Abstract
A recently published macrogenetic dataset of California’s flora and fauna, CaliPopGen, comprehensively summarizes population genetic research published between 1985 and 2020. Integrating these genetic data into the requisite “best available science” upon which conservation professionals rely should facilitate the prioritization of populations based on genetic health. We evaluate the extent to which the CaliPopGen Dataset provides genetic diversity estimates that are 1) unbiased, 2) sufficient in quantity, 3) cover entire species’ ranges, and 4) include potentially adaptive loci. We identified genetic diversity estimates for 4,462 spatially referenced populations of 432 species, confirming California’s rich published history of population genetics research. Most recent studies used microsatellites markers, which have uniquely high levels of variation, and estimates of all genetic metrics varied significantly across marker types. Most studies used less than 10 loci for inferences, rendering parameter estimates potentially unreliable, and covered small spatial extents that include only a fraction of the studied species’ California distribution (median 16.3%). In contrast, the ongoing California Conservation Genomics Project (CCGP) aims to cover the full geographical and environmental breadth of each species’ occupied habitats, and uses a consistent approach based on whole-genome data. However, the CCGP will sequence only 12% of the number of individuals, and covers only about half the evolutionary diversity, of the CaliPopGen Database. There is clearly a place in the evaluation of the genetic health of California for both approaches going forward, especially if differences among studies can be minimized, and overlap emphasized. A complementary use of both datasets is warranted to inform optimal conservation decision-making. Finally, a synopsis of the available population genetic data for California, all other US states and 241 other countries, allows us to identify states and countries for which meaningful data summaries, such as CaliPopGen, could be collated and others, which have limited published data available and are prime targets for future, empirical work.
Keywords: California Conservation Genomics Project, California Floristic Provence, CaliPopGen, CCGP, landscape genomics, multispecies conservation, spatial conservation prioritization
Introduction
Conservation practitioners, from federal and state agencies to local nonprofits, protect and enhance habitats that sustain species over the long term. This requires identifying ecosystems that are necessary to protect, habitats that require restoration, and populations that are at risk of extinction (Soulé 1985). Increasingly, conservation management includes information about the genetic health of populations (Murphy and Weiland 2016), ranging from measures of genetic diversity and heterozygosity, to determining genetic effective population size, to the estimation of the effects of landscape features, both natural and anthropogenic, on levels of gene flow between populations (Holderegger et al. 2019). Integrating genetic data into the requisite “best available science” upon which conservation professionals rely provides a unique benefit to the decision-making process by allowing practitioners to prioritize land for acquisitions based on the genetic health of populations, and to identify source populations for genetic rescue (Frankham et al. 2019). This information is also crucial for establishing baselines for conservation, assessing the success of recovery actions, and identifying barriers to species recovery (Kardos 2021).
Several recent efforts have synthesized genetic metrics for hundreds of species and thousands of spatially georeferenced populations, including the MacroPopGen Database for the Americas (Lawrence et al. 2019), and the CaliPopGen Database for California (Beninde et al. 2022). The CaliPopGen Database provides an extensive database of spatially referenced population genetic data from the peer-reviewed literature published over the last 35 yr for populations in California. This historical database encompasses the shifting baselines of population genetics research from its infancy to the present, an astonishing progression of advances in genomic technologies, statistics, and geospatial analyses, and more generally, tracks our understanding of genomic structure and function over time (Allendorf et al. 2010; Schweizer et al. 2021). As a result of these changes in technology and approach, the “best available science” for one species may be based on entirely different data types and analyses than for another, although both are used to make conservation decisions.
This variability can lead to great disparities in the quality, reliability, and utility of genetic data, although many data users may be unaware that such variation exists or is important (Shafer et al. 2015). For example, it has been demonstrated that population-level estimates of genetic diversity can vary substantially, both quantitatively and in rank order, depending on the marker type used, and markers have gone through several complete turnovers in the last few decades (Allendorf et al. 2010; Beninde et al. 2022). In one thorough example, estimates of expected heterozygosity generated using microsatellites and genome-wide single nucleotide polymorphisms (SNPs) were not significantly correlated in the model plant species Arabidopsis halleri (Fischer et al. 2017). The same study found correlated estimates of FST between microsatellite and SNP markers, although microsatellite estimates were generally higher than those based on SNPs. A similar study in the brown trout, Salmo trutta, revealed that among-population FST estimates were similar between microsatellites and SNPs, as were the rank order of populations for allelic richness and heterozygosity, while effective population size estimates based on the 2 marker types were uncorrelated (Lemopoulos et al. 2019). Similarly, a reexamination of genetic variation in the critically endangered Magdelena River turtle Podocnemis lewyana across its range in Columbia by Gallego-García et al. (2021) found that the long-held inference of the species as an extreme outlier for low genetic diversity based on microsatellites did not hold up when thousands of SNPs were examined: P. lewyana diversity was still low, but not lower than other endangered turtles. What these and many other studies demonstrate is the importance of marker type when evaluating raw estimates of genetic diversity for any given species or population. Even within the same marker types, the number of loci can be important when quantifying spatial population structure using microsatellites (Rosenberg et al. 2002), or SNPs (Willing et al. 2012; McCartney-Melstad et al. 2018).
To best inform conservation decision-making, genetic data need to conform to a number of assumptions and prerequisites. Data should 1) be unbiased and comprehensive with respect to collection locations to allow for accurate spatial prioritization; 2) provide genetic metrics that can be used directly for prioritization of populations, such as heterozygosity, allelic richness, and effective population size; 3) be available across the entire species’ range to capture genetic patterns across all environmental gradients; and 4) include potentially adaptive loci that may inform future genetic rescue efforts.
Here, we make use of the CaliPopGen Database, a comprehensive compilation of studies published on the population genetics of California flora and fauna between 1985 and 2020, and explore the utility of this dataset, and the literature on which it is based, for conservation decision-making. The publication by Beninde et al. (2022) primarily describes the creation and organization of the database for those members of the community who can utilize it for diverse research questions. More detailed analyses of patterns of genetic diversity, the spatial distribution of sampling within and between species across California, as well as more focused comparisons across related taxa and ecosystems, remain to be explored.
At the center of the analyses presented here are the raw estimates of several standard genetic metrics, including heterozygosity and allelic richness, which we assess for their ability to deliver comprehensive and comparable information within and between species. Specifically, we quantify both the total number of population-level estimates of genetic metrics, and the differences among these estimates depending on the genetic marker type used, to summarize our current state of population genetic knowledge across California. We also calculate the proportion of the geographic range for each species that is covered by these earlier studies, the number of listed (under federal and state regulatory acts) species for which genetic data are available, and the distribution of sampling sites by land use type and accessibility. Because California is the most populous state in the United States, has a high density of research institutions, and is located in a biodiversity hotspot, we tested the expectation that California ranks highly in the number of population genetics publications produced among all 50 US states and 241 countries globally. This synopsis of the available population genetic data also allows us to identify states and countries for which meaningful data summaries, such as CaliPopGen, could be collated and other states and countries, which have limited published data available and are prime targets for future work. Finally, we contrast identified shortcomings in the CaliPopGen Database to the aims of the ongoing California Conservation Genomics Project (CCGP, Shaffer et al. 2022) and assess its potential to generate these missing genetic data.
Materials and methods
Genetic metrics and marker types
We compared the estimates of 8 commonly used metrics of genetic diversity summarized in the Population Genetic dataset of CaliPopGen: expected and observed heterozygosity, allelic richness, nucleotide diversity, genetic effective population size, percent polymorphic loci, haplotype diversity, and inbreeding coefficients across the 4 most commonly employed marker types: allozymes (allozy.), mitochondrial DNA sequences (mtDNA), nuclear DNA sequences (nDNA), and microsatellites (microsat.). We restricted our analysis to this subset of marker types because there were data for fewer than 100 populations for each of the other marker types (amplified fragment length polymorphism [AFLP] = 92, chloroplast DNA [cpDNA] = 3, plastid DNA [ptDNA] = 39, random amplified polymorphic DNA [RAPD] = 16, ribosomal DNA [rDNA] = 52, restriction fragment length polymorphism [RFLP] = 5, sex-linked loci = 2, and single-strand conformation polymorphism [SSCP] = 4). We calculated the number of populations, species, and published studies available for each of the marker types, for the full dataset as well as for a subset that only included studies with a minimum of 10 loci to at least partially guard against variance derived from a limited number of loci (Rosenberg et al. 2002). We collapsed the estimates of “allelic richness” and “alleles per locus” as reported in CaliPopGen into one metric because they are highly correlated: comparing data for 174 populations that had both metrics available returned a strongly positive association that was highly significant (Pearson’s correlation r = 0.894, df = 173, P < 0.001). We tested for differences of genetic diversity estimates by marker type using Kruskal–Wallis tests and used Dunn’s tests for pairwise comparisons. All analyses were conducted using R Statistical Software (v4.1.2; R Core Team 2021).
Spatial distribution and extent of studies
Species are very often nonhomogeneous across their entire range, which may limit the utility of local scale landscape genomics studies for making inferences about a species across its broader range (Trumbo et al. 2013). We examined the degree to which study extents of CaliPopGen covered the range of species in California. We first calculated the area of each study extent for a species as the minimum convex polygon (MCP) of all sampling locations in a study via the mcp() function in the r-package “adehabitatHR” (v0.4.19; Calenge 2006). If multiple species were part of a single study, we generated separate study extents for each. Similarly, if a single species was included in 2 or more publications, study extents were generated separately for each of the studies and their areas calculated separately. We then estimated the California range of all species contained in CaliPopGen, as the MCP of georeferenced (≤250 m accuracy), research-grade observations (up to 10,000 per species) from iNaturalist (www.iNaturalist.org), retrieved using the get_inat_obs() function in the “rinat” package (v0.1.9; Barve et al. 2022). We express the fraction of the range of species covered by study extents as the proportion of a study’s MCP divided by the MCP of the full range of the given species in California. We excluded species for which there were less than 4 localities available in either the CaliPopGen Database or iNaturalist (this is the minimum number of points required by the mcp() function). We note that this approach is specifically quantifying the fraction of a species’ range covered by individual studies, rather than the composite of several studies that may focus on a given taxon. To evaluate the efficacy of our approach in estimating the range of a species based on iNaturalist observations, we used Pearson’s correlations to evaluate the potential influence of the number of iNaturalist observations on the area of MCPs. Calculating species ranges from iNaturalist data likely underestimates the full area of the range, as observations on iNaturalist are unlikely to cover the full range of species.
Previous summaries of the ecological literature have demonstrated that sample availability varies with land use type, and sites that are easily accessible, such as those along roads and close to cities, are often overrepresented (Martin et al. 2012; Zizka et al. 2021). To evaluate such spatial biases in the distribution of CaliPopGen localities, we quantified the accessibility of sites as the value of the human influence index at each site. This index summarizes human population size and access infrastructure, including roads, rivers, and railway tracks (Sanderson et al. 2002). We also evaluated the distribution of CaliPopGen localities among USGS land cover classes (U.S. Geological Survey 2014) to determine whether certain land cover classes were over- or underrepresented in the literature. We tallied the observed number of study sites in each land cover class and compared it to the number of expected sites, if sampling sites were distributed in proportion to the relative area of each land cover class in California.
Endangered species in California
We cross-referenced the list of federally listed species and populations (U.S. Fish & Wildlife Service 2022) to the CaliPopGen Database to quantify how many of the 287 listed populations and species have been studied to date using population genetics.
Estimated availability of population genetic data in other US states and countries
Our expectation is that California may be over-studied relative to many other states and countries, given its high human populations size, high concentration of universities and research funding, its location within a biodiversity hotspot (the California Floristic Provence), and its generally proactive approach to environmental stewardship. To estimate how much published information similar to that in the CaliPopGen Database is likely available for other US states and for other countries, we compared the results obtained from the Web of Science (WOS) Core Collection (https://webofknowledge.com/) using the same search criteria as was used to generate CaliPopGen from 1900 to 2022 (topic = (California*) AND topic = (genetic* OR genomic*) AND topic = (species OR taxa* OR population*, see Beninde et al. (2022) for a full description of the search criteria). We replaced “California*” with each of the other US states and with 241 countries (including constituent countries and territories).
We gathered additional information on all 50 US states to explore variation in the number of publications retrieved by the WOS searches, and, by inference, the intensity of past population genetic research efforts, including the number of research institutions (U.S. Department of Education 2020), National Science Foundation funding (NSF 2020), Gross Domestic Product (GDP, U.S. Department of Commerce 2022), electoral votes of states in the 2020 presidential election (Federal Election Commission 2021), and the number of federally listed species under the US Endangered Species Act (U.S. Fish & Wildlife Service 2022). We used random forest models (Breiman 2001), implemented in the r-package “randomForest” (v4.7-1.1; Liaw and Wiener 2022), to quantify the importance of each of these variables as predictors for our primary response variable, the number of publications retrieved by WOS searches. The importance of predictors is quantified by the % increase in the mean squared error of prediction (%IncMSE) after permuting this variable in the out-of-bag cross-validation.
Relevance of the published literature to conservation in California
We quantified how many CaliPopGen studies used the word “conservation” in their title to assess the original intent of studies with respect to conservation questions.
Results
The original intended application of the existing literature to conservation questions was relatively modest, and only 20 publications included the term “conservation” in their title, out of the total 450 publications that constitute the CaliPopGen Database.
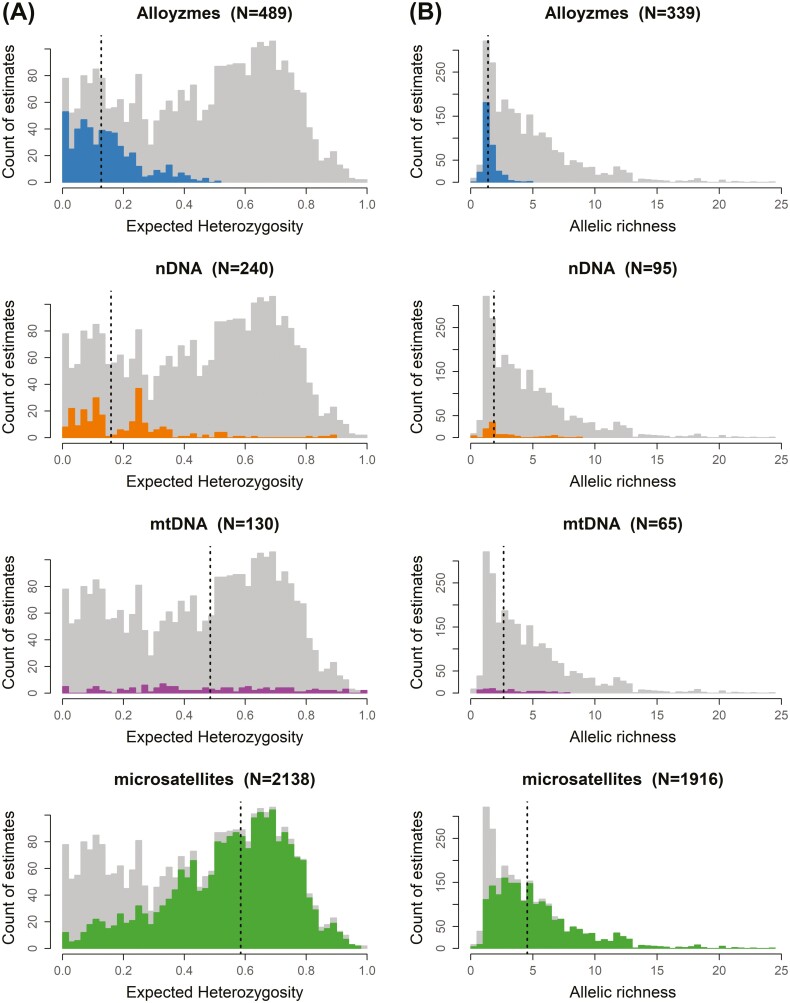
In total, the subset of the CaliPopGen Database explored here contains data on 4,462 populations with information for at least one of the 8 genetic metrics derived from allozymes, nDNA, mtDNA, or microsatellites. The number of population-level estimates available for each marker type and each of the 8 genetic diversity estimates is summarized in Table 1. Estimates for all genetic metrics were significantly different between marker types (Table 2). All pairwise comparisons of expected and observed heterozygosity and allelic richness, the most commonly reported variables, were significantly different among marker types, with the exception of the mtDNA–nDNA comparison of allelic richness (Table 2). Significance of pairwise differences between marker types was more variable for the other genetic metrics (Table 2). For the 2 most commonly reported variables (Fig. 1), median values of expected heterozygosity were 0.13 (allozymes), 0.16 (nDNA), 0.49 (mtDNA), and 0.59 (microsatellites), while median values of allelic richness were 1.4 (allozymes), 1.9 (nDNA), 2.7 (mtDNA), and 4.5 (microsatellites). More than half of all populations were studied using 9 or fewer loci, and the dataset was reduced to 2,069 populations when setting a minimum threshold of 10 loci.
Table 1.
The number of population-level estimates available from the CaliPopGen Database, for all genetic metrics and the most commonly reported marker types, and the total number of populations, species, and studies per marker type.
| Genetic metric | Allozyme | nDNA | mtDNA | Microsatellite |
|---|---|---|---|---|
| Expected heterozygosity | 489 | 240 | 130 | 2,138 |
| Observed heterozygosity | 575 | 155 | 74 | 2,021 |
| Nucleotide diversity | 2 | 173 | 684 | 12 |
| Effective population size | 1 | 19 | 24 | 236 |
| Percent polymorphic loci | 216 | 15 | 16 | 54 |
| Haplotype diversity | 0 | 39 | 367 | 0 |
| Inbreeding coefficient value | 90 | 169 | 5 | 953 |
| Allelic richness | 339 | 95 | 65 | 1,916 |
| Total number of populations | 670 | 450 | 896 | 2,473 |
| Total number of species | 93 | 59 | 164 | 192 |
| Total number of studies | 69 | 42 | 109 | 200 |
Table 2.
Results of Kruskal–Wallis tests of estimates of genetic metrics by marker type (first row per genetic metric, in bold) and results of Dunn’s test for significant differences between pairwise comparisons by marker type (the following rows per genetic diversity metrics, in italics).
| Genetic metric | Chi2/pairwise comparison | df/Z | P adj. |
|---|---|---|---|
| Expected heterozygosity | 1,234.8 | 3 | <0.0001* |
| Expected heterozygosity | allozy.–microsat. | −31.34 | <0.0001* |
| Expected heterozygosity | allozy.–mtDNA | −13.11 | <0.0001* |
| Expected heterozygosity | microsat.–mtDNA | 3.07 | 0.0043* |
| Expected heterozygosity | allozy.–nDNA | −2.66 | 0.0078* |
| Expected heterozygosity | microsat.–nDNA | 20.00 | <0.0001* |
| Expected heterozygosity | mtDNA–nDNA | 9.96 | <0.0001* |
| Observed heterozygosity | 1,105.6 | 3 | <0.0001* |
| Observed heterozygosity | allozy.–microsat. | −32.41 | <0.0001* |
| Observed heterozygosity | allozy.–mtDNA | −7.37 | <0.0001* |
| Observed heterozygosity | microsat.–mtDNA | 5.26 | <0.0001* |
| Observed heterozygosity | allozy.–nDNA | −6.67 | <0.0001* |
| Observed heterozygosity | microsat.–nDNA | 11.13 | <0.0001* |
| Observed heterozygosity | mtDNA–nDNA | 2.16 | 0.0305* |
| Nucleotide diversity | 38.974 | 3 | <0.0001* |
| Nucleotide diversity | allozy.–microsat. | 0.49 | 1.0000 |
| Nucleotide diversity | allozy.–mtDNA | 1.39 | 0.6555 |
| Nucleotide diversity | microsat.–mtDNA | 2.10 | 0.1776 |
| Nucleotide diversity | allozy.–nDNA | 0.69 | 1.0000 |
| Nucleotide diversity | microsat.–nDNA | 0.38 | 0.7038 |
| Nucleotide diversity | mtDNA–nDNA | −5.86 | <0.0001* |
| Effective population size | 47.042 | 3 | <0.0001* |
| Effective population size | allozy.–microsat. | −0.94 | 0.3478 |
| Effective population size | allozy.–mtDNA | −1.79 | 0.2214 |
| Effective population size | microsat.–mtDNA | −4.13 | 0.0002* |
| Effective population size | allozy.–nDNA | −2.24 | 0.1005 |
| Effective population size | microsat.–nDNA | −5.69 | <0.0001* |
| Effective population size | mtDNA–nDNA | −1.54 | 0.2469 |
| Percent polymorphic loci | 30.296 | 3 | <0.0001* |
| Percent polymorphic loci | allozy.–microsat. | −4.37 | 0.0001* |
| Percent polymorphic loci | allozy.–mtDNA | 1.98 | 0.0948 |
| Percent polymorphic loci | microsat.–mtDNA | 4.14 | 0.0002* |
| Percent polymorphic loci | allozy.–nDNA | −2.56 | 0.0314* |
| Percent polymorphic loci | microsat.–nDNA | −0.06 | 0.9482 |
| Percent polymorphic loci | mtDNA–nDNA | −3.33 | 0.0035* |
| Haplotype diversity | 5.0507 | 1 | 0.0246* |
| mtDNA–nDNA | |||
| Inbreeding coefficient | 24.505 | 3 | <0.0001* |
| Inbreeding coefficient | allozy.–microsat. | 2.89 | 0.0154* |
| Inbreeding coefficient | allozy.–mtDNA | 0.67 | 1.0000 |
| Inbreeding coefficient | microsat.–mtDNA | −0.02 | 0.9834 |
| Inbreeding coefficient | allozy.–nDNA | 4.78 | <0.0001* |
| Inbreeding coefficient | microsat.–nDNA | 3.66 | 0.0012* |
| Inbreeding coefficient | mtDNA–nDNA | 0.69 | 1.0000 |
| Allelic richness | 701.6 | 3 | <0.0001* |
| Allelic richness | allozy.–microsat. | −25.19 | <0.0001* |
| Allelic richness | allozy.–mtDNA | −5.71 | <0.0001* |
| Allelic richness | microsat.–mtDNA | 5.64 | <0.0001* |
| Allelic richness | allozy.–nDNA | −4.24 | <0.0001* |
| Allelic richness | microsat.–nDNA | 9.44 | <0.0001* |
| Allelic richness | mtDNA–nDNA | 1.74 | 0.0815 |
Significance of tests is denoted with an asterisk (*); allozy. = allozymes and microsat. = microsatellites.
Fig. 1.
Histograms showing the distribution of estimates of expected heterozygosity (A) and allelic richness (B) by marker type. Gray bars in the background show counts of estimates of all marker types and dashed lines indicate the median estimate, separately for each marker type.
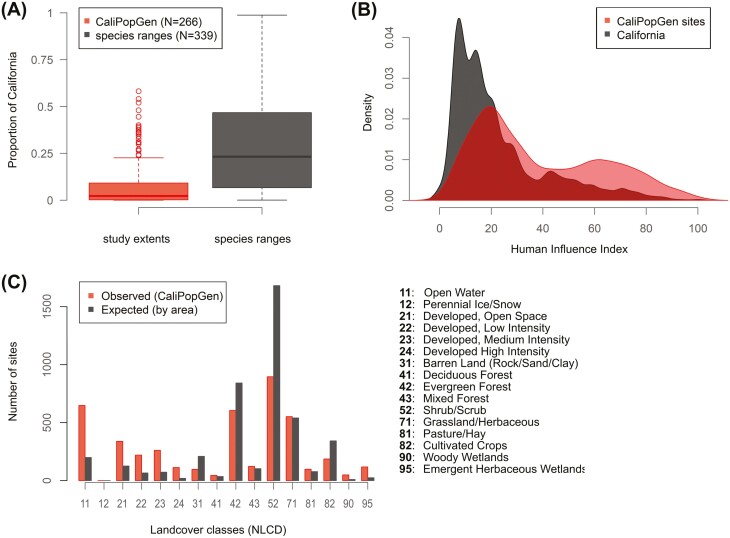
The study extent of the 255 CaliPopGen studies with at least 4 unique sample sites had a median area = 9,365 km2, while the range extent of the 339 species with at least 4 unique occurrence records in iNaturalist had a median area = 98,292 km2, or roughly 10-fold larger (Fig. 2A). Of those studies, 217 corresponded to species we quantified ranges for (as estimated in iNaturalist), and the median proportion of the species range covered by study extents was 16.3%. The genetic study extent of only 10 species covered >100% of the species range, and most of these were from freshwater or marine studies with a low corresponding number of observations on iNaturalist (median iNaturalist occurrences = 10 vs. median iNaturalist occurrences of other species = 261). In addition, the MCP calculated from iNaturalist observations was significantly and positively correlated to the total number of available iNaturalist observations (r = 0.55, df = 335, P < 0.0001), suggesting that range size as estimated by iNaturalist does, to some extent, reflect the number of available observations, and may well be a lower estimate of the true species range for species with few observations.
Fig. 2.
(A) The area of study extents of all CaliPopGen studies (in red) and the size of study species entire ranges within California (dark gray). On average study extents covered 16.3 % of species ranges; (B) Distribution of the human influence index across California (in gray), as a proxy for accessibility of sites, where low values indicate little human influence and high values great human influence, and for CaliPopGen sample sites (in red); (C) the distribution of CaliPopGen sampling sites (in red) compared with their expected frequency (in gray) across 16 land use types.
Studied populations in the California population genetics literature were located in easily accessible areas more frequently than expected, while less accessible areas were under-sampled, clearly indicating some form of access bias present in the dataset (Fig. 2B). Studied populations were also more frequently located in developed land cover classes, open water and wetlands than expected, while shrub/scrub, cultivated crops, barren land, and evergreen forest are underrepresented (Fig. 2C).
Of the 287 federally listed species and populations known to occur in California, 49 (17%) are contained in the CaliPopGen Database. Another 9 listed species are also contained in CaliPopGen, but it is unclear from the records if the data covers the specific, listed population. Thus, at a maximum, 20% (58/287) of the federally listed species in California have received some attention from population genetics research.
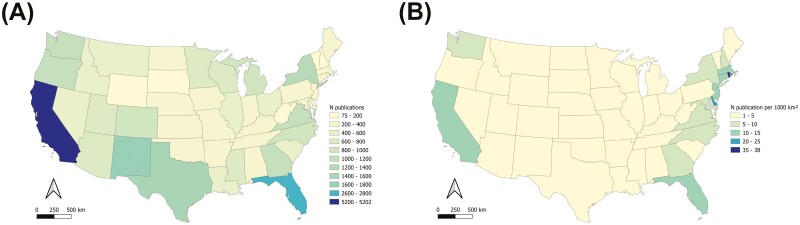
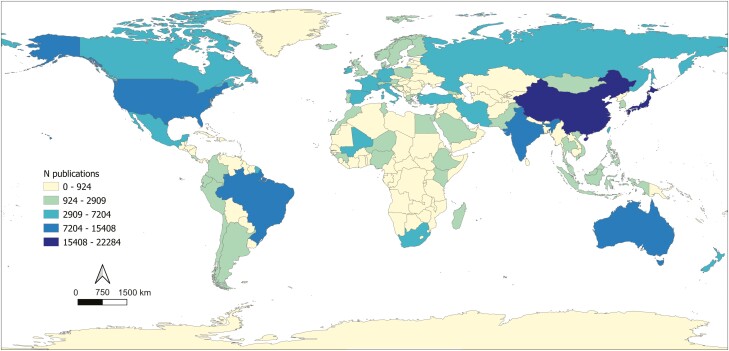
Querying the WOS, California has the highest total number of publications of all US states and ranks 8th among US states in a comparison of the proportion of publications per surface area (Fig. 3; Table 3). In comparison to other countries of similar size (area of California ±20%), California ranks 2nd behind Japan both in the total number of publications and the proportional number of publications per surface area (Table 4). In comparison to all other countries California ranks 10th in the total number of publications (Fig. 4), behind China, Japan, India, Brazil, the United States, Australia, Mali, Mexico, and South Africa, in descending order. The total number of publications retrieved for the United States is 12,003, which is equal to only 34.5% of the sum of all publications of US states queried separately, at 34,797 (Table 3).
Fig. 3.
The number of publications resulting from searching the Web of Science (WOS) using the same search string as used in CaliPopGen, but replacing “California” with each US state: the total number of publications (A) and the number of publications per 1,000 km2 (B). Hawaii and Alaska were omitted from maps. Hawaii ranked 4th by the total number of publications (1,578; see Table 3) and 1st by the number of publications per 1,000 km2 (55.7). Alaska ranked 5th by the total number of publication (1,483) and 48th by the number of publications per 1,000 km2 (0.9).
Table 3.
The 20 US states with the highest number of Web of Science (WOS) publications per surface area, using the same search criteria as for California but replacing for State names (WOS hits = the raw number of results; WOS per 1,000 km2 = the number of WOS hits per 1,000 km2).
| State | Population | Total km2 | WOS hits | WOS per 1,000 km2 | N listed species |
|---|---|---|---|---|---|
| Hawaii | 1,415,872 | 28,313 | 1,578 | 55.7 | 474 |
| Rhode Island | 1,059,361 | 4,001 | 153 | 38.2 | 9 |
| Delaware | 973,764 | 6,446 | 153 | 23.7 | 14 |
| Florida | 21,477,737 | 170,312 | 2,606 | 15.3 | 133 |
| Connecticut | 3,565,278 | 14,357 | 217 | 15.1 | 12 |
| New Jersey | 8,882,190 | 22,591 | 301 | 13.3 | 17 |
| Massachusetts | 6,892,503 | 27,336 | 345 | 12.6 | 18 |
| California | 39,512,223 | 423,967 | 5,202 | 12.3 | 287 |
| Virginia | 8,535,519 | 110,787 | 1,109 | 10.0 | 76 |
| New York | 19,453,561 | 141,297 | 1,239 | 8.8 | 24 |
| Maryland | 6,045,680 | 32,131 | 256 | 8.0 | 23 |
| Georgia | 10,617,423 | 153,910 | 1,032 | 6.7 | 76 |
| North Carolina | 10,488,084 | 139,391 | 899 | 6.4 | 69 |
| Washington | 7,614,893 | 184,661 | 1,153 | 6.2 | 31 |
| New Hampshire | 1,359,711 | 24,214 | 145 | 6.0 | 12 |
| Mississippi | 2,976,149 | 125,438 | 684 | 5.5 | 52 |
| New Mexico | 2,096,829 | 314,917 | 1,640 | 5.2 | 58 |
| South Carolina | 5,148,714 | 82,933 | 416 | 5.0 | 39 |
| Oregon | 4,217,737 | 254,799 | 1,051 | 4.1 | 45 |
| Wisconsin | 5,822,434 | 169,635 | 670 | 3.9 | 24 |
| Ohio | 11,689,100 | 116,098 | 410 | 3.5 | 27 |
| Pennsylvania | 12,801,989 | 119,280 | 340 | 2.9 | 16 |
| Illinois | 12,671,821 | 149,995 | 417 | 2.8 | 33 |
| Texas | 28,995,881 | 695,662 | 1,434 | 2.1 | 105 |
| Alaska | 731,545 | 1,723,337 | 1,483 | 0.9 | 8 |
Additionally, we included States that have a large population (>10 million) and/or have a high number of publications (>1,000). The table is sorted by the number of publications per unit area. California is highlighted in italics for reference.
Table 4.
The number of WOS hits for countries with a comparable size to California (±20% of surface area), using the same search criteria as for California but replacing for country names.
| Countries | Total km2 | WOS hits | WOS per 1,000 km2 | GDP (millions) |
|---|---|---|---|---|
| Japan | 377,976 | 17,412 | 46.1 | 4,937,421.88 |
| California | 423,967 | 5,202 | 12.3 | 3,513,347.50 |
| Spain | 505,992 | 5,112 | 10.1 | 1,425,276.50 |
| Germany | 357,114 | 3,375 | 9.5 | 4,223,116.21 |
| Norway | 385,207 | 2,559 | 6.6 | 482,437.02 |
| Sweden | 450,295 | 2,319 | 5.1 | 627,437.90 |
| Congo | 342,000 | 849 | 2.5 | 12,523.96 |
| Morocco | 446,550 | 1,076 | 2.4 | 132,725.26 |
| Cameroon | 475,442 | 860 | 1.8 | 45,238.61 |
| Papua New Guinea | 462,840 | 728 | 1.6 | 26,594.28 |
| Paraguay | 406,752 | 383 | 0.9 | 38,986.81 |
| Iraq | 438,317 | 411 | 0.9 | 207,889.33 |
| Zimbabwe | 390,757 | 345 | 0.9 | 26,217.73 |
| Uzbekistan | 447,400 | 122 | 0.3 | 69,238.90 |
| Turkmenistan | 488,100 | 83 | 0.2 | 45,231.43 |
The table is sorted by the number of publications per unit area. California is highlighted in italics for reference.
Fig. 4.
The number of publications resulting from searching the WOS using the same search string as used in CaliPopGen, but replacing “California” with each country. This map shows the total number of publications only (not corrected for unit area).
Random forest models explained 33.1% of the variation in the total number of publications obtained by querying the WOS for US states. The most important predictors were the number of listed species (44.7%IncMSE), human population size (35.1%IncMSE), surface area (34.7%IncMSE), amount NSF funding (32.6%IncMSE) and GDP (28.7%IncMSE). Predictors summarizing the number of research institutions, i.e. only R1, R1–R3, all 4-yr program institutions and all institutions, or the political convictions of states were less important (all <16%IncMSE).
Discussion
California has a rich history of research on the population and landscape genetics of its native flora and fauna. Most of the work thus far conducted in the state has focused on a single species, or at most a handful of species (with the exception of Dawson 2001; Kelly and Palumbi 2010), and drew inferences from a variety of marker types and analytical techniques that have evolved over time. The population genetics research community has carried out hundreds of genetic studies for hundreds of taxa in California, but no comprehensive synthesis has yet been produced for this vast collection of studies. Where do we stand with conservation genomic data for California? And what does that data summary imply about the state of knowledge for other states in the US and other countries around the world?
The CaliPopGen Database (Beninde et al. 2022) is a comprehensive collection of population genetic data for studies in California. The full database includes primary population genetic information for 5,453 populations of 448 species from 4 main marker types. In comparison to other macrogenetic databases (Miraldo et al. 2016; Lawrence et al. 2019; Manel et al. 2020; Millette et al. 2020; Theodoridis et al. 2020), when expressed as a proportion of the study area, CaliPopGen contains at least an order of magnitude more species (0.83/1,000 km2), populations (9.59/1,000 km2), and individuals (284.04/1,000 km2) than those previous analyses, making it one of the richest regional data compilations in the world. However, most studies in the CaliPopGen Database were motivated by evolutionary or landscape genetic questions, rather than conservation per se. Only 20 of 450 total publications in the CaliPopGen Database include the term “conservation” in their title. This may explain the low number of estimates of some genetic metrics, especially essential metrics for conservation such as effective population size, which was only reported for 280, or 6.3% of populations (Table 1). It also may explain why so few federally listed species, between 17% and 20% of the 287 in the state, have any population genetic data available in the published literature. In the past, evolutionary and ecological scientists have often eschewed working on threatened species (Britt et al. 2018), potentially due to difficulties in obtaining permits and limitations on sampling imposed by regulatory agencies. Although we lack data on this, our sense is that this is changing—as the biodiversity crisis continues to grow in California and globally, a greater research effort is being focused on threatened and endangered taxa.
The subset of the CaliPopGen Database analyzed here contains genetic information for 4,462 populations of 432 species and most commonly reported genetic metrics were estimates of heterozygosity, both expected and observed, and allelic richness, which each comprised estimates for several hundred populations across multiple marker types. A critically important result from our analysis is that the greatest number of population estimates were derived from microsatellites (55.1%), followed by mtDNA (20%), allozymes (14.9%), and nDNA (10%). Estimates of all genetic metrics were significantly different across marker types and for most pairwise comparisons of marker types (Table 2). Among markers, microsatellite markers, and sometimes mtDNA, were the consistent outliers. Microsatellites yielded higher estimates for both expected heterozygosity and allelic richness, followed, in descending order, by mtDNA, nDNA, and allozymes (Fig. 1). This is in line with previous direct comparisons of estimates from different marker types; microsatellite markers specifically tend to have higher estimates of heterozygosity than nDNA (Fischer et al. 2017; Lemopoulos et al. 2019). While there is nothing inherently wrong with such estimates, the predominance of microsatellite studies, combined with their very high estimates of genetic variation, suggest that great caution should be used when comparing them to studies conducted using other marker types. In a conservation context, these inherent differences could have considerable consequences for prioritization of populations based on levels of standing genetic variation if study-specific differences, such as marker type, are not accounted for—Gallego-García et al. (2021) is a case in point for endangered turtles. A results of studies across marker types is to standardize to a mean of 0 and a variance of 1, which at least places them on a comparable scale of variation (Kort et al. 2021). However, this cannot compensate for differences in spatial scale and sample densities between studies (Leigh et al. 2021), nor for the uniquely high mutation rate of microsatellites that may result in different rank orders of genetic variation across populations and species compared with more standard nuclear DNA markers (Gallego-García et al. 2021).
Despite a higher density of population genetic data available from CaliPopGen than from other macrogenetic dataset, the coverage of species ranges is limited: on average, each study covers 16.3% of a species range in California (Fig. 2A). Again, when the goals of a study are to quantify regional genetic variation on a specific landscape, or learn about realized migration or gene flow, there may not be a need to cover the full range of a species. However, conservation actions benefit from comprehensive, species-wide analyses to prioritize management units, or species, for management actions (Frankham et al. 2019), and the existing data fall short of this objective for most taxa. However, summaries of genetic diversity across species and the entire state are a more achievable goal with the existing data. One of the major impediments to comprehensive analyses, in California and elsewhere, is spatial sampling biases. Across species, we detected considerable sampling bias, similar to other summaries of sample site locations in ecological research (Martin et al. 2012; Zizka et al. 2021). Samples tended to be collected at locations that are easily accessible with a higher-than-expected frequency, and were likely to come from developed land, open water, or wetlands (Fig. 2B and 2C). More remote areas, especially shrub/scrub, barren land and evergreen forests are underrepresented in the existing data, highlighting spatial knowledge gaps. Whether this is driven by the distribution of private and public lands, proximity to urban areas and research institutions, or other factors is still unclear. However, this legacy of working in convenient, rather than more ecologically intact landscapes, is striking and implies that we systematically under sample those landscapes that may tell us the most about populations in least-impacted, more natural settings. Similar to findings for agricultural areas in a global analysis of ecological research activities (Martin et al. 2012), cultivated crops in California (Fig. 2C), were underrepresented in the existing data. This is especially surprising when considering the high levels of human infrastructure and thus accessibility in most agricultural areas, and probably indicates that researchers either are less interested in agricultural areas than in any other accessible land use types, in fact have less access than assumed given that private ownership predominates agricultural landscapes, or that many species simply are no longer found in agricultural areas. However, given the enormous acreage devoted to agriculture (according to the California Department of Food and Agriculture, roughly 43 million of California’s 100 million acres are devoted to agriculture, see https://www.cdfa.ca.gov/agvision/docs/Agricultural_Loss_and_Conservation.pdf), the potential for agricultural landscapes to harbor many components of local species assemblages is an important conservation question that certainly deserves additional, immediate research attention.
In comparison to other US states as well as comparable countries globally, California has among the richest literature on population genetics (Fig. 3; Tables 3 and 4). The state has almost double the number of publications (using our search criteria in WOS) as compared with Florida, the second ranked state. When ranked by the number of publications per unit of land area of each state, California ranks 8th, and all higher-ranking states are considerably smaller in size (0.9% to 40.2% of California; Table 3). In comparison to countries of similar geographic size, California ranks second, with Japan having both the highest absolute number of publications as well as the highest number of publications per unit area (Table 4; Fig. 4). This implies that, from a historical research perspective, California represents essentially a best-case scenario in terms of our current levels of knowledge, and emphasizes the generally sparse spatial coverage of population genetic data across other parts of the United States and many other countries. Economic status of states and countries seems to be linked to the number of publications; GDP accounted for 28.7%IncMSE in analysis of states, and only one of the lowest publishing countries that is similar in size to California (Uzbekistan) does not belong to the UN defined list of countries of the Global South. However, there is also considerable variation in countries of the Global North (Fig. 4) and within the states of the United States. For US states, the number of endangered species present was the most important factor to explain the total number of publications, while, surprisingly, the number of research institutions or political orientation of states played only minor roles. We note that while California does reside in one of the world’s biodiversity hotspots (Mittermeier et al. 2005), there are also well-documented biases associated with listing status among taxonomic groups as well as differences in listing propensities among states, all of which contribute to regional differences in the density of endangered species (Puckett et al. 2016).
CaliPopGen showcases the vast amount of data that can be retrieved from the primary literature and analyzed for current and historical trends. California has among the richest population genetic literature available, both within the United States and globally. Even so, the genetics are not “done” for California, and we have delineated key data gaps that need addressing. Older markers are idiosyncratic, and coverage across groups is spotty at best. There are certainly valuable single-species studies, and with more sophisticated analyses, there may well be statewide patterns to be resolved across taxonomic groups. However, persistent differences in spatial scale and sample numbers/densities across studies probably cannot be corrected for statistically, and going forward, we should also be looking for new opportunities to generate genetic data using consistent, repeatable methodologies and sampling schemes.
The CCGP (Shaffer et al. 2022, www.ccgproject.org), and the Los Angeles Genomics Project (LAG, Beninde et al. unpublished data) are 2 such initiatives that are being spearheaded in California. As described in this issue of the Journal of Heredity, there are a number of distinct advantages of the CCGP over a compilation of historical data as embodied in CaliPopGen, as well as some disadvantages. On the plus side, projects like the CCGP can be planned with common goals and objectives that cannot be achieved with post hoc meta-analyses. Given its goals and objectives, all of the 153 species projects in the CCGP aim to cover the geographical and environmental breadth of occupied habitats, avoiding the problem of partial range coverage that characterizes the existing literature. The CCGP also uses a unitary approach to data collection and analysis—all species have a very high-quality reference genome, all resampling is exclusively with whole-genome resequencing at a target 10× coverage, and all bioinformatics and landscape genomic analyses are run through the same pipelines with the same filters and summary statistics. This uniformity of approach should allow the CCGP to incorporate adaptive genetic diversity into spatial analyses and prioritization recommendations consistently across the state, information that CaliPopGen is lacking. Using the same, whole-genome methodology also allows for unbiased estimates of genetic metrics, which are indispensable for spatially prioritizing populations with high levels of genetic diversity. The disadvantages primarily center on the number of species and populations that can be covered. The CCGP has covered roughly half of the evolutionary diversity contained in the historical database (Toffelmier et al. 2022), and doing so required a level of funding and coordination that has never been available previously.
Finally, the CCGP achieves high geographic coverage of species ranges by implementing an individual-level sampling scheme for all projects. This type of sampling is optimally suited for inferences of landscape connectivity (Prunier et al. 2013) and greatly reduces the number of samples necessary and the field and molecular bench workloads. In total, CCGP aims to sequence about 20,000 individuals from as many unique locations. On the other hand, the population-level sampling schemes employed by studies summarized in CaliPopGen, stem from a more modest 4,462 sites sampled for 168,240 individuals (these numbers are true for the subset of the dataset created for this study, the full CaliPopGen Database contains higher numbers). Population-level samples provide more robust estimates of population genetic diversity as they average across multiple individuals.
In conclusion, there is clearly a place for both single-species analyses at the population level, and synthetic ones like the CCGP going forward, and conservation science needs both. The key is to develop future studies so that multiple lines of evidence can be brought together seamlessly to enhance conservation actions. Doing so requires coordination, across the research community and the agency–university nexus.
Contributor Information
Joscha Beninde, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; IUCN WCPA Connectivity Conservation Specialist Group, Gland, Switzerland.
Erin Toffelmier, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
H Bradley Shaffer, UCLA La Kretz Center for California Conservation Science, Institute of the Environment and Sustainability, University of California, Los Angeles, Los Angeles, CA, United States; Department of Ecology and Evolutionary Biology, University of California, Los Angeles, Los Angeles, CA, United States.
Funding
Funding for JB and ET was provided by the UCLA La Kretz Center for California Conservation Science. ET received funding by a grant provided to the University of California by the State of California, State Budget Act of 2019 [UC Award ID RSI-19-690224]. JB received funding by the German Science Foundation (DFG: BE 6887/1-1).
Competing interests
The authors declare no competing interests.
References
- Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11(10):697–709. [DOI] [PubMed] [Google Scholar]
- Barve V, Hart E, Guillou S. rinat: access “iNaturalist” data through APIs. V0.1.9. 2022. https://cran.r-project.org/web/packages/rinat/rinat.pdf, accessed 8/4/2022
- Beninde J, Toffelmier EM, Andreas A, Nishioka C, Slay M, Soto A, Bueno JP, Gonzalez G, Pham HV, Posta M, et al. A genetic and life history database for the fauna and flora of California. Sci Data. 2022;9(1):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Britt M, Haworth SE, Johnson JB, Martchenko D, Shafer ABA. The importance of non-academic coauthors in bridging the conservation genetics gap. Biol Conserv. 2018;218:118–123. [Google Scholar]
- Calenge C. The package adehabitat for the R software: tool for the analysis of space and habitat use by animals. Ecol Model. 2006;197(3–4):516–519. [Google Scholar]
- Dawson MN. Phylogeography in coastal marine animals: a solution from California? J Biogeogr. 2001;28:723–736. [Google Scholar]
- Federal Election Commission. Official 2020 presidential general election results. General election date: 11/03/2020. State Certificates of Vote, U.S. National Archives and Records Administration; 2021. https://www.fec.gov/resources/cms-content/documents/2020presgeresults.pdf, accessed 07/20/2022. [Google Scholar]
- Fischer MC, Rellstab C, Leuzinger M, Roumet M, Gugerli F, Shimizu KK, Holderegger R, Widmer A. Estimating genomic diversity and population differentiation—an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics. 2017;18:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Ralls K, Eldridge MDB, Dudash MR, Fenster CB, Lacy RC, Sunnucks P. A practical guide for genetic management of fragmented animal and plant populations. With assistance of Karina H. McInnes. 1st ed.Oxford (UK): Oxford University Press; 2019. [Google Scholar]
- Gallego-García N, Caballero S, Shaffer HB. Are genomic updates of well-studied species worth the investment for conservation? A case study of the Critically Endangered Magdalena River turtle. J Hered. 2021;112(7):575–589. [DOI] [PubMed] [Google Scholar]
- Holderegger R, Balkenhol N, Bolliger J, Engler JO, Gugerli F, Hochkirch A, Nowak C, Segelbacher G, Widmer A, Zachos FE. Conservation genetics: linking science with practice. Mol Ecol. 2019;28(17):3848–3856. [DOI] [PubMed] [Google Scholar]
- Kardos M. Conservation genetics. Curr Biol. 2021;31(19):R1185–R1190. [DOI] [PubMed] [Google Scholar]
- Kelly RP, Palumbi SR. Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS One. 2010;5(1):e8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort H, Prunier JG, Ducatez S, Honnay O, Baguette M, Stevens VM, Blanchet S. Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nat Commun. 2021;12(1):516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence ER, Benavente JN, Matte J-M, Marin K, Wells ZRR, Bernos TA, Krasteva N, Habrich A, Nessel GA, Koumrouyan RA, et al. Geo-referenced population-specific microsatellite data across American continents, the MacroPopGen database. Sci Data. 2019;6:14. doi: 10.1038/s41597-019-0024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh DM, van Rees CB, Millette KL, Breed MF, Schmidt C, Bertola LD, Hand BK, Hunter ME, Jensen EL, Kershaw F, et al. Opportunities and challenges of macrogenetic studies. Nat Rev Genet. 2021;11:791–807. doi: 10.1038/s41576-021-00394-0 [DOI] [PubMed] [Google Scholar]
- Lemopoulos A, Prokkola JM, Uusi-Heikkilä S, Vasemägi A, Huusko A, Hyvärinen P, Koljonen M-L, Koskiniemi J, Vainikka A. Comparing RADseq and microsatellites for estimating genetic diversity and relatedness—implications for brown trout conservation. Ecol Evol. 2019;9(4):2106–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. randomForest: Breiman and Cutler’s random forests for classification and regression. 2022. https://cran.rstudio.com/web/packages/randomForest/randomForest.pdf, accessed 5/29/2022.
- Manel S, Guerin P-E, Mouillot D, Blanchet S, Velez L, Albouy C, Pellissier L. Global determinants of freshwater and marine fish genetic diversity. Nat Commun. 2020;11(1):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ, Blossey B, Ellis E. Mapping where ecologists work. Biases in the global distribution of terrestrial ecological observations. Front Ecol Environ. 2012;10(4):195–201. [Google Scholar]
- McCartney-Melstad E, Vu JK, Shaffer HB. Genomic data recover previously undetectable fragmentation effects in an endangered amphibian. Mol Ecol. 2018;27(22):4430–4443. [DOI] [PubMed] [Google Scholar]
- Millette KL, Fugère V, Debyser C, Greiner A, Chain FJJ, Gonzalez A. No consistent effects of humans on animal genetic diversity worldwide. Ecol Lett. 2020;23(1):55–67. [DOI] [PubMed] [Google Scholar]
- Miraldo A, Li S, Borregaard MK, Florez-Rodriguez A, Gopalakrishnan S, Rizvanovic M, Wang Z, Rahbek C, Marske KA, Nogues-Bravo D. An Anthropocene map of genetic diversity. Science. 2016;353(6307):1532–1535. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Gil PR, Hoffman M, Pilgrim J, Brooks T, Mittermeier CG, Lamoreux J, Da Fonseca GA. Hotspots revisited: Earth’s biologically richest and most endangered terrestrial ecoregions: Conservation International. Sierra Madre, Cemex. 2005;315.
- Murphy DD, Weiland PS. Guidance on the use of best available science under the U.S. Endangered Species Act. Environ Manage. 2016;58(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSF. Summary proposal and award information (funding rate) by state and organization. Budget Division; 2020. https://dellweb.bfa.nsf.gov/starth.asp, accessed 07/20/2022. [Google Scholar]
- Prunier JG, Kaufmann B, Fenet S, Picard D, Pompanon F, Joly P, Lena JP. Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations. Towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol Ecol. 2013;22(22):5516–5530. [DOI] [PubMed] [Google Scholar]
- Puckett EE, Kesler DC, Greenwald DN. Taxa, petitioning agency, and lawsuits affect time spent awaiting listing under the US Endangered Species Act. Biol Conserv. 2016;201(1841):220–229. [Google Scholar]
- R Core Team (2021): R: A language and environment. Vienna, Austria: R Foundation for. Available online at http://www.R-project.org/. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. https://www.science.org/doi/pdf/10.1126/science.1078311?download=true, checked on 7/28/2022. [DOI] [PubMed] [Google Scholar]
- Sanderson EW, Jaiteh M, Levy MA, Redford KH, Wannebo AV, Woolmer G. The human footprint and the last of the wild. BioScience. 2002;52(10):891–904. [Google Scholar]
- Schweizer RM, Saarman N, Ramstad KM, Forester BR, Kelley JL, Hand BK, Malison RL, Ackiss AS, Watsa M, Nelson TC, et al. Big data in conservation genomics: boosting skills, hedging bets, and staying current in the field. J Hered. 2021;112(4):313–327. [DOI] [PubMed] [Google Scholar]
- Shafer ABA, Wolf JBW, Alves PC, Bergström L, Bruford MW, Brännström I, Colling G, Dalén L, Meester L de, Ekblom R, et al. Genomics and the challenging translation into conservation practice. Trends Ecol Evol. 2015;30(2):78–87. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, Toffelmier E, Corbett-Detig RB, Escalona M, Erickson B, Fiedler P, et al. Landscape genomics to enable conservation actions: the California Conservation Genomics Project. J Hered. 2022;113(6):577–588. [DOI] [PubMed] [Google Scholar]
- Soulé ME. What is conservation biology? BioScience. 1985;35(11):727–734. [Google Scholar]
- Theodoridis S, Fordham DA, Brown SC, Li S, Rahbek C, Nogues-Bravo D. Evolutionary history and past climate change shape the distribution of genetic diversity in terrestrial mammals. Nat Commun. 2020;11(1):2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffelmier E, Beninde J, Shaffer HB. The phylogeny of California, and how it informs setting multi-species conservation priorities. J. Hered. 2022;113(6):597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbo DR, Spear SF, Baumsteiger J, Storfer A. Rangewide landscape genetics of an endemic Pacific northwestern salamander. Mol Ecol. 2013;22(5):1250–1266. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Commerce. Regional economic accounts: SAGDP tables: annual GDP by state.Bureau of Economic Analysis; 2022. https://apps.bea.gov/regional/downloadzip.cfm, accessed 07/20/2022. [Google Scholar]
- U.S. Department of Education. Integrated Postsecondary Education Data System (IPEDS). Institutional Characteristics Component, National Center for Education Statistics; 2020. https://nces.ed.gov/programs/digest/d20/tables/dt20_317.20.asp, accessed 07/20/2022. [Google Scholar]
- U.S. Fish & Wildlife Service. ECOS. Environmental Conservation Online System: listed species believed to or known to occur in each state. 2022. https://ecos.fws.gov/ecp/report/species-listings-by-state-totals?statusCategory=Listed, accessed 07/20/2022.
- U.S. Geological Survey. NLCD 2011 Percent Developed Imperviousness (2011 Edition, amended 2014)—National Geospatial Data Asset (NGDA) Land Use Land Cover. Sioux Falls, SD: U.S. Geological Survey;2014. [Google Scholar]
- Willing E-M, Dreyer C, van Oosterhout C. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS One. 2012;7(8):e42649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizka A, Antonelli A, Silvestro D. sampbias, a method for quantifying geographic sampling biases in species distribution data. Ecography. 2021;44(1):25–32. [Google Scholar]