Abstract
In multicellular eukaryotes, autophagy is a conserved process that delivers cellular components to the vacuole or lysosome for recycling during development and stress responses. Induction of autophagy activates AUTOPHAGY-RELATED PROTEIN 1 (ATG1) and ATG13 to form a protein kinase complex that initiates autophagosome formation. However, the detailed molecular mechanism underlying the regulation of this protein complex in plants remains unclear. Here, we determined that in Arabidopsis thaliana, the regulatory proteins 14-3-3λ and 14-3-3κ redundantly modulate autophagy dynamics by facilitating SEVEN IN ABSENTIA OF ARABIDOPSIS THALIANA (SINAT)-mediated proteolysis of ATG13a and ATG13b. 14-3-3λ and 14-3-3κ directly interacted with SINATs and ATG13a/b in vitro and in vivo. Compared to wild-type (WT), the 14-3-3λ 14-3-3κ double mutant showed increased tolerance to nutrient starvation, delayed leaf senescence, and enhanced starvation-induced autophagic vesicles. Moreover, 14-3-3s were required for SINAT1-mediated ubiquitination and degradation of ATG13a. Consistent with their roles in ATG degradation, the 14-3-3λ 14-3-3κ double mutant accumulated higher levels of ATG1a/b/c and ATG13a/b than the WT upon nutrient deprivation. Furthermore, the specific association of 14-3-3s with phosphorylated ATG13a was crucial for ATG13a stability and formation of the ATG1–ATG13 complex. Thus, our findings demonstrate that 14-3-3λ and 14-3-3κ function as molecular adaptors to regulate autophagy by modulating the homeostasis of phosphorylated ATG13.
14-3-3λ and 14-3-3κ function redundantly in regulating autophagy initiation by facilitating SINAT1 and SINAT2-mediated degradation of ATG13 proteins.
Introduction
The ubiquitin proteasome system (UPS) and autophagy are the two predominant pathways for protein quality control in eukaryotes (Balchin et al., 2016). In particular, the UPS degrades short-lived proteins marked with ubiquitin (Ub), while autophagy recycles long-lived proteins or cytosolic components (Klionsky and Emr, 2000; Ding et al., 2018). In yeast (Saccharomyces cerevisiae), mammals, and plants, autophagy is a conserved membrane trafficking mechanism that delivers protein aggregates, cytoplasmic components, and damaged organelles to the lysosome or vacuole for breakdown by resident hydrolases (Bassham, 2009; Li and Vierstra, 2012; Zhuang et al., 2015; Marshall and Vierstra, 2018; Avin-Wittenberg, 2019; Qi et al., 2021).
Three distinct types of autophagy have been described in plants: microautophagy, mega-autophagy, and macroautophagy (Marshall and Vierstra, 2018; Qi et al., 2021). Macroautophagy (hereafter referred to as autophagy) is the primary form of autophagy and begins with the initiation and expansion of the phagophore, which elongates and encircles cytoplasmic components to form a double-membrane structure termed the autophagosome. Thereafter, the outer membrane of the mature autophagosome fuses with the tonoplast to release inner single-membrane structures termed autophagic bodies containing the cellular components into the vacuolar lumen, where the sequestered cargo is degraded (Liu and Bassham 2012; Yang and Bassham 2015; Soto-Burgos et al., 2018; Qi et al., 2021).
Over 40 autophagy-related proteins (ATGs) of the core autophagic machinery have been identified in plants (Marshall and Vierstra, 2018; Yoshimoto and Ohsumi, 2018; Zhuang et al., 2018; Avin-Wittenberg, 2019). These proteins assemble four functional protein complexes: (1) the ATG1–ATG13 protein kinase complex; (2) the ATG6–phosphatidylinositol 3-kinase (PI3K) complex; (3) the ATG9 membrane delivery complex; and (4) two Ub-like conjugation systems (ATG5–ATG12 and ATG8–phosphatidylethanolamine). In Arabidopsis (Arabidopsis thaliana), genetic analyses have shown that the inactivation of ATG genes results in conventional autophagy-deficient phenotypes, such as premature leaf senescence, shorter life cycle under normal growth conditions, increased susceptibility to nutrient deprivation, altered biotic and abiotic stress tolerance, and activated innate immunity (Doelling et al., 2002; Xiong et al., 2007; Hayward et al., 2009; Liu et al., 2009; Chen et al., 2015; Liu et al., 2018).
In the autophagic machinery, the ATG1–ATG13 complex is a tightly controlled kinase regulator that initiates autophagosome formation (Suttangkakul et al., 2011). Specifically, posttranslational modifications such as phosphorylation and ubiquitination play a key role in the regulation of autophagy by modulating the activity or stability of components of the ATG1–ATG13 complex (Qi et al., 2021). For example, in yeast and mammals, the kinase TARGET OF RAPAMYCIN (TOR) interacts with its effectors RAPTOR and LETHAL WITH SEC THIRTEEN8 to stimulate its kinase activity, leading to the phosphorylation of ATG13 and diminishing the affinity of the ATG1–ATG13 complex under nutrient-rich conditions. In contrast, starvation or rapamycin treatment inactivates TOR, allowing ATG13 to accumulate in its dephosphorylated form, which promotes the formation of an active ATG1–ATG13 kinase complex for autophagy induction (Kamada et al., 2010; Suttangkakul et al., 2011; Liu and Bassham, 2012; Li et al., 2014; Liu and Xiong, 2022). Moreover, the abundance of UNC-51-LIKE KINASE1 (ULK1), a mammalian homolog of yeast Atg1p, is stabilized by the E3 Ub ligase TUMOR NECROSIS FACTOR RECEPTOR-ASSOCIATED FACTOR6 (TRAF6) through lysine 63 (K63)-linked ubiquitination (Nazio et al., 2013). Furthermore, ULK1 is also governed by K48-linked ubiquitination, which triggers ULK1 degradation by the 26S proteasome pathway to terminate autophagy (Liu et al., 2016a).
Overexpression of Arabidopsis TOR suppresses autophagy, whereas lowering TOR levels via RNA interference constitutively activates autophagy (Liu and Bassham, 2010; Pu et al., 2017), indicating that TOR is a conserved negative regulator of autophagy in plants. Increasing evidence suggests that upon induction of autophagy, dephosphorylated ATG13 proteins interact with ATG1 kinases, increasing their affinity for the accessory protein subunits ATG11 and ATG101 in Arabidopsis (Suttangkakul et al., 2011; Liu and Bassham, 2012; Li et al., 2014). More recently, we showed that members of the RING-type E3 Ub ligase protein family SEVEN IN ABSENTIA OF ARABIDOPSIS THALIANA (SINAT) differentially regulate the stability of the ATG1–ATG13 complex by modulating the proteolysis of ATG13, thereby helping to regulate autophagy (Qi et al., 2020, 2022). These results suggested that, as in mammals, the activity and stability of constituents of the Arabidopsis ATG1–ATG13 complex are strongly affected by phosphorylation and ubiquitination during autophagosome formation. However, the molecular mechanism by which the interplay between ubiquitination and phosphorylation regulates autophagy dynamics remains largely unknown.
In this study, we demonstrated that two Arabidopsis 14-3-3 proteins, 14-3-3λ and 14-3-3κ, function redundantly in the regulation of autophagosome formation by modulating the SINAT1- and SINAT2-mediated stability of ATG13a. Moreover, we revealed that under differential nutrient conditions, the specific association of 14-3-3 proteins with phosphorylated ATG13a acts as a molecular switch for the maintenance of the ATG1–ATG13 complex and autophagosome formation.
Results
SINATs interact with 14-3-3λ and 14-3-3κ in vivo
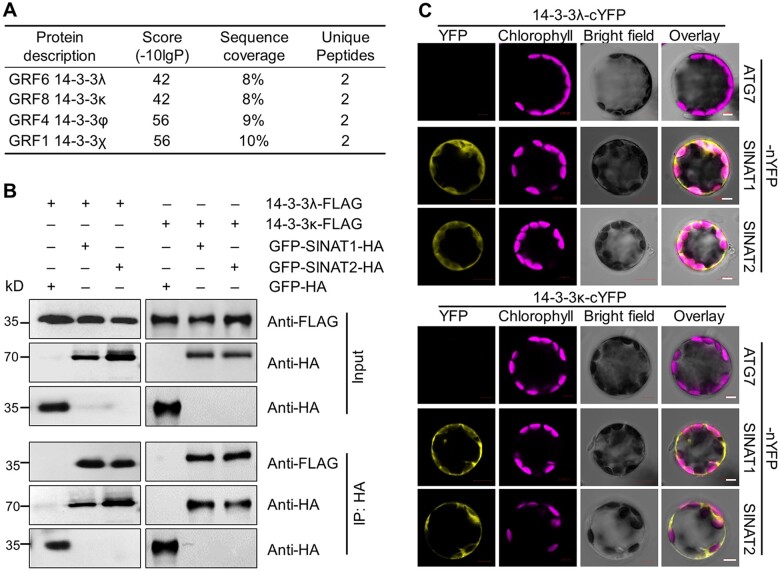
We previously showed that members of the SINAT family of E3 Ub ligases are involved in autophagy via the ubiquitination and degradation of ATG6 (Qi et al., 2017) and ATG13 (Qi et al., 2020) in Arabidopsis. To further investigate the regulatory network of SINATs, we screened for interactors of SINAT1 by immunoprecipitation followed by mass spectrometry (IP-MS) (Xia et al., 2020). In addition to 34 candidate proteins primarily associated with protein degradation systems (Xia et al., 2020), we discovered that four members of the 14-3-3 protein family, 14-3-3λ (also named GENERAL REGULATORY FACTOR 6 [GRF6]), 14-3-3κ (GRF8), 14-3-3φ (GRF4), and 14-3-3χ (GRF1), likely interact with SINAT1 (Figure 1A).
Figure 1.
SINATs interact with 14-3-3λ and 14-3-3κ proteins in vivo. A, Screening for SINAT1-interacting proteins by IP-MS analysis. Total protein was extracted from GFP-SINAT1-HA transgenic seedlings grown on MS medium, immunoprecipitated by anti-GFP agarose beads, and then subjected to in-gel digestion and MS analysis. Transgenic seedlings expressing free GFP-HA were used as a negative control. B, In vivo Co-IP assay validating the association between SINATs (SINAT1 and SINAT2) and 14-3-3s (14-3-3λ and 14-3-3κ). Constructs encoding 14-3-3λ-FLAG or 14-3-3κ-FLAG and GFP-SINAT1-HA or GFP-SINAT2-HA were transiently co-transfected into protoplasts from WT plants (Col-0) and incubated under light conditions for 16 h before immunoprecipitation with HA affinity agarose beads. GFP-HA was co-transfected with 14-3-3λ-FLAG or 14-3-3κ-FLAG as a negative control. Numbers on the left indicate the molecular weight (kDa) of each band. C, BiFC assay of 14-3-3s (14-3-3λ and 14-3-3κ) and SINATs (SINAT1 and SINAT2) in Arabidopsis protoplasts. Constructs encoding the split nYFP fusions SINAT1-nYFP or SINAT2-nYFP and the cYFP fusions 14-3-3λ-cYFP or 14-3-3κ-cYFP were co-transfected in leaf protoplasts and incubated for 16 h under light conditions. The vector pairs 14-3-3λ-cYFP + ATG7-nYFP and 14-3-3κ-cYFP + ATG7-nYFP were co-transfected as negative controls. Confocal micrographs obtained from YFP, chlorophyll autofluorescence, brightfield, and merged images are shown. Scale bars, 5 μm.
To validate the potential protein–protein interaction between SINATs and 14-3-3 proteins, we performed a yeast two-hybrid (Y2H) assay to assess the association of the four 14-3-3 proteins (14-3-3λ, 14-3-3κ, 14-3-3χ, and 14-3-3φ) identified by IP-MS with SINAT proteins, using 14-3-3ω (GRF2) as a control. We included several members and forms of the SINAT family, including SINAT1, SINAT2, SINAT5-S1 (encoded by a splice variant found in the Columbia-0 (Col-0) accession that produces a truncated SINAT5 lacking the RING finger domain; Qi et al., 2017; Zhang et al., 2019), SINAT6, SINAT1C, and SINAT2C (SINAT1 or SINAT2 harboring a mutation of a conserved Cys residue to Ser in the RING finger domain to impair E3 ligase activity; Xia et al., 2020). We determined that among these 14-3-3 proteins, only 14-3-3κ interacts with all SINATs, while 14-3-3λ showed a weak interaction with SINAT5-S1 in yeast cells (Supplemental Figure S1). In contrast, 14-3-3χ, 14-3-3φ, and 14-3-3ω did not interact with SINATs in the Y2H assay (Supplemental Figure S1A).
To further test the possible involvement of 14-3-3 proteins in regulating autophagy, we performed reverse transcription-quantitative PCR (RT–qPCR) to measure the expression levels of five genes encoding 14-3-3s in 1-week-old wild-type (WT) seedlings at various time points (0, 24, 48, or 72 h) during carbon (C) starvation treatment. As shown in Supplemental Figure S1B, the transcripts of 14-3-3λ and 14-3-3κ were significantly upregulated, while those of 14-3-3χ, 14-3-3φ, and 14-3-3ω were significantly downregulated in response to fixed-C starvation. These findings suggest that 14-3-3λ and 14-3-3κ likely function in regulating SINAT-mediated autophagy.
We confirmed the interaction of both 14-3-3λ and 14-3-3κ with SINAT1 and SINAT2 fused to the green fluorescent protein (GFP) and harboring a HA tag, as evidenced by in vivo co-immunoprecipitation (Co-IP) assays with an anti-HA antibody in WT Arabidopsis protoplast cells co-transfected with the relevant constructs (Figure 1B). To test the specificity of the interaction, we co-transfected protoplasts with GFP-HA and 14-3-3-FLAG constructs and failed to co-immunoprecipitate 14-3-3s or SINATs with anti-HA antibodies (Figure 1B).
We also used bimolecular fluorescence complementation (BiFC) analysis to confirm the interaction between 14-3-3λ or 14-3-3κ and SINAT1 or SINAT2 in Arabidopsis cells. We transiently co-transfected the appropriate pairs of constructs into WT protoplasts and incubated them under continuous light for 16 h before assessing fluorescence resulting from the reconstitution of yellow fluorescent protein (YFP). In agreement with the Co-IP results, we observed YFP fluorescence when SINAT1-nYFP and SINAT2-nYFP constructs were transiently co-transfected with 14-3-3λ-cYFP or 14-3-3κ-cYFP, respectively (Figure 1C). In contrast, co-transfection of the constructs 14-3-3s-cYFP and ATG7-nYFP failed to yield YFP signals in Arabidopsis leaf protoplasts, as expected (Figure 1C). These results suggest that 14-3-3λ and 14-3-3κ form a protein complex with SINAT1 and SINAT2 in vitro and in vivo.
14-3-3λ and 14-3-3κ predominately localize to the cytoplasm and the nucleus
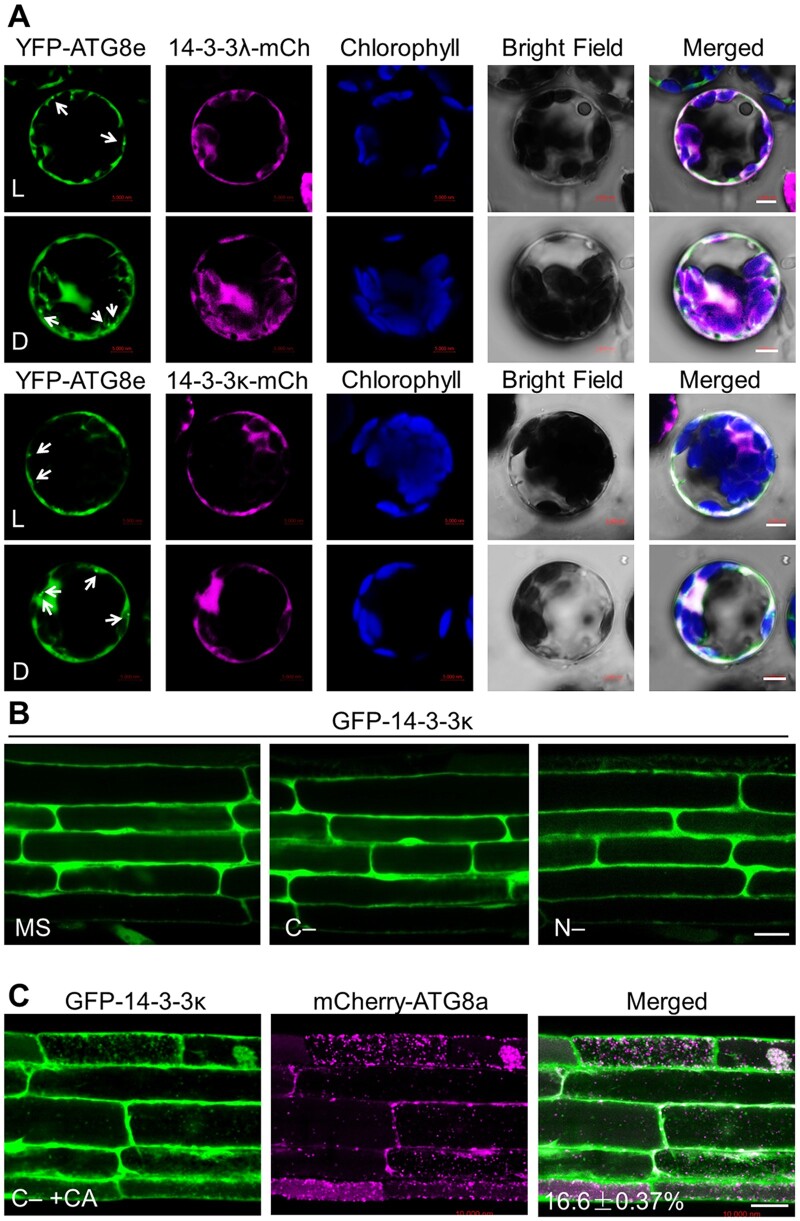
To explore the involvement of 14-3-3s in regulating autophagy, we determined the subcellular localization of GFP-14-3-3 fusion proteins. Confocal microscopy analyses showed that GFP-14-3-3λ and GFP-14-3-3κ predominantly localize to the cytoplasm and the nucleus of transfected Col-0 leaf protoplasts when incubated under constant light conditions (Supplemental Figure S2). Moreover, constant darkness treatment did not change the localization of GFP-14-3-3λ or GFP-14-3-3κ (Supplemental Figure S2). We also co-transfected protoplasts prepared from the WT accession Col-0 with constructs encoding 14-3-3λ-mCherry (a fusion between 14-3-3λ and the red fluorescent protein [RFP] mCherry) or 14-3-3κ-mCherry with the autophagy marker YFP-ATG8e (Qi et al., 2017). While we detected YFP fluorescence in punctate dots marking the site of ATG8e action, neither 14-3-3λ-mCherry nor 14-3-3κ-mCherry colocalized with the mCherry signal under light or dark conditions (Figure 2A).
Figure 2.
Subcellular distribution of 14-3-3 proteins. A, Colocalization of mCherry-tagged 14-3-3λ (14-3-3λ-mCh) or 14-3-3κ (14-3-3κ-mCh) fusions with the autophagy marker YFP-ATG8e in Arabidopsis protoplasts under light (L) or dark (D) conditions. The 14-3-3λ-mCherry or 14-3-3κ-mCherry construct was co-transfected with YFP-ATG8e into Arabidopsis protoplasts and incubated for 16 h, followed by confocal microscopy analysis. Arrows indicate ATG8-labeled autophagic puncta. Confocal images obtained from GFP, mCherry, chlorophyll autofluorescence, brightfield, and merged images are shown. Scale bars, 5 µm. B, Examination of GFP-14-3-3κ subcellular distribution pattern. One-week-old transgenic seedlings expressing GFP-14-3-3κ and grown on MS medium under normal growth conditions (16-h light/8-h dark) were exposed to C- and N-sufficient (MS) or C- or N-deficient (C– or N–) conditions for 16 h. GFP fluorescence was observed by confocal microscopy. Scale bar, 20 µm. C, Evaluation of the colocalization between GFP-14-3-3κ and the autophagy marker mCherry-ATG8a in transgenic plants expressing GFP-14-3-3κ and mCherry-ATG8a. Transgenic seedlings expressing GFP-14-3-3κ and mCherry-ATG8a were grown on MS medium under normal growth conditions for 6 days before being exposed to C-sufficient (MS) or C-deficient conditions in the presence of 1 µM CA (C–+CA) for 16 h. GFP and mCherry fluorescence was then visualized by confocal microscopy. The number in the merged image indicates the colocalization ratio (number of colocalization puncta over total GFP-14-3-3λ puncta). The percentage is the mean ± sd (n = 3) of three independent experiments. For each experiment, three images were used for analysis. Scale bar, 20 µm.
We also generated stable transgenic lines harboring a GFP-14-3-3κ-HA transgene (Supplemental Figure S3). We transferred 1-week-old GFP-14-3-3κ-HA seedlings grown under normal light/dark conditions to sucrose- or nitrogen (N)-deficient medium to induce C (C–) or N (N–) starvation. Confocal laser scanning microscopy of GFP fluorescence showed that the GFP-14-3-3κ-HA fusion localizes to the cytoplasm and the nucleus in both leaf epidermal cells and mature root cells under these conditions (Figure 2B; Supplemental Figure S3C), which was consistent with the results from transient transfections in protoplasts.
The GFP fluorescence pattern above appeared to be different from the puncta typically observed with components of the autophagy complex. Accordingly, we crossed the GFP-14-3-3κ transgenic plants to a transgenic line carrying the autophagy vesicle marker mCherry-ATG8a to generate GFP-14-3-3κ mCherry-ATG8a plants. After a 16-h C starvation treatment in the presence of concanamycin A (CA) (C–+CA), which leads to the accumulation of autophagic bodies, we noticed the marked induction of mCherry-labeled autophagic puncta in 1-week-old GFP-14-3-3κ mCherry-ATG8a root mature cells (Figure 2C). Only 16.6% of GFP-labeled punctate foci colocalized with the mCherry-ATG8 autophagic puncta under the same conditions (Figure 2C). These results suggest that the 14-3-3λ and 14-3-3κ proteins are predominately distributed in the cytosol and the nucleus, and do not appear to be associated with autophagic bodies in Arabidopsis cells.
Plants lacking 14-3-3λ and 14-3-3κ function are more tolerant to nutrient deprivation
To investigate the biological functions of 14-3-3λ and 14-3-3κ proteins, we identified several independent T-DNA insertional mutants (Supplemental Figure S4, A and B) for each 14-3-3 gene. RT–PCR analyses indicated that the full-length transcripts of 14-3-3λ and 14-3-3κ are undetectable in the corresponding 14-3-3λ or 14-3-3κ mutants (Supplemental Figure S4C), indicating that all these lines are likely null mutants.
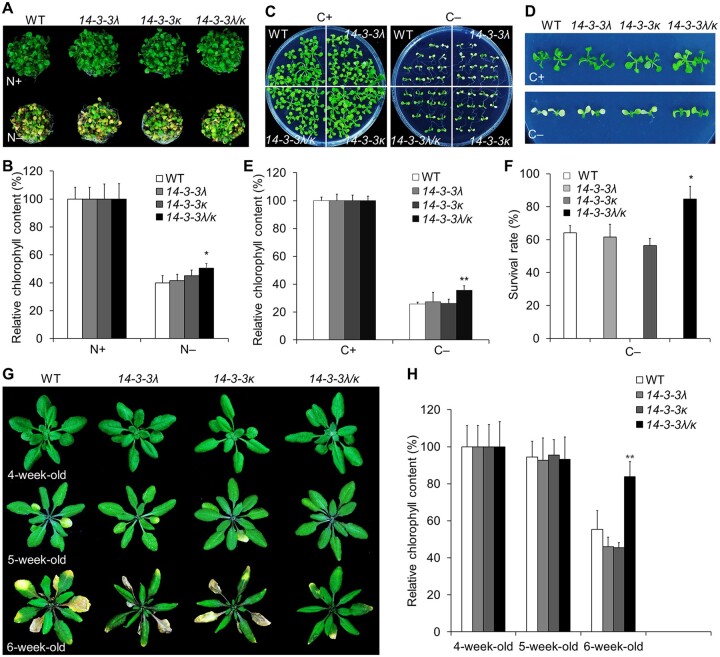
To test whether the 14-3-3 mutants affect processes that involve autophagy, we examined how 14-3-3 mutants responded to nutrient deprivation, as well as their natural leaf senescence. When grown in nutrient-rich or nutrient-limited conditions, all 14-3-3λ (two alleles) and 14-3-3κ (three alleles) homozygous single mutants appeared similar to WT seedlings (Supplemental Figure S5). We also obtained a 14-3-3λ 14-3-3κ double mutant (14-3-3λ/κ; Zhou et al., 2014) to rule out partial redundancy. When grown under nutrient-rich conditions, 14-3-3λ 14-3-3κ double mutant seedlings did not display obvious phenotypic differences compared to the WT or the 14-3-3λ and 14-3-3κ single mutants (Figure 3, A, C, and D). In contrast, the 14-3-3λ 14-3-3κ double mutant showed a substantially higher tolerance when grown in N-free Murashige and Skoog (MS) medium or after fixed C starvation (Figure 3, A–F). The leaves of the 14-3-3λ 14-3-3κ double mutant retained higher relative chlorophyll contents in comparison to WT and the 14-3-3λ and 14-3-3κ single mutant seedlings following N starvation in liquid medium for 4 days (Figure 3, A and B). Similarly, the 14-3-3λ 14-3-3κ double mutant showed enhanced tolerance to fixed C starvation for 7 days after growth in C-rich conditions for 1 week, with green leaves accumulating higher chlorophyll contents compared to those of WT seedlings (Figure 3, C–E). In addition, ∼80% of the 14-3-3λ 14-3-3κ double mutant seedlings survived 1 week after return to normal light/dark conditions, whereas >40% of the WT and 14-3-3λ and 14-3-3κ single mutant seedlings died after the same treatment (Figure 3F).
Figure 3.
Mutations in 14-3-3λ and 14-3-3κ confer increased tolerance to C and N starvation. A, Phenotypes of the 14-3-3λ and 14-3-3κ single mutants and the 14-3-3λ 14-3-3k double mutant (14-3-3λ/κ) in response to N starvation. One-week-old WT, 14-3-3λ-2 (14-3-3λ), 14-3-3κ-3 (14-3-3κ) single mutant, and 14-3-3λ 14-3-3k double mutant seedlings grown on MS medium were transferred to N-rich (N+) or N-deficient (N–) liquid medium and photographed 4 days later. B, Relative chlorophyll contents of WT, 14-3-3λ, 14-3-3κ, and 14-3-3λ 14-3-3k seedlings grown under N+ or N– conditions shown in (A). Chlorophyll contents under N+ conditions were set to 100%. Data are means ± sd calculated from four independent experiments. For each experiment, four technical replicates pooling 15 seedlings each were used per genotype (*P < 0.05 by Student’s t test). C and D, Phenotypes of the 14-3-3λ and 14-3-3κ single mutants and the 14-3-3λ 14-3-3k double mutant in response to fixed C starvation. One-week-old WT, 14-3-3λ-2 (14-3-3λ), 14-3-3κ-3 (14-3-3κ) single mutant, and 14-3-3λ 14-3-3k double mutant seedlings grown on MS medium were transferred to MS plates with sucrose (C+) or without sucrose followed by constant dark treatment (C–) for 7 days. The images were recorded after a 7-day recovery. E and F, Survival rates (E) and relative chlorophyll contents (F) of WT, 14-3-3λ, 14-3-3κ, and 14-3-3λ 14-3-3k seedlings described in (C) and (D) following recovery. Chlorophyll contents under C+ conditions were set to 100%. Data are means ± sd calculated from three independent experiments. For each experiment, four technical replicates pooling 15 seedlings each were used per genotype (*P < 0.05; **P < 0.01 by Student’s t test). G, Images showing the onset of leaf senescence in WT, 14-3-3λ, 14-3-3κ, and 14-3-3λ 14-3-3k plants grown under normal light/dark growth conditions. Photographs were taken at 4, 5, and 6 weeks after germination. H, Relative chlorophyll contents of plants grown under normal light/dark growth conditions for the indicated times. The values from 4-week-old plants were set to 100%. Data are means ± sd calculated from four independent experiments. For each experiment, five entire plants (technical replicates) were used per genotype. Asterisks indicate significant differences from WT (**P < 0.01 by Student’s t test).
To assess naturally occurring leaf senescence, we grew all genotypes for up to 6 weeks on soil under light/dark conditions. Four-week-old 14-3-3λ 14-3-3κ double mutant plants showed few morphological differences relative to the WT or 14-3-3λ or 14-3-3κ single mutant plants (Figure 3G). The 14-3-3λ 14-3-3κ double mutant showed delayed onset of senescence for rosette leaves in 5- and 6-week-old plants compared to the WT or single mutant plants (Figure 3G). Consistent with this observation, 6-week-old 14-3-3λ 14-3-3κ double mutant plants accumulated more chlorophyll than the WT or the 14-3-3λ and 14-3-3κ single mutant plants (Figure 3H). Together, these results indicate that the 14-3-3λ 14-3-3κ double mutant shows enhanced tolerance to nutrient starvation and delayed leaf senescence.
14-3-3λ and 14-3-3κ inhibit starvation-induced formation of autophagic vesicles
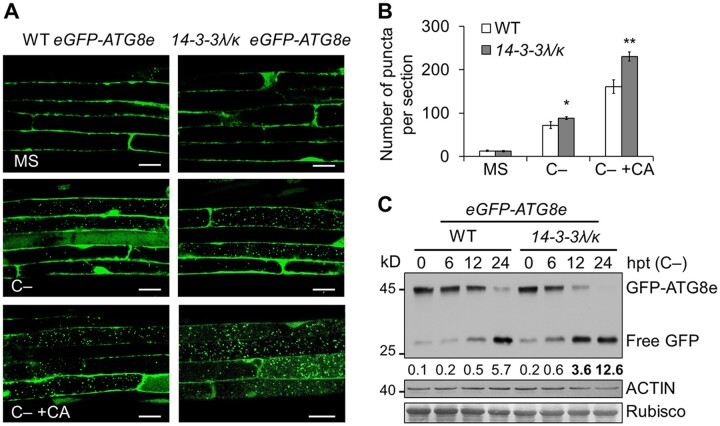
To further assess the role of 14-3-3λ and 14-3-3κ in autophagy, we crossed the 14-3-3λ 14-3-3κ double mutant to the eGFP-ATG8e transgenic line, a well-characterized autophagosome marker line (Xiao et al., 2010; Chen et al., 2017a), to generate the 14-3-3λ 14-3-3κ eGFP-ATG8e line. A 16-h C starvation treatment induced the formation of GFP-labeled autophagic puncta in 1-week-old WT root cells, as determined by confocal microscopy (Figure 4, A and B). Under starvation conditions, significantly more puncta accumulated in the 14-3-3λ 14-3-3κ double mutant background compared to the WT background (Figure 4, A and B). For an independent confirmation of this result, we used the release of free GFP to monitor the autophagic flux and degradation of the GFP-ATG8e reporter into the vacuole (Chung et al., 2010; Chen et al., 2015; Qi et al., 2017). As with the microscopy results, C starvation induced the release of more free GFP in the 14-3-3λ 14-3-3κ double mutant compared to the WT (Figure 4C). Together, these results indicate that 14-3-3λ and 14-3-3κ are involved in regulating starvation-induced autophagic vesicle formation.
Figure 4.
14-3-3 proteins negatively regulate autophagosome formation in response to C starvation. A, Confocal analysis of eGFP-ATG8e and 14-3-3λ 14-3-3k eGFP-ATG8e (14-3-3λ/κ eGFP-ATG8e) lines. One-week-old eGFP-ATG8e and 14-3-3λ 14-3-3k eGFP-ATG8e lines were subjected to C starvation (C–) conditions in the presence or absence of 1 µM CA for 16 h. The formation of autophagosomes was visualized by fluorescence confocal microscopy. Scale bars, 20 µm. B, Number of puncta per root section in root cells from the WT and the 14-3-3λ 14-3-3k double mutant (14-3-3λ/κ) in (A). Data are means ± sd calculated from three independent experiments. For each experiment, 15 sections were used for the calculation for each genotype. Asterisks indicate significant differences from the WT (*P < 0.05; **P < 0.01 by Student’s t test). C, Immunoblot analysis showing the processing of GFP-ATG8e in WT and 14-3-3λ 14-3-3k seedlings in response to C starvation (C–). One-week-old seedlings were collected at 0, 6, 12, and 24-h posttransfer (hpt). Anti-GFP antibodies were used for immunoblotting. The position of GFP-ATG8e fusion and free GFP are indicated on the right. The ratio between free GFP and GFP-ATG8e is shown below. The numbers on the left indicate the molecular weight (kDa) of each size marker. Anti-ACTIN antibodies and Ponceau S-stained membranes are shown below the blot to indicate the amount of protein loaded per lane.
14-3-3λ and 14-3-3κ physically interact with ATG13a
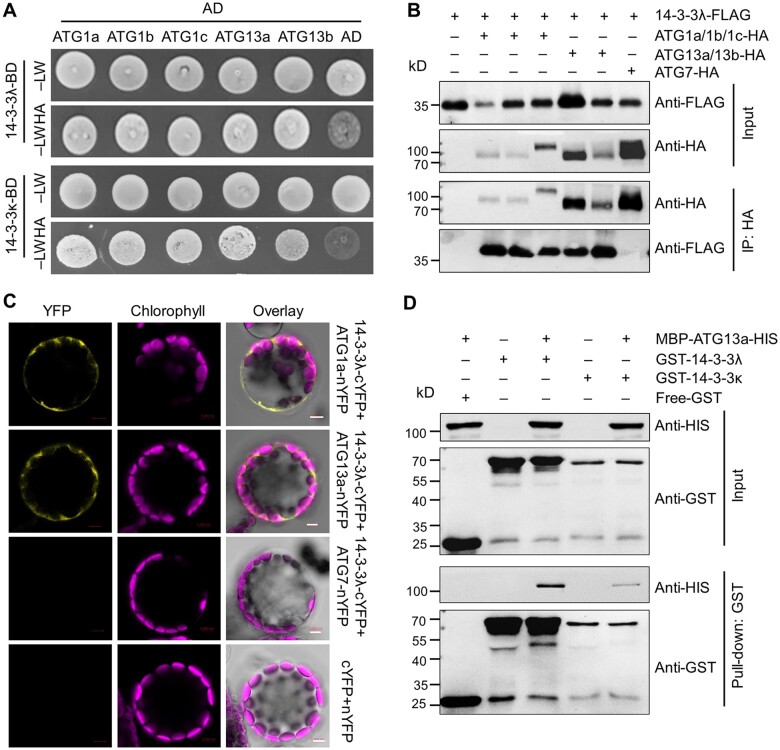
To understand the molecular mechanism underlying the involvement of 14-3-3 proteins in autophagy, we generated 14-3-3λ-FLAG and 14-3-3κ-FLAG transgenic plants (Supplemental Figure S6) to screen for 14-3-3 interacting partners in Arabidopsis by IP-MS analysis. Using 14-3-3λ as a bait pulled down with anti-FLAG antibodies, we determined that the autophagy-associated proteins ATG13a and ATG13b potentially interact with 14-3-3λ (Supplemental Figure S7), suggesting that 14-3-3s may regulate the ATG1–ATG13 kinase complex. To test this possibility, we performed an Y2H assay and confirmed that 14-3-3λ and 14-3-3κ interact with ATG1a, ATG1b, ATG1c, ATG13a, and ATG13b, which are members of the ATG1–ATG13 complex, in yeast cells (Figure 5A).
Figure 5.
14-3-3λ and 14-3-3κ physically interact with ATG13a. A, Y2H assay of the interaction of 14-3-3λ and 14-3-3κ with ATG1/ATG13 proteins (ATG1a, ATG1b, ATG1c, ATG13a, and ATG13b). Full-length 14-3-3λ or 14-3-3κ was fused to the GAL4 DNA-binding domain (BD) and co-transformed with ATG1a-AD, ATG1b-AD, ATG1c-AD, ATG13a-AD, and ATG13b-AD in yeast strain YH109. The positive clones were selected on SD medium lacking Trp, Leu, His, and Ade (–LWHA) containing 5-mM 3-AT. AD indicates the empty AD plasmid. B, In vivo Co-IP assay confirming the association between ATG1/13 (ATG1a, ATG1b, ATG1c, ATG13a, and ATG13b) and 14-3-3λ. Constructs encoding FLAG-tagged 14-3-3λ (14-3-3λ-FLAG) and HA-tagged ATG1/13 (ATG1a-HA, ATG1b-HA, ATG1c-HA, ATG13a-HA, or ATG13b-HA) were transiently co-transfected into Col-0 protoplasts and incubated under light conditions for 16 h, before immunoprecipitation with HA affinity agarose beads. Protoplasts transfected with ATG7-HA and 14-3-3λ-FLAG served as a negative control. C, BiFC assay between ATG1/ATG13 proteins (ATG1a and ATG13a) and 14-3-3λ in Arabidopsis protoplasts. Constructs encoding the split nYFP fusions ATG1a-nYFP, ATG13a-nYFP, or ATG7-nYFP and 14-3-3λ-cYFP were co-transfected in leaf protoplasts and incubated for 16 h under light conditions. The 14-3-3λ-cYFP+ATG7-nYFP and nYFP+cYFP vector pairs were co-transfected as negative controls. Confocal micrographs obtained from YFP, chlorophyll autofluorescence, brightfield, and merged images are shown. Bars = 5 μm. D, Pull-down assay showing the interaction between GST-14-3-3λ or GST-14-3-3κ and MBP-ATG13a-HIS. Recombinant GST-14-3-3λ or GST-14-3-3κ and MBP-ATG13a-HIS were incubated in pull-down assay buffer, followed by incubation with glutathione Sepharose 4B beads (GE Healthcare). The bound proteins were analyzed by immunoblotting using anti-HIS and anti-GST antibodies. Free GST and MBP-ATG13a-HIS were used as negative controls. Numbers on the left indicate the molecular weight (kDa) of each band.
To extend these results in planta, we transiently co-transfected WT leaf mesophyll protoplasts with constructs encoding ATG13a-HA, ATG13b-HA, ATG1a-HA, ATG1b-HA, or ATG1c-HA with 14-3-3λ-FLAG or 14-3-3κ-FLAG. We then incubated all transfected protoplasts under continuous light conditions for 16 h before Co-IP assays. Immunoblot analysis revealed that 14-3-3λ-FLAG and 14-3-3κ-FLAG are immunoprecipitated by ATG13a-HA, ATG13b-HA, ATG1a-HA, ATG1b-HA, and ATG1c-HA, but not by the control ATG7-HA, when protein extracts were incubated with anti-HA antibodies for immunoprecipitation (Figure 5B; Supplemental Figure S8A). We also confirmed the interaction of ATG1s and ATG13s with 14-3-3λ and 14-3-3κ by BiFC assays. For this experiment, we co-transfected protoplasts with constructs encoding ATG1a-nYFP, ATG1b-nYFP, ATG1c-nYFP, ATG13a-nYFP, or ATG13b-nYFP, with 14-3-3λ-cYFP, or 14-3-3κ-cYFP, followed by a 16-h incubation under continuous light or dark conditions, after which we detected YFP fluorescence in the cytoplasm for all combinations and both growth conditions (Figure 5C; Supplemental Figure S8, B and C). In contrast, co-transfection of the negative controls 14-3-3λ-cYFP/ATG7-nYFP, 14-3-3κ-cYFP/ATG7-nYFP, and nYFP/cYFP failed to reconstitute YFP fluorescence, as expected (Figure 5C; Supplemental Figure S8, B and C).
To independently validate the direct association between 14-3-3λ/κ and ATG1/ATG13 proteins, we performed an in vitro pull-down assay. Accordingly, we incubated purified recombinant ATG13a fused to maltose-binding protein (MBP) and a histidine tag (MBP-ATG13a-HIS) with glutathione S-transferase (GST)-tagged 14-3-3s (GST-14-3-3λ and GST-14-3-3κ) or with the negative control GST. We then used glutathione beads to isolate the GST fusion proteins and any interacting proteins, which revealed that GST-14-3-3λ and GST-14-3-3κ can pull down MBP-ATG13a-HIS (Figure 5D). GST did not pull down MBP-ATG13a-HIS (Figure 5D). We conducted a similar assay by incubating recombinant HIS-tagged HIS-14-3-3λ or HIS-14-3-3κ proteins with GST-ATG1a or GST, and isolated the interacting proteins using Ni-NTA resin. Both HIS-14-3-3λ and HIS-14-3-3κ successfully pulled down GST-ATG1a, but not GST (Supplemental Figure S9). Taken together, our findings suggest that 14-3-3λ and 14-3-3κ are likely involved in autophagy by directly associating with the ATG1–ATG13 protein complex.
14-3-3λ and 14-3-3κ are essential for SINAT1-mediated ubiquitination and degradation of ATG13a
We previously suggested that Arabidopsis SINAT1–4 regulate ubiquitination and degradation of ATG13s to help modulate autophagosome formation in response to nutrient starvation (Qi et al., 2020). Given that 14-3-3 proteins interact with both SINATs and ATG13a in vitro and in vivo (Figures 1 and 5) and play a negative role in regulating starvation-induced autophagosome formation in planta (Figure 4), we speculated that 14-3-3λ and 14-3-3κ may act as adaptors for the SINAT-mediated ubiquitination and degradation of ATG13s.
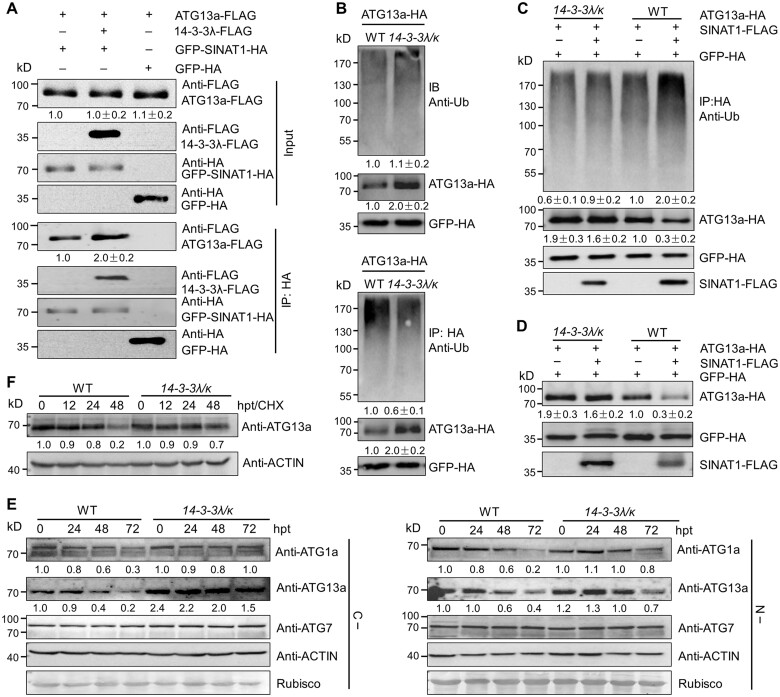
To test this possibility, we compared the interaction of ATG13a and SINATs in the presence or absence of 14-3-3λ. To this end, we co-transformed ATG13a-FLAG and GFP-SINAT1-HA with or without 14-3-3λ-FLAG into protoplasts isolated from WT rosettes. Co-IP analysis revealed that both ATG13a-FLAG and 14-3-3λ-FLAG were immunoprecipitated by GFP-SINAT1-HA, but much more ATG13a-FLAG was immunoprecipitated in the presence of 14-3-3λ-FLAG (Figure 6A), indicating that 14-3-3λ and 14-3-3κ are involved in mediating the interaction between SINAT1 and ATG13a.
Figure 6.
14-3-3λ and 14-3-3κ regulate the SINAT1-mediated ubiquitination and degradation of ATG13a. A, 14-3-3 proteins increase the interaction of SINAT1 and ATG13a. In vivo Co-IP assay showing the interaction between ATG13a-FLAG and GFP-SINAT1-HA in the presence of 14-3-3λ-FLAG. Constructs encoding ATG13a-FLAG and GFP-SINAT1-HA with or without 14-3-3λ-FLAG were transiently co-transfected into Col-0 protoplasts (Col-0) and incubated under light conditions for 16 h before immunoprecipitation with HA affinity agarose beads. GFP-HA was co-transfected with ATG13a-FLAG as negative control. Numbers on the left indicate the molecular weight (kDa) of each band. Relative intensity of the immunoprecipitated ATG13a-FLAG normalized to the GFP-SINAT1-HA is shown below. B, Ubiquitination of ATG13a in the 14-3-3λ 14-3-3k double mutant. ATG13a-HA was transiently transfected into Arabidopsis protoplasts isolated from 4-week-old WT and 14-3-3λ 14-3-3k mutant plants, and the ubiquitination of ATG13a was detected by immunoprecipitation and immunoblot analysis. Proteins were extracted after a 16-h incubation under constant light conditions, followed by the addition of HA affinity agarose beads. The proteins were detected with anti-HA and anti-Ub antibodies. Relative intensity of each protein normalized to the GFP-HA is shown below. The relative intensity of each protein is the mean ± sd calculated from three independent experiments. C, SINAT1-mediated ubiquitination of ATG13a is attenuated in the 14-3-3λ 14-3-3k double mutant. ATG13a-HA and SINAT1-FLAG constructs were transiently co-transfected into Arabidopsis protoplasts prepared from WT or 14-3-3λ 14-3-3k plants and incubated for 16 h under constant light conditions. ATG13a-HA ubiquitination was determined with anti-Ub and anti-HA antibodies. Relative intensity of each protein normalized to the GFP-HA is shown below. The relative intensity of each protein is the mean ± sd calculated from three independent experiments. D, SINAT1-associated degradation of ATG13a is dependent on 14-3-3λ and 14-3-3κ. ATG13a-HA and SINAT1-FLAG constructs were transiently co-transfected into Arabidopsis protoplasts prepared from WT and 14-3-3λ 14-3-3k double mutant plants and incubated for 16 h under continuous light conditions. ATG13a-HA stability was determined with anti-HA antibodies. Relative intensity of each protein normalized to GFP-HA is shown below. The relative intensity of each protein is the mean ± sd calculated from three independent experiments. E, ATG1a, ATG13a, and ATG7 protein abundance in 14-3-3λ 14-3-3k double mutant seedlings. One-week-old WT and 14-3-3λ 14-3-3k mutant seedlings were subjected to C (C–; upper images) or N starvation (N–; lower images) treatment for 24, 48, or 72 h. The blots were probed with anti-ATG1a, anti-ATG13a, and anti-ATG7 specific antibodies. Relative intensity of each protein normalized to the loading control is shown below. F, Stability of ATG13a protein in the 14-3-3λ 14-3-3k double mutant and WT seedlings upon CHX treatment. One-week-old WT and 14-3-3λ 14-3-3k double mutant seedlings were treated with 0.5-mM CHX for 0, 12, 24, or 48 h. Specific anti-ATG13a antibodies were used for immunoblot analysis. Relative intensity of each protein normalized to the loading control is shown below. Numbers on the left indicate the molecular weight (kDa) of each band. GFP-HA was used as control for transfection efficiency. Anti-ACTIN antibodies and Ponceau S-stained membranes are shown below the blots to indicate the amount of protein loaded per lane.
Furthermore, we transiently overexpressed the ATG13a-HA construct in protoplasts prepared from the WT and the 14-3-3λ 14-3-3κ double mutant and determined the resulting levels of ubiquitinated ATG13a. As shown in Figure 6B, total ATG13a ubiquitination levels were largely similar in WT and the 14-3-3λ 14-3-3κ double mutant. However, after immunoprecipitating ATG13a-HA with anti-HA antibodies and probing with anti-Ub antibodies, we observed much lower ubiquitination levels of ATG13a-HA in the 14-3-3λ 14-3-3κ double mutant compared to the WT, which was accompanied by a greater accumulation of ATG13a-HA in the double mutant than in WT (Figure 6B).
We confirmed the reduced levels of ubiquitinated ATG13a-HA in the 14-3-3λ 14-3-3κ double mutant using tandem Ub-binding entities (TUBEs) agarose beads to isolate ubiquitinated proteins, followed by immunoblotting with anti-HA antibodies (Supplemental Figure S10). To determine what effect, if any, SINATs have on ATG13a abundance, we co-transfected ATG13a-HA and SINAT1-FLAG constructs into protoplasts isolated from WT and 14-3-3λ 14-3-3κ double mutant rosettes. We determined that SINAT1-FLAG increases the levels of ATG13a-HA ubiquitination in the WT background (Figure 6C; Qi et al., 2020; Supplemental Figure S11). In contrast, ATG13a-HA ubiquitination appeared much lower in protoplasts prepared from the 14-3-3λ 14-3-3κ double mutant relative to the WT, regardless of the presence of SINAT1-FLAG (Figure 6C; Supplemental Figure S11), suggesting that 14-3-3λ and 14-3-3κ are required for the SINAT-mediated ubiquitination of ATG13a. Moreover, we discovered that transient transfection of SINAT1-FLAG induces the degradation of ATG13a-HA in WT protoplasts, but the degradation was impaired in protoplasts prepared from the 14-3-3λ 14-3-3κ double mutant (Figure 6D; Supplemental Figure S11).
To investigate the effect of 14-3-3λ and 14-3-3κ in regulating the stability of the ATG1 and ATG13 proteins, we examined ATG1a and ATG13a abundance in 1-week-old seedlings from the WT and the 14-3-3λ 14-3-3κ double mutant using anti-ATG1a and anti-ATG13a-specific antibodies. Immunoblot analysis revealed that ATG1a and ATG13a accumulate to higher levels in the 14-3-3λ 14-3-3κ double mutant compared to WT under C- and N-deficient conditions (Figure 6E). As a control, ATG7 levels did not change in the 14-3-3λ 14-3-3κ double mutant relative to the WT under the same conditions (Figure 6E). To monitor the effects of 14-3-3λ and 14-3-3κ loss-of-function mutations on ATG13a stability, we incubated 1-week-old WT and 14-3-3λ 14-3-3κ double mutant seedlings with the protein translation inhibitor cycloheximide (CHX; Qi et al., 2020; Xia et al., 2020). Upon CHX treatment for 12, 24, or 48 h, ATG13a was more stable with a longer half-life in the 14-3-3λ 14-3-3κ double mutant than in WT seedlings, as evidenced by immunoblotting with an anti-ATG13a antibody (Figure 6F). This result supports the notion that 14-3-3λ and 14-3-3κ play a primary role in modulating ATG13a stability.
14-3-3λ and 14-3-3κ interact with phosphorylated ATG13a to modulate formation of the ATG1–ATG13 complex
Previous studies suggested that 14-3-3 proteins typically interact with their partners in a phosphorylation-dependent manner (Camoni et al., 2018). We identified 18 putative phosphorylation sites in ATG13a: S248, S259, S263, S341, S343, S337, Y351, T366, S397, S400, T401, S404, S406, S407, S422, S425, S473, and S528 (Supplemental Figure S12), 15 of which were identified by IP-MS analysis (Supplemental Figure S13), and 3 of which (S366, S400, and T401) were identified by Wang et al. (2022).
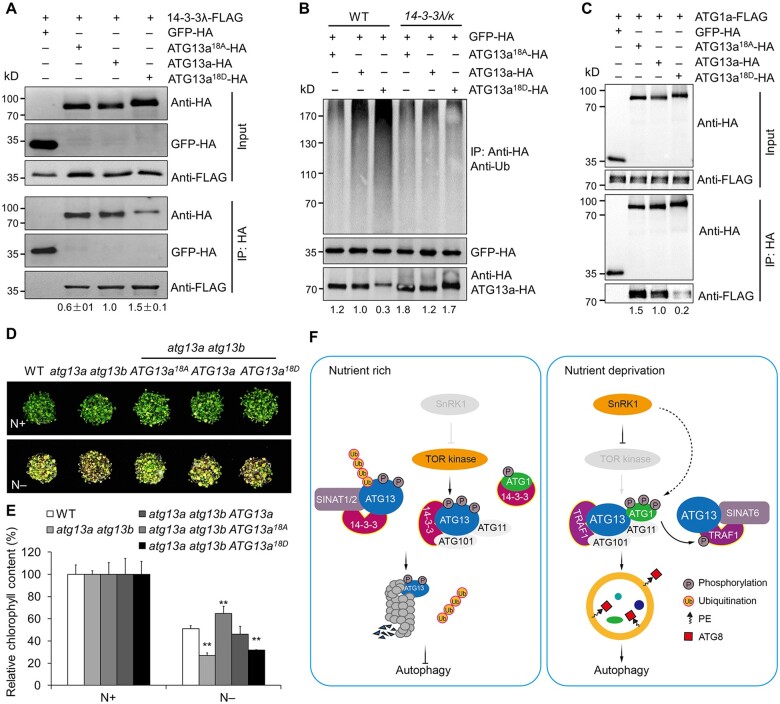
To test whether 14-3-3 proteins associate with phosphorylated ATG13a, we mutated all 18 putative sites to alanine (A), resulting in the nonphosphorylatable protein ATG13a18A, or to aspartic acid (D) to generate the phosphomimic protein ATG13a18D. We tested whether the ATG13a phosphorylation sites are recognized by 14-3-3λ by co-transfecting WT leaf mesophyll protoplasts with the ATG13a-HA, ATG13a18A-HA, or ATG13a18D-HA construct with the 14-3-3λ-FLAG construct, followed by immunoprecipitation with an anti-HA antibody and detection of 14-3-3λ with anti-FLAG antibodies. As shown in Figure 7A, the ATG13a18A variant decreased the interaction between ATG13a and 14-3-3λ, while the ATG13a18D variant increased their association.
Figure 7.
14-3-3s interact with phosphorylated ATG13a to modulate ATG13a degradation and ATG1a–ATG13a complex formation. A, In vivo Co-IP assay of the interaction between 14-3-3λ and ATG13a with different phosphorylation states. Constructs encoding 14-3-3λ-FLAG and HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently co-transfected into protoplasts from WT plants and incubated under light conditions for 16 h, before immunoprecipitation with HA affinity agarose beads. The blots were probed with anti-HA and anti-FLAG antibodies. Relative intensity of the immunoprecipitated 14-3-3λ-FLAG normalized to the different phosphorylation states of ATG13a is shown below. The relative intensity of each protein is the mean ± sd (n = 3) calculated from three independent experiments. B, Ubiquitination of ATG13a in different phosphorylation states in the 14-3-3λ 14-3-3k double mutant. Constructs encoding HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently transfected in Arabidopsis protoplasts isolated from 4-week-old WT and 14-3-3λ 14-3-3k mutant plants and incubated for 16 h under continuous light conditions before the ubiquitination and stability of ATG13a was determined by immunoblot analysis using anti-Ub and anti-HA antibodies. The relative intensity of each protein normalized to GFP-HA is shown below. C, Interaction between ATG13a in different phosphorylation states and ATG1a by Co-IP assay. Constructs encoding FLAG-tagged ATG1a (ATG1a-FLAG) and HA-tagged ATG13a in different phosphorylation states (ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA) were transiently co-transfected into protoplasts from WT plants and incubated under light conditions for 16 h. Total proteins were extracted and immunoprecipitated with HA affinity agarose beads; the blots were probed with anti-HA and anti-FLAG antibodies. Numbers on the left indicate the molecular weight (kDa) of each band. The relative intensity of the immunoprecipitated ATG1a-FLAG normalized to the different phosphorylation states of ATG13a is shown below. D, Sensitivity of the WT, atg13a atg13b double mutant, and atg13a atg13b ATG13a, atg13a atg13b ATG13a18A, atg13a atg13b ATG13a18D transgenic lines to N starvation. Seedlings were grown for 1 week on MS medium, followed by transfer to N-rich (N+) or N deficient (N–) medium for an additional 4 days. E, Relative chlorophyll contents of seedlings with or without N starvation shown in (D). The relative chlorophyll contents were calculated by comparing the values of N–treated versus N+-treated seedlings. Data are means ± sd (n = 3) calculated from three independent experiments. For each experiment, three technical replicates from pools of 15 seedlings each were used per genotype. Asterisks indicate significant differences from the WT (**P < 0.01 by Student’s t test). F, Working model for 14-3-3 family proteins in regulating autophagy in Arabidopsis. Under nutrient-rich conditions, the TOR kinase complex is active and phosphorylates ATG13a to maintain it in a hyperphosphorylated state. The 14-3-3 proteins associate with the phosphorylated form of ATG13a and act as adaptors to recruit the E3 Ub ligases SINAT1 and SINAT2 to form a protein complex that promotes the ubiquitination and degradation of ATG13 via the 26S proteasome pathway. Phosphorylated ATG13 also disassociates from ATG1, suppressing autophagy. Under nutrient starvation, however, SnRK1 inhibits TOR activity, resulting in ATG13 dephosphorylation, which impairs interaction with 14-3-3 proteins and forms a complex with hyperphosphorylated ATG1a mediated by the SnRK1. The scaffold proteins TRAF1s interact with ATG13a together with SINAT6, forming a protein complex that decreases SINAT1- and SINAT2-mediated ubiquitination and degradation of ATG13 to induce autophagy. Furthermore, the ATG1-mediated phosphorylation of TRAF1s in vivo may represent a positive feedback loop that promotes the stability and function of TRAF1 proteins in autophagy induction.
We further tested the association of 14-3-3λ with ATG13a, ATG13a18A, and ATG13a18D proteins by an in vitro pull-down assay. To this end, we individually incubated the recombinant MBP fusion proteins MBP-ATG13a-HIS, MBP-ATG13a18A-HIS, and MBP-ATG13a18D-HIS with GST-tagged 14-3-3λ (GST-14-3-3λ) using GST as a negative control. Immunoblotting suggested that compared to MBP-ATG13a-HIS, the MBP-ATG13a18D-HIS and MBP-ATG13a18A-HIS variants pulled down more or less GST-14-3-3λ protein, respectively (Supplemental Figure S14), confirming the in vivo Co-IP data (Figure 7A). Together, these results indicate that 14-3-3 proteins predominately interact with the phosphorylated form of ATG13a in vitro and in vivo.
To confirm the effects of ATG13a phosphorylation on its ubiquitination and stability, we transiently transfected the ATG13a-HA, ATG13a18A-HA, or ATG13a18D-HA constructs into protoplasts prepared from WT or 14-3-3λ 14-3-3κ double mutant plants. Immunoblot analysis indicated that ATG13a18A shows decreased ubiquitination, while ATG13a18D showed increased ubiquitination in WT protoplasts (Figure 7B). Consistent with this observation, ATG13a abundance was lower in protoplasts transfected with ATG13a18D but higher in protoplasts transfected with ATG13a18A compared to intact ATG13a (Figure 7B). In contrast, the putative phosphomimic variant ATG13a18D appeared much more stable when the relevant construct was transfected into protoplasts prepared from the 14-3-3λ 14-3-3κ double mutant (Figure 7B), confirming that 14-3-3λ and 14-3-3κ are likely involved in regulating SINAT-mediated degradation of ATG13a in a phosphorylation-dependent manner.
In yeast cells, phosphorylated ATG13 has a lower affinity toward ATG1 under nutrient-rich conditions (Kamada et al., 2010; Qi et al., 2021). To test the effect of ATG13 phosphorylation status on the formation of the ATG1–ATG13 complex, we examined the interaction strength of ATG1a with ATG13a18A, ATG13a, and ATG13a18D by transiently co-transfecting Arabidopsis protoplasts with ATG13a18A-HA, ATG13a-HA, or ATG13a18D-HA constructs with the ATG1a-FLAG construct. Following Co-IP with anti-HA antibodies, we detected lower ATG1a-FLAG levels in the immunoprecipitates from protoplasts transfected with ATG13a18D-HA compared to those expressing ATG13a-HA (Figure 7C). These results indicate that the association of 14-3-3s with phosphorylated ATG13a is crucial for the formation of the ATG1–ATG13 complex.
To validate the significance of ATG13a phosphorylation in autophagy-mediated tolerance of nutritional stress in Arabidopsis, we used stable transgenic lines overexpressing YFP-ATG13a (ATG13a-OE), YFP-ATG13a18A, and YFP-ATG13a18D, with comparable ATG13a transcript levels (Supplemental Figure S15A), for further phenotypic analysis in response to nutrient starvation. We grew 1-week-old WT, ATG13a-OE, ATG13a18A, and ATG13a18D seedlings on MS medium before transfer to N-replete liquid MS medium (N+) or N-free liquid MS medium (N–) for 4 days. We determined that ATG13a-OE and ATG13a18A seedlings display a greater tolerance to N starvation than do WT seedlings (Supplemental Figure S15B). Lines ATG13a18A and ATG13a18D exhibited increased and decreased tolerance to N starvation relative to the ATG13a-OE line, respectively, as evidenced by their relative chlorophyll contents (Supplemental Figure S15C).
We further performed a complementation test by introducing YFP-ATG13a, YFP-ATG13a18A, and YFP-ATG13a18D overexpression constructs into the atg13a atg13b double mutant (Suttangkakul et al., 2011) to generate the atg13a atg13b ATG13a, atg13a atg13b ATG13a18A, and atg13a atg13b ATG13a18D transgenic lines. RT–qPCR analysis showed that ATG13a transcript levels of the three variants in representative lines are 23-, 43-, and 48-fold higher than in WT (Supplemental Figure S15D). When 1-week-old WT, atg13a atg13b, atg13a atg13b ATG13a, atg13a atg13b ATG13a18A, and atg13a atg13b ATG13a18D seedlings were subjected to a 4-day N starvation treatment, the atg13a atg13b mutant showed increased sensitivity to nutritional starvation, with significantly lower chlorophyll contents (Figure 7, D and E). The sensitivity of atg13a atg13b mutants to N starvation was fully rescued by the expression of YFP-ATG13T, with relative chlorophyll contents in atg13a atg13b ATG13a plants comparable to those of WT seedlings (Figure 7, D and E). We also observed that atg13a atg13b ATG13a18A and atg13a atg13b ATG13a18D seedlings displayed increased and decreased tolerance, respectively, to N starvation with significantly elevated or reduced relative chlorophyll contents, in comparison to the WT (Figure 7, D and E). These results suggest that phosphorylation of ATG13a is crucial for autophagy-mediated tolerance of nutrient starvation in plants.
Discussion
The 14-3-3 proteins are highly conserved regulatory proteins in eukaryotic organisms (Huber et al., 2002; Wilson et al., 2016; Liu et al., 2016b; Camoni et al., 2018). These proteins are present in multiple isoforms and function as scaffolding molecules by interacting with specific phosphothreonine and phosphoserine residues in their target proteins. They regulate almost all aspects of cellular activities, including growth and development, primary metabolism, phytohormone signaling, and biotic and abiotic stress responses in plants (Denison et al., 2011; Liu et al., 2017; Yang et al., 2019; Zhao et al., 2021). However, whether 14-3-3 proteins regulate autophagy, specifically the ATG1–ATG13 complex, has not been investigated.
In this study, we present several lines of evidence that demonstrate how the two Arabidopsis 14-3-3 proteins, 14-3-3λ and 14-3-3κ, modulate autophagy dynamics by facilitating SINAT-mediated proteolysis of ATG13s. First, 14-3-3λ and 14-3-3κ proteins physically interacted with both SINATs and ATG13s in vitro and in vivo (Figures 1 and 5), to enhance the association between SINAT1 and ATG13a (Figure 6) and help modulate the SINAT-mediated ubiquitination and degradation of ATG13a in planta (Figure 6). Second, GFP-tagged 14-3-3λ and 14-3-3κ fusion proteins localized to the cytoplasm and the nucleus in both transiently transfected protoplasts and stable transgenic lines (Figure 2). Third, knocking out both 14-3-3λ and 14-3-3κ in the 14-3-3λ 14-3-3κ double mutant increased plant tolerance to nutrient starvation, delayed leaf senescence, and enhanced starvation-induced autophagosome formation (Figures 3 and 4). Fourth, the direct interaction of 14-3-3λ and 14-3-3κ with the phosphorylated form of ATG13a is likely essential for ubiquitination of ATG13a and disassociation of the ATG1–ATG13 complex (Figure 7). Furthermore, transgenic Arabidopsis lines overexpressing ATG13a variants at all 18 putative phosphorylation sites showed altered plant sensitivity to nutrient starvation (Figure 7, D and E). Thus, our findings suggest that 14-3-3λ and 14-3-3κ are molecular adaptors that help regulate autophagy by modulating ATG13 stability and the dynamics of the ATG1–ATG13 kinase complex in Arabidopsis.
Our recent studies have highlighted the central roles of Ub modification in regulating ATG1 and ATG13 protein stability during autophagosome formation in plants (Qi et al., 2020, 2022). Under nutrient-rich conditions, the E3 Ub ligases SINAT1 and SINAT2 form a functional protein complex with TRAF1a/1b and ATG13 to ubiquitinate and degrade ATG13 by K48-linked ubiquitination in vivo, thereby maintaining a low level of autophagy (Qi et al., 2020). In response to nutrient starvation, however, SINAT6 competitively interacts with ATG13 to suppress the ubiquitination and degradation of the ATG1–ATG13 complex, leading to the induction of autophagy in plant cells (Qi et al., 2020). Furthermore, upon nutrient deprivation, ATG1a enhances the stability of TRAF1a in vivo, in a phosphorylation-dependent manner, suggesting feedback regulation of autophagy in Arabidopsis (Qi et al., 2020). Together, these results demonstrate that the activity and stability of the Arabidopsis ATG1–ATG13 protein complex are tightly regulated during autophagosome formation. Although ATG1 also undergoes 26S proteasome-dependent degradation under nutrient deprivation conditions (Figure 1; Qi et al., 2020), the molecular mechanism by which SINATs or other E3 Ub ligases induce ATG1a degradation remains to be investigated.
In mammals, 14-3-3 proteins regulate autophagy by binding key components of the autophagy machinery, including Beclin1, human vacuolar protein sorting 34 (hVps34), and Atg9A (Pozuelo-Rubio, 2011, 2012; Wang et al., 2012; Weerasekara et al., 2014). Under physiological conditions, 14-3-3 proteins directly interact with hVps34 in a protein kinase C-dependent phosphorylation manner that inhibits hVps34 kinase activity (Pozuelo-Rubio, 2011). In contrast, starvation or rapamycin treatment promotes the disassociation of 14-3-3 from hVps34 and thus activates hVps34 kinase activity (Pozuelo-Rubio, 2011). Furthermore, 14-3-3 proteins also promote autophagy by interacting with Beclin-1 (Wang et al., 2010; Xiong et al., 2019). In particular, 14-3-3 proteins prevent Beclin1 degradation by associating with a form of Beclin1 phosphorylated on S295, increasing its binding to hVps34 to form a Beclin1–PI3K complex (Wang et al., 2012; Tang et al., 2020). In mammalian cells, 14-3-3 proteins also interact with phosphorylated Atg9A to activate hypoxia-induced autophagy (Weerasekara et al., 2014). Under normoxic conditions, ULK1 and AMP-activated protein kinase (AMPK) mediate the phosphorylation of Atg9A at S761 to maintain its protein abundance at low levels. During hypoxia, however, activated AMPK phosphorylates Atg9A at S761, bypassing the ULK1 kinase complex, which enhances the interaction between 14-3-3 and Atg9A and recruits the Atg9A to the autophagosome (Weerasekara et al., 2014). These studies provide strong support for the idea that 14-3-3 proteins play important roles in autophagosome formation during the nucleation and membrane donation stages.
A previous study observed that phosphorylation of ULK1 at S555 by AMPK promoted its interaction with 14-3-3 proteins in vivo and in vitro, indicating a role for 14-3-3 proteins during autophagy initiation (Pozuelo-Rubio, 2012). Our data presented herein further suggest that 14-3-3 proteins are involved in maintaining the homeostasis of the ATG1–ATG13 kinase complex and contribute to the proper control of autophagy initiation, an early but key stage during autophagosome formation. Thus, our findings may represent a new mechanism for the 14-3-3/SINAT–ATG1/ATG13 module in the regulation of autophagy. A parallel mechanism has not yet been validated in other eukaryotic species.
The Arabidopsis genome harbors thirteen genes encoding 14-3-3 proteins (Rosenquist et al., 2001; Sehnke et al., 2002). The encoded 14-3-3 proteins participate in diverse signal transduction events, such as kinase-mediated signal transduction, response to stress, change of client protein activity, subcellular localization, and posttranslational modifications (Sehnke et al., 2002; Chang et al., 2009; Jaspert et al., 2011; Liu et al., 2017; Yang et al., 2019). Studies of the autophagy system in plants have identified the ATG1–ATG13 kinase complex as a crucial regulator of autophagy and autophagic vesicle assembly via interaction with the regulatory proteins ATG11 and ATG101 (Suttangkakul et al., 2011; Liu and Bassham, 2012; Li et al., 2014). In response to nutrient availability, the core components of the autophagic machinery are controlled by upstream phosphorylation regulators, such as the protein kinases TOR and SUCROSE NONFERMENTING 1-RELATED KINASE 1 (SnRK1) (Qi et al., 2021). In yeast, TOR is a key negative regulator that phosphorylates ATG13 under nutrient-rich conditions and reduces the affinity of ATG1 for ATG13. However, nutrient starvation inactivates TOR, allowing ATG13 to become dephosphorylated and the ATG1–ATG13 complex to form and initiate autophagy (Rabinowitz and White, 2010). In plants, although we lack biochemical evidence directly linking TOR to autophagy, we know that TOR is a conserved negative regulator of autophagy in Arabidopsis (Liu and Bassham, 2010; Pu et al., 2017).
Plant SnRK1 is a highly conserved energy sensor that is activated upon nutrient deprivation (Baena‐González and Sheen, 2008). Overexpressing the gene encoding the Arabidopsis α-subunit of SnRK1, SNF1 KINASE HOMOLOG 10 (KIN10), leads to enhanced tolerance to nutrient starvation, constitutive autophagosome formation, and delayed flowering and leaf senescence, as well as greater levels of phosphorylated ATG1a in Arabidopsis (Baena‐González et al., 2007; Chen et al., 2017b), indicating that KIN10 is likely a positive regulator of autophagy in plants. Furthermore, KIN10 functions upstream of TOR to activate autophagy, suggesting a crosstalk between these two phosphorylation-based regulators of plant autophagy (Soto‐Burgos and Bassham, 2017). To date, however, we do not know whether ATG13a and/or ATG13b proteins are direct substrates for TOR under nutrient-rich conditions in plants. Here, we identified all 18 putative Ser, Thr, and Tyr phosphorylation sites in ATG13a, and showed that mutation of these sites to Ala (ATG13a18A) or Asp (ATG13a18D) altered ATG13a phosphorylation (Figure 7A), suggesting that these 18 residues are potential TOR phosphorylation sites in Arabidopsis. Consistent with this idea, the phosphorylation of these 18 residues in ATG13a was crucial for its association with 14-3-3s, SINAT1-mediated ubiquitination, and ATG1–ATG13 complex formation (Figure 7, A–C). It is therefore conceivable that 14-3-3s are key regulators of SINAT-mediated ubiquitination of phosphorylated ATG13a. Future biochemical investigation of the direct association and phosphorylation of ATG13s by TOR and/or KIN10/11 kinases will deepen our understanding of the mechanisms behind upstream energy signaling in regulating autophagy in plants.
Based on the findings of this study, we propose a working model illustrating the role of 14-3-3 proteins in regulating autophagosome formation (Figure 7F). Under nutrient-rich conditions, the TOR kinase complex is active and phosphorylates ATG13 proteins to keep them hyperphosphorylated. The 14-3-3 proteins associate with phosphorylated ATG13s and act as adaptors to recruit the E3 Ub ligases SINAT1 and SINAT2 to form a protein complex that promotes the ubiquitination and degradation of phosphorylated ATG13s via the 26S proteasome pathway. Phosphorylated ATG13 then disassociates from ATG1s, suppressing autophagy (Figure 7F, left). Under nutrient starvation conditions, however, SnRK1 inhibits TOR activity, resulting in the dephosphorylation of ATG13, which impairs its interaction with 14-3-3 proteins, allowing the formation of a complex with hyperphosphorylated ATG1a mediated by SnRK1. The scaffold proteins TRAF1a and TRAF1b interact with ATG13s together with SINAT6, forming a TRAF1–SINAT6–ATG13 TRAFsome complex that reduces SINAT1- and SINAT2-mediated ubiquitination and degradation of ATG13 to activate autophagy (Figure 7F, right). Furthermore, the ATG-mediated phosphorylation of TRAF1s in vivo may represent a positive feedback loop that promotes the stability and function of TRAF1 proteins upon autophagy induction.
Materials and methods
Plant materials, growth conditions, and treatments
All WT, mutant, and transgenic A. thaliana plants used in this study are in the Col-0 background. The T-DNA insertional mutants described in this study were obtained from the Arabidopsis Biological Resource Center (ABRC), with the names 14-3-3λ-1 (SALK_075218), 14-3-3λ-2 (SALK_075219), 14-3-3κ-1 (SALK_148929), 14-3-3κ-2 (SALK_000496), and 14-3-3κ-3 (SALK_071097). Homozygous mutant plants were identified by PCR using gene-specific primer pairs with a T-DNA border-specific primer (Supplemental Data Set 1). The 14-3-3λ-2 single mutant was crossed to 14-3-3κ-3 to generate the 14-3-3λ 14-3-3κ double mutant (Zhou et al., 2014). The primers used to genotype the single and double mutant plants are listed in Supplemental Data Set 1. The mutants and transgenic lines generated in this study are listed in Supplemental Tables S1 and S2.
All Arabidopsis seeds were surface sterilized in 20% (v/v) bleach containing 0.1% (v/v) Tween-20 for 20 min and washed with sterilized water at least 5 times. The seeds were sown on MS medium (Sigma-Aldrich, St Louis, MO, USA) containing 2% (w/v) sucrose and 0.8% (w/v) agar, followed by stratification at 4°C in the dark for 3 days. The plates were then released at 22°C under a long-day (16-h light/8-h dark) photoperiod and a light intensity of 170 µmol m−2s−1 using fluorescent bulbs (cat. no. F17T8/TL841 17W; Philips, Amsterdam, Netherlands). One week after germination, the seedlings were transferred to soil and incubated under long-day or short-day (8-h light/16-h dark) conditions for further growth and analysis.
For C starvation treatment, 1-week-old MS-grown seedlings were transferred to solid sucrose-free medium and incubated under continuous dark conditions until the seedlings started showing poor growth. After recovery under normal growth conditions for 7 days, seedling phenotypes were recorded as photographs, and the survival rate after C starvation was calculated from 15 seedlings per genotype and defined as the percentage of seedlings with obvious regreening and the appearance of new leaves. For N starvation treatments, 1-week-old seedlings grown on MS medium were transferred to liquid MS or N-free liquid MS medium and grown under normal growth conditions. For biochemical treatments, 1-week-old Arabidopsis seedlings grown on solid MS containing 2% (w/v) sucrose were transferred to sterile 12-well plates with each well containing either liquid MS medium or sucrose-free or N-free liquid MS medium (C– or N–), or containing 1-µM CA or 0.5-mM CHX (cat. no. 2112; Cell Signaling Technology, Danvers, MA, USA) for treatment, followed by total protein extraction and immunoblot analysis.
Plasmid construction
All plasmids used in this study were generated by an In-Fusion method using the ClonExpress II One Step Cloning Kit (catalog no. C112; Vazyme, Nanjing, China). All gene-specific primers with 15-bp extensions homologous to the corresponding vectors are listed in Supplemental Data Set 1. Plasmids for transient expression analyses were derived from the pHBT and pUC119 vectors (Li et al., 2013). For the 14-3-3λ-FLAG, 14-3-3κ-FLAG, ATG1a-FLAG, ATG13a18A-HA, and ATG13a18D-HA constructs, the full-length coding sequences of 14-3-3λ, 14-3-3κ, ATG1a, ATG13a18A, and ATG1318D were inserted into BamHI- and StuI-digested pUC119 plasmids. The full-length coding sequences of 14-3-3λ and 14-3-3κ were individually cloned into StuI- or BamHI-digested pUC119 to generate GFP-14-3-3κ-HA, GFP-14-3-3λ-FLAG, GFP-14-3-3κ-FLAG, 14-3-3λ-mCherry-FLAG, and 14-3-3κ-mCherry-FLAG constructs. The constructs ATG1a-HA, ATG1b-HA, ATG1c-HA, ATG13a-HA, ATG13b-HA, ATG7-HA, ATG13a-FLAG, SINAT1-FLAG, GFP-SINAT1-HA, and GFP-SINAT2-HA were constructed as described previously (Qi et al., 2020).
To generate stable transgenic plants expressing 14-3-3λ-FLAG or 14-3-3κ-FLAG, the UBQ10pro:14-3-3λ-FLAG and UBQ10pro:14-3-3κ-FLAG fragments derived from pUC119 constructs were digested with AscI and cloned into the binary vector pFGC-RCS (Li et al., 2013). For the GFP-14-3-3κ-HA transgenic plants, the UBQ10pro:GFP-14-3-3κ-HA fragment was cloned into the binary vector pFGC-RCS with the UBQ10-NOS expression cassettes of pUC119 vectors (Xia et al., 2020) digested by StuI. These expression cassettes were subsequently introduced into WT Arabidopsis (Col-0) by Agrobacterium (Agrobacterium tumefaciens)-mediated transformation via the floral dip method (Clough and Bent, 1998). The ATG13a-HA transgenic plants were described previously (Qi et al., 2017, 2020).
To generate plasmids for Y2H analysis, the full-length coding sequences of 14-3-3λ, 14-3-3κ, 14-3-3χ, 14-3-3ω, and 14-3-3φ were individually amplified and inserted into pGADT7 or pGBKT7 vectors digested by EcoRI and BamHI to generate the AD or BD vectors for Y2H analysis. The ATG1a-AD, ATG1b-AD, and ATG1c-AD plasmids were described previously (Qi et al., 2017). The ATG13a-AD, ATG13b-AD, and SINAT5-S1-BD plasmids were described previously (Qi et al., 2020). The SINAT1-BD, SINAT2-BD, SINAT6-BD, SINAT1C-BD, and SINAT2C-BD were described previously (Xia et al., 2020).
To generate plasmids for the BiFC assay, the full-length coding sequences of 14-3-3λ and 14-3-3κ were inserted into BamHI-digested pUC-YC or pUC-YN, respectively, placed under the control of the UBQ10 promoter, to generate fusions with nYFP or cYFP. The full-length coding sequences of ATG1a, ATG13a, and ATG7 were inserted into pHBT-YC or pHBT-YN with 35S:PPDK digested with BamHI, respectively, to generate vectors for BiFC assays. SINAT1-nYFP and SINAT2-nYFP were constructed as described previously (Qi et al., 2020).
To generate plasmids for protein production, the full-length coding sequences of 14-3-3λ, 14-3-3κ, and ATG1a were inserted into BamHI- and EcoRI-digested pRSETA or pGEX-6p to generate HIS-14-3-3λ, HIS-14-3-3κ, GST-14-3-3λ, GST-14-3-3κ, and GST-ATG1a fusions. The MBP-ATG13a-HIS, MBP-ATG13a18A, and MBP-ATG13a18D plasmids were constructed by cloning the coding sequence of ATG13a into the BamHI-digested vector pMAL-c5x (New England Biolabs, Ipswich, MA, USA).
Measurements of chlorophyll contents
Measurements of chlorophyll contents were performed as described previously (Porra et al., 1989; Xiao et al., 2010). Total chlorophylls were extracted from Arabidopsis seedlings or leaves after N or C starvation or at different growth stages by immersing the samples in 2-mL (seedlings) or 4-mL (adult leaves) ethanol in continuous dark conditions at 4°C for 48 h. Absorbance was determined at 664 and 647 nm, and total chlorophyll contents were measured and normalized to fresh weight of the samples.
Protein isolation and immunoblot analysis
For total protein extraction, 1-week-old Arabidopsis seedlings grown on MS medium or after nutrient starvation or biochemical treatment were ground in liquid N and homogenized in ice-cold protein extraction buffer (50-mM sodium phosphate pH 7.0, 200-mM NaCl, 10-mM MgCl2, 0.2% [v/v] β-mercaptoethanol, and 10% [v/v] glycerol) containing protease inhibitor cocktail (Roche, Basel, Switzerland; 04693132001). The samples were placed on ice for 30 min and centrifuged at 4°C for 30 min at 12,000 g. The resulting supernatants were transferred to a new microfuge tube containing 5× soldium dodecyl sulfate (SDS) loading buffer and denatured at 95°C for 10 min before electrophoresis. For immunoblot analysis, the denatured samples were subjected to soldium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a Hybond-C membrane (Amersham, Staffordshire, UK). The following specific antibodies were used for immunoblotting analysis: anti-ATG1a (cat. no. AS194274, 1:3,000, Agrisera, Vännäs, Sweden), anti-ATG13a (cat. no. AS194279, 1:3,000, Agrisera), anti-ATG7 (cat. no. ab99001, 1:2,000; Abcam, Cambridge, Uk), anti-HA (cat. no. H6533, Sigma Aldrich, St Louis, MO, USA), anti-FLAG (cat. no. A8592, 1:1,500; Sigma Aldrich), anti-Ub (cat. no. 10201-2-AP, 1:2,000; Proteintech, Rosemont, IL, USA), anti-GFP (cat. no. M20004, 1:3,000; Abmart, Shanghai, China), and anti-ACTIN (cat. no. 58169; 1:1,000; Cell Signaling Technology).
IP-MS assay
Plant materials (10 g) from 10-day-old transgenic seedlings expressing 14-3-3λ-FLAG, ATG13a-HA (Qi et al., 2020) or free GFP and grown in liquid MS medium were ground to powder in liquid N and homogenized in 10 mL 2× IP buffer (100-mM Tris–HCl pH 7.5, 200-mM NaCl, 2-mM EDTA, 20% [v/v] glycerol, 0.5% [v/v] Triton X-100, and protease inhibitor cocktail). After centrifugation twice at 12,000 g for 30 min at 4°C, the supernatant was incubated with anti-FLAG M2 magnetic beads (cat. no. M8823, Sigma Aldrich) for 14-3-3λ-FLAG, or anti-HA affinity agarose beads (cat. no. A2095, Sigma Aldrich) for ATG13a-HA at 4°C overnight with gentle rotation. The beads were then collected and washed with IP buffer containing 0.1% (v/v) Triton X-100 5 times and were eluted with SDS sample buffer. The samples were denatured at 95°C for 10 min and separated on 12% (w/v) SDS–PAGE, followed by Coomassie Brilliant Blue staining. The resulting stained bands were digested with trypsin before liquid chromatography–tandem MS LTQ Orbitrap Elite analysis for identification of 14-3-3λ interacting proteins and ATG13a phosphorylation sites. Seedlings expressing free GFP were used as a blank control for eliminating nonspecific interactions.
Y2H analysis
Y2H assays were conducted using the one-step-transformation method as described previously (Chen et al., 1992) with minor modifications. A volume of 300-μL saturated culture of yeast strain YH109 was centrifuged and resuspended in transformation buffer (10-μL 2 M LiAc, 10-μL 1 M DTT, 20-μg salmon sperm DNA, and 80-μL 50% [w/v] polyethylene glycol 3350) with 300 ng of plasmid DNA for each AD or BD fusion. After a heat shock of 30 min at 45°C, the total suspension was plated directly onto synthetic defined (SD) medium –Leu and Trp (SD–2) and incubated at 30°C for 72 h. Successfully transformed colonies were resuspended in sterile double-distilled H2O and diluted 10-fold and 100-fold, before spotting 10 μL from each dilution onto SD medium –Leu–Trp–His–Ade (SD–4) for 2 days before protein interaction was assessed. To avoid self-activation of the clones, 5-mM 3-amino-1,2,4-triazole (3-AT) was added to the medium.
Co-IP assay
Arabidopsis mesophyll protoplasts were prepared and transfected according to Yoo et al. (2007). Protoplasts isolated from 4-week-old rosettes were transfected with the indicated plasmids and cultured for 16 h before protein extraction. The cells were collected and lysed in 200-μL ice-cold IP buffer with 0.5% (v/v) Triton X-100, followed by centrifugation at 12,000 g for 30 min at 4°C. A 10% aliquot of each supernatant was used for input, and the remainder was incubated with HA-agarose beads (Sigma-Aldrich) or FLAG affinity magnetic beads (Sigma-Aldrich) at 4°C with gentle rotation for 3 h. The beads were then collected and washed 5 times with ice-cold IP buffer containing 0.1% (v/v) Triton X-100, followed by the addition of 5× SDS–PAGE sample buffer and denaturation at 95°C for 10 min before immunoblot analysis with anti-HA and anti-FLAG antibodies. Quantification of the protein immunoblot signal was determined with the software ImageJ.
BiFC assay and microscopy analyses
For BiFC assays, pairs of nYFP and cYFP fusion constructs were co-transfected into leaf protoplasts prepared from WT Arabidopsis plants and incubated for 16 h in constant light or dark conditions. The reconstituted YFP signals indicating a positive protein interaction were detected by confocal microscopy with excitation at 488 nm, and visualized with a band-pass infrared (IR) filter from 500 to 530 nm.
To determine the subcellular localization of 14-3-3s, the plasmids GFP-14-3-3λ or GFP-14-3-3κ and the nucleus localization marker ARF4-RFP (encoding a fusion between AUXIN RESPONSE FACTOR 4 [ARF4] and the RFP), or 14-3-3λ-mCherry or 14-3-3κ-mCherry and YFP-ATG8e (Qi et al., 2017), were co-transfected into 4-week-old WT (Col-0) Arabidopsis mesophyll protoplasts and incubated for 16 h in constant light or dark conditions. The GFP and mCherry/RFP signals were detected by confocal microscopy at 488 nm (GFP) or 516 nm (mCherry/RFP), and visualized with band-pass IR filters from 500 to 530 nm (GFP) or from 560 to 610 nm (mCherry/RFP).
To determine the effect of 14-3-3λ and 14-3-3κ genetic inactivation on autophagosome formation, the 14-3-3λ 14-3-3κ double mutant was crossed to the stable eGFP-ATG8e line (Xiao et al., 2010) to generate the 14-3-3λ 14-3-3κ eGFP-ATG8e reporter line. Three-week-old eGFP-ATG8e and 14-3-3λ 14-3-3κ eGFP-ATG8e seedlings grown on MS medium were transferred to sucrose-free MS medium containing 1-μM CA and incubated in constant dark conditions for fixed-C starvation treatment. Arabidopsis leaf epidermal cells were observed by confocal microscopy with excitation at 488 nm and visualized with a band-pass IR filter between 500 and 530 nm.
Pull-down assay
The full-length coding sequences of ATG13a, ATG13a18A, or ATG13a18D were cloned individually into the vector pMAL-c5x (New England Biolabs) to generate the MBP-ATG13a-HIS, MBP-ATG13a18A-HIS, or MBP-ATG13a18D-HIS fusions. The full-length coding sequences of ATG1a, 14-3-3λ, and 14-3-3κ were cloned into the vector pGEX-6P-1 or pRSETA to generate the GST-ATG1a, GST-14-3-3λ, GST-14-3-3κ, HIS-14-3-3λ, and HIS-14-3-3κ fusions. For production of the HIS-14-3-3κ fusion protein, the culture was harvested without isopropyl β-d-1-thiogalactopyranoside (IPTG) induction. For other proteins produced in this study, IPTG was added to a 250-mL culture to a final concentration of 300 μM when optical density (OD)600 reached 0.6. After an additional 4 h of growth at 28°C (MBP-ATG13a-HIS, MBP-ATG13a18A-HIS, and MBP-ATG13a18D-HIS), or 6 h at 28°C (HIS-14-3-3λ or GST-ATG1a), or 9 h at 37°C (GST-14-3-3λ or GST-14-3-3κ), the cells were harvested and the recombinant proteins purified using a pMAL Protein Fusion and Purification System (cat. no. E8021; New England Biolabs) for MBP-ATG13a-HIS, MBP-ATG13a18A-HIS, MBP-ATG13a18D-HIS, or glutathione Sepharose 4B (cat. no. 17-0756; GE Healthcare, Chicago, IL, USA) for GST-14-3-3λ, GST-14-3-3κ, or GST-ATG1a, or Ni-NTA resin (cat. no. R901-15; Invitrogen, Waltham, MA, USA) for HIS-14-3-3λ or HIS-14-3-3κ following the manufacturer’s instructions. The resulting fusion proteins were desalted and concentrated with IP buffer using Amicon Ultra-4 Centrifugal Filter Devices (cat. no. UFC803024; Millipore, Burlington, MA, USA) and then the levels were examined by immunoblot analysis and Coomassie Brilliant Blue staining using bovine serum albumin as a standard.
For in vitro pull-down assays of 14-3-3λ or 14-3-3κ with ATG1a, 1-μg recombinant HIS-14-3-3λ or HIS-14-3-3κ was mixed with 1-μg recombinant GST-ATG1a or free GST in IP buffer with 0.2% (v/v) Triton X-100. For physical interaction of 14-3-3λ or 14-3-3κ and ATG13a, recombinant GST-14-3-3λ, GST-14-3-3κ, or free GST was mixed with 1-μg recombinant MBP-ATG13a-HIS in IP buffer with 0.2% (v/v) Triton X-100. The above mixtures were incubated at 16°C, with shaking at 1,200 rpm for 2 h, followed by another 2-h incubation after the addition of 10-μL GST resin or Ni-NTA resin, respectively. The resins were washed in IP buffer with 0.2% (v/v) Triton X-100 5 times and then eluted in 5× SDS sample buffer at 95°C for 10 min, followed by immunoblot analysis with anti-HIS and anti-GST antibodies.
In vivo ubiquitination and protein degradation assays
For the in vivo ubiquitination assay, the ATG13a-HA plasmid was transfected alone into Arabidopsis mesophyll protoplasts isolated from WT or the 14-3-3λ 14-3-3κ double mutant or co-transfected with SINAT1-FLAG into WT or 14-3-3λ 14-3-3κ protoplasts. After a 16-h incubation under constant light conditions, the cells were collected and lysed in IP buffer containing 0.5% (v/v) Triton X-100 with vigorous vortexing, followed by centrifugation at 4°C for 30 min at 12,000 g. The supernatants were incubated with HA or TUBE (Cat. no. UM-0401-1000, Agarose) affinity agarose beads before immunoblot analysis. The empty vector pUC119-UBQ10-GFP-HA was co-transfected with other constructs to assess transfection efficiency. The ubiquitination patterns and stability of ATG13a-HA were detected with anti-Ub and anti-HA antibodies.
For protein degradation analysis, 7-day-old 14-3-3λ 14-3-3κ double mutant and WT seedlings grown on solid MS medium were transferred to liquid MS medium with 500-μM CHX for the indicated times before total proteins were extracted for immunoblot analyses. The abundance of ATG13a was detected using anti-ATG13a antibodies and the half-life of the protein determined.
RNA extraction and RT–qPCR analysis
Total RNA was extracted from 1-week-old Arabidopsis seedlings or 3-week-old Arabidopsis rosettes using a HiPure Plant RNA Mini kit (E.Z.N.A.; Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. One microgram of isolated total RNA was reverse transcribed into first-strand cDNA with the HiScript II Q Select RT Super Mix with gDNA Wiper (Vazyme). The cDNA samples were diluted 1:16 in water for qPCR analysis.qPCR was performed in 10-μL reactions with gene-specific primers using ChamQ SYBR Color qPCR Master Mix (Vazyme) on a StepOne Plus Real-time PCR System (Applied Biosystems). qPCR was performed with the following protocol: 95°C for 5 min followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s, and a subsequent standard dissociation protocol to validate the presence of a unique polymerase chain reaction (PCR) product.
For calculation of the relative expression levels of different genes, three technical replicates were performed per reaction. The delta of threshold cycle (ΔCt) values were calculated by subtracting the arithmetic mean Ct values of the targets from the normalizing ACTIN2. The relative transcript level (2−ΔΔCt) was calculated from three independent experiments. The gene-specific primers used for qPCR are listed in Supplemental Data Set 1.
Statistical analyses
Data reported in this study are means ± standard deviation (sd) from three independent experiments unless indicated otherwise. Statistical significance of differences between groups was determined by a two-tailed Student’s t test. The P < 0.05 or <0.01 were considered significant. The two-tailed Student’s t test results are listed in Supplemental Data Set 2.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/European Molecular Biology Laboratory databases under the following accession numbers: 14-3-3λ (At5g10450), 14-3-3κ (At5g65430), 14-3-3φ (At1g35160), 14-3-3χ (At4g09000), 14-3-3ω (At1g78300), SINAT1 (At2g41980), SINAT2 (At3g58040), SINAT5 (At5g53360), SINAT6 (At3g13672), ATG13a (At3g49590), ATG13b (At3g18770), ATG1a (At3g61960), ATG1b (At3g53930), ATG1c (At2g37840), ATG7 (At5g45900), and ARF4 (At5g60450).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. 14-3-3λ and 14-3-3κ interact with SINATs in vitro.
Supplemental Figure S2. Subcellular localization of GFP-14-3-3λ and GFP-14-3-3κ in Arabidopsis protoplasts under constant light or dark conditions.
Supplemental Figure S3. Identification of GFP-14-3-3κ-HA transgenic lines.
Supplemental Figure S4. Identification of 14-3-3λ and 14-3-3κ single mutants.
Supplemental Figure S5. Phenotypic analysis of 14-3-3λ and 14-3-3κ single mutants.
Supplemental Figure S6. Generation of 14-3-3λ and 14-3-3-κ overexpression lines.
Supplemental Figure S7. Screening of 14-3-3λ-interacting proteins by IP-MS analysis.
Supplemental Figure S8. 14-3-3λ and 14-3-3κ interact with ATG1/13 in vivo.
Supplemental Figure S9. Pull-down assay showing the direct interaction of 14-3-3s and ATG1a.
Supplemental Figure S10. Ubiquitination of ATG13a in the 14-3-3λ 14-3-3k double mutant (14-3-3λ/κ).
Supplemental Figure S11. Original images showing the immunoblotting data in Figure 6, C (A) and D (B).
Supplemental Figure S12. Alignment of the ATG13a and ATG13b protein sequences.
Supplemental Figure S13. Mass spectrometry data showing the phosphorylation sites of ATG13a.
Supplemental Figure S14. Pull-down assay showing the direct interaction of 14-3-3λ and ATG13a phosphorylation mutants.
Supplemental Figure S15. Phenotypic analyses of ATG13a phosphorylation mutants.
Supplemental Table S1. Mutants generated in this study.
Supplemental Table S2. Transgenic Arabidopsis plants generated in this study.
Supplemental Data Set 1. Sequence of primers used in this study.
Supplemental Data Set 2. Student’s t test analysis in this study.
Supplementary Material
Acknowledgments
We thank the ABRC for providing the 14-3-3λ and 14-3-3κ mutant seed. We thank Prof. Yan Guo (China Agricultural University) for providing the 14-3-3λ 14-3-3κ double mutant and Prof. Sui-Wen Hou (Lanzhou University) for providing the YFP-ATG13a, YFP-ATG13a18A, YFP-ATG13a18D, atg13a atg13b ATG13a, atg13a atg13b ATG13a18A, and atg13a atg13b ATG13a18D stable transgenic plants.
Funding
This work was supported by the National Natural Science Foundation of China (Project 31725004; 31800217), the Key Realm R&D Program of Guangdong Province (Project 2020B0202090001; 2020B0202080001), the Natural Science Foundation of Guangdong Province (Project 2018A030313210; 2022A1515012500; 2021B1212040008), and Laboratory of Lingnan Modern Agriculture (Project NG2021002; NT2021010; 2022ZD001).
Conflict of interest statement. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Hua Qi, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China; Guangdong Provincial Key Laboratory of Agricultural & Rural Pollution Abatement and Environmental Safety, College of Natural Resources and Environment, South China Agricultural University, Guangzhou, 510642, China.
Xue Lei, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
Yao Wang, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
Shan Yu, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
Ting Liu, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, College of Plant Protection, South China Agricultural University, Guangzhou, 510642, China.
Shun-Kang Zhou, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, College of Plant Protection, South China Agricultural University, Guangzhou, 510642, China.
Jin-Yu Chen, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
Qin-Fang Chen, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
Rong-Liang Qiu, Guangdong Provincial Key Laboratory of Agricultural & Rural Pollution Abatement and Environmental Safety, College of Natural Resources and Environment, South China Agricultural University, Guangzhou, 510642, China.
Liwen Jiang, School of Life Sciences, Centre for Cell & Developmental Biology, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Shatin, Hong Kong, China.
Shi Xiao, Guangdong Laboratory for Lingnan Modern Agriculture, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, 510275, China.
S.X. designed the research. H.Q., X.L., Y.W., S.Y., T.L., S.K.Z., J.Y.C., and Q.F.C. carried out the experiments. S.X., L.J., R.L.Q., and H.Q. analyzed the data and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Shi Xiao (xiaoshi3@mail.sysu.edu.cn).
References
- Avin-Wittenberg T (2019) Autophagy and its role in plant abiotic stress management. Plant Cell Environ 42: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena‐González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353: aac4354. [DOI] [PubMed] [Google Scholar]
- Bassham DC (2009) Function and regulation of macroautophagy in plants. Biochim Biophys Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Camoni L, Visconti S, Aducci P, Marra M (2018) 14-3-3 Proteins in plant hormone signaling: doing several things at once. Front Plant Sci 9: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IF, Curran A, Woolsey R, Quilici D, Cushman JC, Mittler R, Harmon A, Harper JF (2009) Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9: 2967–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT (1992) One-step transformation of yeast in stationary phase. Curr Genet 21: 83–84 [DOI] [PubMed] [Google Scholar]
- Chen L, Liao B, Qi H, Xie LJ, Huang L, Tan WJ, Zhai N, Yuan LB, Zhou Y, Yu LJ, et al. (2015) Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11: 2233–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li F, Xiao S (2017a) Analysis of plant autophagy. Methods Mol Biol 1662: 267–280 [DOI] [PubMed] [Google Scholar]
- Chen L, Su ZZ, Huang L, Xia FN, Qi H, Xie LJ, Xiao S, Chen QF (2017b) The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci 8: 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Denison FC, Paul AL, Zupanska AK, Ferl RJ (2011) 14-3-3 proteins in plant physiology. Semin Cell Dev Biol 22: 720–727 [DOI] [PubMed] [Google Scholar]
- Ding X, Zhang X, Otegui MS (2018) Plant autophagy: new flavors on the menu. Curr Opin Plant Biol 46: 113–121 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Hayward AP, Tsao J, Dinesh-Kumar SP (2009) Autophagy and plant innate immunity: defense through degradation. Semin Cell Dev Biol 20: 1041–1047 [DOI] [PubMed] [Google Scholar]
- Huber SC, MacKintosh C, Kaiser WM (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol Biol 50: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Jaspert N, Throm C, Oecking C (2011) Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front Plant Sci 2: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y (2010) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) AUTOPHAGYRELATED11 plays a critical role in general autophagy-and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marshall RS, Li F (2018) Understanding and exploiting the roles of autophagy in plants through multi-omics approaches. Plant Sci 274: 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, Wu PR, Lin MY, Jiang ST, Tsai TF, et al. (2016a) Cul3‐ KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell 61: 84–97 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang S, Liu B (2016b) 14-3-3 proteins: macro-regulators with great potential for improving abiotic stress tolerance in plants. Biochem Biophys Res Commun 477: 9–13 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2010) TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y (2022) Plant target of rapamycin signaling network: complexes, conservations, and specificities. J Integr Plant Biol 64: 342–370 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66: 117–9128. [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 29: 173–208 [DOI] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self‐association and function through AMBRA1 and TRAF6. Nat Cell Biol 15: 406–416 [DOI] [PubMed] [Google Scholar]
- Pozuelo-Rubio M (2011) Regulation of autophagic activity by 14-3-3ζ proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ 18: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo-Rubio M (2012) 14-3-3 Proteins are regulators of autophagy. Cells 1: 754–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pu Y, Luo X, Bassham DC (2017) TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Li J, Xia FN, Chen JY, Lei X, Han MQ, Xie LJ, Zhou QM, Xiao S (2020) Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 32: 263–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Xia FN, Xiao S (2021) Autophagy in plants: physiological roles and post-translational regulation. J Integr Plant Biol 63: 161–179 [DOI] [PubMed] [Google Scholar]
- Qi H, Xia FN, Xiao S, Li J (2022) TRAF proteins as key regulators of plant development and stress responses. J Integr Plant Biol 64: 431–448 [DOI] [PubMed] [Google Scholar]
- Qi H, Xia FN, Xie LJ, Yu LJ, Chen QF, Zhuang XH, Wang Q, Li F, Jiang L, Xie Q, et al. (2017) TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 29: 890–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E (2010) Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M (2001) Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol 127: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ (2002) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Burgos J, Bassham DC (2017) SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One 12: e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Burgos J, Zhuang X, Jiang L, Bassham DC (2018) Dynamics of autophagosome formation. Plant Physiol 176: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhang Y, Liu S, Sun Z, Wang C, Li L, Zhou W, Cheng S (2020) 14-3-3ζ binds to and stabilizes phospho-beclin 1S295 and induces autophagy in hepatocellular carcinoma cells. J Cell Mol Med 24: 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ling S, Lin WC (2010) 14-3-3τ regulates Beclin 1 and is required for autophagy. PLoS One 5: e10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qin Q, Su M, li N, Zhang J, Liu Y, Yan L, Hou S (2022) Type one protein phosphatase regulates fixed-carbon starvation-induced autophagy in Arabidopsis. Plant Cell https://doi.org/10.1093/plcell/koac251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338: 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL (2014) Metabolic-stress-induced rearrangement of the 14-3-3ζ interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3ζ interaction with phosphorylated Atg9. Mol Cell Biol 34: 4379–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Swatek KN, Thelen JJ (2016) Regulation of the regulators: post-translational modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front Plant Sci 7: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia FN, Zeng B, Liu HS, Qi H, Xie LJ, Yu LJ, Chen QF, Li JF, Chen YQ, Jiang L, et al. (2020) SINAT E3 ubiquitin ligases mediate FREE1 and VPS23A degradation to modulate abscisic acid signaling. Plant Cell 32: 3290–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML (2010) Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong XX, Hu DX, Xu L, Lin H, Zhang Y, Li CY, Chen XQ (2019) Selective 14-3-3γ upregulation promotes beclin-1-LC3-autophagic influx via β-catenin interaction in starved neurons in vitro and in vivo. Neurochem Res 44: 849–858 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2007) Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3: 257–258 [DOI] [PubMed] [Google Scholar]
- Yang X, Bassham DC (2015) New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol 320: 1–40 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang C, Xue Y, Liu X, Chen S, Song C, Yang Y, Guo Y (2019) Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat Commun 10: 1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Ohsumi Y (2018) Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol 59: 1337–1344 [DOI] [PubMed] [Google Scholar]
- Zhang C, Hao Z, Ning Y, Wang GL (2019) SINA E3 ubiquitin ligases: versatile moderators of plant growth and stress response. Mol Plant 12: 610–612 [DOI] [PubMed] [Google Scholar]
- Zhao X, Li F, Li K (2021) The 14-3-3 proteins: regulators of plant metabolism and stress responses. Plant Biol (Stuttg) 23: 531–539 [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26: 1166–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Chung KP, Luo M, Jiang L (2018) Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci 23: 677–692 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Cui Y, Gao C, Jiang L (2015) Endocytic and autophagic pathways crosstalk in plants. Curr Opin Plant Biol 28: 39–47 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.