Abstract
Stem cells play important roles in animal and plant biology, as they sustain morphogenesis and tissue replenishment following aging or injury. In plants, stem cells are embedded in multicellular structures called meristems. The formation of new meristems is essential for the plastic expansion of the highly branched shoot and root systems. In particular, axillary meristems (AMs) that produce lateral shoots arise from the division of boundary domain cells at the leaf base. The CUP-SHAPED COTYLEDON (CUC) genes are major determinants of the boundary domain and are required for AM initiation. However, how AMs get structured and how stem cells become established de novo remain elusive. Here, we show that two NGATHA-LIKE (NGAL) transcription factors, DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (DPA4)/NGAL3 and SUPPRESSOR OF DA1-1 7 (SOD7)/NGAL2, redundantly repress CUC expression in initiating AMs of Arabidopsis thaliana. Ectopic boundary fate leads to abnormal growth and organization of the AM and prevents de novo stem cell establishment. Floral meristems of the dpa4 sod7 double mutant show a similar delay in de novo stem cell establishment. Altogether, while boundary fate is required for the initiation of AMs, our work reveals how it is later repressed to allow proper meristem establishment and de novo stem cell niche formation.
While boundary domains are required for the initiation of meristems, repression of boundary domain fate by the transcription factors DPA4 and SOD7 is required for rapid de novo stem cell formation.
IN A NUTSHELL.
Background: Stem cells function as sources of new cells for growth or replacing injured tissues. In plants, all tissues are descended from stem cells located in their growing tips, within the meristems, which are, therefore, essential for forming the shoot and root systems. Some of these meristems form during embryogenesis, but others form later. For example, the axillary meristems (AMs) that produce lateral shoots arise from a few cells in the boundary domain at the junction between the leaf and the stem. The CUP-SHAPED COTYLEDON (CUC) genes are major determinants of the boundary domain and are required for initiating these AMs. However, little is known about the later steps that lead to well-organized AMs and, in particular, to the de novo establishment of stem cells.
Questions: How are AMs structured during development? How is a new stem cell pool established de novo in an AM?
Findings: We mainly focused on the establishment of cauline AMs (CaAMs) in Arabidopsis thaliana because they are poorly characterized compared to rosette AMs. CaAMs form rapidly after the floral transition and we described the dynamic modification of gene expression that occurs along with their formation. Accordingly, while the CUC genes are required for the early steps of AM initiation, they have to be cleared for meristem establishment and activation of a stem cell population. We showed that two NGATHA-LIKE (NGAL) transcription factors are required to repress CUC expression in the initiating AM (cauline and rosette AMs) to permit de novo establishment of stem cells. Because we observed that stem cell formation is also affected in floral meristems of double mutants for these two NGAL transcription factors, our work reveals a mechanism operating in different types of newly formed meristems.
Next steps: One of the next steps would be to connect the role of the NGAL–CUC interaction during AM formation with other well-known regulators such as cytokinin signaling or HAIRY MERISTEM transcription factors. In addition, it would be interesting to test for a molecular connection between CUC transcription factors and genes controlling stem cell fate.
Introduction
Stem cells play a central role in animal and plant biology, as they are the sources of all cells that form organs and tissues during morphogenesis and allow cells to be replaced following injury or at the end of their lifecycle (Bäurle and Laux, 2003; Morrison and Spradling, 2008; Birnbaum and Alvarado, 2008). In both animals and plants, stem cells are maintained in their undifferentiated and pluripotent state through interactions with the microenvironment, which form a niche (Xie and Spradling, 2000; Dinneny and Benfey, 2008; Janocha and Lohmann, 2018; Pardal and Heidstra, 2021; Comazzetto et al., 2021). However, in contrast to what occurs in animals, plant stem cells cannot move and, as a consequence, stem cell niches have to be formed de novo in plants (Laird et al., 2008). Indeed, de novo stem cell establishment is essential to support the formation of new growth axes (shoots or roots), which allow plants to plastically expand their shape, enabling them to explore their environment.
In plants, stem cells and niches are embedded in multicellular structures called meristems. The shoot apical meristem (SAM), which forms during embryogenesis, is the direct source of the main shoot, forming stem and leaves after germination (Long et al., 1996). The SAM is a dynamic yet organized structure that is maintained through interactions between its different domains. In the apical part of the SAM lies a group of semi-permanent stem cells maintained by an underlying organizing center (OC) that contributes to the stem cell niche function (Laux et al., 1996). In Arabidopsis thaliana, the OC expresses the WUSCHEL (WUS) transcription factor, which travels through cellular connections to the overlying layers to induce stem cell fate (Mayer et al., 1998; Yadav et al., 2011; Daum et al., 2014; Perales et al., 2016; Sloan et al., 2020). In turn, stem cells express the excreted CLAVATA3 (CLV3) peptide that through interactions with different receptor kinases including CLAVATA1 (CLV1) undergoes feedback regulation to repress WUS activity in the OC (Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000; Müller et al., 2008; Schlegel et al., 2021). This core WUS/CLV regulatory feedback circuit is connected to other regulators such as auxin and cytokinin signals or the HAIRY MERISTEM (HAM) transcription factors and their regulatory miRNA, miR171 (Leibfried et al., 2005; Chickarmane et al., 2012; Zhou et al., 2015; Gruel et al., 2016; Ma et al., 2019; Han et al., 2020b). Altogether, this network contributes to proper spatial positioning of the stem cells and their fine tuning to allow meristem activity to respond to environmental signals (Yoshida et al., 2011; Pfeiffer et al., 2016; Landrein et al., 2018).
On the flanks of the meristem, new organ primordia are initiated following a spatial and temporal pattern that is orchestrated by auxin and cytokinin signaling (Reinhardt et al., 2003; Besnard et al., 2014). Proper initiation and separation of the organ primordia require the establishment of an organ boundary domain by multiple factors in which the CUP-SHAPED COTYLEDON (CUC) genes play a prominent role (Aida and Tasaka, 2006; Žádníková et al., 2014). This domain separates the leaf primordium from the meristem and will later give rise to the axillary region that lies on the inner base of the leaf. Multiple factors help coordinate primordium initiation with stem cell activities. For instance, the CUC genes are required for both organ formation and meristem maintenance (Aida et al., 1997), the HD-ZIP III transcription factors contribute to leaf polarity and meristem function (Kim et al., 2008; Caggiano et al., 2017), and auxin and cytokinins regulate both organ initiation and stem cell activity (Reinhardt et al., 2003; Chickarmane et al., 2012; Besnard et al., 2014; Ma et al., 2019).
While the root apical meristem and SAMs that form during embryogenesis generate the primary root and main shoot, respectively, the ramified architectures of the shoot and root systems result from the activity of meristems newly formed during postembryonic development. These lateral root and shoot meristems arise from a group of dividing cells originating from the root pericycle layer and the leaf axillary region, respectively, and acquire an organization and activity similar to the primary embryonic meristems, including a de novo established stem cell population and niche.
The formation of an axillary meristem (AM) between the developing leaf primordia and the SAM can be divided into three steps: the maintenance of a few meristematic cells at the leaf axil, the expansion of this cell population, and the establishment of a functional meristem (Wang et al., 2016; Wang and Jiao, 2018; Cao and Jiao, 2020). Multiple factors regulating these events have been characterized during the formation of the AM in the rosette leaves of A. thaliana. During the maintenance phase, a small group of cells located at the base of the developing leaf retains meristematic features while neighboring cells differentiate (Grbic and Bleecker, 2000; Long and Barton, 2000). Expression of the meristematic gene SHOOT MERISTEMLESS (STM) and the boundary domain genes CUC2 and CUC3 in these cells is required for AM initiation and, accordingly, stm, cuc2, or cuc3 mutants show defective AM formation (Grbic and Bleecker, 2000; Long and Barton, 2000; Hibara et al., 2006; Raman et al., 2008; Shi et al., 2016).
The maintenance of STM expression requires auxin depletion from the axillary region by polar auxin transport (Wang et al., 2014a, 2014b) and involves a self-activation loop facilitated by a permissive epigenetic environment (Cao et al., 2020). These cells can remain latent during a long period of time and, upon receiving proper environmental or endogenous signals, switch to the activation phase, during which their number rapidly increases by cell divisions to generate a small bulge. A strong increase in STM expression is instrumental for the switch to the activation phase (Shi et al., 2016). Multiple transcription factors, such as REVOLUTA, DORNRÖSCHEN, DORNRÖSCHEN LIKE, REGULATOR OF AMS1, 2, and 3 and REGULATOR OF AM FORMATION provide spatial and temporal cues for the local activation of STM expression (Greb et al., 2003; Keller et al., 2006; Müller et al., 2006; Raman et al., 2008; Shi et al., 2016; Zhang et al., 2018; Yang et al., 2012). Furthermore, a local pulse of cytokinin signaling reinforces STM expression to promote the formation of the AM, possibly through a mutual positive feedback loop between the STM and cytokinins (Wang et al., 2014b).
During the establishment phase, the bulge progressively acquires a typical meristem organization with functional sub-domains. Cytokinins promote de novo WUS expression, thus defining the OC (Wang et al., 2017). For this, the type-B Arabidopsis response regulator proteins, which mediate the transcriptional response to cytokinin, directly bind to the WUS promoter. In turn, WUS expression initiates the activation of the stem cell population marked by the expression of CLV3 (Xin et al., 2017). Interestingly, during the initial phase of WUS and CLV3 activation, both genes are expressed in overlapping domains in internal layers of the AM and only later do they shift to their proper expression patterns, with CLV3 expression shifting to the upper layers (Xin et al., 2017). This spatial rearrangement of CLV3 expression requires an apical–basal gradient of the activities of HAM genes, which results in part from the epidermis-specific expression of their negative regulator miR171 (Zhou et al., 2018; Han et al., 2020b, 2020a). Therefore, AM establishment is a gradual process, during which the expanding population of meristematic cells acquires specific identities, including the specification of apical stem cells and an underlying stem cell niche combined with organ boundary domains at the meristem flanks.
Arabidopsis floral meristems are thought to be modified AMs in which the subtending leaf is replaced by a cryptic bract whose development is suppressed (Long and Barton, 2000). Floral meristems also establish de novo a stem cell population marked by the rapid activation of WUS expression in Stage 1 floral meristems (Mayer et al., 1998) and by CLV3 expression by late Stage 2 (Seeliger et al., 2016). However, in contrast to AMs, in which stem cells are maintained, floral meristems are determinate structures with only a transient maintenance of stem cells. Indeed, the C-class floral gene AGAMOUS directly represses WUS by recruiting the Polycomb Repressive Complex 1 factor TERMINAL FLOWER 2 and induces KNUCKLES, which in turn represses WUS and interferes with WUS-mediated CLV3 activation (Lenhard et al., 2001; Liu et al., 2011; Sun et al., 2014; Shang et al., 2021).
Thus, while the molecular mechanisms allowing the preservation and amplification of a pool of meristematic cells leading to AM emergence has started to be deciphered, how the newly formed meristem becomes organized and activated remains far less understood. Here, we analyzed AM establishment in Arabidopsis, concentrating on cauline AMs (CaAMs), which are poorly characterized compared to rosette AMs (RoAMs). We show that CaAMs form rapidly following floral induction and that this is associated with dynamic changes in gene expression. Accordingly, while the CUC genes are required for the maintenance and activation phase, they have to be cleared for meristem establishment and activation of the stem cell population. Indeed, ectopic expression of the CUC boundary genes leads to asynchronous AM development and delayed de novo stem cell formation. We provide a molecular mechanism for this dynamic regulation of the CUC genes by two members of the NGATHA-like (NGAL) family of transcriptional repressors. A similar delay in de novo stem cell establishment is observed during floral meristem formation. Altogether, we reveal a genetic circuit repressing boundary cell fate that is required for de novo stem cell formation.
Results
Dynamic gene expression accompanies CaAM establishment
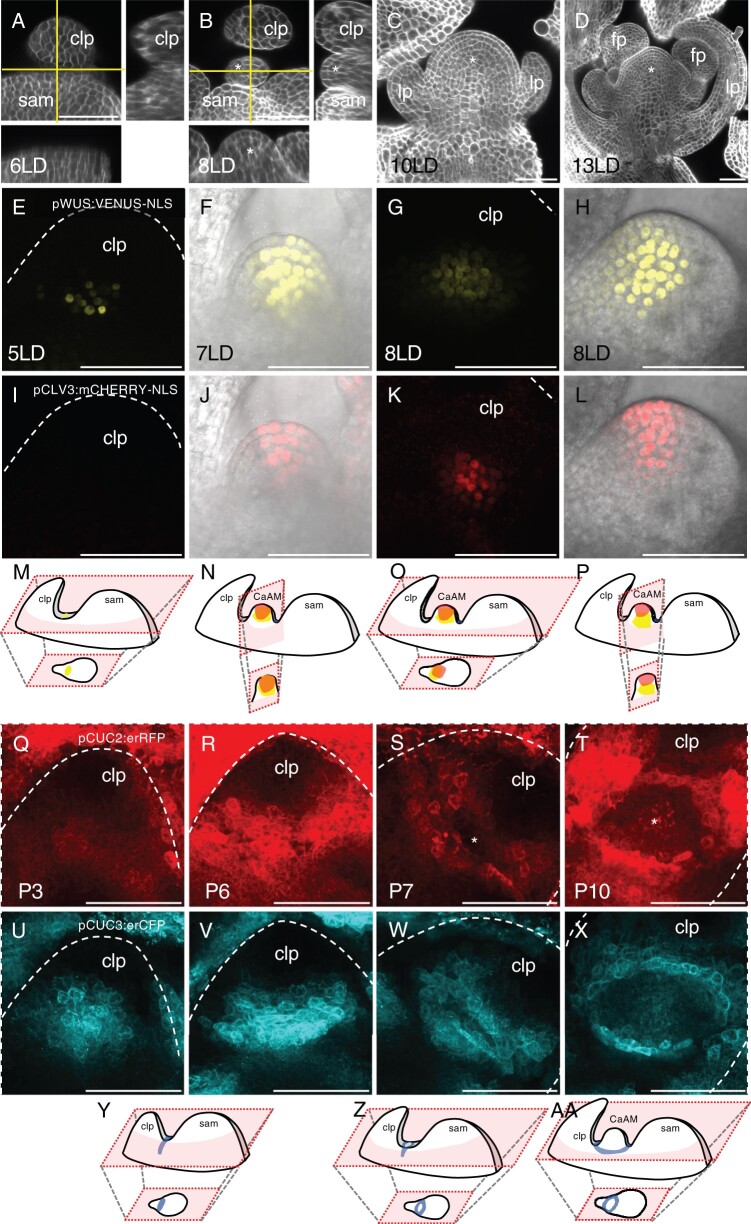
CaAMs rapidly form and grow out following the floral transition (Hempel and Feldman, 1994; Grbic and Bleecker, 2000; Burian et al., 2016). To provide a framework for CaAM formation in A. thaliana, we analyzed morphological changes in calcofluor-stained samples and gene expression dynamics in the leaf axillary region using reporter lines following the shift from short-day (SD) to long-day (LD) conditions (Figure 1). Six days after shifting to LD (6LD), the cauline leaf primordium was separated from the main meristem by a boundary containing small and narrow cells (Figure 1A). At 8LD, a bulge emerged between the cauline leaf primordium and the main meristem, defining the dome stage of the developing AM (Figure 1B). At 10 LD and 13 LD, leaf and flower primordia were formed by the AM (Figure 1, C and D), defining the leaf primordium and flower primordium stages, respectively.
Figure 1.
Rapid morphological changes and dynamic gene expression accompany CaAM formation. A–D, Optical sections of calcofluor-stained WT axillary regions following the SD to LD transition. A and B, Main panel: transverse optical section (with respect to the main stem axis), lower: reconstructed optical tangential section, right: reconstructed optical radial sections. Horizontal and vertical lines mark the position of the tangential and radial sections. C and D, Optical tangential sections. The number of days under LD conditions is indicated. E–L, Maximum projections of transverse (E, G, I, and K) and tangential (F, H, J, and L) optical sections of a line co-expressing pWUS:VENUS-NLS (E–H) and pCLV3:mCHERRY-NLS (I–L) reporters during CaAM formation. (F, J, H, and L) are a merge between reporter fluorescence and transmitted light. The number of days under LD conditions is indicated. M–P, Schemes showing the 3D structures of the apex with the SAM, the cauline leaf primordium and the developing CaAM. Schemes of the expression patterns of WUS and CLV3 are shown. The schemes of the optical sections are shown below. Scheme M corresponds to (E) and (I), scheme N to (F) and (J), and so on. Q–X, Maximum projections of transverse optical sections of a line co-expressing pCUC2:erRFP (Q–T) and pCUC3:erCFP (U–X) reporters during CaAM formation. Positions are numbered according to the rank of the primordium. Primordium number is indicated. Y–AA, Schemes showing the 3D structures of the apex with the SAM, the cauline leaf primordium and the developing CaAM. Schemes of the expression patterns of CUC3 are shown. The schemes of the optical sections are shown below. Scheme Y corresponds to (U) and (V), scheme Z to (W), scheme AA to (X). Scale bars = 50 µm; clp: cauline leaf primordium; asterisks: AM; lp: leaf primordium formed by the AM, fp: flower primordium formed by the AM. The dotted lines correspond to the outlines of the cauline leaf primordia.
To trace back the formation of the OC and stem cells during CaAM establishment, we first analyzed the expression dynamics of WUS and CLV3 transcriptional reporters (Pfeiffer et al., 2016). At 5 LD, pWUS:3xVENUS-NLS expression appeared in a few cells in P7, the seventh youngest visible primordia (Figure 1, E and M). The number of VENUS-expressing cells progressively increased during later stages (Figure 1, F–H and M–P). pCLV3:mCHERRY-NLS expression appeared only later: some CaAMs started to express CLV3 at 7LD, while at 8 LD, most CaAMs expressed CLV3 (Figure 1, I–L). Analysis of longitudinal optical sections showed that at 7 LD, WUS expression expanded from the corpus into the L2 and sometimes L1 layer (Figure 1F). Concomitant with the onset of CLV3 expression (Figure 1L), WUS expression became progressively excluded from the three outermost layers to finally mimic the expression observed in the SAM (Figure 1H). Therefore, like in the RoAMs, during de novo establishment of the stem cell niche in the CaAM, WUS is first activated, while CLV3 is expressed later in a domain contained in the WUS-expressing cells. These two overlapping domains then resolve into an apical CLV3 domain and a central WUS domain. However, whereas in the RoAMs, CLV3 showed a dynamic shift from the central to the apical domain (Xin et al., 2017) (Supplemental Figure S1), in CaAMs, the WUS domain shifted from the apical to the central domain.
Next, we followed the dynamics of CUC2 and CUC3 expression as these genes are redundantly required for CaAM formation (Hibara et al., 2006; Raman et al., 2008). During the early stages (P1–P6), the transcriptional reporters pCUC2:erRFP and pCUC3er:CFP showed a compact domain of expression at the boundary between the cauline leaf primordia and the meristem (Figure 1, Q, U, and Y). These expression domains became progressively more elongated while the groove separating the primordium from the meristem formed (P5–P6, Figure 1, R, V, and Y). Such CUC2 and CUC3 expression dynamics were independent of CaAM formation, as they were observed in the apices of plants regardless of whether they were shifted to LD. However, while in noninduced plants, the expression pattern of CUC2 and CUC3 remained as a compact line, starting 5LD onwards, it split into an eye-shaped structure, leaving a central region with reduced expression in P7–P8 primordia (Figure 1, S, W, and Z). The central domain depleted for CUC2 and CUC3 expression expanded during later stages (P9–P10 in plants at >6 LD), with the expression of the two reporters concentrating into a necklace-shaped structure around the outgrowing meristematic dome (Figure 1, T, X, and AA).
In conclusion, CaAM formation is a rapid process leading to de novo establishment of a novel functional meristem containing an organizing center and stem cell population. CUC2 and CUC3 expression is dynamic during CaAM formation, shifting from expression throughout the meristem to expression restricted around the meristem.
Identification of putative regulators of dynamic CUC gene expression
To identify possible transcriptional regulators of the dynamic expression of the CUC2 and CUC3 genes during AM formation, we performed an enhanced yeast one-hybrid screen using the CUC2 and CUC3 promoter regions as baits, as described in Gaudinier et al. (2011). Thus, we identified SUPPRESSOR OF DA1-1 7 (SOD7)/NGAL2 as a protein that binds to the CUC3 promoter, as revealed by LacZ activation. SOD7/NGAL2 is a member of the small family of NGAL transcription factors (Swaminathan et al., 2008; Romanel et al., 2009). We did not detect any interaction with ABS2/NGAL1, while DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (DPA4)/NGAL3 was not present in the transcription factor collection we screened (see Supplemental Data Set 1). However, because NGAL genes were shown to repress CUC gene expression during leaf and seedling development (Engelhorn et al., 2012; Shao et al., 2020), we next tested whether the NGAL genes could be involved in AM development.
SOD7/NGAL2 and DPA4/NGAL3 are redundantly required for efficient AM formation
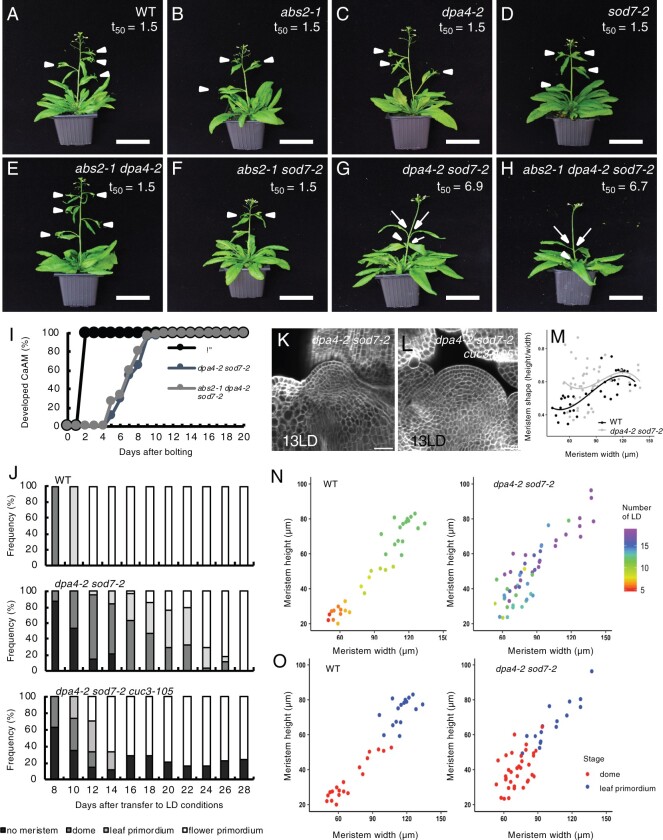
To determine if NGAL genes play a role in AM development, we grew single and multiple ngal mutants for 5 weeks in LD conditions. Undeveloped or delayed CaAMs were frequently observed in the dpa4-2 sod7-2 double mutant and abs1 dpa4-2 sod7-2 triple mutant compared to the CaAMs in wild-type (WT) and other single or double mutants (Figure 2, A–H). To quantify this phenotype more precisely, we examined the kinetics of CaAM development and calculated the time point after bolting at which half of the CaAMs were developed (t50). We observed a delay in the development of CaAMs for dpa4-2 sod7-2 (t50 = 6.9 days) and abs1 dpa4-2 sod7-2 (t50 = 6.6 days) compared to the WT and the other mutants (t50 = 1.5 days) (Figure 2I; Supplemental Figure S2A). Finally, the dpa4-3 sod7-2 double mutant with another dpa4 mutant allele also showed delayed CaAM development (Supplemental Figure S2, B and C). All together, these data show that the NGAL genes are redundantly required for CaAM development and that DPA4 and SOD7 play a major role in this process, while ABS2 has only a minor contribution.
Figure 2.
DPA4 and SOD7 are required for the rapid development of CaAMs. A–H, Inflorescences of WT, single, double, and triple ngal mutants. Plants were grown for 5 weeks in LD conditions. White arrowheads point to developed CaAMs, while the arrows point to delayed CaAMs. The time point after bolting at which half of the CaAMs are developed (t50, in days) is indicated under the genotype. I, Kinetics of CaAM development after bolting. Development of the CaAM is indicated as the percentage of developed branches (≥3 mm) to the total number of cauline leaves (n ≥ 11). J, Kinetics of CaAM development in WT, dpa4-2 sod7-2, and dpa4-2 sod7-2 cuc3-105 grown for 4 weeks in SD and transferred to LD (n ≥ 10). K and L, Tangential optical sections of calcofluor-stained WT of dpa4-2 sod7-2 (K) and dpa4-2 sod7-2 cuc3-105 (L) CaAM at 13LD. The WT control is shown in Figure 1D. M, Evolution of CaAM shape in WT and dpa4-2 sod7-2. N, CaAM height and width as a function of the number of days grown under LD conditions (number of LD; shown on the right) in WT and dpa4-2 sod7-2. O, CaAM height and width as a function of the CaAM stage (dome or leaf primordium) in WT and dpa4-2 sod7-2. Scale bars: (A–H) = 5 cm; (K and L) = 100 µm.
Next, we traced back the origin of the delayed AM development by looking at early stages of CaAMs in the WT and the dpa4-2 sod7-2 mutant. In the WT, all the CaAMs rapidly switched from the dome stage at 8 LD to the leaf primordium stage at 10 LD and at the flower primordium stage at 12 LD (Figure 2J, top plot). In contrast, no meristem was visible in the majority of dpa4-2 sod7-2 cauline leaves at 8 LD, while meristems at the dome stage were present only in approximately half of the axils at 10 LD (Figure 2J, middle plot). The appearance of leaf primordia and flower primordia was also delayed compared to the WT. In addition, confocal observations of dpa4-2 sod7-2 meristems at the dome stage showed that their shape was often abnormal, with a perturbed cellular organization, as divisions in any orientation were observed in the L2 layer (Figure 2K). To quantify the morphology of CaAMs, we measured their width and height and calculated the meristem aspect ratio (height divided by width). Interestingly, small dpa4-2 sod7-2 CaAMs (width < 90 µm) contained a higher meristem, on average, and a more variable shape compared to WT (Figure 2M). Larger meristems tended to regain a normal shape when their size increased. We noticed asynchronous development of the CaAM in dpa4-2 sod7-2; in contrast to the WT, the size of the meristem was not linked to the time spent by the plant under LDs (Figure 2N). Nevertheless, both mutant and WT meristems switched from the dome to the leaf primordium stage at a similar size (Figure 2O). In conclusion, in the dpa4-2 sod7-2 double mutant, CaAM formation is delayed, asynchronous, and associated with an abnormal cellular organization and shape at the dome stage, which reverts to a normal structure at the stage when leaf primordia are initiated.
SOD7/NGAL2 and DPA4/NGAL3 are required for proper CUC2 and CUC3 expression in CaAMs
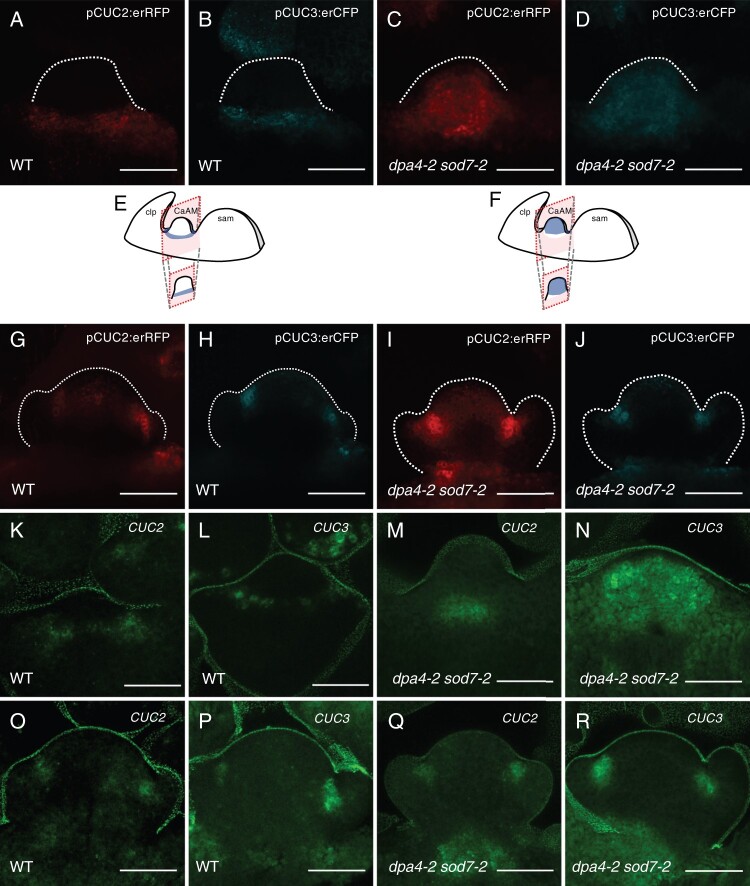
Because NGAL genes are known as negative regulators of CUC gene expression (Engelhorn et al., 2012; Shao et al., 2020), we analyzed CUC2 and CUC3 expression during CaAM development in dpa4-2 sod7-2 and WT by quantitative reverse transcription polymerase chain reaction (RT-qPCR). CUC2 and CUC3 mRNAs levels increased in developing axillary branches of dpa4-2 sod7-2 compared to the WT (Supplemental Figure S3, I and J). To follow CUC2 and CUC3 expression during early stages of CaAM development, we introduced the pCUC2:erRFP and pCUC3:erCFP transcriptional reporters into the dpa4-2 sod7-2 double mutant background. In the WT at the dome stage, pCUC2:erRFP and pCUC3:erCFP reporter expression was excluded from the meristem and was localized to its base (Figure 3, A, B, and E). In contrast, strong and uniform expression of the reporters was observed in dpa4-2 sod7-2 domes (Figure 3, C, D, and F). At the leaf primordium stage, pCUC2:erRFP and pCUC3:erCFP reporters were expressed at the boundary domain of the developing leaf primordia in the WT (Figure 3, G and H). A similar expression pattern was observed in the dpa4-2 sod7-2 mutant, with sometimes weak ectopic expression in the meristem (Figure 3, I and J).
Figure 3.
SOD7 and DPA4 are required for proper CUC2 and CUC3 expression in CaAM. A–D, Maximum projections of tangential optical sections of developing CaAMs of WT and dpa4-2 sod7-2 plants harboring the pCUC2:erRFP and pCUC3:erCFP reporters during CaAM development at the dome stage. E and F, Schemes showing the 3D structures of the apex with the SAM, the cauline leaf primordium (clp) and the developing CaAM. Schemes of the expression patterns of CUC3 are shown. The schemes of the optical sections are shown below. Scheme E corresponds to (B), scheme F to (D). G–J, Maximum projections of tangential optical sections of developing CaAMs expressing the pCUC2:erRFP and pCUC3:erCFP reporters in the WT and dpa4-2 sod7-2 backgrounds during CaAM development at the leaf primordia stage. K–R, Maximum projections of tangential optical sections of whole mount in situ hybridization of CUC2 and CUC3 transcripts in WT and dpa4-2 sod7-2 during CaAM development at the dome stage (K–N) and leaf primordia stage (O–R). Plants were grown for 4 weeks in SD conditions and then shifted to LD. Scale bars = 50 µm. The dotted lines correspond to outlines of the meristems and leaf primordia.
Whole mount in situ hybridization confirmed the similar localization of CUC2 and CUC3 mRNA in the organ primordia boundary domains of both WT and mutant meristems at the “leaf primordium” stage (Figure 3, O–R). CUC3 mRNA was distributed throughout the meristem at the dpa4-2 sod7-2 dome stage, in agreement with the expression pattern of the pCUC3:erCFP reporter (Figure 3N). CUC2 mRNA was observed in the rib zone of dpa4-2 sod7-2 dome stage meristems (Figure 3M), in contrast to the larger expression domain of the pCUC2:erRFP reporter (Figure 3C). Such a reduction of the pattern of CUC2 mRNA may be due to the posttranscriptional regulation of CUC2 by miR164 (Nikovics et al., 2006; Peaucelle et al., 2007; Sieber et al., 2007). The hypothesis that miR164 negatively regulates CUC2 expression during AM development is supported by the observation that the delay in CaAM development in the dpa4-2 sod7-2 (t50 = 5.74 days) mutant was enhanced by the inactivation of MIR164A (t50 = 6.77 days for dpa4-2 sod7-2 mir164a-4), one of the 3 MIR164 genes (Nikovics et al., 2006)(Supplemental Figure S3, A–E). Moreover combining dpa4-2 sod7-2 with the miRNA-resistant version of CUC2, CUC2g-m4 (Nikovics et al., 2006), led to an even stronger phenotype than dpa4-2 sod7-2 mir164a-4, with no development of the CaAM (Supplemental Figure S3, F–H). Together, these data show that DPA4 and SOD7 repress CUC2 and CUC3 expression in the developing AM at the dome stage.
CUC2 and CUC3 are required for the delayed CaAM development of the dpa4-2 sod7-2 double mutant
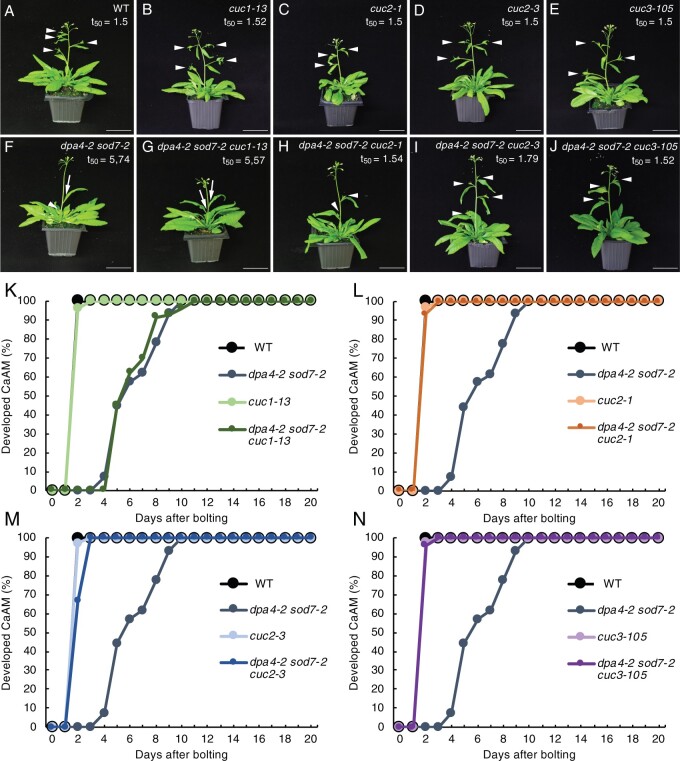
Because ectopic expression of CUC2 and CUC3 coincides with the developmental defects of the dpa4-2 sod7-2 CaAMs, we genetically tested the requirement for CUC genes to delay CaAM development in dpa4-2 sod7-2 (Figure 4). Introducing the cuc2-1 (t50 = 1.54 days) or cuc3-105 null allele (t50 = 1.52 days) into dpa4-2 sod7-2 restored growth of the CaAMs (Figure 4, A–J, L, and N). The cuc2-3 weak allele (t50 = 1.79 days) also led to the restoration of CaAM development, although not as efficiently as the cuc2-1 null allele (Figure 4M). In contrast, introducing the cuc1-13 null allele (t50 = 5.57 days) had no effect on CaAM development (Figure 4, G and K). Observation of early stages of CaAM development showed that an active meristem with proper cellular organization was more rapidly initiated in the dpa4-2 sod7-2 cuc3-105 triple mutant compared to dpa4-2 sod7-2 (Figure 2J, lower plot and Figure 2, K and L). Accelerated meristem development has been reported in mutants affected in the strigolactone pathway or the growth repressor gene BRC1 (Stirnberg et al., 2002; Booker et al., 2004; Aguilar-Martínez et al., 2007). However, introducing a mutant allele of BRC1, MORE AXILLARY BRANCHES2 (MAX2), or MAX3 into the dpa4-2 sod7-2 background led to no or weak restoration of CaAM growth (Supplemental Figure S4), suggesting that DPA4 and SOD7 do not control the strigolactone or BRC1 pathway. Together, these observations suggest that ectopic expression of the CUC2 and CUC3 genes is responsible for defective CaAM organization and delayed activity in dpa4-2 sod7-2.
Figure 4.
CUC2 and CUC3 are required for delayed CaAM development in dpa4-2 sod7-2 mutants. A–J, Inflorescences of WT, single cuc mutants, the dpa4-2 sod7-2 double mutant, and the dpa4-2 sod7-2 cuc triple mutants. Plants were grown for 5 weeks in LD. White arrowheads point to the developed CaAMs, while the arrows point to delayed CaAMs. The time point after bolting at which half of the CaAMs are developed (t50, in days) is indicated under the genotype. K–N, Kinetics of CaAM development in WT (K–N), dpa4-2 sod7-2 (K–N), cuc1-13, and dpa4-2 sod7-2 cuc1-13 (K), cuc1 and dpa4-2 sod7-2 cuc1 (L), cuc3 and dpa4-2 sod7-2 cuc3 (M) and cuc3-105 and dpa4-2 sod7-2 cuc3-105 (N) plants after bolting. Development of the CaAM is indicated as the percentage of developed branches (≥3 mm) to the total number of cauline leaves (n ≥ 7). All data were generated in the same experiments, and therefore the same WT and dpa4-2 sod7-2 data were used in (K–N). Scale bars = 5 cm.
DPA4 and SOD7 are expressed in the boundary domain and transiently in the initiating CaAM
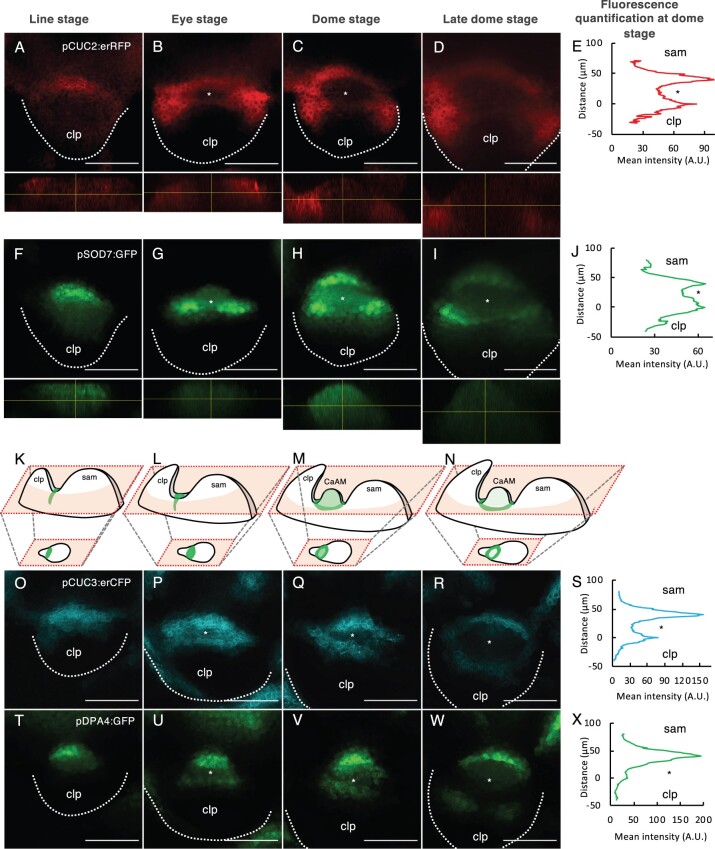
To follow the expression of the DPA4 and SOD7 genes, we generated transcriptional reporters and combined them with the pCUC3:erCFP or pCUC2:erRFP reporters (Figure 5; Supplemental Figure S5). During early stages, pSOD7:GFP and pDPA4:GFP expression overlapped with pCUC3:erCFP and pCUC2:erRFP in an elongated domain between the meristem and the cauline leaf primordium (Figure 5, A, F, K, O, and T). At the “eye” and “dome” stages, pSOD7:GFP and pDPA4:GFP were maintained in the central domain from which the meristem emerged, while pCUC3:erCFP or pCUC2:erRFP disappeared (Figure 5, B, C, G, H, L, M, P, Q, U, and V). At these stages, pDPA4:GFP tended to show higher expression on the SAM side. Fluorescence quantification along a radial axis from the SAM to the leaf primordium confirmed the stronger depletion of pCUC2:erRFP than pSOD7:GFP in the meristematic dome (Figure 5, C, E, H, and J) while pDPA4:GFP showed a peak of expression in the boundary domains closer to the SAM, with weaker expression in the emerging meristem and on the leaf primordium side (Figure 5, Q, S, V, and X). Later, pSOD7:GFP and pDPA4:GFP also became excluded from the meristem and limited to the boundary domain where pCUC3:erCFP is expressed (Figure 5, D, I, N, R, and W). A similar dynamic was observed when we compared the expression of pCUC2:erRFP with pDPA4:GFP or pCUC3:erRFP with pSOD7:GFP (Supplemental Figure S5).
Figure 5.
DPA4 and SOD7 have overlapping expression patterns with CUC2 and CUC3 in the boundary domain and are transiently expressed in the early AM. A–D and F–I, Maximum projections of transverse optical sections of plants co-expressing pCUC2:erRFP (A–D) and pSOD7:GFP (F–I) reporters. Orthoslices through the developing AM are shown below the transverse sections. E and J, Mean fluorescence along the radial axis of CaAM at the dome stage of the pCUC2:erRFP (E) or pSOD7:GFP (J) reporters (n = 6). K–N Schemes showing the 3D structures of the apex with the SAM, the cauline leaf primordium and the developing CaAM. Schemes of the expression patterns of SOD7 are shown. The schemes of the optical sections are shown below. Scheme K corresponds to (F), scheme L to (G), and so on. O–R and T–W, Maximum projections of transverse optical sections of plants co-expressing pCUC3:erCFP (O–R) and pDPA4:GFP (T–W) reporters. S and X, Mean fluorescence along the radial axis of CaAM at the dome stage of the pCUC3:erCFP (S) or pDPA4:GFP (X) reporters. A.U. = arbitrary units (n = 6). CaAMs are at the (A, F, O, and T) line, (B, G, P, and U) eye, (C, H, Q, and V) dome and late dome stage (D, I, R, and W). Scale bars = 50 µm; clp: cauline leaf primordium; asterisks: AM; The dotted lines correspond to the outlines of the cauline leaf primordia.
Disruption of putative NGAL-binding sites in CUC3 is sufficient to phenocopy the delayed secondary stem growth of dpa4-2 sod7-2
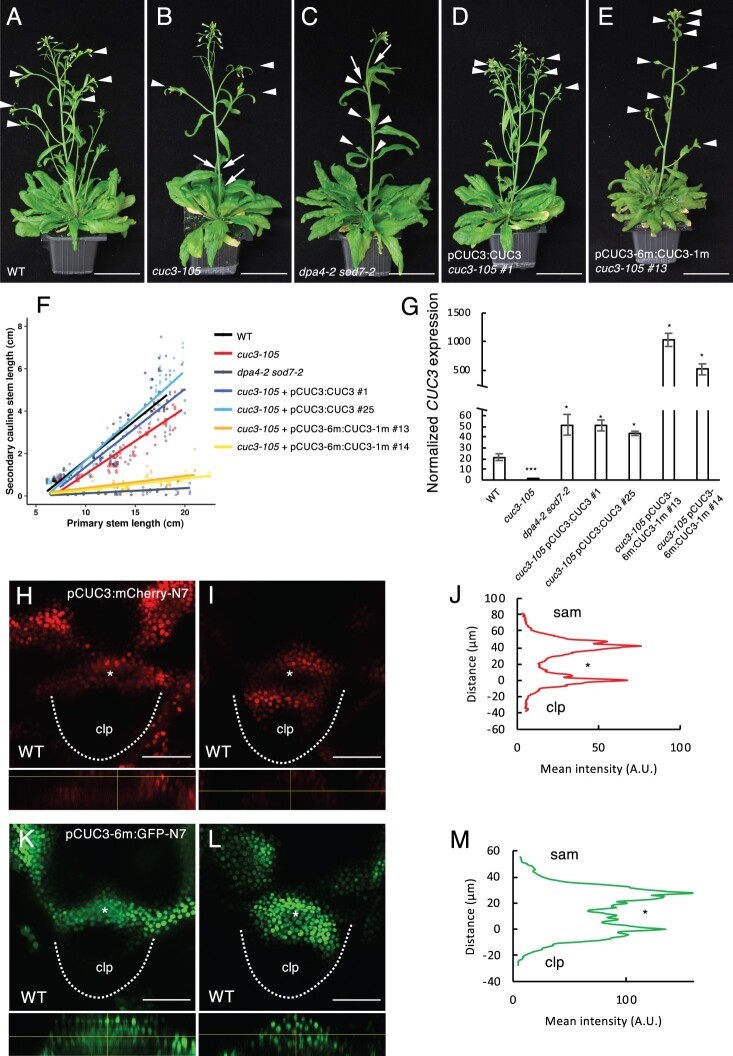
Next, we investigated the molecular interaction between NGAL proteins and CUC genes. We and others have shown that DPA4 and SOD7 repress CUC2 and CUC3 expression and that ABS2/NGAL1 directly binds to the CUC2 promoter (Engelhorn et al., 2012; Shao et al., 2020). It is also known that SOD7 binds to the promoter of the KLUH gene through its CACTTG motif (Zhang et al., 2015), and we found three such motifs in the CUC3 promoter (Supplemental Figure S6A). The DNA-binding matrix of the B3 domain of RAV1, a transcription factor of the same family as DPA4/SOD7, has been determined in vitro and contains a CACCTG consensus site (Yamasaki et al., 2004; Dmytrenko et al., 2015). Using the Jaspar database (Castro-Mondragon et al., 2022), we identified several such putative RAV1-binding sites in CUC3 (motif MA0583.1, Supplemental Table S1). SOD7 was able to bind to a sequence containing the CACCTG motif in the CUC3 promoter in vitro, as determined by electrophoretic mobility shift assay (EMSA; Supplemental Figure S6B). Altogether we identified three of these CACCTG motifs in the CUC3 promoter and one in its coding sequence.
To examine the roles of these motifs in regulating CUC3 expression, we generated a mutated version of CUC3 containing mutations in the three CACTTG and four CACCTG motifs (in pCUC3-6m:CUC3-1m, the mutation in the coding sequence is silent). We introduced pCUC3-6m:CUC3-1m or the pCUC3:CUC3 control construct into the cuc3-105 null mutant background. In contrast to what is observed under LD conditions (Figure 4), cuc3-105 plants shifted from SD to LD conditions showed a strong defect in CaAM initiation (63% of CaAM were not initiated at 32LD, Figure 6, A and B). This CaAM initiation defect was suppressed in two pCUC3:CUC3 cuc3-105 (4% of the CaAMs were not initiated) and two pCUC3-6m:CUC3-1m cuc3-105 lines (all CaAMs were initiated) (Figure 6, D and E), suggesting that a functional CUC3 was produced from both constructs. However, while growth of the secondary stems was similar to the WT in the two complemented pCUC3:CUC3 cuc3-105 lines, the two cuc3-105 lines complemented with the mutated pCUC3-6m:CUC3-1m constructs showed delayed development of secondary stems, similar to dpa4-2 sod7-2 (Figure 6, A–F). Furthermore, we observed a massive increase in CUC3 transcript levels in the pCUC3-6m:CUC3-m1 cuc3-105 lines compared to WT, cuc3-105, and the mutant complemented with pCUC3:CUC3 (Figure 6G). These results suggest that the putative NGAL-binding sites are required to repress CUC3 expression and the CUC3 overexpression resulting from their mutation leads to a delay in CaAM growth, thus partially phenocopying the dpa4-2 sod7-2 double mutant.
Figure 6.
Disruption of putative NGAL binding sites in CUC3 induces ectopic CUC3 expression and a delay in CaAM development. A–E, Inflorescences of WT, cuc3-105, dpa4-2 sod7-2, pCUC3:CUC3 cuc3-105 line #1, and pCUC3-6m:CUC3-1m cuc3-105 line #13. Plants were grown for 4 weeks in SD and then shifted to LD for 3 weeks. White arrowheads point to the developed CaAMs, while the arrows point to delayed CaAMs. F, Secondary cauline stem length as a function of primary length stem for WT, cuc3-105, dpa4-2 sod7-2, two pCUC3:CUC3 cuc3-105 lines (#1 and #25), and two pCUC3-6m:CUC3-1m cuc3-105 lines (#13 and #14). G, Quantification of the transcript level of CUC3 by RT-qPCR on 10 day-old seedlings of WT, cuc3-105, dpa4-2 sod7-2, two pCUC3:CUC3 cuc3-105 lines (#1 and #25) and two pCUC3-6m:CUC3-1m cuc3-105 lines (#13 and #14). Error bars are se. A Student’s t test was performed to compare the expression levels in the mutants compared to the WT (*P < 0.05; **P < 0.01; ***P < 0.001). H, I, K, and L, Maximum projections of transverse optical sections of CaAMs of WT plants harboring the pCUC3:mCherry-N7 (H and I) or pCUC3-6m:GFP-N7 (K and L) reporters during CaAM formation at the eye (H and K) and (I and L) dome stage. Orthoslices through the developing AM are shown below the transverse sections. J and M, Mean fluorescence along the radial axis of CaAMs harboring the pCUC3:mCherry-N7 (L) or pCUC3-6m:GFP-N7 (M) reporters at the dome stage. (n ≥ 5). Scale bars: (A–E) = 5 cm; (H, I, K, and L) = 50 µm. clp: cauline leaf primordium; asterisks: AM; the dotted lines correspond to the edges of the cauline leaf primordia.
Because we observed stronger CUC3 expression in dpa4-2 sod7-2 than the WT, we generated a pCUC3-6m reporter line to follow the pattern of the mutated promoter during CaAM development. The control reporter pCUC3:mCherry-N7 showed a clear depletion of fluorescence in the initiating meristem at the eye and dome stages (Figure 6, H and I), as previously observed with the pCUC3:erCFP reporter (Figure 1). In contrast, the fluorescence of the pCUC3-m6:GFP-N7 reporter remained homogeneous, and no clear depletion was observed at the eye stage (Figure 6K), while ectopic fluorescence remained in the developing meristem at the dome stage (Figure 6L). Quantifications confirmed the diminution of the mean fluorescence intensity inside the dome of the pCUC3:mCherry-N7 line, whereas it remained high in the pCUC3-m6:GFP-N7 line (Figure 6, J and M). At the leaf primordium stage, both WT and mutated reporter constructs showed a similar expression pattern in the boundary domain (Supplemental Figure S6, C and D). This suggests that mutating putative NGAL-binding sites in the CUC3 promoter delays its dynamic repression in the developing meristem. Remarkably, the pCUC3-m6:GFP-N7 reporter showed the same dynamic pattern as observed for the CUC3 transcript or pCUC3:erCFP reporter in dpa4-2 sod7-2. All these results suggest that disrupting putative NGAL-binding sites in CUC3 can induce the ectopic expression of CUC3 in the center of the CaAM, as observed in dpa4-2 sod7-2, which in turn delays secondary branch development.
Repression of the boundary identity is required for stem cell and stem cell niche establishment
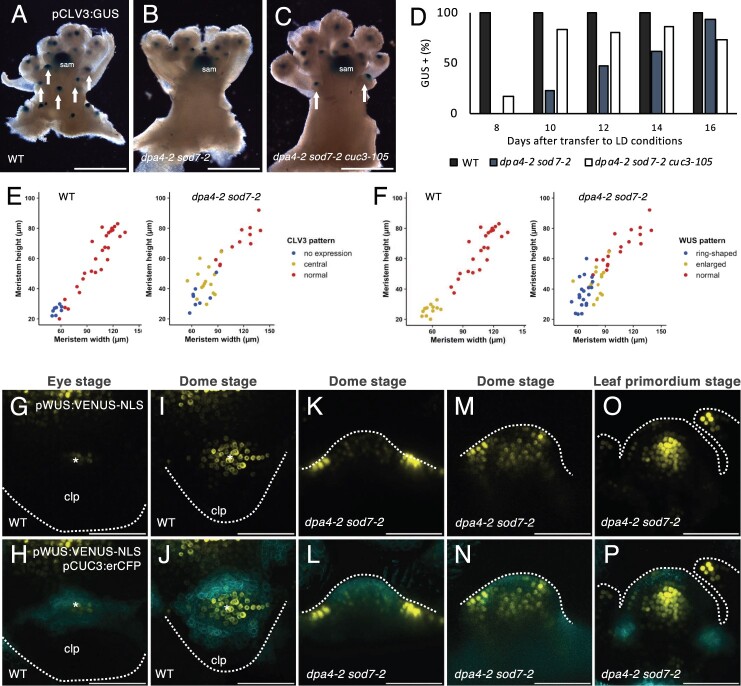
Because AM function is associated with the de novo establishment of stem cells, we next investigated whether stem cell formation is perturbed in dpa4-2 sod7-2 CaAMs. For this, we first followed the dynamics of pCLV3:GUS reporter activation in the CaAM (Figure 7, A–D). Whereas at 8 LD, pCLV3:GUS was expressed in all cauline leaf axils of the WT, none of the dpa4-2 sod7-2 double mutant axils showed visible GUS staining, and at 12LD, only approximately half of the axils of the double mutant expressed the pCLV3:GUS reporter. The dpa4-2 sod7-2 cuc3 triple mutant showed more rapid pCLV3:GUS activation, confirming that CaAM formation was partly restored in this background compared to the dpa4-2 sod7-2 double mutant (Figure 7, C and D).
Figure 7.
Stem cell specification is delayed in dpa4-2 sod7-2 AM. A–C, Expression of the pCLV3:GUS reporter in (A) dpa4-2 sod7-2 (B) dpa4-2 sod7-2 cuc3-105 (C) inflorescences. Plants were grown for 4 weeks in SD and then shifted to LD for 10 days. The arrows point to CaAM with pCLV3:GUS expression, and sam indicate the SAM. D, Quantification of GUS positive CaAM with pCLV3:GUS expression in plants shifted to LD. n ≥ 30. E, CLV3 expression pattern as a function of CaAM width and height in WT and dpa4-2 sod7-2. “normal” is CLV3 expressed in the apical region as shown in Supplemental Figure S7, A, C, D, and E, while “central” is CLV3 expression in the center of the meristem as shown in Supplemental Figure S7F. F, WUS expression pattern as a function of CaAM width and height in WT and dpa4-2 sod7-2. “enlarged” is WUS expressed in the entire meristem, as shown in (J) and (N), “ring-shaped” is WUS expressed at the base of the meristem as in (L), and “normal” is WUS expressed in a few cells in the center of the meristem as in (P). G–P, Maximum projections of transverse (G–J) and tangential (K–P) optical sections of CaAMs of WT and dpa4-2 sod7-2 plants harboring pWUS:VENUS-NLS and pCUC3:erCFP during CaAM development. Plants were grown for 4 weeks in SD conditions and then shifted to LD. Scale bars: (A–C) = 0.5 cm; (G–P) = 50µm; clp: cauline leaf primordium; asterisks: AM; the dotted lines correspond to the outlines of cauline leaf primordia (G–J), AM (K–P).
To further test whether the delayed pCLV3:GUS expression was due to the delayed outgrowth of CaAMs in the double mutant, we compared CLV3 expression in CaAMs of different genotypes at similar morphological stages by whole mount in situ hybridization (Supplemental Figure S7, A–D). Whereas at the dome stage, most WT CaAMs expressed CLV3, only 23% of dpa4-2 sod7-2 CaAMs showed CLV3 expression (n = 17). CLV3 expression was restored in all of the dome stage dpa4-2 sod7-2 cuc3 CaAMs (n = 10). These results suggest that the ectopic expression of CUC3 in the dpa4-2 sod7-2 meristem at the dome stage prevents the activation of CLV3 and that boundary fate needs to be repressed to allow stem cell establishment. Interestingly, while CLV3 expression was again observed in CaAMs of dpa4-2 sod7-2 and dpa4-2 sod7-2 cuc3-105 at the dome stage, its expression pattern was sometimes abnormal, as CLV3 tended to be expressed in the center of the meristem, as was observed during WT RoAM initiation (Supplemental Figure S7, E and F; Figure 1S). The central ectopic expression of CLV3 was observed in 78% and 70% of dpa4-2 sod7-2 (n = 14) and dpa4-2 sod7-2 cuc3-105 (n = 10) CaAMs, respectively. This expression of CLV3 likely represents a transitional phase, as it was mostly observed in small CaAMs in dpa4-2 sod7-2 (width <90 µm), while larger meristems showed a normal apical expression pattern (Figure 7E). Interestingly, the ectopic expression of CLV3 in dpa4-2 sod7-2 corresponds to the perturbed cellular organization observed at the dome stage in dpa4-2 sod7-2 (Figure 2).
Next, because WUS is expressed earlier than CLV3, and WUS and activates this gene, we wanted to know if WUS expression was also delayed in dpa4-2 sod7-2. For this, we compared the dynamics of the pWUS:VENUS-NLS and pCUC3:erCFP reporters in WT and dpa4-2 sod7-2 plants (Figure 7, G–P). In the WT background, at the eye stage, we observed a few cells expressing pWUS:VENUS-NLS in the center of the developing CaAM where pCUC3:erCFP expression started to disappear (Figure 7, G and H). Later during the dome stage, the pWUS:VENUS-NLS expression pattern enlarged and was highest in the part of the meristem where pCUC3:erCFP expression was low (Figure 7, I and J). Interestingly, in the smallest CaAMs of dpa4-2 sod7-2, pWUS:VENUS-NLS signals were very strong in a few cells at the outer base of the meristem, forming a ring-shaped structure, which is complementary to the pattern of pCUC3:erCFP expression inside the whole dome of the CaAM (Figure 7, K and L). Much weaker pWUS:VENUS-NLS expression was detected in a few cells within the meristem. Later on, pWUS:VENUS-NLS expression increased in the meristems of the dpa4-2 sod7-2 mutants (Figure 7, M and N). Lastly, during the leaf primordium stage, a normal pWUS:VENUS-NLS expression pattern was observed in dpa4-2 sod7-2, while pCUC3:erCFP expression returned to the boundary domains (Figure 7, O and P). Whole mount in situ hybridization confirmed the ectopic expression of WUS at the base of the dpa4-2 sod7-2 meristem, while the dpa4-2 sod7-2 cuc3-105 triple mutant showed a WT WUS pattern (Supplemental Figure S7, G–I).
Linking these WUS patterns with meristem size, we confirmed that in the WT, small meristems had an enlarged WUS expression zone, while at later stages, it became restricted to the center of the meristem (Figure 7F). In dpa4-2 sod7-2 mutants, WUS switched from an initial ring-shaped expression pattern around its base to expression throughout the meristem before becoming restricted to the central domain (“normal”; like in the WT; Figure 7F). These results suggest that the ectopic expression of CUC2/CUC3 prevents the activation of WUS in the meristem.
Together, our results suggest a scenario in which the transcription factors DPA4 and SOD7 are essential for the rapid repression of CUC2/CUC3 in the developing AM during the expansion phase (when the number of meristematic cells increases). If such a rapid repression does not occur, ectopic CUC2/CUC3 expression leads to defective meristem growth and organization and the delayed activation of WUS in the meristem, which in turn leads to the delayed activation of CLV3 and hence to defective de novo stem cell niche establishment.
Rosette AM development shows a similar requirement for DPA4/SOD7-mediated regulation of CUC genes to CaAM development
Next, we tested whether the findings specifically described above for CaAMs could be extended to RoAMs. A delay in RoAM outgrowth was observed for the dpa4-2 sod7-2 and abs1 dpa4-2 sod7-2 mutants grown in LD (Supplemental Figure 8SA), and this delay was partially suppressed by the mutation of CUC2 and CUC3 (Supplemental Figure 8SB). RoAM initiation was delayed in dpa4-2 sod7-2 grown in SD compared to the WT, but no modification of growth dynamics was subsequently observed in RoAM, in contrast to CaAMs (Supplemental Figure S8, C–F). The number of RoAMs expressing CLV3 was also reduced in dpa4-2 sod7-2 plants grown in SDs compared to the WT (Supplemental Figure S8G). Finally, cuc3-105 lines expressing pCUC3-6m:CUC3-1m, a CUC3 gene mutated at seven putative DPA4/SOD7 binding sites, showed a delay in RoAM growth (Supplemental Figure S8H). Altogether, these results suggest that the repression of CUC gene expression by DPA4/SOD7 is required for the proper establishment and growth of all AMs in Arabidopsis.
DPA4 and SOD7 facilitate the establishment of stem cells in the floral meristem
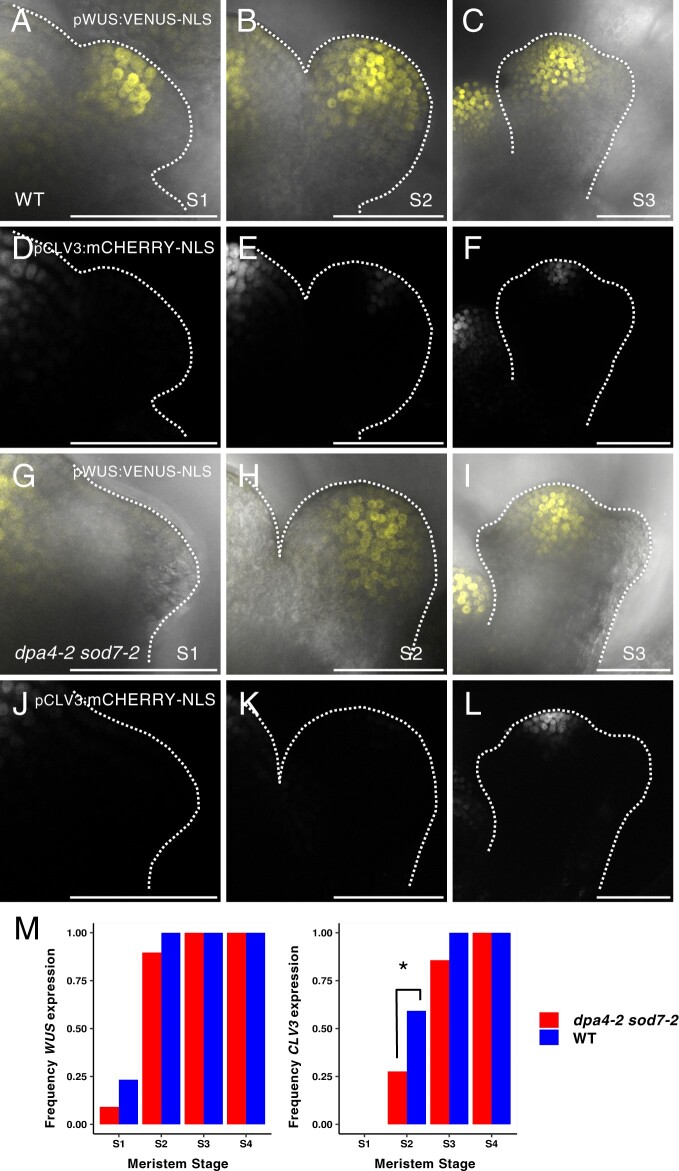
To investigate whether DPA4 and SOD7 play general roles in de novo stem cell formation, we analyzed stem cell establishment in newly formed floral meristems using the pWUS:VENUS-NLS and pCLV3:mCHERRY-NLS reporters (Figure 8). In agreement with previous reports (Mayer et al., 1998), in the WT, pWUS:VENUS-NLS was expressed in a small proportion of floral meristems at Stage 1 and was expressed in all Stage 2 flowers (Figure 8, A, B, and M). Slightly fewer Stage 1 and stage 2 dpa4-2 sod7-2 floral meristems seemed to express pWUS:VENUS-NLS, but this difference was not statistically significant (Figure 8, G, H, and M). WUS activation appeared to be also delayed when the meristems were staged according to their sizes (Supplemental Figure S9A). Interestingly, CLV3 expression was more strongly affected than WUS. Indeed, while 59% of WT Stage 2 floral meristems expressed pCLV3:mCHERRY-NLS (n = 27), only 27% of dpa4-2 sod7-2 Stage 2 floral meristems expressed this reporter (n = 29, Figure 8, E, K, and M, P < 0.05). At Stage 3, all WT meristems expressed the CLV3 reporter (n = 16), while it was absent from 14% of the dpa4-2 sod7-2 meristems (n = 14, Figure 8, F, L, and M). Accordingly, pCLV3:mCHERRY-NLS started to be expressed in dpa4-2 sod7-2 floral meristems that were almost twice as big as the WT (Supplemental Figure S9). Based on these results, we conclude that DPA4 and SOD7 act together to facilitate de novo stem cell establishment in floral meristems.
Figure 8.
Stem cell specification is delayed in dpa4-2 sod7-2 floral meristems. A–L, Maximum projections of developing WT (A–F) and dpa4-2 sod7-2 (G–L) floral meristems at Stage 1, (A, D, G, and J), 2 (B,E,H,K), or 3 (C, F, I, and L) co-expressing the pWUS:VENUS-NLS (A–C, and G–I) and pCLV3:mCHERRY-NLS reporters (D–F, and J–K). M, pWUS:VENUS-NLS and pCLV3:mCHERRY-NLS expression as a function of floral meristem stage. A Fisher’s test was performed to compare the expression levels of the mutants compared to the WT (*P < 0.05). n ≥ 30 for S1, n ≥ 27 for S2, n ≥ 14 for S3 and n ≥ 9 for S4. Scale bars = 50 µm.
Discussion
Stem cells are important throughout the lifecycles of all living organisms. In plants, new populations of stem cells and the meristems that contain them must form throughout the lifecycle to enable continuous growth and branching. Such meristems form in the axils of leaves from boundary domains that maintain meristematic features. Work in recent years has shown that AM initiation requires the maintenance of a meristematic fate by a dense network of interacting transcription factors and phytohormones, in which the CUC boundary genes play a central role, and accordingly, A. thaliana cuc mutants show strong defects in meristem initiation (Hibara et al., 2006; Keller et al., 2006; Müller et al., 2006; Raman et al., 2008; Tian et al., 2014). Here we showed that CUC gene expression must be downregulated in order for the initiating meristem to proceed to the establishment phase and become active. We showed that the NGAL transcription factors DPA4 and SOD7 are required to effectively remove CUC expression from the initiating AM. CUC mis-expression in the developing AM leads to asynchronous and delayed meristem formation, which is associated with abnormal cellular organization. Notably, ectopic expression of these boundary cell fate genes prevented stem cell establishment, which is required for meristem activity. Because we observed that delayed stem cell formation also occurs in floral meristems of the dpa4-2 sod7-2 double mutant, our work reveals a conserved genetic circuit by which the NGAL transcription factors repress the CUC boundary genes to allow de novo stem cell establishment in newly formed meristems.
Arabidopsis can form AMs from both its rosette and cauline leaves, and our work highlights the differences between the development of these two structures. First, while the formation of the RoAMs is a slow process extending over numerous plastochrons, the formation of the CaAM is much faster. It has been proposed that CaAMs are initiated as a result of the floral transition from the axils of the uppermost preexisting leaf primordia (Hempel and Feldman, 1994). Such CaAM induction occurs on leaf primordia at different stages and, accordingly, within successive CaAMs, we observed a trend in which the AM developed even more rapidly relative to the leaf primordium in the upper nodes. CaAM formation occurs rapidly: for instance, WUS expression is initiated in P13 in RoAMs (Wang et al., 2017), while we observed WUS expression as early as P7 in CaAMs. Activation of the floral program at the SAM may be an important factor contributing to the rapid formation of CaAMs. Support for this hypothesis comes from the observation that increased LEAFY (LFY) expression speeds up AM outgrowth (Chahtane et al., 2013). LFY forms a positive feedback loop with the auxin pathway during flower development (Yamaguchi et al., 2013), and while the formation of RoAMs requires local auxin depletion (Wang et al., 2014a, 2014b), high auxin signaling is observed in the young CaAM (Burian et al., 2016). This suggests that positive feedback between auxin and LFY could be active in CaAMs and contribute to their rapid growth.
A second difference between RoAMs and CaAMs is that CaAMs grow out directly after their initiation. As a consequence, mutations in genes inhibiting AM outgrowth, such BRC1 or genes of the strigolactone pathway such as MAX2/3 (Stirnberg et al., 2002; Booker et al., 2004; Aguilar-Martínez et al., 2007) do not further increase CaAM branching. Our genetic analyses indicated that the slow outgrowth of the dpa4-2 sod7-2 CaAMs can be slightly sped-up by mutations in strigolactone pathway components or brc1, suggesting that these pathways may still be active in CaAMs. However, the level of phenotypic restoration observed in these mutants was much lower than that observed in cuc mutants, suggesting that these pathways are not the primary pathways that are affected in the dpa4-2 sod7-2 mutants.
A third difference between CaAMs and RoAMs involves the dynamics of gene activation leading to stem cell establishment. While in both organs, WUS is activated before CLV3, in CaAMs, the WUS domain shifts from an apical to a central position whereas in RoAMs, WUS is already expressed in the central domain. In turn, CLV3 is properly positioned in the apical position from the beginning in CaAMs, while in RoAMs, it moves from a central to an apical position (Xin et al., 2017). Further characterization of cytokinin signaling or HAM gene spatial patterns (which contribute to stem cell establishment in RoAMs) in CaAMs will be necessary to understand these differences (Wang et al., 2017; Zhou et al., 2018; Han et al., 2020b, 2020a).
Our data show that while CUC genes are required for AM formation (Hibara et al., 2006; Raman et al., 2008), likely by preventing cell differentiation and maintaining cells in a meristematic fate, their expression must be negatively regulated to allow proper meristem establishment. Their prolonged, ectopic expression in the meristem is associated with asynchronous growth, abnormal cellular and meristem organization, and delayed organ initiation. These defects can be traced back to some roles of the CUC genes, as these genes have been shown to affect cell proliferation and cell expansion (Peaucelle et al., 2007; Sieber et al., 2007; Larue et al., 2009; Serra and Perrot-Rechenmann, 2020; Kierzkowski et al., 2019), as well as auxin transport and signaling (Bilsborough et al., 2011; Heisler et al., 2005; Maugarny-Calès et al., 2019). However, following an initial phase during which dpa4-2 sod7-2 meristems are misshapen, they recover, restraining CUC2 and CUC3 expression to the boundary. This transition is accelerated in the cuc3 mutant background, suggesting that ectopic CUC activity may be limiting for this. Such a reversion to a recovering meristem could be controlled by genetic factors. For instance, ABS2 (the third NGAL gene) might help exclude CUC expression from the meristem. However, no major differences in AM phenotypes were observed between dpa4-2 sod7-2 double and dpa4-2 sod7-2 abs1 triple mutants, suggesting that the role of ABS2 may be limited. Alternatively, miR164, a well-known repressor of CUC2 expression that acts independently of NGAL genes (Engelhorn et al., 2012), may also be involved in this process. Our genetic analysis with mutations modifying miR164 activity supports such a role.
An alternative hypothesis also emerges from a comparison with the patterning of the leaf margin that leads to tooth formation. In the leaf margin, a pattern with discontinuous CUC expression stripes forms as an emergent property of interconnected feedback loops between CUC activity and auxin transport and signaling (Bilsborough et al., 2011). In addition to the dynamics of these feedback loops, growth is essential for this patterning process, as it generates a cellular template large enough for the feedback loops to be deployed. In such a view, CUC expression patterns would be able to reorganize once the slowly growing meristems of the dpa4 sod7 mutants reach a critical size threshold. Testing such a hypothesis would require further investigations of the interconnections between AM growth and gene expression dynamics, for instance through combined modeling and experimental perturbation of growth.
The final step in meristem formation is the de novo establishment of an active stem cell niche. This is essential for the indeterminate fate of the AM but is also required for proper floral morphogenesis, as a reduction of the inner organs is observed in wus flowers whose stem cell niche is not properly specified (Laux et al., 1996). In both axillary and floral meristems, WUS activation precedes the expression of the stem cell marker CLV3. Here, we showed that in the WT initiating CaAM, WUS expression is rapidly induced in a few cells that are depleted for CUC3 expression. Later, the WUS domain progressively enlarges, occupying most of the developing meristem, a pattern complementary to that of CUC3-expressing cells. In dpa4-2 sod7-2, CUC2 and CUC3 mis-expression during the dome stage profoundly modifies WUS expression patterns, which become mostly restricted to a ring-shaped structure at the base of the meristem and excluded from the meristem itself. Therefore, like in the WT, the expression patterns of the CUC genes and WUS are essentially mutually exclusive in the dpa4 sod7 double mutant. This observation suggests a scenario in which CUC3 represses WUS expression, although alternative scenarios are possible. For instance, it has been suggested that geometrical changes of an emerging meristem might be sufficient for the activation of new WUS and CLV3 domains (Gruel et al., 2016). In such a view, defects in WUS and CLV3 activation in the double dpa4-2 sod7-2 mutant could be a consequence of abnormal meristem growth or shape.
While AMs are initiated from a group of cells expressing the CUC2 and CUC3 organ boundary genes, these boundary domains are located on one side of the initiating floral meristem (Heisler et al., 2005). Indeed, in floral meristems, CUC genes are expressed at Stage 1, forming the boundary between the floral primordia and the SAM until their expression disappears at Stage 4 (Hibara et al., 2006). Despite these differences in the origin of the meristem relative to the boundary domain, the dpa4 sod7 mutants show delayed stem cell specification in both the AM and floral meristems, suggesting that the NGAL/CUC regulatory module controls de novo stem cell formation in all aerial postembryonically formed meristems in a similar manner.
Materials and methods
Plant material and growth conditions
All A. thaliana genotypes used in this study are in the Columbia-0 (Col-0; WT) ecotype background. The cuc2-1 mutant was isolated from the Landsberg erecta ecotype but was backcrossed 5 times to Col-0 (Hasson et al., 2011). The mutant alleles dpa4-2 (Engelhorn et al., 2012), sod7-2 and dpa4-3 (Zhang et al., 2015), abs1 (Shao et al., 2012), cuc1-13, cuc2-3, cuc3-105 (Hibara et al., 2006), brc1-2 (Aguilar-Martínez et al., 2007), max2-1 (Stirnberg et al., 2007), and max3-11 (Booker et al., 2004) were previously described, as were the lines harboring pCUC3:erCFP (Gonçalves et al., 2015), pCUC2:erRFP (Gonçalves et al., 2017), CUC2g-m4 (Nikovics et al., 2006), pCLV3:GUS (Brand et al., 2002), and pCLV3:mCHERRY-NLS/pWUS:3X VENUS-NLS (Pfeiffer et al., 2016).
Seeds were soaked in water at 4°C for 48 h prior to sowing. Plants were grown in soil under LD conditions (2 h dawn [19°C, 65% relative humidity, 80 μmol.m−2.s−1 light, OSRAM LUMILUX T8 fluorescent tubes], 12 h day [21°C, 65% relative humidity, 120 μmol.m-2.s−1 light], 2-h dusk [20°C, 65% relative humidity, 80 μmol.m−2.s−1 light], 16-h dark [18°C, 65% relative humidity, no light]) or SD conditions (1 h dawn [19°C, 65% relative humidity, 80-μmol.m−2.s−1 light], 6-h day [21°C, 65% relative humidity, 120 μmol.m−2.s−1 light], 1-h dusk [20°C, 65% relative humidity, 80 μmol.m−2.s−1 light], 16-h dark [18°C, 65% relative humidity, no light]) and then shifted to LD. Seedlings from Figure 7G were grown in vitro on Arabidopsis medium (Duchefa) under long day conditions (16-h light/8-h dark at 21°C).
Enhanced yeast one-hybrid analysis
The CUC2 and CUC3 promoters were amplified by PCR using promCUC2 Fwd and promCUC2 Rv (3.7 kb) and prCuc3—Fw and prCuc3—R (4.3 kb) (see Primers in Supplemental Table S2). The promoters were recombined with the 5′-TOPO plasmid and then into pMW2 and pMW3 for HIS3 and LACZ reporter selection, respectively. Bait constructs were transformed into yeast as described in Gaudinier et al. (2011) and selected on -His and -Ura dropout medium and for minimal auto-activation in the reporter assays. The prey transcription factor collection used is described in Gaudinier et al. (2011) and Truskina et al. (2021) (see full list in Supplemental Data Set 1). Bait and prey transcription factors were introduced into a diploid yeast colony using the mating method as described in Gaudinier et al. (2011). The interaction between SOD7 and pCUC3 led to LacZ reporter activation but no HIS3 activation.
Generation of transgenic plants
The 2.8-kb promoter of DPA4 was amplified with Pdpa4-2FW and Pdpa4-2RV (Supplemental Table S2) and inserted in front of GFP in the pMDC107 plasmid to generate pDPA4:GFP. The 2.1 kb promoter of SOD7 (Zhang et al., 2015) was amplified with SOD7Profwattb1 and SOD7Prorvattb2 primers and inserted in front of GFP in pMDC107 to generate pSOD7:GFP. The promoters of DPA4 and SOD7 were cloned using a Gateway strategy.
All the parts used in the GoldenBraid version 2.0 strategy (Sarrion-Perdigones et al., 2013) are listed in Supplemental Table S3. The 4.3-kb promoter of CUC3 was amplified with GB_S1pCUC3S2_F and GB_S1pCUC3S2_R and inserted into pUPD2. Three fragments of the CUC3 coding sequence 175, 644, and 273 bp in size, respectively, were amplified with CUC3_S2F and CUC3_dom1R for fragment1, CUC3_dom1F and CUC3_dom2R for fragment2 and CUC3_dom2F and CUC3_S7R for fragment3, combined to obtain a 1-kb fragment, and then inserted into pUPD2. To generate CUC3-1m, we used CUC3 pUPD2 as a matrix and amplified it with CUC3_S2F and CUC3_CDS_PF3_r for the first fragment and with CUC3_CDS_PF3_f and CUC3_S7R for the second fragment to generate a silent mutation into the NGAL-binding site mutation BS3 (Supplemental Figure S7A). A 3.7-kb fragment of the CUC3 promoter with the six binding sites mutated (pCUC3-6m) (Supplemental Figure S7A) was synthesized by Genewiz (https://www.genewiz.com/) in the pUC-GW-Kan vector. The 3.7-kb pCUC3-6m fragment was the excised from pUC-GW-Kan with NsiI–PstI enzymes and inserted into the pCUC3 pUPD2 vector also digested NsiI–PstI enzymes to generate pCUC3-6m pUPD2.
To form the transcriptional unit, the different parts in the pUPD2 vectors were inserted into the pDGB3_α1 binary vector. The different TUs in pDGB3_α1 were combined either with pnos:hygro:tnos pDGB3_α2 or with pCMV:DSRed:tnos pDGB3_α2 into the pDGB3_Ω1 binary vector.
The resulting constructs (pMDC107 or pDGB3_Ω1) were sequence-verified and transferred into Agrobacterium tumefaciens strain GV3101. Plants were transformed by floral dipping. Primary transformants were selected in vitro for their resistance to hygromycin (pMDC107, pDGB3_Ω1) or selected with the red selection marker (pDGB3_Ω1). Several primary transformants were analyzed for their phenotype and for each construct, at least two independent lines were selected based on segregation of the resistance trait.
RNA whole mount in situ hybridization
RNA in situ hybridization was performed as described (L. Chelysheva, H. Morin, A. Nicolas, L. Barel, M. Da Costa, P. Rech, and J.C. Palauqui, unpublished data). Primers used to amplify the probes are indicated in Supplemental Table S2. In situ signal was revealed using a Vector Blue Substrate Kit, Alkaline Phosphatase (Vector Laboratories) and imaged by confocal microscopy (see Supplemental Table S4).
Preparation of CaAMs for confocal imaging
Plants were grown for 4 weeks in SD and shifted to LD. All observations were done in CaAMs between 5 and 16 days after shifting to LD. All observations were performed on fresh samples, except for samples shown in Figures 1, A–D and 2, K and L which were fixed in 4% paraformaldehyde under a vacuum for 1 h and cleared in Clearsee (10% xylitol, 25% urea, and 15% deoxycholate) (Kurihara et al., 2015) and Calcofluor (0.1%) for at least 2 weeks. Hand dissected meristems were mounted between a slide and coverslip with Tris–HCl 10 mM pH = 8.5, Triton 0.01%.
Confocal imaging
Confocal imaging was performed on a Leica SP5 inverted microscope (Leica Microsystems, Wetzlar, Germany). The lenses were Leica 40x HCX PL APO CS. Acquisition parameters are presented in Supplemental Table S4. Imaging was done from above for apices until 10–12 LD, while older apices had to be imaged from the side. Max intensity projections along the Z axis were made using ImageJ (see Supplemental Table S5 for details of projections), and FigureJ (Mutterer and Zinck, 2013) was used to assemble the figures.
Signal normalization and averaging
Fluorescence profiles were computed using Fiji, then spatially normalized and averaged based on the two major signal peaks. First, the localization of each peak was determined along each individual signal profile. For this, the profile was split into two halves on either side of the profile median position. Each of the two peaks was localized as the position of the maximal signal value on the corresponding side. To register several profiles, the resulting peaks were put in correspondence using linear scaling and translation of the profile axis. In the resulting reference axis, the distance between the two peaks can either be arbitrarily chosen, for example, by specifying that the two normalized peaks are separated from each other by a distance of 1 unit, or automatically from input data, for example, using the average distance between peaks computed from the data. After individual data normalization, profiles were averaged to yield the mean signal intensity profile. A script was developed in R for this and used to generate Figures 5, E, J, S, and X and 6, J and M (https://doi.org/10.57745/P6LIN2).
Scanning electron microscopy
Freshly sampled tissues were cooled to −33°C by a Peltier cooling stage (Deben) and observed with a Hirox SH-1500 benchtop scanning electron microscope.
RNA extraction and RT–qPCR expression analysis
Total RNA was isolated from the samples using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions for plant tissue, including DNase treatment. Reverse transcription was performed using RevertAid H Minus M-MuLV Reverse transcriptase (Fermentas), followed by a RNase H treatment for 20 min at 37°C to eliminate DNA–RNA duplexes. RT–qPCR analysis was performed on a Bio-Rad CFX connect machine using SsoAdvanced Universal SYBR Green Supermix following the manufacturer’s instructions. The PCR conditions were as follows: 95°C 3 min; (95°C 10 s; 63°C 10 s; 72C 10 s) × 45 cycles. Primers used for RT-qPCR analysis are listed in Supplemental Table S2. At2g28390 (QREF) and At4g26410 (REFA) (Czechowski et al., 2005) were used to normalize expression levels. Expression data were normalized using the ΔΔCt method (Livak and Schmittgen, 2001).
GUS staining
GUS staining was performed as described (Sessions et al., 1999) in the presence of 0.2-mM potassium ferricyanide and potassium ferrocyanide The reaction was stopped by adding 95% ethanol, which was also used to remove the chlorophyll from the tissues.
Phenotypic analysis
A count of CaAM and RoAM development was carried out over a period of 20 days after bolting (determined when the primary stem >1 mm) on plants grown for 5 weeks on LD. CaAMs and RoAMs were counted every 2 days. A meristem was considered present when it began to grow and be sufficiently visible to the naked eye (>3 mm). In addition, the final number of stem leaves and rosettes was also counted. We calculated the time point after bolting at which half of the CaAMs or RoAMs were developed (t50) using an R script (https://doi.org/10.57745/ZQZVPD).
A count of the stages of CaAM development was carried out over a period of 20 days after bolting (determined when the primary stem >1 mm) on plants grown for 4 weeks in SD and then shifted to LD. Observations were done on CaAMs between 8 and 28 days after shifting in LD under a binocular microscope.
AM height and width were measured on confocal longitudinal sections (either obtained directly during the imaging or reconstructed as orthoslices). The base of the meristem was fixed as a segment joining the junctions between the meristem and the neighboring tissues, and the width of the meristem was measured as the length of this segment and the height of the meristem as the length from this segment to the summit of the meristem. For floral meristems, developmental stages were recognized according to (Smyth et al., 1990). The sizes of floral meristems were measured on transverse sections: meristem length corresponds to the size of the floral meristem along the adaxial/abaxial axis, and the width was measured along the lateral axis.
EMSA
The MBP-SOD7 construct was generated as described previously (Zhang et al., 2015). The plasmid was introduced into Escherichia coli BL21 (DE3). Bacterial cells expressing MBP-SOD7 were lysed using PBS buffer (137-mM NaCl, 2.7-mM KCl, 1.4-mM KH2PO4, 4.3-mM Na2HPO4, pH 7.4) and then sonicated. Amylose resin (New England Biolabs, Ipswich, MA, USA, E8021L) was used to purify the MBP-SOD7 proteins. EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol. The competition experiments were performed by adding 5-, 10-, and 100-fold unlabeled DNA.
Statistical analysis
All values are represented as means and standard errors (SEs). Student’s t tests (Figure 6G, 3SI–J, 8SG) and Fisher’s tests (Figure 8M, 8SG) were performed by using Microsoft Excel 2016 and R as described in the individual figure legends. Results of the tests are shown in Supplemental Data Set 2.
Accession numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (https://www.arabidopsis.org) under the following accession numbers: CUC1 (At3g15170), CUC2 (At5g53950), CUC3 (At1g76420), DPA4/NGAL3 (AT5G06250), SOD7/NGAL2 (AT3G11580), ABS2/NGAL1 (AT2G36080), WUS (AT2G17950), and CLV3 (AT2G27250).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. WUS and CLV3 expression in the CaAM and rosette AM.
Supplemental Figure S2. DPA4 and SOD7 are required for the rapid development of CaAMs.
Supplemental Figure S3. Genetic interaction between MIR164 and DPA4/SOD7 during CaAM development and CUC2/CUC3 mRNA quantification in dpa4-2 sod7-2.
Supplemental Figure S4. Delayed development of dpa4-2 sod7-2 is not restored by mutations in BRC1/MAX genes.
Supplemental Figure S5. DPA4 and SOD7 have overlapping expression patterns with CUC2 and CUC3 in the boundary domain and are transiently expressed in the early AM.
Supplemental Figure S6. Putative NGAL binding sites in CUC3 and pCUC3/pCUC3-6m reporter expression in CaAM.
Supplemental Figure S7. CLV3 and WUS expression patterns in CaAMs.
Supplemental Figure S8. DPA4 and SOD7 are required for the rapid development of rosette AMs.
Supplemental Figure S9. WUS and CLV3 activation is delayed in dpa4-2 sod7-2 floral meristems.
Supplemental Table S1. Putative RAV1 binding sites identified in CUC3 using the Jaspar database.
Supplemental Table S2. Primers used in this work.
Supplemental Table S3. List of domesticated parts and TU in GoldenBraid.
Supplemental Table S4. Acquisition parameters for confocal imaging.
Supplemental Table S5. Projections details for all figures.
Supplemental Data Set 1. Prey transcription factors of the eY1H screen.
Supplemental Data Set 2. Details and results of the statistical tests.
Supplementary Material
Acknowledgments
We thank P. Cubas, C. Rameau, and the NASC for providing seeds. We thank N. Arnaud, N. Bouré, M. Azzopardi, and L. Gissot for providing parts used in the GoldenBraid cloning steps and Anne-Sophie Sarthou for technical help. We thank members of the FTA team at IJPB for discussion and N. Arnaud for comments on the manuscript.
Contributor Information
Antoine Nicolas, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France; Université Paris-Saclay, Orsay, 91405, France.
Aude Maugarny-Calès, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France; Université Paris-Saclay, Orsay, 91405, France.
Bernard Adroher, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France.
Liudmila Chelysheva, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France.
Yu Li, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Jasmine Burguet, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France.
Anne-Maarit Bågman, Department of Plant Biology and Genome Center, University of California, Davis, California 95616, USA.
Margot E Smit, Department of Plant Biology and Genome Center, University of California, Davis, California 95616, USA.
Siobhan M Brady, Department of Plant Biology and Genome Center, University of California, Davis, California 95616, USA.
Yunhai Li, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Patrick Laufs, Université Paris-Saclay, INRAE, AgroParisTech, Institut Jean-Pierre Bourgin (IJPB), Versailles, 78000, France.
Funding
The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). This work has benefited from the support of IJPB’s Plant Observatory technological platforms and financial support from the France Berkeley Fund.
Conflict of interest statement. None declared.
A.N., P.L., and A.M.C. conceived the project and P.L. supervised the project. A.N. performed most of the experiments with the help of P.L., A.M.C., A.M.B., and M.S. performed the Y1H screen under the supervision of S.B. A.M.C. did the preliminary genetic analysis. B.A. contributed to the generation of the double mutant and transgenic lines. L.C. conceived the whole mount in situ protocol and supervised A.N. for this. Yu.L. performed the gel shift experiment under the supervision of Y.L. J.B. wrote the fluorescence average script. A.N. and P.L. wrote the article with inputs of AMC.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Patrick Laufs (patrick.laufs@inrae.fr).
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis Branched1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Tasaka M (2006) Genetic control of shoot organ boundaries. Curr Opin Plant Biol 9: 72–77 [DOI] [PubMed] [Google Scholar]
- Bäurle I, Laux T (2003) Apical meristems: the plant’s fountain of youth. Bioessays 25: 961–970 [DOI] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S et al. (2014) Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421 [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108: 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum KD, Alvarado AS (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science (80) 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Brand U, Grunewald M, Hobe M, Simon R (2002) Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol 129: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian A, Barbier de Reuille P, Kuhlemeier C (2016) Patterns of stem cell divisions contribute to plant longevity. Curr Biol 26: 1385–1394 [DOI] [PubMed] [Google Scholar]
- Caggiano MP, Yu X, Bhatia N, Larsson A, Ram H, Ohno CK, Sappl P, Meyerowitz EM, Jönsson H, Heisler MG (2017) Cell type boundaries organize plant development. Elife 6: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jiao Y (2020) Control of cell fate during axillary meristem initiation. Cell Mol Life Sci 77: 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Wang J, Xiong Y, Yang H, Yang M, Ye P, Bencivenga S, Sablowski R, Jiao Y (2020) A self-activation loop maintains meristematic cell fate for branching. Curr Biol 30: 1893–1904.e4 [DOI] [PubMed] [Google Scholar]
- Castro-Mondragon JA, , Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva PérezN et al. (2022) JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 50: D165–D173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahtane H, Vachon G, Le Masson M, Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M et al. (2013) A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J Cell Mol Biol 74: 678–689 [DOI] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comazzetto S, Shen B, Morrison SJ (2021) Niches that regulate stem cells and hematopoiesis in adult bone marrow. Dev Cell 56: 1848–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Benfey PN (2008) Plant stem cell niches: standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Dmytrenko OP, Brusentsov VA, Pavlenko OL, Kulish MP, Busko TO, Grabovskyi YE, Zabolotny MA, Strelchuk VV, Romaniuk BM (2015) Irradiation-induced polymerization and damages of fullerite С60 under irradiation with Fe ions. Probl At Sci Technol 99: 8–14 [Google Scholar]
- Engelhorn J, Reimer JJ, Leuz I, Göbel U, Huettel B, Farrona S, Turck F (2012) DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 controls leaf margin architecture in Arabidopsis thaliana. Development 139: 2566–2575 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Zhang L, Reece-HoyesJS, , Taylor-Teeples M, Pu L, Liu Z, Breton G, Pruneda-PazJL, , Kim D, KaySA et al. (2011) Enhanced Y1H assays for Arabidopsis. Nat Methods 8: 1053–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves B, Hasson A, Belcram K, Cortizo M, Morin H, Nikovics K, Vialette-Guiraud A, Takeda S, Aida M, Laufs P, et al. (2015) A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J 83: 732–742 [DOI] [PubMed] [Google Scholar]
- Gonçalves B, Maugarny-Calès A, Adroher B, Cortizo M, Borrega N, Blein T, Hasson A, Gineau E, Mouille G, Laufs P, et al. (2017) GDP-L-fucose is required for boundary definition in plants. J Exp Bot 68: 5801–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V, Bleecker AB (2000) Axillary meristem development in Arabidopsis thaliana. Plant J 21: 215–223 [DOI] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruel J, Landrein B, Tarr P, Schuster C, Refahi Y, Sampathkumar A, Hamant O, Meyerowitz EM, Jönsson H (2016) An epidermis-driven mechanism positions and scales stem cell niches in plants. Sci Adv 2: 22–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Geng Y, Guo L, Yan A, Meyerowitz EM, Liu X, Zhou Y (2020a) The overlapping and distinct roles of HAM family genes in Arabidopsis shoot meristems. Front Plant Sci 11: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Yan A, Li L, Zhu Y, Feng B, Liu X, Zhou Y (2020b) A signal cascade originated from epidermis defines apical-basal patterning of Arabidopsis shoot apical meristems. Nat Commun 11: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P (2011) Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ (1994) Bi-directional inflorescence development inArabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286 [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janocha D, Lohmann JU (2018) From signals to stem cells and back again. Curr Opin Plant Biol 45: 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzkowski D, Runions A, Vuolo F, Strauss S, Lymbouridou R, Routier-Kierzkowska A-L, Wilson-Sánchez D, Jenke H, Galinha C, Mosca G, et al. (2019) A Growth-Based Framework for Leaf Shape Development and Diversity. Cell 177: 1405–1418.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T (2015) ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DJ, von Andrian UH, Wagers AJ (2008) Stem cell trafficking in tissue development, growth, and disease. Cell 132: 612–630 [DOI] [PubMed] [Google Scholar]
- Landrein B, Formosa-Jordan P, Malivert A, Schuster C, Melnyk CW, Yang W, Turnbull C, Meyerowitz EM, Locke JCW, Jönsson H (2018) Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc Natl Acad Sci USA 115: 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue CT, Wen J, Walker JC (2009) A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J 58: 450–463 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Liu X, Kim YJ, Müller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X (2011) AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218: 341–353 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K et al. (2019) WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugarny-Calès A, Cortizo M, Adroher B, Borrega N, Gonçalves B, Brunoud G, Vernoux T, Arnaud N, Laufs P (2019) Dissecting the pathways coordinating patterning and growth by plant boundary domains. PLoS Genet 15: e1007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC (2008) Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutterer J, Zinck E (2013) Quick-and-clean article figures with FigureJ. J Microsc 252: 89–91 [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal R, Heidstra R (2021) Root stem cell niche networks: it’s complexed! Insights from Arabidopsis. J. Exp. Bot. 72: 6727–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Morin H, Traas J, Laufs P (2007) Plants expressing a miR164 -resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV (2016) Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc Natl Acad Sci 113: E6298–E6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, Medzihradszky A, Schöne S, Daum G, Suzaki T, Forner J, Langenecker T, Rempel E et al. (2016) Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. Elife 5: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Romanel EAC, Schrago CG, Counago RM, Russo CAM, Alves-Ferreira M, Couñago RM, Russo CAM, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D (2013) Goldenbraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162: 1618–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel J, Denay G, Wink R, Pinto KG, Stahl Y, Schmid J, Blümke P, Simon RG (2021) Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. Elife 10: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Seeliger I, Frerichs A, Glowa D, Velo L, Comelli P, Chandler JW, Werr W (2016) The AP2-type transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE promote G1/S transition. Mol Genet Genomics 291: 1835–1849 [DOI] [PubMed] [Google Scholar]
- Serra L, Perrot-Rechenmann C (2020) Spatiotemporal control of cell growth by CUC3 shapes leaf margins. Development 147: dev183277. [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shang E, Wang X, Li T, Guo F, Ito T, Sun B (2021) Robust control of floral meristem determinacy by position-specific multifunctions of Knuckles. Proc Natl Acad Sci USA 118: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Liu X, Wang R, Zhang G, Yu F (2012) The over-expression of an Arabidopsis B3 transcription factor, ABS2/NGAL1, leads to the loss of flower petals. PLoS One 7: e49861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Meng J, Wang F, Shou B, Chen Y, Xue H, Zhao J, Qi Y, An L, Yu F, et al. (2020) NGATHA-LIKEs Control leaf margin development by repressing cup-shaped cotyledon2 transcription1. Plant Physiol 184: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Zhang C, Tian C, Wang J, Wang Q, Xu T, Xu Y, Ohno C, Sablowski R, HeislerMG et al. (2016) Two-Step Regulation of a Meristematic Cell Population Acting in Shoot Branching in Arabidopsis. PLoS Genet 12: e1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM (2007) Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Sloan J, Hakenjos JP, Gebert M, Ermakova O, Gumiero A, Stier G, Wild K, Sinning I, Lohmann JU (2020) Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL. Nat Commun 11: 2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Sun B, Looi LS, Guo S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T (2014) Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T (2008) The plant B3 superfamily. Trends Plant Sci 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C et al. (2014) An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Han J, Chrysanthou E, Galvan-AmpudiaCS, , Lainé S, Brunoud G, Macé J, Bellows S, Legrand J, Bågman A-M et al. (2021) A network of transcriptional repressors modulates auxin responses. Nature 589: 116–119 [DOI] [PubMed] [Google Scholar]
- Wang J, Tian C, Zhang C, Shi B, Cao X, Zhang TQ, Zhao Z, Wang JW, Jiao Y (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K (2016) Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol 209: 485–498 [DOI] [PubMed] [Google Scholar]
- Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K (2014a) Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiao Y (2018) Axillary meristem initiation — a way to branch out. Curr Opin Plant Biol 41: 61–66 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y (2014b) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC (2000) A niche maintaining germ line stem cells in the Drosophila ovary. Science (80) 290: 328–330 [DOI] [PubMed] [Google Scholar]
- Xin W, Wang Z, Liang Y, Wang Y, Hu Y (2017) Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J Plant Physiol 214: 1–6 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D (2013) A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–82 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y. et al. (2004) Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 16: 3448–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Mandel T, Kuhlemeier C (2011) Stem cell activation by light guides plant organogenesis. Genes Dev 25: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žádníková P, Simon R, Petra Z (2014) How boundaries control plant development. Curr Opin Plant Biol 17: 116–125 [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang J, Wenkel S, Chandler JW, Werr W, Jiao Y (2018) Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du L, Xu R, Cui R, Hao J, Sun C, Li Y (2015) Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 1: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yan A, Han H, Li T, Geng Y, Liu X, Meyerowitz EM (2018) Hairy meristem with wuschel confines clavata3 expression to the outer apical meristem layers. Science (80) 361: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.