Abstract
Background
DBA/1 mice have a higher susceptibility to generalized audiogenic seizures (AGSz) and seizure-induced respiratory arrest (S-IRA) than C57/BL6 mice. The gene expression profile might be potentially related to this difference. This study aimed to investigate the susceptibility difference in AGSz and S-IRA between DBA/1 and C57BL/6 mice by profiling long noncoding RNAs (lncRNAs) and mRNA expression.
Methods
We compared lncRNAs and mRNAs from the brainstem of the two strains with Arraystar Mouse lncRNA Microarray V3.0 (Arraystar, Rockville, MD). Gene Ontology (GO) and pathway analyses were performed to determine the potentially related biological functions and pathways based on differentially expressed mRNAs. qRT–PCR was carried out to validate the results.
Results
A total of 897 lncRNAs and 438 mRNAs were differentially expressed (fold change ≥2, P < 0.05), of which 192 lncRNAs were upregulated and 705 lncRNAs were downregulated. A total of 138 mRNAs were upregulated, and 300 mRNAs were downregulated. In terms of specific mRNAs, Htr5b, Gabra2, Hspa1b and Gfra1 may be related to AGSz or S-IRA. Additionally, lncRNA Neat1 may participate in the difference in susceptibility. GO and pathway analyses suggested that TGF-β signaling, metabolic process and MHC protein complex could be involved in these differences. Coexpression analysis identified 9 differentially expressed antisense lncRNAs and 115 long intergenic noncoding RNAs (lincRNAs), and 2010012P19Rik and its adjacent RNA Tnfsf12-Tnfsf13 may have participated in S-IRA by regulating sympathetic neuron function. The results of the qRT–PCR of five selected lncRNAs (AK038711, Gm11762, 1500004A13Rik, AA388235 and Neat1) and four selected mRNAs (Hspa1b, Htr5b, Gabra2 and Gfra1) were consistent with those obtained by microarray.
Conclusion
We concluded that TGF-β signaling and metabolic process may contribute to the differential sensitivity to AGSz and S-IRA. Among mRNAs, Htr5b, Gabra2, Hspa1b and Gfra1 could potentially influence the susceptibility. LncRNA Neat1 and 2010012P19Rik may also contribute to the different response to sound stimulation. Further studies should be carried out to explore the underlying functions and mechanisms of differentially expressed RNAs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-09016-3.
Keywords: Long noncoding RNAs, AGSz, S-IRA, SUDEP model
Introduction
C57BL/6 and DBA/1 mice are two different inbred strains that show diverse behavioral characteristics [1–4]. Specifically, DBA/1 mice have significantly higher susceptibility to audiogenic generalized seizures (AGSz), followed by seizure-induced respiratory arrest(S-IRA) [5]. This distinct feature may mimic the clinically observed sudden unexpected death in epilepsy (SUDEP) and thus makes DBA/1 mice relevant SUDEP models [6]. The underlying molecular mechanism of S-IRA following AGSz and S-IRA has not yet been clearly illustrated; however, many studies have provided valuable insights into these outcomes.
AGSz and S-IRA can be observed in many mouse strains and have been confirmed to indicate a unique form of seizure that originates in the brainstem. The physiological network in AGSz and S-IRA is thought to be common to different strains of mice, but the susceptibility to such seizures differs and may be influenced by many factors [7]. Considering only genetic background, the DBA mouse family (including DBA/1 and DBA/2 mice) show much higher susceptibility to AGSz than C57BL/6 mice, and several mutations have been found to be related to AGSz [5, 7]. However, genetic mutations cannot wholly explain the differences in susceptibility. Recent findings suggest that the serotonin (5-hydroxytryptamine, 5-HT) system plays an important role in AGSz and S-IRA. Selective serotonin reuptake inhibitors (SSRIs) were found to reduce S-IRA [8–10]. Western blotting also indicated that the tryptophan hydroxylase-2 (TPH 2) level in the DBA/1 mouse brainstem was significantly lower than that in the brainstem of the C57BL/6 J mouse [11]. Another report described that optogenetic activation of 5-HT neurons on the dorsal raphe of the brainstem reduced S-IAR in DBA/1 mice [12].
Other factors, such as time after birth, also influence susceptibility. Naturally, the AGSz and S-IRA of DBA/1 mice only exist in the first 5 weeks of age. However, they can be induced by repeated audio stimulation in early life, which was termed ‘priming’ [6]. Priming endows DBA/1 mice with much greater susceptibility, which may be reestablished in later life via the same stimulation [9]. These studies have suggested that brain development and the environment participate in susceptibility, and thus, epigenetic regulation should be considered.
However, direct evidence for epigenetic regulation is limited. In this study, we aimed to compare the differences in mRNA and long noncoding RNA (lncRNA) expression profiles in the brainstem of DBA/1 and C57BL/6 mice, which may provide insights into the potential epigenetic regulation that mediates AGSz and S-IRA.
Materials and methods
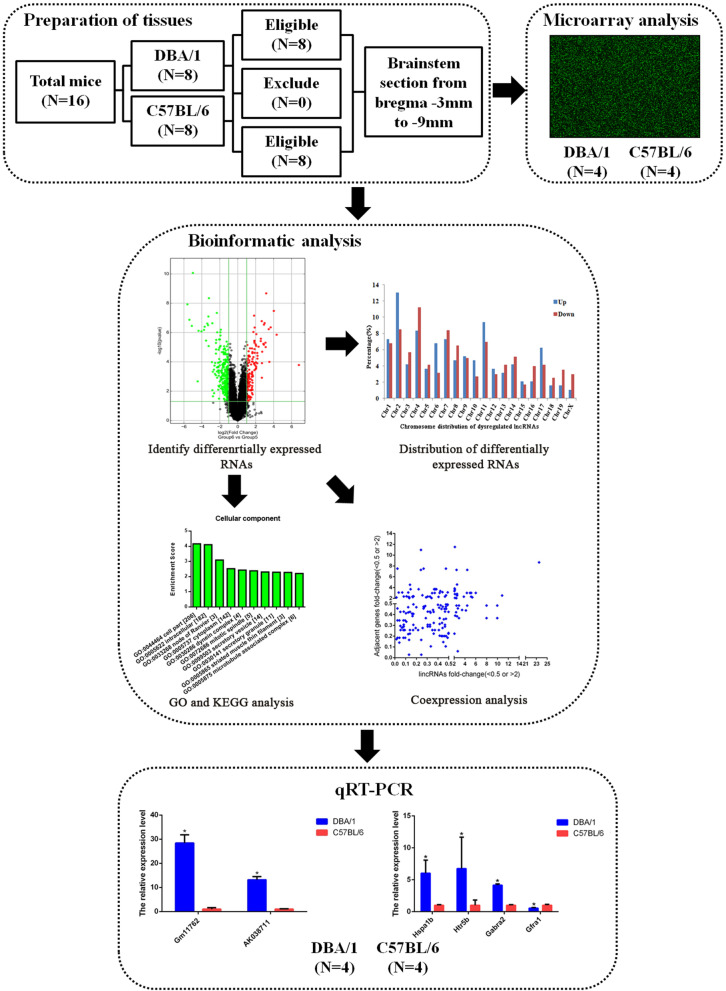
In this study, we established four procedural modules, including modules for the preparation of tissues, a microarray analysis, a bioinformatics analysis, and qRT–PCR (Fig. 1).
Fig. 1.
The flowchart of experimental procedure. The study contained four procedural modules, including preparation of tissues, microarray analysis, bioinformatics analysis, and qRT-PCR
Preparation of tissues
All experimental operations and procedures with animals were performed in accordance with the Guidelines of Animal Care and Use Committee of Sichuan University West China Hospital. The study was not preregistered. Since the AGSz response without priming in DBA/1 mice was quite stable on different postnatal days (PNDs) from 21 to 112 PNDs [13], we purchased PND 28-30 DBA/1JNCrlj and C57BL/6JNifdc male mice from the Charles River Laboratories Experimental Animal Center (Beijing, China). The DBA/1 and C57BL/6 mice were housed in standard laboratory cages (4 mice per cage), and the mice had free access to water and food in a temperature-controlled room (21 °C-25 °C). All animals were maintained under a 12-h light/dark cycle. We evaluated only male DBA/1 mice because 1. previous reports indicated that they are slightly more susceptible to AGSz and S-IRA than females [9] and 2. the susceptibility to seizure may be influenced by ovarian hormones in female mice [14]. Since AGSz and audio stimulation potentially affect RNA expression, these mice were housed in specific pathogen-free (SPF) conditions for 1 week without testing for AGSz. The DBA/1 (n = 8) and C57BL/6 (n = 8) mice were decapitated at the age of 5 weeks under anesthesia with isoflurane inhalation, and the whole brainstem between bregma − 3 mm and bregma − 9 mm was taken as described in a previous study [11]. The brainstem samples were immediately frozen in liquid nitrogen and then stored at − 80 °C for later use. Four samples from each strain were used for the microarray analysis, and the other 4 samples were used for verification by qRT–PCR.
RNA extraction
Total RNA was extracted from DBA/1 and C57BL/6 brainstem tissues using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). RNA quantification and quality were evaluated with a NanoDrop ND-1000 spectrometer (Thermo Fisher Scientific, USA). Standard denaturing agarose gel electrophoresis was applied for the measurement of RNA integrity.
Microarray analysis
An Arraystar mouse lncRNA Microarray V3.0 (Arraystar, Rockville, MD) was used in our study, which could detect about 35,923 lncRNAs and 24,881 coding transcripts. The microarray analysis was performed by KangChen Biotech (Shanghai, China). The lncRNAs were annotated by using authoritative public transcriptome databases (NCBI Refseq 2014, UCSC Known Gene 6.0, Ensembl 38.71 and Genbank) and landmark publications (see Additional file 1), while coding mRNAs were collected from Collaborative Consensus Coding Sequence (CCDS) Project. Sample labeling and array hybridization were carried out on the basis of the Agilent one-color microarray-based gene expression analysis experimental scheme (Agilent Technology). First, we extracted rRNA from total RNA to obtain purified mRNA (RNA-ONLY Eukaryotic RNA Isolation Kit, Epicentre). Second, we used a random primer method to amplify and transcribe each sample into fluorescent cRNA (Arraystar flash RNA labeling kit, Arraystar). Following this, an RNeasy Mini Kit (Qiagen) was utilized to purify the labeled cRNA, the concentration and activity of which were further determined with a NanoDrop ND-1000. Then, the labeled cRNA was hybridized onto microarray slides, and the hybridized arrays were washed, fixed and scanned with an Agilent DNA Microarray Scanner (part number G2505C).
Bioinformatic analysis
The Agilent Feature Extraction software (version 11.0.1.1) and the GeneSpring GX v12.1 software package (Agilent Technologies) were applied to analyze array images and quantile normalization of the raw data, respectively. Quantile normalization was performed as follows: The expression values of specific RNAs were listed in a matrix where each row represents one RNA and each column represents one sample. For each sample, the original values of different RNAs were sorted in ascending order in the column. The mean of the sorted order across each row was obtained, and then, the value of each row was replaced by this mean. Finally, the modified matrix in the previous step was rearranged to follow the same order as the input matrix.
A hierarchical clustering map and volcano plot were created to present the profiles of differentially expressed lncRNAs and mRNAs. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed to identify the potential biological functions and pathways in which the differentially expressed mRNAs were enriched.
We performed coexpression analysis to show the aberrantly expressed antisense lncRNAs with their sense mRNAs and the different expression of long noncoding intergenic RNA (lincRNA) with their nearby coding genes, which was considered important in a bioinformatics analysis [15]. We first subdivided lncRNAs into 6 subgroups, which were defined as follows:
Sense overlapping, the lncRNA exon overlapped a coding transcript exon in the same genomic strand;
Intronic, the lncRNA overlapped an intron of a coding transcript in the same genomic strand;
Natural antisense, the lncRNA was transcribed from the antisense strand and overlapped a coding transcript;
Nonoverlapping antisense, the lncRNA was transcribed from the antisense strand without overlapping an exon;
Bidirectional, the lncRNA was oriented head-to-head with a coding transcript within 1000 bp;
Intergenic, there were no overlapping or bidirectional coding transcripts near the lncRNA.
In the second part of the coexpression analysis, we included all intergenic lncRNAs and set the distance from the lncRNAs to nearby genes to be < 300 kb to identify lincRNAs. As a major lncRNA subtype, a lincRNA is transcribed from intergenic regions and is involved in regulating the expression of adjacent genes [16].
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction (qRT–PCR) were performed to verify different expression patterns of lncRNAs and mRNAs obtained in the microarray analysis by a SYBR green PCR kit and a ViiA 7 Real-time PCR System (Applied Biosystems). All the expression levels of lncRNAs and mRNAs were normalized to the level of the internal reference gene (GAPDH). In this study, dysregulated lncRNAs used for verification were selected primarily on the basis of fold change. We also considered homology between mice and humans and selected some lncRNAs of interest in our study. For the selection of mRNAs, we took the results of GO and KEGG analyses into account and then combined these results with potential known mechanisms that may contribute to differences in AGSz or S-IRA. The sequences of the primers are listed in Table 1.
Table 1.
The sequences of lncRNA and mRNA primers used in the study
| Differential expression | Gene name | Primers (5′-3′) (F=Forward; R = Reverse) |
Amplicon size (bp) |
|---|---|---|---|
| lncRNAs | uc007uzp.1 | F: CACCAAATGGGCTGGACAA | 197 |
| R: GGCTAAAGGCAGACTGGAATC | |||
| NR_045099 | F: GCCATCCAGTTCCATCTTTCT | 143 | |
| R: GCCCCTGTCTGTTCTCCATAA | |||
| NR_015498 | F: CAACGGAGTTACTATGGGTCG | 279 | |
| R: GAGGCTACGGGTGAGGTTAT | |||
| NR_033305 | F: CAAGGAACTTTGGTCGTAGC | 86 | |
| R: AGCAATACAACAATGACTAAGACA | |||
| NR_003513 | F: GGTTGTTTTGTGAGTGTGCTTA | 169 | |
| R: GGGGAGGAAAATGGTTAGTG | |||
| mRNAs | Hspa1b | F: TATAGTCTAGCTGCCCAGTTCC | 75 |
| R: CAGTGCCAAGACGTTTGTTT | |||
| Htr5b | F: GGTGGTGCTCTTCGTCTACT | 179 | |
| R: AGTCTCCGCTTGTCTGGAAG | |||
| Gabra2 | F: ACAGTCCAAGCCGAATGTCC | 138 | |
| R: AACGGAGTCAGAAGCATTGTAAGT | |||
| Gfra1 | F: CCACTCCTGGATTTGCTGAT | 152 | |
| R: CTGAAGTTGGTTTCCTTGCC | |||
| Internal control | GAPDH | F: CACTGAGCAAGAGAGGCCCTAT | 144 |
| R: GCAGCGAACTTTATTGATGGTATT |
Statistical analyses
Differentially expressed lncRNAs and mRNAs were identified as those with a fold change ≥2.0 and a P value < 0.05. We performed false discovery rate (FDR) correction to minimize false-positives. Another analysis based on a fold change ≥2.0 and an FDR < 0.05 was performed as a sensitivity test. The qRT–PCR results are shown as the relative expression levels. We used an independent set of 4 DBA/1 and 4 C57BL/6 mice for qRT–PCR to verify the microarray findings. By setting the expression value of the target genes in the C57BL/6 control group to 1, the expression level in the DBA/1 mouse group is reported the fold change compared with the control group.
Results
Differentially expressed lncRNAs and mRNAs in DBA/1 mice compared with C57BL/6 mice
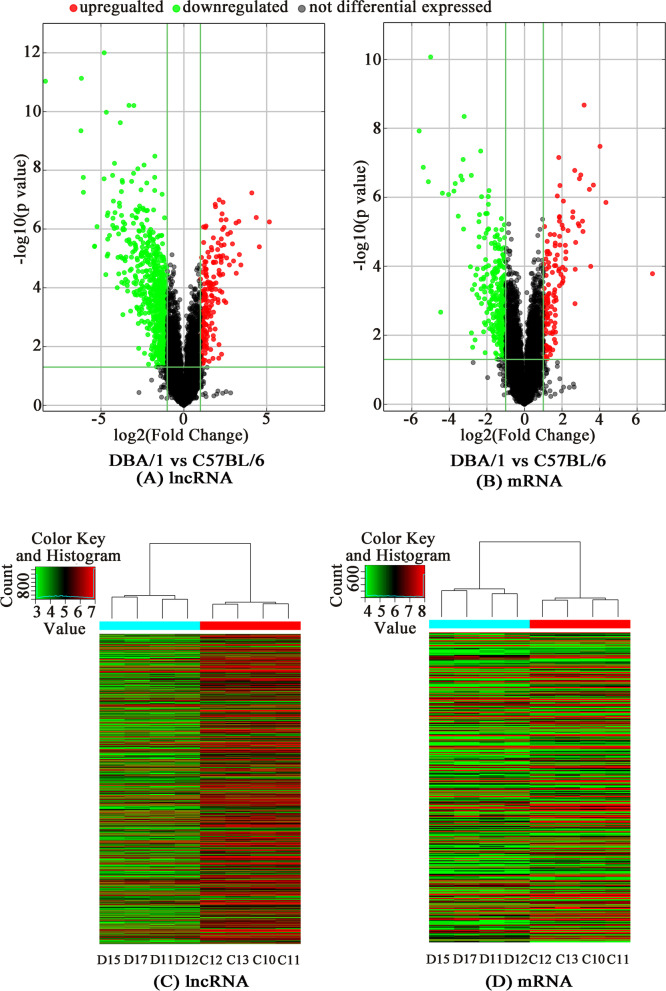
The microarray analysis led to the identification of a total of 897 significantly differentially expressed lncRNAs (192 up- and 705 downregulated) and 438 differentially expressed mRNAs (138 up- and 300 downregulated). The details of all these differentially expressed RNAs are showed in the supplementary material (Additional files 1 and 2). The 20 most differentially expressed lncRNAs and mRNAs between DBA/1 and C57BL/6 mice are listed in Table 2 and Table 3, respectively. Figure 2 shows the volcano plots and hierarchical clustering analysis depicting the expression levels of the distinguishable lncRNAs and mRNAs.
Table 2.
Top 20 differentially expressed lncRNAs between DBA/1 and C57BL/6 mice
| Up-regulated lncRNAs | Down-regulated lncRNAs | ||||||
|---|---|---|---|---|---|---|---|
| Seqname | GeneSymbol | Fold Changea | FDR | Seqname | GeneSymbol | Fold Changea | FDR |
| ENSMUST00000117627 | Gm14201 | 35.75 | 0.0002 | uc009bwo.2 | AK005187 | 326.12 | 0.0000 |
| uc007uzp.1 | AK038711 | 23.50 | 0.0005 | uc008pwq.2 | 1500004A13Rik | 73.85 | 0.0000 |
| NR_045099 | Gm11762 | 20.65 | 0.0001 | NR_015498 | 1500004A13Rik | 72.49 | 0.0000 |
| ENSMUST00000173149 | H2-Bl | 17.14 | 0.0000 | NR_033305 | AA388235 | 66.92 | 0.0000 |
| ENSMUST00000174778 | Gm10499 | 10.86 | 0.0013 | AK047380 | AK047380 | 66.64 | 0.0000 |
| uc008ryr.1 | AK019984 | 10.21 | 0.0008 | AK047372 | AK047372 | 41.87 | 0.0005 |
| ENSMUST00000137728 | AI847159 | 9.97 | 0.0002 | AK143879 | AK143879 | 41.70 | 0.0005 |
| uc008mqp.1 | AK085768 | 8.95 | 0.0021 | AK053631 | AK053631 | 37.72 | 0.0002 |
| ENSMUST00000161336 | Agl | 8.87 | 0.0002 | TCONS_00025043 | XLOC_018501 | 28.09 | 0.0000 |
| ENSMUST00000151051 | Gm14029 | 7.99 | 0.0010 | AK043180 | AK043180 | 27.94 | 0.0000 |
| ENSMUST00000180930 | Gm26793 | 7.63 | 0.0005 | NR_045175 | Smc2os | 26.11 | 0.0022 |
| ENSMUST00000142000 | Ift140 | 7.03 | 0.0003 | uc008pws.2 | 1500004A13Rik | 25.55 | 0.0000 |
| ENSMUST00000129337 | Gm11508 | 6.73 | 0.0013 | AK040275 | AK040275 | 24.70 | 0.0008 |
| ENSMUST00000174018 | Grm7 | 6.25 | 0.0003 | AK053990 | AK053990 | 24.55 | 0.0001 |
| ENSMUST00000176545 | AA465934 | 5.99 | 0.0009 | uc008ouc.1 | AK007174 | 24.15 | 0.0004 |
| AK155705 | AK155705 | 5.85 | 0.0001 | AK157804 | AK157804 | 23.90 | 0.0008 |
| AK017289 | AK017289 | 5.79 | 0.0099 | ENSMUST00000178906 | Gm10593 | 23.88 | 0.0001 |
| ENSMUST00000181014 | D330041H03Rik | 5.40 | 0.0091 | AK047207 | AK047207 | 22.35 | 0.0003 |
| AK136371 | AK136371 | 5.39 | 0.0005 | AK037460 | AK037460 | 19.67 | 0.0000 |
| AK084340 | AK084340 | 5.37 | 0.0002 | AK157092 | AK157092 | 19.58 | 0.0002 |
Notes: lncRNAs, long non-coding RNAs; FDR, false discovery rate. a DBA/1 mice vs. C57BL/6 mice
Table 3.
Top 20 differentially expressed mRNAs between DBA/1 and C57BL/6 mice
| Up-regulated mRNAs | Down-regulated mRNAs | ||||||
|---|---|---|---|---|---|---|---|
| Seqname | GeneSymbol | Fold Changea | FDR | Seqname | GeneSymbol | Fold Changea | FDR |
| NM_001037713 | Xaf1 | 112.48 | 0.0159 | NM_025617 | Tceanc2 | 48.72 | 0.0000 |
| NM_001163810 | Tescl | 20.19 | 0.0008 | NM_010500 | Ier5 | 41.87 | 0.0002 |
| NM_001142938 | AK010878 | 16.23 | 0.0001 | NM_001161411 | Trappc12 | 34.68 | 0.0004 |
| NM_011414 | Slpi | 12.71 | 0.0004 | NM_024472 | Gltpd1 | 31.90 | 0.0000 |
| NM_009247 | Serpinale | 11.51 | 0.0123 | NM_001033149 | Ttc9 | 22.04 | 0.0652 |
| NM_001083918 | Gm13139 | 10.97 | 0.0005 | NM_001039533 | Pdxdc1 | 20.65 | 0.0005 |
| NM_001111119 | Ccnb1ip1 | 9.03 | 0.0000 | NM_001145899 | Slc15a2 | 16. 47 | 0.0006 |
| NM_001001490 | Oxgr1 | 8.66 | 0.0027 | NM_198619 | Zfp933 | 13.51 | 0.0005 |
| NM_053127 | Pcdhb2 | 8.51 | 0.0018 | NM_207533 | Dbx2 | 12.88 | 0.0004 |
| NM_019788 | Bloc1s6 | 7.98 | 0.0003 | NM_011562 | Tdgf1 | 11.52 | 0.0015 |
| NM_022420 | Gprc5b | 7.59 | 0.0003 | NM_032002 | Nrg4 | 10.46 | 0.0003 |
| NM_029865 | Ocel1 | 7.51 | 0.0022 | NM_183167 | AI987944 | 9.91 | 0.0003 |
| NM_001103158 | Gm13242 | 7.27 | 0.0021 | NM_015800 | Crim1 | 9.62 | 0.0002 |
| NM_026645 | Iqcf3 | 6.61 | 0.0044 | NM_145594 | Fgl1 | 9.47 | 0.0024 |
| NM_009244 | Serpina1b | 6.50 | 0.0132 | NM_001130176 | Tnnt2 | 9.25 | 0.0000 |
| NM_175296 | Mael | 6.44 | 0.0469 | NM_025922 | Itpa | 7.34 | 0.0124 |
| NM_175537 | Zbtb38 | 6.38 | 0.0003 | NM_030707 | Fcrls | 7.11 | 0.0003 |
| NM_153568 | Lrrc66 | 5.97 | 0.0014 | NM_019440 | Irgm2 | 7.08 | 0.1372 |
| NM_010478 | Hspa1b | 5.92 | 0.0016 | NM_011723 | Xdh | 6.85 | 0.0292 |
| NM_001127188 | Zfp534 | 4.99 | 0.0026 | NM_001143686 | Apol11b | 6.77 | 0.2280 |
Notes: FDR, false discovery rate. a DBA/1 mice vs. C57BL/6 mice
Fig. 2.
Differentially expressed lncRNAs and mRNAs in DBA/1 mice compared with C57BL/6 mice. The Volcano Plots of lncRNA (a) and mRNA (b) expression; Hierarchical clustering of differentially expressed lncRNA (c) and mRNA (d). ‘red’ indicates high relative expression, and ‘green’ indicates low relative expression. ‘C’ and ‘D’ respectively represent C57BL/6 and DBA/1 group (each group with four mice)
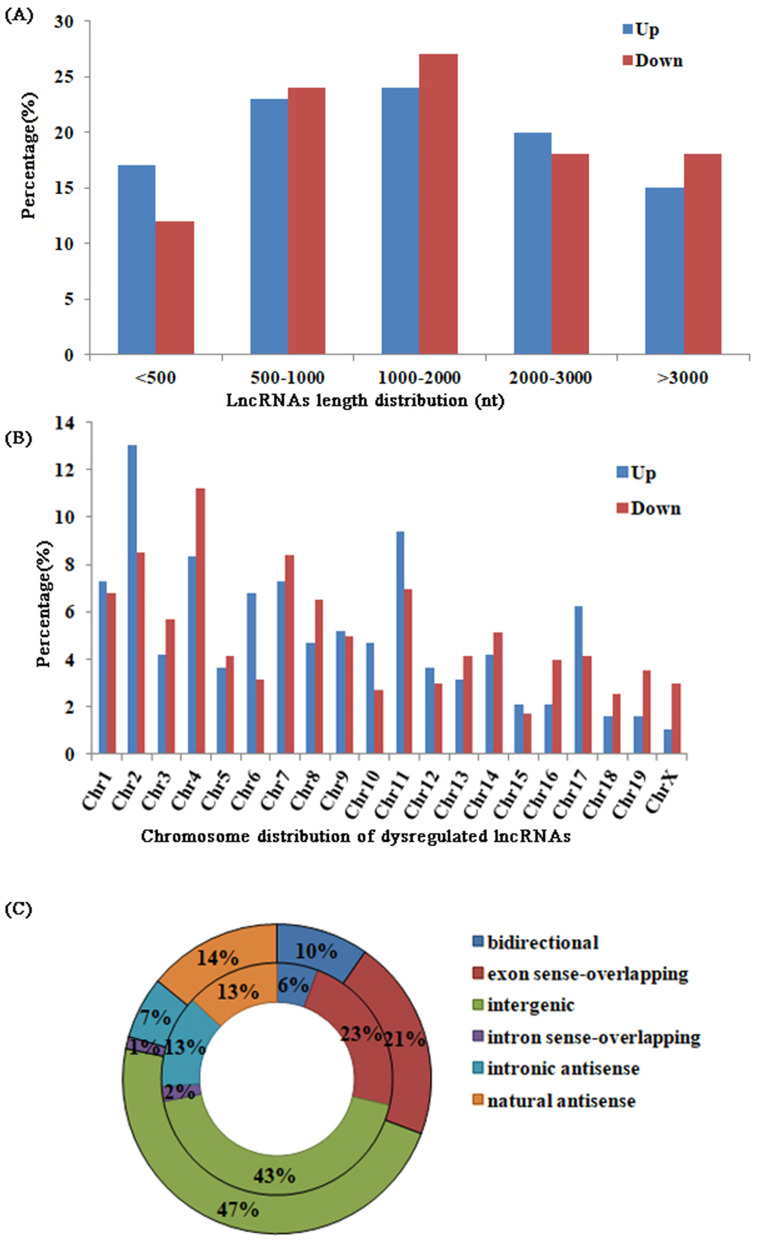
The detailed characteristics of the lncRNAs are also described (Fig. 3): the length distribution of the up- and downregulated lncRNAs was greatest in the 1000-2000 nt bin (24 and 27%, respectively); the most frequent transcriptional locations of up- and downregulated lncRNAs were chromosomes 2 and 4 (13 and 11%, respectively); and 43% of up- and 47% of downregulated lncRNAs were intergenic.
Fig. 3.
a The percentage of the length distribution of differentially expressed lncRNAs; (b) The percentage of the chromosome distribution of differentially expressed lncRNAs; (c) The types of differentially expressed lncRNAs. Upregulated lncRNAs were showed in the inner circle, and downregulated lncRNAs were described in the outer circle
The results of the sensitivity test based on an FDR < 0.05 are available in the supplementary material.
GO and pathway analysis of differentially expressed mRNAs
GO analysis was performed to assess the biological functions of genes and gene products, which were classified into biological processes (BP), cellular components (CC), and molecular function (MF). The most highly enriched GO terms targeted by upregulated genes were negative regulation of peptidase activity (GO: 0010466) in BP, MHC protein complex (GO: 0042611) in CC and serine-type endopeptidase inhibitor activity (GO:0004867) in MF (Fig. 4a). The most enriched GO terms among downregulated genes were transforming growth factor beta receptor signaling pathway (GO: 0007179) in BP, cell part (GO:0044464) in CC, and hydrolase activity (GO: 0016787) in MF (Fig. 4b).
Fig. 4.
GO analysis comparing DBA/1 group with C57BL/6 group. a Top 10 enriched GO terms from upregulated mRNAs in biological process, cellular component, and molecular function. b Top 10 enriched GO terms from downregulated mRNAs in biological process, cellular component, and molecular function
KEGG was performed to identify genes involved in different biological pathways [17–19]. This analysis revealed 28 pathways enriched with significantly differentially expressed genes, including 20 pathways corresponding to upregulated transcripts and 8 corresponding to downregulated transcripts. The most correlated pathway among upregulated genes was Type I diabetes mellitus, and among downregulated genes, it was natural killer cell-mediated cytotoxicity. The details are shown in Table 4.
Table 4.
Pathways identified from comparison between DBA/1 mice and C57BL/6 mice
| lncRNA types | KEGG pathways | Associated genes |
|---|---|---|
| Up-regulated lncRNAs | 1. Type I diabetes mellitus | H2-D1,H2-DMA,H2-Q6,H2-T24,LTA |
| 2. Viral myocarditis | CXADR,H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 3. Herpes simplex virus 1 infection | H2-D1,H2-DMA,H2-Q6,H2-T24,LTA,ZFP457,ZFP51,ZFP658,ZFP961,ZFP963 | |
| 4. Antigen processing and presentation | H2-D1,H2-DMA,H2-Q6,H2-T24,HSPA1B | |
| 5. Cell adhesion molecules | CLDN13,H2-D1,H2-DMA,H2-Q6,H2-T24,MPZL1 | |
| 6. Allograft rejection | H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 7. Graft-versus-host disease | H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 8. Autoimmune thyroid disease | H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 9. Neuroactive ligand-receptor interaction | CORT,GABRA2,GRIN3A,HTR5B,PTAFR,PTGFR,PYY | |
| 10. Staphylococcus aureus infection | DEFA17,DEFA24,H2-DMA,PTAFR | |
| 11. Kaposi sarcoma-associated herpesvirus infection | ANGPT2,GM5741,H2-D1,H2-Q6,H2-T24 | |
| 12. Human T-cell leukemia virus 1 infection | H2-D1,H2-DMA,H2-Q6,H2-T24,LTA | |
| 13. Endocytosis | H2-D1,H2-Q6,H2-T24,HSPA1B,IQSEC1 | |
| 14. Phagosome | H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 15. Nicotine addiction | GABRA2,GRIN3A | |
| 16. Calcium signaling pathway | CAMK4,HTR5B,PTAFR,PTGFR | |
| 17. Serotonergic synapse | GM5741,HTR5B,TPH2 | |
| 18. Epstein-Barr virus infection | H2-D1,H2-DMA,H2-Q6,H2-T24 | |
| 19. Human immunodeficiency virus 1 infection | GM5741,H2-D1,H2-Q6,H2-T24 | |
| 20. Glycerolipid metabolism | DGKB,LPL | |
| Down-regulated lncRNAs | 1. Natural killer cell mediated cytotoxicity | GZMB,KLRA7,NFATC2,PPP3R2,SOS1,TNFSF10 |
| 2. Endocrine and other factor-regulated calcium reabsorption | CLTA,KLK1B3,KLK1B8,KLK1B9 | |
| 3. Starch and sucrose metabolism | GBE1,GPI1,SIS | |
| 4. Renin-angiotensin system | KLK1B3,KLK1B8,KLK1B9 | |
| 5. Glycosphingolipid biosynthesis | KLK1B3,KLK1B8,KLK1B9 | |
| 6. Cell cycle | CHEK2,CUL1,MCM6,ORC6,PTTG1 | |
| 7. Amyotrophic lateral sclerosis (ALS) | MAP3K5,PPP3R2,TNFRSF1B | |
| 8. T cell receptor signaling pathway | 4930544G11RIK,NFATC2,PPP3R2,SOS1 |
The sensitivity test for GO and KEGG was showed in the supplemental files based on a fold change ≥2.0 and an FDR < 0.05.
Coexpression analysis
In this study, nine differentially expressed antisense lncRNAs were found between the DBA/1 group and the C57BL/6 control group. Among these lncRNAs, 2010012P19Rik was the most downregulated lncRNA (fold change = 6.09, FDR < 0.001) with its nearby gene, Tnfsf12-Tnfsf13 (fold change =2.04, FDR = 0.025). Other differentially expressed antisense lncRNAs and their paired sense mRNAs are shown in Table 5.
Table 5.
Differentially expressed antisense lncRNAs and nearby coding gene
| Seqname of lncRNA | Gene symbol | Fold changea (lncRNAs) | Regulation of lncRNA | Genome relationship | Nearby gene seqname | Nearby gene Symbol |
Fold changea (mRNAs) |
Regulation of mRNA |
|---|---|---|---|---|---|---|---|---|
| ENSMUST00000145435 | 2010012P19Rik | 6.0917526 | down | natural antisense | NM_001034097 | Tnfsf12-Tnfsf13 | 2.0415635 | down |
| AK017289 | AK017289 | 5.792853 | up | natural antisense | NM_001267808 | H2-L | 2.8392743 | up |
| AK017289 | AK017289 | 5.792853 | up | natural antisense | NM_010380 | H2-D1 | 2.6094529 | up |
| AK155933 | AK155933 | 4.4353006 | down | intronic antisense | NM_146191 | Lrrk1 | 2.53359 | down |
| AK087052 | AK087052 | 3.4286643 | up | intronic antisense | NM_001035242 | Trpm3 | 2.0411979 | down |
| AK087052 | AK087052 | 3.4286643 | up | intronic antisense | NM_001035243 | Trpm3 | 2.0100955 | down |
| AK149710 | AK149710 | 3.163844 | down | natural antisense | NM_008850 | Pitpna | 2.4984672 | down |
| AK158573 | AK158573 | 2.5693914 | down | natural antisense | NM_028803 | Gbe1 | 2.5839838 | down |
| ENSMUST00000148180 | Gm15396 | 2.5550359 | down | natural antisense | NM_008437 | Napsa | 4.8828493 | down |
| AK007047 | AK007047 | 2.510748 | down | natural antisense | NM_001200023 | Zfp963 | 3.3589015 | up |
| ENSMUST00000124513 | Gm15247 | 2.064782 | down | natural antisense | NM_183151 | Mid1 | 3.2269554 | down |
Notes: lncRNAs, long non-coding RNAs. a DBA/1 mice vs. C57BL/6 mice
A total of 115 long intergenic noncoding RNAs (lincRNAs) were found to be differentially expressed. Among them, approximately 68.5% of the lincRNAs and their adjacent coding genes were changed in the same direction (which means that both lncRNAs and mRNAs were upregulated or that both were downregulated), including 12.9% upregulated and 55.6% downregulated pairs. Eight percent of dysregulated lincRNAs were upregulated and their paired mRNAs were downregulated, while 22.5% of lincRNAs were downregulated, with their nearby mRNAs upregulated. Fold-changes of differential expression among lincRNA-gene pairs are presented in a scatter plot (Fig. 5a). The length distribution of differentially expressed lincRNAs was greatest in the 1000-2000 nt bin (30.4%), and these differential lincRNAs were most frequently transcribed from chromosome 4 (27%). The details are shown in Fig. 5b and Fig. 5c, respectively. The 20 most differentially expressed lincRNAs and their adjacent mRNAs are presented in Table 6.
Fig. 5.
a Fold change of significantly dysregulated lincRNA and their differentially expressed adjacent mRNAs for DBA/1mice vs. C57BL/6 mice; (b) The percentage of the length distribution of differentially expressed lincRNAs; (c) The percentage of the chromosome distribution of differentially expressed lincRNAs
Table 6.
Top 20 differentially expressed lincRNAs and adjacent mRNAs
| Seqname of lncRNA | Gene symbol | Fold changea (lncRNAs) | Regulation of lncRNA | Genome relationship | Nearby gene seqname | Nearby gene Symbol | Fold changea (mRNAs) | Regulation of mRNA |
|---|---|---|---|---|---|---|---|---|
| NR_033305 | AA388235 | 66.9199947 | down | downstream | NM_010386 | H2-Dma | 2.170267 | up |
| NR_033305 | AA388235 | 66.9199947 | down | upstream | NM_019420 | B3galt4 | 5.7973092 | down |
| AK047372 | AK047372 | 41.8748727 | down | downstream | NM_001163042 | Haus8 | 2.1908769 | down |
| AK047372 | AK047372 | 41.8748727 | down | downstream | NM_029865 | Ocel1 | 7.5104853 | up |
| AK143879 | AK143879 | 41.7014068 | down | upstream | NM_001193667 | Gm1987 | 2.8931939 | down |
| AK143879 | AK143879 | 41.7014068 | down | upstream | NM_001277167 | Gm12429 | 5.3971264 | down |
| TCONS_00025043 | XLOC_018501 | 28.0948375 | down | upstream | NM_008861 | Pkd2 | 3.0839338 | up |
| AK053990 | AK053990 | 24.5521064 | down | upstream | NM_030707 | Fcrls | 7.1055169 | down |
| ENSMUST00000178906 | Gm10593 | 23.884969 | down | downstream | NM_001193667 | Gm1987 | 2.8931939 | down |
| ENSMUST00000178906 | Gm10593 | 23.884969 | down | downstream | NM_001277167 | Gm12429 | 5.3971264 | down |
| uc007uzp.1 | AK038711 | 23.496569 | up | downstream | NM_001001490 | Oxgr1 | 8.6586855 | up |
| AK037460 | AK037460 | 19.6676915 | down | upstream | NM_001008232 | Asap3 | 2.3508325 | down |
| AK037460 | AK037460 | 19.6676915 | down | upstream | NM_011542 | Tcea3 | 3.3860996 | down |
| uc029usn.1 | Gm5859 | 19.0222774 | down | upstream | NM_001085530 | Gm13298 | 3.895603 | down |
| AK037363 | AK037363 | 15.2204905 | down | downstream | NM_008911 | Ppox | 3.5903657 | down |
| AK136314 | AK136314 | 14.6596357 | down | upstream | NM_008861 | Pkd2 | 3.0839338 | up |
| NR_040401 | C920006O11Rik | 13.1823016 | down | downstream | NM_025274 | Dppa5a | 3.5029739 | down |
| uc029urz.1 | DQ551946 | 12.8034244 | down | downstream | NM_001085530 | Gm13298 | 3.895603 | down |
| AK141495 | AK141495 | 12.6885049 | down | downstream | NM_001193667 | Gm1987 | 2.8931939 | down |
| AK141495 | AK141495 | 12.6885049 | down | downstream | NM_001277167 | Gm12429 | 5.3971264 | down |
| ENSMUST00000151374 | Snhg3 | 12.1370958 | down | downstream | NM_001081651 | Rab42 | 3.2691229 | down |
| ENSMUST00000151374 | Snhg3 | 12.1370958 | down | upstream | NM_001081211 | Ptafr | 2.3332466 | up |
| ENSMUST00000151374 | Snhg3 | 12.1370958 | down | upstream | NM_001161797 | Phactr4 | 3.4588717 | down |
| ENSMUST00000151374 | Snhg3 | 12.1370958 | down | upstream | NM_026039 | Med18 | 2.3377675 | down |
| ENSMUST00000121728 | Gm13301 | 11.4196096 | down | upstream | NM_001085530 | Gm13298 | 3.895603 | down |
| ENSMUST00000178043 | Gm3892 | 11.2523847 | down | upstream | NM_001085530 | Gm13298 | 3.895603 | down |
| ENSMUST00000174778 | Gm10499 | 10.8643014 | up | downstream | NM_008207 | H2-T24 | 2.5411856 | up |
| ENSMUST00000107991 | Gm3892 | 10.8301037 | down | upstream | NM_001085530 | Gm13298 | 3.895603 | down |
| AK052053 | AK052053 | 10.6618917 | down | downstream | NM_001193667 | Gm1987 | 2.8931939 | down |
| AK052053 | AK052053 | 10.6618917 | down | downstream | NM_001277167 | Gm12429 | 5.3971264 | down |
Notes: lincRNAs, long intergenic noncoding RNAs. a DBA/1 mice vs. C57BL/6 mice
A sensitivity test was also performed on the basis of an FDR < 0.05, and the results are also given in the supplementary material.
Validation of differentially expressed lncRNAs and mRNAs
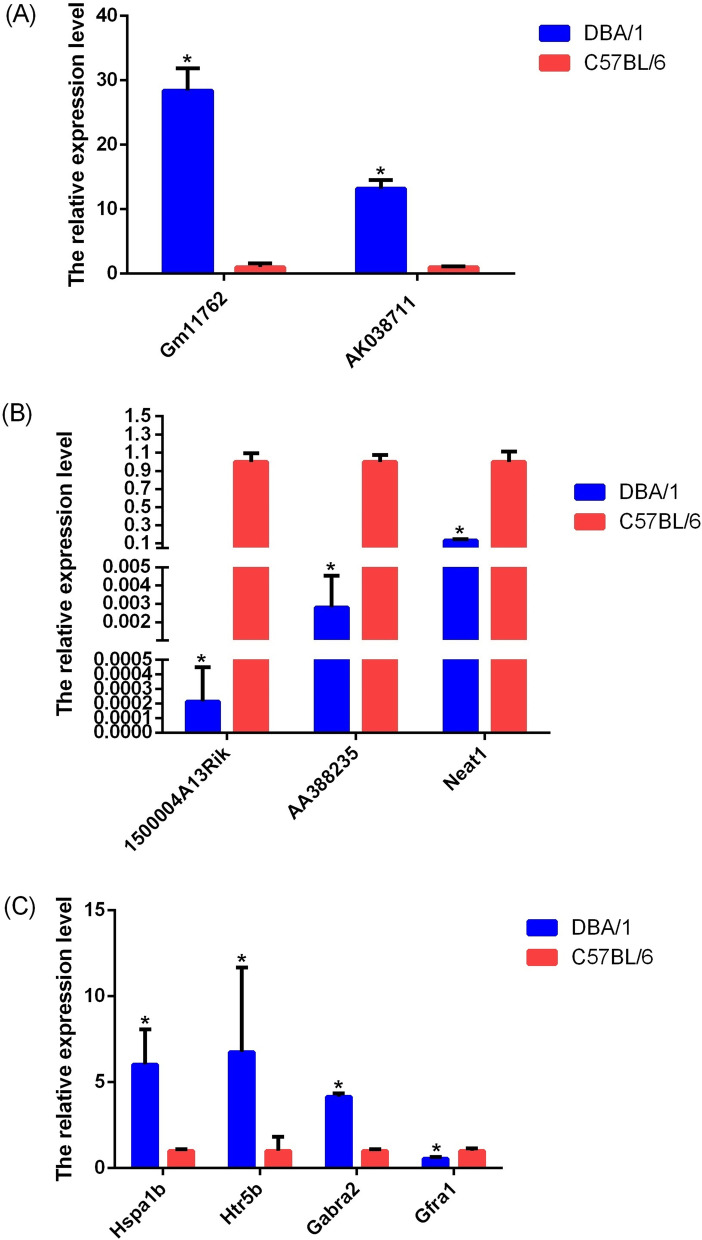
Differentially expressed genes were selected to be analyzed by qRT–PCR, including five lncRNAs (two upregulated and three downregulated) and four mRNAs (three upregulated and one downregulated). According to the qRT–PCRs, the expression of AK038711 (uc007uzp.1) and Gm11762 (NR_045099) was upregulated (Fig. 6a), whereas that of 1500004A13Rik (NR_015498), AA388235 (NR_033305) and Neat1 (NR_003513) was downregulated (Fig. 6b) in DBA/1 mice compared with C57BL/6 mice. This result was consistent with the microarray assay.
Fig. 6.
The qRT-PCR vadidation of differentially expressed lncRNAs and mRNAs between DBA/1 mice with C57BL/6. a The qRT-PCR results of up-regulated lncRNAs; (b) The qRT-PCR results of down-regulated lncRNAs; (c) The qRT-PCR results of differentially expressed mRNAs. By setting the expression value of target genes in C57BL/6 control group at 1, the expression level of which in DBA/1 mice group was the fold change relative to control group. Significant levels were indicated by * (P < 0.05). The results of qRT-PCR were consistent with that in microarray analysis (n = 4 animals/group)
Through a GO and KEGG analysis, we identified 4 potentially related transcripts for validation according to known mechanisms that are potentially related to AGSz or S-IRA. They were Hsp1a, Htr5b, Gabra2 and Gfra1. In our enrichment analysis, Hsp1a was found to be involved in multiple process of regulation on multiple enzyme activity (GO) and stress responses (GO), while Gfra1 was located on axon (GO) and exert molecular function as binding (GO). Both Htr5b and Gabra2 participated in neurotransmitter-related functions (GO) and neuroactive ligand–receptor interactions (KEGG). Compared to the C57BL/6 group, qRT–PCR showed that the expression levels of these genes (Hsp1a, Htr5b and Gabra2) in the DBA/1 group were significantly higher (Fig. 6c), while Gfra1 expression was significantly lower in DBA/1. The results of qRT-PCR were consistent with that in microarray analysis.
Discussion
DBA/1 mice were more susceptible to AGSz and S-IRA than C57BL/6 mice. Previous studies have explored the influence of genetic background [7] but the differences were not completely explained. The gene expression profiles of these mice were not fully understood. In the present study, using microarray analysis, we investigated the potential roles of mRNAs and lncRNAs in the different AGSz and S-IRA susceptibilities of these two strains and identified 897 lncRNAs and 438 mRNAs that were dysregulated in the brainstems of DBA/1 and C57BL/6 mice.
GO and KEGG analyses revealed that the significantly differentially expressed mRNAs were involved in many biological functions. Comparing DBA/1 mice with C57BL/6 mice, the most highly enriched biological process in GO analysis was transforming growth factor beta (TGF-β) receptor signaling pathway for downregulated genes. In this process, the cluster of genes included TDGF1, RASL11B, SNX6, HTRA3, ADAM9, BMPR1B, HPGD, and ARRB. The TGF-β signaling pathway is involved in the dysfunction of neuronal, glial cell and blood–brain barrier (BBB) via alteration of ion channels, adenosine, glutamate, and GABA receptors [20]. Thus, disruption to the TGF-β signaling pathway can lead to changes in neuronal excitability and increase the risk of seizures [21, 22]. TGF-β signaling is also related to inflammation, which plays an important role in epileptogenesis [23]. In this study, the enrichment of downregulated genes in this term seems to contradict the epileptogenesis and thus higher susceptibility to AGSz. However, we found one study reporting that the C57BL/6 strain was resistant to TGF-β- or IL-6-induced seizures [24]. This previous finding hints to the fact that seizure in DBA/1 mice is induced by sound and is not spontaneous, and therefore, we hypothesized that lower expression of TGF-β-related genes in DBA/1 mice might represent instability due to external influences on this biological process, especially sound priming, making these mice vulnerable to AGSz. Further study is still needed.
Another important BP enriched with downregulated genes was metabolic process, with 145 genes differentially expressed. An early study demonstrated that metabolic dysfunction is evident in and may even directly cause epilepsy [25]. More specifically, one study reported that in C57BL/6 mice, a soy protein-containing diet was associated with higher susceptibility to AGSz [7]. Similarly, two recent publications indicated that a high tryptophan or ketogenic diet reduced the risk for S-IRA [26, 27]. Although one of these studies attributed these changes to gut microbes [26], combined with our data, recent evidence strongly suggests that metabolic process differences between these two strains may play an important role in terms of susceptibility to AGSz and S-IRA.
Serine-type endopeptidase inhibitor activity and hydrolase activity showed the highest enrichment score (the former due to upregulated genes and the latter due to downregulated genes) in molecular function in the GO analysis. However, the relation of these results to AGSz or S-IRA is largely unknown.
The most highly enriched cellular component terms were major histocompatibility complex (MHC) class protein complexes, enriched with upregulated genes. MHC molecules participate in negatively regulation of synaptic plasticity [28–31], and overexpression of MHC complex I protein can lead to a decreased ability to form synapses, which has been linked to several central nervous system (CNS) disorders, including autism spectrum disorders (ASDs) and schizophrenia [32, 33]. MHC may exert its effects by interacting with inflammatory cytokines that are involved in epilepsy [33, 34]. These findings suggest that MHC has the potential to be involved in AGSz and S-IRA in DBA/1 mice, although no direct evidence is currently available to support this possibility.
KEGG pathway analysis revealed that Type I diabetes mellitus (T1DM) (upregulated genes) was one of the most differentially expressed pathways. Energy metabolism through glycolysis is related to hereditary susceptibility to epileptic seizures. A previous study reported that maintenance of low blood glucose levels exerted seizure-protecting effects [35]. On the other hand, another study noted that the common mechanism in seizures and T1DM is autoimmune dysfunction [36]. In our study, other immune-mediated pathways were differentially expressed between DBA/1 and C57BL/6 mice, including antigen processing and presentation, allograft rejection and graft-versus-host disease. These findings, combined with the findings from the GO analysis (i.e., TGF-β receptor signaling pathway, MHC, etc.) implied that a broad spectrum of immune pathways might play a role in the susceptibility to AGSz in DBA/1 mice.
When considering the results of GO and KEGG together, we found that the following transcripts required extensive investigation: Htr5b, Gabra2, and Hspa1b.
The Htr5b mRNA level in DBA/1 mice was higher than that in the C57BL/6 mice, with a fold change = 6.76 (P < 0.05). As mentioned, serotonergic neurons were the focus of a recent study on the mechanism of S-IRA and SUDEP. Intervention to activate 5-HT neurons has been proven to reduce S-IRA in DBA/1 mice [8–10]. An electrophysiological study revealed that serotonergic neuron function was suppressed during and after seizures [37]. Since the serotonin system in the brainstem plays a key role in the regulation of breathing and arousal [38, 39], our finding on Htr5b may be related to AGSz-induced S-IRA.
We found that Gabra2 mRNA was more highly expressed in DBA/1 mice than in C57BL/6 mice (fold change = 4.15, P < 0.05). Other studies performed on DBA/2 mice have found that gamma-aminobutyric acid (GABA) and its receptors were related to AGSz and S-IRA. Elevated brain GABA concentrations protected against AGSz [40]. However, DBA/2 mice showed lower K + -induced GABA release on PND 30, which may have been related to susceptibility to AGSz [41]. Thus, we assumed that similar to DBA/2, the increased expression of the GABA receptor might be compensate for the lower GABA level in the brainstem of the DBA/1 mice compared to that in the C57BL/6 mice at certain time point. However, this hypothesis requires further examination. A previous study pointed out that the sequence and expression of the 2 subunits (gamma-1 and alpha-4) of the GABA receptor in DBA/2 mice did not differ from those in the C57BL/6 strain [42, 43]. However, these two studies were performed on the cortex and cerebellum, and the expression of the GABA receptor in DBA/1 mice had not been previously tested. Our findings suggest that, similar to those in the DBA/2 mice, the GABA receptors in the DBA/1 strain may have potentially influenced AGSz and S-IRA.
Hspa1b, which translates into heat shock protein 70 (HSP70), was expressed at significantly higher levels in the DBA/1 group than in the C57BL/6 group (fold change = 6.04, P < 0.05). HSP70 is an indicator of the stress response, which can be induced by various stimuli, including ischemia, traumatic injury and seizure [44–47]. A prolonged stress response is related to cell injury in neurological disease and may lead to damage to the brainstem [48]. Hence, the elevated Hspa1b expression level in DBA/1 mice may represent a high stress state in the DBA/1 strain and may be related to S-IRA susceptibility. A recent proteomic and RNA-seq study on human SUDEP cases indicated increased HSP70-positive neurons in the hippocampus, which was considered to be related to antemortem neuronal injury, such as seizures prior to death [49]. However, in our study, we found higher expression of Hspa1b in DBA/1 mice without seizures than in normal C57BL/6 mice. These results suggest that HSP70, may be more than a biomarker. It may be involved in the mechanism of AGSz, S-IRA and even SUDEP. Studies on stress are potential new prospects for mechanistic research on S-IRA and SUDEP.
In the Leitner et al. study on high-risk SUDEP patients, Gfra1 was identified as the most downregulated mRNA in the hippocampus compared to that in low-risk patients [49]. Similarly, Gfra1 was also expressed at low levels in DBA/1 mice compared to C57BL/6 mice in our study (DBA/1 vs. C57BL/6, fold change = 1.81, downregulated). Gfra1 binds to glial cell-derived neurotrophic factor (GDNF), influencing the survival and differentiation of GABAergic interneurons [50]. Decreased Gfra1 also affects the release of GDNF, resulting in more seizure activities and thus a higher risk for SUDEP [49, 51]. Combined with our findings on Gabra2, these previous findings point to the GABA system as an important modulator for both S-IRA and SUDEP.
Among lncRNAs, nuclear paraspeckle assembly transcript 1 (Neat1) is highly conserved in mammals and participates in various developmental and pathological processes. In our study, we found that the expression of Neat1 in the brainstem of DBA/1 mice was significantly lower than that in the brainstem of C57BL/6 mice (fold change = 7.51, P < 0.001). It has been reported that seizures can lead to a transient downregulation of Neat1, providing a scaffolding function for regulating ion channels and thus was believed to act as a protective mechanism for restoring neuron functionality after seizure [52]. We presume that Neat1 had the ability to participate in forming an electrical barrier against spreading depolarization, which has been found to be closely related to S-IRA and SUDEP [37, 53], by increasing the threshold for propagating. The relatively low Neat1 level in the brainstem of DBA/1 mice represents a deficiency in response to seizures, making these mice vulnerable to postictal electroencephalogram suppression and leading to their high susceptibility to S-IRA.
A number of antisense lncRNAs (n = 9) and lincRNAs (n = 115) were found to be dysregulated in a coexpression analysis. Among the differentially expressed lncRNAs, intergenic lncRNAs were the most common, while exon sense-overlapping lncRNAs ranked second. Among these lncRNAs, Gm14201 (ENSMUST00000117627) was the most markedly upregulated lncRNA, with a fold change = 35.75, and AK005187 (uc009bwo.2) was the most notably downregulated lncRNA, with a fold change of 326.12. The functions of these lncRNAs remain unclear. However, with such marked fold changes, these lncRNAs may have critical functions in the regulation of development and may be potential biomarkers for certain physiological processes.
In our coexpression analysis, 2010012P19Rik was the most differentially expressed antisense lncRNA and was associated with the significantly downregulated adjacent coding gene Tnfsf12-Tnfsf13. Tnfsf12-Tnfsf13 is also known as the TWE-PRIL gene. The gene is, in fact, a hybrid transcript of TWEAK and APRIL. One study reported that knocking down TWE-PRIL enhanced axonal growth of sympathetic neurons [54]. Although sympathetic neuron function in AGSz and S-IRA has not yet been studied in DBA/1 mice, a clinical case reported that SUDEP patients could present with sympathetic hyperactivity [55]. Thus, it is reasonable to deduce that downregulation of Tnfsf12-Tnfsf13 enhanced sympathetic neuron function by stimulating axon growth, leading to hyperactivity related to S-IRA.
A number of differentially expressed lincRNAs were detected. Our analysis showed that a majority of lincRNAs and their nearby coding genes shared the same direction of expression change. The most differentially expressed lincRNA was AA388235, with the upregulated downstream mRNA H2-Dma and the downregulated upstream mRNA B3galt4. The determination of the biological functions and detailed regulatory mechanisms of these lincRNAs requires further exploration.
Limitation
There are several limitations to this research. First, the sample lacked anatomical precision. In this study, the whole brainstem was used to extract RNA, leading to high heterogeneity in cell types and nuclei. Thus, further separation of tissue via anatomical methods or single-cell sequencing of certain nuclei is recommended. Second, the DBA/1 mice were not tested for AGSz or S-IRA. This choice was based on a consideration that seizure itself, as well as S-IRA, which potentially influence the expression of RNA. Notably, the susceptibility of DBA/1 mice to S-IRA increased after priming by daily stimulation with sound [6]. Thus, testing the effect of priming on the RNA expression of DBA/1 mice may provide insight into the acquisition of susceptibility to S-IRA. Third, anesthesia was given according to ethics considerations. Even though we managed to dissect the brainstem as soon as possible after anesthesia, isoflurane inhalation may have influenced the expression of RNA. The determination of the extent to which this drug may have affected RNA requires further study. Additionally, the sample size of this study was small, as only 4 mice from each group were tested by either microarray or qRT–PCR.
Conclusions
Our findings showed that a number of lncRNAs and mRNAs were differentially expressed between the brainstems of DBA/1 and C57BL/6 mice. We found TGF-β signaling and metabolic process may contribute to the differential sensitivity to AGSz and S-IRA. Also, many differentially expressed mRNAs such as Htr5b, Gabra2, Hspa1b and Gfra1 could potentially influence the susceptibility. Finally, current evidence suggested that lncRNA Neat1 and 2010012P19Rik might exert effect on AGSz and S-IRA. These findings provide new directions in the study of AGSz, S-IRA and even SUDEP.
Supplementary Information
Additional file 1. The details of all differentially expressed lncRNAs.
Additional file 2. The details of all differentially expressed mRNAs.
Additional file 3. Supplementary Table based on fold change ≥ 2.0 and FDR<0.05.
Additional file 4. Supplementary Figure based on fold change ≥ 2.0 and FDR<0.05.
Acknowledgements
We thank KangChen Bio-tech, Shanghai, China for providing the microarray and qRT-PCR service.
Abbreviations
- 5-HT
5-hydroxytryptamine
- AGSz
Audiogenic seizures
- ASD
Autism spectrum disorders
- BBB
Blood-brain barrier
- BP
Biological processes
- CC
Cellular components
- CNS
Central nervous system
- FDR
False discovery rate
- GABA
Gamma-aminobutyric acid
- GDNF
Glial cell-derived neurotrophic factor
- GO
Gene ontology
- HSP70
Heat shock protein 70
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lincRNA
Long noncoding intergenic RNA
- lncRNAs
Long non-coding RNAs
- MF
Molecular function
- MHC
Major histocompatibility complex
- NEAT1
Nuclear Paraspeckle Assembly Transcript 1
- PND
Postnatal day
- qRT-PCR
Quantitative Real-time polymerase chain reaction
- S-IRA
Seizure-induced respiratory arrest
- SPF
Specefic pathogen free
- SSRI
Selective Serotonin Reuptake Inhibitor
- SUDEP
Sudden Unexpected Death in Epilepsy
- T1DM
Type I diabetes mellitus
- TGF-β
Transforming growth factor beta
Authors’ contributions
XL and DC performed the experiments. LNZ carried out data analysis and prepared the figures and tables. LNZ and DC wrote the manuscript. LL designed the experiment and critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Health Commission of Sichuan Province [grant number 20ZD005].
Availability of data and materials
The datasets presented in this study can be found in online repositories and have been deposited with the Gene Expression Omnibus (GEO) under the project accession number GSE152931 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152931).
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Sichuan University West China Hospital. All experiments were performed in accordance with relevant guidelines and regulations, which were also in accordance with ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lina Zhu and Deng Chen contributed equally to this work.
References
- 1.Collins RL, Fuller JL. Audiogenic seizure prone (asp): a gene affecting behavior in linkage group 8 of the mouse. Science. 1968;162:1137–1139. doi: 10.1126/science.162.3858.1137. [DOI] [PubMed] [Google Scholar]
- 2.Fuller JL. Effect of drinking schedule upon alcohol preference in mice. Quarterly Journal of Studies on Alcohol. 1967;28:22–26. doi: 10.15288/qjsa.1967.28.022. [DOI] [PubMed] [Google Scholar]
- 3.Randt CT, Barnett BM, McEwen BS, Quartermain D. Amnesic effects of cycloheximide on two strains of mice with different memory characteristics. Exp Neurol. 1971;30:467–474. doi: 10.1016/0014-4886(71)90147-6. [DOI] [PubMed] [Google Scholar]
- 4.Wimer RE, Symington L, Farmer H. Differences in memory processes between inbred mouse strains C57BL/6J AND DBA/2J. J Comp Physiol Psychol. 1968;65:126–131. doi: 10.1037/h0025405. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber RA, Graham JM. Audiogenic priming in DBA/2J and C57 BL/6J mice: interactions among age, prime-to-test interval, and index of seizure. Dev Psychobiol. 1976;9:57–66. doi: 10.1002/dev.420090109. [DOI] [PubMed] [Google Scholar]
- 6.Faingold CL, Randall M, Tupal S. DBA/1 mice exhibit chronic susceptibility to audiogenic seizures followed by sudden death associated with respiratory arrest. Epilepsy Behav. 2010;17:436–440. doi: 10.1016/j.yebeh.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Maxson SC. A genetic context for the study of audiogenic seizures. Epilepsy Behav. 2017;71:154–159. doi: 10.1016/j.yebeh.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Faingold CL, Tupal S, Randall M. Prevention of seizure-induced sudden death in a chronic SUDEP model by semichronic administration of a selective serotonin reuptake inhibitor. Epilepsy Behav. 2011;22:186–190. doi: 10.1016/j.yebeh.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Faingold CL, Randall M. Effects of age, sex, and sertraline administration on seizure-induced respiratory arrest in the DBA/1 mouse model of sudden unexpected death in epilepsy (SUDEP) Epilepsy Behav. 2013;28:78–82. doi: 10.1016/j.yebeh.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Zeng C, Long X, Cotten JF, Forman SA, Solt K, Faingold CL, Feng HJ. Fluoxetine prevents respiratory arrest without enhancing ventilation in DBA/1 mice. Epilepsy Behav. 2015;45:1–7. doi: 10.1016/j.yebeh.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Zhao H, Yang X, Xue Q, Cotten JF, Feng HJ. 5-Hydroxytryptophan, a precursor for serotonin synthesis, reduces seizure-induced respiratory arrest. Epilepsia. 2016;57:1228–1235. doi: 10.1111/epi.13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Zhao H, Zeng C, Van Dort C, Faingold CL, Taylor NE, Solt K, Feng HJ. Optogenetic activation of 5-HT neurons in the dorsal raphe suppresses seizure-induced respiratory arrest and produces anticonvulsant effect in the DBA/1 mouse SUDEP model. Neurobiol Dis. 2018;110:47–58. doi: 10.1016/j.nbd.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin B, Dieuset G, Pawluski JL, Costet N, Biraben A. Audiogenic seizure as a model of sudden death in epilepsy: a comparative study between four inbred mouse strains from early life to adulthood. Epilepsia. 2020;61:342–349. doi: 10.1111/epi.16432. [DOI] [PubMed] [Google Scholar]
- 14.Hom AC, Leppik IE, Rask CA. Effects of estradiol and progesterone on seizure sensitivity in oophorectomized DBA/2J mice and C57/EL hybrid mice. Neurology. 1993;43:198–204. doi: 10.1212/wnl.43.1_part_1.198. [DOI] [PubMed] [Google Scholar]
- 15.Xu JZ, Zhang JL, Zhang WG. Antisense RNA: the new favorite in genetic research. J Zhejiang Univ Sci B. 2018;19:739–749. doi: 10.1631/jzus.B1700594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M. Toward understanding the origin and evolution of cellular organisms., protein Science : a publication of the protein. Society. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vezzani A, Balosso S, Ravizza T. Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy, nature reviews. Neurology. 2019;15:459–472. doi: 10.1038/s41582-019-0217-x. [DOI] [PubMed] [Google Scholar]
- 21.Weissberg I, Wood L, Kamintsky L, Vazquez O, Milikovsky DZ, Alexander A, Oppenheim H, Ardizzone C, Becker A, Frigerio F, Vezzani A, Buckwalter MS, Huguenard JR, Friedman A, Kaufer D. Albumin induces excitatory synaptogenesis through astrocytic TGF-β/ALK5 signaling in a model of acquired epilepsy following blood-brain barrier dysfunction. Neurobiol Dis. 2015;78:115–125. doi: 10.1016/j.nbd.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, Senatorov VV, Morrissey CS, Lippmann K, Vazquez O, Milikovsky DZ, Gu F, Parada I, Prince DA, Becker AJ, Heinemann U, Friedman A, Kaufer D. TGFβ signaling is associated with changes in inflammatory gene expression and perineuronal net degradation around inhibitory neurons following various neurological insults. Sci Rep. 2017;7:7711. doi: 10.1038/s41598-017-07394-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhtar I. Inflammatory and immune mechanisms underlying epileptogenesis and epilepsy: from pathogenesis to treatment target. Seizure. 2020;82:65–79. doi: 10.1016/j.seizure.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Levy N, Milikovsky DZ, Baranauskas G, Vinogradov E, David Y, Ketzef M, Abutbul S, Weissberg I, Kamintsky L, Fleidervish I, Friedman A, Monsonego A. Differential TGF-β signaling in glial subsets underlies IL-6-mediated Epileptogenesis in mice. J Immunol. 2015;195(4):1713–1722. doi: 10.4049/jimmunol.1401446. [DOI] [PubMed] [Google Scholar]
- 25.Kovac S, Kostova ATD, Herrmann AM, Melzer N, Meuth SG, Gorji A. Metabolic and homeostatic changes in seizures and acquired epilepsy—mitochondria, calcium dynamics and reactive oxygen species. Int J Mol Sci. 2017;18. 10.3390/ijms18091935. [DOI] [PMC free article] [PubMed]
- 26.Yue Q, Cai M, Xiao B, Zhan Q, Zeng C. A high-tryptophan diet reduces seizure-induced respiratory arrest and alters the gut microbiota in DBA/1 mice. Front Neurol. 2021;12:762323. doi: 10.3389/fneur.2021.762323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotts MS, Kim Y, Bravo E, Richerson GB, Teran FA. A ketogenic diet protects DBA/1 and Scn1a(R1407X/+) mice against seizure-induced respiratory arrest independent of ketosis. Epilepsy & Behavior: E&B. 2021;124:108334. doi: 10.1016/j.yebeh.2021.108334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estes ML, McAllister AK. Maternal immune activation: implications for neuropsychiatric disorders. Science. 2016;353:772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estes ML, McAllister AK. Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci. 2015;16:469–486. doi: 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glynn MW, Elmer BM, Garay PA, Liu XB, Needleman LA, El-Sabeawy F, McAllister AK. MHCI negatively regulates synapse density during the establishment of cortical connections. Nat Neurosci. 2011;14:442–451. doi: 10.1038/nn.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekar A, Bialas AR, De Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Daly MJ, Carroll MC, Stevens B, McCarroll SA. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang NM, Cowan M, Moonah SN, Petri WA. The impact of systemic inflammation on neurodevelopment. Trends Mol Med. 2018;24:794–804. doi: 10.1016/j.molmed.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerner-Natoli M, Montpied P, Rousset MC, Bockaert J, Rondouin G. Sequential expression of surface antigens and transcription factor NFkappaB by hippocampal cells in excitotoxicity and experimental epilepsy. Epilepsy Res. 2000;41:141–154. doi: 10.1016/s0920-1211(00)00132-7. [DOI] [PubMed] [Google Scholar]
- 35.Greene AE, Todorova MT, Seyfried TN. Perspectives on the metabolic management of epilepsy through dietary reduction of glucose and elevation of ketone bodies. J Neurochem. 2003;86:529–537. doi: 10.1046/j.1471-4159.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 36.Verrotti A, Scaparrotta A, Olivieri C, Chiarelli F. Seizures and type 1 diabetes mellitus: current state of knowledge. Eur J Endocrinol. 2012;167:749–758. doi: 10.1530/EJE-12-0699. [DOI] [PubMed] [Google Scholar]
- 37.Zhan Q, Buchanan GF, Motelow JE, Andrews J, Vitkovskiy P, Chen WC, Serout F, Gummadavelli A, Kundishora A, Furman M, Li W, Bo X, Richerson GB, Blumenfeld H. Impaired serotonergic brainstem function during and after seizures. J Neurosci. 2016;36:2711–2722. doi: 10.1523/JNEUROSCI.4331-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Severson CA, Wang W, Pieribone VA, Dohle CI, Richerson GB. Midbrain serotonergic neurons are central pH chemoreceptors. Nat Neurosci. 2003;6:1139–1140. doi: 10.1038/nn1130. [DOI] [PubMed] [Google Scholar]
- 39.Buchanan GF, Richerson GB. Central serotonin neurons are required for arousal to CO2. Proc Natl Acad Sci U S A. 2010;107:16354–16359. doi: 10.1073/pnas.1004587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schechter PJ, Tranier Y, Jung MJ, Böhlen P. Audiogenic seizure protection by elevated brain GABA concentration in mice: effects of gamma-acetylenic gaba and gamma-vinyl GABA, two irreversible GABA-T inhibitors. Eur J Pharmacol. 1977;45:319–328. doi: 10.1016/0014-2999(77)90270-9. [DOI] [PubMed] [Google Scholar]
- 41.Hertz L, Schousboe A, Formby B, Lennox-Buchthal M. Some age-dependent biochemical changes in mice susceptible to seizures. Epilepsia. 1974;15:619–631. doi: 10.1111/j.1528-1157.1974.tb04034.x. [DOI] [PubMed] [Google Scholar]
- 42.Cestari IN, Liu ZF, Mu W, Burt DR. GABA(a) receptor alpha4 subunit in DBA/2J and C57BL/6J mice. Brain Res Bull. 1998;47:643–647. doi: 10.1016/s0361-9230(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 43.Wang JB, Liu ZF, Kofuji P, Burt DR. The GABA(a) receptor gamma1-subunit in seizure prone (DBA/2) and resistant (C57BL/6) mice. Brain Res Bull. 1998;45:421–425. doi: 10.1016/s0361-9230(97)00348-1. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong JN, Plumier JCL, Robertson HA, Currie RW. The inducible 70,000 molecular/weight heat shock protein is expressed in the degenerating dentate hilus and piriform cortex after systemic administration of kainic acid in the rat. Neuroscience. 1996;74:685–693. doi: 10.1016/0306-4522(96)00164-9. [DOI] [PubMed] [Google Scholar]
- 45.de Freitas MS, Spohr TCLS, Benedito AB, Caetano MS, Margulis B, Lopes UG, Moura-Neto V. Neurite outgrowth is impaired on HSP70-positive astrocytes through a mechanism that requires NF-kappaB activation. Brain Res. 2002;958:359–370. doi: 10.1016/s0006-8993(02)03682-x. [DOI] [PubMed] [Google Scholar]
- 46.Thomas G, Souil E, Richard MJ, Saunier B, Polla BS, Bachelet M. Hyperthermia assists survival of astrocytes from oxidative-mediated necrotic cell death., cellular and molecular biology, vol. 48. Noisy-Le-Grand, France; 2002. p. 191–8. [PubMed]
- 47.Krueger AMR, Armstrong JN, Plumier JC, Robertson HA, Currie RW. Cell specific expression of Hsp70 in neurons and glia of the rat hippocampus after hyperthermia and kainic acid-induced seizure activity. Mol Brain Res. 1999;71:265–278. doi: 10.1016/S0169-328X(99)00198-9. [DOI] [PubMed] [Google Scholar]
- 48.Farley MM, Watkins TA. Intrinsic neuronal stress response pathways in injury and disease, Annual Review of Pathology. Mechanisms of Disease. 2018;13:93–116. doi: 10.1146/annurev-pathol-012414-040354. [DOI] [PubMed] [Google Scholar]
- 49.Leitner DF, Mills JD, Pires G, Faustin A, Drummond E, Kanshin E, Nayak S, Askenazi M, Verducci C, Chen BJ, Janitz M, Anink JJ, Baayen JC, Idema S, van Vliet EA, Devore S, Friedman D, Diehl B, Scott C, Thijs R, Wisniewski T, Ueberheide B, Thom M, Aronica E, Devinsky O. Proteomics and Transcriptomics of the Hippocampus and cortex in SUDEP and high-risk SUDEP patients. Neurology. 2021;96:e2639–e2652. doi: 10.1212/WNL.0000000000011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canty AJ, Dietze J, Harvey M, Enomoto H, Milbrandt J, Ibáñez CF. Regionalized loss of parvalbumin interneurons in the cerebral cortex of mice with deficits in GFRalpha1 signaling., the journal of Neuroscience : the official journal of the society for. Neuroscience. 2009;29:10695–10705. doi: 10.1523/JNEUROSCI.2658-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanter-Schlifke I, Fjord-Larsen L, Kusk P, Angehagen M, Wahlberg L, Kokaia M. GDNF released from encapsulated cells suppresses seizure activity in the epileptic hippocampus. Exp Neurol. 2009;216:413–419. doi: 10.1016/j.expneurol.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 52.Barry G, Briggs JA, Hwang DW, Nayler SP, Fortuna PRJ, Jonkhout N, Dachet F, Maag JLV, Mestdagh P, Singh EM, Avesson L, Kaczorowski DC, Ozturk E, Jones NC, Vetter I, Arriola-Martinez L, Hu J, Franco GR, Warn VM, Gong A, Dinger ME, Rigo F, Lipovich L, Morris MJ, O’Brien TJ, Lee DS, Loeb JA, Blackshaw S, Mattick JS, Wolvetang EJ. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci Rep. 2017;7:40127. doi: 10.1038/srep40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aiba I, Noebels JL. Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med. 2015;7:282ra46. doi: 10.1126/scitranslmed.aaa4050.Spreading. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard L, Wosnitzka E, Okakpu D, White MA, Wyatt S, Davies AM. TWE-PRIL reverse signalling suppresses sympathetic axon growth and tissue innervation. Development (Cambridge). 2018;145. 10.1242/dev.165936. [DOI] [PMC free article] [PubMed]
- 55.Picard RW, Migliorini M, Caborni C, Onorati F, Regalia G, Friedman D, Devinsky O. Wrist sensor reveals sympathetic hyperactivity and hypoventilation before probable SUDEP. Neurology. 2017;89:633–635. doi: 10.1212/WNL.0000000000004208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The details of all differentially expressed lncRNAs.
Additional file 2. The details of all differentially expressed mRNAs.
Additional file 3. Supplementary Table based on fold change ≥ 2.0 and FDR<0.05.
Additional file 4. Supplementary Figure based on fold change ≥ 2.0 and FDR<0.05.
Data Availability Statement
The datasets presented in this study can be found in online repositories and have been deposited with the Gene Expression Omnibus (GEO) under the project accession number GSE152931 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152931).