Abstract
Hepatocellular carcinoma (HCC) is one of the leading lethal malignant tumors worldwide. DEAD-box (DDX) family helicases are implicated in numerous human cancers. However, the role of DDX1 in HCC has not yet been fully elucidated. We downloaded gene expression data and clinical information data of HCC from The Cancer Genome Atlas and International Cancer Genome Consortium (ICGC) database and conducted subsequent analyses using the R package and online portal. The results revealed that HCC tissues had higher DDX1 expression compared with either paired or unpaired normal tissues. The increased DDX1 expression was closely related to the advanced pathological grade and histologic grade of HCC. Further analysis suggested that patients with high DDX1 expression contributed to poor prognosis The Cox regression analysis revealed that the expression level of DDX1 was an independent prognostic factor for HCC. In addition, an ICGC cohort was used for external validation. The cBio-Portal, MethSurv, and UALCAN database were used for evaluating the genomic mechanism. Moreover, the Tumor Immune Estimation Resource dataset and QUANTISEQ algorithm revealed that DDX1 expression positively correlates with immune infiltrating cells. We also identified the DDX1-related differentially expressed genes (DEGs) and explored their biological functions by GO, KEGG, and GSEA analyses, which indicated that DDX1 may regulate the progression of HCC. In general, increased DDX1 expression predicts a poor prognosis and drives the progression of HCC.
Keywords: DDX1 , Hepatocellular carcinoma , Prognostic biomarker , Tumor microenvironment , Infiltration immune cells , Methylation
Introduction
Hepatocellular carcinoma (HCC) is known to be the fifth most common cancer and is the third cause of cancer-related mortalities worldwide. Thus, it is considered a huge threat to people’s health [1]. Surgical resection and liver transplantation are the most suitable methods for treatment of early-stage HCC. However, for non-resectable patients, radiofrequency ablation (RFA) and stereotactic body radiation therapy (SBRT) are deemed as first-line local treatment options [2, 3]. Nonetheless, many patients with HCC treated with resection or local ablation show early relapse after treatment [4]. In the past decade, immunotherapy has become popular for successfully treating various cancers [5–8]. Interestingly, cumulative evidence revealed that immune-mediated mechanisms are deeply involved in the progression of HCC [9]. This suggests the great potential of immune-based therapies for treating patients with HCC. However, few studies report only satisfactory immunotherapy efficacy for HCC. The failure of immunotherapy could be attributed to the complex composition of the tumor microenvironment (TME). Thus, it is important to understand the tumor immune microenvironment (TIME) for identifying pivotal therapeutic targets and prognostic biomarkers for HCC.
DDX1 is a member of the DEAD-box RNA helicase family and participates in various biological processes, such as mRNA translation, tRNA splicing, rRNA processing, microRNA maturation, and repair of DNA double-strand breaks (DSBs) [10–14]. In addition to its role in transcriptional regulation, a recent study reported that DDX1 interacts with eIF3a and eIF4b to decrease insulin translation [15]. Furthermore, some studies found that DDX1 can inhibit viral replication [16, 17]. Moreover, researchers have found that DDX1 promotes tumorigenesis in various carcinomas, such as retinoblastoma, neuroblastoma, testicular carcinoma, colorectal cancer, and breast cancer [18–21]. Nevertheless, the underlying mechanism of DDX1 in the progression of HCC remains obscure. In this research, we tried to revealed the potential role of DDX1 in the TIME of HCC.
Materials and methods
Data collection and analysis
We downloaded the raw RNA sequence data and associated clinical data of patients with HCC from the The Cancer Genome Atlas(TCGA) database. We compared the differential expression of DDX1 between HCC and normal tissues by paired t-test and Mann–Whitney U test. Subsequently, an independent sample t-test was used to analyze the correlation of DDX1 expression with clinicopathologic characteristics, including gender, T stage, pathologic stage, histologic grade, age, alpha fetoprotein (AFP) level and Child-Pugh grade. Analysis and plots were carried out using R packages “rstatix,” “car,” “tidyverse,” “ggplot2,” and “reshape2.” A P value of < 0.05 was considered statistically significant.
Survival analysis and external validation
Based on the average DDX1 expression, the Kaplan Meier (KM) survival curve was used to analyze the prognosis of patients with HCC with different levels of DDX1 mRNA expression. The analyses were conducted using the “survival” and “survminer” packages. Furthermore, we downloaded the RNA-sequencing expression profiles and corresponding clinical information of the liver cancer dataset (RIKEN, JP) from the International Cancer Genome Consortium (ICGC) database. According to the level of DDX1 expression in patient samples, two groups were created. The log-rank test was used to evaluate the difference in overall survival (OS) between the groups. The timeROC analysis was used to compare the predictive accuracy. Log-rank test and Cox regression methods were used to calculate the KM curve P values and hazard ratio (HR) with 95% confidence (CI) intervals.
Genetic mutation and methylation analysis
The cBioPortal (www.cbioportal.org) is an online exploratory tool for visualizing and analyzing cancer genomic data in multidimensional ways [22]. In this study, we used three different datasets (INSERM, Nat Genet 2015; AMC, Hepatology 2014; TCGA, Firehose Legacy) in cBioPortal to characterize the genomic profiles of DDX1. We categorized the patients into two groups (no mutation and mutation of DDX1) and used the Kaplan–Meier survival curves for evaluating the effect of mutations on survival.
Metasurv (https://biit.cs.ut.ee/methsurv/) provides survival analysis based on CpG methylation patterns [23]. To gain a comprehensive understanding of methylation and prognosis, we accessed the survival rate of the CpG methylation sites in DDX1. Survival was expressed using a Kaplan–Meier (KM) curve. In addition, the UALCAN (ualcan.path.uab.edu/index.html) database was employed to access the correlation of promoter methylation level of DDX1 in HCC with clinicopathological characteristics. Statistical significance was defined as a difference of < 0.05.
Correlation between DDX1 expression and immune infiltration
With the use of QUANTISEQ algorithm, we determined the association between DDX1 expression and tumor-infiltrating immune cells (TIICs), which including CD4+ T cells, CD8+ T cells, macrophages and so on. The R package “immunedeconv” was used to reliably estimate immune infiltration [24].
TIMER (https://cistrome.shinyapps.io/timer/) is a public website that analyzes the abundance of TIICs and gene expression across various cancers [25]. According to TIMER dataset, the expression level of DDX1 was associated with the degree of TIICs, including as CD8+ T cells, CD4+ T cells,macrophages and so on. The above results were executed by the Gene module and visualized by scatter plots. We measured the correlation between TIICs and DDX1 expression using Spearman’s correlation.
Biological function enrichment analysis
The differentially expressed mRNAs were screened using the Limma package based on the RNA sequence and associated clinical information downloaded from the TCGA database. To correct the false-positive results, the adjusted P-value was analyzed. We set the thresholds for differentially expressed genes (DEGs) as adjusted P-value of < 0.05 and |log2 fold change| ≥ 1.5. Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.kegg.jp/) is a sophisticated database resource for the systematic analysis of gene functions, which links genomic information with higher order functional information [26–28]. Subsequently, the ClusterProfiler R package was utilized to analyze the pathways enriched in DEGs based on Gene Ontology (GO) and KEGG databases.
Protein interaction screening
STRING (version 11.5) is an online tool that evaluates the interactions of genes [29]. In this study, the STRING dataset was used to search co-expressed genes and construct Protein-Protein Interaction Networks(PPI) networks with an interaction score ≥ 0.15. We identified the top 50 DDX1-interacted proteins. Moreover, using the Similar Genes Detection module of GEPIA (http://gepia.cancer-pku.cn/index.html), we identified the top 100 DDX1-related genes in liver cancer samples from TCGA. We then utilized Online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to carry out an intersection analysis between the top 50 DDX1-binding proteins and the top 100 genes related to DDX1 expression. Finally, we investigated the association of DDX1 with the selected genes via the Correlation Analysis module of GEPIA. Results were presented using scatter plots and Pearson correlation coefficient.
GSEA analysis
RNA-seq data collected from the TCGA database were analyzed using Gene Set Enrichment Analysis (GSEA) to preliminarily classify the genes based on their correlation with DDX1 expression. By GSEA, we carried out a KEGG enrichment analysis to explore the underlying biological roles of DDX1. A false discovery rate (FDR) < 0.05 and a nominal P-value < 0.05 were considered statistically significant.
Results
Baseline and clinical characteristics of patients with HCC
In all, 374 HCC samples with corresponding clinical features were obtained from the TCGA database. The baseline characteristics and clinical features are summarized in Table 1. The study included 253 males (67.6%) and 121 females (32.4%). Among all participants, 47.5% (n = 177) were ≤ 60 years and 52.5% (n = 196) were > 60 years. As for the histologic grade, 55 (14.9%) patients were grade I, 178 (48.2%) patients were grade II, 124 (33.6%) patients were grade III, and only 12 (3.3%) patients were grade IV. In terms of liver function, there were 219 (90.9%) cases of Child-Pugh Grade A, 21 (8.7%) cases of Grade B, and 1 (0.4%) case of Grade C.
Table 1.
Clinical characteristics of the patients with LIHC
| Characteristic | Levels | Overall (%) |
|---|---|---|
| Age, n (%) | ≤ 60 | 177 (47.5) |
| > 60 | 196 (52.5) | |
| Gender, n (%) | Female | 121 (32.4) |
| Male | 253 (67.6) | |
| Race, n (%) | Asian | 160 (44.2) |
| Black or African American | 17 (4.7) | |
| White | 185 (51.1) | |
| Pathologic stage, n (%) | Stage I | 173 (49.4) |
| Stage II | 87 (24.9) | |
| Stage III | 85 (24.3) | |
| Stage IV | 5 (1.4) | |
| T stage, n (%) | T1 | 183 (49.3) |
| T2 | 95 (25.6) | |
| T3 | 80 (21.6) | |
| T4 | 13 (3.5) | |
| N stage, n (%) | N0 | 254 (98.4) |
| N1 | 4 (1.6) | |
| M stage, n (%) | M0 | 268 (98.5) |
| M1 | 4 (1.5) | |
| Histologic grade, n (%) | G1 | 55 (14.9) |
| G2 | 178 (48.2) | |
| G3 | 124 (33.6) | |
| G4 | 12 (3.3) | |
| Adjacent hepatic tissue inflammation, n (%) | None | 118 (49.8) |
| Mild | 101 (42.6) | |
| Severe | 18 (7.6) | |
| AFP(ng/ml), n (%) | ≤ 400 | 215 (76.8) |
| > 400 | 65 (23.2) | |
| Child-Pugh grade, n (%) | A | 219 (90.9) |
| B | 21 (8.7) | |
| C | 1 (0.4) |
Association between DDX1 expression and clinicopathological characteristics in HCC samples
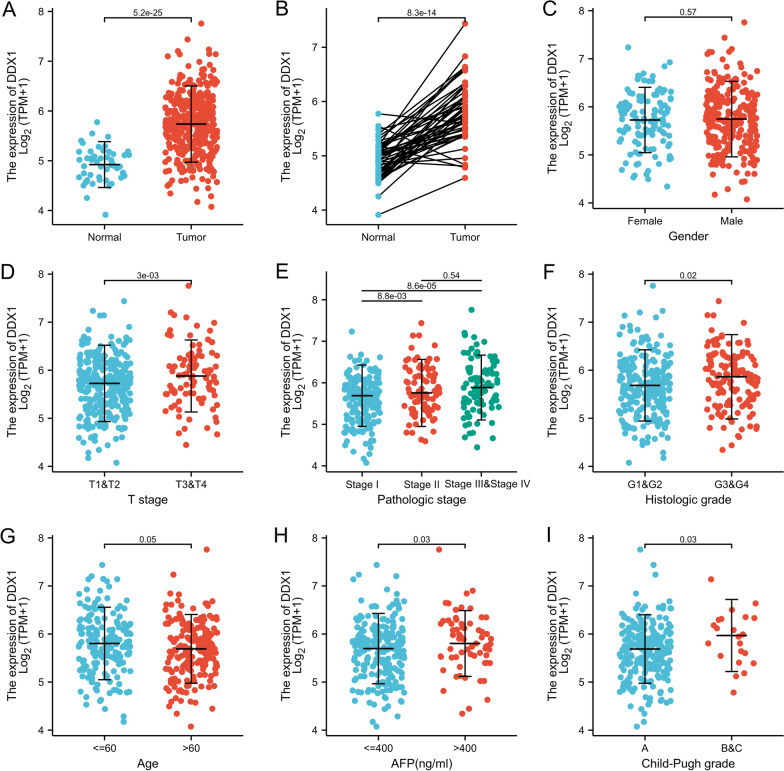
Firstly, we analyzed DDX1 expression in patients with HCC. Both in paired and unpaired samples, DDX1 was expressed at higher levels in tumor samples than in normal samples (Fig. 1A, P < 0.001 and Fig. 1B, P < 0.001). DDX1 expression showed no significant difference between females and males (Fig. 1C, P = 0.574). We found that DDX1 expression was positively associated with the T stage of the tumor (Fig. 1D, P = 0.003), pathologic stage (Fig. 1E, P < 0.001), and histologic grade of HCC (Fig. 1F, P = 0.02). Furthermore, the DDX1 expression was closely linked with age, AFP level and Child-Pugh grade of the patients with HCC (Fig. 1G, P = 0.047; Fig. 1H, P = 0.032; Fig. 1I, P = 0.03).
Fig. 1.
Correlation analysis between DDX1 expression and clinicopathologic features. A DDX1 expression in unpaired samples. B DDX1 expression in paired sample group; C Gender; D T stage; E Pathologic stage; F Histologic grade; G Age; H AFP level; I Child-Pugh grade
High DDX1 expression was an independent prognostic factor for HCC
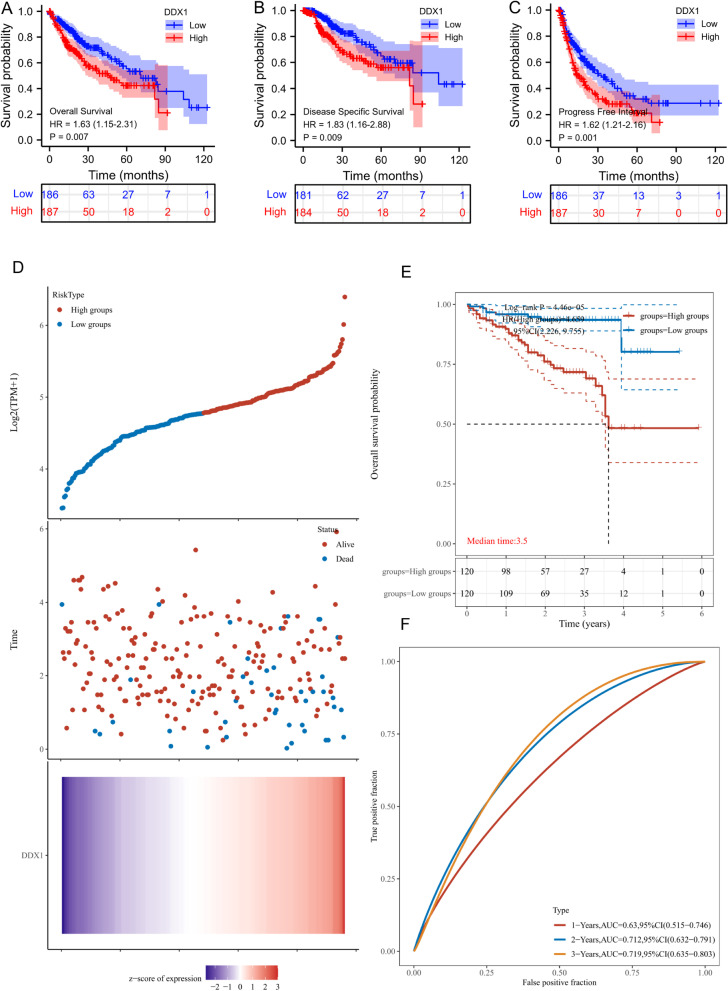
In the TCGA cohort, patients with high-DDX1 expression had significantly shorter OS, disease specific survival(DSS) and progress free interval (PFI) than patients with low-DDX1 expression (Fig. 2A–C). We employed the ICGC database to validate the relation between DDX1 expression and outcome of HCC. According to median DDX1 expression, patients were stratified into high- and low-expression groups. The results revealed that the fatality rate was significantly higher in the high DDX1 expression group compared with the low-expression group (Fig. 2D). In addition, the KM survival analysis indicated that upregulated DDX1 was related to poor patient survival (HR = 4.659, 95% CI = 2.226–9.755, log-rank P = 4.46e-05) (Fig. 2E). We further predicted the 1, 2, and 3 year risk of patients with HCC by estimating the area under the curve (AUC) under the ROC curve (1 year, AUC = 0.63; 2 year, AUC = 0.712; 3 year, AUC = 0.719) (Fig. 2F). Overall, the results indicate that DDX1 expression is closely linked to the outcome of patients with HCC.
Fig. 2.
Prognostic analysis and ICGC dataset validation. A–C Kaplan–Meier survival curve analysis for the effect of DDX1 expression on the prognosis of patients with liver cancer (OS, DSS, and RFI respectively). D The DDX1 expression, and survival analyze of the ICGC dataset. E Kaplan–Meier survival analysis of the DDX1 signature from ICGC dataset. F The ROC curve of the gene
In addition, univariate Cox regression analysis showed significant relationship of pathologic stage, DDX1 expression (hazard ratio = 1.628, 95% CI = 1.145–2.313, P = 0.007), T stage, M stage, and tumor status with poor OS (Table 2). Additionally, multivariate Cox regression analysis identified DDX1 gene expression (HR = 1.822, 95% CI = 1.139–2.915, P = 0.012) and tumor status (HR = 1.873, 95% CI = 1.172–2.995, P = 0.009) as independent risk factors of total survival for patients with HCC.
Table 2.
Univariate and multivariate Cox regression analyses of clinical characteristics associated with overall survival
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 373 | ||||
| ≤ 60 | 177 | Reference | |||
| > 60 | 196 | 1.205 (0.850–1.708) | 0.295 | ||
| Gender | 373 | ||||
| Female | 121 | Reference | |||
| Male | 252 | 0.793 (0.557–1.130) | 0.200 | ||
| Pathologic stage | 349 | ||||
| Stage I & Stage II | 259 | Reference | |||
| Stage III & Stage IV | 90 | 2.504 (1.727–3.631) | < 0.001 | 1.341 (0.182–9.864) | 0.773 |
| Histologic grade | 368 | ||||
| G1&G2 | 233 | Reference | |||
| G3&G4 | 135 | 1.091 (0.761–1.564) | 0.636 | ||
| DDX1 | 373 | ||||
| Low | 186 | Reference | |||
| High | 187 | 1.628 (1.145–2.313) | 0.007 | 1.822 (1.139–2.915) | 0.012 |
| AFP(ng/ml) | 279 | ||||
| ≤ 400 | 215 | Reference | |||
| > 400 | 64 | 1.075 (0.658–1.759) | 0.772 | ||
| T stage | 370 | ||||
| T1&T2 | 277 | Reference | |||
| T3&T4 | 93 | 2.598 (1.826–3.697) | < 0.001 | 1.833 (0.248–13.569) | 0.553 |
| N stage | 258 | ||||
| N0 | 254 | Reference | |||
| N1 | 4 | 2.029 (0.497–8.281) | 0.324 | ||
| M stage | 272 | ||||
| M0 | 268 | Reference | |||
| M1 | 4 | 4.077 (1.281–12.973) | 0.017 | 1.856 (0.427–8.075) | 0.410 |
| Child-Pugh grade | 240 | ||||
| A | 218 | Reference | |||
| B&C | 22 | 1.643 (0.811–3.330) | 0.168 | ||
| Residual tumor | 344 | ||||
| R0 | 326 | Reference | |||
| R1&R2 | 18 | 1.604 (0.812–3.169) | 0.174 | ||
| Tumor status | 354 | ||||
| Tumor free | 202 | Reference | |||
| With tumor | 152 | 2.317 (1.590–3.376) | < 0.001 | 1.873 (1.172–2.995) | 0.009 |
Bold values indicates p < 0.05
DDX1 genetic mutation and methylation in patients with HCC
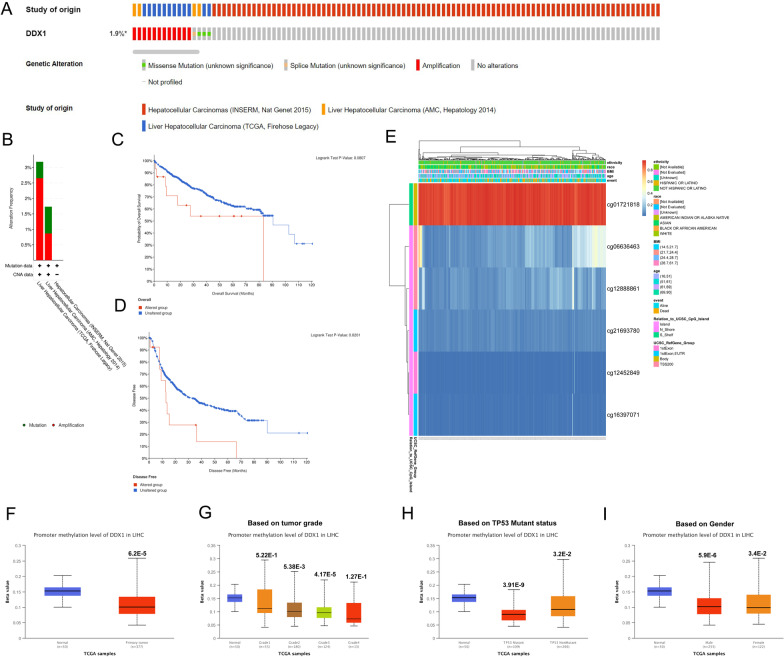
In all, we included data of 916 patients across three datasets (Fig. 3A) and found that the DDX1 gene alteration rate ranged from 1.73 to 1.81% (Fig. 3B). The mutated and not mutated groups showed no difference in OS (log-rank P = 0.0807) (Fig. 3C). However, the DFS rates of the two groups differed significantly (Fig. 3D log-rank P = 0.0201). Next, we detected the DNA methylation levels of DDX1, with the prognostic value of each single CpG by using MethSurv. The headmap graph shown in Fig. 3E showed the methylation of the six CpG sites in LIHC. Of these, cg01721818 methylation was the highest. In addition, the higher the DNA methylation level of cg01721818 and cg21693780, the worse the prognosis (Table 3). Moreover, subgroup analysis of the UALCAN database showed that the promoter methylation of DDX1 was significantly lower in patients with LIHC than in normal controls in subgroups based on grade, gender, and TP53 mutant status (Fig. 3F–I).
Fig. 3.
Genetic alteration and methylation of DDX1 in HCC. A OncoPrint visual summary of alteration in DDX1. B Summary of alterations in DDX1 in HCC based on three datasets. C, D Comparison of the survival prognosis between DDX1 mutation and unmutation groups. E Visualization of six CpG sites on DDX1 with methylation level. F–I Relation between DDX1 promoter methylation in HCC and normal tissues and clinicopathological characteristics: F normal versus primary tumor, G tumor grade, H TP53 mutation status, and I gender
Table 3.
Effect of hypermethylation level on prognosis in LIHC
| CpG | HR | CI | P-value |
|---|---|---|---|
| cg01721818 | 1.948 | (1.209;3.14) | 0.006177134 |
| cg06636463 | 1.079 | (0.765;1.522) | 0.664338648 |
| cg12452849 | 1.19 | (0.842;1.681) | 0.325095456 |
| cg12888861 | 1.544 | (0.984;2.423) | 0.058794328 |
| cg16397071 | 0.87 | (0.617;1.226) | 0.425388277 |
| cg21693780 | 1.519 | (1.071;2.155) | 0.019020876 |
DDX1 is correlated with tumor immune infiltration in HCC
We assessed the expression of 10 immune cell subtypes in the high and low DDX1 expression groups using the QUANTISEQ algorithm (Fig. 4A, B). We found that DDX1 expression was related to CD8+ T cell, B cell, M2 macrophages, Tregs, and monocytes. The immune infiltration scores of M2 macrophages and Tregs were higher in the high DDX1 expression group compared with the low DDX1 expression group. Moreover, we analyzed the relationshipof DDX1 expression the immune infiltration cells via TIMER. As depicted in Fig. 4C, the expression level of DDX1 was positively related with B cells (r = 0.279, P-value = 1.49e-07), CD8+ T cells (r = 0.229, P-value = 1.93e-05), CD4+ T cells (r = 0.285, P-value = 7.86 × 10− 8), macrophages (r = 0.365, P-value = 3.69 × 10− 12), neutrophils (r = 0.398, P-value = 1.54 × 10− 14), and dendritic cells (r = 0.353, P-value = 2.16 × 10− 11). These results indicate that DDX1 plays a pivotal role in immune infiltration in HCC.
Fig. 4.
A, B The different proportions of 10 subtypes of immune cells in tumor sumple expressing high and low level of DDX1. C Association of DDX1 expression with immune infiltration levels
Screening of DEGs and functional enrichment analysis
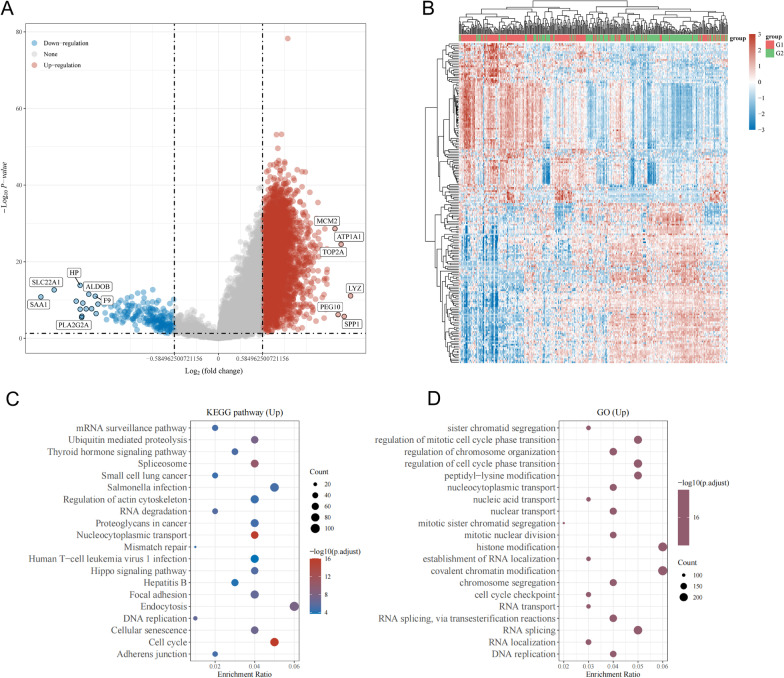
To further investigate the biological functions of DDX1 in HCC, we identified the DEGs. Figure 5A shows the volcano plot for DEGs, of which 4212 were upregulated and 205 were downregulated. Using a hierarchical clustering analysis of DEGs, heatmaps were created to observe the genes in similar samples (Fig. 5B). To explore the potential biological roleof upregulated DEGs in HCC, we used GO and KEGG enrichment analyses. KEGG enrichment results demonstrated that the upregulated DEGs were mainly enriched in the spliceosome, proteoglycans in cancer, focal adhesion, cell cycle, cellular senescence, and endocytosis (Fig. 5C). Next, GO enrichment analysis of upregulated DEGs indicated that they were correlated with cell cycle regulation, RNA editing including RNA splicing, covalent chromatin modification, histone modification, regulation of cell cycle phase transition, and regulation of mitotic cell cycle phase transition (Fig. 5D).
Fig. 5.
Enrichment analysis of DDX1 expression-correlated DEGs in HCC. A Volcano plot of DEGs between high and low DDX1 expression groups. B Clustering analysis heatmap of DDX1 expression-correlated DEGs. C, D KEGG and GO analyses of upregulated DEGs
PPI network construction and correlation analysis
To determine the intrinsic role of DDX1 in HCC tumorigenesis, we used STRING to identify the top 50 DDX1-binding proteins (Fig. 6A). Subsequently, GEPIA was used to identify the top 100 genes correlating with DDX1 expression. We conducted the intersection analysis of the above two groups and found HNRNPU (heterogeneous nuclear ribonucleoprotein U), TARDBP (TAR DNA-binding protein), and HNRNPK (heterogeneous nuclear ribonucleoprotein K) as common factors (Fig. 6B). The DDX1 expression was positively correlated with HNRNPU (Fig. 6C, R = 0.78, P-value = 2.8e-77), TARDBP (Fig. 6D, R = 0.73, P-value = 2e-61), and HNRNPK (Fig. 6E, R = 0.76, P-value = 4.2e-71).
Fig. 6.
DDX1-related gene enrichment analysis and GSEA analysis. A STRING website was used to extract the top 50 DDX1-binding proteins which were supported by available experiments. B Intersection analysis of DDX1-binding and correlated genes. C–E Using the GEPIA approach, we obtained the top 100 DDX1-correlated genes in TCGA and analyzed the expression correlation between DDX1 and selected targets, including HNRNPU, TARDBP, HNRNPK F Pathways in cancer, G cell cycle, H DNA replication, I Wnt signaling pathway, J MAPK signaling pathway, and K TGF-β signaling pathway
Identification of DDX1-related signaling pathways by GSEA analysis
We performed a GSEA analysis to identify the DDX1-related signaling pathways involved in HCC. The results showed that DDX1-associated DEGs were significantly enriched in cancer pathway (Fig. 6F–K), such as Pathways in cancer [normalized enrichment score (NES) = 2.186, adjusted P-value = 0.017, FDR = 0.011], MAPK signaling pathway (NES = 1.704, adjusted P-value = 0.017, FDR = 0.011), Wnt signaling pathway (NES = 2.166, adjusted P-value = 0.017, FDR = 0.011), TGF-β signaling pathway (NES = 1.839, adjusted P-value = 0.021, FDR = 0.013), and DNA replication (NES = 1.935, adjusted P-value = 0.021, FDR = 0.013). Interestingly, DDX1-associated DEGs were associated with cell cycle (NES = 2.448, adjusted P-value = 0.017, FDR = 0.011). These results suggest that DDX1-associated DEGs may participate in these signaling pathways to modulate tumor progression.
Discussion
The DEAD-box RNA helicase family is known to play critical roles in various RNA metabolic processes. Recent studies indicate that DDX members are dysregulated in multiple cancers and function as key players in tumor progression [30]. Hu et al. identified that DDX51 regulates cellular proliferation in esophageal squamous cell carcinoma through the PI3K/AKT/mTOR pathway [31]. Jiang et al. proved that knock-down of DDX46 caused a significant reduction in cell invasion and migration in osteosarcoma [32]. DDX1 activates the transcription of 12p stem cell genes in testicular tumorigenesis [19]. Tanaka et al. proved the role of DDX1 in promoting colorectal tumorigenesis in vitro and in vivo [20]. Researchers have proved that high DDX1 expression is associated with improvement in local control, distant metastatic-free survival, and OS when compared with low DDX1 expression in node-negative and early-stage patients with breast cancer [33]. However, the role of DDX1 in HCC is not yet determined. Therefore, in this study, we focused on determining the potential prognostic value of DDX1 in HCC.
We acquired HCC samples from the TCGA database to explore the role of DDX1 in HCC progression. The results reveal that high DDX1 expression is associated with a poor prognosis of HCC. On analyzing the relationship between DDX1 and clinicopathologic features we found that DDX1 may serve as an adverse prognostic factor in HCC. Patients with high DDX1 expression may present an advanced T stage, histologic grade, and pathological stage.
According to molecular profiles and clinical outcomes, HCC has a highly heterogeneous nature, which present a formidable challenge to an accurate diagnosis and treatment [34]. Luckily, previous studies have proved that molecular subtype stratification could overcome the hurdles caused by tumor heterogeneity [35, 36]. In this research, 240 HCC samples downloaded from ICGC dataset were divided into high and low DDX1-expression subgroups, the KM survival analysis revealed that patients with high levels of DDX1 are at higher risk of suffering a poor prognosis. This results were consistent with the findings from TCGA dataset.
As liver carcinoma is typically associated with high malignancy, it is usually diagnosed at a late stage, posing a challenge for radical surgery. In order to improve the outcome of HCC, early diagnosis and treatment are crucial. As we known, tumorigenesis has been demonstrated to result from multiple gene mutations, patients with gene mutations are known to have a poor prognosis, In this research,we accessed the alteration percentage of DDX1 in HCC. After analyzing three independent datasets, we found the percentage of DDX1 alteration to be 1.9%. Furthermore, no significant differences was observed in OS between the mutation and unmutation DDX1 groups. In addition to gene mutations, numerous epigenetic changes, such as DNA methylation and histone modifications, contribute to tumor development [37–39]. Nowadays, since DNA methylation is tissue-specific and stable, detecting abnormal DNA methylation in liquid biopsies has been shown to be a promising biomarker for cancer diagnosis [40, 41]. A previous study indicated that HCC is caused by DNA methylation [42]. For example, Kuo et al. found that patients with higher IRAK3 methylation had worse prognosis [43]. However, no study has examined the relationship between DDX1 methylation and the oncologic outcome of HCC. Hence, in this study, we carried out methylation analysis and evaluated the correlation of methylation level with prognosis. We identified six CpG sites and two of them with hypermethylation were associated with poor prognosis. Besides, a previous study demonstrated that the hypomethylation status of oncogenes could also promote tumor development [44]. In this study, using UALCAN we found that HCC tissues had lower levels of DDX1 promoter methylation than normal tissues (P < 0.05). Further analysis indicated that the high tumor grade was linked to low promoter methylation levels. Thus, DDX1 methylation examination has the potential to be developed as a screening tool for predicting tumor status and progression; however, further in vivo and in vitro experiments are needed.
TME is a complicated assembly of the tumor, immune, stromal, and extracellular components [45]. Previous studies revealed that TME facilitates the progression of HCC, thus indicating that it could be exploited as a potential therapeutic target [46]. The importance of immune cell infiltration in the TME has been recognized for various solid tumors [47, 48]. In this study, we determined the association between DDX1 expression and immune cell infiltration in HCC through the TIMER database. The results revealed that DDX1 is positively associated with dendritic cells, B cells, macrophages, and T cells. In addition, we verified the positive correlation between DDX1 expression and immune cell infiltration using the QUANTISEQ algorithm. M2 macrophages and Tregs were higher in the high DDX1 expression group compared with the low DDX1 expression group. Several studies indicate that macrophages are essential components of the TME and play key roles in tumor progression [49]. Liu et al. [50] found that M2 macrophages target miR-149-5p/MMP9 signaling pathway thereby facilitating HCC progression. Yeung et al. demonstrated that M2 macrophages are related to the adverse prognosis of liver cancer and promote HCC invasion by promoting Epithelial mesenchymal transformation(EMT) [51]. Another research revealed that M2-polarized macrophages promote EMT of HCC cells and accelerate tumor progression through the TLR4/STAT3 signaling pathway [52]. Similarly, Tregs are also critical components of TME. Jiang et al. proved that Tregs are closely associated with the prognosis of HCC. They secrete TGF-β1 which triggers EMT, thereby enhancing tumor invasiveness [53]. Shen et al. [54] put forth that TGF-β could drive Treg cell polarization to promote the progression of HCC. These results suggest that DDX1 plays a vital role in the TIME of HCC. However, additional experimental and theoretical studies are needed to validate the relationship between DDX1 and tumor infiltration.
To explore the potential biological functions of DDX1, we identified the DEGs in HCC samples and conducted GO and KEGG enrichment analyses. The results showed that DDX1 was mainly related to cell cycle and RNA editing. In addition, since DEAD box proteins function by interacting with other proteins, they are susceptible to being regulated by their partners and their microenvironment [55]. Hence, using STRING and GEPIA database, we extracted 50 DDX1-binding proteins and the top 100 DDX1-related genes. Intersection analysis of the two groups identified HNRNPU, TARDBP, and HNRNPK as the common hits. TARDBP is an RNA-binding protein involved in the cell cycle of HCC tumor cells, and its expression level is related to an advanced stage and high grade of HCC [56]. The CDK2 protein is known as an essential role in cell cycle regulation [57]. Liang et al. have found HNRNPU enhances CDK2 transcription, thereby promoting HCC development [58]. GSEA analysis also identified significant KEGG pathways associated with the cell cycle.
Conclusion
In summary, this study is the first to identify DDX1 as a diagnostic and prognostic biomarker of HCC. The results indicate that DDX1 has an important role in TIME and is involved in the regulation of the cell cycle of HCC. Therefore, targeted DDX1 therapy is a potential treatment strategy for patients with HCC.
Acknowledgements
This work was supported by Shuangshuang Sun.
Author contributions
SC and SS conceived and devised the study and gave aid in writing the manuscript. MY and JX conducted the analysis of data and wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study is funded by Shuangshuang Sun.
Availability of data and materials
All data analyzed in the present study was obtained from TCGA and ICGC.
Declarations
Ethics approval and consent to participate
Our analysis is based on available publicly database, not involving in the problem of ethics.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengping Yuan and Jinyong Xu contributed equally to this work
Contributor Information
Mengping Yuan, Email: ymp6098@163.com.
Jinyong Xu, Email: xujinyong@hotmail.com.
Shuguang Cao, Email: letous202@163.com.
Shuangshuang Sun, Email: 459200804@qq.com.
References
- 1.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Shiina S, et al. Radiofrequency ablation for hepatocellular carcinoma: 10-year outcome and prognostic factors. Am J Gastroenterol. 2012;107(4):569–77. doi: 10.1038/ajg.2011.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahl DR, et al. Outcomes after stereotactic body radiotherapy or radiofrequency ablation for hepatocellular carcinoma. J Clin Oncol. 2016;34(5):452–9. doi: 10.1200/JCO.2015.61.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yegin EG, et al. Progress in surgical and nonsurgical approaches for hepatocellular carcinoma treatment. Hepatobiliary Pancreat Dis Int. 2016;15(3):234–56. doi: 10.1016/s1499-3872(16)60097-8. [DOI] [PubMed] [Google Scholar]
- 5.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TSK, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–30. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 8.Seiwert TY, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–65. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, et al. Immunometabolism: a novel perspective of liver cancer microenvironment and its influence on tumor progression. World J Gastroenterol. 2018;24(31):3500–12. doi: 10.3748/wjg.v24.i31.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pazo A, et al. hCLE/RTRAF-HSPC117-DDX1-FAM98B: a new cap-binding complex that activates mRNA translation. Front Physiol. 2019;10:92. doi: 10.3389/fphys.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popow J, et al. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511(7507):104–7. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki T, et al. A novel all-in-one conditional knockout system uncovered an essential role of DDX1 in ribosomal RNA processing. Nucleic Acids Res. 2021;49(7):e40. doi: 10.1093/nar/gkaa1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han C, et al. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014;8(5):1447–60. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, et al. Role for RIF1-interacting partner DDX1 in BLM recruitment to DNA double-strand breaks. DNA Repair (Amst) 2017;55:47–63. doi: 10.1016/j.dnarep.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, et al. RNA-binding protein DDX1 is responsible for fatty acid-mediated repression of insulin translation. Nucleic Acids Res. 2018;46(22):12052–66. doi: 10.1093/nar/gky867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan X, et al. Foot-and-mouth disease virus infection suppresses autophagy and NF-кB antiviral responses via degradation of ATG5-ATG12 by 3 C(pro) Cell Death Dis. 2017;8(1):e2561. doi: 10.1038/cddis.2016.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Q, et al. The DEAD-Box RNA helicase DDX1 interacts with the viral protein 3D and inhibits foot-and-mouth disease virus replication. Virol Sin. 2019;34(6):610–7. doi: 10.1007/s12250-019-00148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godbout R, et al. Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol. 2007;3(5):575–87. doi: 10.2217/14796694.3.5.575. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka K, et al. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene. 2009;28(21):2142–51. doi: 10.1038/onc.2009.89. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka K, et al. DEAD box protein DDX1 promotes colorectal tumorigenesis through transcriptional activation of the LGR5 gene. Cancer Sci. 2018;109(8):2479–89. doi: 10.1111/cas.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Germain DR, et al. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res Treat. 2011;127(1):53–63. doi: 10.1007/s10549-010-0943-7. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modhukur V, et al. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–88. doi: 10.2217/epi-2017-0118. [DOI] [PubMed] [Google Scholar]
- 24.Sturm G, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–45. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545-d551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605-d612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali MAM. The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int J Clin Oncol. 2021;26(5):795–825. doi: 10.1007/s10147-021-01892-1. [DOI] [PubMed] [Google Scholar]
- 31.Hu DX, et al. Knockdown of DEAD-box 51 inhibits tumor growth of esophageal squamous cell carcinoma via the PI3K/AKT pathway. World J Gastroenterol. 2022;28(4):464–78. doi: 10.3748/wjg.v28.i4.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang F, et al. Knockdown of DDX46 inhibits the invasion and tumorigenesis in osteosarcoma cells. Oncol Res. 2017;25(3):417–25. doi: 10.3727/096504016X14747253292210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taunk NK, et al. DEAD box 1 (DDX1) expression predicts for local control and overall survival in early stage, node-negative breast cancer. Cancer. 2012;118(4):888–98. doi: 10.1002/cncr.26352. [DOI] [PubMed] [Google Scholar]
- 34.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 35.Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: linking molecular mechanisms to disease prognosis. Brief Bioinform. 2015;16(3):461–74. doi: 10.1093/bib/bbu020. [DOI] [PubMed] [Google Scholar]
- 36.Yu F, et al. Breast cancer prognosis signature: linking risk stratification to disease subtypes. Brief Bioinform. 2019;20(6):2130–40. doi: 10.1093/bib/bby073. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(1):R50–9. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 38.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4(12):988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 39.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y, et al. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18(1):1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 41.Yu F, et al. CFEA: a cell-free epigenome atlas in human diseases. Nucleic Acids Res. 2020;48(D1):D40-d44. doi: 10.1093/nar/gkz715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342(2):223–30. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo CC, et al. Methylation of IRAK3 is a novel prognostic marker in hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3960–9. doi: 10.3748/wjg.v21.i13.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N, et al. Analysis of APC mutation in human ameloblastoma and clinical significance. Springerplus. 2016;5:314. doi: 10.1186/s40064-016-1904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Höpken UE, Rehm A. Targeting the tumor microenvironment of leukemia and lymphoma. Trends Cancer. 2019;5(6):351–64. doi: 10.1016/j.trecan.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Muppala S. Significance of the tumor microenvironment in liver cancer progression. Crit Rev Oncog. 2020;25(1):1–9. doi: 10.1615/CritRevOncog.2020034987. [DOI] [PubMed] [Google Scholar]
- 47.Gnjatic S, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44. doi: 10.1186/s40425-017-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilain RE, et al. Dynamic changes in PD-L1 expression and immune infiltrates early during treatment predict response to PD-1 blockade in melanoma. Clin Cancer Res. 2017;23(17):5024–33. doi: 10.1158/1078-0432.CCR-16-0698. [DOI] [PubMed] [Google Scholar]
- 49.Qian BZ, Pollard JW. Macrophage divers enhances tumor progression metastasis cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G, et al. M2 macrophages promote HCC cells Invasion and Migration via miR-149-5p/MMP9 signaling. J Cancer. 2020;11(5):1277–87. doi: 10.7150/jca.35444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung OW, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62(3):607–16. doi: 10.1016/j.jhep.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 52.Yao RR, et al. M2-polarized tumor-associated macrophages facilitated migration and epithelial-mesenchymal transition of HCC cells via the TLR4/STAT3 signaling pathway. World J Surg Oncol. 2018;16(1):9. doi: 10.1186/s12957-018-1312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang AH, et al. Infiltrating regulatory T cells promote invasiveness of liver cancer cells via inducing epithelial-mesenchymal transition. Transl Cancer Res. 2019;8(6):2405–15. doi: 10.21037/tcr.2019.09.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, et al. TGF-β regulates hepatocellular carcinoma progression by inducing treg cell polarization. Cell Physiol Biochem. 2015;35(4):1623–32. doi: 10.1159/000373976. [DOI] [PubMed] [Google Scholar]
- 55.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10(1):121–32. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng B, Tian J, Chen Y. Identification of RNA binding protein interacting with circular RNA and hub candidate network for hepatocellular carcinoma. Aging. 2021;13(12):16124–43. doi: 10.18632/aging.203139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chohan TA, et al. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr Med Chem. 2015;22(2):237–63. doi: 10.2174/0929867321666141106113633. [DOI] [PubMed] [Google Scholar]
- 58.Liang Y, et al. HNRNPU promotes the progression of hepatocellular carcinoma by enhancing CDK2 transcription. Exp Cell Res. 2021;409(1):112898. doi: 10.1016/j.yexcr.2021.112898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed in the present study was obtained from TCGA and ICGC.