Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) produce the characteristic “attaching and effacing” (A/E) lesion of the brush border. Intimin, an outer membrane protein encoded by eae, is responsible for the tight association of both pathogens with the host cell. Several eae have been cloned from different EPEC and EHEC strains isolated from humans and animals. These sequences are conserved in the N-terminal region but highly variable in the last C-terminal 280 amino acids (aa), where the cell binding activity is localized. Based on these considerations, we developed a panel of specific primers to investigate the eae heterogeneity of the variable 3′ region by using PCR amplification. We then investigated the distribution of the known intimin types in a large collection of EPEC and EHEC strains isolated from humans and different animal species. The existence of a yet-unknown family of intimin was suspected because several EHEC strains, isolated from human and cattle, did not react with any of the specific primer pairs, although these strains were eae positive when primers amplifying the conserved 5′ end were used. We then cloned and sequenced the eae present in one of these strains (EHEC of serotype O103:H2) and subsequently designed a PCR primer that recognizes in a specific manner the variable 3′ region of this new intimin type. This intimin, referred to as “ɛ,” was present in human and bovine EHEC strains of serogroups O8, O11, O45, O103, O121, and O165. Intimin ɛ is the largest intimin cloned to date (948 aa) and shares the greatest overall sequence identity with intimin β, although analysis of the last C-terminal 280 aa suggests a greater similarity with intimins α and γ.
All mammals and birds are colonized by Escherichia coli, generally at birth, and these organisms become a permanent part of the normal microflora of the gastrointestinal tract. However, certain E. coli strains have been associated with gastroenteritis, urogenital disease, septicemia, and pleural infections in both humans and animals. Among these strains, enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) constitute a significant risk to human and animal health worldwide.
EHEC strains constitute a subset of serotypes of Shiga toxin (Stx)-producing E. coli (STEC) that has been firmly associated with bloody diarrhea and hemolytic-uremic syndrome (HUS) in industrialized countries (31). Numerous outbreaks of disease have been attributed to EHEC O157:H7 (5, 31), but serotypes other than O157:H7 can be responsible for outbreaks and sporadic cases of human disease (30; A. Caprioli et al., Letter, Emerg. Infect. Dis. 3:578–579, 1997). EHEC strains have been shown to be pathogenic to neonatal calves (12, 49) and are frequently isolated from diarrheic calves (45, 66), though systemic complications, such as HUS, have never been observed. Nevertheless, cattle are above all an important reservoir of EHEC O157, and asymptomatic carriage by young calves and adult cows has been well documented (5, 31, 34). EHEC and other STEC strains have also been detected in the feces of other domestic animals such as sheep (7, 34), pigs (7, 58), and cats and dogs (7) and in the feces of wild animals such as deer (57) and pigeons (14).
In contrast to EHEC, EPEC strains do not produce Stx and are not associated with HUS. Nevertheless, they are a major cause of infant diarrhea in non-industrialized countries (50) and are pathogenic to several animal species. EPEC strains are a serious cause of morbidity and mortality in weaned rabbits (8, 55). They are also pathogenic to neonatal calves (21, 52) and seem to be isolated most frequently in cattle farms with recurrent problems of diarrhea (10). In swine, EPEC strains have been involved in cases of postweaning diarrhea (68), and there is also increasing evidence for a diarrheagenic role of EPEC in dogs (17, 63). Finally, EPEC strains have been isolated from wild and domestic birds (22, 26), although the role of these strains in avian diseases has yet to be defined.
Like all diarrheagenic E. coli strains, EHEC and EPEC must first colonize the intestinal mucosa. Both pathovars produce a characteristic histopathological feature, known as the “attaching-and-effacing” (A/E) lesion, by subverting the intestinal epithelial cell function (recently reviewed in reference 23). This lesion is characterized by the effacement of microvilli and by intimate adherence between the bacteria and the epithelial cell membrane. Marked cytoskeletal changes, including accumulation of polymerized actin, occur directly beneath the adherent bacteria. The formation of A/E lesions is governed by a pathogenicity island known as the locus of enterocyte effacement (LEE), which was first described in the EPEC O127 strain E2348/69 (44). The LEE is present in EPEC and EHEC strains and in other bacterial species, such as Hafnia alvei and Citrobacter rodentium (formerly C. freundii biotype 4280) (for a review, see reference 37). The LEE from the EPEC strain E2348/69 contains 41 genes organized into three major regions, with known function (19). A similar organization has been observed in the LEE from the EHEC O157:H7 strain EDL933, which presents 13 additional genes belonging to a putative P4 family prophage (54). The LEE central region contains the eae (for E. coli attachment effacement) gene encoding a 94- to 97-kDa outer membrane protein known as intimin (36). This protein mediates close contact between the bacteria and the target cell, upon interaction with its translocated receptor Tir (for translocated intimin receptor), encoded by a gene located upstream eae (13, 41). Tir had been initially identified as a 90-kDa tyrosine phosphorylated protein from the target cell membrane and was called Hp90 (59). The role of intimin in human disease has been demonstrated by studies in human volunteers with an isogenic eae null mutant of EPEC E2348/69 (15). In animal models, intimin has been shown to be necessary for EHEC O157:H7 to intensively colonize the intestines and cause diarrhea and A/E lesions in calves and colonic edema and A/E lesions in piglets (11, 16, 47).
E. coli eae genes have been cloned and sequenced from C. rodentium (60) and H. alvei (25) and from different EPEC or EHEC strains isolated from human (36, 67), calf (29), dog (6), pig, and rabbit (4) sources. The overall pattern for these sequences shows high conservation in the N-terminal region and variability in the last C-terminal 280 amino acids of the intimin, where binding to the enterocytes (25) and Tir (33) occurs. Various studies have investigated the heterogeneity of eae among E. coli strains by amplification of the variable 3′ region by PCR and restriction fragment length polymorphism (RFLP) analysis of PCR products. Schmidt et al. (61) designed two primer pairs capable of differentiating the eae genes of EPEC and EHEC strains of serogroup O157. However, Giammanco et al. (28) observed that these primers were not able to amplify the intimin determinants in more than half of a series of eae-positive strains belonging to various serotypes, and Agin and Wolf (3) provided evidence for the existence of at least three immunologically distinct intimin types called α, β, and γ. A multiplex PCR was designed to detect eae and simultaneously identify the specific alleles encoding these three intimin variants (56). In another study, Adu-Bobie et al. (1) used antisera to the C-terminal region and PCR to investigate antigenic variation and classify the cell-binding domain of intimin expressed by A/E lesion-forming bacterial pathogens. By these means, these authors identified four distinct intimin types: intimin α, intimin β, intimin γ, and intimin δ. Nevertheless, some EHEC and EPEC strains still express nontypeable intimins (10, 53, 56).
In the present study, we describe the nucleotide sequence of a fifth intimin type, referred to as “ɛ”, which is present in human and bovine EHEC strains of serotype O103:H2 and designed a PCR primer that recognizes this sequence in a specific manner. A panel of specific primer pairs was used to investigate the distribution of the different intimin types among a collection of eae-positive E. coli strains isolated from humans and different animal species.
MATERIALS AND METHODS
Bacterial strains.
The eae-positive E. coli isolates used in this study were part of the culture collections of the Istituto Superiore di Sanità in Rome, of the Institut für Hygiene und Mikrobiologie of the University of Wurzburg, and of the Ecole Veterinaire in Toulouse. Many of them have been described in previous studies (28, 61). Some human strains were provided by M. A. Karmali (Toronto, Ontario, Canada) and Diana Karpman (Lund, Sweden). EPEC O86:H34 strain ICC95 was kindly provided by G. Frankel, London, United Kingdom. Porcine and canine EPEC strains were kindly provided by Josée Harel, Saint Hyacinthe, Quebec, Canada. We also used the EPEC O127:H6 strain E2348/69, the EHEC O157:H7 strain EDL933, and the EHEC O26:H11 strain H19 as prototypes of the α, γ, and β intimin types. Some of the isolates produced Stx, as assessed by the Vero cell cytotoxicity assay and PCR amplification of stx genes (48).
Sequence analysis.
DNA sequences of the different eae genes were retrieved from GenBank and included human EPEC O127:H6 strain E2348/69 (M58154), human EHEC O157:H7 strain EDL933 (Z11541), human EHEC O103:H2 strain PMK5 (AF116899 and this study), human EHEC O26:H11 strain H19 (U62656), human EHEC O111:H− strain 95NR1 (AF025311), human EPEC O111a,b:H− strain E2430/78 (U62655), human EPEC O86:H34 strain ICC95 (Y13112), rabbit EPEC O15:H− strain RDEC-1 (U60002), rabbit EPEC O103:H2 strain 84/110/1 (U59502), dog EPEC strain 4221 (U66102), human EPEC O55:H7 strain DEC 5d Orskov C586-65 (AF081184), human EPEC O128:H2 strain DEC 11a CDC 2254-75 (AF081186), calf EHEC O26H− strain 413/89-1 (AJ223063), human Hafnia alvei (L29509), and mouse Citrobacter rodentium (L11691). Comparisons were made by using the database at the National Center for Biotechnology Information (National institute of Health, Bethesda, Md.) with the BLAST search algorithm and GCG alignment software and with CLUSTAL W Multiple Sequence Alignment software. The phylogenetic tree was constructed by the neighbor-joining method with a bootstrap 10,000 time.
PCR analysis of eae gene.
The presence of the intimin determinant was assessed in all strains by PCR amplification of the 5′ conserved region by using the eae universal primers SK1 and SK2 (38). The different types of eae genes were identified by using SK1 as the universal forward primer and the reverse primers LP2 and LP3 as described by Schmidt et al. (61), which were able to amplify the determinants related to the sequences of EPEC O127 strain E2348/69 and EHEC O157 strain EDL933, respectively. A new primer, termed LP4, was designed in the hypervariable region of the eae gene of RDEC-1 strain, spanning from nucleotide 2283 to 2309 (Table 1). Another new primer, LP5, was designed in the hypervariable region of the eae gene of EHEC O103 strain PMK5, spanning from nucleotide 2605 to 2630 (Table 1). LP4 and LP5 were used as reverse primers in combination with SK1. PCR conditions were as described by Schmidt et al. (61), and the amplification products were analyzed by electrophoresis on 1% agarose gel.
TABLE 1.
PCR primers used for Intimin gene typing
| Primer | Positiona | Orientation | Sequence | PCR product size with SK1 (bp) |
|---|---|---|---|---|
| SK1 | 26–46 | Forward | 5′-CCCGAATTCGGCACAAGCATAAGC | NAb |
| SK2 | 879–903 | Reverse | 5′-CCCGGATCCGTCTCGCCAGTATTCG | 881 |
| LP2 | 2803–2829 | Reverse | 5′-CCCGAATTCTTATTTTACACAAGTGGC | 2,807 |
| LP3 | 2788–2814 | Reverse | 5′-CCCGAATTCTTATTCTACACAAACCGC | 2,792 |
| LP4 | 2283–2309 | Reverse | 5′-CCCGTGATACCAGTACCAATTACGGTC | 2,287 |
| LP5 | 2605–2630 | Reverse | 5′-AGCTCACTCGTAGATGACGGCAAGCG | 2,608 |
The position of LP2 primer is referred to as the eae sequence of human EPEC O127:H6 strain E2348/69 (M58154); the position of LP3 primer is referred to as the eae sequence of human EHEC O157:H7 strain EDL933 (Z11541); the position of LP4 primer is referred to as the eae sequence of rabbit EPEC O15:H− strain RDEC-1 (U60002); the position of LP5 primer is referred to as the eae sequence of O103:H2 strain PMK5 (AF116899).
NA, not applicable.
Cloning and sequencing of the eae gene from EHEC O103:H2.
The intimin gene of human EHEC O103 strain PMK5, isolated from the stools of a child suffering from HUS (46), was cloned into pCR2-1 vector by PCR. DNA amplification was carried out in a Perkin-Elmer apparatus by using high-fidelity Pfu DNA polymerase (Stratagene) with the primers orfU-upper (5′-TATGATGATCTATGGCGTCTGT-3′) and escD-lower (5′-TATTTTCAAAAAGAATGATGTC-3′). The 3.7-kb PCR amplification product from strain PMK5 was cloned into pCR2.1 vector (Invitrogen). The nucleotide sequence of the amplification product was determined by the dideoxynucleotide triphosphate chain termination method of Sanger, with the Dye Deoxy Terminator Cycle Sequence Kit with an ABI 373A DNA automatic sequencer (Applied Bio-Systems). After the initial sequences were determined by using universal and reverse M13 primers, internal primers were designed to sequence the whole DNA fragment.
F-actin staining (FAS) after interaction between HeLa cells and bacteria.
HeLa cells were seeded at 2 · 104 cells per well on Lab-Tek 8 chamber slides (Nunc) and cultivated for 24 h in Eagle minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS; Gibco) and gentamicin (80 mg · ml−1) at 37°C in a 5% CO2–95% air atmosphere. The interaction was made in 300 ml of MEM buffered with 25 mM HEPES complemented with 5% FCS and 1% mannose, with a starting inoculum of 15 ml of bacterial culture (ca. 103 bacteria per cell). After 4 h of interaction at 37°C, the cells were washed four times with Earle balanced saline solution, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; pH = 7.4) for 30 min at 4°C, and then permeabilized with 0.1% Triton X-100 in PBS for 5 min. F-actin was labelled with rhodamine-phalloidin (Molecular Probes) in accordance with the manufacturer's instructions.
RESULTS
Construction of a specific intimin β primer.
We designed a reverse primer, specific for intimin β, in the hypervariable region of the eae gene from the rabbit EPEC strain RDEC-1 (O15:H−) and the human EHEC strain H19 (O26:H11). This primer, termed LP4, was designed to have no sequences in common at the 3′ end with the corresponding region of the eae genes from strains EDL933 (O157:H7) and E2348/69 (O127:H6), which produce, respectively, intimins γ and α. When used together with SK1, LP4 generated a PCR product of the expected size (2,287 bp) with DNA from EPEC strain RDEC-1 and EHEC strain H19 and no product with templates obtained from EPEC strain E2348/69 or EHEC strain EDL933.
Distribution of the eae gene types among E. coli strains.
The primer pairs SK1 and LP2, SK1 and LP3, and SK1 and LP4 specifically recognized the three types of intimin represented, respectively, by the control strains E2348/69 (intimin α), H19 and RDEC-1 (intimin β), and EDL933 (intimin γ), and these PCRs always generated a product of the expected size (Table 1). The three primer pairs were then used to investigate by PCR the distribution of intimins α, β, and γ among a collection of 189 eae-positive E. coli strains isolated from different sources (i.e., humans, cattle, pigs, rabbits, dogs, and pigeons). The isolates belonged to 22 serogroups and 41 serotypes, and 138 of them produced Stx. Table 2 shows the PCR results obtained with the E. coli strains according to serotype, source of isolation, and capacity to produce Stx.
TABLE 2.
Distribution of intimin types among EPEC and EHEC strains of different serotypes and from different sources
| Intimin type and serotypea | nb | Stx | Source |
|---|---|---|---|
| α1/α2 | |||
| O55:H6 | 1 | − | Human |
| O125:H6 | 3 | − | Human |
| O127:H6 | 2 | − | Human |
| O127:H− | 1 | − | Human |
| O157:H45c | 7 | − | Human |
| O157:H−c | 4 | − | Human |
| NTd | 1 | − | Dog |
| β1/β2 | |||
| O15:H− | 1 | − | Rabbit |
| O18:H− | 2 | + | Pigeon |
| O26:H8 | 1 | − | Human |
| O26:H11 | 12 | + | Human |
| O26:H11 | 4 | + | Cattle |
| O26:H11 | 3 | − | Human |
| O26:H11 | 1 | − | Rabbit |
| O26:H− | 13 | + | Human |
| O26:H− | 4 | − | Human |
| O45:H− | 1 | − | Human |
| O45:Hnde | 2 | + | Pigeon |
| O45:Hnd | 1 | − | Pig |
| O49:Hnd | 1 | − | Dog |
| O86:H34 | 1 | − | Human |
| O86:H− | 1 | − | Human |
| O103:H2 | 2 | − | Rabbit |
| O111:H− | 2 | − | Human |
| O114:H2 | 1 | − | Human |
| O118:H16 | 1 | + | Human |
| O118:H30 | 1 | + | Human |
| O118:H− | 2 | + | Human |
| O123:H11 | 1 | + | Cattle |
| O128:H2 | 4 | − | Human |
| γ1/γ2 | |||
| O55:H7 | 3 | − | Human |
| O86:H40 | 1 | + | Human |
| O111:H− | 16 | + | Human |
| O111:H8 | 1 | + | Human |
| O111:H− | 2 | + | Buffalo |
| O127:H40 | 3 | − | Human |
| O128:H8 | 1 | − | Human |
| O128:H− | 1 | − | Human |
| O145:H− | 15 | + | Human |
| O157:H7 | 19 | + | Human |
| O157:H7 | 15 | + | Cattle |
| O157:H− | 1 | + | Human |
| O157:H−c | 5 | + | Human |
| ɛ | |||
| O8:H2 | 2 | + | Human |
| O11:H2 | 1 | + | Human |
| O45:H2 | 1 | + | Human |
| O103:H2 | 13 | + | Human |
| O103:H18 | 2 | + | Human |
| O103:H− | 2 | + | Human |
| O103:Hnd | 2 | + | Cattle |
| O121:H19 | 1 | + | Human |
| O165:H− | 1 | + | Human |
Serotypes in boldface were those in either α2, β2/δ, or γ2.
Values are numbers of strains examined.
Sorbitol-fermenting EHEC strains belonging to serogroup O157 and previously described by Gunzer and colleagues (32).
NT, nontypeable.
nd, not done.
Intimin α was found in human EPEC strains, which, in addition to the prototype serotype O127:H6, belonged to serotypes O55:H6, O125:H6, O127:H−, O157:H45, and O157:H−; unlike the typical EHEC strains, the O157 EPEC isolates were able to ferment sorbitol. Intimin α was also found in a dog EPEC strain. Intimin γ was mainly found among human and cattle STEC strains, including both the sorbitol-negative and sorbitol-positive (33) EHEC O157 strains, as well as the EHEC isolates of serotypes O86:H40, O111:H−, O111:H8, and O145:H−. Intimin γ was also found in human EPEC O55:H7, O128:H8, O128:H− and O127:H40. Intimin β was found in EPEC strains belonging to classical human (O26, O86, O111, O114, and O128) and rabbit (O15 and O103) serogroups, as well as in EHEC strains of serogroups O26, O118, and O123, in a porcine EPEC strain of serogroup O45, and in a dog EPEC strain of serogroup O49.
As a whole, the panel of PCR primers was able to amplify the eae genes of 164 of the 189 strains tested (87%), and none of the strains reacted with more than one primer pair. The 25 cattle and human EHEC strains that did not react with any of the specific primer pairs belonged to serogroups O8, O11, O45, O103, O121, and O165.
Cloning and sequencing the eae gene of human EHEC O103:H2.
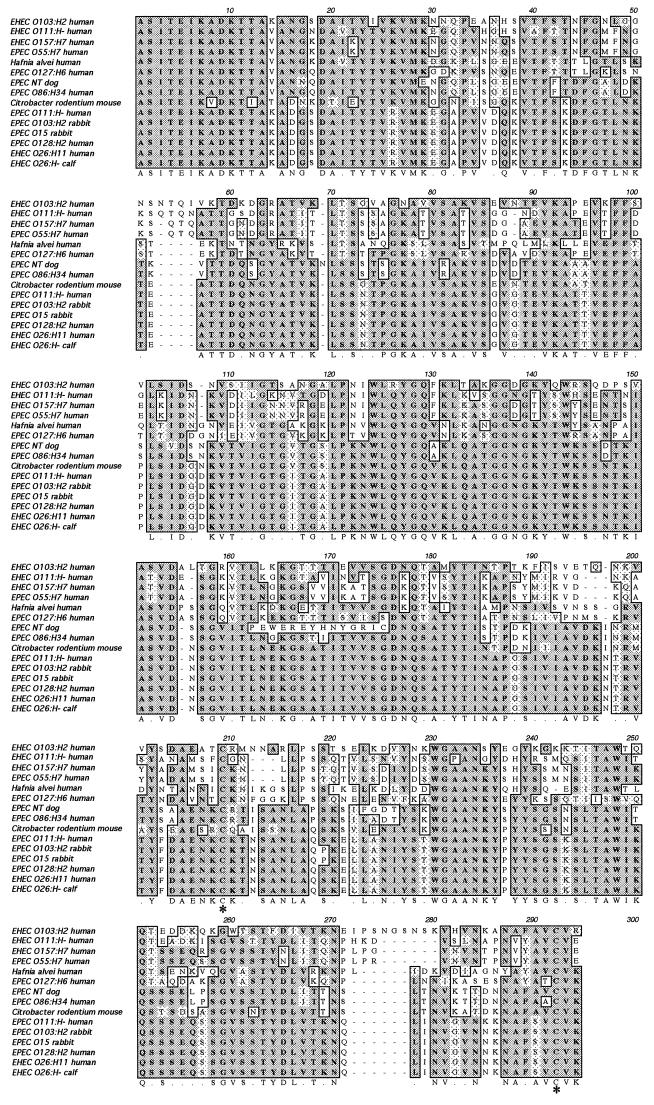
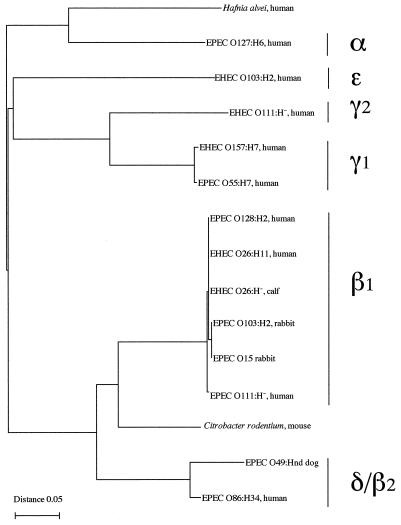
Since several EHEC strains isolated from humans and calves were positive with the universal eae primer pair SK1-SK2 but did not react with any of the type-specific primers, the existence of a yet-unknown type of intimin was suspected. We therefore decided to clone and sequence the eae present in one of these strains. We chose the EHEC strain PMK5 (O103:H2), which had been previously isolated from a patient with HUS in France (46). We first demonstrated that PMK5 produced a functional intimin, since it was able to induce a classical FAS response on human epithelial cells (data not shown). We then cloned the complete eae gene by using primers designed in LEE regions located upstream and downstream of eae. Previous characterizations of the LEE have shown that eae is located between two genes: orfU, which codes for a putative chaperone (19), and escD (also known as Pas) which codes for a member of a type III system required for protein secretion (19). DNA analysis of the orfU and escD genes in LEE from different strains prompted us to design two consensus primers (orfU-upper and escD-lower) that allowed us to clone by PCR a 3.7-kb DNA fragment containing eae (GenBank accession number AF113597). Sequence analysis revealed that the eae gene from EHEC O103:H2 was similar to but larger than those previously described in other EPEC or EHEC strains (948 codons versus 934 to 940 codons). As expected, comparison of the DNA sequences showed a major divergence in the 3′ half of the gene or C terminus, where the activity of binding to the enterocytes is localized. Although larger and quite different, the C terminus of intimin, termed ɛ, contained conserved features, such as the two cysteine residues (Fig. 1) forming a disulfide bond and required for binding activity of the intimin (24). Intimin ɛ shared the greatest overall sequence identity (71%) with intimin β, reaching 98% when the comparison was made with just the first 657 amino acids from the N terminus. However, an analysis of the last C-terminal amino acids (starting with alanine 658) performed with CLUSTAL W suggested a greater similarity with intimins α and γ (Fig. 1). A phylogenetic tree was drawn from this multiple alignment (Fig. 2).
FIG. 1.
CLUSTAL W alignment of intimin ɛ sequence (AF116899) from the EHEC O103:H2 strain PMK5 with α, β, γ and δ intimins cloned from different pathogenic E. coli (EPEC or EHEC), an intimin homologue from H. alvei, and an intimin homologue from C. rodentium. The accession numbers in GenBank for these sequences and the names of the bacterial strains are given in Materials and Methods. Of note, a recent study (35) suggests that the H. alvei strains containing the intimin gene are, in fact, unusual biotypes of E. coli or represent a new species in the genus Escherichia. The multiple alignment was based on the last C-terminal amino acids of the different intimin (starting with alanine 658). The two cysteine residues necessary for the formation of a disulfide bond and the binding activity are indicated by asterisks.
FIG. 2.
Phylogenetic tree based on the last C-terminal amino acids of the different intimin (starting with alanine 658) of different α, β, γ, δ, and ɛ intimins of pathogenic E. coli, an intimin homologue from H. alvei, and an intimin homologue from C. rodentium. The accession numbers in GenBank for these sequences and the names of the bacterial strains are given in Materials and Methods. Of note, a recent study (35) suggests that the H. alvei strains containing the intimin gene are, in fact, unusual biotypes of E. coli or represent a new species in the genus Escherichia.
Construction of a specific intimin ɛ primer.
On the basis of the nucleotide sequence of intimin ɛ, a specific primer, termed LP5, was designed in the hypervariable region of eae. When used together with SK1, LP5 generated a PCR product of the expected size (2,608 bp) with DNA from the EHEC strain PMK5 (O103:H2) as template, but it did not react with DNA from the prototype strains producing intimins α, β, or γ. PCR analysis with LP5 showed that intimin ɛ was present in all of the 25 human or bovine STEC strains that were negative with the other three primers (Table 2).
Subtyping of the intimin determinants.
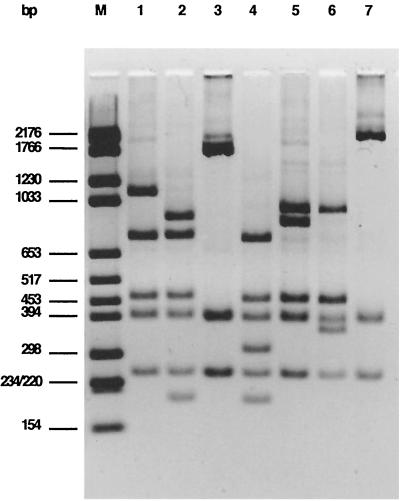
To establish further differentiation within the different types of intimin genes, restriction analysis of the PCR amplification products was performed (Fig. 3) according to the nucleotide sequence analysis of the prototype eae genes. When the α intimin genes of the different serotypes were digested with PstI, the same RFLP pattern (α1) was found in all of the isolates except for the three O125 strains, which shared a second profile termed α2.
FIG. 3.
PstI RFLP analysis of the eae PCR products obtained with the different type-specific primers from E. coli strains representative of the different intimin types. M, molecular size marker; lane 1, type α1 (EPEC O127:H6); lane 2, type α2 (EPEC O125:H6); lane 3, type β1 (EHEC O26:H11); lane 4, type β2 (EPEC O86:H34); lane 5, type γ1 (EHEC O157:H7); lane 6, type γ2 (EHEC O111:H−); lane 7, type ɛ (EHEC O103:H2).
Two other PstI RFLP profiles were observed within the strains possessing the γ intimin type: the first one (γ1) was shared by all the EHEC strains belonging to serogroups O157 and O145, by the Stx-producing strain O86:H40, and by EPEC O55:H7; the second profile (γ2) was represented by all the EHEC O111 strains and by EPEC O127:H40 and O128:H8. Analysis of the β intimin genes with PstI, FokI, and HaeIII showed that all of the isolates harbored the same gene subtype (β1), with the exception of the two human EPEC O86 and the canine EPEC O49 strains, which shared a very different profile, termed β2. The eae gene of one of these isolates (strain ICC95) had been previously classified by Adu-Bobie et al. (1) as the only representative of type δ.
No differences were observed within the intimin ɛ genes, which showed the same digestion profile with PstI, FokI, and HaeIII regardless the serotype of the strains examined. For the intimin γ-positive isolates, the results obtained by RFLP analysis were confirmed by PCR amplification by using the serotype-specific intimin primer pairs previously described by Gannon et al. (27) for EHEC O157 (primers 19 and 20) and by Louie et al. (42) for EHEC O111 (primers P40 and P20). The two primer pairs correctly identified all of the strains carrying the γ1 and γ2 gene subtypes, respectively.
DISCUSSION
Intimin mediates the intimate attachment of the bacteria to the host cell surface and is required for the formation of the characteristic A/E lesion associated with EPEC and EHEC infections. Several studies have shown that a considerable heterogeneity exists within the DNA sequences of the eae genes of different E. coli strains and that the corresponding changes in the amino acid sequence also represent antigenic variations. Using immunological and genetic approaches, Agin and Wolf (3) and Adu-Bobie et al. (1) revealed the existence of four distinct intimin types: intimin α, intimin β, intimin γ, and intimin δ.
We have developed a PCR-RFLP method capable of identifying the four intimins and of further differentiating within the gene types. This method was used to extend the first published observations (1, 3, 27, 42) about the distribution of the different eae genes among human and animal EPEC and EHEC serotypes. In addition, our study revealed the existence of a novel intimin type, termed “intimin ɛ,” which was found in human and bovine STEC strains, including the EHEC serogroups O8, O11, O45, O103, O121, and O165.
By studying a large collection of strains, we observed that intimin α seems to be specifically expressed by human EPEC strains belonging to classical serotypes (O55:H6, O125:H6, O127:H6, O142:H6, and O142:H34), although a nontypeable dog EPEC strain was also shown to express intimin α. Conversely, the novel intimin type ɛ was found only in Stx-producing strains, including two serogroups associated with severe human disease, i.e., O103 (43, 46, 62; A. Caprioli et al., Letter, Emerg. Infect. Dis., 3:578–579, 1997) and O121 (39, 40). Intimin γ is also associated with several EHEC serogroups highly pathogenic to humans: O157, O111, and O145. However, it has also been found in EPEC O55:H7, the likely ancestor of O157:H7 (20), and in nonclassical EPEC serotypes such as O127:H40 and O128:H8. Intimin β appears to be the most ubiquitous type, in that it has been found in both EPEC and EHEC isolates from several animal species: humans, cattle, pigs, rabbits, dogs, and birds. β-Positive strains include important diarrheagenic clones such as human EPEC O26:H11, O111:H2, and O128:H2; rabbit EPEC O15 and O103:H2; and human and bovine EHEC O26:H11. In our study, we also typed the intimin of the O86:H34 EPEC strain ICC95 as β, though Adu-Bobie et al. (1) had previously classified this intimin as type δ. However, this discrepancy was only apparent and may have been due to the fact that the respective primers were designed in different regions of eae. In fact, restriction analysis of the PCR products obtained with our β-specific primer from strain ICC95, from another O86:H− EPEC strain, and from a dog O49 EPEC strain showed a digestion profile, termed β2, different from the β1 pattern shared by all the other intimin β-positive strains. This confirms that there are differences between the eae DNA sequences of O86 EPEC and the other β-type strains and indicates that our β2 subtype corresponds to the δ type of Adu-Bobie et al. (1).
The observation that different intimin types and subtypes are closely associated with different pathogenic E. coli clones could contribute to our understanding of the evolution of intimin genes. It has been suggested that the diversity within the polypeptide cell-binding domain is driven by natural selection, since intimin is highly immunogenic (1). Voss and colleagues (65) and Agin and Wolf (3) have demonstrated differential reactivity of human and rabbit antisera with intimin from different EHEC and EPEC isolates. Recently, Adu-Bobie et al. (2) have confirmed these observations by identifying different immunodominant regions within the C terminus of intimin α and intimin β. Nevertheless, the immune response of the host may not be the sole parameter driving the selection of the different intimin types. The presence of several distinct eae genes could also account for the ability of the intimin-producing strains to colonize different tissue and/or different hosts. This hypothesis is based on the experiment performed by Tzipori and colleagues (64) in a piglet model with EPEC and EHEC strains previously isolated from humans. This experiment showed that an EPEC strain producing intimin α caused A/E lesions in both the small and the large intestine, whereas an EHEC strain producing γ intimin caused A/E lesions only in the large intestine. When the cloned EPEC eae was introduced into the EHEC eae mutant, the hybrid EHEC strain expressing the EPEC intimin caused A/E lesions in both the small and large intestine. The ability to change the site of intestinal colonization by substituting the intimin gene demonstrates that, at least in the piglet model, the intimin protein is essential for specific colonization of the large intestine. Although, EPEC and EHEC host specificity might also lie in the transcriptional regulation of expression of the virulence factors in response to environmental conditions or in the production of intestinal adherence factor distinct from intimin, it is tempting to speculate that intimin type may play a crucial role in the host specificity and/or tissue tropism. Since the α-eae genes seem to be specific for EPEC, whereas the γ-eae genes are mainly found in EHEC, it is conceivable that the expression of different intimin types and the related tissue tropism may have an important role in determining some of the differences in the pathogenesis of EPEC and EHEC infections.
To date, intimin is the only E. coli O157:H7 adherence factor that has been demonstrated to play a role in intestinal colonization in vivo in an animal model. In such models, intimin was shown to be required for EHEC O157:H7 to intensively colonize the intestines and cause diarrhea and A/E lesions in calves and to cause colonic edema and A/E lesions in piglets (11, 16, 47). These results suggest that antiintimin vaccines might interfere with EHEC infections. If used in cattle, such vaccines could help in reducing the level of EHEC intestinal colonization, thus favoring the control of EHEC infections in humans. However, additional research is needed if an intimin γ-based vaccine against E. coli O157 is to be developed. First, domestic animals carry a large variety of non-O157 STEC serotypes, and many of them have been associated with human disease around the world. Reducing the load of E. coli O157 producing intimin γ, without tackling the problem of the non-O157 serogroups producing other intimin types or subtypes, would create an empty niche for these serogroups and could make the problem even worse. Second, the lack of correlation between levels of intimin antibodies in serum and disease severity do not support the hypothesis that an immune response to intimin provides protection against subsequent disease (15). Third, the existence of intestinal adherence factors distinct from intimin is suggested by the isolation of non-O157 STEC strains that lack the eae gene but are still associated with bloody diarrhea or HUS in humans (18, 48). In conclusion, EHEC and EPEC strains possess at least seven different types or subtypes of intimin-coding eae genes: α1, α2, β1, β2 (or δ), γ1, γ2, and ɛ. The novel ɛ subtype is produced by isolates of E. coli O103:H2, which have been associated with HUS in Europe (9, 43, 46), the United States (62), and Canada (51) and can be considered as an emerging EHEC clone. The distribution of intimin types among EHEC and EPEC strains isolated from different hosts (humans, cattle, pigs, rabbits, dogs, and birds) suggests that the host and/or the tissue tropism of the different A/E bacteria may be influenced by the type of intimin they express. Better characterization of the variable 3′ end of eae in a large collection of EHEC and EPEC strains may provide PCR tools for predicting the ability of E. coli strains to cause severe disease in humans.
ACKNOWLEDGMENTS
We thank Fabio Minelli and Barbara Plaschke for their skillful technical assistance. We thank Alain Milon and Jean De Rycke for critical reading of the manuscript.
This study was funded in part by grant BMH4-CT96-0970 from the Commission of the European Communities, in part by a grant from the Région Midi-Pyrénées (grant number 9609691), and in part by a grant from the Institut National de la Recherche Agronomique. This work was also supported by the Concerted Action CT98-3935 from the Commission of the European Communities.
REFERENCES
- 1.Adu-Bobie J, Frankel G, Bain C, Goncalves A G, Trabulsi L R, Douce G, Knutton S, Dougan G. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adu-Bobie J, Trabulsi L R, Carneiro-Sampaio M M, Dougan G, Frankel G. Identification of immunodominant regions within the C-terminal cell binding domain of intimin alpha and intimin beta from enteropathogenic Escherichia coli. Infect Immun. 1998;66:5643–5649. doi: 10.1128/iai.66.12.5643-5649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agin T S, Wolf M K. Identification of a family of intimins common to Escherichia coli causing attaching-effacing lesions in rabbits, humans, and swine. Infect Immun. 1997;65:320–326. doi: 10.1128/iai.65.1.320-326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agin T S, Cantey J R, Boedeker E C, Wolf M K. Characterization of the eaeA gene from rabbit enteropathogenic Escherichia coli strain RDEC-1 and comparison to other eaeA genes from bacteria that cause attaching-effacing lesions. FEMS Microbiol Lett. 1996;144:249–258. doi: 10.1111/j.1574-6968.1996.tb08538.x. [DOI] [PubMed] [Google Scholar]
- 5.Altekruse S F, Cohen M L, Swerdlow D L. Emerging foodborne diseases. Emerg Infect Dis. 1997;3:285–293. doi: 10.3201/eid0303.970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An H, Fairbrother J M, Dubreuil J D, Harel J. Cloning and characterization of the eae gene from a dog attaching and effacing Escherichia coli strain 4221. FEMS Microbiol Lett. 1997;148:239–245. doi: 10.1111/j.1574-6968.1997.tb10295.x. [DOI] [PubMed] [Google Scholar]
- 7.Beutin L, Geier D, Steinrück H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco J E, Blanco M, Blanco J, Mora A, Balaguer L, Mourino M, Juarez A, Jansen W H. O serogroups, biotypes, and eae genes in Escherichia coli strains isolated from diarrheic and healthy rabbits. J Clin Microbiol. 1996;34:3101–3107. doi: 10.1128/jcm.34.12.3101-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.China B, Jacquemin E, Devrin A-C, Pirson V, Mainil J. Heterogeneity of the eae genes in attaching and effacing Escherichia coli from cattle: comparison with human strains. Res Microbiol. 1999;150:323–332. doi: 10.1016/s0923-2508(99)80058-8. [DOI] [PubMed] [Google Scholar]
- 10.China B, Pirson V, Mainil J. Prevalence and molecular typing of attaching and effacing Escherichia coli among calf populations in Belgium. Vet Microbiol. 1998;63:249–259. doi: 10.1016/S0378-1135(98)00237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean-Nystrom E A, Bosworth B T, Moon H W, O'Brien A D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean-Nystrom E A, Bosworth B T, Cray W C, Jr, Moon H W. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect Immun. 1997;65:1842–1848. doi: 10.1128/iai.65.5.1842-1848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deibel C, Kramer S, Chakraborty T, Ebel F. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol Microbiol. 1998;28:463–474. doi: 10.1046/j.1365-2958.1998.00798.x. [DOI] [PubMed] [Google Scholar]
- 14.Dell'Omo G, Morabito S, Quondam R, Agrimi U, Ciuchini F, Macri A, Caprioli A. Feral pigeons as a source of verocytotoxin-producing Escherichia coli. Vet Rec. 1998;142:309–310. doi: 10.1136/vr.142.12.309. [DOI] [PubMed] [Google Scholar]
- 15.Donnenberg M S, Tacket C O, Losonsky G, Frankel G, Nataro J P, Dougan G, Levine M M. Effect of prior experimental human enteropathogenic Escherichia coli infection on illness following homologous and heterologous rechallenge. Infect Immun. 1998;66:52–58. doi: 10.1128/iai.66.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Tzipori S, McKee M L, O'Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drolet R, Fairbrother J M, Harel J, Helie P. Attaching and effacing and enterotoxigenic Escherichia coli associated with enteric colibacillosis in the dog. Can J Vet Res. 1994;58:87–92. [PMC free article] [PubMed] [Google Scholar]
- 18.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 20.Feng P, Lampel K A, Karch H, Whittam T S. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 21.Fischer J, Maddox C, Moxley R, Kinden D, Miller M. Pathogenicity of a bovine attaching effacing Escherichia coli isolate lacking Shiga-like toxins. Am J Vet Res. 1994;55:991–999. [PubMed] [Google Scholar]
- 22.Foster G, Ross H M, Pennycott T W, Hopkins G F, McLaren I M. Isolation of Escherichia coli O86:K61 producing cytolethal distending toxin from wild birds of the finch family. Lett Appl Microbiol. 1998;26:395–398. doi: 10.1046/j.1472-765x.1998.00359.x. [DOI] [PubMed] [Google Scholar]
- 23.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 24.Frankel G, Candy D C, Fabiani E, Adu-Bobie J, Gil S, Novakova M, Phillips A D, Dougan G. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect Immun. 1995;63:4323–4328. doi: 10.1128/iai.63.11.4323-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frankel G, Candy D C A, Everest P, Dougan G. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect Immun. 1994;62:1835–1842. doi: 10.1128/iai.62.5.1835-1842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukui H, Sueyoshi M, Haritani M, Nakazawa M, Naitoh S, Tani H, Uda Y. Natural infection with attaching and effacing Escherichia coli (O103:H−) in chicks. Avian Dis. 1995;39:912–918. [PubMed] [Google Scholar]
- 27.Gannon V P, Rashed M, King R K, Thomas E J. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 1993;31:1268–1274. doi: 10.1128/jcm.31.5.1268-1274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giammanco A, Maggio M, Giammanco G, Morelli R, Minelli F, Scheutz F, Caprioli A. Characteristics of Escherichia coli strains belonging to enteropathogenic E. coli serogroups isolated in Italy from children with diarrhea. J Clin Microbiol. 1996;34:689–694. doi: 10.1128/jcm.34.3.689-694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goffaux F, Mainil J, Pirson V, Charlier G, Pohl P, Jacquemin E, China B. Bovine attaching and effacing Escherichia coli possess a pathogenesis island related to the LEE of the human enteropathogenic Escherichia coli strain E2348/69. FEMS Microbiol Lett. 1997;154:415–421. doi: 10.1111/j.1574-6968.1997.tb12676.x. [DOI] [PubMed] [Google Scholar]
- 30.Goldwater P N, Bettelheim K A. New perspectives on the role of Escherichia coli O157:H7 and other enterohaemorrhagic E. coli serotypes in human disease. J Med Microbiol. 1998;47:1039–1045. doi: 10.1099/00222615-47-12-1039. [DOI] [PubMed] [Google Scholar]
- 31.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 32.Gunzer F, Bohm H, Russmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartland E L, Batchelor M, Delahay R M, Hale C, Matthews S, Dougan G, Knutton S, Connerton I, Frankel G. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol Microbiol. 1999;32:151–158. doi: 10.1046/j.1365-2958.1999.01338.x. [DOI] [PubMed] [Google Scholar]
- 34.Heuvelink A E, van den Biggelaar F L, de Boer E, Herbes R G, Melchers W J, Huis in't Veld J H, Monnens L A. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J Clin Microbiol. 1998;36:878–882. doi: 10.1128/jcm.36.4.878-882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janda J M, Abbott S L, Albert M J. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. J Clin Microbiol. 1999;37:2399–2401. doi: 10.1128/jcm.37.8.2399-2401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaper J B. EPEC delivers the goods. Trends Microbiol. 1998;6:169–172. doi: 10.1016/s0966-842x(98)01266-9. [DOI] [PubMed] [Google Scholar]
- 38.Karch H, Bohm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin producing, sorbitol fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1201–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 40.Karpman D, Hakansson A, Perez M T, Isaksson C, Carlemalm E, Caprioli A, Svanborg C. Apoptosis of renal cortical cells in the hemolytic-uremic syndrome: in vivo and in vitro studies. Infect Immun. 1998;66:636–644. doi: 10.1128/iai.66.2.636-644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 42.Louie M, de Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luzzi I, Tozzi A E, Rizzoni G, Niccolini A, Benedetti I, Minelli F, Caprioli A. Detection of serum antibodies to the lipopolysaccharide of Escherichia coli O103 in patients with hemolytic-uremic syndrome. J Infect Dis. 1995;171:514–515. doi: 10.1093/infdis/171.2.514. [DOI] [PubMed] [Google Scholar]
- 44.MacDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mainil J G, Duchesnes C J, Whipp S C, Marques L R, O'Brien A D, Casey T A, Moon H W. Shiga-like toxin production and attaching effacing activity of Escherichia coli associated with calf diarrhea. Am J Vet Res. 1987;48:743–748. [PubMed] [Google Scholar]
- 46.Mariani-Kurkdjian P, Denamur E, Milon A, Picard B, Cave H, Lambert-Zechovsky N, Loirat C, Goullet P, Sansonetti P J, Elion J. Identification of a clone of Escherichia coli O103:H2 as a potential agent of hemolytic-uremic syndrome in France. J Clin Microbiol. 1993;31:296–301. doi: 10.1128/jcm.31.2.296-301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKee M L, Melton-Celsa A R, Moxley R A, Francis D H, O'Brien A D. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morabito S, Karch H, Mariani-Kurkdjian P, Schmidt H, Minelli F, Bingen E, Caprioli A. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J Clin Microbiol. 1998;36:840–842. doi: 10.1128/jcm.36.3.840-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moxley R A, Francis D H. Natural and experimental infection with an attaching and effacing strain of Escherichia coli in calves. Infect Immun 1986. 1986;53:339–346. doi: 10.1128/iai.53.2.339-346.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pai C H, Ahmed N, Lior H, Johnson W M, Sims H V, Woods D E. Epidemiology of sporadic diarrhea due to verocytotoxin-producing Escherichia coli: a two-year prospective study. J Infect Dis. 1988;157:1054–1057. doi: 10.1093/infdis/157.5.1054. [DOI] [PubMed] [Google Scholar]
- 52.Pearson G R, Watson C A, Hall G A, Wray C. Natural infection with an attaching and effacing Escherichia coli in the small and large intestines of a calf with diarrhoea. Vet Rec. 1989;124:297–299. doi: 10.1136/vr.124.12.297. [DOI] [PubMed] [Google Scholar]
- 53.Pelayo J S, Scaletsky I C, Pedroso M Z, Sperandio V, Giron J A, Frankel G, Trabulsi L R. Virulence properties of atypical EPEC strains. J Med Microbiol. 1999;48:41–49. doi: 10.1099/00222615-48-1-41. [DOI] [PubMed] [Google Scholar]
- 54.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pohl P H, Peeters J E, Jacquemin E R, Lintermans P F, Mainil J G. Identification of eae sequences in enteropathogenic Escherichia coli strains from rabbits. Infect Immun. 1993;61:2203–2206. doi: 10.1128/iai.61.5.2203-2206.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reid S D, Betting D J, Whittam T S. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J Clin Microbiol. 1999;37:2719–2722. doi: 10.1128/jcm.37.8.2719-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rice D H, Hancock D D, Besser T E. Verotoxigenic Escherichia coli O157 colonisation of wild deer and range cattle. Vet Rec. 1995;137:524. doi: 10.1136/vr.137.20.524. [DOI] [PubMed] [Google Scholar]
- 58.Rios M, Prado V, Trucksis M, Arellano C, Borie C, Alexandre M, Fica A, Levine M M. Clonal diversity of chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J Clin Microbiol. 1999;37:778–781. doi: 10.1128/jcm.37.3.778-781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenshine I, Donnenberg M S, Kaper J B, Finlay B B. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schauer D B, Falkow S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt H, Russmann H, Karch H. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect Immun. 1993;61:4894–4898. doi: 10.1128/iai.61.11.4894-4898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarr P I, Fouser L S, Stapleton A E, Wilson R A, Kim H H, Vary J C, Jr, Clausen C R. Hemolytic-uremic syndrome in a six-year-old girl after a urinary tract infection with Shiga-toxin-producing Escherichia coli O103:H2. N Engl J Med. 1996;335:635–638. doi: 10.1056/NEJM199608293350905. [DOI] [PubMed] [Google Scholar]
- 63.Turk J, Maddox C, Fales W, Ostlund E, Miller M, Johnson G, Pace L, Turnquist S, Kreeger J. Examination for heat-labile, heat-stable, and Shiga-like toxins and for the eaeA gene in Escherichia coli isolates obtained from dogs dying with diarrhea: 122 cases (1992–1996) J Am Vet Med Assoc. 1998;212:1735–1736. [PubMed] [Google Scholar]
- 64.Tzipori S, Gunzer F, Donnenberg M S, de Montigny L, Kaper J B, Donohue-Rolfe A. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect Immun. 1995;63:3621–3627. doi: 10.1128/iai.63.9.3621-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voss E, Paton A W, Manning P A, Paton J C. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect Immun. 1998;66:1467–1472. doi: 10.1128/iai.66.4.1467-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wieler L H, Schwanitz A, Vieler E, Busse B, Steinruck H, Kaper J B, Baljer G. Virulence properties of Shiga toxin-producing Escherichia coli (STEC) strains of serogroup O118, a major group of STEC pathogens in calves. J Clin Microbiol. 1998;36:1604–1607. doi: 10.1128/jcm.36.6.1604-1607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 68.Zhu C, Harel J, Jacques M, Desautels C, Donnenberg M S, Beaudry M, Fairbrother J M. Virulence properties and attaching-effacing activity of Escherichia coli O45 from swine postweaning diarrhea. Infect Immun. 1994;62:4153–4159. doi: 10.1128/iai.62.10.4153-4159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]