Abstract
With the development of novel prognostic tools derived from omics technologies, transplant medicine is entering the era of precision medicine. Currently, there are no established predictive biomarkers for post-transplant kidney function. A total of 270 deceased donor pretransplant kidney biopsies were collected and posttransplant function was prospectively monitored. This study first assessed the utility of pretransplant gene expression profiles in predicting 24-month outcomes in a training set (n=174). Nearly 600 differentially expressed genes were associated with 24-month graft function. Grafts that progressed to low function at 24-months exhibited upregulated immune responses and downregulated metabolic processes at pre-transplantation. Using penalized logistic regression modeling, a 55 gene model AUROC for 24-month graft function was 0.994. Gene expression for a subset of candidate genes was then measured in an independent set of pretransplant biopsies (n=96) using qPCR. The AUROC when using 13 genes with 3 donor characteristics (age, race, BMI) was 0.821. Subsequently, a risk score was calculated using this combination for each patient in the validation cohort, demonstrating the translational feasibility of using gene markers as prognostic tools. These findings support the potential of pretransplant transcriptomic biomarkers as novel instruments for improving posttransplant outcome predictions and associated management.
Introduction
Kidney transplantation (KT) significantly improves overall quality-of-life and survival for patients with end-stage renal disease (ESRD), however, sustaining long-term allograft survival remains an ongoing challenge.1 The donor organ shortage has resulted in the increased use of marginal donor kidneys, further complicating the evaluation of organ quality without objective markers.2–4
Currently, the evaluation of donor organ quality largely depends on the Kidney Donor Profile Index (KDPI), a numerical score that combines 10 donor characteristics, and on histological evaluations of core biopsies collected prior to transplantation.5,6 The use of histology to predict short-term function was introduced nearly two decades ago when investigators reported that severe glomerulosclerosis increases the risk of delayed graft function (DGF) and poor 6-month outcomes.7 However, histological scores at transplant time showed no correlation to long-term allograft survival. Histological evaluation has been widely disputed due to concerns related to bias and inter-observer discrepancies, yet this practice continues to be a standard of care in most US medical centers.6 Thus far, clinical characteristics and histological findings have not allowed for the robust prediction of post-transplant function.2,5–8
Recent advancements in transcriptomic technology have improved the diagnosis and management of human diseases, presenting a unique opportunity for molecular evaluations to assist in KT outcome prediction. A transcriptomic profile serves as a snapshot of the temporary cell state and thus, its analysis can provide detailed and personalized information on the biological responses to injury.9
There is a critical need for molecular tools that can accurately predict functional outcomes for kidney transplant patients.8 Over the past decade, many teams have explored differentially expressed genes (DEGs) associated with post-transplant events and outcomes, with very few reporting predictive scores for direct clinical translation.
This prospective multicenter study developed and validated a multivariable model, combining baseline clinical characteristics and transcriptomic (biological) data, that predicts post-transplant function and can be easily transferred to clinical settings. The prediction of long-term outcomes has the potential to allow for early interventions to prevent or ameliorate progression to graft dysfunction, revealing a critical opportunity for transcriptomics to become a canon of contemporary transplant medicine.
2. Materials and Methods
2.1. Patients and Samples.
A total of 295 consecutive deceased donor (DD) KT recipients were enrolled from 4 transplant centers in the US, including 1) Virginia Commonwealth University (VCU) Medical Center, 2) University of Virginia (UVA) Medical Center, 3) Montefiore Medical Center, and 4) University of Tennessee Health Science Center (UTHSC). The study protocol was approved by the Institutional Review Board (IRB #HP-00092097). The clinical and research activities being reported are consistent with the Principles of the Declaration of Istanbul as outlined in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism. Written informed consent was obtained from KT recipients at transplantation time. Living donor recipients, re-transplant recipients, pregnant women, recipients <18 years old, HIV+ recipients, and recipients with previous history of malignancy were excluded from the study.
Tissue was obtained shortly before transplantation (back-bench biopsies) using an 18-gauge biopsy needle and immediately suspended in RNAlater (Ambion, Austin, USA). Patients received triple immunosuppression with calcineurin inhibitors, mycophenolate mofetil, and steroids; as for induction therapies, either anti-thymocyte globulin or basiliximab were administered.
Samples collected from UVA and VCU were included as part of the training set, while samples collected from Montefiore and UTHSC were included as part of the external validation set. Out of the 295 patients enrolled, a total of 25 were excluded due to follow-up loss, death with graft function, microarray quality control criteria, and biopsy RNA integrity. The patient flow diagram is shown in Figure S1.
2.2. Pre-processing Methods.
Total RNA was isolated from renal biopsies using TRIzol reagent (Invitrogen, Waltham, USA). RNA quality and integrity were evaluated using the Agilent RNA 6000 Nano Kit on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). Samples with an RNA integrity number of <8 were excluded from the analysis.
Gene expression of biopsies from the training set was measured using Affymetrix GeneChip microarrays (HG-U133A 2.0) (access: GSE147451) (Thermo Fisher Scientific, Waltham, USA). The Affymetrix Detection Call algorithm was used to determine whether probe sets were present, marginally present, or absent in each sample. Quality control was performed as previously published.10 To obtain probe set expression summaries, we used the robust multiarray average method.11 Prior to statistical analysis, the gene expression data matrix was filtered to exclude probe sets called absent in all samples and control probe sets, leaving 19,380 probe sets remaining for statistical analysis.
2.3. Study Design.
Estimated Glomerular Filtration Rate (eGFR) was calculated using the abbreviated Modification of Diet in Renal Disease (MDRD) formula.12 Study endpoints were defined as graft function at 24-months post-transplant (mean=24.3±1.2 months). Categorically, patients were considered to have low graft function with a 24-month eGFR <45 mL/min/1.73m2, while an eGFR of ≥45 mL/min/1.73m2 represented the high function group, corresponding to the chronic kidney disease KDIGO guidelines (www.kidney-international.org). Additionally, patients who experienced graft failure prior to 24-months were included in the low-functioning group. Linear mixed-effects models that included eGFR recorded at all time points (1-, 6-, 9-, 12-, 18-, 21-, and 24-months post-KT) were fit to demonstrate how continuous eGFR differed by this dichotomous categorization. To assess long-term outcomes, graft/patient survival was calculated as the time from 24-month post-transplant until the date of graft failure or date of death, censoring for those alive without graft failure at their last follow-up date. Only patients alive at 24-months were included in the survival analysis.
2.4. Statistical Methods.
The Kaplan-Meier method was used to estimate graft/patient survival and the log-rank test was used to compare the high vs. low eGFR groups. Descriptive statistics (mean and standard deviation (SD)) were applied to summarize continuous variables, while frequencies and percentages were used to summarize categorical variables.
To identify differentially expressed genes (DEGs) associated with outcome group, probe set level linear models were fit with high vs. low graft function group assignments as the predictor variable adjusting for the surrogate variable representing batch effect, using the limma Bioconductor package of the open-source R software for statistical computing and graphics (R Foundation for Statistical Computing, Vienna, Austria). All resulting p-values were adjusted for multiple hypothesis testing using Benjamini and Hochberg’s false discovery rate (FDR) method.13
Penalized logistic regression models were applied to simultaneously perform automatic variable selection and outcome prediction for high-dimensional covariate spaces. First, the gene expression data matrix was filtered to retain differentially expressed probe sets having an FDR <0.05. Thereafter, repeated 10-fold cross-validation (CV) was used to identify the optimal tuning parameters for fitting a penalized logistic regression model predicting outcome (high vs. low graft function). The repeated 10-fold CV procedure was performed using the caret package14 with glmnet15 in the R programming environment. Gene expression data was applied to derive a multivariable model. A grid search was performed to optimize the two tuning parameters required by elastic net, the penalty term λ, and the proportion of the penalty associated with the LASSO versus ridge regression, αLASSO. The combination of DEGs that optimized the area under the receiver operating characteristic curve (AUROC) from the repeated 10-fold CV procedure was selected for fitting the gene expression model. Significant demographic/clinical characteristics (p-value<0.05) were combined with DEGs to develop a gene expression + clinical data model. Two additional models were fit for performance comparison: one using all significant clinical characteristics and another that included the patient’s numerical KDPI value as the sole predictor.16
2.5. Pathway Analyses.
GO and KEGG pathway enrichment analyses were performed using enrichGO and enrichKEGG functions which adjust the estimated significance level to account for multiple hypothesis testing (FDR≤0.05). Finally, Metascape (https://metascape.org) was used for functional enrichment, interactome analysis, gene annotation, cell enrichment, and protein-protein interactions (PPIs).17 The Molecular Complex Detection (MCODE) algorithm was applied to the PPI network to identify densely connected networks.
2.6. QPCR Validation.
An initial set of genes was selected for further validation based on i) statistical significance, ii) high predictive performance in final models, and iii) association with relevant biological pathways. Individual predesigned TaqMan™ assays (ThermoFisher Scientific, Waltham, USA) were used for qPCR reactions. Gene expression results were expressed as ΔCt values normalized by a dual reference gene combination (ACTB and GAPDH).18 Univariable logistic regression models were fit for each gene to identify whether gene expression was significantly associated with 24-month outcome. Thereafter, multivariable logistic regression models were fit for each gene to determine significance after adjusting for important clinical covariates identified in the training set, and the AUROC and associated 95% confidence intervals (CI) were estimated.
2.7. Risk Score Equation.
The estimated regression coefficients (b) for each independent variable (X) in the multivariable regression model were used to form the linear risk score equation:
The optimal threshold which maximizes both specificity and sensitivity (Youden’s index) was used to predict whether the subjects would have low or high eGFR at 24 months. Lastly, the linear predictor (risk score) for each patient was converted into a probability scale (0.0–1.0) using the following equation:
3. Results
3.1. Clinical markers discriminating 24-month eGFR outcomes.
Among the 174 KT recipients in the training set, 67 (38.5%) subjects had low graft function and 107 (61.5%) had high function based on the criteria described in Materials and Methods. Clinical characteristics and demographics are shown in Table 1. On average, the high functioning group was composed of younger donor kidneys (37±16 years) compared to the low graft function group (48±14 years) (p<0.001). The groups also differed with respect to donor race (p=0.006), and BMI (p<0.001). No recipient variables were significantly different between groups. A spaghetti plot separated by high vs. low graft function with lowess smooths overlaid and the linear mixed-effects model demonstrated the difference between the eGFR trajectories over time (Figure S2). Regarding the individual eGFR courses, there was a significant difference (p<0.001) between the two groups across each timepoint throughout the 24-month period of observation. The high-functioning group showed a stable positive eGFR slope of 0.067 ml/min/month (0.81 ml/min/year), while the low-functioning group had a negative slope of −0.53 ml/min/month (−6.36 ml/min/year).
Table 1.
Characteristics of donor and recipients sub-stratified based on eGFR at 24-month post kidney transplant in the training set (n=174).
| Clinical Characteristic | Category | High eGFR (n=107) |
Low eGFR (n=67) |
p-value |
|---|---|---|---|---|
| Donor Characteristics | ||||
| Donor age, years (avg ± SD) | 37.12 ±15.97 | 48.49 ± 13.79 | <0.001 | |
| Donor gender n (%) | Male | 66 (61.7) | 36 (53.7) | 0.38 |
| Female | 41 (38.3) | 31 (46.3) | ||
| Donor race n (%) | American Indian | 1 (0.9) | 0 (0.0) | 0.006♦ |
| Asian | 2 (1.9) | 0 (0.0) | ||
| African American | 24 (22.4) | 30 (44.8) | ||
| Caucasian | 79 (73.8) | 37 (55.2) | ||
| Hispanic | 1 (0.9) | 0 (0.0) | ||
| DCD, n (%) | 16 (15.0) | 12 (17.9) | 0.761 | |
| Donor cause of death n (%) | Anoxia | 33 (30.8) | 20 (29.9) | 0.113♦ |
| Head trauma | 36 (33.6) | 13 (19.4) | ||
| Stroke | 34 (31.8) | 32 (47.8) | ||
| Other/Unknown | 4 (3.7) | 2 (3.0) | ||
| Delayed graft function n (%) | 34 (31.8) | 28 (41.8) | 0.238 | |
| Donor BMI (avg ± SD) | 26.57 ±5.83 | 31.10 ± 9.07 | <0.001 | |
| CIT, hours (avg ± SD) | 19.48 ±9.01 | 19.73 ± 6.65 | 0.837 | |
| WIT, min (avg ± SD) | 30.79 ±7.33 | 31.79 ± 6.82 | 0.367 | |
| Pump used, n (%) | 53 (49.5) | 44 (65.7) | 0.054 | |
| Pump time hours (avg ± SD) | 7.05 ± 8.06 | 7.84 ± 7.16 | 0.497 | |
| Last donor creatinine mg/dL (avg ± SD) | 1.24 ±0.87 | 1.25 ± 0.55 | 0.903 | |
| Donor HBV cAb n (%) | Negative | 93 (86.9) | 61 (91.0) | 0.613♦ |
| Positive | 9 (8.4) | 3 (4.5) | ||
| N/A | 5 (4.7) | 3 (4.5) | ||
| Donor HCV Ab n (%) | Positive | 12 (11.2) | 7 (10.4) | 1.00 |
| Donor CMV, n (%) | Positive | 63 (58.9) | 42 (62.7) | 0.734 |
| KDPI (avg ± SD) | 49.46 ± 27.40 | 69.93 ± 22.00 | <0.001 | |
| KDRI (avg ± SD) | 1.07 ± 0.36 | 1.34 ± 0.40 | <0.001 | |
| Histological Evaluation of Pretransplant Biopsies | ||||
| Pretransplant glomerulosclerosis (gsc) n (%) | Absent | 62 (57.9) | 46 (68.6) | 0.603♦ |
| Mild | 17 (15.9) | 8 (11.9) | ||
| Moderate | 2 (1.9) | 1 (1.5) | ||
| Severe | 0 (0.0) | 0 (0.0) | ||
| N/A | 26 (24.3) | 12 (17.9) | ||
| Pretransplant interstitial fibrosis (if) n (%) | Absent | 25 (23.4) | 10 (14.9) | 0.160♦ |
| Mild | 52 (48.6) | 39 (58.2) | ||
| Moderate | 4 (3.7) | 6 (9.0) | ||
| Severe | 0 (0.0) | 0 (0.0) | ||
| N/A | 26 (24.3) | 12 (17.9) | ||
| Pretransplant tubular atrophy (ta) n (%) | Absent | 46 (43.0) | 26 (38.8) | 0.263♦ |
| Mild | 34 (31.8) | 26 (38.8) | ||
| Moderate | 1 (0.9) | 3 (4.5) | ||
| Severe | 0 (0.0) | 0 (0.0) | ||
| N/A | 26 (24.3) | 12 (17.9) | ||
| Recipient Characteristics | ||||
| Recipient age (avg ± SD) | 51.98 ± 12.62 | 53.09 ± 11.06 | 0.556 | |
| Recipient gender n (%) | Male | 64 (59.8) | 40 (59.7) | 1.00 |
| Female | 43 (40.2) | 27 (40.3) | ||
| Recipient race n (%) | Asian/Pacific Islander | 1 (0.9) | 0 (0.0) | 0.898♦ |
| African American | 79 (73.8) | 50 (74.6) | ||
| Caucasian | 22 (20.6) | 16 (23.9) | ||
| Hispanic | 4 (3.7) | 1 (1.5) | ||
| Other/Unknown | 1 (0.9) | 0 (0.0) | ||
| Recipient BMI, (avg ± SD) | 27.92 ± 5.19 | 28.48 ± 4.86 | 0.479 | |
| Recipient HCV, n (%) | Positive | 13 (12.1) | 6 (9.0) | 0.621♦ |
| Negative | 94 (87.9) | 61 (91.0) | ||
| CMV disease, n (%) | Positive | 2 (1.9) | 4 (6.0) | 0.206♦ |
| Recipient CMV n (%) | Positive | 82 (76.6) | 51 (76.1) | 1.00 |
| Pretransplant diagnosis n (%) | DM | 20 (18.7) | 15 (22.4) | 0.516♦ |
| DM/HTN | 24 (22.4) | 8 (11.9) | ||
| HTN | 37 (34.6) | 25 (37.3) | ||
| FSGS | 8 (7.5) | 5 (7.5) | ||
| Other | 18 (16.8) | 14 (20.9) | ||
| Matched sex, n (%) | 49 (45.8) | 41 (61.2) | 0.068 | |
| Months on dialysis pretransplant (avg ± SD) | 40.37 ± 34.62 | 45.95 ± 37.51 | 0.333 | |
| AR episodes within 12 months posttransplant n (%) | 10 (9.3) | 10 (14.9) | 0.330♦ | |
| HLA mismatch (avg ± SD) | 4.38 ± 1.33 | 4.41 ± 1.21 | 0.768 | |
| PRA | >80% | 30 (28.0) | 22 (32.8) | 0.501 |
| dnDSA, n (%) | Positive | 8 (7.5) | 10 (14.9) | 0.131♦ |
A two-sample t-test was computed for continuous variables, while categorical variables were compared using a Chi-square test (or Fisher’s exact test when there were small cell sizes).
Fisher’s exact test used due to small expected cell sizes.
AR: acute rejection; BMI: body mass index; CIT: cold ischemia time; CMV: cytomegalovirus; DCD: donation after circulatory death; DM: diabetes mellitus; dnDSA: de novo donor specific antibody, FSGS: focal segmental glomerulosclerosis; HBV: hepatitis B virus; HCV: Hepatitis C virus; HLA: human leukocyte antigen; HTN: hypertension; KDPI: Kidney Donor Profile Index; KDRI: Kidney Donor Risk Index; SCD: standard criteria donor; WIT: warm ischemia time.
Patients with low 24-month graft function experienced significantly poorer long-term survival outcomes than patients with high 24-month graft function (p=0.03) (Figure S3). Using the combined analytical approaches, it was evident that the two groups were significantly different throughout follow-up.
3.2. Molecular markers discriminating 24-month eGFR outcomes.
A total of 595 unique genes (corresponding to 699 probe sets) were differentially expressed (FDR<0.05) in pretransplant donor organs, of which 408 were upregulated and 187 were downregulated in low function kidneys (Table S1). A volcano plot showing for all DEGs is displayed in Figure 1A. A heatmap displaying the top shared and unique pretransplant biological pathways in low-function kidneys is depicted in Figure 1B. These pretransplant biopsies are highly enriched in genes inducing innate (e.g., ADAM8, C1QA, CCL5, CD68, CLEC7A, HLA-F, NCKAP1L TYROBP,) and adaptive (e.g., C1QB, CD3D, CD6, CD48, CD84, GPR183, IGLL5, HLA-DQA1, HLA-DQB1, HLA-DQB2, IL7R) immune responses. Cell-type enrichment analyses identified dendritic, monocytes, myeloid, and natural killer cells as the main cell sources for the upregulated genes in pretransplant biopsies with low 24-month function (Figure S4). In contrast, downregulated genes such as CTNND1, DLAT, ENO1, FH, GOT1, IDH2, PDS5A, RFC3, and PGK1 are involved in metabolic processes (carbon/glucose metabolism, TCA cycle), gluconeogenesis, and cell-cell adhesion, and are associated with low 24-month function.
Figure 1.
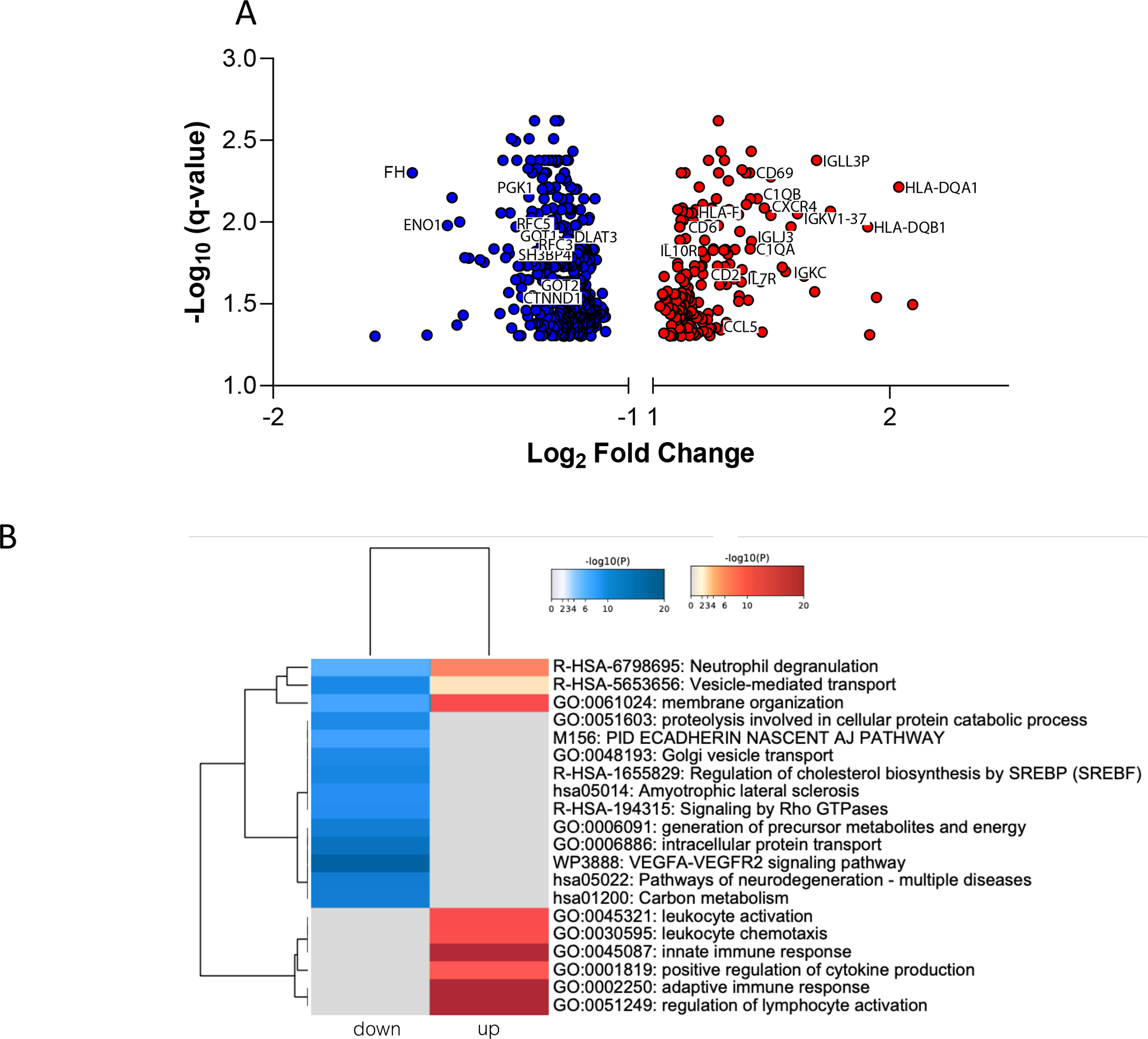
A. Volcano plot showing fold changes and the adjusted p-values for all differentially expressed genes between groups at pre-transplantation. The red dots represent down-regulated genes and blue dots represent up-regulated genes in low-functioning kidneys. B. Heatmap of top enriched biological pathways in low-functioning kidneys, colored by p-values. Grey values indicate no detected expression patterns.
The PPIs between down- and up-regulated DEGs are displayed in Figures S5 and S6. Kidneys with low 24-month function exhibited many downregulated biological processes at pre-transplantation including the metabolism of cholesterol, carbon, and carbohydrates, DNA damage recognition, regulation of intrinsic apoptotic signaling, and cell cycle regulation (Figure S5). These same kidneys showed upregulated PPI networks related to dendritic cell migration, regulation of chemotaxis, interferon gamma (IFN-γ) signaling, and the Fc epsilon receptor 1 (FCER1) pathway (Figure S6).
3.3. 24-month multivariable models
(1). Gene Expression (GE) model.
When searching over the grid of parameters, the optimal value from our repeated 10-fold CV procedure was λ=0.02 and αLASSO=1. When applying gene expression data (FDR≤0.05) to predict 24-month function, there were 55 significant probe sets in the penalized model. A plot of these 55 probe sets by their variable importance is displayed in Figure 2A. The AUROC using gene expression data (55 genes) was 0.994 (95% CI: 0.986, 1.0). When performing N-fold CV on the GE model, the AUROC was 0.767 (0.696, 0.838).
Figure 2.
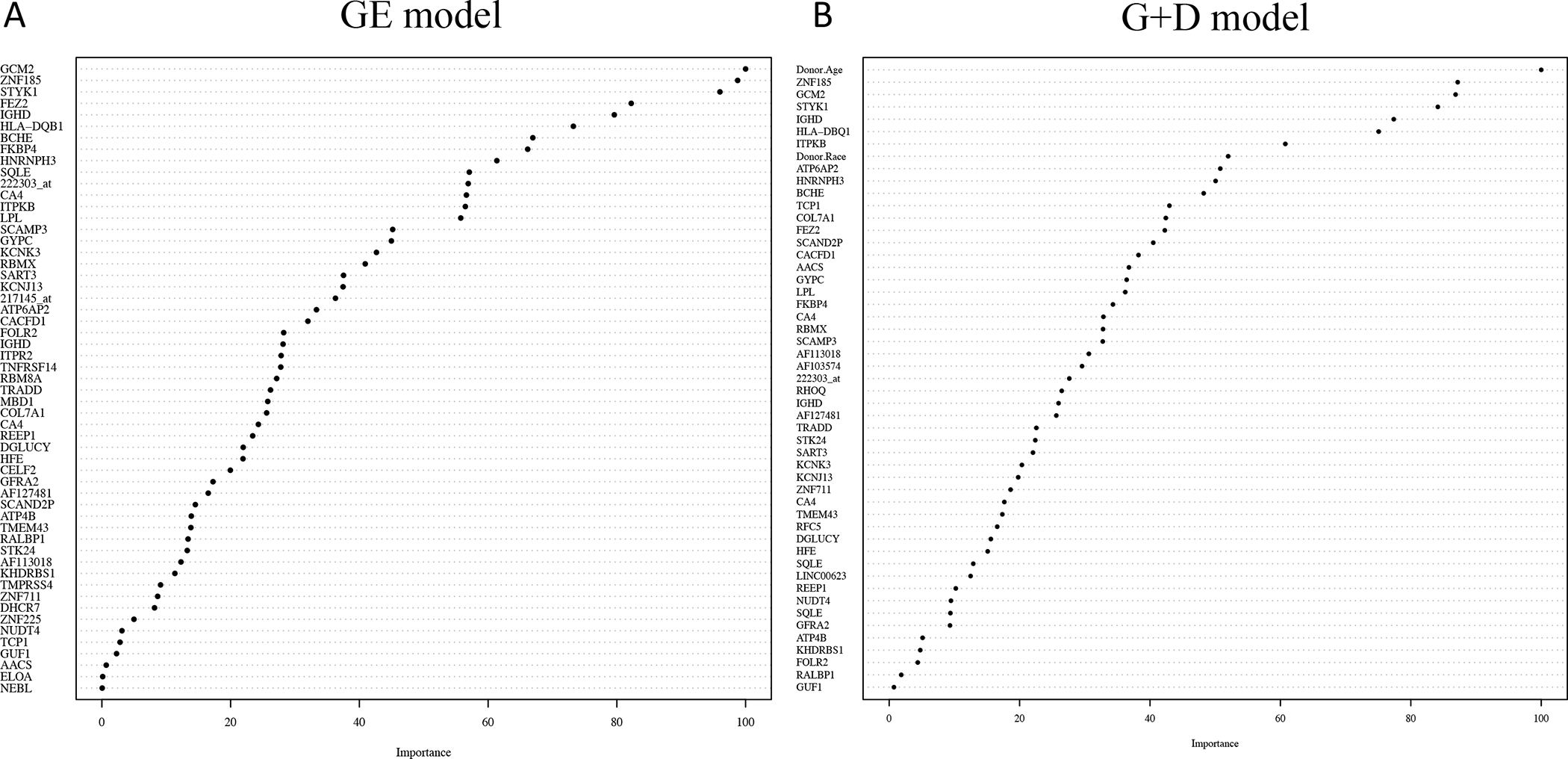
A. Plot of the 55 genes listed by their variable importance in predicting 24-month function for the gene expression (GE) model. B. Plot of the 52 variables (49 genes + 3 donor characteristics) in order of variable importance used in predicting 24-month function for the gene expression + donor characteristics (G+D) model.
(2). Donor Characteristics (DC) model.
Donor age, race, and BMI were the only clinical characteristics significantly different when comparing the high vs. low eGFR groups (p<0.05) (Table 1). Parameter estimates, standard errors, and p-values from the DC logistic regression model are shown in Table S2. The AUROC for the training data using the 3 characteristics with statistical significance (donor age, race, BMI) was 0.754 (95% CI: 0.680, 0.828). The N-fold CV for the donor age, race, and BMI model is 0.727 (0.649, 0.805).
(3). Gene Expression + Donor Characteristics (G+D) model.
When searching over the grid of parameters, the optimal values from our repeated 10-fold CV procedure were also λ=0.02 and αLASSO=1. When fitting the model there were 49 probe sets in the final model when donor age, race, and BMI were included. A plot of the 52 variables (49 probe sets and 3 donor characteristics) in order of their variable importance is displayed in Figure 2B. The AUROC for the G+D model was 0.996 (95% CI: 0.990, 1.0). When performing the N-fold CV the AUROC was 0.809 (0.744, 0.875).
(4). KDPI model.
The KDPI for each patient was calculated using 10 donor characteristics (donor age, height, weight, race, cause of death, HCV status, serum creatinine, DCD criteria, history of hypertension, and history of diabetes). Resulting numerical KDPI scores were used for the predictive model. The AUROC for the training data was 0.718 (95% CI: 0.642, 0.794). The AUROC for the N-fold CV is 0.705 (0.627, 0.782).
The respective AUROC curves for the 4 models in the training set are shown in Figure 3.
Figure 3.
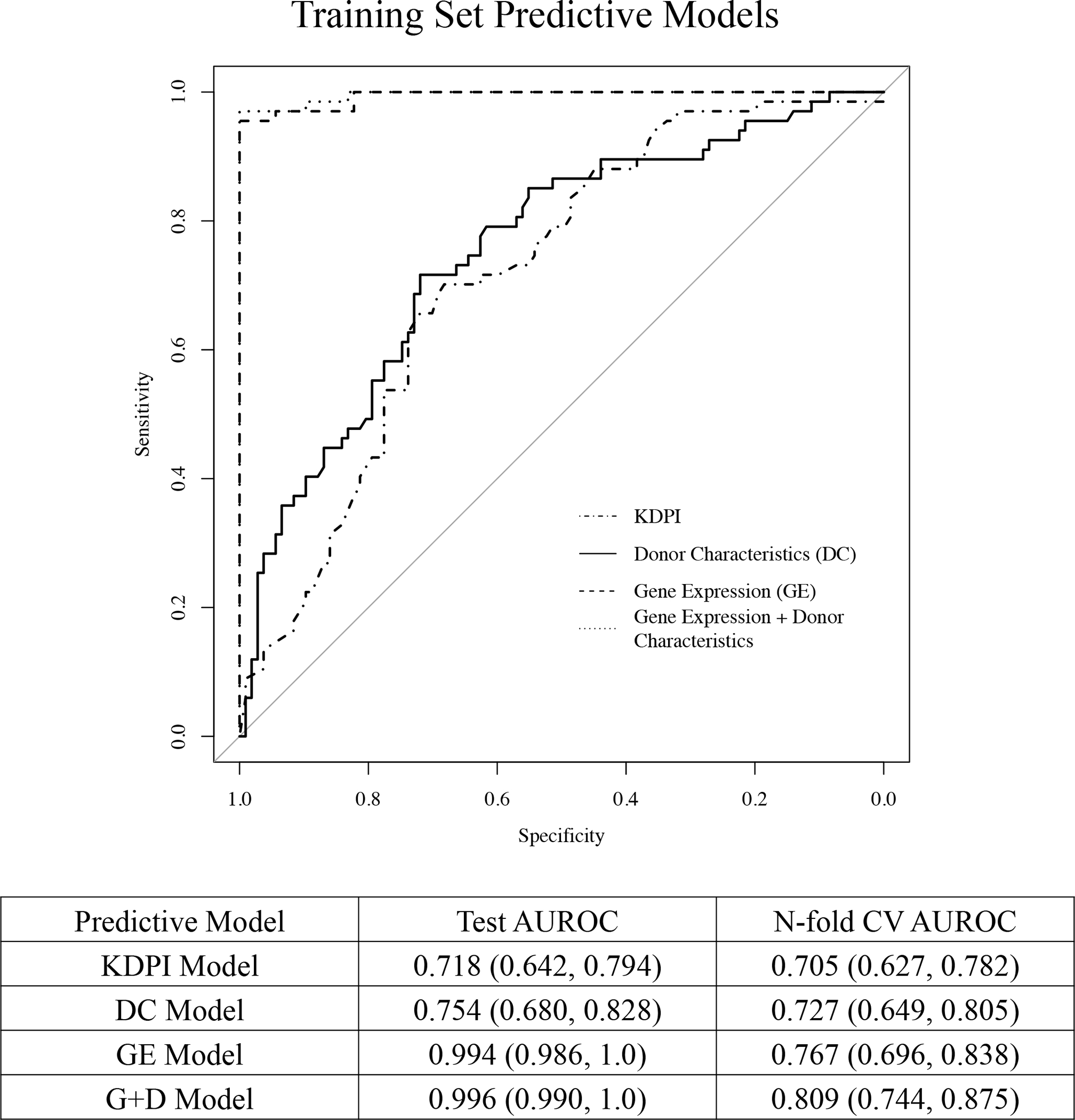
Area under the receiver operating characteristic (AUROC) curves for the training data for the donor characteristics (DC) model, gene expression (GE) model, gene expression + donor characteristics (G+D) model, and the KDPI model in predicting high vs. low eGFR group 24-months posttransplant. The diagonal line represents performance of a chance model.
3.4. External Validation using qPCR.
The validation set included 96 KT recipients, of which 36 (37.5%) had low eGFR and 60 (62.5%) had high eGFR at 24-months post-transplant (Table 2). The AUROC for the donor characteristics model (age, BMI, race) is 0.691 (95% CI: 0.584–0.797). The KDPI model calculated using 10 donor characteristics yielded the same point estimate for AUROC = 0.691 (95% CI: 0.585–0.797). The 13 genes that were validated from the final models (GE and G+D) included BCHE, FKBP4, GYPC, HLA-DQB1, HNRNPH3, IGHD, NUDT4, RBM8A, RHOQ, SQLE, STK24, TRADD, and ZNF185 (assay IDs provided in Table S3). The combined model (13 genes + 3 donor characteristics) showed an AUROC of 0.821 (95% CI: 0.733, 0.909) for 24-month function. The respective AUROC curves for the 4 models after the 10-fold CV procedure are shown in Figure 4.
Table 2.
Characteristics of donor and recipients sub-stratified based on eGFR at 24-month post kidney transplant in the validation set (n=96).
| Clinical Characteristics | Category | High eGFR (n=60) |
Low eGFR (n=36) |
p-value |
|---|---|---|---|---|
| Donor Characteristics | ||||
| Donor age (avg ± SD) | 38.22 ±12.65 | 46.33±13.50 | 0.004 | |
| Donor gender, n (%) | Male | 26 (43.3) | 15 (41.7) | 0.570 |
| Female | 33 (55.0) | 19 (52.8) | ||
| Unknown | 1 (1.7) | 2 (5.6) | ||
| Donor race, n (%) | Asian | 0 (0.0) | 1 (2.8) | 0.359 |
| African American | 11 (18.3) | 7 (19.4) | ||
| Caucasian | 44 (73.3) | 22 (61.1) | ||
| Hispanic | 3 (5.0) | 2 (5.6) | ||
| Other | 2 (3.3) | 4 (11.1) | ||
| DCD, n (%) | 4 (6.7) | 6 (16.7) | 0.227 | |
| Donor cause of death, n (%) | Anoxia | 26 (43.3) | 12 (33.3) | 0.241 |
| Head trauma | 11 (18.3) | 6 (16.7) | ||
| Stroke | 17 (28.3) | 17 (47.2) | ||
| Other/Unknown | 6 (10.0) | 1 (2.9) | ||
| Delayed graft function, n (%) | 26 (43.3) | 16 (44.4) | 1.000 | |
| Donor BMI (avg ± SD) | 29.85 (9.06) | 36.47 (50.16) | 0.320 | |
| CIT, hours (avg ± SD) | 21.49 ±10.80 | 20.99 ±8.02 | 0.816 | |
| WIT, min (avg ± SD) | 35.42 (5.23) | 33.79 (5.83) | 0.169 | |
| Pump used, n (%) | 31 (51.7) | 16 (44.4) | 0.635 | |
| Pump Time min (avg ± SD) | 261.25 (356.61) | 261.83 (383.20) | 0.994 | |
| Last Donor Creatinine mg/dL (avg ± SD) | 1.83 (1.71) | 1.38 (1.18) | 0.172 | |
| Donor HBV cAb, n (%) | Positive | 4 (6.7) | 2 (5.6) | 0.718 |
| Negative | 55 (91.7) | 34 (94.4) | ||
| N/A | 1 (1.7) | 0 (0.0) | ||
| Donor HCV Ab, n (%) | Positive | 19 (31.7) | 10 (27.8) | 0.863 |
| Donor CMV, n (%) | Positive | 29 (48.3) | 21 (58.3) | 0.237 |
| Negative | 31 (51.7) | 14 (38.9) | ||
| N/A | 0 (0.0) | 1 (2.8) | ||
| KDPI (avg ± SD) | 51.68 (23.34) | 67.36 (20.08) | 0.001 | |
| KDRI (avg ± SD) | 1.08 (0.33) | 1.33 (0.46) | 0.003 | |
| Recipient Characteristics | ||||
| Recipient Age (avg ± SD) | 53.33 (12.16) | 50.64 (11.43) | 0.290 | |
| Recipient Gender n (%) | Female | 21 (35.0) | 9 (25.0) | 0.571 |
| Male | 38 (63.3) | 26 (72.2) | ||
| Unknown | 1 (1.7) | 1 (2.8) | ||
| Recipient Race n (%) | African American | 42 (70.0) | 21 (58.3) | 0.698 |
| Caucasian | 11 (18.3) | 9 (25.0) | ||
| Hispanic | 3 (5.0) | 3 (8.3) | ||
| Unknown | 4 (6.7) | 3 (8.3) | ||
| Recipient BMI, (avg ± SD) | 39.20 (38.37) | 45.48 (62.18) | 0.546 | |
| Recipient HCV, n (%) | Positive | 3 (5.0) | 6 (16.7) | 0.108 |
| Negative | 47 (78.3) | 27 (75.0) | ||
| N/A | 10 (16.7) | 3 (8.3) | ||
| CMV disease, n (%) | Positive | 6 (10.0) | 2 (5.6) | 0.726 |
| Negative | 50 (83.3) | 31 (86.1) | ||
| N/A | 4 (6.7) | 3 (8.3) | ||
| Recipient CMV, n (%) | Positive | 29 (48.3) | 20 (55.6) | 0.165 |
| Negative | 30 (50.0) | 13 (36.1) | ||
| N/A | 1 (1.7) | 3 (8.3) | ||
| Pretransplant diagnosis n (%) | DM | 11 (18.3) | 6 (16.7) | 0.676 |
| DM/HTN | 10 (16.7) | 8 (22.2) | ||
| HTN | 14 (23.3) | 5 (13.9) | ||
| FSGS | 5 (8.3) | 3 (8.3) | ||
| Other | 18 (30.0) | 14 (38.9) | ||
| Unknown | 2 (3.3) | 0 | ||
| Matched sex, n (%) | 21 (36.2) | 16 (47.1) | 0.421 | |
| Months on dialysis pretransplant (avg ± SD) | 45.44 ±24.58 | 54.22 ±50.52 | 0.262 | |
BMI: Body Mass Index; CIT: Cold Ischemia Time; CMV: Cytomegalovirus; DCD: Donation after Circulatory Death; DM: Diabetes Mellitus; FSGS: Focal Segmental Glomerulosclerosis; HBV: Hepatitis B Virus; HCV: Hepatitis C Virus; HTN: Hypertension; KDPI: Kidney Donor Profile Index; KDRI: Kidney Donor Risk Index; SCD: Standard Criteria Donor; SD Standard Deviation; WIT: Warm Ischemia Time.
Figure 4.
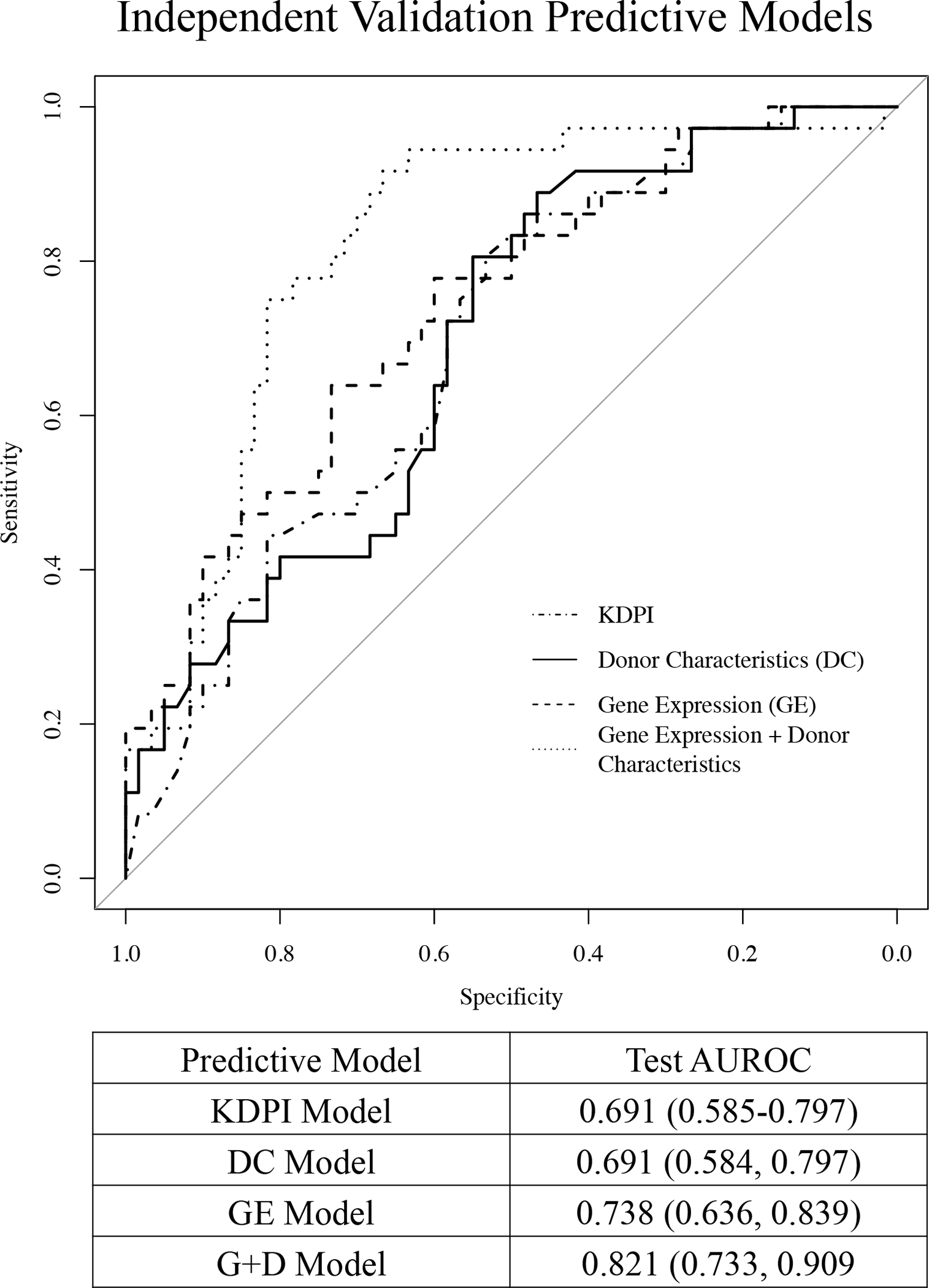
Area under the receiver operating characteristic (AUROC) curves for the validation set for the KDPI, donor characteristics (age, race, BMI), 14 genes alone, and 14 genes + 3 donor characteristics in predicting high vs. low eGFR group 24-months post-transplantation. The diagonal line represents performance of a chance model.
3.5. Risk Score Calculation.
A 24-month graft function risk score was calculated for each patient in the independent validation cohort (n=96) based on the combined model (13 genes + 3 donor characteristics). Regression coefficients, confidence intervals, and p-values are described in Table 3. Gene expression values and donor characteristics were linearly combined into a risk score as follows:
Table 3.
Regression coefficients for the logistic regression model that includes 13 genes and 3 donor characteristics (age, BMI, and race).
| Coefficient | Lower bound | Upper bound | P-value | |
|---|---|---|---|---|
| Intercept | −4.544 | −13.485 | 4.397 | 0.319 |
| Donor Age | 0.057 | 0.015 | 0.1 | 0.009 |
| Donor Race | 0.586 | −0.62 | 1.792 | 0.341 |
| Donor BMI | 0.004 | −0.014 | 0.023 | 0.628 |
| BCHE | 0.29 | −0.001 | 0.581 | 0.051 |
| FKBP4 | 0.023 | −1.535 | 1.582 | 0.977 |
| GYPC | −0.981 | −1.993 | 0.032 | 0.058 |
| HLA-DQB1 | −0.105 | −0.222 | 0.012 | 0.08 |
| HNRNPH3 | −0.327 | −1.982 | 1.328 | 0.698 |
| IGHD | 0.039 | −0.128 | 0.207 | 0.647 |
| NUDT4 | 0.975 | 0.131 | 1.818 | 0.024 |
| RBM8A | 0.717 | −1.522 | 2.956 | 0.53 |
| RHOQ | −2.182 | −3.885 | −0.478 | 0.012 |
| SQLE | 0.112 | −0.583 | 0.808 | 0.752 |
| STK24 | 1.073 | −0.201 | 2.346 | 0.099 |
| TRADD | 0.171 | −0.865 | 1.207 | 0.746 |
| ZNF185 | 0.378 | −0.783 | 1.539 | 0.523 |
Lower and upper bounds of the 95% Confidence Intervals and adjusted p-values for each regression coefficient.
Risk score = −4.544 + 0.29 (ΔCt BCHE) + 0.023 (ΔCt FKBP4) − 0.981 (ΔCt GYPC) − 0.105 (ΔCt HLA-DQB1) − 0.327 (ΔCt HNRNPH3) + 0.039 (ΔCt IGHD) + 0.975 (ΔCt NUDT4) + 0.717 (ΔCt RBM8A) − 2.182 (ΔCt RHOQ) + 0.112 (ΔCt SQLE) + 1.073 (ΔCt STK24) + 0.171 (ΔCt TRADD) + 0.378 (ΔCt ZNF185) + 0.057 (donor age) + 0.004 (donor BMI) + 0.586 (donor race indicator variable).
Donor race was converted to a dichotomous variable, with Caucasian = 0 and all other races = 1. The risk equation was then converted to a probability scale (0.0–1.0). The probability of low-graft function for each patient is plotted in Figure 5A and the KDPI score for each patient is plotted in Figure 5B. Youden’s index was calculated for both the probability score and the KDPI, with y=0.306 and y=52 as the respective thresholds that maximize specificity and sensitivity for the validation set. When using KDPI to predict low 24-month function, the sensitivity was 80.6% and the specificity was 53.3%. When using the risk probability score, the sensitivity was 88.9% and the specificity was 66.6% (Figure 5C).
Figure 5.
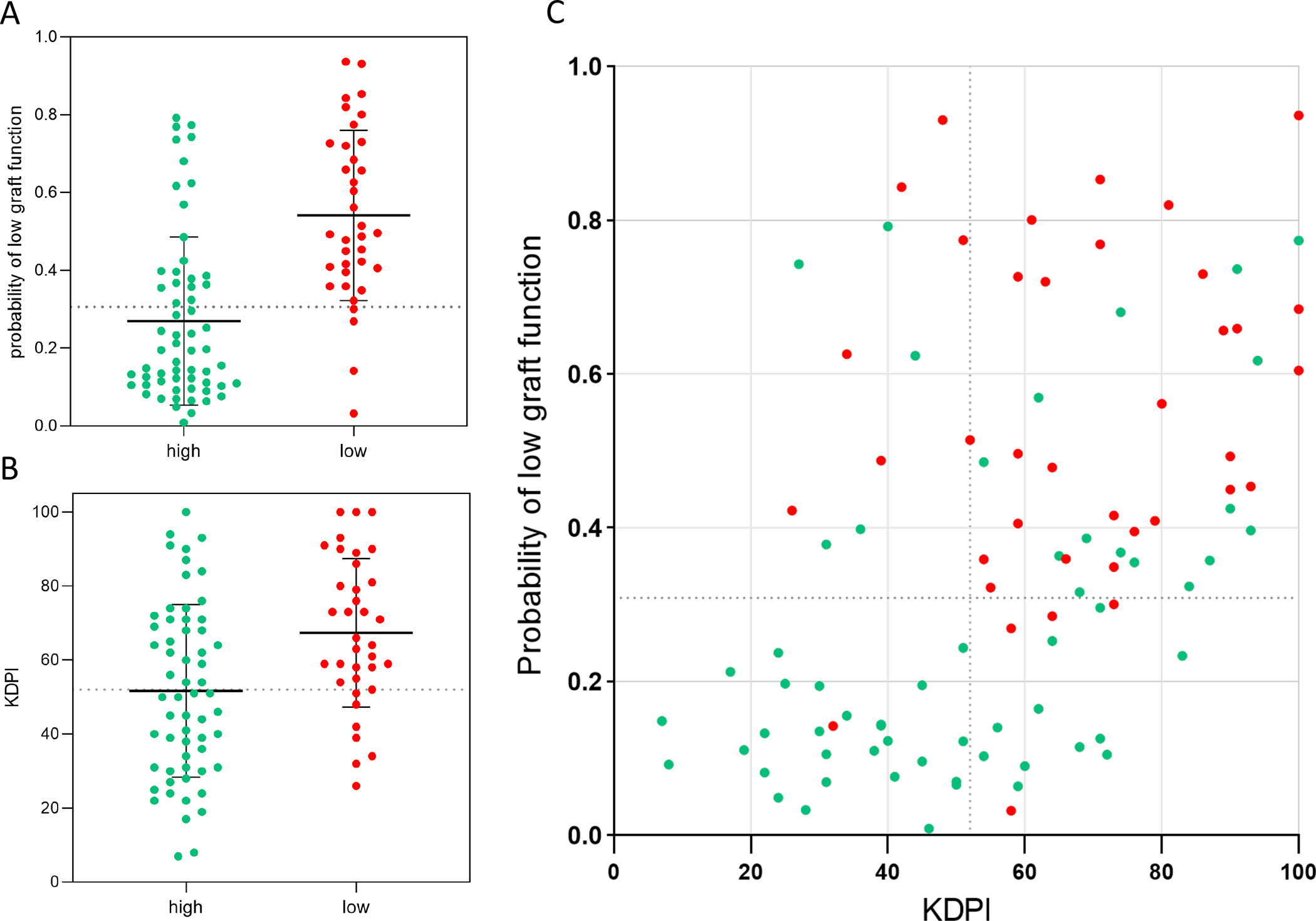
A. Probability score (derived from predictive equation) of each patient in the validation set (n=96) separated by 24-month outcome group. Dotted horizontal line at 0.306 represents Youden’s index. Mean and standard deviation bars displayed. Green represents high and red represents low 24-month function. B. KDPI score for each patient in the validation set separated by 24-month outcome group. Dotted horizontal line at 52 represents Youden’s index (where specificity and sensitivity are maximized). Mean and standard deviation bars displayed. C. KDPI and probability score of each patient plotted with Youden’s indices depicted for each axis.
4. Discussion
The field of transplantation is in critical need of more accurate tools to predict allograft outcomes.19–21 Current in-use clinical scores and histological assessments have only demonstrated modest predictive accuracy for short-term outcomes.22–25 Over the last decade, transcriptomic profiling has emerged as a powerful approach for revealing unbiased biological information useful for posttransplant management.
Our study represents the largest high-throughput transcriptomic analysis of pretransplant donor kidneys predicting 24-month outcomes. Herein, we present a predictive risk score, which combines donor age, race, BMI, and donor quality gene markers, that can be calculated prior to transplantation to predict graft function. We also identified differential pretransplant transcriptional profiles between kidneys with low and high function at 24-months, providing a deeper insight into the early biological processes leading to graft dysfunction.
This prospective study has three critical features to consider: i) the inclusion of 270 patients from 4 transplant centers, ii) high-throughput genome-wide approaches, and iii) a well-characterized external validation cohort. Furthermore, our unique patient cohort includes a broad spectrum of kidney donor organs (i.e., aged, DCD, HCV+, pumped, and AKI donors), and a significant number of African American recipients (70.8%).
Thus far, a limited number of peer-reviewed pretransplant kidney gene expression studies have been conducted in the field.26–34 Of these studies, only 2 evaluated graft outcomes beyond 1 year (both of which had small sample sizes and used targeted gene approaches).30,34 Critically, none of the previous studies included external validations, which are necessary to determine the reproducibility and generalizability of results in different patient populations.
Additionally, the majority of predictive transcriptomic studies in kidney transplantation have focused on delayed graft function (DGF) as a surrogate marker, without being able to predict longer-term outcomes (>12 months).28,29,31–36 We found that DGF was not significantly associated with 24-month function (p=0.238), explaining why gene sets associated with DGF have poor predictive value.8,37 Furthermore, most transcriptomic studies have utilized post-reperfusion biopsies, which are less likely to capture intrinsic organ quality due to the ‘transcriptional noise’ induced by reperfusion injury, surgical procedures, recipient immune infiltration, and immunosuppressive medications.8,28,30,38–40 Our results indicate that the use of pre-reperfusion biopsies allows for a more accurate evaluation of donor organ quality.8,41–43
Our results showed that grafts with low function at 24 months displayed upregulated innate and adaptive immune responses (e.g., B cell proliferation, positive regulation of phagocytes, dendritic cell migration) prior to transplantation. This finding is in concordance with our previous studies, which reported an upregulated donor immune signature associated with short-term graft function.29,31,44 We also recently reported that pretransplant donor biopsies from grafts progressing to chronic allograft dysfunction presented differentially methylated epigenetic profiles related to an activated immune state.45
Moreover, the downregulation of fundamental biological processes such as metabolic function (e.g., metabolism of cholesterol, carbon, and carbohydrates) further exacerbates the degree of injury posttransplant in kidneys with low 24-month function. Metabolic dysfunction in native kidney tissue (involving oxidative phosphorylation, fatty acid oxidation, cellular respiration) is associated with impaired repair mechanisms in kidney disease, 46–50 which may contribute to the progressive decline of graft function.
Overall, increased immune responses and decreased metabolic activity prior to transplantation disrupt graft homeostasis and result in the gradual loss of kidney function over time. These results are independent of cold ischemia time and other pre-/peri-transplant factors, reflecting the importance of evaluating the inherent donor mechanisms responsible for triggering and likely, sustaining post-transplant injury.
Although many genes have been identified to play important roles in kidney disease progression and pathophysiology, they do not inherently serve as reliable predictors of post-transplant graft function and disease state.51 This study serves as one of the first computational studies to integrate experimental and clinical data to identify novel markers of graft function.
All clinical and demographic characteristics from both the donors and recipients were analyzed, and statistically significant variables were used to develop a multivariable predictive model. As expected, donor age was the most predictive clinical variable, 8,29 followed by BMI and race. Current models including KDPI use less accessible/objective donor characteristics such as “history of hypertension” and “history of diabetes.” Interestingly, no recipient characteristics (including age, rejection events, or donor-specific antibodies) correlated with 24-month outcomes, demonstrating the prevailing importance of donor organ quality in predicting graft function.
We found that 24-month graft function was more accurately predicted by the transcriptomic profile of preimplantation biopsies (GE model AUROC = 0.994) than by significant donor characteristics (DC model AUROC = 0.754) or by KDPI scores (KDPI model AUROC = 0.718) (p<0.001). The same was true of the combined gene and donor characteristic (G+D) model (AUROC=0.996) (p<0.001).
To confirm the generalizability of these results, a small set of genes from the final models were tested in an independent cohort of patients (G+D model AUROC=0.821). This model more accurately predicted 24-month function than the KDPI (AUROC=0.691) and DC models (AUROC=0.691) (p=0.026). In the same patients, qPCR results and clinical characteristics were combined to develop a predictive equation quantifying patient risk for decreased 24-month graft function.
Challenges associated with the statistical modeling of biomedical datasets include high-dimensionality, incompleteness, bias, heterogeneity, overfitting, and background noise.52,53,54,55 However, the use of an external validation set largely mitigates these concerns. While our analyses exceed the outcome measurements performed by nearly all pretransplant biomarker studies, predictive markers beyond 24-months need to be identified.8 Nonetheless, this study lays the groundwork for future clinical trials aiming to implement molecular markers in post-transplant management. While this study showed high accuracy of outcome prediction using 13 genes, a predesigned 96-well qPCR plate can be loaded with all the genes from the final models (GE and G+D models), which will further improve the predictive accuracy of this approach.
Defining surrogate endpoints, standards for outcome reporting, and statistical strategies to appropriately analyze differences between outcome groups is critical in biomarker discovery research.56 Currently, there is a great deal of complexity associated with patient classification approaches in kidney transplantation. A reliable classification of kidney function and progression is needed but not yet achieved. Thus, when designing this study, we considered multiple different patient classification approaches that utilized one or more of the following parameters: overall eGFR slope, Y-intercepts, final eGFR as a continuous outcome, and multiple eGFR measurements. We chose to analyze eGFR as a dichotomous outcome to enable the reporting of clinically meaningful statistics that frequently accompany diagnostic/prognostic assays, such as the AUROC. Ultimately, this eGFR categorization (supported by significant differences in long-term graft survival) allows for significant statistical power to detect important differences across primary endpoints for direct clinical translation.56,57
In conclusion, this is the first genome-wide large-cohort study to demonstrate that the donor kidney transcriptome, prior to implantation, captures intrinsic organ quality and carries significant predictive weight for 24-month transplant function. Our findings shift the paradigm of understanding longer-term kidney transplant outcomes away from recipient factors/post-transplant events (e.g., DGF) and towards the intrinsic donor organ quality, which can be captured by molecular techniques. Notably, we demonstrate that a combined predictive equation using both clinical and biological data can more accurately predict 24-month outcomes as compared to the current established scoring system (KDPI) in an external patient cohort. Such findings transform our understanding of graft quality, relevant biological pathways, and potential therapeutic targets, offering more personalized criteria for posttransplant management.
Supplementary Material
Figure S1. Patient flow diagram. A total of 295 patients were enrolled from 4 transplant centers (n=195 training set, n=100 validation set). Purple boxes represent exclusions. 21 patients were excluded from the training set due to follow-up loss, death with graft function, and microarray quality control criteria. 4 patients were excluded from the validation set due to low RNA integrity. The remaining 270 patients were included in the final training (n=174) and validation (n=96) sets. QC: Quality control; RIN: RNA integrity number.
Figure S3. Kaplan-Meier estimates for time until graft failure or death showing graft/patient survival after 24-months, separated by 24-month graft function group with log-rank test comparing the two groups. Only patients who were alive at 24-months were included in the analyses, with 24-months as time-zero. NA: not available.
Figure S4. Bar chart visualizing the top enriched cell-types for the upregulated DEGs (in low-functioning kidneys) and their associated q-values.
Figure S2: A. Spaghetti plot separated by high and low graft function group at 24 months with lowess smooths overlaid. Smoothed eGFR post-transplant (black line) and fitted linear mixed effects model (white dotted line) with equation. B. Mean eGFR (corresponding to black line) and standard deviation at each timepoint separated by high and low 24-month graft function.
Figure S5. Downregulated protein-protein interaction (PPI) networks encoded by differentially expressed genes (represented by nodes). Lines represent interaction relationships between nodes. Color-coded clusters were selected from PPI network using Molecular Complex Detection (MCODE). Downregulated pretransplant biological pathways associated with each MCODE subnetwork and adjusted p-values are listed.
Figure S6. Upregulated protein-protein interaction (PPI) networks encoded by differentially expressed genes (represented by nodes). Lines represent interaction relationships between nodes. Color-coded clusters were selected from PPI network using Molecular Complex Detection (MCODE). Upregulated pretransplant biological pathways associated with each MCODE subnetwork and adjusted p-values are listed.
Table S1. List of differentially expressed probe sets (n=699) with gene symbols listed in order of descending fold change (FC) (472 upregulated and 227 downregulated). Probe sets map to 595 unique genes. Unmapped probe sets (n=6) labeled as “N/A.” False discovery rate (FDR) <0.05.
Table S2. Regression coefficients, confidence intervals, and p-values from donor characteristics (DC) logistic regression model.
Table S3. Gene expression assays used for qPCR validation.
Acknowledgements/Funding:
The authors would like to thank the consenting patients, clinical staff, and research coordinators of Montefiore Medical Center, Northwestern Memorial Hospital, University of Tennessee Health Science Center, University of Virginia Medical Center, and Virginia Commonwealth University Medical Center for providing, collecting and processing invaluable samples used for this multicenter study.
Research reported in the publication was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health under awards numbers: R01DK109581 (VM/KJA) and R01DK122682 (VM).
Abbreviations:
- AKI
acute kidney injury
- AUROC
area under the receiver operating curve
- BMI
body mass index
- CI
confidence interval
- CT
cycle threshold
- DC
donor characteristics
- DCD
donor after cardiac death
- DD
deceased donor
- DEGs
differentially expressed genes
- DGF
delayed graft function
- eGFR
estimated glomerular filtration rate
- ESRD
end-stage renal disease
- FDR
false discovery rate
- GE
gene expression
- G+D
gene expression and donor characteristics
- GO
gene ontology
- HCV
hepatitis C virus
- IPA
Ingenuity Pathway Analysis
- KDIGO
Kidney Disease Improving Global Outcomes
- KDPI
kidney donor profile index
- KDRI
kidney donor risk index
- MDRD
modification of diet in renal disease study
- qPCR
quantitative polymerase chain reaction
Footnotes
Disclosure: The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting information statement: Additional Supporting Information (Tables S1–S3 and Figures S1–S4) may be found online in the supporting information tab for this article.
Data Availability Statement:
Raw data from all Affymetrix GeneChip microarrays is available in the Gene Expression Omnibus public repository (GSE147451) which can be viewed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147451.
References:
- 1.Hart A, Lentine KL, Smith JM, et al. OPTN/SRTR 2019 Annual Data Report: Kidney. Am J Transplant. 2021;21 Suppl 2:21–137. doi: 10.1111/ajt.16502 [DOI] [PubMed] [Google Scholar]
- 2.Ojo AO. Expanded criteria donors: process and outcomes. Semin Dial. 2005;18(6):463–468. doi: 10.1111/j.1525-139X.2005.00090.x [DOI] [PubMed] [Google Scholar]
- 3.Filiopoulos V, Boletis JN. Renal transplantation with expanded criteria donors: Which is the optimal immunosuppression? World J Transplant. 2016;6(1):103–114. doi: 10.5500/wjt.v6.i1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao PS, Ojo A. The alphabet soup of kidney transplantation: SCD, DCD, ECD--fundamentals for the practicing nephrologist. Clin J Am Soc Nephrol. 2009;4(11):1827–1831. doi: 10.2215/CJN.02270409 [DOI] [PubMed] [Google Scholar]
- 5.Dahmen M, Becker F, Pavenstädt H, Suwelack B, Schütte-Nütgen K, Reuter S. Validation of the Kidney Donor Profile Index (KDPI) to assess a deceased donor’s kidneys’ outcome in a European cohort. Sci Rep. 2019;9(1):11234. doi: 10.1038/s41598-019-47772-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lentine KL, Kasiske B, Axelrod DA. Procurement Biopsies in Kidney Transplantation: More Information May Not Lead to Better Decisions. J Am Soc Nephrol. 2021;32(8):1835–1837. doi: 10.1681/ASN.2021030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaber LW, Moore LW, Alloway RR, Amiri MH, Vera SR, Gaber AO. Glomerulosclerosis as a determinant of posttransplant function of older donor renal allografts. Transplantation. 1995;60(4):334–339. doi: 10.1097/00007890-199508270-00006 [DOI] [PubMed] [Google Scholar]
- 8.von Moos S, Akalin E, Mas V, Mueller TF. Assessment of Organ Quality in Kidney Transplantation by Molecular Analysis and Why It May Not Have Been Achieved, Yet. Front Immunol. 2020;11:833. doi: 10.3389/fimmu.2020.00833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supplitt S, Karpinski P, Sasiadek M, Laczmanska I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int J Mol Sci. 2021;22(3):1422. doi: 10.3390/ijms22031422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supplitt S, Karpinski P, Sasiadek M, Laczmanska I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int J Mol Sci. 2021;22(3):1422. Published 2021 Jan 31. doi: 10.3390/ijms22031422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 12.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002 [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 14.Kuhn M Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28(1):1–26. doi: 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- 15.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Harrell FE. Multivariable Modeling Strategies. In: Harrell Frank E Jr, ed. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. Springer Series in Statistics. Springer International Publishing; 2015:63–102. doi: 10.1007/978-3-319-19425-7_4 [DOI] [Google Scholar]
- 17.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. Published 2019 Apr 3. doi: 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herath S, Dai H, Erlich J, et al. Selection and validation of reference genes for normalisation of gene expression in ischaemic and toxicological studies in kidney disease. PLoS One. 2020;15(5):e0233109. doi: 10.1371/journal.pone.0233109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reese PP, Harhay MN, Abt PL, Levine MH, Halpern SD. New Solutions to Reduce Discard of Kidneys Donated for Transplantation. J Am Soc Nephrol. 2016;27(4):973–980. doi: 10.1681/ASN.2015010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moeckli B, Sun P, Lazeyras F, et al. Evaluation of donor kidneys prior to transplantation: an update of current and emerging methods. Transpl Int. 2019;32(5):459–469. doi: 10.1111/tri.13430 [DOI] [PubMed] [Google Scholar]
- 21.Stegall MD, Gaston RS, Cosio FG, Matas A. Through a glass darkly: seeking clarity in preventing late kidney transplant failure. J Am Soc Nephrol. 2015;26(1):20–29. doi: 10.1681/ASN.2014040378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae S, Massie AB, Thomas AG, et al. Who can tolerate a marginal kidney? Predicting survival after deceased donor kidney transplant by donor-recipient combination. Am J Transplant. 2019;19(2):425–433. doi: 10.1111/ajt.14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husain SA, Shah V, Alvarado Verduzco H, et al. Impact of Deceased Donor Kidney Procurement Biopsy Technique on Histologic Accuracy. Kidney Int Rep. 2020;5(11):1906–1913. Published 2020 Aug 14. doi: 10.1016/j.ekir.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall IE, Parikh CR, Schröppel B, et al. Procurement Biopsy Findings Versus Kidney Donor Risk Index for Predicting Renal Allograft Survival. Transplant Direct. 2018;4(8):e373. doi: 10.1097/TXD.0000000000000816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Y, Schaubel DE, Kalbfleisch JD, Ashby VB, Rao PS, Sung RS. Reevaluation of the Kidney Donor Risk Index. Transplantation. 2019;103(8):1714–1721. doi: 10.1097/TP.0000000000002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84(3):353–361. doi: 10.1038/labinvest.3700037 [DOI] [PubMed] [Google Scholar]
- 27.Kainz A, Perco P, Mayer B, et al. Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation. 2007;83(8):1048–1054. doi: 10.1097/01.tp.0000259960.56786.ec [DOI] [PubMed] [Google Scholar]
- 28.Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8(1):78–85. doi: 10.1111/j.1600-6143.2007.02032.x [DOI] [PubMed] [Google Scholar]
- 29.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85(4):626–635. doi: 10.1097/TP.0b013e318165491f [DOI] [PubMed] [Google Scholar]
- 30.Bodonyi-Kovacs G, Putheti P, Marino M, et al. Gene expression profiling of the donor kidney at the time of transplantation predicts clinical outcomes 2 years after transplantation. Hum Immunol. 2010;71(5):451–455. doi: 10.1016/j.humimm.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mas VR, Scian MJ, Archer KJ, et al. Pretransplant transcriptome profiles identify among kidneys with delayed graft function those with poorer quality and outcome. Mol Med. 2011;17(11–12):1311–1322. doi: 10.2119/molmed.2011.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goncalves-Primo A, Mourão TB, Andrade-Oliveira V, et al. Investigation of apoptosis-related gene expression levels in preimplantation biopsies as predictors of delayed kidney graft function. Transplantation. 2014;97(12):1260–1265. doi: 10.1097/01.TP.0000442579.12285.e8 [DOI] [PubMed] [Google Scholar]
- 33.McGuinness D, Mohammed S, Monaghan L, et al. A molecular signature for delayed graft function. Aging Cell. 2018;17(5):e12825. doi: 10.1111/acel.12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Snijders MLH, Haasnoot GW, et al. Elevated intragraft expression of innate immunity and cell death-related markers is a risk factor for adverse graft outcome. Transpl Immunol. 2018;48:39–46. doi: 10.1016/j.trim.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 35.Guerrieri D, Re L, Petroni J, et al. Gene expression profile in delay graft function: inflammatory markers are associated with recipient and donor risk factors. Mediators Inflamm. 2014;2014:167361. doi: 10.1155/2014/167361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferdinand JR, Hosgood SA, Moore T, et al. Cytokine absorption during human kidney perfusion reduces delayed graft function-associated inflammatory gene signature. Am J Transplant. 2021;21(6):2188–2199. doi: 10.1111/ajt.16371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller TF, Solez K, Mas V. Assessment of kidney organ quality and prediction of outcome at time of transplantation. Semin Immunopathol. 2011;33(2):185–199. doi: 10.1007/s00281-011-0248-x [DOI] [PubMed] [Google Scholar]
- 38.Cippà PE, Liu J, Sun B, Kumar S, Naesens M, McMahon AP. A late B lymphocyte action in dysfunctional tissue repair following kidney injury and transplantation. Nat Commun. 2019;10(1):1157. Published 2019 Mar 11. doi: 10.1038/s41467-019-09092-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreepala C, Famulski KS, Chang J, Halloran PF. Comparing molecular assessment of implantation biopsies with histologic and demographic risk assessment. Am J Transplant. 2013;13(2):415–426. doi: 10.1111/ajt.12043 [DOI] [PubMed] [Google Scholar]
- 40.Kamińska D, Kościelska-Kasprzak K, Mazanowska O, et al. Pretransplant Immune Interplay Between Donor and Recipient Influences Posttransplant Kidney Allograft Function. Transplant Proc. 2018;50(6):1658–1661. doi: 10.1016/j.transproceed.2018.03.129 [DOI] [PubMed] [Google Scholar]
- 41.Lim MA, Bloom RD. Medical Therapies to Reduce Delayed Graft Function and Improve Long-Term Graft Survival: Are We Making Progress?. Clin J Am Soc Nephrol. 2020;15(1):13–15. doi: 10.2215/CJN.13961119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kayler LK, Sokolich J, Magliocca J, Schold JD. Import kidney transplants from nonmandatory share deceased donors: characteristics, distribution and outcomes. Am J Transplant. 2011;11(1):77–85. doi: 10.1111/j.1600-6143.2010.03359.x [DOI] [PubMed] [Google Scholar]
- 43.Le Meur Y, Aulagnon F, Bertrand D, et al. Effect of an Early Switch to Belatacept Among Calcineurin Inhibitor-Intolerant Graft Recipients of Kidneys From Extended-Criteria Donors. Am J Transplant. 2016;16(7):2181–2186. doi: 10.1111/ajt.13698 [DOI] [PubMed] [Google Scholar]
- 44.Scian MJ, Maluf DG, Archer KJ, et al. Identification of biomarkers to assess organ quality and predict posttransplantation outcomes. Transplantation. 2012;94(8):851–858. doi: 10.1097/TP.0b013e318263702b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bontha SV, Maluf DG, Archer KJ, et al. Effects of DNA Methylation on Progression to Interstitial Fibrosis and Tubular Atrophy in Renal Allograft Biopsies: A Multi-Omics Approach. Am J Transplant. 2017;17(12):3060–3075. doi: 10.1111/ajt.14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kingsmore KM, Bachali P, Catalina MD, et al. Altered expression of genes controlling metabolism characterizes the tissue response to immune injury in lupus. Sci Rep. 2021;11(1):14789. Published 2021 Jul 20. doi: 10.1038/s41598-021-93034-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afshinnia F, Rajendiran TM, Soni T, et al. Impaired β-Oxidation and Altered Complex Lipid Fatty Acid Partitioning with Advancing CKD. J Am Soc Nephrol. 2018;29(1):295–306. doi: 10.1681/ASN.2017030350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hallan S, Afkarian M, Zelnick LR, et al. Metabolomics and Gene Expression Analysis Reveal Down-regulation of the Citric Acid (TCA) Cycle in Non-diabetic CKD Patients. EBioMedicine. 2017;26:68–77. doi: 10.1016/j.ebiom.2017.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma K, Karl B, Mathew AV, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24(11):1901–1912. doi: 10.1681/ASN.2013020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grayson PC, Eddy S, Taroni JN, et al. Metabolic pathways and immunometabolism in rare kidney diseases. Ann Rheum Dis. 2018;77(8):1226–1233. doi: 10.1136/annrheumdis-2017-212935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He JC, Chuang PY, Ma’ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81(1):22–39. doi: 10.1038/ki.2011.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, Hoffman MM. Machine Learning for Integrating Data in Biology and Medicine: Principles, Practice, and Opportunities. Inf Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14 [DOI] [PubMed] [Google Scholar]
- 54.Demšar J, Zupan B. Hands-on training about overfitting. PLoS Comput Biol. 2021;17(3):e1008671. Published 2021 Mar 4. doi: 10.1371/journal.pcbi.1008671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lever J, Krzywinski M, Altman N. Model selection and overfitting. Nat Methods. 2016;13(9):703–704. doi: 10.1038/nmeth.3968 [DOI] [Google Scholar]
- 56.Maggiore U, Leventhal J, Cravedi P. Rethinking clinical endpoints in kidney transplant trials. Curr Opin Organ Transplant. 2020;25(1):1–7. doi: 10.1097/MOT.0000000000000719 [DOI] [PubMed] [Google Scholar]
- 57.Baek CH, Kim H, Yang WS, Han DJ, Park SK. A postoperative 1-Year eGFR of More Than 45 ml/min May be the Cutoff Level for a Favorable Long-Term Prognosis in Renal Transplant Patients. Ann Transplant. 2016;21:439–447. Published 2016 Jul 15. doi: 10.12659/aot.897938 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Patient flow diagram. A total of 295 patients were enrolled from 4 transplant centers (n=195 training set, n=100 validation set). Purple boxes represent exclusions. 21 patients were excluded from the training set due to follow-up loss, death with graft function, and microarray quality control criteria. 4 patients were excluded from the validation set due to low RNA integrity. The remaining 270 patients were included in the final training (n=174) and validation (n=96) sets. QC: Quality control; RIN: RNA integrity number.
Figure S3. Kaplan-Meier estimates for time until graft failure or death showing graft/patient survival after 24-months, separated by 24-month graft function group with log-rank test comparing the two groups. Only patients who were alive at 24-months were included in the analyses, with 24-months as time-zero. NA: not available.
Figure S4. Bar chart visualizing the top enriched cell-types for the upregulated DEGs (in low-functioning kidneys) and their associated q-values.
Figure S2: A. Spaghetti plot separated by high and low graft function group at 24 months with lowess smooths overlaid. Smoothed eGFR post-transplant (black line) and fitted linear mixed effects model (white dotted line) with equation. B. Mean eGFR (corresponding to black line) and standard deviation at each timepoint separated by high and low 24-month graft function.
Figure S5. Downregulated protein-protein interaction (PPI) networks encoded by differentially expressed genes (represented by nodes). Lines represent interaction relationships between nodes. Color-coded clusters were selected from PPI network using Molecular Complex Detection (MCODE). Downregulated pretransplant biological pathways associated with each MCODE subnetwork and adjusted p-values are listed.
Figure S6. Upregulated protein-protein interaction (PPI) networks encoded by differentially expressed genes (represented by nodes). Lines represent interaction relationships between nodes. Color-coded clusters were selected from PPI network using Molecular Complex Detection (MCODE). Upregulated pretransplant biological pathways associated with each MCODE subnetwork and adjusted p-values are listed.
Table S1. List of differentially expressed probe sets (n=699) with gene symbols listed in order of descending fold change (FC) (472 upregulated and 227 downregulated). Probe sets map to 595 unique genes. Unmapped probe sets (n=6) labeled as “N/A.” False discovery rate (FDR) <0.05.
Table S2. Regression coefficients, confidence intervals, and p-values from donor characteristics (DC) logistic regression model.
Table S3. Gene expression assays used for qPCR validation.
Data Availability Statement
Raw data from all Affymetrix GeneChip microarrays is available in the Gene Expression Omnibus public repository (GSE147451) which can be viewed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147451.


