Abstract
The promyelocytic leukemia zinc finger transcription factor (PLZF) is essential for nearly all of the unique, innate-like functions and characteristics of natural killer T cells (NKT cells). It is not known, however, if the activity of PLZF is regulated by other factors. Here we show that the function of PLZF is completely dependent on the transcription factor Ying Yang 1 (YY1). Mouse NKT cells expressing wild type levels of PLZF, but deficient for YY1 had developmental defects, lost their characteristic “pre-formed” mRNA for cytokines and failed to produce cytokine protein upon primary activation. Immunoprecipitation experiments showed that YY1 and PLZF were co-associated. Taken together these biochemical and genetic data show that the broadly expressed transcription factor, YY1, is required for the cell specific “master regulator” functions of PLZF.
Introduction
Classic adaptive immunity requires antigen specific activation of T or B cells, followed by cell differentiation into discrete effector subtypes. Only after secondary stimulation are the effector functions and cytokines elaborated, enabling these lymphocytes to fully participate in the immune response. Therefore, adaptive immune responses are rather slow to develop and somewhat limited in terms of function. Natural Killer T cells (NKT cells), a unique subset of α T cells, are, on the other hand, poised for immediate response to activation (1). Minutes after activation, NKT cells produce large amounts of a wide array of cytokines. The breadth of the response allows NKT cells to participate in diverse responses including immunity to pathogens, protection from autoimmunity, tumor immunity and regulation of obesity-induced inflammation, among others (2). Indeed, triggering of NKT cells impacts the activation status of nearly every other cell type of the immune system. The control of NKT cells clearly straddles a very fine line between tolerance and fulminant response.
The BTB-ZF transcription factor PLZF (Zbtb16) is the signature regulator of the functional and phenotypic dimorphism between NKT cells and conventional T cells. PLZF deficient NKT cells lose their normal activated phenotype and do not produce cytokines upon primary activation (3, 4). Ectopic expression of PLZF in conventional T cells, on the other hand, results in the acquisition of innate T cell-like characteristics such as an activated phenotype and the rapid secretion of Th1 and Th2 cytokines in response to primary activation (5). Interestingly, although PLZF is broadly required for all innate-like effector functions, NKT cells are stratified into distinct effector subgroups. PLZF activity, therefore, must be modified to direct its subgroup specific effects. Activation of T cells via the TCR, TCR plus SLAM signaling or even by Type 1 inflammation induced by bacterial infection does not induce detectable levels of PLZF expression in non-innate T cells (6). Suppression of PLZF expression in conventional T cells occurs early in T cell development and appears to be stably maintained (6). Although initially thought to be expressed continuously in all NKT cells, it recently was reported that adipose tissue resident NKT cells (arNKT cells) express little or no PLZF (7). arNKT cells appear to be a distinct lineage that is selected in the thymus (8). Interestingly, the lack of PLZF expression correlates with the acquisition of regulatory functions including an increased capacity to produce IL-10 (7).
Interest in PLZF has continued to increase since it is now known that it also controls the development of innate lymphoid cells (9), MAIT cells (10) and human NK cells (11). Indeed, the interest in the BTB-ZF family of transcription factors, in general, has grown enormously over the last few years as it has been shown that they are often the “toggle switch” that controls key inflection points during development, such as the commitment of T versus B cells or CD4 versus CD8 T cells (12).
Here, we show that major functions of PLZF are completely dependent upon the expression of the transcription factor, Yin Yang 1 (YY1) in NKT cells. YY1 belongs to the polycomb protein family, which is a subfamily of zinc-finger transcription factors with four typical zinc-finger DNA binding motifs (13). YY1 has been shown to be essential in diverse biological processes such as hematopoiesis (14), cancer development and progression (15, 16), and heart development (17). YY1 modulates chromatin structure and thus gene expression by mediating the formation of enhancer-promoter loops and by recruiting HAT and HDAC proteins to specific loci (18). Conditional deletion of YY1 at the DN stage results in a major block in thymocyte development (19). Interestingly, however, the loss of YY1 in thymocytes is compensated for by simultaneous deletion of p53 (19). Therefore, despite the roles reported for YY1 in some mature T cell lineages (20, 21), the function of YY1 during the complex process of T cell development appears only to be the control of p53’s apoptotic activity.
We show that in the absence of YY1, NKT cells acquired the typical activated/memory phenotype associated with this T cell lineage, but failed to undergo normal expansion. Furthermore, distinct from conventional thymocytes, development of YY1 deficient NKT cells was not rescued by deletion of p53. YY1 deficient NKT cells, however, had a near complete defect in their ability to produce cytokines. Remarkably, the profound failure of development and acquisition of effector functions occurred despite normal PLZF expression levels. Interestingly, we show the two proteins are in complex, raising the possibility that YY1 may be a cofactor necessary for the function of PLZF. Overall, our data show that the broadly expressed, multifaceted YY1 transcription factor plays a specific role in NKT cells, which is to enable the function of the “master regulator” functions of PLZF for the control of the development and effector functions of NKT cells.
Experimental Procedures
Animals
YY1 flx.flx mice were obtained from the Jackson Laboratory (Bar Harbor, ME) (22) and, also, from a colleague (Dr. Michael Verzi) at Rutgers University. CD4-cre mice and p53 flx.flx mice were obtained from Jackson (23, 24). PLZF-Cre mice were developed in our lab as reported (7). PLZF knockout mice were reported previously and were fully backcrossed to C57Bl/6 (4) Lck.PLZF transgenic mice were reported previously (5). All mouse strains were bred and maintained in the animal facility of the Child Health Institute of New Jersey. All experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee of the Child Health Institute of New Jersey. Animal Care and experimental procedures were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Child Health Institute of New Jersey and the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Preparation of Single Cell Suspensions from Lymphoid and Non-lymphoid Tissues
Whole tissues were dissociated between glass slides and filtered through a 40-μm mesh. Spleen, lymph node and thymus samples were treated with Red Blood Cell Lysing Buffer (Sigma). Cells were then counted on a hemacytometer. Liver homogenate was suspended in Percoll (Sigma) to a final concentration of 30% Percoll. Liver homogenates were then centrifuged for 20 min at 2,000 r.p.m. in a Sorvall RTH-750 rotor. Lymphocytes were then collected from the buffy layer, washed once with FACS buffer, treated with Red Blood Cell Lysing Buffer (Sigma) and counted on a hemacytometer.
Flow cytometry, antibodies, automacs enrichment, and cell sorting
Single cell suspensions were blocked in FACS buffer with 2% normal mouse serum, 0.1% anti-Fcγ antibody, and 0.1ug/ml streptavidin for 15 minutes at 4C followed by staining with primary antibodies for 20 minutes at 4C. Intracellular staining for transcription factors and cytokines was accomplished using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). All intracellular staining was performed at room temperature. The following antibodies were used in this study: anti-CD3 (500A2), Anti-CD4 (RM4–5), anti-CD8α (53–6.7), anti–IL-17A (Ebio17B7), anti-CD19 (1D3), anti-CD24 (M1/69), anti-CD44 (IM7), CD45.2 (104), anti-CD69 (H1.2F3), anti-IFN-γ(XMG1.2), anti-IL4 (11B11), anti-MHCII (212.A1), anti-NK1.1 (PK136), anti-PLZF (Mags.21F7), anti-RORγT (B2D), anti-T-bet (eBio4B10), anti-YY1 (sc-7341), anti-TCRvB7 (TR310), anti-TCRvB8.1/8.2 (MR5–2). The PBS57-loaded CD1d-tetramer was provided by the National Institutes of Health tetramer core facility. Dead cells were excluded when possible, by DAPI staining and doublet events were eliminated by comparing FSC-W to FSC-H and SSC-W to SSC-H. Events were acquired on a LSRII cytometer (BD Biosciences, San Jose, CA), and the data were analyzed with the FlowJo software (TreeStar, Ashland, OR).
NKT cell enrichment was performed using an AutoMACS (Miltenyi Biotec). Cells were stained with the PE-labeled CD1d tetramer, incubated with anti-PE microbeads (Miltenyi Biotec), and separated on the AutoMacs (Miltenyi Biotec). Samples were at least 80% pure following enrichment.
Cell sorting was performed at the Flow Cytometry/Cell Sorting & Confocal Microscopy Core Facility at Rutgers EOSHI or at the Cancer Institute of NJ.
Co-IP and Western blot: Lck.PLZF transgenic mice
30 million thymocytes were lysed in 1 mL of hypotonic lysis buffer (0.05% NP-40, 10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 5 mM EDTA), and complete protease inhibitor cocktail (Roche), and incubated on ice for 30 min. The cytosolic fraction was discarded. Cell nuclei were incubated on ice for 1 hr in a nuclear digestion buffer (20 mM HEPES, 300 mM NaCl, 20 mM KCl), supplemented with an EDTA-free complete protease inhibitor cocktail (Roche). Following lysis, nuclear lysates were sonicated for 3 seconds at 30% amplitude using a Branson Digital Sonifier. Nuclear lysates were combined with 1ug of antibody, either anti-PLZF(D9) or anti-YY1(sc-7341), and 50 uL of Protein G Agarose Beads (Sigma). Samples were incubated for 2 hr with rotation at 4°C. Beads were washed (5x) with ice-cold PBS. Bound proteins were eluted by boiling for 5 min in reducing sample buffer. The eluted samples were loaded on Criterion pre-cast TGX gels (Criterion) and separated. Proteins were transferred to PVDF Transfer Membranes (Immobilon). Primary antibodies used for the Western Blot were: anti-YY1 (sc1703) and anti-PLZF (D9).
Co-IP and Western blot: In vivo expanded NKT cells
Mice were injected intravenously via the retro orbital route with 60 ug of α-GalCer. 72 hours after injection, mice were sacrificed, and single cell suspensions were prepared from spleens. 30 million splenocytes were lysed in 1 mL of hypotonic lysis buffer (0.05% NP-40, 10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 5 mM EDTA), and complete protease inhibitor cocktail (Roche), and incubated on ice for 30 min. The cytosolic fraction was discarded. Cell nuclei were incubated on ice for 1 hr in a nuclear digestion buffer (20 mM HEPES, 300 mM NaCl, 20 mM KCl), supplemented with an EDTA-free complete protease inhibitor cocktail (Roche). Following lysis, nuclear lysates were sonicated for 3 seconds at 30% amplitude using a Branson Digital Sonifier. Nuclear lysates were pre-cleared by incubation with Protein G Agarose Beads (sigma) for 30 minutes at 4°C. Beads were discarded and the pre-cleared lysates were combined with Protein G Agarose Beads and 5ug of antibody, either anti-PLZF(D9), anti-YY1(sc-7341), or non-specific Immunoglobulin. Mock IPs were also conducted, in which anti-PLZF(D9) or anti-YY1(sc-7341) antibodies were incubated with Protein G Agarose Beds in the absence of lysate. Samples were incubated for 2 hr with rotation at 4°C. Beads were washed (5x) with ice-cold PBS. Bound proteins were eluted by boiling for 5 min in reducing sample buffer. The eluted samples were loaded on Criterion pre-cast TGX gels (Criterion) and separated. Proteins were transferred to P Transfer Membranes (Immobilon). Primary antibodies used for the Western Blot were: anti-YY1 (sc1703) and anti-PLZF (D9).
In vivo activation
Mice were injected intravenously via the retro orbital route with 10 ug of the NKT cell specific agonist α-galactosylceramide (α-GalCer). 90 minutes after injection, splenocytes were harvested.
In vitro activation
NKT cells were incubated in vitro with 50ng/mL PMA (Sigma) and 500 ng/ML Ionomycin (Sigma) for 5 hours. After one hour in culture, Brefeldin A (Biolegend) was added to the media at a final concentration of 5 ug/mL.
RNA Isolation and Quantitative PCR Analysis
Sorted NKT cells were lysed in Trizol solution at a concentration of 1×106 cells per mL. RNA was isolated from the trizol solution using the Direct-zol RNA MicroPrep Kit (Zymo Research). Reverse transcription was performed using the GoScript Reverse Transcription System (Promega). The resultant cDNA was utilized to perform TaqMan (Life Technologies) based qPCR. Taqman Universal PCR Master Mix No AmpErase UNG (Life Technologies) was used. The following TaqMan probes were used: B2M (Mm00437762_m1), IL-4(Mm00445259_m1), IFN-γ (Mm01168134_m1. Samples were run on a QuantStudio6 Flex Real Time PCR System(Life Technology). Relative expression of IL-4 and IFN-γ was calculated by using the ΔΔCT method, using B2M as the internal housekeeping control gene.
General experiment design and statistical analysis
Data from at least three samples in three or more independent tests were collected as detailed in the figure legends. Statistical analysis was performed using Microsoft Excel or GraphPad Prism. All data was subjected to analysis with Mann Whitney U-Test or One-Way Anova.
Results
YY1 is required for normal development of NKT cells
Deletion of YY1 in NKT cells was induced by crossing mice with an allele of YY1 flanked by flox recombination sites (YY1 flx.flx) (22) with a mouse line that expresses the cre recombinase in PLZF-expressing cells (PLZF-Cre) (7, 25–27). PLZF expression is induced very early during NKT cell development, but is not expressed in conventional T cells (4, 6, 28). Consistent with this, we found that YY1 expression was ablated in NKT cells, but expressed in conventional, non-PLZF expressing thymocytes and T cells (Figure 1A). Deletion of the target gene in PLZF-Cre mice might also occur in undefined hematopoietic stem cells (25–27), however, if this occurred for YY1, these cells did not contribute to the general T cell population in a measurable way. A small subset of NKT cells in PLZF-Cre YY1 flx.flx mice continued to express YY1 indicating that the gene had not been deleted in those cells (Figure 1A).
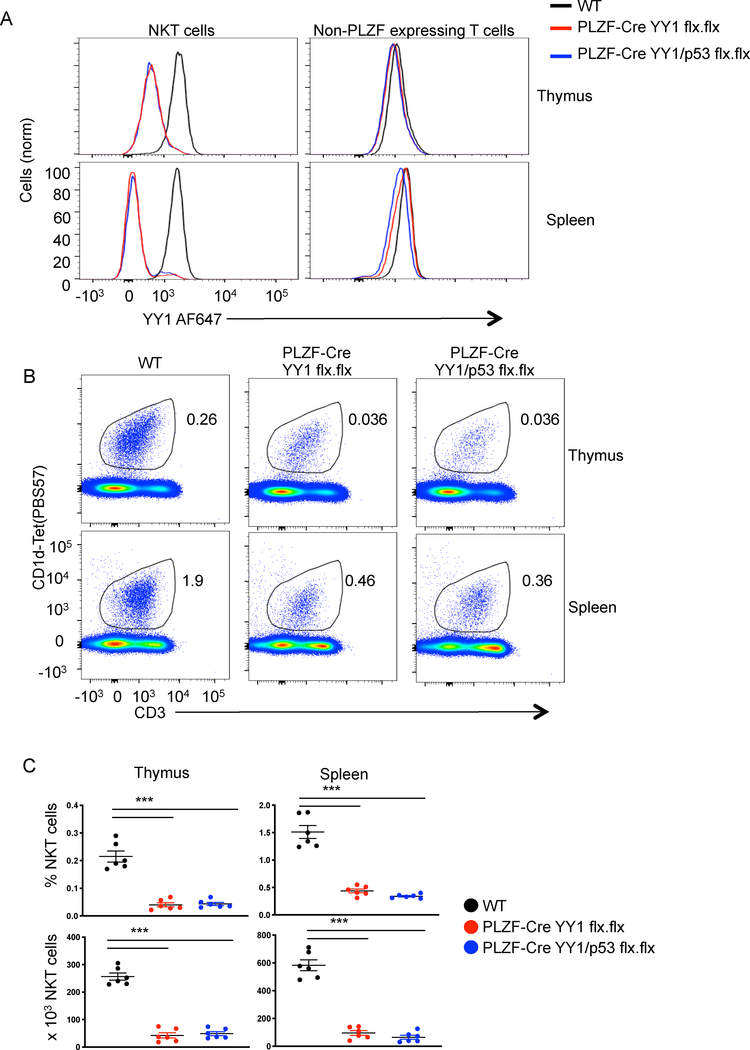
Figure 1. Cell specific deletion of YY1 causes a decrease in the number of NKT cells.
The indicated tissues were collected from C57BL/6 (WT) mice, PLZF-Cre YY1 flx.flx mice or PLZF-Cre YY1/p53 flx.flx mice and analyzed by FACS with the indicated antibodies. (A) YY1 expression was compared between WT NKT cells (MHCII−, CD3+, CD1d tet+, CD24−), PLZF-Cre YY1 flx.flx NKT cells, and PLZF-Cre YY1/p53 flx.flx NKT cells (left) isolated from the thymus (top) or spleen (bottom). YY1 expression was also compared between conventional T cells (MHCII−, CD3+, CD1dtet−, PLZF−) (right) isolated from WT mice, PLZF-Cre YY1 flx.flx mice, and PLZF-Cre YY1/p53 flx.flx mice. Data are representative of 5 or more independent experiments. (B, C) The frequency and absolute numbers of NKT cells (MHCII−, CD3+, CD1dtet+) in the thymus and spleen of WT mice, PLZF-Cre YY1 flx.flx mice, and PLZF-Cre YY1/p53 flx.flx mice were determined by FACS. Representative plots are shown in (B) and pooled data concerning the frequency (C, top) and absolute number (C, bottom) are in (C). FACS plots show representative results from at least 5 mice analyzed in at least 5 independent experiments (A, B). Graphs (C) show compiled data from 6 mice, examined in 5 independent experiments. The horizontal lines indicate the mean (±s.e.m.). ***P<.001 determined by One-Way Anova.
The loss of YY1 expression in NKT cells resulted in an ~85% reduction in both the percentage and absolute numbers of the cells in the thymus (Figures 1B, 1C). A similar reduction in the number of NKT cells was also found in peripheral tissues including the spleen, lymph nodes and liver (Figures 1B, 1C, S1A and S1B). In a previous study, we showed that deletion of YY1 with CD2-Cre reduced total thymic cellularity to ~1% of wild type, while deletion at the early double positive stage with Lck-Cre reduced cellularity to ~30% of WT (19). p53, a gene known to be inhibited by YY1 (29), was substantially upregulated (19). Normal thymocyte development was restored by a secondary deletion of p53, which showed that YY1 was necessary for protection against apoptosis (19). To test if the same mechanism was involved in NKT cell development, we generated mice with conditional “floxed” alleles of both YY1 and TP53 (p53). We then introduced the PLZF-Cre BAC transgene (PLZF-Cre YY1/p53 flx.flx). This approach induced simultaneous deletion of both genes soon after positive selection of NKT cells. In contrast to what was reported for conventional T cell development, deletion of p53 in YY1 deficient NKT cells did not restore NKT cell development (Figure 1B). Both the percentage and total number of NKT cells was not restored in the thymus as compared to PLZF-Cre YY1flx.flx mice or in the spleen. (Figure 1C). Therefore, the need for YY1 during NKT cell development is distinct from its control of apoptosis mediated by p53 during conventional T cell development.
Similar results were obtained with conditional YY1 and p53 alleles crossed to CD4-Cre (CD4-Cre YY1/p53 flx.flx), which induces deletion early during the double positive stage of development (Figure S2). Once again, loss of p53 expression did not restore the frequency or number of NKT cells in the thymus as compared to CD4-Cre YY1flx.flx mice (Figure S2B). Distinct from previous results with CD2-Cre and LCK-Cre mediated deletion of YY1, deletion with CD4-Cre did not result in a consistent reduction in the cellularity or development of thymocytes (Figure S2C). Simultaneous deletion of p53 and YY1, therefore, did not impact development of conventional thymocytes when deleted later in development (Figure S2D).
Overall, these data show that deletion of YY1 either early in thymocyte development (with CD4-Cre) or post positive selection (with PLZF-Cre) resulted in a similar disruption of NKT cell development. Most importantly, the PLZF-Cre data show that the impact of YY1 on NKT cell development is cell intrinsic and, since NKT cells can be detected with the CD1d-tetramer, the recombination event required for the production of the invariant TCR is intact in the absence of YY1. Finally, the mechanism for reduced NKT cells development is not apoptosis as a consequence of upregulation of p53.
NKT cell maturation is only partially compromised by loss of YY1
After positive selection, NKT cells progress through a developmental program during which they acquire an activated/memory phenotype. Following downregulation of CD24, discrete development steps are typically demarked first by expression of CD44 and than NK1.1. These steps include Stage 1 (CD44-, NK1.1-), upregulation of CD44 (Stage 2: CD44+, NK1.1-), followed by expression of NK1.1 (Stage 3: CD44+, NK1.1+) (Figure 2A). In the absence of PLZF, NKT cells fail to acquire the characteristic activated phenotype (3, 4). Most notably, PLZF deficient NKT cells do not fully upregulate CD44 or NK1.1, consistent with a Stage 1 phenotype (Figure 2A).
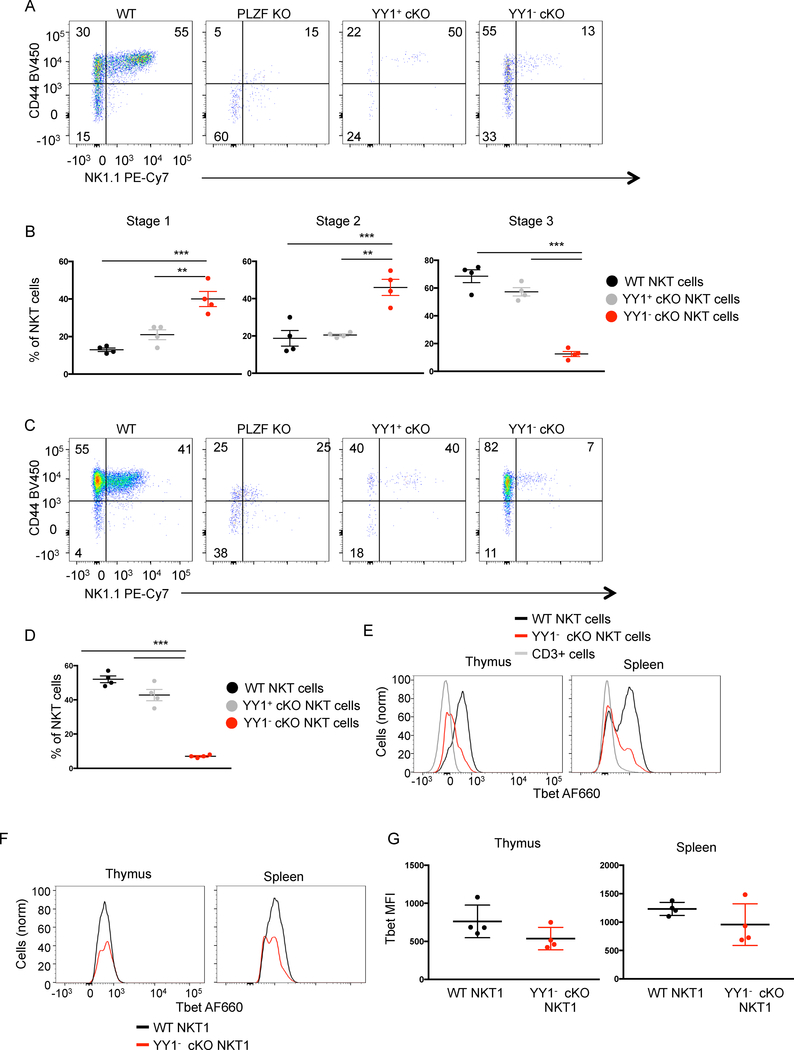
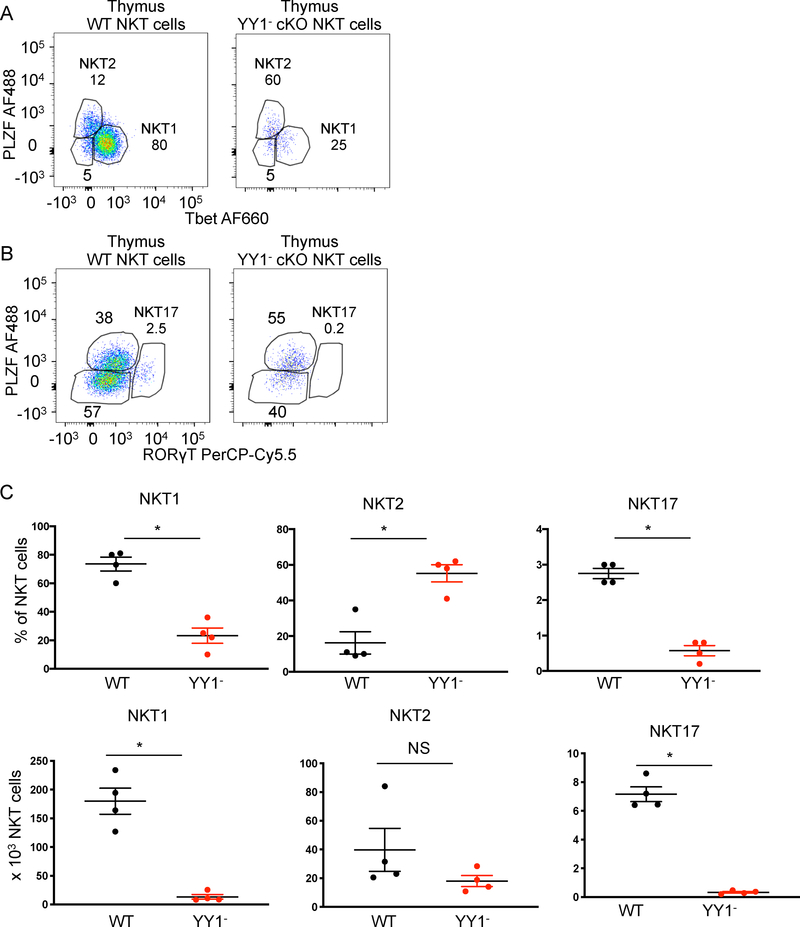
Figure 2. YY1 deficient NKT cells acquire an effector/memory phenotype but fail to reach stage 3 of development.
The indicated tissues were collected from C57BL/6 (WT) mice, PLZF KO mice or PLZF-Cre YY1 flx.flx mice and analyzed by FACS with the indicated antibodies. NKT cells (MHCII−, CD3+, CD1dtet+, CD24−) isolated from PLZF-Cre YY1 flx.flx mice were stained for YY1. The NKT cells from the PLZF-Cre YY1 flx.flx mice that stained YY1- are denoted as YY1− cKO NKT cells. The NKT cells from the PLZF-Cre YY1 flx.flx mice that stained YY1+ are denoted as YY1+ cKO NKT cells. (A, B) NKT cells isolated from the thymus of four week old WT mice, PLZF KO mice and PLZF-Cre YY1 flx.flx mice were stained for CD44 and NK1.1. Representative FACs plots are shown in (A). Pooled data concerning the distribution of NKT cells in each stage are shown in (B). The stages were defined as follows: Stage 1 (CD24-, CD44-, NK1.1-), Stage 2 (CD24-, CD44+, NK1.1-), and Stage 3 (CD24-, CD44+, NK1.1+). (C, D) Splenic NKT cells isolated from WT mice, PLZF-Cre YY1 flx.flx mice, and PLZF KO mice were stained for CD44 and NK1.1. A representative FACs plot is shown in (C). Pooled data concerning the frequency of splenic NK1.1+ NKT cells are depicted in (D). (E) Tbet expression was compared between WT NKT cells and YY1− cKO NKT cells from the thymus (left) and spleen (right). (F-G) Tbet expression levels were compared between Tbet+ WT NKT cells and Tbet+ YY1− cKO NKT cells. A representative histogram is shown in (F) and cumulative data from 4 mice are shown in (G). FACS plots show typical results from indicated tissues and graphs show compiled data from 4 mice, examined in 3 or more independent experiments. The horizontal lines indicate the mean (±s.e.m.). **P<0.01, ***P<.001 determined by One Way Anova (B, D) or Mann-Whitney U Test (G).
In contrast, to PLZF deficient NKT cells, the majority of YY1 deficient NKT cells from PLZF-Cre YY1 flx.flx (denoted as YY1−cKO) acquired the activated CD44 high phenotype (Figure 2A). NKT cells from PLZF-Cre YY1 flx.flx that continued to express YY1 due to failed deletion of the gene (denoted as YY1+cKO), similarly upregulated CD44, as did NKT cells from wild type mice. NK1.1 expression was, however, reduced in the YY1−cKO NKT cells. Therefore, based on upregulation of CD44, YY1− cKO NKT cell development progressed to a stage 2 like phenotype, but continued development to stage 3 was impeded (Figure 2B). The YY1 sufficient NKT cells (YY1+ cKO NKT cells) from the PLZF-Cre YY1 flx.flx mice displayed no developmental defects as compared to wild type NKT cells, indicating the YY1− cKO NKT cell developmental defect was cell intrinsic. NKT cells leave the thymus at stage 2 and upregulate NK1.1 expression in peripheral tissues such as the spleen (30). Here too, YY1− cKO NKT cells were found to upregulate CD44 similarly to YY1 sufficient NKT cells. Further differentiation, based on a failure to upregulate NK1.1 was, however, disrupted (Figure 2C, 2D).
One potential explanation for the failure to progress to the NK1.1+ stage of development would be the inability to upregulate Tbet (31). Although fewer YY1-cKO NKT cells express, Tbet, the transcription factor is clearly induced in the absence of YY1 both in thymus and spleen NKT cells (Fig. 2E). Comparison of only the Tbet expressing NKT cells shows that the level of expression is very similar (Fig. 2F), with no statistically significant difference between the mean fluorescent intensities (MFI) (Fig. 2G). Overall, these data indicate YY1 is necessary for some, but not all aspects of NKT cell development.
YY1 deficient NKT cells express wild type levels of PLZF
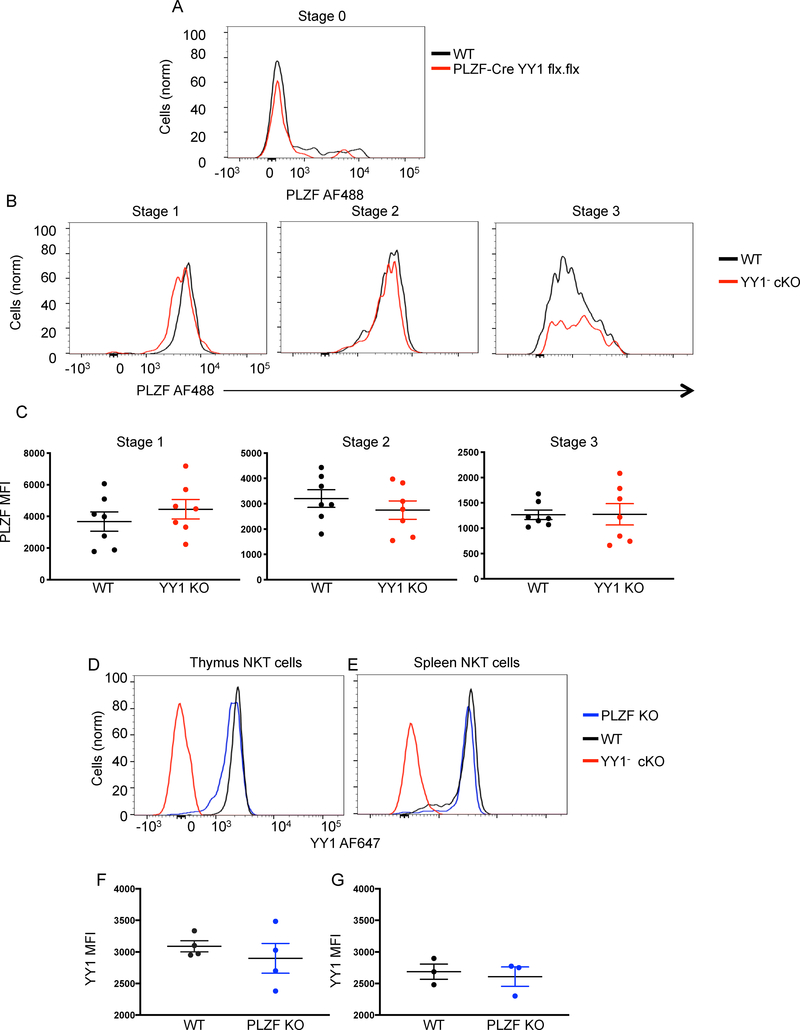
The failure of YY1 deficient NKT cells to develop normally could be due to a concurrent loss of PLZF expression (32). PLZF expression levels were compared by flow cytometry between wild type NKT cells and YY1− cKO NKT cells from PLZF-Cre YY1 flx.flx mice. Since PLZF expression levels change as NKT cells develop (4), we compared expression within the discrete stages defined by CD44 and NK1.1 expression. The level of PLZF expression was found to be statistically indistinguishable between wild type NKT cells and YY1− cKO NKT cells at all stages of development (Figure 3A–3C). These data showed that YY1 was not required for PLZF expression and, more importantly, that development of NKT cells was disrupted despite normal PLZF expression. PLZF expression in YY1 deficient NKT cells from CD4-Cre YY1 flx/flx mice was also found to be similar to wild type (Figure S3A). YY1 deficiency induced by CD4 did, however, result in an increase in CD24+ Stage 0 cells (Figure S3B), which, as expected, have not yet upregulated expression of PLZF. A previous report suggested that PLZF was not expressed in YY1 deficient NKT cells (32), however, the accumulation of PLZF negative Stage 0 cells was not appreciated.
Figure 3. PLZF and YY1 expression are independent.
The indicated tissues were collected from C57BL/6 (WT) mice, PLZF-Cre YY1 flx.flx mice or PLZF KO mice and analyzed by FACS with the indicated antibodies. (A) PLZF expression was determined by flow cytometry and compared between WT Stage 0 NKT cells (MHCII−, CD3+, CD1dtet+, CD24+) and Stage 0 NKT cells isolated from PLZF-cre YY1 flx.flx mice. (B, C) PLZF expression was determined by flow cytometry and compared between WT NKT cells and YY1− cKO at the same developmental stage. Representative histograms are shown in (B). Cumulative MFI data for PLZF are plotted in (C). NKT cell stages were defined as follows: Stage 1 (CD24−, CD44−, NK1.1−) (left), Stage 2 (CD24−, CD44+, NK1.1−) (middle), and Stage 3 (CD24−, CD44+, NK1.1+) (right). NKT cells isolated from the thymus (D, F) and spleen (E, G) of WT mice and PLZF KO mice were stained for YY1. Representative histograms of YY1 expression are shown in (D and E). Pooled data concerning the MFI of YY1 are shown in (F and G). Each symbol (C, F, G) represents an individual mouse. Data are from 3 or more independent experiments representing (C) 7, (F) 4, or (G) 3 biological replicates. The horizontal lines indicate the mean (±s.e.m.). P values determined by Mann Whitney U-Test (C, F, G).
Finally, to determine if PLZF impacted the expression of YY1, wild type NKT cells and PLZF KO NKT cells were compared by flow cytometry. Wild type and PLZF deficient NKT cells isolated from the thymus (Figure 3D) and the spleen (Figure 3E) expressed statistically indistinguishable levels of YY1 (Figure 3F, 3G).
Collectively these data indicate that these two transcription factors do not directly regulate each other’s protein expression levels. Furthermore, since PLZF is expressed at wild type levels, yet NKT cells fail to fully develop, these data raise the possibility that YY1 is either directly or indirectly required for the function of PLZF.
YY1 deficient NKT cells do not produce cytokines
Wild type and PLZF-cre YY1 flx.flx mice were intravenously injected with 10 μg of the NKT cell specific agonist ligand, α-GalCer. 90 minutes after injection, mice were sacrificed, spleens were removed and NKT cells were enriched by sorting with magnetic beads. The cells were made permeable and stained for IL-4 and IFN-γ. YY1 expressing NKT cells from the PLZF-cre YY1 flx.flx (YY1+cKO) mice produced both cytokines at levels that were indistinguishable from NKT cells from wild type mice (Figure 4A, 4B). In sharp contrast, almost none of the YY1− cKO NKT cells produced cytokine (Figure 4A, 4B).
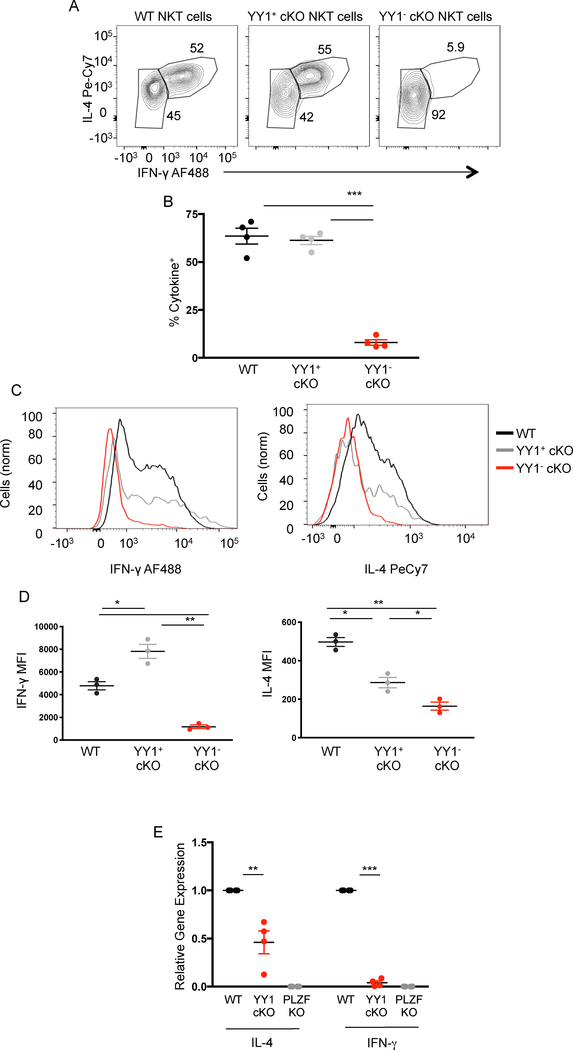
Figure 4. YY1 deficient NKT cells do not produce cytokines upon primary activation.
(A, B) C57BL/6 (WT) and PLZF-cre YY1 flx.flx (YY1 cKO mice) were injected I.V. with α-GalCer (10ug) suspended in saline. 90 minutes after injection, splenic NKT were harvested and stained for IL-4 and IFN-γ. Representative FACs plot depicting IL-4 and IFN-γ expression in WT NKT cells (MHCII−, CD3+, CD1dtet+, CD24−), YY1− cKO NKT cells, and YY1+ cKO NKT cells are depicted in (A) and cumulative data from 4 experiments are shown in (B). (C, D) NKT cells isolated from the spleens of WT and YY1 cKO mice were activated in vitro for 5 hours by culture with PMA (50ng/mL) and Ionomycin (500n/mL). Activated NKT cells were stained for IL-4, IFN-γ, and YY1. Representative histograms (C) and aggregate data (D), show the production of IFN-γ (left) and IL-4 (right) by WT NKT cells, YY1+ cKO NKT cells, and YY1− cKO NKT cells following in vitro activation. (E) NKT cells were FACs sorted from WT, PLZF KO, and YY1 cKO mice. RNA was prepared and cDNA was generated. qPCR was used to measure the presence of IL-4 and IFN-γ mRNA in the sorted NKT cell populations. Data are representative of 4 (B, E) or 3 (D) independent experiments and biological replicates. The horizontal lines indicate the mean (±s.e.m.). *P<0.05 **P<0.01, ***P<.001 determined by one-way ANOVA (B, D, E).
YY1 deficient NKT cells may have a deficiency in migration or localization that could impact their in vivo response to injected ligands. Alternatively, the loss of YY1 might impede TCR signaling. To bypass these concerns, we isolated NKT cells from wild type and PLZF-cre YY1 flx.flx mice and activated them in vitro by use of PMA/ionomycin. Similar to in vivo activation, YY1−cKO NKT cells did not produce either IFNγ or IL-4 (Figure 4C, 4D). Both wild type NKT cells and YY1+ cKO NKT cells produced both cytokines under the same activation conditions.
Unlike conventional T cells, the mRNA for IL-4 and other cytokines is present in NKT cells, even prior to activation (Figure 4E). This “pre-formed” mRNA likely plays a direct role in the rapid production of cytokines by NKT cells. PLZF deficient NKT cells do not have pre-formed cytokine mRNA, however, which presumably is a major reason for why PLZF deficient NKT cells cannot produce cytokine upon primary activation (4). The failure of YY1 deficient NKT cells to produce cytokines suggested that a similar mechanism might be involved. Total NKT cells (both YY1− cKO and YY1+ cKO) were sorted from wild type, PLZF deficient and PLZF-cre YY1 flx.flx mice. RNA was isolated and levels of specific cytokine message were measured by qPCR. As expected, mRNA for IL-4 and IFN-γ were readily detected in wild type NKT cells, but were absent from PLZF deficient NKT cells (Figure 4D). Despite the presence of some YY1 sufficient NKT cells, the NKT cells from the PLZF-cre YY1 flx.flx mice showed a substantial reduction in the level of these cytokine mRNAs. Therefore, despite wild type expression levels, in the absence of YY1, PLZF was unable to induce an open configuration at these gene loci, thereby preventing continuous mRNA transcription, which prevents the rapid release of cytokines.
Overall, these experiments show that despite expressing wild type levels of PLZF, YY1 deficient NKT cells could not produce their characteristic burst of cytokines in response to activation. These data imply that as in development of NKT cells, YY1 expression is necessary for the function of PLZF. Therefore, the lineage specific master regulator of NKT cell effector functions, PLZF, requires co-expression of the ubiquitously expressed YY1 transcription factor for its activity.
Subset development
The propensity of NKT cell subsets to be have particular effector functions can be defined by the expression of Tbet, PLZF and RORγt (33). Therefore, to further explore the impact of the loss of YY1, we determined if expression of these transcription factors was altered in YY1 deficient NKT cells. NKT1 cells express PLZF and Tbet, NKT2 cells express high levels of PLZF, but no Tbet and NKT17 cells express intermediate levels of PLZF and RORγt. Consistent with the known functions of these transcription factors, NKT1, NKT2 and NKT17 cells are skewed towards producing INFγ, IL-4 or IL-17, respectively (34).
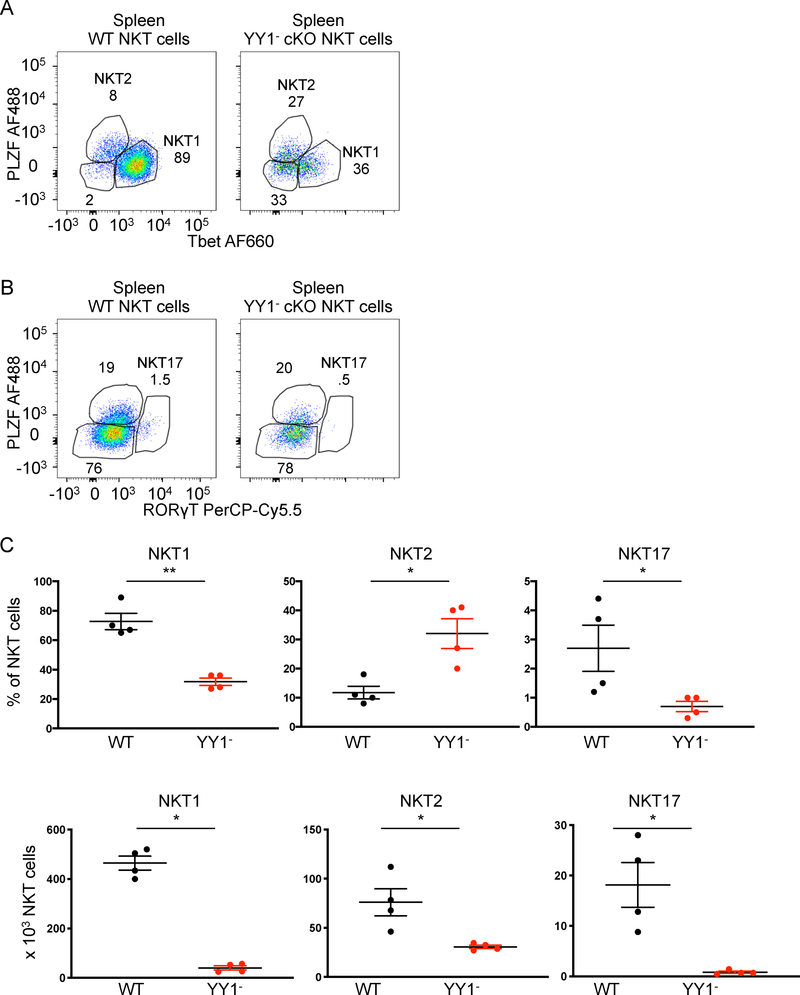
Most wild type thymic NKT cells belong to the NKT1 subpopulation (Figure 5A). In contrast, the majority of thymic YY1− cKO NKT cells had a NKT2 –like profile (Figure 5A). The NKT17 subpopulation was substantially diminished in the YY1− cKO NKT cell population compared to wild type (Figure 5B). While the percentage of NKT2 cells was increased, the absolute number of NKT2 cells was statistically unchanged (Figure 5C). Reflecting the thymus, spleen YY1−cKO cells also had an increased NKT2 population, but decreased frequencies of NKT1 and NKT17 cells (Figure 6A, 6B and 6C). The increased percentage of NKT2 cells did not, however, reflect increased numbers of the cells as compared to wild type (Figure 6C). Finally, we tested the possibility that changes in subset distribution might reflect altered TCR usage, which can impact the strength of signal received during selection (35–37). Approximately 70% of YY1+cKO and YY1-cKO NKT cells expressed Vβ8 (Figure S3C). The frequency of the less common Vβ7 expressing NKT cells was, however, altered approximately 2-fold (10% YY1+cKO versus 20% YY1-cKO). Vβ2, which typically represents less than 5% of total NKT cells, was notably changed (Figure S3C). Therefore, TCR usage was similar suggesting that the strength of signaling during development was not substantially altered. Overall, although substantially altered in frequency, based on transcription factor expression, all three NKT cell effector subtypes develop in the absence of YY1.
Figure 5. NKT cell subset development in the absence of YY1.
FACs analysis was used to determine the expression of key transcription factors associated with NKT cell effector functions in YY1 deficient thymus NKT cells. NKT cells isolated from the thymuses of C57BL/6 (WT) mice and PLZF-Cre YY1 flx.flx mice were stained for the transcription factors PLZF, Tbet and ROR γT. Gating schemes and representative FACS data for the (A) NKT1, NKT2, and (B) NKT17 populations are shown. (C) The frequency and absolute number of thymic WT NKT cells and thymic YY1− cKO NKT cells in these subpopulations are summarized. Data are from 4 or more independent experiments representing 4 biological replicates. The horizontal lines indicate the mean (±s.e.m.). *P<0.05determined by Mann Whitney U-Test (C).
Figure 6. NKT cell subsets in the spleen in the absence of YY1.
FACs analysis was used to determine the expression of key transcription factors associated with NKT cell effector functions in YY1 deficient spleen NKT cells. Splenic NKT cells isolated from WT and PLZF-Cre YY1 flx.flx mice were stained for PLZF, Tbet, and ROR γT (A, B). The frequency and absolute number of splenic WT NKT cells and YY1 deficient NKT cells present in the NKT1, NKT2, and NKT17 subpopulations are summarized in (C). Data are from 4 or more independent experiments representing 4 biological replicates. The horizontal lines indicate the mean (±s.e.m.). *P<0.05, **P<0.01, determined by Mann Whitney U-Test (C).
Interaction of YY1 with PLZF
To determine if YY1 might be directly influencing the function of PLZF, we tested if the two transcription factors were complexed by carrying out co-immunoprecipitation (co-IP) experiments (Figure 7). Reported mass spectrometry analysis suggested that such an association might occur (38). We used cells from mice carrying a transgene for PLZF under the control of the T cell specific LCK promoter (PLZF tg) in which all T cells express PLZF (5). We have previously shown that conventional T cells from these PLZF Tg mice take on innate-like T cell characteristics, suggesting that PLZF functions similarly in this system as it does in NKT cells. Importantly, the expression of PLZF in T cells from these mice was shown to be similar to endogenous PLZF expression in NKT cells (5). Therefore, this is ectopic expression, but not overexpression.
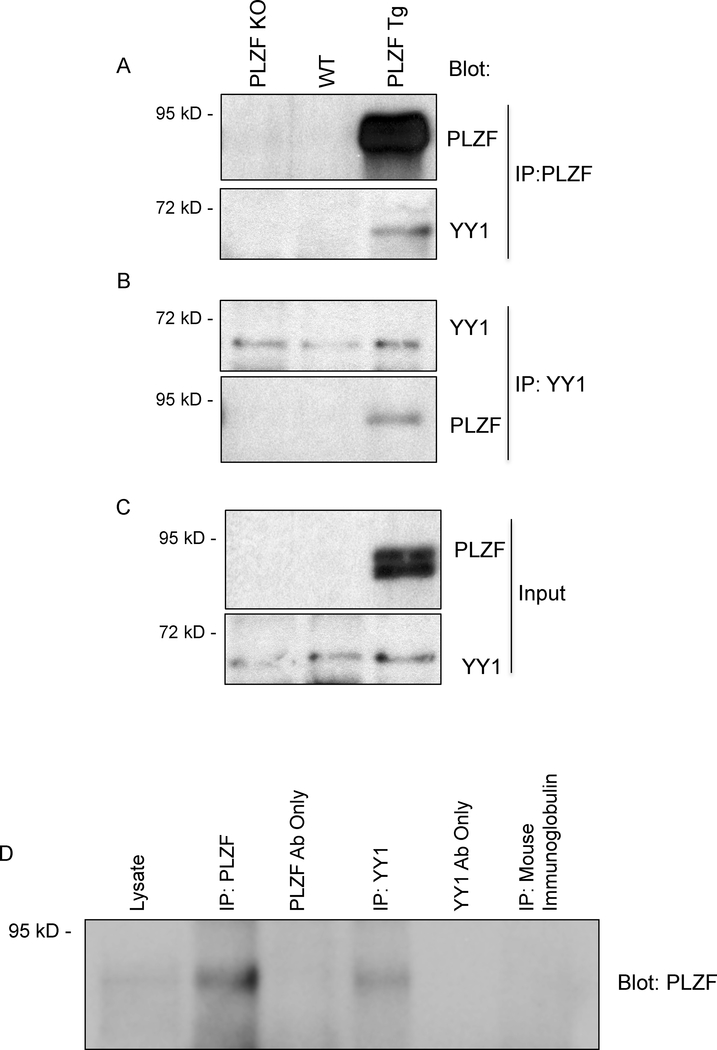
Figure 7. Co-Immunoprecipitation of PLZF and YY1 demonstrates a molecular interaction between the two transcription factors.
(A-C) Nuclear lysates were generated from PLZF deficient (PLZF KO), C57BL/6 (WT), and Lck.PLZF transgenic (PLZF TG) thymocytes. Lystates were immunoprecipitated (IP) with (A) PLZF or (B) YY1 specific antibodies followed by Western blot with antibodies against YY1 or PLZF to test for the co-association of the two transcription factors. (C) Western blot of thymocyte lysates (input) from the same lysastes as (A,B) were probed with the indicated antibody (anti-PLZF, top; anti-YY1, bottom). Data are representative of 4 independent experiments. (D) Nuclear lysates were generated from the spleens of α-GalCer treated mice. Total lysate (lane 1), lysate imunoprecipitated with a PLZF specific antibody (lane 2), YY1 specific antibody (lane 4), or non-specific immunoglobulin (lane 6). Mock PLZF IP (lane 3) and mock YY1 IP (lane 5) were loaded in the indicated lanes. Western blot of the samples were probed with anti-PLZF antibody. Data are representative of 4 independent experiments.
To determine if PLZF and YY1 interact, PLZF complexes were captured from nuclear lysates isolated from thymocytes from PLZF tg mice or from control wild type or PLZF deficient (PLZF KO) thymocytes followed by western blotting for YY1 (Figure 7A). The results showed that YY1 was specifically detected in association with PLZF only in PLZF Tg thymocytes and not in negative control PLZF deficient or wild type thymocytes. Importantly, these results were confirmed by capturing YY1 from the thymocytes followed by Western blotting for PLZF (Figure 7B). Western blotting of the input nuclear lysates showed that YY1 was present in thymocytes from all three mice, however, PLZF was only detected in thymocytes isolated from PLZF Tg mice (Figure 7C). As expected, PLZF was not detected in total thymocytes from wild type mice due the very low frequency of PLZF expressing T cells.
Next, we confirmed this result in primary NKT cells. The scarcity of NKT cells in wild type mice makes direct biochemical assays such as this difficult. To overcome this, we first expanded NKT cells in vivo by injection of the α-GalCer ligand. Three days post activation approximately 20 million NKT cells could be obtained from the spleen, representing an ~20 fold increase (Figure S4A). Importantly, NKT cells post activation continue to express both YY1 and PLZF (Figure S4B, S4C). Nuclear isolates from purified NKT cells were immunoprecipitated with anti-YY1 antibody followed by Western blot with anti-PLZF. Associated PLZF protein was specifically detected only in the lane with precipitated YY1 (Figure 7D). Overall, these data showed that YY1 is in complex with PLZF suggesting that YY1 might directly control the function of PLZF in NKT cells.
Discussion
T cell progenitors, derived from bone marrow hematopoietic stem cells, migrate to the thymus and are signaled to become early thymocyte progenitors (ETPs). These early thymocytes undergo a well-regulated, stepwise development involving TCR rearrangement, proliferation, culling via apoptosis, migration, and interaction with thymic stromal cells, which ultimately results in a self-referential, self-tolerant and functionally heterogeneous T cell repertoire (39–41). Disruption of any of these steps leads to aberrant or blocked T cell development. Nearly all the factors that control this process for conventional T cells are also essential for NKT cell development. Furthermore, disruption of positive or negative selection owing to defects in the TCR due to loss of TCRβ and/or TCRα chain rearrangement (42), or loss of key proteins required for TCR signaling pathways, such as Lck, ZAP-70 (43, 44), LAT (45), SLP-76 (46), PLCγ1 (47) or VAV1 (48), also block NKT cell development. Therefore, it is clear that much of NKT cell development is similar to the development of conventional T cells.
A number of genes, however, have been shown to be uniquely essential for NKT development. For example, loss of expression of either the tyrosine kinase, Fyn (49) or its adapter protein, SAP (50) cause the specific loss of NKT cells, while conventional T cell development is not perturbed. This is potentially due to a specific need for SLAM family member signaling (51, 52). Also, since NKT cells express a canonical TCRα chain, encoded by the Vα14 gene segment joined to the distally located Jα18 gene segment, disruption of the lifespan of the double positive thymocytes impacts NKT cell development, such as is the case in the absence of RORγt (53) or HEB (42).
Although multiple gene deficiencies are now known to impact NKT cell development, the BTB-ZF transcription factor, PLZF, appears to be the primary, if not only, known transcription factor essential for the development of the characteristic NKT innate-like effector functions and phenotype (3, 5). Importantly, among T cells, PLZF is only expressed in innate-like T cells (NKT cells, γδ NKT cells and MAIT cells) and is not induced in conventional T cells (6). The full mechanism by which PLZF endows the innate phenotype in T cells is, however, unknown.
We generated mice with a deletion of YY1 that was largely restricted to NKT cells. Deletion of YY1 resulted in a sharp reduction in the total number of NKT cells in the thymus, spleen, liver and, in contrast to PLZF deficiency, the lymph nodes. A similar, albeit moderately more severe phenotype is found using thymocyte specific expression of Cre controlled by the CD4 promoter (32), which deletes YY1 earlier in development.
Of most interest, however, is that YY1 deficient NKT cells lost the capacity to produce cytokines following primary activation. Loss of rapid cytokine production is likely caused by a failure to produce “pre-formed” mRNA, which also occurs in the absence of PLZF. These data suggest that YY1 is required to establish the genetic program that is ultimately controlled by PLZF. However, PLZF clearly can direct some gene expression activity in the absence of YY1. For example, upregulation of CD44, which is a characteristic of nearly all PLZF expressing lymphocytes (54, 55), still occurs. The CD44 gene has been shown by chromatin immunoprecipitation (CHIP) to be a direct target of PLZF (56). These data show that YY1 is only necessary for certain aspects of PLZF function and, therefore, it is possible that PLZF and YY1 control both complementary and independent aspects of NKT cell development and function.
Despite the loss of “pre-formed” cytokine mRNA, the complete disruption of cytokine production and the failure to accumulate in the thymus and liver, YY1 deficient NKT cells expressed wild type levels of PLZF. These data show that PLZF function, but not expression, is dependent on co-expression of YY1. YY1, however, clearly is not sufficient for NKT cell effector functions, since PLZF deficient NKT cells express wild type levels of the transcription factor. This co-dependency implies there is a direct, functional coordination. Our finding that the two proteins are in a complex suggests that YY1 might directly modulate the function of PLZF in NKT cells. Understanding this interplay will be an important step towards understanding the transcriptional programming of NKT cells.
This is the first report of the requirement for a co-expression 0f YY1 and a BTB-ZF transcription factor for cellular function. BTB-ZF transcription factors, for example, ThPOK, LRF, Bcl6, and MAZR regulate many of the key inflection points during the development of the immune system and, also, basic biological functions such as male germ cell homeostasis, axial-skeletal patterning, liver development, brain function, etc. The molecular mechanisms by which BTB-ZF transcription factors execute their functions remain poorly described. It is possible molecular coordination with YY1 may be necessary for the function of BTB-ZF transcription factors in these other settings, in addition to its function in NKT cells. YY1 itself has been shown to play a role in the development of the immune system. Perhaps some functions which are attributed to YY1 are actually a consequence of the gross dysfunction of BTB-ZF transcription factors in the absence of YY1.
In summary, we show that complete loss of YY1 in NKT cells leads to a loss of the expansion and function of NKT cells. These disrupted functions occur despite wild type expression levels of PLZF. Combined these data identify a new function for YY1, which is to mediate specific functions of PLZF.
Supplementary Material
Key Points.
YY1 deficient NKT cells display cell intrinsic developmental and functional defects.
YY1 deficient NKT cells maintain wildtype levels of PLZF expression.
YY1 and PLZF protein associate with one another.
Acknowledgements
We’d like to thank Joshua Vieth from the Child Health Institute of New Jersey for critical analysis of the data and/or help with experimental procedures and Arthur Roberts, from the CINJ Flow Cytometry Core Facility for cell sorting.
These studies were supported by NIH NIAID R01 AI083988 and AI059739 (to D.B.S.), the Robert Wood Johnson Foundation (grant #67038) to the Child Health Institute of New Jersey, NCI Comprehensive Core Grant (P30CA072720), the Aresty Undergraduate Research Fellowships (to P.W.D.) and the New Jersey Health Foundation Excellence in Research Award (to D.B.S.).
References
- 1.Bendelac A, Savage PB, and Teyton L 2007. The Biology of NKT Cells. Annu Rev Immunol 25: 297–336. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay K, Marrero I, and Kumar V 2016. NKT cell subsets as key participants in liver physiology and pathology. Cell Mol Immunol 13: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, and Sant’Angelo DB 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology 9: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, and Sant’Angelo DB 2010. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. Journal of immunology 184: 6746–6755. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Laouar A, Denzin LK, and Sant’Angelo DB 2015. Zbtb16 (PLZF) is stably suppressed and not inducible in non-innate T cells via T cell receptor-mediated signaling. Sci Rep 5: 12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, Godfrey DI, Leadbetter EA, Sant’Angelo DB, von Andrian U, and Brenner MB 2015. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nature immunology 16: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieth JA, Das J, Ranaivoson FM, Comoletti D, Denzin LK, and Sant’Angelo DB 2017. TCRalpha-TCRbeta pairing controls recognition of CD1d and directs the development of adipose NKT cells. Nat Immunol 18: 36–44. [DOI] [PubMed] [Google Scholar]
- 9.Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A 2014. A committed precursor to innate lymphoid cells. Nature 508: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, Goodnow CC, McCluskey J, Rossjohn J, Uldrich AP, Pellicci DG, and Godfrey DI 2015. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med 212: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, Yuan J, Wolchok JD, Horsthemke B, Wieczorek D, and Sant’Angelo DB 2011. Altered development of NKT cells, gammadelta T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS One 6: e24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu AM, and Sant’Angelo DB 2011. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. Journal of immunology 187: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Seto E, Chang LS, and Shenk T 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67: 377–388. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z, Hong CC, Kong G, Assumpcao A, Ong IM, Bresnick EH, Zhang J, and Pan X 2018. Polycomb Group Protein YY1 Is an Essential Regulator of Hematopoietic Stem Cell Quiescence. Cell Rep 22: 1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE, McCubrey JA, Canevari S, and Libra M 2009. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell cycle 8: 1367–1372. [DOI] [PubMed] [Google Scholar]
- 16.Patten DK, Corleone G, Gyorffy B, Perone Y, Slaven N, Barozzi I, Erdos E, Saiakhova A, Goddard K, Vingiani A, Shousha S, Pongor LS, Hadjiminas DJ, Schiavon G, Barry P, Palmieri C, Coombes RC, Scacheri P, Pruneri G, and Magnani L 2018. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morikawa Y, Leach J, and Martin JF 2013. Yin-Yang 1, a new player in early heart development. Circulation research 112: 876–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, and Young RA 2017. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171: 1573–1588 e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Foreman DP, Sant’Angelo DB, and Krangel MS 2016. Yin Yang 1 Promotes Thymocyte Survival by Downregulating p53. J Immunol 196: 2572–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, Lee W, Kim J, Souabni A, Busslinger M, and Lee GR 2013. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc Natl Acad Sci U S A 110: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang SS, Jang SW, Kim MK, Kim LK, Kim BS, Kim HS, Kim K, Lee W, Flavell RA, and Lee GR 2016. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat Commun 7: 10789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, and Shi Y 2006. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 26: 3565–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino S, Vooijs M, van Der Gulden H, Jonkers J, and Berns A 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14: 994–1004. [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, and Wilson CB 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774. [DOI] [PubMed] [Google Scholar]
- 25.Thapa P, Chen MW, McWilliams DC, Belmonte P, Constans M, Sant’Angelo DB, and Shapiro VS 2016. NKAP Regulates Invariant NKT Cell Proliferation and Differentiation into ROR-gammat-Expressing NKT17 Cells. J Immunol 196: 4987–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thapa P, Romero Arocha S, Chung JY, Sant’Angelo DB, and Shapiro VS 2017. Histone deacetylase 3 is required for iNKT cell development. Sci Rep 7: 5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin MN, Sultana DA, Lorentsen KJ, Cho JJ, Kirst ME, Brantly ML, Califano D, Sant’Angelo DB, and Avram D 2016. Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival. Proc Natl Acad Sci U S A 113: 7608–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosokawa H, Romero-Wolf M, Yui MA, Ungerback J, Quiloan MLG, Matsumoto M, Nakayama KI, Tanaka T, and Rothenberg EV 2018. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol 19: 1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, and Shi Y 2004. Yin Yang 1 is a negative regulator of p53. Cell 117: 859–872. [DOI] [PubMed] [Google Scholar]
- 30.Benlagha K, Kyin T, Beavis A, Teyton L, and Bendelac A 2002. A thymic precursor to the NK T cell lineage. Science 296: 553–555. [DOI] [PubMed] [Google Scholar]
- 31.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, and Glimcher LH 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20: 477–494. [DOI] [PubMed] [Google Scholar]
- 32.Ou X, Huo J, Huang Y, Li YF, Xu S, and Lam KP 2018. Transcription factor YY1 is essential for iNKT cell development. Cell Mol Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology 14: 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monteiro M, and Graca L 2014. iNKT cells: innate lymphocytes with a diverse response. Crit Rev Immunol 34: 81–90. [DOI] [PubMed] [Google Scholar]
- 35.Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, Dragone LL, Lefferts A, Halluszczak C, Riemondy K, Hesselberth JR, Rao A, O’Connor BP, Marrack P, Scott-Browne J, and Gapin L 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Commun 9: 2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M, Svensson MND, Venken K, Chawla A, Liang S, Engel I, Mydel P, Day J, Elewaut D, Bottini N, and Kronenberg M 2018. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat Commun 9: 2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathew R, Seiler MP, Scanlon ST, Mao AP, Constantinides MG, Bertozzi-Villa C, Singer JD, and Bendelac A 2012. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature 491: 618–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrie HT, and Zuniga-Pflucker JC 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol 25: 649–679. [DOI] [PubMed] [Google Scholar]
- 40.Sant’Angelo DB, Lucas B, Waterbury PG, Cohen B, Brabb T, Goverman J, Germain RN, and Janeway CA Jr. 1998. A molecular map of T cell development. Immunity 9: 179–186. [DOI] [PubMed] [Google Scholar]
- 41.Krangel MS 2007. T cell development: better living through chromatin. Nature immunology 8: 687–694. [DOI] [PubMed] [Google Scholar]
- 42.D’Cruz LM, Knell J, Fujimoto JK, and Goldrath AW 2010. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nature immunology 11: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negishi I, Motoyama N, Nakayama K, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan AC, and Loh DY 1995. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature 376: 435–438. [DOI] [PubMed] [Google Scholar]
- 44.Wiest DL, Ashe JM, Howcroft TK, Lee HM, Kemper DM, Negishi I, Singer DS, Singer A, and Abe R 1997. A spontaneously arising mutation in the DLAARN motif of murine ZAP-70 abrogates kinase activity and arrests thymocyte development. Immunity 6: 663–671. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, Long EO, Love PE, and Samelson LE 1999. Essential role of LAT in T cell development. Immunity 10: 323–332. [DOI] [PubMed] [Google Scholar]
- 46.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt FW, and Geha RS 1998. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell 94: 229–238. [DOI] [PubMed] [Google Scholar]
- 47.Fu G, Chen Y, Yu M, Podd A, Schuman J, He Y, Di L, Yassai M, Haribhai D, North PE, Gorski J, Williams CB, Wang D, and Wen R 2010. Phospholipase C{gamma}1 is essential for T cell development, activation, and tolerance. J Exp Med 207: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner M, Mee PJ, Walters AE, Quinn ME, Mellor AL, Zamoyska R, and Tybulewicz VL 1997. A requirement for the Rho-family GTP exchange factor Vav in positive and negative selection of thymocytes. Immunity 7: 451–460. [DOI] [PubMed] [Google Scholar]
- 49.Eberl G, Lowin-Kropf B, and MacDonald HR 1999. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol 163: 4091–4094. [PubMed] [Google Scholar]
- 50.Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, and Latour S 2005. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med 201: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borowski C, and Bendelac A 2005. Signaling for NKT cell development: the SAP-FynT connection. J Exp Med 201: 833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y, Zhong MC, Qian J, Calderon V, Cruz Tleugabulova M, Mallevaey T, and Veillette A 2019. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat Immunol 20: 447–457. [DOI] [PubMed] [Google Scholar]
- 53.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, and Littman DR 2005. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22: 705–716. [DOI] [PubMed] [Google Scholar]
- 54.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, and Sant’Angelo DB 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 9: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, and Sant’Angelo DB 2010. Development of promyelocytic zinc finger and ThPOK-expressing innate gammadelta T cells is controlled by strength of TCR signaling and Id3. J Immunol 184: 1268–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, and Bendelac A 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A 113: 7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.