Abstract
Basophils are innate immune cells associated with Type 2 immunity, allergic reactions and host defense against parasite infections. In this study, we show that the transcription factor PLZF, which is known for its essential role in the function and development of several innate lymphocyte subsets, is also important for the myeloid-derived basophil lineage. PLZF-deficient mice had decreased numbers of basophil progenitors in the bone marrow and mature basophils in multiple peripheral tissues. Functionally, PLZF-deficient basophils were less responsive to IgE activation and produced reduced amounts of IL-4. The altered function of basophils resulted in a blunted Th2 T cell response to a protein allergen. Additionally, PLZF deficient basophils had reduced expression of the IL-18 receptor, which impacted migration to lungs. PLZF, therefore, is a major player in controlling Type 2 immune responses mediated not only by innate lymphocytes but also by myeloid derived cells.
Introduction
Despite representing less than 1% of circulating leukocytes in the peripheral blood of mammals, basophils have been shown to play multiple roles in the immune response, including regulating innate and the adaptive immune responses, protecting against parasitic infection and contributing to immunological disorders such as allergy and autoimmunity (1–8). Activated basophils can produce a range of Th2 cytokines including IL-4, IL-6 and IL-13 that can help promote Th2 cell differentiation. Therefore, basophils impact immune responses such as the clearance of parasites, ectoparasites and the initiation of allergic responses(9) (10, 11).
Like natural killer T cells (NKT cells), basophils have the unusual feature of continuous transcription of the IL-4 gene (12–14). The continuous presence of “pre-formed” mRNA for IL-4 presumably plays a role in enabling basophils to rapidly release IL-4 after activation(15–17). Depletion of basophils or disrupting IL-4 production by basophils leads to impaired differentiation of T cells into the Th2 lineage (17) (15) and altered ILC2 responses (18). Basophils are reported to express MHCII as well as the co-stimulation molecules CD40, CD80 and CD86 and have been shown to interact with lymphocytes (19). Therefore, basophils might also influence lymphocyte activation via contact-dependent mechanism (20–22). Beyond activating ILC2s and T cells, basophils also express IL13, B cell-activating factor (BAFF), and a proliferation-inducing ligand (APRIL), which can lead to B cell activation and class-switch recombination to produce IgD, IgG, IgA and IgE (23–27).
Basophils primarily mature in the bone marrow before they enter the periphery where they have a life span of only ~60 hours (28). Basophils are currently reported to arise from the common myeloid progenitor (CMP), which gives rise to granulocyte/macrophage progenitors (GMPs). GMPs are reported to have the potential to differentiate into neutrophils, eosinophils, basophils and mast cells. Along the basophil/mast cell lineage, GMPs develop into the pre-basophil and mast cell progenitors (pre-BMPs) in the bone marrow or, alternatively, into basophil-mast cell progenitors (BMCPs) in the spleen (29), (30). Both pre-BMPs and BMCPs can differentiate into basophil progenitors (BaPs) and mast cell progenitors (MCPs), which ultimately differentiate into mature basophils and mast cells, respectively (31).
Studies have demonstrated that basophil development is dependent on the expression of CCATT enhancer-binding proteins C/EBPα and GATA2. GATA2 expression is induced by IRF8. The ordered expression of these two proteins determines whether GMPs develop into basophils, mast cells or eosinophils (32, 33). GMPs that first downregulate and then upregulate C/EBPα expression while remaining high for GATA2 expression will differentiate into basophil progenitors. In contrast, GMPs will differentiate into eosinophils if C/EBPα expression remains high throughout development, but into mast cell progenitors if C/EBPα remains low (8). The commitment to the mast cell lineage is also thought to rely on the transcription factor MITF (34, 35). C/EBPα and MITF suppress each other’s expression, which secures the basophil and mast cell lineages (29).
The BTB-ZF (broad-complex, tramtrack and bric-à-brac, zinc finger) genes are a family of transcription factors that are characterized by a N-terminal BTB domain that is involved in protein-protein interactions, and a domain with C-terminal Krüppel-type zinc fingers enabling binding of DNA (36–38). It is now appreciated that BTB-ZF transcription factors play key roles for the development and/or function of multiple immune cell lineages. For example, the BTB-ZF transcription factor, promyelocytic leukemia zinc finger (PLZF; zbtb16) is essential for the normal function of several types of innate lymphoid derived cells including αβ NKT and γδ NKT cells (39–42), mucosal associated invariant T cells (MAIT cells) (43) and type 2 innate lymphocytes (ILC2) (44–46). In the absence of PLZF, NKT cells do not acquire their characteristic activated phenotype and fail to produce cytokines upon primary activation (41, 42). Instead, these cells require secondary stimulation to be fully activated and carry out effector functions, making them similar to conventional T cells (41). In humans, in addition to NKT and MAIT cells, PLZF is expressed in a population of CD161hi CD4 and CD8 T cells, all γδ T cells, and all natural killer (NK) cells (47). Blood samples from a PLZF-deficient person showed that there was an impact on the frequency and/or phenotype of all of these lymphocyte populations (47).
Ectopic expression of PLZF spontaneously induces conventional T cells to be more sensitive to antigenic stimulation and able to produce pro-inflammatory cytokines such as IL-17, INF-γ and TNF upon primary activation (48–50). Tight regulation of endogenous PLZF expression would, therefore, appear important to prevent aberrant T cell responses that might lead to allergy or autoimmune diseases. Consistent with this, our studies showed that PLZF expression among lymphocytes is restricted to innate-like cells and that it cannot be induced in non-innate T cells (51). Strong TCR signaling in developing thymocytes or in mature T cells, for example, does not induce PLZF (51) and expression in non-innate lymphocytes was also not found in response to inflammatory conditions resulting from bacterial infection. Analysis of the chromatin structure of Zbtb16 (the gene encoding PLZF) using ChIP for H3K4me3 and H3K27me3 also suggests that PLZF expression is suppressed in Th1, Th2, Th17, Tregs and naïve CD4 T cells (51, 52). Repression of expression of PLZF was recently reported to be dependent on Bcl11b and occurs early during thymocyte development (53).
The specificity and tight control of PLZF expression in lymphocytes suggests that other immune cells expressing PLZF might also be dependent on the transcription factor for essential effector functions. To explore this possibility, we examined PLZF expression within the myeloid compartment. Basophils, we find, express PLZF both early in development and as mature cells. Genetic deletion of PLZF showed that the transcription factor plays a role during development and, most interestingly, for the full elaboration of basophil effector functions. PLZF deficient basophils fail to fully respond to a protein allergen, papain, resulting in blunted Th2 CD4 T cell responses. Therefore, PLZF is important for Type 2 responses by directly controlling the function of innate lymphocytes and, also, indirectly, through the control of basophil functions.
Materials and Methods
General experiment design and statistical analysis
This study was designed to study the role of the transcription factor PLZF in basophil development and function in Th2 immunity. Age-matched (8–12 week old) WT and PLZF-KO mice were used for comparisons in this study. Basophil frequency in different mouse tissues was analyzed. We used both in vivo and in vitro system to study basophil functions. Each experiment contained at least three biological replicates. Data are shown as the mean ± SEM. Statistical analyses were performed using GraphPad Prism 10 software. All samples were analyzed using unpaired, two-tailed Student t test. P values less than 0.05 were considered significant.
Mice
Rag1 KO, CD1d KO, IL-18R KO, PLZF-IRES-GFPCre, and Rosa26-tdTomato mice were purchased from Jackson Laboratories. The PEG reporter and PLZF-cre mouse lines were generated in our lab as previously described(51, 54). PLZF-deficient mice were previously described(55). All mouse strains were bred and maintained in the animal facility of the Child Health Institute of New Jersey. All experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee of the Child Health Institute of New Jersey. Animal care and experimental procedures were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Child Health Institute of New Jersey and the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Flow Cytometry
Cells were harvested from the tissues and pre-incubated with the anti-FcγR mAb and normal mouse serum to block nonspecific antibody binding for 15 min at 4C. Surface staining was performed in FACS buffer (PBS with 1% heat-inactivated FBS) for 25 min at 4C using the indicated surface Abs. Dead cells were excluded by DAPI staining and doublet events were eliminated by comparing the FCS-W and FCS-H to the SSC-W and SSC-H. IL-4 intracellular staining was performed using the BD Fixation/Permeabilization Solution Kit (BD Biosciences). Intracellular staining of PLZF was performed by fixing cells in 4% paraformaldehyde in PBS for 30 min at 4C. Cells were then permeabilized for 15 min at 4C in commercially-available Permeabilization Buffer (Invitrogen), and incubated with anti-PLZF antibody in permeabilization buffer for 1 hour at 4C. Samples were washed 3X with permeabilization buffer and 1X with FACS buffer prior to analysis. Events were acquired on an LSRII cytometer (BD Biosciences) and the data were analyzed with the FlowJo software.
Antibodies
The following Ab clones were used: anti-CD3 (500A2), anti-CD4 (RM45), anti-CD19 (1D3), anti-CD49b (DX5), anti-CD34 (RAM34), anti-CD117 (ACK2), anti-CD200R (OX110), anti-CD40L (MR1), anti-CD62L (MEL-14), anti-CCR2 (SA203G11), anti-CCR7 (4B12), anti-CCR9 (CW-1.2), anti-MHCII (212.A1), anti-ST2 (RMST2–2), IL-18Rα (P3TUNYA), anti-FcεRIα (MAR-1), anti-PLZF (Mags.21F7), anti-GATA3 (TWAJ), anti-IL-4 (11b11). Abs were used with different fluorochrome conjugations: FITC-, PE-, PerCP-Cy5.5-, PE-Cy7-, APC-, APC-Cy7-, AF700-, and Pacific Blue.
Quantitative RT-PCR
Total cellular RNA from cells was isolated by mirVana miRNA isolation kit (ThermoFisher). cDNA was generated with reverse transcription using oligo-dT, random primers and reverse transcriptase (Promega). Quantitative PCR of the cDNA was performed on QuantStudio 6 Flex Real-Time PCR System (ThermoFisher) and the following TaqMan gene expression primer sets (ThermoFisher): Hprt (Mm03024075_m1), zbtb16 (Mm01176868_m1). Gene expression was analyzed using Hprt as an endogenous control in each sample.
In vivo basophil activation
Mice were immunized subcutaneously in the rear footpad with 50 ug of papain (Calbiochem) dissolved in sterile PBS. As control, same amount of heat-inactivated papain was injected into the other side of the footpad. To heat-inactivate, papain was dissolved in PBS and placed in boiled water for 10 minutes. Mice were sacrificed either 3 days (for basophil staining) or 4 days (for GATA3 staining in T cells) after injection and analyzed by flow cytometry.
In vitro basophil activation
100 ul of peripheral blood was drawn from the mice and mixed with 100 ul of RPMI 1640. The cells were activated with different concentration of IgE or PMA (50 ng/ml)/Ionomycin (500ng/ml) for 3 hours total. Brefeldin A was added at 1 hour after the start of the activation. After activation, the cells were washed two times with PBS and spun at 500g for 5 minutes. Then the cells were treated with red blood cell lysis buffer to remove red blood cells. The cells were then blocked and stained with surface markers, followed by intracellular cytokine staining.
Results
Mapping PLZF expression during the development of myeloid lineage cells.
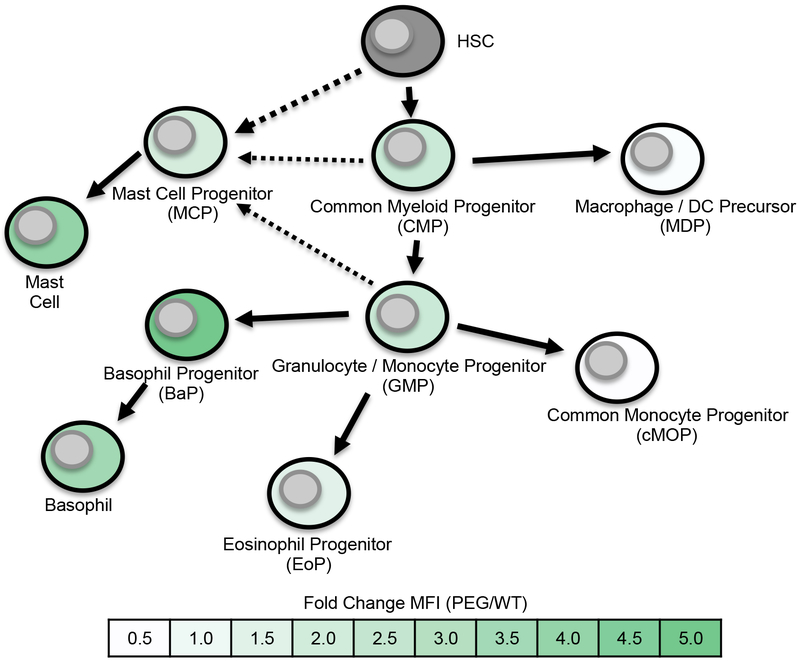
Using our previously described PLZF-eGFP (PEG) reporter mice (51, 54, 56), we evaluated PLZF expression during early myeloid lineage development. In contrast to PLZF expression in lymphocytes, which has been shown to be largely restricted to specific cell types (51), we found expression of PLZF in many myeloid precursor populations (Fig. 1). As indicated by fold change as compared to matched wild type cells (Fig. 1) and as shown in FACS histograms (Fig. S1), PLZF expression levels ranged substantially depending on the subset examined, which were identified as shown in Figs. S2, S3 and S4 (30, 57–60). Expression patterns of PLZF in the different populations were consistent from mouse to mouse (Fig. S1). Low levels of expression of the transcription factor, as defined by eGFP expression, were found in common myeloid progenitors (CMP), mast cell progenitors (MCP) and granulocyte/monocyte progenitors (GMP) (Fig. 1). Eosinophil progenitors (EoP) also expressed low levels of PLZF, however, macrophage/DC precursors (MDP) and common monocyte precursors (cMOP) were negative. The most pronounced expression of PLZF was in mast cells and basophils, as well as the immediate precursors of basophils, BaP. Basophil progenitors (BaP) expressed higher levels of PLZF as compared to mature basophils. Downregulation of PLZF as basophils matured is similar to what has previously been observed during NKT cell development (41).
Fig. 1. PLZF expression during myeloid lineage cell development.
Myeloid lineage cells were stained and identified in bone marrow from PLZF-eGFP reporter mice (PEG) as shown in Supplementary Figs. 2–3. PLZF expression, as defined by eGFP expression, was determined by comparison of the identical population obtained from wild type mice (Fig. S1). The progression of development was based on publications, as indicated in the text. Color-coding in the figure represents mean fluorescence intensity (MFI) values for each subset of cells, based on eGFP expression (MFI of eGFP signal divided by the background fluorescence of the identical cell population from a matched control analyzed in the same experiment). Fold change of MFI key is shown in figure.
We chose to focus on a potential role for PLZF in basophils since these cells are known to rapidly express IL-4 similarly to NKT cells. The burst of IL-4 produced by NKT cells shortly after activation is completely dependent upon PLZF (41, 42). We found that PLZF was expressed in all basophils in the spleen, peripheral blood, bone marrow and lung (Fig. 2A). Expression levels were, however, lower than that found in NKT cells (Fig. 2B). PLZF expression was distinctly higher in BaPs in the bone as compared to mature bone marrow basophils (Fig. 2C). Importantly, intracellular staining with anti-PLZF antibody verified protein expression in basophils (Fig. 2D). The increased concentrations of paraformaldehyde that we found were necessary for reliable retention of proteins in the permeabilized basophils, however, resulted in reduced signal intensity both in the basophils and NKT cells that were stained via the same protocol. Therefore, the conclusions concerning the absolute amounts of PLZF protein in the cells should not be drawn.
Fig. 2. PLZF is expressed in basophils.
(A) eGFP expression in basophils from spleen, bone marrow, blood and lung of PEG mice. Basophils were identified as DAPI−CD3−CD19−FcεRI+CD49b+ cells. (B) Comparison of eGFP expression in basophils, and NKT cells. Wild type basophils are shown as a fluorescence control. (C) Intracellular staining of PLZF in splenic basophils, with CD8 T cells and NKT cells as negative and positive controls, respectively. Quantification of PLZF MFI (fold change over CD8 T cells) is shown (N=3 biological replicates from 2 individual experiments). (D) eGFP expression in basophil progenitors (BaPs) in the bone marrow in PEG mice. BaPs were identified as DAPI−CD3−CD19−CD34+FcεRI+CD49b+cKit− cells. (E) ZsGreen and tdTomato expression in basophils from the bone marrow and spleen in PCre x ZsGreen and PLZF-IRES-GFPCre x tdTomato mice, respectively. Representative FACS plots from 1 of 3 independent experiments are shown.
To further confirm PLZF expression in basophils, we carried out “fate-mapping” experiments. We used both our PLZF-Cre BAC transgenic mice (Pcre), as well as PLZF-IRES-GFPCre “knock-in” mice (44). These two strains were both crossed to mice that have an allele of Rosa 26 modified with a gene that encodes a fluorescent protein, either ZsGreen or tdTomato, that is preceded by a loxP flanked STOP cassette. In these mice, Cre expression from the PLZF BAC or from the knock-in allele, even if it is transient, will permanently label cells with the fluorescent protein. Both mouse lines showed uniform expression of the fluorescent protein in basophils from the bone marrow and spleen (Fig. 2E). It should be noted that direct expression of GFP in basophils from the PLZF-IRES-GFP/Cre “knock-in” construct was not detectable. This presumably was due either to lower expression levels of GFP from the knock-in construct as compared to the BAC transgene or, possibly, alterations in expression resulting from the insertion of the IRES-GFP/Cre cassette in the 3’ UTR of the zbtb16 gene. Indeed, microRNA target sites within the 3’ UTR have been shown to impact PLZF expression (61). Importantly, we found that use of the floxed-stop ROSA26 ZsGreen allele proved to give results that are more consistent with known PLZF expression patterns, as compared to the previously reported floxed-stop ROSA26 tdTomato allele(44, 62). The mice carrying the ZsGreen version of the allele showed that, as expected, nearly 100% of NKT cells in the thymus and spleen were labeled, but that the previously reported and unexpected labeling of thymocytes, B cells and T cells was eliminated or greatly reduced (Fig. S4). For example, B cells and DP thymocytes were essentially negative for ZsGreen expression, indicating that “leaky” expression of the PLZF-Cre in hematopoietic stem cells, as previously suggested, was unlikely. Overall, these data demonstrate that PLZF is expressed both in basophil progenitors in the bone marrow and in mature basophils in multiple mouse tissues.
PLZF-deficient mice have reduced numbers of basophils.
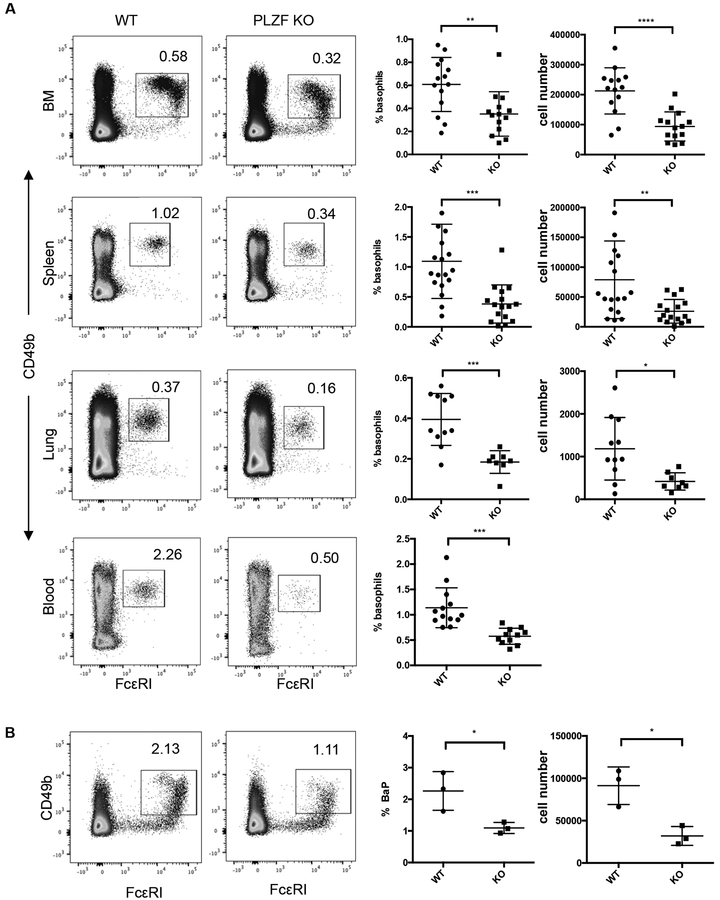
The expression of PLZF in basophil progenitors and mature basophils raised the possibility that, like for NKT cells, basophils require the transcription factor for development. To study this, we compared the frequency and absolute numbers of basophils in C57Bl/6 and age matched PLZF-deficient mice. FACS analysis was done by first excluding lymphocytes by use of anti-CD3 and anti-CD19 and then identifying basophils by expression of CD49b and FcεRI (Fig. 3A). PLZF deficiency resulted in a ~50–60% percent decrease of basophils in multiple tissues, including the bone marrow, spleen, lung and peripheral blood (Fig. 3A). The absolute numbers of basophils were reduced by a similar degree. Consistent with the reduced number of basophils in all tissues, the basophil progenitor population in the bone marrow showed an approximate 70% decrease as compared to age matched wild type mice (Fig. 3B). Therefore, similar to NKT cells, albeit to a lesser degree, PLZF-deficient mice have reduced frequencies of basophils. Unlike NKT cells, PLZF deficiency results in the reduction of basophils is found in all tissues. Combined these data suggest that PLZF has a direct impact on basophil development.
Fig. 3. Reduced numbers of basophils PLZF deficient mice.
(A) Basophil numbers in the bone marrow, spleen, lung and blood of PLZF-deficient and WT mice. Cells shown are DAPI−CD3−CD19−CD34− population and basophils were identified as CD49b+FCεR1+. Numbers indicate the percentage of events within the gated area. Percentage and absolute number of basophils are shown in scatter plot graphs. Each symbol indicates a single mouse. N=14 with ~5 independent experiments for each tissue. (B) Basophil progenitor numbers in the bone marrow of PLZF-deficient and WT mice. Cells shown are DAPI−CD3−CD19−CD34+ population. Numbers in dot plots show the percentage of events in the gated region. Percentage and absolute number of basophil precursors are shown in scatter plot graphs. Each symbol indicates a single mouse. N=3 with 3 independent experiments. The horizontal lines indicate the mean (±s.e.m.). *P<0.05, **P<0.01, ***P<.001 determined by Student’s t-test.
Papain-induced basophil migration is impaired in PLZF-deficient mice
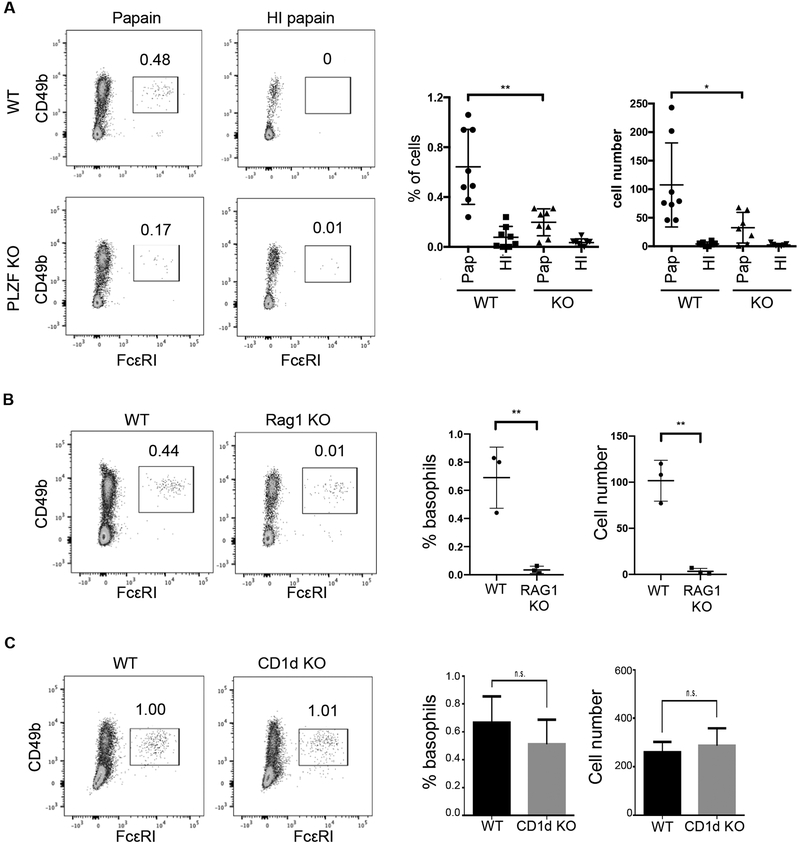
Proteolytic activities that are associated with some allergens and parasite secretions are thought to be directly involved in the activation of basophils (63–66). An excellent model of this type of immune response is the subcutaneous injection of the cysteine protease, papain. We injected 50μg of papain into one rear footpad of mice and the same amount of heat-inactivated (HI) papain into the other footpad as negative control. Three days after injection, the draining lymph nodes were removed and analyzed by FACS. Papain injection resulted in a basophil response that comprised approximately 0.6% of the CD3−, CD19− cells in the draining lymph nodes (Fig. 4A, top). In contrast, very few basophils were recruited to the lymph nodes in response to the allergen in PLZF-deficient mice (Fig. 4A, bottom). Although the number of basophils present in PLZF-deficient mice is reduced, the near complete absence of the cells following treatment implies that a migration deficiency is in effect. Heat inactivated papain did not result in basophil recruitment in either mouse strain.
Fig. 4. Papain-induced basophil-mediated Th2 responses were decreased in PLZF KO mice.
(A) 50μg of papain was injected into the footpads of PLZF knock out (KO) and wild type (WT) mice. The same amount of heat inactivated (HI) papain was injected into the other footpad as control. 3 days after injection, the draining lymph nodes were removed and stained for basophils. Cells shown were gated are DAPI−CD3−CD19−. Frequency of basophils, identified as CD49b+FCεR1+ is shown in the dot plot and for multiple experiments in the scatter plot graphs. Each symbol indicates a single mouse. N=8 with 3–5 independent experiments. (B-C) 50ug of papain or HI papain was injected into (B) RAG1 KO (N=3; 3 independent experiments) or (C) CD1d KO (N=5; 3 independent experiments) mice and WT controls. Basophil frequency was determined as in (A). The horizontal lines in the scatter plots indicate the mean (±s.e.m.). *P<0.05, **P<0.01, ***P<.001, n.s. (not significant) was determined by Student’s t-test.
Basophil responses (both their population expansion and recruitment) to most stimuli including helminth parasite and papain are dependent on IL-3-IL-3R signaling (67–69). Further, T cells are reported to be the main source of IL-3 required for basophil responses to be initiated (70). Consistent with these data, basophil activation and recruitment are disrupted in lymphocyte deficient mice as a result of IL-3 deficiently (70). Consistent with these previous studies, injection of RAG1-deficient mice with papain resulted in limited basophil migration to the draining lymph nodes (Fig. 4B). This result raised the possibility that the failure of basophil migration was due to loss of functional NKT cells in the PLZF-deficient mice. We tested this by injecting CD1d-deficient mice, which are devoid of invariant NKT cells, with papain. The results showed that there was no difference in the frequency of recruited basophils between CD1d KO and WT mice (Fig. 4C). Combined, these data show that PLZF expression in basophils is important for their response to an allergic stimulus.
After papain injection, naïve CD4+ T cells in the lymph nodes have been shown to respond to the IL-4 produced by basophils and undergo Th2 differentiation, which requires the transcription factor, GATA3. Therefore, the frequency of GATA3+ T cells indirectly reflects the extent of the Th2 response mediated by basophils (17). Results showed that in wild type mice, ~6% of T cells in the lymph nodes expressed GATA3 after papain immunization (Fig. 5A). The frequency of GATA3-positive CD4 T cells in the draining lymph nodes following papain injection of PLZF-deficient mice was, however, reduced by nearly half (Fig. 5A). PLZF expressing NKT cells do not play a role in this Th2 differentiation response since there was no difference in GATA3-positive CD4 T cells in the lymph nodes of CD1d-KO mice following papain injection (Fig. 5B).
Fig. 5. Differentiation of Th2 T cells is reduced in the absence of PLZF.
(A) 50μg of papain was injected into the footpads of WT or PLZF KO mice. 4 days after papain injection, the draining lymph nodes were removed. Th2 differentiation, as measured by intracellular staining for GATA3, in CD3+ CD4+ T cells, with the percent positive cells shown in the dot plots. Scatter plots show the frequency of GATA3+ CD4+ T cells, with an N = 5 and 4 independent experiments. (B) 50μg of papain was injected into the footpads of WT or CD1d KO mice. The frequency of GATA3+ CD4+ T cells is shown in the dot plots. FACS is representative of 3 independent experiments. The horizontal lines in the scatter plots indicate the mean (±s.e.m.). *P<0.05, **P<0.01, ***P<.001, n.s (not significant) was determined by Student’s t-test.
Functional analysis of basophils in PLZF-deficient mice in vitro
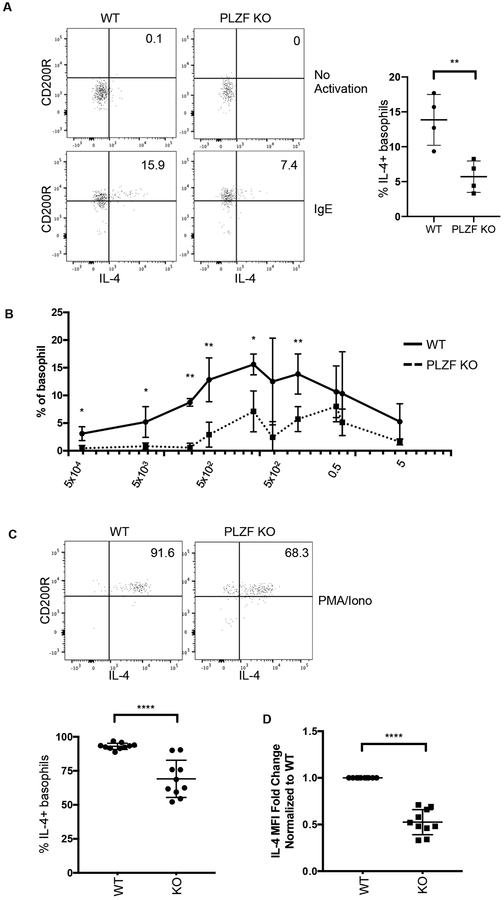
The reduced basophil-mediated Th2 response in PLZF-deficient mice was a consequence of either the decreased number of basophils or, alternatively, reduced basophil effector functions. We considered the possibility that like NKT cells, PLZF deficiency in basophils might result in disrupted IL-4 production. To test this, peripheral blood from mice was activated in vitro with either IgE or PMA/Ionomycin. Intracellular staining for cytokines in basophils was done three hours after activation. Incubation with 0.05ng/ml of IgE resulted in activation of basophils as measured by upregulation of CD200R expression (Fig. 6A). Approximately 15% of wild type basophils produced IL-4 under these conditions. In contrast, only ~7% of PLZF-deficient basophils produced IL-4 (Fig. 6A). Next, we used titrated concentrations of IgE to determine if the sensitivity of PLZF-deficient basophils to activating signals was reduced. The percentage of responding basophils that were deficient for PLZF was significantly reduced at nearly every concentration of IgE. Furthermore, we found that the dose of IgE needed to activate PLZF-deficient basophils was approximately 100-times higher than that needed for wild type cells (Fig. 6B).
Fig. 6. Function of PLZF deficient basophils.
(A) Peripheral blood from PLZF KO and WT mice was activated with 0.05ng/ml of IgE for 3 hours. Brefeldin A (BFA) was added for the last two hours of activation. The cells were made permeable and stained for intracellular IL-4. CD200R was analyzed to measure activation. Cells shown are CD3−CD19−FcεRI+CD49b+. Representative FACS plots from 1 of 4 independent experiments are shown. (B) Peripheral blood from WT or PLZF KO mice was activated with increasing concentrations of IgE for 3 hours. The percentage of basophils expressing IL-4 is shown. (C) Peripheral blood samples from PLZF KO and WT mice were activated with PMA/Ionomycin for 3 hours. BFA was added for the last two hours of activation. The cells were then stained for intracellular IL-4. Basophils were identified as the CD3−CD19−FcεRI+CD49b+ population. The scatter plot shows the frequency of basophils that produced IL-4 under these conditions. (D) Fold change of IL-4 Mean Fluorescence Intensity (MFI) of the cytokine producing basophils from (C), normalized to WT. N=10 with 5 independent experiments.
PMA/Ionomycin was used to determine the maximal IL-4 production capacity of wild type and PLZF-deficient basophils. Fewer PLZF KO basophils responded, as measured by intracellular staining of IL-4 (Fig. 6C). The mean fluorescence intensity (MFI) of IL-4 was also determined as a measure of the amount of cytokine produced by the responding cells and was found to be about 2-fold lower (Fig. 6D). Overall, these data show that in the absence of PLZF, fewer basophils produced IL-4 and those that did produce the cytokine, made less.
PLZF-deficient basophils have reduced IL-18R expression
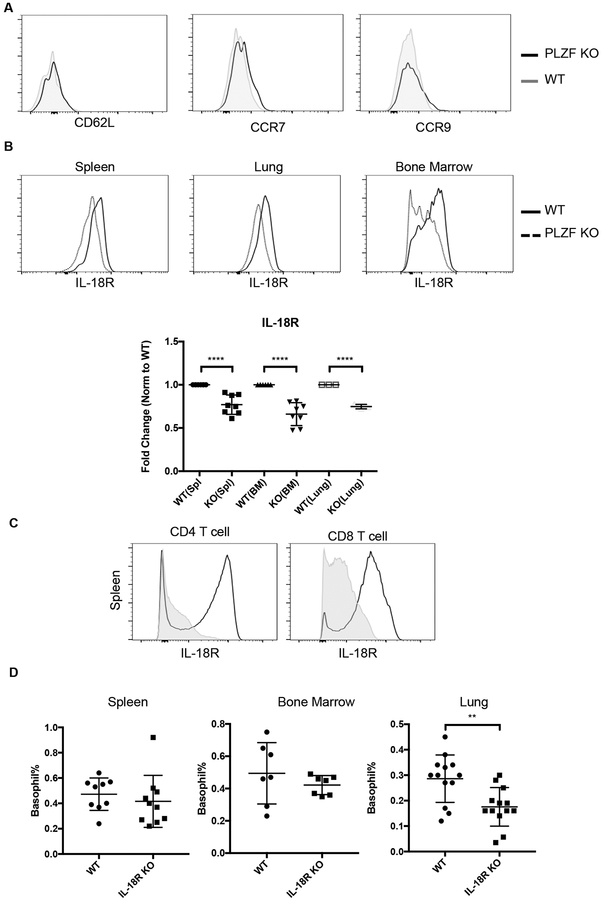
To investigate the potential mechanism for reduced migration of PLZF-deficient basophils after papain immunization, we stained various cytokine and chemokine receptors on basophils. Loss of PLZF did not impact the expression levels of CD62L, CCR7 or CCR9, all of which have been implicated in basophil migration (Fig. 7A) (17, 22, 71, 72). There also were no differences in the expression of other cell surface proteins such as ST2, MHCII or CD200R in the absence of PLZF (data not shown). Interestingly, however, PLZF-deficient basophils were found to express lower levels of the IL-18 receptor (Fig. 7B). These differences were relatively small, but were highly consistent in multiple tissues including the spleen, lung and bone marrow (Fig. 7B). Altered expression of IL-18R is consistent with data showing that PLZF binds to and, possibly, directly regulates expression of Il18r1 (73, 74). Furthermore, ectopic expression of PLZF in conventional T cells resulted in upregulation of IL-18R expression (Fig. 7C).
Fig. 7. PLZF impacts IL-18R expression on basophils and their migration to the lung.
(A) Expression of the indicated proteins was not altered in PLZF deficient spleen basophils. (B) IL-18R expression on basophils from spleen, lung and bone marrow was reduced on PLZF deficient basophils. Representative FACS plots from 1 of 4 independent experiments is shown. The fold change on basophils from each of these tissues is shown in the scatter plot. N=8 with 4 independent experiments. (C) IL-18R expression is shown for spleen T cells (CD3+CD8+ or CD3+CD4+) taken from WT mice (grey filled) or from mice ectopically expressing PLZF in all T cells. (D) The frequency of basophils in the lung, spleen and bone marrow from IL-18R KO and WT mice. N=7–12 mice with 3–5 independent experiments.
IL-18R-deficient mice were analyzed to determine if altered expression of this receptor impacted basophil migration. The frequency (Fig. 7D) and phenotype of the cells from the spleen and bone marrow was not notably altered. In the lung, however, there was a ~30% decrease in the frequency of basophils. Therefore, PLZF control of IL-18R may influence the ability of basophils to migrate to some tissues.
Together, we show that expression of PLZF during myeloid lineage development is necessary for full population of the basophil compartment. Furthermore, PLZF deficient basophils display decreased migration, reduced IL-4 production upon activation, and lower surface levels of IL-18R, indicating that PLZF plays an important role in basophil trafficking and function.
Discussion
The importance of the expression of PLZF in “innate-like” lymphocytes such as NKT cells, Vγ1.1;Vδ6.3 γδ NKT cells, ILCs, and MAIT cells has been well established (75). Less is known, however, about the transcription factor’s function in myeloid lineage subsets. PLZF has been shown to impact the early development of the myeloid lineage cells in humans (76), but its expression and potential function in mature myeloid lineage cell types is largely unexplored. In this study, we found that PLZF is expressed in basophils where it plays an important for role both their development and function.
PLZF was found to be expressed in basophils from all tissues including the bone marrow, spleen, lung and peripheral blood. Like during NKT cell development, PLZF was downregulated as basophil progenitors matured in the bone marrow. In the absence of PLZF, we found a ~50–60% percent decrease of the cells. More interestingly, PLZF-deficient basophils proved to have a reduced capacity to migrate and produce IL-4. Nearly 100-times the dose of IgE was required to induce IL-4 release in the absence of PLZF. Furthermore, maximal IL-4 production, as determined by activation with PMA/ionomycin, was reduced. The reduced IL-4 release, therefore, correlated with a diminished Th2 immune response to a protein allergen. It has been reported that IL-13 production by PLZF deficient ILC2 harvested from the lungs of mice treated with papain for 3 days produce somewhat less IL-13 (45). Therefore, it is possible that reduced function of IL2s in the absence of PLZF also impacts Th2 T cell differentiation. Overall, data suggest that PLZF impacts the lymphocyte contribution in type 2 immune responses both by cell-intrinsic and cell-extrinsic means.
In humans with fatal asthma there often is an increased infiltration of basophils into the lung(77). In mouse asthma models, basophils have been shown to mediate early-phase airway obstruction (78). Therefore, the control of basophil migration and activation by PLZF might play a role in the pathogenesis of asthma. In support of this, our results showed that PLZF-deficient basophils have reduced levels of IL-18R and that loss of this receptor impacted basophil migration to the lung. IL-18 is a pleiotropic cytokine produced by macrophages, Kupffer cells as well as epithelial cells such as keratinocytes (79, 80). Previous studies have shown that IL-18, combined with IL-3, stimulates large amounts of IL-4 and IL-13 production by basophils, in vitro (79). Injection of IL-18 in mice has been shown to increase IL-4 production by basophils, recruit antigen-induced eosinophils, and increase Th2 cytokines in the lung (12, 81, 82). Recently, it was shown that IL-18 signaling is necessary for the response of PLZF expressing NKT cells to influenza infection in the lung (83). Perhaps, therefore, activation by IL-18 signaling is a common feature of lung NKT cells and basophils. This interpretation may, however, not apply to humans where it has been shown that, although basophils express high levels of IL18R, in vitro, they do not obviously respond to IL-18 (84).
In the absence of PLZF, basophil numbers were significantly decreased. PLZF has been shown to regulate development of human cord blood-derived myeloid progenitor cells by transcriptional modulation of key transcription factors including GFI-1, C/EBPα, LEF-1, DUSP6 and ID2 (38, 85). The maturation of basophils also requires the C/EBPα, suggesting a possible mechanism for the control of basophil development by PLZF.
Collectively, our data showed that the transcription factor PLZF regulates the development and function of basophils. Although, there is not a complete loss of basophil effector functions, the reduction in IL-4 production in response to a protein allergen is sufficient to blunt Th2 immunity. With a cell intrinsic role in controlling the NKT cells, type 2 innate-like lymphocytes (ILC2) and now a cell extrinsic role that impacts Th2 cell development, PLZF is emerging as a major regulator of the overall type 2 immune response. Interestingly, data show that variations in the expression levels of PLZF have major impacts on cell function (62, 86). Therefore, subtle variations in PLZF expression in the human population could be associated with the efficiency of clearing various pathogens and parasites. In addition, the control of IL-18R expression, which we show influences migration of basophils to the lung, implies that variations in PLZF expression might also impact susceptibility to asthma. Overall, our results further elucidate the transcriptional regulation of basophil functions, which helps for better understanding of the role of basophils in the prevention and control of disease.
Supplementary Material
Key points:
PLZF deficient mice have reduced numbers of basophil progenitors and mature basophils
PLZF deficient basophils respond poorly to the protein allergen, papain;
PLZF deficient basophils are refractory to activation and produce less IL-4;
Acknowledgments:
We thank A. Rabson for insightful discussions; L. Osorio for technical assistance; A. Roberts for cell sorting
These studies were supported by NIH NIAID R01 AI083988 and AI059739 (to D.B.S.), the Robert Wood Johnson Foundation (grant #67038) to the Child Health Institute of New Jersey, the Dr. James J. O’Connell Fund (to S.Z.) and the New Jersey Health Foundation Excellence in Research Award (to D.B.S.).
References and Notes:
- 1.Schroeder JT 2009. Basophils beyond effector cells of allergic inflammation. Adv Immunol 101: 123–161. [DOI] [PubMed] [Google Scholar]
- 2.Mack M, Schneider MA, Moll C, Cihak J, Bruhl H, Ellwart JW, Hogarth MP, Stangassinger M, and Schlondorff D. 2005. Identification of antigen-capturing cells as basophils. J Immunol 174: 735–741. [DOI] [PubMed] [Google Scholar]
- 3.Kubo M 2018. Mast cells and basophils in allergic inflammation. Curr Opin Immunol 54: 74–79. [DOI] [PubMed] [Google Scholar]
- 4.Rigoni A, Colombo MP, and Pucillo C. 2018. Mast cells, basophils and eosinophils: From allergy to cancer. Semin Immunol 35: 29–34. [DOI] [PubMed] [Google Scholar]
- 5.Siracusa MC, Kim BS, Spergel JM, and Artis D. 2013. Basophils and allergic inflammation. The Journal of allergy and clinical immunology 132: 789–801; quiz 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasuyama H, Mukai K, Obata K, Tsujimura Y, and Wada T. 2011. Nonredundant roles of basophils in immunity. Annu Rev Immunol 29: 45–69. [DOI] [PubMed] [Google Scholar]
- 7.Min B 2008. Basophils: what they ‘can do’ versus what they ‘actually do’. Nat Immunol 9: 1333–1339. [DOI] [PubMed] [Google Scholar]
- 8.Siracusa MC, Perrigoue JG, Comeau MR, and Artis D. 2010. New paradigms in basophil development, regulation and function. Immunol Cell Biol 88: 275–284. [DOI] [PubMed] [Google Scholar]
- 9.Falcone FH, Haas H, and Gibbs BF. 2000. The human basophil: a new appreciation of its role in immune responses. Blood 96: 4028–4038. [PubMed] [Google Scholar]
- 10.Sokol CL, and Medzhitov R. 2010. Role of basophils in the initiation of Th2 responses. Current opinion in immunology 22: 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min B, Brown MA, and Legros G. 2012. Understanding the roles of basophils: breaking dawn. Immunology 135: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohrs K, Wakil AE, Killeen N, Locksley RM, and Mohrs M. 2005. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 23: 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min B, Prout M, Hu-Li J, Zhu J, Jankovic D, Morgan ES, Urban JF Jr., Dvorak AM, Finkelman FD, LeGros G, and Paul WE. 2004. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J Exp Med 200: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gessner A, Mohrs K, and Mohrs M. 2005. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J Immunol 174: 1063–1072. [DOI] [PubMed] [Google Scholar]
- 15.Oh K, Shen T, Le Gros G, and Min B. 2007. Induction of Th2 type immunity in a mouse system reveals a novel immunoregulatory role of basophils. Blood 109: 2921–2927. [DOI] [PubMed] [Google Scholar]
- 16.Hida S, Tadachi M, Saito T, and Taki S. 2005. Negative control of basophil expansion by IRF-2 critical for the regulation of Th1/Th2 balance. Blood 106: 2011–2017. [DOI] [PubMed] [Google Scholar]
- 17.Sokol CL, Barton GM, Farr AG, and Medzhitov R. 2008. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol 9: 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, Noti M, Tait Wojno ED, Fung TC, Kubo M, and Artis D. 2014. Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol 193: 3717–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan BM, Liang HE, Bando JK, Wu D, Cheng LE, McKerrow JK, Allen CD, and Locksley RM. 2011. Genetic analysis of basophil function in vivo. Nat Immunol 12: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokol CL, Chu NQ, Yu S, Nish SA, Laufer TM, and Medzhitov R. 2009. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat Immunol 10: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrigoue JG, Saenz SA, Siracusa MC, Allenspach EJ, Taylor BC, Giacomin PR, Nair MG, Du Y, Zaph C, van Rooijen N, Comeau MR, Pearce EJ, Laufer TM, and Artis D. 2009. MHC class II-dependent basophil-CD4+ T cell interactions promote T(H)2 cytokine-dependent immunity. Nat Immunol 10: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimoto T, Yasuda K, Tanaka H, Nakahira M, Imai Y, Fujimori Y, and Nakanishi K. 2009. Basophils contribute to T(H)2-IgE responses in vivo via IL-4 production and presentation of peptide-MHC class II complexes to CD4+ T cells. Nat Immunol 10: 706–712. [DOI] [PubMed] [Google Scholar]
- 23.Gauchat JF, Henchoz S, Mazzei G, Aubry JP, Brunner T, Blasey H, Life P, Talabot D, Flores-Romo L, Thompson J, and et al. 1993. Induction of human IgE synthesis in B cells by mast cells and basophils. Nature 365: 340–343. [DOI] [PubMed] [Google Scholar]
- 24.Yanagihara Y, Kajiwara K, Basaki Y, Ikizawa K, Akiyama K, and Saito H. 1997. Induction of human IgE synthesis in B cells by a basophilic cell line, KU812. Clinical and experimental immunology 108: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagihara Y, Kajiwara K, Basaki Y, Ikizawa K, Ebisawa M, Ra C, Tachimoto H, and Saito H. 1998. Cultured basophils but not cultured mast cells induce human IgE synthesis in B cells after immunologic stimulation. Clinical and experimental immunology 111: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K, Xu W, Wilson M, He B, Miller NW, Bengten E, Edholm ES, Santini PA, Rath P, Chiu A, Cattalini M, Litzman J, J BB, Huang B, Meini A, Riesbeck K, Cunningham-Rundles C, Plebani A, and Cerutti A. 2009. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat Immunol 10: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merluzzi S, Betto E, Ceccaroni AA, Magris R, Giunta M, and Mion F. 2015. Mast cells, basophils and B cell connection network. Mol Immunol 63: 94–103. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz C, Eberle JU, and Voehringer D. 2016. Basophils in inflammation. Eur J Pharmacol 778: 90–95. [DOI] [PubMed] [Google Scholar]
- 29.Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, Chabon J, Graham B, Ohmori K, Li Y, and Huang H. 2013. Antagonistic regulation by the transcription factors C/EBPalpha and MITF specifies basophil and mast cell fates. Immunity 39: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, and Akashi K. 2005. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A 102: 18105–18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki H, Kurotaki D, and Tamura T. 2016. Regulation of basophil and mast cell development by transcription factors. Allergol Int 65: 127–134. [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki H, and Akashi K. 2007. Myeloid lineage commitment from the hematopoietic stem cell. Immunity 26: 726–740. [DOI] [PubMed] [Google Scholar]
- 33.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, and Akashi K. 2006. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes & development 20: 3010–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura Y, Morii E, Jippo T, and Ito A. 2002. Regulation of mast cell phenotype by MITF. Int Arch Allergy Immunol 127: 106–109. [DOI] [PubMed] [Google Scholar]
- 35.Takemoto CM, Lee YN, Jegga AG, Zablocki D, Brandal S, Shahlaee A, Huang S, Ye Y, Gowrisankar S, Huynh J, and McDevitt MA. 2008. Mast cell transcriptional networks. Blood Cells Mol Dis 41: 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beaulieu AM, and Sant’Angelo DB. 2011. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J Immunol 187: 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siggs OM, and Beutler B. 2012. The BTB-ZF transcription factors. Cell Cycle 11: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suliman BA, Xu D, and Williams BR. 2012. The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Front Oncol 2: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, and Sant’Angelo DB. 2010. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol 184: 1268–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, and von Boehmer H. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A 106: 12453–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, and Sant’Angelo DB. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 9: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, Russ BE, Nold-Petry CA, Nold MF, Bedoui S, Chen Z, Corbett AJ, Eckle SB, Meehan B, d’Udekem Y, Konstantinov IE, Lappas M, Liu L, Goodnow CC, Fairlie DP, Rossjohn J, Chong MM, Kedzierska K, Berzins SP, Belz GT, McCluskey J, Uldrich AP, Godfrey DI, and Pellicci DG. 2016. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol 17: 1300–1311. [DOI] [PubMed] [Google Scholar]
- 44.Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A. 2014. A committed precursor to innate lymphoid cells. Nature 508: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verhoef PA, Constantinides MG, McDonald BD, Urban JF Jr., Sperling AI, and Bendelac A. 2016. Intrinsic functional defects of type 2 innate lymphoid cells impair innate allergic inflammation in promyelocytic leukemia zinc finger (PLZF)-deficient mice. J Allergy Clin Immunol 137: 591–600 e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, and Bendelac A. 2015. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci U S A 112: 5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, Yuan J, Wolchok JD, Horsthemke B, Wieczorek D, and Sant’Angelo DB. 2011. Altered development of NKT cells, gammadelta T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS One 6: e24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, and Sant’Angelo DB. 2010. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol 184: 6746–6755. [DOI] [PubMed] [Google Scholar]
- 49.Savage AK, Constantinides MG, and Bendelac A. 2011. Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling. J Immunol 186: 5801–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, and Ellmeier W. 2008. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A 105: 17919–17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S, Laouar A, Denzin LK, and Sant’Angelo DB. 2015. Zbtb16 (PLZF) is stably suppressed and not inducible in non-innate T cells via T cell receptor-mediated signaling. Sci Rep 5: 12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, and Zhao K. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosokawa H, Romero-Wolf M, Yui MA, Ungerback J, Quiloan MLG, Matsumoto M, Nakayama KI, Tanaka T, and Rothenberg EV. 2018. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol 19: 1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu L, Qiao Y, Choi ES, Das J, Sant’angelo DB, and Chang CH. 2013. A transgenic TCR directs the development of IL-4+ and PLZF+ innate CD4 T cells. J Immunol 191: 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barna M, Hawe N, Niswander L, and Pandolfi PP. 2000. Plzf regulates limb and axial skeletal patterning. Nat Genet 25: 166–172. [DOI] [PubMed] [Google Scholar]
- 56.Thapa P, Das J, McWilliams D, Shapiro M, Sundsbak R, Nelson-Holte M, Tangen S, Anderson J, Desiderio S, Hiebert S, Sant’angelo DB, and Shapiro VS. 2013. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun 4: 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, Sigvardsson M, and Jacobsen SE. 2005. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3-short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood 105: 2717–2723. [DOI] [PubMed] [Google Scholar]
- 58.Akashi K, Traver D, Miyamoto T, and Weissman IL. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197. [DOI] [PubMed] [Google Scholar]
- 59.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, and Akashi K. 2005. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med 201: 1891–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, and Feuerer M. 2013. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 14: 821–830. [DOI] [PubMed] [Google Scholar]
- 61.Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, Kimura MY, Sharrow SO, Guinter TI, Feigenbaum L, and Singer A. 2015. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol 16: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, Godfrey DI, Leadbetter EA, Sant’Angelo DB, von Andrian U, and Brenner MB. 2015. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol 16: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenstein RK, Bezbradica JS, Yu S, and Medzhitov R. 2014. Signaling pathways activated by a protease allergen in basophils. Proc Natl Acad Sci U S A 111: E4963–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyake K, and Karasuyama H. 2017. Emerging roles of basophils in allergic inflammation. Allergol Int 66: 382–391. [DOI] [PubMed] [Google Scholar]
- 65.Schroeder JT 2011. Basophils: emerging roles in the pathogenesis of allergic disease. Immunol Rev 242: 144–160. [DOI] [PubMed] [Google Scholar]
- 66.Karasuyama H, Wada T, Yoshikawa S, and Obata K. 2011. Emerging roles of basophils in protective immunity against parasites. Trends Immunol 32: 125–130. [DOI] [PubMed] [Google Scholar]
- 67.Lantz CS, Boesiger J, Song CH, Mach N, Kobayashi T, Mulligan RC, Nawa Y, Dranoff G, and Galli SJ. 1998. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature 392: 90–93. [DOI] [PubMed] [Google Scholar]
- 68.Voehringer D 2013. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol 13: 362–375. [DOI] [PubMed] [Google Scholar]
- 69.Kim S, Karasuyama H, Lopez AF, Ouyang W, Li X, Le Gros G, and Min B. 2013. IL-4 Derived from Non-T Cells Induces Basophil- and IL-3-independent Th2 Immune Responses. Immune Netw 13: 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen T, Kim S, Do JS, Wang L, Lantz C, Urban JF, Le Gros G, and Min B. 2008. T cell-derived IL-3 plays key role in parasite infection-induced basophil production but is dispensable for in vivo basophil survival. Int Immunol 20: 1201–1209. [DOI] [PubMed] [Google Scholar]
- 71.Iikura M, Ebisawa M, Yamaguchi M, Tachimoto H, Ohta K, Yamamoto K, and Hirai K. 2004. Transendothelial migration of human basophils. J Immunol 173: 5189–5195. [DOI] [PubMed] [Google Scholar]
- 72.Tang H, Cao W, Kasturi SP, Ravindran R, Nakaya HI, Kundu K, Murthy N, Kepler TB, Malissen B, and Pulendran B. 2010. The T helper type 2 response to cysteine proteases requires dendritic cell-basophil cooperation via ROS-mediated signaling. Nat Immunol 11: 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, and Bendelac A. 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A 113: 7602–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gleimer M, von Boehmer H, and Kreslavsky T. 2012. PLZF Controls the Expression of a Limited Number of Genes Essential for NKT Cell Function. Front Immunol 3: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alonzo ES, and Sant’Angelo DB. 2011. Development of PLZF-expressing innate T cells. Curr Opin Immunol 23: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dick JE, and Doulatov S. 2009. The role of PLZF in human myeloid development. Ann N Y Acad Sci 1176: 150–153. [DOI] [PubMed] [Google Scholar]
- 77.Kepley CL, McFeeley PJ, Oliver JM, and Lipscomb MF. 2001. Immunohistochemical detection of human basophils in postmortem cases of fatal asthma. Am J Respir Crit Care Med 164: 1053–1058. [DOI] [PubMed] [Google Scholar]
- 78.Nabe T, Matsuya K, Akamizu K, Fujita M, Nakagawa T, Shioe M, Kida H, Takiguchi A, Wakamori H, Fujii M, Ishihara K, Akiba S, Mizutani N, Yoshino S, and Chaplin DD. 2013. Roles of basophils and mast cells infiltrating the lung by multiple antigen challenges in asthmatic responses of mice. Br J Pharmacol 169: 462–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshimoto T, Tsutsui H, Tominaga K, Hoshino K, Okamura H, Akira S, Paul WE, and Nakanishi K. 1999. IL-18, although antiallergic when administered with IL-12, stimulates IL-4 and histamine release by basophils. Proc Natl Acad Sci U S A 96: 13962–13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sims JE, and Smith DE. 2010. The IL-1 family: regulators of immunity. Nature reviews. Immunology 10: 89–102. [DOI] [PubMed] [Google Scholar]
- 81.Kumano K, Nakao A, Nakajima H, Hayashi F, Kurimoto M, Okamura H, Saito Y, and Iwamoto I. 1999. Interleukin-18 enhances antigen-induced eosinophil recruitment into the mouse airways. Am J Respir Crit Care Med 160: 873–878. [DOI] [PubMed] [Google Scholar]
- 82.Wild JS, Sigounas A, Sur N, Siddiqui MS, Alam R, Kurimoto M, and Sur S. 2000. IFN-gamma-inducing factor (IL-18) increases allergic sensitization, serum IgE, Th2 cytokines, and airway eosinophilia in a mouse model of allergic asthma. J Immunol 164: 2701–2710. [DOI] [PubMed] [Google Scholar]
- 83.Gaya M, Barral P, Burbage M, Aggarwal S, Montaner B, Warren Navia A, Aid M, Tsui C, Maldonado P, Nair U, Ghneim K, Fallon PG, Sekaly RP, Barouch DH, Shalek AK, Bruckbauer A, Strid J, and Batista FD. 2018. Initiation of Antiviral B Cell Immunity Relies on Innate Signals from Spatially Positioned NKT Cells. Cell 172: 517–533 e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pecaric-Petkovic T, Didichenko SA, Kaempfer S, Spiegl N, and Dahinden CA. 2009. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood 113: 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doulatov S, Notta F, Rice KL, Howell L, Zelent A, Licht JD, and Dick JE. 2009. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes & development 23: 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Constantinides MG, Picard D, Savage AK, and Bendelac A. 2011. A naive-like population of human CD1d-restricted T cells expressing intermediate levels of promyelocytic leukemia zinc finger. J Immunol 187: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.