Abstract
The efficacy and specificity of protein, DNA, and RNA-based drugs make them popular in the clinic; however, these drugs are often delivered via injection, requiring skilled medical personnel, and producing biohazardous waste. Here, we report an approach that allows for their controlled delivery, affording either a burst or slow release without altering the formulation. We show that when encapsulated within zeolitic-imidazolate framework eight (ZIF-8), the biomolecules are stable in powder formulations and can be inoculated with a low-cost, gas-powered “MOF-Jet” into living animal and plant tissues. Additionally, their release profiles can be modulated through judicious selection of the carrier gas used in the MOF-Jet. Our in vitro and in vivo studies reveal that when CO2 is used, it creates a transient and weakly acidic local environment that causes a near-instantaneous release of the biomolecules through an immediate dissolution of ZIF-8. Conversely, when air is used, ZIF-8 biodegrades slowly, releasing the biomolecules over a week. This is the first example of controlled-biolistic delivery of biomolecules using ZIF-8, which provides a powerful tool for fundamental and applied science research.
We can alter the release kinetics of highly stabilized biomacromolecules in both skin and plant tissues to allow for either instant release or slow release simply by changing the typeof gas used in a pneumatic delivery jet.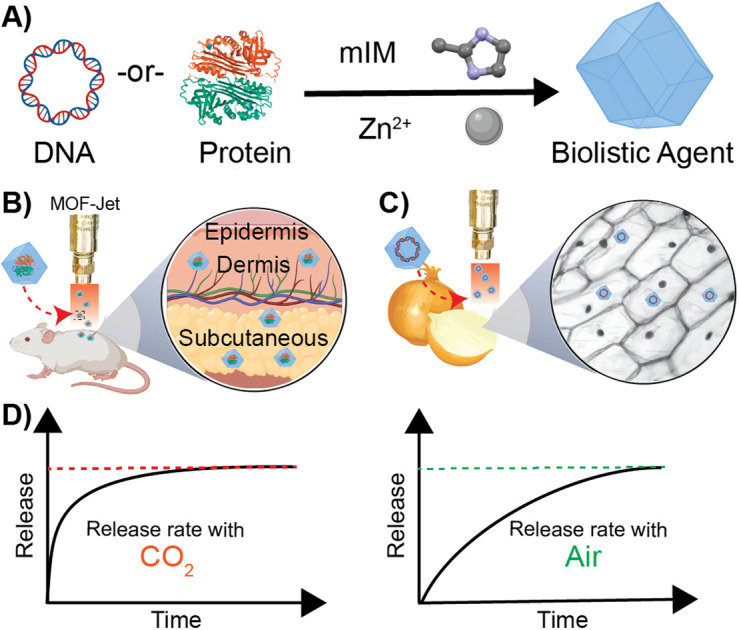
1. Introduction
Biomacromolecule-based therapeutics have been clinically transformative owing to their affinity, specificity, and efficacy in treating diseases.1 Advancements in biotechnology have led to the mass production and adaption of drugs based on proteins or DNA. Indeed, four of the five top-selling drugs in the US are biomacromolecules.2,3 With the development of COVID-19 vaccines that run the range of DNA-based viral vectors to mRNA-based lipid nanoparticles, it is now reasonable to say that most people have been treated and spared serious illness thanks to biomacromolecular agents. While their efficacy and specificity in treating diseases are typically considered exceptional, most biomacromolecular drugs are not stable under ambient conditions without modifications.4,5 Further, these drugs are susceptible to degradation or denaturation at body temperature, complicating controlled-release implantable depot systems within the body.6,7 Recent advances in drug delivery platforms have emerged to control the delivery of these drugs.7 For instance, microneedles create micron-sized channels to transport therapeutics directly into the targeted tissue when pressed on the skin. Recent advances in 3D printing and clever molding with biodegradable and thermoplastic polymers have created opportunities to expand this technology to stabilize proteins.8,9 The advancements here are exciting, with comprehensive studies underway to address issues that have stalled broad adoption.10 Liquid Jet Injectors (JIs) or jet guns that use mechanical energy to create a high-pressure stream of a liquid capable of penetrating the skin were once popular in the mass delivery of liquid vaccine formulations.11 Through the 2000s, JIs were used to deliver millions of intramuscular and subcutaneous vaccine doses per year. However, safety concerns arose from the use of discharging high-pressure liquid, which would bounce off the skin and spray back onto the jet injector, which has recently limited their usage to veterinary applications.11 In recent years, solid-particle delivery using high-pressure compressed gas—or so-called biolistic delivery—has emerged as a non-invasive way of delivering payloads into both cells and tissues in plants and animals without concerns for spray-back.12 In this approach, biomacromolecules are adsorbed onto heavy micron-sized metal particles, such as gold or tungsten, and then accelerated to high speeds via pressurized gas flow and bombarded directly into the targeted tissue.12,13 This simplicity has allowed biolistic methods to progress steadily over traditional methods like direct DNA injection or Agrobacterium-mediated gene transfection in plants.13–15 While this technique is already widely used in plants, recent work has found this approach to be very effective in delivering DNA, RNA, and protein therapeutics to animals.12,16,17 Even though biolistic delivery has become routine for some applications, it has several drawbacks. A typical commercial gene gun is quite expensive, with costs exceeding US$30 000 18 and coating the particles with DNA or proteinaceous material is a cumbersome task, requiring its own apparatus and drying of particles.19 Further, the metal microparticles are non-biodegradable, which means they will persist within the organism indefinitely.20 Finally, once the genetic material is loaded onto the metal microparticle surface, it is highly susceptible to nuclease and enzymatic degradation, meaning they would not overcome issues with biotherapeutic degradation in vivo.21 Beyond all that, a major disadvantage of this current technology is that there is no control over the release profile of the therapeutic material after it is delivered into cells or tissue as these biomacromolecules are merely surface coated and weakly physisorbed onto the metal particle surface. Here, the active biomolecules are instantaneously released in the physiological or cellular environment, leaving their metal particle carrier behind.22
Since the mid-90s, a class of hybrid micro and mesoporous materials called metal–organic frameworks (MOFs), made from interconnected metal nodes and organic linkers, has attracted ever-growing attention in a broad range of applications.23–28 Recently, MOFs have emerged as a method to protect biomacromolecules from degradation or thermal denaturation by being protected in a thermodynamically stable crystalline shell.29,30 ZIF-8 is a highly crystalline MOF composed of the organic ligand methylimidazole (mIM) interconnected by Zn2+ cations. Recent work by us and others have shown that ZIF-8 crystals will nucleate and grow on the surface of biomacromolecules via a “biomimetic” mechanism resulting in their complete encapsulation and protection—mimicking the natural biomineralization process of soft-body bearing living organisms (Scheme 1A). Specifically, biomimetic mineralization with ZIF-8 is effective in enabling long-term room temperature storage of many macromolecules while also allowing for slow-release in vivo for vaccine and cancer therapeutics.7,31–33 A critical issue, however, is that the release rate is governed by the kinetics of metal competition in vivo. So far, there have been no reports on controlling the rate of degradation when ZIF-8 is implanted. Consequently, while MOFs may make great shells to protect biomacromolecules, if they are injected into the skin there is no way to alter the release rate—it is essentially fixed by the chemistry of the MOF.32,34
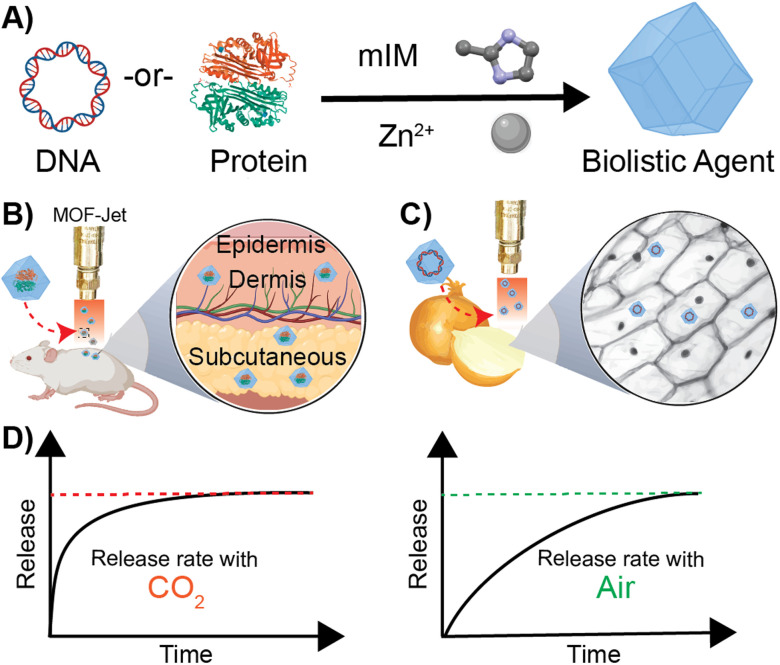
Scheme 1. Schematic representation of biomimetic mineralization with ZIF-8 (A) synthesis of DNA-or-Protein loaded ZIF-8 crystals from 2-methyl imidazole and zinc acetate dihydrate in aqueous conditions at room temperature. (B) Biolistic delivery of protein-loaded ZIF-8 crystals into animal skin. (C) Biolistic delivery of DNA-loaded ZIF-8 into plant tissue. (D) Respective release profiles of the encapsulated biomolecule when biolistically delivered with CO2 and air as the propellent.
Degradation of ZIF-8 in tissue—as distinct from what happens following endocytosis by a cell—is the result of competition for the zinc centers by inorganic phosphates and proteins such as albumin, which exploit the kinetic lability of the bond between Zn2+ and 2-methylimidazole. In contrast, following endocytosis by a cell, ZIF-8 is rapidly dissolved. The endosome acidifies over time, causing the disintegration of the ZIF-8 and releasing the encapsulated molecules within a cell. Although normal tissue and cytoplasm are typically slightly above neutral pH, we realized it should be possible to induce the rapid dissolution of ZIF-8 using CO2 as a reactive carrier gas during biolistic delivery. CO2 has been shown to acidify the interior of MOFs35,36 and, since ZIF-8 is capable of modest adsorption of CO2,37,38 we hypothesized that bombarding dried ZIF-8-based biomacromolecules out of a pneumatic gun using high pressures of CO2 would result in a local and rapid pH drop once the CO2 encounters the water within the cells or tissues. CO2 reacts very rapidly with moisture to create carbonic acid, which should rapidly dissolve the ZIF-8, subsequently releasing the encapsulated therapeutics. On the other hand, high-pressure air, which contains only 0.04% CO2, should deliver a slow-releasing depot of ZIF-8 coated biomolecules. In this paper, we show that we can change the release of thermally and enzymatically protected DNA and proteins encapsulated in ZIF-8 by using CO2 as a pneumatic carrier gas. Specifically, we have developed a method to change the kinetics of releasing biomacromolecules via biolistic delivery between “instant” and “slow-release.” These findings are important because changes in release kinetics typically require significant changes in how a drug is formulated, which must be tuned to whatever tissue or cell type is targeted. Our approach is exceptionally generalizable, and we demonstrate so in two very different proof-of-principle experiments. Specifically, we show effective control of gene delivery into plant cells and protein delivery in animal tissue by shooting a powder formulation with either air or CO2 as the propellant.
2. Results and discussion
2.1. Carrier gas delivery
Our central hypothesis is that we can alter the release profile of biomacromolecules from ZIF-8 by changing the carrier gas from air to CO2. Once dispersed in water, CO2 lowers the pH in the media, causing the dissolution of ZIF materials. We systematically progressed to investigate changes in ZIF morphology caused by using different gas formulations. We designed an apparatus—which we call a MOF-Jet—to discharge ZIF particles at high speed with commercially obtained carrier gases. Our MOF-Jet (Fig. 1A) was inspired by a previously disclosed design39 with specific modifications to the firing tip (Fig. 1B) to allow us to fire dry MOF powder. This was accomplished with a thin piece of parafilm sandwiched between two metal washers (Fig. 1C) and seated snugly inside the tip (complete schematics and part numbers are in the ESI†). ZIF powder was set onto the top of this thin parafilm membrane and resided there until fired. We initially assessed solution changes following delivery of pristine ZIF-8 crystals, as illustrated in the inset of Fig. 1A. In this experiment, ZIF-8 was bombarded into water using either 100% CO2 or compressed ambient air, which contains 0.04% CO2.
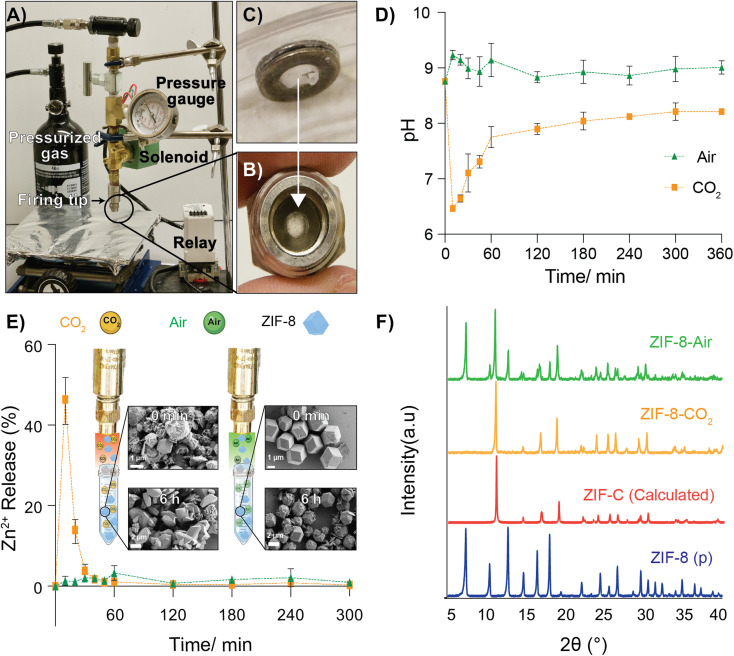
Fig. 1. In vitro release profiles of ZIF-8 delivered via MOF-jet using CO2 and air along with post shooting characterization of ZIF-8. (A) Photographs of MOF-Jet, and individual components are highlighted. Pressurized gas is released by triggering the green electronic solenoid. A relay controlling the solenoid is triggered by an electronic firing push button. The DNA is inoculated from the firing tip onto a sample below. (B) Close up of the firing tip containing the loading washers. (C) Close up of the loading washers showing the sandwiched parafilm design. (D) pH Profiles of solutions after ZIF-8 was fired using compressed air or CO2. Data shows transient mild acidification with a peak at t = 10 min. (E) Zn2+ release profile as measured by ICP-MS after shooting ZIF-8 into DI water with either CO2 or compressed air. The sudden burst of Zn2+ reaches its maximum at t = 10 min whereas little free Zn2+ is observed in the air sample. Following dissolution, a new phase of ZIF (ZIF-C) is formed. SEM images were obtained soon after shooting of ZIF-8 with CO2 and compressed air and after 6 h with CO2 and compressed air. (F) PXRD patterns of the ZIF-8 crystals after discharging them into water with compressed air or CO2 propellants after 6 h.
Time-resolved measurements of free Zn2+ in solution, as well as pH changes, were recorded. Briefly, 25 mg of pristine ZIF-8 was delivered into a tube containing 18 MΩ water (initial pH 8.76) with either CO2 or compressed air. We used unbuffered water to avoid known complications arising from many buffering salts.34 After shooting ZIF-8 into the water, the Zn2+ concentration was monitored in time intervals for up to 6 h using inductively coupled plasma mass spectroscopy (ICP-MS) and the pH of the solution was monitored using a micro-pH probe connected to a customized controller (schematics and controller coding included in ESI†). As shown in Fig. 1E, when using CO2 as the carrier gas, there was a massive increase of Zn2+ in solution, peaking at 10 min post-delivery. This peak alone accounts for nearly 45% of the original Zn2+ ions present in the ZIF-8 crystals. After about 10 min, the Zn2+ concentration gradually decreased, indicating Zn2+ was leaving the solution. Correspondingly, we noted that at the time the Zn2+ concentration was at its highest, the pH was at its lowest (6.0–6.2) and then gradually returned to the starting pH value over the next 6 h. In contrast, the pH of the solution of ZIF-8 delivered with compressed air did not show significant changes (Fig. 1D). This difference in pH changes between carrier gas can be explained by CO2 dissolution in water making weakly acidic carbonic acid, which yields CO32− and HCO3− anions. The increase of the pH after 10 min can be explained by the gradual decarbonization of the water as the CO2 reenters the air. After shooting with CO2, SEM data obtained from samples of ZIF isolated several minutes after shooting (Fig. 1E) shows a different morphology compared to the ZIF-8 bombarded with compressed air, which still looks like the as-synthesized ZIF-8 crystals. Even more profound changes were observed if the solution was allowed to rest for 6 h after shooting.
The PXRD data (Fig. 1F) obtained from the recovered powder following delivery with CO2 or compressed air both shows prominent new reflections, a particularly large one at 11° 2Θ in both samples, which corresponds to the previously reported ZIF polymorph ZIF-CO3-1 40 (also reported as ZIF-C).41 When CO2 was used, pure ZIF-CO3-1 was formed; however, when air was used, a significant fraction of the original ZIF-8 remained. This result is in perfect accord with our hypothesis—prior literature has shown that carbonated water substantially increases the CO2 adsorption of ZIF-8, but affects its crystal structure.42 This phenomenon has been explained based on an irreversible two-step reaction that happens when ZIF-8 and carbonated water mix, initially dissolving into zinc carbonate and 2-methylimidazole and promoting the loss of the ZIF-8 structure and reformation of ZIF-C.40,42 We believe the sudden “loss” of Zn2+ in solution is from the formation of ZIF-C, which appears to begin in earnest at t = 10 m after firing. The addition of buffers and serum seemed to inhibit the formation of ZIF-C in this experiment and complicated pH measurements greatly. We believed that, though buffered solutions did not reliably show changes in bulk aqueous pH, it was still very likely that local pH around the ZIF was being affected, so we next turned to study our release in plant and animal tissues with model biotherapeutics.
2.2. Synthesis and characterization of biomaterials-loaded ZIF-8
We next sought to assess the release of biomolecules from the ZIF-8 framework using different carrier gases. Biomolecules including enzymes,43 genetic material,44 bacteria and yeast,33,45,46 whole viruses,45,47 and intact liposomes48 have been encapsulated within ZIF-8, through a process known as biomimetic mineralization. The rigidity of the MOF structure reduces molecular motions, which enhances the thermal stability compared to its free form.23,24,49 The MOF shell also serves as a physical barrier against proteases and nucleases.23,24 The low toxicity of the ZIF family of MOFs, which biodegrade back into their discrete zinc metal ions and imidazolate linkers, have made them attractive “sheddable” protective coatings for highly sensitive therapeutic biomolecules. Further, ZIF particle size can be controlled by altering the concentrations of the starting biomacromolecule, zinc, and imidazole solutions yielding particles ranging in size between several nanometers to micrometers.
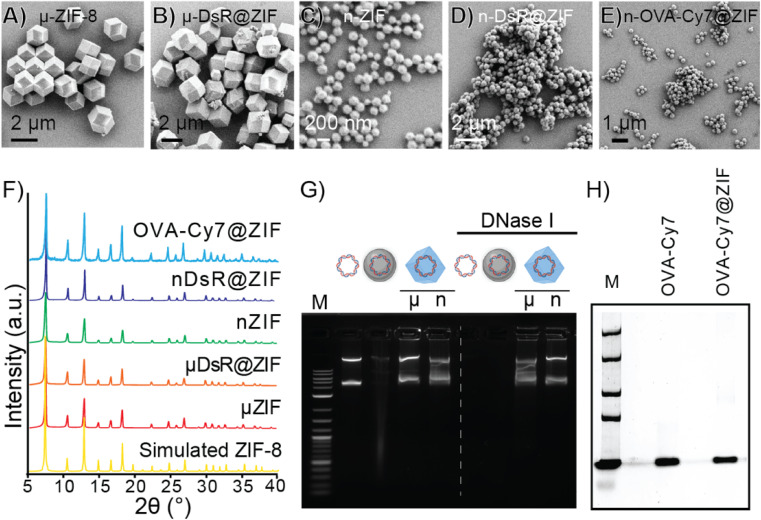
The first of our two proof-of-principle applications will involve the delivery of DNA into plants. This proof-of-principle is significant because transient genetic modification of plants is considered a potential approach to creating pest resistance without germ-line modifications, which require extensive regulatory approval and consumers have found such “GMO” foods undesirable. We selected the pEGB35S:DsRed DNA plasmid, which encodes for a red fluorescent protein derived by Discosoma sp. (Scheme S3†) in planta. In our second proof-of-principle application, we will deliver an intact protein into the subcutaneous layer of the skin of a mouse. Subcutaneous delivery of slow-release proteins is challenging yet is a highly promising avenue for enhancing vaccine performance as well as reducing the frequency of injection of vaccines and therapies (e.g., insulin). To that end, we synthesized a fluorescently tagged ovalbumin protein (OVA-Cy7) for use in animals. We then systematically identified synthetic conditions needed to quantitatively encapsulate the DsRed plasmid and OVA-Cy7 within ZIF-8 to form DsR@ZIF and OVA-Cy7@ZIF respectively. DNA transfection requires very little material (2.5 μg of DNA per 1.0 mg of ZIF-8) and we were able to optimize the metal to ligand ratio to produce both micro (1.02 ± 0.03 μm) and nano (355.00 ± 48.38 μm) sized crystals of pristine ZIF-8 (Fig. 2A and C respectively) and DNA loaded ZIF-8 (Fig. 2B and D). The composites were analyzed by powder X-ray diffraction (PXRD), which confirmed the sodalite ZIF-8 topology (Fig. 2F). Scanning electron microscopy (SEM) shows uniform rhombic dodecahedral DsR@ZIF and OVA-Cy7@ZIF (Fig. 2E) crystals and confirms that the encapsulation of the DNA plasmid and protein does not affect the crystal morphology of ZIF-8. Characterization by thermogravimetric analysis (TGA) and N2 adsorption data show minimal differences between pristine ZIF-8 and DsR@ZIF (Fig. S5A and S5B†). The EDAX (Fig. S5C–F†) data supports DNA encapsulation, as indicated by faint signals for the presence of phosphorus in DsR@ZIF.
Fig. 2. Synthesis and characterization of the ZIF encapsulated DNA plasmid and protein. SEM micrographs of (A) micro ZIF-8, (B) micro DsR@ZIF, (C) nano ZIF-8, (D) nano DsR@ZIF and (E) nano OVA-Cy7@ZIF. (F) PXRD of nano and micro formulations of DsR@ZIF compared to synthesized and simulated ZIF-8. (G) 1% agarose gel before (left) and after (right) DNase I treatment of DsRed plasmid, DsR@W, and DsR@ZIF-8. (H) 10% SDS gel characterization of pristine OVA-Cy7 and exfoliated OVA-Cy7@ZIF with sodium acetate buffer (pH = 3).
While ZIF-8 has been used to deliver DNA and mRNA into mammalian cells by endocytosis50,51 where the acidity of the late endosome dissolves the ZIF, plant cells do not have a comparable endocytosis route so nanoparticle delivery is much more challenging and requires harsh manual (rubbing, grinding, or shooting) delivery. The protection of genetic materials against nuclease degradation is an important aspect of efficient gene transfection, particularly in agricultural settings. The stability of the encapsulated DNA against nuclease degradation was investigated in the nano and micro DsR@ZIF formulations. Metal-based microparticles are the current state-of-the-art carrier in gene delivery for plants, thus we tested DsR@ZIF against DsR coated on tungsten metal microparticles (DsR@W) as well as pristine plasmid (DsR). The samples were incubated with DNase I for 10 min. The DsR@ZIF formulations were removed from the solution and the ZIF shell was dissolved in 0.5 M ethylenediaminetetraacetic acid (EDTA). The solutions were run on an agarose gel, as shown in Fig. 2G. The left of the agarose gel shows the electrophoretic mobility of the DNA plasmid without any nuclease treatment. On the right of the gel, the bands corresponding to free plasmid and DsR@W are notably absent, indicative of DNA digestion. These results show that only the DsR@ZIF prevented nuclease degradation because the DNA is on the inside of the ZIF-8, and therefore inaccessible to nucleases.
In contrast to DNA plasmid encapsulation, for which there are very few examples, ZIF-8 growth and its ability to protect proteins has been demonstrated extensively.52–54 In accord with previously published reports, we found that ZIF-8 growth is quantitative on ovalbumin and it is possible to re-isolate the OVA following ZIF digestion (Fig. 2H) using sodium acetate (pH 5.0).55
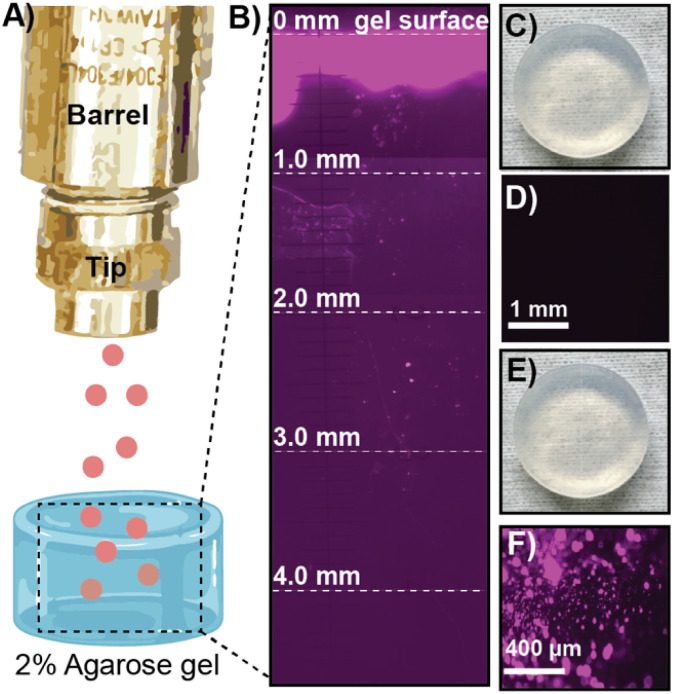
After confirming our cargo was encapsulated and protected within ZIF-8, we moved to optimize the delivery of the microparticles into soft tissue. We optimized the biolistic delivery parameters—operating pressure and distance between the gun nozzle and the targeted tissue—for the gun by firing ZIF-8 particles loaded with the fluorescent dye Cy-5 into homemade ballistic gel, as schematically illustrated in Fig. 3A. Agarose gel (2%, 2 cm × 2 cm) emulates soft tissues in the body12 and allows for the imaging of penetration via fluorescence microscopy, which helps calculate the distance traveled by the fluorescently labeled ZIF-8 particles (Fig. 3B). We found that using the MOF-Jet at a 1400 kPa and 1 cm away from the target, the ZIF-8 particles were principally lodged within the first 0.3 mm of the gel with several particles traveling as far as 3 mm from the gel surface. Importantly, the firing of the MOF gun did not damage the gel surface as shown by the photograph and brightfield images before (Fig. 3C and D) and after firing (Fig. 3E and F). Based on these results, we selected the pressure of 1400 kPa and the tip-to-sample distance of 1 cm for the gene delivery in planta experiment. The optimized shooting pressure and distance for the protein delivery in vivo were obtained as 3450 kPa and 1 cm, according to the same optimization method with the exception that mouse skin was placed over the 2% agarose gel (Fig. S4†) to simulate shooting through the stratum corneum—the hard outermost layer skin. Our full optimization strategy is included in Table S3.†
Fig. 3. Optimization of shooting parameters using 2% agarose ballistic gel model which emulate soft tissue in vivo. (A) Schematic of shooting Cy-5 labeled ZIF-8 into 2% agarose gel. (B) Cross sectional fluorescence image of the gel showing particle penetration. (C) Photograph of gel before shooting Cy-5@ZIF. (D) Fluorescence image of the gel before shooting Cy-5@ZIF. (E) Photograph of gel after shooting Cy-5@ZIF. (F) Fluorescence image of gel after shooting Cy-5@ZIF.
2.3. Gene delivery in planta
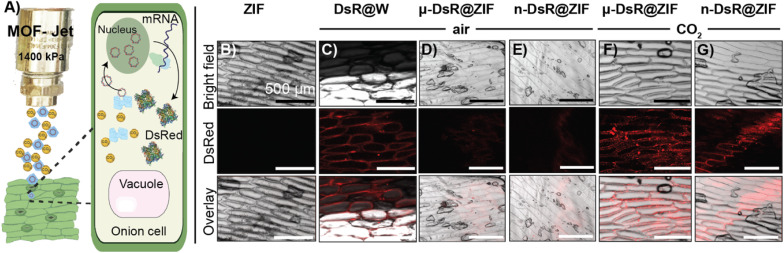
For our first proof-of-principle experiment, gene transfection was done using white onions as the test plant as they have large cells with well-defined structures. Indeed, they were the plant tested in the original biolistic gene delivery paper.56,57 For gene expression to occur, DsR@ZIF must enter the cell, the ZIF shell must dissolve, and then the genetic payload must go to the nucleus (Fig. 4A). The DsRed plasmid encodes mRNA for the synthesis of a red fluorescence protein, which can be monitored through confocal laser scanning microscopy (CLSM). In our first experiment, DsR@ZIF, DsR@W (positive control), and pristine ZIF-8 (negative control) were delivered using compressed air (0.04% CO2) as the propellant (1400 kPa, 1 cm) into an onion bulb, which was then incubated in the dark for 24 h (Fig. S6A†). The negative ZIF control produced no fluorescence while the “classical” metal-based DsR@W produced obvious fluorescence (Fig. 4B and C, respectively). We then tested our ZIF formulations. When compressed air was used there was little fluorescence, indicating low gene transfection in either the micro- or nano-particle formulations of DsR@ZIF (Fig. 4D and E respectively). Shooting ZIF directly into the cytosol of cells, which have pH closer to neutral, is unlikely to induce rapid dissolution and plasmid release, thus these results are in line with the in vitro experiments discussed above. In contrast, when we switched to 100% CO2, we observed gene expression from both nano and micro DsR@ZIF formulations (Fig. 4F and G). These results show the importance of inducing “burst release” in gene delivery with ZIF-8—which is not something that is possible unless this compressed-gas delivery mechanism is utilized.
Fig. 4. Evaluation of gene delivery and gene transfection in planta. (A) Schematic of the expected gene transfection process after shooting DsR@ZIF into plant cells. Confocal micrographs of post gene transfection with (B) ZIF-8 (negative control) and (C) DsR@W (positive control). Confocal micrographs of (D) micro DsR@ZIF and (E) nano DsR@ZIF shooting with compressed air. (F and G) Gene expression is seen with micro and nano DsR@ZIF respectively when CO2 was used as the particle propellant.
2.4. Protein delivery in vivo
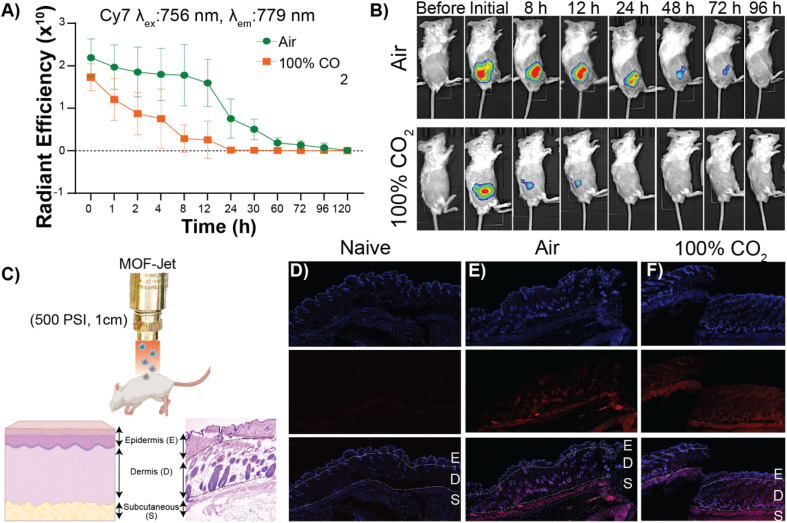
In our second proof-of-principle experiment, we extended this controlled biolistic delivery to study the release profile of protein therapeutics in the skin of a mouse model. Plant cells are large and tightly packed, and live cells are located directly on the surface of plants. In contrast, the epidermis is coated in a thick layer of dead cells and, under that, the cells that make up the skin are soft, and not as tightly packed as those in a plant. In general, delivery of microparticles into the skin is thought to result in these particles being trapped in the interstitium—or spaces between the cells.58 One reason drugs are delivered into the skin—as is the case for vaccines and insulin—is that delivery into the interstitium provides a means of slow-release into circulation. Until now, however, controlling that release from the interstitium required specific polymer formulations that had tuned decay times.59 We will show here that, using the exact same material, we can change release kinetics just by changing the propulsion gas used in our MOF-Jet. Ovalbumin (OVA) is an established model protein/antigen owing to its well-documented properties.55 As discussed before, OVA was fluorescently labeled with near-infrared Cy7 dye (OVA-Cy7) for the purpose of monitoring its tissue residency over time. OVA-Cy7 was then encapsulated in ZIF-8 (OVA-Cy7@ZIF) and dried to get a solid formulation. We then progressed to fire these into mice flanks. For our experiments, eight BALB/c mice (6–8 weeks old) were divided into two groups (n = 4). The same amount of material (0.5 mg dry weight) was inoculated into the left flank of each mouse with 3450 kPa pressure and a 1 cm distance between the gun nozzle and the skin surface. After shooting with compressed air (0.04% CO2) and 100% CO2, the tissue residency of the OVA-Cy7@ZIF was monitored until fluorescence decreased to their baseline levels. The obtained results were in agreement with the data obtained for the plant gene transfection study. According to the tissue residency data reported in Fig. 5A, OVA-Cy7@ZIF was delivered using 100% CO2 cleared out from the flank tissue environment within 12 h (t1/2 ∼ 3 h). In contrast, the fluorescence of OVA-Cy7@ZIF delivered using compressed air remained at the tissue site for almost 4 days (t1/2 ∼ 18 h). These observations are in line with our hypothesis that when CO2 is used as the carrier gas, ZIF dissolution promoted by media acidification results in burst release.
Fig. 5. Evaluation of protein delivery in vivo. (A) The radiant efficiency changes of the OVA-Cy7@ZIF over time with compressed air (0.04% CO2) and pure CO2 (100% CO2). (B) Time-resolved fluorescence images showing the tissue residency of biolistically bombarded Cy7 labeled OVA in ZIF-8 (OVA-Cy7@ZIF) into the left flank of BALB/c mice with compressed air (top) or pure CO2 (bottom) as the carrier gas. (C) (Top) A schematic representation biolistically bombarded ZIF into the mouse using smURFP@ZIF. (Bottom left) The layers of the skin are labeled schematically. (Bottom right) An H&E-stained skin cross-section obtained from a male BALB/c mouse to show the actual physiology of the skin. Skin section from (D) naïve mice, (E) mice inoculated with smURFP@ZIF propelled by air, and (F) mice inoculated with smURFP@ZIF propelled by CO2. The fluorescent micrographs show DAPI-stained nuclei (top) and the location of the smURFP/Cy5 particles (middle) and their respective overlay images (bottom) with the skin sections labeled as (E)pidermis, (D)ermis, and (S)ubcutaneous. The majority of the red fluorescence is in the subcutaneous area.
Next, we analyzed how deep ZIF particles traveled through tissue when inoculated into a mouse flank. For that purpose, we encapsulated small-ultra red fluorescent protein (smURFP) in ZIF-8 (smURFP@ZIF). smURFP was selected because of its stability, brightness, and compatibility with common filter sets on scanning confocal microscopes. Male BALB/c mice were inoculated with 0.5 mg of smuRFP@ZIF into the left flank, mice were immediately sacrificed to minimize the dissipation of smURFP in the CO2 sample, and organs/tissue were collected for further analysis. Fluorescence and histological examination of the skin and underlying tissues showed that most of the particles had been logged in the dermis and subcutaneous layer (Fig. 5D, E and F). Importantly, our results show that either choice of gas resulted in the particles entering the same layers of skin (Fig. 5D and F).
3. Conclusion
In the present work, we demonstrate a facile method for biomolecule biolistic delivery using ZIF-8 as a scaffold. We show that qualitatively, ZIF-8 performs as well as tungsten microparticles in the transfection of onion cells following gene gun delivery, demonstrating for the first time the successful transfection of plant cells using a MOF-based material. We further demonstrated that the encapsulated therapeutics can be released either slowly or in a burst release simply by changing the carrier gas from compressed air to CO2. We show, for the first time, that the release of biomacromolecules can be altered without changing the underlying substrate, rather, we need merely change the carrier gas used in the delivery process. When neat (100%) CO2 is used, the ZIF carrier brings with it all that is necessary for its fast dissolution and rapid transfection in the plant and to release the therapeutics faster. This latter observation has considerable promise to promote new pathways for controllable biolistic delivery in a broad range of organisms for various therapeutic and gene transfection applications. Therefore, the lower gene transfection efficiency when air is used can be attributed to the fact that, without the help of CO2, the ZIF-8 does not dissolve or degrade quickly, and the encapsulated DNA and therefore released slowly. This slower rate of release may ultimately be useful as the “time-delayed” aspect of ZIF-8 dissolution has recently been used to promote better immune responses to bacterial infections in vivo and it might be applicable to gene transfection in vivo and in planta as well.
4. Experimental
4.1. Materials and methods
All reagents here described were used without further purification. Ethylenediaminetetraacetic acid (EDTA), 2-methylimidazole, and zinc acetate dihydrate were purchased from Sigma-Aldrich. pEGB 35S:DsRed:Tnos (GB0361) was gifted from Diego Orzaez (Addgene plasmid ## 68220; http://n2t.net/addgene:68220; RRID:Addgene_68220) was gifted from Addgene (USA). Propidium iodide and nuclease-free water were purchased from Thermo Fisher Scientific (Waltham, MA, USA). DNase I and SYBR Gold DNA stain were purchased from New England Biolabs. Ultrapure water was obtained from an ELGA PURELAB Flex 2 system with resistivity measured at least 18.2 MΩ cm−1. White onions were purchased from the local grocery (Walmart). PureYield™ Plasmid Maxiprep System for plasmid purification and extraction was purchased from Promega (USA). Ovalbumin was purchased from Worthington biochemicals. Lumiprobe sulfocyanine7 NHS ester was purchased from Thermo Fisher Scientific.
4.2. DsRed plasmid purification
Kanamycin-supplied LB agar plates were streaked with pEGB 35S:DsRed transformed Escherichia coli (purchased from Addgene) using a sterilized loop and incubated overnight at 37 °C. Single colonies were added to 5 mL of LB kanamycin-containing media and incubated overnight at 37 °C with continuous shaking. Further, the cells were amplified in 1000 mL of LB media, incubated at 37 °C for 19 h, and their optical density monitored. At an optical density value of 0.9, cells were harvested by centrifugation at 5000 × g for 10 min, and the plasmid was isolated using a Promega Maxiprep kit. Steps for plasmid isolation and purification are described elsewhere.60 Purified pEGB 35S:DsRed was collected in DNase-free water and stored at −20 °C for further use. The concentration of the DNA was determined by NanoDrop™ UV-vis spectroscopy.
4.3. Synthesis of pEGB 35S:DsRed@ZIF-8 micro and nanocomposites
Two formulations were used for the encapsulation of the DsRed plasmid. The micro-DNA-containing ZIF formulation was prepared as follows: 25 μL of 1 mg mL−1 sDsRed plasmid were added to a solution of 752 μL of nuclease-free water and 213 μL of 3 M 2-methylimidazole. Then, 10 μL of 1 M zinc acetate dihydrate was added. The resulting solution was mixed thoroughly, and almost immediately following the addition of zinc, the mixture went from colorless to cloudy. The reaction proceeded for 18 h at RT, and the DNA@ZIF crystals were harvested by centrifugation 10 000 × g for 15 min. The resulting supernatant was collected and used for encapsulation efficiency determination. Two additional water washes removed excess precursors, and crystals were allowed to dry at RT for use as is.
Similarly, the micro pristine ZIF-8 solution was prepared by mixing 777 μL of nuclease-free water, 213 μL of 3 M 2-methylimidazole, and 10 μL of 1 M zinc acetate dihydrate. The nanoformulation was prepared following a protocol described elsewhere.50 Briefly, 95 mg of 2-methylimidazole and 7 mg of zinc acetate dihydrate were dissolved separately in 500 μL of nuclease-free water. 25 μL of 1 mg mL−1 DsRed plasmid was added to the 2-methylimidazole solution. Next, the zinc solution was added, and the solution was thoroughly mixed. The encapsulation proceeded for 15 min at RT, and the DNA-loaded ZIF-8 crystals were harvested by centrifugation 10 000 × g for 15 min. The supernatant was collected and used for the assessment of encapsulation efficiency. Accordingly, the nano pristine ZIF-8 crystals were afforded by reacting 95 mg of 2-methylimidazole and 7 mg of zinc acetate dihydrate in a final volume of 1000 μL of nuclease-free water for 15 min at RT. Unreacted precursors were removed by water washes (2×) and dried in the open air at RT.
4.4. Synthesis of Cy5@ZIF
Synthesis of Cy5@ZIF was done following the same procedure used to synthesize nano DsR@ZIF. Briefly, 95 mg of 2-methylimidazole and 7 mg of zinc acetate dihydrate were dissolved separately in 500 μL of water. Then, 25 μL of 1 mg mL−1 Cy5 stock was added to the 2-methylimidazole solution and mixed well. Once thoroughly mixed, zinc acetate dihydrate solution (500 μL) was added to Cy5, 2-methylimidazole mixture and mixed well. The mixture was allowed to react for 15 min at RT. Blue Cy5@ZIF powder was harvested by centrifugation at 17 000 × g for 15 min. The Cy5@ZIF was washed twice with water and dried at RT.
4.5. Synthesis of OVA-Cy7
Protein solution – Ovalbumin 10 mg was dissolved in 810 μL of H2O. This Ovalbumin solution was added to 90 μL of 1 M NaHCO3 buffer (pH = 8.4). Dye solution-63 μL of Cy7NHS ester was dissolved in 37 μL of DMSO. Then, the protein and dye solutions were mixed well and incubated overnight at 4 °C overnight on a rotisserie. The resulted OVA-Cy7 was washed several times with water in a 10 k spin column to remove all the non-reacted Cy7 dye.
4.6. Synthesis of OVA-Cy7@ZIF and nanocomposites
OVA-Cy7@ZIF was synthesized following the same procedure as nano DsRed@ZIF by adding 3.125 μL of OVA-Cy7 (32 mg mL−1) to encapsulate 100 μg of OVA-Cy7 in 1 mL of ZIF-8 final reaction mixture. Then to confirm the successful encapsulation of OVA in ZIF, OVA-Cy7@ZIF was exfoliated using 1 M sodium acetate buffer (pH = 3) and run on 10% SDS gel, 100 V for 45 min.
4.7. Synthesis of smURFP and smURFP@ZIF and nanocomposites
smURFP protein was synthesized according to a previously reported protocol.61 smURFP@ZIF was synthesized using the same method used for Cy5@ZIF using smURFP instead of Cy5.
4.8. Fluorescence spectroscopy
We used fluorescence spectroscopy to determine the encapsulation efficiency. Briefly, propidium iodide (1 mg mL−1) was added to the supernatant of the DsR@ZIF. A 1 mg mL−1 pristine DNA solution was used as a control. The reaction proceeded at RT for 10 min (covered in foil). Fluorescence spectra were then collected using a Tecan Spark 20 M plate reader (Ex λmax = 535 nm and Em λmax = 617 nm). Data collected revealed an encapsulation efficiency of 95.5% for micron formulation and 94.8% for nanoformulation.
4.9. Material characterization
Surface morphology was investigated using a Zeiss Supra 40 scanning electron microscope at 1 kV and a working distance of 6 mm. Surface area measurements were carried out on a Micrometrics ASAP 2020 surface area analyzer by nitrogen adsorption under 77 K. Surface area was independently analyzed using the Brunauer–Emmett–Teller (BET) method, and pore size was determined under the non-localized density functional theory with a carbon slit pore model 18. Sample activation took place before analysis. All samples were soaked in MeOH and dried under a high vacuum overnight. The crystals were then washed in dichloromethane overnight. Lastly, the dichloromethane was removed by high vacuum overnight. Sample degassing was done at 120 °C under a vacuum for 12 h. PXRD data was measured using a Rigaku SmartLab X-ray diffractometer equipped with CuKα (1.54060 Å) at 30 mA and 40 kV. All samples reported here were activated before analysis. Data for each PXRD collected was uploaded into the Global fit software and analyzed from 5° to 40° (2θ). Thermogravimetric analysis of each sample was done in a TA Instruments SDT Q600 Analyzer. The temperature was ramped up from 30 to 800 °C, under an N2 atmosphere, under a constant heating rate of 5 °C min−1.
4.10. DNase I assay
DNase I treatment was used to evaluate the protection of DNA from nuclease degradation through encapsulation inside ZIF-8. The treatment was done using TURBO™ DNase (Invitrogen) following the manufacturer's protocol. In brief, DNase I (1 μL of 2 U μL−1) was added to both micro and nano DsR@ZIF, pristine DsRed plasmid DNA, and DsR@W. The reaction was allowed to occur for 10 min at 37 °C. As controls, untreated pristine DsRed plasmid DNA, micro DsR@ZIF, nano DsR@ZIF, and DsRed@W were used. Next, 1 μL of 0.5 M EDTA was added to terminate the enzymatic reaction by denying the divalent cations needed for the degradation reaction. The treated DsR@ZIF samples were centrifuged, and the supernatants were removed. Finally, 100 μL of 0.5 M EDTA was added to dissolve the ZIF-8 crystals, releasing the encapsulated DNA. The solutions were run on an agarose gel. It was noted that the bands corresponding to free plasmid and DsR@W are notably absent, indicative of DNA digestion. On the other hand, the DsR@ZIF formulations still contain DNA, showing they prevented nuclease degradation. In other words, the DNA is on the inside of the ZIF-8 and therefore inaccessible to nucleases.
4.11. Optimization of shooting parameters of the particle delivery system for plants
Cy-5 fluorescent dye-labeled ZIF-8 was inoculated at different distances with different pressure values ranging from 700 kPa to 2100 kPa to obtain the maximum penetration depth conditions. This process was carried out using 2 × 2 cm 2% agarose gels made using a silicon mold. 2% agarose gels were used for this optimization as it has previously been reported as a model for emulating soft tissue.12 After shooting fluorescently labeled ZIF-8 into the gel, the penetration depth of ZIF-8 particles inside the gel was measured by epifluorescence microscopy using a microscope calibration slide (visible behind the gel in Fig. 2). X denotes a broken agarose gel. The process and results are shown in Fig. S6. and Table S3.†
4.12. Biolistic delivery of DsRed@ZIF-8 into onion tissues
The synthesized micro and nano-size crystals of DsRed@ZIF-8 were dried at RT before being used for bombardment. The optimized bombardment parameters for the onion epidermis were 1400 kPa, 1 cm distance between the gun nozzle and tissue. Bombardment of onion bulbs was performed according to the lab safety protocols inside a fume hood. After bombardment, the onion bulbs were placed inside a culture plate on a napkin wetted with DI water. The plates were covered with aluminum foil and incubated for 24 h. The epidermis tissue layer was then carefully excised from the bulb and imaged using confocal laser microscopy and epifluorescence microscopy to evaluate the DsRed fluorescence protein expression. Air and CO2 gas were used as particle propellants, and both data sets were analyzed for comparison.
4.13. Study of ZIF-8 release profile in vitro with CO2 and compressed air
25 mg of pristine ZIF-8 was delivered directly into 10 mL of DI water using CO2 and compressed air as particle propellants, respectively. After delivering ZIF-8 into DI water, the falcon tubes were capped and allowed to mix on a rotisserie properly. 0.1 mL aliquots of the supernatant were collected at each time interval and replaced with DI water. The pH of the solution at each time point was recorded using a micro-pH probe connected to a customized controller. To obtain Zn2+ concentrations, 9.9 mL of 2% HNO3 acid was added to 0.1 mL of the supernatant and allowed to react for 48 h before ICP-MS measurements. The standard curve for Zn2+ was prepared by dissolving/digesting 25 mg of pristine ZIF-8 in 10 mL of 2% HNO3 acid. Both experiments were done in triplicate (n = 3).
4.14. Study of ZIF-8 release profile without shooting, under standard RT conditions
To study the ZIF-8 stability/releasing profile without shooting, 25 mg of pristine ZIF-8 in powder form was dispersed and resuspended in 10 mL of DI water in falcon tubes. Then the falcon tubes were capped and allowed to mix in a rotisserie properly. 0.1 mL aliquots of the supernatants were collected at each time interval and replaced the solution with DI water. The pH of the solution at each time point was also recorded using a micro-pH probe connected to a customized controller. Then, 9.9 mL of 2% HNO3 acid was added to 0.1 mL of the supernatant and allowed to digest for 48 h before ICP-MS measurements of Zn2+. The standard curve for Zn2+ was prepared by dissolving/digesting 25 mg of pristine ZIF-8 in 10 mL of 2% HNO3 acid. Both experiments were done in triplicate (n = 3). Finally, the obtained powders were analyzed by PXRD, and the ZIF phase analysis was done by (https://rapps.tugraz.at/apps/porousbiotech/ZIFphaseanalysis/) named ZIF phase analysis. By uploading diffraction patterns collected using Cu Kα radiation, this web application allows for rapid identification of the crystalline phases of ZIF. Data is shown in Fig. S7.†
4.15. Optimization of the amount of OVA-Cy7 to be encapsulated in ZIF-8 for each flank shooting
Different amounts of OVA-Cy7 ranging from 6.25–100 μg was used to determine the optimum fluorescence intensity through the live animal imager (IVIS Lumina III- PerkinElmer, Waltham, MA, USA) as shown in Fig. S3.†
4.16. In vivo release study by tissue residency of OVA-Cy7@ZIF after shooting into mouse flank
Cy7 labeled ovalbumin 25 μg was encapsulated in ZIF-8 (OVA-Cy7@ZIF) in 250 μL final volume for one-inoculation (∼0.5 mg of OVA-Cy7@ZIF) and was dried to get a solid formulation of the protein. Then each formulation was inoculated into the left flank of male BALB/c mice (n = 4) model with 3450 kPa of gas pressure and a 1 cm distance between the gun nozzle and the skin surface. After shooting with compressed air (0.04% CO2) and 100% CO2, the tissue residency of the OVA-CY7@ZIF in two groups was monitored over several days using the live animal imager.
4.17. Skin penetration studies of smURFP@ZIF
The same experiment mentioned above in 16 was performed using smURFP@ZIF. A total of four BALB/c male mice were divided into two groups (n = 2) as 100% CO2 and air. 100 μg of smURFP protein was encapsulated in ZIF-8 according to the same protocol mentioned above. Then each formulation was inoculated into the left flank of male BALB/c mice models with 3450 kPa of gas pressure and a 1 cm distance between the MOF-jet nozzle and the skin surface. Crossed sections of the skin inoculated with smURFP@ZIF and skin samples from the naïve mice (n = 2) were collected into 4% PFA in 1X PBS for fluorescence analysis.
4.18. smURFP fluorescence analysis of the skin tissues through confocal microscopy
Soon after shooting, mice were euthanized, and skin cross-sections of the inoculated area intact with the muscle were fixed in 4% PFA in 1X PBS, processed, and embedded in paraffin wax. The tissue samples were sectioned at 4 μm sized using a Leica rotary microtome. The sections were collected and stained with DAPI to stain the nuclei. The fixed samples were analyzed through a confocal microscope for smURFP fluorescence and DAPI to determine the smURFP@ZIF particle penetration depth.
4.19. Histological analysis of the skin and other major organs after shooting experiments
Major organs, skin surface, and cross-sectioned tissue samples were collected into 4% PFA in 1XPBS. Then the organs were fixed in 4%PFA for 48 h under shaking conditions. After that, the organs and tissues were washed with 70% ethanol, processed, and embedded in paraffin wax. Organs and tissue samples were sectioned at 4 μm thickness using a Leica rotary microtome. For pathological analysis, the sections were collected and stained with hematoxylin and eosin (H&E). The results shown in Fig. S8.†
Ethical statement
Female and male BALB/c mice were obtained from Charles River Lab (Wilmington, Ma). All animal studies including biolistic delivery were performed in strict accordance with the protocol #19-06 approved by the University of Texas at Dallas Institutional Animal Care and Use Committee (IACUC).
Data availability
Data are avaliable upon request.
Author contributions
Conceptualization: JJG, YHW, methodology: YHW, FCH, investigation: YHW, FCH, OT, IT, ORB, SK, TH, CEB, AS, SDD, SDP, SAC, JPV, KJB, RAS, NDN, JJG, supervision: JJG, writing—original draft: YHW, JJG, writing—review & editing: YHW, JJG.
Conflicts of interest
The authors declare that they have no competing interests.
Supplementary Material
Acknowledgments
R. A. S thanks the, Army Research Laboratory (W911NF-18-2-0035). J. J. G. acknowledges support from the National Science Foundation (DMR-2003534). Welch Foundation (AT-1989-20190330). University of Texas at Dallas lab animal resource center (LARC) for their assistance in animal care. N. J. D thanks the Welch Foundation (AT-2030-20200401).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc04982a
References
- Tyagi P. Santos J. L. Drug Discovery Today. 2018;23:1053–1061. doi: 10.1016/j.drudis.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Bruno B. J. Miller G. D. Lim C. S. Ther. Delivery. 2013;4:1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowacz M., Krans A., Wallén C., Martinez A. and Mohammadi N., Student thesis, Independent thesis Basic level (degree of Bachelor), Uppsala University, Disciplinary Domain of Science and Technology, 2017 [Google Scholar]
- Crommelin D. J. A. Anchordoquy T. J. Volkin D. B. Jiskoot W. Mastrobattista E. J. Pharm. Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami F. Vatanara A. Park E. J. Na D. H. Pharmaceutics. 2018;10(3):131. doi: 10.3390/pharmaceutics10030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Hong S. Liu M. D. Yu W. Y. Zhang M. K. Zhang L. Zeng X. Zhang X. Z. J. Controlled Release. 2020;320:159–167. doi: 10.1016/j.jconrel.2020.01.038. [DOI] [PubMed] [Google Scholar]
- Luzuriaga M. A. Shahrivarkevishahi A. Herbert F. C. Wijesundara Y. H. Gassensmith J. J. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2021:e1735. doi: 10.1002/wnan.1735. [DOI] [PubMed] [Google Scholar]
- Luzuriaga M. A. Berry D. R. Reagan J. C. Smaldone R. A. Gassensmith J. J. Lab Chip. 2018;18:1223–1230. doi: 10.1039/C8LC00098K. [DOI] [PubMed] [Google Scholar]
- Ray S. Wirth D. M. Ortega-Rivera O. A. Steinmetz N. F. Pokorski J. K. Biomacromolecules. 2022;23:903–912. doi: 10.1021/acs.biomac.1c01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly R. F. Raj Singh T. R. Woolfson A. D. Drug Delivery. 2010;17:187–207. doi: 10.3109/10717541003667798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger B. G. Papania M. J. Vaccines. 2013:1200–1231. [Google Scholar]
- Zilony N. Tzur-Balter A. Segal E. Shefi O. Sci. Rep. 2013;3:2499. doi: 10.1038/srep02499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J. A. Lummis S. C. Nat. Protoc. 2006;1:977–981. doi: 10.1038/nprot.2006.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aps L. Tavares M. B. Rozenfeld J. H. K. Lamy M. T. Ferreira L. C. S. Diniz M. O. J. Biotechnol. 2016;228:58–66. doi: 10.1016/j.jbiotec.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Crichton M. L. Muller D. A. Depelsenaire A. C. I. Pearson F. E. Wei J. Coffey J. Zhang J. Fernando G. J. P. Kendall M. A. F. Sci. Rep. 2016;6:27217. doi: 10.1038/srep27217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Hoai T. Hohn O. Vu M. D. Baldenhofer G. Sayed Ahmed M. S. Dörken B. Norley S. Lipp M. Pezzutto A. Westermann J. Cancer Gene Ther. 2012;19:880–887. doi: 10.1038/cgt.2012.78. [DOI] [PubMed] [Google Scholar]
- O'Brien J. A. Lummis S. C. R. BMC Biotechnol. 2011;11:66. doi: 10.1186/1472-6750-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.-Y. Huang S.-H. Lin C.-M. Plant Biotechnol. Rep. 2008;2:267–270. doi: 10.1007/s11816-008-0067-2. [DOI] [Google Scholar]
- Zhang D. Das D. B. Rielly C. D. Drug Delivery. 2014;21:571–587. doi: 10.3109/10717544.2013.864345. [DOI] [PubMed] [Google Scholar]
- Russell J. A. Roy M. K. Sanford J. C. Plant Physiol. 1992;98:1050–1056. doi: 10.1104/pp.98.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods G. Zito K. J. Visualized Exp. 2008;12:675. doi: 10.3791/675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefi O. Simonnet C. Baker M. W. Glass J. R. Macagno E. R. Groisman A. J. Neurosci. Res. 2006;26:6119–6123. doi: 10.1523/JNEUROSCI.1237-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H. Cordova K. E. O'Keeffe M. Yaghi O. M. Science. 2013;341:1230444. doi: 10.1126/science.1230444. [DOI] [PubMed] [Google Scholar]
- Liang K. Ricco R. Doherty C. M. Styles M. J. Bell S. Kirby N. Mudie S. Haylock D. Hill A. J. Doonan C. J. Falcaro P. Nat. Commun. 2015;6:7240. doi: 10.1038/ncomms8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire C. V. Forgan R. S. Chem. Commun. 2015;51:5199–5217. doi: 10.1039/C4CC04458D. [DOI] [PubMed] [Google Scholar]
- Abanades Lazaro I. Wells C. J. R. Forgan R. S. Angew. Chem., Int. Ed. Engl. 2020;59:5211–5217. doi: 10.1002/anie.201915848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. Liang K. Adv. Funct. Mater. 2020:30. [Google Scholar]
- Drout R. J. Robison L. Farha O. K. Coord. Chem. Rev. 2019;381:151–160. doi: 10.1016/j.ccr.2018.11.009. [DOI] [Google Scholar]
- Wu M. X. Yang Y. W. Adv. Mater. 2017;29(23):1606134. doi: 10.1002/adma.201606134. [DOI] [PubMed] [Google Scholar]
- Chen H. Yang J. Sun L. Zhang H. Guo Y. Qu J. Jiang W. Chen W. Ji J. Yang Y. W. Wang B. Small. 2019;15:e1903880. doi: 10.1002/smll.201903880. [DOI] [PubMed] [Google Scholar]
- Riccò R. Liang W. Li S. Gassensmith J. J. Caruso F. Doonan C. Falcaro P. ACS Nano. 2018;12:13–23. doi: 10.1021/acsnano.7b08056. [DOI] [PubMed] [Google Scholar]
- Luzuriaga M. A. Welch R. P. Dharmarwardana M. Benjamin C. E. Li S. Shahrivarkevishahi A. Popal S. Tuong L. H. Creswell C. T. Gassensmith J. J. ACS Appl. Mater. Interfaces. 2019;11:9740–9746. doi: 10.1021/acsami.8b20504. [DOI] [PubMed] [Google Scholar]
- Luzuriaga M. A. Herbert F. C. Brohlin O. R. Gadhvi J. Howlett T. Shahrivarkevishahi A. Wijesundara Y. H. Venkitapathi S. Veera K. Ehrman R. Benjamin C. E. Popal S. Burton M. D. Ingersoll M. A. De Nisco N. J. Gassensmith J. J. ACS Nano. 2021;15:17426–17438. doi: 10.1021/acsnano.1c03092. [DOI] [PubMed] [Google Scholar]
- Luzuriaga M. A. Benjamin C. E. Gaertner M. W. Lee H. Herbert F. C. Mallick S. Gassensmith J. J. Supramol. Chem. 2019;31:485–490. doi: 10.1080/10610278.2019.1616089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldone R. A. Forgan R. S. Furukawa H. Gassensmith J. J. Slawin A. M. Z. Yaghi O. M. Stoddart J. F. Angew. Chem., Int. Ed. 2010;49:8535. doi: 10.1002/anie.201004618. [DOI] [PubMed] [Google Scholar]
- Gassensmith J. J. Kim J. Y. Holcroft J. M. Farha O. K. Stoddart J. F. Hupp J. T. Jeong N. C. J. Am. Chem. Soc. 2014;136:8277–8282. doi: 10.1021/ja5006465. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Xian S. Xi H. Wang H. Li Z. Chem. Eng. Sci. 2011;66:4878–4888. doi: 10.1016/j.ces.2011.06.051. [DOI] [Google Scholar]
- Chen R. Yao J. Gu Q. Smeets S. Baerlocher C. Gu H. Zhu D. Morris W. Yaghi O. M. Wang H. Chem. Commun. 2013;49:9500–9502. doi: 10.1039/C3CC44342F. [DOI] [PubMed] [Google Scholar]
- Lin H.-J., Huang K.-J., Yang H.-T. and Chen W.-C., U.S.Pat., US6436709B1, 2002
- Basnayake S. A. Su J. Zou X. Balkus K. J. Inorg. Chem. 2015;54:1816–1821. doi: 10.1021/ic5027174. [DOI] [PubMed] [Google Scholar]
- Carraro F. Velásquez-Hernández M. d. J. Astria E. Liang W. Twight L. Parise C. Ge M. Huang Z. Ricco R. Zou X. Villanova L. Kappe C. O. Doonan C. Falcaro P. Chem. Sci. 2020;11:3397–3404. doi: 10.1039/C9SC05433B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Guo P. Regueira T. Wang Z. Du J. Chen G. J. Phys. Chem. C. 2016;120:13287–13294. doi: 10.1021/acs.jpcc.6b03772. [DOI] [Google Scholar]
- Liang W. Xu H. Carraro F. Maddigan N. K. Li Q. Bell S. G. Huang D. M. Tarzia A. Solomon M. B. Amenitsch H. Vaccari L. Sumby C. J. Falcaro P. Doonan C. J. J. Am. Chem. Soc. 2019;141:2348–2355. doi: 10.1021/jacs.8b10302. [DOI] [PubMed] [Google Scholar]
- Poddar A. Conesa J. J. Liang K. Dhakal S. Reineck P. Bryant G. Pereiro E. Ricco R. Amenitsch H. Doonan C. Mulet X. Doherty C. M. Falcaro P. Shukla R. Small. 2019;15:e1902268. doi: 10.1002/smll.201902268. [DOI] [PubMed] [Google Scholar]
- Liang K. Richardson J. J. Doonan C. J. Mulet X. Ju Y. Cui J. Caruso F. Falcaro P. Angew. Chem., Int. Ed. Engl. 2017;56:8510–8515. doi: 10.1002/anie.201704120. [DOI] [PubMed] [Google Scholar]
- Li S. Zhou X. Chen Z. Herbert F. C. Jayawickramage R. Panangala S. D. Luzuriaga M. A. Alahakoon S. B. Diwakara S. D. Meng X. Fei L. Ferraris J. Smaldone R. A. Gassensmith J. J. ACS Appl. Mater. Interfaces. 2020;12:11884–11889. doi: 10.1021/acsami.9b15667. [DOI] [PubMed] [Google Scholar]
- Li S. Dharmarwardana M. Welch R. P. Benjamin C. E. Shamir A. M. Nielsen S. O. Gassensmith J. J. ACS Appl. Mater. Interfaces. 2018;10:18161–18169. doi: 10.1021/acsami.8b01369. [DOI] [PubMed] [Google Scholar]
- Herbert F. C. Abeyrathna S. S. Abeyrathna N. S. Wijesundara Y. H. Brohlin O. R. Carraro F. Amenitsch H. Falcaro P. Luzuriaga M. A. Durand-Silva A. Diwakara S. D. Smaldone R. A. Meloni G. Gassensmith J. J. Nat. Commun. 2021;12:2202. doi: 10.1038/s41467-021-22285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W. Ricco R. Maddigan N. K. Dickinson R. P. Xu H. Li Q. Sumby C. J. Bell S. G. Falcaro P. Doonan C. J. Chem. Mater. 2018;30:1069–1077. doi: 10.1021/acs.chemmater.7b04977. [DOI] [Google Scholar]
- Li Y. Zhang K. Liu P. Chen M. Zhong Y. Ye Q. Wei M. Q. Zhao H. Tang Z. Adv. Mater. 2019;31:e1901570. doi: 10.1002/adma.201901570. [DOI] [PubMed] [Google Scholar]
- Orellana-Tavra C. Haddad S. Marshall R. J. Abánades Lázaro I. Boix G. Imaz I. Maspoch D. Forgan R. S. Fairen-Jimenez D. ACS Appl. Mater. Interfaces. 2017;9:35516–35525. doi: 10.1021/acsami.7b07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddigan N. K. Tarzia A. Huang D. M. Sumby C. J. Bell S. G. Falcaro P. Doonan C. J. Chem. Sci. 2018;9:4217–4223. doi: 10.1039/C8SC00825F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J. Young A. P. Tsung C.-K. Small. 2017;13:1700880. doi: 10.1002/smll.201700880. [DOI] [PubMed] [Google Scholar]
- Liu J. Wen Q. Zhou B. Yuan C. Du S. Li L. Jiang L. Yao S. Q. Ge J. ACS Chem. Biol. 2022;17:32–38. doi: 10.1021/acschembio.1c00872. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Wang F. Ju E. Liu Z. Chen Z. Ren J. Qu X. Adv. Funct. Mater. 2016;26:6454–6461. doi: 10.1002/adfm.201600650. [DOI] [Google Scholar]
- Klein T. M. Wolf E. D. Wu R. Sanford J. C. Nature. 1987;327:70–73. doi: 10.1038/327070a0. [DOI] [Google Scholar]
- Lacroix B. Citovsky V. Methods Mol. Biol. 2020;2124:125–139. doi: 10.1007/978-1-0716-0356-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow T. W. Grice J. E. Lin L. L. Faye R. Butler M. Becker W. Wurm E. M. T. Yoong C. Robertson T. A. Soyer H. P. Roberts M. S. Adv. Drug Delivery Rev. 2011;63:470–491. doi: 10.1016/j.addr.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Welch R. P. Lee H. Luzuriaga M. A. Brohlin O. R. Gassensmith J. J. Bioconjugate Chem. 2018;29:2867–2883. doi: 10.1021/acs.bioconjchem.8b00483. [DOI] [PubMed] [Google Scholar]
- Traustadóttir G. Á. Jensen C. H. Garcia Ramirez J. J. Beck H. C. Sheikh S. P. Andersen D. C. Int. J. Biol. Macromol. 2017;97:460–467. doi: 10.1016/j.ijbiomac.2017.01.067. [DOI] [PubMed] [Google Scholar]
- Herbert F. C. Brohlin O. R. Galbraith T. Benjamin C. Reyes C. A. Luzuriaga M. A. Shahrivarkevishahi A. Gassensmith J. J. Bioconjugate Chem. 2020;31:1529–1536. doi: 10.1021/acs.bioconjchem.0c00190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are avaliable upon request.