Abstract
The automated construction of datasets has become increasingly relevant in computational chemistry. While transition-metal catalysis has greatly benefitted from bottom-up or top-down strategies for the curation of organometallic complexes libraries, the field of organocatalysis is mostly dominated by case-by-case studies, with a lack of transferable data-driven tools that facilitate both the exploration of a wider range of catalyst space and the optimization of reaction properties. For these reasons, we introduce OSCAR, a repository of 4000 experimentally derived organocatalysts along with their corresponding building blocks and combinatorially enriched structures. We outline the fragment-based approach used for database generation and showcase the chemical diversity, in terms of functions and molecular properties, covered in OSCAR. The structures and corresponding stereoelectronic properties are publicly available (https://archive.materialscloud.org/record/2022.106) and constitute the starting point to build generative and predictive models for organocatalyst performance.
A database of thousands of experimentally-derived or combinatorially enriched organocatalysts and fragments to navigate chemical space and optimize reaction properties.
Introduction
Constructing extensive yet tailored databases is crucial for the successful development and application of data-driven tools in catalysis and materials science.1,2 The way datasets are generated largely reflects how chemists think about the structure of a catalyst. In turn, this not only influences the way improved molecular systems are searched, but also how their structure is manipulated, for example through trial-and-error,3 fine-tuning according to mechanistic insight,4–7 or generating compound libraries for activity/selectivity screening.8–11
Transition-metal catalysts are naturally viewed in a modular fashion as a combination of active metal centre and ligands, which are further decomposed into metal-coordinating groups, backbone/bridging units, and substituents.12 This simple, yet powerful fragment-based strategy has enabled tremendous advancements in computer-aided catalyst design,13,14 from the exploration of the chemical space of inorganic species curated through bottom-up or top-down approaches,15–20 the construction of ligand databases with associated steric and electronic descriptors,21–27 to the development of algorithms for the assembly of metal complexes from fragments and evolutionary experiments.28–30 Modularity is even more apparent in biocatalysts,31 which combine a limited number of building blocks, the amino acids; inspired by natural evolution, strategies such as combinatorial backbone assembly32 have allowed to generate libraries of structurally diverse enzymes with altered catalytic properties.
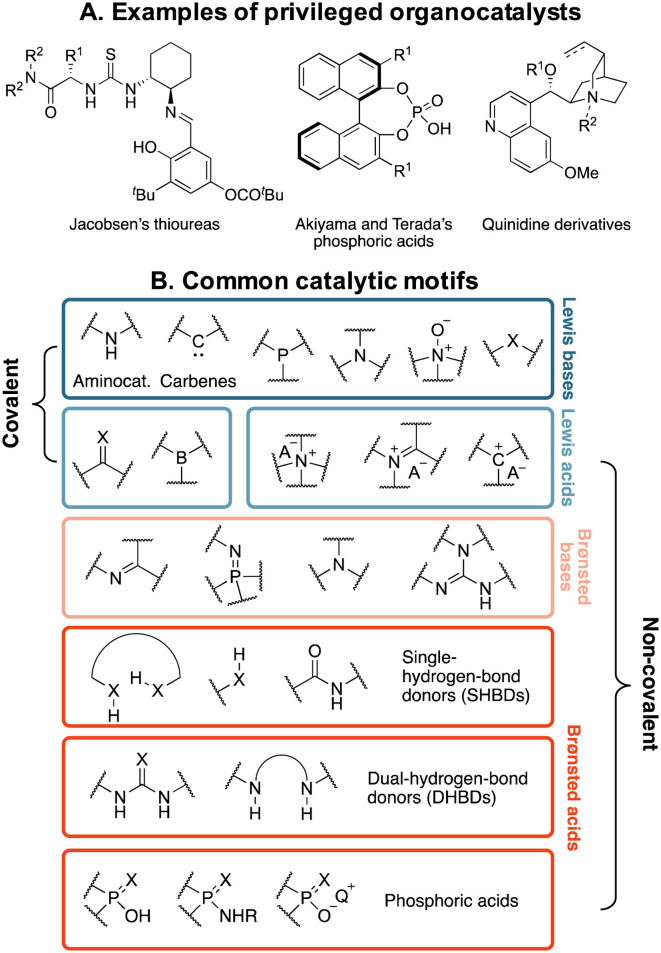
Organocatalysts are far less frequently classified according to fragment-based schemes. Instead, they are typically grouped into families of “privileged catalysts”,33,34 or according to the functional components that encapsulate their catalytic power (Fig. 1).35 Privileged catalysts are those species possessing certain chiral scaffolds that have proven to be effective at inducing high levels of enantioselectivity across a wide range of mechanistically unrelated reactions.33,34 Some effort has been made to summarize these catalytic motifs,36 however their comprehensive enumeration across all of chemical space is challenging due to the large possible variations in functionalities. This problem is exacerbated by the fact that organocatalysts are essentially a subclass of organic molecules, whose space is estimated to exceed 1060,37,38 and chemical expertise is required to evaluate whether an organic molecule could function as a catalyst in a reaction. Therefore, de novo organocatalyst design is a formidable, seldomly approached task, primarily due to the lack of robust ways of defining and assembling their building blocks,39,40 and reaction optimization is dominated by testing closely related analogues of a known privileged catalyst.41
Fig. 1. (A) Prototypical privileged chiral frameworks for asymmetric catalysis. (B) Classification of organocatalysts according to their catalytic motifs (X = O, S).
Similarly, the field of data-driven organocatalysis has been dominated by efforts, either on the automation side42 (e.g., AARON43 or ACE/Virtual Chemist44,45) or on the development of statistical models for enantioselectivity prediction,46–52 that have focused on specific reaction classes or structurally related catalysts. There is currently a dearth of general strategies and platforms for organocatalysts comparison, fragmentation into building blocks, and assembly across a wide region of catalyst space, encompassing functionally and chemically diverse molecules with a multitude of catalytic functions.
In this work, we propose a solution in the form of OSCAR (Organic Structures for CAtalysis Repository), a database of experimentally derived or combinatorially enriched organocatalysts and of the corresponding molecular fragments that are extracted from them. Not only OSCAR constitutes a map to navigate organocatalyst space and potentially enable informed catalyst design, but the modular strategy behind its construction paves the way to a multitude of data-driven and fragment-based reaction optimization methods.53,54 Herein, we show how such a dataset is curated and augmented with crystallographically determined structures using a combination of top-down and bottom-up approaches, and how the fragments are assembled in a combinatorial fashion to generate thousands of species. In its current forms, OSCAR contains 4000 catalysts, whose use has either been documented in the literature for organic synthesis or with chemically analogous structure reported in the Cambridge Structural Database (CSD), spanning various catalytic functions (Lewis/Brønsted acids and bases), and two exemplary enriched combinatorial supersets, OSCAR!(NHC) and OSCAR!(DHBD). The former consists of over 8000 carbenes for covalent catalysis, the latter contains ca. 1.5 million non-covalent dual-hydrogen-bond donors. The approaches used to generate these combinatorial databases (vide infra) are however transferable to other classes, implying the possibility of further extending OSCAR. A selection of stereoelectronic molecular descriptors, including reactivity indices derived from conceptual DFT, are provided and may help establishing structure–reactivity relationships for reaction optimization. All structures and properties are publicly available on the Materials Cloud for interactive visualization with Chemiscope (https://doi.org/10.24435/materialscloud:gy-3h).55 They could serve as the starting point to define the combinatorial space for evolutionary experiments,56 as well as the basis for dataset curation to train machine learning models for applications in organic synthesis.54
Results and discussion
Database curation
No comprehensive repository of organocatalysts' structures covering all of the functionalities summarized in Fig. 1B currently exists. Most frequently, they are reported in the literature in 2D format (i.e., ChemDraw pictures) with associated experimental characterization data in the ESI† (NMR and IR spectra and, less often, crystal structure information), but molecular geometries are not easily accessible. To construct OSCAR, we followed a five-step protocol (Fig. 2), which starts with the manual collection of catalysts (as 2D objects) from reviews,35,57–68 journal articles,69–72 books,73–77 and commercial catalogues78,79 into a “seed” database (step 1). Each of the 1000 2D entries in this library is labelled according to the classes in Fig. 1 and converted into a 1D (i.e., SMILES strings) and subsequently 3D (i.e., optimized XYZ geometry) structure (see the Computational methods). Given that more than ∼1500 publications on organocatalysis are published each year,80 it is virtually impossible to curate an exhaustive library of all existing catalysts. Nonetheless, the seed database aims at covering the chemical diversity observed across all of organocatalyst space in terms of chemical functionalities, catalytic motifs and scaffolds/substituents, with the added bonus of each structure either being commercially available or synthetically accessible, having being mined from the literature.
Fig. 2. Graphical summary of the steps followed for the curation of OSCAR.
This top-down approach ensures that only organic molecules that have been reported to display, or be tested for, catalytic activity are included in the database. However, it is a slow, human error-prone process that cannot be automated and might either introduce in the repository erroneous or mislabelled structures or lead to chemically interesting ones being excluded. Existing crystallographic databases (e.g., CSD,81,82 COD83) offer the most comprehensive collection of organic (and inorganic) molecules that have been synthetised. Although it not possible to filter out a priori those compounds that have not been tested as organocatalysts, CSD offers the chance to significantly augment the seed database with more, chemically diverse structures, provided that the right chemical motifs, which might make a molecule catalytically active, are searched. To achieve this goal we enumerated, in 1D format, 64 “function-based fragments” included in the seed database (step 2 Fig. 2 and S1 and S2†). Although not exhaustive, they represent the most common catalytic motifs and ensure that the species that contain them are relevant to the task at hand. In step 3, these fragments are searched in CSD and the corresponding whole molecules extracted. After retrieving the 3D geometries from the cif files with the cell2mol software,84 3010 compounds are added to the seed database, yielding a total of 4000 entries (after filtering out identical ones, see the ESI†). All 3D entries are then converted into 1D format for subsequent fragmentation and recombination (steps 4 and 5, vide infra).
With respect to the catalytic motifs (cf.Fig. 1B), the distribution of the CSD-extracted structures changes significantly from the seed database (see the two histograms in Fig. 3A). In OSCAR, the majority of species (40%) are classified as dual-hydrogen-bond donors; their large increase in number upon CSD extraction is likely due to the popularity of the (thio)urea moiety as pharmacophore85–87 and for anion recognition.88 The second most popular class (24%) is aminocatalysts based on the pyrrolidine motif: in the early days of organocatalysis, the vast majority of reactions were indeed amine-based59,89 and five-membered (polycyclic) secondary amines are widely encountered in natural products, as well as being a preferred scaffold in pharmaceutical science and drug design.90 The other classes are more or less equally represented (∼5–6%, Fig. 3B), with a slight predominance of Lewis bases (11%), given the large variety of N(O)-, P(O)-, and S(O)-nucleophilic organocatalysts. If we consider the increase in type of heteroatoms from the seed to the CSD-extracted database (Fig. 3C), sulphur and nitrogen are the most abundant due to the predominance of the thiourea and pyrrolidine catalytic motifs. The amount of P, Si, X, and especially B atoms increases to a significantly lesser extent. In the case of phosphorous, even though we seek to augment the quantity of P-containing motifs, only a limited number of phosphoric acids (ca. 25) are extractable from CSD. On the other hand, no catalytic unit that specifically contains halogens, silicon or boron is searched. An exhaustive description of the functional groups present in OSCAR is given in the ESI (Table S2 and Fig. S4†). Finally, the catalysts in the two datasets have a similar distribution of molecular weights (Fig. 3D), with the seed database containing on average slightly larger molecules (∼430 u) and a displaying smoother decrease in occurrences as their size increases.
Fig. 3. (A) Distribution histograms of catalytic motifs in the seed database and in the CSD-extracted structures. (B) Pie chart showing percentages of catalytic motifs in the seed and CSD-extracted datasets. (C) Distribution histograms of heteroatom types (X = halogens), and (D) molecular weight in the seed and in the CSD-extracted sets.
Structure and property maps
The chemical and structural diversity contained in OSCAR is visualized in Fig. 4A with a 2D t-SNE map91 based on FCHL1992 of the 4000 organocatalysts from the seed and CSD databases. Alternative representations and dimensionality reduction can be found in the ESI (Fig. S5–S7†). Although the two axes (dimensions) of this structure map have no formal physical meaning, it is possible to establish a qualitative relationship between them and chemical properties. In particular, species found higher in the map are bigger (higher molecular weight/surface area), whereas the degree of conjugation and the presence of aromatic scaffolds and substituents decreases left to right. For example, diol-based catalysts,93 which act as single-hydrogen-bond donors, and phosphoric acids94 are found along the upper edge of the map, with the fully aromatic BINOL derivatives on the left, the H8-BINOL core in the middle, and BAMOLs on the right. Dual-HBDs, especially diaryl (thio)ureas, occupy the lower left corner of the map, while simple proline derivatives the bottom right, with larger and more complex aminocatalysts in the upper left region. Other noticeable clusters correspond to the ketone epoxidation catalysts developed by Shi and Shu (covalent Lewis acidic carbohydrate derivatives),95,96 and to iminophosphorane Brønsted bases.97
Fig. 4. (A) 2D t-SNE map of OSCAR on the basis of the FCHL19 representation.92 Each point represents an organocatalyst, coloured by the corresponding catalytic motif. Each cluster contains catalysts with similar structure, with some examples being shown. R = alkyl group; Ar = aromatic group; PTC = phase-transfer catalyst. (B) Property map: computed (ωB97X-D/Def2-TZVP//B97-D/Def2-TZVP) nucleophilicity (Nrel) vs. electrophilicity (E-index) parameters.98 A zoom-in of the map is provided on the right hand side.
The structure map is complemented by a “property map” (Fig. 4B) in which the organocatalysts are evaluated in terms of their DFT-computed global electro/nucleophilicity indices (see the Computational methods), which assume that, when these catalysts react, they do so cumulatively and simultaneously at all their atomic sites.99 The largest influence on the descriptors is exerted by the total molecular charge, and three regions are found (four if the green point corresponding to the phosphorylated sulfonimidamide100 with −1 charge is considered). E-index increases with the charge, while Nrel decreases. Highly electrophilic and charged species include phase transfer catalysts101 (PTCs, non-covalent Lewis acids) and azolium ions, which are the conjugate acid precursors of carbene organocatalysts.102 The zoom-in on the right hand side of Fig. 4B shows the spread of E-index and Nrel values for neutral organocatalysts. Among the most nucleophilic species, Brønsted bases, in particular iminophosphoranes, and phosphoramidite Lewis bases are found towards the top of the map ( , σ = 0.6 eV), while ketone epoxidation electrophiles are at the bottom (
, σ = 0.6 eV), while ketone epoxidation electrophiles are at the bottom ( , σ = 0.2 eV,
, σ = 0.2 eV,  , σ = 0.5 eV). Some families of catalysts, such as DHBDs containing the thiourea motif and aminocatalysts, cover a wide range of values (0.4 < E-index,DHBD < 2.1 eV), indicating that their electronic properties are highly dependent on the nature of the substituents bound to the catalytic motif. Although it is unlikely that these simple reactivity indices can accommodate a robust and universal scale for electrophilicity and nucleophilicity of such diverse molecules with a varied range of structural, electronic, and bonding properties, the property map in Fig. 4B and the set of descriptors provided with OSCAR may supplement existing structure–reactivity scales in organocatalysis,103–109 such as the ones developed by Mayr et al.110–113
, σ = 0.5 eV). Some families of catalysts, such as DHBDs containing the thiourea motif and aminocatalysts, cover a wide range of values (0.4 < E-index,DHBD < 2.1 eV), indicating that their electronic properties are highly dependent on the nature of the substituents bound to the catalytic motif. Although it is unlikely that these simple reactivity indices can accommodate a robust and universal scale for electrophilicity and nucleophilicity of such diverse molecules with a varied range of structural, electronic, and bonding properties, the property map in Fig. 4B and the set of descriptors provided with OSCAR may supplement existing structure–reactivity scales in organocatalysis,103–109 such as the ones developed by Mayr et al.110–113
Combinatorial databases
OSCAR currently covers a significant part of organocatalyst space and a large pool of chemically and functionally diverse catalytic motifs. However, given the nearly infinite number of possible derivatives of each catalyst, only relatively few examples are included. Harnessing the fragment-based strategy used to enrich the seed database with structures from CSD in a bottom-up fashion, we exponentially increase the size of OSCAR by building combinatorial databases from molecular fragments. The exact nature of the fragments depends on the family of organocatalysts, but they can be grouped into two categories: catalytic motifs (i.e., the chemical groups that contain the reactive components) and structural substituents (which modulate their stereoelectronic properties). If the catalytic motif is easily distinguishable from the rest of the molecule (e.g., for dual-hydrogen-bond donors, vide infra), it is extracted as a subgraph of the whole catalyst, and the rest handled as structural substituents. If the catalytic motif exhibits larger chemical diversity and substitution patterns (e.g., carbenes, vide infra), the possible functional units and substituents are curated manually based on chemical expertise. Herein, we show how to do this for two types of covalent and non-covalent organocatalysts, specifically N-heterocyclic carbenes [OSCAR!(NHC)] and dual-hydrogen-bond donors [OSCAR!(DHBD)]. In the first case, a relatively “small” database (8622 catalysts) is curated by carefully selecting catalytic motifs and substituents found in OSCAR. In the second, we adopt a graph-based approach to generate 1 573 015 DHBDs.
In the first example (Fig. 5, top), 17 cores/scaffolds are extracted from the seed and CSD libraries (Fig. 4A and S9,† most central ring system generated with DataWarrior114); based on structural features reported in the literature,115–118 60 substituents grouped into three categories (R1–3, Fig. S10–S12†) and appropriate substitution patterns are defined. They are then translated into flexible SMILES strings (Table S4†), written in such a way that different R1–3 in each core can easily be introduced and exchanged. Finally, 3D structures are generated from the SMILES and fully optimized, yielding a database of 8622 species. In the second example (Fig. 5, bottom), all the organocatalysts containing one DHBD unit in the seed and CSD-extracted datasets (1593) are interpreted as molecular graphs119 (i.e., undirected multigraphs with RDKit) and fragmented into the central catalytic motif and the two substituents on either side (R1,2), affording a combinatorial space of 7 × 6942 groups. After duplicate removal and recombination with RDKit, they yield a total of 1 573 015 species (all optimized at the xTB level); 1000 structures per each DHBD motif are selected and optimized with DFT, and 6994 are shown in Fig. 6B.
Fig. 5. Graphical summary of the steps followed to generate the combinatorial databases OSCAR!(NHC) (top) and OSCAR!(DHBD) (bottom). X = O/S.
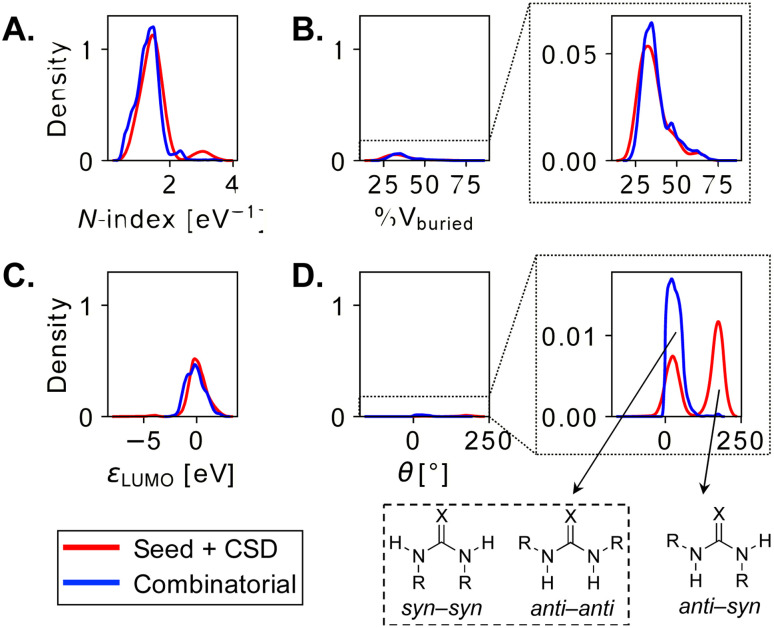
Fig. 6. (A) Percentage buried volume vs. N-index of combinatorial NHC organocatalysts. N-index is found to scale linearly with known experimental pKa values of azolium ions (Fig. S8†). (B) HNNH dihedral angle (θ) vs. LUMO energy (ωB97X-D/Def2-TZVP//B97-D/Def2-TZVP) of dual-hydrogen-bond donor species. Good linear correlation between εLUMO and the pKa's of DHBDs has been found (Fig. S13†).
The two combinatorial datasets are visualized with chemical space maps (Fig. 6),120 which are typically constructed from steric and electronic molecular descriptors. Based on their popularity and chemical meaningfulness, the percentage of buried volume121,122 %Vburied and nucleophilicity N-index (see the Computational methods) are the parameters chosen for OSCAR!(NHC), while the LUMO energy εLUMO and the HNNH dihedral angle of the HBD unit (θ) are plotted for OSCAR!(DHBD). The electronic descriptors provide an indirect estimate of the catalysts' Brønsted acidity/basicity: analysis of the experimental equilibrium acidities of 23 NHCs123 shows that the pKa values of their precursors (the azolium ions) are directly proportional to the N-index of the carbene (R2 = 0.80, 2σ = 0.72, Fig. S8†), while the LUMO energies of 74 DHBDs69 scale linearly (R2 = 0.92, 2σ = 2.32, Fig. S13†) with their experimental pKa's (as previously noted by Sigman for a smaller subset).50 %Vburied and θ quantify the steric influence exerted by the catalysts' core and substituents.
The NHCs in Fig. 6A are coloured according to common structural features. The N-substituent (R1 in Fig. 5 and S10†) has the greatest effect on nucleophilicity, with catalysts bearing electron-donating alkyl groups [i.e., Me, Et, iPr, Cy, and C(Me)Cy] having the highest N-index (blue points). These species are predicted to be the most reactive towards electrophilic attack, however their precursors have pKa's over 20,123 meaning that relatively strong bases must be used for active catalyst generation. The steric demand of the carbene is mostly influenced by R3 (Fig. S12†): l-pyroglutamic acid-derived bicyclic NHCs124 with diaryl- and diaryl(hydroxy)methyl substituents125,126 (red and purple points) are located towards the top of the map (large %Vburied). Despite their ability to enforce a rigid asymmetric environment, which could be beneficial in enantioselective reactions, these catalysts are poorly nucleophilic and predicted to be less reactive. Green and orange species, based on the tetracyclic amino-indanol-derived core developed by Rovis and Bode127,128 and on morpholine- and pyrrolidine-based triazoliums, have more balanced steric and electronic properties and indeed are among the most popular and versatile NHCs used in organocatalysis.102 Analysis of the descriptors provided with the 8622 carbenes in OSCAR!(NHC) could eventually be used to tune the catalyst's composition for performance improvement in specific reactions, as outlined in structure–activity–stereoselectivity studies using similar physical organic parameters.129–131 For example, Rovis, Lee, and co-workers found correlations between the computed gas-phase acidity of a series of triazolium cations and their selectivity in two Umpolung reactions,132 while Wei and Lan developed a linear model to predict the chemoselectivity of an NHC-catalyzed ester functionalization based on the global nucleophilicity and electrophilicity indices of the species involved in the product-determining step.133
In Fig. 6B, each point is coloured according to the nature of the central DHBD unit. Based on εLUMO, and in agreement with pKa measurements,134,135 croconamides and thiosquaramides (purple and red species) are more acidic than thioureas, ureas, and deltamides (yellow, blue, and light blue). Sulfamides (orange points) cover a relatively wider range of εLUMO values, implying that the higher electron-withdrawing ability of the sulfonyl group, which should result in stronger acidity of the N–H bonds compared to ureas,136 is significantly modulated by the substituents. The rapid estimation and comparison of the acidity of various DHBDs is useful for reaction optimization, as dual-hydrogen-bond donors with lower pKa's have been found to give better enantioselectivities and faster reaction times.137 Sulfamides are also the most flexible species, as indicated by the large number of catalysts with θ > 80°. In OSCAR!(DHBD), the majority of structures generated and selected for DFT optimization are in the anti–anti or syn–syn conformation (θ < 80°, Fig. 7D and S17†),138 the former being the most relevant to catalysis, since the hydrogens point in the same direction.139
Fig. 7. Distribution plots (y-axis: normalized probability density) of molecular descriptors for NHCs (A and B) and DHBDs (C and D) in the seed + CSD-extracted (red curves) and combinatorial databases (blue). X = O/S.
If we compare the distribution of θ values in the “original” and combinatorial datasets (Fig. 7D), we see that many CSD-extracted DHBDs adopt the anti–syn conformation (θ > 80°). In a comprehensive study of diaryl(thio)ureas from CSD, Paton et al. found that the majority (99%) of ureas exist as anti–anti conformers, whereas about 60% thioureas are in the anti–syn form.140 These results agree with our own, with thioureas extracted from CSD having large θ's (Fig. 7D). The “original” and combinatorial sets are more similarly distributed in terms of the other molecular descriptors (Fig. 7A–C, N-index, %Vburied, and εLUMO), suggesting that the recombination of the same fragments does not significantly alter the property space covered; instead, the combinatorial strategy provides more instances/structures for each property value.
Conclusions
We have introduced OSCAR (Organic Structures for CAtalysis Repository), a database of 4000 organocatalysts mined from the literature and CSD and enriched with several thousand species generated from fragments in a combinatorial fashion. We have developed a transferable fragment-based strategy for dataset generation, which exploits the modularity of organocatalysts by defining function-based catalytic motifs and structural substituents. OSCAR covers a wide region of catalyst space with incomparable chemical diversity, and includes a selection of steric and electronic molecular descriptors useful for catalytic properties estimation and performance prediction. All content (geometries, stereoelectronic parameters) is publicly available on the Materials Cloud for interactive visualization with Chemiscope55 (https://doi.org/10.24435/materialscloud:gy-3h) and fully searchable and interoperable with chemoinformatics software (e.g., RDKit, SMILES-based tools); the corresponding chemical space maps could be used for many potential applications, including data and training set curation, organocatalyst inverse design through evolutionary experiments,56 and mechanistic understanding. We expect OSCAR, and its future extensions and refinements, to assist in the establishment of data-driven and fragment-based reaction optimization methods in organic synthesis.53
Computational methods
Quantum chemistry
All DFT computations were performed with the Gaussian16 software package.141 Geometry optimizations were carried out at the B97-D/Def2-TZVP level142–144 in the gas-phase applying density fitting techniques. ωB97X-D/Def2-TZVP single-point energies145 were computed in the gas-phase at the B97-D geometries. The ionization potential and electron affinity of a subset 2060 organocatalysts from the seed and CSD datasets were also computed at the IP/EA-EOM-DLPNO-CCSD146/cc-pVTZ level as implemented in Orca 5.0.147 All coupled cluster computations used the RIJCOSX approximation148 with the cc-pVTZ/C and the Def2/J auxiliary basis sets for correlation and resolution of identity. This high-level data is available and can be used for the training of ML models. The structures in the combinatorial databases were pre-optimized with the semiempirical GFN2-xTB Hamiltonian149 in the gas-phase, followed by DFT optimizations and single-points, as described above.
The initial set of Cartesian coordinates for each organocatalyst was either obtained by converting SMILES formats150 into three-dimensional structures with the 3D structure generator operation (i.e., gen3d operation) implemented in the OpenBabel software,151 or applying cell2mol84 on selected CSD entries exported with ConQuest (version 5.42), included in the CCSD software, from the CSD database updated to May 2021. The t-SNE map91 for the 4000 catalysts in OSCAR was computed on the basis of the FCHL19 representation92 of each molecule. The perplexity used to generate the structure map was set to 20 and the maximum number of optimization iterations was fixed at 5000.
Open shell single-point computations (n − 1 and n + 1 electrons) were also performed at the optimized n-electron B97-D geometries and uωB97X-D/Def2-TZVP level for the 4000 catalysts in the seed + CSD dataset and for the 8622 carbenes in OSCAR!(NHC). These energies provide an alternative way of estimating the organocatalysts' ionization potential [IP = E(n − 1) − E(n)] and electron affinity [EA = E(n) − E(n + 1)] (see the ESI† for further details).152
Reaction indices
The organocatalysts' ionization potential (IP) and electron affinity (EA) were estimated from the frontier molecular orbital energies (FMOs) of the n-electron species (in the gas-phase, at the ωB97X-D/Def2-TZVP level) using Koopman's theorem153 within a Hartree–Fock scheme and used to calculate the conceptual DFT descriptors98,154,155 chemical potential (μ), hardness (η), E-index, N-index, and relative nucleophilicity (Nrel) as follows:
 |
1 |
 2
2
 |
3 |
 |
4 |
| Nrel = εHOMO − εHOMO(TCNE) | 5 |
where TCNE is tetracyanoethylene. Note that, based on the different formalisms for defining nucleophilicity,156 a distinction has been made between N-index (the reciprocal of the E-index) and relative nucleophilicity (Nrel).157
Data availability
The structures of the the organocatalysts and their stereoelectronic descriptors are publicly available on the Materials Cloud for interactive visualization with Chemiscope (https://archive.materialscloud.org/record/2022.106).
Author contributions
S. G. and C. C. conceived the project. S. G. performed DFT computations, curated the data, and analysed the results, with help from P. v. G. R. L. developed and implemented scripts for the combinatorial databases generation and analysis. S. V. developed and implemented cell2mol. A. F. provided support with the coupled cluster computations and t-SNE plot generation. S. G. and C. C. wrote the manuscript with help and feedback from all authors. C. C. provided supervision throughout.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
S. G. acknowledges the European Research Council (ERC, Grant Agreement No. 817977) within the framework of European Union's H2020 for financial support. The National Centre of Competence in Research (NCCR) “Sustainable chemical process through catalysis (Catalysis)” of the Swiss National Science Foundation (SNSF, grant number 180544) is acknowledged for financial support of P. v. G. and R. L. S. V. and A. F. acknowledge the National Centre of Competence in Research (NCCR) “Materials' Revolution: Computational Design and Discovery of Novel Materials (MARVEL)” of the Swiss National Science Foundation (SNSF, grant number 182892). The authors also acknowledge support from EPFL. Dr Guillaume Fraux is acknowledged for his help with Chemiscope.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc04251g
References
- Bo C. Maseras F. López N. The role of computational results databases in accelerating the discovery of catalysts. Nat. Catal. 2018;1:809–810. doi: 10.1038/s41929-018-0176-4. [DOI] [Google Scholar]
- Nandy A. Duan C. Kulik H. J. Audacity of huge: overcoming challenges of data scarcity and data quality for machine learning in computational materials discovery. Curr. Opin. Chem. Eng. 2022;36:100778. doi: 10.1016/j.coche.2021.100778. [DOI] [Google Scholar]
- McNally A. Prier C. K. MacMillan D. W. C. Discovery of an α-Amino C–H Arylation Reaction Using the Strategy of Accelerated Serendipity. Science. 2011;334:1114–1117. doi: 10.1126/science.1213920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumori S. Zhang H. Ha-Yeon Cheong P. Houk K. N. Tanaka F. Barbas C. F. Direct Asymmetric anti-Mannich-Type Reactions Catalyzed by a Designed Amino Acid. J. Am. Chem. Soc. 2006;128:1040–1041. doi: 10.1021/ja056984f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming E. M. Quigley C. Rozas I. Connon S. J. Computational Study-Led Organocatalyst Design: A Novel, Highly Active Urea-Based Catalyst for Addition Reactions to Epoxides. J. Org. Chem. 2008;73:948–956. doi: 10.1021/jo702154m. [DOI] [PubMed] [Google Scholar]
- Gammack Yamagata A. D. Datta S. Jackson K. E. Stegbauer L. Paton R. S. Dixon D. J. Enantioselective Desymmetrization of Prochiral Cyclohexanones by Organocatalytic Intramolecular Michael Additions to α,β-Unsaturated Esters. Angew. Chem., Int. Ed. 2015;54:4899–4903. doi: 10.1002/anie.201411924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iribarren I. Trujillo C. Improving phase-transfer catalysis by enhancing non-covalent interactions. Phys. Chem. Chem. Phys. 2020;22:21015–21021. doi: 10.1039/D0CP02012E. [DOI] [PubMed] [Google Scholar]
- Rosales A. R. Wahlers J. Limé E. Meadows R. E. Leslie K. W. Savin R. Bell F. Hansen E. Helquist P. Munday R. H. Wiest O. Norrby P.-O. Rapid virtual screening of enantioselective catalysts using CatVS. Nat. Catal. 2019;2:41–45. doi: 10.1038/s41929-018-0193-3. [DOI] [Google Scholar]
- Doney A. C. Rooks B. J. Lu T. Wheeler S. E. Design of Organocatalysts for Asymmetric Propargylations through Computational Screening. ACS Catal. 2016;6:7948–7955. doi: 10.1021/acscatal.6b02366. [DOI] [Google Scholar]
- Gerosa G. G. Spanevello R. A. Suárez A. G. Sarotti A. M. Joint Experimental, in Silico, and NMR Studies toward the Rational Design of Iminium-Based Organocatalyst Derived from Renewable Sources. J. Org. Chem. 2015;80:7626–7634. doi: 10.1021/acs.joc.5b01214. [DOI] [PubMed] [Google Scholar]
- Gerosa G. G. Marcarino M. O. Spanevello R. A. Suárez A. G. Sarotti A. M. Re-Engineering Organocatalysts for Asymmetric Friedel–Crafts Alkylation of Indoles through Computational Studies. J. Org. Chem. 2020;85:9969–9978. doi: 10.1021/acs.joc.0c01256. [DOI] [PubMed] [Google Scholar]
- Foscato M. Jensen V. R. Automated in Silico Design of Homogeneous Catalysts. ACS Catal. 2020;10:2354–2377. doi: 10.1021/acscatal.9b04952. [DOI] [Google Scholar]
- Houk K. N. Liu F. Holy Grails for Computational Organic Chemistry and Biochemistry. Acc. Chem. Res. 2017;50:539–543. doi: 10.1021/acs.accounts.6b00532. [DOI] [PubMed] [Google Scholar]
- Falivene L. Cao Z. Petta A. Serra L. Poater A. Oliva R. Scarano V. Cavallo L. Towards the online computer-aided design of catalytic pockets. Nat. Chem. 2019;11:872–879. doi: 10.1038/s41557-019-0319-5. [DOI] [PubMed] [Google Scholar]
- Nandy A. Duan C. Taylor M. G. Liu F. Steeves A. H. Kulik H. J. Computational Discovery of Transition-metal Complexes: From High-throughput Screening to Machine Learning. Chem. Rev. 2021;121:9927–10000. doi: 10.1021/acs.chemrev.1c00347. [DOI] [PubMed] [Google Scholar]
- Janet J. P. Duan C. Nandy A. Liu F. Kulik H. J. Navigating Transition-Metal Chemical Space: Artificial Intelligence for First-Principles Design. Acc. Chem. Res. 2021;54:532–545. doi: 10.1021/acs.accounts.0c00686. [DOI] [PubMed] [Google Scholar]
- Nandy A. Zhu J. Janet J. P. Duan C. Getman R. B. Kulik H. J. Machine Learning Accelerates the Discovery of Design Rules and Exceptions in Stable Metal–Oxo Intermediate Formation. ACS Catal. 2019;9:8243–8255. doi: 10.1021/acscatal.9b02165. [DOI] [Google Scholar]
- Gugler S. Janet J. P. Kulik H. J. Enumeration of de novo inorganic complexes for chemical discovery and machine learning. Mol. Syst. Des. Eng. 2020;5:139–152. doi: 10.1039/C9ME00069K. [DOI] [Google Scholar]
- Liu F. Duan C. Kulik H. J. Rapid Detection of Strong Correlation with Machine Learning for Transition-Metal Complex High-Throughput Screening. J. Phys. Chem. Lett. 2020;11:8067–8076. doi: 10.1021/acs.jpclett.0c02288. [DOI] [PubMed] [Google Scholar]
- Hueffel J. A. Sperger T. Funes-Ardoiz I. Ward J. S. Rissanen K. Schoenebeck F. Accelerated dinuclear palladium catalyst identification through unsupervised machine learning. Science. 2021;374:1134–1140. doi: 10.1126/science.abj0999. [DOI] [PubMed] [Google Scholar]
- Fey N. Tsipis A. C. Harris S. E. Harvey J. N. Orpen A. G. Mansson R. A. Development of a Ligand Knowledge Base, Part 1: Computational Descriptors for Phosphorus Donor Ligands. Chem.–Eur. J. 2006;12:291–302. doi: 10.1002/chem.200500891. [DOI] [PubMed] [Google Scholar]
- Jover J. Fey N. Harvey J. N. Lloyd-Jones G. C. Orpen A. G. Owen-Smith G. J. J. Murray P. Hose D. R. J. Osborne R. Purdie M. Expansion of the Ligand Knowledge Base for Monodentate P-Donor Ligands (LKB-P) Organometallics. 2010;29:6245–6258. doi: 10.1021/om100648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey N. Harvey J. N. Lloyd-Jones G. C. Murray P. Orpen A. G. Osborne R. Purdie M. Computational Descriptors for Chelating P,P- and P,N-Donor Ligands1. Organometallics. 2008;27:1372–1383. doi: 10.1021/om700840h. [DOI] [Google Scholar]
- Durand D. J. Fey N. Computational Ligand Descriptors for Catalyst Design. Chem. Rev. 2019;119:6561–6594. doi: 10.1021/acs.chemrev.8b00588. [DOI] [PubMed] [Google Scholar]
- Durand D. J. Fey N. Building a Toolbox for the Analysis and Prediction of Ligand and Catalyst Effects in Organometallic Catalysis. Acc. Chem. Res. 2021;54:837–848. doi: 10.1021/acs.accounts.0c00807. [DOI] [PubMed] [Google Scholar]
- Fey N. Koumi A. Malkov A. V. Moseley J. D. Nguyen B. N. Tyler S. N. G. Willans C. E. Mapping the properties of bidentate ligands with calculated descriptors (LKB-bid) Dalton Trans. 2020;49:8169–8178. doi: 10.1039/D0DT01694B. [DOI] [PubMed] [Google Scholar]
- Gensch T. dos Passos Gomes G. Friederich P. Peters E. Gaudin T. Pollice R. Jorner K. Nigam A. Lindner-D'Addario M. Sigman M. S. Aspuru-Guzik A. A Comprehensive Discovery Platform for Organophosphorus Ligands for Catalysis. J. Am. Chem. Soc. 2022;144:1205–1217. doi: 10.1021/jacs.1c09718. [DOI] [PubMed] [Google Scholar]
- Foscato M. Venkatraman V. Occhipinti G. Alsberg B. K. Jensen V. R. Automated Building of Organometallic Complexes from 3D Fragments. J. Chem. Inf. Model. 2014;54:1919–1931. doi: 10.1021/ci5003153. [DOI] [PubMed] [Google Scholar]
- Foscato M. Occhipinti G. Venkatraman V. Alsberg B. K. Jensen V. R. Automated Design of Realistic Organometallic Molecules from Fragments. J. Chem. Inf. Model. 2014;54:767–780. doi: 10.1021/ci4007497. [DOI] [PubMed] [Google Scholar]
- Chu Y. Heyndrickx W. Occhipinti G. Jensen V. R. Alsberg B. K. An Evolutionary Algorithm for de Novo Optimization of Functional Transition Metal Compounds. J. Am. Chem. Soc. 2012;134:8885–8895. doi: 10.1021/ja300865u. [DOI] [PubMed] [Google Scholar]
- Thorpe T. W. Marshall J. R. Harawa V. Ruscoe R. E. Cuetos A. Finnigan J. D. Angelastro A. Heath R. S. Parmeggiani F. Charnock S. J. Howard R. M. Kumar R. Daniels D. S. B. Grogan G. Turner N. J. Multifunctional biocatalyst for conjugate reduction and reductive amination. Nature. 2022;604:86–91. doi: 10.1038/s41586-022-04458-x. [DOI] [PubMed] [Google Scholar]
- Lapidoth G. Khersonsky O. Lipsh R. Dym O. Albeck S. Rogotner S. Fleishman S. J. Highly active enzymes by automated combinatorial backbone assembly and sequence design. Nat. Commun. 2018;9:2780. doi: 10.1038/s41467-018-05205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T. P. Jacobsen E. N. Privileged Chiral Catalysts. Science. 2003;299:1691–1693. doi: 10.1126/science.1083622. [DOI] [PubMed] [Google Scholar]
- Strassfeld D. A. Algera R. F. Wickens Z. K. Jacobsen E. N. A Case Study in Catalyst Generality: Simultaneous, Highly-Enantioselective Brønsted- and Lewis-Acid Mechanisms in Hydrogen-Bond-Donor Catalyzed Oxetane Openings. J. Am. Chem. Soc. 2021;143:9585–9594. doi: 10.1021/jacs.1c03992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seayad J. List B. Asymmetric Organocatalysis. Org. Biomol. Chem. 2005;3:719–724. doi: 10.1039/B415217B. [DOI] [PubMed] [Google Scholar]
- Kenny R. Liu F. Trifunctional Organocatalysts: Catalytic Proficiency by Cooperative Activation. Eur. J. Org. Chem. 2015;2015:5304–5319. doi: 10.1002/ejoc.201500179. [DOI] [Google Scholar]
- Kirkpatrick P. Ellis C. Chemical space. Nature. 2004;432:823. doi: 10.1038/432823a. [DOI] [Google Scholar]
- Reymond J.-L. The Chemical Space Project. Acc. Chem. Res. 2015;48:722–730. doi: 10.1021/ar500432k. [DOI] [PubMed] [Google Scholar]
- Schneider G. Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discovery. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- Liu T. Naderi M. Alvin C. Mukhopadhyay S. Brylinski M. Break Down in Order To Build Up: Decomposing Small Molecules for Fragment-Based Drug Design with eMolFrag. J. Chem. Inf. Model. 2017;57:627–631. doi: 10.1021/acs.jcim.6b00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betinol I. O. Kuang Y. Reid J. P. Guiding Target Synthesis with Statistical Modeling Tools: A Case Study in Organocatalysis. Org. Lett. 2022;24:1429–1433. doi: 10.1021/acs.orglett.1c04134. [DOI] [PubMed] [Google Scholar]
- Reid J. P. Ermanis K. Goodman J. M. BINOPtimal: a web tool for optimal chiral phosphoric acid catalyst selection. Chem. Commun. 2019;55:1778–1781. doi: 10.1039/C8CC09344J. [DOI] [PubMed] [Google Scholar]
- Guan Y. Ingman V. M. Rooks B. J. Wheeler S. E. AARON: An Automated Reaction Optimizer for New Catalysts. J. Chem. Theory Comput. 2018;14:5249–5261. doi: 10.1021/acs.jctc.8b00578. [DOI] [PubMed] [Google Scholar]
- Weill N. Corbeil C. R. De Schutter J. W. Moitessier N. Toward a computational tool predicting the stereochemical outcome of asymmetric reactions: Development of the molecular mechanics-based program ACE and application to asymmetric epoxidation reactions. J. Comput. Chem. 2011;32:2878–2889. doi: 10.1002/jcc.21869. [DOI] [PubMed] [Google Scholar]
- Burai Patrascu M. Pottel J. Pinus S. Bezanson M. Norrby P.-O. Moitessier N. From desktop to benchtop with automated computational workflows for computer-aided design in asymmetric catalysis. Nat. Catal. 2020;3:574–584. doi: 10.1038/s41929-020-0468-3. [DOI] [Google Scholar]
- Zahrt A. F. Henle J. J. Rose B. T. Wang Y. Darrow W. T. Denmark S. E. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science. 2019;363:eaau5631. doi: 10.1126/science.aau5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. P. Sigman M. S. Holistic prediction of enantioselectivity in asymmetric catalysis. Nature. 2019;571:343–348. doi: 10.1038/s41586-019-1384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. P. Simón L. Goodman J. M. A Practical Guide for Predicting the Stereochemistry of Bifunctional Phosphoric Acid Catalyzed Reactions of Imines. Acc. Chem. Res. 2016;49:1029–1041. doi: 10.1021/acs.accounts.6b00052. [DOI] [PubMed] [Google Scholar]
- Shoja A. Zhai J. Reid J. P. Comprehensive Stereochemical Models for Selectivity Prediction in Diverse Chiral Phosphate-Catalyzed Reaction Space. ACS Catal. 2021;11:11897–11905. doi: 10.1021/acscatal.1c03520. [DOI] [Google Scholar]
- Werth J. Sigman M. S. Connecting and Analyzing Enantioselective Bifunctional Hydrogen Bond Donor Catalysis Using Data Science Tools. J. Am. Chem. Soc. 2020;142:16382–16391. doi: 10.1021/jacs.0c06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexa K. W. Belyk K. M. Henle J. Xiang B. Sheridan R. P. Denmark S. E. Ruck R. T. Sherer E. C. Application of Machine Learning and Reaction Optimization for the Iterative Improvement of Enantioselectivity of Cinchona-Derived Phase Transfer Catalysts. Org. Process Res. Dev. 2022;26:670–682. doi: 10.1021/acs.oprd.1c00155. [DOI] [Google Scholar]
- Gallarati S. Fabregat R. Laplaza R. Bhattacharjee S. Wodrich M. D. Corminboeuf C. Reaction-based machine learning representations for predicting the enantioselectivity of organocatalysts. Chem. Sci. 2021;12:6879–6889. doi: 10.1039/D1SC00482D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallarati S. Laplaza R. Corminboeuf C. Harvesting the fragment-based nature of bifunctional organocatalysts to enhance their activity. Org. Chem. Front. 2022;9:4041–4051. doi: 10.1039/D2QO00550F. [DOI] [Google Scholar]
- Tsuji N. Sidorov P. Zhu C. Nagata Y. Gimadiev T. Varnek A. List B. Predicting Highly Enantioselective Catalysts Using Tunable Fragment Descriptors. ChemRxiv. 2022 doi: 10.26434/chemrxiv-2022-bsmdl. [DOI] [PubMed] [Google Scholar]
- Fraux G. Cersonsky R. K. Ceriotti M. Chemiscope: interactive structure–property explorer for materials and molecules. J. Open Source Softw. 2020;5:2117. doi: 10.21105/joss.02117. [DOI] [Google Scholar]
- Laplaza R. Gallarati S. Corminboeuf C. Genetic Optimization of Homogeneous Catalysts. Chem.: Methods. 2022:e202100107. [Google Scholar]
- Holland M. C. Gilmour R. Deconstructing Covalent Organocatalysis. Angew. Chem., Int. Ed. 2015;54:3862–3871. doi: 10.1002/anie.201409004. [DOI] [PubMed] [Google Scholar]
- Dondoni A. Massi A. Asymmetric Organocatalysis: From Infancy to Adolescence. Angew. Chem., Int. Ed. 2008;47:4638–4660. doi: 10.1002/anie.200704684. [DOI] [PubMed] [Google Scholar]
- Dalko P. I. Moisan L. In the Golden Age of Organocatalysis. Angew. Chem., Int. Ed. 2004;43:5138–5175. doi: 10.1002/anie.200400650. [DOI] [PubMed] [Google Scholar]
- Dalko P. I. Moisan L. Enantioselective Organocatalysis. Angew. Chem., Int. Ed. 2001;40:3726–3748. doi: 10.1002/1521-3773(20011015)40:20<3726::AID-ANIE3726>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Maji R. Mallojjala S. C. Wheeler S. E. Chiral phosphoric acid catalysis: from numbers to insights. Chem. Soc. Rev. 2018;47:1142–1158. doi: 10.1039/C6CS00475J. [DOI] [PubMed] [Google Scholar]
- Enders D. Niemeier O. Henseler A. Organocatalysis by N-Heterocyclic Carbenes. Chem. Rev. 2007;107:5606–5655. doi: 10.1021/cr068372z. [DOI] [PubMed] [Google Scholar]
- Wong O. A. Shi Y. Organocatalytic Oxidation. Asymmetric Epoxidation of Olefins Catalyzed by Chiral Ketones and Iminium Salts. Chem. Rev. 2008;108:3958–3987. doi: 10.1021/cr068367v. [DOI] [PubMed] [Google Scholar]
- McGarrigle E. M. Myers E. L. Illa O. Shaw M. A. Riches S. L. Aggarwal V. K. Chalcogenides as Organocatalysts. Chem. Rev. 2007;107:5841–5883. doi: 10.1021/cr068402y. [DOI] [PubMed] [Google Scholar]
- Marcelli T. Hiemstra H. Cinchona Alkaloids in Asymmetric Organocatalysis. Synthesis. 2010:1229–1279. doi: 10.1055/s-0029-1218699. [DOI] [Google Scholar]
- Benaglia M. Rossi S. Chiral phosphine oxides in present-day organocatalysis. Org. Biomol. Chem. 2010;8:3824–3830. doi: 10.1039/C004681G. [DOI] [PubMed] [Google Scholar]
- Liu X. Lin L. Feng X. Chiral N,N′-Dioxides: New Ligands and Organocatalysts for Catalytic Asymmetric Reactions. Acc. Chem. Res. 2011;44:574–587. doi: 10.1021/ar200015s. [DOI] [PubMed] [Google Scholar]
- Wei Y. Shi M. Applications of Chiral Phosphine-Based Organocatalysts in Catalytic Asymmetric Reactions. Chem.–Asian J. 2014;9:2720–2734. doi: 10.1002/asia.201402109. [DOI] [PubMed] [Google Scholar]
- Yang Q. Li Y. Yang J.-D. Liu Y. Zhang L. Luo S. Cheng J.-P. Holistic Prediction of the pKa in Diverse Solvents Based on a Machine-Learning Approach. Angew. Chem., Int. Ed. 2020;59:19282–19291. doi: 10.1002/anie.202008528. [DOI] [PubMed] [Google Scholar]
- Yang C. Xue X.-S. Li X. Cheng J.-P. Computational Study on the Acidic Constants of Chiral Brønsted Acids in Dimethyl Sulfoxide. J. Org. Chem. 2014;79:4340–4351. doi: 10.1021/jo500158e. [DOI] [PubMed] [Google Scholar]
- Christ P. Lindsay A. G. Vormittag S. S. Neudörfl J.-M. Berkessel A. O'Donoghue A. C. pKa Values of Chiral Brønsted Acid Catalysts: Phosphoric Acids/Amides, Sulfonyl/Sulfuryl Imides, and Perfluorinated TADDOLs (TEFDDOLs) Chem.–Eur. J. 2011;17:8524–8528. doi: 10.1002/chem.201101157. [DOI] [PubMed] [Google Scholar]
- Walvoord R. R. Huynh P. N. H. Kozlowski M. C. Quantification of Electrophilic Activation by Hydrogen-Bonding Organocatalysts. J. Am. Chem. Soc. 2014;136:16055–16065. doi: 10.1021/ja5086244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano A., in Stereoselective Organocatalysis, John Wiley & Sons, Ltd, 2013, pp. 11–80 [Google Scholar]
- Dalko P. I., in Enantioselective Organocatalysis, John Wiley & Sons, Ltd, 2007, pp. 1–17 [Google Scholar]
- Chi Y. R. Comprehensive Enantioselective Organocatalysis. Edited by Peter I. Dalko. Angew. Chem., Int. Ed. 2014;53:6858. doi: 10.1002/anie.201403631. [DOI] [Google Scholar]
- List B., Asymmetric Organocatalysis, Springer, 2009 [Google Scholar]
- Pihko P., in Hydrogen Bonding in Organic Synthesis, John Wiley & Sons, Ltd, 2009, pp. 1–4 [Google Scholar]
- Asymmetric Organocatalysis, ChemFiles, Sigma-Aldrich, 2006, vol. 6, pp. 1–16 [Google Scholar]
- Organocatalysis, ChemFiles, Sigma-Aldrich, 2007, vol. 7, pp. 1–24 [Google Scholar]
- Xiang S.-H. Tan B. Advances in asymmetric organocatalysis over the last 10 years. Nat. Commun. 2020;11:3786. doi: 10.1038/s41467-020-17580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom C. R. Allen F. H. The Cambridge Structural Database in Retrospect and Prospect. Angew. Chem., Int. Ed. 2014;53:662–671. doi: 10.1002/anie.201306438. [DOI] [PubMed] [Google Scholar]
- Groom C. R. Bruno I. J. Lightfoot M. P. Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gražulis S. Chateigner D. Downs R. T. Yokochi A. F. T. Quirós M. Lutterotti L. Manakova E. Butkus J. Moeck P. Le Bail A. Crystallography Open Database – an open-access collection of crystal structures. J. Appl. Crystallogr. 2009;42:726–729. doi: 10.1107/S0021889809016690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela S. Laplaza R. Cho Y. Corminboeuf C. Cell2mol: encoding chemistry to interpret crystallographic data. npj Comput. Mater. 2022;8:188. doi: 10.1038/s41524-022-00874-9. [DOI] [Google Scholar]
- Garuti L. Roberti M. Bottegoni G. Ferraro M. Diaryl Urea: A Privileged Structure in Anticancer Agents. Curr. Med. Chem. 2016;23:1528–1548. doi: 10.2174/0929867323666160411142532. [DOI] [PubMed] [Google Scholar]
- Anil S. M. Rajeev N. Kiran K. R. Swaroop T. R. Mallesha N. Shobith R. Sadashiva M. P. Multi-pharmacophore Approach to Bio-therapeutics: Piperazine Bridged Pseudo-peptidic Urea/Thiourea Derivatives as Anti-oxidant Agents. Int. J. Pept. Res. Ther. 2020;26:151–158. doi: 10.1007/s10989-019-09824-4. [DOI] [Google Scholar]
- Azeem S. Ataf Ali A. Ashfaq Mahmood Q. Amin B. Thiourea Derivatives in Drug Design and Medicinal Chemistry: A Short Review. J. Drug Des. Med. Chem. 2016;2:10–20. [Google Scholar]
- Bregović V. B. Basarić N. Mlinarić-Majerski K. Anion binding with urea and thiourea derivatives. Coord. Chem. Rev. 2015;295:80–124. doi: 10.1016/j.ccr.2015.03.011. [DOI] [Google Scholar]
- Xu L.-W. Luo J. Lu Y. Asymmetric catalysis with chiral primary amine-based organocatalysts. Chem. Commun. 2009:1807–1821. doi: 10.1039/B821070E. [DOI] [PubMed] [Google Scholar]
- Li Petri G. Raimondi M. V. Spanò V. Holl R. Barraja P. Montalbano A. Pyrrolidine in Drug Discovery: A Versatile Scaffold for Novel Biologically Active Compounds. Top. Curr. Chem. 2021;379:34. doi: 10.1007/s41061-021-00347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L. Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Christensen A. S. Bratholm L. A. Faber F. A. Anatole von Lilienfeld O. FCHL revisited: Faster and more accurate quantum machine learning. J. Chem. Phys. 2020;152:044107. doi: 10.1063/1.5126701. [DOI] [PubMed] [Google Scholar]
- Nguyen T. N. Chen P.-A. Setthakarn K. May J. A. Chiral Diol-Based Organocatalysts in Enantioselective Reactions. Molecules. 2018;23:2317. doi: 10.3390/molecules23092317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar D. Sugiono E. Raja S. Rueping M. Complete Field Guide to Asymmetric BINOL-Phosphate Derived Brønsted Acid and Metal Catalysis: History and Classification by Mode of Activation; Brønsted Acidity, Hydrogen Bonding, Ion Pairing, and Metal Phosphates. Chem. Rev. 2014;114:9047–9153. doi: 10.1021/cr5001496. [DOI] [PubMed] [Google Scholar]
- Shu L. Shi Y. An Efficient Ketone-Catalyzed Epoxidation Using Hydrogen Peroxide as Oxidant. J. Org. Chem. 2000;65:8807–8810. doi: 10.1021/jo001180y. [DOI] [PubMed] [Google Scholar]
- Denmark S. E. Wu Z. The Development of Chiral, Nonracemic Dioxiranes for the Catalytic, Enantioselective Epoxidation of Alkenes. Synlett. 2000;1999:847–859. doi: 10.1055/s-1999-3123. [DOI] [Google Scholar]
- Formica M. Rozsar D. Su G. Farley A. J. M. Dixon D. J. Bifunctional Iminophosphorane Superbase Catalysis: Applications in Organic Synthesis. Acc. Chem. Res. 2020;53:2235–2247. doi: 10.1021/acs.accounts.0c00369. [DOI] [PubMed] [Google Scholar]
- Chakraborty D. Chattaraj P. K. Conceptual density functional theory based electronic structure principles. Chem. Sci. 2021;12:6264–6279. doi: 10.1039/D0SC07017C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. Yoo J. Kang K. Predicting the chemical reactivity of organic materials using a machine-learning approach. Chem. Sci. 2020;11:7813–7822. doi: 10.1039/D0SC01328E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patureau F. W. Worch C. Siegler M. A. Spek A. L. Bolm C. Reek J. N. H. SIAPhos: Phosphorylated Sulfonimidamides and their Use in Iridium-Catalyzed Asymmetric Hydrogenations of Sterically Hindered Cyclic Enamides. Adv. Synth. Catal. 2012;354:59–64. doi: 10.1002/adsc.201100692. [DOI] [Google Scholar]
- Xiang B. Belyk K. M. Reamer R. A. Yasuda N. Discovery and Application of Doubly Quaternized Cinchona-Alkaloid-Based Phase-Transfer Catalysts. Angew. Chem., Int. Ed. 2014;53:8375–8378. doi: 10.1002/anie.201404084. [DOI] [PubMed] [Google Scholar]
- Flanigan D. M. Romanov-Michailidis F. White N. A. Rovis T. Organocatalytic Reactions Enabled by N-Heterocyclic Carbenes. Chem. Rev. 2015;115:9307–9387. doi: 10.1021/acs.chemrev.5b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mood A. Tavakoli M. Gutman E. Kadish D. Baldi P. Van Vranken D. L. Methyl Anion Affinities of the Canonical Organic Functional Groups. J. Org. Chem. 2020;85:4096–4102. doi: 10.1021/acs.joc.9b03187. [DOI] [PubMed] [Google Scholar]
- Kadish D. Mood A. D. Tavakoli M. Gutman E. S. Baldi P. Van Vranken D. L. Methyl Cation Affinities of Canonical Organic Functional Groups. J. Org. Chem. 2021;86:3721–3729. doi: 10.1021/acs.joc.0c02327. [DOI] [PubMed] [Google Scholar]
- Kaupmees K. Tolstoluzhsky N. Raja S. Rueping M. Leito I. On the Acidity and Reactivity of Highly Effective Chiral Brønsted Acid Catalysts: Establishment of an Acidity Scale. Angew. Chem., Int. Ed. 2013;52:11569–11572. doi: 10.1002/anie.201303605. [DOI] [PubMed] [Google Scholar]
- Jakab G. Tancon C. Zhang Z. Lippert K. M. Schreiner P. R. (Thio)urea Organocatalyst Equilibrium Acidities in DMSO. Org. Lett. 2012;14:1724–1727. doi: 10.1021/ol300307c. [DOI] [PubMed] [Google Scholar]
- Ni X. Li X. Wang Z. Cheng J.-P. Squaramide Equilibrium Acidities in DMSO. Org. Lett. 2014;16:1786–1789. doi: 10.1021/ol5005017. [DOI] [PubMed] [Google Scholar]
- Li Z. Li X. Ni X. Cheng J.-P. Equilibrium Acidities of Proline Derived Organocatalysts in DMSO. Org. Lett. 2015;17:1196–1199. doi: 10.1021/acs.orglett.5b00143. [DOI] [PubMed] [Google Scholar]
- Li Y. Zhang L. Luo S. Bond Energies of Enamines. ACS Omega. 2022;7:6354–6374. doi: 10.1021/acsomega.1c06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji B. Breugst M. Mayr H. N-Heterocyclic Carbenes: Organocatalysts with Moderate Nucleophilicity but Extraordinarily High Lewis Basicity. Angew. Chem., Int. Ed. 2011;50:6915–6919. doi: 10.1002/anie.201102435. [DOI] [PubMed] [Google Scholar]
- Mayr H. Lakhdar S. Maji B. Ofial A. R. A quantitative approach to nucleophilic organocatalysis. Beilstein J. Org. Chem. 2012;8:1458–1478. doi: 10.3762/bjoc.8.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F. Maji B. Min E. Ofial A. R. Mayr H. Basicities and Nucleophilicities of Pyrrolidines and Imidazolidinones Used as Organocatalysts. J. Am. Chem. Soc. 2020;142:1526–1547. doi: 10.1021/jacs.9b11877. [DOI] [PubMed] [Google Scholar]
- Maji B. Joannesse C. Nigst T. A. Smith A. D. Mayr H. Nucleophilicities and Lewis Basicities of Isothiourea Derivatives. J. Org. Chem. 2011;76:5104–5112. doi: 10.1021/jo200803x. [DOI] [PubMed] [Google Scholar]
- Sander T. Freyss J. von Korff M. Rufener C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- Enders D. Balensiefer T. Nucleophilic Carbenes in Asymmetric Organocatalysis. Acc. Chem. Res. 2004;37:534–541. doi: 10.1021/ar030050j. [DOI] [PubMed] [Google Scholar]
- Hopkinson M. N. Richter C. Schedler M. Glorius F. An overview of N-heterocyclic carbenes. Nature. 2014;510:485–496. doi: 10.1038/nature13384. [DOI] [PubMed] [Google Scholar]
- Biju A. T. Kuhl N. Glorius F. Extending NHC-Catalysis: Coupling Aldehydes with Unconventional Reaction Partners. Acc. Chem. Res. 2011;44:1182–1195. doi: 10.1021/ar2000716. [DOI] [PubMed] [Google Scholar]
- Ryan S. J. Candish L. Lupton D. W. Acyl anion free N-heterocyclic carbene organocatalysis. Chem. Soc. Rev. 2013;42:4906–4917. doi: 10.1039/C3CS35522E. [DOI] [PubMed] [Google Scholar]
- RDKit: Open-Source Chemoinformatics and Machine Learning. https://www.rdkit.org
- Dotson J., van Dijk L., Timmerman J., Grosslight S., Walroth R., Püntener K., Gosselin F., Mack K. and Sigman M. S., Data-driven multi-objective optimization tactics for catalytic asymmetric reactions, ChemRxiv, 2022 [DOI] [PMC free article] [PubMed]
- Clavier H. Nolan S. P. Percent buried volume for phosphine and N-heterocyclic carbene ligands: steric properties in organometallic chemistry. Chem. Commun. 2010;46:841–861. doi: 10.1039/B922984A. [DOI] [PubMed] [Google Scholar]
- Gómez-Suárez A. Nelson D. J. Nolan S. P. Quantifying and understanding the steric properties of N-heterocyclic carbenes. Chem. Commun. 2017;53:2650–2660. doi: 10.1039/C7CC00255F. [DOI] [PubMed] [Google Scholar]
- Li Z. Li X. Cheng J.-P. An Acidity Scale of Triazolium-Based NHC Precursors in DMSO. J. Org. Chem. 2017;82:9675–9681. doi: 10.1021/acs.joc.7b01755. [DOI] [PubMed] [Google Scholar]
- Chen X.-Y. Gao Z.-H. Ye S. Bifunctional N-Heterocyclic Carbenes Derived from l-Pyroglutamic Acid and Their Applications in Enantioselective Organocatalysis. Acc. Chem. Res. 2020;53:690–702. doi: 10.1021/acs.accounts.9b00635. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-R. He L. Wu X. Shao P.-L. Ye S. Chiral N-Heterocyclic Carbene Catalyzed Staudinger Reaction of Ketenes with Imines: Highly Enantioselective Synthesis of N-Boc β-Lactams. Org. Lett. 2008;10:277–280. doi: 10.1021/ol702759b. [DOI] [PubMed] [Google Scholar]
- He L. Zhang Y.-R. Huang X.-L. Ye S. Chiral Bifunctional N-Heterocyclic Carbenes: Synthesis and Application in the Aza-Morita-Baylis-Hillman Reaction. Synthesis. 2008;2008:2825–2829. doi: 10.1055/s-2008-1067216. [DOI] [Google Scholar]
- Kerr M. S. Read de Alaniz J. Rovis T. A Highly Enantioselective Catalytic Intramolecular Stetter Reaction. J. Am. Chem. Soc. 2002;124:10298–10299. doi: 10.1021/ja027411v. [DOI] [PubMed] [Google Scholar]
- He M. Struble J. R. Bode J. W. Highly Enantioselective Azadiene Diels–Alder Reactions Catalyzed by Chiral N-Heterocyclic Carbenes. J. Am. Chem. Soc. 2006;128:8418–8420. doi: 10.1021/ja062707c. [DOI] [PubMed] [Google Scholar]
- Wang N. Xu J. Lee J. K. The importance of N-heterocyclic carbene basicity in organocatalysis. Org. Biomol. Chem. 2018;16:8230–8244. doi: 10.1039/C8OB01667D. [DOI] [PubMed] [Google Scholar]
- Li Z. Li X. Cheng J.-P. Recent Progress in Equilibrium Acidity Studies of Organocatalysts. Synlett. 2019;30:1940–1949. doi: 10.1055/s-0037-1611890. [DOI] [Google Scholar]
- Gadekar S. C. Dhayalan V. Nandi A. Zak I. L. Mizrachi M. S. Kozuch S. Milo A. Rerouting the Organocatalytic Benzoin Reaction toward Aldehyde Deuteration. ACS Catal. 2021;11:14561–14569. doi: 10.1021/acscatal.1c04583. [DOI] [Google Scholar]
- Niu Y. Wang N. Muñoz A. Xu J. Zeng H. Rovis T. Lee J. K. Experimental and Computational Gas Phase Acidities of Conjugate Acids of Triazolylidene Carbenes: Rationalizing Subtle Electronic Effects. J. Am. Chem. Soc. 2017;139:14917–14930. doi: 10.1021/jacs.7b05229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Xu J. Li S.-J. Qu L.-B. Li Z. Chi Y. R. Wei D. Lan Y. Prediction of NHC-catalyzed chemoselective functionalizations of carbonyl compounds: a general mechanistic map. Chem. Sci. 2020;11:7214–7225. doi: 10.1039/D0SC01793K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. Zwicker V. E. Yuen K. K. Y. Jolliffe K. A. Quantum Chemical Prediction of Equilibrium Acidities of Ureas, Deltamides, Squaramides, and Croconamides. J. Org. Chem. 2017;82:10732–10736. doi: 10.1021/acs.joc.7b02083. [DOI] [PubMed] [Google Scholar]
- Zwicker V. E. Yuen K. K. Y. Smith D. G. Ho J. Qin L. Turner P. Jolliffe K. A. Deltamides and Croconamides: Expanding the Range of Dual H-bond Donors for Selective Anion Recognition. Chem.–Eur. J. 2018;24:1140–1150. doi: 10.1002/chem.201704388. [DOI] [PubMed] [Google Scholar]
- Zhang X. Liu S. Li X. Yan M. Chan A. S. C. Highly enantioselective conjugate addition of aldehydes to nitroolefins catalyzed by chiral bifunctional sulfamides. Chem. Commun. 2009:833–835. doi: 10.1039/B818582D. [DOI] [PubMed] [Google Scholar]
- Li X. Deng H. Zhang B. Li J. Zhang L. Luo S. Cheng J.-P. Physical Organic Study of Structure-Activity-Enantioselectivity Relationships in Asymmetric Bifunctional Thiourea Catalysis: Hints for the Design of New Organocatalysts. Chem.–Eur. J. 2010;16:450–455. doi: 10.1002/chem.200902430. [DOI] [PubMed] [Google Scholar]
- Sandler I. Larik F. A. Mallo N. Beves J. E. Ho J. Anion Binding Affinity: Acidity versus Conformational Effects. J. Org. Chem. 2020;85:8074–8084. doi: 10.1021/acs.joc.0c00888. [DOI] [PubMed] [Google Scholar]
- Wittkopp A. Schreiner P. R. Metal-Free, Noncovalent Catalysis of Diels–Alder Reactions by Neutral Hydrogen Bond Donors in Organic Solvents and in Water. Chem.–Eur. J. 2003;9:407–414. doi: 10.1002/chem.200390042. [DOI] [PubMed] [Google Scholar]
- Luchini G. Ascough D. M. H. Alegre-Requena J. V. Gouverneur V. Paton R. S. Data-mining the diaryl(thio)urea conformational landscape: understanding the contrasting behavior of ureas and thioureas with quantum chemistry. Tetrahedron. 2019;75:697–702. doi: 10.1016/j.tet.2018.12.033. [DOI] [Google Scholar]
- Frisch M., Trucks G., Schlegel H., Scuseria G., Robb M., Cheeseman J., Montgomery J., Vreven T., Kudin K., Burant J., Millam J., Iyengar S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J., Hratchian H., Cross J., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R., Yazyev O., Austin A., Cammi R., Pomelli C., Ochterski J., Ayala P., Morokuma K., Voth G., Salvador P., Dannenberg J., Zakrzewski V., Dapprich S., Daniels A., Strain M., Farkas O., Malick D., Rabuck A., Raghavachari K., Foresman J., Ortiz J., Cui Q., Baboul A., Clifford S., Cioslowski J., Stefanov B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R., Fox D., Keith T., Laham A., Peng C., Nanayakkara A., Challacombe M., Gill P., Johnson B., Chen W., Wong M., Gonzalez C. and Pople J., Gaussian 16, Revision C.01, Wallingford, CT, 2016 [Google Scholar]
- Becke A. D. Density-functional thermochemistry. V. Systematic optimization of exchange–correlation functionals. J. Chem. Phys. 1997;107:8554–8560. doi: 10.1063/1.475007. [DOI] [Google Scholar]
- Grimme S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006;27:1787–1799. doi: 10.1002/jcc.20495. [DOI] [PubMed] [Google Scholar]
- Weigend F. Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005;7:3297–3305. doi: 10.1039/B508541A. [DOI] [PubMed] [Google Scholar]
- Chai J.-D. Head-Gordon M. Systematic optimization of long-range corrected hybrid density functionals. J. Chem. Phys. 2008;128:084106. doi: 10.1063/1.2834918. [DOI] [PubMed] [Google Scholar]
- Haldar S. Riplinger C. Demoulin B. Neese F. Izsak R. Dutta A. K. Multilayer Approach to the IP-EOM-DLPNO-CCSD Method: Theory, Implementation, and Application. J. Chem. Theory Comput. 2019;15:2265–2277. doi: 10.1021/acs.jctc.8b01263. [DOI] [PubMed] [Google Scholar]
- Neese F. Wennmohs F. Becker U. Riplinger C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020;152:224108. doi: 10.1063/5.0004608. [DOI] [PubMed] [Google Scholar]
- Neese F. Wennmohs F. Hansen A. Becker U. Efficient, approximate and parallel Hartree–Fock and hybrid DFT calculations. A ‘chain-of-spheres’ algorithm for the Hartree–Fock exchange. Chem. Phys. 2009;356:98–109. doi: 10.1016/j.chemphys.2008.10.036. [DOI] [Google Scholar]
- Bannwarth C. Ehlert S. Grimme S. GFN2-xTB—An Accurate and Broadly Parametrized Self-Consistent Tight-Binding Quantum Chemical Method with Multipole Electrostatics and Density-Dependent Dispersion Contributions. J. Chem. Theory Comput. 2019;15:1652–1671. doi: 10.1021/acs.jctc.8b01176. [DOI] [PubMed] [Google Scholar]
- Weininger D. Weininger A. Weininger J. L. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 1989;29:97–101. doi: 10.1021/ci00062a008. [DOI] [Google Scholar]
- O'Boyle N. M. Banck M. James C. A. Morley C. Vandermeersch T. Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. Roy D. R. Subramanian V. Chattaraj P. K. Are strong Brønsted acids necessarily strong Lewis acids? J. Mol. Struct.: THEOCHEM. 2007;812:13–24. doi: 10.1016/j.theochem.2007.02.013. [DOI] [Google Scholar]
- Koopmans T. Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den Einzelnen Elektronen Eines Atoms. Physica. 1934;1:104–113. doi: 10.1016/S0031-8914(34)90011-2. [DOI] [Google Scholar]
- Domingo L. R. Ríos-Gutiérrez M. Pérez P. Applications of the Conceptual Density Functional Theory Indices to Organic Chemistry Reactivity. Molecules. 2016;21:748. doi: 10.3390/molecules21060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings P. De Proft F. Langenaeker W. Conceptual Density Functional Theory. Chem. Rev. 2003;103:1793–1874. doi: 10.1021/cr990029p. [DOI] [PubMed] [Google Scholar]
- Domingo L. R. Pérez P. The nucleophilicity N index in organic chemistry. Org. Biomol. Chem. 2011;9:7168–7175. doi: 10.1039/C1OB05856H. [DOI] [PubMed] [Google Scholar]
- Domingo L. R. Chamorro E. Pérez P. Understanding the Reactivity of Captodative Ethylenes in Polar Cycloaddition Reactions. A Theoretical Study. J. Org. Chem. 2008;73:4615–4624. doi: 10.1021/jo800572a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures of the the organocatalysts and their stereoelectronic descriptors are publicly available on the Materials Cloud for interactive visualization with Chemiscope (https://archive.materialscloud.org/record/2022.106).