Abstract
Summary
We present a flexible, user-friendly R package called protti for comprehensive quality control, analysis and interpretation of quantitative bottom-up proteomics data. protti supports the analysis of protein-centric data such as those associated with protein expression analyses, as well as peptide-centric data such as those resulting from limited proteolysis-coupled mass spectrometry analysis. Due to its flexible design, it supports analysis of label-free, data-dependent, data-independent and targeted proteomics datasets. protti can be run on the output of any search engine and software package commonly used for bottom-up proteomics experiments such as Spectronaut, Skyline, MaxQuant or Proteome Discoverer, adequately exported to table format.
Availability and implementation
protti is implemented as an open-source R package. Release versions are available via CRAN (https://CRAN.R-project.org/package=protti) and work on all major operating systems. The development version is maintained on GitHub (https://github.com/jpquast/protti). Full documentation including examples is provided in the form of vignettes on our package website (jpquast.github.io/protti/).
1 Introduction
With novel and evolving proteomics methods such as limited proteolysis coupled to mass spectrometry (LiP-MS; Feng et al., 2014), N-terminomics, phosphoproteomics and other post-translational modification-centric mass spectrometry-based experiments, all of which require peptide-centric data analysis, there is a growing need for flexible software tools to analyse bottom-up proteomics data. Data structures of such bottom-up proteomics experiments are diverse, and users often require specific workflows. Existing bottom-up proteomics R packages such as MSstats (Choi et al., 2014), DEP (Zhang et al., 2018), msmsEDA (Gregori et al., 2021), MSnbase (Gatto et al., 2021), TPP (Childs et al., 2021), HaDeX (Puchala et al., 2020) and NormalyzerDE (Willforss et al., 2019) either offer fixed analysis pipelines (e.g. packages of the MSstats family) and are therefore not easily implementable for specific user needs or are not suited for comprehensive peptide-level analysis, especially of LiP-MS data, since they offer a limited set of functions (DEP, msmsEDA, MSnbase, TPP, HaDeX, NormalyzerDE). Finally, few available tools offer functions for quality control, data analysis and data interpretation in one package.
To fill this gap, we developed protti, a user-friendly label-free bottom-up proteomics R package that facilitates the analysis of data-dependent, data-independent and targeted mass spectrometry data on the protein, peptide, precursor or fragment level. protti provides flexible functions for quality control, data filtering, differential quantification and dose-response analysis that can be tailored to a user’s needs, as well as supporting biological interpretation such as functional enrichment and network analysis. protti follows the design principles of the tidyverse package family (Wickham et al., 2019), which makes it accessible and easy to modify even for inexperienced R users; the code is written with the novice user in mind, and is carefully documented. Due to its modular design, most functions can be used independently of each other and can be applied to input data from many sources. Although protti does not provide a graphical user interface, unlike proteomics tools such as proteosign (Efstathiou et al., 2017), Perseus (Tyanova and Cox, 2018), Prostar (Wieczorek et al., 2017), DAnTE (Karpievitch et al., 2009), PIQMIe (Kuzniar and Kanaar, 2014), StatQuant (van Breukelen et al., 2009), LFQ-Analyst (Shah et al., 2020), ProtExA (Minadakis et al., 2020) and MSqRob (Goeminne et al., 2018) this aids the seamless implementation of protti into any R data analysis workflow.
2 Overview
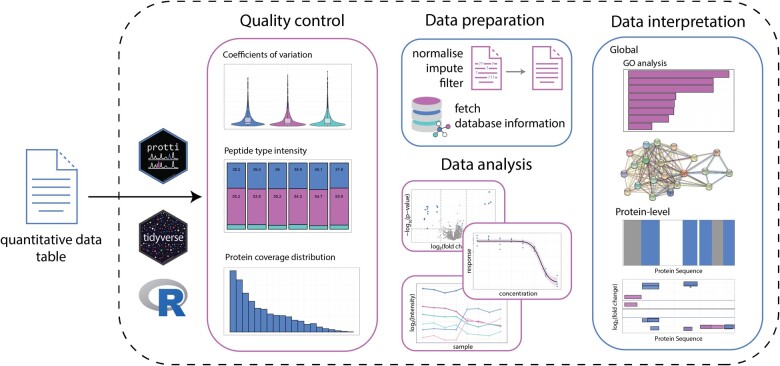
protti provides a wide range of functions (Fig. 1). Briefly, the user can assess and visualize data quality and perform median normalization to correct for unequal sample amounts. Protein abundances can be inferred from precursor or peptide intensities based on methods adapted from the MaxLFQ algorithm (Cox et al., 2014) and as previously implemented in the R package iq (Pham et al., 2020). Differential abundance calculation in conjunction with statistical testing can be applied to any case-control dataset. Statistical testing can either be performed using a standard Welch's t-test, ANOVA, a moderated t-test based on the limma R package (Ritchie et al., 2015) or the proDA R package (Ahlmann-Eltze, 2020). Dose-response experiments can be analysed by fitting four-parameter log-logistic regression models to the data based on functions from the drc R package (Ritz et al., 2015).
Fig. 1.
Overview of protti functions. protti can be used on the output of any software package for quantitative analysis of bottom-up proteomics experiments and provides a flexible set of functions for quality control, as well as data pre-processing, data analysis and data interpretation
Lists of significantly altered proteins obtained with any of the described workflows, can be further analysed using functional (Ashburner et al., 2000) or pathway enrichment analyses, as well as a network analysis tool based on the STRINGdb R package (Szklarczyk et al., 2019). Furthermore, to obtain additional information on proteins, including PTMs, subcellular location, PDB structures, and interaction partners, data can be annotated using information imported from several databases (UniProt (The UniProt Consortium, 2021), KEGG (Kanehisa and Goto, 2000), MobiDB (Piovesan et al., 2021), ChEBI (Hastings et al., 2016)). With the respective fetch functions, the user can load this information directly into R. Functions for the analysis of peptide-centric data from LiP-MS experiments allow visualiztion of differentially abundant peptides along the protein sequence.
2.1 Applications
protti can be applied to any bottom-up label-free proteomics dataset. In principle, the output of any table-formatted proteomic quantitative data analysis software is supported if it contains information on sample, condition, intensity, protein ID (UniProt ID), and the feature the measured intensity reports on (fragment, precursor, peptide) if different from protein intensity. The quality control, data analysis and data interpretation modules of protti can be used serially or independently.
2.2 Implementation
The modularity of protti enables its use in conjunction with user-defined workflows and functions. protti is written as simply as possible using tidyverse packages (Wickham et al., 2019), making its source code easy to understand and adapt or extend, even for inexperienced R users.
3 Conclusions
We describe protti, a flexible, user-friendly R package for analysis of various bottom-up proteomics datasets. protti provides functions for quality control, data analysis on protein, peptide, precursor or fragment levels, as well as data interpretation within a single, easy-to-use package. We include examples of data analysis workflows as vignettes to illustrate the utility of the different functions for the analysis of protein- or peptide-centric datasets.
Acknowledgements
The authors thank all members of the Picotti lab for testing protti and providing feedback. In addition, we thank Natalie de Souza for providing valuable feedback on documentation and the manuscript.
Funding
This work was supported by the Promedica Stiftung, Chur and the European Research Council [grant agreement number 866004].
Conflict of Interest: P.P. is a scientific advisor for the company Biognosys AG (Zurich, Switzerland) and an inventor of a patent licensed by Biognosys AG that covers the LiP-MS method mentioned in this manuscript.
References
- Ahlmann-Eltze C. (2020) Differential Abundance Analysis of Label-Free Mass Spectrometry Data. R package version 1.4.0.
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs D. et al. (2021) TPP: Analyze Thermal Proteome Profiling (TPP) Experiments. R package version 3.20.1.
- Choi M. et al. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics, 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- Cox J. et al. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou G. et al. (2017) ProteoSign: an end-user online differential proteomics statistical analysis platform. Nucleic Acids Res., 45, W300–W306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. et al. (2014) Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol., 32, 1036–1044. [DOI] [PubMed] [Google Scholar]
- Gatto L. et al. (2021) MSnbase, efficient and elegant R-based processing and visualization of raw mass spectrometry data. J. Proteome Res., 20, 1063–1069. [DOI] [PubMed] [Google Scholar]
- Goeminne L.J.E. et al. (2018) Experimental design and data-analysis in label-free quantitative LC/MS proteomics: a tutorial with MSqRob. J. Proteomics, 171, 23–36. [DOI] [PubMed] [Google Scholar]
- Gregori J. et al. (2021) msmsEDA: Exploratory Data Analysis of LC-MS/MS Data by Spectral Counts. R package version 1.30.0.
- Hastings J. et al. (2016) ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res., 44, D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpievitch Y. et al. (2009) A statistical framework for protein quantitation in bottom-up MS-based proteomics. Bioinformatics, 25, 2028–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A., Kanaar R. (2014) PIQMIe: a web server for semi-quantitative proteomics data management and analysis. Nucleic Acids Res., 42, W100–W106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minadakis G. et al. (2020) ProtExA: a tool for post-processing proteomics data providing differential expression metrics, co-expression networks and functional analytics. Comput. Struct. Biotechnol. J., 18, 1695–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.V. et al. (2020) iq: an R package to estimate relative protein abundances from ion quantification in DIA-MS-based proteomics. Bioinformatics, 36, 2611–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D. et al. (2021) MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res., 49, D361–D367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchala W. et al. (2020) HaDeX: an R package and web-server for analysis of data from hydrogen-deuterium exchange mass spectrometry experiments. Bioinformatics, 36, 4516–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E. et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz C. et al. (2015) Dose-response analysis using R. PLoS One, 10, e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.D. et al. (2020) LFQ-analyst: an easy-to-use interactive web platform to analyze and visualize label-free proteomics data preprocessed with MaxQuant. J. Proteome Res., 19, 204–211. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D. et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res., 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res., 49, D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Cox J. (2018) Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol., 1711, 133–148. [DOI] [PubMed] [Google Scholar]
- van Breukelen B. et al. (2009) StatQuant: a post-quantification analysis toolbox for improving quantitative mass spectrometry. Bioinformatics, 25, 1472–1473. [DOI] [PubMed] [Google Scholar]
- Wickham H. et al. (2019) Welcome to the tidyverse. J. Open Source Softw., 4, 1686. [Google Scholar]
- Wieczorek S. et al. (2017) DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics, 33, 135–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willforss J. et al. (2019) NormalyzerDE: online tool for improved normalization of omics expression data and high-sensitivity differential expression analysis. J. Proteome Res., 18, 732–740. [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. (2018) Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc., 13, 530–550. [DOI] [PubMed] [Google Scholar]