Abstract
Cell Painting is a high-throughput, phenotypic profiling assay that uses fluorescent cytochemistry to visualize a variety of organelles and high-content imaging to derive a large number of morphological features at the single cell level. Most Cell Painting studies have used the U-2 OS cell line for chemical or functional genomics screening. The Cell Painting assay can be used with many other human-derived cell types, given that the assay is based on the use of fluoroprobes that label organelles that are present in most (if not all) human cells. Questions remain, however, regarding the optimization steps required and overall ease of deployment of the Cell Painting assay to novel cell types. Here, we used the Cell Painting assay to characterize the phenotypic effects of sixteen phenotypic reference chemicals in concentration-response screening mode across six biologically diverse human-derived cell lines (U-2 OS, MCF7, HepG2, A549, HTB-9, ARPE-19). All cell lines were labeled using the same cytochemistry protocol and the same set of phenotypic features were calculated. We found it necessary to optimize image acquisition settings and cell segmentation parameters for each cell type but did not adjust the cytochemistry protocol. For some reference chemicals, similar subsets of phenotypic features corresponding to a particular organelle were associated with the highest effect magnitudes in each affected cell type. Overall, for certain chemicals the Cell Painting assay yielded qualitatively similar biological activity profiles across a group of diverse, morphologically distinct human-derived cell lines without the requirement for cell-type specific optimization of cytochemistry protocols.
Introduction
The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency (i.e. US EPA CompTox Blueprint)1 advocates the use of non-targeted, high-throughput profiling assays for initial characterization of the biological activity of environmental chemicals. Ideally, such profiling assays should: 1) be capable of being deployed in a high-throughput, concentration-response screening format and 2) provide high-content data that be used to identify potency thresholds for perturbation of cellular biology and provide information on putative mechanisms of toxicity. To date, two high-throughput profiling assays have been identified that meet this criteria: high-throughput transcriptomics (HTTr) using whole transcriptome-targeted RNA-Seq2 and imaging-based, high-throughput phenotypic profiling (HTPP) using Cell Painting3. Both of these assays are currently being operationalized at US EPA for use in a tiered toxicity testing framework based on New Approach Methodologies (NAMs): i.e. technologies, methodologies, approaches or combinations thereof that can be used to provide information on chemical hazard and risk assessment that avoids the use of intact animals4.
A third criterion that is desirable for high-throughput profiling assays is compatibility for use across a wide variety of human-derived cell types. Different cell types express different complements of genes / proteins both in vivo and when cultured in vitro. As such, it is widely recognized in the field of toxicology that no single cell type or cell-based in vitro test system can capture the diversity of human biology that may be perturbed by the diverse universe of chemicals that may be present in the environment. As such, the US EPA CompTox Blueprint recommends using high-throughput profiling assays to screen environmental chemicals across multiple biologically-diverse cell types that represent different aspects of human biology1. Over the next several years, it is anticipated that the HTTr and HTPP approaches will be applied to multiple human-derived cell types, thereby providing broad, complementary coverage of molecular and phenotypic responses across a much larger swath of biological space than the existing ToxCast and Tox21 assay portfolios5. The compatibility of targeted RNA-Seq with multiple human-derived cell types has been demonstrated in the scientific literature6–9. Many questions remain, however, regarding the general compatibility and portability of the Cell Painting assay among different cell types, as well as what optimization steps are needed for adapting the assay to novel cell models. These topics were briefly discussed at a recent colloquium entitled “Cell Painting in Drug Discovery” at the annual Society for Biomolecular Imaging and Informatics (SBI2) conference in 2019. These topics remain an open research question as, to date, published examples of the compatibility of the Cell Painting assay with multiple human-derived cell types is scarce.
In one of the first Cell Painting publications, Gustafsdottir et al. presented a qualitative comparison of fluorophore labeling patterns in a collection of morphologically-distinct human-derived cell types (U-2 OS, MCF7, HTB-9, A549, 3T3)10. This qualitative comparison indicated that the fluoroprobe labeling and imaging procedures used in the Cell Painting assay are compatible with multiple cell types. Likewise, Bray et al. (2016) note that their research group and collaborators have successfully applied the Cell Painting assay to variety of different cell types and provide qualitative images of three human-derived cell types (U-2 OS, A549 and human umbilical vein endothelial cells)3. Absent from these reports, however, were quantitative data, information on cell type-specific optimization steps, information on feature extraction or comparison among cell lines of phenotypic responses due to treatment. More recently, Warchal et al. have used the Cell Painting assay to screen libraries of pharmacological compounds in a set of biologically and morphologically distinct breast cancer cell lines and have developed computational approaches to quantify and visualize the degree of dissimilarity in phenotypic response profiles across this cell line panel11–13. These studies were conducted in the context of personalized medicine: i.e. identification of drugs that produce distinct phenotypes in cell lines with a particular clinical subtype. Each cell line in the breast cancer cell line panel responded similarly to a majority of active chemicals tested whereas as a smaller subset of chemicals produced a dissimilar phenotype in at least one of the cell lines within the panel11. Chemicals that produced a dissimilar phenotype in a particular cell line were identified as the most interest for potential personalized medicine applications. In contrast, chemicals that reliably produce similar phenotypes across diverse cell lines would be of interest to researchers aiming to identify sets of Cell Painting reference treatments (i.e. assay controls) for incorporation into high-throughput screening of biologically diverse cell line panels and for tracking of Cell Painting assay performance.
In pursuit of this objective, we used the Cell Painting assay to quantitatively characterize the phenotypic effects of sixteen reference chemicals in concentration-response screening mode (n = 7 test concentrations / chemical) across six biologically-diverse and morphologically-distinct human-derived cell lines (U-2 OS, MCF7, HepG2, A549, HTB-9, ARPE-19) grown in monolayer. To assess general compatibility and ease of portability across cell lines, the same cytochemistry protocol was used and the same set of phenotypic features were extracted for each cell line. Image acquisition parameters, including z-offsets, laser power and acquisition times for confocal imaging, as well as cell segmentation were optimized for each cell line. In addition to qualitative and quantitative comparisons of phenotypic response profiles, concentration-response modeling of the data was performed to assess variation in potencies associated with changes in cellular morphology across the cell line panel. Overall, for certain chemicals the Cell Painting assay yielded similar biological activity profiles across the cell line panel without the requirement for cell-type specific optimization of cytochemistry protocols. For a majority of chemicals tested, potency thresholds associated with changes in cellular morphology did not vary by more than one order of magnitude. Chemicals that produce similar phenotypic profiles at similar potencies across the diverse cell line space are the most promising candidates for selection as plate-based assay controls for screening of biologically diverse cell line panels using the Cell Painting assay.
Materials and Methods
Materials and Test Chemicals
A549 human lung carcinoma cells (CCL-185™), ARPE-19 human retinal pigmented epithelial cells (CRL-2302™), HepG2 human hepatocellular carcinoma cells (HB-8065™), 5637 human urinary bladder carcinoma cells (HTB-9™), MCF7 human breast adenocarcinoma cells (HTB-22™), and U-2 OS human osteosarcoma cells (HTB-96™) were purchased from ATCC (Manassas, VA). Dulbecco′s Modified Eagle′s Medium (DMEM, Cat-No: 10–013-CV), conical tubes, and culture flasks of various sizes (T25, T75, T225) were purchased from Corning® (Corning, NY). Heat-inactivated fetal bovine serum (HI-FBS) was purchased from Sigma-Aldrich (St. Louis, MO). CellEvent® Caspase 3/7, 10x Gibco® penicillin-streptomycin-glutamine (PSG, Cat-No: 10378–016), TrypLE Select (Cat-No: 125663–011), 0.4% trypan blue solution, Countess® cell counting chamber slides and MicroAmp® optical adhesive film were purchased from ThermoFisher Scientific (Waltham, MA). Solutions of 16% paraformaldehyde (PFA) were purchased from Electron Microscopy Sciences (Hatfield, PA). CellCarrier-384 Ultra microplates were purchased from PerkinElmer (Waltham, MA). Echo qualified 384-polypropylene (384PP) and 384-well low dead volume (384LDV) plates were purchased from LabCyte (San Jose, CA). Alexa Fluor™ 568 Phalloidin (Cat-No: A12380), Caspase3/7 (Cat-No: C10423), Concanavalin A (Cat-No: C11252), Hoechst-33342 (Cat-No: H3570), MitoTracker DeepRed (Cat-No: M22426), Propidium Iodide (Cat-No: P3566), SYTO14 (Cat-No: S7576), and Wheat Germ Agglutinin Alexa Fluor™ 555 Conjugate (Cat-No: W32464) were purchased from ThermoFisher Scientific.
Concentration-response screening was performed using a set of 14 phenotypic reference chemicals found to produce morphological phenotypes in U-2 OS cells using the Cell Painting assay10. Four additional chemicals were also screened, two of which were not expected to illicit phenotypic responses in any of the six cell lines at or below 100 μM (saccharin and sorbitol). Two cytotoxic chemicals (staurosporine and ionomycin) were also included at a single concentration as cell viability positive controls. All 14 reference chemicals, negative controls, and cytotoxic chemicals can be found in Nyffeler et al. (2019) and the EPA chemical dashboard (https://comptox.epa.gov/dashboard/chemical_lists/HTPP2019_REFSET).
Cell Culture
Once acquired from ATCC, each of the six cell lines were stored in vapor phase liquid nitrogen, thawed one at a time, and cultured in the same growth media (DMEM + 10% HI-FBS + 1x PSG). Cell lines were detached from the flask via incubation with 1X TrypLE Select for 7 minutes (ARPE-19), 8 minutes (A549, HepG2, MCF7, U-2 OS), and 10 minutes (HTB-9). Passaging and maintenance proceeded for each cell line as described.14 Appropriate seeding densities for use during the expansion phase were determined for each cell line in order to keep a consistent passaging schedule (4 days in vitro, DIV4) among all cell types. Each cell line was passaged and expanded individually with cryopreservation occurring at P4 (HepG2 & HTB-9), P5 (A549), P6 (MCF7 & U-2 OS), and P11 (ARPE-19) using the manufacturer’s recommended cryopreservation media.
For concentration-response screening experiments, a single vial of each cell line was thawed, cultured in growth media, and expanded up to three consecutive passages for each experimental replicate in order to achieve enough cell mass for 384-well plating. Cell yields and viabilities were determined using a Countess™ II automated cell counter (ThermoFisher Scientific) via the manufacturer′s protocol prior to seeding in CellCarrier-384 Ultra microplates. Cell seeding densities for 384-well plating were optimized for each cell type to allow analysis of at least 300 cells/well across less than 9 individual fields-of-view and not exceed 50% confluency at the time of sampling. Each cell line was seeding at the following densities and passage numbers: A549 (P7-P9) and ARPE-19 (P13-P15) at 500 cells/well, HepG2 (P6-P8) and HTB-9 (P5-P7) at 1000 cells/well, MCF7 (P8-P10) at 2500 cells/well, and U-2 OS (P8-P10) at 400 cells/well. Total culture volume for each cell type was 40 μL per well. Each test plate was placed into a 37°C humidified incubator with 5% CO2 atmosphere for a 24-hour recovery period prior to dosing.
Chemical Treatment
All 14 phenotypic reference chemicals, along with the two negative controls and two cytotoxic compounds, were solubilized in dimethylsulfoxide (DMSO) at concentrations ranging from 4 to 20 mM to create chemical stock solutions and stored at −20°C. These chemical stocks were pipetted into an Echo qualified 384-well polypropylene (384PP) plate and were used in dose plate preparation to make dilution series as described.14 The resulting Echo qualified 384-well low dead volume (384LDV) plates containing reference chemicals at 200x testing concentration were sealed with an adhesive aluminum film and stored at −80°C until needed. At 24 hours post-plating, a LabCyte Echo 550 acoustic dispenser was used to dispense 200 nL of 200x concentrated chemical treatment into randomized test wells.
The 14 reference chemicals and two negative controls were tested at seven concentrations (approximate half-log spacing) with three technical replicates per plate. Concentration ranges are listed in Table 1. The two cytotoxic compounds were tested at a single concentration (30 μM Ionomycin, 1 μM Staurosporine) with six technical replicates per plate. Each cell line used in this experiment had a total of three independent biological replicates with cultures established on different days. Cells were exposed to chemicals 24 hours after initial plating and sampled 48 hours after chemical treatment.
Table 1.
Chemical identifiers and test concentration ranges.
| Group | Chemical Namea | CASRN | DTXSID | Minimum Test Concentration (μM) | Maximum Test Concentration (μM) |
|---|---|---|---|---|---|
| Reference chemical | Amperozide | 75558-90-6 | DTXSID6048416 | 0.1 | 100 |
| Berberine Chloride | 633-65-8 | DTXSID8024602 | 0.1 | 100 | |
| Ca-074-Me | 147859-80-1 | DTXSID50881386 | 0.1 | 100 | |
| Etoposide | 33419-42-0 | DTXSID5023035 | 0.1 | 100 | |
| Fenbendazole | 43210-67-9 | DTXSID0040672 | 0.02 | 20 | |
| Fluphenazine | 69-23-8 | DTXSID2023068 | 0.1 | 100 | |
| Latrunculin B | 76343-94-7 | DTXSID10881387 | 0.05 | 50 | |
| Metoclopramide | 364-62-5 | DTXSID6045169 | 0.1 | 100 | |
| 5-Nitro-2-(3-phenylpropylamino)benzoic acid (NPPD) | 107254-86-4 | DTXSID90147978 | 0.1 | 100 | |
| Oxibendazole | 20559-55-1 | DTXSID5045625 | 0.02 | 20 | |
| Rapamycin | 53123-88-9 | DTXSID5023582 | 0.1 | 100 | |
| Rotenone | 83-79-4 | DTXSID6021248 | 0.01 | 10 | |
| Paclitaxel (Taxol) | 33069-62-4 | DTXSID9023413 | 0.0005 | 0.5 | |
| Tetrandrine | 518-34-3 | DTXSID10178062 | 0.025 | 25 | |
| Negative control | Saccharin | 81-07-2 | DTXSID5021251 | 0.1 | 100 |
| Sorbitol | 50-70-4 | DTXSID5023588 | 0.1 | 100 | |
| Cytotoxicity Control | Ionomycin | 56092-81-0 | DTXSID2040521 | 30 | 30 |
| Staurosporine | 62996-74-1 | DTXSID6041131 | 1 | 1 |
Chemical vendors and catalog number described in Nyffeler et al. (2020).
Cell Viability (CV) Assay and Image Processing
Assay plates were live-labeled with a solution of Hoechst-33342 (H-33342), CellEvent Caspase 3/7 (Casp), and propidium iodide (PI) in growth media to assess cytostatic and cytotoxic chemical effects. A Certus FLEX micro dispenser equipped with a 0.10 / 0.03 Gyger microvalve was utilized to dispense 2 μL of staining solution into each assay well. Final concentrations of each fluorescent label per well were 8.1 μM of H-33342, 5 μM of Casp, and 3.75 μM of PI. Plates were incubated at 37°C for 30 minutes prior to being fixed with 12 μL of 16% PFA using a MultiFlo FX Microplate Dispenser with a 1 μL peri-pump cassette. Fixation was followed by a 10 min incubation period at room temperature protected from light. Plates were washed four times with 1X PBS using the same MultiFlo FX Microplate Dispenser with the final wash remaining in the wells as a storage buffer. Plates were sealed with optical adhesive tape and stored at 4°C. Plates were removed from 4°C storage at least an hour prior to image acquisition to give each plate time to equilibrate to room temperature.
Fluorescent images were captured and processed using an Opera Phenix High Content Screening System (PerkinElmer) and Harmony software (v4.8, PerkinElmer). Images were acquired using a 5x objective and one field per well (2536 μm × 2536 μm). Excitation / emission pairings for imaging of H-33342, CASP and PI were 405 nm / 435–480 nm, 488 nm / 500–550 nm and 561 nm / 570–630 nm, respectively. Images were prefiltered via the H-33342 channel to exclude regions containing debris and/or staining irregularities. This same channel was used for segmentation of the nuclei and valid nuclei were selected based on intensity and size. The mean intensity of each valid nuclei was measured for both the CASP and PI channels. Cell-level data for each plate were exported for downstream analysis using R statistical software (v3.4.1). This process is described in further detail in Nyffeler et al. (2020)14 with both the CASP and PI channels being analyzed using the same methodology. Within each plate, cells with labeling intensity exceeding the median + 3*SD of vehicle control cells in either the CASP channel or PI channel were scored as non-viable cells (i.e. responders). For both CASP and PI, percent responder was then calculated for each well by dividing the total number of responders by the total number of all cells. A benchmark concentration was then determined for CASP and PI and the cytotoxic benchmark concentration (BMC) was identified as the minimum of the CASP and PI BMCs. Cytostatic effects were determined via the effective concentration 50 (EC50) of the normalized cell count. The in vitro point-of-departure (POD) for the cell viability assay (CV POD) was calculated using the minimum of the cytostatic EC50 and cytotoxic BMC values. The no observable effect concentration (NOEC) and lowest observable effect concentration (LOEC) were determined as the highest tested concentration below and lowest tested concentration above the CV POD, respectively.
Cell Painting (CP) Assay and Image Processing
Assay plates were fluorescently labeled using the adapted ‘Cell Painting’ procedure as described.14 Phenotypic profiling of cellular organelles (nucleus, nucleoli, endoplasmic reticulum (ER), actin cytoskeleton, golgi, and plasma membrane (AGP), mitochondria) was performed to examine morphological responses to chemical treatments across a variety human-derived cell lines (A549, ARPE-19, HepG2, HTB-9, MCF7, and U-2 OS). Each plate was first live-cell labeled with MitoTracker Deep Red, followed by a fixation and washing step utilizing a MultiFlo FX Microplate Dispenser. Cells were permeabilized with 0.1% Triton X-100 and washed prior to staining with a 1% bovine serum albumin in 1X PBS solution containing the remaining labeling reagents (Alexa Fluor™ 568 Phalloidin, Concanavalin A Alexa Fluor 488 conjugate, Hoechst-33342, SYTO14, Wheat Germ Agglutinin Alexa Fluor™ 555 Conjugate). In the final wash step 80 μL 1X PBS was dispensed into each well as a storage buffer. Plates were then sealed with optical adhesive tape and stored at 4°C until image acquisition. Each plate was removed from 4°C and allowed to equilibrate to room temperature for at least one hour prior to image acquisition.
An Opera Phenix High Content Screening System was used to acquire images of fluorescently labeled cells that were subsequently analyzed with Harmony® software (v4.8). Images for each cell line were obtained using an 20x water immersion objective in confocal mode using 2×2 pixel binning. Channel specific Z-offsets, exposure times, and laser power settings were optimized for each individual cell line (Supplementary Table 1) using a combination of randomly selected wells and vehicle-treated wells. These settings remained constant for each cell line throughout experiments. Images for each cell line were acquired with 8 or 9 unique fields per well, covering an area up to 1908 μm × 1908 μm per well.
Image processing was performed via Harmony® software using a combination of cell segmentation and defining of cellular compartments (nucleus, membrane, cytoplasm, perinuclear space). Nuclear and cell segmentation was optimized for each cell line, with the Hoechst-33342 channel (nuclei) acting as a seed. A total of 1300 features (texture, intensity, etc.) were measured for each individual cell. The approximate number of cells analyzed for each cell line is listed in Supplementary Table 1. Images were screened for wells containing <50 cells in order to exclude these from downstream analysis. Cell-level data corresponding to each plate were exported and data reduction was performed using R statistical software. Median absolute deviation (MAD) was used to normalize cell-level data to vehicle controls within each plate for each endpoint.3 The median of the normalized data was calculated for each well to derive well-level aggregate values. Well level median data was then Z-transformed and used as input for concentration-response modeling with the BMDExpress software package15. Highly cytotoxic treatments were excluded from BMC modeling of CP data by removing tested concentrations above the CV LOEC. Standardization and BMC modeling of the well-level data are as described in detail.14 Features considered responsive to chemical treatment were those with a BMC below the highest tested concentration. Each of the 1300 features were grouped using channel, module, and compartment into 49 different categories as described.14 BMC values for each feature were used to calculate a median BMC for each category and categories with ≥30% responsive features were ranked in ascending order by median BMC. The CP POD was defined as the category with the lowest potency values.
Comparison of Phenotypic Profiles
Well-level data corresponding to DMSO controls and the cell viability NOEC for each chemical by cell type combination were analyzed using a variation of the Theta Comparative Cell Scoring (TCCS) method described by Warchal et al.11, 13. In instances where a cell viability NOEC could not be determined, the highest tested concentration was retained in the analysis. A total of n = 9 treatment wells per chemical x cell line combination and n = 72 DMSO control wells per cell line were included in the TCCS analysis. In brief, features with a z-transformed well level value less than 1.5 were imputed with a value of 0. Principal components analysis (PCA) was then performed using the prcomp function of the R stats package. The results from the first ten principal components (PC) for each sample (capturing >75% of total variance in the data) were retained and centered to DMSO controls (regardless of cell type). The Euclidean norm of a vector containing values for PC1 through PC10 was calculated as a metric of “distance” (i.e. magnitude of phenotypic change) from control and the angular degree of each vector in comparison to a reference vector (i.e. theta) was calculated as metric of similarity of phenotypic response profiles.
Results
Morphological Heterogeneity of Human-Derived Cell Lines
For this study, the Cell Painting assay as implemented in Nyffeler et al. (2020)14 was used to evaluate chemical effects across six biologically diverse and morphologically distinct human-derived cell lines. In a first step, the morphology of the different cell lines was investigated in the absence of chemical treatment to qualitatively assess whether the fluorescent probes labeled organelles in a similar pattern to those characteristically observed in U-2 OS cells. Fluorescent labeling of the organelles of interest was apparent in all six cell lines and were generally consistent with the sub-cellular localization patterns previously observed in U-2 OS cells (Figure 1)10, 14. Qualitative differences in cell size, compactness, and organelle symmetry were apparent when examining fluorescent channels, independently, across cell lines. U-2 OS, A549 and ARPE-19 cells tended to be larger in total area and to have higher granularity in the perinuclear region within the ER/RNA channel compared to other cell lines. In U-2 OS, A549 and ARPE-19 cells, ER/RNA fluorescent intensity diminished with increasing distance from the center of the cell. This pattern was less prominent in cell lines with more compact cytoplasm (i.e. HepG2, HTB-9, MCF7). Nucleoli were prominently labeled in the nucleus region of each cell type. Labeling patterns in the actin cytoskeleton/Golgi/plasma membrane (AGP) channel had the highest degree of qualitative dissimilarity across cell lines. Prominent labeling of the cytoskeleton (visualized as striations) were observable in U-2 OS, A549 and ARPE-19 cells. In contrast, AGP labeling in the cytoplasm of more compact cell lines (HepG2, HTB-9, MCF7) appeared more diffuse. The prominence of perinuclear AGP labeling varied both across cell lines and from cell-to-cell within a cell line. HTB-9 cells had the most prominent labeling of the plasma membrane amongst all cell types. Asymmetric perinuclear localization of mitochondria was apparent in U-2 OS, A549 and ARPE-19 cells while more symmetric and diffuse mitochondrial labeling was apparent in HepG2, HTB-9 and MCF7 cells. Overall, organelles of interest appear to be reliably labeled in each of the respective cell types using a uniform staining protocol.
Figure 1: Examples of Cell Painting labeling patterns in six different human-derived cell lines.
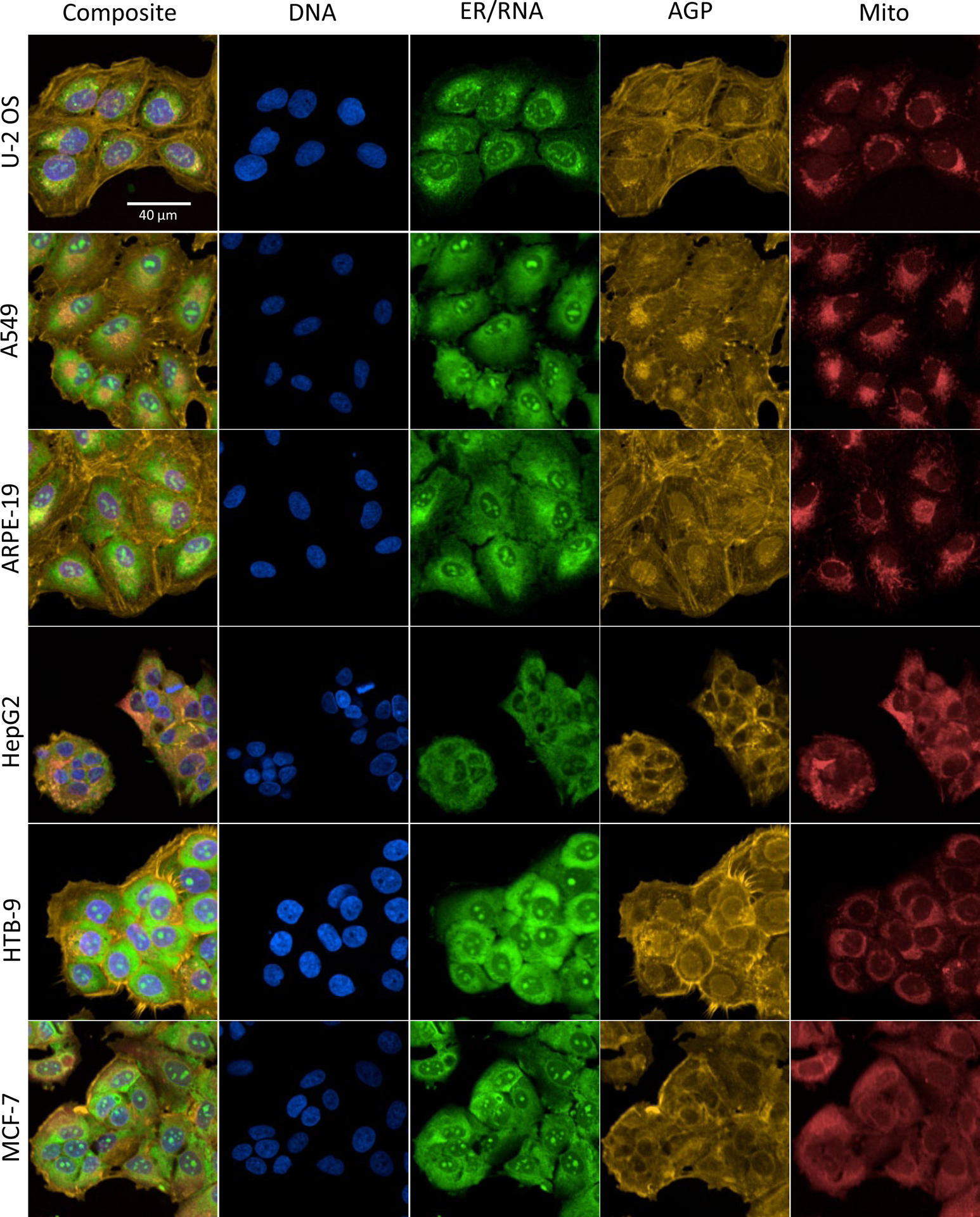
Representative images of DMSO control wells, live-labeled for mitochondria, fixed, permeabilized, and labeled with the remaining fluorescent probes. All images were acquired at 20x magnification. A composite image for each cell line is shown along with the four individual channels corresponding to a total of seven different cellular organelles. Similar organelle labeling patterns can be observed across all six cell lines along with variations cell size as well as organelle localization, compactness, and granularity. Scale bar = 40 μm.
Comparison of Chemical-Induced Phenotypic Profiles Across Cell Lines
Next, a set of 14 reference chemicals along with two chemicals not expected to produce phenotypic changes in any of the cell lines up to 100 μM (saccharin, sorbitol) were screened in concentration-response mode. The chemicals were tested in seven-point concentration-response across all cell lines with an exposure duration of 48 hours as described in previous studies10, 14. For several of the tested chemicals, visual changes in cellular morphology were obvious in multiple cell lines, were qualitatively similar across affected cell lines and were consistent with findings from previous studies in U-2 OS cells10, 14. For example, Figure 2 illustrates the effects of treatment with 30 μM berberine chloride across the various cell types. Effects on mitochondria morphology were observed in each cell line (Fig. 2). The effects manifested as an increase in mitochondrial compactness. In addition, treatment with 3 μM Ca-074-Me produced changes in AGP-labeling patterns in each cell line with the most prominent increases in intensity perinuclear labeling observed in cell lines with comparatively larger cytoplasmic area (i.e. U-2 OS, A549, ARPE-19) (Fig S1). Lastly, 0.3 μM etoposide treatment resulted in increases in overall cell size in most cell lines (U-2 OS, A549, ARPE-19, HepG2, MCF7, but not HTB-9). The increase in cell size corresponded to effects in a variety of organelles and phenotypic features.
Figure 2: Examples of chemical specific phenotypes across six human-derived cell lines.
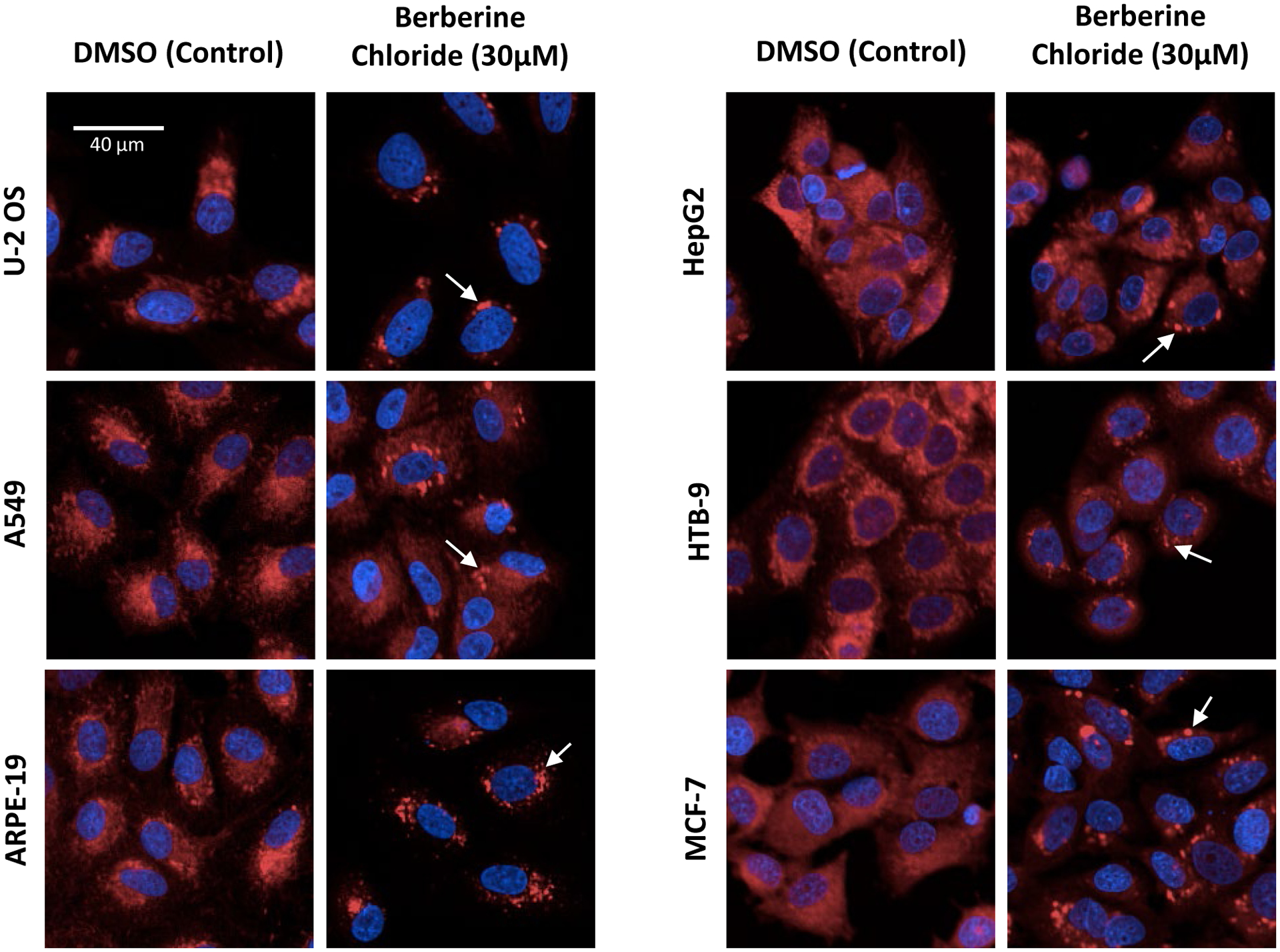
Each cell line was treated for 48 hr and comparisons between DMSO control and berberine chloride (30μM) are shown within the DNA and mitochondria channels. All images were acquired at 20x magnification. Effects on mitochondria compactness and texture between DMSO controls and cells treated with berberine chloride were observed (arrows). Phenotypic responses appear qualitatively similar across cell lines. Scale bar = 40 μm.
Concentration-response modeling of phenotypic profiling data was performed on all six cell lines. Results were visualized using potency-magnitude and accumulation plots as described.14 Representative results for berberine chloride are shown in Figure 3 and results for the other 15 chemicals in the study are shown in Supplementary Figure 2. Each point on the potency magnitude plots (Figure 3, left two columns) is a phenotypic feature represented by the potency (i.e. benchmark concentration, BMC) and effect size (i.e. normalized magnitude) from the concentration-response modeling results, coded by fluorescent channel, cellular compartment, and feature type. Each point on the accumulation plots (Figure 3, right two columns) represents the median feature-level potency for phenotypic categories where ≥ 30 % of constituent features were affected by chemical treatment and are also coded by fluorescent channel, cellular compartment and feature type. Grey and red vertical dotted lines represent the cytostatic EC50 and cell viability BMC, respectively. For all 14 reference chemicals phenotypic effects occurred below the threshold for cytostatic effects or cytotoxicity in each of the six cell lines (Figure 3, Fig S2). Sorbitol and saccharin affected none or very few phenotypic features in each of the six cells lines (Fig S2A–B). There was only one instance where an in vitro point-of-departure (i.e. CP POD) was determined for either of these chemicals (saccharin in A549 cells) (Fig S2B).
Figure 3: Concentration-response modeling of phenotypic profiling data following berberine chloride treatment in six human-derived cell lines.
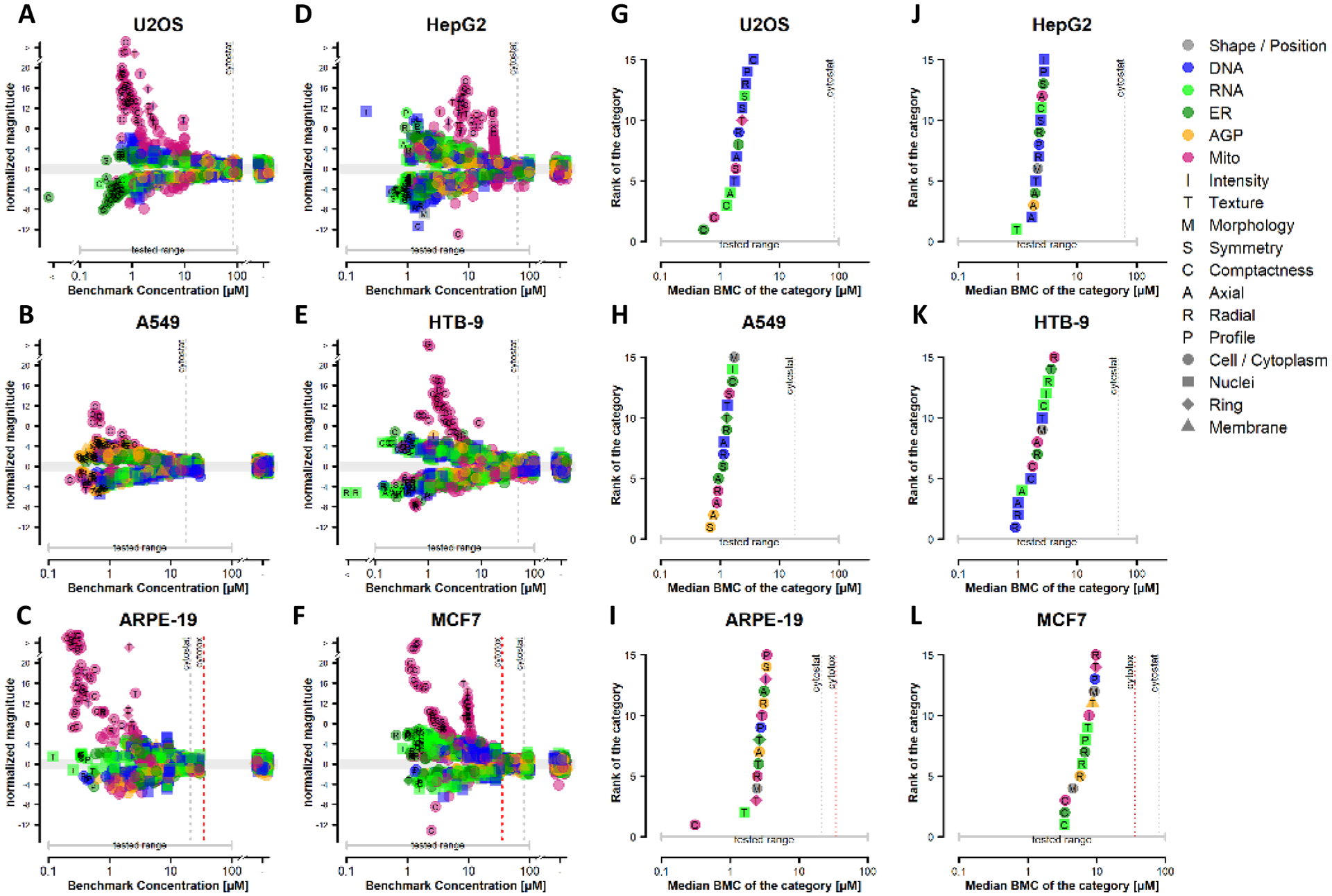
(A-F) Potency-magnitude plots corresponding to an individual cell line treated with berberine chloride. Each point on the potency-magnitude plots refers to a phenotypic feature represented by the BMC on the x-axis and the normalized effect magnitude on the y-axis. Features are coded by fluorescent channel (color), compartment (shape) and feature type (letter). The gray shaded area in the potency-magnitude plots shows −1 < magnitude < 1 and represents the threshold for a marked response from DMSO control. (G-L) Accumulation plots corresponding to an individual cell line treated with berberine chloride. Feature-level BMCs were grouped into 49 categories. Categories where ≥30% of the constituent features were concentration-responsive were ranked in ascending order according to the median BMC of the category. The 15 most sensitive categories for each cell line are displayed and coded as described in panels A-F. The median feature-level BMC for each category is represented on the x-axis and the category rank represented on the y-axis. In all panels, the onset of cytotoxicity and cytostatic effects are marked by red and grey vertical dotted lines, respectively. Absence of vertical lines indicates that cytotoxicity or cytostatic effects were not observed within the concentration range tested.
Comparison of potency-magnitude plots for the exemplar chemical berberine chloride demonstrated that phenotypic features associated with mitochondrial texture, compactness and intensity were associated with the largest response magnitudes in each cell line (Figure 3A – 3F). This is consistent with the qualitative observations in Figure 2. Of note, the most sensitive features and the most sensitive phenotypic category (i.e. those having the highest potency) for berberine chloride were not always associated with changes in mitochondrial morphology across the cell line panel. For instance, in each cell type, individual features associated with DNA, ER/RNA or AGP morphology were affected at lower concentrations than features associated with mitochondrial morphology (Figure 3A – 3F) albeit with much lower response magnitudes. Further, the most sensitive category (i.e. lowest median BMC) corresponded to a different channel / compartment / feature type for each cell type. For ARPE-19 cells, the most sensitive phenotypic category was mitochondrial compactness and the potency value for this category was left-shifted compared with the next most sensitive categories (Figure 3I). For other cell lines, the potency of the first mitochondria-associated category was within one half-log unit of the most sensitive phenotypic category. Additional comparisons of phenotypic effects across cell lines based on review of concentration-response modeling results can be found in Supplementary Figure 2.
In order to more easily identify patterns of phenotypic effects in the profiling data across the large variety of cell types and chemicals tested, a heatmap was constructed that illustrated normalized effect magnitudes of the highest non-cytotoxic test concentration for each chemical x cell type combination (Fig 4A). If a chemical did not produce cytotoxicity, the highest tested concentration was retained. In addition, an analysis using a modification of the theta comparative cell scoring method (TCCS) (Warchal et al. 2016) was performed in order to quantify and compare the robustness and similarity of phenotypic response profiles across chemicals and cell types. Results are presented as a series of radial plots (Fig 4B). Points relatively further from the origin indicate relatively more robust phenotypic effects and points positioned along similar angular degrees are indicative of similarity in response profiles.
Figure 4: Similarity in phenotypic response profiles across six human-derived cell lines.
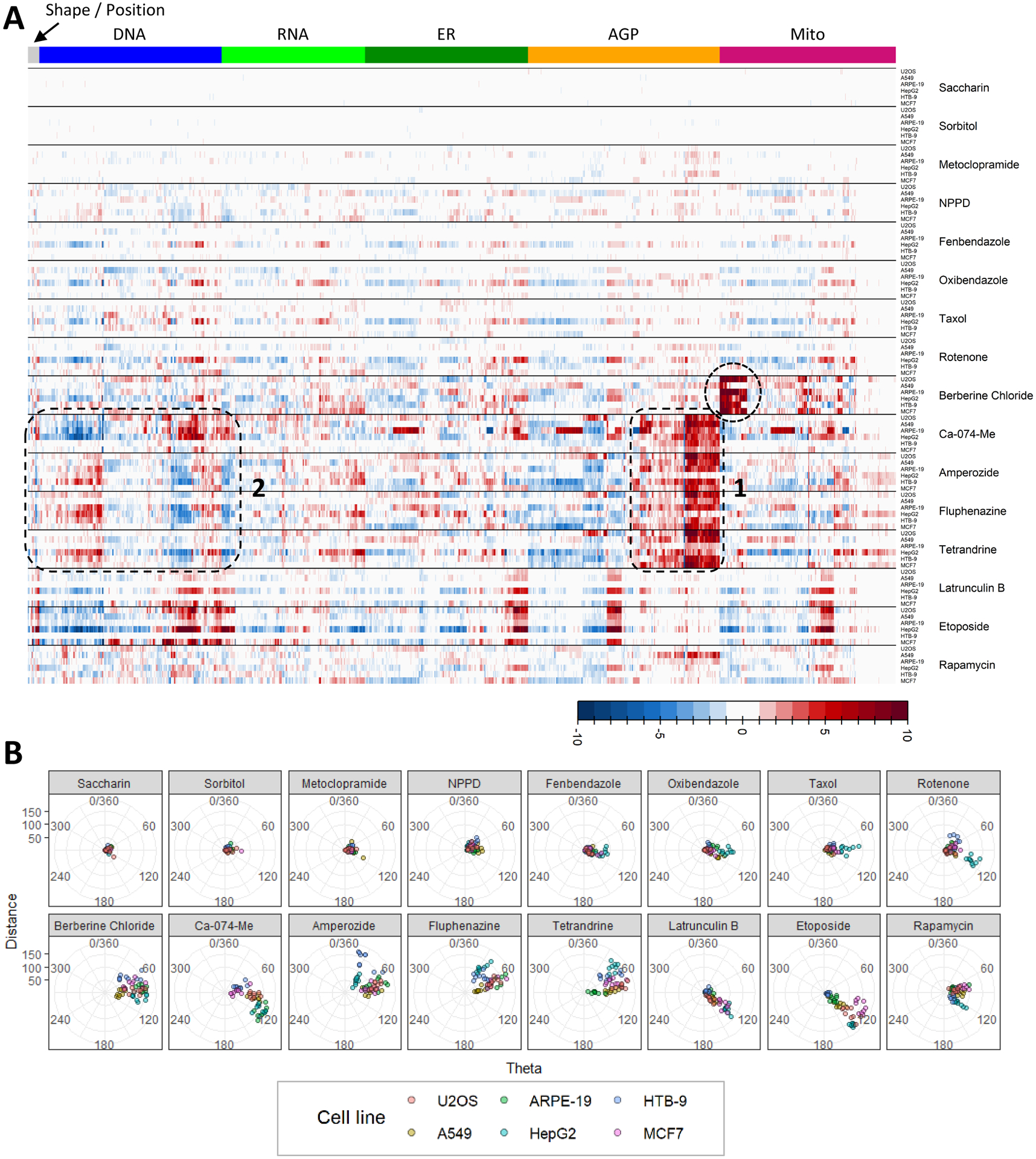
(A) Summary of phenotypic effects for the highest non-cytotoxic test concentration of each chemical. Well level feature data were normalized and scaled per plate, then averaged. The columns of the heatmap correspond to the 1300 phenotypic features, organized by fluorescent channel. Rows correspond to different cell types treated with the chemicals listed to the right of each heatmap clade. Colors in the heatmap represent the magnitude of increase or decrease in a measured feature with respect to DMSO control. Dotted circles or boxes denote areas or similarity or dissimilarity in phenotypic response profiles as described in the text. (B) Theta cell comparative scoring (TCCS) radial plots, faceted by chemical. Data from the highest non-cytotoxic concentration of each test chemical were analyzed using a modification of TCCS analysis (Warchal et al. 2016). Each point represents and individual test well (n = 9 test wells / chemical / cell type). Points are color coded by cell type. Points relatively further from the origin (distance) indicate relatively more robust phenotypic effects and points positioned along similar angular degrees (theta) are indicative of relative similarity in response profiles.
The highest tested concentrations of saccharin, sorbitol and metoclopramide affected none or very few phenotypic features across the different cell types (Fig 4A). Therefore, points on the TCCS plots are tightly packed around the origin (Fig 4B). NPPD affected a variety of different phenotypic features throughout different cell types, albeit with low magnitudes of effect (Fig 4A). Points on the TCSS plot are slightly more spread from the origin as compared to three aforementioned chemicals, however, no discernable similarity in response profiles across cell types is apparent for NPPD (Fig 4B). Fenbendazole, oxibendazole, taxol and rotenone affected a variety of different phenotypic features throughout different cell types, albeit with low magnitudes of effect (Fig 4A). Of note, for each of these chemicals many more features were affected in HepG2 cells as compared to the other cell types tested. This observation is manifest in the TCCS plots, as points corresponding to HepG2 samples are shifted away from the origin (along a similar angular degree) as compared to points associated with the other cell types (Figure 4B). Rotenone also produced a response in HTB-9 cells that was dissimilar to that observed in HepG2 cells.
As shown in Figure 2 and Figure 3, berberine chloride produced changes in mitochondrial morphology in each cell type. Those effects were clearly visible and consistent in the heatmap (Fig 4A, circle) with the exception of A549 cells where a smaller number of affected mitochondrial features was observed upon comparison to other cell types. In the TCCS plot, all points were shifted away from the origin with A549 cells located closest to the origin.
The test chemicals Ca-074-Me, amperozide, fluphenazine and tetrandrine produced pronounced changes in AGP labeling (Fig 4A, rounded box 1) and produced a variety of effects in other channels and feature types, the identity of which varied across cell types. In general, the pattern of changes in many features within the DNA channel are in opposite directions when comparing Ca-074-Me and amperozide, fluphenazine or tetrandrine, respectively (Fig 4A, rounded box 2). The differential characteristics of the phenotypic responses of these chemicals are apparent upon comparison of individual TCCS plots (Fig 4B). Cells treated with Ca-074-Me tend to be off-shifted from center in the lower right direction while cells treated with amperozide, fluphenazine or tetrandrine tend be off-shifted from center in the upper right direction. The similar angular degree amongst amperozide, fluphenazine and tetrandrine is indicative of quantitative similarity in phenotypic profiles across cell types with tetrandrine covering the narrowest range of angular degrees amongst these three chemicals.
Latrunculin B produced effects in a subset of ER, AGP and mitochondrial features that are consistent in terms of identity and directionality across U-2 OS, A549, HepG2 and MCF7 cell lines (Fig 4A). Many additional features were affected in HepG2 and MCF7 cells, but not the other two cell types. Samples associated with all four cells types were markedly off shifted from center along a narrow range of angular degrees in TCCS analysis (Fig 4B), indicating some quantitative similarity in response of these cell types. Responses to latrunculin B in the remaining two cell types (ARPE-19 and HTB-9) were not robust. Etoposide also produced effects in a sub-set of features from each channel except RNA that are consistent in terms of identity and directionality across all cell lines except HTB-9 (Fig 4A). Samples associated with all cell types except HTB-9 were markedly off-shifted from the origin along a narrow range of angular degrees in TCCS analysis, indicating some quantitative similarity in response of these cell types. Lastly, rapamycin affected a variety of different phenotypic features across the various cell types with little consistency in identity, magnitude or direction of effects (Fig 4A). No discernable similarity in response profiles across cell types is apparent for rapamycin using these approaches (Fig 4B).
In summary, some degree of quantitative similarity in phenotypic response profiles across biologically diverse cell types was observed with berberine chloride, Ca-074-Me, amperozide, fluphenazine, tetrandrine, latrunculin B and etoposide. In some instances, chemicals that produce robust responses in a particular cell type did not produce robust responses in others. This underscores the need to identify and rigorously characterize putative reference chemicals prior to use as plate-based controls for the Cell Painting assay in novel cell types.
Comparison of Potencies for Chemical Bioactivity Across Cell Lines
In addition to evaluating the similarity/dissimilarity of chemical-induced phenotypic profiles across cell types, we also compared potencies for chemical bioactivity using concentration-response modeling. The PODs corresponding to the Cell Painting assay (i.e. CP POD), cytotoxicity BMC, and cytostatic EC50 were plotted for comparison of each chemical treatment and cell type (Fig. 5). Concentration-response curves for normalized cell count (nCC), cell viability and apoptosis for each chemical by cell type combination are shown in Supplementary Figure 3. No response in cell viability, cytostatic (i.e. nCC) or Cell Painting assays was observed for sorbitol in any cell type. No response in cell viability or cytostatic assays was observed for saccharin. Saccharin was active in the Cell Painting assay in ARPE-19 cells only and this finding was associated with effects in a single phenotypic category (Fig S2B).
Figure 5. CP PODs of fourteen reference chemicals and two negative control chemicals across six human-derived cell lines.
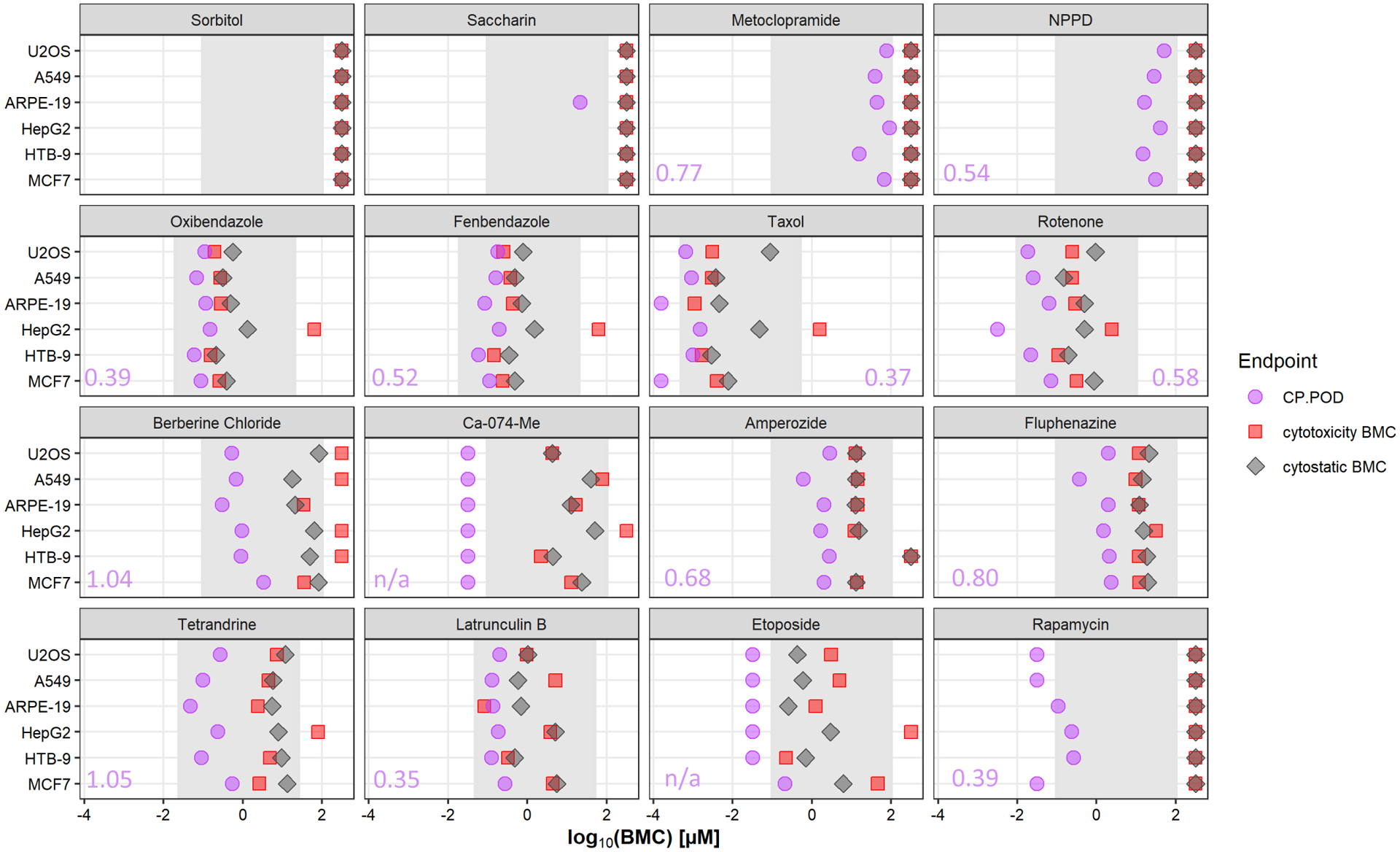
PODs for the Cell Painting assay (CP POD) (purple cicles), cytotoxicity BMC (red squares) and cytostatic BMC (gray diamonds) were compared across six cells types. Gray shading represents the range of tested concentrations for each chemical. Values in purple are the range of potencies, in log10 units, between the maximum and minimum CP POD observed across cell types for each chemical. Only chemical x cell type combinations where a CP POD could be determined within the tested concentration range are included in this comparison.
All 14 reference chemicals were active in the Cell Painting assay in each of the cell lines tested, often at concentrations markedly below the threshold for cytotoxicity or cytostatic effects. Chemicals with the largest difference between the threshold for bioactivity as determined by the Cell Painting assay and thresholds for cytotoxicity or cytostatic effects were Ca-074-Me, berberine chloride and rapamycin. Of note, accurate CP PODs for taxol in ARPE-19 and MCF7 cells, rotenone in HepG2 cells, Ca-074-Me in any cell type, etoposide in all cell types except MCF7 and rapamycin in U-2 OS, A549 and MCF7 cells could not be determined. This was due to changes in cellular phenotypes occurring at the lowest concentration tested in each of these instances. These instances are illustrated in Figure 5 as purple points to the left of the gray shaded regions that represents the concentration range tested for each chemical.
For the remaining instances where a CP POD could be reliably determined within the tested concentration range, the difference in CP PODs across the cell lines varied by less than approximately one order of magnitude (i.e. log10 unit). The largest ranges of CP PODs across cell types was observed with tetrandrine and berberine chloride (1.04 and 1.05 log10 units, respectively). The smallest ranges of CP PODs across cell types was observed with latrunculin B and taxol (0.35 and 0.37 log10 units, respectively). This consistency in CP PODs was apparent despite the observation that the most sensitive phenotypic features or phenotypic categories varied across cell types for any given chemical (except Ca-074-Me where effects in the AGP channel were always the most sensitive) (Fig S2). Overall, these data indicate that for the chemical set tested in the present study, bioactivity estimates (i.e. potency) are generally consistent across cell lines even when dissimilarity in phenotypic profiles is observed. It is unclear if this observation is applicable to other / larger chemical sets.
Discussion
The Cell Painting assay can be used as a non-targeted high-throughput profiling assay for characterizing the biological activity of environmental chemicals1. Questions remain, however, regarding the general compatibility and portability of the Cell Painting assay throughout different human-derived cell types and what optimization steps may be required for deploying the assay in a novel cell model. To investigate this topic, we established Cell Painting sample preparation, image acquisition, feature extraction and data analysis workflows (described in Nyffeler et al. (2020)14) and applied these workflows to concentration-response screening of fourteen phenotypic reference chemicals (and two negative control chemicals) across six human-derived cell types (U-2 OS, A549, ARPE-19, HepG2, HTB-9, MCF7). A uniform cytochemistry protocol with no cell type-specific optimization steps was used to prepare samples of each cell type for high-content imaging. In contrast, we found it necessary to optimize parameters associated with image acquisition (i.e. z-offset, laser power, exposure duration) as well as those associated with nucleus and cell segmentation on a per cell type basis. A second objective of this study was to compare phenotypic profiles and potency estimates for perturbation of cellular biology across the biologically diverse cell lines to inform strategies for selection of reference chemicals (i.e. assay controls) for high-throughput screening. For several of the chemicals tested, similar subsets of phenotypic features corresponding to a particular organelle were associated with the highest effect magnitudes in each affected cell type. For other chemicals, more pronounced effects were observed in one particular cell type upon comparison to other cell types in the panel. Lastly, potencies for biological activity of the tested chemicals were comparable (i.e. less than or approximating one log10 unit of potency) across cell types even though the identity of the most sensitive phenotypic feature categories or most highly affected phenotypic features tended to vary across cell types. Variations in the response of different cell types to chemicals underscores the need to identify and rigorously characterize reference chemicals for the Cell Painting assay on a per cell type basis as active chemicals do not always produce similar phenotypes in different cellular contexts.
The U-2 OS cell line was included in the present study given that this was the cell type used for initial development of Cell Painting and is widely considered the “gold standard” for this assay3, 10, 14, 16, 17. Three of the other cell lines included in the present study (MCF7, HTB-9 and A549) were used in the qualitative comparison of fluorophore labeling patterns presented in the initial Cell Painting publication10. The remaining two cell lines (HepG2, ARPE-19) were selected based on previous use at EPA in various toxicological research areas18, 19. Overall, this panel of cell lines is derived from a diverse set of human tissues, have diverse baseline gene expression patterns20, 21 and are morphologically heterogenous with regards to cellular size, nucleus-to-cytoplasm ratios and tightly-packed versus dispersed growth patterns when cultured in 2-D monolayers. Despite this morphological heterogeneity, the uniform cytochemistry protocol employed in the present study labeled organelles of interest within each cell line with patterns characteristic of what is observed in U-2 OS cells. This was to be expected given that the fluorescent probes used in the Cell Painting assay bind to molecular target sites that are present in most (if not all) human-derived cell types. The characteristic patterns we observed did not indicate to us a need to optimize the cytochemistry protocol for each individual cell type before proceeding to development of image acquisition and analysis workflows. However, it should not be concluded that deployment of the Cell Painting assay to novel human-derived cell types will never require optimization of cytochemistry protocols. For example, Warchal et al (2016)11 noted a sensitivity of the MDA-MB-231 cell line to live cell staining with mitotracker in the context of the Cell Painting assay. Upon addition of the live cell label, MDA-MB-231 cells assumed a rounded appearance (N. Carragher, personal communication) that may have been indicative of an adverse effect on the overall health of these cells. This finding prompted the authors to change the sequential addition of “live-cell” (i.e. mitotracker) and “fixed-cell” (i.e. all other probes) reagents recommended in previous studies3, 10 to a single addition after cells had been fixed with paraformaldehyde. This eliminated the artifactual effect observed in MDA-MB-231 cells. Of note (and similar to the present study), a uniform cytochemistry protocol was applied to all eight breast cancer cell types, not just the one where the adverse response to the live cell labeling reagent was observed. These observations support the need to critically evaluate novel cell lines for compatibility with Cell Painting sample preparation protocols before proceeding to downstream studies and adjust where necessary to maintain consistency within a study. In the case of the present study, further optimization of cytochemistry protocols beyond those described previously14 was unnecessary and portability of the sample preparation procedures across cell types was straightforward.
In the present study, we did find it necessary to optimize image acquisition parameters on a per cell type basis to account for differences in the size and three-dimensional shape of the respective cell types when cultured in monolayers. The present study uses single z-plane confocal microscopy with lasers as light sources and a 16-bit large-format sCMOS camera for image acquisition. During optimization, the focal height, laser power and integration time for image acquisition was adjusted for each channel for each cell line based on inspection of randomly selected treatment wells and vehicle control wells. The goal in each case was to identify a focal plane where the organelle(s) of interest were in sharp focus while keeping the maximal pixel intensities of the image within the lower quarter of the dynamic range of the 16-bit camera. Even in monolayer, different objects (i.e. organelles) of interest within a cell can be present at different heights in relation to apical or basal surface of the cell22, 23, thereby necessitating different focal heights for obtaining the sharpest images when using single plane confocal imaging. In the context of the Cell Painting assay, multiplexed labeling of endoplasmic reticulum (ER) with Alexa Fluor®-conjugated concanavalin A and nucleoli provide good examples of this phenomenon. Even though these stains are multiplexed within the same fluorescent channel in the Cell Painting assay, the optimal setting for imaging of nucleoli was consistently identified at a higher focal plane (relative to the default reference point determined by the Opera Phenix High Content Screening System using CellCarrier-384 Ultra microplates) as compared to the ER or any other organelle (Supplementary Table 1). This necessitated use of two acquisitions with the 488 nm laser at different focal heights to obtain sharp images of the ER and nucleoli, respectively. Similarly, in most cell lines (except HepG2), the optimal setting for imaging of mitochondria was at a lower focal plane compared to that for any other organelle. The use of maximum projections from 3D confocal microscopy would eliminate the need for identifying an optimal focal height for imaging each channel, assuming an appropriate Z-range has been determined that spans the thickness of the cell of interest. This approach is more computationally intensive, however, and it would increase the overall duration of image acquisition but not eliminate the need to optimize laser power and image acquisition times for different fluorescent channels and different cell types. The focal offsets and image acquisition settings in Supplementary Table 1 are particular to the high-content screening system and the cell culture vessels that were used in the present study, and would need to be adjusted for other imaging platforms and/or plate types. Our conclusion was that the Cell Painting sample preparation protocol could be deployed throughout the cell types used in the present study with ease (i.e., without modification), while the image acquisition phase of experimentation required detailed, cell type–specific adjustments prior to screening chemicals.
Comparison of chemical-induced phenotypic profiles demonstrated that some chemicals can produce qualitatively similar effects in biologically diverse cell types. Similarities in response profiles were evaluated in several different ways including representative images (Fig 2), feature level potency-magnitude plots of concentration-response modeling results (Fig 3, Supp Fig 2), heatmaps (Fig 4A) and through use of the theta comparative cell scoring (TCCS) method (Fig 4B)11. In all cell types, berberine chloride produced marked changes primarily in features related to mitochondrial morphology. This is consistent with the biological activity of this chemical as an inhibitor of the mitochondrial respiratory complex I24. In all cell types, the chemicals amperozide, fluphenazine and tetrandrine produced marked changes in textural features associated within the AGP channel and changes in a variety of different feature types in the DNA channel. Ca-074-Me also produced marked changes in AGP textural features and a variety of different feature types in the DNA channel. Ca-074-Me induced changes in the AGP channel had similar directionality as compared to the aforementioned three chemicals while Ca-074-Me-induced changes in the DNA channel tended to be in the opposite direction. Dissimilarity in the profiles of Ca-074-Me and the other three chemicals is illustrated in the TCCS radial plots in Fig 4B. Samples associated with Ca-074-Me treatment tended to shift from the origin along a different range of radial degrees as compared to the other three chemicals. The underlying reason for the similarity in phenotypic response profiles for amperozide, fluphenazine and tetrandrine is not obvious given that the primary molecular mechanisms of these chemicals is diverse: amperozide is a 5-HT2A serotonin receptor antagonist25, fluphenazine and is D2 dopamine receptor antagonist26 and tetrandrine is a calcium channel blocker27. It could be that some promiscuous or off-target pharmacological activity of one, some or all of these chemicals may underlie the similarity in response profiles. Follow-up studies would be needed to determine this. Regardless of the underlying molecular mechanisms it is clear that certain chemicals can produce similar quantitative phenotypic response profiles in the Cell Painting assay in different biologically diverse cell types when a uniform sample preparation and feature extraction algorithm is applied. This supports the general extensibility of the Cell Painting assay across a variety of human-derived cell types and use of this assay as a bioactivity screening approach.
There were also instances where chemicals produced a pronounced phenotypic effect in one cell line that was not recapitulated in other cell lines. This was most notable with the HepG2 cell model upon treatment with a variety of chemicals whose typical uses are associated with biocidal activity28–30. Fenbendazole, oxibendazole and taxol (i.e. paclitaxel) all bind β-tubulin and affect microtubule stability (albeit in different ways) to produce cytotoxicity in rapidly dividing cells31, 32. Rotenone is a mitochondrial toxicant with common use as a piscicide33. The identity and directionality of features affected in HepG2 cells by these chemicals is markedly similar as evidenced by visual comparison of heatmap results (Fig 4A) and similar angular degree of HepG2 samples in TCCS analysis (Fig 4B). Features in the DNA channel had the largest magnitude of effects in HepG2 cells for each of these chemicals (Supp Fig2I–L). In addition, for each of these chemicals the differences in potency between Cell Painting effects and cytotoxicity / cytostatic effects was greater in HepG2 cells than in any other cell type although CP PODs did not vary by to a great extent across cell types. Comparison of the cytotoxicity BMC, cytostatic EC50 and CP PODs is indicative of a more gradual transition toward cytotoxicity in HepG2 cells as compared to other cell types for this group of chemicals. The reason for differing sensitivity of HepG2 cells as compared to other cell types is unknown but may be due to differences in basal metabolism, replication rate, expression of pharmacological targets, chemical transport, metabolism or overall robustness across the cell line panel.
Another finding of this study is that for most chemicals, the threshold potency for phenotypic effects as determined by the Cell Painting assay did not vary by a substantial amount across cell lines. This was despite the fact that for any particular chemical (except Ca-074-Me) different phenotypic features or phenotypic categories were identified as the most sensitive across cell types. One interpretation of this finding is that while the concentration where chemicals begin to perturb cellular biology is consistent the manner in which these effects initially manifest is dependent upon the biological context of the cells. Of note, in many instances the CP POD for phenotypic effects occurred at concentrations of test chemicals that did not affect cell counts, indicating that the most sensitive changes in phenotype were not associated with changes in cell density (Figure 5, Supplementary Figure 3). It is unknown whether the consistency in bioactive potency is generalizable across a larger diversity of chemical or biological space (i.e. more cell lines).
The results of the present study inform strategy for selection of reference chemicals in situations that involve the use of the Cell Painting assay across multiple cell types and where harmonization of reference chemicals across cell types is desired. In high-throughput screening, assay performance is assessed by the use of positive controls and/or reference chemicals that produce a predictable and reproducible response in the endpoint(s) of interest34. Confirmation that a culture or an assay plate responds to the reference chemical as predicted is indicative that the assay was conducted properly and increases confidence that data from test substances is of acceptable quality. As compared to traditional high-throughput screening (HTS) assays, positive controls / reference chemicals are more difficult to identify for high-throughput profiling assays due the multiplexed nature of the assay readout35. Previously, we used a set of four chemicals (i.e. berberine chloride, Ca-074-Me, etoposide, rapamycin) as reference chemicals for the Cell Painting assay in the U-2 OS model14. These chemicals produced robust profiles of phenotypic effects that were dissimilar across the chemical set (spanning nearly every category measured in the Cell Painting assay) but highly reproducible for each chemical across the many assay plates used in the study. The reproducibility was observed in terms of the phenotypic signature these chemicals produced as well as the potency for which phenotypic effects occurred. These chemicals were evaluated and selected for use as reference chemicals specifically in the context of the U-2 OS cell model. Extensibility to other cell types was not evaluated at the time. Data from the present study indicate that it is possible to identify reference chemicals that produce similar (but not identical) phenotypes across multiple biologically diverse cell types and are complementary to one another in terms of producing robust effects in different fluorescent channels / organelles. The most robust phenotypic effects of three of the reference chemicals from the previous study14 (i.e. berberine chloride, Ca-074-Me, etoposide) are consistent across the cell line panel, the lone exception being etoposide in HTB-9 cells. Using a “channel-centric“ approach one could identify several chemicals, each of which produce robust effects in a different fluorescent channel or organelle across a collection of cell lines and use them in a complementary manner to track assay performance while providing coverage the many features measured as part of the Cell Painting assay.
In summary, we conclude that the Cell Painting assay is capable of being deployed to novel human-derived cell types grown in monolayer in a straightforward manner, with minimal (or no) adjustment to sample preparation protocols. However, cell-type specific optimization of image acquisition and feature extraction parameters is required. The degree of qualitative and quantitative similarity in phenotypic response profiles across biologically diverse cell types will vary from chemical to chemical, with some chemicals producing the most robust effects in the same organelle / fluorescent channel regardless of biological context.
Supplementary Material
Acknowledgements
The author would like to thank Terri Fairley, Dan Hallinger and Sandra Roberts for operations support activities during conduct of this research. The authors would also like to thank Drs. Keith Houck, Timothy Shafer, Brian Chorley and Maureen Gwinn for their insightful comments during review of this manuscript.
Funding Information
The U.S. Environmental Protection Agency through its Office of Research and Development provided funding for this research.
Abbreviations
- BMC
Benchmark Concentration
- CP
Cell Painting
- CV
Cell Viability
- DMEM
Dulbecco′s Modified Eagle Medium
- DMSO
Dimethyl sulfoxide
- EC50
Effective Concentration 50
- LOEC
Lowest Observable Effect Concentration
- NAM
New Approach Methodologies
- NOEC
No Observable Effect Concentration
- PC
Principal Component
- PCA
Principal Components Analysis
- PI
Propidium Iodide
- POD
Point-of-Departure
- RNA-Seq
Ribonucleic Acid Sequencing
- SBI2
Society for Biomolecular Imaging and Informatics
- sCMOS
scientific Complementary Metal–Oxide–Semiconductor
- TCCS
Theta Comparative Cell Scoring
- Tox21
Toxicity Testing in the 21st Century
- ToxCast
Toxicology Forecaster
- US EPA
United States Environmental Protection Agency
Footnotes
Conflict of Interest
This manuscript does not necessarily reflect U.S. EPA policy nor does mention of trade names or products constitute endorsement for use. The authors declare no conflict of interest regarding this manuscript. The U.S. Environmental Protection Agency has provided administrative review and has approved this manuscript for publication. The views expressed in this manuscript are those of the authors and do not necessarily reflect the view or policies of the U.S. Environmental Protection Agency. Reference to commercial products or services does not constitute endorsement.
Bibliography
- 1.Thomas RS; Bahadori T; Buckley TJ; et al. The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicol Sci 2019, 169, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeakley JM; Shepard PJ; Goyena DE; et al. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One 2017, 12, e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray MA; Singh S; Han H; et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc 2016, 11, 1757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.USEPA. Strategic Plan to Promote the Development and Implementation of Alternative Test Methods Within the TSCA Program. U.S. Environmental Protection Agency, O. o. C. S. a. P. P., Washington, DC., Ed. 2018. [Google Scholar]
- 5.Richard AM; Judson RS; Houck KA; et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol 2016, 29, 1225–51. [DOI] [PubMed] [Google Scholar]
- 6.Ramaiahgari SC; Auerbach SS; Saddler TO; et al. The Power of Resolution: Contextualized Understanding of Biological Responses to Liver Injury Chemicals Using High-throughput Transcriptomics and Benchmark Concentration Modeling. Toxicol Sci 2019, 169, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon JM; Paranjape SR; Wolter JM; et al. High-throughput screening and classification of chemicals and their effects on neuronal gene expression using RASL-seq. Sci Rep 2019, 9, 4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limonciel A; Ates G; Carta G; et al. Comparison of base-line and chemical-induced transcriptomic responses in HepaRG and RPTEC/TERT1 cells using TempO-Seq. Arch Toxicol 2018, 92, 2517–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm FA; Iwata Y; Sirenko O; et al. A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives. Green Chem 2016, 18, 4407–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustafsdottir SM; Ljosa V; Sokolnicki KL; et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS One 2013, 8, e80999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warchal SJ; Dawson JC; Carragher NO Development of the Theta Comparative Cell Scoring Method to Quantify Diverse Phenotypic Responses Between Distinct Cell Types. Assay Drug Dev Technol 2016, 14, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warchal SJ; Dawson JC; Carragher NO Evaluation of Machine Learning Classifiers to Predict Compound Mechanism of Action When Transferred across Distinct Cell Lines. SLAS Discov 2019, 24, 224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warchal SJ; Dawson JC; Shepherd E; et al. High content phenotypic screening identifies serotonin receptor modulators with selective activity upon breast cancer cell cycle and cytokine signaling pathways. Bioorg Med Chem 2020, 28, 115209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nyffeler J; Willis C; Lougee R; et al. Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol Appl Pharmacol 2020, 389, 114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips JR; Svoboda DL; Tandon A; et al. BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics 2019, 35, 1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bray MA; Gustafsdottir SM; Ljosa V; et al. A dataset of images and morphological profiles of 30,000 small-molecule treatments using the Cell Painting assay. Gigascience 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohban MH; Singh S; Wu X; et al. Systematic morphological profiling of human gene and allele function via Cell Painting. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah I; Setzer RW; Jack J; et al. Using ToxCast Data to Reconstruct Dynamic Cell State Trajectories and Estimate Toxicological Points of Departure. Environ Health Perspect 2016, 124, 910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zucker RM; Ortenzio J; Degn LL; et al. Biophysical comparison of four silver nanoparticles coatings using microscopy, hyperspectral imaging and flow cytometry. PLoS One 2019, 14, e0219078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M; Fagerberg L; Hallstrom BM; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 21.Barretina J; Caponigro G; Stransky N; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teiten MH; Bezdetnaya L; Morliere P; et al. Endoplasmic reticulum and Golgi apparatus are the preferential sites of Foscan localisation in cultured tumour cells. Br J Cancer 2003, 88, 146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanueva J; Viniegra S; Gimenez-Molina Y; et al. The position of mitochondria and ER in relation to that of the secretory sites in chromaffin cells. J Cell Sci 2014, 127, 5105–14. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y; Ye J Mitochondrial inhibitor as a new class of insulin sensitizer. Acta Pharm Sin B 2012, 2, 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svartengren J; Simonsson P Receptor binding properties of amperozide. Pharmacol Toxicol 1990, 66 Suppl 1, 8–11. [DOI] [PubMed] [Google Scholar]
- 26.Meltzer HY; Matsubara S; Lee JC Classification of typical and atypical antipsychotic drugs on the basis of dopamine D-1, D-2 and serotonin2 pKi values. J Pharmacol Exp Ther 1989, 251, 238–46. [PubMed] [Google Scholar]
- 27.Xie QM; Tang HF; Chen JQ; et al. Pharmacological actions of tetrandrine in inflammatory pulmonary diseases. Acta Pharmacol Sin 2002, 23, 1107–13. [PubMed] [Google Scholar]
- 28.Matthews JB Anthelmintic resistance in equine nematodes. Int J Parasitol Drugs Drug Resist 2014, 4, 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melo KM; Oliveira R; Grisolia CK; et al. Short-term exposure to low doses of rotenone induces developmental, biochemical, behavioral, and histological changes in fish. Environ Sci Pollut Res Int 2015, 22, 13926–38. [DOI] [PubMed] [Google Scholar]
- 30.Jordan MA; Wilson L Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004, 4, 253–65. [DOI] [PubMed] [Google Scholar]
- 31.Naaz F; Haider MR; Shafi S; et al. Anti-tubulin agents of natural origin: Targeting taxol, vinca, and colchicine binding domains. Eur J Med Chem 2019, 171, 310–331. [DOI] [PubMed] [Google Scholar]
- 32.Page SW Chapter 10 - Antiparasitic drugs. In Small Animal Clinical Pharmacology (Second Edition); Maddison JE; Page SW; Church DB, Eds.; Elsevier: 2008. [Google Scholar]
- 33.Xiong N; Long X; Xiong J; et al. Mitochondrial complex I inhibitor rotenone-induced toxicity and its potential mechanisms in Parkinson’s disease models. Crit Rev Toxicol 2012, 42, 613–32. [DOI] [PubMed] [Google Scholar]
- 34.Buchser W; Collins M; Garyantes T; et al. Assay Development Guidelines for Image-Based High Content Screening, High content Analysis and High Content Imaging. In Assay Guidance Manual; Sittampalam GS; Grossman A; Brimacombe K, Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, 2012. [PubMed] [Google Scholar]
- 35.Harrill J; Shah I; Setzer RW; et al. Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Current Opinion in Toxicology 2019, 15, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


