Abstract
Group A streptococcal M proteins are type-specific virulence factors that inhibit phagocytosis. We used two M proteins, M5 and Emm22, to analyze the influence of genetic background on the properties of M proteins. Mutant strains, engineered to lack these M proteins, were complemented with genes encoding the homologous or heterologous M protein, and the complemented strains were analyzed for phagocytosis resistance. Neither the M5 nor the Emm22 protein conferred phagocytosis resistance in the heterologous background, but they did do so in the homologous background. This was not due to lack of surface expression in the heterologous background. Moreover, the M5 and Emm22 proteins expressed in heterologous background appeared to have normal structure, since they were not affected in their ability to bind different human plasma proteins. In particular, M5 or Emm22 had normal ability to bind human complement inhibitors, a property that has been implicated in phagocytosis resistance. Results similar to those obtained with M5 and Emm22 were obtained in experiments with the M6 and Emm4 proteins. Together, these data suggest that the surface expression of M protein alone may not be sufficient to confer phagocytosis resistance and consequently that strain-specific factors other than M and Emm proteins may contribute to the ability of group A streptococci to resist phagocytosis.
Group A streptococcus (Streptococcus pyogenes) is the cause of tonsillitis and impetigo and also causes severe diseases, including necrotizing fasciitis and the streptococcal toxic shock syndrome (2). In addition, group A streptococcal infections are sometimes complicated by one of the postinfectious sequelae, rheumatic fever and glomerulonephritis.
The M proteins of group A streptococci are major virulence factors that confer resistance to phagocytosis (14). The N-terminal sequences of the M proteins are highly variable, giving rise to the existing ∼100 serotypes. Protection against phagocytosis is usually serotype specific. It should be noted that a large fraction of all group A streptococcal strains encode three proteins with structural features typical for M proteins (12, 18). These proteins, designated Mrp, Emm, and Enn, are encoded by adjacent genes on the chromosome and are regulated by a common positive regulatory gene element now designated mga (7, 20). At least two of the gene products (Mrp and Emm) from a single strain can contribute to resistance against phagocytosis, although Emm appears to be more important (22, 32). Strains with three genes encoding M-like proteins differ from those with a single gene in that they usually express opacity factor (OF), a lipoproteinase that is both surface bound and secreted (25, 26). Recent evidence indicates that OF contributes to group A streptococcal virulence (4).
Streptococcal mutants lacking M protein(s) are readily phagocytosed. However, the antiphagocytic property can be restored by complementation with the homologous M protein (19). Moreover, there is evidence that introduction of DNA encoding heterologous M proteins can provide phagocytosis resistance (5, 23, 27). For example, phagocytosis resistance was restored following integration of the gene encoding the M5 protein in the chromosome of an M protein-negative isolate derived from a serotype 24 strain (5). In contrast, the Emm4 protein (Arp4), derived from an OF+ type 4 strain, was unable to complement the phagocytosis resistance of an OF− type 6 strain deleted of its M protein-encoding gene, leading to the conclusion that the expression of Emm4 is not sufficient to confer phagocytosis resistance (8). On the other hand, gene inactivation experiments suggest that Emm4 and similar proteins have antiphagocytic properties (22, 32). Taken together, these data suggested to us that the ability of M and Emm proteins to provide the antiphagocytic property might be limited by the genetic background of the strain in which it is expressed. To resolve this issue, we have analyzed the ability of different proteins to confer phagocytosis resistance in heterologous strains. We find that the ability of M proteins to provide protection against phagocytosis is indeed highly restricted by the genetic background of the strain. This finding suggests that phagocytosis resistance may require cooperation between M proteins and other strain-specific streptococcal component(s).
MATERIALS AND METHODS
Bacterial strains and plasmids.
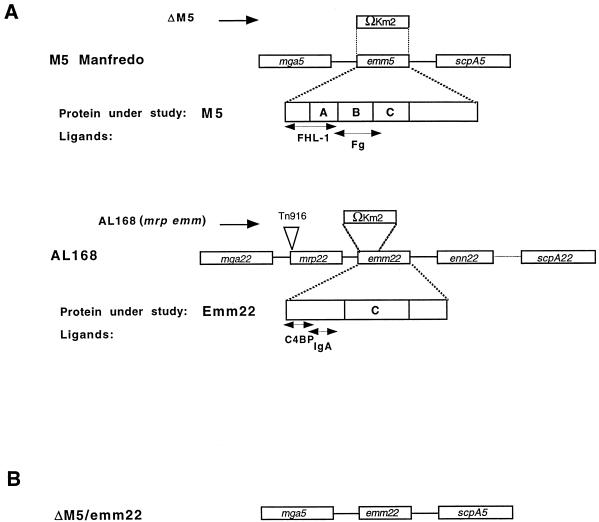
Strain M5 Manfredo was kindly provided by Michael Kehoe, University of Newcastle-upon-Tyne. The type 22 strain AL168 has been described previously (28). The type 6 strain JRS4 was from June R. Scott (Emory University, Atlanta, Ga.). The plasmid pLZ12Spec is an Escherichia coli-Streptococcus shuttle vector carrying a spectinomycin resistance marker (8), whereas derivatives of the temperature-sensitive suicide vector pJRS233 (21) used here carry resistance markers for erythromycin and kanamycin. Deletion of the emm5 gene in M5 Manfredo was achieved by homologous recombination by using a derivative of pJRS233 (10). This strain is referred to as ΔM5. The mrp22 gene of strain AL168 was inactivated by insertional mutagenesis by using the conjugative transposon Tn916, whereas inactivation of the emm22 gene in the mrp22 negative strain was performed by homologous recombination by using the pJRS233 vector as described elsewhere (32).
Culture conditions.
Streptococci were grown in Todd-Hewitt broth (TH), in 5% CO2, at 37°C. Streptococci transformed with pLZ12Spec, or derivatives thereof, were selected on medium supplemented with spectinomycin at 100 mg/liter. For isolation of streptococci transformed with derivatives of pJRS233, erythromycin and kanamycin were used at concentrations of 1.0 and 200 mg/liter, respectively. E. coli LE392 was grown in Luria-Bertani medium, supplemented with spectinomycin at 20 mg/liter, if transformed with derivatives of pLZ12Spec, or with erythromycin at 500 mg/liter and kanamycin at 50 mg/liter, if transformed with derivatives of pJRS233.
Binding of radiolabeled ligands to group A streptococci.
For binding studies, overnight cultures of streptococci were harvested by centrifugation, washed twice in PBSA (0.15 M NaCl, 0.03 M phosphate, 0.02% sodium azide; pH 7.2), and were then resuspended in PBSA supplemented with 0.05% Tween 20 (PBSAT). The binding of radiolabeled proteins to bacteria, at different dilutions, was measured in a total volume of 250 μl of PBSAT. After incubation for 1 h at 20°C, the samples were centrifuged (4,000 × g, 10 min), the supernatant was discarded, and the remaining pellet was washed with 2 ml of PBSAT. After centrifugation and subsequent removal of the supernatant, the radioactivity associated with the pellet was measured in a gamma counter.
Bactericidal assay.
Resistance to phagocytosis was analyzed in a bactericidal assay as described elsewhere (15, 21, 32). Briefly, an overnight culture of bacteria in TH, supplemented with 100 mg of spectinomycin per liter, was diluted 1:50 in TH and grown without agitation to an A620 of 0.15 at 37°C. The bacteria were then diluted by 104 in TH, and 100 μl of the suspension, containing ∼50 CFU, was added to 1.0 ml of human blood supplemented with sodium heparin at 20 U/ml in acid-cleaned glass tubes (100 by 10 mm). The tubes were incubated with rotation at 37°C for 3 h. For some experiments, heparin was replaced with hirudin at 140 U/ml (Calbiochem). For experiments with hirudin, 2.2-ml polypropylene tubes were used instead of glass tubes, and 250 μl of fresh hirudinized blood was mixed with ∼10 CFU of log-phase bacteria in TH. CFU in the samples were counted before and after incubation by using the pour plate method.
Emm protein and OF-encoding shuttle plasmids.
Construction of pJRS264, an emm4 (arp4) gene containing derivative of pLZ12Spec has been described previously (8). A 2.1-kb EcoRI/SphI fragment of pKEJ1 harboring the emm5 gene (11) was inserted into pLZ12Spec, resulting in plasmid pLZemm5. For construction of pLZemm22, a 2.2-kb HindIII fragment of Sir2202 carrying the emm22(Sir22) gene (29), was ligated with HindIII-digested pLZ12Spec to generate pLZemm22. To construct pLZemm6, the emm6 gene was amplified by PCR by using chromosomal DNA from strain JRS4 as template and the synthetic oligonucleotides 5′-GGCTGGATCCTTAATAGCATTTAGGTC-3′ and 5′-GCTAGGCATGCATAAAAGAGAGAACCG-3′ as primers. Restriction sequences for BamHI and SphI were introduced by the primers. The amplified fragment was digested with BamHI and SphI and was subsequently ligated with BamHI/SphI-digested pLZ12Spec. To construct pLZsof22, the gene encoding opacity factor from type 22 strain AL168 was amplified by PCR by using the synthetic oligonucleotides 5′-GTCCCATGGCGGATCCTTAATTTTTATCTCACC-3′ and 5′-GCAAGCTTGGCATGCTTAAA-GCCAAAGGCTTAGGG-3′ as primers. Primer sequences were selected according to the published sequence for sof22 (25). The resulting fragment was digested with BamHI and SphI, for which restriction sequences were introduced by the primers and subsequently ligated with pLZ12Spec to generate pLZsof22.
Chromosomal replacement of emm5 by emm22 in strain M5 Manfredo.
Integration of the emm22 gene into the chromosome of the ΔM5 strain was achieved by homologous recombination. A 1,356-bp DNA fragment with the entire emm22 gene was amplified by PCR with the synthetic oligonucleotides 5′-CGAAGGATCCAAAAAAAGAGGAAGCCCCTTCC-3′ and 5′-GGTCTGCATGCGATTGTTAGTTACTTAGCC-3′ as primers and with chromosomal DNA from the AL168 strain as a template. The PCR fragment was digested with BamHI and SphI, recognition sequences for which had been introduced through the primers. The fragment was subsequently used to replace the ΩKm2 cassette in a derivative of pJRS233 containing this cassette flanked by sequences surrounding the emm5 gene (10). In the resulting plasmid, the emm22 gene was flanked by two sequences (1,200 and 1,000 bp, respectively) that are derived from sequences upstream and downstream of the emm5 gene in the chromosome of M5 Manfredo. This plasmid was electroporated into ΔM5. The transformed bacteria were first grown for 48 h at 30°C in the presence of erythromycin. Individual erythromycin-resistant bacteria were then used to inoculate 10 ml of TH and were incubated at 37°C, a temperature that does not allow pJRS233 or its derivatives to replicate in streptococci (21). Mutants in which ΩKm2 in ΔM5 had been replaced by emm22 were selected by screening streptococci able to grow at 37°C for the loss of kanamycin resistance. Integration of emm22 in the correct chromosomal location was verified by PCR.
Recombinant DNA techniques.
Standard recombinant DNA techniques were used. Ligase and restriction enzymes were purchased from Promega. Transformation of E. coli was performed by the CaCl2 method. Electroporation of streptococci was carried out as described previously (3).
Other methods and reagents.
Human C4BP (complement factor 4b-binding protein) was a kind gift from Björn Dahlbäck, Malmö General Hospital, Lund University, Malmö, Sweden, and FHL-1 (complement factor H-like protein 1) was kindly provided by Peter Zipfel, Bernhard-Nocht Institute of Tropical Medicine, Hamburg, Germany. Polyclonal human serum immunoglobulin A (IgA) was from Cappel. Human fibrinogen and fibronectin was from Sigma. Proteins were labeled with 125I by using the chloramine-T method (6). Measurement of lipoproteinase activity was made according to the method of Maxted et al. (16).
RESULTS
Construction of strains expressing homologous and heterologous M proteins.
To study the ability of different M proteins to confer phagocytosis resistance to strains with different background, the well-defined M5 and M22 systems were used. The M5 Manfredo strain is OF− and expresses a single M protein (17, 33) (Fig. 1), whereas the OF+ M22 strain AL168 contains an mga regulon encoding three M-like proteins (32) (Fig. 1), of which at least two (Mrp and Emm) are expressed on the streptococcal surface and confer phagocytosis resistance. The genes encoding M5 and Emm22 were cloned separately in the shuttle vector pLZ12Spec. The resulting plasmids, designated pLZemm5 and pLZemm22, respectively, were used to transform ΔM5 (10), an M5-deficient derivative of M5 Manfredo, or AL168(mrp emm), a phagocytosis-sensitive derivative of AL168 lacking expression of the Mrp22 and Emm22 proteins (32). As a result, four strains were obtained, in which M5 and Emm22 were expressed in either a homologous or a heterologous background. The four strains were designated ΔM5/pLZemm5, ΔM5/pLZemm22, AL168(mrp emm)/pLZemm5, and AL168(mrp emm)/pLZemm22.
FIG. 1.
Schematic representation of the mga5 and mga22 regulons and the mutants used in this study. (A) Organization of the mga regulons of the type 5 strain M5 Manfredo and the type 22 strain AL168. Replacement of the emm5 gene with a gene encoding kanamycin resistance (Ωkm2) by homologous recombination gave rise to strain ΔM5 (10). In the AL168(mrp emm) derivative, the mrp22 gene has been disrupted by insertion of the conjugative transposon Tn916, whereas a large part of the emm22 gene has been replaced by a kanamycin casette (32). The mga gene encodes the multigene activator of group A streptococcus, and the mrp, emm, and enn genes encode proteins in the M protein family, whereas the scpA gene encodes the streptococcal C5a peptidase. The regions in the proteins marked A, B, and C are distinct domains containing three to five different repeat units. (B) Organization of the mga regulon of the ΔM5/emm22 derivative of the M5 Manfredo strain, in which the emm5 gene has been replaced by the emm22 gene. Protein ligands that are relevant for this study and their binding sites in M5 (Fg [fibrinogen] and FHL-1) and Emm22 (IgA and C4BP) are indicated.
Binding properties of streptococcal strains expressing homologous and heterologous M proteins.
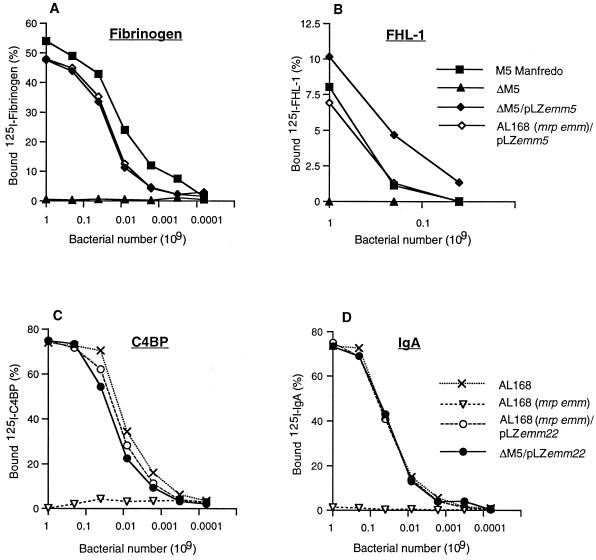
To analyze whether the surface expression of the M5 and Emm22 proteins was similar in the different bacterial backgrounds, we determined the ability of the complemented streptococci to bind radiolabeled ligands known to specifically interact with the M5 and Emm22 proteins, respectively (Fig. 2). Surface expression of the M5 protein was monitored by using the capacity of the streptococci to bind FHL-1 and fibrinogen, both of which bind to M5, but not to Emm22 (10, 13). Similarly, C4BP and IgA, both of which interact with Emm22 but not with M5 were used to determine the expression level of the Emm22 protein (11, 31). The two strains expressing M5 in homologous and heterologous backgrounds showed similar FHL-1 and fibrinogen-binding capacity. The slight differences noted are unlikely to reflect different M5 protein expression levels, particularly since for fibrinogen the binding capacity of ΔM5/pLZemm5 and AL168(mrp emm)/pLZemm5 was somewhat reduced as compared to M5 Manfredo, whereas for FHL-1 the binding capacity of ΔM5/pLZemm5 was slightly improved over the other two strains (Fig. 2A and B). The two strains expressing Emm22 in homologous and heterologous background showed C4BP- and IgA-binding levels that were practically identical (Fig. 2C and D). No difference in the binding patterns was noted between complemented isolates that had been cultured with or without antibiotic selection (data not shown), suggesting that the plasmids were not highly unstable. Finally, the complemented strains all possessed the hair-like surface structures that are typical for M proteins (30), as determined by transmission electron microscopy (data not shown).
FIG. 2.
Binding of purified ligands to streptococci expressing the M5 and Emm22 proteins. The binding of 125I-labeled ligands to the indicated group A streptococcal strains was measured as a function of bacterial number. Fibrinogen (A) and FHL-1 (B) are known to interact specifically with the M5 protein, whereas C4BP (C) and IgA (D) bind to the Emm22 protein. The data presented are based on three independent experiments with triplicate samples. The variation was <5% of the binding, and therefore error bars have not been introduced.
Ability of M proteins to confer antiphagocytic property in a homologous or heterologous background.
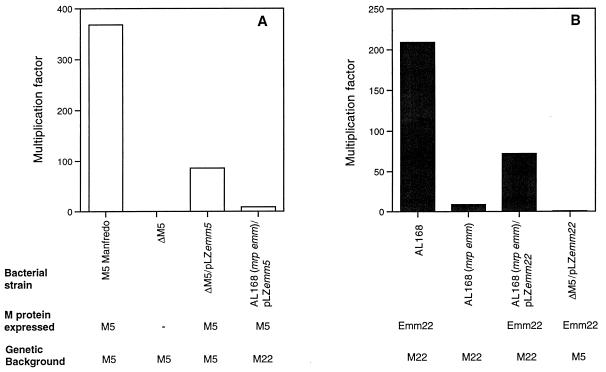
The ability of the four complemented strains to resist phagocytosis was compared with the parental M protein-expressing strains, and with the corresponding M negative strains, in the bactericidal assay in whole blood by using heparin as the anticoagulant (15). Ten independent experiments with blood from six different blood donors were carried out (Fig. 3 and Table 1). There was a donor-dependent interexperimental variation, but the outcome of each individual experiment showed a similar pattern, namely, that M5 and Emm22 provided phagocytosis resistance in the homologous but not in the heterologous background. Similar results were obtained with hirudin, a specific inhibitor of thrombin, as the anticoagulant (data not shown). Together, these data show that the ability of M proteins to provide protection against phagocytosis is restricted by the genetic background of the strain.
FIG. 3.
Growth of group A streptococcal strains in human blood. The multiplication factor indicated on the ordinate represents the factor of the increase in number of CFU during a 3-h incubation in rotating tubes. (A) Experiments with strains expressing the M5 protein or no M protein (strain ΔM5). (B) Experiments with strains expressing the Emm22 protein or no M protein (AL168mrp−emm−). The data are also shown in Table 1.
TABLE 1.
Survival of group A streptococcal strains, expressing homologous and heterologous M proteins, in a phagocytosis assaya
| Bacterial strain | M protein expressed | Genetic background | Multiplication factor (range) |
|---|---|---|---|
| M5 Manfredo | M5 | M5 | 368 (52–666) |
| ΔM5 | M5 | 0.15 (0–0.5) | |
| ΔM5/pLZemm5 | M5 | M5 | 86 (11–275) |
| AL168(mrp emm)/ pLZemm5 | M5 | M22 | 9 (0–31) |
| AL168 | Emm22, Mrp22 | M22 | 209 (60–276) |
| AL168(mrp emm) | M22 | 9 (0–40) | |
| AL168(mrp emm)/ pLZemm22 | Emm22 | M22 | 72 (19–220) |
| ΔM5/pLZemm22 | Emm22 | M5 | 0.57 (0–3.25) |
The data are based on the results from 10 independent experiments with six different donors.
The differences in antiphagocytic properties of the strains were not due to different growth rates, since the growth curves (in TH) were similar for all eight strains used in these experiments (data not shown). Nevertheless, a strong selective pressure favoring isolates cured from plasmid could influence the results obtained in the phagocytosis assay. However, bacteria surviving phagocytosis experiments still grew when replated on agar supplemented with spectinomycin, making this explanation unlikely. Moreover, the binding properties of rescued streptococci were similar to those displayed by the parent strains, i.e., surviving AL168(mrp emm)/pLZemm5 bound FHL-1 and fibrinogen as efficiently as M5 Manfredo, whereas surviving ΔM5/pLZemm22 showed C4BP- and IgA-binding levels comparable to those obtained with wild-type AL168 bacteria (data not shown).
To analyze whether the results were dependent on the extrachromosomal location of the complementing gene, the emm22 gene was inserted into the region of the M5 Manfredo chromosome normally occupied by emm5. The resulting strain, designated ΔM5/emm22, bound C4BP and IgA as efficiently as ΔM5/pLZemm22 or AL168 (Fig. 4A and Fig. 2C and D), indicating that the strain expressed the intact M22 protein in an amount that corresponds to the levels expressed by AL168 itself. However, like ΔM5/pLZemm22, strain ΔM5/emm22 failed to grow in heparinized blood (Table 2), indicating that the results obtained with the complemented isolates were not due to the location of the M protein gene on a plasmid.
FIG. 4.
Binding of host ligands to an M-negative type 5 expressing Emm22 and OF22. The emm5 gene of M5 Manfredo was replaced by emm22, resulting in strain ΔM5/emm22. (A) This strain expresses Emm22 on its surface, as shown by its ability to bind C4BP and IgA. The gene encoding the opacity factor (sof) was cloned from the type M22 strain AL168 and inserted in pLZ12Spec. The resulting plasmid was then introduced into ΔM5/emm22, producing strain ΔM5/emm22(pLZsof22). (B) The ability of ΔM5/emm22(pLZsof22) to bind fibronectin, a ligand for OF, was compared with the fibronectin-binding capacity of the M5 Manfredo, ΔM5, and ΔM5/emm22 strains.
TABLE 2.
Survival of group A streptococci expressing different M proteins in a phagocytosis assaya
| Bacterial strain | M protein expressed | Genetic background | Multiplication factor (range) |
|---|---|---|---|
| ΔM5/emm22 | Emm22 | M5 | 0.36 (0.07–0.9) |
| ΔM5/emm22(pLZsof22) | Emm22 | M5 | 0.79 (0.08–2) |
| ΔM5/pJRS264 | Emm4 | M5 | 0.22 (0.03–0.45) |
| ΔM5/pLZemm6 | M6 | M5 | 72 (24–125) |
| AL168(mrp emm)/pJRS264 | Emm4 | M22 | 79.2 (12.6–121) |
| AL168(mrp emm)/pLZemm6 | M6 | M22 | 4.8 (1.4–20) |
The data are based on counts from three independent experiments with different donors.
The inability of M proteins to confer the antiphagocytic property to heterologous strains is not limited to the M5 and M22 systems.
To analyze whether strain-specific protection against phagocytosis is limited to the M5-M22 system, ΔM5 and AL168(mrp emm) were complemented with the emm6 gene cloned in pLZ12Spec. Binding analysis with fibrinogen and FHL-1 showed that the resulting heterologous strains expressed the M6 protein at levels comparable to those of the wild-type strain (data not shown). The complemented strains were analyzed in the bactericidal assay. Expression of the M6 protein in ΔM5 supported growth in blood, whereas its expression in AL168(mrp emm) did not (Table 2). It has been shown previously that expression of Emm4 (Arp4) failed to protect M6-deficient streptococci against phagocytosis (8). In agreement with this finding, ΔM5 streptococci complemented with the emm4 gene on the pLZ plasmid (pJRS264) were readily phagocytosed (Table 2). In contrast, when pJRS264 was used to complement AL168(mrp emm), the resulting isolate became resistant against phagocytosis (Table 2), showing that the Emm4 protein does indeed have the functional characteristics of an M protein.
The opacity factor is unable to restore the antiphagocytic property of Emm22 in a heterologous background.
The restriction in the ability of M proteins to provide protection against phagocytosis in different genetic background suggests that other strain-specific components affect the ability of M proteins to confer phagocytosis resistance. One such protein is the OF. To analyze whether coexpression of OF allows Emm22 to confer phagocytosis resistance in a heterologous background, we introduced the plasmid pLZsof22 encoding the OF22 protein into ΔM5/emm22. The resulting strain, ΔM5/emm22(pLZsof22), had lipoproteinase activity (data not shown) and bound fibronectin (Fig. 4B), a characteristic property of OF (25). However, expression of OF22 did not rescue ΔM5/emm22(pLZsof22) streptococci in the bactericidal assay (Table 2). Other factors that may affect the strain-specific restriction must therefore be analyzed.
DISCUSSION
Our observations clearly suggest that M proteins are restricted in their ability to provide group A streptococci with resistance against phagocytosis. This conclusion is based on the finding that the M5 and M6 proteins, originating from isolates of the OF− lineage, conferred phagocytosis resistance in the background of an OF− strain of type 5 but failed to do so in the OF+ strain of type 22. Similarly, the Emm4 and Emm22 proteins, both originating from OF+ strains, only conferred phagocytosis resistance to the OF+ strain.
Several explanations for the surprising phenomenon described here can be imagined. A trivial reason would be that M protein encoding plasmids are unstable in some strains. However, several lines of evidence indicated that the plasmids used here were not lost during the course of the experiments. More importantly, such an explanation does not account for the failure of ΔM5 bacteria carrying the emm22 gene inserted into the chromosome to grow in blood. A second alternative could be that certain M proteins are not properly surface expressed in a heterologous background. Our data do not support this explanation, since the ability of the complemented strains to bind different ligands were similar to that of the parent strains. However, it seems possible that there exist lineage specific systems necessary for correct folding, processing, or cell surface distribution of M proteins and that these systems influence the ability of M proteins to confer phagocytosis resistance. For example, it has been demonstrated that the streptococcal cysteine proteinase cleaves proteins belonging to the M protein family, thereby providing a mechanism for modulating the surface expression of these proteins (1, 24). It remains to be investigated whether such protease activity can show the substrate specificity required to explain the present findings. The binding data with C4BP and FHL-1, which bind to the N-terminal surface-exposed portion of Emm22 and M5, respectively (10, 11), do not support this explanation since the complemented strains had the expected binding properties, showing that their M proteins are not processed in any extensive way. The failure of emm5 to provide the AL168(mrp emm) strain with the capacity to survive in blood could partially be explained by the requirement for expression of both the mrp22 and emm22 gene products. However, this explanation is not sufficient, since emm22 by itself provides the mutant strain with phagocytosis resistance (Fig. 3) (32).
Until now, there has been no evidence that M proteins would require additional factors to exert their antiphagocytic effect. However, this may well be the explanation behind the present findings. The opacity factor is one such factor, but the finding that simultaneous expression of Emm22 and OF22 did not provide the OF− ΔM5 strain with the antiphagocytic property indicates that OF is not a likely candidate. Moreover, a requirement for OF production would not explain the failure of the M5 and M6 proteins to provide protection in the OF+ background. Other factors should therefore be evaluated. Among known surface proteins, the T proteins are perhaps the most interesting candidates, since they appear to be variable and since expression of a specific T protein is usually associated with expression of certain M serotypes (9).
Regardless of the molecular explanation, the present findings show that there are functional barriers between group A streptococci of different serotypes. In addition, the data imply that great care must be taken when interpreting functional data derived from experiments involving the introduction of M and Emm protein-encoding genes into different strains of group A streptococci.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Medical Research Council (grants 9490 and 9926), the Axson Johnson Trust, the Crafoord Trust, the Johan and Greta Kock Trust, the Wiberg Trust, the Royal Physiographic Society in Lund, the Österlund Trust, and Actinova Ltd.
We are very grateful to Björn Dahlbäck and Peter Zipfel for the donation of valuable reagents.
REFERENCES
- 1.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 2.Bisno A L, Stevens D L. Streptococcal infections of skin and soft tissues. N Engl J Med. 1996;334:240–245. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- 3.Caparon M G, Scott J R. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 4.Courtney H S, Hasty D L, Li Y, Chiang H C, Thacker J L, Dale J B. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol Microbiol. 1999;32:89–98. doi: 10.1046/j.1365-2958.1999.01328.x. [DOI] [PubMed] [Google Scholar]
- 5.Courtney H S, Liu S, Dale J B, Hasty D L. Conversion of M serotype 24 of Streptococcus pyogenes to M serotypes 5 and 18: effect on resistance to phagocytosis and adhesion to host cells. Infect Immun. 1997;65:2472–2474. doi: 10.1128/iai.65.6.2472-2474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwood F C, Hunter W M, Glover J S. The preparation of (125I)-labeled human growth hormone of high specific activity. Biochem J. 1963;89:114–123. doi: 10.1042/bj0890114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haanes E J, Cleary P P. Architecture of the vir regulons of group A streptococci parallels opacity factor phenotype and M protein class. J Bacteriol. 1992;174:4967–4976. doi: 10.1128/jb.174.15.4967-4976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husmann L K, Scott J R, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson D R, Kaplan E L. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J Med Microbiol. 1993;38:311–315. doi: 10.1099/00222615-38-5-311. [DOI] [PubMed] [Google Scholar]
- 10.Johnsson E, Berggård K, Kotarsky H, Hellwage J, Zipfel P F, Sjöbring U, Lindahl G. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161:4894–4901. [PubMed] [Google Scholar]
- 11.Johnsson E, Thern A, Dahlbäck B, Heden L O, Wikstrom M, Lindahl G. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J Immunol. 1996;157:3021–3029. [PubMed] [Google Scholar]
- 12.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 13.Kotarsky H, Hellwage J, Johnsson E, Skerka C, Svensson H G, Lindahl G, Sjöbring U, Zipfel P F. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J Immunol. 1998;160:3349–3354. [PubMed] [Google Scholar]
- 14.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 15.Lancefield R C. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med. 1957;106:525–544. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxted W R, Widdowson J P, Fraser C A. Antibody to streptococcal opacity factor in human sera. J Hyg. 1973;71:35–42. doi: 10.1017/s0022172400046180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller L, Gray L, Beachey E, Kehoe M. Antigenic variation among group A streptococcal M proteins. Nucleotide sequence of the serotype 5 M protein gene and its relationship with genes encoding types 6 and 24 M proteins. J Biol Chem. 1988;263:5668–5673. [PubMed] [Google Scholar]
- 18.Navarre W W, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Casal J, Caparon M G, Scott J R. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res Microbiol. 1992;143:549–558. doi: 10.1016/0923-2508(92)90112-2. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Casal J, Price J A, Maguin E, Scott J R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 22.Podbielski A, Schnitzler N, Beyhs P, Boyle M D. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 23.Poirier T P, Kehoe M A, Whitnack E, Dockter M E, Beachey E H. Fibrinogen binding and resistance to phagocytosis of Streptococcus sanguis expressing cloned M protein of Streptococcus pyogenes. Infect Immun. 1989;57:29–35. doi: 10.1128/iai.57.1.29-35.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raeder R, Woischnik M, Podbielski A, Boyle M D P. A secreted streptococcal cysteine protease can cleave a surface-expressed M1 protein and alter the immunoglobulin binding properties. Res Microbiol. 1998;149:539–548. doi: 10.1016/s0923-2508(99)80001-1. [DOI] [PubMed] [Google Scholar]
- 25.Rakonjac J V, Robbins J C, Fischetti V A. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect Immun. 1995;63:622–631. doi: 10.1128/iai.63.2.622-631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saravani G A, Martin D R. Opacity factor from group A streptococci is an apoproteinase. FEMS Microbiol Lett. 1990;56:35–39. doi: 10.1111/j.1574-6968.1990.tb04118.x. [DOI] [PubMed] [Google Scholar]
- 27.Scott J R, Guenthner P C, Malone L M, Fischetti V A. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenberg L, O'Toole P, Lindahl G. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol Microbiol. 1992;6:1185–1194. doi: 10.1111/j.1365-2958.1992.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 29.Stenberg L, O'Toole P W, Mestecky J, Lindahl G. Molecular characterization of protein Sir, a streptococcal cell surface protein that binds both immunoglobulin A and immunoglobulin G. J Biol Chem. 1994;269:13458–13464. [PubMed] [Google Scholar]
- 30.Swanson J, Hsu K, Gotschlich E C. Electron microscopic studies on streptococci. I. M antigen. J Exp Med. 1969;130:1063–1091. doi: 10.1084/jem.130.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thern A, Stenberg L, Dahlbäck B, Lindahl G. Ig-binding surface proteins of Streptococcus pyogenes also bind human C4b-binding protein (C4BP), a regulatory component of the complement system. J Immunol. 1995;154:375–386. [PubMed] [Google Scholar]
- 32.Thern A, Wästfelt M, Lindahl G. Expression of two different antiphagocytic M proteins by Streptococcus pyogenes of the OF+ lineage. J Immunol. 1998;160:860–869. [PubMed] [Google Scholar]
- 33.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]