Abstract
Rotifers of the class Bdelloidea are microscopic animals notorious for their long-term persistence in the apparent absence of sexual reproduction and meiotic recombination. This evolutionary paradox is often counterbalanced by invoking their ability to repair environmentally induced genome breakage. By studying the dynamics of DNA damage response in the bdelloid species Adineta vaga, we found that it occurs rapidly in the soma, producing a partially reassembled genome. By contrast, germline DNA repair is delayed to a specific time window of oogenesis during which homologous chromosomes adopt a meiotic-like juxtaposed configuration, resulting in accurate reconstitution of the genome in the offspring. Our finding that a noncanonical meiosis is the mechanism of germline DNA repair in bdelloid rotifers gives previously unidentified insights on their enigmatic long-term evolution.
Discovering meiosis coupled to DNA repair in a bdelloid rotifer demystifies the success of this asexual clade.
INTRODUCTION
The class Bdelloidea is the largest and oldest metazoan taxon, composed of microscopic free-living animals, in which no males or hermaphrodites have been conclusively described (1). Bdelloid females are thought to develop from unfertilized and unreduced eggs resulting from the ameiotic maturation of primary oocytes (2, 3). This mode of reproduction, known as mitotic parthenogenesis (or apomixis), is considered an evolutionary dead-end, which contrasts with the 460 morphospecies and the 60-Ma evolution of Bdelloidea (4, 5). However, recent signatures of genetic exchanges and recombination have been reported in the model bdelloid species Adineta vaga and more recently in Macrotrachela quadricornifera, but the underlying mechanisms are unknown (6–9). Therefore, the ancient asexuality of bdelloid rotifers, often reported as a notorious exception in textbooks, remains a mystery. Environmental stresses affecting genome stability could be central to bdelloid rotifers evolutionary success (10). These organisms resist complete desiccation, a stress frequently encountered in their limno-terrestrial environments such as lichens and mosses. This desiccation tolerance seems to correlate with their ability to survive extreme doses of ionizing radiation (i.e., >0.5 kGy). Such high level of resistance is coupled to an exceptional capacity at repairing DNA double-strand breaks (DSBs) that accumulate during genotoxic stresses (11–13). Repeated cycles of chromosome breakage and reassembly could thus contribute to their genome plasticity and adaptability, by promoting DNA recombination (10). Our recent work has revealed that the genome of A. vaga is composed of six pairs of homologous chromosomes with signatures of homologous recombination (14). To maintain such genome organization in an ancient asexual thriving in habitats where genome breakage is frequent, faithful repair of DNA lesions must occur in the germ line to ensure complete chromosome reconstruction and transmission to the offspring. How and when this takes place in the apparent lack of meiosis remain an open question.
RESULTS
To address this question, DNA repair was investigated at the cellular level by exposing desiccated A. vaga individuals to proton radiation (PR). Such treatment induced dose-dependent DNA damage (fig. S1A) without markedly altering their survival and egg-laying activity (fig. S1, B and C). DNA lesions affected both somatic and germline nuclei as assessed by pulsed-field gel electrophoresis (PFGE) analysis on whole animals (fig. S1A) and terminal deoxynucleotidyl transferase (TdT)–mediated deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) assay on primary oocytes nuclei (fig. S1, D and E). However, fertility, i.e., the ability of eggs to produce a viable progeny, was reduced at a PR dose of 0.4 kGy and above (fig. S1F), confirming that A. vaga germline cells are more sensitive to ionizing radiation-induced damage than somatic cells (11, 13).
PFGE analysis performed after digesting genomic DNA of 0.4-kGy irradiated animals with the rare-cutting enzyme Sbf I confirmed that PR produced massive DNA breakage, generating a smear of low–molecular weight DNA fragments (Fig. 1A, 0 hours). Within 48 hours of recovery, the genome was partially reassembled with specific fragments of the initial restriction pattern, obtained before treatment, being recovered (Fig. 1A). In contrast, the nonirradiated parental PFGE restriction profile was fully restored in the progeny of irradiated individuals, suggesting that the germline genome was completely and accurately reassembled during oogenesis and/or early embryogenesis (Fig. 1B). Somatic and germline cells are therefore distinguished not only by their tolerance to PR-induced damage at the phenotypic level but also by their molecular response to DNA damage.
Fig. 1. Resolution of DNA damage after PR in A. vaga.
(A) Kinetics of DSB repair monitored by comparing the PFGE profiles of Sbf I–digested chromosomal DNA from control (Unirrad.) and from 0.4-kGy irradiated animals either immediately (0 hours) or at different recovery time points. This PFGE analysis detects DNA repair mainly in the somatic nuclei of adult A. vaga. (B) PFGE profiles of Sbf I–digested chromosomal DNA from unirradiated A. vaga (P) and three F1 generation clones (C1 to C3) derived from 0.5-kGy irradiated animals. (C) (2′S)-2′-Deoxy-2′fluoro-5-ethynyluridine (F-ara-EdU)–labeling scheme of A. vaga. Positive control consists of A. vaga individuals exposed to F-ara-EdU (green dot and arrow) since the mitotic phase of their embryogenesis. In the other conditions, adults were rehydrated in the presence of F-ara-EdU after a short desiccation period (white dot) directly followed or not by 1.28-kGy PR (red dot). (D) Visualization of F-ara-EdU incorporation in the nuclear DNA of representative controls (positive control and unirradiated control) and 48 hours after 1.28-kGy PR (irradiated A. vaga). F-ara-EdU was detected after coupling the fluorescent 6-FAM-azide by click reaction (green), and DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (red). For each condition, a micrograph of a confocal Z section from a whole individual (left scale bars, 50 μm) and a higher magnification of the inset focusing on the gonads (right scale bars, 10 μm) are shown. Arrows point to the pool of primary oocytes, arrowheads indicate several nurse nuclei, and the other nuclei belong to somatic cells.
Because DNA synthesis is a common step of virtually all repair pathways, DNA damage response was examined in both somatic and germline cells by incubating irradiated and nonirradiated A. vaga individuals with (2′S)-2′-deoxy-2′fluoro-5-ethynyluridine (F-ara-EdU) (Fig. 1C). This thymidine analog was subsequently labeled at the sites of de novo DNA synthesis by click reaction with 6-fluorescein (FAM)–azide. As expected, when unirradiated eggs were exposed to F-ara-EdU during the mitotic phase of development, fluorescent labeling was observed in all embryonic and adult nuclei, including those of primary oocytes (fig. S2A and Fig. 1D, left). Labeling was sensitive to hydroxyurea treatment but not to the RNA polymerase II inhibitor α-amanitin, demonstrating that it is specific to DNA synthesis (fig. S2B). In contrast, only the large nurse nuclei of the ovaries were labeled when unirradiated adults were exposed to F-ara-EdU. This is consistent with the eutelic nature of bdelloid rotifers and the arrest of DNA replication in postmitotic adult cells, except for nurse nuclei that undergo multiple cycles of postdevelopmental endoreplication to ensure yolk production during oogenesis (Fig. 1D, middle).
The pattern of F-ara-EdU incorporation was markedly altered after PR showing, 48 hours later, an increased labeling of somatic nuclei (Fig. 1D, right) as a function of the PR dose (fig. S3). The level of F-ara-EdU labeling followed the kinetics of genome reassembly as determined by PFGE, with the highest labeling observed when the analog is added directly upon PR recovery (0 hours) and no labeling beyond 48 hours after PR (fig. S4). These results are consistent with de novo DNA repair synthesis being triggered in somatic nuclei immediately after DNA damage.
The damaged chromatin of primary oocytes remained negative to F-ara-EdU labeling whichever the PR dose tested (as exemplified for the highest 1.28-kGy exposure in Fig. 1D, right). This suggests that contrarily to somatic cells, primary oocytes in A. vaga do not activate DNA repair as an immediate response to DNA lesions and that recovery of genome integrity must occur at a later stage of germline maturation.
Confirming this idea, specifically tracking F-ara-EdU incorporation in nuclear DNA of maturing oocytes showed that DNA repair synthesis occurred only in the developing egg of irradiated females after the onset of oogenesis when the chromosomes are individualized into discrete condensed structures, while primary oocytes remained unlabeled (Fig. 2, right). This spatially and temporally localized F-ara-EdU labeling was specific to heavily irradiated animals and was not detected in the absence of PR (Fig. 2, middle). The results therefore suggest that primary oocytes are kept in a temporary cell cycle arrest and that genome repair in the germ line is delayed to a specific time window that coincides with oogenesis resumption.
Fig. 2. Repair of PR-induced DNA damage during oocyte maturation in A. vaga.
Maximum intensity projections of confocal Z sections from DAPI-stained positive control and unirradiated or irradiated (1.28 kGy) individuals bearing a maturing oocyte and exposed to F-ara-EdU according to Fig. 1C. Dashed lines outline the developing egg. Scale bars, 10 μm. Insets show a close-up view of the nucleus from the single maturing oocyte (top, delineated by a circle) and from several primary oocytes (bottom, indicated by arrows). Scale bars, 2 μm.
This finding gave us an impetus to reexamine in detail the cytology of A. vaga oogenesis. Flow cytometry and cytofluorometric measurements of nuclei DNA content revealed that, except for nurse nuclei, the bulk of A. vaga somatic cells are in the G1 phase of the cell cycle, while contrarily to what was previously published (15), primary oocytes are arrested in the G2 phase (fig. S5 and Supplementary Text). This means that each oocyte contains duplicated copies of the 12 chromosomes forming the diploid genome of A. vaga (i.e., 4c DNA content).
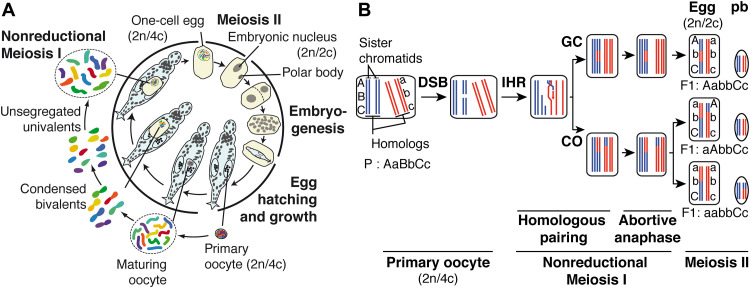
Standard confocal microscopy coupled to fluorescent in situ hybridization (FISH) was used to characterize the main stages of A. vaga oogenesis based on chromosome dynamics (Fig. 3). FISH probes were designed to track the localization of two distinct, nonoverlapping regions of a chromosome pair. The data suggest that primary oocyte maturation starts with the condensation of typically long and thin thread-like prophase chromosomes that gradually become shorter, thicker, and well individualized (Fig. 3, B to D). At a certain stage, homologous chromosomes were shown to adopt a tightly juxtaposed and condensed organization equivalent to bivalents that are formed during canonical meiosis I prophase (Fig. 3E). Homologs could be connected by chiasmata (fig. S6A), and their pairing appears to take place when chromosomes are clumped into a dense tangle showing fused or nearly fused FISH signals (Fig. 3, D and E). Homologous chromosomes then separate into two groups to form a typical anaphase arrangement, each containing a FISH-tagged homolog (Fig. 3F). However, this step was never followed by a reductional nuclear division. Rather, the 12 condensed chromosomes remain together and are transmitted as a complete diploid set to the developing egg, with no polar body being produced (Fig. 3G).
Fig. 3. Modified meiosis in A. vaga.
(A and Ba to La) Maximum intensity projections of confocal Z sections from DAPI-stained adult animals (A and Ba to Ga) and deposited eggs (Ha to La) at different stages of oogenesis (A and Ba to Ia) or early embryogenesis (Ja to La). Dashed lines outline the eggshell once visible (F to L). Dotted lines delineate blastomeres (J to L). Arrows point to the single polar body (I to L). Scale bars, 50 μm. Insets show a close-up view of the gonad (A) and of the nucleus either from the developing egg (B to I) or from mitotic blastomeres in early embryos (Ja to La). When possible, chromosome entities were randomly numbered. Scale bars, 2 μm. (Bb to Lb) Maximum intensity projections of confocal Z sections from DAPI-stained nuclei (gray, left; blue, right) and from FISH identifying two regions of a chromosome pair (green and red, right) in maturing oocyte nucleus (Bb to Ib) or in nuclei of early embryos (Jb to Lb). Scale bars, 2 μm.
The A. vaga egg is laid at the one-cell stage whose chromosomes are still composed of two sister chromatids (2n and 4c) (Fig. 3H and fig. S6B). Oogenesis ends shortly after oviposition with an unequal cellular division that produces a large cell and a polar body, each inheriting one chromatid per chromosome (i.e., 2n and 2c) (Fig. 3I). This equational division must be equivalent to meiosis II except that it results in two cells containing the parental number of chromosomes.
The larger cell then undergoes first cleavage, and blastomeres further divide by regular mitosis, while the polar body remains along the furrow (Fig. 3J). During embryogenesis, homologous chromosomes remain separated throughout the cell cycle (Fig. 3, I to L) generating up to four FISH signals during anaphase when sister chromatids segregate (Fig. 3K).
DISCUSSION
As reported in textbooks, the reproductive mode of bdelloid rotifers has long been considered as mitotic parthenogenesis (16, 17). Our findings here rather support a meiotic-derived oogenesis with an incomplete or abortive meiosis I during which homologous chromosomes associate and separate but do not segregate into haploid nuclei (Fig. 4A; see Supplementary Text). A nonreductional first meiosis, known as first division restitution (18), has been described in several parthenogenetic species (19–22) and probably also applies to the asexual phase of the monogonont rotifers reproductive cycle (see Supplementary Text). This suggests that the modified meiotic mode of reproduction that we describe here is not species specific and has been acquired by bdelloid rotifers independently of their desiccation sensitivity.
Fig. 4. A model of parthenogenesis and its genomic consequences in bdelloid rotifer A. vaga.
(A) During oocyte maturation, the six pairs of duplicated homologous chromosomes condense and form bivalents but do not segregate into haploid cells. Sister chromatids then separate during a single division analogous to meiosis II, which generates a diploid embryonic nucleus and an extruded diploid polar body. Cell division cycles end after embryogenesis; the hatched individual is eutelic. (B) A single pair of duplicated homologous chromosomes in the germ line is shown in blue and red. Following accidental or meiotic programmed DNA DSBs, they can engage into distinct mechanisms of interhomolog recombination (IHR), involving for example gene conversion (GC) or crossing over (CO). This generates chimeric chromosomes and results in some loss of heterozygosity in the egg after the independent segregation of recombinant chromatids in meiosis II. One set ends up in a polar body (pb) that does not contribute to the future embryo. For convenience, only some possible genomic outcomes of abortive meiosis I are depicted.
The overall maintenance of two distinct haplotypes in A. vaga genome (14) corroborates with the cytological mechanism of oogenesis proposed here, in which the maternal allelic diversity is retained unless interhomolog recombination (IHR), an almost universal feature of the onset of meiosis (23, 24), occurs (Fig. 4B). The signatures of IHR found in specific regions of A. vaga genome (14) or in natural A. vaga populations (8) thus likely result from mechanisms such as crossing over (CO) and/or gene conversion that take place during the meiotic pairing of homologs (Fig. 4B). Supporting this idea, most genes encoding the classical actors of the meiotic machinery are found in the genome of bdelloid rotifers (25).
In A. vaga, the repair of oocyte DNA damage caused by PR appears to be delayed to oogenesis when chromosomes initiate the meiotic-like process. We hypothesize that the temporal relationship between the two events reflects their functional link. Repair of accidental DNA DSBs in the germ line would use the meiotic recombination machinery to promote the complete genome reassembly without major chromosomal rearrangements as we observe. Thus, the nonreductional meiotic process in bdelloid rotifers was likely evolutionary maintained to serve primarily for DNA repair to safeguard the genetic information of the species, especially when thriving in semiterrestrial environments where DNA DSBs do accumulate during prolonged periods of desiccation (12).
IHR is probably not restricted to the repair of accidental DNA DSBs, but it may also result from meiotic programmed DSB because its genomic signatures are observed in hydrated lines of A. vaga that did not undergo any environmental stress (14). Our observation of structures resembling chiasmata, i.e., the cytological sites of meiotic CO in unirradiated animals, also supports this hypothesis (fig. S6A). The nondetection of DNA synthesis activity during oogenesis in unirradiated A. vaga (Fig. 2) is therefore possibly due to the sensitivity of the F-ara-EdU–based assay that would only reveal relatively high levels of DNA synthesis and thus of DNA damage.
On the other hand, the immediate response of G0/G1 somatic nuclei to PR-caused DNA DSBs is unlikely to involve homology-directed mechanisms in the absence of sister chromatids and DNA replication. Therefore, somatic DSBs are most likely to be sealed by nonhomologous end joining (NHEJ), a pathway that generally predominates in nondividing cells (26, 27). Specific genes encoding critical actors of NHEJ-related mechanisms are particularly well represented in the genome of A. vaga, and many of them were found to be highly expressed or up-regulated during desiccation (14, 28). The same is true for other DNA repair pathways like base excision repair or nucleotide excision repair that are expected to act on other radiation-induced DNA lesions such as abasic sites, altered bases, or single-strand breaks (29). Each of these different mechanisms involves some extent of de novo DNA synthesis, which may contribute to the dose-dependent incorporation of F-ara-EdU that we observed in somatic nuclei upon irradiation.
The incomplete DNA DSB repair in the somatic nuclei, potentially including genetic alterations at the sequence level, could be tolerated due to their noncycling, postmitotic status in eutelic bdelloid rotifers and/or the absence of signaling of unrepaired DNA DSBs, which generally leads to apoptosis (30). The detected level of somatic DNA repair seems therefore sufficient to maintain gene expression and cellular functions to ensure survival of adult individuals under severe genotoxic conditions.
Discovering meiosis-specific cytological events coupled to DNA repair activity during oogenesis in the model species A. vaga demystifies the success of what remained the most notorious evolutionary scandal as it strongly suggests that parthenogenesis in bdelloid rotifers is not synonym to strict clonality. Regardless of the origin of DNA DSBs (programmed DSBs during the meiotic-derived oogenesis or accidental DSBs due to genotoxic stresses), IHR in the germ line can efficiently and accurately reconstruct broken chromosomes while shuffling the allelic content and creating offspring that are genetically diverse from their mother. After the independent segregation of putative recombinant chromatids in meiosis II, one set ends up in a polar body that does not contribute to the future embryo (Fig. 4B). In the absence of syngamy, by breaking up interactions between linked loci, IHR also contributes to eliminate deleterious mutations by reducing genetic hitchhiking and background selection (31). However, the frequency of IHR events and the mutation rate that would be required to limit the detrimental effects of loss of heterozygosity over generations in bdelloid lineages as well as the role of horizontal gene transfer (14) and interindividual genetic exchanges (6, 7) in their evolution remain to be determined.
MATERIALS AND METHODS
Bdelloid rotifer culture
All experiments were performed using isogenic A. vaga clones derived from a single individual from the Meselson Laboratory. The cultures were maintained hydrated in petri dishes with spring water, at 25°C, and fed with lettuce filtrate.
Desiccation and exposure to PR
A. vaga was starved overnight before being desiccated for 3 days as previously described (12) and submitted to a homogeneous broad proton beam defocused over 1-cm2 area produced by a 2-MV Tandem accelerator (High Voltage Engineering Europa). They were exposed to 1.7-MeV proton, with a linear energy transfer of 25 keV/μm, ranging from 0.16 to 1.28 kGy. A description of the experimental setup and the irradiation procedure is given in (32). After PR exposure, samples were processed immediately or stored at −80°C.
Survival and fertility
Twenty-four desiccated A. vaga individuals submitted or not to different doses of PR were individually isolated in a 96-well plate containing 0.12 ml of spring water and lettuce filtrate and placed at 25°C. During 1 week, which covered one filial generation, survival of rotifers was determined each day by monitoring their activity. At the same time, the number of eggs and daughters (recognizable by their small size) present in each well was scored. Their cumulative number reflects the egg-laying capacity of irradiated animals, while the number of active daughters was used to determine population growth. At least two independent experiments were performed, and data are presented as means ± 1 SD.
Analysis of genomic integrity by PFGE
The accumulation of DNA DSBs induced by PR and the DNA repair kinetics were screened using PFGE according to the protocol described in (12). When required, genomic DNA was digested with Sbf I restriction enzyme (New England Biolabs, UK) as follows. Agarose plugs containing A. vaga DNA were washed three times in 1× tris-EDTA (TE;tris-HCl, 10 mM; EDTA, 1 mM) and immersed in 100 μl of CutSmart buffer before 4 hours of incubation with 10 U of Sbf I at 37°C. Plugs were then rinsed three times in 1× TE and two times in 0.5× tris borate EDTA (TBE; Bio-Rad, 1610733) and stored in 0.5× TBE at 4°C until they were loaded in a 0.8 or 1% agarose gel (Lonza, Rockland, ME, USA) along with Saccharomyces cerevisiae chromosomes as ladder (Bio-Rad, Hercules, CA, USA). Migration was performed on a Bio-Rad CHEF Mapper XA system according to the following parameters: 22 hours of migration time, 14°C, 5.5 V/cm, 120° switch angle, and 60- to 185-s switch times with a linear ramp. These parameters provide resolution over a size range from 225 to 1600 kb. They were adapted for Fig. 1B with 50- to 150-s switch times to improve resolution of restriction fragments. Gels were stained with SYBR Gold (Invitrogen, Carlsbad, CA, USA) and scanned with a Bio-Rad Chemidoc XRS camera.
TUNEL on primary oocytes
DNA damage in primary oocytes was monitored by using the In Situ Cell Death Detection Kit, Fluorescein (Roche, 11684795910). This assay is based on the ability of the enzyme TdT to incorporate fluorescein-dUTP to 3′-OH termini accessible at DNA breaks. The TUNEL reaction was performed on cells and nuclei extruded from A. vaga. Briefly, rotifers previously irradiated by 1.28-kGy PR or not were transferred on Superfrost microscope slides and covered with a coverslip. They were squashed to release cells and nuclei from rotifers’ cuticle. Samples were then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; 137 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4) and permeabilized with 0.5% Triton X-100/0.5% Saponin in PBS before treatment with proteinase K (25 μg/ml; Thermo Fisher Scientific, EO0491) in PBS. Each incubation was performed for 15 min at room temperature (RT) and was followed by three 5-min washes with PBS. TUNEL reaction mix was then prepared according to the manufacturer’s protocol, and 100 μl was added on samples that were subsequently covered by a coverslip and incubated in a dark humidified atmosphere for 1 hour at 37°C. After three 5-min washes in PBS, slides were incubated 15 min in 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; Sigma-Aldrich, D9542) in PBS, rinsed in PBS, and mounted with Mowiol. A negative control consisted in proton-irradiated A. vaga cells was prepared as described here above, except that the TUNEL reaction mix did not contain the TdT enzyme. A positive control was obtained by pretreating unirradiated cells at 25°C for 15 min with 100 U of deoxyribonuclease I (Invitrogen, 18047019) in 50 mM tris (pH 7.5)/bovine serum albumin (1 mg/ml) before the TUNEL reaction. Confocal microscopy was performed on a Leica SP5 (Leica Microsystems, Wetzlar, Germany) by keeping constant parameters. The Integrated Density (IntDen) function of ImageJ software (v.1.51 for Mac) was used to calculate frequency distribution of TUNEL normalized to DAPI fluorescence intensity. Each histogram plot is a representative result from at least two independent TUNEL assays where more than 245 primary oocytes (at least 24 rotifers) from four independent PR experiments were examined. The data are presented as means ± 1 SD.
F-ara-EdU incorporation and DNA labeling
F-ara-EdU was used to label newly synthesized DNA with high sensitivity in A. vaga. Desiccated rotifers, either proton-irradiated or not, were rehydrated in spring water containing lettuce filtrate and 50 μM F-ara-EdU (Sigma-Aldrich, T511293) from a 10 mM stock solution in dimethyl sulfoxide. They were incubated at 25°C for 48 hours (Fig. 1D) or until oocyte maturation (Fig. 2). In the case of experimental controls, early embryos were incubated with 50 μM F-ara-EdU for several hours before egg hatching in the presence or not of 100 mM hydroxyurea (Sigma-Aldrich, H8617) or α-amanitin (10 μg/ml; Sigma-Aldrich, A2263) (fig. S2). Alternatively, exposure of embryos to 50 μM F-ara-EdU in the absence of any drug continued after hatching (Fig. 1D, left). For the F-ara-EdU kinetics experiments, the thymidine analog was added in the medium either directly (0 hours), 24, 48, 72, or 96 hours after rehydration. After its metabolic incorporation into newly synthesized DNA, F-ara-EdU was detected by coupling to fluorescein 6-FAM-azide. Briefly, adults or embryos were harvested by a 2-min centrifugation at 10,000 rpm, spotted on Superfrost microscope slides, and covered with a coverslip after removal of excess liquid. Slides were then inverted onto a pad of Kimtech Science Kimwipes, submitted to a gentle pressure, and placed in liquid nitrogen for 10 s before coverslips were removed with a razor blade. The samples were then fixed and permeabilized as described for the TUNEL assay, and F-ara-EdU was coupled to 6-FAM-azide by click reaction using the EdU-Click Kit (Baseclick BCK-EdU488) according to the manufacturer’s instructions. Nuclei were counterstained with DAPI (1 μg/ml) in PBS for 15 min. Following PBS washes, slides were mounted with Mowiol for confocal microscopy. The data presented are representative results of at least three independent experiments.
Oogenesis
A. vaga showing early signs of oogenesis, namely, a swollen gonad with single nucleus isolated from the cluster of primary oocytes, was selectively pipetted onto a new petri dish with spring water and lettuce filtrate placed at 15°C. At regular intervals, several rotifers were transferred into a 1.5-ml tube on ice, pelleted by a 2-min centrifugation at 10,000 rpm, and deposited onto a new plate. After removal of excess water, they were covered with methanol–acetic acid (3:1) fixative for 20 min at RT and harvested by quick centrifugation. The pellet was dropped on Superfrost microscope slides and air-dried, and nuclei were stained with DAPI (1 μg/ml) in PBS before imaging by confocal microscopy. Data are representative of at least three independent experiments.
DNA FISH
A. vaga adults at different stages of oogenesis were selectively harvested into a 1.5-ml tube on ice, pelleted by a 2-min centrifugation at 10,000 rpm, and spotted on Superfrost microscope slides. Animals were freeze-cracked in liquid nitrogen, fixed, and permeabilized as described in the F-ara-EdU labeling procedure. Preparation of embryos after oviposition was performed according to the protocol reported in (14). The FISH probe library used on adults and embryos targeted two regions of chromosome pairs 2 and 5, respectively. They were described in (14), prepared, and used accordingly.
Nuclear DNA content
To prepare A. vaga nuclei for DNA content estimation by flow cytometry, cultures were starved for 24 hours before 5000 rotifers were collected in a 1.5-ml tube on ice and washed twice in stock solution [0.4 mM trisodium citrate dihydrate, Nonidet P-40 at 0.1% (v/v), 1.5 mM sperminetetrahydrochloride, and 0.5 mM trishydroxymethyl-aminomethane (pH 7.6)]. They were then transferred into a 1-ml Dounce tissue homogenizer and lysed on ice with 400 strokes to free individual nuclei. The homogenate was filtered through a 30-mm mesh filter and digested for 10 min at 37°C by addition of 100 ml of 0.003% trypsin in stock solution. Next, 75 ml of 0.05% trypsin inhibitor/0.01% ribonuclease A (RNase A) was added, and samples were incubated at 37°C for another 10 min. Last, they were stained with propidium iodide (PI; 10 mg/ml; Sigma-Aldrich, P4864) for 15 min in the dark. PI-stained samples were kept at 4°C and subjected to flow cytometry on a FACSVerse (BD Biosciences). Chicken erythrocyte nuclei, used as an internal standard, were added extemporaneously. Data are representative of three independent experiments. For cytofluorometry measurement of nuclear DNA content, 7- to 10-day-old A. vaga was harvested, fixed in methanol–acetic acid (3:1), and spotted on Superfrost microscope slides. Slides were stained with PI (10 mg/ml) in PBS supplemented with 0.01% RNase A for 30 min. Slides were washed in PBS, mounted with Mowiol, and subjected to confocal microscopy analyses. ImageJ software was used to quantify the PI fluorescence intensity in primary oocytes, somatic nuclei, and nurse nuclei. Data are representative at least three independent experiments, each analyzing at least 350 primary oocytes, 600 somatic nuclei, and 200 nurse nuclei from at least 50 rotifers.
Acknowledgments
The MORPH-IM platform and the Research Unit for Analysis by Nuclear Reactions (LARN) of UNamur are acknowledged for technical help. M.T. thanks A. Houtain and P. Simion for constructive discussion.
Funding: This work was supported by grant agreement 725998 (RHEA) from the European Research Council to K.V.D., by a grant “Action de Recherche Concertée” from the Fédération Wallonie-Bruxelles to B.Ha. and K.V.D., by the PRODEX program to B.He. and K.V.D. in support of European Space Agency selected ILSRA-2014-0106 project, and by a postdoctoral research fellowship to M.T. from F.R.S.-FNRS (2014-2017). E.N. obtained a “Fonds spécial pour la recherche” from the Fédération Wallonie-Bruxelles/UNamur.
Author contributions: Conceptualization: M.T. Methodology: M.T. Validation: M.T. and E.N. Formal analysis: M.T., E.N., B.He., B.Ha., and K.V.D. Investigation: M.T., E.N., B.He., L.H., J.V., C.D., and A.-C.H. Visualization: M.T. Supervision: M.T., B.Ha., and K.V.D. Writing—original draft: M.T., B.Ha., and K.V.D. Writing—review and editing: M.T., B.Ha., and K.V.D. Funding acquisition: B.Ha. and K.V.D.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S6
References
REFERENCES AND NOTES
- 1.Birky C. W. Jr, Positively negative evidence for asexuality. J. Hered. 101 (Suppl. 1), S42–S45 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Hsu W. S., Oogenesis in Habrotrocha tridens (Milne). Biol. Bull. 111, 364–374 (1956). [Google Scholar]
- 3.Hsu W. S., Oogenesis in the Bdelloidea rotifer Philodina roseola Ehrenberg. Cellule 57, 283–296 (1956). [Google Scholar]
- 4.Segers H., Annotated checklist of the rotifers (Phylum Rotifera), with notes on nomenclature, taxonomy and distribution. Zootaxa 1564, 1–104 (2007). [Google Scholar]
- 5.Tang C. Q., Obertegger U., Fontaneto D., Barraclough T. G., Sexual species are separated by larger genetic gaps than asexual species in rotifers. Evolution 68, 2901–2916 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Signorovitc A., Hur J., Gladyshev E., Meselson M., Allele sharing and evidence for sexuality in a mitochondrial clade of bdelloid rotifers. Genetics 200, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debortoli N., Li X., Eyres I., Fontaneto D., Hespeels B., Tang C. Q., Flot J. F., Van Doninck K., Genetic exchange among bdelloid rotifers is more likely due to horizontal gene transfer than to meiotic sex. Curr. Biol. 26, 723–732 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Vakhrusheva O. A., Mnatsakanova E. A., Galimov Y. R., Neretina T., Gerasimov E. S., Naumenko S. A., Ozerova S. G., Zalevsky A. O., Yushenova I. A., Rodriguez F., Arkhipova I. R., Penin A. A., Logacheva M. D., Bazykin G. A., Kondrashov A. S., Genomic signatures of recombination in a natural population of the bdelloid rotifer Adineta vaga. Nat. Commun. 11, 6421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laine V. N., Sackton T. B., Meselson M., Genomic signature of sexual reproduction in the bdelloid rotifer Macrotrachella quadricornifera. Genetics 220, iyab221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gladyshev E. A., Arkhipova I. R., Genome structure of bdelloid rotifers: Shaped by asexuality or desiccation? J. Hered. 101 (Suppl. 1), S85–S93 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Gladyshev E., Meselson M., Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. 105, 5139–5144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hespeels B., Knapen M., Hanot-Mambres D., Heuskin A. C., Pineux F., Lucas S., Koszul R., Van Doninck K., Gateway to genetic exchange? DNA double-strand breaks in the bdelloid rotifer Adineta vaga submitted to desiccation. J. Evol. Biol. 27, 1334–1345 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Hespeels B., Penninckx S., Cornet V., Bruneau L., Bopp C., Baumlé V., Redivo B., Heuskin A. C., Moeller R., Fujimori A., Lucas S., Van Doninck K., Iron ladies - how desiccated asexual rotifer Adineta vaga deal with X-rays and heavy ions? Front. Microbiol. 11, 1792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simion P., Narayan J., Houtain A., Derzelle A., Baudry L., Nicolas E., Arora R., Cariou M., Cruaud C., Gaudray F. R., Gilbert C., Guiglielmoni N., Hespeels B., Kozlowski D. K. L., Labadie K., Limasset A., Llirós M., Marbouty M., Terwagne M., Virgo J., Cordaux R., Danchin E. G. J., Hallet B., Koszul R., Lenormand T., Flot J. F., Van Doninck K., Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Sci. Adv. 7, eabg4216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mark Welch D. B., Meselson M., Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual Class Bdelloidea. Biol. J. Linn. Soc. 79, 85–91 (2003). [Google Scholar]
- 16.N. H. Barton, D. E. G. Briggs, J. A. Eisen, D. B. Goldstein, N. H. Patel, Evolution of genetic systems, in Evolution (Cold Spring Harbor Laboratory Press, ed. 1, 2007), pp. 257–694. [Google Scholar]
- 17.D. B. Mark Welch, C. Ricci, M. Meselson, Bdelloid rotifers: Progress in understanding the success of an evolutionary scandal, in Lost Sex (Springer, ed. 1, 2009), pp. 259–279. [Google Scholar]
- 18.Mirzaghaderi G., Horandl E., The evolution of meiotic sex and its alternatives. Proc. Biol. Sci. 283, 20161221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narbel-Hofstetter M., Cytologie comparée de l’espèce parthenogénétique Luffia ferchaultella Steph. et de l’espèce bisexuée L. lapidella Goeze (Lepidoptera, Psychidae). Chromosoma 12, 505–552 (1961). [DOI] [PubMed] [Google Scholar]
- 20.Beukeboom L. W., Pijnacker L. P., Automictic parthenogenesis in the parasitoid Venturia canescens (Hymenoptera: Ichneumonidae) revisited. Genome 43, 939–944 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Belshaw R., Quicke D. L. J., The cytogenetics of thelytoky in a predominantly asexual parasitoid wasp with covert sex. Genome 46, 170–173 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Hiruta C., Nishida C., Tochinai S., Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res. 18, 833–840 (2010). [DOI] [PubMed] [Google Scholar]
- 23.N. Hunter, Meiotic recombination, in Molecular Genetics of Recombination (Springer, ed. 1, 2007), pp. 381–442. [Google Scholar]
- 24.Zickler D., Kleckner N., Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Nowell R. W., Almeida P., Wilson C. G., Smith T. P., Fontaneto D., Crisp A., Micklem G., Tunnacliffe A., Boschetti C., Barraclough T. G., Comparative genomics of bdelloid rotifers: Insights from desiccating and nondesiccating species. PLOS Biol. 16, e2004830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyama T., Wilson D. M. III, DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 12, 620–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H. H. Y., Pannunzio N. R., Adachi N., Lieber M. R., Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecox-Lea B. J., Mark Welch D. B., Evolutionary diversity and novelty of DNA repair genes in asexual Bdelloid rotifers. BMC Evol. Biol. 18, 177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.E. C. Friedber, G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, T. Ellenberger, DNA Repair and Mutagenesis (American Society for Microbiology Press, ed. 2, 2006). [Google Scholar]
- 30.Vermezovic J., Stergiou L., Hengartner M. O., d’Adda di Fagagna F., Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Diff. 19, 1847–1855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartfield M., Keightley P. D., Current hypotheses for the evolution of sex and recombination. Integr. Zool. 7, 192–209 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Wéra A. C., Heuskin A. C., Riquier H., Michiels C., Lucas S., In vitro irradiation station for broad beam radiobiological experiments. Nucl. Instrum. Methods Phys. Res., Sect. B 269, 3120–3124 (2011). [Google Scholar]
- 33.Zelinka C., Studien uber Raderthiere. III. Zur entwicklungsgeschichte der Raderthiere nebst Bemerkungen uber ihre Anatomie und BiologieIII. Zur entwicklungsgeschichte der Raderthiere nebst Bemerkungen uber ihre Anatomie und Biologie. Zeitschrift fur wissenschaftliche Zoologie 53, 1–159 (1891). [Google Scholar]
- 34.Boschetti C., Ricci C., Sotgia C., Fascio U., The development of a bdelloid egg: A contribution after 100 years. Hydrobiologia 546, 323–331 (2005). [Google Scholar]
- 35.Mark Welch J. L., Mark Welch D. B., Meselson M., Cytogenetic evidence for asexual evolution of bdelloid rotifers. Proc. Natl. Acad. Sci. 101, 1618–1621 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenssen D., Contribution à l’étude du développement et de la maturation des oeufs chez l’Hydatina Senta. Zoologischer Anzeige 21, 617–621 (1898). [Google Scholar]
- 37.Whitney D. D., Observations on the maturation stages of the parthenogenetic and sexual eggs of Hydatina senta. J. Exp. Zool. Phil. 6, 137–146 (1909). [Google Scholar]
- 38.Shull A. F., Chromosomes and the life cycle of Hydatina senta. Biol. Bull. 41, 55–61 (1921). [Google Scholar]
- 39.Whitney D. D., The chromosome cycle in the rotifer Asplanchna amphora. J. Morphol. 47, 415–433 (1929). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S6
References