Abstract
Innate immunity provides an ever-present or rapidly inducible initial defense against microbial infection. Among the effector molecules of this defense in many species are broad-spectrum antimicrobial peptides. Tracheal antimicrobial peptide (TAP) was the first discovered member of the β-defensin family of mammalian antimicrobial peptides. TAP is expressed in the ciliated epithelium of the bovine trachea, and its mRNA levels are dramatically increased upon stimulation with bacteria or bacterial lipopolysaccharide (LPS). We report here that this induction by LPS is regulated at the level of transcription. Furthermore, the transfection of reporter gene constructs into tracheal epithelial cells indicates that DNA sequences in the 5′ flanking region of the TAP gene, within 324 nucleotides of the transcription start site, are responsible in part for mediating gene induction. This region includes consensus binding sites for NF-κB and nuclear factor interleukin-6 (NF IL-6) transcription factors. Gel mobility shift assays indicate that LPS induces NF-κB binding activity in the nuclei of these cells, while NF IL-6 binding activity is constitutively present. The gene encoding human β-defensin 2, a human homologue of TAP with similar inducible expression patterns in the airway, was cloned and found to have conserved NF-κB and NF IL-6 consensus binding sites in its 5′ flanking region. Previous studies of antimicrobial peptides from insects indicated that their induction by infectious microbes and microbial products also occurs via activation of NF-κB-like and NF IL-6-like transcription factors. Together, these observations indicate that a strategy for the induction of peptide-based antimicrobial innate immunity is conserved among evolutionarily diverse organisms.
Innate immunity provides animals with a dynamic first-line host defense against microbes. Epithelial cells lining the mammalian airway are a crucial site in host defense of the respiratory tract. One of the innate host defense responses of these cells is the inducible production of β-defensins, a class of homologous antibiotic peptides whose members have been found in leukocytes and epithelial cells in a wide distribution of animals, including birds, rodents, ruminants and humans (8). Together with clearance mechanisms, barrier properties of epithelial surfaces, and additional antimicrobial factors, β-defensins are proposed to help maintain the respiratory and other mucosal surfaces free from infection (10).
Tracheal antimicrobial peptide (TAP) is the first described β-defensin. TAP is a 38-amino-acid peptide with broad-spectrum antimicrobial activity isolated from the bovine tracheal mucosa (12). The TAP gene is expressed in vivo in the ciliated airway epithelium (9), and its expression levels are dramatically increased following experimentally induced bacterial infection (45). In vitro incubation of bovine tracheal epithelial cells (TEC) with heat-killed bacteria or bacterial lipopolysaccharide (LPS) markedly increased TAP mRNA levels (11). This response was shown to be mediated by CD14, a well-characterized mammalian coreceptor for LPS (11). Although initially characterized as a cell surface marker for cells of the monocyte/macrophage lineages, CD14 is also expressed by epithelial cells and likely provides these cells with the capacity to detect and respond to bacteria at their luminal surface (11, 16, 49). These findings suggest that certain mucosal epithelial cells can autonomously detect bacteria and then responsively mount a direct antimicrobial action.
Studies of the innate immune response in Drosophila and other insects have led to the understanding that upon challenge with microbes, there is an induction of a collection of antimicrobial factors, including antimicrobial peptides (5, 6, 23). This induction is mediated at the transcriptional level and involves proteins homologous to the mammalian toll-like receptors (TLRs), interleukin receptor-associated kinase, IκB, NF-κB, and nuclear factor interleukin-6 (NF IL-6) (14, 25, 28, 30, 34, 39, 50). In mammals, these factors are integral to a variety of immune and inflammatory pathways, suggesting a striking similarity between certain insect and mammalian host defense responses (22, 34, 38).
The insights from the studies with Drosophila and the discovery of inducible antimicrobial peptide expression in mammals suggest that parallels may extend to regulatory mechanisms of host defense gene expression in epithelial cells. Accordingly, we addressed potential mechanisms which regulate expression of TAP in TEC. Examination of the TAP gene indicated the presence of consensus binding sites for NF-κB and NF IL-6 (9), two transcription factors implicated in a wide array of inducible immune and inflammatory responses. Here, we examined the possible role of these transcription factors in the expression of TAP in response to challenge of TEC by LPS.
MATERIALS AND METHODS
General methodology.
All reagents and general methods were as previously described (11), unless otherwise noted. LPS was obtained from either Sigma Chemical Co., St. Louis, Mo. (no. L-8643) or List Biological Laboratories, Inc., Cambell, Calif. My4 was obtained from Biogenex Laboratories, San Ramon, Calif., and mouse immunoglobulin G2b was obtained from Antigenix America, Inc., Franklin Square, N.Y.
Primary culture of bovine TEC.
Cells were cultured by the method of Wu et al. (51, 52) as described previously (11). The epithelial cells were plated on petri dishes (5 × 105 cells/35-mm-diameter dish) containing a collagen gel (Vitrogen 100; Collagen Biomedical, Palo Alto, Calif.) in a defined growth medium. The cells maintain their epithelial characteristics, including the presence of active cilia on many of the cells (11, 51, 52). Cells were cultured in 49% Dulbecco modified Eagle medium–49% F-12–2% Ultroser G supplemented with antibiotic-antimycotic (Life Technologies) and gentamicin (50 μg/ml). The complete medium was determined to be free of bacterial endotoxin as tested by the Limulus amebocyte lysate-gel clot assay (sensitivity = 0.125 endotoxin unit/ml) (Associates of Cape Cod).
Northern blot analysis.
For harvesting, cells were washed with phosphate-buffered saline (PBS) and then incubated with collagenase (type 2; Worthington Biochemical, Freehold, N.J.) (10 mg/ml) at 37°C for 10 min. The suspended cells were pelleted by centrifugation at 150 × g for 5 min at 4°C, resuspended in 3 ml of PBS, and then repelleted. Total RNA was extracted from harvested cells, and the RNA samples were then prepared for Northern blot analysis as described previously (11). The probe for TAP was TAP48a (5′-CCAAGCAGACAGGACCAGGAAGAGGAGCGCGAGGAGCAGGTGATGGAGCCTCAT-3′). Labeled probes were hybridized overnight to immobilized RNA in 37.5% (vol/vol) formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt's solution–1% (vol/vol) sodium dodecyl sulfate (SDS) at 42°C and then washed at high stringency in 0.1× SSC–0.1% SDS at 52°C for 30 min (43). A bovine α-tubulin oligonucleotide probe (11) was used as a control for RNA integrity and relative amounts. The hybridization and wash conditions for this probe were modified such that hybridization buffer contained 25% formamide and the final stringency wash with was 2× SSC–0.1% SDS at 65°C. Signal intensities of Northern blots were quantitated by Phosphorimager analysis (Molecular Dynamics, Sunnyvale, Calif.). In all cases the signal was normalized for the relative RNA amount by using the signal intensity of the control probe.
EMSA.
Cells were harvested with collagenase and rinsed twice in PBS. Extracts were prepared by the method of Dignam et al. (13). Protein concentrations were adjusted to 0.2 mg/ml. Electrophoretic mobility shift assays (EMSA) were carried out in the presence of poly(dI/dC) by a standard protocol (2) with 6% nondenaturing polyacrylamide gel electrophoresis in 0.25× Tris-borate-EDTA. In certain experiments, anti-human p65 or p50 antibodies (400 ng/reaction) (Santa Cruz Biochemicals) were added to the incubation reaction prior to electrophoretic analysis. Complementary oligonucleotides containing either the NF-κB or NF IL-6 sequence were annealed to create double-stranded molecules and labeled with [γ-32P]ATP and T4 polynucleotide kinase. The sequence of the sense NF-κB oligonucleotide (TAP/NF32) is 5′-AGCTTTTTCTGGGGTTTTCCCCAGCCTCAT-3′ (the consensus sequence is underlined), and that of the sense NF IL-6 oligonucleotide (TAP/NFIL30) is 5′-TAAGCGAAGGTTCAGCAAGAAGTCTGTGCC-3′. The nonspecific oligonucleotide used for competition experiments is a mutant form of the NF-κB oligonucleotide (TAP/NFmut32), with the sequence 5′-AGCTTTTTCTCTCATTTTCCCCAGCCTCAT-3′ (sense).
Nuclear run-on assay.
Epithelial cells (5 × 105 cells/35-mm-diameter dish) were cultured as described above and treated with bacterial LPS (100 ng/ml). Control cultures were grown in parallel and given only vehicle (PBS). After 16 h of culture incubation, the nuclei were isolated from the cells (2). The nuclei from 10 dishes (in each group) were pooled, and the transcription that had been initiated in the intact cells was allowed to complete by incubating the nuclei in the presence of [α-32P]UTP by a method described previously (19). RNA was isolated from the nuclei and then was hybridized to target DNA sequences immobilized on Hybond nylon membranes (Amersham). The target sequences were TAP, i.e., a 1.9-kb EcoRI genomic restriction fragment that encompasses both exons of the TAP gene, the single intron, and proximal portions of the 5′ and 3′ flanking regions, and (ii) tubulin, i.e., a 1.2-kb cDNA clone (9). After high-stringency washing, specific hybridization of RNA was assessed by autoradiography and quantitated by phosphorimager analysis.
Plasmids and transfections.
Sequences from the upstream region of the TAP gene were isolated by PCR amplification and subcloned into the multiple cloning site of the pGL-2 luciferase reporter plasmid (Promega). The transcription start site of the TAP gene (9) is designated +1. Plasmid p1S contains bases −324 to +1, p3S contains bases −294 to +1, and plasmid p4S contains bases −180 to +1. Early-passage bovine TEC were grown on plastic coated with a 1:75 dilution of Vitrogen and transfected with 1.8 μg of plasmid DNA by using Lipofectamine (Life Technologies), followed by cultivation for 48 h before reporter gene analysis. Cells were cotransfected with 0.2 μg of pB-gal (Promega) as an internal control for transfection efficiency. After 18 h of incubation with either LPS (100 ng/ml) or buffer vehicle, the cells were lysed and assayed for enzymatic activity according to the manufacturer's instructions. Luciferase activity was measured with the Gene-light assay kit (Promega) in an LKB luminometer, and data were normalized to β-galactosidase activity, measured by using the β-gal assay kit (Promega).
Gene cloning.
The human β-defensin 2 (HBD-2) gene was cloned from a human genomic library (lambda-FIX phage vector [no. 944201]; Stratagene) with oligonucleotide probes based on the published human cDNA (20) by standard methods as described previously (35). Phage insert DNA from positive clones was subcloned into pBluescript II SK(+) plasmid, and purified plasmid DNA was sequenced from both strands by using a thermal cycling method with fluorescent dye-labeled dideoxynucleotide terminators.
Nucleotide sequence accession number.
The GenBank accession number for the sequence determined in this work is AF071216.
RESULTS
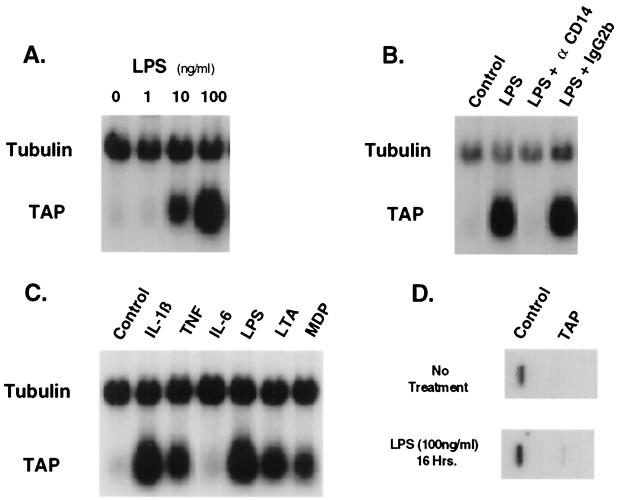
To investigate regulation of TAP gene expression, TEC were studied in primary culture under serum-free conditions. Northern blot analysis revealed that TAP mRNA levels in TEC are elevated in response to LPS (10.3-fold maximal response in this representative experiment), compared with levels in control cultures. The induction was dependent on the LPS concentration, with a half-maximal response seen at approximately 10 ng/ml (Fig. 1A). A blocking anti-CD14 monoclonal antibody, My4, abrogated the effects of LPS on TAP mRNA levels in the TEC, while an isotype control antibody had no effect at equivalent concentrations (Fig. 1B). These results are consistent with previous studies (11) and demonstrate that TAP mRNA is inducible in TEC in response to challenge with LPS via a CD14-dependent mechanism.
FIG. 1.
Induction of TAP mRNA levels in TEC by LPS and other inflammatory mediators. (A) Concentration dependence of LPS on TAP mRNA levels in bovine TEC. Total RNA was isolated from primary cultures of TEC (2 × 105 cells/culture) after incubation for 16 h with no additions (lane 1) or in the presence of purified LPS from Pseudomonas aeruginosa (1 ng/ml [lane 2], 10 ng/ml [lane 3], or 100 ng/ml [lane 4]). Probes for Northern blot analysis were TAP48a (TAP) and α-tubulin cDNA (Tubulin). (B) Effect of My4, an anti-CD14 blocking monoclonal antibody, on LPS-inducible changes in TAP mRNA levels. Cultures of TEC were incubated without LPS (lane 1) or with LPS (100 ng/ml) (lanes 2 to 4) for 16 h. The anti-CD14 (αCD14) mouse monoclonal antibody My4 (500 ng/ml) (lane 3) or an equivalent concentration of immunoglobulin G2b (IgG2b), an isotype-specific control antibody (lane 4), was coincubated as indicated. Northern blot analysis was as described for panel A. (C) Effects of inflammatory cytokines and bacterial products on TAP mRNA levels. RNA was isolated from TEC after incubation for 16 h with no additions (lane 1) or in the presence of TNF-α (100 ng/ml) (lane 2), IL-1β (100 ng/ml) (lane 3), IL-6 (500 U/ml) (lane 4), LPS (100 ng/ml) (lane 5), lipoteichoic acid (LTA) (10 μg/ml) (lane 6), or muramyl dipeptide (MDP) (100 μg/ml) (lane 7). Northern blot analysis was as described for panel A. (D) Nuclear run-on transcription of TAP in TEC. TEC were untreated (Control) or treated with bacterial LPS (100 ng/ml) for 16 h. Nuclei from 5 × 106 cells were harvested and used for nuclear run-on analysis as described in Materials and Methods. Nylon filter blots containing cDNA encoding TAP and tubulin were hybridized under high-stringency conditions with 32P-labeled primary RNA transcripts. Specific hybridization of RNA was assessed by autoradiography and quantitated by phosphorimager analysis.
TAP mRNA levels were also analyzed in TEC exposed to the inflammatory cytokines IL-1β, tumor necrosis factor alpha (TNF-α), and IL-6 (Fig. 1C). TAP mRNA levels were induced in TEC exposed to 100 ng of either IL-1β or TNF-α per ml for 16 h, compared to the control cells, but not in cells challenged with IL-6 (500 U/ml). In addition, elevated TAP mRNA levels were induced in response to the bacterial products lipoteichoic acid and muramyl dipeptide (Fig. 1C). Control experiments found that the addition of polymyxin B to cultures effectively blocked the response to LPS but had little or no effect on the response to lipoteichoic acid, muramyl dipeptide, or cytokines (data not shown). These data demonstrate that when challenged by bacterial membrane and cell wall components or certain inflammatory cytokines, TEC respond (in part) by raising steady-state levels of TAP mRNA. We saw no similar response of an increase in β-defensin mRNA levels in primary alveolar macrophages (42) or in a bovine turbinate epithelial cell line (data not shown).
In order to determine whether the LPS-induced increase in mRNA levels is due to an increase in the rate of transcription, we performed nuclear run-on experiments. Bovine TEC were incubated in the presence or absence of 100 ng of LPS per ml, and nascent run-on transcription was analyzed by hybridization of 32P-labeled nuclear RNA to TAP and α-tubulin probes. The results from pools of 2 × 106 TEC per group indicate that the elevated levels of TAP mRNA in LPS-challenged cells compared to controls correlate with an increase in TAP transcription (Fig. 1D). There was an approximately 9-fold induction of TAP nascent transcripts detectable, which compares favorably with the approximately 8- to 13-fold increase in steady-state mRNA levels observed when cells are challenged with 100 ng of LPS per ml (Fig. 1A) (11, 41). Additional experiments showed that the rates of decay of TAP mRNA levels were essentially the same (half-life of approximately 4 h) in TEC after removal of LPS stimulation and in LPS-stimulated TEC upon addition of actinomycin D, a blocker of transcription (data not shown). Together these data support that the principal mechanism for induction of TAP mRNA levels by LPS stimulation is via increased transcription, with little significant effect attributable to prolongation of the TAP mRNA half-life.
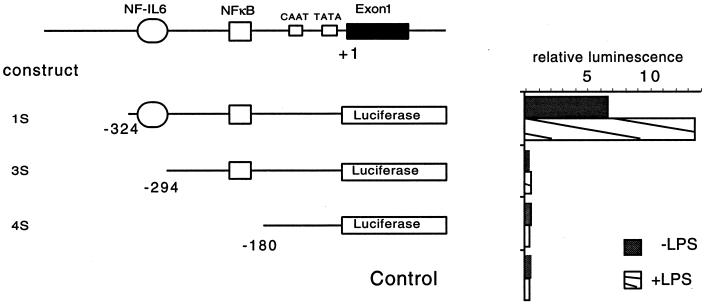
An examination of the 5′ flanking sequence of the TAP gene reveals the presence of several potential transcription factor binding sites involved in LPS-mediated induction of host defense genes (Fig. 2). Several members of the NF-κB (Rel) family of transcription factors are known to mediate the transcriptional induction of numerous genes in immune and inflammatory reactions (31), including the induction of antimicrobial peptides in insects (6, 25, 34). The presence of an NF-κB consensus site 177 bp upstream from the transcriptional start site of the TAP gene suggested that this transcription factor may be involved in the induction of TAP mRNA. The transcription factor NF IL-6 has been shown to participate in activation of numerous innate immune responses, often through interactions with NF-κB (1, 4, 28, 36, 47). The presence of a consensus binding site for NF IL-6 adjacent to the NF-κB site suggested that this factor may also participate in the regulated expression of TAP. In order to assess the functional participation of these sites in the regulated expression of the TAP gene, we transfected reporter gene constructs into TEC in primary culture. A luciferase-reporter gene construct with the 5′ flanking region of the TAP gene containing the NF-κB and the NF IL-6 sites (−324 to +1; p1S), another with just the NF-κB (−294 to +1; p3S), and a third that included neither of these sites but included simply the more proximal CAAT and TATA boxes (−180 to +1; p4S) were transfected into these cells and tested for luciferase activity. Figure 2 indicates that only p1S, the construct containing both the NF-κB and the NF IL-6 sites, was capable of expressing the reporter gene. Furthermore, only this construct allowed for induced expression when coincubated with LPS. These data support that sequences necessary and sufficient for inducible transcription of TAP in TEC are present in the 324 nucleotides of 5′ flanking sequence contained in the p1S reporter plasmid and further indicate the involvement of the NF-κB and NF IL-6 sites in this response.
FIG. 2.
Promoter analysis of the upstream region of the TAP gene. A map of the putative promoter region of the TAP gene is shown at the top. Segments of the 5′ flanking region of the TAP gene were ligated into a promoterless luciferase reporter expression vector. Plasmid constructs containing the putative NF-κB and NF IL-6 sites are shown to the left, along with the control vector containing no promoter. Plasmid constructs were transfected into cultured primary TEC, followed by stimulation with LPS. The promoter activity for each plasmid is exhibited as relative luminescence of each transfection, normalized to cotransfected β-galactosidase, and is shown to the right of the respective plasmid.
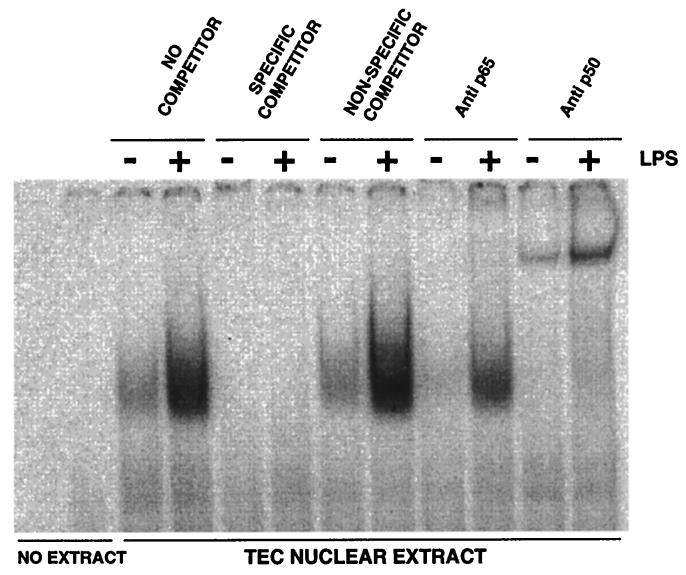
To analyze the binding of transcription factors to these regions of the TAP gene in response to LPS, nuclear extracts from cultured TEC were assayed by EMSA. A 32-bp double-stranded oligonucleotide (ds-TAP/NF32), whose sequence encompassed the NF-κB consensus binding sequence in the 5′ flanking region of the TAP gene (nucleotides −165 to −197), was radioactively labeled and incubated with nuclear lysates. The complexes were fractionated by gel electrophoresis under nondenaturing conditions and analyzed by autoradiography (Fig. 3). Coincubation of extracts with unlabeled specific (ds-TAP/NF32) and nonspecific (ds-TAP/NFmut32) competitors indicated that TEC nuclear extracts contained a low level of NF-κB. When TEC were incubated with LPS, NF-κB binding activity in nuclear extracts was increased. EMSA with coincubation of specific antibodies to the p50 and p65 subunits of NF-κB show a distinct retardation in the mobility with the p65 antibody and a reduction in the intensity of the shifted band with the p50 antibody. These results support that a predominant factor which binds to the NF-κB site of the TAP gene in LPS-stimulated TEC is the p50-p65 heterodimer.
FIG. 3.
Mobility shift analysis of NF-κB in bovine TEC nuclear extracts. A 32P-labeled, double-stranded oligonucleotide probe containing the NF-κB sequence flanking the TAP gene (ds-TAP/NF32) was incubated with nuclear extracts from bovine TEC treated with LPS (100 ng/ml) or untreated. Specificity of binding is shown by competition with unlabeled specific (ds-TAP/NF32) and nonspecific (ds-TAP/NFmut32) double-stranded oligonucleotides, as described in Materials and Methods. The composition of the NF-κB complex was analyzed by including either anti-p50 or anti-p65 antibodies in the binding reaction. Inclusion of the anti-p50 antibody causes retarded migration of the binding complex, whereas anti-p65 antibody decreases the signal intensity of the binding complex.
For EMSA with NF IL-6 sequences, a 30-bp double-stranded oligonucleotide (ds-TAP/NFIL30), whose sequence encompassed the NF IL-6 consensus binding sequence (nucleotides −329 to −300), was used. Specific binding to the NF IL-6 site of the TAP gene was observed in the TEC nuclear extracts. However, unlike the NF-κB binding, there was no detectable increase in response to LPS (data not shown). These data indicate that NF IL-6 binding activity is constitutively present in TEC nuclei. Together with the reporter gene studies described above, which indicated the requirement of DNA encompassing both NF IL-6 and NF-κB sites for inducible TAP transcription, the EMSA data suggest that binding of both NF IL-6 and NF-κB transcription factors to the flanking region of the TAP gene may be required for its transcriptional induction in response to LPS. In total, these results suggest that TAP gene expression in TEC is induced by LPS at the transcriptional level through a pathway common to other host defense response genes.
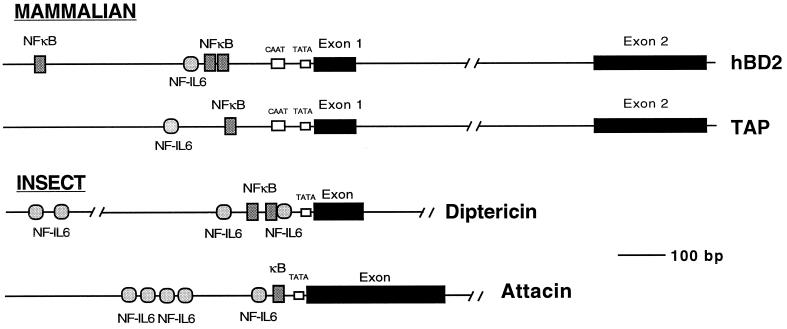
Recent publications described the discovery of a human homologue of TAP which was expressed in the airway and whose expression was observed to be inducible upon stimulation with bacteria and inflammatory cytokines (3, 20, 37). This suggests that a similar pathway may be involved in host defense of the human and bovine upper airways. To initiate the studies that will further address this possibility, we screened a human genomic library for the HBD-2 gene by using the published cDNA sequence (20). A single phage clone was obtained, which contained the entire HBD-2-coding sequence along with flanking regions. The genomic sequence indicated that the HBD-2 gene encompassed two exons separated by an intron of 1.6 kB (Fig. 4). This structure is similar to that of the genes which encode TAP (9) and enteric β-defensin (48). In comparing the promoter regions, we observed that the NF-κB site in the TAP promoter was conserved in its location. In addition, two additional putative NF-κB sites were found in the HBD-2 gene. While the NF IL-6 site was not conserved in location, a putative site was found approximately 70 bp downstream of the one in the TAP promoter.
FIG. 4.
Structural organization of the HBD-2 gene and other selected antimicrobial peptide genes. The HBD-2 gene was cloned from a human genomic bacteriophage library. The sequence of 4.8 kb of DNA encompassing the gene was determined from each complementary strand. Intron-exon boundaries were defined based on comparison with the published cDNA sequence. Putative enhancer binding sites were identified by using MacVector software. The organization of the HBD-2 gene was compared with those of the TAP gene (9) and two inducible antimicrobial peptide genes from insects, acidic attacin (46) and diptericin (28).
The conservation of these sites is striking when compared with the antibacterial response in insects. There, numerous antibacterial and antifungal peptides have been identified, and their genes are induced at the transcriptional level in response to infectious agents through defined pathways which involve NF-κB-like and NF IL-6-like factors (15, 27, 28, 34, 40). In Fig. 4, the upstream regions of two such genes (attacin from Hyalophora cecropia and diptericin from Drosophila melanogaster) are schematically aligned with the flanking regions of the mammalian TAP and HBD-2 genes. The conservation of these sites in inducible antimicrobial peptide genes highlights a dramatic parallel of innate immune responses in distantly related species of the animal kingdom.
DISCUSSION
Innate immunity is thought to provide the host with a defense system capable of effectively dealing with the continuous challenge by a wide array of microbes at surface epithelia. A hallmark of the innate immune system is that it remains ever present or immediately inducible. This property serves to distinguish it from the lymphocyte-mediated acquired immune system, in which the antigen-specific response is developed over a period of days to weeks (26).
Emerging evidence supports that many aspects of innate immune defenses are conserved in the animal kingdom, including the use of antimicrobial peptides capable of killing a wide range of microbes (5, 6, 22). Such peptides are expressed both in circulating phagocytic cells and in epithelial cells of mammals and many invertebrate species. In insect model systems, where provocative testing has been possible, antimicrobial peptides have proven to be critically important to an effective innate host defense response (21, 23, 33, 34). Insects lack lymphocytes and therefore rely solely on their innate defense system for protection. In mammals, the abundance and in vitro activity of antimicrobial peptides in phagocytic leukocytes and at mucosal epithelia support a similar key role in innate immunity (18, 32, 53), but further testing of this hypothesis is warranted.
A distinctive feature of insect antimicrobial peptides is that their production is inducible upon challenge with microbes and microbial outer membrane components (6, 23). This induction is regulated at the transcriptional level and has been shown to be mediated by transcription factors homologous to the NF-κB and NF IL-6 families of mammalian transcription factors (14, 25, 28, 30, 34, 39, 50). Inducible expression was observed in numerous epithelial cells, including those of the insect trachea (17). Studies of TAP have indicated that a remarkable parallel exists in the regulated expression of this mammalian molecule in TEC and that of insect antimicrobial peptides (8, 9, 11).
TAP mRNA levels are increased in TEC upon challenge with heat-killed bacteria (11), bacterial outer membrane components (Fig. 1A to C) (11), and certain inflammatory cytokines (Fig. 1C) (41). Because bovine epithelial cell lines that express TAP are not available, studies with primary cultures of TEC were used as a model system to examine mechanisms governing TAP expression (Fig. 1 to 3). These cells show expression patterns (Fig. 1A to C) (11) that appear to recapitulate those of TEC cells in vivo (45) but impose significant technical challenges that limit the breadth of mechanistic studies. Nevertheless, the studies reported here show that like that of the insect peptides, the induction of TAP in response to LPS is regulated at the transcriptional level (Fig. 1D). Sequences necessary for the inducible response are present in the 324 nucleotides of the proximal 5′ flanking sequence of the TAP gene (Fig. 2). This region contains consensus binding sites for the NF-κB and NF IL-6 transcription factors (Fig. 2). EMSA analysis indicates that LPS induces NF-κB binding activity in the nuclei of these cells (Fig. 3). Experiments using specific antibodies to the p50 and p65 subunits of NF-κB indicate that these two subunits are included in a significant fraction of NF-κB in TEC (Fig. 3). NF IL-6 binding activity is detectable in nuclei of control TEC and does not increase upon LPS stimulation (data not shown). Together, these data support the conclusions that (i) a response of TEC to challenge by LPS is the induction of TAP mediated at the transcriptional level, (ii) the transcriptional induction of TAP depends on sequences in the 5′ flanking region of the gene, and (iii) the mechanism of induction likely involves the binding of NF-κB and NF IL-6 transcription factors to their respective sites in the TAP gene.
Our data suggest that CD14-mediated induction of TAP in response to LPS and other microbial products exemplifies a mechanism by which mammals can recognize microbes at epithelial surfaces and transcriptionally induce local antibiotic peptides. We speculate that these peptides, together with other antimicrobial factors, are capable of effectively dealing with the continuous challenge by a wide array of microbes at surface epithelia. The lower respiratory tracts of mammals are considered to be relatively free of microbes (39). A current hypothesis holds that the defensive responses of epithelial cells are important to host defense because they prevent colonization and/or subsequent infection (8, 24). In vivo evidence of this response was provided by the studies of Stolzenberg et al., in which TAP mRNA levels were dramatically induced within 4 h following experimentally induced bacterial infection of the bovine airway (45). The kinetics of this inducible response suggests that it could contribute to host defense prior to the development of lymphocyte-mediated adaptive immune responses. Similarly, the expression in epithelial cells of a homologous β-defensin, HBD-2, in response to bacterial LPS indicates that a similar response is found in humans (3, 20, 44). This gene was cloned and found to have conserved binding sites for NF-κB and NF IL-6 transcription factors. We have recently found that NF-κB is activated in human TEC upon stimulation with IL-1β (M. Becker, G. Diamond, M. Verghese, and S. Randell, unpublished data). In the same study, we have identified the expression of both CD14 and LTRs in the human TEC. At least two of the identified TLRs have been implicated as functioning in conjunction with CD14 to transduce the signal from microorganisms (7, 29). Bovine TEC express at least one homologous TLR (D. Legarda and G. Diamond, unpublished data), indicating a conservation of this induction pathway. In light of these mammalian responses in epithelial cells and the aforementioned analogous responses in insects, it appears that transcriptional induction of antimicrobial peptides via pathways utilizing NF-κB and NF IL-6 transcription factors is phylogenetically conserved in the animal kingdom.
ACKNOWLEDGMENTS
We thank Thomas Hamilton and Jennifer Major for technical advice.
G.D. was supported by grants from the NIH (HL53400), the USDA, and the Cystic Fibrosis Foundation. C.L.B. was supported by grants from the NIH (AI32738 and AI32234).
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a c/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley; 1994. [Google Scholar]
- 3.Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson J M. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Invest. 1998;102:874–880. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts J C, Cheshire J K, Akira S, Kishimoto T, Woo P. The role of NF-κB and NF-IL6 transactivating factors in the synergistic activation of human serum amyloid A gene expression by interleukin-1 and interleukin-6. J Biol Chem. 1993;268:25624–25631. [PubMed] [Google Scholar]
- 5.Boman H G. Gene-encoded peptide antibiotics and the concept of innate immunity: an update review. Scand J Immunol. 1998;48:15–25. doi: 10.1046/j.1365-3083.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Boman H G. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 7.Chow J C, Young D W, Goldenbock D T, Christ W J, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 8.Diamond G, Bevins C L. Beta-defensins: endogenous antibiotics of the innate host defense response. Clin Immunol Immunopathol. 1998;88:221–225. doi: 10.1006/clin.1998.4587. [DOI] [PubMed] [Google Scholar]
- 9.Diamond G, Jones D E, Bevins C L. Airway epithelial cells are the site of expression of a mammalian antimicrobial peptide gene. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond, G., D. Legarda, and L. K. Ryan. The innate immune response of the respiratory epithelium. Immunol. Rev., in press. [DOI] [PubMed]
- 11.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a novel cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dushay M S, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engstrom Y, Kadalayil L, Sun S, Smakovlis C, Hultmark D, Faye I. κB-like motifs regulate the induction of immune genes in Drososphila. J Mol Biol. 1993;232:327–333. doi: 10.1006/jmbi.1993.1392. [DOI] [PubMed] [Google Scholar]
- 16.Fearns C, Kravchenko V V, Ulevitch R J, Loskutoff D J. Murine CD14 gene expression in vivo: extramyeloid synthesis and regulation by lipopolysaccharide. J Exp Med. 1995;181:857–866. doi: 10.1084/jem.181.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrandon D, Jung A C, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann J A. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganz T, Lehrer R I. Antimicrobial peptides of vertebrates. Curr Opin Immunol. 1998;10:41–44. doi: 10.1016/s0952-7915(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 20.Harder J, Bartels J, Christophers E, Schroder J-M. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann J A. Innate immunity of insects. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann J A, Reichart J-M. Drosophila immunity. Trends Cell Biol. 1997;7:309–316. doi: 10.1016/S0962-8924(97)01087-8. [DOI] [PubMed] [Google Scholar]
- 24.Huttner K M, Bevins C L. Antimicrobial peptides as mediators of epithelial host defense. Ped Res. 1999;45:785–794. doi: 10.1203/00006450-199906000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Ip Y T, Reach M, Engstrom Y, Kadalayil L, Cai H, González-Crespo S, Tatei K, Levine M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 26.Janeway C A, Jr, Travers P. Immunobiology: the immune system in health and disease. 3rd ed. London, United Kingdom: Current Biology, Ltd.; 1997. [Google Scholar]
- 27.Kadalayil L, Petersen U M, Engstrom Y. Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res. 1997;25:1233–1239. doi: 10.1093/nar/25.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kappler C, Meister M, Lagueux M, Gateff E, Hoffmann J A, Reichhart J-M. Insect immunity. Two 17 bp repeats nesting a κB-related sequence confer inducibility to the diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. EMBO J. 1993;12:1561–1568. doi: 10.1002/j.1460-2075.1993.tb05800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirschning C J, Wesche H, Merrill Ayres T, Rothe M. Human Toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi A, Matsui M, Kubo T, Natori S. Purification and characterization of a 59-kilodalton protein that specifically binds to NF-κB-binding motifs of the defense protein genes of Sarcophagia peregina (the flesh fly) Mol Cell Biol. 1993;13:4049–4056. doi: 10.1128/mcb.13.7.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp E B, Ghosh S. NF-κB and Rel proteins in innate immunity. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer R I, Bevins C L, Ganz T. Defensins and other antimicrobial peptides. In: Ogra P L, Mestecky J, Lamm M E, Strober W M, Bienstock J, editors. Mucosal immunology. 2nd ed. New York, N.Y: Academic Press; 1998. pp. 89–99. [Google Scholar]
- 33.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J-M, Hoffmann J A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in Drosophila host defense. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffmann J A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls potent antifungal response in drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 35.Mallow E B, Harris A, Salzman N, Russell J P, DeBerardinis J R, Ruchelli E, Bevins C L. Human enteric defensins: gene structure and developmental expression. J Biol Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- 36.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCray P B, Bentley L. Human airway epithelia express a beta-defensin. Am J Respir Cell Mol Biol. 1997;16:343–349. doi: 10.1165/ajrcmb.16.3.9070620. [DOI] [PubMed] [Google Scholar]
- 38.Meng X, Khanuja B S, Ip Y T. Toll receptor-mediated drosophila immune response requires dif, an NF-kappaB factor. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas E, Reichhart J M, Hoffmann J A, Lemaitre B. In vivo regulation of the IkappaB homologue cactus during the immune response of Drosophila. J Biol Chem. 1998;273:10463–10469. doi: 10.1074/jbc.273.17.10463. [DOI] [PubMed] [Google Scholar]
- 40.Petersen U M, Bjorklund G, Ip Y T, Engstrom Y. The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J. 1995;14:3146–3158. doi: 10.1002/j.1460-2075.1995.tb07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan L K, Rhodes J, Bhat M, Diamond G. Expression of beta-defensin genes in bovine alveolar macrophages. Infect Immun. 1998;66:878–881. doi: 10.1128/iai.66.2.878-881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Singh P K, Jia H P, Wiles K, Hesselberth J, Liu L, Conway B A D, Greenberg E P, Valore E V, Welsh M J, Ganz T, Tack B F, McCray P B., Jr Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stolzenberg E D, Anderson G M, Ackermann M R, Whitlock R H, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun S-C, Lindstrom I, Lee J-Y, Faye I. Structure and expression of the attacin genes in Hyalophora cecropia. Eur J Biochem. 1991;196:247–254. doi: 10.1111/j.1432-1033.1991.tb15811.x. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Yoshida N, Kishimoto T. Targetted disruption of the NF-IL6 gene discloses its essential role in bacterial killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 48.Tarver A P, Clark D P, Diamond G, Russell J P, Erdjument-Bromage H, Tempst P, Cohen K S, Jones D E, Sweeney R W, Wines M, Hwang S, Bevins C L. Enteric β-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verghese, M. W., G. Diamond, M. N. Becker, and S. H. Randell. 1998. Upregulation of human β-defensin 2 by LPS and TNF-α in cultured human bronchial epithelial cells. Role of epithelial-cell expressed CD14. Ped. Pulmonol. Suppl., A585.
- 50.Wu L P, Anderson K V. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 51.Wu R, Nolan E, Turner C. Expression of tracheal differentiated functions in serum-free hormone-supplemented medium. J Cell Physiol. 1985;125:167–181. doi: 10.1002/jcp.1041250202. [DOI] [PubMed] [Google Scholar]
- 52.Wu R, Yankaskas J, Cheng E, Knowles M R, Boucher R. Growth and differentiation of human nasal epithelial cells in culture. Am Rev Respir Dis. 1985;132:311–320. doi: 10.1164/arrd.1985.132.2.311. [DOI] [PubMed] [Google Scholar]
- 53.Zasloff M. Antibiotics peptides as mediators of innate immunity. Curr Opin Immunol. 1992;4:3–7. doi: 10.1016/0952-7915(92)90115-u. [DOI] [PubMed] [Google Scholar]