Abstract
In an effort to better understand genetic and cellular factors that influence innate immunity, we examined host and bacterial factors involved in the nonopsonic phagocytosis and killing of Escherichia coli K-12 by mouse macrophages. Unelicited (resident) peritoneal macrophages from five different mouse strains, BALB/c, C57BL/6, CD-1, C3H/HeJ, and C3H/HeN, were employed. Additional macrophage populations were obtained from CD-1 mice (bone marrow-derived macrophages). Also, for BALB/c and C57BL/6 mice, peritoneal macrophages elicited with either thioglycolate or proteose peptone, bone marrow-derived macrophages, and macrophage-like cell lines derived from the two strains were employed. Two E. coli K-12 strains that differed specifically in their abilities to produce type 1 pili containing the adhesive protein FimH were examined. The parameters used to assess macrophage bacteriocidal activity were (i) the killing of internalized (gentamicin-protected) E. coli during the approximately 4-h assay and (ii) the initial rate at which internalized E. coli were eliminated. Data on these parameters allowed the following conclusions: (i) unelicited or proteose peptone-elicited peritoneal macrophages were significantly better at eliminating internalized bacteria than thioglycolate-elicited peritoneal macrophages, bone marrow-derived macrophages, or macrophage cell lines; (ii) the host genetic background had no significant effect upon the ability of unelicited peritoneal macrophages to kill E. coli (even though the mouse strains differ widely in their in vivo susceptibilities to bacterial infection); and (iii) the FimH phenotype had no significant effect upon E. coli survival once the bacterium was inside a macrophage. Additionally, there was no correlation between the bacteriocidal effectiveness of a macrophage population and the number of bacteria bound per macrophage. However, macrophage populations that were the least bacteriocidal tended to bind higher ratios of FimH+ to FimH− E. coli. The effect of gamma interferon, fetal calf serum, and the recombination proficiency of E. coli were examined as factors predicted to influence intracellular bacterial killing. These had no effect upon the rate of E. coli elimination by unelicited peritoneal macrophages.
The mechanism by which bacteria are taken up and killed by macrophages and other cells of the reticuloendothelial system in the absence of normal or immune serum components has been under study for a number of years (39). This process has been referred to as nonopsonic phagocytosis. Nonopsonic processes differ in a number of respects from the opsonic mechanisms. One important difference is an increased rate of killing of opsonized internalized bacteria (3, 54, 59). Nonopsonic phagocytosis has been characterized as a primitive holdover from protozoal ingestion mechanisms (40). However, since some antibodies, particularly those directed against bacterial adhesins, actually prevent phagocytosis (62), it may be fortunate that this poorly understood mechanism is still in place.
One well-described bacterial ligand that mediates nonopsonic phagocytosis is the type 1 pilus (8). These pili are produced by many members of the Enterobacteriaceae (12) and promote bacterial adherence to the mucosal surfaces of a wide variety of hosts through a mannose-sensitive interaction with receptors on eucaryotic cells (12). This adherence is thought to allow the colonization of a number of host compartments (42) and promote interindividual spread (4). Type 1 pili also mediate adherence to phagocytic cells (39). The interactions between type 1 pili and neutrophils (58), mast cells (30–32), macrophages (3), and other leukocytes (45) have been some of the more carefully examined interactions between bacteria and phagocytes.
Early and more recent work on the nature of the interaction of type 1 piliated Escherichia coli cells with macrophages indicates that one minor component of the pili, the product of the fimH gene (FimH), is responsible for adherence (25). Whereas this adherence, in effect, tethers the piliated bacteria to macrophages and effectively increases the number of bacteria bound compared to the number of fimH mutants bound (25) and induces an oxidative burst (5, 15, 29, 41), reports on whether type 1 piliation actually results in an increased rate of killing compared to that for nonpiliated or FimH− cells, under defined in vitro conditions, are conflicting (5, 15, 16, 22, 25, 29). Reports agree that leukocyte-bound type 1 piliated cells are better protected against killing than opsonized FimH− E. coli cells (3, 16, 55). This protection is due, at least in part, to a difference in the compartmentalization of opsonized E. coli versus that of FimH+ E. coli in bone marrow-derived macrophages (3). Direct comparisons of otherwise isogenic FimH+ and FimH− bacteria (both unopsonized) suggest that there is a modest but statistically significant increase in the survivability of FimH+ over that of FimH− E. coli in microphages (25).
In order to better understand factors affecting innate host susceptibility to bacterial diseases and also the role of type 1 pili in the nonopsonic phagocytic process, we examined resident peritoneal macrophages from five different mouse strains for their abilities to kill E. coli that were phenotypically either FimH+ or FimH−. Also, for some mouse strains, additional elicitation methods, anatomical sources, and derivation methods were examined to see if these factors affected macrophage function.
MATERIALS AND METHODS
Mouse strains and cell lines.
Male BALB/c, C3H/HeN, C3H/HeJ, C57BL/6, and CD-1 mice 8 to 12 weeks of age were used in these experiments. The mice were purchased from either Charles River Laboratories (Wilmington, Mass.) or Taconic Farms (Germantown, N.Y.). Mice were maintained under pathogen-free husbandry conditions and were fed food and water ad libitum. Relevant genotypic and phenotypic differences in the mouse strains and macrophage-like cell lines are listed in Table 1.
TABLE 1.
Mouse and bacterial strains and cell lines used in this study
| Organism or Cell line | Description and/or relevant phenotype | Source or referencea |
|---|---|---|
| Mice | ||
| BALB/c | Inbred; ityS lpsNN; sensitive to Y. enterocolitica, L. monocytogenes, and S. enterica serovar Typhimurium | 11, 17, 48 |
| C57BL/6 | Inbred; ityS lpsN; resistant to Y. enterocolitica, L. monocytogenes, and Legionella pneumophila | 11, 17, 65 |
| CD-1 | Outbred; ityR (presumed) lpsN; resistant to S. enterica serovar Typhimurium and L. monocytogenes | 1, 2, 38 |
| C3H/HeJ | Inbred; ityR lpsD; susceptible to S. enterica serovar Typhimurium and Francisella tularensis | 21, 44, 61 |
| C3H/HeN | Inbred; ityR lpsN; resistant to S. enterica serovar Typhimurium and F. tularensis | 21, 48 |
| Mouse cell lines | ||
| J774 | Macrophage-like cell line derived from BALB/c reticulum cell sarcoma | 47a |
| IC-21 | Macrophage cell line derived from simian virus 40 transformation of C57BL/6 peritoneal macrophages | 34 |
| Bacteria | ||
| E. coli | ||
| ORN115 | thr-1 leuB thi-1 Δ(argF-lac)U169 malA1 xyl-7 ara-13 mt1-2 gal-6 rpsL fhuA2 supE44 pilG Pil+ Hag+ (FimH+) Mal− | 56 |
| ORN133 | Same as ORN115 except fimH::Kan Hag− (FimH−) Mal− | 35 |
| ORN175 | Same as ORN115 except Mal+ | 25 |
| ORN204 | Same as ORN133 except Mal+ | P1 transduction from ORN109 (19) |
| ORN205 | Same as ORN115 except recA13 Rec− | P1 transduction from E. coli EC901 and EC931 (43) |
| ORN172 | Same as ORN115 except Δfim(BEACDEFGH) Pil− Rec+ | 63 |
| ORN201 | Same as ORN172 except recA13 Rec− | P1 transduction from E. coli EC901 and EC931 (48) |
| L. monocytogenes | ||
| EGD 1/2a | Presumed wild type | 37 |
References for mouse strains refer to their resistant properties. More-comprehensive listings of resistances and derivations of mouse strains can be found at the Jackson Laboratory, Bar Harbor, Maine.
Bacterial strains and growth conditions.
Bacterial strains used in this study are shown in Table 1. All bacteria were grown in L broth (36) overnight with shaking to stationary phase. Prior to the bacteriocidal assay, bacteria were harvested by centrifugation (6,000 × g for 10 min), resuspended, and diluted to ca. 2 × 108 cells/ml in phosphate-buffered saline (PBS). Media used to assess bacterial numbers both before and after macrophage killing assays consisted of maltose tetrazolium agar (53) for E. coli (25) and L agar (36) for Listeria monocytogenes.
Isolation and cultivation of peritoneal macrophages.
Peritoneal cells were isolated by intraperitoneal (i.p.) injection of 7 to 8 ml of RPMI 1640 medium (Gibco BRL, Grand Island, N.Y.) containing 5% fetal bovine serum (FBS), gentamicin (5 μg/ml), and heparin (5 U/ml) into mice that had been killed by cervical dislocation. Following i.p. injection, the mice were shaken to dislodge peritoneal cells and the lavage fluids were removed by syringe. The peritoneal cells were centrifuged (430 × g for 5 min), the resulting cell pellet was suspended in the aforementioned medium lacking heparin at a concentration of 5 × 105 cells/ml, and 0.5 ml of the cell suspension was placed into each well of a 48-well cell culture cluster plate (Costar, Cambridge, Mass.). Two hours later, the medium was removed and the plastic adherent cells were washed three times with 0.5 ml of Hanks balanced salt solution (HBSS) and then incubated in 0.5 ml of RPMI 1640 medium containing 5% FBS without antibiotics at 37°C in a humidified 5% CO2 incubator. Eighteen to twenty-four hours later, the culture medium was aspirated and the cells were washed once with HBSS and then incubated with 0.25 ml of the above culture medium lacking antibiotics. Elicitation of proteose peptone- or thioglycolate-elicited inflammatory macrophages was done by injecting mice i.p. with 2 ml of either sterile 10% proteose peptone (Difco, Detroit, Mich.) or thioglycolate broth (Remel, Kansas City, Mo.). Three days later, the peritoneal cells were harvested as described above.
In some experiments, macrophages were treated with gamma interferon (IFN-γ) prior to use in the bacteriocidal assays. Immediately following the 2-h plastic adherence procedure, macrophages were incubated in medium containing 10 antiviral units of homogeneously pure recombinant mouse IFN-γ/ml (8.0 × 106 U/mg of protein) for 18 to 24 h as previously reported (26, 47) and then used in the bacteriocidal assays. The recombinant mouse IFN-γ (lot 2271-54-F2) was the kind gift of Genentech, Inc. (South San Francisco, Calif.).
Cultivation of bone marrow-derived macrophages and macrophage cell lines.
Bone marrow-derived macrophages were cultured as previously reported (20). IC-21 cells were grown in RPMI 1640–10% FBS. J774 cells were grown in Dulbecco minimal essential medium–Ham's F-12 mixture (1:1) with 10% FBS.
Macrophage bacteriocidal assay.
Macrophages in 48-well tissue culture plates (∼105 to 2 × 105 cells/well) containing 0.25 ml of RPMI 1640 medium were exposed to approximately 106 bacteria added in 50 μl of PBS for 10 min at 37°C. After incubation, wells were washed four times (each wash was with 1 ml of PBS). After the final wash, 0.5 ml of prewarmed RPMI 1640 was added to each well. Gentamicin (2.5 μl of a 1-mg/ml stock) was added to selected wells to produce a final concentration of 5 μg of gentamicin/ml. The addition of 0.1 ml of 1.0% Triton X-100 lysed the macrophages and defined the end of the incubation. One to two minutes after the Triton X-100 additions, the contents of the wells were diluted and plated. The brief exposure of bacteria to gentamicin and Triton X-100 had no effect on bacterial viability. At each time point, the contents of (typically) four wells were plated: two wells contained macrophages, with each well having a mixture of FimH+ and FimH− E. coli cells added at approximately a 1:1 ratio. (Sometimes the two wells were duplicates; occasionally one well had some other treatment.) The two remaining wells did not have macrophages but were treated as if they did, both before and after the addition of bacteria. In one of the wells, gentamicin was added. In the last well gentamicin was omitted. In pilot experiments, we found that a small number of bacteria remained bound to the plastic of the wells without macrophages (1 to 10% of the macrophage-bound bacteria). The bacteria in each of these wells were used to assess the bacteriocidal kinetics of gentamicin when bacteria were “unprotected” and bacterial growth in the absence of gentamicin. Time points were spaced according to data from pilot experiments, which indicated that >90% of unprotected bacteria were eliminated after 20 to 25 min of exposure to gentamicin (establishing the basis for the first time point) and that the numbers of internalized bacteria were constant or tending upward after 4 h (establishing the basis for the end of the assay). The number of bacteria initially bound to macrophages was determined as described above but just prior to the addition of gentamicin.
Maltose tetrazolium agar allowed the convenient identification of FimH+ and FimH− strains, which were genetically marked by their abilities to utilize maltose (25) (Table 1). The ratio of FimH+ to FimH− E. coli cells obtained after macrophage exposure was normalized to the starting FimH+/FimH− ratio to determine if there was any difference in the rate of macrophage killing based upon the FimH phenotype. FimH+ and FimH− cells did not differ in their gentamicin sensitivities (tested in the control well lacking macrophages but with gentamicin) and did not differ noticeably in their growth rates (determined in wells lacking macrophages and gentamicin). Plating efficiencies on maltose tetrazolium agar, previously noted as slightly different depending upon the Mal phenotype (25), were not appreciably different under the present conditions.
Statistical analysis.
Each experiment was typically performed with at least duplicate sets of wells. Standard deviations of the means of at least two separate experiments were calculated with the aid of the Microsoft Excel STDEV function. Standard error was calculated as the standard deviation divided by the square root of the number of experiments. All tabulated or illustrated values are the averages of at least two independent experiments. Regression analysis of the bacteriocidal curves was performed with the aid of the Microsoft Excel, version 5, trend line generator feature. Significant differences between means were determined by either Student's t test or analysis of variance. Both tests were provided by the Microsoft Excel, version 4, statistics package. The F statistic was also used to determine the probability of an accidental association of two variables in generating a trend line (Microsoft Excel, version 4). Statistically significant differences were defined as P < 0.05.
RESULTS
Analysis of macrophage killing curves.
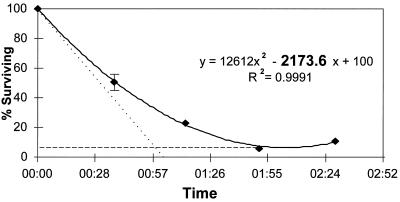
In order to compare the bacteriocidal activities of distinct macrophage populations from different anatomical sites of different mouse strains, a standard was needed. Pilot experiments revealed that the most consistent results came from conditions that produced a final ratio (after washing off unbound bacteria) of approximately 1 bound bacterium per 10 macrophages. This low multiplicity allowed direct comparisons between FimH+ and FimH− bacteria in a single well under noncompetitive binding conditions and reduced potentially confounding effects associated with high multiplicities of infection (e.g., endotoxin effects). At 20 to 25 min after gentamicin addition, we began measuring the rate of killing. At least 90% of the bacteria still viable at this time had been internalized by macrophages (and were thus protected from gentamicin), but had not yet been killed. The remaining ca. 10% were external bacteria that had not been killed by gentamicin in the 25-min period. A considerable fraction (84 ± 41% of the total originally bound; 10 experiments averaged) of the FimH+ and FimH− E. coli cells initially bound (i.e., present immediately after washing) were still viable after 20 to 25 min of gentamicin exposure, even in the most bacteriocidal macrophage populations (unelicited peritoneal macrophages from BALB/c mice; see below). The fate of this “gentamicin-protected” population (set as 100%) over the next 4 h constituted our bacteriocidal curves. An example of one such curve, along with two parameters associated with the trend line through the points, is shown in Fig. 1. In this example, the points on killing curves represent values averaged from both FimH+ and FimH− E. coli.
FIG. 1.
Curve denoting the percentage of gentamicin-protected E. coli surviving over time. The trend line through the points (diamonds) describes a parabolic curve represented by the top equation. The instantaneous slope of the curve (dy/dx at x = 0) is depicted by the dotted line, and the numerical value is highlighted in the top equation. The y value at the vertex (dy/dx = 0) of the curve is illustrated by the dashed line. The R2 statistic (coefficient of determination) denotes the degree to which the values of the experimentally derived points match those predicted by the equation shown. (R2 values range from 0 to 1.0, with 1.0 being a perfect match.) Vertical bars represent standard errors from an experiment performed in triplicate.
Parameters employed to assess differences in bacteriocidal activity between macrophage populations.
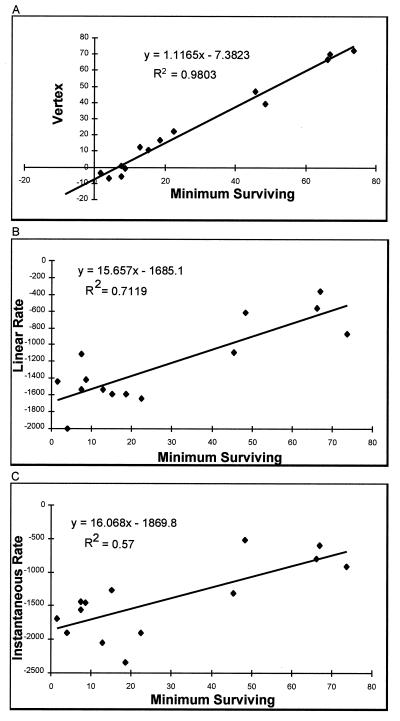
Regression analysis of linear, exponential, and parabolic trend lines showed that the shape of most bacteriocidal curves most closely matched that of a parabola. This is reflected in the R2 statistic (coefficient of determination), which gave the highest average value (a perfect correlation produces an R2 value of 1.0) when 18 killing curves (employing unelicited peritoneal macrophages from different mouse strains and with values for FimH+ and FimH− E. coli averaged) were compared (parabolic, R2 = 0.80 ± 0.17; linear (first three points), R2 = 0.78 ± 0.24; exponential, R2 = 0.74 ± 0.16). For the linear curve, only the first three time points were considered because the killing curves departed from linearity relatively rapidly. Data compiled from 15 different macrophage populations and 33 individual experiments revealed that the ordinate value of the vertex of the parabolic trend line accurately and precisely predicted the actual recorded minimum percentage of surviving E. coli cells (FimH+ and FimH− cells averaged), further indicating the natural parabolic shape of the killing curves (Fig. 2A). Initial rate measurements (of both linear and parabolic curves) were less predictive of the ability of a macrophage population to eliminate internalized E. coli (reflected in the lower R2 statistic; Fig. 2B and C). Nevertheless, rate measurements were linearly related to the minimal percent surviving. This linear relationship was confirmed by calculating the significance of the F statistic, which indicates the probability of erroneously concluding that a linear relationship exists (in all cases P < 0.05).
FIG. 2.
Comparison of curve parameters for their abilities to predict the minimum percent of internalized bacteria surviving in a bacteriocidal assay. (A) Points represent the vertex values of curves describing the bacteriocidal assay versus the actual minimal percentage of bacteria surviving in the bacteriocidal assay. (B) Points represent the initial linear rate of elimination (in units of percentage of internalized bacteria eliminated per 24-h period) versus the actual minimal percentage of bacteria surviving the bacteriocidal assay. (C) Points represent the instantaneous rate of bacterial elimination based upon a parabolic trend line best describing the points of the bacteriocidal assay (in arbitrary units of percentage of internalized bacteria eliminated per 24-h period) versus the actual minimal percentage of bacteria surviving in the bacteriocidal assay. Negative values on the ordinates in panels B and C reflect the negative slopes of the bacteriocidal assays. The top equation in each panel describes the curve derived from linear regression analysis. The R2 value denotes the coefficient of determination as defined in the text.
For the purpose of comparing killing curve parameters (Fig. 2), we excluded twelve experiments because a killing curve was not generated. That is, in these experiments, the ingested E. coli grew, generating a positively sloped linear trend line. While this exclusion of these curves was justified in drawing the conclusions about killing curve shape, this limited the use of values calculated from the parabolic curve (e.g., vertex) since not all experimental results could be analyzed this way. In view of this shortcoming, two curve parameters, percent eliminated and the initial linear rate of elimination, were used for all subsequent measurements. Values for both of these parameters could be derived from any curve, and the parameters could be applied to any macrophage population.
Comparison of the relative abilities of distinct macrophage populations from different mouse strains to eliminate internalized FimH+ and FimH− E. coli.
Peritoneal, bone marrow, and cell line macrophages derived from BALB/c and C57BL/6 mice were initially compared for their abilities to eliminate internalized FimH+ and FimH− E. coli. When results indicated that elicitation did not enhance the rate or degree of killing of either FimH type of E. coli, elicitation methods were not employed for subsequent mouse strains examined (CD-1, C3H/HeJ, and C3H/HeN). For C3H/He strains, only resident peritoneal macrophages were tested. In all macrophage populations tested, there was no statistically significant difference in the degree or rate at which FimH+ and FimH− E. coli cells were eliminated. Consequently, average measurements of killing effectiveness include both FimH+ and FimH− E. coli.
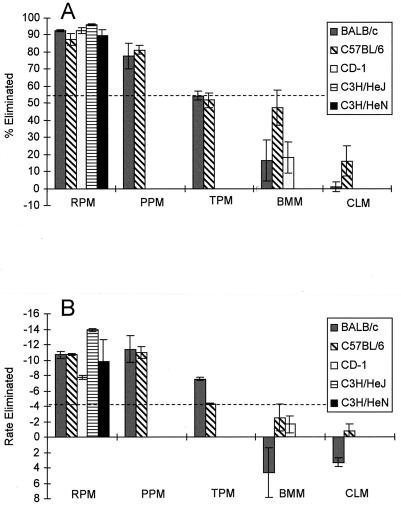
For BALB/c and C57BL/6 mice, unelicited and proteose peptone-elicited peritoneal macrophages were superior to thioglycolate-elicited macrophages, bone marrow-derived macrophages, and cell line macrophages in their ability to eliminate internalized E. coli (Fig. 3). This was especially apparent when the maximal number of ingested bacteria eliminated was used as a measure (Fig. 3A). Nevertheless, the rate measurement produced the same trend (Fig. 3B). One noticeable difference between the statistically significant groupings in Fig. 3A and B was that the BALB/c thioglycolate-elicited peritoneal macrophages showed a high initial rate of E. coli elimination that was not indicative of the relatively low total percentage of E. coli cells eliminated. Also, in some of the macrophage populations that killed poorly, E. coli actually grew (initially) before being reduced in number later in the assay (Fig. 3B). The outbred CD-1 strain exhibited the same general trend as the BALB/c and C57BL/6 mice, indicating the superiority of the resident peritoneal macrophages over bone marrow-derived macrophages (Fig. 3). The killing capacities of resident peritoneal macrophages from all five mouse strains tested were statistically indistinguishable regardless of the type of measurement employed (statistical analysis of data shown in Fig. 3).
FIG. 3.
Capacities of macrophage populations to eliminate internalized E. coli. (A) Percentages of internalized E. coli cells eliminated by macrophages from the mouse strains. (B) Initial rate of E. coli elimination in units of percent internalized bacteria eliminated per 10-min period. Negative values indicate a decrease in the internalized population; positive values denote an increase. RPM, resident (unelicited) peritoneal macrophages; PPM, proteose peptone-elicited peritoneal macrophages; TPM, thioglycolate-elicited peritoneal macrophages; BMM, bone marrow-derived macrophages; CLM, cell line macrophages. The cell line macrophages constitute the J774 line for BALB/c mice and IC-21 for C57BL/6 mice. Only unelicited peritoneal macrophages and bone marrow-derived macrophages were tested for CD-1 mice, and only unelicited peritoneal macrophages were tested for C3H/HeJ and C3H/HeN mice. Vertical lines represent the standard errors of the means. The dashed line in each panel divides two statistically distinct groupings: results extending above the line were (as a group) statistically distinct from those grouped below the line.
E. coli binding characteristics of macrophage populations in comparison to their bacteriocidal abilities.
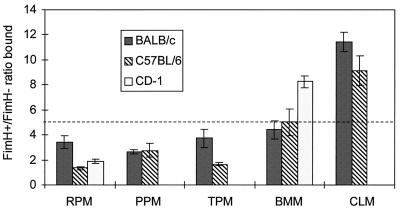
Whereas we could find no difference in the abilities of macrophage populations to eliminate FimH+ E. coli relative to FimH− E. coli, they did differ in the binding of FimH+ and FimH− cells (Fig. 4). The macrophage populations that bound the highest ratios of FimH+ to FimH− bacteria (bone marrow and cell line macrophages) were the populations least able to kill E. coli once ingested. For BALB/c and C57BL/6 mice, this trend was statistically significant only in distinguishing the cell lines from the peritoneal and bone marrow-derived macrophages. For the outbred CD-1 mice, where just bone marrow and unelicited peritoneal macrophages were examined, there was a statistically significant difference between these two macrophage populations with respect to binding. There was no correlation between the absolute numbers of E. coli cells bound per macrophage and the bacteriocidal effectiveness. That is, the macrophage populations most effective at killing internalized E. coli did not bind significantly more of them. This was established in tests comparing unelicited macrophages (used as one statistical grouping) and the other macrophage populations (data not shown).
FIG. 4.
Preferential binding of FimH+ E. coli over FimH− E. coli by macrophages from different host strains, anatomical locations, and derivations. The ratio of FimH+ to FimH− E. coli cells bound is shown on the vertical axis. The macrophage populations are as defined in the legend for Fig. 3. Only unelicited peritoneal macrophages and bone marrow-derived macrophages were tested for CD-1 mice. Vertical lines represent the standard errors of the means. The dashed line divides two statistically distinct groupings: results extending above the line are (as a group) statistically distinct from those grouped below the line.
Examination of additional variables for macrophage bacteriocidal ability.
The effect of serum in the assay, the recombination proficiency of the ingested bacteria (their ability to repair DNA), and the effect of IFN-γ treatment on macrophage bacteriocidal activity were examined. We tested the effect of FBS on macrophage bacteriocidal activity because E. coli antibodies, if elicited in utero, had the potential to serve as opsonizing antibodies. However, experiments done in the absence of serum had no significant effect upon the binding or the subsequent rate or degree of killing of FimH+ or FimH− E. coli. This was true whether the serum was left out during the absorption period only or left out of the macrophage preparation protocol and killing assay entirely (data not shown).
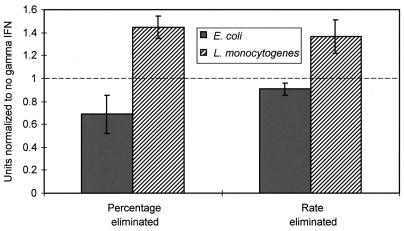
Our results for E. coli recA mutants (we tested both fimbriate and nonfimbriate recA mutants) indicated that the rate and degree to which they were eliminated by BALB/c mouse unelicited macrophages did not differ significantly from those for the parent (data not shown). Similarly, IFN-γ had no effect upon the rate or degree to which FimH+ and FimH− E. coli cells were killed by unelicited macrophages from the BALB/c and C57BL/6 strains (Fig. 5). IFN-γ did have a significant effect upon the degree to which L. monocytogenes was killed by unelicited peritoneal macrophages (the lowest point in the killing curve); consequently the IFN-γ did appear to be having an effect on the macrophages. This effect of IFN-γ on macrophage killing of L. monocytogenes did not produce a difference in the initial elimination rate of these microorganisms.
FIG. 5.
Effect of the addition of 10 U of IFN-γ/ml to unelicited peritoneal macrophages prior to the bacteriocidal assay. IFN-γ was added as described in the text. The percentages of internalized E. coli and L. monocytogenes cells eliminated and the initial rates of elimination were recorded in three experiments for L. monocytogenes and four experiments for E. coli. In each experiment, the differences in degree and rate of elimination were normalized to the values obtained with no IFN-γ addition and then averaged (standard errors are denoted by the vertical lines). The dashed line denotes the normalized value (1) with no IFN-γ addition.
DISCUSSION
In the experiments described herein, we compared the innate abilities of different macrophage populations from mice of different genetic backgrounds to kill internalized E. coli. Bacteriocidal activity was measured by two parameters (i) the initial elimination rate and (ii) the maximal percentage of internalized E. coli eliminated. FimH+ and FimH− E. coli cells were used to assess the effects of different bacterial cell surface interactions upon the binding and elimination of the bacteria once internalized. Additional assays that examined physiological and genetic factors that might influence the process were employed.
Quantitative bacteriocidal measurements which involved measuring the percentage of internalized bacteria eliminated were developed. Employing this measurement effectively normalized the results so that data from macrophages with different binding and uptake kinetics could be compared. We chose to present two measurements, (i) the initial rate and (ii) the degree to which an internalized population was eliminated. Of these two, the measurement of maximal percentage eliminated was found to be the more reproducible. In particular, we often found that high initial elimination rates often did not result in a high percentage of internalized bacteria eliminated. A similar observation was made by van Dissel et al. (60) with opsonized Salmonella enterica serovar Typhimurium.
Using regression analysis, we found that the ordinate value of the vertex of a parabola describing the killing curves predicted the maximal percentage of internalized bacteria eliminated quite well. In contrast, van Dissel et al. (60) found that opsonized S. enterica serovar Typhimurium underwent an exponential decrease after being taken up by macrophages over the 90-min period of their assays. An exponential decrease would be expected if pure probability determined elimination rate (i.e., the macrophages acting as a simple bacteriocidal agent). We expect that the parabolic shape coincidentally best described a dynamic state in which most ingested bacteria were being killed by macrophages in an exponential fashion but in which other ingested bacteria were growing, protected from the gentamicin by macrophages incapable of killing them (or not killing all of them). As time progressed, the replicative power of the protected E. coli began to be witnessed as the upward slope of the parabola. Consequently, the parabolic shape may be simply indicative of a generally inefficient process. However, the killing curve shape is not a widely analyzed feature of bacteriocidal assays. Future and retrospective attention to the shapes of killing curves may reveal unappreciated mechanistic relationships.
Both the initial linear elimination rate and the degree to which internalized E. coli cells were eliminated supported the same order of macrophage bacteriocidal effectiveness, with resident and proteose peptone-elicited peritoneal macrophages being consistently the best, followed by, in declining order of effectiveness, thioglycolate-elicited peritoneal macrophages, bone marrow-derived macrophages, and macrophage-like cell lines. The killing curves of macrophage populations that eliminated approximately 60% (or less) of the ingested bacteria had significantly lower R2 values (for linear or parabolic curves), and the curve parameters had higher standard deviations, than those of macrophage populations killing greater than 90% of the ingested bacteria. We found it difficult to draw any conclusions about the killing capacities of macrophage populations in this low-level killing category. The inability of thioglycolate-elicited macrophages to efficiently kill bacteria has been often reported (7, 18, 27, 57). Likewise, our observation that macrophage cell lines and bone marrow-derived macrophages were inefficient at killing internalized bacteria, compared to unelicited peritoneal macrophages, was consistent with those of others (9, 46–48). However, bone marrow-derived macrophages have been frequently used in bacterial phagocytosis assays (3, 46). When bone marrow-derived macrophages were examined with FimH+ E. coli (3), the degree to which FimH+ E. coli cells were eliminated was found to be similar to that reported here (FimH− E. coli cells were not tested previously [3]).
Mouse genetic background had no effect on the in vitro bacteriocidal capacity of macrophages even though the strains of mice differ rather dramatically in their susceptibilities to gram-negative and gram-positive bacterial pathogens (33) (Table 1). We were somewhat surprised that there was no effect. However, susceptibility to bacterial infection, as it relates to macrophage bacteriocidal effectiveness, depends upon a number of factors, among them the type of bacteria under investigation (17, 51) and whether the bacteria are opsonized or not (1, 28, 60). Our results indicate that the innate susceptibility of mice to a variety of bacterial infections does not correlate with the ability of host macrophages to take up or kill unopsonized E. coli.
Whereas previous experiments have shown statistically significant differences between the survival of FimH+ E. coli and that of FimH− E. coli in resident peritoneal macrophages (25), we witnessed no such differences here. The assay conditions, the criteria used to assess killing, and the magnitude of the differences shown in this earlier report leave open the possibility that there may be little difference between the killing of FimH+ and FimH− E. coli once internalization has occurred. Consequently, our results support observations that piliation has little effect on the outcome of nonopsonic phagocytosis as far as killing of ingested bacteria is concerned (5, 6, 16, 22). Our results leave open the possibility that FimH+ and FimH− E. coli cells are directed to different vacuolar compartments once internalized (analogous to FimH+ and opsonized E. coli) (3) but indicate that this hypothetical difference in trafficking makes no effective difference in terms of killing, at least as measured by the methods we employed.
Whereas the killing of ingested bacteria was not influenced by the FimH+ or the FimH− phenotype, macrophage populations were found to have distinctly different binding properties when the ratios of FimH+ to FimH− E. coli bound were compared. We attribute these binding differences to different FimH receptor densities (3, 13) on the various macrophage populations relative to the densities of receptors that simply bind FimH− E. coli via another mechanism or mechanisms (e.g., via lipopolysaccharide [64]). Interestingly, the macrophage populations least effective in killing ingested E. coli exhibited the highest relative binding of FimH+ E. coli. The biological significance of this is unknown. Since a number of different receptors have been proposed to have a role in binding FimH on phagocytic cells (3, 14, 24, 52), it may be that certain macrophage populations vary in the expression of receptor type as well as density.
Two factors that have been shown to affect the fate of bacteria internalized by macrophages are (i) recombination proficiency, shown to be a factor in the survival of (opsonized) S. enterica serovar Typhimurium (10) and (ii) the exogenous addition of IFN-γ, shown to affect the fate of unopsonized L. monocytogenes (47) and opsonized S. enterica serovar Typhimurium (23, 48), as well as the phagocytosis of unopsonized E. coli (49, 50; killing was not measured in these reports). Neither of these factors appears to affect the rate or degree to which unopsonized E. coli cells (FimH+ or FimH−) were killed as measured by our methods, although control experiments showed that IFN-γ did significantly improve the degree to which unopsonized L. monocytogenes was eliminated, as has been previously reported (47). Understanding the reason for the differential effect of IFN-γ may aid in understanding the molecular mechanisms by which certain bacteria are able to thwart macrophage killing. Under our assay conditions, the addition of heat-inactivated 5% FBS to unelicited peritoneal macrophages had no significant effect upon the binding or subsequent killing of FimH+ E. coli as has been previously reported (3). It may be that FBS effects are manifested only under certain conditions or with certain macrophage populations.
There are many complex biochemical events involved in the internalization and killing of bacteria by phagocytes. In vitro assays to quantify bacterial killing by macrophages can provide useful insights into host-pathogen interactions. However, there are numerous variables in such assays that make comparisons from different laboratories difficult. In the present study, we systematically examined several of the principal assay variables for their effects on the fate of phagocytized, unopsonized E. coli. We found that several factors previously reported to influence the microbicidal activity of macrophages, such as the presence or absence of serum, activation of the macrophage by IFN-γ, recombination proficiency of the bacteria, FimH phenotype, and the mouse strain background of the macrophage population had no effect. Of relevance for future studies was our finding that the anatomical source of macrophages and their derivation significantly influenced bacteriocidal activity.
ACKNOWLEDGMENTS
We thank Craig Altier for a critical reading of the manuscript and helpful suggestions.
This work was supported by grant AI 222223 from the Public Health Service and the State of North Carolina.
REFERENCES
- 1.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson J A, Miller S I. Spacious phagosome formation within mouse macrophages correlates with Salmonella serotype pathogenicity and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archinal W A, Wilder M S. Susceptibility of HRS/J mice to listeriosis: macrophage activity. Infect Immun. 1988;56:613–618. doi: 10.1128/iai.56.3.613-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baorto D M, Gao Z, Malaviya R, Dustin M L, van der Merwe A, Lublin D M, Abraham S N. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature. 1997;389:636–639. doi: 10.1038/39376. [DOI] [PubMed] [Google Scholar]
- 4.Bloch C A, Stocker B A D, Orndorff P E. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol. 1992;6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 5.Blumenstock F, Jann K. Adhesion of piliated Escherichia coli strains to phagocytes: differences between bacteria with mannose-sensitive pili and mannose-resistant pili. Infect Immun. 1982;35:264–269. doi: 10.1128/iai.35.1.264-269.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boner G, Mhashilkar A M, Rodriguez-Ortega M, Sharon N. Lectin-mediated, nonopsonic phagocytosis of type 1 Escherichia coli by human peritoneal macrophages of uremic patients treated by peritoneal dialysis. J Leukoc Biol. 1989;46:239–245. doi: 10.1002/jlb.46.3.239. [DOI] [PubMed] [Google Scholar]
- 7.Briles D E, Lehmeyer J, Forman C. Phagocytosis and killing of Salmonella typhimurium by peritoneal exudate cells. 1981. Infect Immun. 1981;33:380–388. doi: 10.1128/iai.33.2.380-388.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinton C C., Jr The structure, function, synthesis and genetic control of bacterial pili and a molecular model of DNA and RNA transport in gram negative bacteria. Trans NY Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchmeier N A, Lipps C J, So M Y, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 11.Cheers C, McKenzie I C. Resistance and susceptibility of mice to bacterial infection: genetics of listeriosis. Infect Immun. 1978;19:755–762. doi: 10.1128/iai.19.3.755-762.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duguid J P, Old D C. Adhesive properties of Enterobacteriaceae. In: Beachey E H, editor. Bacterial adherence: receptors and recognition. Vol. 6. London, United Kingdom: Chapman and Hall; 1980. pp. 187–217. [Google Scholar]
- 13.Gbarah A, Gahmberg C G, Boner G, Sharon N. The leukocyte surface antigens CD11b and CD18 mediate the oxidative burst activation of human peritoneal macrophages induced by type 1 fimbriated Escherichia coli. J Leukoc Biol. 1993;54:111–113. doi: 10.1002/jlb.54.2.111. [DOI] [PubMed] [Google Scholar]
- 14.Gbarah A, Gahmberg C G, Ofek I, Jacobi U, Sharon N. Identification of the leukocyte adhesion molecules CD11 and CD18 as receptors for type 1-fimbriated (mannose-specific) Escherichia coli. Infect Immun. 1991;59:4524–4530. doi: 10.1128/iai.59.12.4524-4530.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gbarah A, Mirelman D, Sansonetti P J, Verdon R, Bernhard W, Sharon N. Shigella flexneri transformants expressing type 1 (mannose-specific) fimbriae bind to, activate, and are killed by phagocytic cells. Infect Immun. 1993;61:1687–1693. doi: 10.1128/iai.61.5.1687-1693.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz M B, Kuriyama S M, Silverblatt F J. Phagolysosome formation by polymorphonuclear neutrophilic leukocytes after ingestion of Escherichia coli that express type 1 pili. J Infect Dis. 1987;156:229–233. doi: 10.1093/infdis/156.1.229. [DOI] [PubMed] [Google Scholar]
- 17.Hancock G E, Schaedler R W, MacDonald T T. Yersinia enterocolitica infection in resistant and susceptible strains of mice. Infect Immun. 1986;53:26–31. doi: 10.1128/iai.53.1.26-31.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington-Fowler L, Henson P M, Wilder M S. Fate of Listeria monocytogenes in resident and activated macrophages. Infect Immun. 1981;33:11–16. doi: 10.1128/iai.33.1.11-16.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris S L, Elliott D A, Blake M C, Must L M, Messenger M, Orndorff P E. Isolation and characterization of mutants that have lesions affecting receptor-binding of type 1 pili in Escherichia coli. J Bacteriol. 1990;172:6411–6418. doi: 10.1128/jb.172.11.6411-6418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havell E A, Spitalny G L. The induction and characterization of interferon from pure cultures of murine macrophages. Ann NY Acad Sci. 1980;350:413–421. doi: 10.1111/j.1749-6632.1980.tb20643.x. [DOI] [PubMed] [Google Scholar]
- 21.Hernychova L, Kovarova H, Macela A, Kroca M, Krocova Z, Stulik J. Early consequences of macrophage-Francisella tularensis interaction under the influence of different genetic background in mice. Immunol Lett. 1997;57:75–81. doi: 10.1016/s0165-2478(97)00063-1. [DOI] [PubMed] [Google Scholar]
- 22.Iwahi T, Imada A. Interaction of Escherichia coli with polymorphonuclear leukocytes in pathogenesis of urinary tract infection in mice. Infect Immun. 1988;56:947–953. doi: 10.1128/iai.56.4.947-953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagaya K, Watanabe K, Fukazawa Y. Capacity of recombinant gamma interferon to activate macrophages for Salmonella killing activity. Infect Immun. 1989;57:609–615. doi: 10.1128/iai.57.2.609-615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson A, Carlsson S R, Dahlgren C. Identification of the lysosomal membrane glycoprotein Lamp-1 as a receptor for type-1-fimbriated (mannose-specific) Escherichia coli. Biochem Biophys Res Commun. 1996;219:168–172. doi: 10.1006/bbrc.1996.0200. [DOI] [PubMed] [Google Scholar]
- 25.Keith B R, Harris S L, Russell P W, Orndorff P E. Effect of type 1 piliation on in vitro killing of Escherichia coli by mouse peritoneal macrophages. Infect Immun. 1990;58:3448–3454. doi: 10.1128/iai.58.10.3448-3454.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koestler T P, Badger A M, Rieman D J, Greig R, Poste G. Induction by immunomodulatory agents of a macrophage antigen recognized by monoclonal antibody 158.2 and correlation with macrophage function. Cell Immunol. 1985;96:113–125. doi: 10.1016/0008-8749(85)90344-2. [DOI] [PubMed] [Google Scholar]
- 27.Leijh P C, van Zwet T L, ter Kuile M N, van Furth R. Effect of thioglycolate on phagocytic and microbicidal activities of peritoneal macrophages. Infect Immun. 1984;46:448–452. doi: 10.1128/iai.46.2.448-452.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lissner C R, Weinstein D L, O'Brien A D. Mouse chromosome I Ity locus regulates microbicidal activity of isolated peritoneal macrophages against a diverse group of intracellular and extracellular bacteria. J Immunol. 1985;135:544–547. [PubMed] [Google Scholar]
- 29.Lock R, Dahlgren C, Linden M, Stendahl O, Svensbergh A, Ohman L. Neutrophil killing of two type 1 fimbria-bearing Escherichia coli strains: dependence on respiratory burst activation. Infect Immun. 1990;58:37–42. doi: 10.1128/iai.58.1.37-42.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malaviya R, Ikeda T, Ross E, Abraham S N. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381:77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 31.Malaviya R, Ross E, Jakschik B A, Abraham S N. Mast cell degranulation induced by type 1 fimbriated Escherichia coli in mice. J Clin Investig. 1994;93:1645–1653. doi: 10.1172/JCI117146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malaviya R, Ross E A, MacGregor J I, Ikeda T, Little J R, Jakschik B A, Abraham S N. Mast cell phagocytosis of FimH-expression enterobacteria. J Immunol. 1994;152:1907–1914. [PubMed] [Google Scholar]
- 33.Malo D, Skamene E. Genetic control of host resistance to infection. Trends Genet. 1994;10:365–371. doi: 10.1016/0168-9525(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 34.Mauel J, DeFendi V. Infection and transformation of mouse peritoneal macrophages by simian virus 40. J Exp Med. 1971;134:335–350. doi: 10.1084/jem.134.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurer L M, Orndorff P E. Identification and characterization of genes determining receptor binding and pilus length of Escherichia coli type 1 pili. J Bacteriol. 1987;169:640–645. doi: 10.1128/jb.169.2.640-645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Murray E G D, Webb R A, Swann M B R. A disease of rabbits characterized by large mono-nuclear leukocytosis caused by a heretofore undescribed bacillus Bacterium monocytogenes. J Pathol Bacteriol. 1926;29:407–412. [Google Scholar]
- 38.O'Brien A D. Innate resistance of mice to Salmonella typhi infection. Infect Immun. 1982;38:948–952. doi: 10.1128/iai.38.3.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ofek I, Goldhar J, Keisari Y, Sharon N. Nonopsonic phagocytosis of microorganisms. Annu Rev Microbiol. 1995;49:239–276. doi: 10.1146/annurev.mi.49.100195.001323. [DOI] [PubMed] [Google Scholar]
- 40.Ofek I, Sharon N. Lectinophagocytosis: a molecular mechanism of recognition between cell surface sugars and lectins in the phagocytosis of bacteria. Infect Immun. 1988;56:539–547. doi: 10.1128/iai.56.3.539-547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohman L, Hed J, Stendahl O. Interaction between human polymorphonuclear leukocytes and two different strains of type-1 fimbriae-bearing Escherichia coli. J Infect Dis. 1982;146:751–757. doi: 10.1093/infdis/146.6.751. [DOI] [PubMed] [Google Scholar]
- 42.Orndorff P E, Bloch C A. The role of type 1 pili in the pathogenesis of Escherichia coli infections: a short review and some new ideas. Microb Pathog. 1990;9:75–79. doi: 10.1016/0882-4010(90)90081-z. [DOI] [PubMed] [Google Scholar]
- 43.Orndorff P E, Spears P A, Schauer D, Falkow S. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J Bacteriol. 1985;164:321–330. doi: 10.1128/jb.164.1.321-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 45.Ponniah S, Abraham S N, Endres R O. T-cell-independent stimulation of immunoglobulin secretion in resting human B lymphocytes by the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1992;60:5197–5203. doi: 10.1128/iai.60.12.5197-5203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Portnoy D A, Jacks P S, Hinrichs D J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portnoy D A, Schreiber R D, Connelly P, Tilney L G. Gamma interferon limits access of Listeria monocytogenes to the macrophage cytoplasm. J Exp Med. 1989;170:2141–2146. doi: 10.1084/jem.170.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Ralph P, Nakoinz I. Phagocytosis by a macrophage tumor and its clonal cell line. Nature. 1975;257:393–394. doi: 10.1038/257393a0. [DOI] [PubMed] [Google Scholar]
- 48.Riikonen P, Makela P H, Saarilahti H, Sukupolvi S, Taira S, Rhen M. The virulence plasmid does not contribute to growth of Salmonella in cultured murine macrophages. Microb Pathog. 1992;13:281–291. doi: 10.1016/0882-4010(92)90038-p. [DOI] [PubMed] [Google Scholar]
- 49.Rollag H, Degre M. Effect of interferon preparations on the uptake of non-opsonized Escherichia coli by mouse peritoneal macrophages. Acta Pathol Microbiol Scand Sect B. 1981;89:153–159. doi: 10.1111/j.1699-0463.1981.tb00169_89b.x. [DOI] [PubMed] [Google Scholar]
- 50.Rollag H, Degre M, Sonnenfeld G. Effects of interferon-alpha/beta and interferon-gamma preparation on phagocytosis by mouse peritoneal macrophages. Scand J Immunol. 1984;20:149–155. doi: 10.1111/j.1365-3083.1984.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 51.Sapru K, Stotland P K, Stevenson M M. Quantitative and qualitative differences in bronchoalveolar inflammatory cells in Pseudomonas aeruginosa-resistant and -susceptible mice. Clin Exp Immunol. 1999;115:103–109. doi: 10.1046/j.1365-2249.1999.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauter S L, Rutherfurd S M, Wagener C, Shively J E, Hefta S A. Identification of the specific oligosaccharide sites recognized by type 1 fimbriae from Escherichia coli on nonspecific cross-reacting antigen, a CD66 cluster granulocyte glycoprotein. J Biol Chem. 1993;268:15510–15516. [PubMed] [Google Scholar]
- 53.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. p. 102. [Google Scholar]
- 54.Silverblatt F J, Dreyer J S, Schauer S. Effect of pili on susceptibility of Escherichia coli to phagocytosis. Infect Immun. 1979;24:218–223. doi: 10.1128/iai.24.1.218-223.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverblatt F J, Ofek I. Interaction of bacterial pili and leukocytes. Infection. 1983;11:235–238. doi: 10.1007/BF01641208. [DOI] [PubMed] [Google Scholar]
- 56.Spears P A, Schauer D, Orndorff P E. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J Bacteriol. 1986;168:179–185. doi: 10.1128/jb.168.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spitalny G L. Dissociation of bactericidal activity from other functions of activated macrophages in exudates induced by thioglycolate medium. Infect Immun. 1981;34:274–284. doi: 10.1128/iai.34.1.274-284.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tewari R, MacGregor J I, Ikeda T, Little J R, Hultgren S J, Abraham S N. Neutrophil activation by nascent FimH subunits of type 1 fimbriae purified from the periplasm of Escherichia coli. J Biol Chem. 1993;268:3009–3015. [PubMed] [Google Scholar]
- 59.Valenti-Weigand P, Benkel P, Rohde M, Chhatwal G S. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect Immun. 1996;64:2467–2473. doi: 10.1128/iai.64.7.2467-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Dissel J T, Leijh P C J, van Furth R. Differences in initial rate of intracellular killing of Salmonella typhimurium by resident peritoneal macrophages from various mouse strains. J Immunol. 1985;134:3404–3410. [PubMed] [Google Scholar]
- 61.Weinstein D L, Lissner C R, Swanson R N, O'Brien A D. Macrophage defect and inflammatory cell recruitment dysfunction in Salmonella susceptible C3H/HeJ mice. Cell Immunol. 1986;102:68–77. doi: 10.1016/0008-8749(86)90326-6. [DOI] [PubMed] [Google Scholar]
- 62.Weinstein R, Silverblatt F J. Antibacterial mechanisms of antibody to mannose-sensitive pili of Escherichia coli. J Infect Dis. 1983;147:882–889. doi: 10.1093/infdis/147.5.882. [DOI] [PubMed] [Google Scholar]
- 63.Woodall L D, Russell P W, Harris S L, Orndorff P E. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli using a chromosomal lacUV5 promoter. J Bacteriol. 1993;175:2770–2778. doi: 10.1128/jb.175.9.2770-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright S D, Jong M T C. Adhesion-promoting receptors on human macrophages recognize Escherichia coli by binding to lipopolysaccharide. J Exp Med. 1986;164:1876–1888. doi: 10.1084/jem.164.6.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida S, Goto Y, Mizuguchi Y, Nomoto K, Skamene E. Genetic control of natural resistance in mouse macrophages regulating intracellular Legionella pneumophila multiplication in vitro. Infect Immun. 1991;59:428–432. doi: 10.1128/iai.59.1.428-432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]