SUMMARY
Somatic adult stem cell lineages in high-turnover tissues are under tight gene regulatory control. Like its mammalian counterpart, the Drosophila intestine precisely adjusts the rate of stem cell division with the onset of differentiation based on physiological demand. Although Notch signaling is indispensable for these decisions, the regulation of Notch activity that drives the differentiation of stem cell progenies into functional, mature cells is not well understood. Here, we report that commitment to the terminally differentiated enterocyte (EC) cell fate is under microRNA control. We show that an intestinally enriched microRNA, miR-956, fine-tunes Notch signaling activity specifically in intermediate, enteroblast (EB) progenitor cells to control EC differentiation. We further identify insensitive mRNA as a target of miR-956 that regulates EB/EC ratios by repressing Notch activity in EBs. In summary, our study highlights a post-transcriptional gene-regulatory mechanism for controlling differentiation in an adult intestinal stem cell lineage.
Graphical Abstract
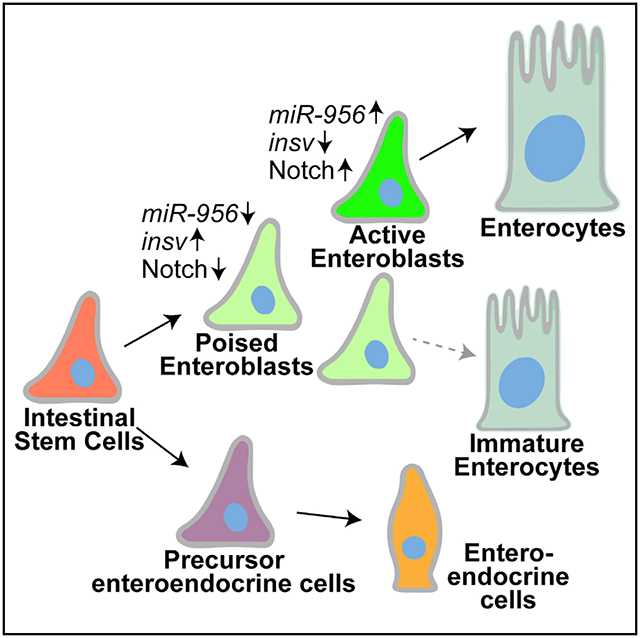
In brief
The regulatory mechanisms governing the choice between stem cell fate retention and the onset of differentiation is not well understood. Here, Mukherjee et al. describe that this decision is undertaken by a tissue enriched microRNA, miR-956, in the Drosophila midgut by suppressing the Notch inhibitor insensitive in stem cell progenies.
INTRODUCTION
Precise regulation of somatic stem cell activity is essential for the maintenance of tissue homeostasis, which, in turn, is crucial for ensuring fitness and long-term survival of an organism. Stem cell populations display remarkable plasticity that reflects a sensitive balance in their rates of self-renewal and differentiation (Morrison and Spradling, 2008). To retain constancy in cell numbers during tissue homeostasis, many adult stem cells divide asymmetrically to give rise to one stem cell that self-renews and another transient cell type that undergoes terminal differentiation (Knoblich, 2008). However, the mechanisms whereby stem cells dynamically regulate gene expression to maintain this balance between stem cell fate retention and terminal differentiation commitment are still not well understood. Understanding these basic mechanisms of stem cell biology can provide fundamental insights into tissue dysplasias that arise from aging as well as diseases including cancer (Clarke and Fuller, 2006).
The adult Drosophila intestinal epithelium has emerged as a premier model system for studying tissue homeostasis because it harbors a population of somatic stem cells with striking similarity to mammalian intestinal stem cells as well as a relatively simple, genetically tractable lineage (Ohlstein and Spradling, 2006; Casali and Batlle, 2009). The Drosophila intestine is maintained by a pool of mitotically active intestinal stem cells (ISCs) that give rise to two differentiated cell types, absorptive enterocytes (ECs) and secretory enteroendocrine cells (EEs) through distinct mechanisms (Biteau and Jasper, 2014; Wang et al., 2015). ECs are generated via the intermediate enteroblast (EB) progenitor cell type in a Notch-dependent manner (Ohlstein and Spradling, 2006; Micchelli and Perrimon, 2006), whereas EEs are generated directly from a separate population of stem cells called pre-EEs that express Prospero (Pros) (Bardin et al., 2010; Biteau and Jasper, 2014; Wang et al., 2015; Zeng and Hou, 2015). ISCs express the transmembrane protein Delta (Dl), which activates the Notch receptor in EBs to promote their terminal differentiation. Repression of Notch signaling in ISCs maintains stem cell fate and prevents differentiation, whereas high Notch activity in EBs promotes differentiation. While mechanisms that regulate ISC fate and EE specification have been extensively analyzed, the molecular mechanisms that regulate Notch signaling in EBs to drive EC differentiation are still unclear.
Recent studies report that EBs exhibit plasticity by coordinating their lineage differentiation rate to meet local demand (Antonello et al., 2015; Tang et al., 2021). In some cases, EBs delay terminal differentiation by undergoing a mesenchymal-to-epithelial transition (MET) that initiates a “paused” state. EBs are then activated to differentiate in response to local damage cues via miR-8, which antagonizes the transcription factor escargot (esg) and reverses the MET (Antonello et al., 2015). These dynamic “differentiation-poised” versus “differentiation-activated” differences in EB status have been demonstrated through intravital image tracing of the adult ISC lineage (Tang et al., 2021). Control of EB status likely involves Klumpfuss, a Notch-induced transcription factor that directs cell fate toward the EC lineage (Korzelius et al., 2019). EBs can also be culled via Diap1-induced apoptosis in order to maintain gut homeostasis (Reiff et al., 2019). These studies have shed light on how EB plasticity maintains overall tissue robustness in a high-turnover tissue such as the gut. However, the regulatory mechanisms that drive EC commitment in EBs during homeostasis are not well defined.
Here, we have characterized the function of an intestinally enriched microRNA (miRNA), miR-956, in regulating the onset of EB differentiation. We found that miR-956 is enriched in the progenitor cell population and maintains the balance of progenitor versus differentiated cell types by affecting EB-to-EC differentiation. We further show that miR-956 regulates EB-to-EC differentiation by suppressing Notch signaling in EBs via the target mRNA insensitive (insv). Thus, we identify a miRNA-mediated mechanism of regulating the commitment to EC differentiation and thereby tissue homeostasis in the Drosophila adult intestine.
RESULTS
miR-956 is highly enriched in the adult intestinal epithelium
We previously profiled small RNAs that were enriched in the Drosophila intestine and identified miR-956 as one of the most highly expressed intestinal miRNAs (Mukherjee et al., 2021). To verify the enrichment of miR-956 in the intestine, we used TaqMan real-time qPCR analysis to measure mature miR-956 levels in dissected intestinal tissues relative to intact whole animals. We also performed parallel analysis on adult “carcass” tissue, which we defined as the tissue remaining after the gastro-intestinal tract was removed. miR-956 displayed significantly higher expression in the intestinal tissue compared with whole animals and was hardly detectable in the carcass (Figure 1A). In contrast, the miR-210 control was absent from the intestine but detected in the carcass (Figure 1A), consistent with previous analysis (Weigelt et al., 2019).
Figure 1. miR-956 is enriched in Drosophila intestinal tissue.
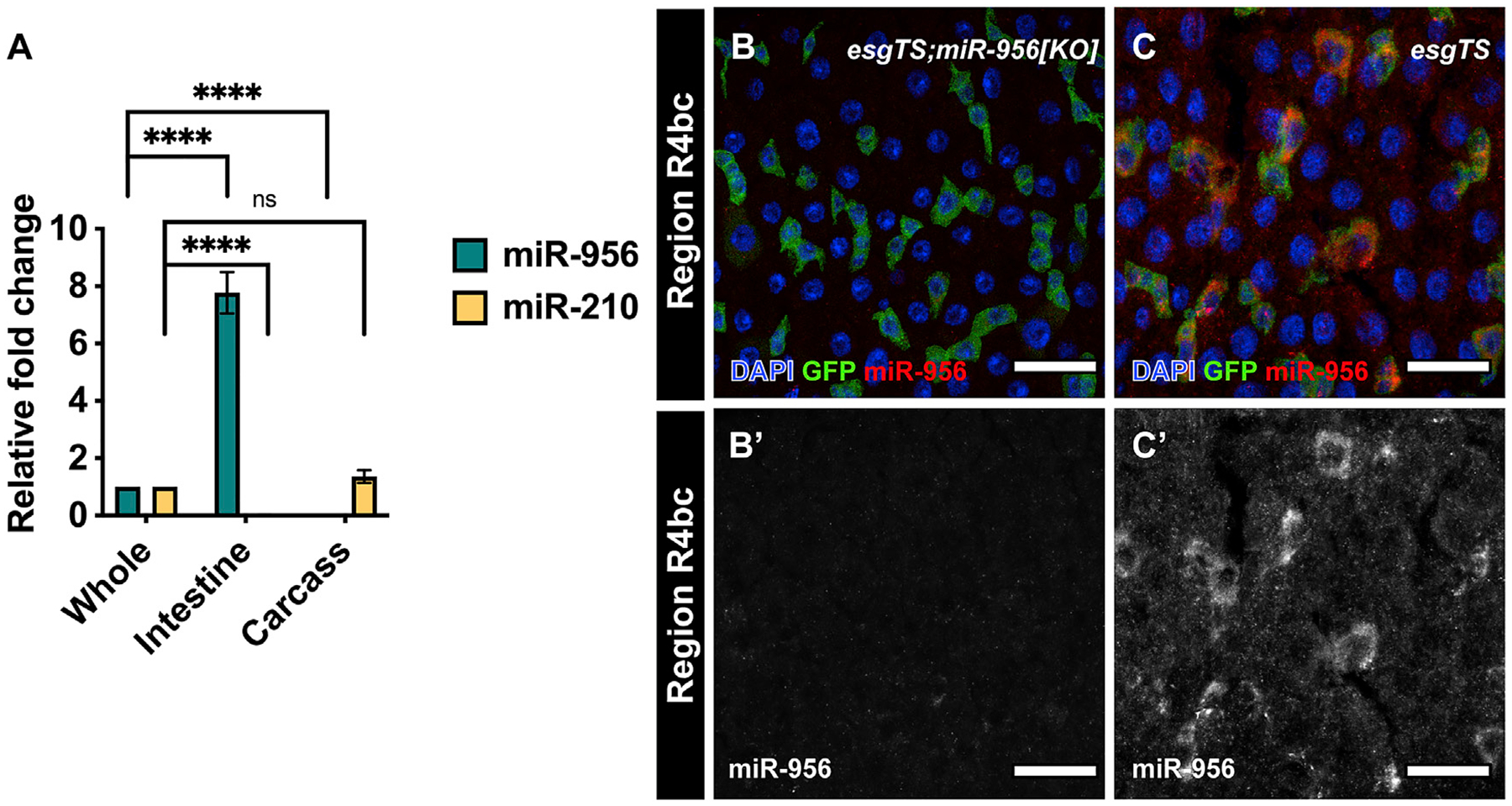
(A) qPCR analysis of miR-956 and miR-210 levels in intestinal tissues and carcass relative to whole animals. For each experiment, samples were collected from three separate animals in triplicates. Statistical significance of the difference in miRNA levels in intestinal and carcass samples relative to whole tissue is indicated. (B and C) esgTS-labeled progenitor cells (green) in (B) esgTS; miR-956[KO] mutants, which were used as a control, and (C) wild-type esgTS animals, with counterstaining of all cell nuclei (DAPI in blue) and miR-956 using RNA in situ probes (red). (B′–C′) Grayscale images of miR-956 RNA in situ. Scale bar, 25 μm. Data shown as mean ± SEM. Significance values: ns, not significant; ****p < 0.0001.
Despite its high expression levels in the Drosophila intestine, the role of miR-956 has not been investigated. To assess where it functions in the intestine, we analyzed its expression pattern with RNA in situ hybridization using a miR-956 probe. Signal for this probe was undetectable in the intestines of a miR-956 [KO] loss-of-function allele (Chen et al., 2014), which also lacked miR-956 RNA according to TaqMan qPCR analysis (Figures 1B, S1A, and S1B). While in situ analysis detected very high miR-956 levels in all cell types in most regions of control midguts (Figure S1C), we noticed that miR-956 was particularly enriched in a subset of cells in posterior midgut regions R4bc and R5. To identify this cell population, we performed in situ analysis on intestines in which progenitor cells were marked with GFP driven by a conditional combination of the progenitor-specific P {GawB}NP5130 driver and the ubiquitously expressed, temperature-sensitive GAL4 repressor P{tubP-GAL80[ts]}, referred to as esg-GAL4TS (Micchelli and Perrimon, 2006). In situ and GFP signals strongly overlapped, indicating that miR-956 was enriched in progenitor cells including those in region R4bc (Figure 1C). For this and future experiments involving esg-GAL4TS-driven GFP, we refer to GFP-negative cells as differentiated cells. Since stem cells in the R4bc region have been previously shown to be more active than those in other regions (Buchon et al., 2013), this expression pattern suggested that miR-956 may be particularly important in responding to proliferation and/or differentiation cues.
miR-956 maintains the balance between progenitor and differentiated cells
To determine the function of miR-956 in progenitor cells, we knocked it down using esg-GAL4TS and an available miR-956 sponge strain (Fulga et al., 2015). The miR-956 sponge strain, referred to as UAS-miR-956sp, harbors two copies of an mCherry-encoding transgene that contains 20 3′ UTR-located, antisense miR-956 sequences and is under upstream activation sequence (UAS) control. Esg-GAL4TS-driven sponge expression led to a significant increase in the proportion of GFP-positive progenitor cells and a concomitant decrease in the proportion of GFP-negative differentiated cells (Figures 2A, 2B, 2D, and 2E). These proportions were quantified as the ratio of the number of each individual cell type per the total number of cells per field of view. We also quantified the proportion of EEs and ECs using Pros and Pdm1 antibodies, respectively, and saw no significant change in EE numbers but a significant reduction in EC cell numbers (Figure S2). In contrast, we found that overexpression of miR-956 in the progenitors had the opposite effect: it decreased the number of GFP-positive progenitor cells and increased the number of GFP-negative differentiated cells (Figures 2A, 2C, 2D, and 2E).
Figure 2. miR-956 balances the proportion of progenitor cells to differentiated cells.
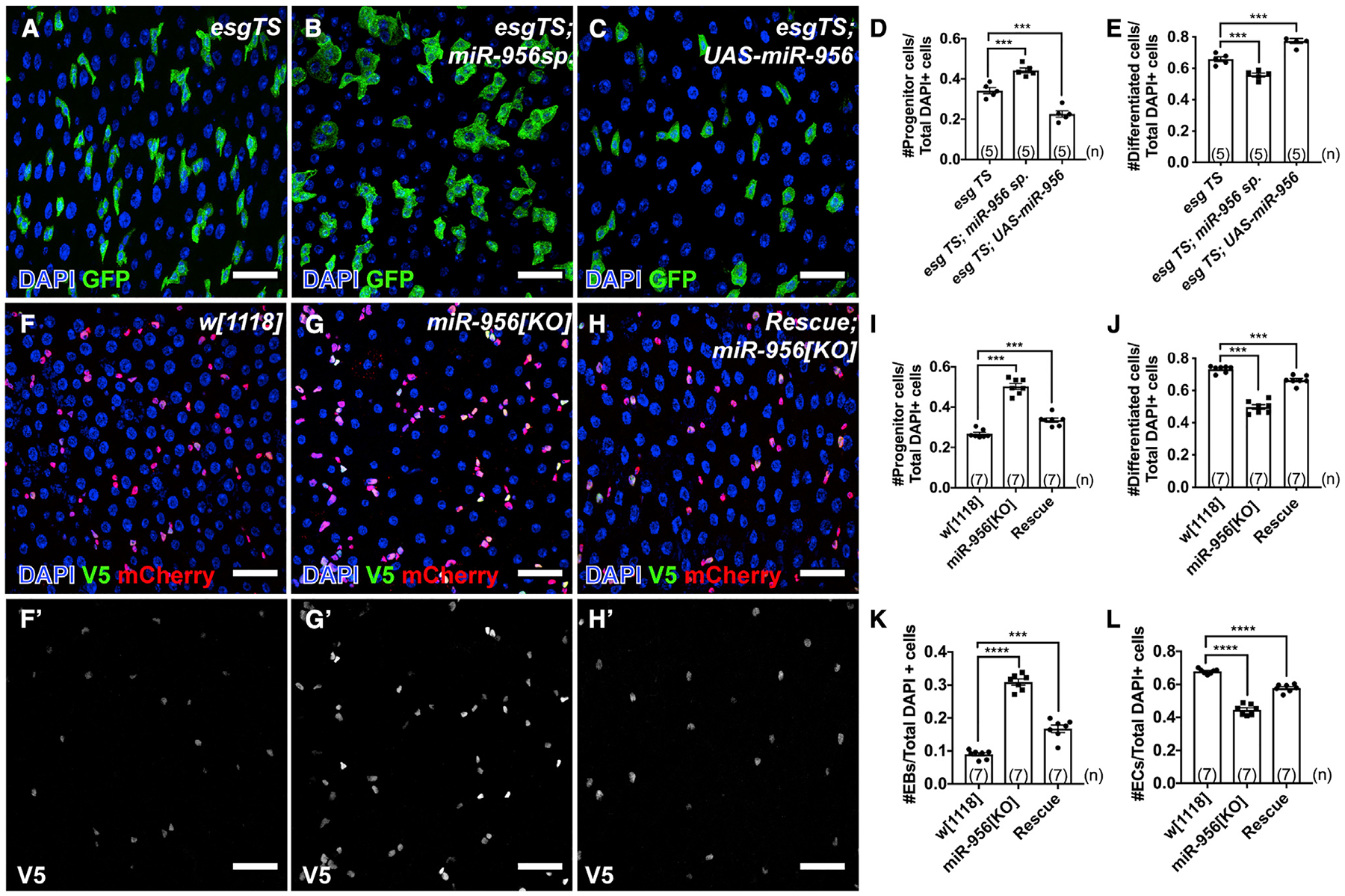
(A–C) esgTS-labeled progenitor cells (green) in (A) esgTS, (B) esgTS; UAS-miR-956sp, and (C) esgTS; UAS-miR-956 midguts counterstained for all cell nuclei (DAPI in blue).
(D) Quantification of progenitor cell numbers in esgTS, esgTS; miR-956sp and esgTS, UAS-miR-956 animals.
(E) Quantification of GFP-negative differentiated cell numbers in esgTS, esgTS; miR-956sp and esgTS; UAS-miR-956 animals.
(F–H) Midguts in (F) wild-type(w[1118]), (G) miR-956[KO], and (H) mutants rescued with miR-956 rescue transgene counterstained for progenitors (anti-mCherry in red), EBs marked using Su(H)-GBE-V5 (anti-V5 in green), and all cell nuclei (DAPI in blue). (F′–H′) Grayscale images of indicated channels from (F)–(H).
(I–L) Quantification of (I) progenitor cells, (J) mCherry-negative differentiated cells, and (K) EB and (L) EC numbers in miR-956[KO] mutants compared with wild type and rescued controls.
Data shown as mean ± SEM. Significance values: ***p < 0.001; ****p < 0.0001. Scale bar, 25 μm. n values in the graphs indicate the number of intestines.
To verify these results, we looked at the proportion of cell types in the intestinal epithelium of homozygous miR-956[KO] mutants that survived to adulthood. Consistently, miR-956[KO] mutants displayed a significant increase in the number of progenitor cells, which were quantified using the mira-His2A. mCherry.HA (Miller et al., 2020) reporter (Figures 2F, 2G, and 2I). To determine whether progenitor subtypes were differentially affected by this expansion, we quantified both ISC and EB numbers in wild type and miR-956[KO] mutants. EBs were identified by the presence of both the V5-epitope tagged, EB-specific marker 3Xgbe-smGFP.V5.nls (Buddika et al., 2020) and mira-His2A.mCherry.HA, while ISCs were identified by the presence of mira-His2A.mCherry.HA and the absence of 3Xgbe-smGFP.V5.nls. We found that the number of EBs, but not ISCs, was significantly increased in miR-956[KO] mutants (Figures 2F, 2G, 2K, and S3A). These 3Xgbe-smGFP.V5.nls-positive cells did not express the ISC-specific marker Dl, confirming that they were EBs and not ISCs with increased Notch activity (Figure S3B). miR-956[KO] mutants also had an associated decrease in the number of mira-His2A.mCherry.HA-negative, differentiated cells (Figures 2F, 2G, and 2J). To characterize this cell class, we distinguished ECs and EEs based on their ploidy and Pros expression and found that only the EC numbers were significantly decreased (Figures 2F, 2G, 2L, and S3C–S3E). While the nuclei of mutant ECs were larger than progenitor cells and therefore easily identifiable, we also noted that mutant EC nuclei were smaller than controls (Figures S3F–S3H). We verified that these cells with smaller nuclei were indeed ECs by staining for the EC-specific marker Pdm1 and also found that they displayed a significant reduction in DAPI fluorescence intensity relative to controls, suggesting a reduction in ploidy (Figure S3I). We further verified that none of the 3Xgbe-smGFP.V5.nls-positive EB cells showed ectopic Pdm1 signal in wild type and miR-956 [KO] mutants (Figures S3F and S3G). These miR-956[KO] mutant phenotypes were substantially rescued by a miR-956 rescue transgene that harbored a 3-kb fragment that spanned the miR-956 gene locus (Figures 2F–2L, S1A, and S3A). The increase in EB number together with the decrease in EC number and size suggested that loss of miR-956 disrupted the EB-to-EC transition.
miR-956 promotes EB-to-EC differentiation
To test whether miR-956 acted in progenitor cells to control EB number, we depleted miR-956 in progenitor cells and quantified the number of EBs. To do so, we used a second conditional progenitor GAL4-driver, the I-KCKT-GAL4TS driver (Buddika et al., 2021), that, like esgTS, is specific to adult progenitor cells but was already combined with 3Xgbe-smGFP.V5.nls to enable EB quantification. The I-KCKT system consists of a progenitor-specific Gal4 driver and a UAS-GFP responder that can be activated by an independent transgene (Figure 3A). Driven by I-KCKT-GAL4TS, UAS-miR-956sp led to a significant increase in the number of EBs (Figures 3B–3D). Confirming that this expansion was cell autonomous, UAS-miR-956sp driven specifically in adult EBs using a temperature-sensitive EB-specific Gal4 driver called Gbe-GAL4TS (Zeng et al., 2010) led to a significant expansion in EB numbers (Figures 3E–3G).
Figure 3. miR-956 promotes EB-to-EC differentiation.
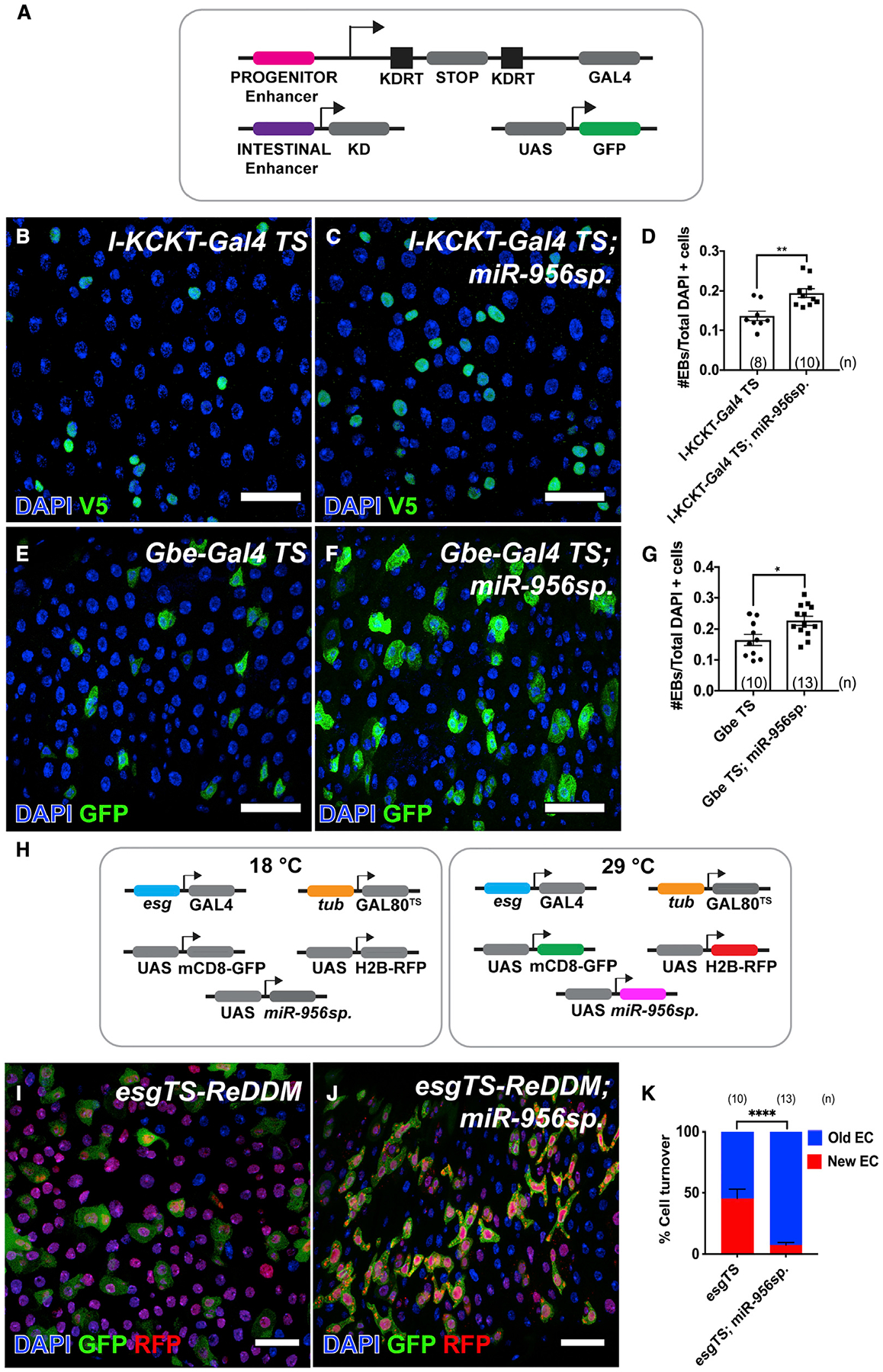
(A) Schematic of the I-KCKT system.
(B and C) Midguts showing EBs marked using Su(H)-GBE-V5 (anti-V5 in green) in (B) I-KCKT-Gal4 TS, and (C) I-KCKT-Gal4TS; UAS-miR-956sp animals counterstained for all cell nuclei (DAPI in blue).
(D) Quantification of EB numbers in I-KCKT-Gal4 TS and I-KCKT-Gal4TS; UAS-miR-956.sp midguts.
(E and F) Gbe-Gal4 TS labeled EBs (green) in (E) Gbe-Gal4 TS, and (F) Gbe-Gal4 TS; UAS-miR-956sp midguts counterstained for all cell nuclei (DAPI in blue).
(G) Quantification of EB numbers in Gbe-Gal4 TS and Gbe-Gal4TS; UAS-miR-956sp midguts.
(H) Schematic of the ReDDM system.
(I and J) EC turnover analysis using (I) esgTS-ReDDM and (J) esgTS-ReDDM; UAS-miR-956sp midguts counterstained for all cell nuclei (DAPI in blue); GFP and RFP reporters in green and red, respectively.
(K) Quantification of percentage of cell turnover (red ECs/unlabeled DAPI + ECs) in esgTS-ReDDM and esgTS-ReDDM; UAS-miR-956sp midguts.
Data shown as mean ± SEM. Significance values: *p < 0.1; **p < 0.01; ****p < 0.0001. Scale bar, 25 mm. n values in the graphs indicate the number of intestines.
In order to track the process of differentiation in miR-956 mutants, we used a fluorescence reporter-based lineage tracing analysis method called repressible dual differential stability cell marker (ReDDM) (Antonello et al., 2015). This method relies on the esgReDDM strain that combines the conditional esg-GAL4TS driver with two UAS-responsive fluorescence reporters with short (mCD8-GFP) and long (H2B-RFP) half-lives (Figure 3H). The esg-GAL4TS driver can be activated in adult ISCs and their intermediate EB progeny by a temperature shift that label progenitor cells in both red and green. Although the GAL4 driver is not transcribed in differentiated ECs and EEs, the red H2B-RFP reporter persists in these newly differentiated cell populations because of its longer half-life, whereas the short-lived green mCD8-GFP does not. Older differentiated cells that arose before GAL4 activation can also be distinguished by the absence of RFP. After GAL4 activation, therefore, the ratio of red-labeled to unlabeled cells is a readout of the rate of differentiation of progenitor into EC cells and EEs. We combined esgReDDM with UAS-miR-956sp and quantified cell turnover as the ratio of newly RFP-labeled ECs to older, unlabeled differentiated ECs at 7 days post-induction and performed the same analysis in esgReDDM control midguts. UAS-miR-956sp displayed both an accumulation of dual GFP/RFP-positive clusters of progenitor cells and a decrease in the proportion of newly generated differentiated cells relative to control (Figures 3I–3K). This decrease in newly differentiated ECs combined with the expansion in the EB population suggested that loss of miR-956 inhibits EB-to-EC differentiation. Collectively, our results indicated that miR-956 acted in EBs to drive their differentiation into ECs.
miR-956 regulates notch signaling in EBs
We next sought to determine how miR-956 promoted the EB-to-EC transition. The central pathway that dictates EB-to-EC differentiation is the highly conserved Notch signaling pathway (Micchelli and Perrimon, 2006). ISCs express the Notch ligand Dl, which activates Notch via juxtracrine signaling in nearby EBs (Ohlstein and Spradling, 2006). The level of Notch signaling is asymmetric in ISC/EB pairs: high in EBs and low in ISCs (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Low Notch activity is maintained in ISCs by transcriptional repression of Notch target genes via Hairless, whereas high Notch activity in EBs promotes EC differentiation (Bardin et al., 2010; Ohlstein and Spradling, 2006). Loss of Notch has also been associated with the formation of tumor-like clones containing both ISCs and EEs and reduced ECs, suggesting differential cell-type-specific roles in ISCs versus EBs (Ohlstein and Spradling, 2006). In miR-956 mutant animals, we did not observe an increase in ISC or EE numbers but rather an expansion in EBs and reduced ECs, which could be due to defects in Notch signaling specifically in EBs.
To determine whether the Notch signaling pathway was specifically affected in EBs, we compared Notch activity in control and miR-956[KO] mutants using 3Xgbe-smGFP.V5.nls. This reporter is not only an EB cell-type marker but also a Notch signaling reporter because it contains a Notch-responsive enhancer (NRE) element. We simultaneously marked ISC/EBs by staining with antibodies against horseradish peroxidase (HRP), known to mark intestinal progenitor cells as well as neural tissue (Haase et al., 2001; Jan and Jan, 1982; O’Brien et al., 2011). To rigorously compare reporter expression between genotypes, we combined wild-type and mutant tissues in the same tube for fixation and processing. Resulting tissues harbored cells with a range of low-to-high smGFP.V5 expression levels, reflecting EBs with varying levels of Notch activity (Figures 4A and 4B). Average fluorescence intensity of smGFP.V5 levels was significantly lower in miR-956[KO] mutants relative to control animals (Figure 4C). This indicated that miR-956 promoted Notch signaling in EBs, raising the possibility that reduced Notch activity caused the changes in EB/EC cell numbers in miR-956 mutants. To address whether miR-956 directly promoted Notch activity, we compared miR-956 expression levels, detected using RNA in situ, with the levels of the NRE GFP reporter. We found that most cells with high miR-956 expression displayed weak GFP expression (Figure S4A, yellow arrowheads), and vice versa (Figure S4A, white arrowheads).
Figure 4. miR-956 regulates the Notch signaling pathway via insv.
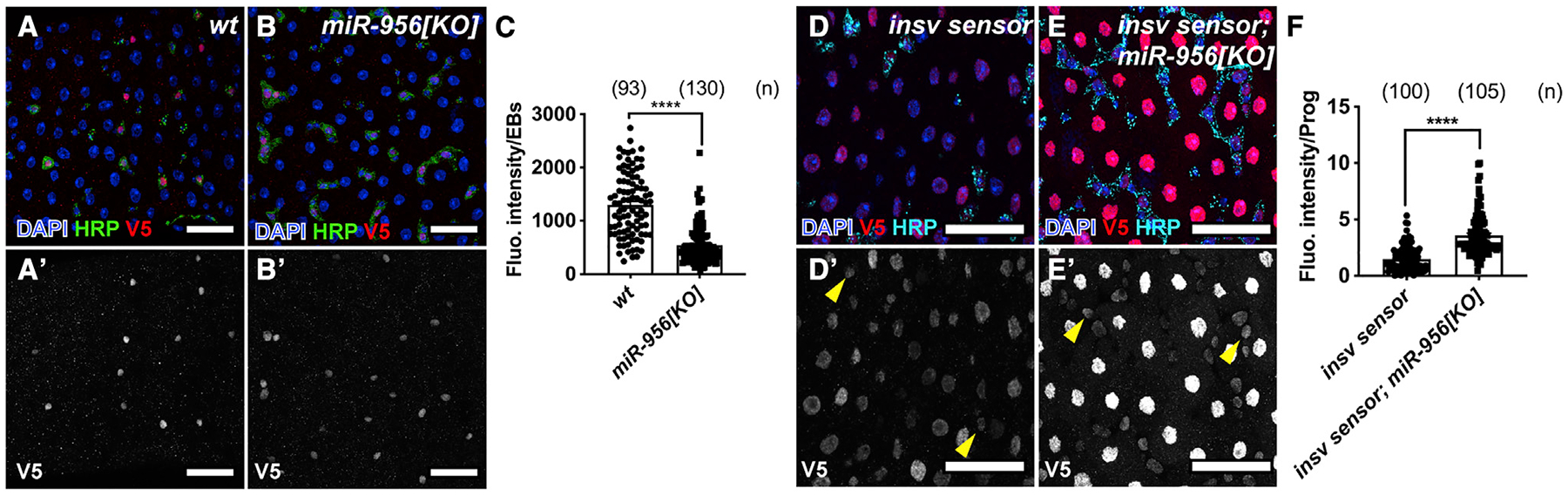
(A and B) Notch signaling reporter expression (anti-V5 in red) in (A) wild type and (B) miR-956[KO] mutants counterstained for progenitors in HRP (green) and all cell nuclei (DAPI in blue). (A′–B′) Grayscale images of Notch signaling reporter in wild type versus miR-956[KO] mutants.
(C) Fluorescence intensity of Notch reporter expression in EBs in wild type versus miR-956[KO] mutants.
(D and E) Midguts from (D) control or (E) miR-956[KO] mutants stained for smGFP.V5.insv 3′UTR (red), progenitors in HRP (cyan), and all cell nuclei (DAPI in blue).
(D′–E′) Grayscale images of indicated channels from (D) and (E). Progenitor cells are labeled with yellow arrowheads.
(F) Fluorescence intensity of V5 reporter expression in progenitors of control and miR-956[KO] mutants stained for smGFP.V5.insv 3′UTR (red).
Data shown as mean ± SEM. Significance values: ****p < 0.0001. Scale bar, 25 mm. n values in the graphs indicate the number of cells quantified from at least five intestines.
To confirm that Notch activity was affected by loss of miR-956, we measured the expression of downstream Notch targets encoded by the Enhancer of split-Complex (E(spl)-C) in miR-956[KO] mutants relative to control animals using qRT-PCR. Of the seven bHLH repressors that are directly activated during Notch signaling by Su(H) (Bailey and Posakony, 1995; Lecourtois and Schweisguth, 1995) that we tested, six were significantly downregulated in the mutants (Figure S4B). Thus, the combined reporter, in situ analysis, and qRT-PCR results indicated that loss of miR-956 led to downregulation of Notch signaling activity.
miR-956 regulates Notch signaling via insv
To identify mRNA targets of miR-956 that might be relevant to Notch signaling, we used TargetScan (Agarwal et al., 2015) to search for mRNAs with predicted miR-956 binding sites. Of the 87 predicted targets that are conserved across Drosophila species, insv stood out because it is known to downregulate Notch signaling (Reeves and Posakony, 2005; Duan et al., 2011). Insv is a member of the BAN, E5R, and NAC1 (BEN) domain-containing family of proteins and acts with Su(H) to repress Notch signaling during peripheral nervous system development (Duan et al., 2011). Ectopic Insv has been associated with multiple Notch loss-of-function phenotypes and represses an array of E(spl)-C target genes (Duan et al., 2011).
To investigate whether miR-956 promoted Notch signaling in EBs by repressing insv, we evaluated Insv protein levels in miR-956[KO] animals using an anti-Insv antibody (Duan et al., 2011). Insv protein levels were higher in the miR-956 mutant relative to controls (Figures S4C and S4D), indicating that miR-956 repressed Insv under wild-type conditions. Furthermore, esg-GAL4TS-driven expression of an UAS-insv transgene led to phenotypes that were very similar to those associated with miR-956 loss, including an increase in the proportion of GFP-positive progenitor cells and a decrease in the proportion of GFP-negative differentiated cells (Figures S5A–S5D). In order to distinguish between the role of elevated Insv in ISCs versus EBs, we drove UAS-insv in either ISCs or EBs using cell-specific Gal4 driver ISC-KCKT-GAL4TS (Buddika et al., 2021) or Gbe-GAL4TS (Zeng et al., 2010), respectively. Expression of insv in ISCs did not lead to any significant changes in ISC/EB numbers, whereas expression in EBs led to a significant expansion in EBs and a concomitant reduction in EC numbers (Figures S5E–S5L). This further suggested that insv specifically acted in EBs to regulate EB-to-EC differentiation.
To test whether miR-956 regulated insv via the predicted binding sites in its 3′ UTR, we prepared an insv 3′ UTR sensor containing the 3′ UTR downstream of a V5-tagged nuclear protein expressed under the control of an intestine-specific enhancer (Nern et al., 2015; Buddika et al., 2021). V5 staining intensity was significantly higher in the miR-956[KO] mutant intestine compared with control intestines (Figures 4D and 4E). To investigate whether miR-956 regulated insv in progenitor cells, we quantified V5 intensity in HRP-positive progenitor cells and found that it was significantly higher in the mutant background (Figure 4F). We also observed higher reporter expression in ECs in the mutant, suggesting that, despite the low levels of miR-956 detected in ECs by in situ hybridization, it is also active in those cells. To test whether insv plays a role in ECs, we crossed an insv RNAi line to the EC-specific P{GawB}Myo31DFNP0001 driver (Jiang and Edgar, 2009) and found no effect on EC numbers (Figures S5M–S5O). Additionally, UAS-miR-956 expression in ECs suppressed insv 3′ UTR reporter expression (Figures S4E–S4G). Overall, our data suggested that miR-956 promoted Notch signaling activity in progenitor cells by repressing insv.
miR-956 promotes EB-to-EC differentiation by regulating Notch signaling activity via insv
These results suggested that miR-956 mutant defects in EB-to-EC differentiation were caused by inappropriately elevated insv levels. We therefore tested whether reduction in insv gene dosage could suppress miR-956 mutant defects. We modified insv dosage by incorporating one copy of the verified loss-of-function insv23B allele (Reeves and Posakony, 2005) into the miR-956 homozygous mutant background. Loss of one copy of insv dominantly suppressed three miR-956 phenotypes: (1) the increase in the number of EBs (Figure 5A), (2) the decrease in number of ECs (Figure 5B), and (3) the reduction in the level of the Notch activity reporter (Figures 5C and S6). These results indicated that elevated Insv levels in the miR-956 mutant repressed Notch signaling and regulated EB/EC ratios.
Figure 5. miR-956 promotes EB-to-EC differentiation by regulating the Notch signaling activity.
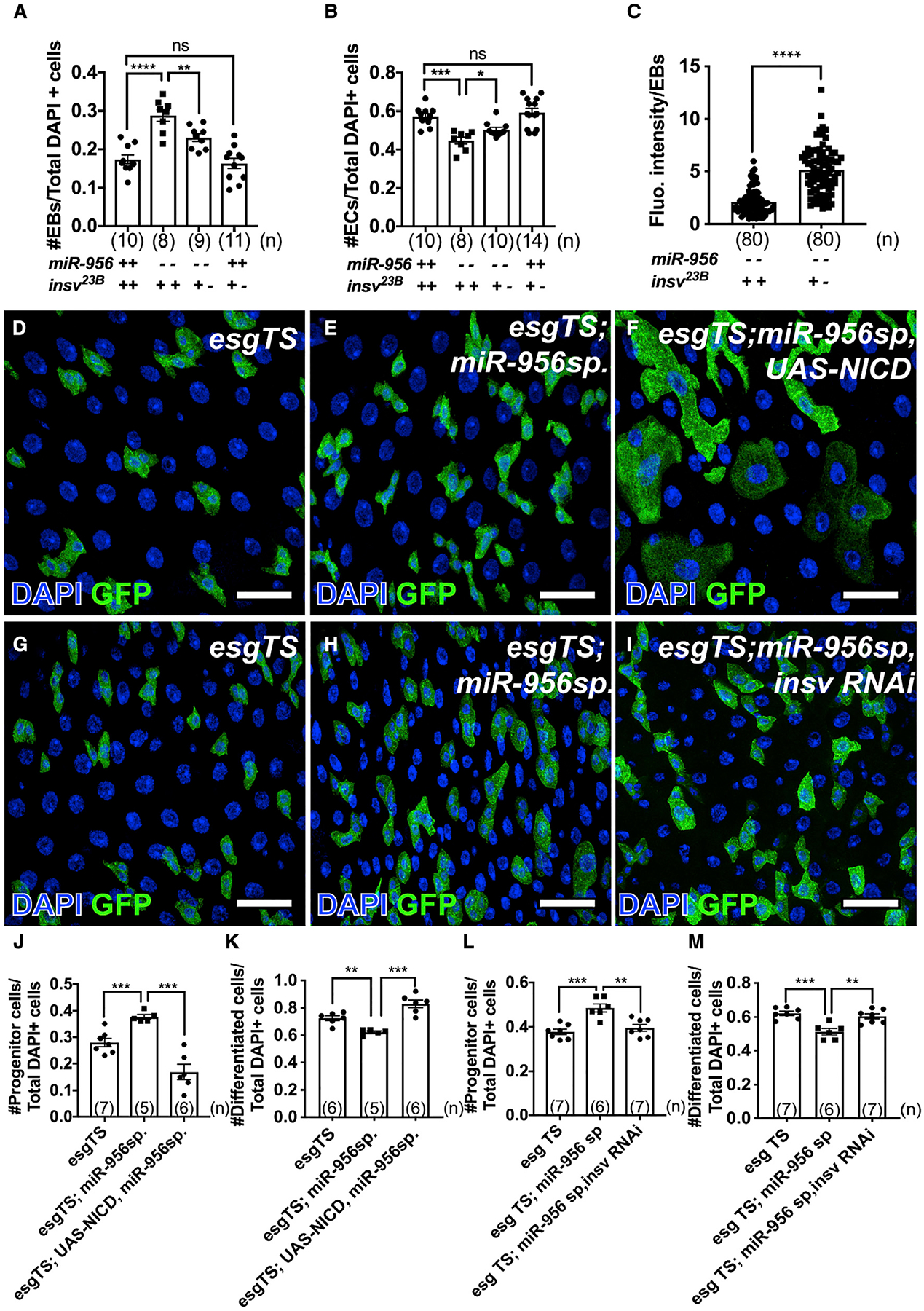
(A) Quantification of EB numbers in control or miR-956[KO] mutants that harbor two or one wild-type alleles of insv.
(B) Quantification of EC numbers in control or miR-956[KO] mutants that harbor two or one wild-type alleles of insv.
(C) Fluorescence intensity of Notch reporter expression in EBs in miR-956[KO] mutants that harbor two versus one wild-type alleles of insv.
(D–F) esgTS-labeled progenitor cells (green) in (D) esgTS, (E) esgTS; UAS-miR-956sp, and (F) esgTS; UAS-miR-956sp, UAS-NICD midguts counterstained for all cell nuclei (DAPI in blue).
(G–I) esgTS-labeled progenitor cells (green) in (G) esgTS, (H) esgTS; UAS-miR-956sp, and (I) esgTS; UAS-miR-956sp, insv RNAi midguts counterstained for all cell nuclei (DAPI in blue).
(J and K) Quantification of (J) progenitor cell numbers and (K) GFP-negative differentiated cell numbers in esgTS, esgTS, UAS-miR-956sp and esgTS, UAS-miR-956sp, UAS-NICD animals.
(L and M) Quantification of (L) progenitor cell numbers and (M) GFP-negative differentiated cell numbers in esgTS, esgTS, UAS-miR-956sp and esgTS, UAS-miR-956sp, insv RNAi animals.
Data shown as mean ± SEM. Significance values: n.s., not significant; *p < 0.1; **p < 0.01; ***p < 0.001; ****p < 0.0001. Scale bar, 25 mm. n values in the graphs indicate the number of intestines.
These results suggested that miR-956, insv, and Notch interacted to promote differentiation and thereby regulated tissue homeostasis. To address that question, we first tested whether activation of Notch signaling in progenitors that also lacked miR-956 activity could override the differentiation defects that was observed in animals expressing miR-956sp. We found that activation of Notch via the expression of Notch intracellular domain (NICD), the active form of Notch, in progenitors could force the progenitors to differentiate (Figures 5D–5F, 5J, and 5K). This result is consistent with our hypothesis that differentiation defects in miR-956 loss-of-function conditions was due to reduced Notch activity. Finally, to further corroborate whether miR-956 acted through insv to promote differentiation, we knocked down insv in progenitor cells that also lacked miR-956 activity and found that it suppressed miR-956sp defects (Figures 5G–5I, 5L, and 5M). Together, these experiments showed that suppression of insv increased Notch signaling activity in miR-956 mutants and that de-repression of Notch signaling rescued miR-956-associated defects. Thus, this study showed that miR-956 was crucial for adjusting Notch signaling activity in intestinal progenitor cells via insv and thereby regulating the timing of differentiation during intestinal tissue homeostasis.
DISCUSSION
In this study, we have uncovered that a tissue-enriched miRNA, miR-956, promotes Notch signaling in EBs and thereby regulates EB-to-EC differentiation in the Drosophila adult intestine. Our data showed that miR-956 is highly enriched in the active progenitor cells of the posterior midgut region and boosts Notch signal in EBs, most likely via repression of insv. In the absence of miR-956, this balance is perturbed due to elevated Insv, which is known to interact with Su(H) to downregulate Notch signaling (Duan et al., 2011). Thus, miR-956 contributes to the decision between the retention of progenitor fate and the differentiation into a terminal cell type and is indispensable for maintaining tissue homeostasis.
Although miR-956 is enriched in both ISCs and EBs in the posterior midgut region, miR-956 does not appear to regulate ISC cell fate. To account for this, we propose a model whereby miR-956 also regulates insv in ISCs but is dispensable for maintaining ISC fate due to functional redundancy with Hairless, a critical inhibitor of Notch signal in ISCs (Figure 6). Under wild-type conditions, Hairless binds to its cofactor Su(H) and transcriptionally represses Notch target genes (Bardin et al., 2010). Hairless potentially compensates for lack of Insv activity in ISCs, enabling the retention of ISC fate and self-renewal activity. This is in accordance with a previous study, which showed that elevated Insv could rescue hairless-null phenotypes and antagonize Notch independently of Hairless (Duan et al., 2011). We predict that in the absence of miR-956 activity, high levels of Insv supplements Hairless function in ISCs to ensure low Notch activity and maintain ISC cell fate.
Figure 6. Model.
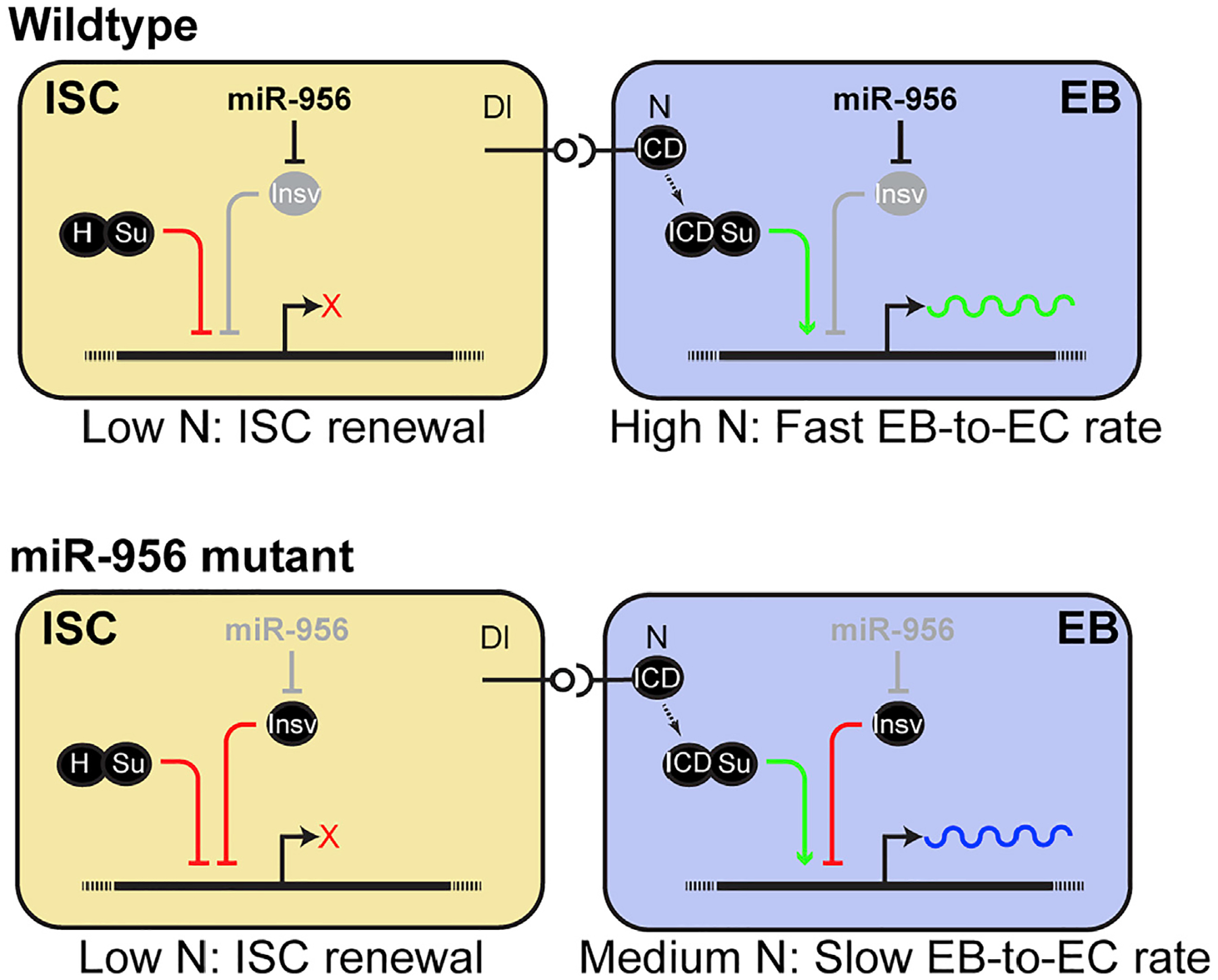
miR-956 represses insv to promote Notch signaling and modulates EB differentiation.
See discussion for details.
On the other hand, there are high levels of Notch activity in EBs due to the activation of Notch signaling in these cells. Previous work indicated that, upon activation, Notch is cleaved and that its active form, NICD, binds to Su(H), replacing Hairless, in EBs (Bardin et al., 2010). Our model predicts that under wild-type conditions, miR-956 reinforces high Notch activity in EBs by repressing a Notch inhibitor, Insv (Figure 6). High Notch activity in EBs ensures faster EB-to-EC turnover. We propose that in the absence of miR-956 activity, high levels of Insv in EBs outcompetes NICD for binding to its cofactor Su(H) and thereby represses Notch target genes. Moderate Notch activity in EBs, in turn, slows down EB-to-EC differentiation, resulting in aberrant EB/EC ratios. This is consistent with the observation that miR-956 expression is higher in the more active progenitor cells of the posterior midgut region that are presumably differentiating faster (Buchon et al., 2013). miR-956 function is thus crucial for maintaining EB plasticity by buffering Notch signaling levels and thereby rate of EC turnover.
We also noticed weak expression of miR-956 in ECs, which in turn possibly regulates Insv expression in ECs. However, the functional relevance of miR-956 in ECs is unclear. In addition, we also observed that in miR-956[KO] mutants, ECs have significantly smaller nuclei. Reduced EC nuclear size in miR-956 [KO] mutants might reflect the Notch signaling defects in EBs that prevent them from fully differentiating into mature ECs. Alternatively, it could also be due to a cell-autonomous effect of miR-956 activity in endocycling ECs that results in lower DNA content and nuclear size. We predict that miR-956-mediated repression of insv in ECs is not a major regulatory mechanism because (1) miR-956 levels are much lower in ECs compared with progenitor cells, and (2) Insv directly interacts with Su(H) to modulate Notch activity, which is absent in differentiated ECs.
The Notch signaling pathway is highly dose sensitive, and mild changes in signal levels can have profound developmental consequences (Lai, 2004). miRNA-mediated regulation of Notch signaling effectors has been shown to impact cell fate transitions in many Drosophila tissues such as ovaries, wings, and the nervous system (Caygill and Brand, 2017; Kavaler et al., 2018; Lai et al., 2005; Yatsenko and Shcherbata, 2018). Our study, however, has identified a miRNA-mediated circuit in the highly adaptive intestinal tissue that functions in a more context-dependent manner rather than being hardwired during development. We showed that subtle changes in insv can suppress strong miR-956 loss-of-function phenotypes; this miR-956/insv circuit is likely used as a mechanism by the tissue to adjust its rate of EC turnover to meet tissue demand. Thus, miR-956 may have evolved as an intestine-specific adaptation to control Notch pathway and thereby maintain overall tissue robustness.
The role of miRNAs in the Drosophila intestinal tissue are only beginning to be understood and has so far been mostly studied in the context of adaptive tissue responses (Antonello et al., 2015; Foronda et al., 2014; Mukherjee et al., 2021). Another miRNA, miR-305, is also known to target a Notch inhibitor, Hairless, but it specifically acts in ISCs (Foronda et al., 2014). Collectively, these studies have outlined cell-type-specific modulation of Notch signaling in the intestinal lineage and thus highlights the importance of miRNA-mediated regulation in actively regenerating intestinal tissue. Like the Drosophila intestine, the evolutionarily conserved Notch signaling pathway is also the predominant differentiation cue in the mammalian ISC lineage, and thus understanding Notch signaling dynamics has future potential in colon cancer therapies (Qiao and Wong, 2009; Vinson et al., 2016).
Limitations of the study
Here, we have used miR-956[KO] whole-animal mutants as well as UAS-miR-956 sp. transgene for analysis of miR-956 function in the intestinal tissue. However, due to technical limitations, we were unable to use lineage tracing methods such as mosaic analysis with a repressible cell marker (MARCM) to look at individual stem cell lineages that lack miR-956 activity. In addition, in this study we focused on miR-956 function in the tissue, which we found acts mainly in EBs by suppressing insv mRNA. However, functional studies of insv and genetic interaction between insv and Su(H) in ISCs still awaits future experimentation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nicholas S. Sokol (nssokol@gmail.com).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact without restriction.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila strains and husbandry
Age-matched, 5–7 days old female flies were used in all experiments. All flies used in this study were reared on standard Bloomington Drosophila Stock Center (BDSC) food and maintained inside a 25°C incubator set for a 12-h light/dark cycle and 65% humidity. For temperature-sensitive experiments, crosses were set at 18°C incubators and 2–3 days old progenies were shifted to 29°C for 5–7 days to activate Gal4 expression. Drosophila strains used in this study are listed in the key resources table. Unless otherwise indicated, w[1118] was used as the wildtype control.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-V5 | Bio-Rad | Cat# MCA1360GA; RRID:AB_567249 |
| Rabbit anti-GFP | Thermo Fisher Scientific | Cat# A11122; RRID:AB_221569 |
| Rabbit anti-mCherry | BioVision | RRID:AB_5993-100 |
| Goat Alexa Fluor 488-conjugated anti-rabbit | Thermo Fisher Scientific | RRID:AB_2576217 |
| Goat Alexa Fluor 568-conjugated anti-mouse | Thermo Fisher Scientific | RRID:AB_144696 |
| Goat Alexa Fluor 647-conjugated anti-Horseradish Peroxidase | Jackson ImmunoResearch | RRID:AB_2338967 |
| Anti-DIG POD | Millipore Sigma | Cat# 11207733910 |
| Rabbit Anti-Insv | (Duan et al., 2011) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde solution | Electron Microscopy Sciences | Cat# 15714 |
| TRIzol® LS reagent | Ambion | Cat# 10296028 |
| 10X PBS pH 7.4, RNase-free | Invitrogen | Cat# AM9624 |
| Heptane | Sigma-Aldrich | Cat# 246654 |
| Triton X-100 | Sigma-Aldrich | Cat# 11332481001 |
| Vectashield | Vector Laboratories | Cat# H-1000 |
| ProLong Diamond Antifade | Invitrogen | Cat# P36965 |
| SUPERasIn RNase Inhibitor | Invitrogen | Cat# AM2696 |
| cOmplete Protease Inhibitor | Roche | Cat# 1183617001 |
| Turbo DNase | Thermo Fisher Scientific | Cat# AM2239 |
| Superscript III | Thermo Fisher Scientific | Cat# 56575 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621L |
| Western Block Solution | Millipore Sigma | Cat# 11921673001 |
| Critical commercial assays | ||
| TSA Cyanine 3 System Kit | Akoya BioSciences | Cat#: NEL704A001KT |
| miRCURY LNA miRBA ISH Buffer Set | Qiagen | Cat# 339450 |
| miR-956 LNA in situ probes | Qiagen | Cat# 339111 |
| miR-956 Taqman probe | Thermo Fisher Scientific | Cat# PN4440887 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: P{CG10116-smGFP.V5.nls-insv3′UTR}attP40 | This study | N/A |
| D. melanogaster: P{miR-956 rescue3kb}attP40 | This study | N/A |
| D. melanogaster: esg-Gal4 UAS-GFP tubGal80 ts | (Micchelli and Perrimon, 2006) | N/A |
| D. melanogaster: gbe-smGFP::V5::nls | (Buddika et al., 2021) | N/A |
| D. melanogaster: w; Su(h)Gbe-Gal4, UAS-mCD8GFP/CyO; tubGal80 ts /TM6B | B. Edgar | N/A |
| D. melanogaster: P{insv[23B]} | (Duan et al., 2011) | N/A |
| D. melanogaster: P{UAS-NICD} | (Go et al., 1998) | N/A |
| D. melanogaster: TI{GAL4}mir-956[KO], P{GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, | BDSC | RRID:BDSC_58941 |
| D. melanogaster: PBac{UAS-mir-956.S}VK00037 | BDSC | RRID:BDSC_60607 |
| D. melanogaster: P{UAS-mCherry.mir-956.sponge.V2}attP40; P{UAS-mCherry.mir-956.sponge.V2}attP2 | BDSC | RRID:BDSC_61442 |
| D. melanogaster: P{mira(KDRT.stop)GAL4}attP40, P{tubP-GAL80[ts]}20; P{CG10116-KDR.PEST} attP2 Note: Also referred to as I-KCKT-GAL4ts | BDSC | RRID:BDSC_91410 |
| D. melanogaster: P{GD10842}v34494 Note: Also referred to as insv-RNAi | VDRC | RRID:VDRC_34494 |
| D. melanogaster: M{UAS-insv.ORF.3xHA.GW} ZH-86Fb Note: Also referred to as UAS-insv | BDSC | RRID:FlyORF_002980 |
| Oligonucleotides | ||
| List of oligo sequences | This study | See Table S1 |
| Software and algorithms | ||
| Adobe Photoshop and Adobe Illustrator Software | Adobe | N/A |
| Leica LAS-X | Leica | N/A |
| Prism | GraphPad Software | N/A |
| Other | ||
| StepOnePlus Real-time PCR machine | Life Technologies | N/A |
| Leica SP8 Scanning Confocal microscope | Leica | N/A |
METHOD DETAILS
Transgenes
miR-956 rescue
To rescue miR-956, first a 3,021 bp PCR fragment was amplified from genomic DNA with oligonucleotide pair 3989/4024. It was then cloned into a pATTB-based transformation plasmid that includes mini-white using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs E2621L). Transgenesis of the plasmid yielded {miR-956 rescue3kb}attP40. Oligo sequences are listed in Table S1 insv 3′UTR sensor: To generate insv 3′UTR sensor, first a 440-bp PCR fragment was amplified from genomic DNA with oligonucleotide primer pair 4845/4855. It was then cloned into a homemade XbaI/Pme1-digested transformation plasmid that contained the intestine-specific CC10116 enhancer fragment (Buddika et al., 2021) upstream of the smGFP.V5.nls ORF (Buddika et al., 2020). Transgenesis of the plasmid yielded {CG10116-smGFP.V5.nls-insv3′UTR}attP40. Oligo sequences are listed in Table S1.
Immunostaining
For all experiments, intestinal tissues were dissected from 5 to 7 days old female adult flies in ice-cold 1× phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (PF). For some experiments where control and mutant tissue were processed in the same tube, we made distinct incisions during the dissection step by cutting off the anterior portion of the gut from wildtype intestines and leaving the mutant guts intact. Tissues were then fixed on a nutator for 45 min at room temperature. After fixation, samples were washed in 1XPBT (1× PBS, 0.1% Triton X-100) solution for at least an hour by changing washes every 15 min. After washes, tissue samples were then blocked in 0.5% BSA in 1XPBT for 30 min at room temperature. Samples were incubated with primary antibody solution at 4°C overnight. The primary antibody was removed, and the samples were then washed in 1XPBT solution before incubating them with secondary antibody and 5 μg/mL DAPI in 1XPBT for 2 to 3 h at room temperature. Next the samples were washed for an hour by changing the wash solution at least thrice and mounted on Vectashield mounting medium. Primary and secondary antibodies used in this study have been listed in the key resources table.
Imaging and image analysis
Intestinal samples were imaged using Leica HC PL APO CS2 63×/1.40 objective (Leica Type F Immersion Liquid, n = 1.518) on Leica SP8 Scanning Confocal microscope. To maintain consistency and minimize variations due to regional differences in the gut, all images were acquired from the R4bc region of the adult posterior midgut as defined in a previous study (Buchon et al., 2013). The images were processed using Adobe Photoshop CC software. For quantifying proportion of cell numbers, count tool was used to quantify the ratio of individual cell types to total cells counted per field of view. The number of replicates for each experiment are indicated as n values shown in graphs. All experiments have been repeated at least three times. For comparing protein levels across samples, both control and experimental samples were dissected and stained in the same tube and imaged under identical settings. Fluorescence intensity was measured using ImageJ and quantified as corrected total cell fluorescence (CTCF) using the following formula: CTCF = integrated density (area of selected cell 3 mean fluorescence of background readings). For each experiment, we quantified fluorescence intensity in cells from at least five intestines. Final images were assembled in Adobe Illustrator CC.
RNA in situ hybridization
The RNA in situ hybridization protocol used for detecting miR-956 expression in the gut was adapted from the miRCURY LNA hand-book provided by the vendor of the in situ probes (Qiagen) as well as a previous study (Kucherenko et al., 2012). Briefly, intestinal tissues were dissected in ice-cold 1X PBS solution, fixed in 4% PF solution, and washed in 1X PBS three times. The samples were then dehydrated in 3:1, 1:1, 1:3 PBT/ethanol mix for 10 minutes each followed by a 10-minute wash in 100% ethanol. Next, they were rehydrated in 1:3, 1:1, 3:1 PBT/EtOH mix for 10 minutes each and again transferred to 1X PBS wash solution. Following three quick washes, the samples were next washed with 0.5X HYB buffer in 1X PBS and then prehybridized in 1X HYB buffer at 59°C shaker. The samples were incubated with preheated miRCURY LNA probe for miR-956 in 40 nM final concentration and left overnight at 59°C shaker.
Following hybridization, the samples were washed in HYB solution for 20 minutes followed by five quick washes in PBT at 59°C. Next, the samples were blocked in Western Block (WB) solution for 1 hour and incubated with antibodies against DIG conjugated to HRP diluted 1:2000 in WB solution for 2 hours at room temperature. For signal detection, we used the TSA Cyanine 3 System kit. The samples were washed in PBT: WB solution six times and then incubated with streptavidin-HRP solution for 1 hour. They were again washed in PBT: WB solution six times followed by two subsequent washes in PBT and 1X PBS. Next the samples were incubated in solution containing Cyanine 3 Tyramide diluted 1:50 in Amplification Dilutent for 2 hours at the room temperature protected from the light. The samples were washed in 1X PBS six times before mounting them in Prolong Diamond.
qRT-PCR Assays
For all analysis, total RNA was extracted from whole animals or dissected intestinal samples separated from the carcass collected in triplicates using TRIsol reagent. For TaqMan qRT-PCR analysis, all RNA samples were first diluted to 25 ng/μL concentration and then reverse transcribed with SuperScript III (Life Technologies) to make complementary DNA (cDNAs). The cDNAs were then further diluted to 1:25 and used as a template for TaqMan qRT-PCR. For each reaction, cDNA from individual samples were mixed with TaqMan Universal PCR master mix and a miR-956 Taqman probe (Life Technologies) in triplicates and run with the qRT-PCR setup on a StepOnePlus Real time PCR machine (Life Technologies). Relative miRNA abundance was calculated using the Pfaffl method and normalized to 2s rRNA levels. For other qPCR analysis, reaction was set up using the PowerUp SYBR Green Master Mix (Life Technologies) instead and mRNA abundance values were normalized to RP49 levels. qPCR oligos are listed in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
All graphs shown in main and supplementary figures were plotted using Prism (GraphPad, version 7.0). All data have been represented as Mean ± Standard Error of Mean (SEM) and n values in the graphs indicate number of intestines or number of cells used for quantification as mentioned in the figure legends. Significance values for each dataset were calculated using a two-tailed un-paired t test with Welch’s correction. Significance values are indicated as follows: n.s., not significant; *, p < 0.1; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Supplementary Material
Highlights.
Tissue enriched miR-956 is highly expressed in intestinal progenitor cells
miR-956 acts in stem cell progenies to promote enterocyte differentiation
miR-956 regulates differentiation by promoting Notch activity in enteroblasts
miR-956 mediates its function by suppressing insensitive mRNA
ACKNOWLEDGMENTS
We thank Norbert Perrimon, Bruce Edgar, Eric Lai, Spyros Artavanis-Tsakonas, the Bloomington Drosophila Stock Center (supported by grant NIH-P4OOD018537), and the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) for reagents; the Light Microscopy Imaging Center (supported by grant NIH1S10OD024988-01) for access to the SP8 confocal; our colleagues at Indiana University for helpful discussions; and the National Institutes of Health for financial support (award R01GM124220 to N.S.S. and B.R.C.). We are also grateful to Claude Desplan for valuable feedback on the manuscript and supporting S.M. in his lab during revisions and to Yi Ting Huang for handling reagents used in this manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111495.
DECLARATION OF INTERESTS
No competing interest declared.
REFERENCES
- Agarwal V, Bell GW, Nam J-W, and Bartel DP (2015). Predicting effective microrna target sites in mammalian mrnas. Elife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonello ZA, Reiff T, Ballesta-Illan E, and Dominguez M (2015). Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by mir-8-escargot switch. EMBO J. 34, 2025–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AM, and Posakony JW (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to notch receptor activity. Gene Dev. 9, 2609–2622. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Perdigoto CN, Southall TD, Brand AH, and Schweisguth F (2010). Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, and Jasper H (2014). Slit/robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 7, 1867–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FPA, Fang HY, Boquete JP, Deplancke B, and Lemaitre B (2013). Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738. [DOI] [PubMed] [Google Scholar]
- Buddika K, Ariyapala IS, Hazuga MA, Riffert D, and Sokol NS (2020). Canonical nucleators are dispensable for stress granule assembly in intestinal progenitors. J. Cell Sci 133. Jcs243451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddika K, Xu J, Ariyapala IS, and Sokol NS (2021). I-Kckt allows dissection-free rna profiling of adult Drosophila intestinal progenitor cells. Development 148. dev196568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A, and Batlle E (2009). Intestinal stem cells in mammals and Drosophila. Cell Stem Cell 4, 124–127. [DOI] [PubMed] [Google Scholar]
- Caygill EE, and Brand AH (2017). Mir-7 buffers differentiation in the developing Drosophila visual system. Cell Rep. 20, 1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Song S, Weng R, Verma P, Kugler JM, Buescher M, Rouam S, and Cohen SM (2014). Systematic study of Drosophila microrna functions using A collection of targeted knockout mutations. Dev. Cell 31, 784–800. [DOI] [PubMed] [Google Scholar]
- Clarke MF, and Fuller M (2006). Stem cells and cancer: two faces of eve. Cell 124, 1111–1115. [DOI] [PubMed] [Google Scholar]
- Duan H, Dai Q, Kavaler J, Bejarano F, Medranda G, Nègre N, and Lai EC (2011). Insensitive is A corepressor for suppressor of hairless and regulates Notch signalling during neural development. EMBO J. 30, 3120–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D, Weng R, Verma P, Chen YW, and Cohen SM (2014). Coordination of insulin and Notch pathway activities by microrna mir-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 28, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Mcneill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, Deluca T, et al. (2015). A transgenic resource for conditional competitive inhibition of conserved Drosophila micrornas. Nat. Commun 6, 7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, and Artavanis-Tsakonas S (1998). Cell proliferation control by Notch signaling in Drosophila development. Development 125, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Haase A, Stern M, Wächtler K, and Bicker G (2001). A tissue-specific marker of ecdysozoa. Dev. Gene. Evol 211, 428–433. [DOI] [PubMed] [Google Scholar]
- Jan LY, and Jan YN (1982). Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA 79, 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, and Edgar BA (2009). Egfr signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 136, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaler J, Duan H, Aradhya R, De Navas LF, Joseph B, Shklyar B, and Lai EC (2018). Mirna suppression of A Notch repressor directs non-neuronal fate in Drosophila mechanosensory Organs. J. Cell Biol 217, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583–597. [DOI] [PubMed] [Google Scholar]
- Korzelius J, Azami S, Ronnen-Oron T, Koch P, Baldauf M, Meier E, Rodriguez-Fernandez IA, Groth M, Sousa-Victor P, and Jasper H (2019). The Wt1-like transcription factor Klumpfuss maintains lineage commitment of enterocyte progenitors in the Drosophila intestine. Nat. Commun 10, 4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherenko MM, Barth J, Fiala A, and Shcherbata HR (2012). Steroid-induced microrna let-7 acts as A spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31, 4511–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC (2004). Notch signaling: control of cell communication and cell fate. Development 131, 965–973. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, and Rubin GM (2005). Pervasive regulation of Drosophila Notch target genes by gy-box-brd-box-and K-Box-Class micrornas. Genes Dev. 19, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, and Schweisguth F (1995). The neurogenic suppressor of hairless dna-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9, 2598–2608. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, and Perrimon N (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479. [DOI] [PubMed] [Google Scholar]
- Miller DE, Kahsai L, Buddika K, Dixon MJ, Kim BY, Calvi BR, Sokol NS, Hawley RS, and Cook KR (2020). Identification and characterization of breakpoints and mutations on Drosophila melanogaster balancer chromo-somes. G3 10, 4271–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, and Spradling AC (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Paricio N, and Sokol NS (2021). A stress-responsive mirna regulates bmp signaling to maintain tissue homeostasis. Proc. Natl. Acad. Sci. USA 118. e2022583118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien LE, Soliman SS, Li X, and Bilder D (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell 147 (3), 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474. [DOI] [PubMed] [Google Scholar]
- Qiao L, and Wong BCY (2009). Role of Notch signaling in colorectal cancer. Carcinogenesis 30, 1979–1986. [DOI] [PubMed] [Google Scholar]
- Reeves N, and Posakony JW (2005). Genetic programs activated by proneural proteins in the developing Drosophila pns. Dev. Cell 8, 413–425. [DOI] [PubMed] [Google Scholar]
- Reiff T, Antonello ZA, Ballesta-Illán E, Mira L, Sala S, Navarro M, Martinez LM, and Dominguez M (2019). Notch and egfr regulate apoptosis in progenitor cells to ensure gut homeostasis in Drosophila. EMBO J. 38, E101346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Qin P, Liu X, Wu S, Yao R, Cai G, Gao J, Wu Y, and Guo Z (2021). Intravital imaging strategy flyvab reveals the dependence of Drosophila enteroblast differentiation on the local physiology. Commun. Biol 4, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson KE, George DC, Fender AW, Bertrand FE, and Sigounas G (2016). The Notch pathway in colorectal cancer. Int. J. Cancer 138, 1835–1842. [DOI] [PubMed] [Google Scholar]
- Wang C, Guo X, Dou K, Chen H, and Xi R (2015). Ttk69 acts as A master repressor of enteroendocrine cell specification in Drosophila intestinal stem cell lineages. Development 142, 3321–3331. [DOI] [PubMed] [Google Scholar]
- Weigelt CM, Hahn O, Arlt K, Gruhn M, Jahn AJ, Eßer J, Werner JA, Klein C, Büschges A, Grönke S, and Partridge L (2019). Loss of mir-210 leads to progressive retinal degeneration in Drosophila Melanogaster. Life Sci. Alliance 2. E201800149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko AS, and Shcherbata HR (2018). Stereotypical architecture of the stem cell niche is spatiotemporally established by mir-125-dependent coordination of Notch and steroid signaling. Development 145. Dev159178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Chauhan C, and Hou SX (2010). Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis 48, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, and Hou SX (2015). Enteroendocrine cells are generated from stem cells through A distinct progenitor in the adult Drosophila posterior midgut. Development 142, 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


