Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sublineages have escaped most receptor-binding domain (RBD)-targeting therapeutic neutralizing antibodies (NAbs), which proves that previous NAb drug screening strategies are deficient against the fast-evolving SARS-CoV-2. Better broad NAb drug candidate selection methods are needed. Here, we describe a rational approach for identifying RBD-targeting broad SARS-CoV-2 NAb cocktails. Based on high-throughput epitope determination, we propose that broad NAb drugs should target non-immunodominant RBD epitopes to avoid herd-immunity-directed escape mutations. Also, their interacting antigen residues should focus on sarbecovirus conserved sites and associate with critical viral functions, making the antibody-escaping mutations less likely to appear. Following these criteria, a featured non-competing antibody cocktail, SA55+SA58, is identified from a large collection of broad sarbecovirus NAbs isolated from SARS-CoV-2-vaccinated SARS convalescents. SA55+SA58 potently neutralizes ACE2-utilizing sarbecoviruses, including circulating Omicron variants, and could serve as broad SARS-CoV-2 prophylactics to offer long-term protection, especially for individuals who are immunocompromised or with high-risk comorbidities.
Keywords: SARS-CoV-2, sarbecovirus, omicron, antibody, cocktail, broad-spectrum, prophylactics
Graphical abstract
Cao et al. describe a strategy to identify a broad sarbecovirus-neutralizing antibody cocktail that would be hard to escape for future SARS-CoV-2 variants using high-throughput epitope mapping. The resulting antibody cocktail, named SA55+SA58, exhibits high neutralizing potency and breadth against ACE2-utilizing sarbecoviruses and efficiently protects mice from BA.1 and BA.5 infection.
Introduction
Over 2 years after its emergence, the COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is still spreading. Neutralizing antibodies (NAbs) play a critical role in the prevention and treatment of COVID-19.2 , 3 , 4 , 5 However, the constant emergence of new variants has caused large-scale evasion of NAbs, posing severe challenges to SARS-CoV-2 NAb drugs.1 , 6 , 7 , 8 , 9 Given their prophylactic and therapeutic efficacy, the clinical development of NAb drugs that are difficult for future variants to escape is still in high demand,10 , 11 especially for individuals who are immunocompromised or with high-risk comorbidities.
All currently approved SARS-CoV-2 NAb drugs target the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein, such as LY-CoV016+LY-CoV555 (bamlanivilab + etesevimab)12 , 13; REGN10933+REGN10987 (casirivimab + imdevimab)10 , 14; S309 (sotrovimab)15; AZD1061+AZD8895 (cilgavimab + tixagevimab, AZD7442, Evusheld)16; and LY-CoV1404 (bebtelovimab).17 Unfortunately, the majority of them have already been escaped by the Omicron variants.1 , 8 , 18 , 19 Most of the NAbs were selected for clinical development mainly based on two criteria: (1) the antibody candidates were potent against the circulating variants at the time,10 , 12 , 13 , 14 , 15 , 16 , 17 , 20 and (2) the antibody candidates should ideally form a non-competing antibody cocktail.10 , 16 , 21 Given that high potency could lead to lower dosage and that non-overlapping cocktails would reduce the chance of being completely escaped, the strategy is reasonable but insufficient, considering the repeated evolution of multiple mutations on the RBD that could escape both antibodies in a cocktail simultaneously.22 , 23 High potency does not guarantee good neutralization breadth against rapidly emerging SARS-CoV-2 variants. Better broad NAb drug selection strategies, beyond simply picking the most potent ones to form cocktails, are needed to counter the fast-evolving virus.
One potential broad SARS-CoV-2 NAb drug selection strategy is to choose NAbs that exhibit broad sarbecovirus neutralizing activity.4 , 5 These broad sarbecovirus NAbs (bsNAbs) target epitopes located on sarbecovirus conserved regions, which may represent functional constraints and thus hardly mutate. Various bsNAbs targeting spike protein’s RBD or S2 region have been discovered.24 , 25 , 26 , 27 S2-targeting bsNAbs indeed displayed exceptional SARS-CoV-2 neutralization breadth, but their potency is rather low for NAb drug development. On the other hand, anti-RBD bsNAbs, such as S309,15 ADG-2,28 DH1047,29 S2X259,5 and S2K146,30 could exhibit high neutralization potency and are good candidates for NAb drugs; however, the majority of them are escaped by Omicron BA.2 or BA.4/BA.5.1 , 18 This suggests that NAbs with broad sarbecovirus neutralizing capability are not necessarily broad SARS-CoV-2 NAbs, and a better strategy to select broad NAb drugs is still needed.
In this article, we describe a rational approach for identifying RBD-targeting broad SARS-CoV-2 NAb cocktails with high potency that are strongly resistant to both current and potential future mutants. The resulting featured antibody cocktail, SA55+SA58, exhibits broad sarbecovirus neutralizing activity and is highly potent against current Omicron variants, including BA.1, BA.2, BA.2.12.1, BA.4/BA.5, and the latest evasive variants with convergent mutations, such as BQ.1.1 and XBB, making it a valuable bsNAb drug candidate.31
Results
A rational strategy for identifying potent bsNAb cocktails
We believe that rational identification of broad SARS-CoV-2 NAb drug candidates relies on the accurate estimation of antibodies’ neutralization breadth; however, neutralization breadth is not only governed by the biochemical properties of the NAbs but also depends on the virus mutation pattern and evolution direction. Based on the study of SARS-CoV-2 variants, the evolution of the virus RBD seems to comply with the following patterns: SARS-CoV-2 is more likely to evolve RBD mutations on “hot” epitopes that are frequently targeted by NAbs elicited by SARS-CoV-2 convalescents and vaccinees to achieve efficient humoral immunity evasion1 , 23 , 32; also, the virus is less likely to evolve mutations that could disrupt its conserved functions, such as ACE2 binding or RBD folding.33 On the basis of these observations, we propose two additional criteria for identifying RBD-targeting broad SARS-CoV-2 NAb drugs: (3) the candidate NAbs should target a rare RBD epitope such that NAbs with a similar epitope are not abundant in SARS-CoV-2 convalescents and vaccinees. This way, the RBD mutations that escape the candidate NAbs are not aligned with those that heavily evade herd humoral immunity and would have a lower chance of appearing. (4) The candidate NAbs should target sarbecovirus conserved epitopes on the RBD that are also associated with critical viral functions, such as ACE2 binding or glycosylation. These NAbs would naturally exhibit broad sarbecovirus binding capability, and their escaping mutations are less likely to prevail.30
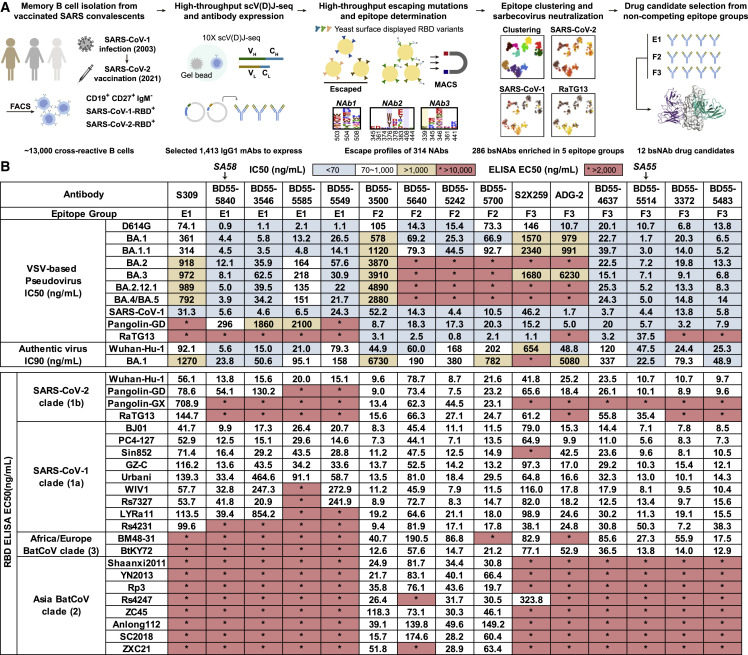
However, several difficulties exist for identifying NAb candidates that meet these four criteria. First, a substantially large antibody library is needed to contain a collection of rare anti-RBD NAbs that target sarbecovirus conserved regions. To reduce the size of the antibody library needed, we choose SARS-CoV-2-vaccinated SARS convalescents as the antibody source since their memory B cells are more likely to encode NAbs that target sarbecovirus conserved epitopes.34 Second, a high-throughput method is needed to determine the binding epitopes and antibody-escaping mutations of each NAb in a large library. Previously, we presented a high-throughput unsupervised epitope mapping technology based on deep mutational scanning (DMS), which can be employed for epitope distribution and escaping mutation analyses at a much higher efficiency than traditional epitope binning techniques.23 , 35 By combining high-throughput DMS with droplet-based single-cell V(D)J sequencing (scVDJ-seq) or single-cell reverse transcription PCR (RT-PCR) of RBD-specific memory B cells,20 , 21 , 36 , 37 an efficient pipeline can be built to isolate a large panel of NAbs and, at the same time, identify their binding epitopes, solving the above difficulties1 (Figure 1A).
Figure 1.
Identification of broad sarbecovirus neutralizing antibodies
(A) Schematic for the identification and characterization of bsNAbs, and selection of bsNAb drug candidates from SARS-CoV-2-vaccinated SARS convalescents.
(B) Neutralizing IC50 against SARS-CoV-2 variants VSV-based pseudoviruses and authentic viruses, and ELISA EC50 against sarbecovirus RBD by identified bsNAbs and other published antibodies in groups E1, E3, and F3. Background colors of grids indicate the following: blue, IC50 ≤ 70 ng/mL; white, 70 ng/mL < IC50 < 1,000 ng/mL; yellow, IC50 ≥ 1,000 ng/mL. Asterisks with red backgrounds indicate that the corresponding values were larger than the limits of detection. Neutralization assays and ELISA were conducted in at least two biological replicates.
We successfully obtained a large bsNAb collection from SARS-CoV-2-vaccinated SARS convalescents with each antibody’s epitope and escape profiles determined (Figure 1A). Specifically, we used fluorescence-activated cell sorting (FACS) and collected ∼13,000 CD19+CD27+IgM− memory B cells that can cross-bind to both the SARS-CoV-2 RBD and the SARS-CoV-1 RBD from 28 individuals who recovered from SARS in 2003 and received 2 doses of SARS-CoV-2 ancestral strain (referred to as wild type [WT]) inactivated vaccine (CoronaVac) and 1 booster dose of RBD-based protein subunit vaccine (ZF2001) in 202138 , 39 (Figure S1; Table S1). A total of 2,838 heavy-light chain-paired antibody sequences were recovered from those cross-reactive memory B cells by high-throughput scVDJ-seq.20 Among them, 1,413 antibody sequences contain immunoglublin G1 (IgG1) heavy-chain constant regions and were selected to express in vitro as monoclonal antibodies (mAbs) (Table S2). The neutralizing activity of the mAbs against SARS-CoV-2 was further screened using VSV-based pseudovirus (D614G). We then selected NAbs (D614G IC50 < 10 μg/mL) for further epitope mapping and successfully obtained the escape profiles of 314 NAbs using DMS for RBD, among which 286 displayed sarbecovirus neutralizing activity (SARS-CoV-1 IC50 < 10 μg/mL)1 , 23 (Figure 1A; Table S2).
These bsNAbs mainly appeared in five epitope groups (E1, E3, F1, F2, and F3), which were determined by unsupervised clustering of DMS results as previously presented1 , 23 (Figure 2A). In general, group E1 (S309 epitope) antibodies bind to the front of the RBD and react with all clade 1a/1b sarbecoviruses15 (Figure 1B). F3 (ADG-2 epitope) NAbs bind to the back of the RBD and could bind to all ACE2-utilizing sarbecovirus clades, including BtKY72 and BM48-31 in clade 328 (Figure 1B). Group E3 (S2H97 epitope), F1 (S304 epitope), and F2 (BD55-1239 epitope, near the linoleic acid binding pocket) antibodies exhibited broad specificity to sarbecoviruses in all clades1 , 15 , 27 , 40 (Figure 1B); however, antibodies from cryptic epitope E3 and F1 exhibited low neutralizing activities and are not suitable for NAb drugs. In comparison, E1 and F3 NAbs displayed high neutralizing activity, while F2 demonstrated moderate potency but better breadth (Figure 1B). A total of 49 E1, 109 F2, and 57 F3 bsNAbs were identified (Table S2).
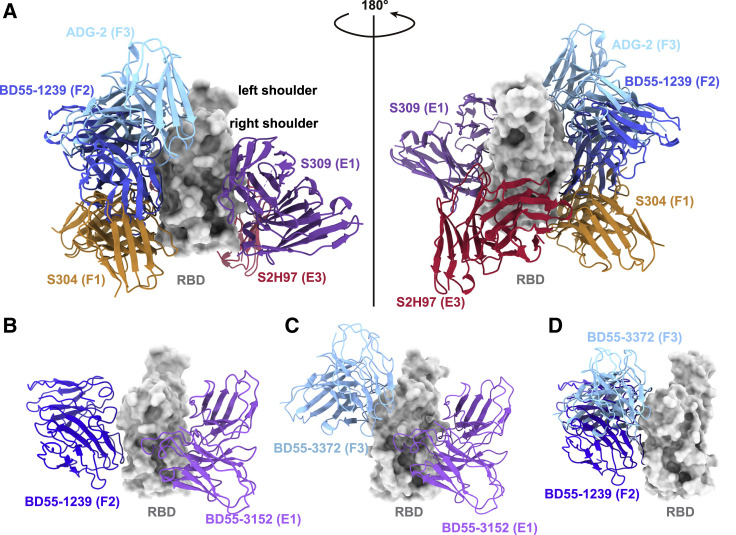
Figure 2.
Proposed strategies for non-overlapping bsNAb combination
(A) Aligned structures of representative antibodies in epitope groups with broad reactivity against sarbecoviruses are shown, including S309 in E1 (PDB: 6WPS), S2H97 in E3 (PDB: 7M7W), and S304 in F1 (PDB: 7JW0); BD55-1239 in F2 (PDB: 7WRL); and ADG-2 in F3 (PDB: 7U2D). Structural alignments were based on their RBD part.
(B and C) Aligned structures of BD55-3152 (group E1, PDB: 7WR8) together with (B) BD55-1239 (group F2, PDB: 7WRL) or (C) BD55-3372 (group F3, PDB: 7WRO) in complex with SARS-CoV-2 BA.1 RBD.
(D) Aligned structure shows BD55-1239 (PDB: 7WRL) in F2 and BD55-3372 (PDB: 7WRO) in F3 could not form non-overlapping cocktails.
According to criterion 1, we selected the top four most potent NAbs from each of the three epitope groups, E1, F2, and F3, as bsNAb drug candidates based on the pseudovirus neutralizing activity against SARS-CoV-1, SARS-CoV-2 D614G, and Omicron BA.1, which is the dominant variant at the time of experiment. The majority of bsNAb candidates showed high somatic hypermutation (SHM) rates resulting from long-term antibody affinity maturation since SARS infection in 2003 (Table S3). Among the four candidates in group E1, BD55-5840 (SA58) showed the highest authentic virus neutralization activity against both the SARS-CoV-2 ancestral strain (IC90 = 5.6 ng/mL) and Omicron BA.1 (IC90 = 23.8 ng/mL), surpassing an archetypical E1 antibody, S309 (Figure 1B). In group F3, BD55-5514 (SA55) exhibited the highest activity against BA.1 authentic virus (IC90 = 22.5 ng/mL), which again surpassed the potency of ADG-2 (Figure 1B). The overall authentic virus neutralization of NAbs from group F2 is indeed much lower than that of E1 and F3 (Figure 1B).
Next, we examined the possible cocktail pairing strategies among these three epitope groups. We aligned the RBD structures of three representative antibody Fab:RBD complexes: SA58 for E1, BD55-1239 for F2, and BD55-3372 for F3 (Figures 2B–2D).1 Results demonstrated that E1 antibodies could generally form non-overlapping cocktails with antibodies in either group F2 or F3 (Figures 2B and 2C), while F2 and F3 antibodies covered overlapping epitopes (Figure 2D). Thus, E1+F2 and E1+F3 are the two practical strategies to build non-competing bsNAb cocktails. However, we previously showed that F2 antibodies are abundant in SARS-CoV-2WT convalescents/vaccinees, while E1 and F3 are generally rare.1 Therefore, E1/F3 cocktails are better than E1/F2 cocktails, according to criterion 3, in that the rarity of these NAbs would make their escaping RBD mutations less likely to prevail. Also, the epitopes of E1 NAbs are centered around N343 glycan (Figure 3A), and F3 NAbs’ escaping mutations mainly focus on the binding interface of ACE2 (Figure 3B), while the epitopes of F2 NAbs are not linked to any known critical viral functions. Indeed, almost all F2 NAbs were escaped by Omicron BA.2 subvariants mainly due to D405N and R408S mutations located in their epitopes, despite these two sites being highly conserved among sarbecoviruses (Figure 1B).23 This exemplifies criterion 4: that antibodies targeting sarbecovirus conserved epitopes can also be easily escaped unless their major escaping mutations directly overlap with sites that associate with critical viral functions.
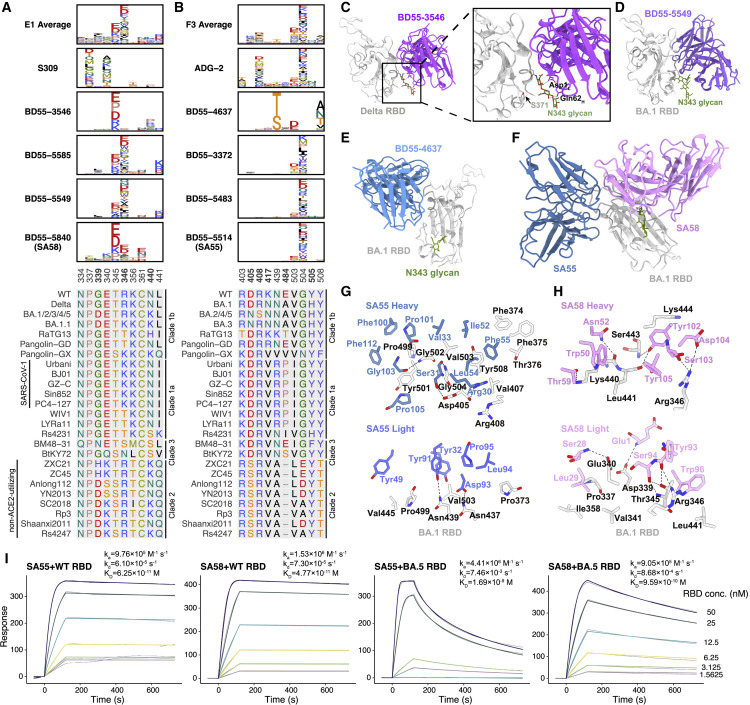
Figure 3.
Escape maps and binding modes of representative E1 and F3 antibodies
(A and B) Average escape scores of antibodies in groups E1 (A) and F3 (B), and escape maps of selected drug-candidate bsNAbs in groups E1 (A) and F3 (B). Colors are assigned according to the type of amino acids, and the heights of the letters in the logo plot indicate escape scores. Multiple sequence alignment on the corresponding escaping hotspot residues is shown. Residues are numbered based on SARS-CoV-2 numbering.
(C–H) Structures of mAbs in complex with RBD. Colors of text in each panel are the same as that of the corresponding structural models.
(C) Cryo-EM structure of BD55-3546 bound to RBDDelta.
(D) X-ray crystallographic structure of BD55-5549 bound to RBDBA.1.
(E) Cryo-EM structure of BD55-4637 bound to RBDBA.1.
(F) Cryo-EM of SA55 and SA58 bound together to RBDBA.1.
(G and H) Interactions between the heavy-chain and light-chain SA55 (G) or SA58 (H) and RBDBA.1. Residues of RBD, SA55, and SA58 are black, blue, and pink, respectively.
(I) SPR sensorgrams for the measurement of the binding kinetics of SA55 and SA58 to WT and BA.5 RBD. Gray curves are raw responses, and colored curves are results fitted with a 1:1 binding model. Representative results of two replicates are shown.
Structures and RBD-binding of the bsNAb drug candidates
To decide the final bsNAb cocktail composition, we further examined the escape profiles and structural properties of the E1 and F3 NAb candidates. Most E1 NAbs are susceptible to mutations of T345 and R346 (Figure 3A). The four E1 bsNAbs displayed three different escape profiles, suggesting three different RBD-binding modes. BD55-3546 is mainly escaped by T345 and N440 mutations. BD55-5585 and BD55-5549 have similar escape profiles and are mainly escaped by R346, T345, and L441 mutations. SA58 is less affected by R346 mutations compared with BD55-5585 and BD55-5549. To analyze their detailed molecular interactions with RBD, we determined the crystal structure of BD55-5549/RBDBA.1 and the cryoelectron microscopy (cryo-EM) structures of BD55-3546/S6PDelta (Figures 3C, 3D, and S2A; Tables S4 and S5). Previously, we also solved the cryo-EM structures of SA58/S6PBA.1 and SA58/S6PBA.2.1 Similar to S309, the epitopes of these antibodies all encompass the N343 glycan, which plays a critical role in modulating RBD conformation. Thus, critical mutations that can alter the position of the glycan motif could reduce the activities of the E1 antibodies, such as S371F, which emerged in BA.2.1 BD55-3546 is more affected by this mutation than SA58, with 5- to 10-fold decreased activities toward the S371F-containing Omicron variants (Figure 1B). In the BD55-3546/S6PDelta structure, Q62H and D1L pack intimately with the N343 glycan (Figures 3C and S3A), rationalizing the sensitivity of BD55-3546 to its displacement. BD55-3546 is also sensitive to mutations of N440, as N440 is nestled in a pocket formed by W50H, N52H, T55H, I57H, and T59H and forms extensive van der Waals and hydrogen-bond interactions with these residues (Figure S3A). By comparison, BD55-5549 is not affected by the changes of N440. Indeed, the N440K site is not involved in interacting with BD55-5549 (Figures 3D and S3B). On the other hand, L441 is surrounded by Y110H, Y110H, and F111H, and BD55-5549 could be escaped by the L441 mutations (Figure 3A). Contrarily, SA58 is not susceptible to the substitutions of 440-441 as the side chain of neither site is closely targeted by SA5 and is only slightly affected by S371F.1 Mutations of T345 could lead to prominent escapes from SA58 since T345 is enclosed by Y105H, L94L, and W96L (Figures 3A and 3H); however, T345 substitutions are unlikely to emerge, as it is essential for the proper glycosylation of N343 that is critical for RBD function.33 Together, considering the prevalence of R346 (BA.1.1) and S371 (BA.2) mutations, we selected SA58 as the final E1 bsNAb for clinical development due to its higher potency and lesser sensitivity to glycan displacement and R346 substitutions.
F3 NAbs are mainly escaped by mutations of V503 and G504, as well as the D405 mutation found in BA.2 (Figure 3B). BD55-4637 has a rather unique escape profile, while BD55-3372, BD55-5483, and SA55 share highly similar profiles, suggesting similar interactions with RBD (Figure 3B). BD55-4637 can be escaped by the N439T/S mutations, as these mutations would lead to the glycosylation of N437, which packs against W55H, revealed by the cryo-EM structure of BD55-4637/S6PBA.1 (Figures 3E, S2B, and S3C). BD55-4637 is also sensitive to mutations of Y508 due to the interaction between Y508 and D102H; however, Y508 is essential to ACE2 binding and could hardly be evolved33 (Figure S4C). In contrast, BD55-3372, BD55-5483, and SA55 are not susceptible to the changes of these two sites but are sensitive to the changes on V503 and G504 (Figure 3B). Nevertheless, G504 is also critical for ACE2 binding as shown by DMS, and both V503 and G504 are conserved among sarbecoviruses,33 , 41 making it difficult for variants with mutations on these sites to prevail. Based on these characteristics, all four F3 bsNAbs are good drug candidates since their escaping mutations could not easily appear, and SA55 is chosen as the final F3 bsNAb for clinical development due to its highest neutralization potency (Figure 1B).
SA55 indeed can bind to the RBD together with SA58 in a largely non-interfering manner, as demonstrated by the SA58/SA55/S6PBA.1 ternary complex structure (Figures 3F and S2C). Interestingly, a close examination of the SA55/S6PBA.1 structure reveals that the S373P and S375F mutations could promote the interaction with SA55, consistent with the pseudovirus results (Figure 1B). F374 is flipped out because of these two mutations and robustly packs with F55CDRH2 in SA55 (Figure 3G). P373BA.1 also forms a hydrophobic interaction with L94CDRL3 (Figure 3G). Although T376, D405, and R408 are involved in the interaction with SA55, they are all located at the periphery of SA55’s epitope, and the activities of SA55 toward BA.2/BA.3/BA.2.12.1/BA.4/BA.5 are only slightly decreased when compared with BA.1 (Figures 1B and 3G). Most importantly, SA58 and SA55 could simultaneously bind and neutralize the virus in a complementary way. SA58 does not directly impede ACE2 but can engage both the up and down RBDs, whereas SA55 only binds to the up RBDs but promptly blocks ACE2 (Figure S3D).
We also determined the binding kinetics of SA55 and SA58 to WT or BA.5 RBD using surface plasma resonance (SPR) assays. The equilibrium dissociation constants (KD) of SA55 and SA58 against BA.5 RBD are 1.7 and 0.96 nM, respectively, which are increased by 20- and 27-fold compared to the KD against WT RBD for SA55 and SA58, respectively. However, their neutralization activities were almost not affected by mutations carried by BA.5 (Figure 1B). The reductions in affinities are mainly attributed to the increased rate of dissociation, and the rate of association did not change significantly (Figure 3I). The non-competing binding of SA55 and SA58 is also validated by SPR competition assays (Figure S3E). Together, SA55+SA58 is rationally selected as the final non-competing bsNAb cocktail and is expected to exhibit exceptional SARS-CoV-2 neutralization breadth and potency.
SA55+SA58 display broad resilience against RBD single substitutions
To further explore possible escape mutants of SARS-CoV-2 against SA55/SA58, we screened for escape mutations by infesting Vero cells with replication-competent recombinant VSV (rVSV)-SARS-CoV-2-SBA.1 at a gradient of NAb concentration to simulate the evolutionary stress caused by NAbs.42 , 43 After 4 days of co-culture with different concentrations of antibodies and rVSV-SARS-CoV-2-SBA.1, GFP+ wells reflect significant viral replication and antibody evasion (Figure 4A). The supernatant and cell layer of the well that has the highest antibody concentration among the GFP+ wells were collected. Total RNA was extracted and subjected to reverse transcription, amplification, and deep sequencing. Next-generation sequencing reads were aligned to a codon-optimized Omicron BA.1 spike nucleotide sequence to determine enriched substitutions (Figure 4A).
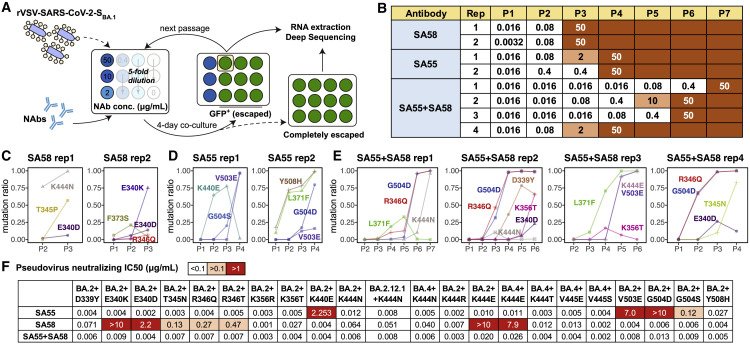
Figure 4.
SA55+SA58 cocktail robustly neutralizes SARS-CoV-2 escaping mutants
(A) Schematic of in vitro escape mutation screening using SARS-CoV-2BA.1-spike-pseudotyped rVSV. The virus was passed in the presence of antibody dilutions for 4 days on Vero cells. Cells were screened for virus replication by monitoring GFP+.
(B) Antibody concentration of dilutions that are escaped, passed, and sequenced of each passage and each antibody. SA55 and SA58 alone were tested in two biological replicates, respectively. SA55+SA58 cocktail was tested in four biological replicates.
(C–E) The ratio of enriched escape mutations of each replicate and each passage by deep sequencing (C) using SA58 only, (D) using SA55 only, or (E) using the SA55+SA58 cocktail.
(F) Neutralizing activities of SA55, SA58, and SA55+SA58 against pseudoviruses of selected SARS-CoV-2 variants harboring mutations that could potentially escape SA55 or SA58. Neutralization assays were conducted in two biological replicates.
In both replicates, SA58 was escaped in passage 3 (P3), but the evasion could be attributed to different mutations (Figure 4B). In replicate 1 (rep1), SA58 showed a decreased activity in P2 due to K444N and completely escaped by the combination of K444N and T345P in P3 (Figure 4C). Compared with rep1, the evasion of SA58 in rep2 was dominated by mutations on E340 instead of K444. Specifically, a slight enrichment of E340D in P2 resulted in a mild drop in potency, and the sharp enrichment of E340K in P3 completely escaped SA58 (Figure 4C). Slightly better than using SA58 alone, SA55 alone was not escaped until P4 in both replicates. In SA55 rep1, K440E was enriched in P2 and P3 but disappeared in P4 when it was replaced by G504S and V503E (Figure 4D). In SA55 rep2, the enrichment of Y508H in P2/P3 resulted in a drop of activity, and the final evasion was caused by G504D/V503E in P4, similar to rep1. Interestingly, L371F was identified since P2 in SA55 rep2, which is far from its binding interface and not an escaping mutation against SA55, given that SA55 could potently neutralize BA.2, which harbors 371F. This may be due to the potential advantage of 371F in cell infection compared with 371L.
In comparison with using SA55 or SA58 alone, inhibition by the SA55+SA58 cocktail could survive more passages, ranging from P4 to P7 in four replicates (Figure 4B). In rep1, the neutralization by the SA55+SA58 cocktail was not affected until the co-occurrence of G504D (which escaped SA55) and R346Q (which affected SA58) in P6, and the remaining activity of SA58 was finally eliminated by K444N in P7. Co-occurrence of G504D and R346Q was also observed at rep2 in P3–P4 and rep4 in P2–P3, while the remaining activity of SA58 was eliminated by D339Y/E340D in rep2 and T345N/E340D in rep4, respectively (Figure 4E). In rep3, L371F was observed again, which slightly affected the binding affinity of SA58 as reported previously.1 The final evasion was caused by the combination of SA58-escaping K444E and SA55-escaping V503E. These results demonstrate the enhanced ability of the SA55+SA58 cocktail to resist escaping mutations compared with their individual usage. Similar to most existing antibody cocktails, at least two escaping substitutions are required to escape SA55+SA58.
To further validate the identified escaping mutations and the broad activity of the SA55+SA58 cocktail, we constructed multiple VSV-based pseudoviruses harboring SARS-CoV-2 BA.2/BA.2.12.1/BA.5 spike with additional substitutions on RBD, including 339Y, 340K/D, 345N, 346Q/T, 356R/T, 440E, 444N/R/E/T, 445E/S, 503E, 504D/S, or 508H, and determined the neutralizing activity of SA55, SA58, and SA55+SA58 cocktails against them. The results correspond well with the above DMS, structural analyses, and rVSV-based mutation screening. Specifically, the efficacy of SA55 was slightly affected by Y508H; moderately affected by G504S; strongly affected by K440E; and escaped by V503E and G504D, and SA58 was slightly affected by K356T, D339Y, and K444N; moderately affected by T345N, R346Q, and R346T; strongly affected by E340D; and escaped by E340K and K444E (Figures 4F and S4A). However, most of the identified escaping mutations would lead to changes in surface electrostatic potential located on sites that are conserved across ACE2-utilizing sarbecoviruses (Figures 3A and 3B) and are not observed in circulation. Notable exceptions are R346T and K444N/T, which are not conserved and have emerged recently in circulation (Figures S4B and S4C). Sequences with D339H are also emerging, which is mainly contributed by BA.2.75 and its sublineages.44 , 45 Besides the pressure caused by NAbs in group E1, the emergence of these mutations can be promoted by NAbs in groups D1/D2, which are also affected by R346 and K444 mutations.1 Fortunately, R346T only causes a moderate drop in SA58 activity. The accumulation of mutations on R346 and K444 may further affect the neutralization of SA58, but SA55 could efficiently neutralize such mutants, including recently emerging variants with convergent mutations on RBD such as BQ.1, BQ.1.1, and XBB.31 SA55+SA58 potently neutralizes all tested SARS-CoV-2 Omicron mutants, demonstrating its noteworthy breadth and resistance against escaping mutations.
SA55+SA58 demonstrate high viral clearance efficacy in vivo
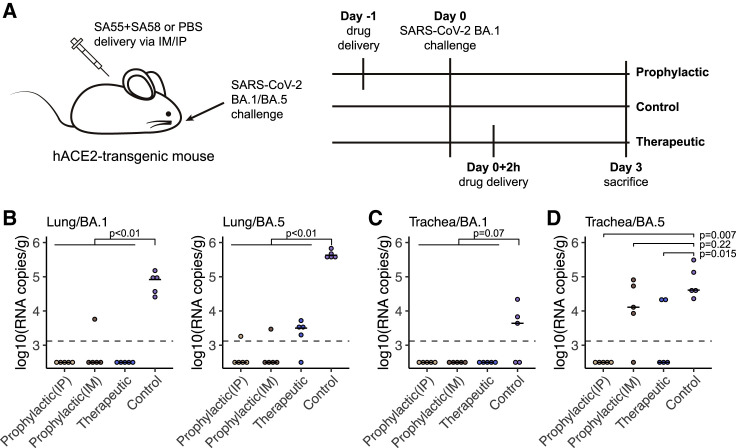
To evaluate the therapeutic and prophylactic efficacy of the SA55+SA58 cocktail against SARS-CoV-2 in vivo, human ACE2 (hACE2)-transgenic mice were challenged by an infectious SARS-CoV-2 BA.1 or BA.5 isolate before or after treatment by SA55+SA58. In prophylactic and therapeutic groups, 20 mg/kg 1:1 SA55 and SA58 was given to the hACE2-transgenic mice via intraperitoneal (i.p.) injection 24 h pre-infection or 2 h post-infection, respectively (Figures 5A and S5A). To test the robustness of different antibody delivery approaches, an additional prophylactic group of mice received SA55+SA58 via intramuscular (IM) injection. A control group of mice that received phosphate-buffered saline (PBS) instead of SA55+SA58 was also included. Each group consisted of five mice (Figure S5A). The body weight of each mouse in each group was monitored and recorded daily. All mice were sacrificed at day 3, and the lungs and trachea were collected for viral load analyses via qPCR.
Figure 5.
SA55/SA58 demonstrate potent protection efficacy in mice
(A) Schematic of the design for the SARS-CoV-2 BA.1 or BA.5 challenge experiment in mice. In prophylactic groups, 20 mg/kg SA55+SA58 was administered intraperitoneally or intramuscularly and challenged by SARS-CoV-2 BA.1 or BA.5 intranasally 1 day later. In therapeutic groups, antibodies were given intraperitoneally 2 h after virus challenge. Each group consists of 5 mice.
(B) Viral load in lungs of mice in each group.
(C and D) Viral load in trachea of mice challenged by BA.1 (C) or BA.5 (D) in each group. Each point corresponds to samples from a mouse. p values were calculated using two-tailed Wilcoxon rank-sum test. The limit of detection is about 1.3 × 103 copies/g and shown as the dashed line.
In general, mice in the control group exhibited significantly higher viral RNA copies in lungs compared with those that received SA55+SA58 (p < 0.01). Specifically, all ten mice in the control group exhibited a viral load of >104 RNA copies per gram in lung, while all 30 mice treated with SA55+SA58 before or after BA.1 or BA.5 challenge exhibited a viral load lower than 104 (Figure 5B).
Considering that Omicron variants infect more cells in the upper, rather than the lower respiratory, tract, we also measured the viral load in trachea.46 In the trachea of mice challenged by BA.1, virus was only detected in three of the five mice in the control group (Figure 5C). Although the difference between mice that received SA55+SA58 and PBS is not very significant (p = 0.07), we did not detect any viral RNA in the trachea of all 15 mice in prophylactic or therapeutic groups challenged by BA.1 (Figure 5C). In contrast, all 5 mice challenged by BA.5 in the control group exhibited >104 viral RNA copies per gram in their trachea (Figure 5D). No viral RNA was detected in trachea of all mice treated with SA55+SA58 before BA.5 challenge via i.p. (p = 0.007), but four mice treated before challenge via IM and two mice treated after challenge exhibited considerable viral load (p = 0.22 and 0.015, respectively) (Figure 5D). Notably, BA.1 and BA.5 infection only caused mild diseases that do not lead to obvious changes in body weight (Figure S5B). Together, SA55+SA58 displayed strong viral clearance and blocking efficacy in vivo, especially in lungs and when delivered via i.p.
Discussion
In this study, we proposed a rational strategy to discover broad NAb drugs that are strongly resistant to SARS-CoV-2 RBD mutations. We hypothesized that a good broad NAb candidate should avoid targeting the public immunodominant epitopes such that the virus will be less likely to evolve mutations on the susceptible residues of these NAb drug candidates; also, the drug candidates should target conserved residues that are associated with critical functions, such as receptor binding or antigen folding, which will enable stronger resistance to viral mutations. By integrating high-throughput DMS with scVDJ-seq, we successfully obtained a large collection of bsNAbs from SARS-CoV-2-vaccinated SARS convalescents and rationally selected a pair of featured bsNAbs, SA55+SA58, to form a non-competing cocktail that demonstrated exceptional neutralizing breadth and potency against SARS-CoV-2 variants of concern (VOCs) and related sarbecoviruses. Also, SA55+SA58 exhibited strong resistance to RBD mutations and may be resistant to escape by future SARS-CoV-2 variants. We believe this rational strategy can also be applied to discover broadly reactive antibody drugs against other rapidly evolving viruses.
SA55, which is from group F3, is indeed extremely rare and unique in SARS-CoV-2 vaccinees and convalescents, and its escape mutations, mainly V503X and G504X, did not exhibit any trends to prevail.1 , 31 In contrast, group E1 NAbs, including SA58, are not as rare as group F3, and they share some escape mutations, such as K444X and R346X, with groups D1/D2, promoting recent emergence of these mutations.31 , 47 , 48 , 49 The accumulation of such mutations may affects the neutralization of SA58, although group E1 is the only choice to form a cocktail with SA55, and SA58 is the best one among NAbs in group E1 that we could find.1 Despite this, the principle of our strategy still holds, and SA55+SA58 as a cocktail still neutralizes all tested SARS-CoV-2 variants, including BA.2.75, BA.2.75.2, BF.7, BQ.1, BQ.1.1, CH.1.1, XBB, and XBB.1, which accumulated evasive convergent mutations.31 , 44
Due to antibodies’ prolonged half-life, NAb drugs may serve as long-term prophylactics, where small-molecule drugs, such as Paxlovid, could not.50 , 51 When engineered on the Fc region, such as YTE modification, the half-life of NAbs can be further extended to nearly 90 days, suggesting that NAb drugs can stay effective for around half a year.50 Therefore, Fc-engineered SA55+SA58 could serve as long-term broad SARS-CoV-2 prophylactic NAb drugs, similar to Evusheld, which has been approved to be used as pre-exposure prophylaxis for the prevention of SARS-CoV-2 symptomatic infections. Compared with Evusheld, SA55+SA58 is more difficult for SARS-CoV-2 to escape, making it an advantageous drug candidate to afford long-lasting protection against current and future sarbecovirus mutants. Prophylactic SA55+SA58 would be especially valuable for elderly and immunocompromised individuals who cannot produce enough effective antibodies or are not suitable for vaccination. SA55+SA58 is scheduled to enter phase 1 clinical trial in December 2022.
Limitations of the study
DMS data used in this study were obtained based on a mutant library of SARS-CoV-2 ancestral strain RBD, but the mutations carried by Omicron have led to significant antigenic drift. Due to potential epistasis, there may be escape mutations based on Omicron that were not captured by DMS and that limit the neutralizing breadth of the selected NAbs against future variants as the accumulation of mutations. The neutralization mechanisms of group E1 NAbs, including SA58, that do not directly block ACE2 have not been clearly resolved. A recent study reported an NAb targeting an epitope similar to SA58 and demonstrated that it blocked viral-host membrane fusion.52 Although the strategy proposed in this study may be applied to other fast-evolving viruses in principle, this study only focus on identifying a NAb cocktail against sarbecoviruses, especially SARS-CoV-2 variants. Also, the efficacy of SA55+SA58 has only be tested in vitro and in mice. Further clinical studies are necessary to evaluate its safety and prophylactic/therapeutic efficacy against COVID-19 in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-human CD19 antibody | BioLegend | Cat#392508; RRID: AB_2750099 |
| FITC anti-human CD20 antibody | BioLegend | Cat#302304; RRID: AB_314252 |
| Brilliant Violet 421™ anti-human CD27 antibody | BioLegend | Cat#302824; RRID: AB_11150782 |
| PE/Cyanine7 anti-human IgM antibody | BioLegend | Cat#314532; RRID: AB_2566485 |
| FITC-conjugated chicken anti-cMyc antibody | Immunology Consultants Laboratory, Inc. | Cat#CMYC-45F |
| BD series SARS-CoV-2 RBD antibodies | This study | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 Wuhan-Hu-1 (SARS-CoV-2/human/CHN/CN1/2020) | This study | MT407649 |
| SARS-CoV-2 Omicron BA.1 (SARS-CoV-2/human/CHN/Omicron-1/2021) | This study | OM095411 |
| SARS-CoV-2 Omicron BA.5 (SARS-CoV-2/human/CHN/GD-5/2022) | This study | OP678016 |
| DH10Bac E. coli | Invitrogen | Cat#10359-016 |
| Cell lines | ||
| Saccharomyces cerevisiae strain EBY100 | ATCC | MYA-4941 |
| 293T | ATCC | CRL-3216 |
| HEK293F | ThermoFisher | Cat#R79007 |
| Huh-7 | JCRB | Cat#0403 |
| 293T-hACE2 | SinoBiological | N/A |
| Vero | ATCC | CCL-81 |
| HEK293F (in CD293) | ThermoFisher | Cat#11625019 |
| Sf21 | Invitrogen | Cat#B821-01 |
| High Five | Invitrogen | Cat#B855-02 |
| Chemicals, peptides, and recombinant proteins | ||
| X-tremeGENE 9 DNA Transfection Reagent | Roche | Cat#19129300 |
| Polyethylenimine | Polysciences | Cat#23966-2 |
| DMEM | ThermoFisher | Cat#11965092 |
| HEPES | ThermoFisher | Cat#15630080 |
| Iodoacetamide | Sigma Aldrich | Cat#I1149 |
| SMM 293-TI medium | Sino Biological | Cat#M293TI |
| SIM SF medium | Sino Biological | Cat#MSF1 |
| SIM HF medium | Sino Biological | Cat#MHF1 |
| PE-conjugated streptavidin | ThermoFisher | Cat#S866 |
| 3% Glutamine | Gibco | Cat#21051 |
| 199 culture medium | BasalMedia | Cat#X103A6 |
| SARS-CoV-2 (BA.4/BA.5) Spike RBD Protein (His Tag) | Sino Biological | Cat#40592-V08H130 |
| SARS-CoV-2 (WT) Spike RBD Protein (His Tag) | Sino Biological | Cat#40592-V08H |
| Critical commercial assays | ||
| MoFlo Astrios EQ Cell Sorter | Beckman Coulter | N/A |
| EasySep™ Human CD19 Positive Selection Kit II | BioLegend | Cat#392508 |
| Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1.1 | 10x Genomics | PN-1000165 |
| Chromium Single Cell 5′ Library Construction Kit | 10x Genomics | PN-1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B Cell | 10x Genomics | PN-1000016 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | PN-1000120 |
| NovaSeq 6000 S4 Reagent Kit v1.5 | Illumina | Cat#20028312 |
| NovaSeq XP 4-Lane Kit v1.5 | Illumina | Cat#20043131 |
| QIAamp Viral RNA Mini Kit | Qiagen | Cat#52904 |
| PrimeScript RT reagent Kit with gDNA Eraser | TaKaRa | Cat#RR047B |
| PrimeSTAR GXL DNA Polymerase | TaKaRa | Cat#R050B |
| Superdex 200 Increase 10/300 GL | Cytiva | Cat#28990944 |
| Superose 6 Increase 10/300 GL | Cytiva | Cat#29091596 |
| Biacore 8K | Cytiva | N/A |
| Series S Sensor Chip Protein A | Cytiva | Cat#29127556 |
| Series S Sensor Chip CM5 | Cytiva | Cat#29149603 |
| His Capture Kit | Cytiva | Cat#29234602 |
| Recombinant DNA | ||
| pVSV-SARS-CoV-2-S (Delta) | This study | encoding eGFP gene and codon-optimized SARS-CoV-2 Delta spike gene (GISAID: EPI_ISL_1911250) in place of VSV G |
| Plasmids expressing T7 polymerase and N, P, M and L of VSV, pcDNA3.1 | Li et al., 202042 | N/A |
| SARS-CoV-2 Omicron/Delta S gene, residues 1-1208, 6P and furin cleavage mutation, T4 fibritin trimerization motif, 8xHisTag, pcDNA | This study | N/A |
| BD55-5514 Fab, pcDNA | This study | N/A |
| BD55-5840 Fab, pcDNA | This study | N/A |
| BD55-3546 Fab, pcDNA | This study | N/A |
| BD55-4637 Fab, pcDNA | This study | N/A |
| BD55-5549 Fab, pcDNA | This study | N/A |
| Deposited data | ||
| Local refinement of BD55-4637 Fab in complex of BA.1 RBD | This study | PDB ID 7WRJ, EMD-32728 |
| Local refinement of BD55-3546 Fab in complex of Delta RBD | This study | PDB ID 7WRY, EMD-32737 |
| Local refinement of SA55 and SA58 Fab in complex of BA.1 RBD | This study | PDB ID 7Y0W, EMD-33552 |
| BD55-5549 Fab in complex of BA.1 RBD | This study | PDB ID 7Y0V |
| Software and algorithms | ||
| PyMOL (version 2.6.0a0) | Schrödinger, LLC. | https://pymol.org |
| dms_variants (version 0.8.9) | The Bloom lab | https://github.com/jbloomlab/dms_variants |
| ggplot2 (version 3.3.3) | Wickham, 201653 | https://ggplot2.tidyverse.org |
| FlowJo™ (version 10.8) | BD Biosciences | https://www.flowjo.com |
| logomaker (version 0.8) | Tareen and Kinney, 201954 | https://github.com/jbkinney/logomaker |
| hisat2 (version 2.2.1) | Kim et al., 201955 | http://daehwankimlab.github.io/hisat2/ |
| HKL2000 | HKL Research | https://www.hkl-xray.com/ |
| PHENIX | Liebschner et al., 201956 | https://www.phenix-online.org/ |
| COOT | Emsley et al., 201057 | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| SerialEM software | Mastronarde, 200558 | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | Zheng et al., 201759 | https://emcore.ucsf.edu/ucsf-software |
| RELION | Zivanov et al., 201860 | https://www2.mrc-lmb.cam.ac.uk/relion |
| UCSF Chimera | Pettersen et al., 200461 | https://www.cgl.ucsf.edu/chimera |
| UCSF ChimeraX | Pettersen et al., 202162 | https://www.cgl.ucsf.edu/chimerax |
| Biacore Insight Evaluation Software (version 4.0.8) | Cytiva | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents could be directed to and will be fulfilled by the Lead Contact, Xiaoliang Sunney Xie (sunneyxie@biopic.pku.edu.cn).
Materials availability
All reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cells
For rVSV-based mutant screening assays, HEK293T and Vero cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The cultures were maintained at 37°C in an incubator supplied with 5% CO2.
For neutralization assays, HEK293T and Huh-7 cells were cultured in DMEM supplemented with 10% FBS, maintained at 37°C in an incubator supplied with 8% CO2.
SARS-CoV-2 VSV-based pseudovirus
The SARS-CoV-2 pseudovirus was constructed as previously described using VSV pseudotyped virus (G∗ΔG-VSV).63 Pseudovirus carrying spikes of SARS-CoV-2 D614G (SARS-CoV-2 spike (GenBank: MN908947) +D614G), BA.1 (A67V, H69del, V70del, T95I, 142-144del, Y145D, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F), BA.2 (GISAID: EPI_ISL_7580387, T19I, del24–26, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K), BA.4/BA.5 (T19I, L24S, del25-27, del69-70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K) was constructed and used, as described previously.1
Authentic SARS-CoV-2 virus
SARS-CoV-2 Omicron BA.1 (SARS-CoV-2/human/CHN/Omicron-1/2021, GenBank: OM095411) was supplied from ILAS, PUMC. SARS-CoV-2 Omicron BA.5 (SARS-CoV-2/human/CHN/GD-5/2022, GenBank: OP678016) was kindly provided by Guangdong provincial center for disease control and prevention, China. To identify the stocks of the virus, the plaque-purified viral isolate was amplified as described previously. The virus titer of the supernatant was determined using a standard 50% tissue culture infection dose TCID50 assay.
Transgenic mice
Specific-pathogen-free, 6- to 8-week-old hACE2 mice (17–19g) were supplied by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Science, Peking Union Medical College. Transgenic mice were generated by microinjection of the mouse Ace2 promoter driving the human ACE2 coding sequence into the pronuclei of fertilized ova from ICR mice, and then human ACE2 integrated was identified by PCR as previously described.64
Method details
Antigen-specific cell sorting, V(D)J sequencing, and data analysis
Plasma samples were isolated from SARS-CoV-2-vaccinated SARS convalescents as described previously.1 Written informed consent was obtained from each participant in accordance with the Declaration of Helsinki. CD27+ IgM− B cells cross-binding to SARS-CoV-1 and SARS-CoV-2 RBD were sorted from plasma samples from SARS-CoV-2-vaccinated SARS convalescents with MoFlo Astrios EQ Cell Sorter (Beckman Coulter). Briefly, CD19+ B cells were enriched from PBMC with EasySep Human CD19 Positive Selection Kit II (STEMCELL, 17,854). CD19+ B cells were then stained with FITC anti-human CD19 antibody (BioLegend, 392,508), FITC anti-human CD20 antibody (BioLegend, 302,304), Brilliant Violet 421 anti-human CD27 antibody (BioLegend, 302,824), PE/Cyanine7 anti-human IgM antibody (BioLegend, 314,532), biotinylated Ovalbumin (SinoBiological) conjugated with Brilliant Violet 605 Streptavidin (BioLegend, 405,229), SARS-CoV-1 biotinylated RBD protein (His & AVI Tag) (SinoBiological, 40,634-V27H-B) conjugated to PE-streptavidin (BioLegend, 405,204), SARS-CoV-2 biotinylated RBD protein (His & AVI Tag) (SinoBiological, 40,592-V27H-B) conjugated to APC-streptavidin (BioLegend, 405,207). After washed twice, 7-AAD (Invitrogen, 00-6993-50) were added. 7-AAD-, CD19/CD20+, CD27+, IgM−, OVA−, SARS-CoV-1 RBD+, and SARS-CoV-2 RBD+ were sorted.
Isolated SARS-CoV-1 and SARS-CoV-2 RBD cross-binding B cells were then subjected to Chromium Next GEM Single Cell V(D)J Reagent Kits v1.1 following the manufacturer’s user guide (10x Genomics, CG000208). Briefly, cells were resuspended to an appropriated concentration after centrifugation. Cells were processed with 10X Chromium Controller to obtain gel beads-in-emulsion (GEMs) and then subjected to reverse transcription (RT). RT products were subjected to clean up, preamplification and purification with SPRIselect Reagent Kit (Beckman Coulter, B23318). Paired V(D)J sequence were enriched with 10X primers. After library preparation, libraries were sequenced by Novaseq 6000 platform running Novaseq 6000 S4 Reagent Kit v1.5 300 cycles (Illumina, 20,028,312) or NovaSeq XP 4-Lane Kit v1.5 (Illumina, 20,043,131).
Sequenced V(D)J raw data were mapped to CRCh38 V(D)J reference by Cell Ranger (v6.1.1). We filtered out non-productive contigs and kept the cell with only one heavy chain and one light chain. With IMGT reference, antibody region and gene annotation were performed by IgBlast (v1.17.1). Mutations were identified by the different nucleotide numbers between antibody and corresponding germline reference.
High-throughput yeast display-based mutation escape profile
Escape mutation profiling assays were performed based on the previously constructed deep mutational scanning libraries of SARS-CoV-2 RBD.1 , 23 , 65
Briefly, yeast display libraries were first thawed and amplified in SD-CAA media overnight. Then, RBD expression was induced in SG-CAA media at room temperature with mild agitation for 16-18h. The induced libraries were washed with PBST buffer (PBS with 0.02% Tween 20) and ready for further magnetic-activated cell sorting (MACS)-based mutation escape profiling. Protein A magnetic beads (Thermo Fisher, 10008D) were first conjugated with antibodies by 30min incubation at room temperature, then the Protein A-antibody conjugated beads were washed and incubated with prepared yeast libraries. To obtain pure result, two round of sequential above selections were conducted. The third-round selection was performed using the anti-c-Myc magnetic beads (Thermo Fisher, 88,843) to bring down background noise. Yeast cells finally obtained were grown overnight with SD-CAA media and proceed to plasmid extraction with 96 Well Plate Yeast Plasmid Preps Kit (Coolaber, PE053). The extracted plasmid products were used as the template for PCR amplification to detect N26 barcodes as described in.33 Final PCR products were further purified and submitted to Illumina Next 550 sequencing.
All sequencing data of yeast display deep mutational scanning are processed using the previously described pipeline.23 In brief, PacBio SMRT reads were mapped to a reference sequence containing SARS-CoV-2 RBD sequence, constant spacer region, and barcode N26, using the Python package dms_variants (v0.8.9), to construct the barcode-variant map. Sequenced barcodes from antibody-selected samples and their corresponding reference samples were also parsed using dms_variants (v0.8.9). Escape scores of each barcode X were defined as F×(nX,ab/Nab)/(nX,ref/Nref), where n and N are number of detected barcode X and total barcodes in antibody-selected (ab) or reference (ref) samples, respectively, and F is a scale factor to normalize the scores to 0-1 range. Escape scores of each substitution on RBD were estimated using epistasis models as described previously. For each antibody, only the mutations or residues whose escape scores were at least two times higher than the median score of all mutations or residues were retained. Logo plots for visualization of escape maps were generated using the R package ggseqlogo (v0.1).
Protein expression and purification for structural analysis
The SARS-CoV-2 S6P (F817P, A892P, A899P, A942P, K986P, and V987P) expression construct that encodes the spike ectodomain (residues 1-1208) with a “GSAS” substitution at the furin cleavage site (residues 682–685) was previously described.21 Delta mutations (T19R, G142D, 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N) and Omicron BA.1 mutations (A67V, H69del, V70del, T95I, G142D, V143del, Y144del, Y145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) were further engineered based on this construct. These plasmids were transiently transfected into HEK293F cells using polyethylenimine (Polysciences) to express the corresponding proteins. Culture supernatants were harvested at 96 h post-transfection, concentrated, and exchanged into the binding buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl). The proteins were first isolated using the Ni-NTA affinity method, and then further purified using a Superose 6 increase column in the final buffer (20 mM HEPES, pH 7.2, 150 mM NaCl). The antibody Fabs were expressed and purified as previously described.21
Cryo-EM data collection, processing, and structure building
Four microliter S6P protein (0.9 mg/mL) was mixed with the same volume of indicated Fabs (1 mg/mL each), and the resulting mixtures were applied onto glow-discharged Quantifoil holey carbon Au R1.2/1.3 grids using a Vitrobot (Mark IV), at 4°C and 100% humidity.37 After ∼3 s, the grids were plunged into the liquid ethane for vitrification. Data were collected using a Titan Krios (operating at 300 kV) equipped with a K3 direct detection camera (Gatan), and processed using cryoSPARC.66 To improve the local density for the Fab/RBD interface, UCSF Chimera61 and Relion67 were used to generate masks, and local refinements were further performed using cryoSPARC. COOT57 and PHENIX68 were used for structural modeling and the real-space refinement, respectively. Structural figures were generated using USCF ChimeraX62 and Pymol (Schrödinger, LLC.).
Crystallization and structure determination
The BD55-1403/BA.1 RBD and BD55-5549/BA.1 RBD complexes were formed by mixing the corresponding protein components at equimolar ratios and incubated on ice for 2 h. The resulting complexes were then further purified using a Superose 200 increase column in the final buffer. Purified Fab/RBD complexes were concentrated to ∼8 mg/mL and subjected to crystal screens. The BD55-1403/BA.1 RBD crystals were obtained by the vapor diffusion method at 18°C, and the reservoir solution contains 2% v/v Tacsimate (pH 5.0), 0.1 M Sodium citrate tribasic dihydrate (pH 5.6), and 16% w/v Polyethylene glycol 3,350. The BD55-5549/BA.1 RBD crystals were obtained in 0.1 M HEPES (pH 7.25), 10% w/v 2-Propanol, and 18% w/v Polyethylene glycol 4,000. For cryo-protection, crystals were soaked in the reservoir solutions supplemented with 17-19% v/v ethylene glycol, and then flash-cooled in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (beamline BL10U2), and processed then using HKL2000 (HKL Research). The structures were solved by molecular replacement using PHASER,69 adjusted in COOT and refined using PHENIX.
Pseudotyped virus neutralization assay
SARS-CoV-2 Spike pseudotyped virus was prepared based on a vesicular stomatitis virus (VSV) pseudotyped virus packaging system as previously reported.1 Pseudovirus neutralization assays were performed using the Huh-7 cell line (Japanese Collection of Research Bioresources [JCRB], 0403) (for D614G, BA.1, BA.1.1, BA.2, BA.3, BA.2.12.1, BA.4/BA.5, and SARS-CoV-1 pseudotyped virus neutralization assay) or 293T cells overexpressing human angiotensin-converting enzyme 2 (293T-hACE2) (Sino Biological Company) (for Pangolin-GD and RaTG13 pseudotyped virus neutralization assay). Antibodies were serially diluted in DMEM (Hyclone, SH30243.01) and mixed with pseudotyped virus and incubated for 1 h in a 37°C incubator with 5% CO2. Digested Huh-7 cells or 293T-hACE2 cells were dispensed to the antibody-virus mixture. After 22-26 h cells culture in 5% CO2, 37°C incubator, the supernatant was discarded and D-luciferin reagent (PerkinElmer, 6,066,769) was added, and plates were incubated in darkness for 2 min to allow complete cell lysis. Lysis was transferred to chemiluminescence detection plate and the luminescence value was detected with a microplate spectrophotometer (PerkinElmer, HH3400). IC50 was determined by a four-parameter logistic regression model.
Authentic virus neutralization CPE assay
Neutralization assay for authentic SARS-CoV-2 and its mutant strains using a cytopathic effect (CPE) assay. Monoclonal antibodies or plasma samples were serially diluted in DMEM, mixed with the same volume of the virus, and incubated in a 37°C incubator with 5% CO2. The mixture was added to a monolayer of Vero cells (ATCC, CCL-81) in a 96-well plate and cultured for 5 days. All wells were examined under a microscope and the CPE effect of each well was recorded. All experiments were performed in a biosafety level 3 (BSL-3) facility.
Generation of replication-competent rVSV-SARS-CoV-2-SBA.1
Replication-competent rVSV-SARS-CoV-2-SBA.1 was generated as described previously, expressing the eGFP reporter gene and the codon-optimized SARS-CoV-2 BA.1 strain spike protein (GISAID: EPI_ISL_6590782.2).42 , 43 Briefly, 293T cells were transfected with the rVSV backbone plasmid and five support plasmids encoding the T7 polymerase, N, P, M, and L of VSV using the calcium phosphate method. Recovery of the virus was determined by cytopathic effects and eGFP expression. Viruses in the supernatant were harvested and passaged on Vero cells to obtain viral stocks.
Selection of escape mutations
The rVSV-SARS-CoV-2-SBA.1 mutants that evade mAb neutralization were screened according to a previous report. Briefly, rVSV-SARS-CoV-2-SBA.1 (5 x 105 FFU) was preincubated with 5-fold serial dilution of monoclonal antibody at RT for 30 min. Then the antibody-virus mixtures were added to monolayers of Vero cells and incubated at 37°C. After four days, the supernatants from the wells containing the highest concentration of mAbs that showed extensive GFP-positive foci were harvested. For the next round of selection, the supernatants were passaged on fresh Vero cells in the presence of the same concentrations of mAbs as before. The passages continued until 50 μg/mL mAbs could no longer neutralize the mutant viruses.
Viral RNA was extracted from the supernatants of each passage using the QIAamp Viral RNA Mini Kit (Qiagen, US). Purified RNAs were reverse transcribed using PrimeScript RT reagent Kit with gDNA Eraser (Takara, JP) and the S gene was amplified using PrimeSTAR GXL DNA Polymerase (Takara, JP) for next-generation sequencing.
Identification of enriched escape substitutions
Illumina sequencing adapters were first trimmed from raw sequencing reads using trim_galore (default parameters, version 0.6.7, cutadapt version 1.18). Trimmed reads were mapped to the nucleotide sequence of SARS-CoV-2 BA.1 spike glycoprotein (codon-optimized for rVSV) using hisat2 (default parameters, v.2.2.1). PCR duplicates were removed from mapped reads using Picard MarkDuplicates (REMOVE_DUPLICATES = TRUE, version 2.18.29). SNP information was extracted from deduplicated BAM files using bcftools mpileup (-d 900000 -C 50 -Q 30 -q 5, version 1.8). Detected nucleotide mutations at each position were then parsed using custom Python scripts to remove all insertions or deletions, and converted into the occurrence ratio of residue substitutions.
Surface plasma resonance
SPR experiments were performed using Biacore 8K (Cytiva). SA55 or SA58 (human IgG1) was captured by a Sensor Chip Protein A (Cytiva), and SARS-CoV-2 WT or BA.5 RBD (His Tag, Sino Biological) of various concentration (1.5625 nM, 3.125 nM, 6.25 nM, 12.5 nM, 25 nM, and 50 nM) were injected. The response was recorded at room temperature, and the raw data curves were fitted to a 1:1 binding model using Biacore Insight Evaluation Software (Cytiva, v4.0.8).
For competition assays, SARS-CoV-2 WT or BA.5 RBD (His Tag, Sino Biological) was captured by a Sensor Chip CM5 (Cytiva) with immobilized anti-His using His Capture Kit (Cytiva), and SA55 or SA58 (200 nM) was injected. After equilibrium, the other mAb was also added (SA58+SA55, 200 nM each). The response was recorded by Biacore Insight Evaluation Software (Cytiva, v4.0.8) at room temperature. For control, we also determined the responses using SA55 or SA58 (200 nM) only.
In vivo SARS-CoV-2 challenge in mice
Murine studies were performed in an animal biosafety level 3 (ABSL3) facility using HEPA-filtered isolators. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Science, Peking Union Medical College (BLL22007). Mice were inoculated intranasally with authentic SARS-CoV-2 BA.1 (SARS-CoV-2/human/CHN/Omicron-1/2021, GenBank: OM095411) or SARS-CoV-2 BA.5 (SARS-CoV-2/human/CHN/GD-5/2022, GenBank: OP678016) stock virus at 1 × 105 TCID50. The infected mice were observed daily to record body weights and were sacrificed at 3 days post infection, and the lungs and tracheas were collected for viral load detection.
Viral load analysis was performed by qRT-PCR. The total RNA of the lungs and trachea was extracted with the RNeasy Mini Kit (Qiagen). Lung and trachea homogenates were prepared by using an electric homogenizer. The reverse transcription was processed with PrimerScript RT Reagent Kit (TaKaRa) according to the manufacturers’ instructions. qRT-PCR reactions were performed using the PowerUp SYBG Green Master Mix Kit (Applied Biosystems), according to following cycling protocol: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles at 95°C for 15 s and 60°C for 30 s, and then 95°C for 15 s, 60°C for 1 min, 95°C for 45 s. Forward primer 5′-TCGTTTCGGAAGAGACAGGT-3′ and reverse primer 5′-GCGCAGTAAGGATGGCTAGT-3′ were used in qRT-PCR. Standard curves were constructed by using 10-fold serial dilutions of recombinant plasmids with known copy numbers (from 1.47 × 109 to 1.47 × 101 copies/μl).
Quantification and statistical analysis
SPR assays were performed in two biological replicates. SPR data were fitted to a 1:1 binding model by Biacore Insight Evaluation Software. Neutralization assays were performed in at least two biological replicates. IC50 values were determined by a four-parameter logistic regression model. Two-tailed Wilcoxon rank-sum tests were used to examine the significant differences in viral load between groups, as described in Figures 5B–5D.
Acknowledgments
We thank Sino Biological for the technical assistance on mAb and RBD expression. We thank J. Luo and H. Lv for the help with flow cytometry. We thank Singlomics Biopharmaceuticals for providing technical help on antibody screening. We thank Changwen Ke from the Guangdong Provincial Center for Disease Control and Prevention for providing SARS-CoV-2 BA.5 as a gift. This project is financially supported by Beijing Municipal Science and Technology Project (Z211100002521018), the Ministry of Science and Technology of the People's Republic of China and Changping Laboratory (2021A0201 and 2021D0102), and the National Natural Science Foundation of China (32222030).
Author contributions
Y.C. and X.S.X. designed the study. Y.C., F.J., and X.S.X. wrote the manuscript with input from all authors. Q.G. proofed the manuscript. Y.C. and F.S. coordinated the expression and characterization of the neutralizing antibodies. J.W., F.J., and Y.C. performed and analyzed the yeast display screening experiments. Y.Y., T.X., P.W., R.A., Yao Wang, and X.N. performed the neutralizing antibody expression and characterization, including pseudovirus neutralization assays and ELISA. Y.Y. and Youchun Wang prepared the VSV-based SARS-CoV-2 pseudovirus. F.Y., Yuhang Zhang, and A.Z. conceived and performed the rVSV-EGFP-SARS-CoV-2-based mutant screening assays, and F.J. analyzed the data. A.Y., Yao Wang, S.Y., R.A., and W.S. performed and analyzed the antigen-specific single B cell VDJ sequencing. S.D., Ying Zhang, P.L., Z.Z., and J.X. performed the structural analyses. Y.H. and W.Y. performed and coordinated the authentic virus neutralization cytopathic effect (CPE) assay. L.B. and C.Q. performed animal experiments. X.H. and R.J. recruited the SARS convalescents.
Declaration of interests
X.S.X. and Y.C. are the inventors of the provisional patent applications for BD series antibodies, which includes BD55-5840 (SA58) and BD55-5514 (SA55). X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals. W.Y. and Y.H. are the CEO and vice president of Sinovac Biotech, respectively. SA58 and SA55 have been transferred to Sinovac Biotech for clinical development.
Published: December 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111845.
Supplemental information
Data and code availability
Sequences of the mAbs involved in this study are available in Table S2, and processed mutation escape scores reported previously could be downloaded from https://github.com/jianfcpku/SARS-CoV-2-RBD-DMS-broad, which have been described previously.1 Processed rVSV-based mutant screening data and custom scripts for analyses of sequencing data from rVSV-based mutant screening assays are available at https://github.com/jianfcpku/escaping-mutants-screening.
Cryo-EM density maps have been deposited in the Electron Microscopy DataBank with accession codes EMDB: EMD-32728, EMD-32737, EMD-33552. Structural coordinates have been deposited in the Protein DataBank with accession codes PDB: 7WRJ, 7WRY, 7Y0W, 7Y0V.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 3.Cromer D., Steain M., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Kent S.J., Triccas J.A., Khoury D.S., Davenport M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet. Microbe. 2022;3:E52–E61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon-Loriere E., Schwartz O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022;20:187–188. doi: 10.1038/s41579-022-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 8.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184:3949–3961.e11. doi: 10.1016/j.cell.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougan M., Azizad M., Chen P., Feldman B., Frieman M., Igbinadolor A., Kumar P., Morris J., Potts J., Baracco L., et al. Bebtelovimab, alone or together with bamlanivimab and etesevimab, as a broadly neutralizing monoclonal antibody treatment for mild to moderate, ambulatory COVID-19. medRxiv. 2022 doi: 10.1101/2022.03.10.22272100. Preprint at. [DOI] [Google Scholar]
- 12.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 13.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 16.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park Y.J., Pinto D., Walls A.C., Liu Z., De Marco A., Benigni F., Zatta F., Silacci-Fregni C., Bassi J., Sprouse K.R., et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. bioRxiv. 2022 doi: 10.1101/2022.05.08.491108. Preprint at. [DOI] [PubMed] [Google Scholar]
- 20.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 23.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dacon C., Tucker C., Peng L., Lee C.C.D., Lin T.H., Yuan M., Cong Y., Wang L., Purser L., Williams J.K., et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022;377:728–735. doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Low J.S., Jerak J., Tortorici M.A., McCallum M., Pinto D., Cassotta A., Foglierini M., Mele F., Abdelnabi R., Weynand B., et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W.T., Musharrafieh R., Song G., Dueker K., Tse L.V., Martinez D.R., Schäfer A., Callaghan S., Yong P., Beutler N., et al. Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS-like viruses. Nat. Immunol. 2022;23:960–970. doi: 10.1038/s41590-022-01222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez D.R., Schäfer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Stalls V., Janowska K., et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 2022;14:eabj7125. doi: 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y.J., De Marco A., Starr T.N., Liu Z., Pinto D., Walls A.C., Zatta F., Zepeda S.K., Bowen J.E., Sprouse K.R., et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science. 2022;375:449–454. doi: 10.1126/science.abm8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Zhang N., Wang Y., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaney A.J., Starr T.N., Bloom J.D. An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain. Virus Evol. 2022;8:veac021. doi: 10.1093/ve/veac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan C.W., Chia W.N., Young B.E., Zhu F., Lim B.L., Sia W.R., Thein T.L., Chen M.I.C., Leo Y.S., Lye D.C., Wang L.F. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du S., Liu P., Zhang Z., Xiao T., Yasimayi A., Huang W., Wang Y., Cao Y., Xie X.S., Xiao J. Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants. Cell Res. 2021;31:1130–1133. doi: 10.1038/s41422-021-00555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An Y., Li S., Jin X., Han J.B., Xu K., Xu S., Han Y., Liu C., Zheng T., Liu M., et al. A tandem-repeat dimeric RBD protein-based covid-19 vaccine zf2001 protects mice and nonhuman primates. Emerg. Microbes Infect. 2022;11:1058–1071. doi: 10.1080/22221751.2022.2056524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toelzer C., Gupta K., Yadav S.K.N., Borucu U., Davidson A.D., Kavanagh Williamson M., Shoemark D.K., Garzoni F., Staufer O., Milligan R., et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starr T.N., Greaney A.J., Stewart C.M., Walls A.C., Hannon W.W., Veesler D., Bloom J.D. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. bioRxiv. 2022 doi: 10.1101/2022.09.20.508745. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Zhao C., Zhang Y., Yuan F., Zhang Q., Shi X., Zhang L., Qin C., Zheng A. Establishment of replication-competent vesicular stomatitis virus-based recombinant viruses suitable for SARS-CoV-2 entry and neutralization assays. Emerg. Microbes Infect. 2020;9:2269–2277. doi: 10.1080/22221751.2020.1830715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Zhang Y., Li D., Deng Y.Q., Xu H., Zhao C., Liu J., Wen D., Zhao J., Li Y., et al. Enhanced protective immunity against SARS-CoV-2 elicited by a VSV vector expressing a chimeric spike protein. Signal Transduct. Target. Ther. 2021;6:389. doi: 10.1038/s41392-021-00797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30:1527–1539.e5. doi: 10.1016/j.chom.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Anraku Y., Kimura I., Ito J., Yamasoba D., Nasser H., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Cell Host Microbe. 2022;30:1540–1555.e15. doi: 10.1016/j.chom.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu P., Evans J.P., Faraone J., Zheng Y.-M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Distinct neutralizing antibody escape of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. bioRxiv. 2022 doi: 10.1101/2022.10.19.512891. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jian F., Yu Y., Song W., Yisimayi A., Yu L., Gao Y., Zhang N., Wang Y., Shao F., Hao X., et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect. Dis. 2022;22:1535–1537. doi: 10.1016/s1473-3099(22)00642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Li Z., Ho J., Guo Y., Yeh A.Y., Liu M., Wang M., Yu J., Sheng Z., Huang Y., et al. Resistance of SARS-CoV-2 Omicron subvariant BA.4.6 to antibody neutralization. bioRxiv. 2022 doi: 10.1101/2022.09.05.506628. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loo Y.M., McTamney P.M., Arends R.H., Abram M.E., Aksyuk A.A., Diallo S., Flores D.J., Kelly E.J., Ren K., Roque R., et al. The SARS-CoV-2 monoclonal antibody combination, AZD7442, is protective in nonhuman primates and has an extended half-life in humans. Sci. Transl. Med. 2022;14:eabl8124. doi: 10.1126/scitranslmed.abl8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfizer Pfizer shares top-line results from phase 2/3 EPIC-PEP study of PAXLOVID™ for post-exposure prophylactic use. 2022. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-shares-top-line-results-phase-23-epic-pep-study
- 52.Luo S., Zhang J., Kreutzberger A.J.B., Eaton A., Edwards R.J., Jing C., Dai H.Q., Sempowski G.D., Cronin K., Parks R., et al. An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion. Sci. Immunol. 2022;7:eadd5446. doi: 10.1126/sciimmunol.add5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham H. Springer-Verlag New York; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 54.Tareen A., Kinney J.B. Logomaker: beautiful sequence logos in Python. Bioinformatics. 2020;36:2272–2274. doi: 10.1093/bioinformatics/btz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.-W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 65.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 67.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of the mAbs involved in this study are available in Table S2, and processed mutation escape scores reported previously could be downloaded from https://github.com/jianfcpku/SARS-CoV-2-RBD-DMS-broad, which have been described previously.1 Processed rVSV-based mutant screening data and custom scripts for analyses of sequencing data from rVSV-based mutant screening assays are available at https://github.com/jianfcpku/escaping-mutants-screening.
Cryo-EM density maps have been deposited in the Electron Microscopy DataBank with accession codes EMDB: EMD-32728, EMD-32737, EMD-33552. Structural coordinates have been deposited in the Protein DataBank with accession codes PDB: 7WRJ, 7WRY, 7Y0W, 7Y0V.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.