Abstract
Recently, MDS with mutated SF3B1 and blast count <5% was proposed as distinct entity with favorable prognosis by the international working group for the prognosis of MDS (IWG-PM), the 5th edition of the WHO classification and the International Consensus Classification. To further characterize this entity with respect to the genomic landscape, AML transformation rate and clinical outcome, we analyzed 734 MDS patients by whole genome sequencing. SF3B1 mutations were identified in 31% (n = 231), most frequently accompanied by TET2 mutations (29%). 144/231 (62%) SF3B1mut samples fulfilled entity criteria proposed by IWG-PM (SF3B1ent). These cases were associated with longer survival, lower AML transformation rate, normal karyotypes and harbored less accompanying mutations compared to SF3B1mut samples not falling into the proposed SF3B1 entity (SF3B1nent). Of SF3B1mut cases 7% (15/231; SF3B1ent: 3/144 [2%]; SF3B1nent: 12/87 [14%]) progressed to AML compared to 15% SF3B1 wild-type patients (75/503). Of these 15 SF3B1mut cases, 10 (67%) showed RUNX1 mutations at MDS or AML stage. Multivariate analysis revealed that del(5q) and RUNX1 mutations were independent negative prognostic factors for overall survival, while blast count >5% was not. In conclusion, SF3B1mut MDS has a favorable prognosis independent of blast count if karyotype and RUNX1 mutations are considered.
Subject terms: Myelodysplastic syndrome, Cancer genomics
Introduction
Myelodysplastic neoplasms (MDS) are clonal disorders characterized by peripheral cytopenias, morphologic dysplasia in hematopoietic cells and ineffective hematopoiesis [1, 2]. The currently used revised 4th edition of the WHO classification (WHO 2017) in MDS is mainly based on the number of cytopenias, dysplastic lineages, and the percentage of ring sideroblasts (RS) and blasts detected in bone marrow and peripheral blood samples [3]. Within the last years, the use of next generation sequencing (NGS) enabled the identification of driver genes in MDS providing insights into the underlying heterogeneous genetic landscape [4–6]. In this line, about half of MDS patients harbor somatic mutations in splicing pathway genes. Of these, SF3B1 is the most commonly mutated gene and if mutated shown to be associated with RS, higher white blood cell counts and lower bone marrow blasts [6–9]. Moreover, SF3B1 mutations define a distinct MDS subset showing favorable prognosis and indolent disease course [10]. Thus, in the WHO 2017 the SF3B1 mutation is integrated into the diagnosis of MDS-RS (diagnostic criteria: RS ≥ 15% or RS ≥ 5% if SF3B1mut) [1].
Following up on this, the international working group for the prognosis of MDS (IWG-PM) proposed MDS with mutated SF3B1 as a distinct entity if certain criteria are fulfilled (Supplementary Table S1) [11]. These criteria included: (1) cytopenia as defined by standard hematologic values, (2) somatic SF3B1 mutation, (3) morphologic dysplasia (with or without RS), (4) bone marrow blasts <5% and peripheral blood blasts <1%, and (5) WHO 2017 criteria for MDS 5q-, MDS/MPN-RS-T, or other MDS/MPN or MPN are not met. Further exclusion criteria were: (1) poor-risk cytogenetics comprising monosomy 7, inv(3) or abnormalities of chromosome 3q26, and complex karyotype (≥3 chromosomal abnormalities); and (2) accompanying mutations in RUNX1 and/or EZH2. The presence of JAK2V617F, CALR, or MPL mutations would strongly support the diagnosis of MDS/MPN-RS-T.
The upcoming 5th edition of the WHO Classification (WHO 2022) emphasizes a genetic basis for defining diseases and has now categorized MDS into morphologically defined MDS and MDS with defining genetic abnormalities (DGA) while largely abandoning the blast cut-off between MDS and AML if AML DGA are present [2]. It has further incorporated many of the proposed IWG-PM criteria into the newly introduced entity “MDS with low blasts and SF3B1 mutation” [2]. However, according to WHO 2022 only biallelic TP53 inactivations are excluded besides certain cytogenetic abnormalities (Supplementary Table S1). In contrast to the WHO 2022, the International Consensus Classification (ICC) requires an SF3B1 variant allelic frequency (VAF) ≥ 10% in the absence of certain cytogenetic abnormalities, RUNX1 and multi-hit TP53 (Supplementary Table S1) [12]. It further sets the blast cut-off for AML-DGA to 10%, while cases with 10–19% blasts without DGA are assigned as a new category MDS/AML. In this study, we defined the SF3B1 entity (SF3B1ent) based on the first publication proposed by the IWG-PM, but also discuss the changes in classification according to 5th edition of the WHO classification and ICC.
The aim of the study was to analyze the SF3B1 mutation and the proposed SF3B1 entity in a large cohort of 734 MDS patients with respect to the incidence, genomic landscape, AML transformation rate and clinical outcome.
Material and methods
Patients cohort and samples
For this analysis, we selected 734 MDS samples with material available to perform whole genome sequencing sent to the MLL Munich Leukemia Laboratory between 09/2005 and 12/2019. Diagnoses (from peripheral blood and bone marrow) were made based on cytomorphology, cytogenetics and molecular genetics as previously published [13–15]. All cases were classified into specific subgroups according to WHO 2017 [16]. For abbreviations of entities, see Table 1. Therapy-related MDS were excluded from this study. The MDS cohort comprised 310 (42%) female and 424 (58%) male cases with a median age of 73 years (range: 23–93 years) and a median follow-up of 9.3 years. All patients gave their written informed consent for genetic analyses and to the use of laboratory results as well as clinical data for research purposes according to the Declaration of Helsinki. The study was further approved by the laboratory´s institutional review board.
Table 1.
WHO entities and SF3B1 classification of the MDS cohort.
| WHO 2017 Diagnosis | Number of samples, n (%) | SF3B1wt, n (%) | SF3B1mut, n (%) | SF3B1ent, n (% of SF3B1mut) | SF3B1nent, n (% of SF3B1mut) |
|---|---|---|---|---|---|
| MDS with single lineage dysplasia (MDS-SLD) | 22 (3) | 21 (95) | 1 (5) | 1 (100) | 0 (0) |
| MDS with multilineage dysplasia (MDS-MLD) | 105 (14) | 104 (99) | 1 (1) | 1 (100) | 0 (0) |
| MDS with single lineage dysplasia with ring sideroblasts (MDS-RS-SLD) | 51 (7) | 8 (16) | 43 (84) | 37 (84) | 6 (16) |
| MDS with multilineage dysplasia with ring sideroblasts (MDS-RS-MLD) | 149 (20) | 21 (14) | 128 (86) | 105 (82) | 23 (18) |
| MDS with isolated del(5q) (MDS 5q-) | 107 (15) | 86 (80) | 21 (20) | 0 (0) | 21 (100) |
| MDS with excess blasts (MDS-EB-1) | 149 (20) | 124 (83) | 25 (17) | 0 (0) | 25 (100) |
| MDS with excess blasts (MDS-EB-2) | 151 (21) | 139 (92) | 12 (8) | 0 (0) | 12 (100) |
| MDS total | 734 | 503 (69) | 231 (31) | 144 (62) | 87 (38) |
wt wild-type, mut mutated, SF3B1ent proposed SF3B1 entity, SF3B1nent SF3B1 mutated cases not meeting SF3B1 entity criteria.
Whole genome sequencing (WGS) and variant filtering
WGS analysis was performed for all patients. For this, total genomic DNA was extracted from lysed cell pellet of bone marrow or peripheral blood using the MagNA Pure 96 with DNA and Viral Nucleic Acid Large Volume Kit and Cellular RNA Large Volume Kit (Roche, Basel, Switzerland). Library preparation and sequencing as well as calling and filtering of single nucleotide variants, structural variants and somatic copy number variations (CNVs) were performed as previously described [17, 18]. Copy neutral loss of heterozygosity (CN-LOH) was assessed using HadoopCNV.
Mutational analysis
In this study, we evaluated mutations in 73 genes associated with myeloid neoplasms for all patients from WGS data only or from combined WGS and targeted NGS panels (see supplementary material). Out of all 734 cases, 605 samples were additionally analyzed by targeted sequencing within a recent study [6] and 87 cases were analyzed by targeted NGS during routine diagnostics [19]. WGS data confirmed all mutations detected by targeted NGS panels and was further consulted for completing the mutational analysis of the 73 genes. The presence of FLT3-ITD and KMT2A-PTD were retrieved from WGS data only.
Statistical analysis
Statistical analyses were performed using SPSS version 19.0 (IBM Corporation, Armonk, NY). Analyses for overall survival (OS) and cumulative incidence (CI) of disease progression were performed according to Kaplan-Meier and compared using two-sided log rank tests. The OS was calculated as time from diagnosis to death or last follow-up. For the CI of disease progression death was considered as a competing event. Between different groups numerical variables were compared using the Mann–Whitney-U-Test, and dichotomous variables using chi-square test. Cox proportional hazards regression model was used to identify the impact of different variables on OS or AML transformation. All results were considered significant at p < 0.05.
Results
Incidence and prognostic impact of SF3B1 mutations
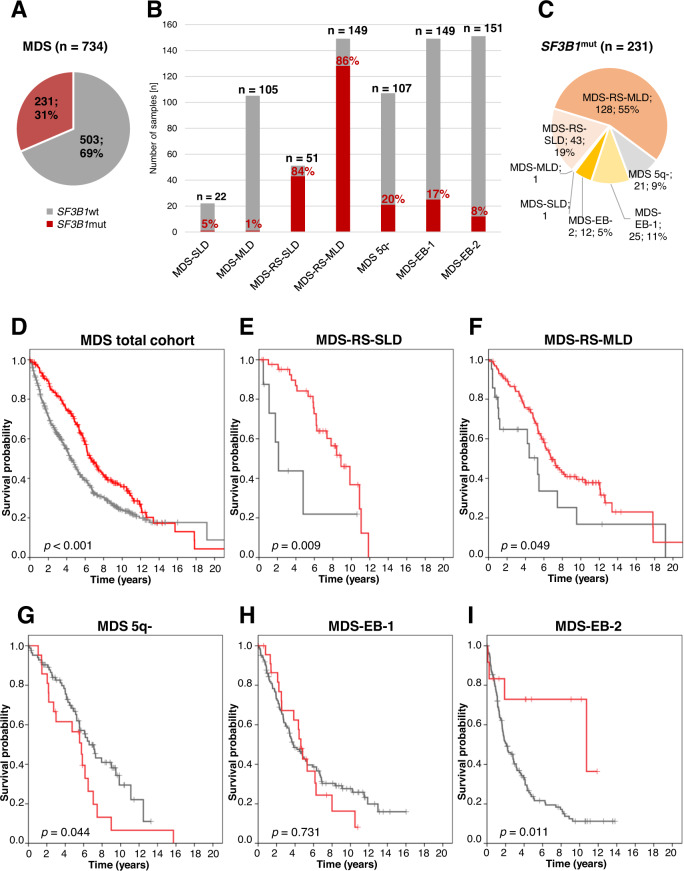
SF3B1 mutations were identified in 231 of 734 (31%) MDS patients and were mainly found in MDS-RS (171/200; 86%; MDS-RS-SLD: 43/51, 84%; MDS-RS-MLD: 128/149; 86%) resulting in 74% (171/231) of all SF3B1mut cases (Table 1; Fig. 1A–C). In addition, 13% (37/300) of MDS with excess blasts (MDS-EB-1/2) and 20% (21/107) of MDS 5q- harbored SF3B1 mutations together accounting for 25% (58/231) of all SF3B1mut cases (Table 1; Fig. 1B, C). The remaining 1% of SF3B1mut cases were an MDS-SLD and an MDS-MLD sample. Of note, SF3B1 mutations were most frequently found in patients with blast count <5% (192/419; 46%).
Fig. 1. Distribution and OS of SF3B1 mutations in MDS.
A Frequency of SF3B1 mutations in the entire MDS cohort; wt wild-type, mut mutated. B Proportion of SF3B1mut cases within different MDS entities (red: mutated; gray: wild-type). C WHO 2017 entities of SF3B1mut MDS. D OS of patients with mutated (n = 231; red) vs. wild-type (n = 503; gray) SF3B1 within the entire MDS cohort. E OS of MDS-RS-SLD patients with mutated (n = 43; red) vs. wild-type (n = 8; gray) SF3B1. F OS of MDS-RS-MLD patients with mutated (n = 128; red) vs. wild-type (n = 21; gray) SF3B1. G OS of MDS 5q- patients with mutated (n = 21; red) vs. wild-type (n = 86; gray) SF3B1. H OS of MDS-EB-1 patients with mutated (n = 25; red) vs. wild-type (n = 124; gray) SF3B1. I OS of MDS-EB-2 patients with mutated (n = 12; red) vs. wild-type (n = 139; gray) SF3B1.
In the total MDS cohort SF3B1 mutations were associated with better OS (median: 79 vs. 53 months; p < 0.001; Fig. 1D). Within the different MDS entities SF3B1 mutations were favorable in MDS-RS-SLD (median OS: 106 vs. 25 months; p = 0.009), MDS-RS-MLD (median: 82 vs. 64 months; p = 0.049) and MDS-EB-2 (median: 129 vs. 25 months; p = 0.011), but were associated with a shorter OS in MDS 5q- (median: 69 vs. 79 months; p = 0.044) (Fig. 1E–I). Irrespective of the SF3B1 mutation status, MDS-RS-SLD patients showed the best OS within the entire MDS cohort, while in contrast MDS with excess blasts was associated with the shortest OS (Supplementary Fig. S1A). A similar pattern was observed when focusing on SF3B1 mutated (SF3B1mut) patients (Fig. S1B). However, the unusual long OS for SF3B1 mutated MDS-EB-2 might be affected by therapy, in this regard allogeneic stem cell transplantation (SCT) received by 3/12 SF3B1mut MDS-EB-2 patients (Supplementary Table S2).
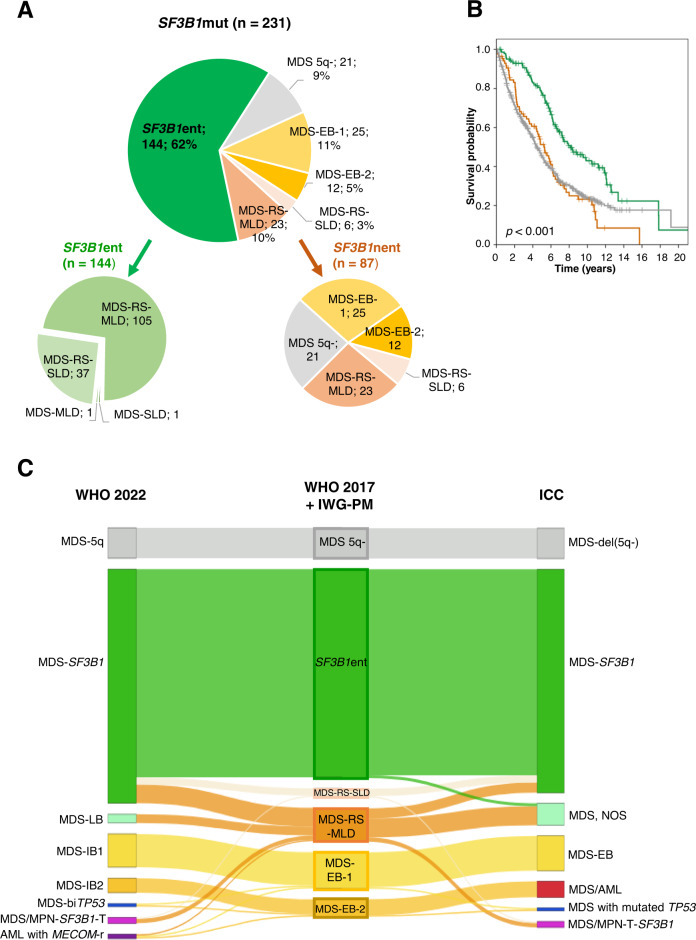
Within the SF3B1mut cohort 144/231 (62%) samples fulfilled the criteria proposed by IWG-PM (SF3B1ent) (Table 1; Fig. 2A; Supplementary Fig. S2A). SF3B1ent cases had a longer OS compared to SF3B1mut samples not falling into the proposed SF3B1 entity (SF3B1nent) (Fig. 2B; median: 97 vs. 63 months; p < 0.001). However, no positive effect of SF3B1 mutations on OS was observed within MDS-RS-SLD or MDS-RS-MLD if SF3B1 non-entity mutated cases were compared to wild-type cases (Supplementary Fig. S3A, B). Of note, SF3B1 mutations were associated with the presence of RS in both groups (SF3B1ent and SF3B1nent), showing median percentages of 63 and 49, respectively (Supplementary Fig. S2B; p < 0.001).
Fig. 2. Categorization and OS of SF3B1mut samples.
A WHO 2017 entities of SF3B1mut samples and classification into the IWG-PM proposed SF3B1 entity (SF3B1ent) or non-SF3B1 entity (SF3B1nent). B OS of patients with mutated SF3B1 fulfilling criteria for proposed SF3B1 entity (n = 144; green) or not (n = 87; brown) vs. wild-type SF3B1 (n = 503; gray) (p < 0.001). C Comparison of SF3B1mut MDS diagnoses based on the currently used revised 4th edition of the WHO (WHO 2017) and the IWG-PM criteria (middle) to the corresponding MDS diagnoses considering the upcoming 5th edition of WHO (WHO 2022; left) and the International Consensus Classification (ICC; right).
Differences in the defining criteria for the SF3B1 entity between WHO 2022, ICC and IWG-PM lead to changes in assignment of 18 and 14 SF3B1mut cases, respectively (Supplementary Table S1; Fig. 2C). A detailed analysis of the changes is described in the supplement.
Recurrent SF3B1 mutations
Within the entire cohort, 25 different SF3B1 mutations, most frequently affecting amino acid K700 (53%, 123/231), were detected with a mean VAF ranging from 22% to 48% (Supplementary Fig. S4). In 5/231 patients two different SF3B1 mutations were detected resulting in 236 SF3B1 mutations in total (Table in Supplementary Fig. S4B). The VAF of each SF3B1 mutation did not exceed 50% (range: 4–50%) (Fig. 3A). Of all SF3B1 mutations 77% (182/236) showed a VAF > 30% with SF3B1ent accounting for 66% (120/182). Moreover, 17% (39/236) showed a VAF between 15% and 29%, mainly belonging to SF3B1ent (25/39, 64%). SF3B1 VAFs <15% were seen in 15 cases, rarely in SF3B1ent (20%, 3/15). However, two of those 15 cases (one SF3B1ent; one MDS-RS-MLD) showed a second SF3B1 mutation with a VAF > 20% (Supplementary Fig. S4B). Of the remaining cases having SF3B1 mutations with VAFs <15% (n = 13), 5/13 (39%) samples were MDS 5q-, 6 (46%) MDS-EB-1/2 and 2 (15%) were SF3B1ent (Fig. 3B). Of note, no CNVs or CN-LOHs overlapping with SF3B1 were found.
Fig. 3. Variant allelic frequencies (VAFs) of 231 SF3B1mut samples.
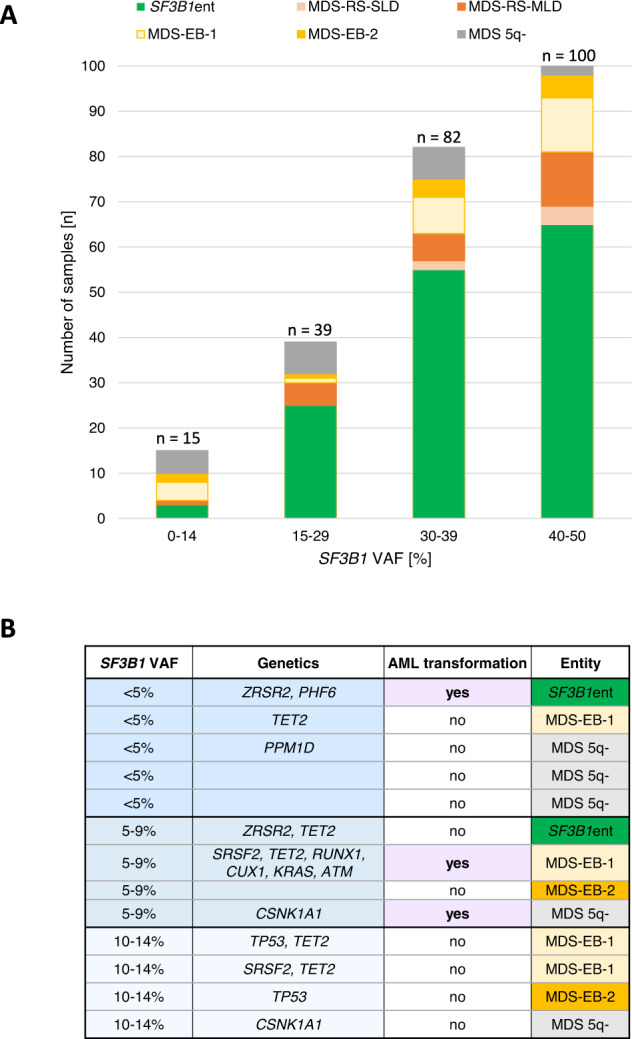
A SF3B1 VAFs with respect to the different entities; n (mutations) = 236. B Characteristics of cases having only one SF3B1 mutation and a VAF below 15%.
Genomic landscape of SF3B1mut patients
Regarding cytogenetic abnormalities, 69/231 (30%) SF3B1mut samples showed aberrant karyotypes (SF3B1ent: 27/144, 19%; SF3B1nent: 42/87, 48%; p < 0.001; Supplementary Fig. S5A). Notably, cytogenetic risk groups poor and very poor according to the Revised International Prognostic Scoring System (IPSS-R) were found in 11 SF3B1nent but in none of SF3B1ent cases (Supplementary Fig. S5B).
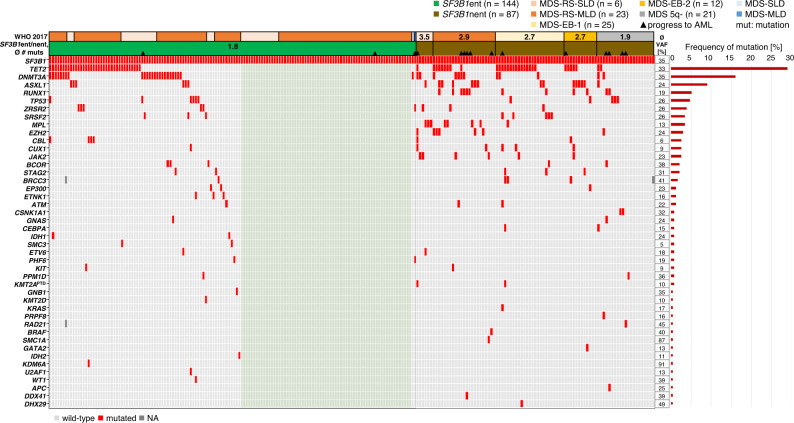
Within SF3B1ent 47% (67/144) did not harbor any additional mutation in 73 analyzed genes resulting in an average of 1.8 mutations (including SF3B1) in this group (Fig. 4), while 53% (77/144) harbored one to four additional mutations. SF3B1nent patients showed on average 2.6 mutations (MDS with isolated del(5q): 1.9; MDS-EB: 2.7; MDS-RS: 3.1; Fig. 4). Although SF3B1ent samples showed in total few mutations, additional mutations (if present) were detected in 27 different genes (Supplementary Fig. S6A). Additional mutations in SF3B1nent samples were found in 9 to 20 different genes depending on the respective entity (Supplementary Fig. S6B–F).
Fig. 4. Molecular characterization of SF3B1mut MDS patients.
Illustration of all 231 samples, each column represents one patient. Genes (gray: wild-type; red: mutated) as well as the WHO entity (incl. SF3B1ent) are given for each patient. Light green: SF3B1ent patients with isolated SF3B1 mutation; VAF variant allelic frequency.
The most frequent additional mutations in all SF3B1mut patients were TET2 (29%), DNMT3A (16%) and ASXL1 (9%) (Fig. 4; Supplementary Fig. S7A, B). The mutational frequencies of RUNX1, MPL, EZH2, and JAK2 in the total SF3B1mut cohort were 5% (RUNX1) and 3% (MPL, EZH2, JAK2) and were present due to the entity criteria only in SF3B1nent in 12, 8, 7 and 6 cases, respectively. Of note, compared to SF3B1 wild-type samples mutations in ASXL1, RUNX1, TP53, ZRSR2, SRSF2 and STAG2 were significantly less frequent in SF3B1mut patients, while DNMT3A mutations were more frequent (Supplementary Fig. S7A). Interestingly, within SF3B1mut samples TP53 mutations (n = 11) were most frequently seen within MDS 5q- (3/21, 14%). However, mutated TP53 was also seen in SF3B1ent (6/144; 4%), MDS-EB-1 (1/25; 4%) and MDS-EB-2 (1/12; 8%) (Supplementary Fig. S7B). Notably, 82% (9/11) of TP53 mutations were monoallelic events. In two samples (MDS-EB-1/2) both a mutation and deletion were detected affecting the TP53 gene (biallelic inactivation). In 17 SF3B1mut samples, additional spliceosome mutations were found, namely ZRSR2 (n = 9) and SRSF2 (n = 8) (Fig. 4; Supplementary Figs. S6, S7). In SF3B1ent patients spliceosome mutations were found in 9 cases (ZRSR2: n = 6, mean VAF: 27% vs. 26% of SF3B1; SRSF2: n = 3, mean VAF: 16% vs. 47% of SF3B1), whereas within SF3B1nent 8 samples showed additional ZRSR2 (n = 3, mean VAF: 26% vs. 37% of SF3B1) or SRSF2 (n = 5, mean VAF: 31% vs. 29% of SF3B1) mutations (Supplementary Fig. S8). Interestingly, additional spliceosome mutations were not detected in MDS 5q- (Fig. 4; Supplementary Fig. S7). In 5/17 (29%) cases the SF3B1 VAF was lower than the VAF of additional spliceosome mutations (SRSF2: n = 2, all MDS-EB-1; ZRSR2: n = 3, all SF3B1ent; Supplementary Fig. S8). In 4 of those samples the SF3B1 VAF was lower than 15% and therefore accounted to the 13 samples of the entire MDS cohort showing only one SF3B1 mutation with a low VAF (<15%; Fig. 3B). Thus, in 11/13 patients with a low SF3B1 VAF either deletions on chromosome 5 (n = 5), additional spliceosome (n = 4) or TP53 mutations (n = 2) were identified at MDS diagnosis.
Prognostic impact of additional aberrations in SF3B1mut MDS
Next, we analyzed the prognostic contribution of additional gene mutations and other risk factors to OS in SF3B1mut patients. Within all SF3B1mut patients the number of mutations showed a significant impact on OS (Supplementary Fig. S9A; p = 0.040). However, within SF3B1ent cases, OS was not affected by the presence of additional mutations (Supplementary Fig. S9B). In univariate analyses bone marrow blasts <5% was a good prognostic marker (hazard ratio HR: 0.616; p = 0.033), while RUNX1 mutations (HR: 4.347; p < 0.001), ASXL1 mutations (HR: 1.836, p = 0.023) and del(5q) (HR: 1.977; p = 0.008) were poor prognostic markers (Table 2). Of note, the poor prognostic impact on OS of complex karyotypes did not reach statistical significance (p = 0.063) presumably due to the small samples size (n = 7). Further, del(5q) was not restricted to isolated del(5q) cases but comprised all cases with deletions on chromosome 5, and thus included also cases with complex karyotypes. In multivariate analysis only RUNX1 mutations (HR: 3.581; p < 0.001) and del(5q) (HR: 2.146; p = 0.003) were independent prognostic factors. Patients with SF3B1mut having either del(5q) or RUNX1 mutations (n = 31) showed shorter OS compared to SF3B1mut patients not having these abnormalities (n = 200; median OS: 43 vs. 88 months; p < 0.001; Supplementary Fig. S9C).
Table 2.
Cox proportional hazards ratio analyses of variables in SF3B1 mutated MDS prognostic of OS.
| Risk factor | Hazard ratio (HR) | 95% CI | P |
|---|---|---|---|
| Univariate analysis | |||
| Sex | 1.080 | 0.764–1.528 | 0.663 |
| SF3B1 VAF, < 15% vs. ≥15% | 0.705 | 0.345–1.443 | 0.339 |
| Bone marrow blast count, <5% vs. ≥5% | 0.616 | 0.395–0.961 | 0.033 |
| Bone marrow blast count, <10% vs. ≥10% | 1.901 | 0.605–5.980 | 0.272 |
| RUNX1 | 4.347 | 2.325–8.128 | <0.001 |
| EZH2 | 1.381 | 0.564–3.381 | 0.480 |
| ASXL1 | 1.836 | 1.085–3.105 | 0.023 |
| DNMT3A | 0.920 | 0.582–1.454 | 0.720 |
| TET2 | 0.979 | 0.682–1.405 | 0.907 |
| JAK2 | 1.115 | 0.411–3.022 | 0.831 |
| MPL | 1.520 | 0.709–3.257 | 0.282 |
| TP53 | 1.401 | 0.652–3.008 | 0.387 |
| del(5q) | 1.977 | 1.198–3.262 | 0.008 |
| Complex karyotype (≥3 abnormalities) | 2.986 | 0.944–9.448 | 0.063 |
| MECOM rearrangement | 0.486 | 0.069–3.556 | 0.497 |
| Other cytogenetic abnormalities | 1.428 | 0.930–2.194 | 0.104 |
| Multivariate analysis | |||
| Bone marrow blast count, <5% vs. ≥5% | 0.693 | 0.418–1.148 | 0.154 |
| RUNX1 | 3.581 | 1.769–7.249 | <0.001 |
| ASXL1 | 1.157 | 0.618–2.164 | 0.649 |
| del(5q) | 2.146 | 1.289–3.574 | 0.003 |
OS overall survival, CI confidence interval, VAF variant allelic frequency.
p-values in bold: statistical significance (p < 0.05).
Molecular genetics of SF3B1mut patients transforming to AML
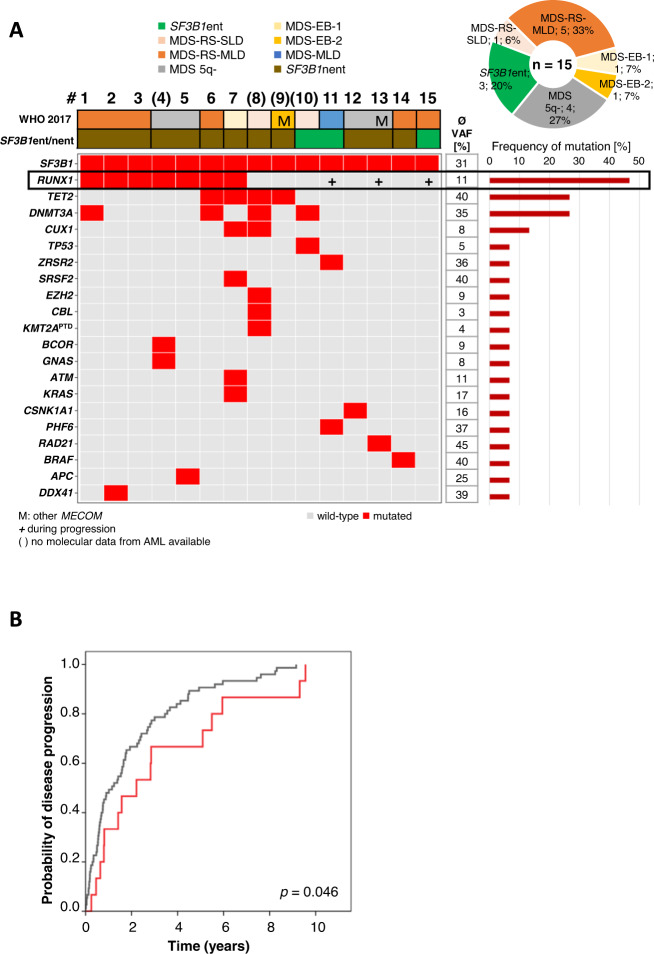
Of SF3B1mut patients 7% (15/231) progressed to AML compared to 15% (75/503) of SF3B1 wild-type patients (median follow-up: 9.3 years; Fig. 5A). In addition, time to AML was shorter in SF3B1 wild-type compared to SF3B1mut patients (median: 14 vs. 27 months, p = 0.046; Fig. 5B). Notably, an AML transformation rate of 14% (12/87) was seen in SF3B1nent and 2% (3/144) in SF3B1ent (median follow-up: 122 and 112 months; Fig. 5A). A trend for a longer time to AML transformation was observed for SF3B1ent compared to SF3B1nent, however not reaching statistical significance (71 vs. 17 months, p = 0.0825). Eleven of 15 SF3B1mut MDS cases were also analyzed for the presence of molecular mutations at their diagnosis of AML (Fig. 5A, further details are provided in the supplement). Regarding the prognostic contribution of additional gene mutations and other risk factors to AML transformation in SF3B1mut patients univariate analyses revealed bone marrow blasts <5% to be associated with lower risk (hazard ratio HR: 0.097; p = 0.021) and RUNX1 mutations (HR: 3.518; p = 0.05) with higher risk for AML transformation (Supplementary Table S3).
Fig. 5. Genetics of MDS patients with mutated SF3B1 progressing to AML.
A Molecular characterization of SF3B1mut patients progressing to AML at MDS stage (n = 15). Each column represents one patient, numbered 1–15. Number in brackets indicate that molecular data at AML stage is not available. Genes (gray: wild-type; red: mutated), WHO 2017 entities and SF3B1ent/nent are given for each patient. ent entity, nent non-entity, VAF variant allelic frequency. B Cumulative incidence of AML transformation of SF3B1 mutated (n = 15; red) vs. wild-type (n = 75; gray) patients.
Discussion
SF3B1 mutations are frequently detected within MDS and associated with favorable prognosis [5–7]. In our WGS-based cohort of 734 MDS patients we identified 231/734 (31%) cases with SF3B1 mutations verifying known hotspots in K700, K666 and H662 [4, 7, 8, 20, 21] and confirming a heterozygous SF3B1 mutation status with high median VAFs (35%) across all entities. VAFs >30% were observed in 77% of SF3B1mut samples. SF3B1 mutations persisted over the entire disease courses in many AML-transforming patients supporting that SF3B1 mutations are early events in MDS. However, SF3B1mut samples with low VAFs (<15%) were mainly found in SF3B1 non-entity cases showing excess blasts, del(5q) or TP53 mutations but also in two SF3B1ent samples harboring other spliceosome mutations. In line with previous reports, SF3B1 mutations were predominantly found in MDS-RS-SLD/MLD supporting the association of SF3B1 with RS [7, 10]. Moreover, we confirmed that SF3B1 mutations in MDS were favorable with regard to OS and AML transformation [7, 10, 11].
Recently, the IWG-PM suggested MDS with mutated SF3B1 as a distinct entity [11]. In this study, we evaluated the IWG-PM proposed SF3B1 entity criteria. We confirmed the favorable clinical outcome of SF3B1 entity similar to recently published studies [21, 22]. Additionally, in line with Komrokji et al. we observed a significantly longer OS of SF3B1 entity patients compared to SF3B1 non-entity patients, in contrast to Venable et al. who did not observe significant differences in OS between SF3B1ent and SF3B1nent presumably due to the small cohort size [21].
In contrast to Malcovati et al. [11], we observed that SF3B1 mutations were associated with significantly shorter OS within MDS 5q- concordant with previous reports [19, 22, 23] highlighting the adverse prognostic impact of mutated SF3B1 within this entity. Within our SF3B1mut cohort, only 5% (11/231; all SF3B1nent) had poor or very poor cytogenetic risk groups concordant with a previous report [11] adding to the reasons for the favorable prognosis of SF3B1 mutations. In this line, the lately published IPSS-M, a unique risk score, improves the risk stratification of MDS patients by including molecular genetics into their model [24], in contrast to the IPSS-R, which considers only morphological features and cytogenetics [25]. The IPSS-M model further incorporates SF3B1 mutations with different weights depending on co-abnormalities (i.e. isolated del(5q) or BCOR, BCORL1, RUNX1, NRAS, STAG2, SRSF2 mutations).
With regard to the mutational landscape of SF3B1mut cases the most frequent additional mutations were DNMT3A, TET2, and ASXL1 (DTA) similar to previous reports showing that epigenetic and histone modifiers are commonly mutated in MDS, but also in aging individuals [5, 21, 26–28]. The number of additional mutations significantly impacted on OS in all SF3B1mut patients. Within SF3B1ent the number of co-mutations did not affect OS as shown in IWG-PM results [11], however the number of co-mutations was low compared to SF3B1nent cases.
In line with previous studies [5, 10], we confirmed that progression to AML occurs at a relatively low frequency in SF3B1mut patients (7%; 15/231). Furthermore, AML transformation was less frequent in SF3B1ent compared to SF3B1nent (2% vs. 14%). Progression of MDS to AML is suggested to be driven by cooperating genetic lesions [28, 29]. In this regard, we found that AML-transforming patients harbored on average more mutations than non-progressing patients (3.2 vs. 2.0). We further demonstrated that at MDS diagnosis 47% (7/15) of AML-transforming patients showed RUNX1 mutations, significantly more frequent in AML-transforming compared to non-transforming patients. Moreover, during disease progression chromosomal aberrations were gained in two cases whereas most frequently RUNX1 mutations were acquired (n = 3) highlighting the role as potential driver gene and confirming the strong adverse prognostic value of RUNX1 mutations [11] associated with worse OS and a higher AML transformation rate within SF3B1mut patients as also shown by Komrokji et al. [22].
Recently, the 5th edition of WHO and the ICC introduced MDS with mutated SF3B1 as a new entity [2, 12]. The entity criteria proposed by the WHO 2022 and ICC mainly follow those suggested by the IWG-PM but differ in excluding mutations. Further, it is stated that the diagnostic criteria of MDS 5q- remain and that an SF3B1 mutation does not per se override this diagnosis. This is supported by our data as SF3B1 mutations show a negative impact on OS in MDS 5q- and do not seem to be the defining mutation in this setting, as suggested by the frequently low SF3B1 VAF. In contrast to the IWG-PM proposal, both WHO 2022 and ICC guidelines exclude biallelic TP53 inactivations from the SF3B1 entity. In our SF3B1mut cohort, only two samples (2/231; <1%) harbored biallelic TP53 inactivations (both with blast count >5%). Concordant with the IWG-PM, ICC also excludes RUNX1 mutations from the SF3B1 entity supported by our data showing RUNX1 mutations as independent negative prognostic factors for OS and AML transformation.
In our univariate analysis, we confirmed the negative prognostic impact of del(5q) on OS of SF3B1mut cases and additionally found a negative impact of blast count >5% as well as RUNX1 and ASXL1 mutations. However, our multivariate analysis could not confirm the independent prognostic impact of blast count >5%, but showed del(5q) and RUNX1 mutations as independent prognostic markers. Thus, based on our data the threshold of <5%, which is used by IWG-PM, ICC and WHO 2022, is not required if presence of del(5q) and RUNX1 mutation are exclusion criteria for the SF3B1 entity. Of note, studies from Malcovati et al. showed a significant impact of excess blasts on the survival of SF3B1mut patients [10, 11], however, RUNX1 mutations were not included in their multivariate analysis.
In conclusion, SF3B1 mutations are associated with good clinical outcome. Patients fulfilling the criteria of the SF3B1 entity proposed by the IWG-PM show an even better prognosis (longer OS, lower AML transformation rate). Our data suggest that the identification of the good prognostic subset within SF3B1mut patients can be achieved by excluding only cases with del(5q) and/or RUNX1 mutations, however completely independent of blast count.
Supplementary information
Acknowledgements
The authors would like to thank all co-workers at the MLL Munich Leukemia Laboratory for their dedicated work. The authors would also like to thank all physicians for providing samples and caring for patients as well as collecting data.
Author contributions
SH and CH designed the study, SH interpreted the data, SH wrote the manuscript. CH was responsible for chromosome banding and FISH analyses, MM, CB, GH, and HS for molecular and bioinformatic analyses, WK for immunophenotyping and TH for cytomorphologic analyses. All authors read and contributed to the final version of the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
CH, WK, and TH declare part ownership of Munich Leukemia Laboratory (MLL). SH, HS, MM, GH, and CB are employed by the MLL.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01728-5.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–19. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasserjan RP, Orazi A, Brunning RD, Le Beau MM, Porwit A, Baumann I, et al. Myelodysplastic syndromes. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Myelodysplastic syndromes. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2017:97–120.
- 4.Cazzola M, la Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–2034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, P VL, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of Genetic Lesions in 944 Patients with Myelodysplastic Syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 9.Swoboda DM, Lee J-H, Chan O, Komrokji RS, Al Ali N, Padron E, et al. Marrow ring sideroblasts are highly predictive for TP53 mutation in MDS with excess blasts. Blood. 2019;134:4244. doi: 10.1182/blood-2019-124772. [DOI] [PubMed] [Google Scholar]
- 10.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–41. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood. 2020;136:157–70. doi: 10.1182/blood.2020004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka H-M, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16:53–9. doi: 10.1038/sj.leu.2402329. [DOI] [PubMed] [Google Scholar]
- 14.Haferlach T, Kern W, Schoch C, Hiddemann W, Sauerland MC. Morphologic dysplasia in acute myeloid leukemia: importance of granulocytic dysplasia. J Clin Oncol. 2003;21:3004–5. doi: 10.1200/JCO.2003.99.091. [DOI] [PubMed] [Google Scholar]
- 15.Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004;104:3078–85. doi: 10.1182/blood-2004-03-1036. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2017.
- 17.Höllein A, Twardziok SO, Walter W, Hutter S, Baer C, Hernandez-Sanchez JM, et al. The combination of WGS and RNA-Seq is superior to conventional diagnostic tests in multiple myeloma: Ready for prime time? Cancer Genet. 2020;242:15–24. doi: 10.1016/j.cancergen.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Stengel A, Baer C, Walter W, Meggendorfer M, Kern W, Haferlach T, et al. Mutational patterns and their correlation to CHIP-related mutations and age in hematological malignancies. Blood Adv. 2021;5:4426–34. doi: 10.1182/bloodadvances.2021004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meggendorfer M, Haferlach C, Kern W, Haferlach T. Molecular analysis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5 reveals a specific spectrum of molecular mutations with prognostic impact: a study on 123 patients and 27 genes. Haematologica. 2017;102:1502–10. doi: 10.3324/haematol.2017.166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, la Porta MG, Pascutto C, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–46. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venable ER, Chen D, Chen CP, Bessonen KR, Nguyen PL, Oliveira JL, et al. Pathologic spectrum and molecular landscape of myeloid disorders harboring SF3B1 mutations. Am J Clin Pathol. 2021;156:679–90. doi: 10.1093/ajcp/aqab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komrokji R, Volpe V, Chan O, Al Ali N, Swoboda D, Kuykendall A, et al. Validation of the international working group proposal for SF3B1 mutant myelodysplastic syndromes. Blood. 2021;138:989–92. doi: 10.1182/blood.2021010831. [DOI] [PubMed] [Google Scholar]
- 23.Chan O, Ali NA, Sallman D, Padron E, Lancet J, Komrokji R. Therapeutic outcomes and prognostic impact of gene mutations including TP53 and SF3B1 in patients with Del(5q) myelodysplastic syndromes (MDS) Clin Lymphoma Myeloma Leuk. 2022;22:e467–e76. doi: 10.1016/j.clml.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Ossa JEA, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;0:EVIDoa2200008. doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, et al. Revised International Prognostic Scoring System (IPSS-R) for myelodysplastic syndromes. Blood. 2012;120:2454–65. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwind S, Jentzsch M, Kubasch AS, Metzeler KH, Platzbecker U. Myelodysplastic syndromes: Biological and therapeutic consequences of the evolving molecular aberrations landscape. Neoplasia. 2021;23:1101–9. doi: 10.1016/j.neo.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meggendorfer M, de AA, Nadarajah N, Alpermann T, Kern W, Steuer K, et al. Karyotype evolution and acquisition of FLT3 or RAS pathway alterations drive progression of myelodysplastic syndrome to acute myeloid leukemia. Haematologica. 2015;100:e487–90. doi: 10.3324/haematol.2015.127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.