To the Editor:
In AML SF3B1 mutations are recurrently found, most frequently in AML-MRC [1] and were shown to be highly specific for secondary AML (s-AML) arising post MDS or MDS/MPN [2]. Thus, the presence of SF3B1 mutations is considered as diagnostic criteria for AML-MR according to the 5th edition of the WHO classification (WHO 2022; [3]). Here, we address the prognostic impact of SF3B1 mutations in AML and evaluate the genetic landscape of SF3B1 mutated patients at AML diagnosis and during follow-up.
Based on the revised 4th edition of the WHO classification (WHO 2017), AML are classified into AML with recurrent genetic abnormalities, AML with myelodysplasia-related changes (AML-MRC) or AML, not otherwise specified (AML-NOS) [4]. Several changes are announced in the WHO 2022 [3] incorporating more genetically defined entity criteria. For example, AML with mutated RUNX1 is no longer recognized as distinct entity, AML-MRC is replaced by AML-MR considering gene mutations while removing morphologic criteria and AML sub-groups with rearranged KMT2A or MECOM are extended including all partner genes.
Within the last years, many prognostically relevant driver genes in AML have been identified including also spliceosome genes [5]. In myeloid malignancies, SF3B1 is most frequently mutated in MDS or MDS/MPN and associated with a favorable prognosis and an indolent disease course [6–8]. More recent data by Bernard et al. indicate that the favorable outcome is restricted to those patients lacking co-mutations in BCOR, BCORL1, NRAS, RUNX1, SRSF2, STAG2 and del(5q) [9].
For this analysis we selected 735 AML samples with material available to perform whole genome sequencing sent to the MLL Munich Leukemia Laboratory between 09/2005 and 01/2020. Therapy-related AML were excluded from this study. Within the cohort 89% (652/735) were de novo AML cases and 11% (83/735) s-AMLs. For further details on cohort and statistics see Supplementary Methods. All cases were classified into specific sub-groups according to the currently used WHO 2017 [4]. SF3B1mut cases were further classified according to WHO 2022 [3] and the International Consensus Classification (ICC; [10]). For abbreviations of entities, see Supplementary Table S1. All patients gave their written informed consent for genetic analyses and to the use of laboratory results and clinical data for research purposes according to the Declaration of Helsinki. The study was further approved by the laboratory´s institutional review board. All samples were subjected to whole genome and targeted panel sequencing (Supplementary Methods).
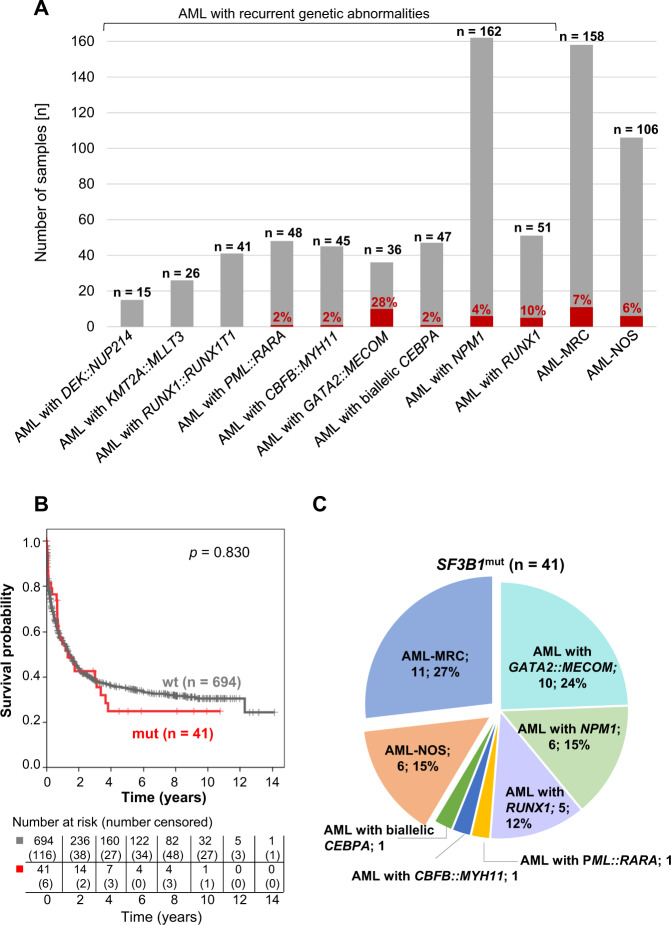
We identified SF3B1 mutations in a small fraction (6%; 41/735) of AML patients (Fig. 1A and Supplementary Table S1) in line with published results [5, 11]. Based on WHO 2017, SF3B1 mutations were found in AML with recurrent genetic abnormalities (24/471; 5%), AML-MRC (11/158; 7%) and AML-NOS (6/106; 6%) (Fig. 1A). Within the entire AML cohort, comprising samples from 16 different entities, SF3B1 mutations were detected in eight different AML entities (Supplementary Table S1), most frequently within AML with GATA2::MECOM (10/36; 28%), thereby confirming the association of SF3B1 mutations with GATA2::MECOM rearrangements as previously published [12]. Notably, within AML-NOS SF3B1 mutations were exclusively found in samples diagnosed with AML with maturation (Supplementary Table S1). The presence of ring sideroblasts in SF3B1mut AML is described in the Supplementary Results. SF3B1 mutations did not affect OS in the total AML cohort (median: 16 vs. 17 months; p = 0.830; Fig. 1B). Within all 41 SF3B1mut cases AML-MRC (11/41; 27%) and AML with GATA2::MECOM (10/41; 24%) were most frequent (Fig. 1C). When stratified for AML sub-entities, there was also no impact of SF3B1 mutations on OS within each sub-entity (Supplementary Fig. S1A–E), however OS was different within SF3B1mut AML if stratified according to WHO entities (Supplementary Fig. S1F, G). Thus, the prognosis of the SF3B1mut AML seems to be dominated by the sub-entity, concordant with a previous report showing that splicing mutations (including SF3B1) per se are not prognostic in AML [13].
Fig. 1. Distribution and OS of SF3B1 mutations in AML.
A SF3B1 mutation status within different AML entities (red: mutated; gray: wild-type). B OS of patients with mutated (n = 41; red) vs. wild-type (n = 694; gray) SF3B1 within the entire AML cohort. C WHO 2017 entities of SF3B1 mutated AML (n = 41).
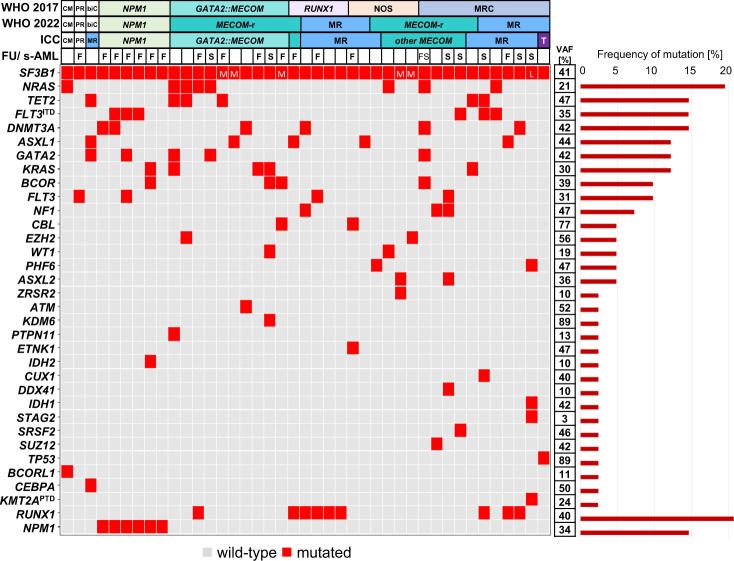
In the total cohort, SF3B1 mutations showed a mean variant allelic frequency (VAF) of 41% and those mutations affecting amino acids K666 and K700 were found most frequently (Supplementary Results and Supplementary Fig. S2) similar to previous studies [13, 14]. On average, SF3B1mut patients harbored 3.3 mutations (AML-NOS: 2.5; AML with RUNX1: 2.8; AML with GATA2::MECOM: 3.3; AML-MRC: 3.6; AML with NPM1: 3.7; Fig. 2). The most frequent additional mutations in SF3B1mut patients were RUNX1 (9/41; 22%) and NRAS (8/41; 20%). NPM1, TET2, or DNMT3A mutations or FLT3-ITD were detected in 15% (6/41) each. RUNX1 mutations were present besides within AML with RUNX1 mutation, also in AML-MRC (n = 3) and AML with GATA2::MECOM (n = 1). Interestingly, 37% (15/41) of SF3B1mut patients harbored at least one mutation in a DTA gene (DNMT3A, TET2, ASXL1). Additional mutations were found in 5 to 21 different genes depending on the respective entity (Supplementary Fig. S3A–E). Within SF3B1mut patients 10 cases showed MECOM rearrangements (MECOM-r) with a different partner gene than GATA2. This resulted in 49% (20/41) of SF3B1mut patients harboring a MECOM-r (Fig. 2). Conversely, 31% (20/64) of all AML with MECOM-r showed an SF3B1 mutation, which was thus the second most frequent mutation within this AML entity after NRAS mutations (36%; 23/64). SF3B1 mutations were significantly associated with MECOM-r (31% [20/64] vs. 3% [21/671]; p < 0.001). In summary, the majority (78%, 32/41) of SF3B1mut AML were either AML with MECOM-r (n = 20) or AML-MR (n = 12), underpinning the strong association of SF3B1 mutations with these two entities (Fig. 2; further details on the classification of SF3B1mut cases are provided in the Supplementary Results). A prior history of MDS or MDS/MPN was documented in 20% (8/41) of SF3B1mut patients harboring on average 4.3 mutations at AML diagnosis (Fig. 2 and Supplementary Fig. S4A). Thereof, 63% (5/8) had a MECOM-r and 25% showed RUNX1, DNMT3A, GATA2, NRAS, BCOR mutations or FLT3-ITD when AML was diagnosed. The SF3B1mut was already present in the prior MDS stage in 4/5 patients with available MDS data (Supplementary Results and Supplementary Fig. S4).
Fig. 2. Molecular characterization of AML patients with mutated SF3B1.
Illustration of all 41 samples, each column represents one patient. Genes (gray: wild-type; red: mutated) as well as the WHO and ICC entities are given for each patient. Secondary AMLs (s-AMLs) are marked with “S” and those with available follow-up (FU) data with “F”. VAF variant allelic frequency (mean), CM CBFB::MYH11, PR PML::RARA, biC biallelic CEBPA, NOS not otherwise specified, MR(C) myelodysplasia-related (changes), MECOM-r MECOM rearrangement, T mutated TP53, L low VAF (0–14%), M medium VAF (15–29%); Remaining cases showed SF3B1 VAFs ≥30%.
In AML with NPM1 or RUNX1 mutations the SF3B1 VAFs exceeded 30% in all cases and were similar to or higher than the VAFs of NPM1 or RUNX1 mutations in 11/11 cases (Fig. 2 and Supplementary Figs. S3F and S5A, B). A comparable pattern was seen in the remaining cases with SF3B1 VAFs higher than 15% (n = 29; Fig. 2 and Supplementary Fig. S5C). In the one AML-MRC patient with a low SF3B1 VAF (6%), other mutations showed higher VAFs (IDH1: 42%; KMT2A-PTD: 24%; Supplementary Fig. S5C). In total, in 40/41 (98%) SF3B1mut cases similar or higher SF3B1 VAFs were observed compared to other co-mutations or aberrations, indicating that SF3B1 mutations are rather primary than secondary mutations during leukemogenesis. This is in line with a previous report, showing that SF3B1 mutations are acquired early in MDS and that splicing mutations are early evolutionary events in myeloid malignancies [14]. In 16/41 (39%) SF3B1mut cases molecular follow-up data was available (Fig. 2). In 1/16 patients, an AML patient with mutated NPM1, the SF3B1 mutation (VAF: 40%) remained detectable, despite complete hematologic remission and undetectable NPM1 mutation (Supplementary Fig. S6B). In 15/16 (94%) cases the SF3B1 VAFs paralleled the VAFs of co-mutations during the entire disease course, even during relapse (for details see Supplementary Results and Supplementary Fig. S6).
As shown, mutations in RUNX1 or DTA genes were found among the most frequent co-mutations of SF3B1mut AML samples, concordant with previously published studies [5, 14]. Notably, DTA genes are frequently mutated in MDS [6] and the most common mutations in clonal hematopoiesis of indeterminate potential (CHIP) [11]. In our study, a prior history of MDS or MDS/MPN has been documented in some SF3B1mut AML patients (8/41). However, it might be unidentified in others. Alternatively, SF3B1mut CHIP or CCUS (clonal cytopenia with undetermined significance) may represent relevant precursor lesions of SF3B1mut AML, in line with Venable et al., showing that SF3B1mut cases comprise the full pathologic spectrum of myeloid disorders from CCUS to AML [14]. In SF3B1mut s-AML patients, we frequently detected MECOM-r and RUNX1 mutations, both known AML driver genes [5]. These two genetic abnormalities were also frequently found within the remaining SF3B1mut AML patients, where no MDS or MDS/MPN history had been reported. In this line, we previously showed that SF3B1mut MDS patients harboring RUNX1 mutations frequently progressed to AML and that RUNX1 mutations and MECOM-r were gained during AML transformation [15]. Thus, our data suggests an MDS/CCUS pre-phase in SF3B1mut AML without antecedent clinical documentation and further supports the guidelines of the WHO 2022 showing that SF3B1mut AML is diagnosed as AML-MR without knowing the patient’s clinical history.
In summary, SF3B1 mutations are found in a small fraction of AML patients, are enriched in poor risk AML subtypes and are strongly associated with MECOM rearrangements and myelodysplasia-related changes. The persistently high VAF of SF3B1 mutations in AML patients suggests that SF3B1 mutations are acquired early in a pre-leukemic clone and may be indicative of an MDS pre-phase.
Supplementary information
Acknowledgements
The authors would like to thank all co-workers at the MLL Munich Leukemia Laboratory for their dedicated work. The authors would also like to thank all physicians for providing samples and caring for patients as well as collecting data.
Author contributions
SH and CH designed the study, SH and GH interpreted the data, SH wrote the manuscript. CH was responsible for chromosome banding and FISH analyses, MM, CB and StH for molecular and bioinformatic analyses, WK for immunophenotyping and TH for cytomorphologic analyses. All authors read and contributed to the final version of the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
CH, WK and TH declare part ownership of Munich Leukemia Laboratory (MLL). SH, StH, MM, GH and CB are employed by the MLL.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01734-7.
References
- 1.Fuhrmann I, Lenk M, Haferlach T, Stengel A, Hutter S, Baer C, et al. AML, NOS and AML-MRC as defined by multilineage dysplasia share a common mutation pattern which is distinct from AML-MRC as defined by MDS-related cytogenetics. Leukemia. 2022;36:1939–42. doi: 10.1038/s41375-022-01631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–76. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2017.
- 5.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangaonkar AA, Lasho TL, Finke C, Ketterling RP, Reichard KK, McCullough K, et al. SF3B1-mutant myelodysplastic syndrome/myeloproliferative neoplasms: a unique molecular and prognostic entity. Haematologica. 2022;107:1189–92. doi: 10.3324/haematol.2021.280463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood. 2020;136:157–70. doi: 10.1182/blood.2020004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard E, Tuechler H, Greenberg Peter L, Hasserjian Robert P, Arango Ossa Juan E, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;1:EVIDoa2200008. doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 10.Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka H-M, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valent P, Kern W, Hoermann G, Milosevic Feenstra JD, Sotlar K, Pfeilstöcker M, et al. Clonal hematopoiesis with oncogenic potential (CHOP): separation from CHIP and roads to AML. Int J Mol Sci. 2019;20:789. [DOI] [PMC free article] [PubMed]
- 12.Awada H, Kerr CM, Rogers HJ, Maciejewski JP, Visconte V. Molecular and clinical aspects of acute myeloid leukemia with Inv(3)(q21q26)/t(3;3)(q21;q26) carrying spliceosomal mutations. Blood. 2020;136:7–8. doi: 10.1182/blood-2020-139953. [DOI] [Google Scholar]
- 13.Bamopoulos SA, Batcha AMN, Jurinovic V, Rothenberg-Thurley M, Janke H, Ksienzyk B, et al. Clinical presentation and differential splicing of SRSF2, U2AF1 and SF3B1 mutations in patients with acute myeloid leukemia. Leukemia. 2020;34:2621–34. doi: 10.1038/s41375-020-0839-4. [DOI] [PubMed] [Google Scholar]
- 14.Venable ER, Chen D, Chen CP, Bessonen KR, Nguyen PL, Oliveira JL, et al. Pathologic spectrum and molecular landscape of myeloid disorders harboring SF3B1 mutations. Am J Clin Pathol. 2021;156:679–90. doi: 10.1093/ajcp/aqab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber S, Hutter S, Meggendorfer M, Hoermann G, Walter W, Baer C, et al. The role of SF3B1 mutations in myelodysplastic syndromes. EHA Libr. 2022;357597:P735. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.