Abstract
Background
Castor (Ricinus communis L.) is cultivated for seed oil and to feed (leaves) Eri silkworm, Samia ricini (Donovan) Hutt. Alternaria blight affects castor cultivation resulting substantial yield loss (∼30%). Uses of synthetic fertilizers and agrochemicals for disease management have serious concerns as the castor leaves are fed to eri silkworms for rearing. Application of plant growth promoting rhizobacteria for disease suppression and to enhance plant growth will be a healthier choice in castor cultivation. The aim of this study was to assess the efficacy of Alternaria blight disease suppression by native rhizobacteria isolated from wasteland castor and their ability on plant growth promotion.
Methodology
We isolated 50 bacterial antagonists from castor rhizosphere using the dilution plate method and evaluated their antagonistic activity against the castor blight pathogen, Alternaria ricini. Based on antimicrobial bioassay and plant growth promotion (PGP) traits (phosphate solubilization, ACC deaminase activities, production of IAA, GA3, HCN, NH3 and siderophore), salt and acid tolerance; we have chosen ten potential isolates and identified them through 16SrRNA gene sequencing and analysis. Disease suppression and plant growth studies were evaluated in pot experiments.
Results and conclusion
Three isolates namely, Enterobacter hormaechei (LRP-2), Bacillus mycoides (HF-1) and B. aryabhattai (UR-6) showed potential antagonistic activities and PGP traits which were selected for disease suppression and PGP studies. Application of PGPR consortia (LRP-2+HF-1) could suppress the plants from A. ricini infection in challenged inoculation. Mix inoculation of LRP-2 and UR-6 showed synergistic effect and enhanced plant growth in pot experiments. Combinations of E. hormaechei (LRP-2), B. mycoides (HF-1) and B. aryabhattai (UR-6) can be applied as bio-control and bio-fertilizer formulation to protect castor from Alternaria blight and also to enhance plant growth.
Keywords: Castor, Alternaria blight, Antagonists, Plant growth promoting rhizobacteria (PGPR), Wasteland
Highlights
-
•
We isolated native rhizobacteria of castor growing in cultivated and wastelands.
-
•
Ten PGPR were selected for disease suppression and plant growth experiments.
-
•
Inoculation of native PGPR could suppress Alternaria blight disease in Castor.
-
•
Co-inoculation of PGPR strains also enhanced plant growth and biomass.
Castor; Alternaria blight; antagonists; Plant growth promoting rhizobacteria (PGPR); wasteland.
1. Introduction
Castor (Ricinus communis L., Family: Euphorbiaceae) is a highly valued economically important crop plant for its non-edible seed oil used for industrial and biofuel applications and leaf biomass for feeding Eri silkworm, one of the popular silk producing insects [1, 2]. Worldwide, 1.30 million hectares of land are being used for castor cultivation [3]. About 95% of the world's castor production is shared by India, China and Brazil and mostly cultivated by marginal farmers [4]. There is an increasing demand for castor leaf biomass as well as seed for both the industries. In this context, a deeper understanding on growth and development of castor plant is necessary to promote its productivity in per unit area.
Castor plant is prone to various diseases in different agro-climatic conditions especially when cultivated in the subtropical and tropical regions. Leaf blight disease of castor is widely scattered in the tropical and sub-tropical regions while cultivated in farm lands causing serious damage to quality of leaves [5]. Alternaria ricini, the causal organism infects about 36.80–68.30% castor plants in agricultural lands [1, 6]. Ecologically, castor is recognized as a wasteland plant and normally grown for the restoration of disturbed and uncultivated soil [7]. Biomass production of individual plants in wasteland is higher compared to cultivated land which might be due to plant-microbe interactions as this interaction governs many processes in the rhizosphere and help in plant fitness under stressed conditions [8]. Alterations in the microbial consortia particularly plant growth promoting rhizobacteria (PGPR) in cultivated castor might alter various processes those are responsible for enhanced plant growth and protection of plant from various stress burdens including diseases [9].
Usually, synthetic fungicides are used for management of disease caused by phyto-pathogenic fungi [10]. However, application of these products have higher risk for public health due to bio-magnification, as the leaves are fed to eri silkworm and the pupae are eaten with or without cooking [1]. Instead, application of bacterial and fungal antagonists and PGPRs are a good choice for the control of fungal diseases as well as plant growth enhancement [11, 12, 13]. Kumar et al. [14] opined that bacterial antagonists were often better in controlling fungal diseases, if applied alone and sometimes in combination with other fungicides. However, successful utilization of PGPR is dependent on its survival in soil, the compatibility with the crop on which it is inoculated, the interaction ability with indigenous microflora in soil, and environmental factors [15].
Castor grows abundantly in uncultivated wastelands and contaminated sites (road or urban dumpsites) and disease incidences in those plants are comparatively less (unpublished field survey data). There might be certain groups of PGPR which colonize in plant rhizosphere promoting plant growth, abiotic stress tolerance and nutrient fixation for easy uptake producing plant growth regulators, siderophores, volatile organic compounds and protective enzymes such as chitinase, glucanase and ACC-deaminase [16, 17, 18, 19]. Native rhizobacteria interact with the plant promoting seed germination, root-shoot growth, delayed leaf senescence and tolerance to stresses [20]. These groups of bacteria protect the plant from the diseases and increase plant growth promotion (PGP) through various mechanisms [21]. It is practical to isolate and test the native rhizobacteria and re-inoculate them in commercial castor cultivation which may acclimatize themselves in castor root environment easily and would be more effective in combating the stress than exotic PGPR strains.
In this study, we have isolated some potential native bacterial antagonists and PGPR from the castor growing in wastelands and re-inoculated those strains in cultivated castor to screen the potential isolate which can effectively suppress the Alternaria blight disease of castor and to evaluate their PGP activities in pot experi-ments.
2. Materials and methods
2.1. The study area
The experiments were carried out at Central Muga Eri Research and Training Institute (CMER&TI), Assam (India). The North-eastern part of India is the native place of Eri silkworms and people grow castor plant for feeding to the Eri silkworms. Castor also grows abundantly in wastelands and unutilized lands in the region. The region is surrounded by the Himalayan foothills and flanked by the river Brahmaputra and Barak valleys and lies between 25.5736° N latitude and 93.2473° E longitude and the altitude varies from sea level to 23,000 ft above sea level. The region experiences very hot-humid weather during summer with an average temperature of 30 °C (7–38.5 °C). The annual rainfall ranges between 1500 -10,000 mm with moderate humidity (75%). Large parts of the area are covered by forests but falling of a constant danger of denudation and deforestation due to the large felling of trees for timber, firewood, annual flood, habitat destruction, etc. [22]. Due to its diverse climatic and topographic conditions, the forests receive abundant rainfall and support a vast variety of floral and faunal biodiversity.
2.2. Soil sampling and isolation of castor rhizobacteria
Rhizospheric soil samples of castor growing in wastelands, contaminated soils (municipality and urban dumpsites), roadsides and farmlands were collected from different parts of Northeast India viz., Kokrajhar, Guwahati, Jorhat, Golaghat, Dibrugarh and foothills of Meghalaya, Nagaland and Arunachal Pradesh. Five healthy plants were randomly selected from each different site mentioned above and roots and root-adhered soil samples were collected. Root soil samples (five) from each block were mixed into one composite sample, collected in polypropylene bags and kept in ice boxes before carrying to the laboratory [18, 23].
Castor rhizobacteria were isolated from the rhizospheric soil samples through the serial dilution method. One gram of soil was suspended in 100 mL of 0.8% salt solution (NaCl) and kept for 30 min at 200 rpm in a shaker incubator (Remi, India). Later, the samples were mixed thoroughly and serial dilutions were made up to 10−10 dilutions using sterile saline water. 100 μL of each dilution was evenly spread on different enriched agar media (make - HiMedia, Mumbai) plates namely Tryptic Soy Agar (TSA), Nutrient Agar (NA), Lauria Britani Agar (LBA), Pseudomonas Agar (PA), Bacillus Agar, Azospirillum Agar (AS) and Azotobacter media (AM). The plates were incubated at 30 °C for 1–2 days after inoculation. Each different bacterial colony that appeared on the plates was counted in every 12 h interval. The population density of rhizobacteria was determined by the total viable colony count method and expressed as colony forming unit (cfu. g−1). Morphologically and visually distinct colonies based on shape, size, elevation, opacity and margin were picked up from the plates and prepared the pure cultures (Supplementary Fig. S1-A). The selected isolates were further purified and re-suspended in 40% glycerol vials and stored in -20 °C for future use. The isolates were further subjected to biochemical characterization using readymade biochemical test kits manufactured by Hi-media Ltd., Mumbai (India) (Supplementary Fig. S1-B). The results were compared with Bergey's Manual of Systematic Bacteriology for identification.
2.3. Isolation of fungal pathogen from infected castor leaf
The infected leaf samples were brought to the laboratory and cut into small pieces (1cm2), surface sterilized for 5 min using sodium hypochlorite solution NaOCl (0.01%) followed by three successive washing with sterilized distilled water [24]. The leaf pieces were air dried and placed on Petri dishes (90 mm) containing Potato Dextrose Agar (PDA, Himedia) medium with streptomycin sulphate (50 mgL-1). Plates were incubated at 25 ± 2 °C for a period of three days. The colonies obtained were isolated and examined under Phase Contrast Microscope (Leica, Germany) for morphological characterization (viz., mycelium, conidiophores, conidia etc.) using cotton blue (5%) as staining reagent. The causal organism of the disease was confirmed through Koch Postulates and identified according to morphological and reproductive characteristics with the help of standard manuals and references [25, 26].
2.4. Antimicrobial bioassay
In vitro antimicrobial bioassay of rhizobacterial isolates was evaluated by dual plate culture technique on PDA medium. The 24 h old bacterial broth cultures (108 cfu mL−1) were streaked on a peptone glucose agar (PGA) plate as circular or semi-circular pattern. Then, the fungal agar plug (5 mm mycelial disc of A. ricini) of 3 days old culture grown on the PDA plate was subsequently inoculated at the center on the PGA plates [27]. Inoculation only with the pathogen (A. ricini) was taken as control. The plates (n = 3) were kept for incubation at 30 ± 2 °C for 5 days and diameter of colony growth (mm) was measured after 72 h intervals. Antagonistic activity i.e. percent inhibition was calculated by formula C-T/C×100, where C and T were the mycelial growth (diameter) on control and the test plate respectively [27]. From antimicrobial bioassay, ten potential bacterial antagonists were selected for the screening of growth promoting traits.
2.5. Qualitative and quantitative assay for PGP traits
2.5.1. Phosphate solubilization
The selected bacterial antagonists were spot inoculated on National Botanical Research Institute Phosphate (NBRIP) medium (Glucose 10 gL-1, Ca3 (PO4)2 5 gL-1, MgCl2 6H2O 5 gL-1, MgSO4. 7H2O 0.25 gL-1, KCl 0.2 gL-1, (NH4)2SO4 0.1 gL-1, Agar 15 gL-1 and at pH 7.0) and allowed to grow for 48 h at 32 °C [28]. Halozones diameters developed around the colonies and their solubilization indexes (SI) were calculated [18]. We also estimated the available phosphate by Vanadomolybdate colorimetric assay method [29]. In this case, the isolates were inoculated in Nutrient Broth (Himedia) and the seed cultures (108 cfu mL−1) were prepared by incubating the broth cultures in rotary shaker at 32 °C for 24 h (Supplementary Fig. S1-D).
Thereafter, the seed cultures were transferred to NBRIP broth and incubated at 32 °C for 96 h by continuous shaking. Phosphate solubilizer strain Bacillus subtilis NCIM2063 available at the institute was taken as positive control with un-inoculated NBRIP broth served as negative control. The pH drop during the log phage at 96 h was recorded with subsequent cell numbers counted by the standard plate count method [18]. Culture aliquots of 10 mL per isolate were taken along with the control, incubated and further centrifuged at 10,500g for 15 min at 4 °C. The supernatants obtained were incubated for 30 min at 30 °C and 5 ml of the supernatant was transferred to fresh test tubes. Later, 2 ml of Vanadomolybdate reagent was added and measured spectrophotometrically (Systronics 2202) at 420 nm. The available phosphorus was finally estimated by calibrating with a standard curve of KH2PO4.
2.5.2. Nitrate reductase, IAA and GA3 production
Qualitative screening of nitrate reductase activities of the bacterial antagonists were carried out as per the Griess test. The isolates were inoculated in nitrate reduction broth (Beef extract 3.0 g. L−1, Gelatin peptone 5.0 g. L−1, KNO3 1.0 g. L−1 and pH 7.0) for 48 h at 37±2 °C. Few drops of sulfanilic acid and α-naphthylamine solutions were added to the broths. Change of colour from pink to red indicated nitrate reduction activity. Quantitative estimation of Indole 3-acetic acid (IAA) was carried out according to tge standard protocol of Wohler [30]. Overnight broth cultures were centrifuged at 23000g for 5 min. Later, the cell pellets were suspended in 3 mL of phosphate buffer (pH 7.5) having 1% of glucose and tryptophan respectively and were further incubated for 24 h at 37 °C. The solutions were then filtered using Whatman paper no. 2 after adding 2 ml of 5% Trichloroacetic acid and 1 mL of 0.5 M CaCl2. Salper solution (2 mL) (2 mL of 0.5 M FeCl3 and 98 mL 35% Perchloric acid) was added to 3 mL of the filtrate, incubated in dark for 30 min at room temperature after vortexing for 2 min. The absorbances of the solutions were measured at 535 nm using a spectrophotometer (Systronics 2202) and the final estimation of IAA was calculated against a standard curve prepared from the graded concentration of IAA (Hi-media Ltd., Mumbai).
We also estimated Gibberellic acid (GA3) production after obtaining the filtrates as mentioned above [31]. The filtrates (3 mL) along with 2 mL of Zinc acetate were taken and 2 mL of Potassium ferrocyanide was added to the solution after 2 min followed by centrifugation at 1750g for 5 min. A total of 5 mL of the supernatant was obtained and incubated for 2 h at 20 °C after adding 5 mL of 30% HCl solution. Finally, the GA3 production was estimated by calibrating with a standard curve of commercial grade Gibberellic acid (Hi-media Ltd., Mumbai) with the absorbance measured at 240 nm wavelength using a UV-vis-spectrophotometer (Systronics 2202).
2.5.3. ACC deaminase
One day old LB cultures of the bacterial antagonists were centrifuged at 17500g for 5 min and the pellets were re-suspended in 5 mL Tris HCl (0.1 mol L−1; pH 7.6). Entire solution was then centrifuged again at 26,500g for 5 min and the resultant pellets were re-suspended in 2.0 mL Tris HCl (0.1 molL-1; pH 8.5). Toluene (30 μL) and 1- aminocyclopropane1-carboxylate (ACC) (20 μL, 0.5 mol L−1) was added to each mixture, vortexed and incubated for 15 min at 32 °C. One milliliter of HCl (0.56 mol L−1) was added to each solution and centrifuged for 5 min at 26,500g immediately after incubation. One mL of HCl (0.56 mol L−1) and 2.0 mL of 2,4-dinitrophenylhydrazine reagents were added to the supernatants, vortexed and incubated at 30 °C for 30 min. Thereafter, 2 mL of NaOH (2 mol.L−1) were added to the suspensions and their absorbances were measured at 540 nm. Serial concentrations of α-ketobutyrate were taken for the preparation of the standard curve and accordingly, the ACC deaminase activity was calculated [32].
2.5.4. Ammonia (NH3), hydrogen cyanide (HCN) and siderophore production
For determination of NH3 production ability, bacterial isolates were inoculated in 10 mL of peptone broth (4%) and allowed to incubate at 32 °C for 48 h. After incubation, 0.5 mL of Nessler's reagent was added to each tube. Change of colour from yellow to brown/red indicates the presence of NH3 [33]. HCN production was measured by the method of Ahmad et al. [34] with slight modification. The Nutrient broth was amended instead of Nutrient agar with glycine 4.4 gL-1 and the bacterial isolates were subsequently added to the broth and incubated for 48 h. Further, 1 mL of Na2CO3 (2%) and 1 mL of Picric acid (0.5%) solutions were mixed to the culture and incubated at 30 °C for 96 h in a shaker. HCN production was confirmed on appearance of red colour in the solutions. Siderophore production was estimated as described by Schwyn and Neilands [35]. The isolates were inoculated on Chrome Azurol S broth (Chrome Azurol S: 60.5 mgL-1, Iron solution: 10 mLL-1, HDTMA: 72.9 mgL-1, PIPES: 30.24 gL-1, NaOH:12 gL-1, Agar: 18 gL-1) and incubated at 32 °C for 72 h. Appearance of yellow or orange colour indicates siderophore production (+ve) by the bacterial antagonists (Supplementary Fig. S1-C).
2.5.5. Salt and pH tolerance
Salt and pH tolerance activities were estimated following the methodologies as described by Romdhane et al. [36] and Kucuk et al. [37] with a slight modification. The bacterial antagonists were streaked on Nutrient agar plates with three different concentrations of NaCl (1.0, 2.0 and 3.0%) along with three different pH (4.0, 6.0 and 9.5) respectively. The capability of the isolates to grow under stress was determined after incubation for 48 h at 32 °C. NaCl of 0.1% and pH 7.0 on nutrient agar medium was taken as control (Supplementary Fig. S1-E).
2.6. Identification of bacterial antagonists and fungal pathogen
Selected bacterial antagonists were identified through 16S rDNA homology study and analysis. The bacterial genomic DNA was isolated by using the genomic DNA extraction kit manufactured by Hi-Media Ltd., Mumbai (India). Universal forward primer 16SF (5-AGAGTTTGATCCTGGCTCAG-3) and reverse primer 16SR (5՛-ACGGCTACCTTGTTACGACTT-3) was used for PCR amplification. PCR amplification was carried out with initial denaturation for 2 min at 95 °C followed by 35 cycles consisting of 95 °C, 55 °C and 72 °C for 1, 1 and 1.5 min respectively followed by final extension of 5 min at 72 °C. Purification of the PCR products was done using a QIA quick PCR purification kit (Qiagen). PCR purified products were sequenced in Bioserve Biotechnologies (India) Private Limited, Hyderabad, (India). The Sequence was subjected to BLAST analysis using NCBI Blast server database. Later, the sequences were submitted to NCBI Gene Bank with accession numbers (Table 1).
Table 1.
Identity of selected rhizobacteria based on the sequence analysis of 16S rRNA genes and their gene bank accession numbers.
| Sl. No. | Strain Code | Identified bacterial strains | NCBI Gene Bank accession no. | Base pair length (bp) |
|---|---|---|---|---|
| 1 | HF-1 | Bacillus mycoides | MH157940.1 | 1011 |
| 2 | KB-4 | B. stratosphericus | MH157939.1 | 1435 |
| 3 | KB-6 | B. cereus | MH157938.1 | 1036 |
| 4 | KM-2 | B. wiedmannii | MH157937.1 | 1435 |
| 5 | KRP-3 | B. aryabhattai | MH157936.1 | 1438 |
| 6 | KRP-6 | B. aryabhattai | MH157935.1 | 1442 |
| 7 | LR-5 | B. megaterium | MH157934.1 | 1453 |
| 8 | LR-7 | B. aryabhattai | MH157933.1 | 1442 |
| 9 | UR-6 | B. aryabhattai | MH157932.1 | 1449 |
| 10 | LRP-2 | Enterobacter hormaechei | MH157931.1 | 1404 |
Further, genomic DNA of fungal pathogen (previously identified as A. ricini) was extracted by the method suggested by Moller et al. [38]. The nuclear ribosomal DNA ITS1 and ITS2 region of the fungal strain were amplified using previously designed primer sets [39] and sequenced in Bioserve Biotechnologies (India) Private Limited, Hyderabad (India). The fungal strain was identified through BLAST search of the sequences in NCBI database. The sequence was aligned with the closest homology of sequences obtained from NCBI database and a phylogenetic tree was constructed using ‘MEGA 7.0’ [40]. The identified sequence was submitted to NCBI database and an accession number was obtained (Accession no MK652889).
2.7. PGPR for disease suppression and growth promotion
From the above experiments, two most effective bacterial antagonists were selected for pot experiments to study the disease suppression under greenhouse condition. Bacterial strains showing promising PGP traits were selected for in vivo study based on their performances in qualitative and quantitative screening for growth promoting traits. These sets of bacteria were prepared to the desired concentration after compatibility test and applied separately in pot experiments.
2.7.1. Screening for disease suppression
Bacterial antagonists namely LRP-2 and HF-1 were selected for disease suppression study in pot experiment. Bacterial isolates were multiplied in LB broth (HiMedia, India) at 30 ± 2 °C for 48 h (continuous shaking @ 200 rpm). The liquid cultures were centrifuged at 17,500g for 15 min and the cell pellets were thinned in sterile distilled water to get the required concentration of 108−109 cfu mL−1. Pot experiments were conducted with eight different treatment combinations for analyzing the percent disease severity and incidence with triplicates for three different seasons [41]. Seeds of non-bloomy red (NBR-1) variety of castor were collected from the Host Plant Division of the institute for in vivo experiments, as this is the most preferred and recommended variety of castor for eri silkworm farming [1]. The seeds were rinsed with 0.2 % (v/v) NaOCl for 10 min followed by sterile water. Seeds were sown in pots (3–4 seeds per pot, diameter 30 cm; height 45 cm) with soil loaded in it under greenhouse condition (28 °C with 75% RH).
Later, only one healthy seedling was kept in the pot and after one month the plants were ready for treatment. For challenged group, 10 g of 5 days old mycelia of A. ricini was mixed in 100 mL distilled water and the suspension was inoculated into the pots (foliar application). Eight sets of treatments were designed namely T1: sterile water (Negative control), T2: only pathogen (A. ricini); T3: Pathogen + fungicide (Indofil M45) next day, T4: Bacterial isolate LRP-2 + pathogen next day, T5: LRP-2 + pathogen applied on the same day, T6: Pathogen + LRP-2 next day, T7: HF-1 + pathogen applied on the same day and T8: LRP-2 + HF-1 + Pathogen applied on the same day. Earlier, seeds were dipped in PGPR suspensions for 1 h before sowing in PGPR treated pots (T4, T5 T6 T7 T8). The results were recorded in alternative days for a month. The appearance of disease in the plants were recorded and analyzed.
2.7.2. Screening of PGPR on plant growth promotion
Pot experiments were also conducted with five treatment combinations including PGPR for analyzing the growth parameters (no. of leaves per plant, shoot length, leaf moisture and biomass). As mentioned above, non-bloomy red (NBR-1) variety seeds of castor were treated with 0.5 % (v/v) NaOCl for 10 min and washed with sterile water. Top layer of soil (up to 15 cm depth; sandy clay loam) was collected from the institute farm for pot experiments. Soil samples were grounded, air dried, and passed through 8-mm sieve. We selected two bacterial strains namely LRP-2 and UR-6 for this study with the inoculums concentrations of 108 cfu mL−1. Five treatments were designed; PGPT1: Agricultural soil (control), PGPT2: Farm yard manure (FYM), PGPT3: Bacterial antagonist (LRP-2), PGPT4: LRP-2 and UR-6 (B. aryabhattai), PGPT5: LRP-2 and UR-6 along with 50% of recommended NPK dose (as per CMER&TI, Central Silk Board). Calculated amounts of FYM, NPK and PGPR (20 mL per plant; 108 cfu mL−1) were well mixed with the soil in each pot [42].
In the PGPR treated pots; seeds were dipped in PGPR suspensions for 1 h before sowing (PGPT3, PGPT4 and PGPT5). Treated and non-treated castor seeds (3–4) were sown into sterilized pots containing 12 kg soil per pot (diameter 30 cm; height 45 cm) which were thinned to one plant after 15 days of germination [42]. Booster doses of PGPR were given at 15, 30 and 45 days after sowing. At maturity stage and data regarding yield and yield contributing parameters were collected and analyzed statistically. The agronomical parameters viz., shoot length (m), leaves per plant (nos.), leaf biomass (g) and leaf moisture (g) contents were recorded after 45 days of treatments (1st harvesting time) (Supplementary Fig. S2). FYM, inorganic fertilizers and PGPR strains were supplemented in the pots for analysis of plant growth parameters of castor. The fertilizer inputs except PGPR were given as per the recommendation given in Soil Health Card (Agronomy and Soil Science Division, CMER&TI, Lahdoigarh).
2.8. Data analysis
All experiments were replicated and the data including antagonistic activity, biochemical parameters, rearing and growth parameters were recorded in Microsoft Excel and analysed by Statistical Analysis System (SAS), Origin Pro 8 statistical software. Standard error and significant differences between values were determined using Duncan's multiple range test (P < 0.01), following one-way ANOVA. Regression, graphs and diagrams were presented with the help of statistical software program ‘Origin Pro 8’.
3. Results
3.1. Isolation and identification of castor rhizobacteria
From the 30 soil samples collected from different castor growing regions of Northeast India, we isolated fifty morphologically distinct rhizobacterial pure cultures by serial dilution plate technique and subsequently, their gram staining reactions and other morphological characters were recorded. The bacterial populations in the castor rhizosphere were ranged from 1.5 × 104 to 2.5 × 106 cfug−1. Most of the isolates were gram positive, small to large rod shaped bacteria and a few of them were gram negative rods arranged in different orientations. The potential bacterial antagonists were identified up to species level by polyphasic approach (morphological, biochemical and molecular characterization). Soil pH and soil texture data were also recorded. Further, selected bacterial isolates were identified through sequence analysis of 16S rRNA gene. Nine isolates were belonged to the genus Bacillus. The most potential isolate LRP-2 was identified as Enterobacter hormaechei (Figure 1). The other isolates were identified as Bacillus mycoides (HF-1), B. stratosphericus (KB-4), B. cereus (KB-6), B. wiedmannii (KM-2), B. aryabhattai (KRP-3), B. aryabhattai (KRP-6), B. megaterium (LR-5), B. aryabhattai (LR-7) and B. aryabhattai (UR-6). The details of the isolates and their identity are given in Table 1. The 16S rRNA gene sequences of the strains were submitted to NCBI database and accordingly their accession numbers were obtained (MH157931.1 to MH157940.1).
Figure 1.
Bacterial 16SrRNA sequences based homology study of the selected bacterial sequences indicating the position of strain LRP-2, HF-1 and UR-6 (antagonist and plant growth promoting strains) using Neighbour Joining method.
3.2. Identification of fungal pathogen
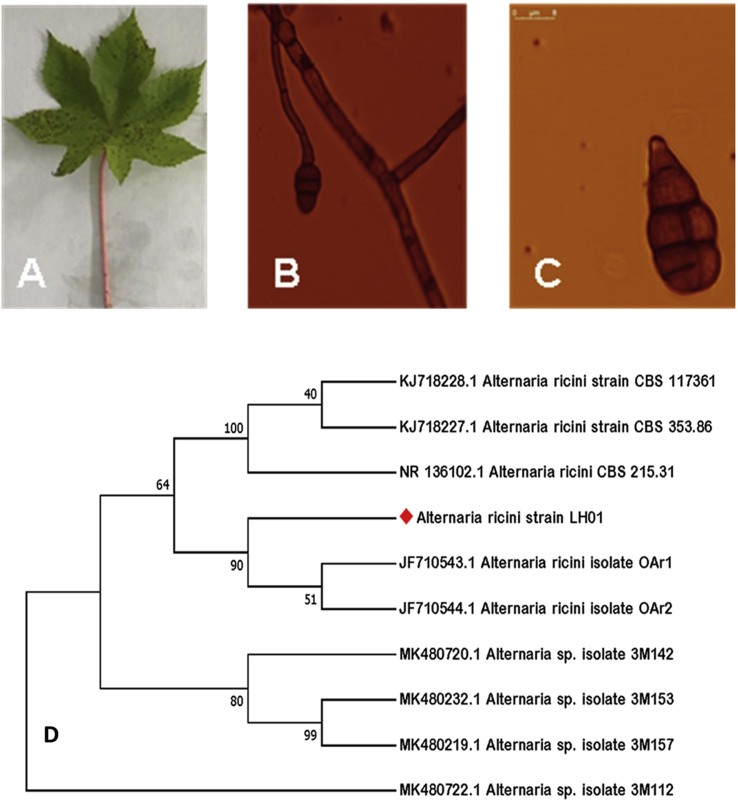
Four fungal purified colonies were isolated from the infected leaves and morphologically characterized. Conidia were typically produced in chains on the conidiophore. Hyphae were hyaline, septate and branched. The colour of the conidia was dark brown with horizontal and vertical septations (Figure 2A-C). Based on the colony coulour and morphology, mycelial and spore character, the pathogen was identified as A. ricini. The ITS1 and ITS2 region of the fungal strain was amplified and sequenced. The sequence was analyzed in NCBI database through BLAST search and identified as A. ricini strain LH01 (Figure 2D). Later, a phylogenetic tree was constructed using ‘MEGA 7.0’ [40]. The sequences were submitted to NCBI database (Accession no MK652889).
Figure 2.
Identification of fungal pathogen (LH01) of castor (Alternaria ricini). A-C, Infected castor leaf, fungal mycelium and septate spore of the fungus; D – Dendrogram was drawn using the homology study among the closest sequencess through BLAST search in NCBI database (using MEGA).
3.3. Antimicrobial bioassay
Antifungal activity of the rhizobacterial isolates were screened against A. ricini strain LH01. Ten out of 20 isolates (50%) exhibited potential antagonistic activity against the fungus (Figure 3) and later, these isolates were selected for further studies. The isolate LRP-2 (E. hormaechei) was the most potential strain and could reduce the colony growth up to 85.4% followed by B. mycoides (HF-1). All other isolates having potential antagonistic activity belongs to the genus Bacillus other than E. hormaechei. Based on antimicrobial bioassay, bacterial antagonist strains LRP-2 (E. hormaechei) and HF-1 (B. mycoides) were selected for pot experiments for disease suppression study under greenhouse condition.
Figure 3.
Antimicrobial bioassay of the selected bacterial antagonists against A. ricini (dual culture method). Values followed by a different letter are significantly different (P ≤ 0.01). Error bars represent standard deviations.
3.4. Screening of bacterial antagonists for PGP traits
3.4.1. Phosphate solubilization and nitrate reductase activity
Among the ten most potential rhizobacteria, phosphate solubilization index (PSI) was highest (1.72) in E. hormaechei (LRP-2) at 96 h of incubation and the least in B. mycoides (HF-1) (0.8). This result was proved in the quantitative assay (ability to solubilize inorganic phosphate in selected medium). In quantitative analysis, isolate LRP-2 (E. hormaechei) could solubilize highest amount of phosphate (63.52 μmol. L−1) whereas lowest in HF-1 (B. mycoides) (26.67 μmol. L−1) (Table 2). There were significant differences (p ≤ 0.01) on phosphate solubilization activities among the isolates. The pH drop was noticed up to 5.4 in isolate LRP-2 followed by 5.6 in KB-4 (B. stratosphericus), 5.9 in KRP-6 (B. aryabhattai) and 5.9 in LR-5 (B. megaterium) respectively at 96 h of incubation with an initial pH of 7.0. Isolate KB-6 (B. cereus) and KRP-6 (B. aryabhattai) were capable of producing nitrate reductase whereas all other isolates showed negative results against the control (Supplementary Table S1).
Table 2.
Qualitative and quantitative estimation of phosphate solubilization activity of the bacterial antagonists.Superscripts values with different alphabets differ significantly (P < 0.05) ± Standard deviation (SD).
| Isolate no. | Solubilization index (S.I.) | Soluble phosphate concentration (μmol/L)± SD | Initial pH | Final pH ± SD |
|---|---|---|---|---|
| Control | 0 | 0 ± 0.00 | 7.0 | 7.0 ± 0.45 |
| HF-1 | 0.80a | 26.67 ± 6.18a | 7.0 | 6.2 ± 0.36a |
| KB-4 | 1.57b | 55.72 ± 2.09b | 7.0 | 5.6 ± 0.42b |
| KB-6 | 1.40b | 48.83 ± 3.13c | 7.0 | 6.0 ± 0.28a |
| KM-2 | 1.10a | 48.96 ± 5.24c | 7.0 | 6.1 ± 0.19a |
| KRP-3 | 1.30a | 54.47 ± 2.33b | 7.0 | 6.3 ± 0.57a |
| KRP-6 | 1.66b | 51.89 ± 2.37bc | 7.0 | 5.9 ± 0.39b |
| LR-5 | 1.11a | 52.06 ± 4.39bc | 7.0 | 5.9 ± 0.44b |
| LR-7 | 1.32a | 56.50 ± 3.44b | 7.0 | 6.2 ± 0.29a |
| UR-6 | 1.26a | 53.74 ± 3.63bc | 7.0 | 6.5 ± 0.33b |
| LRP-2 | 1.72d | 63.52 ± 4.09d | 7.0 | 5.4 ± 0.51c |
3.4.2. IAA, GA3 and ACC deaminase activity
Rhizobacterial antagonists produced a range of IAA ranging between 9.42 ± 1.22 μg mL−1 to 30.62 ± 1.08 μg mL−1. The maximum production of IAA was found in isolate LRP-2 (30.62 ± 1.08 μg mL−1) which was very significant (p ≤ 0.01) whereas the lowest production was recorded in KRP-3 (9.42 ± 1.22 μg mL−1). All the selected 10 isolates produced IAA and the differences were significant among the strains. Similarly, LRP-2 produced highest amount of GA3 (18.96 ± 1.11 μg mL−1) among the antagonists and the least was produced by KB-4 (B. stratosphericus) (4.47 ± 0.57 μgmL−1) (Figure 4). Saleemi et al. ([43]) reported that PGPR exhibited production of IAA and GA3 ranging from 5.5 to 30.6 and 10.0–14.8 μg mL−1 which included Bacillus species dominantly. Moreover, UR-6 (B. aryabhattai) was on top in degrading ACC into α-ketobutyrate (5.43 ± 0.15 μg mL−1) followed by LR-7 (B. aryabhattai) (4.73 ± 0.12 μg mL−1) and KB-6 (B. cereus) (3.29 ± 0.14 μg mL−1) (Figure 5). Significant differences were observed among the isolates having ACC-deaminase activities (p ≤ 0.01).
Figure 4.
Indole-3-Acetic Acid (IAA) and Gibberellic acid (GA3) production by the selected bacterial antagonists. Values followed by a different letter are significantly different (P ≤ 0.01). Error bars represent standard deviations.
Figure 5.
ACC (1-aminocyclopropane-1-carboxylic acid) deaminase activity of the selected bacterial antagonists. Values followed by a different letter are significantly different (P ≤ 0.01). Error bars represent standard deviations.
3.4.3. NH3, HCN, siderophore production and salt tolerance
All these selected bacterial antagonists showed positive reaction for production of HCN and NH3 in qualitative screening (Supplementary Table S1). NH3 is the source of nitrogen and it plays a vital role in plant promoting shoot and root growth and biomass production [33]. Siderophore production was observed by LRP-2 (E. hormaechei) and UR-6 (B. aryabhattai). The rhizobacterial isolates were grown in different salt concentrations i.e. 1%, 2% and 3% NaCl. Except KRP-3 (B. aryabhattai), all other isolates were able to grow in these salt concentrations. Moreover, five isolates viz., KRP-3, KRP-6, LR-5, LR-7 and LRP-2 were able to grow at pH 4.0. At alkaline pH (9.5), seven isolates (HF-1, KB-4, KRP-3, KRP-6, LR-7, UR-6 and LRP-2) showed profuse growth leaving behind three isolates (KB-6, KM-2 and LR-5) with no growth at all (Supplementary Table S1).
3.5. In vivo disease suppression of bacterial antagonists
Among the eight different treatment combinations, the percent disease severity (PDS) was the highest in the treatments T2 and T4 (13.33%) followed by T6 whereas the lowest PDS was observed in treatment T8. Similarly, percent disease incidence (PDI) was the highest (100%) in the control treatment T2 (pathogen only) and in T6 where LRP-2 was inoculated as treatment regime. Further, PDI was lowest (33.3%) in treatments T4, T5 and T8 respectively where bacterial antagonists were inoculated before the pathogen or as mix inoculation (Table 3). From these results, it was apparent that the bacterial antagonists LRP-2 and HF-1 when applied before the fungal infection could protect the plant from Alternaria blight disease. Even, when LRP-2 and pathogens were applied at the same time, the disease incidence was very less. The effect of LRP-2 was at par with commercial fungicide (Indofil M45). But, the synergistic effect of LRP-2 and HF-1 was better and this treatment could protect the castor plant from fungal infection.
Table 3.
Percent disease severity and disease incidence after application of different treatment combinations. Values having different superscripts in the column differ significantly (P < 0.05).
| Treatment name | Treatment combinations | Disease severity index (%) | Disease incidence (%) |
|---|---|---|---|
| T1 | Sterile water (Negative control) | 0 | 0 |
| T2 | Pathogen (Positive control) | 13.33a | 100a |
| T3 | Pathogen + Fungicide (next day) | 6.66b | 66.66b |
| T4 | LRP-2 + Pathogen (next day) | 13.33a | 33.33c |
| T5 | LRP-2 + Pathogen (same day) | 8.33b | 33.33c |
| T6 | Pathogen + LRP-2 (next day) | 10.66c | 100a |
| T7 | HF-1 + Pathogen (same day) | 8.22b | 66.66b |
| T8 | LRP-2 + HF-1 + Pathogen (same day) | 3.33d | 33.33c |
3.6. Plant growth promotion assay
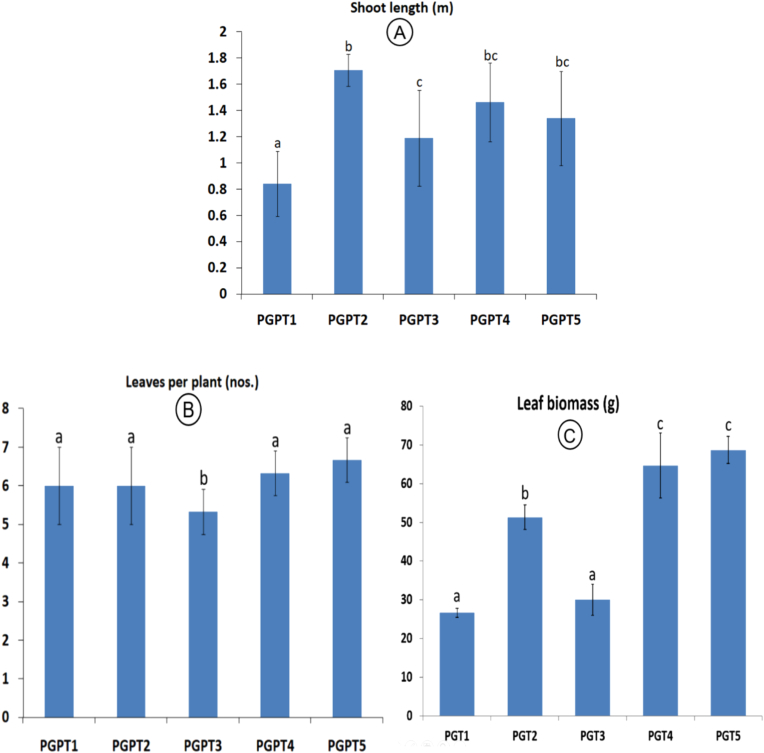
For plant growth promotion assay, we selected two promising bacterial isolates i.e., LRP-2 (E. hormaechei) and UR-6 (B. aryabhattai) and applied in different treatments. The height of the plant i.e., shoot length (1.71 ± 0.12 m) was maximum when treated with FYM (PGPT2) followed by application of combination of LRP-2 and UR-6 (PGPT4: 1.46 ± 0.30 m) and combinations of LRP-2, UR-6 and 50% of recommended NPK dose (PGPT5: 1.34 ± 0.36 m). However, number of leaves per plant (6.67 ± 0.21) and total leaf biomass (68.7 ± 3.5 g) was higher in the treatment PGPT5 which are the most important growth parameters (Figure 6). Overall, PGPR application along with the 50 % dose of NPK showed promising results in biomass yield. Even though, the plants were treated with LRP-2 and UR-6 (PGPT5), leaf moisture content was higher in the plants treated with FYM (PGPT2) and LRP-2 + UR-6 (PGPT4) i.e. 64.28 % and 63.92% respectively.
Figure 6.
Plant growth parameters of castor in different treatment combinations (PGPT1: Normal soil, PGPT2: Cow dung, PGPT3: LRP-2, PGPT4: LRP-2 + UR-6, PGPT5: LRP-2 + UR-6 + 50% of recommended NPK). A - Shoot length (m), B - Leaves per plant (nos.), C - Leaf biomass (g). Values followed by a different letter are significantly different (P ≤ 0.01). Error bars represent standard deviations.
4. Discussion
Ecologically, castor is known as wasteland colonizer and has gained the biological resource status because of the multiple uses of seeds as well as leaves (silkworm feeding) [44]. The plant is ideal for restoring disturbed soil as well as colonization and modification of wastelands [45, 46]. Biomass production of individual castor plant is higher when compared to other wasteland plant species like Achyranthes aspera, Abutilon indicum, Cassia occidentalis etc [7]. The yield parameters of castor are dependent on many factors including environmental stress [47]. Most importantly, multifactorial nature of plant-microbial interactions play a significant role on adaptation of plant in stressed environment and also protect from pathogens. Over the period of time, application of PGPR in plant abiotic stress management has been comprehensively studied which can even neutralize the toxic effect of heavy metals, improve leaf water status under salinity and other abiotic stress conditions [48, 49, 50].
The native rhizobacteria, we isolated from castor growing in the wasteland and selected for this study showed potential antagonistic activities against the fungal pathogen (A. ricini) and PGP activities enhancing plant growth. Among the three selected isolates, E. hormaechei (LRP-2) exhibited promising in vitro antifungal activity against A. ricini. In the pot experiments, the bacterial antagonist LRP-2 alone and in combination with B. mycoides (HF-1) could protect the plant from A. ricini infection which was at par with the commercial fungicides (Indofil M45) if applied before the symptoms appeared and at the same time with the pathogenic fungus (challenged inoculation). These isolates had potential PGP traits like ACC deaminase and phosphate solubilization as well as production of IAA, NH3 and siderophores. Previously, Enterobacter spp. and Bacillus spp. were reported to have a wide range of PGP characteristics by many workers [23, 51, 52, 53]. The homologies shown by the bacterial isolates with E. hormaechei (LRP-2) in phylogenetic studies have different function and were not evaluated their PGP and antagonistic activity. The biological functions of the other isolates showing homologies with B. mycoides (HF-1) and B. aryabhattai (UR-6) in the dendrogram are not well described. However, these two species exhibit genomic and phenotypic diversities and are widely used biocontrol agents. B. mycoides has already been used in a foliar spray preparation as a fungicide for disease prevention in plants and B. aryabhattai has been used as biocontrol agent of Heterodera glycines in soybean [54, 55, 56]. Bendaha and Belaouni [57] reported E. hormaechei as a potential antagonist against Fusarium oxysporum f.sp. radices lycopersici (FORL). These native rhizobacteria associated in castor root environment in wasteland might have helped the plants to uptake the nutrients from soils and protected from the infective microbes.
IAA mediated plant development and plant growth is augmented by exogenous IAA. Low amount of IAA stimulates root elongation, while higher amount of it decreases root length, increases root hair formation, and stimulates the formation of lateral roots [58, 59]. PGPR strains LRP-2, UR-6 and HF-1could produce IAA and GA3 which had the significant role for plant growth and development. These strains could increase both the root surface area and length so that the plants had greater access to soil nutrients [59]. Earlier, it was reported that inoculation of GA3 producing Sphingomonas sp. in tomato plants had significant effect on shoot elongation, seed germination and emergence, floral induction, fruit development as well as stem and leaf growth [60]. Additionally, E. hormaechei (LRP-2) and B. aryabhattai (UR-6) were the two candidates having potential phosphate solubilization activities. There are previous reports about PGPR strains those can solubilize phosphate by producing low molecular weight organic acids [61].
The highest ACC deaminase activity was showed by the bacterium B. aryabhattai (UR-6) among the isolates and used as one of the potential PGPR in this study. Microorganisms having ACC-deaminase activities have the ability to improve the plant growth under stressed environments like cold, drought, flooding, infections of pathogens and the presence of heavy metals [62]. ACC deaminase serves as the precursor of plant hormone ethylene synthesized in plant tissues during stressful conditions and alleviates biotic and abiotic stress conditions [62, 63]. Due to the presence of such rhizobacteria, castor might grow luxuriantly under stressed conditions even in the degraded wastelands.
We also found that all the selected PGPR showed positive reactions for production of HCN and NH3 which were the sources of nitrogen promoting shoot and root growth including biomass production [33]. Bacterial strains namely LRP-2 and UR-6 were also positive for siderophore production. Siderophore producing PGPR bind Fe3+ in the plant rhizospheres and prevent bacterial and fungal pathogens by depriving them from available iron [64, 65]. These bacterial strains could grow easily in acidic and saline soil. Raaijmakers et al. and Glick [61, 66] also reported that Bacillus spp. produce lipopeptide biosurfactants which is an advantage in competitive interactions with other organisms e.g. bacteria, fungi, protozoa and plants.
After inoculation with PGPR in pot experiments, yield related agronomical parameters like shoot length, leaf biomass and leaf moisture content etc. were recorded in different treatment combinations. Total leaf biomass production was higher when the plants were treated with E. Hormaechei (LRP-2) and B. aryabhattai (UR-6) (PGPT4) compared to control (un-inoculated soil) and other treatments. Castor grows in different habitats and as such, it encounters different soil microbial communities leading to complex plant microbe interactions [67]. Such interactions alter different plant growth parameters in various crops as reported earlier [68, 69]. Dutta and Thakur [23] previously reported that inoculation of E. lignolyticus strain TG1 in tea enhanced plant growth in greenhouse condition which was isolated from tea rhizosphere. As such, the native PGPR strains isolated from castor rhizosphere inoculated in pot experiments with high inoculum densities might have synergistic effect on plant growth.
5. Conclusion
Bacterial antagonists and PGPR strains isolated and identified in this study could protect castor from fungal infection, enhanced plant growth and also developed physiological mechanisms to withstand stress and survive in adverse conditions competing with the surrounding environment. In this study, we have highlighted the significance of native PGPR for plant growth and disease suppression through in-vitro screening and pot experiments. Such comprehensive screening followed by field testing helps in identifying rhizobacterial strains adaptable to diverse environment and soil conditions. These PGPR strains may be used as a possible alternative and can be included into appropriate nutrient and disease management practices of castor.
Declarations
Author contribution statement
Sosaka Protim Sandilya: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jeevan B.: Conceived and designed the experiments; Performed the experiments.
Kironta Dutta: Performed the experiments.
Gangavarapu Subrahmanyam, Nabanita Bhattacharyya: Performed the experiments; Analyzed and interpreted the data.
Vijay N.: Analysed and interpreted the data.
Mahananda Chutia: Conceived and designed the experiments; Analysed and interpreted the data; Wrote the paper.
Funding statement
Dr. Mahananda Chutia was supported by Central Silk Board, Bangalore [PRP-5880].
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors are thankful to the Director, Central Muga Eri Research & Training Institute (CMER&TI), Central Silk Board, Lahdoigarh, Jorhat (Assam) for providing necessary support and guidance to carry out the experiments. The authors are also thankful to the ‘Advanced Level Institutional Biotech Hub’ of CMER&TI for proving logistic support. We are also thankful to Bioserve Biotechnologies (India) Private Limited, Hyderabad for sequencing and analysis of microbial samples.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Sarmah M.C., Chutia M., Neog K., Das R., Rajkhowa G., Gogoi S.N. Evaluation of promising castor genotype in terms of agronomical and yield attributing traits, biochemical properties and rearing performance of Eri silkworm, Samia ricini (Donovan) Ind. Crop. Prod. 2011;31:192–196. [Google Scholar]
- 2.Lima J.R.D.S., Antonino A.C.D., Souza E.S.D., Lira C.A.B.D.O., Silva I.D.F.D. Seasonal and inter-annual variations of evapotranspiration, energy exchange, yield and water use efficiency of castor grown under rainfed conditions in North-eastern Brazil. Ind. Crop. Prod. 2013;50:203–211. [Google Scholar]
- 3.Dharajiya D.T., Shah A., Galvadiya B.P., Patel M.P., Srivastava R., Pagi N.K., Solanki S.D., Parida S.K., Tiwari K.K. Genome-wide microsatellite markers in castor (Ricinus communis L.): identification, development, characterization, and transferability in Euphorbiaceae. Ind. Crop. Prod. 2020;151 [Google Scholar]
- 4.Sailaja M., Tarakeswari M., Sujatha M. Stable genetic transformation of castor (Ricinus communis L.) via particle gun-mediated gene transfer using embryo axes from mature seeds. Plant Cell Rep. 2008;27:1509–1519. doi: 10.1007/s00299-008-0580-3. [DOI] [PubMed] [Google Scholar]
- 5.Das N., Kayastha A.M., Srivastava P.K. Purification and characterization of urease from de husked pigeon pea (Cajanus cajan L.) seeds. Phytochemistry (Oxf.) 2002;61(5):513–521. doi: 10.1016/s0031-9422(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 6.Nagaraja O., Krishnappa M. Leaf blight of Castor caused by Alternaria ricini: detection and pathogenicity in Castor (Ricinus communis L) Seed. Int. J. Agric. Env. Sci. 2016;3(4):6–10. [Google Scholar]
- 7.Mehmood F., Khan A.U.H., Khan Z. Appraisal of ecological significance of Ricinus communis Linn. in the wasteland of Lahore, Pakistan. Biologia (Pakistan) 2011;57:97–103. [Google Scholar]
- 8.Rajkumar M., Sandhya S., Prasad M.N.V., Freitas H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012;30:1562–1574. doi: 10.1016/j.biotechadv.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Jones D.L., Dennis P.G., Owen A.G., van Hees P.A.W. Organic acid behaviour in soils—misconceptions and knowledge gaps. Plant Soil. 2003;248:31–41. [Google Scholar]
- 10.Thilagam R., Hemalatha N. Plant growth promotion and chilli anthracnose disease suppression ability of rhizosphere soil actinobacteria. J. Appl. Microbiol. 2019;126:1835–1849. doi: 10.1111/jam.14259. [DOI] [PubMed] [Google Scholar]
- 11.Berg G., Fritze A., Roskot N., Smalla K. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahlia Kleb. J. Appl. Microbiol. 2001;91:963–971. doi: 10.1046/j.1365-2672.2001.01462.x. [DOI] [PubMed] [Google Scholar]
- 12.Palaniyandi S.A., Yang S.H., Cheng J.H., Meng L., Suh J.W. Biological control of anthracnose (Colletotrichum gloeosporioides) in yam by Streptomyces sp. MJM5763. J. Appl. Microbiol. 2011;111:443–455. doi: 10.1111/j.1365-2672.2011.05048.x. [DOI] [PubMed] [Google Scholar]
- 13.Chandra H., Kumari P., Bisht R., Prasad R., Yadav S. Plant growth promoting Pseudomonas aeruginosa from Valeriana wallichii displays antagonistic potential against three phytopathogenic fungi. Mol. Biol. Rep. 2020;7(8):6015–6026. doi: 10.1007/s11033-020-05676-0. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A., Patel J.S., Meena V.S., Ramteke P.W. Plant growth promoting rhizobacteria: strategies to improve abiotic stresses under sustainable agriculture. J. Plant Nutr. 2019;42(11-12):1402–1415. [Google Scholar]
- 15.Martinez-Viveros O., Jorquera M.A., Crowley D.E., Gajardo G., Mora M.L. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010;10:293–319. [Google Scholar]
- 16.Choudhary D.K., Sharma K.P., Gaur R.K. Biotechnological perspectives of microbes in agro-ecosystems. Biotechnol. Lett. 2011;33:1905–1910. doi: 10.1007/s10529-011-0662-0. [DOI] [PubMed] [Google Scholar]
- 17.García-Fraile P., Menéndez E., Rivas R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015;2:183–205. [Google Scholar]
- 18.Sandilya S.P., Bhuyan P.M., Gogoi D.K., Kardong D. Phosphorus solubilization and plant growth promotion ability of rhizobacteria of Ricinus communis L. growing in Assam, India. Proc. Natl. Acad. Sci. India B Biol. Sci. 2016;88:959–966. [Google Scholar]
- 19.Sandilya S.P., Bhuyan P.M., Vijay N., Gogoi D.K., Kardong D. Impact of Pseudomonas aeruginosa strain MAJ PIA03 affecting the growth and phytonutrient production of castor, a primary host-plant of Samia ricini. J. Soil Sci. Plant Nutr. 2017;17(2):499–515. [Google Scholar]
- 20.Habib S.H., Kausar H., Saud H.M. Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/6284547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinesha R., Anandaraja M., Kumarb A., Binia Y.K., Subilaa K.P., Aravind R. Isolation, characterization, and evaluation of multi-trait plant growth promoting rhizobacteria for their growth promoting and disease suppressing effects on ginger. Microbiol. Res. 2015;173:34–43. doi: 10.1016/j.micres.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharyya N., Sarma S. Assessment of availability, ecological feature, and habitat preference of the medicinal herb Houttuynia cordata Thunb. in the Brahmaputra Valley of Assam, India. Environ. Monit. Assess. 2010;160(1-4):277–287. doi: 10.1007/s10661-008-0694-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta J., Thakur D. Evaluation of multifarious plant growth promoting traits, antagonistic potential and phylogenetic affiliation of rhizobacteria associated with commercial tea plants grown in Darjeeling, India. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0182302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das R., Chutia M., Das K., Jha D.K. Factors affecting sporulation of Pestalotiopsis disseminata causing grey blight disease of Persea bombycina Kost., the primary food plant of muga silkworm. Crop Protect. 2010;29(9):963–968. [Google Scholar]
- 25.Dugan F.M. APS Press; US: 2006. The Identification of Fungi: an Illustrated Introduction with Keys, Glossary and Guide to Literature. [Google Scholar]
- 26.Gilman J.C. third ed.s. Biotech Books; New Delhi: 2008. A Manual of Soil Fungi. [Google Scholar]
- 27.El-Sayed W.S., Akhkha A., El-Naggar M.Y., Elbadry M. In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 2014;5:651. doi: 10.3389/fmicb.2014.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islama T., Deoraa A., Hashidokoa Y., Rahmana A., Itoa T., Taharaa S. Isolation and identification of potential phosphate solubilizing bacteria from the rhizoplane of Oryza sativa L. cv. BR29 of Bangladesh. Z. Naturforsch., C: J. Biosci. 2007;62:103–110. doi: 10.1515/znc-2007-1-218. [DOI] [PubMed] [Google Scholar]
- 29.Gupta A.P., Neue H.U., Singh V.P. Phosphorus determination in rice plants containing variable manganese content by the phospho-molybdo-vanadate (yellow) and phosphomolybdate (blue) colorimetric methods. Commun. Soil Sci. Plant Anal. 1993;24:1309–1318. [Google Scholar]
- 30.Wohler I. Auxin-indole derivatives in soils determined by a colorimetric method and by high performance liquid chromatography. Microbiol. Res. 1997;152:399–405. [Google Scholar]
- 31.Paleg L.G. Physiological effects of gibberellins. Annu. Rev. Plant Physiol. 1965;16:291–322. [Google Scholar]
- 32.Marques A.P.G.C., Pires C., Moreira H., Rangel A.O.S.S., Castro P.M.L. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol. Biochem. 2010;42:1229–1235. [Google Scholar]
- 33.Cappuccino J.C., Sherman N. A Laboratory Manual; New York: 1992. Microbiology; pp. 125–179. [Google Scholar]
- 34.Ahmad F., Ahmad I., Khan M.S. Screening of free living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008;163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Schwyn B., Neilands J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 36.Romdhane S., Nasr H., Samba-Mbaye R., Neyra M., Ghorbel M.H. Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J. Appl. Microbiol. 2006;100:436–444. doi: 10.1111/j.1365-2672.2005.02765.x. [DOI] [PubMed] [Google Scholar]
- 37.Kucuk C., Kivanc M., Kinaci E. Characterization of Rhizobium sp. isolated from bean. Turk. J. Biol. 2006;30:127–132. [Google Scholar]
- 38.Moller E.M., Bahnweg G., Sandermann H., Geiger H.H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992;20(22):6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Op De Beeck M., Lievens B., Busschaert P., Declerck S., Vangronsveld J., Colpaert J.V. Comparison and validation of some ITS primer pairs useful for fungal meta barcoding studies. PLoS One. 2014;9(6):e97629. doi: 10.1371/journal.pone.0097629. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N., Nie M. The Neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Pandey K.K., Pandey P.K., Kallo G., Banerjee M.K. Resistance to early blight of tomato with respect to various parameters of disease epidemics. J. Plant Pathol. 2003;69:364–371. [Google Scholar]
- 42.Masood S., Naz T., Javed M.T., Ahmed I., Ullah H., Iqbal M. Effect of short-term supply of farmyard manure on maize growth and soil parameters in pot culture. Arch. Agron Soil Sci. 2014;60(3):337–347. [Google Scholar]
- 43.Saleemi M., Kiani M.Z., Sultan T., Khalid A., Mahmood S. Integrated effect of plant growth promoting rhizobacteria and phosphate solubilizing microorganisms on growth of wheat (Triticum aestivum L.) under rainfed condition. Agric. Food Secur. 2017;6:46. [Google Scholar]
- 44.James T.A., Harden G.J. National herbarium. NSW. Royal Botanic Garden; Sydney, Australia: 1990. Flora of New South Wales. [Google Scholar]
- 45.Bhattacharyya N. Improvement of soil quality of wasteland by growing Ricinus communis L. under agronomic supplements. Delve. 2013;2(II):101–107. [Google Scholar]
- 46.Boda R.K., Majeti N.V.P., Suthari S. Ricinus communis L. (castor bean) as a potential candidate for revegetating industrial waste contaminated sites in peri-urban Greater Hyderabad: remarks on seed oil. Environ. Sci. Pollut. Res. Int. 2017;24:19955–19964. doi: 10.1007/s11356-017-9654-5. [DOI] [PubMed] [Google Scholar]
- 47.Severino L.S., Auld D.L., Vale L.S., Marques L.F. Plant density does not influence every castor plant equally. Ind. Crop. Prod. 2017;107:588–594. [Google Scholar]
- 48.Ahmad M., Zahir Z.A., Khalid M. Efficacy of Rhizobium and Pseudomonas strains to improve physiology, ionic balance and quality of mung bean under salt-affected conditions on farmer’s fields. Plant Physiol. Biochem. 2013;63:170–176. doi: 10.1016/j.plaphy.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 49.Naveed M., Hussain M.B., Zahir Z.A., Mitter B., Sessitsch A. Drought stress amelioration in wheat through inoculation with Burkholderia phytofirmans strain PsJN. Plant Growth Regul. 2014;73:121–131. [Google Scholar]
- 50.Vurukonda S.S.K.P., Vardharajula S., Shrivastava M., SkZ A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016;184:13–24. doi: 10.1016/j.micres.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Jha C.K., Aeron A., Patel B.V., Maheshwari D.K., Saraf M. In: Bacteria in Agrobiology: Plant Growth Responses. Maheshwari D., editor. Springer; Berlin, Heidelberg: 2011. Enterobacter: role in plant growth promotion. [Google Scholar]
- 52.Gupta M., Kiran S., Gulati A., Singh B., Tewari R. Isolation and identification of phosphate solubilizing bacteria able to enhance the growth and aloin-A biosynthesis of Aloe barbadensis Miller. Microbiol. Res. 2012;167:358–363. doi: 10.1016/j.micres.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 53.Mohamed I., Eid K.E., Abbas M.H., Salem A.A., Ahmed N., Ali M., Shah G.M., Fang C. Use of plant growth promoting rhizobacteria (PGPR) and mycorrhizae to improve the growth and nutrient utilization of common bean in a soil infected with white rot fungi. Ecotoxicol. Environ. Saf. 2019;171:539–548. doi: 10.1016/j.ecoenv.2018.12.100. [DOI] [PubMed] [Google Scholar]
- 54.Guetsky R., Shtienberg D., Elad Y., Fischer E., Dinoor A. Improving biological control by combining biocontrol agents each with several mechanisms of disease suppression. Phytopathology. 2002;92(9):976–985. doi: 10.1094/PHYTO.2002.92.9.976. [DOI] [PubMed] [Google Scholar]
- 55.Farhaoui A., Adadi A., Tahiri A., Alami N.E., Khayi S., Mentag R., Ezrari S., Radouane N., Mokrini F., Belabess Z., Lahlali R. Biocontrol potential of plant growth-promoting rhizobacteria (PGPR) against Sclerotiorum rolfsii diseases on sugar beet (Beta vulgaris L.) Physiol. Mol. Plant Pathol. 2022;119 [Google Scholar]
- 56.Zhao J., Liu D., Wang Y., Zhu X., Chen L., Duan Y. Evaluation of Bacillus aryabhattai Sneb517 for control of Heterodera glycines in soybean. Biol. Control. 2020;142 [Google Scholar]
- 57.Bendaha M.E.A., Belaouni H.A. Tomato growth and resistance promotion by Enterobacter hormaechei subsp. steigerwaltii EB8D. Arch. Phytopathol. Plant Protect. 2019;52:318–332. [Google Scholar]
- 58.Spaepen S., Vanderleyden J., Remans R. In: FEMS Microbiology Reviews. Unden F., editor. Blackwell Publishing Ltd.; New York, NY, USA: 2007. Indole-3-acetic acid in microbial and microorganism-plant signalling; pp. 425–448. [DOI] [PubMed] [Google Scholar]
- 59.Vacheron J., Desbrosses G., Bouffaud M.L., Touraine B., Moënne-Loccoz Y., Muller D. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013;4 doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan A.L., Waqas M., Kang S.M. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014;52:689–695. doi: 10.1007/s12275-014-4002-7. [DOI] [PubMed] [Google Scholar]
- 61.Glick B.R. Hindawi Publishing Corporation; Scientifica: Waterloo, Canada: 2012. Plant Growth-Promoting Bacteria: Mechanisms and Applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glick B.R., Cheng Z., Czarny J., Duan J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 2007;119:329–339. [Google Scholar]
- 63.Chandra D., Srivastava R., Gupta V.V.S.R., Franco C.M.M., Sharma A.K. Evaluation of ACC-deaminase producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 2019;65:387–403. doi: 10.1139/cjm-2018-0636. [DOI] [PubMed] [Google Scholar]
- 64.Kloepper J.W., Leong J., Teintze M., Schroth M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature. 1980;286:885–886. [Google Scholar]
- 65.Vansuyt G., Robin A., Briat J.F., Curie C., Lemanceau P. Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana, mol. Plant-Microbe Interact. 2007;2:441–447. doi: 10.1094/MPMI-20-4-0441. [DOI] [PubMed] [Google Scholar]
- 66.Raaijmakers J.M., de Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 67.Li X., Feng C., Chen L., Liu F., Wang L., Luo K., Pang Y. Cultivable rhizobacteria improve castor bean seedlings root and plant growth in Pb–Zn treated soil. Rhizosphere. 2021;19 [Google Scholar]
- 68.Trabelsi D., Mhamdi R. Microbial inoculants and their impact on soil microbial communities: a review. BioMed Res. Int. 2013 doi: 10.1155/2013/863240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Essalimi B., Esserti S., Rifai L.A., Koussa T., Makroum K., Belfaiza M., Rifai S., Venisse J.S., Faize L., Alburquerque N., Burgos L., Jadoumi S.E., Faize M. Enhancement of plant growth, acclimatization, salt stress tolerance and Verticillium wilt disease resistance using plant growth-promoting rhizobacteria (PGPR) associated with plum trees (Prunus domestica) Sci. Hortic. 2022;291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.