Abstract
SARS-CoV-2 is a betacoronavirus with single-stranded positive-sense RNA, which is a serious global threat to human health. Understanding the molecular mechanism of viral replication is crucial for the development of antiviral drugs. The synthesis of viral polyproteins is a crucial step in viral progression. The synthesis of viral polyproteins in coronaviruses is regulated by the 5ʹ-untranslated region (UTR); however, the detailed regulatory mechanism needs further investigation. The present study demonstrated that the RNA binding protein, RBM24, interacts with the RNA genome of SARS-CoV-2 via its RNA recognition submotifs (RNPs). The findings revealed that RBM24 recognizes and binds to the GUGUG element at stem-loop 4 (SL4) in the 5ʹ-UTR of SARS-CoV-2. The interaction between RBM24 and 5ʹ-UTR prevents 80S ribosome assembly, which in turn inhibits polyproteins translation and the replication of SARS-CoV-2. Notably, other RNA viruses, including SARS-CoV, MERS-CoV, Ebolavirus, rhinovirus, West Nile virus, Zika virus, Japanese encephalitis virus, yellow fever virus, hepatitis C virus, and human immunodeficiency virus-1 also contain one or several G(U/C/A)GUG sequences in the 5ʹ-UTR, which is also targeted by RBM24. In conclusion, the present study demonstrated that RBM24 functions by interacting with the 5ʹ-UTR of SARS-CoV-2 RNA, and elucidated that RBM24 could be a host restriction factor for SARS-CoV-2 and other RNA viruses.
Keywords: SARS-CoV-2, RBM24, Polyproteins, Translation, RNA viruses
1. Introduction
SARS-CoV-2 belongs to the Betacoronavirus genus, which contain a positive-sense single-stranded RNA ((+)ssRNA) genome with a 5ʹ-cap and a poly(A) tail in the 3ʹ-untranslated region (UTR). Host cell entry is mediated via the spike (S) protein of SARS-CoV-2, which attaches to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface (Zhou et al., 2020), following by triggering membrane fusion, viral entry, and infection. Then the uncoated genomic RNA is translated into pp1a and pp1ab polyproteins, which are then cleaved into a total of 16 nonstructural proteins (Nsps) and subsequently assembled into the transcription/replication machinery. The replication and transcription complex assembled by Nsps and virus-induced double-membrane vesicles (DMVs) replicates and synthesizes a nested set of subgenomic RNA species by genomic transcription, which encoded structural proteins and some accessory proteins. The newly formed virus particles subsequently undergo assembly, budding, and release into the extracellular compartment (Ashour et al., 2020; Mohamadian et al., 2021).
The RNA genome of SARS-CoV-2 is less than 30 kb in length and includes 14 open reading frames (ORFs) that encode Nsps, structural proteins and other accessory proteins (Krichel et al., 2020; Naqvi et al., 2020). ORF1a/b is translated into the pp1a (Nsp1–11) or pp1ab (Nsp1–16) polyproteins, which undergo proteolytic cleavage by the proteases such as PLpro and 3CLpro to generate a total of 16 Nsps (Kadam et al., 2021). The polyproteins are essential for the translation of structural proteins (Mohamadian et al., 2021). The 5ʹ-end of the SARS-CoV-2 genome contains a leader sequence and an UTR with several stem-loop structures that are necessary for the translation, replication, and transcription of viral RNA. As the RNA genome of SARS-CoV-2 is capped at the 5ʹ-end, it is suggested that the initiation of translation is canonical and cap-dependent. Based on the experimental SHAPE structure of the 5ʹ-UTR of mouse hepatitis virus (MHV) (Yang and Leibowitz, 2015), the epsilon (ε) structure of hepatitis B virus (HBV) (Yao et al., 2018), and the secondary structure of the 5ʹ-UTR of SARS-CoV-2, it is assumed that the 5ʹ-UTR potentially forms a translation initiation complex in proximity of the 5ʹ-cap. RNA structures in proximity of the 5ʹ-cap are known to regulate RNA translation (Babendure et al., 2006; Pestova and Kolupaeva, 2002). Previous structural studies on the 5ʹ-UTRs of other coronaviruses have demonstrated the presence of hairpin structures in the proximity of the cap structure, and the efficiency of translational initiation is significantly modulated by these secondary structures (Babendure et al., 2006). Miao et al. have proved the 5ʹ-UTR of SARS-CoV-2 is highly structured with five simple hairpin structures, similar with other coronaviruses. Among the stem-loops (SLs), the SL2 hairpin domain is expected to be involved in the replication complex formation, the SL3 hairpin is known to encompass the leader TRS (TRS-L) sequence, the SL4 has a long hairpin and is a relatively stable, and the SL5 has a more complex four-way junction structure (Miao et al., 2021).
SARS-CoV-2 relies on host RNA-binding proteins (RBPs) for the infection progression, multiple RBPs have been identified involving in SARS-CoV-2 RNA-host protein interaction (such as PABPC1, DDX3X, EIF4B, etc.) by comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) (Flynn et al., 2021). RBPs contain RNA recognition motifs (RRMs) and bind to single or double-strand regions of RNA molecules, and play key roles in the post-transcriptional regulation of RNA. Host RBPs can be recruited for viral replication (Li and Nagy, 2011). RNA-binding protein 24 (RBM24) with two RNA recognition sub-motifs (RNP1 and RNP2) is necessary for cardiovascular development and myogenesis (Grifone et al., 2014; Jiang et al., 2014; Zhang et al., 2016). RBM24 can post-transcriptionally regulate the stability of the members of the p53 transcription factor family (Jiang et al., 2014; Xu et al., 2014), which suggests that RBM24 plays a crucial role in RNA biology. Our previous studies have indicated that RBM24 can modulate the translation and replication of HBV and hepatitis C virus (HCV) by targeting the secondary structure of the viral 5ʹ-UTRs (Cao et al., 2018; Yao et al., 2018). RBM24 is characterized by its single and invariant RRM domain, which binds to the G(U/C/A)GUG element or stem-loop regions (Kuwasako et al., 2014; Yao et al., 2019). The sequence analyses shown that the “GUGUG” elements were located at SL4 and SL5 in the 5ʹ-UTR, implying that RBM24 could be involved in the translation and replication of SARS-CoV-2 polyproteins by targeting the 5ʹ-UTR.
In order to test this hypothesis, we investigated the function of RBM24 in the translation of polyproteins and replication of SARS-CoV-2. The data presented in this study demonstrate that RBM24 inhibits the translation of polyproteins and replication of SARS-CoV-2 via its RNA-binding domains. The findings demonstrated that RBM24 specifically binds to the SL4 GUGUG element in the 5ʹ-UTR of SARS-CoV-2, and subsequently inhibits the formation of the 80S translation initiation complex. We also observed that RBM24 targets and inhibits the translation of viral RNA in several RNA viruses containing the G(U/C/A)GUG sequence in the 5ʹ-UTR. The findings therefore demonstrate that RBM24 may serve as a novel host limiting factor in SARS-CoV-2 and other RNA viruses.
2. Results
2.1. RBM24 inhibits the replication of SARS-CoV-2
Our previous study demonstrated that RBM24 directly binds to the 5ʹ-UTR of HCV RNA (Cao et al., 2018) and the 5ʹ-ε stem-loop of HBV pgRNA via RNA structure-dependent manner (Yao et al., 2018, 2019), and subsequently inhibits viral replication and translation. We therefore assume that RBM24 may be involved in regulating SARS-CoV-2 replication by interfering with translation of viral proteins. In order to evaluate our hypothesis, we determined the functions of RBM24 in SARS-CoV-2. The expression of Nsp3 was determined as a measure of polyprotein expression. As depicted in Fig. 1 , the expression of SARS-CoV-2 Nsp3 protein, N protein, and RNA increased following RBM24 knockdown, and correspondingly decreased following RBM24 overexpression in both H1299-ACE2 (Fig. 1B and C) and A549-ACE2 cells (Fig. 1D and E). In order to further confirm the antiviral effect of RBM24, the infectious titers were measured. As shown in Fig. 1F, overexpression of RBM24 leaded to a decrease in infectious titers in supernatant. Taken together, these results indicated that RBM24 inhibited the replication of SARS-CoV-2.
Fig. 1.
RBM24 inhibits the replication of SARS-CoV-2. (A) Schematic illustration of the experimental protocol. H1299-ACE2 cells or A549-ACE2 cells were seeded in 24-well plates at −1 day post transfection (dpt), transfected at 0 dpt, infected with SARS-CoV-2 at 1 dpt, and harvested at 2 dpt. (B–C) H1299-ACE2 cells were transfected with siNC, siRBM24, pcDNA3.1, or pRBM24, and infected with SARS-CoV-2 at MOI of 0.1. The protein and RNA samples were harvested from the cell lysates. (B) The expression of the Nsp3 and N proteins of SARS-CoV-2 and RBM24 was detected by Western blotting, using actin as the loading control. (C) SARS-CoV-2 RNA was detected by real-time PCR, and the levels were normalized to that of actin RNA; the levels of SARS-CoV-2 RNA in the siNC and pcDNA3.1 groups were considered to be 1. (D–E) A549-ACE2 cells were transfected with siNC, siRBM24, pcDNA3.1, or pRBM24, and infected with SARS-CoV-2 at MOI of 0.1. The protein and RNA samples were harvested from the cell lysates. (D) The Nsp3 and N proteins of SARS-CoV-2 and RBM24 were detected by Western blotting, using actin as the loading control. (E) SARS-CoV-2 RNA was detected by real-time PCR, and the levels were normalized to that of actin RNA; the levels of SARS-CoV-2 RNA in the siNC and pcDNA3.1 groups were considered as 1. (F) H1299-ACE2 cells were transfected with pcDNA3.1 or pRBM24 and infected with SARS-CoV-2. The supernatants were harvested at 16 dpi and subjected to the plaque assay using VeroE6 cells (left panel), then the infectious titers were counted (right panel).
2.2. The RNP domains of RBM24 are essential for the inhibition of SARS-CoV-2
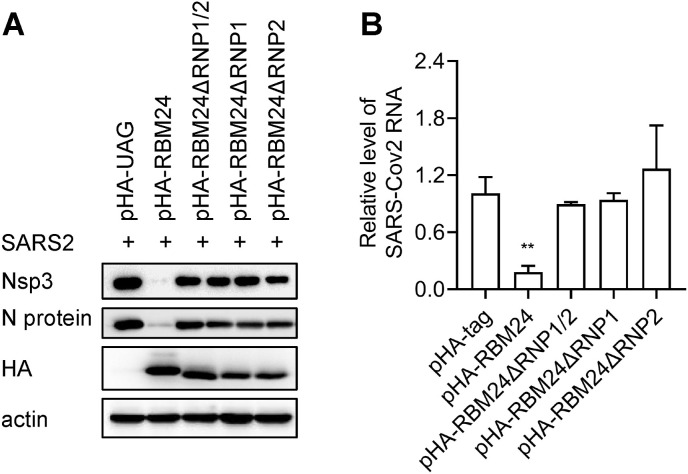
RBM24 mainly functions by binding to RNA via its two putative RNA-binding sub-domains, RNP1 and RNP2 (Yao et al., 2018). In order to verify whether the RNP domains are critical for the inhibitory effects of RBM24 on SARS-CoV-2, H1299-ACE2 cells were transfected with wild-type or truncated forms of RBM24 following infection with SARS-CoV-2. The results demonstrated that the levels of Nsp3 protein, N protein, and viral RNA were significantly reduced only following transfection with the wild-type RBM24; however, their levels remained unaltered following transfection with truncated RBM24 (Fig. 2 A and B). These results confirmed that the RNP1 and RNP2 domains are essential for the inhibitory effect of RBM24 on SARS-CoV-2.
Fig. 2.
The RNP domains of RBM24 are essential for the inhibitory effect of SARS-CoV-2. (A–B) H1299-ACE2 cells were transfected with pHA-RBM24, pHA-tag, pHA-RBM24ΔRNP1/2, pHA-RBM24ΔRNP1, or pHA-RBM24ΔRNP2, and infected with SARS-CoV-2 at MOI of 0.1. The protein and RNA samples were subsequently harvested from the cell lysates. (A) The expression of the Nsp3 and N proteins of SARS-CoV-2 and RBM24 was detected by Western blotting, using actin as the loading control. (B) SARS-CoV-2 RNA was detected by real-time PCR, and the RNA levels were normalized to that of actin RNA; the levels of SARS-CoV-2 RNA in the pHA-tag group were regarded as 1.
2.3. RBM24 directly binds to the 5ʹ-UTR of SARS-CoV-2 RNA
As both RNP1 and RNP2 are RNA recognition domains and truncated RBM24 lacking the RNP1 and RNP2 domains has no influence on the replication of SARS-CoV-2, we speculated that RBM24 may interact with SARS-CoV-2 RNA via the RNP domains. The RNA-IP assay revealed that only wild-type RBM24 co-immunoprecipitated with SARS-CoV-2 RNA (Fig. 3 A), suggesting that RBM24 interacts with SARS-CoV-2 RNA, and the interaction is primarily mediated via the RNP domains.
Fig. 3.
RBM24 directly binds to the 5′-UTR of SARS-CoV-2 RNA. (A) H1299-ACE2 cells were transfected with pHA-RBM24, pHA-tag, pHA-RBM24ΔRNP1/2, pHA-RBM24ΔRNP1, or pHA-RBM24ΔRNP2, and infected with SARS-CoV-2 at MOI of 0.1. The RNA samples were subsequently harvested from the cell lysates. The input or co-immunoprecipitated RNA of SARS-CoV-2 was detected by real-time PCR. The relative levels of the co-immunoprecipitated RNA of SARS-CoV-2 was normalized to that of the input RNA. (B) Schematic illustration of the 5′-UTR of SARS-CoV-2. (C) HEK293T cells were transfected with pRBM24 and the cell lysates were harvested at 48 hpt. The expression of input RBM24 and actin was detected by Western blotting. (D) Lysates of cells (400 μg) transfected with pRBM24 or 50 pM of rhRBM24 were incubated with 2 μg of biotin-labeled HBV ε, biotin-labeled 5′-UTR of SARS-CoV-2, or yeast tRNA. Unlabeled 5′-UTR RNA (0 × , 10 × , and 20 × ) was added in excess for competing with RBM24 in binding to the biotin-labeled 5′-UTR fragment, followed by pulldown with streptavidin beads and assessment by Western blotting. (E) HEK293T cells were transfected with pHA-RBM24, pHA-tag, pHA-RBM24ΔRNP1/2, pHA-RBM24ΔRNP1, or pHA-RBM24ΔRNP2. The cell lysates were harvested at 48 hpt, and 400 μg of cell lysates were incubated with 2 μg of the biotin-labeled 5′-UTR fragments of SARS-CoV-2 RNA, followed by pulldown assays with streptavidin beads and assessment with Western blotting.
RBM24 interacts with RNA in a structure- or sequence-dependent manner. SARS-CoV-2 contains a stable 5′UTR structure which also contains the “GUGUG” element in SL4 and SL5 (Fig. 3B, black asterisks) (Miao et al., 2021). We further determined the role of RBM24 in binding to the 5ʹ-UTR. RBM24 overexpression lysates or purified recombinant RBM24 protein was incubated with the biotin-labeled 5ʹ-UTRs of SARS-CoV-2 RNA. The RNA-protein complex pulled down using streptavidin beads was detected by Western blotting. In the absence or presence of other cellular proteins, RBM24 was capable of binding to the ε element of HBV (Fig. 3D, lane 2, positive control) and the 5ʹ-UTR of SARS-CoV-2 (Fig. 3D, lane 3). RBM24 incubated with yeast tRNA was used as the negative control (Fig. 3D, lane 1). In order to confirm the binding specificity between RBM24 and the 5ʹ-UTR of SARS-CoV-2, the binding of RBM24 to the biotin-labeled 5ʹ-UTR of SARS-CoV-2 was competitively inhibited by adding an excess of the unlabeled 5ʹ-UTR probe of SARS-CoV-2. The results further confirmed that RBM24 specifically binds to the 5ʹ-UTR of SARS-CoV-2 (Fig. 3D, lanes 4–6).
The findings revealed that both RNP1 and RNP2 were essential for the binding of RBM24 to SARS-CoV-2 RNA (Fig. 3A). We further verified whether the RNP domains are essential for the binding of RBM24 to the 5ʹ-UTR of SARS-CoV-2. As depicted in Fig. 3E, only wild-type RBM24 was pulled down by the biotin-labeled 5ʹ-UTR fragments. Truncated RBM24 with deletions in any of the RNP domains were not pulled down by the 5ʹ-UTR fragments, indicating that the both the RNP domains, RNP1 and RNP2, are essential for the binding of RBM24 to the 5ʹ-UTR.
2.4. RBM24 binds to the GUGUG sequence in the 5ʹ-UTR of SARS-CoV-2
The biotin-labeled 5ʹ-UTRs of truncated RNAs, namely, SL1+SL2, SL3+SL4, SL4.5+SL5d, and SL5a-c, were synthesized based on the structure of the 5ʹ-UTR of SARS-CoV-2 (Fig. 4 A). RBM24 lysates or purified recombinant RBM24 were incubated with the wild-type or truncated 5ʹ-UTRs, and subjected to the pulldown assay. As depicted in Fig. 4B, the binding ability of RBM24 to SL1+SL2, SL4.5+SL5d, and SL5a-c were reduced compared to that of the full length 5ʹ-UTR (Fig. 4B, lanes 3, 4, 6, and 7). However, SL3+SL4 displayed RBM24 binding activity (Fig. 4B, lane 5), indicating that SL3+SL4 may play a key role in the binding of RBM24 in the absence or presence of other cellular proteins.
Fig. 4.
RBM24 binds to the GUGUG sequence in the 5′-UTR of SARS-CoV-2. (A) Schematic illustration of the structure of the 5′-UTR of SARS-CoV-2. (B) The biotin-labeled SL1+SL2, SL3+SL4, SL4.5+SL5d, and SL5a-c truncated 5′-UTRs were constructed based on the structure of the 5′-UTR of SARS-CoV-2. Lysates of cells (400 μg) transfected with pRBM24 or 50 pM of rhRBM24 were incubated with 2 μg of yeast tRNA, biotin-labeled 5′-UTR fragments, or its truncated fragments, followed by pulldown with streptavidin beads and assessment by Western blotting. (C) The biotin-labeled SL3, SL4, 5′-UTRΔSL3+SL4, 5′-UTRΔSL3, and 5′-UTRΔSL4 truncated UTRs were constructed based on the structure of the 5′-UTR of SARS-CoV-2. Lysates of cells (400 μg) transfected with pRBM24 or 50 pM of rhRBM24 were incubated with 2 μg of yeast tRNA, biotin-labeled 5′-UTR fragments, or its truncated fragments, followed by pulldown with streptavidin beads and assessment by Western blotting. (D) Schematic illustration of the wild-type SL4 and mutated SL4 (SL4 mut). The GUGUG sequence was mutated to GAGAG and the CACGCA complementary sequence was mutated to CTCGCA in the SL4 mut mutant. (E) The biotin-labeled SL4 and truncated SL4 mut were constructed based on the structure of the 5′-UTR of SARS-CoV-2. Lysates of cells (400 μg) transfected with pRBM24 or 50 pM of rhRBM24 were incubated with 2 μg of yeast tRNA, biotin-labeled 5′-UTR, SL4, or SL4 mut fragment, followed by pulldown with streptavidin beads and assessment by Western blotting.
The substructural domain of SL3+SL4 was subsequently mapped and we observed that SL4 but not SL3 co-precipitated with RBM24 (Fig. 4C, lanes 3–4). Based on this finding, biotin-labeled 5ʹ-UTRs with deletions of SL3+SL4, SL3, or SL4 were constructed. The truncated 5ʹ-UTR with SL3 deletion was pulled down with RBM24, while there were no clear signals for the truncated 5ʹ-UTRs with deletions of SL3+SL4 or SL4 (Fig. 4C, lanes 5–7), indicating that the SL4 domain in the 5ʹ-UTR is critical for the binding of RBM24 to the 5ʹ-UTR.
The GUGUG element is present in the SL4 domain, and to further confirm whether the GUGUG element in the 5ʹ-UTR of RNA is critical for the binding of RBM24, we synthesized wild-type and mutated SL4 RNAs (substitution at 2U→A, 3U→A, and pairing bases 2A→T), and evaluated their binding potential (Fig. 4D). The findings revealed that the binding capability of RBM24 to mutated SL4 was significantly reduced compared to the binding ability of RBM24 to the wild-type 5ʹ-UTR and SL4 RNA (Fig. 4E). We also identified that the SL5b stem-loop contains a GUGUG element; however, SL5b was incapable of binding to RBM24 (Fig. 4B). This finding demonstrated that the GUGUG element in SL4 in the 5ʹ-UTR of SARS-CoV-2 is critical for the binding of RBM24 to the 5ʹ-UTR.
2.5. RBM24 inhibits mRNA translation via the 5ʹ-UTR by inhibiting 80S ribosome assembly
The 5ʹ-UTR fragment of SARS-CoV-2 is essential for the translational initiation of ORF1a/b (Schubert et al., 2020). RBM24 knockdown apparently enhanced the expression of Nsp3, while the overexpression of RBM24 suppressed Nsp3 expression (Fig. 1B and D). It has been reported that RBM24 inhibits the translation of HBV and HCV by binding to the viral 5ʹ-UTRs (Cao et al., 2018; Yao et al., 2018). These results implied that RBM24 inhibited the translation of polyproteins in SARS-CoV-2 by targeting the 5ʹ-UTR. In order to validate this speculation, the 5ʹ-UTR sequence of SARS-CoV-2 was inserted upstream of the luciferase ORF for generating the pSARS-CoV-2 5ʹ-UTR-luc (pSARS2 5′-UTR-luc) plasmid, which was subsequently co-transfected with pcDNA3.1 or pRBM24. The relative luciferase activity (RLA) was normalized with that of pRBM24 to pcDNA3.1. Compared with the vector plasmid pluc, the insertion of the 5ʹ-UTR of SARS-CoV-2 and the 5ʹ-UTR of HBV significantly reduced the RLA (Fig. 5 B). In order to further verify the effect of RBM24 on the translation of SARS-CoV-2 polyproteins, pluc, pSARS-CoV-2 5ʹ-UTR-luc, and pHBV-5ʹUTR-luc transcripts were generated in vitro and incubated with rhRBM24 or BSA in rabbit reticulocyte lysate (RRL). We observed that RBM24 significantly reduced the RLA following the insertion of the 5ʹ-UTR of SARS-CoV-2 and the 5ʹ-UTR of HBV (Fig. 5C). These results suggested that RBM24 could inhibit the translation of polyproteins by interacting with the 5ʹ-UTR of SARS-CoV-2.
Fig. 5.
RBM24 inhibits the translation of mRNA via the 5′-UTR by preventing 80S ribosome assembly. (A) Schematic illustration of the constructed pluc, pHBV-ε-luc, and pSARS2-5′-UTR-luc plasmids. (B) HEK293T cells were co-transfected with pluc, pHBV-ε-luc, or pSARS2-5′-UTR-luc, along with pcDNA3.1 or pRBM24. Luciferase activity was determined using a Steady-Glo® luciferase assay system. The values of RLA were calculated and normalized with those of the pRBM24 to pcDNA3.1 groups. (C) The luc, HBV-ε-luc, and SARS2-5′-UTR-luc RNAs were transcribed in vitro, and incubated with rhRBM24 or BSA in RRL, and the luciferase activity was determined using a Steady-Glo® luciferase assay system. The values of RLA were calculated and normalized with those of the rhRBM24 to BSA groups. (D) The biotin-labeled 5′-UTR fragments of RNA were incubated with rhRBM24 or BSA in RRL for 15 min at 30 °C. The ribosomal complexes were separated by sucrose density gradient ultracentrifugation. The distribution of biotinylated-RNA fragments was detected by dot-blot assay using streptavidin-HRP and analyzed by densitometry.
Ribosome assembly assay was performed in an RRL system for elucidating the mechanism underlying the inhibitory effect of RBM24 on the 5ʹ-UTR-mediated translation of SARS-CoV-2. The formation of the 80S ribosome was observed in the control group that was incubated with BSA; however, the intensity of the 80S peak was significantly decreased in the presence of rhRBM24, and the majority of RNAs were retained in the 40/48S peaks (Fig. 5D). These findings indicated that the inhibitory effect of the RBM24–5ʹ-UTR interaction on the translation of polyproteins was attributed to the inhibition of 80S ribosome assembly on SARS-CoV-2 RNA.
2.6. Broad-spectrum inhibition of viral translation by RBM24
The G(U/C/A)GUG sequence is widely found in the 5ʹ-UTRs of RNA viruses, including SARS-CoV-2, SARS, MERS, Ebola, rhinovirus, West Nile virus, Zika, JEV, yellow fever virus, HCV, and HIV-1 (Fig. 6 A). RBM24 inhibits the replication of HBV (Yao et al., 2018), HCV (Cao et al., 2018), and SARS-CoV-2 (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5) by inhibiting the translation of viral proteins, which implies that RBM24 may act as an antiviral effector protein. In order to validate this speculation, 11 kinds of viral 5ʹ-UTRs (Fig. 6A) were inserted upstream of the luciferase ORF for generating the pSARS2 5ʹ-UTR-luc, pSARS 5ʹ-UTR-luc, pMERS 5ʹ-UTR-luc, pEbola 5ʹ-UTR-luc, prhinovirus 5ʹ-UTR-luc, pWest Nile virus 5ʹ-UTR-luc, pZika 5ʹ-UTR-luc, pJEV 5ʹ-UTR-luc, pyellow fever virus 5ʹ-UTR-luc, pHCV 5ʹ-UTR-luc, and pHIV-1 5ʹ-UTR-luc recombinant plasmids, which were subsequently co-transfected with pcDNA3.1 or pRBM24. The RLA was normalized with that of pRBM24 to pcDNA3.1 and compared with that of the vector plasmid pluc. The results demonstrated that all the viral 5ʹ-UTRs significantly reduced the RLA in HEK293T (Fig. 6B) and Huh7 cells (Fig. 6C). These findings implied that RBM24 inhibited viral translation by targeting with the viral 5ʹ-UTR, and may act as a broad-spectrum antiviral effector for RNA viruses containing the G(U/C/A)GUG element at the 5ʹ-UTR. The efficacy of RBM24 as a host restriction factor of SARS-CoV-2 is presently being investigated.
Fig. 6.
Broad-spectrum inhibition of viral translation by RBM24. (A) Schematic illustration of the viral 5′-UTR sequences; the specific G(U/C/A)GUG binding sequence of RBM24 has been highlighted. (B–C) The sequences of the viral 5′-UTRs were inserted at the 5′-end of the ORF of luciferase in pluc, to generate the pSARS2 5′-UTR-luc, pSARS 5′-UTR-luc, pMERS 5′-UTR-luc, pEbola 5′-UTR-luc, prhinovirus 5′-UTR-luc, pWest Nile virus 5′-UTR-luc, pZika 5′-UTR-luc, pJEV 5′-UTR-luc, pyellow fever virus 5′-UTR-luc, pHCV 5′-UTR-luc, and pHIV 5′-UTR-luc recombinant vectors. The viral 5′-UTR luciferase plasmids were co-transfected with pRBM24 or pcDNA3.1 into HEK293T cells (B) or Huh7 cells (C). Luciferase activity was determined using a Steady-Glo® luciferase assay system. The values of RLA values were normalized with those of the pRBM24 to pcDNA3.1 groups; the RLA values of the pluc group were considered to be 1.
3. Discussion
Following its release into the cytoplasm, the uncoated genomic RNA of SARS-CoV-2 is first used as a template for the synthesis of the polyproteins, pp1a and pp1ab, following which the synthesized polyproteins mediate the translation of structural proteins and the replication of viral RNA (Ashour et al., 2020; Mohamadian et al., 2021). The detailed mechanism of polyprotein translation remains to be elucidated. In this study, we identified a novel cellular factor, RBM24, which inhibits the translation of SARS-CoV-2 polyproteins by targeting the 5ʹ-UTR. RBM24 directly interacts with the GUGUG element in the SL4 stem-loop in the 5ʹ-UTR of SARS-CoV-2, inhibits 80S ribosome assembly, and suppresses the efficiency of polyproteins translation. RBM24 represents a limiting factor in the replication of SARS-CoV-2, and may also act as a broad-spectrum antiviral effector protein in other RNA viruses containing the G(U/C/A)GUG element in the 5ʹ-UTR.
Coronaviruses have evolved specialized mechanisms for hijacking the host gene expression machinery and utilizes the cellular resources of the host for regulating the translation of viral proteins. The 5ʹ- and 3ʹ-UTRs contain cis-acting sequences that are functionally important for RNA-RNA interactions and for the binding of viral RNA and cellular proteins during RNA replication (Satija and Lal, 2007; Yang and Leibowitz, 2015). In particular, the 5ʹ-UTR of SARS-CoV-2 also efficiently promotes translational initiation (Schubert et al., 2020). The pp1a and pp1ab polyproteins are translated from SARS-CoV-2 RNA and subsequently cleaved into Nsp1–16 (Kadam et al., 2021). The results of this study demonstrated that the RBM24 can directly bind to the 5ʹ-UTR of SARS-CoV-2 (Fig. 3D) to inhibit the translation of SARS-CoV-2 RNA into Nsp3 (Fig. 1B and D). Altogether, the findings demonstrated that RBM24 inhibits the translation of polyproteins (pp1a and pp1ab). The viral proteins, especially Nsp3, Nsp9, Nsp10, Nsp15, and Nsp16 play critical roles in viral replication (Mohamadian et al., 2021), and we also observed that the replication of SARS-CoV-2 were significantly decreased when RBM24 was overexpressed (Fig. 1A–F).
It is postulated that the translation of SARS-CoV-2 RNA into polyproteins is initiated by ribosomal scanning, in which the 43S-preinitiation complex binds near the 5′-cap and scans in the 3ʹ-direction until it encounters the AUG codon. The 60S subunit then joins to form a complete 80S ribosome, which is followed by translational initiation (Sonenberg and Hinnebusch, 2009). RBM24 is a strong inhibitor of viral translation and binds to the 5ʹ-UTR of SARS-CoV-2 to inhibit 80S ribosome assembly, which subsequently reduces the pool of available ribosomes that can engage in translation (Fig. 5D). The results of the in vivo and in vitro assays demonstrated that RBM24 overexpression inhibited the translation of luciferase initiated by the 5ʹ-UTR of SARS-CoV-2 (Fig. 5B and C). Furthermore, the ribosome assembly assay revealed that RBM24 did not affect the binding of the 40S subunit to the mRNA containing the 5ʹ-UTR of SARS-CoV-2; however, RBM24 affected the formation of the 80S ribosome assembly complex on mRNAs containing the 5ʹ-UTR of SARS-CoV-2 (Fig. 5D). These findings led us to speculate that RBM24 possibly binds to the 5ʹ-UTR of SARS-CoV-2 to prevent the 40S ribosomal subunit in scanning for the AUG codon, which leads to the failure of assembly of the 80S ribosome, and subsequently inhibits polyproteins translation. The hypothesis was supported by the fact that RBM24 also hampers ribosome assembly during protein translation in HBV and HCV by interacting with the 5ʹ-UTR of HBV and 5ʹ-UTR of HCV, respectively (Cao et al., 2018; Yao et al., 2018). In a previous report neither silencing nor overexpression of RBM24 causes changes in the total protein level (Zhang et al., 2018). And our previous results of WST-1 analysis showed that the down-regulation of RBM24 did not affect cell viability (Yao et al., 2018). Therefore, the observed antiviral effects of RBM24 were probably due to direct inhibition on SARS-CoV-2 translation. However, we could not totally exclude that RBM24 may affect other cellular proteins and indirectly regulates SARS-CoV-2 translation. We also notice the limitations of relatively artificial binding and translation in vitro assays, further assays such as in vivo translation and RNA fluorescence in situ hybridization (FISH) should be performed in the further.
For the function of regulating virus replication, RBM24 is more like a switch than a host limiting factor. On the one hand, RBM24 inhibits the translation of HBV and HCV, and on the other hand, enhances the replication of the HBV and HCV genome (Cao et al., 2018; Yao et al., 2018, 2019, 2022). Whether or not RBM24 works as switch modulates the protein translation and RNA replication during other virus infection is an opening question now. Though our results suggest RBM24 as an anti-SARS-CoV-2 target, the detail mechanism of RBM24 in virus replication needs more deep investigation. RBM24 was supposed to be a potential therapeutic target for cancer treatment (Shi, 2022), And different efforts such as small molecule inhibitors, siRNA, peptides and CRISPR-based therapies had been made to target RBP functions for cancer therapy (Mehta et al., 2022). However, how to deliver the drugs to specific tissues with minimal cytotoxicity to normal cells is the main question, which is the same for the antivirus purpose. A possible strategy is to block the interaction of RBM24 with its viral specific partner.
RBM24 is characterized by its single and invariant RRM domain, which specifically binds to RNA via a G(U/C/A)GUG sequence-dependent or RNA structure-dependent manner (Kuwasako et al., 2014; Yao et al., 2019). The SARS-CoV-2 RNA contains a 5ʹ-UTR with several stem-loop structures (Manfredonia et al., 2020) and the GUGUG element in SL4 (Fig. 3, Fig. 4D). RBM24 binds to the ε element in the pgRNA of HBV via a structure-dependent manner (Yao et al., 2018, 2019), while in SARS-CoV-2, RBM24 recognizes the 5ʹ-UTR in a sequence-dependent manner, via the GUGUG sequence in SL4 (Fig. 4E). It's still unclear why the “GUGUG”element in SL5b cannot bind to RBM24 (Fig. 4B). Other sequences than the core sequence may also be important for RBM24 binding. More likely, in addition to the sequence, the secondary or even higher structure determines the binding of RBM24 and viral genome. The G(U/C/A)GUG element is present the 5ʹ-UTRs of numerous RNA viruses, including SARS-CoV-2, SARS-CoV, MERS-CoV, Ebola virus, rhinovirus, West Nile virus, Zika virus, Japanese encephalitis virus, yellow fever virus, HCV, and HIV-1 (Fig. 6A). We further evaluated whether RBM24 affects the translation of these 11 viruses by comparing the luciferase activity to that of the vector luciferase. The results demonstrated that RBM24 may suppress the translational efficiency of the luciferase ORF following the insertion of these viral 5ʹ-UTRs (Fig. 6B and C). However, these results haven't shown directly that RBM24 inhibits the translation of these viruses, but suggested that RBM24 may have a common antiviral activity against RNA viruses by inhibiting viral translation. Thus, this hypothesis needs to be evaluated using infection systems. Also, it was not possible to rule out whether RBM24 has inhibitory effects on other viruses that lacking the G(U/C/A)GUG element, and further studies are necessary in this regard.
Altogether, the present study demonstrated that the RBM24 host factor, with RNA-binding activity, targets the GUGUG element in the SL4 stem-loop in the 5ʹ-UTR of SARS-CoV-2 to inhibit the translation of polyproteins and suppress the replication of SARS-CoV-2. The findings also indicated that RBM24 may have a common antiviral activity against several RNA viruses in general. Inducing RBM24 overexpression in vivo or delivering RBM24 to the target site may be the novel antiviral therapies. Taken together, the results provide insights into understanding the mechanism of replication of SARS-CoV-2 and aid in the development of novel antiviral targets for RNA viruses.
4. Materials and methods
4.1. Cell culture and transfection
The H1299-ACE2 cells used in this study were a generous gift from Prof. Peng Li, and were cultured in Gibco RP Basic MI 1640 medium (C11875500BT; Gibco). The stable A549-ACE2 cell line expressing human the ACE2 receptor was generated from A549 cells and cultured in Dulbecco's modified Eagle's medium (DMEM, 12100-046; Gibco). Huh7 (YB–H3110; ATCC) and HEK293T cells (HEMCL-032; ATCC) were also cultured in DMEM. The culture media (1640 Basic medium and DMEM) were supplemented with 10% fetal bovine serum (FBS, 10099-141; Gibco) and 100 U/ml penicillin-streptomycin (15140-122; Gibco), and the cells were cultured at 37 °C in an atmosphere of 5% CO2.
4.2. Plasmids
The plasmids expressing the wild-type RBM24 protein, namely, pRBM24 and pHA-RBM24, as well as the plasmids expressing the truncated RBM24 protein, including pHA-RBM24ΔRNP1/2, pHA-RBM24ΔRNP1, and pHA-RBM24ΔRNP2, were used as previously described (Yao et al., 2018). In addition, pluc and pHBV-TR-luc (pTR-luc) were used as previously described (Yao et al., 2018). The fragments containing the viral 5ʹ-UTR sequences were synthesized and cloned into pluc, which was located at the 5ʹ-end of the ORF of luciferase, to generate the pSARS2 5ʹ-UTR-luc, pSARS 5ʹ-UTR-luc, pMERS 5ʹ-UTR-luc, pEbola 5ʹ-UTR-luc, prhinovirus 5ʹ-UTR-luc, pWest Nile virus 5ʹ-UTR-luc, pZika 5ʹ-UTR-luc, pJEV 5ʹ-UTR-luc, pyellow fever virus 5ʹ-UTR-luc, pHCV 5ʹ-UTR-luc, and pHIV 5ʹ-UTR-luc recombinant vectors.
4.3. Transfection and infection
Plasmids and siRNAs were transfected into H1299-ACE2, A549-ACE2 and Huh7 cells using Lipofectamine® 3000 Reagent (L3000-015; Invitrogen™) or into HEK293T cells using Lipofectamine® 2000 Reagent (11668-019; Invitrogen™) according to the manufacturer's instructions. The cells in the control group were transfected with control siRNA (siNC, SI03650318; QIAGEN), while the cells in the experimental group were transfected with RBM24 siRNA (siRBM24, SI03030195; QIAGEN).
For SARS-CoV-2 infection, the cells were infected with the viral preparation at a multiplicity of infection (MOI) of 0.1, and the cells were harvested 24 h post infection (hpi). Briefly, the infected cells were lysed in TRIzol (15596018; Invitrogen) and the RNA was extracted according to the manufacturer's instructions. The RNA was amplified by real-time polymerase chain reaction (PCR) using the primers for actin (forward: 5ʹ-TGGAATCCTGTGGCATCCATGAAAC-3ʹ; reverse: 3ʹ-TAAAACGCAGCTCAGTAACAGTCCG-5ʹ) and the N protein of SARS-CoV-2 (forward: 5ʹ-GGGGAACTTCTCCTGCTAGAAT-3ʹ; reverse: 3ʹ-CAGACATTTTGCTCTCAAGCTG-5ʹ). The relative N RNA levels were normalized with the actin RNA. The cells were lysed in 2X Laemmli buffer and subjected to Western blotting.
4.4. The plaque assay
Viral supernatants were 5-fold serially diluted in DMEM, 200 μl of each serially diluted supernatant was added to the VeroE6 cells. After 1 h, the inoculums were removed and the 500 μl of 2% Methyl cellulose medium overlay was added to each well for 4 days at 37 °C, 5% CO2. The cells were then fixed with formalin and stained with crystal violet. The number of plaques were counted and the viral titers were expressed in the logarithm of plaque-forming units (PFU) per milliliter.
4.5. Western blotting
Western blotting was performed as previously described (18). The following antibodies were used for Western blotting: anti-actin (66009-1; ProteinTech), anti-RBM24 (ab94567; Abcam), rabbit anti-HA (3724S; Cell Signaling Technology), rabbit anti-Nsp3 (GTX135614; GeneTex), rabbit anti-SARS-CoV-2 nucleocapsid (anti-N) (28769-1-AP; ProteinTech), mouse anti-Flag (F1804; Sigma), streptavidin HRP (CT353; U-cytech), anti-mouse secondary antibodies (115-035-146; Jackson), and anti-rabbit secondary antibodies (111-035-003; Jackson).
4.6. RNA immunoprecipitation (RIP) assays
RIP assays were performed using the previously described method (Cao et al., 2018; Keene et al., 2006). Briefly, H1299-ACE2 cells were transfected with pHA-tag, pHA-RBM24, pHA-ΔRNP1/2, pHA-ΔRNP1, or pHA-ΔRNP2, and infected with SARS-CoV-2 at MOI of 0.1, following which the cell lysates were harvested at 48 hpi. Then, 800 μg of the cell lysates containing the total protein was incubated with protein G precoated with mouse anti-HA (H9658; Sigma) at 30 °C for 4 h. After washing 5 times with washing buffer, the precipitated RNA and 5% of the total RNA (input RNA) extracted from the cell lysates were detected by real-time PCR. The relative level of the precipitated RNA of SARS-CoV-2 was normalized to that of the input RNA.
4.7. Streptavidin pulldown assay
The streptavidin pulldown assay was performed as previously described (Cao et al., 2018; Yang et al., 2012), and the recombinant human RBM24 (rhRBM24) was prepared by prokaryotic expression and purification as previous reported (Cao et al., 2018). Briefly, the biotinylated-RNA fragments were dissolved in RNA folding buffer (20 mM HEPES (pH 7.6), 100 mM KCl, and 5 mM MgCl2) and folded by incubation at 70 °C for 2 min. Then 400 μg of the total protein and 50 pmol of purified rhRBM24 were separately incubated with 2 μg of biotinylated-RNA fragments, 30 μg of yeast tRNA (AM7119; Ambion), and 40 U of RNasin at 30 °C for 30 min. Dynabeads M-280 Streptavidin (11205D; Invitrogen) was resuspended with 300 μL of the RNA binding buffer (100 mM KCl, 20 mM HEPES (pH 7.6), 5 mM MgCl2, 10% glycerol, 0.1% IGEPAL®, 1 mM DTT, and 400 μM RVC). The aforementioned protein-RNA mixtures were subsequently added to the beads and incubated at 30 °C for 2 h. The beads were washed 6 times with RNA binding buffer, and subjected to Western blotting.
4.8. Luciferase reporter assay
The luciferase reporter assay was performed as previously described (Cao et al., 2014). Briefly, HEK293T or Huh7 cells were co-transfected with the indicated plasmids. The activity of firefly luciferase was measured at 48 h post transfection (hpt) with a luciferase reporter assay system (E2940; Promega). All the reporter assays were repeated at least three times.
4.9. In vitro translation and ribosome assembly assay
The capped RNAs from pluc, pHBV-TR-luc, and pSARS2-5′UTR-luc were generated by in vitro transcription using a mMESSAGE mMACHINE® Kit (AM1344; Invitrogen). Biotin-11-UTP (AM8450; Invitrogen) was added to the reaction mixture for producing biotin-labeled capped RNAs. The in vitro translation and ribosome assembly assays were performed as previously described (Locker et al., 2007). Briefly, the template RNAs and recombinant human RBM24 protein (rhRBM24) (Cao et al., 2018) or a non-specific control protein (BSA), were incubated in an RRL (L4960; Promega)-based translation reaction system. The luciferase activity assay was subsequently performed using a Steady-Glo® luciferase assay system (E2520; Promega) (Cao et al., 2014).
For the ribosome assembly assay, 1 μg of the biotin-labeled 5ʹ-UTRs of SARS-CoV-2 RNA and 12.5 pmol of rhRBM24 or BSA were added to an RRL-based ribosome assembly mixture. The mixtures were incubated for 15 min at 30 °C and subjected to 10–40% sucrose density gradient ultracentrifugation. The gradients were fractionated into 21 fractions and blotted onto an Amersham Hybond™-N+ membrane with a Whatman® Minifold® I 96 well dot-blot array system. The membrane was crosslinked with a HL-2000 Hybrilinker, and the blotted biotinylated-RNA was detected with Streptavidin-HRP (CT353; U-Cytech).
4.10. Statistical analyses
The data were analyzed using two-tailed unpaired t-test. Statistical significance was set at * (p < 0.05), ** (p < 0.01), or *** (p < 0.001).
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31900138, to Y. Y.), the China Postdoctoral Science Foundation (grant number 2019M662851, to Y. Y.), and the National Basic Research Priorities Program of China (grant number 2018YFA0507201, to X. C.).
Declaration of competing interest
The authors declare that there is no conflict of interest in the publication of this study.
Acknowledgments
The authors would like to acknowledge Dr. Ding Gao, Ms. Anna Du, and Ms. Juan Min of The Core Facility and Technical Support of Wuhan Institute of Virology for their valuable technical support.
Data availability
Data will be made available on request.
References
- Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9 doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babendure J.R., Babendure J.L., Ding J.H., Tsien R.Y. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Zhao K., Yao Y., Guo J., Gao X., Yang Q., Guo M., Zhu W., Wang Y., Wu C., Chen J., Zhou Y., Hu X., Lu M., Chen X., Pei R. RNA binding protein 24 regulates the translation and replication of hepatitis C virus. Protein Cell. 2018;9:930–944. doi: 10.1007/s13238-018-0507-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Zhu W., Han Q., Pei R., Chen X. Construction of a chimeric hepatitis C virus replicon based on a strain isolated from a chronic hepatitis C patient. Virol. Sin. 2014;29:61–70. doi: 10.1007/s12250-014-3408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C., Schmitz C.O., Parker K.R., Woo E., Chang H.Y., Horvath T.L., Carette J.E., Bertozzi C.R., Wilen C.B., Satpathy A.T. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184:2394–2411. doi: 10.1016/j.cell.2021.03.012. e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R., Xie X., Bourgeois A., Saquet A., Duprez D., Shi D.L. The RNA-binding protein Rbm24 is transiently expressed in myoblasts and is required for myogenic differentiation during vertebrate development. Mech. Dev. 2014;134:1–15. doi: 10.1016/j.mod.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Jiang Y.Q., Zhang M., Qian Y.J., Xu E.S., Zhang J., Chen X.B. Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. J. Biol. Chem. 2014;289:3164–3175. doi: 10.1074/jbc.M113.524413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S.B., Sukhramani G.S., Bishnoi P., Pable A.A., Barvkar V.T. SARS-CoV-2, the pandemic coronavirus: molecular and structural insights. J. Basic Microbiol. 2021;61:180–202. doi: 10.1002/jobm.202000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D., Komisarow J.M., Friedersdorf M.B. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K., Takahashi M., Unzai S., Tsuda K., Yoshikawa S., He F., Kobayashi N., Guntert P., Shirouzu M., Ito T., Tanaka A., Yokoyama S., Hagiwara M., Kuroyanagi H., Muto Y. RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing. Nat. Struct. Mol. Biol. 2014;21:778–786. doi: 10.1038/nsmb.2870. [DOI] [PubMed] [Google Scholar]
- Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8:305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N., Easton L.E., Lukavsky P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M., Incarnato D. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020;48:12436–12452. doi: 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M., Raguraman R., Ramesh R., Munshi A. RNA binding proteins (RBPs) and their role in DNA damage and radiation response in cancer. Adv. Drug Deliv. Rev. 2022;191 doi: 10.1016/j.addr.2022.114569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Tidu A., Eriani G., Martin F. Secondary structure of the SARS-CoV-2 5'-UTR. RNA Biol. 2021;18:447–456. doi: 10.1080/15476286.2020.1814556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadian M., Chiti H., Shoghli A., Biglari S., Parsamanesh N., Esmaeilzadeh A. COVID-19: Virology, biology and novel laboratory diagnosis. J. Gene Med. 2021;23 doi: 10.1002/jgm.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija N., Lal S.K. The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Muhlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Shi D.L. RBM24 in the post-transcriptional regulation of cancer progression: anti-tumor or pro-tumor activity? Cancers. 2022;14 doi: 10.3390/cancers14071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E.S., Zhang J., Zhang M., Jiang Y.Q., Cho S.J., Chen X.B. RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Mol. Cancer Res. 2014;12:359–369. doi: 10.1158/1541-7786.MCR-13-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3' and 5' ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Bi J., Xue X., Zheng L., Zhi K., Hua J., Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- Yao Y., Yang B., Cao H., Zhao K., Yuan Y., Chen Y., Zhang Z., Wang Y., Pei R., Chen J., Hu X., Zhou Y., Lu M., Wu C., Chen X. RBM24 stabilizes hepatitis B virus pregenomic RNA but inhibits core protein translation by targeting the terminal redundancy sequence. Emerg. Microb. Infect. 2018;7:86. doi: 10.1038/s41426-018-0091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Yang B., Chen Y., Huang D., Liu C., Sun H., Hu X., Zhou Y., Wang Y., Chen J., Pei R., Wen Z., Chen X. RNA-Binding motif protein 38 (RBM38) mediates HBV pgRNA packaging into the nucleocapsid. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105249. [DOI] [PubMed] [Google Scholar]
- Yao Y., Yang B., Chen Y., Wang H., Hu X., Zhou Y., Gao X., Lu M., Niu J., Wen Z., Wu C., Chen X. RNA-binding motif protein 24 (RBM24) is involved in pregenomic RNA packaging by mediating interaction between hepatitis B virus polymerase and the epsilon element. J. Virol. 2019;93 doi: 10.1128/JVI.02161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang Y., Xu E., Mohibi S., de Anda D.M., Jiang Y., Zhang J., Chen X. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 2018;25:1118–1130. doi: 10.1038/s41418-017-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Lin Y., Liu J., Zhang Z.G., Fu W., Guo L.Y., Pan L., Kong X., Zhang M.K., Lu Y.H., Huang Z.R., Xie Q., Li W.H., Xu X.Q. Rbm24 regulates alternative splicing switch in embryonic stem cell cardiac lineage differentiation. Stem Cell. 2016;34:1776–1789. doi: 10.1002/stem.2366. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.