Abstract
Background:
Inflammatory linear verrucous epidermal nevus (ILVEN) is a rare skin disease characterized by pruritic erythematous scaly plaques distributed along the lines of Blaschko. Two cases of ILVEN with CARD14 mutations and 1 case with a GJA1 mutation have been previously reported
Objective:
To elucidate the genetic cause of a cohort of patients diagnosed based on clinical and histopathological evaluation with ILVEN.
Methods:
We recruited patients diagnosed with ILVEN based on clinical and histopathological criteria. Exome sequencing of affected skin with or without blood was performed and germline and somatic pathogenic variants were identified.
Results:
Five patients were enrolled. All had skin lesions from birth or early childhood. Two patients developed psoriasis vulgaris after the diagnosis of ILVEN. The first had a germline heterozygous CARD14 mutation and a post-zygotic hotspot mutation in KRT10. Histopathological evaluation did not show epidermolytic hyperkeratosis. The second had a post-zygotic hotspot mutation in HRAS. Her ILVEN became itchy once psoriasis developed. One patient was re-diagnosed with linear porokeratosis based on a germline mutation in PMVK and a post-zygotic second-hit mutation. Two patients were re-diagnosed with CHILD nevus based on germline NSDHL mutations.
Conclusion:
ILVEN is a clinical descriptor for a heterogenous group of mosaic inflammatory disorders. Genetic analysis has the potential to more precisely categorize ILVEN and permits pathogenesis-directed therapies in some cases.
Keywords: genetic skin diseases, inflammatory linear verrucous epidermal nevus, epidermal nevus, mosaicism, CHILD syndrome
INTRODUCTION
Inflammatory linear verrucous epidermal nevus (ILVEN) is a benign nevus characterized by pruritic erythematous papules and plaques distributed along the lines of Blaschko. ILVEN classically presents at birth or within the first year of life. It usually extends over the skin within Blaschko lines for months to years, finally stabilizing in early childhood.1 The classic histology of ILVEN is psoriasiform acanthosis with overlying alternating parakeratosis and orthokeratosis and corresponding hypo- and hypergranulosis underneath.1,2 A superficial perivascular lymphocytic infiltrate is seen within the dermis. However, the histology is not always characteristic and rare histological subtypes have been described.3 There has also been much debate about the clinical and histopathological similarities with linear psoriasis as well as CHILD (congenital hemidysplasia with ichthyosiform nevus and limb defects) syndrome. The variability in histology described in ILVEN, together with the clinical and histopathological similarities with other blaschkolinear inflammatory disorders raise the question of whether ILVEN is a distinct clinical entity.
The linear/blaschkoid distribution of ILVEN suggests that a somatic mutation in keratinocyte precursor cells during embryogenesis underlies its pathogenesis. To date, mosaicism of CARD14 mutations has been reported in two cases, and a GJA1 mutation in one case.4,5
We aimed to identify the genetic cause of five cases referred for a genetic analysis under the diagnosis of ILVEN through whole exome sequencing.
METHODS
The Yale Human Investigation Committee approved the study protocol. Patients were evaluated clinically by a board-certified dermatologist and referred for genetic analysis with a suspected diagnosis of ILVEN. All patients or their guardians provided verbal and written informed consent. Prior to genetic analysis, the medical records and hematoxylin and eosin-stained slides of included patients were reviewed. Biopsies from affected skin and blood/saliva were collected from included patients for genetic analysis.
DNA extraction, sequencing and bioinformatic analysis.
Paired exome sequencing was performed by the Yale Center for Genome Analysis (YCGA) using sheared genomic DNA (gDNA) isolated from saliva (Saliva DNA Isolation Reagent Kit, Norgen) and primary keratinocytes (Allprep, Qiagen) or 1 mm cores from the epidermis of formalin-fixed, paraffin-embedded specimens (QIAamp DNA FFPE Tissue Kit, Qiagen). We used primary keratinocytes from patients 2 and 5. When fresh affected tissue was not available we used cores from FFPE affected tissue. Unpaired analysis of affected tissue was performed for patient 1 since normal tissue was not available. Resultant reads were aligned to the hg19 human reference genome using the Burrows-Wheeler Aligner (BWA-MEM) and trimmed with PCR duplicates removal using Picard (Broad Institute) prior to recalibration with GATK 6. Haplotype caller was used to identify germline variants7 and Mutect2 was used to identify somatic variants 8. Variants were annotated using ANNOVAR 9. Missense, nonsense, indels and splice site mutations were filtered to exclude variants with an allele frequency >2% in GnomAD.10 Aligned reads were examined with Integrative Genomics Viewer software (IGV, Broad Institute) to remove mismapped reads and calling errors 11. Relevant variants were verified by Sanger sequencing.
Cell cultures.
Skin biopsies from affected skin of patients 2 and 5 were transported in DMEM (Invitrogen) with 1% penicillin/streptomycin and incubated overnight in 1X dispase (ZenBio). Primary keratinocytes were isolated from the resulting epidermal sheet using 0.05% trypsin-2% EDTA and were plated onto a feeder layer of mitomycin C treated 3T3 cells in DMEM/F12 medium. Cells were subsequently propagated in Epilife (Invitrogen).
RESULTS
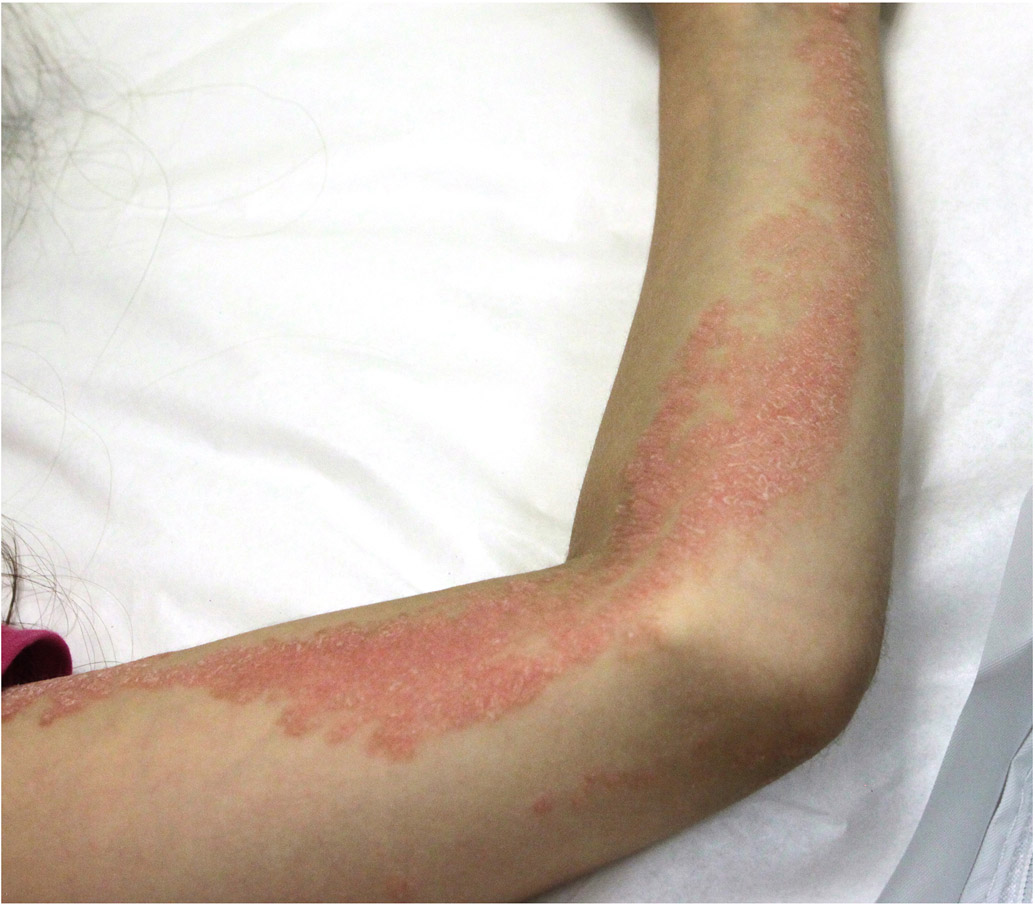
All patients were female and were diagnosed with ILVEN as infants or in early childhood. Clinical presentations included blaschkoid psoriasiform plaques of variable extension (Figure 1). None had bone defects. Histopathological presentation varied (Figure 1).
Figure 1. Clinical and histologic features of inflammatory linear verrucous epidermal nevus (ILVEN).
a) Patient 1 had a linear verrucous plaque over her right foot and ankle that became itchy once she developed generalized plaque psoriasis. b) Histology demonstrated alternating orthokeratosis and parakeratosis acanthosis and spongiosis. c) Patient 2 had itchy blaschkoid psoriasiform plaques over her left chest and arm. d) Histopathologic evaluation showed psoriasiform epidermal hyperplasia and areas of alternating orthokeratosis and parakeratosis. Some of the keratinocytes showed reticular degeneration with balloon cell morphology. e) Patient 3 presented with extensive whorls of pink plaques, some covered with yellowish crusts. f) Histopathologic evaluation showed alternate parakeratosis overlying hypogranulosis and orthokeratosis, acanthosis and papillomatosis with perivascular and interstitial lymphocytic infiltrate in the superficial dermis. The parakeratosis overlying hypogranulosis corresponds to wide cornoid lamellae. g) Patient4 had a small linear verrucous plaque over her left groin. h) Histopathological evaluation showed psoriasiform hyperplasia with hyperkeratosis and parakeratosis, thin supra-papillary plate and dense lymphohistiocytic infiltrate in the papillary dermis. i) Patient 5 has psoriasiform and warty plaques in a blaschkoid distribution over the left side of her body, including the left groin. Several fingernails had longitudinal leukonychia and the second nail was partially absent. j) Histopathology showed marked parakeratosis and acanthosis and foamy macrophages in the papillary dermis.
Patient 1 presented with a linear hyperkeratotic plaque over her right foot and ankle that was first noticed in early infancy (Figure 1a). The plaque became pruritic during adolescence once she developed generalized plaque psoriasis. There was no evidence for superimposed fungal infection. Histopathology was compatible with ILVEN (Figure 1b). The plaque partially responded to potent topical corticosteroids. Paired exome sequencing showed a post-zygotic HRAS c.38G>T, p.G13V mutation. This variant has been reported as a somatic mutation in various types of cancer and as a germline mutation in Costello syndrome.12 Glycine 13 is one of the most mutated amino acid in keratinocytic epidermal nevus 13 There were no germline variants in CARD14.
Patient 2 presented with itchy linear and whorled psoriasiform plaques over her left chest and arm that were noticed since she was born and gradually became thicker (Figure 1c). At the age of 13 months she developed psoriasis vulgaris. A biopsy obtained from a linear plaque showed ILVEN features with psoriasiform epidermal hyperplasia and areas of alternating orthokeratosis and parakeratosis. Some of the keratinocytes showed reticular degeneration with balloon cell morphology (Figure 1d). Paired exome analysis revealed a post-zygotic KRT10 c.467G>A, p.R156H mutation, a frequent mutation in epidermolytic ichthyosis, and a germline CARD14 c. 2044C>T, p.R682W mutation which has been previously reported in psoriasis and pityriasis rubra pilaris (PRP).14,15
Patient 3 was included in previous publications of our group.16,17 She was born with extensive whorled-linear erythematous scaly plaques that progressed to involve the left side of her body. The plaques were pruritic with frequent secondary bacterial infections (Figure 1e). Initial review of her biopsy sections was consistent with ILVEN (Fig 1f), but later genetic analysis showed germline heterozygous PMVK c.79G>T p.E27X and somatic PMVK c.379C>T, p. Q127X mutations and a post-genetic analysis diagnosis of linear porokeratosis was made. Indeed, cornoid lamellae were noticed in serial sections of the skin biopsy that were not present in the initial sections. Based on the alteration of genes in the cholesterol biosynthesis pathway, the patient was treated with lovastatin/cholesterol cream with significant improvement.16
Patient 4 was born with small blaschkoid erythematous plaques over her left groin (Fig1g). Her daughter had similar lesions. Histopathological evaluation showed psoriasiform hyperplasia with hyperkeratosis and parakeratosis (Figure 1h). While somatic analysis showed no pathogenic mutations, germline genetic analysis revealed a novel heterozygous NSDHL mutation predicted to be damaging by functional prediction scores (Polyphen2 and SIFT), and absent in gnomAD leading to a diagnosis of CHILD syndrome.
Patient 5 was born with psoriasiform warty plaques in a blaschkoid distribution over the left side of her body. The blaschkoid psoriasiform plaque over her dorsal left hand and second finger ended in nail dystrophy (Figure 1i). She also had longitudinal leukonychia over her 2nd through 4th left fingernails. Histopathological evaluation showed parakeratosis, acanthosis and foamy macropahges within the papillary dermis (Figure 1j). Based on genetic analysis that revealed a novel heterozygous NSDHL mutation predicted to be damaging and absent in gnomAD, she was diagnosed with CHILD syndrome.
DISCUSSION
ILVEN is a rare mosaic disorder defined by clinical and histological criteria. Since first described by Altman and Mehregan,2 cases of ILVEN with various histological variants have been reported.3 Here we show that ILVEN is not a well-defined monogenic mosaic disorder but rather a heterogenous group of blaschko-linear inflammatory conditions. In our cohort, some nevi looked clinically and histopathologically as “ILVEN” but genetic analysis revealed a more precise diagnosis of CHILD nevus and linear porokeratosis. Others were epidermal nevi with inflamed clinical appearance presumably due to an inflammatory genetic background. The variability in mutations and genes that cause “ILVEN” lead to differences in clinical presentation and histopathology in different individuals.
Two patients who presented with ILVEN in early infancy developed psoriasis vulgaris later in life. Genetic analysis showed post-zygotic mutations in HRAS p.G13V and KRT10 p.R156H, both predicted to cause keratinocytic epidermal nevus (EN) and epidermolytic EN, respectively. Histopathology was compatible with ILVEN and there was no clue for epidermolytic hyperkeratosis in the latter. While the HRAS p.G13V ILVEN became itchy once generalized psoriasis appeared, the KRT10 p.R156H ILVEN was itchy before psoriatic plaques developed. The erythematous nature of these lesions, which clinically resembled ILVEN rather than other forms of epidermal nevi, could be explained by having a pro-inflammatory genetic background that modifies the clinical and histopathological phenotype. Indeed, the patient with the KRT10 ILVEN had a heterozygous CARD14 p.R682W variant that has previously been published as associated with sporadic psoriasis and pityriasis rubra pilaris.14,15
Although case 3 had a typical clinical appearance of ILVEN, meticulous histologic evaluation revealed a cornoid lamella that could easily be appreciated as alternating ortho- and parakeratosis. Finding a germline first-hit and a post-zygotic second hit in PMVK, a gene known to be mutated in porokeratosis rendered the diagnosis as linear porokeratosis. Cornoid lamellae are not pathognomonic for porokeratosis and have been found in ILVEN and in 1.2% of 167 epidermal nevi in studies that did not include genetic evaluation.3,18 Our case suggests that a fraction of individuals thought to have ILVEN may, in fact, have linear porokeratosis and emphasizes the importance of histopathologic evaluation of blaschkoid lesions. Genetic analysis revealing the biallelic PMVK mutations enabled a pathogenesis-based therapy with topical lovastatin/cholesterol, emphasizing the role of genetic evaluation in tailoring therapy for mosaic disorders.16
Patients 4 and 5 clinically showed ILVEN, but in fact had CHILD syndrome with minimal clinical change (so-called CHILD nevus). The histopathologic characteristics of CHILD nevus include verrucous acanthosis of the epidermis, hyperkeratosis with focal parakeratosis, and foamy xanthoma cells in the dermal papillae.19 The latter may be absent as in patient 4. Several cases of ILVEN have noted other affected females in the family and ILVEN lesions have been described with superimposed verruciform xanthoma, which is frequently seen in CHILD nevus, but these reports did not include genetic evaluation.20 Evaluating the cases retrospectively, the presence of an affected daughter in case 4, the intertriginous localization (ptychotropism) in both cases, and the foamy xanthoma cells in case 5 could have hinted at the diagnosis of CHILD nevus.
Taken together, the present study reveals the diversity of genetic causes underlying ILVEN, expands the phenotypic spectrum of known genes, and underscores the power of genetic analysis alongside clinical and histopathological evaluation in characterization and recategorization of rare skin diseases. Our findings also emphasize the power of genetic evaluation of mosaic disorders in tailoring specific therapeutic interventions.
Table 1:
clinical and genetic characteristics of ILVEN cases
| Subject # | Age at diagnosis |
Sex | Anatomic location | Psoriasis vulgaris | # of reads in blood |
# of reads in affected skin |
Germline mutation | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | Non ref. |
Ref. | Non ref. | |||||||
| 1 | 0 | F | Right foot and ankle | Yes | HRAS c.38G>T (p.G13V) | N/A | N/A | 30 | 10 | |
| 2 | 9 months | F | Left chest and arm | Yes | KRT10 c.467G>A (p.R156H) | 68 | 0 | 222 | 11 | CARD14 c.2044C>T (p.R682W) |
| 3 | 0 | F | Widespread blaschkoid plaques over the left side of the body | No | PMVK c.379C>T (p.Q127X) | 113 | 0 | 119 | 34 | PMVK c.79G>T (p.E27X) |
| 4 | 0 | F | Left groin | No | NSDHL c.1008C>G (p.H336Q) | |||||
| 5 | 0 | F | Left trunk, left upper and lower limbs | No | NSDHLc.148G>A (p.G50R) | |||||
N/A= not applicable; ref=reference
Key Message:
ILVEN is a heterogenous group of mosaic inflammatory disorders.
Acknowledgement
We thank the patients and their families for contributing to this paper.
The patients in this manuscript have given written informed consent to publication of their case details.
Funding Sources
This study was supported in part by the National Institutes of Health (R01 AR071491) to Dr. Choate and the Yale Center for Mendelian Genomics U54 HG006504). The funders had no role in design and conduct of the study, collection, management, analysis, and interpretation of the data, preparation, review, or approval of the manuscript, and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Morag C, Metzker A. Inflammatory linear verrucous epidermal nevus: report of seven new cases and review of the literature. Pediatr Dermatol. 1985;3(1):15–18. [DOI] [PubMed] [Google Scholar]
- 2.Altman J, Mehregan AH. Inflammatory linear verrucose epidermal nevus. Arch Dermatol. 1971;104(4):385–389. [PubMed] [Google Scholar]
- 3.Tiwary A, Mishra D. A unique porokeratotic variant of inflammatory linear verrucous epidermal nevus. Indian J Paediatr Dermatol. 2017;18(3):237–240. doi: 10.4103/2319-7250.206088 [DOI] [Google Scholar]
- 4.Umegaki-Arao N, Sasaki T, Fujita H, et al. Inflammatory Linear Verrucous Epidermal Nevus with a Postzygotic GJA1 Mutation Is a Mosaic Erythrokeratodermia Variabilis et Progressiva. J Invest Dermatol. 2017;137(4):967–970. doi: 10.1016/j.jid.2016.11.016 [DOI] [PubMed] [Google Scholar]
- 5.Riachi M, Polubothu S, Stadnik P, et al. Molecular Genetic Dissection of Inflammatory Linear Verrucous Epidermal Naevus Leads to Successful Targeted Therapy. J Invest Dermatol. Published online June 8, 2021. doi: 10.1016/j.jid.2021.02.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017:201178 (November). doi: 10.1101/201178 [DOI] [Google Scholar]
- 8.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VCV000180848.6 - ClinVar - NCBI. Accessed July 4, 2022. https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000180848.6
- 13.Luo S, Tsao H. Epidermal, Sebaceous, and Melanocytic Nevoid Proliferations Are Spectrums of Mosaic RASopathies. J Investig Dermatol. 2014;134(10):2493–2496. doi: 10.1038/jid.2014.244 [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Chung HJ, Ross N, et al. Analysis of CARD14 Polymorphisms in Pityriasis Rubra Pilaris: Activation of NF-κB. J Invest Dermatol. 2015;135(7):1905–1908. doi: 10.1038/jid.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan CT, Cao L, Roberson EDO, et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. Am J Hum Genet. 2012;90(5):796–808. doi: 10.1016/j.ajhg.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atzmony L, Lim YH, Hamilton C, et al. Topical cholesterol/lovastatin for the treatment of porokeratosis: A pathogenesis-directed therapy. J Am Acad Dermatol. 2020;82(1):123–131. doi: 10.1016/j.jaad.2019.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atzmony L, Khan HM, Lim YH, et al. Second-Hit, Postzygotic PMVK and MVD Mutations in Linear Porokeratosis. JAMA Dermatol. 2019;155(5):548–555. doi: 10.1001/jamadermatol.2019.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su WP. Histopathologic varieties of epidermal nevus. A study of 160 cases. Am J Dermatopathol. 1982;4(2):161–170. [DOI] [PubMed] [Google Scholar]
- 19.Gantner S, Rütten A, Requena L, Gassenmaier G, Landthaler M, Hafner C. CHILD syndrome with mild skin lesions: histopathologic clues for the diagnosis. J Cut Pathol. 2014;41(10):787–790. doi: 10.1111/cup.12377 [DOI] [PubMed] [Google Scholar]
- 20.Singh S, Khandpur S, Agarwal S. Verruciform xanthoma overlying inflammatory linear verrucous epidermal nevus and in broad segmental distribution. BMJ Case Rep. 2018;2018. doi: 10.1136/bcr-2018-225964 [DOI] [PMC free article] [PubMed] [Google Scholar]