Abstract
Background
The path to childhood asthma is thought to initiate in utero and be further promoted by postnatal exposures. However, the underlying mechanisms remain underexplored. We hypothesized that prenatal maternal immune dysfunction associated with increased childhood asthma risk (revealed by low IFN‐γ:IL‐13 secretion during the third trimester of pregnancy) alters neonatal immune training through epigenetic mechanisms and promotes early‐life airway colonization by asthmagenic microbiota.
Methods
We examined epigenetic, immunologic, and microbial features potentially related to maternal prenatal immunity (IFN‐γ:IL‐13 ratio) and childhood asthma in a birth cohort of mother–child dyads sampled pre‐, peri‐, and postnatally (N = 155). Epigenome‐wide DNA methylation and cytokine production were assessed in cord blood mononuclear cells (CBMC) by array profiling and ELISA, respectively. Nasopharyngeal microbiome composition was characterized at age 2–36 months by 16S rRNA sequencing.
Results
Maternal prenatal immune status related to methylome profiles in neonates born to non‐asthmatic mothers. A module of differentially methylated CpG sites enriched for microbe‐responsive elements was associated with childhood asthma. In vitro responsiveness to microbial products was impaired in CBMCs from neonates born to mothers with the lowest IFN‐γ:IL‐13 ratio, suggesting defective neonatal innate immunity in those who developed asthma during childhood. These infants exhibited a distinct pattern of upper airway microbiota development characterized by early‐life colonization by Haemophilus that transitioned to a Moraxella‐dominated microbiota by age 36 months.
Conclusions
Maternal prenatal immune status shapes asthma development in her child by altering the epigenome and trained innate immunity at birth, and is associated with pathologic upper airway microbial colonization in early life.
Keywords: childhood asthma, DNA methylation, maternal prenatal immunity, nasal microbiome, trained innate immunity
Through an integrated, inter‐generational approach, we characterized maternal and early‐life epigenetic, immunologic and microbial features that influence asthma development during childhood. We demonstrate that, in children of non‐asthmatic mothers, maternal prenatal immune status (IFN‐γ:IL‐13) shapes the child's path to asthma by altering the epigenome and trained innate immunity in the neonate, and promoting pathologic upper airway microbial colonization in early life. These results provide a novel framework for mechanistic research linking maternal prenatal health to childhood asthma pathogenesis. Abbreviations: CBMC, cord blood mononuclear cells; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cells; rRNA, ribosomal RNA

Abbreviations
- CBMC
cord blood mononuclear cells
- CI
confidence interval
- ConA
concanavalin A
- DMC
differentially methylated CpG site
- FDR
false discovery rate
- IIS
Infant Immune Study
- IFN
interferon
- IL
interleukin
- LPS
lipopolysaccharide
- OR
odds ratio
- PBMC
peripheral blood mononuclear cells
- PC
principal coordinate
- PMA
phorbol 12‐myristate 13‐acetate
- SV
sequence variant
- TLR
Toll‐like receptor
- TRQ
turquoise
- WGCNA
weighted gene co‐expression network analysis
1. INTRODUCTION
Although asthma is the most prevalent chronic disease of childhood, 1 our understanding of its inception mechanisms remains limited. Maternal prenatal traits and exposures [asthma, 2 smoking, 3 , 4 diet, 5 antibiotic use, 6 infections 7 , 8 ] as well as mode of delivery 9 and postnatal diet 10 influence asthma susceptibility in children, suggesting that the path to the disease initiates in utero and progresses under the influence of postnatal exposures. 1 , 11 , 12 , 13 , 14
The role of maternal prenatal immune status in shaping this path was recently highlighted by our work in the Infant Immune Study (IIS), a birth cohort of mother–child dyads sampled pre‐, peri‐, and postnatally over a decade. 15 A decreased ratio of interferon (IFN)‐γ to interleukin (IL)‐13 (IFN‐γ:IL‐13) secretion by mitogen‐stimulated peripheral blood mononuclear cells (PBMC) isolated during the third trimester of pregnancy was associated with increased prevalence of childhood asthma. 16 This relation was limited to pregnancy, suggesting this period may provide a temporal window for intrinsic cellular events involved in childhood asthma inception. Moreover, the association between childhood asthma risk and maternal prenatal IFN‐γ:IL‐13 was specific to nonasthmatic mothers, 16 pointing to a distinct, inter‐generational path to asthma development in this group.
Because children born to non‐asthmatic mothers account for the largest absolute number of asthmatic children in the population at large, 16 mechanisms that lead to disease development in this under‐studied group require elucidation. Our epigenome‐wide study in a subset of IIS neonates revealed distinct DNA methylation profiles in cord blood mononuclear cells (CBMC) of children who did or did not become asthmatic by age 9 years, 17 suggesting that perinatal epigenetic regulation of immunity acts as a gatekeeper for the asthma trajectory from birth to childhood. 18 Early life, and particularly the period between birth and age 3 years, represents a critical period of immune functional development and microbiota establishment. 19 , 20 , 21 Cross‐sectional and longitudinal studies have linked early‐life gut and airway microbiota perturbations and altered microbial developmental trajectories over the first three years of life with increased risk of asthma and atopy in childhood. 22 , 23 , 24 , 25 In infancy, colonization of the upper airways by microbiota dominated by putatively pathogenic genera (Moraxella, Streptococcus, or Haemophilus) increased risk of lower airway infection and development of asthma in later childhood. 26 , 27
Here, we integrated these heretofore independent lines of research and tested the hypothesis that the relationship between maternal prenatal immune dysfunction (revealed by decreased IFN‐γ:IL‐13) and childhood asthma development reflects altered epigenetic regulation of neonatal innate immune responsiveness to microbial stimuli which, we propose, results in early‐life airway colonization by asthmagenic microbiota.
2. METHODS
2.1. Study population
The IIS, a birth cohort of 482 mother–child dyads, was designed to identify immune maturation patterns in early life and their impact on asthma risk, as well as maternal influences shaping these patterns. Pregnant women were recruited between 1996 and 2004 during a prenatal visit. Any healthy newborn was eligible for IIS provided that her/his mother spoke English and planned to use a local, participating pediatrician. Subjects were only excluded from the study if the mother was diagnosed with an immune deficiency disorder or had plans to leave Tucson. 15 Parents completed a respiratory health history questionnaire at enrollment. Enrolled children were monitored for respiratory and immune phenotypes associated with asthma and allergies within the first decade of life. This study included all mother–child dyads for whom maternal prenatal IFN‐γ:IL‐13 was measured, maternal and child (2–9 years) asthma status was known, and CBMC DNA samples were available (N = 182). Nasopharyngeal swabs were longitudinally collected from children at 2–36 months of age and stored at −80°C in fetal calf serum until DNA was extracted. 16S rRNA amplicon sequencing was performed on 149 nasal samples from 73 participants, 43 of whom provided samples at more than one time point (Figure S1). Informed consent or assent was obtained from the parents of all participants and the study was approved by the University of Arizona Institutional Review Board.
Further details about maternal and child characteristics are provided as online supporting information.
2.2. DNA methylation studies
CBMC DNA was isolated with the DNeasy Blood & Tissue kit (Qiagen), quantified using Qubit, and provided (~1.6 μg) to Illumina (San Diego, CA) for epigenome‐wide DNA methylation profiling on the EPIC platform, which surveys 853,307 CpG sites. 28 Analysis was limited to 722,677 sites/probes after excluding probes that map to sex chromosomes, overlap loci with common SNPs (minor allele frequency ≥0.05), have been previously identified as cross‐reactive, 29 or provided signals indistinguishable from background (detection p‐value > .01 in ≥75% of samples). Methylation data were processed using the minfi R package, 30 and raw probe intensities were corrected for background and color imbalance by control‐probe normalization. Infinium type I and II probe bias was corrected using the SWAN algorithm, 31 signals were quantile‐normalized, and methylation data (M‐values) were extracted.
Proportions of CBMC populations (CD4+ and CD8+ T cells, B cells, NK cells, and monocytes) were estimated using the minfi package. 32 Principal component analysis was performed to identify chip effects and potential confounding variables (minfi‐estimated cell proportions, gestational age, mode of delivery, child ethnicity, and child sex). The ComBat function [sva 33 ] was used to adjust for chip effects while preserving the effects of maternal immune status and childhood asthma. Estimated cell proportions, mode of delivery, and child sex remained significantly associated with at least one of the first ten principal components and were selected as covariates. Two additional covariates were identified by latent factor analysis [CorrConf 34 ] and included in subsequent analyses. Linear regression (limma) was used to test for associations between maternal prenatal IFN‐γ:IL‐13 and neonatal DNA methylation at individual CpG sites (M‐values). Analyses were performed in R (v3.4.4).
2.3. Weighted gene co‐expression network analysis (WGCNA)
Networks of differentially methylated CpG sites (DMCs) in CBMCs were constructed by hierarchical clustering through WGCNA. 35 , 36 , 37 , 38 Linear regression was used to adjust M‐values for confounding variables (minfi‐estimated cell proportions, child sex, mode of delivery, and latent factors) identified in the differential methylation analysis. The power threshold was set at 2 to meet scale‐free topology, and co‐methylated networks (modules) were constructed using the unsigned network algorithm. Branch cutting was performed using a “deepSplit” value of 2 with “pamStage” turned off. Minimum module size was set to 5 CpGs. All other settings were at default values. 35 Methylation data for each module were summarized using the eigengene vector, that is, the first principal component of the module. Spearman correlation was used to test for associations between eigengene vectors and traits of interest. The association between Turquoise (TRQ) module DMCs and asthma was further analyzed using linear regression (limma).
Additional details about DMC annotation and bioinformatic analyses, 16S rRNA sequencing, and other statistical approaches are provided as online supporting information.
3. RESULTS
3.1. Maternal prenatal immune status relates to the neonatal immune methylome and childhood asthma risk
We initially examined IIS mother–child dyads with complete data on maternal prenatal IFN‐γ:IL‐13, maternal and child (age 2–9 years) asthma status, and CBMC DNA for methylation analyses (N = 182; Figure S1). Consistent with our original findings, 16 this population exhibited a robust inverse association between maternal prenatal IFN‐γ:IL‐13 and childhood asthma risk (OR 0.2, 95% CI 0.07–0.7, p = .008), which became even stronger when analyses were limited to nonasthmatic mothers and their children (OR 0.1, 95% CI 0.03–0.4, p = .001; p = .025 for the interaction between maternal immune status and maternal asthma, Figure S2A). Therefore, all subsequent studies focused on nonasthmatic mother–child dyads (N = 155). Table S1 shows the characteristics of study participants. We next compared childhood asthma prevalence across quartiles of maternal prenatal IFN‐γ:IL‐13 (Figure S2B). While a significant trend was found across the quartiles (trend chi‐square p = .001, 1 degree of freedom), this trend was driven by the relationship between Q1 and asthma: logistic regression analysis revealed no significant differences in asthma risk among Q2, Q3, and Q4—only between Q1 and the other three quartiles (Table S2). Children of mothers with the lowest prenatal IFN‐γ:IL‐13 (Q1) were significantly more likely to develop asthma during childhood compared to all other quartiles combined (Q2‐4; OR 5.3, 95% CI 2.0–13.8, p = .0008, Figure S2B). Therefore, all subsequent analyses compared children born to Q1 mothers (N = 39) with children born to Q2‐4 mothers (N = 116).
To assess how prenatal maternal immunity influences the path to childhood asthma, we initially determined whether maternal prenatal IFN‐γ:IL‐13 related to the epigenetic profiles of paired fresh‐frozen, untreated CBMCs. An epigenome‐wide comparison of DNA methylation in neonates born to mothers with low (Q1) or high (Q2‐4) prenatal IFN‐γ:IL‐13 identified 2316 DMCs (FDR <0.05: Figure 1A and Table S3). Interestingly, top hits among CpG sites hypermethylated in at‐risk infants born to mothers with low (Q1) IFN‐γ:IL‐13 mapped to genes involved in innate immune responses to microbes. ZFYVE9/SARA is a TGF‐β pathway member 39 that regulates macrophage responses to intracellular pathogenic microbes 40 ; ANXA5 binds directly to LPS, dampening cytokine responses to gram‐negative bacteria 41 ; CCR1, a chemokine receptor, is involved in leukocyte migration to sites of inflammation and infection. 42 These findings provided the first hint that maternal prenatal IFN‐γ:IL‐13 and the epigenetic landscape at birth influence neonatal responses to microbes.
FIGURE 1.
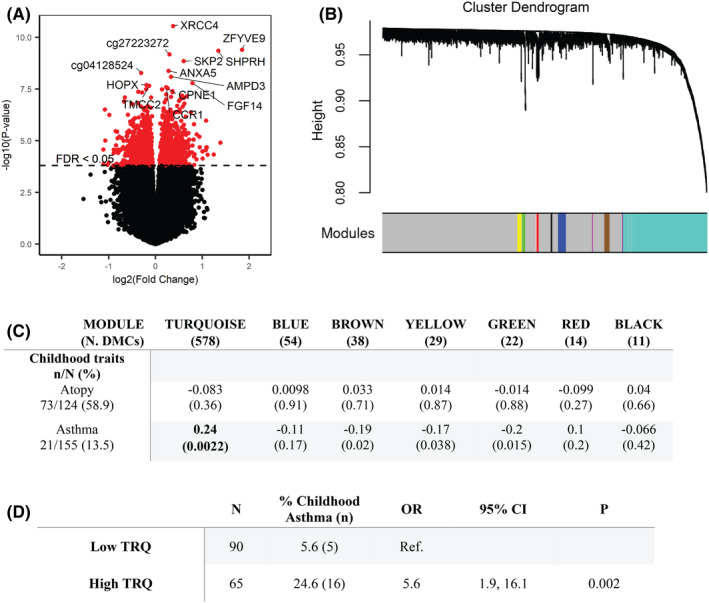
Maternal prenatal IFN‐γ:IL‐13 relates to differences in the CBMC methylome. (A) Differentially methylated CBMC CpGs associated with Q1 versus Q2‐4. The x‐ and y‐axes represent the log2(fold change) and significance, respectively, for differential methylation at each CpG. The dashed line marks the threshold of significance [false discovery rate (FDR) < 0.05]. The most significant DMCs are annotated with their gene symbol (or CpG identifier). (B) Modules identified by hierarchical clustering of maternal prenatal IFN‐γ:IL‐13 Q1 vs. Q2‐4‐associated DMCs (n = 2316) using WGCNA. Distinct modules are denoted by colors represented in the bar below the dendrogram. Gray represents DMCs that did not cluster into a module. (C) Relationships between maternal prenatal IFN‐γ:IL‐13–associated methylation modules at birth and asthma‐related phenotypes in childhood. The table shows Spearman correlation coefficients and p‐values for relationships between module eigengene vectors and asthma‐associated clinical traits. Only modules with >10 CpGs are shown. Bolded associations remained significant after Bonferroni correction (p = .05/14 = .0036). (D) Asthma risk among children with high or low TRQ module eigenvalues at birth (divided at “0”), as determined by logistic regression.
Next, we performed WGCNA to determine which of the 2316 DMCs that distinguish neonates born to mothers with low (Q1) or high (Q2‐4) IFN‐γ:IL‐13 were associated with childhood asthma and/or atopy at age 5–9 years. Ten co‐methylation modules were identified (Figure 1B), and key features (eigengene vectors representing the first principal component of each module) were used to assess module/phenotype relationships. The largest module, Turquoise (TRQ, 578 DMCs: Table S4) showed no association with atopy but correlated significantly with childhood asthma (Spearman's ρ = 0.24, p = .0022, Figure 1C). High TRQ module methylation at birth significantly increased childhood asthma risk (OR 5.6, 95% CI 1.9–16.1, p = .002, Figure 1D), suggesting that maternal prenatal immune status shapes the path to childhood asthma through epigenetic mechanisms.
3.2. Immune training at birth influences asthma risk during childhood
Because TRQ module methylation in CBMCs was associated with childhood asthma (Figure 1C,D), exploring the functional potential of DMCs in this module might reveal early‐life mechanisms of childhood asthma development. Indeed, we found that 410 of the 578 TRQ module DMCs mapped to genomic elements with regulatory potential (promoters or enhancers). Of these regulatory DMCs, 116 were differentially methylated (p < .05) in neonates who did or did not develop asthma by age 9 years. As many as 59% of these asthma‐associated regulatory DMCs resided in elements known to modify their chromatin configuration in responses to stimulation by microbes or their products 43 , 44 , 45 , 46 , 47 , 48 , 49 (Figure 2, Tables S5 and S6).
FIGURE 2.
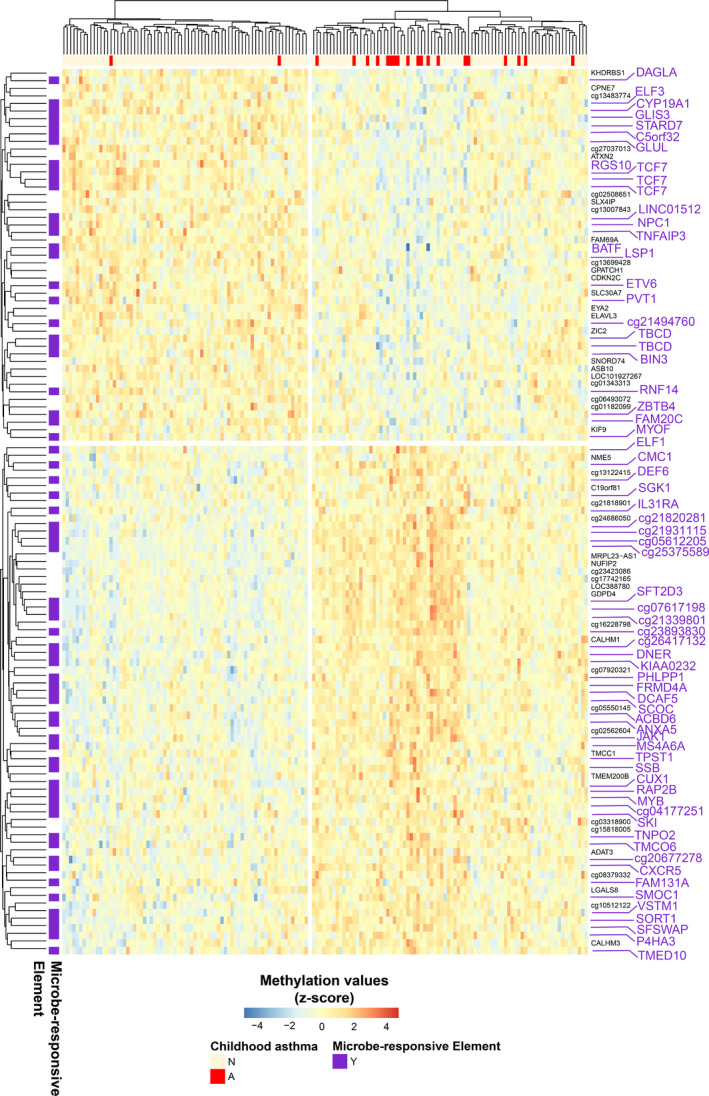
Asthma‐associated differential methylation in the TRQ module co‐localizes with microbe‐responsive elements. Heatmap of TRQ module DMCs (rows) that mapped to promoters/enhancers (as assessed by Roadmap Epigenomics consortium chromatin state annotations) and were significantly (p < .05) associated with asthma. The color bar at the top denotes individual neonates (columns) who did or did not develop asthma during childhood. Microbe‐responsive elements (purple text in the right side legend) were defined as DMCs mapping to genes exhibiting dynamic expression, or putative enhancers exhibiting dynamic changes in chromatin marks (DNaseI/ATAC accessibility, H3K27ac, H3K4me1) in response to microbes or their products. 43 , 44 , 45 , 46 , 47 , 48 , 49 Statistics and annotations for these DMCs are provided in Table S5.
Combined with our initial analyses of neonatal differential methylation associated with maternal prenatal IFN‐γ:IL‐13 (Figure 1A), these findings led us to hypothesize that maternal prenatal immune status leverages epigenetic mechanisms to train neonatal innate immunity to microbes and susceptibility to asthma. To test this hypothesis, we explored the relationship between maternal prenatal IFN‐γ:IL‐13 and concentrations of innate or adaptive cytokines secreted by CBMCs stimulated with LPS or mitogens (Concanavalin A, ConA/phorbol 12‐myristate 13‐acetate, PMA). A significant positive association (Spearman's ρ = 0.23, p = .009) was found between maternal prenatal IFN‐γ:IL‐13 and neonatal LPS‐induced IL‐6 production. TNF secretion also exhibited a trending positive relationship. In contrast, no other cytokine responses, including those triggered by mitogens, were significantly associated with maternal IFN‐γ:IL‐13 after correction for multiple testing (Table 1). LPS‐induced IL‐6 production likely derived from monocytes because it was strongly and positively correlated with the estimated proportion of these cells (Spearman's ρ = 0.30, p = 3.6e−04), but not with other CBMC cell types. These relations point to decreased myeloid cell responsiveness to microbial stimuli —a reflection of inefficient innate immune training 50 —in neonates born to mothers with prenatal immune dysfunction.
TABLE 1.
Maternal prenatal IFN‐γ:IL‐13 is related to LPS‐ but not ConA/PMA‐stimulated cytokine production by CBMC
| LPS | ConA/PMA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IL‐1β | IL‐6 | TNF | IL‐12 | IL‐10 | IFN‐γ | IL‐4 | IL‐5 | IL‐13 | IL‐10 | |
| Spearman's ρ | −0.067 | 0.225 | 0.210 | 0.092 | 0.149 | −0.036 | −0.046 | 0.031 | −0.015 | −0.055 |
| N | 133 | 133 | 133 | 133 | 133 | 149 | 149 | 149 | 149 | 149 |
| p | .447 | .009 | .015 | .294 | .088 | .665 | .581 | .709 | .852 | .509 |
Note: Cytokine levels were measured by ELISA in supernatants of LPS‐ or ConA/PMA‐stimulated CBMC cultures. Data for each cytokine were z‐scored prior to testing for associations, and maternal prenatal IFN‐γ:IL‐13 was coded as a dichotomous variable (0 = Q1 and 1 = Q2‐4). Bolded p‐values remained significant after Bonferroni correction (p = .05/5 = .01).
We next took a stepwise approach to analyze the relationship between neonatal DNA methylation, neonatal LPS‐induced IL‐6 production, and childhood asthma. We detected a significant inverse correlation between TRQ module methylation (i.e., TRQ module eigengene vector) and LPS‐induced IL‐6 responses at birth (Spearman's ρ = −0.34, p = 6e−05). We then found that neonates who developed asthma during childhood clustered in a group characterized by low LPS‐induced IL‐6 production and high TRQ module methylation at birth (Figure 3A). Notably, TRQ methylation levels did not affect asthma prevalence in neonates with high IL‐6 production but had a profound effect on asthma prevalence among neonates with low IL‐6 (p for interaction = .12, Figure 3B). Indeed, only neonates with high TRQ and low IL‐6 values had substantially higher risk for asthma (OR 7.8, 95% CI 2.0–30.0, p = .003, Figure 3C). In combination, these data suggest that low maternal prenatal IFN‐γ:IL‐13 enhances the neonate's risk of asthma by altering epigenetic CBMC programming and impairing innate immune responsiveness to microbial signals.
FIGURE 3.
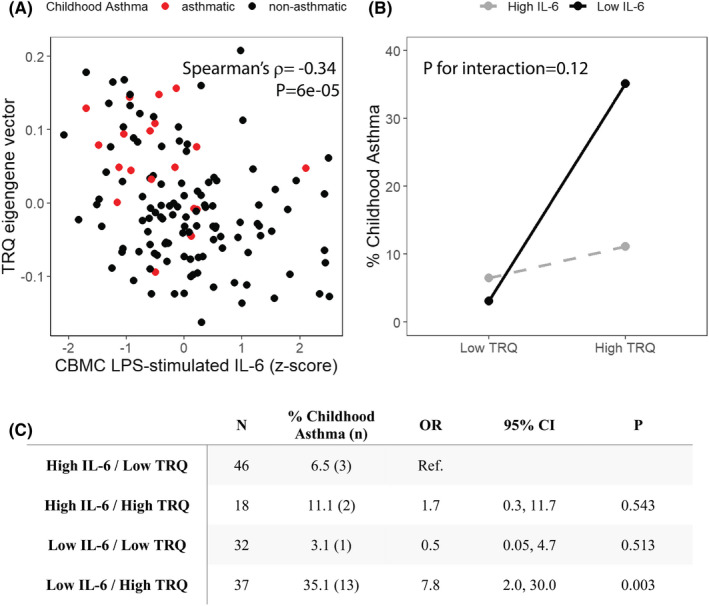
CBMC TRQ module and LPS‐stimulated IL‐6 production co‐associate with childhood asthma development. (A) CBMC TRQ eigengene values and IL‐6 responses to LPS stimulation at birth are significantly correlated. Red and black dots represent neonates with or without a diagnosis of asthma during childhood, respectively. (B) Interaction between TRQ module methylation (eigengene value), LPS‐induced IL‐6 responses at birth, and prevalence of asthma during childhood. p for interaction was assessed using logistic regression. (C) The risk of childhood asthma is related to CBMC TRQ methylation status and IL‐6 responses. Odds ratios were calculated by logistic regression. For panels B and C, TRQ eigengene vector and z‐scored LPS‐stimulated IL‐6 production were each categorized into “High” and “Low” values, divided at zero.
3.3. Prenatal immune training relates to early‐life nasal microbiota development and asthma
Because we found that infants at high‐risk for asthma exhibited impaired capacity to respond to bacterial products at birth, we hypothesized that this impairment favors the development of a pathogenic airway microbiota enriched for asthmagenic bacteria. To test this hypothesis, we assessed repeated nasopharyngeal microbiota profiles (N = 149) collected over the first three years of life from a subset of IIS children (N = 73). Though smaller than our study cohort, associations between maternal prenatal immunity, CBMC methylome and immune function, and childhood asthma development identified in the entire dataset (N = 155) replicated in this smaller population (Table S7). To examine relationships between early‐life nasal microbiota development and maternal prenatal or neonatal features associated with asthma development, we deconstructed upper airway microbiota compositional trajectories over the first three years of life into their first three principal coordinates (PC1‐3, explaining 30.1% of variance). Consistent with previous findings, 51 , 52 the dominant bacterial genus present was significantly related to the first principal coordinate (Weighted UniFrac; Linear Mixed Effects; PC1; p < .001; Table S8), explaining the largest proportion of microbiota variance over time. Maternal prenatal IFN‐γ:IL‐13 quartiles, TRQ module methylation, CBMC IL‐6 responses to LPS, and childhood asthma also explained variance in microbiota development on PC2 and PC3 during this period (Table S8). This suggests that prenatal maternal immune status and early‐life innate immune training and function relate to features of upper airway microbiota development over the first three years of life. To identify these specific microbiological features, we next assessed airway microbiota development in children born to mothers in the highest (Q1) or lower (Q2‐4) risk groups who exhibited significantly distinct trajectories of upper airway microbiota development on PC3 (LME interaction ANOVA, p = .01; Figure 4A). Variance on PC3 was primarily driven by Haemophilus, Dolosigranulum, and Moraxella (Figure 4B); the upper airways of children born to Q1 mothers were colonized in very early life (~first 12 months) by Haemophilus and in later childhood (by 36 months of age) by Moraxella, suggesting that this pattern of upper airway colonization is linked to both prenatal immune status and childhood asthma development. Samples from children who received antibiotics or asthma medications within 1 week of sample collection (n = 4) were not distinct with respect to nasal microbiome diversity or composition. In addition, these relationships were not driven by the children who developed asthma before the 36‐month sample collection (n = 6). Removing these participants did not reduce the significance of the interactions we observed.
FIGURE 4.
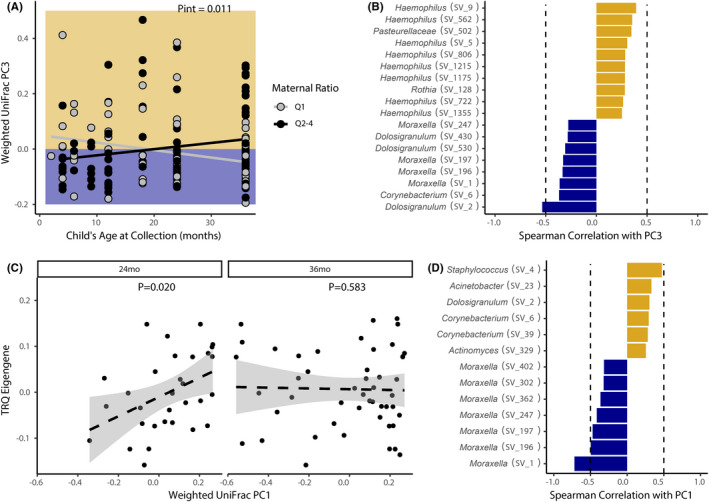
Early‐life upper airway microbiota development is influenced by maternal IFN‐γ:IL‐13 and asthma‐associated DNA methylation in the TRQ module. (A) Upper airway microbiota development over the first three years of life relates to maternal IFN‐γ:IL‐13 status. Lines indicate the slopes from linear mixed effects models. Background represents directionality from panel B. (B) Variance in principal coordinate (PC) 3 is driven by Haemophilus (Spearman's ρ > 0.25), and Dolosigranulum and Moraxella (ρ < −0.25; dotted lines represent ρ of 0.5 or − 0.5, respectively). (C) Weighted UniFrac PC1 correlates with the TRQ eigengene at 24 months, but not 36 months of age (line represents linear model and standard error). (D) Weighted UniFrac PC1 is primarily driven by differences in Moraxella decreases (ρ < −0.25) and Staphylococcus and Corynebacterium (ρ > 0.25) abundances.
Next, to determine duration of effect of neonatal immune training on microbiota development, we examined relationships between the TRQ methylation module and nasal microbiota composition at 24 and 36 months. TRQ module methylation was significantly associated with variance in nasal microbiota composition at 24 months (N = 31) but not at 36 months (N = 48) of age (Linear Model; PC1; p = .020 and p = .583, respectively; Figure 4C), suggesting that CBMC epigenetic modifications may exert the greatest influence on microbiota development over the first 2 years of life. Bacteria whose abundance correlated with PC1 at 24 months included those previously found to dominate the upper airways in early life and relate to childhood asthma susceptibility (Moraxella) or protection (Corynebacterium; Figure 4D). 27 , 53 This suggested that hypo‐ or hyper‐methylation in specific CpGs at birth associates with the presence of these bacteria in the upper airways at 24 months of age. To further deconstruct these relationships, we examined correlations between bacteria on PC1 and asthma‐associated DMCs within the TRQ module (identified in Figure 2 and Table S5). Multiple Moraxella were negatively correlated with hyper‐methylation at genes involved in vesicle‐mediated transport (CUX1, SFT2D3), lipid binding (ACBD6), and regulation of myeloid cell effector function (VSTM1; Figure S3). In contrast, genera including Corynebacterium correlated positively with some of these genes (e.g., SFT2D3 and ACBD6), suggesting that methylation at these sites influences the presence of bacterial species associated with development of, or protection against, childhood asthma. Collectively, these data suggest that differences in neonatal immune training associated with maternal prenatal immune status and immune responsiveness to microbes in very early life promote distinct microbiota developmental trajectories that relate to asthma development in childhood.
4. DISCUSSION
Childhood asthma is a complex disease characterized by distinct endotypes 54 and a multifactorial pathogenesis. Previous work examined the role of individual factors—pre‐ or postnatal, genetic, epigenetic, immunologic, microbial—in the development of the disease. In contrast, we took an integrated, inter‐generational approach and sought to characterize maternal and early‐life epigenetic, immunologic, and microbial features that influence childhood asthma development to identify tractable targets and temporal windows for disease‐preventing interventions. To this end we relied on a unique birth cohort of mother–child dyads carefully monitored pre‐, peri‐, and postnatally for a decade. In this cohort, maternal prenatal immune status, specifically IFNγ:IL‐13, predicts asthma during childhood. 16 Here, we demonstrate that the prenatal immune status of nonasthmatic mothers influences their newborns' epigenetic landscape and innate immune responsiveness to microbes, and the subsequent development of their upper airway microbiome in early life. Each of these components independently and interdependently related to childhood asthma development.
The selective association between neonatal methylation and childhood asthma but not atopy points to a distinct epigenetic path to asthma that is independent of allergy—a notion supported by our previous epidemiological work. 16 As importantly, the epigenetic network associated with asthma development in at‐risk neonates born to mothers with low IFN‐γ:IL‐13 comprised over 100 differentially methylated CpG sites, more than half of which mapped to microbe‐responsive elements. Consistent with a role of these epigenetic modifications in innate immune function, monocyte‐derived IL‐6 and TNF production in those neonates was reduced upon stimulation with bacterial LPS. The implications of these findings are novel and profound. Innate cytokine responses to microbial pro‐inflammatory triggers are a hallmark of trained immunity, a de facto innate immune memory that enhances subsequent myeloid cell responsiveness following an initial infectious or sterile pro‐inflammatory challenge. 50 , 55 Unlike adaptive immune memory, which depends on de novo rearrangements of antigen receptor genes in lymphocytes, trained immunity relies on long‐term functional reprogramming of innate myeloid cells (primarily monocytes and dendritic cells) through epigenetic mechanisms. 50 , 55 Post‐translational histone modifications in trained immune cells have been well characterized, 56 but DNA methylation patterns also discriminate individuals who do or do not develop trained immunity following a primary innate stimulation. 57 , 58 Our results further support the role of DNA methylation in trained immunity, providing targets for future mechanistic analyses. Most importantly, our data provide the first explicit, population‐based link between maternal prenatal immunity, trained immunity at birth and childhood asthma by identifying a network of epigenetically modified genes that associate with decreased innate immune responsiveness to microbes, asthmagenic upper airway bacterial colonization during the critical first three years of life, and increased risk of asthma during childhood. Because innate cytokine responses to microbes were measured at birth, our findings also imply that exposures influencing neonatal innate training and asthma risk occurred in utero. Indeed, recent evidence indicates that fetal innate immunity can be trained by microbial infections 59 , 60 or maternal vaccination during pregnancy, 61 implicating maternal prenatal microbiome‐immune interactions in these processes.
The mechanisms underpinning in utero training of fetal innate immunity remain unclear. While direct movement of maternal microbes to the fetal intestine can occur and modulate immune cell function, 62 , 63 data from mice 64 , 65 , 66 and humans 67 also suggest that metabolites released by maternal prenatal microbiota are sensed by fetal bone marrow progenitors with long‐lasting effects on the progeny's immune function. To the extent that delayed immune maturation in early life promotes type‐2 responses and contributes to asthma and allergy risk, 1 , 63 effective immune training in utero may avert this risk by enabling rapid responses to maturation‐inducing microbial signals. Asthma protection in children raised in traditional, microbe‐rich farm environments 68 provides a case in point. CBMCs from these children demonstrated prenatal priming evidenced by upregulation of microbial‐sensing TLRs 68 and innate cytokines, 69 reiterating that maternal prenatal microbial exposures calibrate the development and maturation of immune responses in the offspring both quantitatively and temporally, ultimately enhancing asthma resistance. 70 , 71 , 72 , 73
Reduced capacity to respond to microbes in the postnatal period undoubtedly influences microbial colonization trajectories, though this had not been previously investigated in human populations. Indeed, children born to mothers with lower IFN‐γ:IL‐13 exhibited microbial successional states in the upper airways dominated by Haemophilus in early infancy, and Moraxella in early childhood. Several independent studies have linked respiratory microbiota dominated by these genera in early life with increased risk of acute respiratory infection and febrile illness in the lower respiratory tract, 26 and with asthma development and susceptibility to pulmonary exacerbations later in childhood. 26 , 52 , 74 Species within both genera, specifically nontypeable Haemophilus influenzae and Moraxella catarrhalis, chronically colonize the upper airways and can survive in host cells. 75 , 76 , 77 , 78 , 79 Hence, inter‐generational transmission of compromised host immunity to intracellular microbial pathogens appears as a keystone of asthma development in children of nonasthmatic mothers.
Although novel and potentially significant, our results should be interpreted with caution. The size of the IIS population was limited. Therefore, despite the suggestive temporal architecture of our data that span the prenatal, perinatal, and early‐life periods, performing formal statistical mediation analyses would have been unwise. Moreover, the unique sample collection schedule in the IIS population prevented direct replication of our findings in other cohorts. Thus, the conceptual framework we propose needs to be tested in future studies, some of which are already ongoing. 73 Moreover, samples appropriate for chromatin and gene expression analyses were not collected, and thus histone and/or transcriptional signatures of trained immunity related to asthma status later in life 56 could not be surveyed in neonatal immune cells. These limitations were partially mitigated by the availability of epigenome‐wide DNA methylation data and extensive information about cytokine protein secretion in response to diverse signals. The biological underpinnings of maternal prenatal IFN‐γ:IL‐13 also remain unclear, even though this study further highlighted its persistent, trans‐generational impact. Finally, sample availability dictated that microbiome composition in early life could be studied in the children's upper airways but not in their gut, and only in a subset of subjects. Therefore, despite their potential implications, these analyses should be considered purely exploratory.
Despite these limitations, our findings appear to suggest that maternal prenatal immune status shapes the child's path to asthma by altering the epigenome and trained innate immunity in the neonate, and then promoting pathologic upper airway microbial colonization in early life. This scenario provides a novel, robust framework for further mechanistic research linking maternal prenatal health to childhood asthma pathogenesis.
AUTHOR CONTRIBUTIONS
Donata Vercelli and Susan V. Lynch designed and oversaw the study, data generation, analyses, and interpretation. Avery DeVries performed DNA methylation studies and statistical analyses with Debra A. Stern. Douglas Fadrosh generated 16S rRNA data. Kei E. Fujimura and Kathryn McCauley performed microbiota analyses. Avery DeVries, Kathryn McCauley, Debra A. Stern, Susan V. Lynch, and Donata Vercelli wrote and revised the final manuscript.
FUNDING INFORMATION
This work was funded by the National Institutes of Health (R21AI133765 to DV and SVL; P01AI148104 to SVL and DV; R21AI144722 to DV; U19AI125357 to DV; T32 ES007091 to ADV). Support was also provided by The BIO5 Institute (to ADV). The Infant Immune Study was funded by the National Institutes of Health (R01AI042268)
CONFLICT OF INTEREST
ADV, KM, DAS, KEF, DF, and DV declare no competing interests. SVL is a co‐founder, board member and consultant of Siolta Therapeutics Inc., and holds stock in this company, she also consults for Solarea Bio.
Supporting information
Appendix S1
Table S1‐S8
ACKNOWLEDGMENTS
The authors thank Anne L. Wright, PhD and Marilyn Halonen, PhD for their advice and comments about this manuscript. They also acknowledge Kole Lynch for his initial work on this project.
DeVries A, McCauley K, Fadrosh D, et al. Maternal prenatal immunity, neonatal trained immunity, and early airway microbiota shape childhood asthma development. Allergy. 2022;77:3617‐3628. doi: 10.1111/all.15442
Avery DeVries, Kathryn McCauley, Debra A. Stern, Susan V. Lynch, and Donata Vercelli contributed equally to this article.
Contributor Information
Susan V. Lynch, Email: susan.lynch@ucsf.edu.
Donata Vercelli, Email: donata@arizona.edu.
REFERENCES
- 1. Martinez FD, Vercelli D. Asthma. Lancet. 2013;382(9901):1360‐1372. doi: 10.1016/s0140-6736(13)61536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lim RH, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta‐analysis. PLoS One. 2010;5(4):e10134. doi: 10.1371/journal.pone.0010134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6(8):660‐662. doi: 10.1513/pats.200907-065DP [DOI] [PubMed] [Google Scholar]
- 4. Neuman A, Hohmann C, Orsini N, et al. Maternal smoking in pregnancy and asthma in preschool children: a pooled analysis of eight birth cohorts. Am J Respir Crit Care Med. 2012;186(10):1037‐1043. doi: 10.1164/rccm.201203-0501OC [DOI] [PubMed] [Google Scholar]
- 5. Chatzi L, Torrent M, Romieu I, et al. Mediterranean diet in pregnancy is protective for wheeze and atopy in childhood. Thorax. 2008;63(6):507‐513. doi: 10.1136/thx.2007.081745 [DOI] [PubMed] [Google Scholar]
- 6. Stensballe LG, Simonsen J, Jensen SM, Bonnelykke K, Bisgaard H. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. Research Support, Non‐U.S. Gov't. J Pediatr. 2013;162(4):832‐838.e3. doi: 10.1016/j.jpeds.2012.09.049 [DOI] [PubMed] [Google Scholar]
- 7. Calvani M, Alessandri C, Sopo SM, et al. Infectious and uterus related complications during pregnancy and development of atopic and nonatopic asthma in children. Allergy. 2004;59(1):99‐106. [DOI] [PubMed] [Google Scholar]
- 8. Xu B, Pekkanen J, Jarvelin MR, Olsen P, Hartikainen AL. Maternal infections in pregnancy and the development of asthma among offspring. Int J Epidemiol. 1999;28(4):723‐727. [DOI] [PubMed] [Google Scholar]
- 9. Stokholm J, Thorsen J, Blaser MJ, et al. Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci Transl Med. 2020;12(569):eaax9929. doi: 10.1126/scitranslmed.aax9929 [DOI] [PubMed] [Google Scholar]
- 10. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222‐227. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kindlund K, Thomsen SF, Stensballe LG, et al. Birth weight and risk of asthma in 3‐9‐year‐old twins: exploring the fetal origins hypothesis. Thorax. 2010;65(2):146‐149. doi: 10.1136/thx.2009.117101 [DOI] [PubMed] [Google Scholar]
- 12. Martino D, Prescott S. Epigenetics and prenatal influences on asthma and allergic airways disease. Research Support, Non‐U S Gov't Review. Chest. 2011;139(3):640‐647. [DOI] [PubMed] [Google Scholar]
- 13. Ober C, Vercelli D. Gene‐environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27(3):107‐115. doi: 10.1016/j.tig.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prescott SL. Early origins of allergic disease: a review of processes and influences during early immune development. Curr Opin Allergy Clin Immunol. 2003;3(2):125‐132. doi: 10.1097/00130832-200304000-00006 [DOI] [PubMed] [Google Scholar]
- 15. Su Y, Rothers J, Stern DA, Halonen M, Wright AL. Relation of early antibiotic use to childhood asthma: confounding by indication? Clin Exp Allergy. 2010;40(8):1222‐1229. doi: 10.1111/j.1365-2222.2010.03539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rothers J, Stern DA, Lohman IC, et al. Maternal cytokine profiles during pregnancy predict asthma in children of mothers without asthma. Am J Respir Cell Mol Biol. 2018;59(5):592‐600. doi: 10.1165/rcmb.2017-0410OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeVries A, Wlasiuk G, Miller SJ, et al. Epigenome‐wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J Allergy Clin Immunol. 2017;140(2):534‐542. doi: 10.1016/j.jaci.2016.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeVries A, Vercelli D. The neonatal methylome as a gatekeeper in the trajectory to childhood asthma. Epigenomics. 2017;9(4):585‐593. doi: 10.2217/epi-2016-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539‐544. doi: 10.1126/science.aad9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1(4):367‐382. doi: 10.3920/bm2010.0027 [DOI] [PubMed] [Google Scholar]
- 21. Jain N. The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut Microbes. 2020;12(1):1824564. doi: 10.1080/19490976.2020.1824564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arrieta MC, Arevalo A, Stiemsma L, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 2018;142(2):424‐434.e10. doi: 10.1016/j.jaci.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- 24. Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187‐1191. doi: 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durack J, Kimes NE, Lin DL, et al. Delayed gut microbiota development in high‐risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun. 2018;9(1):707. doi: 10.1038/s41467-018-03157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704‐715. doi: 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teo SM, Tang HHF, Mok D, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe. 2018;24(3):341‐352.e5. doi: 10.1016/j.chom.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389‐399. doi: 10.2217/epi.15.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole‐genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aryee MJ, Jaffe AE, Corrada‐Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363‐1369. doi: 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maksimovic J, Gordon L, Oshlack A. SWAN: Subset‐quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11(5):1‐9. doi: 10.1080/15592294.2016.1161875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118‐127. doi: 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 34. McKennan C, Nicolae D. Accounting for unobserved covariates with varying degrees of estimability in high‐dimensional biological data. Biometrika. 2019;106(4):823‐840. doi: 10.1093/biomet/asz037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morin A, McKennan CG, Pedersen CT, et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J Allergy Clin Immunol. 2020;146(6):1358‐1366. doi: 10.1016/j.jaci.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nicodemus‐Johnson J, Myers RA, Sakabe NJ, et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight. 2016;1(20):e90151. doi: 10.1172/jci.insight.90151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicodemus‐Johnson J, Naughton KA, Sudi J, et al. Genome‐Wide Methylation Study Identifies an IL‐13‐induced Epigenetic Signature in Asthmatic Airways. Am J Respir Crit Care Med. 2016;193(4):376‐385. doi: 10.1164/rccm.201506-1243OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang WB, Ling GH, Sun L, Liu FY. Smad anchor for receptor activation (SARA) in TGF‐beta signaling. Front Biosci (Elite Ed). 2010;2:857‐860. doi: 10.2741/e147 [DOI] [PubMed] [Google Scholar]
- 40. Reed SG. TGF‐beta in infections and infectious diseases. Microbes Infect. 1999;1(15):1313‐1325. doi: 10.1016/s1286-4579(99)00252-x [DOI] [PubMed] [Google Scholar]
- 41. Rand JH, Wu X‐X, Lin EY, Griffel A, Gialanella P, McKitrick JC. Annexin A5 binds to lipopolysaccharide and reduces its endotoxin activity. mBio. 2012;3(2):e00292‐11. doi: 10.1128/mBio.00292-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7(5):a016303. doi: 10.1101/cshperspect.a016303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cirovic B, de Bree LCJ, Groh L, et al. BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe. 2020;28(2):322‐334.e5. doi: 10.1016/j.chom.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fong ON, Chan KY, Leung KT, et al. Expression profile of cord blood neutrophils and dysregulation of HSPA1A and OLR1 upon challenge by bacterial peptidoglycan. J Leukoc Biol. 2014;95(1):169‐178. doi: 10.1189/jlb.0413219 [DOI] [PubMed] [Google Scholar]
- 45. Jesus Iglesias M, Reilly S‐J, Emanuelsson O, et al. Combined chromatin and expression analysis reveals specific regulatory mechanisms within cytokine genes in the macrophage early immune response. PLoS One. 2012;7(2):e32306. doi: 10.1371/journal.pone.0032306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kariuki SN, Blischak JD, Nakagome S, Witonsky DB, Di Rienzo A. Patterns of transcriptional response to 1,25‐dihydroxyvitamin D3 and bacterial lipopolysaccharide in primary human monocytes. G3 (Bethesda). 2016;6(5):1345‐1355. doi: 10.1534/g3.116.028712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luan L, Patil NK, Guo Y, et al. Comparative transcriptome profiles of human blood in response to the toll‐like receptor 4 ligands lipopolysaccharide and monophosphoryl lipid A. Sci Rep. 2017;7(1):40050. doi: 10.1038/srep40050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Novakovic B, Habibi E, Wang SY, et al. β‐glucan reverses the epigenetic state of LPS‐induced immunological tolerance. Cell. 2016;167(5):1354‐1368.e14. doi: 10.1016/j.cell.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saeed S, Quintin J, Kerstens HHD, et al. Epigenetic programming of monocyte‐to‐macrophage differentiation and trained innate immunity. Science (New York, NY). 2014;345(6204):1251086. doi: 10.1126/science.1251086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Netea MG, Domínguez‐Andrés J, Barreiro LB, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375‐388. doi: 10.1038/s41577-020-0285-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Durack J, Huang YJ, Nariya S, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. 2018;6(1):104. doi: 10.1186/s40168-018-0487-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCauley K, Durack J, Valladares R, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. 2019;144(5):1187‐1197. doi: 10.1016/j.jaci.2019.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang HHF, Lang A, Teo SM, et al. Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J Allergy Clin Immunol. 2021;147(5):1683‐1691. doi: 10.1016/j.jaci.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Conrad LA, Cabana MD, Rastogi D. Defining pediatric asthma: phenotypes to endotypes and beyond. Pediatr Res. 2021;90(1):45‐51. doi: 10.1038/s41390-020-01231-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Netea MG, Joosten LAB, Latz E, et al. Trained immunity: a program of innate immune memory in health and disease. Science (New York, NY). 2016;352(6284):aaf1098. doi: 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fanucchi S, Dominguez‐Andres J, Joosten LAB, Netea MG, Mhlanga MM. The Intersection of Epigenetics and Metabolism in Trained Immunity. Immunity. 2021;54(1):32‐43. doi: 10.1016/j.immuni.2020.10.011 [DOI] [PubMed] [Google Scholar]
- 57. Das J, Verma D, Gustafsson M, Lerm M. Identification of DNA methylation patterns predisposing for an efficient response to BCG vaccination in healthy BCG‐naive subjects. Epigenetics. 2019;14(6):589‐601. doi: 10.1080/15592294.2019.1603963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verma D, Parasa VR, Raffetseder J, et al. Anti‐mycobacterial activity correlates with altered DNA methylation pattern in immune cells from BCG‐vaccinated subjects. Sci Rep. 2017;7(1):12305. doi: 10.1038/s41598-017-12110-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Apostol AC, Jensen KDC, Beaudin AE. Training the fetal immune system through maternal inflammation‐A layered hygiene hypothesis. Front Immunol. 2020;11:123. doi: 10.3389/fimmu.2020.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katzmarski N, Domínguez‐Andrés J, Cirovic B, et al. Transmission of trained immunity and heterologous resistance to infections across generations. Nat Immunol. 2021;22(11):1382‐1390. doi: 10.1038/s41590-021-01052-7 [DOI] [PubMed] [Google Scholar]
- 61. Mawa PA, Webb EL, Filali‐Mouhim A, et al. Maternal BCG scar is associated with increased infant proinflammatory immune responses. Vaccine. 2017;35(2):273‐282. doi: 10.1016/j.vaccine.2016.11.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Halkias J, Rackaityte E, Hillman SL, et al. CD161 contributes to prenatal immune suppression of IFNgamma‐producing PLZF+ T cells. J Clin Invest. 2019;129(9):3562‐3577. doi: 10.1172/JCI125957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rackaityte E, Halkias J, Fukui EM, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. 2020;26(4):599‐607. doi: 10.1038/s41591-020-0761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thorburn AN, McKenzie CI, Shen S, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320 [DOI] [PubMed] [Google Scholar]
- 65. Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159‐166. doi: 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 66. Kimura I, Miyamoto J, Ohue‐Kitano R, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367(6481):eaaw. doi: 10.1126/science.aaw8429 [DOI] [PubMed] [Google Scholar]
- 67. van Dijk SJ, Zhou J, Peters TJ, et al. Effect of prenatal DHA supplementation on the infant epigenome: results from a randomized controlled trial. Clin Epigenetics. 2016;8:114. doi: 10.1186/s13148-016-0281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ege MJ, Bieli C, Frei R, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school‐age children. J Allergy Clin Immunol. 2006;117(4):817‐823. doi: 10.1016/j.jaci.2005.12.1307 [DOI] [PubMed] [Google Scholar]
- 69. Pfefferle PI, Buchele G, Blumer N, et al. Cord blood cytokines are modulated by maternal farming activities and consumption of farm dairy products during pregnancy: the PASTURE Study. J Allergy Clin Immunol. 2010;125(1):108‐115.e1‐3. doi: 10.1016/j.jaci.2009.09.019 [DOI] [PubMed] [Google Scholar]
- 70. Ober C, Sperling AI, von Mutius E, Vercelli D. Immune development and environment: lessons from Amish and Hutterite children. Curr Opin Immunol. 2017;48:51‐60. doi: 10.1016/j.coi.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. von Mutius E, Smits HH. Primary prevention of asthma: from risk and protective factors to targeted strategies for prevention. Lancet. 2020;396(10254):854‐866. doi: 10.1016/S0140-6736(20)31861-4 [DOI] [PubMed] [Google Scholar]
- 72. Holt PG, Strickland DH, Custovic A. Targeting maternal immune function during pregnancy for asthma prevention in offspring: Harnessing the “farm effect”? J Allergy Clin Immunol. 2020;146(2):270‐272. doi: 10.1016/j.jaci.2020.04.008 [DOI] [PubMed] [Google Scholar]
- 73. Lynch SV, Vercelli D. Microbiota, epigenetics, and trained immunity. convergent drivers and mediators of the asthma trajectory from pregnancy to childhood. Am J Respir Crit Care Med. 2021;203(7):802‐808. doi: 10.1164/rccm.202010-3779PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou Y, Jackson D, Bacharier LB, et al. The upper‐airway microbiota and loss of asthma control among asthmatic children. Nat Commun. 2019;10(1):5714. doi: 10.1038/s41467-019-13698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morey P, Cano V, Martí‐Lliteras P, et al. Evidence for a non‐replicative intracellular stage of nontypable Haemophilus influenzae in epithelial cells. Microbiology (Reading). 2011;157(Pt 1):234‐250. doi: 10.1099/mic.0.040451-0 [DOI] [PubMed] [Google Scholar]
- 76. Craig JE, Nobbs A, High NJ. The extracytoplasmic sigma factor, final sigma(E), is required for intracellular survival of nontypeable Haemophilus influenzae in J774 macrophages. Infect Immun. 2002;70(2):708‐715. doi: 10.1128/iai.70.2.708-715.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Slevogt H, Seybold J, Tiwari KN, et al. Moraxella catarrhalis is internalized in respiratory epithelial cells by a trigger‐like mechanism and initiates a TLR2‐ and partly NOD1‐dependent inflammatory immune response. Cell Microbiol. 2007;9(3):694‐707. doi: 10.1111/j.1462-5822.2006.00821.x [DOI] [PubMed] [Google Scholar]
- 78. Spaniol V, Heiniger N, Troller R, Aebi C. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis . Microbes Infect. 2008;10(1):3‐11. doi: 10.1016/j.micinf.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 79. de Vries SP, Bootsma HJ, Hays JP, Hermans PW. Molecular aspects of Moraxella catarrhalis pathogenesis. Microbiol Mol Biol Rev. 2009;73(3):389‐406. doi: 10.1128/mmbr.00007-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Table S1‐S8


