Abstract
Human accelerated regions (HARs) are the fastest-evolving sequences in the human genome. When HARs were discovered in 2006, their function was mysterious due to scant annotation of the noncoding genome. Diverse technologies, from transgenic animals to machine learning, have consistently shown that HARs function as gene regulatory enhancers with significant enrichment in neurodevelopment. It is now possible to quantitatively measure the enhancer activity of thousands of HARs in parallel and model how each nucleotide contributes to gene expression. These strategies have revealed that many human HAR sequences function differently than their chimpanzee orthologs, though individual nucleotide changes in the same HAR may have opposite effects, consistent with compensatory substitutions. To fully evaluate the role of HARs in human evolution, it will be necessary to experimentally and computationally dissect them across more cell types and developmental stages.
Keywords: human accelerated region, enhancer, reporter assay, epigenetics, machine learning, evolution
INTRODUCTION
Comparative genomics has identified thousands of human accelerated regions (HARs), evolutionarily conserved sequences with an unexpected number of nucleotide changes on the human lineage (reviewed in 2, 20, 28, 44, 59). This intriguing signature suggests a functional change unique to humans (Figure 1), making HARs exciting sequences for understanding the basis of human-specific traits and diseases (19, 51). But when HARs were first described in 2006, we lacked the tools and data necessary to decode their ancestral function, let alone to predict how human substitutions altered function. Most HARs lie outside protein-coding genes in what was once called junk DNA due to limited functional annotations. Today, each HAR is decorated with dozens of genomic experiments and computational predictions—too much data for manual interpretation of every HAR.
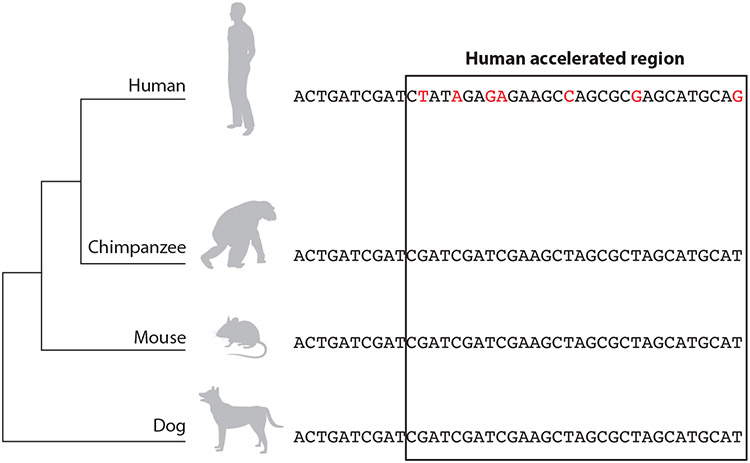
Figure 1.
Human accelerated regions have acquired many nucleotide substitutions (red) in the human genome since their divergence from the common ancestor with chimpanzees, but they are highly conserved in other vertebrates. This sequence signature suggests a constrained function during vertebrate evolution that was lost or changed in humans.
In this review, we describe how the initial hypothesis that most HARs function as developmental enhancers has gained support through a series of technological advances, including epigenetic profiling, massively parallel reporter assays (MPRAs), and machine learning. We integrate recently published data and identify those HARs most likely to function as enhancers in the most studied context, brain development, as well as in other tissues. Our analysis of the literature also pinpoints specific variants with the strongest evidence for altering HAR enhancer activity during human evolution. Equipped with deep learning models and genome editing tools, researchers can now dissect each HAR at the single-nucleotide level to understand its role in human biology.
THE DEVELOPMENTAL ENHANCER HYPOTHESIS
When HARs were first described, it was surprising that nearly all of them fell outside protein-coding exons. We expected the fastest-evolving regions of the human genome to be in genes, even given that mammalian genomes were found to be ~98% noncoding, because the importance of genes was well understood. After it was established that HARs are mostly noncoding, later studies designed to expand upon the initial set of HARs filtered out coding regions or analyzed them separately with models that account for codon evolution (3, 6, 24, 38, 39, 47, 54, 56). Initially, the primary evidence that noncoding HARs were functionally important was their extreme sequence conservation up until the human–chimpanzee ancestor, despite lying in genomic regions with normal mutation rates, which indicates strong negative selection (35, 54). Motivated by King & Wilson’s (36) 1975 discovery that human and chimpanzee blood proteins harbor very few amino acid differences, researchers hypothesized that this conserved function was gene regulation.
To explore the idea that HARs are enhancers and to decipher what pathways they might regulate, the first strategy connected HARs to nearby genes and made guilt-by-association inferences based on the roles of these genes. This leveraged scientists’ much better understanding of proteins compared to regulatory elements at that time. The analyses showed a clear pattern that has held up over the years: HARs are significantly enriched near genes involved in transcription, cell adhesion, development, and disease, with a tissue bias toward activity in the brain (7, 10, 24, 54, 56, 73). This pattern suggested that sequence changes in HARs during human evolution could have altered the expression of important genes that themselves regulate gene networks, potentially explaining anatomical and physiological features unique to our species. But the only evidence supporting this hypothesis was genomic proximity. More data were needed.
The following sections are organized around a series of technologies used to generate these data over the course of the past fifteen years. We include experimental strategies as well as analytical methods for integrating data to test the HAR enhancer hypothesis.
TRANSGENIC ANIMALS: INDIVIDUAL HUMAN ACCELERATED REGIONS FUNCTION AS ENHANCERS IN VIVO
Researchers noted early on that reporter assays in transgenic mice and fish could be used to characterize expression patterns driven by individual HARs (51). This approach continues to be important because it can capture spatiotemporal enhancer activity in whole animals (7, 62). Limitations include cost, throughput, studying primate enhancers in nonprimates, and generating qualitative data.
Integrating results across studies, we find that 74 HARs have been tested with transgenic reporter assays at specific developmental stages in mice and zebrafish (Supplemental Table 1). Activity was observed in at least one tissue for 50 HARs (68%), with 19 being active brain enhancers (71). Notable examples of HAR enhancers characterized in transgenic animals include 2 in introns of AUTS2, which is associated with autism and other neurological disorders (53), and 11 in the NPAS3 locus, which is associated with neurodevelopment, epilepsy, and schizophrenia (7, 33). Thus, transgenics have confirmed that HARs regulate important developmental genes in vivo.
Of the in vivo validated HAR enhancers, 27 have been assayed using both the human and the chimpanzee sequence. Qualitative expression differences between the 2 alleles were shown in 9 (32%) of them (Table 1). Examples that have been further linked to specific genes and phenotypes include HAR2/HACNS1, a Gbx2 enhancer in chondrogenic mesenchyme during limb development (17); 2xHAR.20, an EN1 enhancer in keratinocytes influencing eccrine sweat gland density (1); 2xHAR.238, a Gli2 enhancer in testis Leydig cells influencing male typical behavior (52); and HARE5, an Fzd8 enhancer in neural progenitor cells influencing cell cycle acceleration and brain size (4). These represent the HARs that have been most closely related to human-specific traits.
Table 1.
Human accelerated regions (HARs) where the human and chimpanzee sequences are differentially active in transient transgenic reporter assays
| HARs | Active in mice? | Active in fish? | Active in either? | Human–chimp differences? |
Reference(s) |
|---|---|---|---|---|---|
| HAR2/2xHAR.3/HACNS1 | Yes | NT | Yes | Yes | 57, 69 |
| 2xHAR.20 | Yes | NT | Yes | Yes | 1, 7 |
| 2xHAR.114 | Yes | NT | Yes | Yes | 7 |
| 2xHAR.142 | Yes | Yes | Yes | Yes | 33, 34 |
| 2xHAR.164 | Yes | NT | Yes | Yes | 7 |
| 2xHAR.170 | Yes | NT | Yes | Yes | 7 |
| HAR202 | NT | Yes | Yes | Yes | 7 |
| 2xHAR.238 | Yes | NT | Yes | Yes | 7, 53, 69 |
| HARE5/ANC516 | Yes | NT | Yes | Yes | 4 |
Abbreviation: NT, not tested.
INDUCED PLURIPOTENT STEM CELLS: NONHUMAN PRIMATE DATA
Chimpanzee cells are important for understanding how HARs might have functioned in the human–chimpanzee common ancestor and throughout human evolution. But sampling and research use of tissues from chimpanzees and other apes across the life span is largely forbidden. This means that all of the initial studies of HAR function were performed using mice, fish, and human cell lines. Induced pluripotent stem cell (iPSC) technology (65) changed this by allowing pluripotent cells to be generated from chimpanzee fibroblasts and lymphoblasts, which can be acquired without invasive procedures and are commercially available. HAR researchers quickly adopted this strategy and demonstrated that chimpanzee iPSCs could be reprogrammed into neural progenitors, cardiomyocytes, neural crest cells, and other previously inaccessible cell types (reviewed in 59). As described in the next section, this platform has been used for comparative epigenetic profiling of various cell types from humans and chimpanzees (57, 71). iPSC-derived cells are also employed to directly test HAR enhancer function with MPRAs (70, 71), including in chimpanzee neuronal cells (71), and they could be leveraged for genome editing experiments.
EPIGENETIC AND EXPRESSION PROFILES: MOST HUMAN ACCELERATED REGIONS ARE IN ACTIVE CHROMATIN
The advent of methods to probe the biochemical activity of genome sequences via sequencing was a boon for understanding HAR enhancer function. These techniques include chromatin immunoprecipitation sequencing (ChIP-seq) for binding of transcription factors and modified histones, open chromatin assays [e.g., DNase I hypersensitive sites sequencing (DNase-seq), assay for transposase-accessible chromatin using sequencing (ATAC-seq)], and transcription measurements [e.g., RNA sequencing (RNA-seq), cap analysis of gene expression (CAGE)]. The first use of functional genomics to predict the function of a HAR was when Sanger sequencing of cloned complementary DNAs, called expressed sequence tags, led to the discovery that HAR1 is a long noncoding RNA (55).
As compendia of epigenetic profiles for different human tissues and cell types grew, HARs without annotation became the exception rather than the rule. Today, a typical HAR overlaps dozens of epigenetic marks (Figure 2). Studies consistently have shown that HARs are enriched with marks of active enhancers, such as DNase hypersensitive sites, transcription factor and histone ChIP-seq peaks, and enhancer RNA (22, 42, 57, 67). HARs are particularly enriched in brain data sets, concordant with their genomic proximity to neurodevelopmental genes (7, 22). Leveraging the tissue-specific nature of functional genomics data, researchers further observed that the epigenetic profiles of HARs correlate with expression and functional annotations of nearby genes, providing a link to specific pathways and tissues regulated by individual HARs (57, 60, 67). Evidence for such links grew further with the introduction of chromatin conformation capture data, which have been used to measure three-dimensional proximity of HARs and gene promoters (4, 71, 72).
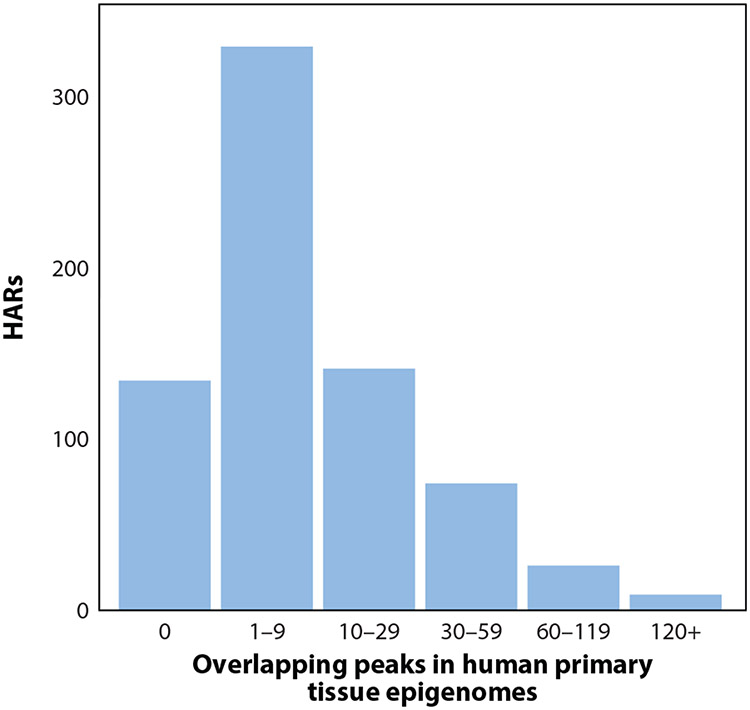
Figure 2.
Human accelerated regions (HARs) are marked with dozens of epigenetic features. This histogram shows the number of epigenetic marks overlapping HARs (71). Less than 20% of HARs (134/713) overlap no peaks, and the top 10% of HARs overlap more than 40 peaks each. This analysis focuses on 5% irreproducible discovery rate peak calls from primary tissues. Including peaks from cell lines and/or less conservative peak calls would increase the number of overlaps.
In addition to functionally annotating HARs, epigenetic data have been used to study the evolution of human gene regulation in two other ways. First, candidate regulatory elements can be generated from human data and subsequently analyzed for human variants and positive selection (15, 23, 31, 32). A substantial minority of the resulting elements overlap previously identified HARs, but many new fast-evolving enhancers have been discovered with this strategy. Similar to HARs, they are enriched for activity in neuronal tissues and cell lines (15). A second related approach is to generate functional genomics data from tissues or cell lines derived from chimpanzees, monkeys, and/or mice and compare these to human data in order to identify human-gained and human-lost enhancers (or promoters) (11, 57, 60, 67). Researchers found that some of these are diverged in sequence, similar to HARs, but many are not. Compared to HARs, they also tend to be less conserved across species (66). Thus, epigenetics-first strategies complement the approach that has been used with HARs, where identifying acceleration precedes assessing enhancer potential.
MACHINE LEARNING: MODELS CAN DECODE HUMAN ACCELERATED REGION ENHANCER FUNCTION
Spurred by the rapid growth of functional genomics data a decade ago, researchers began applying machine learning models to assess the enhancer potential of HARs in different cell types and tissues. HARs may overlap enhancer annotations derived from unsupervised learning, such as genome segmentations (46), or they can be scored for enhancer-like properties with supervised learning models. Such models encode rules about how sequence and/or epigenetic features relate to enhancer activity measured, for example, by transgenic experiments or other reporter assays (18). In an early study implementing both of these strategies (7), segmentations labeled nearly two-thirds of HARs as enhancers, whereas supervised learning trained on the VISTA Enhancer Browser database of developmental enhancer experiments (68) predicted about one-third of HARs to be enhancers. Each method uses different algorithms, cell types, developmental time points, and thresholds to call enhancers, as well as different gold standards for enhancers themselves (e.g., epigenetic signature versus in vivo reporter activity). A strength of both strategies is that dozens or even hundreds of data sets are integrated into tissue-specific enhancer predictions, making them more accurate than using individual epigenetic data sets.
A related approach is to build machine learning models that predict enhancer-associated epigenetic marks from DNA sequence alone (21). There has been an explosion of deep learning approaches to this problem, many of which make tissue-specific predictions (9, 45, 50, 58, 69). Similar to other enhancer prediction approaches, these models can be used to score HARs, in this case based on having enhancer-like sequences. Because the only input is sequence, these models also can be utilized to predict the effects of sequence variants on enhancer activity. This strategy was recently used to identify variants in human-gained enhancers that have large effects on embryonic neocortical enhancer predictions, providing a potential mechanism to explain epigenetic marks present in human but not in macaque samples (45). This work illustrates the ability of deep learning to dissect the sequence basis for lineage-specific enhancers at single-nucleotide resolution.
To extend this approach to HARs, we scored all Single Nucleotide Polymorphism Database (dbSNP) variants (37) that overlapped a HAR with the Sei model (9). We found variants that alter HAR enhancer activity predictions consistently across tissues, as well as some with tissue-specific effects (Figure 3a). As expected for evolutionarily conserved sequences, HARs harbor many human polymorphisms that are predicted to increase or decrease enhancer activity to a greater degree than known disease mutations (Figure 3b). Most HAR variants also disrupt binding sites of tissue-specific transcription factors and/or chromatin loop anchors (Figure 3c). We envision extending this methodology to quantify the effects of human–chimpanzee fixed differences in HARs. Such an analysis would perform the equivalent of millions of reporter assays on the computer in just a few hours, prioritizing specific HAR variants and variant combinations for experimental characterization.
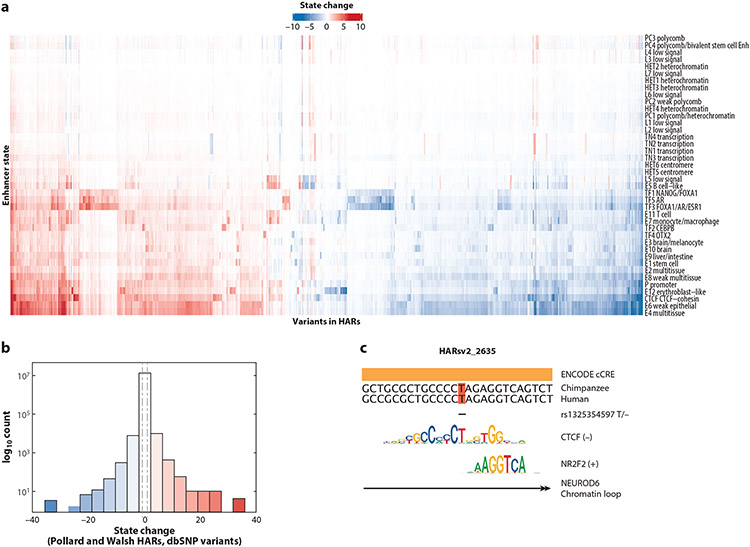
Figure 3.
Deep learning analysis of human variants in human accelerated regions (HARs). All single-nucleotide polymorphisms (SNPs) included on the SNP Database (dbSNP) that overlap with a HAR were scored for their effects on tissue-specific enhancer state predictions using the model Sei (9). This analysis includes all SNPs in all HARs tested in three massively parallel reporter assay (MPRA) studies (22, 66, 71). (a) Increases (red) and decreases (blue) in predicted enhancer state (rows) for all SNPs (columns). (b) Distribution of effects in panel a. Many SNPs in HARs have effect sizes greater than those of known human disease variants [vertical dashed lines represent the median of all SNPs in the Human Gene Mutation Database (HGMD), as reported in Reference 9]. (c) Example of a SNP (rs1325354597) in HARsv2_2635 (22) where the minor allele is predicted to substantially decrease the brain enhancer state and the CTCF state. This variant overlaps an annotated candidate regulatory element (ENCODE cCRE) and motifs of CTCF and NR2F2 as well as other neurological transcription factors (8). The SNP deletes an important nucleotide (T) in the CTCF motif. Consistent with CTCF’s role in loop extrusion, this genomic element has a significant chromatin loop with the promoter of the transcription factor NEUROD6 in cells carrying the major allele (63).
MASSIVELY PARALLEL REPORTER ASSAYS: QUANTIFYING HUMAN ACCELERATED REGION ENHANCER ACTIVITY EN MASSE
Another technology that has vastly increased the throughput of HAR functional characterization is MPRAs (29). In MPRA experiments, thousands of reporter constructs, each with a unique barcode and candidate enhancer, are assayed together in cell lines via RNA-seq (Figure 4). Constructs may be plasmids or integrated into the genome with Lentivirus. DNA sequencing enables normalization of RNA read counts by the abundance of each construct, producing a measure of enhancer activity that is more quantitative than reporter gene staining in transgenic animals but lacks spatiotemporal information due to being performed in vitro. MPRAs have been used in three independent studies to compare human and chimpanzee HAR sequences (22, 66, 71). In several cases, HARs that were prioritized based on MPRA activity have led to identification of gene regulatory differences between humans and nonhuman primates [e.g., PPP1R17 and cell cycle regulation in neural progenitor cells (22)]. MPRAs have also been used to investigate introgressed Neanderthal variants (30), modern human-specific variants (70), human-gained enhancers (66), and autism-associated variants in HARs (13).
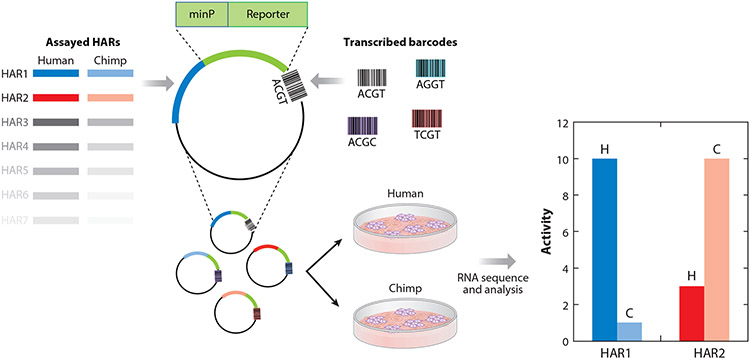
Figure 4.
Comparing enhancer activity between human and chimpanzee human accelerated region (HAR) sequences. Massively parallel reporter assay studies involve cloning HAR sequences into reporter vectors along with barcodes that uniquely identify each tested sequence. These vectors are inserted into cell lines, such as neural progenitor cells, using molecular tools such as lentiviruses. They randomly insert into the cell line’s genome. HAR enhancer activity is measured with RNA sequencing of the transcribed barcodes. By associating each tested sequence with many barcodes, activity can be averaged across genomic integration points, providing a robust measurement.
Similarities and Differences Between Massively Parallel Reporter Assay Studies
Motivated by the enrichment of HARs in neurodevelopmental loci, all of these studies used neuronal cells, which in several cases were derived from iPSCs. Therefore, we do not yet have a comprehensive understanding of HAR enhancer activity in other cell types and developmental stages, but we can now evaluate the consistency of findings across neurodevelopmental studies. This is important because MPRAs are challenging experiments in neuronal cells. Consequently, replicate concordance can be fairly low within studies (Supplemental Figure 1). Furthermore, these MPRA studies used different subsets of HARs, sequence variants, vectors, delivery strategies, cell lines, analysis tools, and statistical thresholds (Table 2).
Table 2.
Comparison of human accelerated region (HAR) massively parallel reporter assay (MPRA) studies
| Study | MPRA type |
Human cells |
Mouse cells |
Primate cells |
Statistical methods |
HARs | HGEs | Variants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active | DE | Total | Active | DE | Total | Active | DE | Total | ||||||
| Doan (14) | Plasmid | SH-SY5Y | Neuro2A | NT | t-test | 43% | NA | 335 | NT | NT | NT | NA | 35% | 343 rare ASD:WT |
| Girskis (23) | Plasmid | SH-SY5Y | Neuro2A | NT | t-test | 49% | 61% (H:C) | 3,132 | NT | NT | NT | NT | NT | NT |
| Weiss (72) | 5′ Lentivirus | ESC, NPC, osteoblasts | NT | NT | MPRAnalyze | NT | NT | NT | NT | NT | NT | 13% | 3% | 14,042 modern: archaic human |
| Uebbing (68) | Plasmid | NSC | NT | NT | t-test | 12% | 28% (H:C) | 1,363 | 34% | 35% | 3,027 | NA | NA | 32,776 human: nonhuman primate |
| Jagoda (31) | Plasmid | K562 | NT | NT | DESeq2 | NT | NT | NT | NT | NT | NT | 48% | 6% | 5,353 Neanderthal introgressed: not |
| Ryu (64) | 3′ Lentivirus | HS1, NPC, HS1, GPC, WTC, NPC, WTC, GPC | NT | Pt2A NPC, Pt2A GPC, Pt5C NPC, Pt5C GPC | limma | 41% | 22% (H:C) | 714 | NT | NT | NT | 28% | NA | 736 human: chimp for 7 HARs |
Abbreviations: HAR, human accelerated region; MPRA, massively parallel reporter assays; NA, not available; NT, not tested.
Consistently Active Human Accelerated Region Enhancers
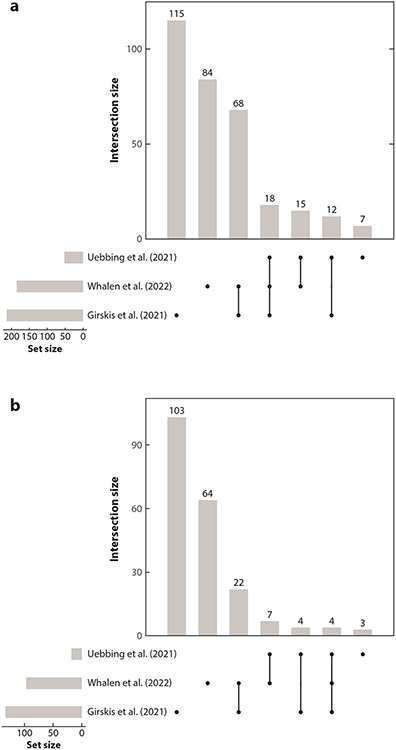
Given the heterogeneity of MPRA approaches, it is not surprising that agreement between studies regarding which HARs are neurodevelopmental enhancers is moderate (Supplemental Table 2). Nonetheless, out of 441 HARs tested in the 3 MPRA studies that compared human and chimpanzee alleles (22, 66, 71), we identified 113 that are active in at least 2 studies and 18 that are active in 3 (Figure 5a). These can be regarded as high-confidence HAR enhancers and a lower bound on how many HARs regulate neurodevelopment. Supporting this idea, the 2 HARs active in 3 studies and also tested in transgenic embryos (2xHAR.114, 2xHAR.548) were both active in vivo (Supplemental Table 1).
Figure 5.
Massively parallel reporter assay (MPRA) studies converge on some of the same active and differentially active human accelerated regions (HARs). The 441 HARs that have been tested in three MPRA studies were compared for consistency of results [Uebbing et al. (66), Girskis et al. (22), Whalen et al. (71)]. (a) Counts of HARs that were active in one, two, or all three studies. (b) Counts of HARs where the human and chimpanzee alleles were differentially active in one, two, or all three studies.
Pinpointing Individual Variants that Alter Human Accelerated Region Enhancer Activity
Comparing sequence variants of HARs is a powerful use of MPRAs because different alleles can be assayed side by side in the same experiment, alleviating much of the technical variability that confounds comparisons across experiments. This powerful strategy has been used to compare human versus chimpanzee homologs (22, 71), individual human-derived nucleotides (fixed or polymorphic) (13, 66, 71), and permutations of human-derived nucleotides (66, 71). Each HAR MPRA study identified hundreds of differentially active HARs, also known as species-biased HAR enhancers. These results vastly increase the number of HARs with strong evidence that human-specific variants altered their enhancer activity in neurodevelopment.
In contrast to within-experiment comparisons, comparisons of differential activity across studies are challenging due to the biological and technical differences described above. However, we identified 37 HARs that are consistently species biased in two studies (Figure 5b; Supplemental Table 3). Four of these HARs (2xHAR.9, 2xHAR.10, 2xHAR.63, and 2xHAR.548) were species biased in all three studies, making them high priority for further functional characterization. In fact, 2xHAR.548, which is in a neural progenitor cell chromatin domain with the transcription factor FOXP1, has already been validated as an ear enhancer in mouse embryos with suggestive differences between the human and chimpanzee sequences that merit further investigation (71). Chromatin domains in neural progenitor cells also support a link between 2xHAR.10 and PAX8, as well as between 2xHAR.63 and the genes BHLHE40 and ITPR1. It will be exciting to see if differential activity in MPRAs pinpoints HARs that function differently in humans compared to other mammals.
Quantifying Interactions Between Variants in Human Accelerated Regions
Several MPRA studies tested individual variants or subsets of the variants in each HAR (13, 66, 71). These strategies are the first data that can be used to dissect how the multiple human-specific nucleotides in each HAR affect its enhancer function. These analyses showed that some individual variants change enhancer activity relative to the chimpanzee sequence as much or more than the full set of human variants does. Another intriguing finding is that variants in the same HAR frequently interact to amplify or dampen each other’s effects on HAR enhancer activity (66, 71). This functional readout suggests that the rapid evolution of HARs may be due in part to compensatory evolution.
WHAT ROLE DID HUMAN ACCELERATED REGIONS PLAY IN HUMAN EVOLUTION?
Since the discovery of HARs, the evolutionary forces that created them and their contribution to human trait evolution have been investigated intensely. Molecular evolutionary and population genetic modeling has shown that most HARs have variant patterns consistent with positive selection, but some appear to have evolved through GC-biased gene conversion or loss of constraint (16, 35, 41, 54). From studies of ancient DNA, we have learned that most human–chimpanzee substitutions in HARs predate our common ancestor with Neanderthals and other archaic hominins, although a handful are unique to modern humans (5, 12, 28, 70). In addition, we know that having accelerated regions is not a human-specific trait. Chimpanzees and other primates have their own lineage-specific accelerated regions, with roughly similar numbers and genomic distributions to HARs (40, 56). While accelerated regions rarely overlap between primates, they cluster near each other in loci linked to neurodevelopment and disease (33, 40, 56). Diverse species beyond primates also have accelerated regions, though their genomic distributions and functional associations differ (25-27, 56). Collectively, a great deal has been revealed about HAR evolution.
However, much of this knowledge does not specifically account for HARs functioning as developmental enhancers. It is therefore a good time to revisit some fundamental questions about HAR evolution. For example, why did HARs evolve so rapidly after millions of years of extreme sequence conservation? MPRAs suggest that compensatory evolution to maintain enhancer activity levels may be an underlying mechanism (66, 71). They have also identified a role for adaptive introgression from Neanderthals (30). As individual HAR nucleotides begin to be dissected computationally and experimentally, we can also ask which variants most affect enhancer function. These investigations point to the importance of known transcription factor–binding sites (66, 70, 71), as well as some large effects that remain to be decoded functionally. With this knowledge, we can start to ask if each HAR evolved through gain-of-function (4), loss-of-function (64), or compensatory evolution to maintain function. Finally, some recent MPRA studies examined how cellular environment (e.g., cell type or species) interacts with sequence variation in HARs, showing few trans effects when comparing HAR enhancer activity in human versus chimpanzee (71) or mouse (22) cells. This is consistent with the high similarity of human and chimpanzee proteomes, other MPRAs in human versus mouse cells (48), and transgenic experiments in mice versus fish (61). However, the domination of trans effects by cis effects remains to be fully tested with an alternative technology.
HOW DOES HUMAN ACCELERATED REGION ENHANCER FUNCTION AFFECT OUR UNDERSTANDING OF DISEASE?
One of the first discoveries about HARs was their genomic proximity to disease genes. Indeed, nearby psychiatric disorder genes such as AUTS2 and NPAS3 inspired researchers to prioritize HARs for functional studies. With strong evidence that many HARs are enhancers, this genomic association takes on new meaning: Sequence changes in HARs are likely to perturb disease gene expression (13). Since many HAR-associated genes are well-known regulators and hubs in transcriptional networks (10, 73), their differential expression would affect many other genes and cellular processes, suggesting outsized effects caused by noncoding HAR mutations. Supporting this idea, rare polymorphisms in HARs may account for 5% of consanguineous autism cases (13). Thus, HAR enhancers are helping researchers to discover the genetic basis for disease (10). Conversely, medical genetics can help to functionally characterize HARs by revealing which HARs and HAR variants are pathogenic (14). Further extending this paradigm, drug target data have been used to map morbidities to HARs via their nearby genes (10). Taken together, these investigations underscore the utility of HARs for discovering new enhanceropathies and the power of disease biology for linking HARs to pathways and phenotypes. As more human and nonhuman primate genomes are sequenced, this promises to be an increasingly fruitful approach.
CONCLUSIONS
We are at an exciting moment for HAR biology. It is clear that many HARs function as enhancers. Machine learning and epigenetic data predict enhancer function for the majority of HARs. MPRAs have rapidly increased the rate at which candidate HAR enhancers can be tested in cells. With only a few developmental stages and cell types interrogated so far, these strategies have already prioritized HAR enhancers for in vivo functional characterization (e.g., with transgenic cells and animals) and for nucleotide-level experiments.
Evidence from genomic location, chromatin interactions, epigenetic signatures, sequence content, and machine learning increasingly suggests that HARs are biased toward neurodevelopment. The question of whether this bias is driven by better annotation and/or more data for neurological loci is important. While some tissues do have less information, others (e.g., developing heart) are similarly well characterized, suggesting that the brain enrichment of HARs is not purely an artifact of knowledge bias.
With this base of recent discoveries, the time is right to revisit questions about HARs that have been challenging to address before now. For example, what forces drove the rapid evolution of HARs? Which polymorphisms, fixed differences, and variants never seen in people affect HAR enhancer function? How many of these are deleterious? Which HAR variants interact with each other and their trans environment, either positively, to amplify their effects, or negatively, as in compensatory evolution? Interrogating the functions of HARs that are not enhancers to determine if they are repressors, insulators, RNA genes, splicing regulators, or protein-binding domains in messenger RNAs will also be interesting.
Addressing these questions will require new strategies. We envision performing MPRAs in more cell types and species with different permutations of HAR variants. Single-cell genomics, and the prospect of single-cell MPRAs, promise to resolve HAR function even further. This would expand the catalog of HAR enhancers and provide more data on interactions among and between variants and the trans environment. CRISPR-Cas genome editing provides a complementary technology for dissecting HARs (43, 59), which we expect will propel studies of individual HAR loci via humanized mice or cell lines, as well as large-scale screens of many HARs with CRISPR activation and interference (34). Beyond HARs, applying these strategies to human-specific deletions (hCONDELs) (49), as well as human-gained and human-lost enhancers (32, 59), will also be exciting. Cell lines and organoids differentiated from iPSCs are likely to remain a powerful system for these investigations (59, 62) by enabling researchers to work in the cellular environment of chimpanzees and other apes and to generate cells from difficult-to-sample tissues and developmental time points. Looking ahead, we predict that machine learning will drive the prioritization of HARs, variant combinations, and trans environments for these experiments.
Supplementary Material
SUMMARY POINTS.
Human accelerated regions (HARs) possess the intriguing evolutionary signature of rapid evolution in the human lineage but strong conservation in other species.
HARs are largely noncoding, and they had no known function when initially discovered in 2006.
Technological development has made it clear that many HARs function as gene regulatory enhancers, with enrichment in neurodevelopment.
Machine learning and massively parallel reporter assays (MPRAs) enable many HAR variants to be screened together for their effects on enhancer activity.
MPRA studies are only moderately concordant, but collectively they identify HARs where human-derived variants confidently alter enhancer activity.
Individual variants in HARs interact, suggesting that compensatory evolution may have driven rapid divergence since the human–chimpanzee ancestor.
Genetic variation in HARs, both natural and engineered, is a promising tool for elucidating the role of HARs in human evolution and disease.
ACKNOWLEDGMENTS
We gratefully acknowledge Nadav Ahituv’s contribution to Figure 4. S.W. and K.S.P. were funded by Gladstone Institutes, Chan Zuckerberg Biohub, and National Institute of Mental Health awards R01MH109907 and U01MH116438.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aldea D, Atsuta Y, Kokalari B, Schaffner SF, Prasasya RD, et al. 2021. Repeated mutation of a developmental enhancer contributed to human thermoregulatory evolution. PNAS 118:e2021722118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae BI, Jayaraman D, Walsh CA. 2015. Genetic changes shaping the human brain. Dev. Cell 32:423–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird CP, Stranger BE, Liu M, Thomas DJ, Ingle CE, et al. 2007. Fast-evolving noncoding sequences in the human genome. Genome Biol. 8:R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd JL, Skove SL, Rouanet JP, Pilaz LJ, Bepler T, et al. 2015. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol 25:772–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbano HA, Green RE, Maricic T, Lalueza-Fox C, de la Rasilla M, et al. 2012. Analysis of human accelerated DNA regions using archaic hominin genomes. PLOS ONE 7:e32877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush EC, Lahn BT. 2008. A genome-wide screen for noncoding elements important in primate evolution. BMC Evol. Biol 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capra JA, Erwin GD, McKinsey G, Rubenstein JL, Pollard KS. 2013. Many human accelerated regions are developmental enhancers. Philos. Trans. R. Soc. B 368:20130025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, et al. 2022. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50:D165–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen KM, Wong AK, Troyanskaya OG, Zhou J. 2021. A sequence-based global map of regulatory activity for deciphering human genetics. bioRxiv 454384. 10.1101/2021.07.29.454384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu XY, Quan Y, Zhang HY. 2020. Human accelerated genome regions with value in medical genetics and drug discovery. Drug Discov. Today 25:821–27 [DOI] [PubMed] [Google Scholar]
- 11.Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, et al. 2013. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154:185–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crisci JL, Wong A, Good JM, Jensen JD. 2011. On characterizing adaptive events unique to modern humans. Genome Biol. Evol 3:791–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, et al. 2016. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167:341–54.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan RN, Shin T, Walsh CA. 2018. Evolutionary changes in transcriptional regulation: insights into human behavior and neurological conditions. Annu. Rev. Neurosci 41:185–206 [DOI] [PubMed] [Google Scholar]
- 15.Dong X, Wang X, Zhang F, Tian W. 2016. Genome-wide identification of regulatory sequences undergoing accelerated evolution in the human genome. Mol. Biol. Evol 33:2565–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duret L, Galtier N. 2009. Comment on “Human-specific gain of function in a developmental enhancer”. Science 323:714. [DOI] [PubMed] [Google Scholar]
- 17.Dutrow EV, Emera D, Yim K, Uebbing S, Kocher AA, et al. 2022. Modeling uniquely human gene regulatory function via targeted humanization of the mouse genome. Nat. Commun 13:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erwin GD, Oksenberg N, Truty RM, Kostka D, Murphy KK, et al. 2014. Integrating diverse datasets improves developmental enhancer prediction. PLOS Comput. Biol 10:e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franchini LF, Pollard KS. 2015. Genomic approaches to studying human-specific developmental traits. Development 142:3100–12 [DOI] [PubMed] [Google Scholar]
- 20.Franchini LF, Pollard KS. 2017. Human evolution: the non-coding revolution. BMC Biol. 15:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghandi M, Mohammad-Noori M, Ghareghani N, Lee D, Garraway L, Beer MA. 2016. gkmSVM: an R package for gapped-kmer SVM. Bioinformatics 32:2205–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girskis KM, Stergachis AB, DeGennaro EM, Doan RN, Qian X, et al. 2021. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 109:3239–51.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gittelman RM, Hun E, Ay F, Madeoy J, Pennacchio L, et al. 2015. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 25:1245–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haygood R, Babbitt CC, Fedrigo O, Wray GA. 2010. Contrasts between adaptive coding and noncoding changes during human evolution. PNAS 107:7853–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holloway AK, Begun DJ, Siepel A, Pollard KS. 2008. Accelerated sequence divergence of conserved genomic elements in Drosophila melanogaster. Genome Res. 18:1592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holloway AK, Bruneau BG, Sukonnik T, Rubenstein JL, Pollard KS. 2016. Accelerated evolution of enhancer hotspots in the mammal ancestor. Mol. Biol. Evol 33:1008–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z, Sackton TB, Edwards SV, Liu JS. 2019. Bayesian detection of convergent rate changes of conserved noncoding elements on phylogenetic trees. Mol. Biol. Evol 36:1086–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubisz MJ, Pollard KS. 2014. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev 29:15–21 [DOI] [PubMed] [Google Scholar]
- 29.Inoue F, Ahituv N. 2015. Decoding enhancers using massively parallel reporter assays. Genomics 106:159–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagoda E, Xue JR, Reilly SK, Dannemann M, Racimo F, et al. 2022. Detection of Neanderthal adaptively introgressed genetic variants that modulate reporter gene expression in human immune cells. Mol. Biol. Evol 39:msab304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y, Gittelman RM, Lu Y, Liu X, Li MD, et al. 2018. Evolution of DNAase I hypersensitive sites in MHC regulatory regions of primates. Genetics 209:579–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones B 2015. Becoming human—identifying human accelerated regulatory DNA. Nat. Rev. Genet 16:439. [DOI] [PubMed] [Google Scholar]
- 33.Kamm GB, Pisciottano F, Kliger R, Franchini LF. 2013. The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol. Biol. Evol 30:1088–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kampmann M 2020. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol 16:465–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. 2010. GC-biased evolution near human accelerated regions. PLOS Genet. 6:e1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–16 [DOI] [PubMed] [Google Scholar]
- 37.Kitts A, Sherry S. 2011. The Single Nucleotide Polymorphism Database (dbSNP) of nucleotide sequence variation. In The NCBI Handbook, ed. McEntyre J, Ostell J. Bethesda, MD: US Natl. Cent. Biotechnol. Inf. [Google Scholar]
- 38.Kosiol C, Vinař T, da Fonseca RR, Hubisz MJ, Bustamante CD, et al. 2008. Patterns of positive selection in six mammalian genomes. PLOS Genet. 4:e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostka D, Hahn MW, Pollard KS. 2010. Noncoding sequences near duplicated genes evolve rapidly. Genome Biol. Evol 2:518–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kostka D, Holloway AK, Pollard KS. 2018. Developmental loci harbor clusters of accelerated regions that evolved independently in ape lineages. Mol. Biol. Evol 35:2034–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostka D, Hubisz MJ, Siepel A, Pollard KS. 2012. The role of GC-biased gene conversion in shaping the fastest evolving regions of the human genome. Mol. Biol. Evol 29:1047–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee KS, Bang H, Choi JK, Kim K. 2020. Accelerated evolution of the regulatory sequences of brain development in the human genome. Mol. Cells 43:331–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KS, Chatterjee P, Choi EY, Sung MK, Oh J, et al. 2018. Selection on the regulation of sympathetic nervous activity in humans and chimpanzees. PLOS Genet. 14:e1007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levchenko A, Kanapin A, Samsonova A, Gainetdinov RR. 2018. Human accelerated regions and other human-specific sequence variations in the context of evolution and their relevance for brain development. Genome Biol. Evol 10:166–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Hannenhalli S, Ovcharenko I. 2021. De novo human brain enhancers created by single nucleotide mutations. bioRxiv 451055. 10.1101/2021.07.04.451055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Libbrecht MW, Chan RCW, Hoffman MM. 2021. Segmentation and genome annotation algorithms for identifying chromatin state and other genomic patterns. PLOS Comput. Biol. 17:e1009423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478:476–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattioli K, Oliveros W, Gerhardinger C, Andergassen D, Maass PG, et al. 2020. Cis and trans effects differentially contribute to the evolution of promoters and enhancers. Genome Biol. 21:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minnoye L, Taskiran II, Mauduit D, Fazio M, Van Aerschot L, et al. 2020. Cross-species analysis of enhancer logic using deep learning. Genome Res. 30:1815–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noonan JP. 2009. Regulatory DNAs and the evolution of human development. Curr. Opin. Genet. Dev 19:557–64 [DOI] [PubMed] [Google Scholar]
- 52.Norman AR, Ryu AH, Jamieson K, Thomas S, Shen Y, et al. 2021. A human accelerated region is a Leydig cell GLI2 enhancer that affects male-typical behavior. bioRxiv 428524. 10.1101/2021.01.27.428524 [DOI] [Google Scholar]
- 53.Oksenberg N, Stevison L, Wall JD, Ahituv N. 2013. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLOS Genet. 9:e1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLOS Genet. 2:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443:167–72 [DOI] [PubMed] [Google Scholar]
- 56.Prabhakar S, Noonan JP, Paabo S, Rubin EM. 2006. Accelerated evolution of conserved noncoding sequences in humans. Science 314:786. [DOI] [PubMed] [Google Scholar]
- 57.Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, et al. 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163:68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quang D, Xie X. 2016. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 44:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reilly SK, Noonan JP. 2016. Evolution of gene regulation in humans. Annu. Rev. Genom. Hum. Genet 17:45–67 [DOI] [PubMed] [Google Scholar]
- 60.Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, et al. 2015. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347:1155–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritter DI, Li Q, Kostka D, Pollard KS, Guo S, Chuang JH. 2010. The importance of being cis: evolution of orthologous fish and mammalian enhancer activity. Mol. Biol. Evol 27:2322–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silver DL. 2016. Genomic divergence and brain evolution: how regulatory DNA influences development of the cerebral cortex. Bioessays 38:162–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song M, Pebworth M-P, Yang X, Abnousi A, Fan C, et al. 2020. Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587:644–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sumiyama K, Saitou N. 2011. Loss-of-function mutation in a repressor module of human-specifically activated enhancer HACNS1. Mol. Biol. Evol 28:3005–7 [DOI] [PubMed] [Google Scholar]
- 65.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–76 [DOI] [PubMed] [Google Scholar]
- 66.Uebbing S, Gockley J, Reilly SK, Kocher AA, Geller E, et al. 2021. Massively parallel discovery of human-specific substitutions that alter enhancer activity. PNAS 118:e2007049118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, et al. 2016. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat. Neurosci 19:494–503 [DOI] [PubMed] [Google Scholar]
- 68.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35:D88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Jaime-Lara RB, Roy A, Sun Y, Liu X, Joseph PV. 2021. SeqEnhDL: sequence-based classification of cell type-specific enhancers using deep learning models. BMC Res. Notes 14:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss CV, Harshman L, Inoue F, Fraser HB, Petrov DA, et al. 2021. The cis-regulatory effects of modern human-specific variants. eLife 10:e63713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whalen S, Inoue F, Ryu H, Fair T, Markenscoff-Papadimitriou E, et al. 2022. Machine-learning dissection of Human Accelerated Regions in primate neurodevelopment. bioRxiv 256313. 10.1101/256313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Won H, Huang J, Opland CK, Hartl CL, Geschwind DH. 2019. Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat. Commun 10:2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu K, Schadt EE, Pollard KS, Roussos P, Dudley JT. 2015. Genomic and network patterns of schizophrenia genetic variation in human evolutionary accelerated regions. Mol. Biol. Evol 32:1148–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.