Abstract
Introduction
A newly identified SARS-CoV-2 variant, VOC202012/01 originating lineage B.1.1.7, recently emerged in the United Kingdom. The rapid spread in the UK of this new variant has caused other countries to be vigilant.
Material and methods
We based our initial screening of B.1.1.7 on the dropout of the S gene signal in the TaqPath assay, caused by the 69/70 deletion. Subsequently, we confirmed the B.1.1.7 candidates by whole genome sequencing.
Results
We describe the first three imported cases of this variant from London to Madrid, subsequent post-arrival household transmission to three relatives, and the two first cases without epidemiological links to UK. One case required hospitalization. In all cases, drop-out of gene S was correctly associated to the B.1.1.7 variant, as all the corresponding sequences carried the 17 lineage-marker mutations.
Conclusion
The first identifications of the SARS-CoV-2 B.1.1.7 variant in Spain indicate the role of independent introductions from the UK coexisting with post-arrival transmission in the community, since the early steps of this new variant in our country.
Keywords: COVID-19, SARS-CoV-2, B.1.1.7, Importation, Transmission, WGS
Abstract
Introducción
Recientemente, ha surgido en Reino Unido una nueva variante de SARS-CoV-2, VOC202012/01, que origina el linaje B.1.1.7. Su rápida distribución en Reino Unido ha alertado a otros países a vigilar su presencia.
Material y métodos
El rastreo inicial de la variante B.1.1.7 se basó en la ausencia de amplificación del gen S en el ensayo TaqPath, causado por la deleción 69/70. Todos los casos candidatos de corresponder a la variante B.1.1.7 con este criterio fueron posteriormente confirmados por secuenciación de genoma completo.
Resultados
Describimos los primeros 3 casos importados de esta variante, desde Londres hasta Madrid, con la posterior transmisión domiciliaria de uno de estos casos a 3 familiares y, adicionalmente, los 2 primeros casos con la variante sin vínculo epidemiológico con Reino Unido. Uno de los casos requirió hospitalización. En todos los casos el criterio de no amplificación del gen S identificó con precisión la variante B.1.1.7, como demostró posteriormente la presencia de las 17 mutaciones marcadoras de este linaje.
Conclusión
Las primeras identificaciones de la variante B.1.1.7 de SARS-CoV-2 indican un papel solapante de las introducciones independientes desde Reino Unido, con eventos de transmisión comunitaria, incluso desde los primeros momentos de la presencia de esta variante en nuestro país.
Palabras clave: COVID-19; SARS-CoV-2; B.1.1.7, Importación; Transmisión; WGS
A newly identified SARS-CoV-2 variant, VOC202012/01 originating lineage B.1.1.7, recently emerged in the UK, is responsible for 85% of all cases in south east England1 at the time of writing this manuscript. This variant has also been reported in other European countries and in most distant locations, e.g., Japan, Australia, or Singapore. B.1.1.7 has higher transmissibility in comparison to other SARS-CoV-2 lineages.1 It carries an unusually high number of specific mutations, 17, most of which are non-synonymous and eight concentrate in the S gene.2 Among the mutations mapping in the S gene, some are linked to relevant functional roles, e.g., immune response evasion, (69/70 deletion)3 or increased affinity to the ACE2 receptor (N501Y).4, 5 These findings have raised the alarm of having to face a new variant with the potential ability to accelerate the spread of the pandemic, although no association was has found initially between the new variant and greater COVID-19 severity or more frequent hospitalizations6, a recent report finds a realistic possibility (as per PHIA probability yardstick) that B.1.1.7 is associated with an increased risk of death compared to infection with no-B.1.1.7 variants.7
From a diagnostic perspective, the presence of the 69/70 deletion allows a rapid screening of the B.1.1.7 variant, as it impairs the hybridization of the S probe in the TaqPath assay (ThermoFisher, Waltham, USA). It causes a drop-out in the S signal, accompanied by a normal positive signal for the other two genes targeted by the assay.8
Here, we communicate the first eight confirmed SARS-CoV-2 B.1.1.7 variants identified in Spain, all diagnosed in Madrid, with epidemiological support to the UK for six of the cases: three cases who recently travelled from London to Madrid (December 19–20, 2020) and three household members related to one of these cases. The other two cases had no known epidemiological relationship with the UK, neither trips nor contact with people coming from that country. One of the cases required hospitalization and the others were indicated to keep quarantine and isolation measures along with their close contacts.
All except the two cases without links with the UK and the hospitalized case were positive for the COVID-19 rapid antigen test (Panbio™ COVID-19 Ag rapid test device, Abbott) and all showed an S gene target failure with the TaqPath RT-PCR COVID-19 kit. The B.1.17 variant was confirmed by identifying its 17 specific mutations by whole genome sequencing (WGS). WGS for Cases 2, 3, 7, and 8 was performed with the Artic_nCov-2019_V3 panel of primers (Integrated DNA Technologies, Inc., Coralville, Iowa, USA) (artic.network/ncov-2019). Libraries were prepared using the Nextera Flex DNA Library Preparation Kit (Illumina lnc, California, USA). WGS for Cases 1, 4–6 followed the MinION procedure (Oxford Nanopore Technologies, Oxford, United Kingdom). Sequences were deposited at GISAID (EPI_ISL_756271, 756272, 756273 and 756274).
Imported cases
Case 1. A 33-year-old male London resident, who returned to Madrid from the UK on December 19, 2020 started with fever and myalgia on December 20, and the next day with cough and diarrhoea. He consulted his general practitioner and had his first positive RT-PCR for which he was prescribed symptomatic treatment. On December 28, he went to the emergency room due to dyspnoea. Physical examination revealed tachypnea, bilateral crackles, and hypoxemia, and the chest X-ray showed multilobar bilateral infiltrates. Lymphocyte count, D-dimers, and ferritin were normal and C-reactive protein value was 16 mg/L (normal < 5). Patient was treated with oxygen, dexamethasone, and prophylactic heparin with favourable clinical evolution. On December 30, 2020, the patient had a second positive RT-PCR result. By that time, his two household contacts were negative.
Case 2. A 30-year-old female without any comorbidity who travelled from London to Madrid on December 20, 2020 and started with fever asthenia, cough, myalgia, and dyspnoea the night of her arrival. She went to the emergency department on December 22 where an antigenic COVID-19 rapid test and RT-PCR were performed, both with positive result; furthermore, unilateral pneumonia was diagnosed.
Transmission event
Case 3 (imported index Case). A 27-year-old male with a past medical history of protein C deficiency who arrived to Madrid from London on December 19, 2020 and presented two days later to the emergency department with a chief complaint of fever, generalized headache, and mild non-productive cough the night before. The SARS-CoV-2 RT-PCR was positive after testing negative on December 16. Physical examination and vital signs were normal and therefore symptomatic treatment and prophylactic anticoagulation regimen were indicated.
Cases 4–6 (household contacts). Three household contacts of the imported index case went to the emergency department on December 23, 2020 due to fever and general malaise and had positive SARS-CoV-2 RT-PCRs. Two female aged 61 and 31 years and a 55-year-old male. None had underlying diseases nor clinical or radiographic signs of pneumonia.
Community cases without epidemiological link to UK
Case 7. A 36-year-old male without any relevant medical history who attended the emergency department on December 27, 2020 after contact with a friend who tested positive for SARS-CoV-2 on December 23. The patient complained of headache and productive cough that had started 24 h before. He was tested positive result for SARS-CoV-2 RT-PCR.
Case 8. A 30-year-old female without any relevant clinical history, who went to the emergency department on December 27, 2020 after close contact with her mother-in-law and partner, who were SARS-CoV-2 positive. The patient presented asthenia, non-productive cough, and diarrhoea, and positive SARS-CoV-2 RT-PCR.
Comparative analysis of strains
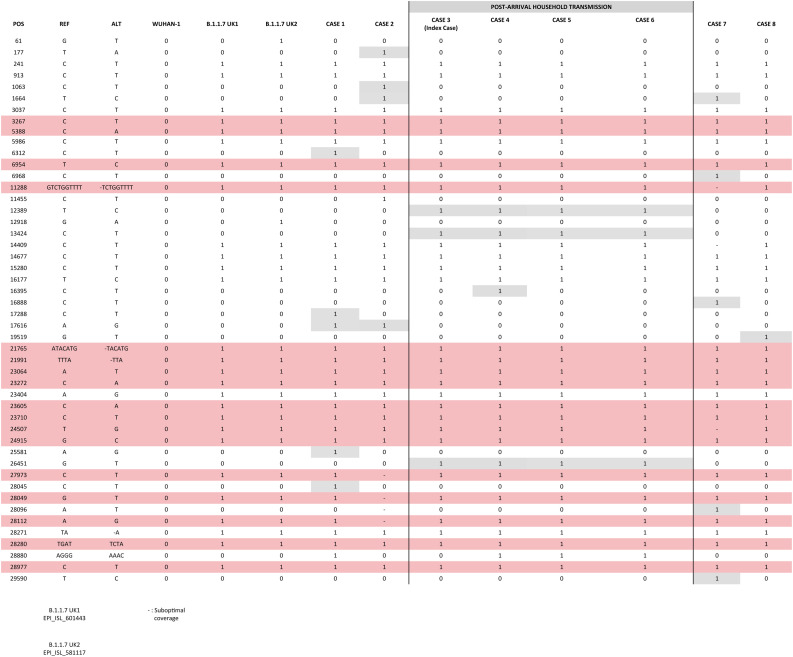
We compared the sequences obtained from the cases identified in Spain; among them and against two UK original B.1.1.7 consensus references (GISAID: EPI_ISL_601443 and 581117; 20 and 21 September 2020) (Fig. 1 ). 3–7 SNPs were observed between our cases and the UK references, consistent with the time since the new variant has been circulating in UK and its import to Spain. Nearly identical sequences (differing in 1 SNP in one of the cases) were obtained for the cases involved in the household transmission. The remaining cases showed 1–4 private SNPs each, consistent with independent importations.
Fig. 1.
Comparison of the sequences obtained from the cases identified in Spain; among them and against two UK original B.1.1.7 consensus references (GISAID: EPI_ISL_601443 and 581117; 20 and 21 September 2020).
Conclusions
We report three independent imported cases from the UK into Spain of the new SARS-CoV-2 B.1.1.7 variant and a rapid household transmission linked to one of the cases. We also communicate the two first identifications of the B.1.1.7 variant in cases without any epidemiological suspicion supporting it, which indicates its circulation in the community. Only one case required hospitalization.
At the moment of closing the final revision of this manuscript, B.1.1.7 variant constitutes around 15–20% of the cases in our population. This makes meaningless to apply a criterion of epidemiological suspect and makes unaffordable to perform continuous confirmatory sequence analysis of all new candidate cases with a drop out signal in the RT-PCR. Further efforts are currently being made in order to constantly update the magnitude of the presence of the B.1.1.7 variant in our population, based on the S dropout feature. Updatings are either population-based (in those laboratories systematically applying the TaqPath test), or on a random subsampling of positive specimens pretested by another diagnostic RT-PCR.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by Instituto de Salud Carlos III (Ref COV20/00140: SeqCOVID – Consorcio para la epidemiología genómica de SARS-CoV-2 en España) and by Consejo Superior de Investigaciones Científicas (CSIC) (PTI Salud Global). We are grateful to Dainora Jaloveckas (cienciatraducida.com) for editing and proofreading assistance.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.eimce.2021.02.013
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Volz E., Mishra S., Chand M., Barret J., Johnson R., Geidelberg L., et al. MRC Centre for Global Infectious Disease Analysis Report; 2020. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data; p. 42. [Google Scholar]
- 2.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. ARTIC Netw. 2020 [Google Scholar]
- 3.Kemp S., Harvey W., Datir R., Collier D., Ferreira I., Carabelli A., et al. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. BioRxiV. 2020 [Google Scholar]
- 4.Chand M., Hopkins S., Dabrera G., Achison C., Barclay W., Ferguson N., et al. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01. Public Health England. 2020 [Google Scholar]
- 5.Chen J., Wang R., Wang M., Wei G. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horby P., Davies N., Edmunds J., Ferguson N., Medley G., Hayward A., et al. NERVTAG note on B.1.1.7 severity. New and Emerging Respiratory Virus Threats Advisory Group NERVTAG. 2021 [Google Scholar]
- 7.ECDC . 2020. Risk related to spread of new SARS-CoV-2 variants of concern in the EU/EEA. [Google Scholar]
- 8.ECDC . 2020. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom THREAT ASSESSMENT BRIEF. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.