Abstract
Gibbons are the most speciose family of living apes, characterized by a diverse chromosome number and rapid rate of large-scale rearrangements. Here we performed single-cell template strand sequencing (Strand-seq), molecular cytogenetics, and deep in silico analysis of a southern white-cheeked gibbon genome, providing the first comprehensive map of 238 previously hidden small-scale inversions. We determined that more than half are gibbon specific, at least fivefold higher than shown for other primate lineage-specific inversions, with a significantly high number of small heterozygous inversions, suggesting that accelerated evolution of inversions may have played a role in the high sympatric diversity of gibbons. Although the precise mechanisms underlying these inversions are not yet understood, it is clear that segmental duplication–mediated NAHR only accounts for a small fraction of events. Several genomic features, including gene density and repeat (e.g., LINE-1) content, might render these regions more break-prone and susceptible to inversion formation. In the attempt to characterize interspecific variation between southern and northern white-cheeked gibbons, we identify several large assembly errors in the current GGSC Nleu3.0/nomLeu3 reference genome comprising more than 49 megabases of DNA. Finally, we provide a list of 182 candidate genes potentially involved in gibbon diversification and speciation.
A fundamental question in evolutionary biology is to establish how Hominina diverged from apes, starting from a common ancestor. It is common knowledge that the mammalian karyotype has been quite conserved during the evolution of eutherian chromosomes (Wienberg 2004; Kim et al. 2017; Damas et al. 2021). However, gibbons (Hylobatidae), also called lesser apes, represent an exception to this widely accepted conclusion because their genome evolution has been accelerated by shuffling events that gave rise to some highly derivative karyotypes (Carbone et al. 2006). This chromosomal instability might be responsible for the diversity of gibbons, the most heterogeneous group of living apes with almost 20 existing species (Fan et al. 2017; Nie et al. 2018). They are classified in four genera, which feature different numbers of chromosomes and extensive chromosome rearrangements that allow us to separate them into Hoolock (2n = 38), Hylobates (2n = 44), Symphalangus (2n = 50), and Nomascus (2n = 52) (Capozzi et al. 2012).
Because of their close evolutionary relationship with humans and great apes (Matsudaira and Ishida 2010) and the high genetic diversity between them, gibbons represent a unique perspective on evolution. Previous studies based on extensive cytogenetic analyses focused on large-scale chromosomal rearrangements (on average, 22 megabases in length) and allowed precise definition of the synteny block organization of the four genera with respect to the reconstructed Hylobates ancestral karyotype and to humans (Roberto et al. 2007; Misceo et al. 2008). However, not much is known about smaller variants within these megabase-large synteny blocks.
Among structural variants (SVs), inversions were first proposed to contribute to speciation because they suppress recombination in the inverted regions when in the heterozygous state (Rieseberg 2001; Navarro and Barton 2003). As a consequence, inversions are powerful forces in diversification because suppressed recombination caused by inversions allows mutations to accumulate independently between ancestral and derived arrangements (Noor et al. 2001; Kirkpatrick and Barton 2006; Fuller et al. 2019). Moreover, inversions have the potential to disrupt genes or associations with regulatory elements or relocate them closer or further away from heterochromatic regions, thus inducing a position effect as previously described in Drosophila, yeast, mouse, and human (Tham and Zakian 2002; Pedram et al. 2006; Elgin and Reuter 2013; Puig et al. 2015; McBroome et al. 2020). Inversions can be maintained over thousands or millions of years and often involve large genomic regions that may contain important genes for intraspecific divergence and speciation (Wellenreuther and Bernatchez 2018).
Despite the important role of inversions in genome evolution, they are difficult to detect and analyze in mammalian genomes owing to their balanced nature and the fact that their breakpoints (BPs) are often embedded within repetitive sequences such as segmental duplications (SDs). Even the latest long-read sequencing and optical mapping technologies often miss inversions or generate a high rate of false positives (Chaisson et al. 2019), leading to a gap in inversion discovery and understanding, as well as errors in sequenced genome assemblies (Kidd et al. 2008; Sanders et al. 2016). To date, only single-cell template strand sequencing (Strand-seq) has been successfully applied to detect inversions in human and nonhuman primate genomes (Sanders et al. 2016; Chaisson et al. 2019; Maggiolini et al. 2020; Porubsky et al. 2020a; Ebert et al. 2021; Porubsky et al. 2022). Strand-seq has the potential to determine the ancestral and derived states of every single inversion, thereby assigning mutational events to different time points during primate evolution, providing remarkable new information about the evolution and variation of primate genomes. Strand-seq generates directional libraries that, once sequenced and aligned against a reference genome, allow investigators to distinguish different orientation variants and to locate them at a single-cell level (Sanders et al. 2016). Although Strand-seq coverage is low, by pooling data from several sequenced cells, each homologous chromosome can be uniquely characterized, and events up to 1 kbp can be detected. Moreover, because in Strand-seq, inversions are detected just by a segmental change in read alignment orientation within the inverted segment, the presence of large SDs at the inversion boundaries does not affect their detection, and the false positive rate has been shown to be as low as 0.003%, even within the most complex regions of the genome (Maggiolini et al. 2020).
Here we applied this newly developed strategy to generate a comprehensive map of small-scale inversions within large synteny blocks in a Nomascus siki (NSI), providing a detailed and fine-structure picture of small-scale, previously hidden, inversions comprising genes potentially important for gibbon intraspecific divergence and speciation.
Results
Strand-seq data analysis
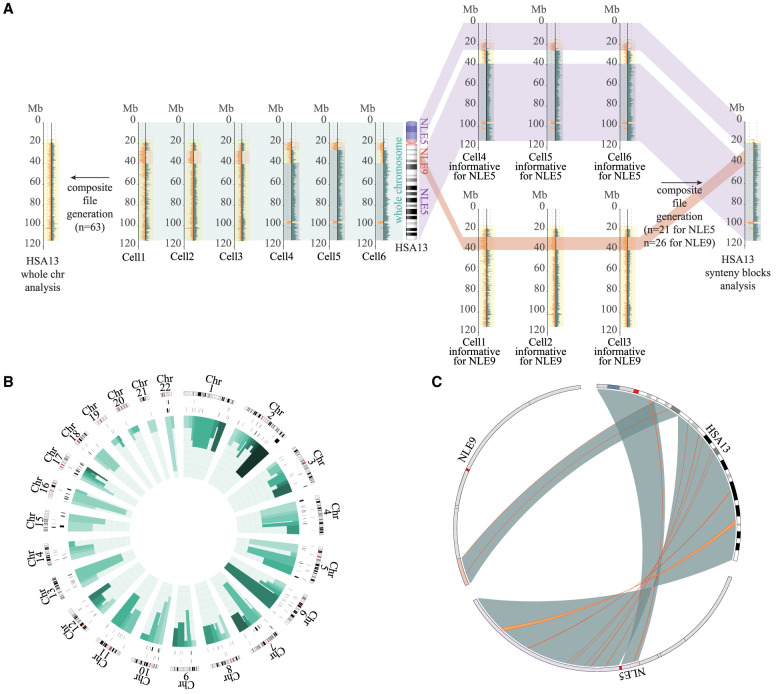
To detect inversions between the genomes of human and gibbon, we sequenced an NSI individual using Strand-seq. We selected the same individual from the genus Nomascus whose genome was well characterized using extensive fluorescence in situ hybridization (FISH) analyses to create a synteny block backbone as part of the NLE genome sequencing project (GGSC Nleu3.0/nomLeu3) (Carbone et al. 2014). Because the gibbon karyotype is highly rearranged compared with that of the human, we developed a new strategy to map and identify inversions between human and gibbon genomes. First, we generated high-quality single-cell Strand-seq libraries, taking advantage of the directionality of single-stranded DNA molecules, which are distinguished as either Crick (C; forward, or plus strand) or Watson (W; reverse, or minus strand) based on their 5′–3′ orientation (Sanders et al. 2017); then we mapped the sequencing data against the gibbon GGSC Nleu3.0/nomLeu3 reference and selected 62 informative libraries for which Strand-seq was successful. The NLE alignment was used to select the specific Strand-seq libraries that were informative for inversion calling for each chromosome (i.e., inherited in a WW or CC orientation, because homozygous inversions are masked on chromosomes that are inherited in the WC orientation) (Sanders et al. 2017). Afterward, in order to map the reads against the human reference genome (GRCh38/hg38), we considered each one of the 119 human–gibbon synteny blocks as a separate chromosome whose boundaries have been assessed using published cytogenetics data (Roberto et al. 2007; Capozzi et al. 2012; http://www.biologia.uniba.it/gibbon/) and further refined using gibbon Strand-seq data (i.e., observing a complete switch of reads from W to C or vice versa) (Supplemental Table S1). Hence, we assigned informative libraries for each synteny block, based on previously selected NLE-informative chromosomes (Fig. 1A). In this way, we generated high-coverage and directional files for each human–gibbon synteny block that were BED-formatted into a unique composite file, and uploaded it as a custom track on UCSC Genome Browser (GRCh38/hg38 release). Using this NSI composite file, we manually scanned all the chromosomes, looking for changes in the directionality of reads and identified 444 events that are >7 kb and have a minimum of 10 reads mapping within the region (Fig. 1B,C; Supplemental Table S2). This threshold was set in order to increase our confidence in detecting real inversions.
Figure 1.
Strand-seq data analysis. (A) Schematic view of the method used to generate the Chr 13 composite file for the detection of the inversions. In the middle, an ideogram of human Chromosome 13 (HSA13) and the corresponding synteny blocks relative to Nomascus (NLE5 or NLE9) is shown. (Left) If single-cell Strand-seq data are pooled directly (Strand-seq output for six single cells is shown as an example) without taking into account the human–gibbon synteny block information, the resulting composite file will show no informative strand-state information (reads will map to both Watson and Crick strands equally, so every chromosome appears as WC). (Right) The method used in the present study considers human–gibbon synteny blocks individually to select libraries for generating a composite file separately for each syntenic region. Only informative libraries for each synteny block are selected and merged to generate a composite file of the whole chromosome. (B) Circos diagram (Krzywinski et al. 2009) reporting detected inversions for each human chromosome ideogram, with heterozygous inversions in the external circle and homozygous inversions in the inner circle. Green bars indicate the number of inversions detected for each synteny block (shown in different shades of green), where the height is proportional to the number of inversions detected for that block (min = 0; max = 9). (C) Circos diagram (Krzywinski et al. 2009) exemplifying the results of the analysis for a single human chromosome. Human–gibbon synteny blocks for human Chr 13 (large green highlights) and the inversions detected within each block (thin orange lines) are shown.
Inversion analysis
By manually analyzing the NSI composite file on the UCSC Genome Browser, we initially identified a total of 444 events showing a switch in strand orientation that can be either inversions or copy-variable regions, which likely reflect intrachromosomal duplications, inverted duplications, unannotated SD/repeat elements, and other reference artifact events. Indeed, Strand-seq can distinguish between simple and nested inversions, as well as copy-variable regions. With simple inversions, the reads within an inverted region appear in the opposite orientation with respect to the flanking reads in the synteny block where they map (Supplemental Fig. S1A); whereas when the inversions map within a larger inverted region, the reads appear to be in direct orientation by Strand-seq and are classified as nested inversions (Supplemental Fig. S1B). Moreover, inversions can be homozygous when both the homologs are inverted and are visualized as a complete switch in the directionality of the reads with respect to the synteny block (Supplemental Fig. S1A,B) or heterozygous when only one homolog is inverted and inversions appear as a partial change in the directionality of the reads, resulting in a mix of green and orange reads (Supplemental Fig. S1C). Copy-variable region events can show a switch in strand orientation by Strand-seq, as well as an increase in read depth (Supplemental Fig. S1D). To distinguish between inversions and copy-variable regions, we then analyzed the sequencing read depth. Because NSI-specific copy-variable regions are represented more than once along the gibbon genome with respect to a single-copy region, we excluded from our analysis 202 regions that had a read depth at least one and a half times higher than the average depth of the synteny block in which they were embedded. To validate our selection filter, we randomly tested by FISH three of these regions (Supplemental Fig. S2) that were confirmed to be segments with variable copy numbers and locations. Following literature interrogation, we also excluded from our inversions list six regions whose call was an artifact owing to misassemblies or to the annotation of minor alleles in the human reference genome (Supplemental Table S3; Sanders et al. 2016; Vicente-Salvador et al. 2017; Catacchio et al. 2018; Audano et al. 2019; Chaisson et al. 2019). Furthermore, we manually integrated our list with two more regions that are polymorphic in human and for which the minor allele is annotated on the GRCh38 human assembly; indeed, Strand-seq appears to be direct but is actually inverted (Supplemental Table S4; Sanders et al. 2016). Overall, we obtained a final list of 238 inversions (Supplemental Table S5). Among these, 231 are simple inversions and seven are nested, because they are included in larger inversions. Moreover, ∼73% of the inversions are homozygous (175 out of 238), whereas 27% (63 out of 238) were heterozygous and therefore likely polymorphic in the NSI population. Worthy of note, homozygous inversions are larger in size than are inversions in the heterozygous state (Supplemental Fig. S3A).
Validation of simple inversions
We selected nine inversions >500 kbp and tested them by FISH in the same NSI for which Strand-seq was generated. In detail, eight out of nine were tested in interphase nuclei by three-color FISH, whereas one larger inversion (Chr2_inv16) was tested on metaphase by two-color FISH (Supplemental Table S6). All the inversions detected by Strand-seq were confirmed by FISH, with two out of nine in heterozygous state in the tested NSI as predicted by Strand-seq (Supplemental Fig. S4).
As another means of validation of the inversion calls, we used BAC-end sequence (BES) pair-mapping data from the same NLE individual used to generate the gibbon reference genome assembly. Because the BAC library belongs to an NLE and the Strand-seq was generated from an NSI, we expected some discrepancies for inversions that could be polymorphic between the two species. First, the BAC ends were mapped against the human reference genome, and the 238 regions were scanned in order to evaluate the presence of BACs spanning the inversions. A total of 97 regions had BACs spanning either one of the inversion BPs and were further analyzed for validation. BAC ends supported an inversion if pairs spanning an inversion BP mapped abnormally far apart and were incorrectly oriented when mapped to the human reference genome (Tuzun et al. 2005). For 74 out of 97 regions (one heterozygous and 73 homozygous), BAC-end mapping profiling supported the inversion, and there was a total concordance between NLE and NSI; for 22, the BES supported a direct orientation in NLE, whereas NSI was heterozygous (n = 16) or inverted in homozygous state (n = 6) by Strand-seq; finally, for one region, NLE was heterozygous, whereas NSI inverted in homozygous state (Supplemental Table S7), suggesting that these regions are polymorphic between NLE and NSI.
Evolutionary and phylogenetic analysis
Next, our goal was to reconstruct the evolutionary history of each region comparing human and nonhuman primate genomes. To accomplish this aim, we took advantage of recently published Strand-seq data for great apes (one chimpanzee, one bonobo, one gorilla, and one orangutan) (Porubsky et al. 2020a) and macaque genomes (Maggiolini et al. 2020). Additional information regarding the orientation and the polymorphic state of the inverted regions in human and nonhuman primates was retrieved from previous literature (Sanders et al. 2016; Vicente-Salvador et al. 2017; Catacchio et al. 2018; Chaisson et al. 2019; Giner-Delgado et al. 2019; Maggiolini et al. 2019; Puig et al. 2020), and using, when necessary, marmoset and squirrel monkey net alignment as outgroups, we reconstructed the evolutionary history of 189 out of 238 inversions (∼79%) (Supplemental Table S5). The remaining inversions included regions with a complex architecture or regions for which the orientation of outgroup species was not available.
Previous studies had already defined the evolutionary history for 32 regions out of 189. However, for 11 out of 32 of these regions, we added additional information. The evolution is described here for the first time for 157 out of 189 (83%) inversions, with 10 events for which we provided two possible alternative evolutionary scenarios (Supplemental Table S5).
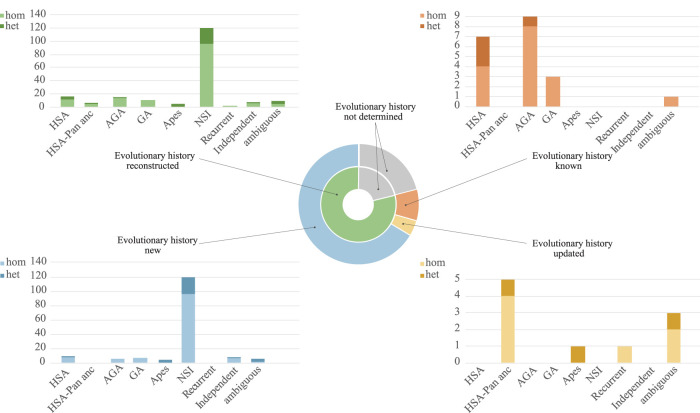
Excluding these 10 regions, the evolutionary history was described with a high-confidence level for the remaining 179 regions as follows: 14 (7.8%) of them are human specific, five (2.7%) occurred in the human-pan ancestor, 16 (8.9%) occurred in the African great ape ancestor, 10 (5.5%) occurred in the great ape ancestor, four (2.2%) occurred in the ape ancestor, 121 (67.4%) are gibbon specific, nine (5%) are inversions that occurred independently in two different lineages (two occurred in gorilla and gibbon, four occurred in orangutan and gibbon, one occurred in the human-pan ancestor and gibbon, one occurred in human and macaque), and one region (0.5%) is a case of recurrent inversion in African great apes and gibbon (Supplemental Table S5; Fig. 2).
Figure 2.
Evolutionary history of inversions. The pie chart in the middle reports a summary of the evolutionary analyses for all the inversions detected by Strand-seq. The four histograms report the number of inversions that occurred in each lineage, with detail on what was already known from previous literature and what has been determined for the first time in the present study.
Finally, we explored the shared inversion pattern for 536 regions (including all the 238 inversions identified in the current study), which were identified as inverted across at least one of six different species (chimpanzee, bonobo, gorilla, orangutan, gibbon, and macaque) compared with human using Strand-seq (current study and previous literature) (Maggiolini et al. 2020; Porubsky et al. 2020a). For each inversion pair, we iteratively considered them as shared when they overlap for at least 50% of the maximum length. This approach is less conservative in assigning shared inversions, and therefore, numbers slightly differ compared with those estimated in the above analysis.
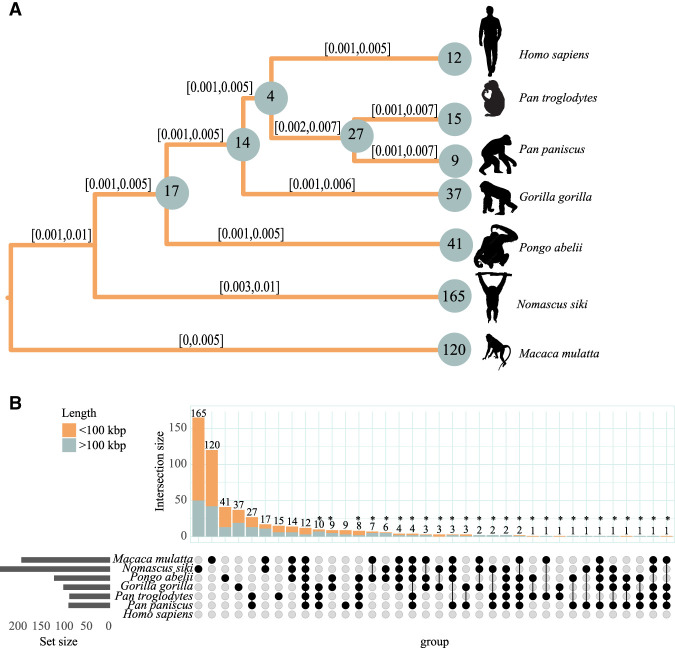
When the species-specific inversions were investigated, we observed a high number of private inversions for NSI, even higher than the one observed for macaque, despite the fact that the latter is the phylogenetic outgroup (Supplemental Fig. S5). Specifically, we observed 165 unique inversions for NSI and 120 for macaque. Furthermore, we identified 12 human-specific inversions, for which the inverted haplotype relative to the human reference genome (GRCh38/hg38) was observed for all the nonhuman species investigated. We estimated the inversion rate as approximately six (5.86) autosomal inversions for a million years, similar to previous estimates reported for great apes (Porubsky et al. 2020a). We constructed a Bayesian evolutionary tree and observed a median rate ranging from 0.0025 to 0.0037 inversion per locus for all the investigated primates with the exception of NSI, which showed a substantially higher (1.7- to 2.5-fold) branch rate (median = 0.0063, 95% confidence interval = 0.0028, 0.0097) (Fig. 3A). Recent surveys suggested that variation at inversion loci in gibbons is mostly owing to long (>100-kbp) variants, with a minor role played by shorter ones (Roberto et al. 2007; Carbone et al. 2014). However, we show that 116 out of 165 gibbon-specific inversions are <100 kbp in size.
Figure 3.
High inversion rates in Nomascus revealed by distance-based and Bayesian analysis. (A) Evolutionary tree based on Markov chain Monte Carlo based on a set of 536 inversions considering both homozygous and heterozygous presence (see Methods). For each branch, the 95% posterior density confidence interval is reported. Numbers on nodes refer to shared inversion presence on the underlying subtrees, and numbers on branches refer to private inversions. (B) Upset plot of a set of 536 inversions. The upper barplot refers to the number of inversions observed in the species indicated in the intersection matrix. The left barplot refers to the total number of inversions identified for each species. Bar colors refer to the length of the inversions, as shown in the legend. Inversions putatively assigned to incomplete lineage sorting and/or recurrence were annotated with an asterisk. The silhouette of the chimpanzee is created by T. Michael Keesey and Tony Hisgett (PhyloPic; http://phylopic.org/; image is under a Creative Commons Attribution 3.0 unported license); silhouettes of bonobo and gorilla are from PhyloPic under a Public Domain Dedication 1.0 license, and the silhouette of the macaque is from PhyloPic under a Public Domain Mark 1.0 license.
Chromosome distributions and genomic features
Inversions ranged in size from 7 kbp to 17.7 Mbp, with heterozygous events being smaller in size than homozygous inversions (Supplemental Fig. S3A). Inversions were distributed among all chromosomes, and generally, the number of inversions showed a positive correlation with the human chromosome size (R² = 0.3673) (Supplemental Fig. S3B). An exception is Chromosome 17, with the highest number of inversions (n = 16) and a relatively small chromosome size. Taking into account only the gibbon-specific inversions and their density along the gibbon chromosomes, the correlation between the number of inversions and chromosome size is less strong (R² = 0.3379) (Supplemental Fig. S3C). Furthermore, comparing the density data of our inversions with the large-scale rearrangement rate for the human–gibbon synteny blocks, there is no evidence of a correlation between the most rearranged chromosomes at the karyotype level and those with the highest number of small-scale inversions identified here (Supplemental Fig. S3C). A similar result was obtained reproducing the analysis in great apes and macaque using previously published data on small-scale inversions (Maggiolini et al. 2020; Porubsky et al. 2020a) and large chromosomal rearrangements (Supplemental Fig. S3D,E; Ventura et al. 2007, 2011).
Next, considering the SD content at inversion BPs, we found that 57 out of 238 inversions (23.5%) have human SDs mapping at both BPs. Moreover, our data show that inversions flanked by SDs at both BPs are significantly (P = 7.18 × 10−6) larger than those without SDs (Supplemental Fig. S6). Of note, considering only the gibbon-specific inversions, only 9.7% are flanked by SDs (Supplemental Fig. S3F).
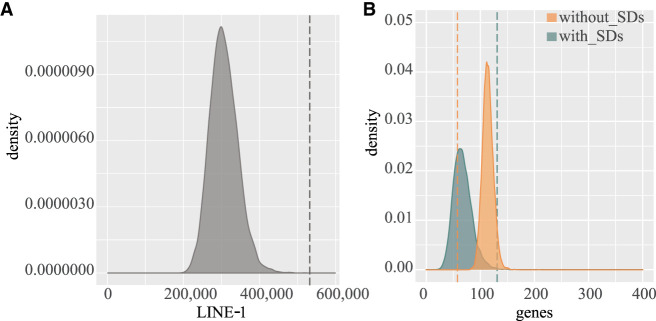
Next, we analyzed the repeat content of the regions harboring the BPs for the remaining gibbon-specific inversions devoid of SDs. Because our inversion coordinates are relative to the human reference genome, we attempted to convert the inversion genomic coordinates between the human GRCh38 and gibbon Nleu3.0/nomLeu3 assemblies. Unfortunately, the draft state of the unfinished gibbon reference genome prevented us from successfully resolving this task. We therefore analyzed the repeat content of the BP regions in human, and we found that they are enriched in two classes of repeats, class I of transposable long interspersed nuclear elements (LINE-1) and long terminal repeat (LTR) of retrotransposon families, with high statistical significance (P < 0.001). The same regions are instead depleted of other classes of repeats, that is, class II of transposable long interspersed nuclear elements (LINE-2) (Fig. 4A; Supplemental Fig. S7).
Figure 4.
Gene density and repeat content at inversion breakpoints. (A) The panel shows data comparisons for LINE-1 between 10,000 random clusters of shuffled regions (gray area) and gibbon-specific BP regions devoid of SDs (dashed line) in human GRCh38. (B) The panel shows the gene content of 10,000 random clusters of shuffled regions, chosen in genomic segments with (green area) and without (orange area) SDs, compared with the gene content at gibbon-specific inversion BPs with (green dashed line) and without (orange dashed line) SDs.
Another interesting question to ask is how many genes map within inverted regions. We identified a total of 2013 human RefSeq-curated genes inside inversions, of which 891 map within gibbon-specific events (Supplemental Table S8) and may have contributed to divergence and speciation. Moreover, because inversions can influence species evolution by disrupting genes or creating new fusion genes, we searched for genes mapping at their BPs. We found a total of 119 out of 238 inversions (50%) that overlap human RefSeq genes for at least one BP, and we obtained a list of 586 genes potentially affected by inversion events (Supplemental Table S9). Taking into account these genes, we performed a Gene Ontology analysis applying the ToppFun default parameters on the ToppGene portal. Our analysis revealed a high percentage of genes involved in the olfactory response (P-value = 4.48 × 10−21) (Supplemental Table S10). Considering only gibbon-specific inversions, we found that 49 out of 121 interrupt 182 human genes (Supplemental Table S9). We performed a Gene Ontology analysis on these genes, highlighting an enrichment in those involved in the perception of smell, detection of stimulus, and nervous system processes (Supplemental Tables S11, S12).
By comparing gibbon-specific inversion BP regions with clusters of regions that were randomly selected in the genome, the regions harboring inversion BPs within SDs showed a significant enrichment in gene content (P-value = 8.15 × 10−5), whereas a depletion of genes was observed within BPs devoid of SDs (P-value = 1.08 × 10−8) (Fig. 4B).
Interspecies inversion polymorphisms and reference errors
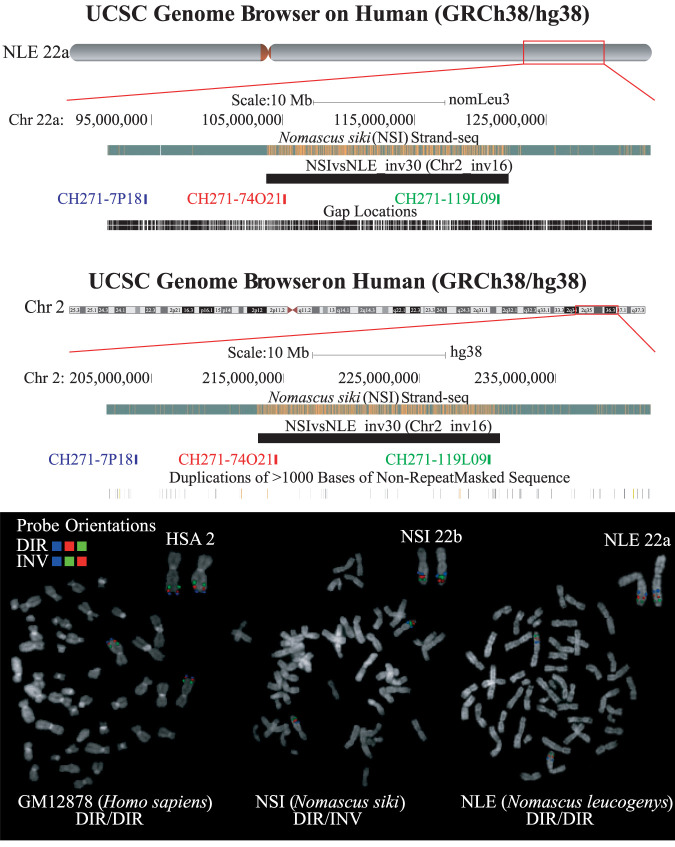
Given the high interspecific variation in gibbons, we analyzed a southern and a northern white-cheeked gibbon, NSI and NLE, respectively. The NSI is the individual that was strand sequenced, and the NLE is the one that was sequenced as part of the gibbon genome sequencing project and for which the reference genome is available (GGSC Nleu3.0/nomLeu3). We first merged 62 Strand-seq selected libraries into a high-coverage and directional composite file that we aligned against the GGSC Nleu3.0/nomLeu3 reference genome. Next, we manually scanned the Strand-seq data with the intent to identify inverted regions between the two gibbon genomes. We focused on regions >500 kbp that are testable by FISH. Using this approach, we initially identified 29 regions potentially inverted between NSI with respect to the NLE reference (Supplemental Table S13). Of note, 13 out of 29 were also detected in the comparison between NSI and the human reference genome so, apparently, the NLE reference individual shared the same orientation with human for those regions. For three out of 13 (Chr18_inv2, Chr15_inv8, and Chr2_inv16), the human-like orientation in NLE was also supported by BES pair mapping (Supplemental Table S7). We selected 15 out of 29 regions and tested them by FISH in human, NLE, and NSI (Fig. 5; Supplemental Table S6; Supplemental Fig. S8). Among these, 12 out of 15 (80%) resulted to be artifacts owing to misassemblies in the gibbon reference (Supplemental Fig. S8). Ten of these regions appeared as homozygous inversions and were simply assembled in the wrong orientation, whereas two appeared directly compared with the human reference and heterozygous compared with the gibbon reference. In the last two cases, the regions are annotated on a wrong chromosome in the gibbon reference; that is, NSIvsNLE_inv9 inversion maps on chromosome NLE6 but is annotated on NLE7, and NSIvsNLE_inv25 inversion is mapping on chromosome NLE10 but is annotated on NLE17 (Supplemental Table S13). Moreover, comparing cytogenetics data reported by http://www.biologia.uniba.it/gibbon/index.html (Carbone et al. 2014) with the gibbon assembly, we found one additional assembly error (NSIvsNLE_inv13): This synteny block maps downstream with respect to the centromere and not upstream as shown in the Nleu3.0 assembly. Finally, three out of 15 regions are real polymorphisms between NLE and NSI, and of note, all of them are heterozygous in our Strand-seq NSI individual (Fig. 5).
Figure 5.
FISH validation of a 17-Mbp inversion. NSI Strand-seq view of NLE Chromosome 22a and HSA Chromosome 2 shows the incomplete switch in the orientation of 17 Mbp against both reference genomes, suggesting the presence of a heterozygous inversion (Chr2_inv16). Metaphase FISH experiments in human, NSI, and NLE show that the region is inverted in heterozygous state only in NSI and is in direct orientation in both human and NLE. (HSA) Homo sapiens; (NSI) Nomascus siki; (NLE) Nomascus leucogenys.
Discussion
Gibbons have a fundamental role in hominoid evolutionary studies because they are the most diverse group of living apes, characterized by different numbers of chromosomes and extensive genome reshuffling both within and among genera. Previous studies have suggested an increased chromosomal rearrangement rate in the white-cheeked gibbon (Nomascus leucogenys), making it an ideal model to study structural variation in primates (Müller et al. 2003) and provide insight into rearrangement formation mechanisms and identify genes that potentially contribute to the evolution of gibbons. Recently, single-cell template strand sequencing was used to successfully discover inversions in great apes and macaque (Maggiolini et al. 2020) genomes. In this work, we challenged the ability of Strand-seq to discover inversions in the southern white-cheeked gibbon and showed the power of this method even in highly shuffled genomes. Unlike great apes and macaque, gibbon genomes are extensively rearranged, with more than 100 large synteny blocks relative to human, and localized on different chromosomes (Roberto et al. 2007; http://www.biologia.uniba.it/gibbon/chromosomes/Fig_3_NLE_synteny.html). By adapting Strand-seq data analysis, under extensive guidance of cytogenetic data (Roberto et al. 2007), and considering each synteny block as an individual chromosome when mapping directional reads to the human reference genome, we were able to create the most comprehensive inversion map between human and gibbon to date, consisting of 238 inversions ranging in size between 7 kbp and 17.7 Mbp. All the inversions have been described here for the first time in the Nomascus genome (Fig. 1B), and 80 of them were previously shown to be inverted in other primate genome analyses (Supplemental Table S5).
The 238 inversions are distributed along all human autosomes, and their density is positively related to the chromosome size, with the exception of Chromosome 17, which shows a high number of inversions despite its small size (Supplemental Fig. S3B). Chromosome 17 is highly enriched in human protein-coding genes (with the second highest gene density in the genome), as well as in SD and repetitive elements (Zody et al. 2006), suggesting that these genomic features may predispose this chromosome to structural events such as inversions. Taking into account only Nomascus-specific inversions, we analyzed their distribution along the autosomes, showing that there is not a significant correlation between large-scale rearrangements and the number of inversions for each chromosome (Supplemental Fig. S3C). This suggests that the mechanisms underlying small-scale structural variation and gibbon karyotype instability might be different. Extending the analysis to previously published small-scale lineage-specific inversions, we confirmed this trend also in macaque and great apes (Supplemental Fig. S3D,E).
To reconstruct the lineage specificity of inversions, we took advantage of published Strand-seq data from great ape and macaque genomes (Maggiolini et al. 2020; Porubsky et al. 2020a) and determined that 121 inversions are gibbon specific (Fig. 2), a number fivefold and 12-fold higher than the ones reported in similar studies for macaque and bonobo, respectively (Maggiolini et al. 2020; Mao et al. 2021). Because the impact of SDs in inversion onset is well documented, we analyzed their content at the inversion BPs (Supplemental Fig. S6). In contrast to inversions that occurred in human and great ape lineages, where 31% (51 out of 165) map to regions of SDs (Maggiolini et al. 2020), our study shows that only 9% (11 out of 121) of overall gibbon-specific inversion BPs map to sites of SDs and are therefore likely mediated by nonallelic homologous recombination (NAHR) (Maggiolini et al. 2020; Porubsky et al. 2020a). This feature is shared with Old World monkeys (10 out of 184, i.e., 5.4%, of macaque inversions mediated by SDs) in contrast to the rest of Hominidae (with 31% of inversions having SDs at the BPs) (Maggiolini et al. 2020). NAHR is clearly less active in macaque and lesser apes than in great apes and correlates with SD architectural differences within their genomes (Marques-Bonet et al. 2009; Carbone et al. 2014; Warren et al. 2020). In search of an explanation for the abundance of structural changes in gibbons, we analyzed the repeat content of the white-cheeked gibbon-specific BP sites present in GRCh38. Breakage regions were found to colocalize with high statistical significance with LINE-1 and LTR elements, suggesting that microhomology-mediated mechanisms might underlie gibbon genome instability. This is consistent with previous evidence, uncovered by the first gibbon genome assembly, that the appearance of lesser ape–specific transposable elements (i.e., LAVA elements) might be associated with the accelerated karyotype evolution in the gibbon lineage (Carbone et al. 2014). Nevertheless, the incomplete nature of the current Nomascus reference genome hampered an investigation of gibbon-specific sequence motifs; therefore, conclusions must be tempered until a higher-quality genome assembly is available.
Among the 121 gibbon-specific inversions, 22% are heterozygous and indicative of inversion polymorphisms in Nomascus (Supplemental Fig. S3A), a number significantly higher than the one reported for great apes (11%) (Porubsky et al. 2020a) and macaque (4%) (Maggiolini et al. 2020). Of note, 70% of the heterozygous inversions are <50 kbp compared with 40% of the homozygous inversions (P-value = 0.0002). It is well known that large heterozygous inversions can be detrimental because they can lead to the loss of unbalanced gametes resulting from recombination. Small inversions in the heterozygous state might not synapse regularly in meiosis, reducing the risk of deleterious crossovers (Torgasheva and Borodin 2010). Consequently, small inversions might have a limited negative impact on the fitness while contributing to gibbon differentiation and speciation. We attempted to gain more insights into the polymorphic state of the detected inversions in N. siki by mapping Strand-seq data against the N. leucogenys reference genome (GGSC Nleu3.0/nomLeu3), another species of the same genus. However, cytogenetic validation of 15 identified inversions showed that 80% of these regions are artifacts owing to errors in the Nomascus reference assembly. In total, we identify 49 Mbp of DNA (Supplemental Table S13), comprising 381 genes, where the sequence has been assembled in the wrong orientation in the gibbon reference genome, highlighting the importance of improving the quality of primate assemblies in order to facilitate additional comparative analyses that might have important applications for evolutionary and biomedical studies.
Because of the dynamic nature of the gibbon genome, we analyzed the distribution of inversions across different species. Taking advantage of great apes and macaque Strand-seq published data (Maggiolini et al. 2020; Porubsky et al. 2020a), we considered a set of 536 nonredundant inversions identified in these species and highlighted a substantially high number of inversions specific to gibbon compared with the other primates, suggesting a complex evolution of structural rearrangements in the investigated species (Supplemental Fig. S5). This excess is mirrored in the inversion rate estimated using a Bayesian phylogenetic reconstruction approach, for which an approximate twofold higher inversion rate is shown for Nomascus (Fig. 3A). Of note, in contrast to what was previously described (Roberto et al. 2007; Girirajan et al. 2009; Carbone et al. 2014), we showed that the higher inversion rate of the gibbon genome with respect to other primates is mirrored also for small-scale inversions (<100 kbp) (Fig. 3B). All these observations point toward a high evolutionary rate for inversions, suggesting that accelerated evolution of these events may have played a role in the high sympatric species of the genus.
Out of the 536 inversions analyzed, 461 are consistent with the known phylogenetic relationship among the analyzed species, whereas 75 could be instances of incomplete lineage sorting or recurrence. However, it is also important to take into account the fact that our sample size (n = 2) prevents the identification of polymorphic inversions. Therefore, it might be possible that some inconsistent inversions are polymorphic in some species, but we failed to observe them. In this context, it is also relevant that we are analyzing a presence/absence pattern using a conservative approach (similar to Porubsky et al. 2020a), and it might be possible that we are pooling together evolutionarily different inversions. In the future, combining high-confidence long-read sequencing and Strand-seq analysis from multiple individuals will ultimately allow fine-grain analyses of single inversions.
An important question is whether inversions occur in gene regions rather than in intergenic regions, thus potentially affecting gene function and resulting in phenotypic diversity in gibbons. In searching for genes that could be altered by the 121 gibbon-specific inversions, we found that 49 events have BPs overlapping 182 human gene orthologs, with 51 genes mapping within single-copy regions and 131 to sites of SDs. The functional redundancy of the latter might make these inversions more tolerable as opposed to disruptions of the 47 unique genes. Gene Ontology analysis on the overall 182 human orthologs reveals the presence of genes involved in sensory perception of smell, detection of stimulus, and nervous system processes (Supplemental Tables S11, S12). Furthermore, density analyses show a nonrandom distribution of inversion BPs with an enrichment for repetitive elements and duplicated genes (Fig. 4A,B, respectively).
Together, these results contribute to our knowledge on structural variation in primate genomes and are exceptionally informative in terms of what genes are potentially involved in the evolution of gibbons. We believe this work will lay the ground for future functional studies and will eventually contribute to our understanding on the mechanisms responsible for gibbon diversification and adaptation. In the future, high-quality and complete reference genome assemblies and gene annotations, as well as long-read sequencing of several individuals, will eventually provide resources to address the molecular mechanisms underlying inversion formation in gibbons and test the effects of inversions and their role in speciation.
Methods
Raw data processing and inversion definition
Strand-seq libraries were prepared for the NSI cell line as previously described (Sanders et al. 2017) and were sequenced on NextSeq 5000 (MID-mode, 75-bp paired-end protocol). Raw data from single-cell sequencing were demultiplexed based on barcodes and converted to FASTQ files using Illumina standard software (bcl2fastq, version 1.8.4). FASTQ reads were mapped to the human (GRCh38/hg38) and NLE (GGSC Nleu3.0/nomLeu3) reference genomes using BWA aligner (version 0.7.15) (Li and Durbin 2009). BAM files were sorted using SAMtools (version 1.3.1-foss-2016b) (Li and Durbin 2009), and duplicate reads were marked using biobambam2 (version 2.0.76) (Tischler and Leonard 2014).
We performed inversion analyses as previously described (Sanders et al. 2016, 2017; Porubsky et al. 2020b). Briefly, we filtered BAM files for low mapping quality reads (mapq < 10) by means SAMtools view, and we selected chromosomal regions that inherited Watson (W; “−”) and Crick (C; “+”) strands. To assess inversions, we generated a directional “composite” file from multiple single cells combining the reverse complement of the reads in the WW BAM files with the reads in CC BAMs for the corresponding chromosome. To perform this step, WW BAM files were first converted to BED format and then back again to BAM files using BEDTools bamtobed and bedtobam, respectively (version v2.29.0) (Quinlan and Hall 2010).
To bioinformatically identify inversions, “composite” merged and sorted files by means SAMtools merge and sort, respectively, were used as input for the R package BreakpointR (daewoooo, n.d.-a) (Porubsky et al. 2020b), with a “windowsize” defined by a size of 50 kbp.
Read-depth analysis
Read-depth analysis was performed using DELLY (version 0.8.7) (Rausch et al. 2012). In detail, read-depth profile for each synteny block was defined starting from the corresponding final composite BAM file: reads in 3-kbp mappable windows (-i 3000) were counted, and the coverage was normalized using the human (GRCh38 based) mappability map (supplied by the software) and genome FASTA files (-m and -g options, respectively). The obtained cov.gz outputs were intersected with the BED coordinates of 444 regions for which Strand-seq showed a switch in read directionality, and the average copy number value for each interval was calculated. All the regions with a copy number 1.5-fold higher than the average copy number of the synteny block were considered as copy-variable regions.
FISH analysis
Metaphase chromosomes and interphase nuclei were obtained from lymphoblastoid cell lines of one human (Coriell Cell Repository, GM12878), one N. leucogenys (Asia), and one N. siki kindly provided by S. Muller (Munich). Three-color FISH experiments were performed using human fosmid (n = 3) and BAC (n = 9) clones, as well as gibbon BAC (n = 33) clones (Supplemental Table S6) directly labeled by nick-translation with Cy3-dUTP (PerkinElmer), Cy5-dUTP (PerkinElmer), and fluorescein-dUTP (Enzo) as previously described (Lichter et al. 1990), with minor modifications. Briefly, 300 ng of labeled probe was used for the FISH experiments; hybridization was performed at 37°C in 2 × SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, and 3 mg sonicated salmon sperm DNA in a volume of 10 mL. Posthybridization washing was at 60°C in 0.1 × SSC (three times, high stringency, for hybridizations on gibbon and human when using species-specific clones) or at 42°C in 2 × SSC (four times each for cross-species hybridizations). Nuclei were simultaneously DAPI stained. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5, and fluorescein fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software. FISH on interphase nuclei was tested through three-color FISH using two probes within the predicted inversion and a reference probe outside. FISH on metaphase chromosomes was performed using two probes within the inverted region.
BES sequence paired mapping
Gibbon BESs (CHORI-271) reported by Carbone et al. (2006) and used to generate the NLE reference genome assembly were aligned against the human reference genome (GRCh38/hg38) as part of a three-step process (recruitment, quality rescoring, and pairing) optimized and published by Tuzun et al. (2005). BACs spanning regions in the same orientation as in human are concordant in size and orientation of the ends, whereas clones spanning inversion BPs are discordant because they have end pairs that are incorrectly oriented and map abnormally far apart when mapped to the human reference genome sequence. BES sequence profiling of 102 BAC clones was used to study the orientation of 238 predicted inversions (Supplemental Table S7).
Gene Ontology analysis
Genes at the inversion BPs were extracted from the curated subset of the RefSeq track from the UCSC Genome Browser. The obtained gene list has been analyzed using the ToppGene portal (Chen et al. 2009; https://toppgene.cchmc.org/), which is a one-stop portal for gene list enrichment analysis and candidate gene prioritization based on functional annotations and protein interaction networks. In particular, the ToppFun function has been used to detect functional enrichment of genes based on transcriptome, proteome, regulome (TFBS and miRNA), ontologies (GO, pathway), phenotype (human disease and mouse phenotype), pharmacome (drug–gene associations), literature cocitation, and other features.
Phylogenetic analysis
To explore the inversion evolutionary pattern among primates, we integrated the inversion data for NSI with that available for Macaca mulatta (Maggiolini et al. 2020) and great apes (Porubsky et al. 2020a). In doing so, we considered evolutionarily shared inversions if their overlap was at least 50% of the longest possible inversion. We then constructed a binary matrix of presence (one) or absence (zero) for each inversion across the seven considered species, without considering heterozygosity. Similar to Porubsky et al. (2020a), we estimated the Manhattan distance (Porubsky et al. 2020a) and performed complete hierarchical clustering relying on the presence/absence of inversions.
Evaluating the abundance of small inversions
To compare the number of small inversions in NSI with those observed for other primates, we constructed a matrix of shared inversions using the same approach as before, but restricting our analysis only to inversions <100 kbp. The resulting matrix was used to build an upset plot, shown in Figure 3B.
Estimating inversion rates
We performed two different inversion rate estimates previously described (Porubsky et al. 2020a). First, we estimated the mean fixation rate of simple inversions per million years as the total number of simple specific inversions multiplied by the sum of divergence times among species. Second, we harnessed the Lewis–Markov k model, as implemented in BEAST v2.6.6. Single inversions were modeled as discrete features coded as 0,1,2 and indicating the homozygous human reference, were inverted in just one allele, and were homozygous inverted. We used the same settings and priors used by Porubsky et al. (2020a) with slight modifications. In detail, given that we were interested in the inversion rate inference and that the phylogenetic relationships among species are established, we defined macaque as outgroup and the Homo-pan clade as unilinear. Specifically, we set the TMRCA prior for macaque as a lognormal with M = 2.9 and S = 0.1, whereas the same distribution for TMRCA for the Homo-pan clade was set with M = 1.61 and S = 0.1. To achieve an effective sample size higher than 200, we performed 1 × 109 iterations, recording every 1000 samples. Convergence was tested using Tracer 1.7, and the maximum clade credibility tree was obtained summarizing the 10,000 obtained trees using treeAnnotator. The resulting tree was plotted using FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Repeats and gene density analyses
A cluster of regions, including all gibbon-specific inversion BPs devoid of SDs in human GRCh38/hg38, was generated. For BPs <2 kbp, a surrounding region of this size was instead considered. Ten thousand random clusters of matching-size regions were extracted from the human reference genome using the BEDTools shuffle function, excluding SDs, centromeres, telomeres, and sequencing gaps. The density in base pair of repeats was evaluated for each cluster using BEDTools intersect function. ggplot2 in R (R Core Team 2022) was used to draw the data in Figure 4A and Supplemental Figure S7.
Similarly, a second cluster of regions, including all gibbon-specific inversion BPs with SDs in human GRCh38/hg38, was generated and used to generate 10,000 random clusters of matching-size regions. The density in the number of genes was individually evaluated for both clusters (with and without SDs) using BEDTools intersect function. ggplot2 in R (R Core Team 2022) was used to draw the data in Figure 4B.
Data access
The Strand-seq library sequence data generated from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA828390.
Supplementary Material
Acknowledgments
This work was supported by a “Fondi di Ateneo, University of Bari” grant (grant number CUP H92F17000190005) to F.A.
Author contributions: F.A. and F.A.M.M. designed the study. A.D.S. and P.H. performed single-cell libraries construction. A.L. performed sequencing and bioinformatic data analysis. P.D.A. and A.L. performed statistical analysis. F.M. performed phylogenetic analysis. D.P. and L.M. performed FISH experiments. F.A., F.A.M.M., L.M., A.D.S., M.V., C.R.C., and J.O.K. contributed to data interpretation. F.A. and F.A.M.M. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276960.122.
Competing interest statement
The authors declare no competing interests.
References
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Carbone L, Stanyon RR, Marra A, Yang F, Whelan CW, de Jong PJ, Rocchi M, Archidiacono N. 2012. A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons. Genome Res 22: 2520–2528. 10.1101/gr.138651.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BF, Zhu B, Osoegawa K, Mootnick A, Kofler A, Wienberg J, Rogers J, Humphray S, et al. 2006. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet 2: e223. 10.1371/journal.pgen.0020223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B, et al. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature 513: 195–201. 10.1038/nature13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catacchio CR, Maggiolini FAM, D'Addabbo P, Bitonto M, Capozzi O, Lepore Signorile M, Miroballo M, Archidiacono N, Eichler EE, Ventura M, et al. 2018. Inversion variants in human and primate genomes. Genome Res 28: 910–920. 10.1101/gr.234831.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10: 1784. 10.1038/s41467-018-08148-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Corbo M, Lewin HA. 2021. Vertebrate chromosome evolution. Annu Rev Anim Biosci 9: 1–27. 10.1146/annurev-animal-020518-114924 [DOI] [PubMed] [Google Scholar]
- Ebert P, Audano PA, Zhu Q, Rodriguez-Martin B, Porubsky D, Bonder MJ, Sulovari A, Ebler J, Zhou W, Serra Mari R, et al. 2021. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372: eabf7117. 10.1126/science.abf7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Reuter G. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan PF, He K, Chen X, Ortiz A, Zhang B, Zhao C, Li YQ, Zhang HB, Kimock C, Wang WZ, et al. 2017. Description of a new species of Hoolock gibbon (Primates: Hylobatidae) based on integrative taxonomy. Am J Primatol 79: e22631. 10.1002/ajp.22631 [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Koury SA, Phadnis N, Schaeffer SW. 2019. How chromosomal rearrangements shape adaptation and speciation: case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol 28: 1283–1301. 10.1111/mec.14923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner-Delgado C, Villatoro S, Lerga-Jaso J, Gayà-Vidal M, Oliva M, Castellano D, Pantano L, Bitarello BD, Izquierdo D, Noguera I, et al. 2019. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat Commun 10: 4222. 10.1038/s41467-019-12173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK, et al. 2009. Sequencing human–gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res 19: 178–190. 10.1101/gr.086041.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64. 10.1038/nature06862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Farré M, Auvil L, Capitanu B, Larkin DM, Ma J, Lewin HA. 2017. Reconstruction and evolutionary history of eutherian chromosomes. Proc Natl Acad Sci 114: E5379–E5388. 10.1073/pnas.1702012114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. 1990. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247: 64–69. 10.1126/science.2294592 [DOI] [PubMed] [Google Scholar]
- Maggiolini FAM, Cantsilieris S, D'Addabbo P, Manganelli M, Coe BP, Dumont BL, Sanders AD, Pang AWC, Vollger MR, Palumbo O, et al. 2019. Genomic inversions and GOLGA core duplicons underlie disease instability at the 15q25 locus. PLoS Genet 15: e1008075. 10.1371/journal.pgen.1008075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiolini FAM, Sanders AD, Shew CJ, Sulovari A, Mao Y, Puig M, Catacchio CR, Dellino M, Palmisano D, Mercuri L, et al. 2020. Single-cell strand sequencing of a macaque genome reveals multiple nested inversions and breakpoint reuse during primate evolution. Genome Res 30: 1680–1693. 10.1101/gr.265322.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Catacchio CR, Hillier LW, Porubsky D, Li R, Sulovari A, Fernandes JD, Montinaro F, Gordon DS, Storer JM, et al. 2021. A high-quality bonobo genome refines the analysis of hominid evolution. Nature 594: 77–81. 10.1038/s41586-021-03519-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Girirajan S, Eichler EE. 2009. The origins and impact of primate segmental duplications. Trends Genet 25: 443–454. 10.1016/j.tig.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira K, Ishida T. 2010. Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol Phylogenet Evol 55: 454–459. 10.1016/j.ympev.2010.01.032 [DOI] [PubMed] [Google Scholar]
- McBroome J, Liang D, Corbett-Detig R. 2020. Fine-scale position effects shape the distribution of inversion breakpoints in Drosophila melanogaster. Genome Biol Evol 12: 1378–1391. 10.1093/gbe/evaa103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D, Capozzi O, Roberto R, Dell'oglio MP, Rocchi M, Stanyon R, Archidiacono N. 2008. Tracking the complex flow of chromosome rearrangements from the Hominoidea ancestor to extant Hylobates and Nomascus gibbons by high-resolution synteny mapping. Genome Res 18: 1530–1537. 10.1101/gr.078295.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Hollatz M, Wienberg J. 2003. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113: 493–501. 10.1007/s00439-003-0997-2 [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution (N Y) 57: 447–459. 10.1111/j.0014-3820.2003.tb01537.x [DOI] [PubMed] [Google Scholar]
- Nie WH, Wang JH, Su WT, Hu Y, He SW, Jiang XL, He K. 2018. Species identification of crested gibbons. Zool Res 39: 356–363. 10.24272/j.issn.2095-8137.2018.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci 98: 12084–12088. 10.1073/pnas.221274498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, Murnane JP. 2006. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol 26: 1865–1878. 10.1128/MCB.26.5.1865-1878.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky D, Sanders AD, Höps W, Hsieh P, Sulovari A, Li R, Mercuri L, Sorensen M, Murali SC, Gordon D, et al. 2020a. Recurrent inversion toggling and great ape genome evolution. Nat Genet 52: 849–858. 10.1038/s41588-020-0646-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky D, Sanders AD, Taudt A, Colomé-Tatché M, Lansdorp PM, Guryev V. 2020b. breakpointR: an R/Bioconductor package to localize strand state changes in Strand-seq data. Bioinformatics 36: 1260–1261. 10.1093/bioinformatics/btz681 [DOI] [PubMed] [Google Scholar]
- Porubsky D, Höps W, Ashraf H, Hsieh P, Rodriguez-Martin B, Yilmaz F, Ebler J, Hallast P, Maria Maggiolini FA, Harvey WT, et al. 2022. Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders. Cell 185: 1986–2005.e26. 10.1016/j.cell.2022.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Casillas S, Villatoro S, Cáceres M. 2015. Human inversions and their functional consequences. Brief Funct Genomics 14: 369–379. 10.1093/bfgp/elv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Lerga-Jaso J, Giner-Delgado C, Pacheco S, Izquierdo D, Delprat A, Gayà-Vidal M, Regan JF, Karlin-Neumann G, Cáceres M. 2020. Determining the impact of uncharacterized inversions in the human genome by droplet digital PCR. Genome Res 30: 724–735. 10.1101/gr.255273.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358. 10.1016/s0169-5347(01)02187-5 [DOI] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, Mardis ER, Lomiento M, Tuzun E, Cheng Z, Mootnick AR, Archidiacono N, Rocchi M, et al. 2007. Molecular refinement of gibbon genome rearrangements. Genome Res 17: 249–257. 10.1101/gr.6052507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Hills M, Porubský D, Guryev V, Falconer E, Lansdorp PM. 2016. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res 26: 1575–1587. 10.1101/gr.201160.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Falconer E, Hills M, Spierings DCJ, Lansdorp PM. 2017. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat Protoc 12: 1151–1176. 10.1038/nprot.2017.029 [DOI] [PubMed] [Google Scholar]
- Tham WH, Zakian VA. 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21: 512–521. 10.1038/sj.onc.1205078 [DOI] [PubMed] [Google Scholar]
- Tischler G, Leonard S. 2014. biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol Med 9: 13. 10.1186/1751-0473-9-13 [DOI] [Google Scholar]
- Torgasheva AA, Borodin PM. 2010. Synapsis and recombination in inversion heterozygotes. Biochem Soc Trans 38: 1676–1680. 10.1042/BST0381676 [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732. 10.1038/ng1562 [DOI] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246. 10.1126/science.1140615 [DOI] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. 2011. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res 21: 1640–1649. 10.1101/gr.124461.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Salvador D, Puig M, Gayà-Vidal M, Pacheco S, Giner-Delgado C, Noguera I, Izquierdo D, Martínez-Fundichely A, Ruiz-Herrera A, Estivill X, et al. 2017. Detailed analysis of inversions predicted between two human genomes: errors, real polymorphisms, and their origin and population distribution. Hum Mol Genet 26: 567–581. 10.1093/hmg/ddw415 [DOI] [PubMed] [Google Scholar]
- Warren WC, Harris RA, Haukness M, Fiddes IT, Murali SC, Fernandes J, Dishuck PC, Storer JM, Raveendran M, Hillier LW, et al. 2020. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science 370: eabc6617. 10.1126/science.abc6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol 33: 427–440. 10.1016/j.tree.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Wienberg J. 2004. The evolution of eutherian chromosomes. Curr Opin Genet Dev 14: 657–666. 10.1016/j.gde.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al. 2006. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440: 1045–1049. 10.1038/nature04689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.