Abstract
Hepatocellular carcinoma (HCC) is dangerous cancer that often evades early detection because it is asymptomatic and an effective detection method is lacking. For people with chronic liver inflammation who are at high risk of developing HCC, a sensitive detection method for HCC is needed. In a meta-analysis of The Cancer Genome Atlas pan-cancer methylation database, we identified a CpG island in the USP44 promoter that is methylated specifically in HCC. We developed methylation-sensitive high-resolution melting (MS-HRM) analysis to measure the methylation levels of the USP promoter in cell-free DNA isolated from patients. Our MS-HRM assay correctly identified 40% of patients with early-stage HCC, whereas the α-fetoprotein test, which is currently used to detect HCC, correctly identified only 25% of early-stage HCC patients. These results demonstrate that USP44 MS-HRM analysis is suitable for HCC surveillance.
Keywords: Hepatocellular carcinoma, High-resolution melting analysis, Liquid biopsy, Methylation, USP44
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most dangerous cancers and is difficult to detect at early stages (1, 2). Although chronic inflammation caused by factors such as alcohol consumption, hepatitis viral infection, and non-alcoholic steatohepatitis can drive carcinogenesis (3-7), it is challenging to detect HCC in the setting of chronic liver disease. Ultrasonography and the α-fetoprotein (AFP) test have been used to detect HCC, but their sensitivities for early-stage HCC are not high enough to provide sufficient surveillance power for patients with an increased risk of HCC (2, 8).
Analysis of cell-free DNA (cfDNA) obtained from a blood-based liquid biopsy is emerging as an alternative method for early cancer detection. Next-generation sequencing (NGS) analysis of cfDNA for tumor-specific mutations or methylations can successfully identify circulating tumor DNA (ctDNA) in diverse cancer types (9-12). Mutations are highly associated with tumors, but low allele frequencies hamper their detection in the blood (13, 14). Although aberrant methylations are usually shared in the same tumor types, resulting in high tumor associations (15), their associations are not limited to tumors but are often shared with other cell types, causing background noise in blood-based assays. To improve detection, it is essential to find methylation markers with high specificity to the cancer of interest, along with techniques that can differentiate small methylation changes in cfDNA caused by increased ctDNA in the blood. NGS analysis of cfDNA methylation provides good statistical power to address the above requirements, but its cost and technical hurdles hinder its use in routine cancer surveillance.
To develop a liquid biopsy assay for early detection of HCC, we selected HCC-specific methylation markers (HMMs) from a meta-analysis of The Cancer Genome Atlas (TCGA) pan-cancer methylome data: https://www.cancer.gov/tcga. We found that cg22538054, located in the promoter region of ubiquitin-specific peptidase 44 (USP44), is hypermethylated only in HCC, and we confirmed this hypermethylation in HCC using pyrosequencing of tumor and paired normal liver tissue samples obtained from the same patients. We developed a methylation-sensitive high-resolution melting (MS-HRM) assay for the USP44 promoter and tested its feasibility in analysis of liquid biopsy samples to detect early-stage HCC. Clinical validation using tissue and blood samples demonstrated that MS-HRM analysis of a single HMM (cg22538054) can detect HCC with high sensitivity and specificity. In particular, the cg22538054 MS-HRM assay showed 40% sensitivity and 95% specificity for early-stage HCC, higher than the sensitivity of the AFP test (25%). A combined MS-HRM and AFP assay showed higher sensitivity (57% of early HCC and 76% of all HCC) that is sufficient for detecting early-stage HCC in a high-risk group.
RESULTS
Identification of HMMs
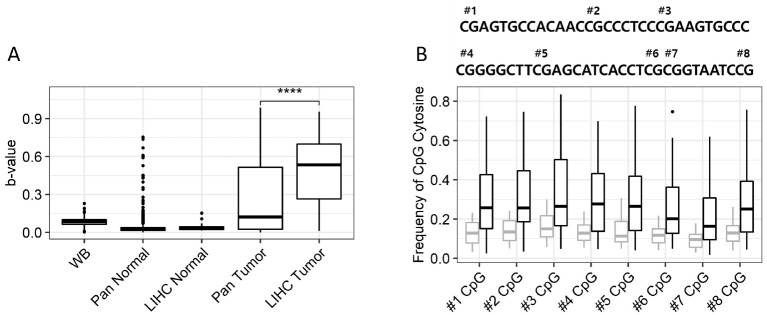
To identify HMMs with minimal cross-reactivity with other tissue types, we examined the methylation levels in non-HCC tissue types of CpG sites that are hypermethylated in HCC using TCGA and Gene Expression Omnibus (GEO) pan-cancer data. In addition, whole blood methylome data from the Korean National Institute of Healthy (KNIH) were used to filter out CpGs with methylation levels of more than 10% in blood. Among the hypermethylated CpGs in HCC, cg22538054 showed less than 10% methylation in all normal tissues, including whole blood cells (Fig. 1A). Although some tumors showed elevated methylation of cg22538054, the methylation levels were considerably lower (<15%) than that seen in HCC. To verify the methylation levels of cg22538054 and the neighboring CpG sites in tumor tissues, we performed bisulfite pyrosequencing of the region using 47 HCC and paired normal tissues. All CpG sites sequenced were significantly hypermethylated in tumor tissues (Fig. 1B). Therefore, we considered hypermethylation of the USP44 promoter to be a good indicator of HCC and developed a PCR-based assay to measure the region’s methylation level in the blood.
Fig. 1.
Methylation of cg22538054 in normal and tumor tissues. (A) The b-value of cg22538054 in public data collected from TCGA, GEO, and KNIH. WB; whole blood, LIHC; liver hepatocellular carcinoma. ****P < 0.0001. (B) Results of pyrosequencing analysis of eight CpG sites including cg22538054 at the USP44 promoter. The sequence of the USP44 promoter region, with CpG sites labeled “#number”, is shown at the top. Boxplot shows the cytosine frequencies of normal (gray) and tumor (black) tissues at CpG sites corresponding to the “#number” label shown above. ****P < 0.0001.
Development of MS-HRM analysis for the USP44 promoter region
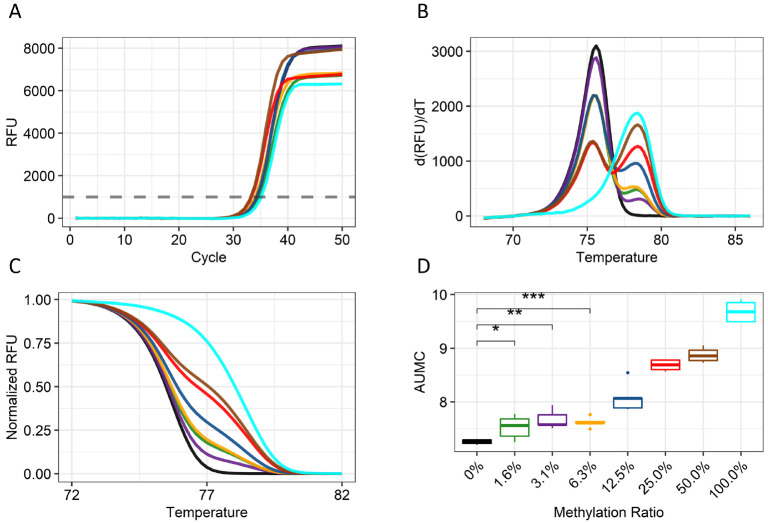
In healthy people, cfDNA are mostly derived from blood cells. When liver cancer develops, DNA fragments are released from the cancer cells and mixed with DNA fragments from other tissues in blood. Therefore, a sensitive, quantitative assay is needed to detect tumor DNA fragments in the cfDNA pool. We designed primers to amplify the promoter region regardless of its methylation status and then measured the methylation levels of the amplified fragments using MS-HRM analysis. Serially diluted bisulfite-treated genomic DNA (gDNA) of Huh-1 liver cancer cells and peripheral blood mononuclear cells (PBMCs) was examined using the assay. The quantification cycle (Cq) of the PCR indicated that all the samples showed similar amplification efficiencies independent of their methylation status (Fig. 2A). Subsequent HRM analysis revealed two distinct peaks, one at 76°C and the other at 78°C, representing unmethylated fragments of the USP44 promoter region from PBMCs and methylated fragments of the USP44 promoter region from Huh-1 cells, respectively (Fig. 2B). The peak size of the higher temperature increased gradually with the percentage of Huh-1 DNA in the sample. Similarly, the slope of the melting curve of the amplified fragments decreased as the amount of Huh-1 DNA increased (Fig. 2C). Quantification of the area under the methylation curve (AUMC) was sensitive enough to detect 1.6% methylation in 1 ng of input DNA, representing a few copies of methylated DNA in 300 genome equivalents of human DNA (Fig. 2D). A significant correlation between methylation level and AUMC value was also observed. These results demonstrate that MS-HRM analysis can quantitatively measure the methylation level of a tumor marker.
Fig. 2.
Melting peak and curve analysis of the cg22538054 MS-HRM assay. (A) Relative fluorescence units (RFUs) values of the amplification for samples with increasing levels of methylation. (B) Melting peak plot of each serially diluted sample at the indicated temperature. The melting peak is derived by taking a negative number to the differential value of the melting curve. (C) A normalized melting curve plot of each serially diluted sample is shown. (D) A boxplot of the AUMC values of the curves of serially diluted samples based on the normalized melting curve. The experiment was done a total of five times. The color codes for the methylated templates are as follows: 0%, black; 1.56%, green; 3.12%, dark orchid; 6.25%, orange; 12.5%, blue; 25.0%, red; 50.0%, brown; and 100%, cyan.
Clinical validation of the HRM assay
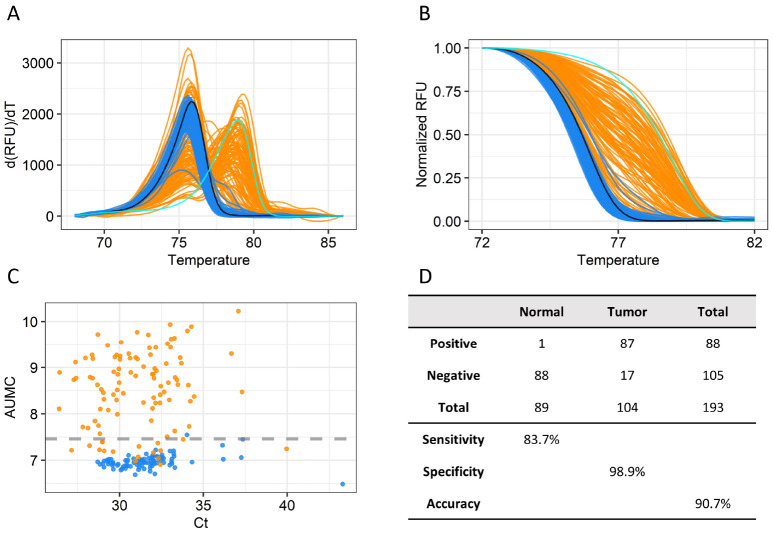
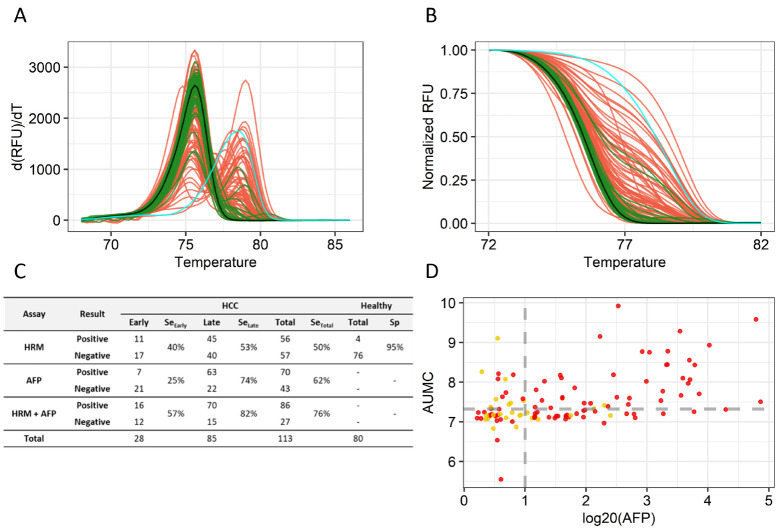
We first analyzed clinical samples (104 HCC tumor tissues and 89 paired normal liver tissues) to examine the accuracy of the cg22538054 MS-HRM assay (Supplementary Table 1). The melting peaks and curves of the amplified fragments from normal tissues overlapped with those of PBMCs, indicating that the USP44 promoter region was not methylated in normal tissues (Fig. 3A). However, the melting peaks and curves of most of the amplified fragments from HCC samples were strongly shifted toward those of the methylated promoter region. The AUMC values of tumor samples were significantly higher than those of normal tissues (Fig. 3B). We set the 99th percentile (7.463) of AUMC values of normal tissues as the cut-off distinguishing normal and tumor samples and observed that 83.7% of tumor tissues had AUMC values above this cut-off (Fig. 3C). The high accuracy of the assay in detecting tumors regardless of HCC subtypes with only one methylation marker encouraged us to test its ability to detect cancer DNA in blood samples from HCC patients and healthy people. We collected plasma from 80 healthy people and 113 HCC patients and analyzed the samples using our MS-HRM assay (Supplementary Table 2). Like the tissue samples, cfDNA from plasma of healthy people showed melting curves and peaks similar to those of the unmethylated control, whereas that of HCC patients showed varying shifts in melting peaks and curves toward those of the methylated control (Fig. 4A, B). Using a cut-off set to the 95th percentile of AUMC values in healthy people, 40% of early-stage and 53% of late-stage HCC patients were correctly identified as having HCC. The sensitivity of the MS-HRM assay (40%) for early-stage cancer (40%) (Barcelona Clinic Liver Cancer [BCLC] stage A) was higher than that of the AFP test (25%) (Fig. 4C), and many of the HCC patients who received negative results on the AFP test received positive results with the MS-HRM analysis (Fig. 4D). Combining the two tests achieved 57% sensitivity for early-stage cancer and 76% sensitivity for all HCC (Fig. 4C). This result indicates that the high accuracy of the USP44 MS-HRM analysis is a suitable blood-based test for HCC surveillance.
Fig. 3.
MS-HRM assay of HCC tumor and paired normal tissue samples. (A) Melting peak plot of NAT and tumor tissue samples in the indicated temperature range. The color codes are as follows: negative control, black; positive control, cyan; NAT, blue; and tumor tissue, orange. (B) Normalized melting curve plot of NAT and tumor tissue samples. (C) Scatter plots for Cq and AUMC of each tissue sample. The gray dotted line indicates the 99th percentile of AUMC values of NATs. (D) Accuracy of the MS-HRM assay. Sensitivity and specificity are based on a cut-off set to the 99th percentile of AUMC values (7.463) of NATs.
Fig. 4.
MS-HRM assay for blood samples from 80 healthy people and 113 HCC patients. (A) Melting peak plot. The color codes are as follows: negative control, black; positive control, cyan; healthy people, green; and HCC patients, red. (B) Normalized melting curve plot. (C) Diagnostic performance of the MS-HRM assay and AFP test. (D) Scatter plots of AFP and AUMC values for tumor stage (early-stage: BCLC 0, A (yellow dot); late-stage: BCLC B, C, D (red dot)). AFP values were converted to log20. The gray dotted line on the y-axis represents the 95th percentile (7.325) of AUMC values of healthy people, and the gray dotted line on the x-axis sets indicates the tumor cut-off point where log20 (AFP) is 1.
DISCUSSION
Analysis of liquid biopsy samples has emerged as a promising method to detect cancer (16). Cancer-specific biomarkers in the blood are good targets for such analysis. The systemic nature of blood samples makes it difficult to identify the source of cancer-associated markers unless they are particular to a certain cancer type. To develop an assay to detect HCC in blood samples, we performed a meta-analysis of methylome databases containing more than 8,500 patient methylome data for various cancers and normal tissues to discover HMMs. We filtered the HMMs using blood-based methylome data to select those without detectable methylation levels in the blood. Hypermethylation of cg22538054 at the CpG island of the USP44 promoter was primarily associated with HCC. Hypermethylation of the USP44 promoter correlates with its downregulation in HCC as was shown in other tumors (17). This result indicates the suitability of USP44 MS-HRM as an effective analysis of blood-based liquid biopsy samples to detect HCC.
Another hurdle in blood-based cancer detection is the small amount of cancer-specific DNAs in the blood (18). Methylation-specific PCR is typically chosen for this analysis but often gives false-positive results due to nonspecific binding of methylation-specific primers to unmethylated sites when the real binding sites for the primers are limiting (19, 20). To avoid this problem, we developed an assay that can amplify DNA fragments of cancer biomarkers regardless of their methylation status (21). Then we measured differences in their melting characteristics caused by the replacement of C/G pairing with A/T pairing in the unmethylated sites after bisulfite conversion of the cfDNA. The AUMC values of all healthy blood and normal liver tissue samples were low, indicating that there were no false-positive results caused by imperfect analytical specificity of the assay. But our MS-HRM assay was sensitive enough to detect small changes in methylation levels in blood samples from HCC patients.
In summary, we developed an assay that addresses two significant hurdles for liquid biopsy to test for HCC: specific biomarkers and high analytical accuracy. Our MS-HRM assay has high sensitivity and specificity in detecting early-stage HCC in blood. Therefore, the USP44 MS-HRM assay could be used for HCC surveillance in high-risk patients.
MATERIALS AND METHODS
Marker discovery from public data
All public data used for analysis were downloaded from TCGA and GEO (HumanMethylation450 bead array). For raw data from whole blood produced in-house, normalization and batch effects were corrected using ‘Minfi’ R package (v.3.4.3). The signal intensity of raw data was measured and converted into a methylation b-value ranging from 0 (unmethylated) to 1 (fully methylated), and the genome was mapped using the hg19 reference genome.
First, to facilitate blood-based liquid biopsy, hypermethylated differentially methylated probes with b-values of 0.3 or higher in tumor compared to normal tissue were selected. Next, unmethylated probes with b-values of 0.1 or less in more than 90% of normal samples were selected. For the selected probes, HCC diagnostic markers were discovered using the ‘randomForest’ R package (v.3.4.3). For modeling, the data were randomly divided; 90% of the data was used for training and the remaining 10% for testing (with 1,000 iterations). The prediction performance of markers was calculated as the area under the curve (data not shown).
Confirmation of analytical sensitivity using control samples
We used DNA from the Huh-1 liver cancer cell line and human blood gDNA (Roche) in fold-dilution series to determine the limit of detection (LOD) of the MS-HRM assay developed for the USP44 promoter region. Control samples with eight different methylation levels (0%, 1.6%, 3.1%, 6.3%, 12.5%, 25%, 50%, and 100%) were prepared and the LOD experiment using the diluted samples was done five times. AUMC values were derived according to the methylation level of each sample, and t-tests were performed to compare the AUMC values in the 0% sample and the 1.6%, 3.1%, and 6.3% samples.
Clinical samples
Normal adjacent tissues (NATs) and tumor tissues were collected from 47 HCC patients for pyrosequencing. Of the 47 HCC patients, 39 had T1- or T2-stage tumors, and 27 had AFP levels higher than 20.
The performance of the tissue-based MS-HRM assay was confirmed using 89 NATs and 104 tumor tissues from HCC patients. Of the 104 tumor tissues, 96 were at stage T1 or T2. In addition, 60 patients had AFP levels higher than 20, accounting for 57.1% of all HCC patients.
Whole blood from 80 healthy people and 113 HCC patients was collected using Alphaliquid TubesTM and PAXgene ccfDNA Tubes. For later processes, cfDNA was preserved in PAXgene ccfDNA Tubes to stabilize cfDNA and reduce the loss of cellular components like gDNA. Whole blood samples were centrifuged at 1,900 g for 15 min, and then the supernatant was transferred to a new 15-ml conical tube and centrifuged at 1,900 g for 15 minutes to obtain plasma, from which as much debris as possible was removed. Tissue and blood samples approved by the Seoul National University Hospital Institutional Review Board (H-1602-016-739 and H-1809-088-974) were used for pyrosequencing and HRM analysis.
Nucleic acid extraction and bisulfite conversion
gDNA of Huh-1 cells or Human Genomic DNA from human blood (Roche) was extracted using the QIAmp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. cfDNA from plasma of HCC patients and healthy volunteers was extracted using the MagListoTM cfDNA Extraction Kit (Bioneer) according to the manufacturer’s instructions. The extracted gDNA and cfDNA were bisulfite-converted using the EZ DNA Methylation-Lightning Kit (Zymo Research) according to the manufacturer’s instructions and then used in an assay to check the methylation level of each sample.
Pyrosequencing
To amplify a region containing eight CpGs, including cg22538054, we used forward primer 5’-GGGAATGGTTTTAGGAAGTTGA-3’ and reverse primer 5’-biotin-AACCCCATCCCTCCACCCTC-3’. Thermocycling was conducted in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) under the following conditions: initial denaturation at 95°C for 15 min; 45 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 min; and a final extension at 72°C for 10 min. Sequencing was completed using a PyroMark Q48 Autoprep System (Qiagen) according to the manufacturer’s instructions from sequencing providers (Macrogen). We performed pyrosequencing on 47 tumor samples and paired normal tissue samples and compared their methylation levels at eight target CpG sites using t-tests.
MS-HRM analysis
MS-HRM was conducted using a separate reverse primer that can bind to methylated and unmethylated templates to detect trace amounts of ctDNA in cfDNA. The forward primer was designed to enable semi-quantification using the methylation independent primer used in the existing MS-HRM. USP44 promoter PCR amplification was performed in a 25-μl reaction containing 20× AccuPowerⓇ Plus DualStarTM qPCR MasterMix (Bioneer), 2× EvaGreenⓇ (Biotium), 0.4 μM common forward primer (5’-GATGGAGAGAAGGCGGGTAAGA-3’), 0.4 μM methylated reverse primer (5’-CGAAATAATACTCGAAACCCCG-3’), unmethylated reverse primer (5’-CACAAAATAATACTCAAAA CCCCA-3’), and nuclease-free water. For PCR, we used samples corresponding to 0.25 ml of plasma. We calculated the sample volume by dividing the elution volume used in the final bisulfite conversion by the plasma volume used in cfDNA extraction. For MS-HRM analysis, thermocycling was conducted in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) under the following conditions: initial denaturation at 95°C for 5 min; 50 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s; and a final extension at 72°C for 5 min. After PCR amplification, HRM analysis spanned a temperature range from 60°C to 95°C, with a ramp rate of 0.2°C/10 s, and used the fluorescence acquisition setting.
Melting curve normalization and AUMC
To remove exponential background noise from the total fluorescence value according to the measured temperature, we selected temperatures (each TL; 68°C, TR; 86°C) with constant MS-HRM fluorescence change rates within the sections below and above the melting point and measured the fluorescence value at each temperature. The parameters of the exponential background noise were estimated from the rate of change. The exponential background noise values were calculated from the estimated parameters and removed from the total fluorescence values at each temperature point to calculate the fluorescence data function M(T) proportional to the amount of residual double-stranded DNA. Next, the methylation level was derived from the value of the area under the melting curve within each sample’s [68°C, 86°C] section.
ACKNOWLEDGEMENTS
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF), funded by the Ministry of Science & ICT (grant number: 2017M3A9A7050614).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–1718. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang I, Kim JA, Kim J, Lee JH, Kim MJ, Ahn JK. Hepatitis B virus X protein promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells by regulating SOCS1. BMB Rep. 2022;55:220. doi: 10.5483/BMBRep.2022.55.5.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Intino PE, Morselli-Labate AM, et al. Trevisani F', author. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34:570–575. doi: 10.1016/S0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 5.Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jepsen P, Ott P, Andersen PK, et al. Risk for hepatocellular carcinoma in patients with alcoholic cirrhosis: a Danish nationwide cohort study. Ann Intern Med. 2012;156:841–847. doi: 10.7326/0003-4819-156-12-201206190-00004. [DOI] [PubMed] [Google Scholar]
- 7.Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman M. Limitations of screening for hepatocellular carcinoma. Hepat Oncol. 2014;1:161–163. doi: 10.2217/hep.13.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labgaa I, Villacorta-Martin C, D'Avola D, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene. 2018;37:3740–3752. doi: 10.1038/s41388-018-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang P, Chan CW, Chan KA, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:1317–1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin CQ, Yuan CH, Qu Z, et al. Liquid biopsy of hepatocellular carcinoma: circulating tumor-derived biomarkers. Dis Markers. 2016;2016:1427849. doi: 10.1155/2016/1427849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh S, Jo Y, Jung S, Yoon S, Yoo KH. From genome sequencing to the discovery of potential biomarkers in liver disease. BMB Rep. 2020;53:299. doi: 10.5483/BMBRep.2020.53.6.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC-challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15:577–586. doi: 10.1038/s41571-018-0058-3. [DOI] [PubMed] [Google Scholar]
- 14.Stetson D, Ahmed A, Xu X, et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis Oncol. 2019;3:1–9. doi: 10.1200/PO.18.00191. [DOI] [PubMed] [Google Scholar]
- 15.Lee SM, Kim-Ha J, Choi WY, et al. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics. 2016;8:993–1005. doi: 10.2217/epi-2016-0027. [DOI] [PubMed] [Google Scholar]
- 16.von Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut. 2020;69:2025–2034. doi: 10.1136/gutjnl-2019-320282. [DOI] [PubMed] [Google Scholar]
- 17.Kang X, Sun L, Guo K, et al. Serum protein biomarkers screening in HCC patients with liver cirrhosis by ICAT-LC-MS/MS. J Cancer Res Clin Oncol. 2010;136:1151–1159. doi: 10.1007/s00432-010-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJ. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18:1174–1196. doi: 10.1039/C8LC00100F. [DOI] [PubMed] [Google Scholar]
- 19.Herman JG, Graff JR, Myöhänen SBDN, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan VTT, Ha NT, Uyen NQ, et al. Standardization of the methylation‑specific PCR method for analyzing BRCA1 and ER methylation. Mol Med Rep. 2014;9:1844–1850. doi: 10.3892/mmr.2014.1990. [DOI] [PubMed] [Google Scholar]
- 21.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.