Abstract
Circular RNAs are single-stranded RNAs with a covalently closed structure formed by the process of back-splicing. Aberrant expression of circular RNAs contributes to the pathogenesis of a wide range of cancers. Pancreatic cancer is one of the most lethal cancers due to diagnostic difficulties and limited therapeutic options. Circular RNAs are emerging as novel diagnostic biomarkers and therapeutic targets for pancreatic cancer. Moreover, recent advances in the therapeutic application of engineered circular RNAs have provided a promising approach to overcoming pancreatic cancer. This review discusses the roles of circular RNAs in the pathogenesis of pancreatic cancer and in potential treatment applications and their usefulness as diagnostic biomarkers.
Keywords: circular RNA, circRNA, pancreatic cancer, biomarker, treatment
1 Introduction
Pancreatic cancer is the seventh leading cause of cancer-related death worldwide, responsible for approximately 470 000 deaths per year (Sung et al., 2021). The number of deaths related to pancreatic cancer is increasing, and the prognosis remains poor, with a 5-year survival rate of only approximately 11% (Siegel et al., 2022). This poor prognosis is because most patients are diagnosed at advanced stages, and there is a lack of effective treatment options. In fact, the 5-year survival rates of patients with stage 0 and stage IV pancreatic cancer are 85.8% and 2.7%, respectively (Egawa et al., 2012). To improve the prognosis of pancreatic cancer, it is necessary to elucidate the molecular mechanisms underlying the pathogenesis of pancreatic cancer, to enable the development of methods for early diagnosis and effective treatment.
Recent progress in research on circular RNAs (circRNAs), which are single-stranded RNAs with a covalently closed structure, has shown that many circRNAs are dysregulated and involved in the pathogenesis of cancers (Vo et al., 2019). The presence of circRNAs was first reported in pathogens, such as plant viroids (Sanger et al., 1976) and hepatitis delta virus (Kos et al., 1986), followed by the discovery in the 1990s of circRNAs among human transcripts (Nigro et al., 1991; Cocquerelle et al., 1992, 1993; Capel et al., 1993). Over the last decade, advances in RNA sequencing technology and analytical methods have begun to reveal the overall picture of circRNAs in the human transcriptome. To date, more than 180 000 circRNAs have been identified, and their expression patterns have been shown to vary according to tissue or cell type and disease state (Jeck et al., 2013; Memczak et al., 2013; Salzman et al., 2013; Rybak-Wolf et al., 2014; Gao et al., 2015; Dong et al., 2018).
With clarification of the diversity of circRNAs in the human transcriptome, their biological functions are also being elucidated. This review discusses the roles of circRNAs in the pathogenesis of pancreatic cancer and potential therapeutic applications and their utility as diagnostic markers for pancreatic cancer.
2 Roles of circRNAs in the pathogenesis of pancreatic cancer
Although circRNAs have identical exon sequences as those of their linear cognate mRNAs, they have distinct cellular functions. circRNAs can interact with DNA, RNA, and proteins to modulate transcription, act as decoys for miRNAs and proteins, enhance mRNA stability, provide scaffolds for protein complex formation, and serve as templates for translation. In pancreatic cancer, many circRNAs are dysregulated and play important roles in various aspects of cancer progression. Most pathogenic circRNAs function as miRNA decoys, and several circRNAs have been reported to interact with proteins to stabilize mRNA, inhibit post-translational modifications, and provide scaffolds for formation of protein complexes (Figure 1). This chapter focuses on representative circRNAs involved in the pathogenesis of pancreatic cancer for which the detailed mechanisms have been elucidated, and those that could not be introduced in the text are summarized in Table 1.
FIGURE 1.
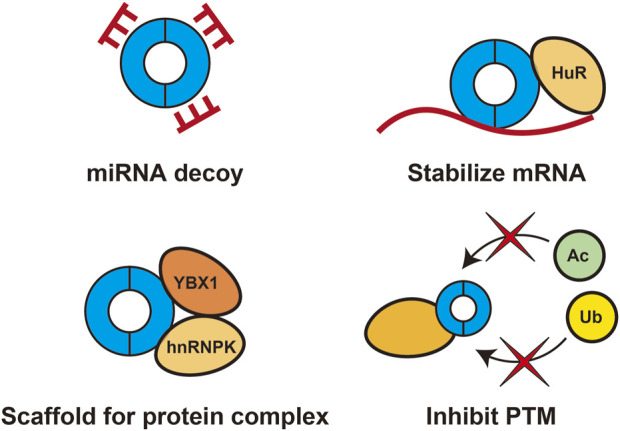
Roles of pathogenic circRNAs in pancreatic cancerPathogenic circRNAs in pancreatic cancer function as miRNA decoys and interact with proteins to stabilize mRNA, inhibit post-translational modifications (PTMs), and provide scaffolds for protein complex formation.
TABLE 1.
Pathogenic circRNAs in pancreatic cancer.
| circRNA | Mechanism | Target | Biological function | Reference |
|---|---|---|---|---|
| circRNA_100782 | miRNA decoy | miR-124/IL6R | Promotes proliferation | Chen et al. (2017) |
| circIARS | miRNA decoy | miR-122/RhoA | Increases endothelial permeability | Li et al. (2018a) |
| circPDE8A | miRNA decoy | miR-338/MACC1 | Promotes invasion | Li et al. (2018b) |
| circZMYM2 | miRNA decoy | miR-335-5p/JMJD2C | Promotes proliferation and invasion; inhibits apoptosis | An et al. (2018) |
| circADAM9 | miRNA decoy | miR-217/PRSS3 | Promotes proliferation and invasion | Xing et al. (2019) |
| circASH2L | miRNA decoy | miR-34a/Notch1 | Promotes proliferation, invasion, and angiogenesis | Chen et al. (2019) |
| circLDLRAD3 | miRNA decoy | miR-137-3p/PTN | Promotes proliferation, invasion, and migration | Yao et al. (2019) |
| hsa_circRNA_0007334 | miRNA decoy | miR-144-3p and miR-577/MMP7 and COL1A1 | Promotes migration | Yang et al. (2019) |
| circ_0000977 | miRNA decoy | miR-153/HIF1A | Helps HIF1A-mediated immune escape | Ou et al. (2019) |
| ciRS-7 (CDR1as) | miRNA decoy | miR-7 | Promotes proliferation and invasion | Liu et al. (2019) |
| circ_0030235 | miRNA decoy | miR-1253 and miR-1294 | Promotes proliferation and invasion; inhibits apoptosis | Xu et al. (2019) |
| circRHOT1 | miRNA decoy | miR-26b, miR-125a, miR-330 and miR-382 | Promotes proliferation, invasion, and migration | Qu et al. (2019) |
| circ_0007534 | miRNA decoy | miR-625 and miR-892b | Promotes proliferation, invasion, and migration | Hao et al. (2019) |
| circRNA_000864 | miRNA decoy | miR-361-3p/BTG2 | Suppresses migration and invasion | Huang et al. (2020) |
| circ_0075829 | miRNA decoy | miR-1287-5p/LAMTOR3 | Promotes proliferation, invasion, and migration | Zhang et al. (2020b) |
| chr7:154954255-154998784+ | miRNA decoy | miR-4459/KIAA0513 | Promotes proliferation | Shao et al. (2020) |
| circ_0013912 | miRNA decoy | miR-7-5p | Promotes proliferation and metastasis | Guo et al. (2020a) |
| hsa_circRNA_001587 | miRNA decoy | miR-223/SLC4A4 | Inhibits migration, invasion, and angiogenesis | Zhang et al. (2020a) |
| circRHOT1 | miRNA decoy | miR-125a-3p/E2F3 | Promotes proliferation and invasion, inhibit apoptosis | Ling et al. (2020) |
| circBFAR | miRNA decoy | miR-34b-5p/MET/Akt | Promotes proliferation and migration | Guo et al. (2020b) |
| circNFIB1 | miRNA decoy | miR-486-5p/PIK3R1/VEGF-C | Inhibits lymphangiogenesis and lymphatic metastasis | Kong et al. (2020) |
| circFOXK2 | miRNA decoy and protein interaction | miR-942/ANK1, GDNF, PAX6 and scaffold for YBX1 and hnRNPK | Promotes proliferation, invasion, and migration | Wong et al. (2020) |
| hsa-circ-001653 | miRNA decoy | miR-377/HOXC6 | Promotes cell-cycle progression, in vitro angiogenesis, and invasion | Shi et al. (2020) |
| circHIPK3 | miRNA decoy | miR-330-5p/RASSF1 | Promotes gemcitabine resistance | Liu et al. (2020) |
| circSEC24A | miRNA decoy | miR-606/TGFBR2 | Promotes proliferation, invasion, and migration | Chen et al. (2021) |
| circRNF13 | miRNA decoy | miR-139-5p/IGF1R | Promotes proliferation, invasion, and migration | Liu et al. (2021c) |
| circ_0092367 | miRNA decoy | miR-1206/ESRP1 | Inhibits EMT and gemcitabine resistance | Yu et al. (2021) |
| circ_0013587 | miRNA decoy | miR-1227/E-Cadherin | Inhibits resistance to erlotinib | Xu et al. (2021a) |
| hsa_circ_0006117 | miRNA decoy | miR-96-5p/KRAS/MAPK | Promotes proliferation, invasion, and migration | Liu et al. (2021a) |
| circPTPN22 | Protein interaction | Inhibit STAT3-SIRT1 interaction | Attenuates immune microenvironment | He et al. (2021) |
| circEYA3 | miRNA decoy | miR-1294/c-Myc | Increases energy production via ATP synthesis | Rong et al. (2021a) |
| circRHOBTB3 | miRNA decoy | miR-600/NACC1/Akt/mTOR | Promotes autophagy and proliferation | Yang et al. (2021) |
| circ_0099999 | miRNA decoy | miR-330-5p/FSCN1 | Promotes proliferation, invasion,, migration, and glycolysis | Wang et al. (2021) |
| circZNF91 | miRNA decoy | miR-23b-3p/SIRT1 | Exosomal transmission to induce chemoresistance | Zeng et al. (2021) |
| circCCT3 | miRNA decoy | miR-613/VEGFA/VEGFR2 | Promotes proliferation and invasion, inhibit apoptosis | Hou et al. (2021) |
| circ-0005105 | miRNA decoy | miR-20a-3p/COL11A1 | Promotes EMT | Ma et al. (2021) |
| circSLIT2 | miRNA decoy | miR-510-5p/c-Myc/LDHA | Promotes glycolysis and proliferation | Guan et al. (2021) |
| circ_0066147 | miRNA decoy | miR-326/E2F2 | Promotes proliferation and invasion, inhibit apoptosis | Zhang and Zhang, (2021) |
| circ_0001666 | miRNA decoy | miR-1251/SOX4 | Promotes EMT and invasion | Zhang et al. (2021a) |
| circPCDH10 | miRNA decoy | miR-338-3p/hTERT | Promotes proliferation and invasion | Zhang et al. (2021b) |
| circ_0030167 | miRNA decoy | miR-338-5p/Wif1/Wnt8/β-catenin | Exosomal transmission to inhibit proliferation, invasion, and migration | Yao et al. (2021) |
| circ_03955 | miRNA decoy | miR-3662/HIF-1α | Inhibits apoptosis and promotes Warburg effect | Liu and Xu, (2021) |
| circ_0092314 | miRNA decoy | miR-671/S100P | Promotes EMT | Shen et al. (2021b) |
| circ-MBOAT2 | miRNA decoy | miR-433-3p/GOT1 | Promotes proliferation and invasion; inhibits apoptosis | Zhou et al. (2021) |
| circNEIL3 | miRNA decoy | miR-432-5p/ADAR1 | Promotes EMT | Shen et al. (2021a) |
| CDR1as | miRNA decoy | miR-432-5p/E2F3 | Promotes proliferation and invasion | Xiong et al. (2021) |
| circRNA_000684 | miRNA decoy | miR-145/KLF5 | Promotes proliferation and invasion, and migration | Liu et al. (2021b) |
| hsa_circ_0071036 | miRNA decoy | miR-489 | Promotes proliferation and invasion; inhibits apoptosis | Han et al. (2021) |
| circRNA_102049 | miRNA decoy | miR-455-3p/CD80 | Promotes proliferation and invasion; inhibits apoptosis | Zhu et al. (2021) |
| circEIF6 | miRNA decoy | miR-557/SLC7A11/PI3K/AKT | Promotes proliferation and invasion; inhibits apoptosis | Zhang et al. (2021c) |
| hsa_circ_0050102 | miRNA decoy | miR-1182/NPSR1 | Promotes proliferation and invasion; inhibits apoptosis | Hua et al. (2021) |
| circSFMBT1 | miRNA decoy | miR-330-5p/PAK1 | Promotes proliferation and invasion | Xu et al. (2021c) |
| circ-MTHFD1L | miRNA decoy | miR-615-3p/RPN6 | Promotes gemcitabine resistance | Chen et al. (2022b) |
| circ_0047744 | miRNA decoy | miR-21/SOCS5 | Suppresses metastasis | Xie et al. (2022) |
| circATG7 | miRNA decoy and protein interaction | miR-766-5p/ATG7 and HuR interaction | Promotes autophagy and proliferation | He et al. (2022) |
| circCUL2 | miRNA decoy | miR-203a-5p/MyD88/NFkB/IL6 | Induces inflammatory cancer-associated fibroblast | Zheng et al. (2022) |
| circANAPC7 | miRNA decoy | miR-373/PHLPP2/AKT/TGFb | Promotes proliferation and muscle wasting | Shi et al. (2022) |
| circ_0072008 | miRNA decoy | miR-545-3p/SLC7A11 | Promotes proliferation, invasion, and glycolysis | Sun et al. (2022) |
| hsa_circ_0050102 | miRNA decoy | miR-218-5p/PPME1 | Promotes proliferation, invasion, and angiogenesis | Feng et al. (2022) |
| circFARP1 | miRNA decoy and protein interaction | miR-660-3p/LIF and CAV1 interaction | Promotes gemcitabine resistance | Hu et al. (2022) |
| circRTN4 | miRNA decoy and protein interaction | miR-497-5p/HOTTIP/HOXA13 and RAB11FIP interaction | Promotes proliferation and invasion; inhibits apoptosis | Wong et al. (2022) |
| circUHRF1 | miRNA decoy | miR-1306-5p/ARL4C | Promotes proliferation, migration, and EMT, inhibits apoptosis | Liu et al. (2022) |
| hsa_circ_0074298 | miRNA decoy | miR-519d/SMOC2 | Promotes proliferation and gemcitabine resistance | Hong et al. (2022a) |
2.1 Decoys for miRNAs
As first demonstrated with CDR1as, some circRNAs function as decoys that bind to miRNAs and prevent them from binding to their target mRNAs (Hansen et al., 2013; Memczak et al., 2013). Hansen et al. also demonstrated that CDR1as reduced miR-7 activity, whereas the cognate linear RNA of CDR1as had little effect on miR-7 activity, suggesting that circRNAs have unique biological functions that differ from linear RNAs despite having the same sequence. In pancreatic cancer, many pathogenic circRNAs have been reported to function as miRNA decoys, leading to cell proliferation, inhibition of apoptosis, metastasis, chemotherapy resistance, and metabolic reprogramming. For example, circBFAR was identified as an upregulated circRNA in pancreatic cancer, and its overexpression was shown to be correlated with poor prognosis (Guo X. et al., 2020). Mechanistically, circBFAR binds to miR-34b-5p and decreases MET expression, thereby activating the MET/PI3K/Akt signaling pathway and promoting the progression of pancreatic cancer cells. In another example, circSLIT2 was shown to upregulate c-Myc expression by binding to miR-510-5p and to facilitate aerobic glycolysis in pancreatic cancer cells (Guan et al., 2021). In addition, many other circRNAs have been reported to contribute to pancreatic cancer progression in a cell-autonomous manner by disrupting miRNA function.
The circRNAs that are dysregulated in cancer cells not only increase their malignant potential in a cell-autonomous manner but also contribute to the progression of pancreatic cancer in a non-cell-autonomous manner by altering the tumor microenvironment (Figure 2). circNFIB1 was shown to be downregulated and to be negatively associated with lymph node metastasis in pancreatic cancer patients (Guan et al., 2021). circNFIB1 functions as a decoy for miR-486-5p and antagonizes the miR-486-5p-mediated suppression of PIK3R1 expression. As PIK3R1 acts as an inhibitor of the PI3K/Akt signaling pathway, circNFIB1 inhibits the PI3K/Akt pathway and downregulates its downstream target, VEGF-C, resulting in suppression of lymphangiogenesis and lymph node metastasis.
FIGURE 2.
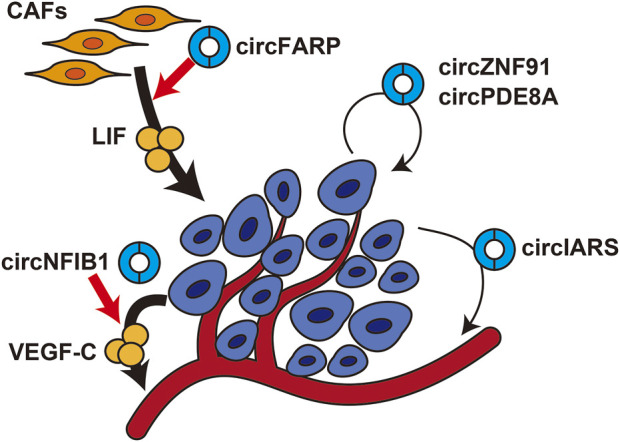
Effects of circRNAs on the tumor microenvironment in pancreatic cancercircFARP enhances LIF expression in cancer-associated fibroblasts. circNFIB1 enhances VEGF-C expression in pancreatic cancer cells. circZNF91 and circPDE8A are transmitted from pancreatic cancer cells to nearby cancer cells. circIARS is transmitted from pancreatic cancer cells to endothelial cells.
Cancer-associated fibroblasts (CAFs) are the predominant cell type in the stroma of pancreatic cancer and play an important role in chemotherapy resistance. circFARP was identified as a circRNA specifically upregulated in gemcitabine-resistant CAFs, and high circFARP expression was shown to be correlated with poor progression-free survival (Guan et al., 2021). When pancreatic cancer cells were cultured in conditioned medium derived from circFARP-overexpressing CAFs, they acquired gemcitabine resistance. Mechanistically, circFARP enhances leukemia inhibitory factor (LIF) expression by decoying miR-660-3p, and LIF secreted from CAFs induces gemcitabine resistance in pancreatic cancer cells by activating the LIF/STAT3 pathway. Interestingly, circFARP inhibits CAV1 degradation by interacting directly with CAV1, which promotes caveola-mediated exocytosis of LIF. Thus, circFARP functions as both a miRNA decoy and protein decoy.
Recent studies have shown that circRNAs are enriched in exosomes and may mediate intercellular communication between cancer cells and the tumor microenvironment to promote cancer progression (Seimiya et al., 2020). For example, circPDE8A was identified as a circRNA enriched in exosomes secreted from liver metastatic pancreatic cancer cells (Li Z. et al., 2018). circPDE8A functions as a decoy for miR-338 to upregulate MACC1 and stimulates invasive growth via the MACC/MET/ERK or AKT pathway. Exosomes derived from circPDE8A-overexpressing cancer cells activate the MACC/MET/ERK or AKT pathway and promote epithelial–mesenchymal transition. In clinical practice, circPDE8A was also detected in exosomes from the plasma of pancreatic cancer patients, and high exosomal circPDE8A expression was shown to be correlated with TNM stage and poor prognosis. circIARS was also enriched in exosomes from liver metastatic pancreatic cancer cells (Li J. et al., 2018). Exosomal circIARS decoyed miR-122 to increase RhoA expression, which in turn increased the permeability of human umbilical vein endothelial cells and facilitated invasion of pancreatic cancer cells. Another study showed that hypoxia-induced exosomal circZNF91 promoted chemoresistance in normoxic pancreatic cancer (Zeng et al., 2021). circZNF91 was upregulated in exosomes derived from pancreatic cancer cells under hypoxic conditions compared with normoxic conditions. When hypoxia-induced exosomal circZNF91 was transmitted to normoxic tumor cells, circZNF91 bound to miR-23b-3p and upregulated SIRT1 expression. Consequently, SIRT1 stabilized HIF-1α protein, which enhanced glycolysis and promoted chemoresistance in recipient cells. These results suggest that exosomal circRNAs may function as decoys for miRNAs in recipient cells and play important roles in cancer progression by mediating signal transmission between pancreatic cancer cells and the tumor microenvironment.
Although there is accumulating evidence that circRNAs are involved in the progression of pancreatic cancer, caution is required when considering whether a given circRNA has a measurable effect on cellular functions by acting as an miRNA decoy. The most well-studied circRNA with miRNA decoy function is CDR1as, which has over 70 binding sites for miR-7 (Hansen et al., 2013). Another well-known circRNA has 16 miR-138 target sequences (Hansen et al., 2013). Compared with these well-known circRNAs, most circRNAs have few miRNA-binding sites and low expression levels (Guo et al., 2014). Therefore, when investigating the potential of a circRNA of interest to act as a miRNA decoy and contribute to the pathogenesis of pancreatic cancer, careful consideration of whether the expression of that circRNA and the number of miRNA-binding sites are sufficient to achieve a measurable effect is needed.
2.2 Inhibiting post-translational modifications
Some circRNAs interact directly with proteins and stabilize them by inhibiting ubiquitin–proteasomal degradation. For example, circFARP1, which is upregulated in CAFs, interacts with CAV1 and blocks the interaction between CAV1 and the E3 ubiquitin ligase ZNRF1 to inhibit CAV1 degradation (Hu et al., 2022). As CAV1 mediates caveola-mediated exocytosis, CAV1 stabilization enhances LIF exocytosis from CAFs and activates the JAK–STAT signaling pathway in pancreatic cancer cells. As another example, circRTN4 interacts with RAB11FIP, which plays an important role in epithelial–mesenchymal transition, and protects it from ubiquitin–proteasomal degradation (Wong et al., 2022). Prediction of the structure of the circRTN4–RAB11FIP1 complex revealed that circRTN4 blocked the Lys578 ubiquitination site of RAB11FIP1. These results suggest that circRNAs bind to proteins and enhance their stability by inhibiting ubiquitin–proteasomal degradation.
It has been suggested that circPTPN22 inhibits protein–protein interactions and promotes STAT3 acetylation (He et al., 2021). circPTPN22 is upregulated in pancreatic cancer, and it interacts directly with STAT3 to block the interaction between STAT3 and SIRT1. In a xenograft model, knockdown of circPTPN22 enhanced intratumoral T cell infiltration and inhibited tumor growth. These results suggest that circPTPN22 inhibits STAT3 deacetylation by SIRT1 and promotes immune evasion of pancreatic cancer.
2.3 Scaffolds for protein complexes
circRNAs also serve as scaffolds for mRNA–protein complex formation to enhance expression of the mRNA. circFOXK2 is highly expressed in pancreatic cancer and promotes cell proliferation, migration, and invasion (Wong et al., 2020). Pulldown of circRNAs followed by mass spectrometry revealed that circFOKX2 interacts with YBX1 and hnRNPK. The YBX1–hnRNPK complex is reportedly involved in cancer progression. To determine the direct targets of the YBX1–hnRNPK complex, a circFOKX2 RNA pulldown assay and RNA immunoprecipitation assay of YBX1 and hnRNPK were performed, and NUF2 and PDXK mRNA were identified as direct targets. Knockdown of circFOXK2 downregulated the expression of NUF2 and PDXK mRNA and inhibited the interactions of YBX1 and hnRNPK with NUF2 and PDXK mRNA. These results suggest that circFOXK2 provides a scaffold for the YBX1–hnRNPK complex to promote expression of NUF2 and PDXK. Another study showed that circATG7 interacted with HuR and increased the mRNA level of ATG7 (He et al., 2022). circATG7 exists in both the cytoplasm and nucleus; cytoplasmic circATG7 acts as a decoy for miR-766-5p to decrease the expression of its target gene, ATG7, while nuclear circATG7 interacts with HuR and increases the stability of ATG7 mRNA in a HuR-dependent manner.
Taken together, these observations suggest that pathogenic circRNAs in pancreatic cancers function as miRNA decoys and/or interact with proteins. However, given that the exact mechanisms of action of many circRNAs remain unknown, that circRNAs acting as templates for protein translation have not been identified in pancreatic cancer, and that a single circRNA can have multiple functions, the impact of circRNAs on the progression of pancreatic cancer may be much greater than known at present. Further studies are needed to elucidate the overall impact of circRNAs on the pathogenesis of pancreatic cancer.
3 circRNAs as diagnostic biomarkers
3.1 Pancreatic cancer
Although early diagnosis is important to improve the prognosis of pancreatic cancer, most pancreatic cancer cases are diagnosed at an advanced stage. One reason for this is that pancreatic cancer biomarkers are ineffective for early diagnosis. For example, serum pancreatic enzymes, such as amylase, are elevated in only 20–50% of patients with pancreatic cancer, and serum CA19-9, which is routinely used as a biomarker for pancreatic tumors, is elevated in 70–90% of cases (Sharma et al., 2011). However, the sensitivity of CA19-9 for detecting pancreatic cancer lesions ≤2 cm decreases to approximately 50% (Liu et al., 2012), which is insufficient for early diagnosis. Therefore, the development of novel diagnostic biomarkers is required.
circRNAs have a half-life of 18.8–23.7 h, which is significantly longer than that of their cognate mRNAs (4.0–7.4 h) (Enuka et al., 2016). In addition, circRNAs exhibit characteristic expression patterns in different types of cancer (Vo et al., 2019). Due to their remarkable stability and specificity, circRNAs are regarded as potential diagnostic biomarkers for cancers. To date, five studies have reported the utility of serum circRNAs as diagnostic biomarkers for pancreatic cancer (Table 2). That is, circLDLRAD3, circPDAC, hsa_circ_0013587, and circ_001569 were reported to be aberrantly expressed in the serum of pancreatic cancer patients (Yang et al., 2017; Xu K. et al., 2021; Shen X. et al., 2021; Seimiya et al., 2021), and another study suggested that the combination of hsa_circ_0006220 and hsa_circ_0001666 can improve diagnostic performance compared with each circRNA alone (Hong L. et al., 2022). These studies demonstrated that circRNAs have a sensitivity of 0.45–0.76 and specificity of 0.70–0.90 for diagnosis of pancreatic cancer.
TABLE 2.
CircRNAs as diagnostic biomarkers for pancreatic cancer.
| circRNA | Sensitivity | Specificity | Mechanism/Target/Biological function | Reference |
|---|---|---|---|---|
| circ-LDLRAD3 | 0.57 | 0.70 | miRNA decoy/miR-137-3p/Promotes proliferation, invasion, and migration | Yang et al. (2017) |
| circPDAC | 0.45 | 0.90 | Unknown | Seimiya et al. (2021) |
| hsa_circ_0013587 | 0.76 | 0.76 | miRNA decoy/miR-1227/Inhibits resistance to erlotinib | Xu et al. (2021b) |
| circ_001569 | 0.63 | 0.74 | Unknown | Shen et al. (2021c) |
| hsa_circ_0006220 and hsa_circ_0001666 | 0.74 | 0.87 | Unknown | Hong et al. (2022b) |
3.2 Intraductal papillary mucinous neoplasm
It has been suggested that circRNAs may be useful for diagnosis of not only pancreatic cancer but also intraductal papillary mucinous neoplasm (IPMN), which is a precancerous lesion of pancreatic cancer (Seimiya et al., 2021). It was shown that circPDAC is highly expressed in pancreatic cancer, and that high circPDAC expression is associated with lymph node metastasis and cancer stage. This circRNA is enriched in exosomes and can be detected in the blood of patients with pancreatic cancer, suggesting that measurement of circPDAC levels in blood may be useful for the diagnosis of pancreatic cancer. Interestingly, circPDAC was also detected in the blood of patients with IPMN. As there are currently no effective biomarkers for diagnosis of IPMN, circPDAC may serve as a novel diagnostic biomarker for this disease. Recently, pancreatic surveillance of high-risk individuals with genetic and familial risk factors for pancreatic cancer was shown to lead to early diagnosis and better long-term survival (Dbouk et al., 2022). In the same way, pancreatic surveillance in patients with IPMN may also reduce pancreatic cancer mortality through early detection.
A number of issues remain to be resolved for the routine application of circRNAs as diagnostic biomarkers for pancreatic cancer or IPMN in real-world clinical practice. First, because of the relatively small number of cases in the studies discussed above, it will be necessary to evaluate the diagnostic performance in larger cohorts including patients with early-stage pancreatic cancer. In addition, PCR is commonly used to measure the expression levels of circRNAs in blood, but PCR is labor-intensive and requires multiple steps, including RNA extraction from blood, reverse transcription, and cDNA amplification. Therefore, it is necessary to develop a simpler and more sensitive measurement method. Future research will resolve these issues and provide hope for early diagnosis of pancreatic cancer.
4 Therapeutic applications of circular RNAs
4.1 circRNAs as therapeutic molecules
As circRNAs have unique biological functions and molecular characteristics, they have potential applications as new nucleic acid therapeutic agents. One possible application is to use circRNAs as stable molecules for RNA interference therapeutics. For example, engineered circRNA molecules encoding miR-34a-3p and -5p sequences, called db34a RNA, significantly inhibited angiogenesis of pancreatic cancer cells (Gnanamony et al., 2021). This is the only study to date that has examined the potential application of circRNAs as therapeutic molecules in pancreatic cancer, but a variety of applications are being investigated in other diseases, which have been reviewed elsewhere (Liu and Chen, 2022). Briefly, based on the stability and biological functions of circRNAs, various applications of these RNAs are being investigated, including as decoys for miRNAs and proteins, stable antisense RNAs, boosters of innate immune responses, inhibitors of innate immune responses, and templates for protein translation. These various circRNA-based therapies may also be applicable in pancreatic cancer.
4.2 Targeting pathogenic circRNAs
As the roles of circRNAs in the pathogenesis of pancreatic cancer become clearer, new therapeutic approaches targeting pathogenic circRNAs are also expected. For example, circRTN4 is highly expressed in pancreatic cancer and is correlated with liver metastasis (Wong et al., 2022). Knockdown of circRTN4 in pancreatic cancer cell lines inhibits their proliferation, migration, and invasion. Furthermore, circRTN4 knockdown significantly suppressed tumor growth and metastasis to the liver in a mouse xenograft model. In addition, many pathogenic circRNAs have also been reported to be potential therapeutic targets.
Although many circRNAs are expected to be therapeutic targets, several challenges remain for clinical application of circRNA-targeted therapies. Antisense oligonucleotides, siRNAs, shRNAs, and CRISPR–Cas systems can be used to target RNAs. Clinical application of CRISPR–Cas systems is being intensively evaluated, and several oligonucleotide drugs have been approved by the FDA and are clinically available (Roberts et al., 2020; Modell et al., 2022). To apply these molecules to circRNA-targeted therapies, they should specifically recognize back-splice junctions. However, because circRNAs have identical sequences to those of their cognate linear mRNAs, in cases where the back-splice junction has a similar nucleotide sequence to that of a canonical splicing junction, it may not be possible to engineer therapeutic molecules that specifically recognize circRNAs. Even with proper design, care is required regarding off-target effects, especially those on cognate linear mRNAs. A number of processes are involved in the regulation of circRNA expression, including transcription, splicing, nuclear export, and degradation (Chen, 2020). Elucidating these molecular mechanisms may provide new therapeutic targets for regulating circRNA expression levels.
5 Concluding remarks
With the development of RNA sequencing methods for circRNAs, a great deal of progress in the research of these molecules has been made over the past decade. Accordingly, some circRNAs have been shown to play important roles in a variety of pathological conditions, including pancreatic cancer (Rong et al., 2021b; Chen Q. et al., 2022). However, the functional significance of many circRNAs is largely unknown. They may be meaningless byproducts generated during the splicing process, but they may have important biological functions that remain to be elucidated. It is hoped that the currently unknown pathological roles of circRNAs will be further elucidated, and new therapeutic strategies for pancreatic cancer using circRNAs will be developed in the future.
Author contributions
TS, MO, and MF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#22K15390 and # 22H02828) (to TS and MO), by JST CREST (to MO, #JPMJCR19H5), and by the Practical Research for Innovative Cancer Control Program and by the Program for Basic and Clinical Research on Hepatitis from AMED (to MO, #JP20ck0106557 and #JP22fk0210092).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- An Y., Cai H., Zhang Y., Liu S., Duan Y., Sun D., et al. (2018). circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell. Physiol. biochem. 51, 2224–2236. 10.1159/000495868 [DOI] [PubMed] [Google Scholar]
- Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030. 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- Chen G., Shi Y., Zhang Y., Sun J. (2017). CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco. Targets. Ther. 10, 5783–5794. 10.2147/OTT.S150678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490. 10.1038/S41580-020-0243-Y [DOI] [PubMed] [Google Scholar]
- Chen Q., Li J., Shen P., Yuan H., Yin J., Ge W., et al. (2022a). Biological functions, mechanisms, and clinical significance of circular RNA in pancreatic cancer: A promising rising star. Cell Biosci. 12, 97. 10.1186/S13578-022-00833-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li Z., Zhang M., Wang B., Ye J., Zhang Y., et al. (2019). Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 38, 466. 10.1186/s13046-019-1436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu S., Liu X., Jiang X., Jiang J. (2021). CircSEC24A upregulates TGFBR2 expression to accelerate pancreatic cancer proliferation and migration via sponging to miR-606. Cancer Cell Int. 21, 671. 10.1186/s12935-021-02392-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-W., Hu J.-F., Wang Z.-W., Liao C.-Y., Kang F.-P., Lin C.-F., et al. (2022b). Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 41, 153. 10.1186/s13046-022-02343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C., Daubersies P., Majerus M. A., Kerckaert J. P., Bailleul B. (1992). Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11, 1095–1098. 10.1002/J.1460-2075.1992.TB05148.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. (1993). Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160. 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- Dbouk M., Katona B. W., Brand R. E., Chak A., Syngal S., Farrell J. J., et al. (2022). The multicenter cancer of pancreas screening study: Impact on stage and survival. J. Clin. Oncol. 40, 3257–3266. 10.1200/JCO.22.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Ma X. K., Li G. W., Yang L. (2018). CIRCpedia v2: An updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinforma. 16, 226–233. 10.1016/j.gpb.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa S., Toma H., Ohigashi H., Okusaka T., Nakao A., Hatori T., et al. (2012). Japan pancreatic cancer registry; 30th year anniversary: Japan pancreas society. Pancreas 41, 985–992. 10.1097/MPA.0B013E318258055C [DOI] [PubMed] [Google Scholar]
- Enuka Y., Lauriola M., Feldman M. E., Sas-Chen A., Ulitsky I., Yarden Y. (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383. 10.1093/nar/gkv1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N., Jiao Z., Zhang Y., Yang B. (2022). Hsa_circ_0050102 regulates the pancreatic cancer development via miR-218-5p/PPME1 axis. J. Clin. Lab. Anal. 36, e24247. 10.1002/jcla.24247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zhao F. (2015). Ciri: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16, 4–16. 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanamony M., Demirkhanyan L., Ge L., Sojitra P., Bapana S., Norton J. A., et al. (2021). Circular dumbbell miR-34a-3p and -5p suppresses pancreatic tumor cell-induced angiogenesis and activates macrophages. Oncol. Lett. 21, 75. 10.3892/ol.2020.12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H., Luo W., Liu Y., Li M. (2021). Novel circular RNA circSLIT2 facilitates the aerobic glycolysis of pancreatic ductal adenocarcinoma via miR-510-5p/c-Myc/LDHA axis. Cell Death Dis. 12, 645. 10.1038/s41419-021-03918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Agarwal V., Guo H., Bartel D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15, 409. 10.1186/S13059-014-0409-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Zhao L., Wei G., Liu P., Zhang Y., Fu L. (2020a). Blocking circ_0013912 suppressed cell growth, migration and invasion of pancreatic ductal adenocarcinoma cells in vitro and in vivo partially through sponging miR-7-5p. Cancer Manag. Res. 12, 7291–7303. 10.2147/CMAR.S255808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhou Q., Su D., Luo Y., Fu Z., Huang L., et al. (2020b). Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer 19, 83. 10.1186/s12943-020-01196-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Fang Y., Chen P., Xu Y., Zhou W., Rong Y., et al. (2021). Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle 20, 369–382. 10.1080/15384101.2021.1874684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hao L., Rong W., Bai L., Cui H., Zhang S., Li Y., et al. (2019). Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J. Cell. Biochem. 120, 3780–3789. 10.1002/jcb.27658 [DOI] [PubMed] [Google Scholar]
- He Y., Han P., Chen C., Xie S., Zhang H., Song Y., et al. (2021). circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation. Cancer Gene Ther. 10.1038/s41417-021-00382-w [DOI] [PubMed] [Google Scholar]
- He Z., Cai K., Zeng Z., Lei S., Cao W., Li X. (2022). Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. 13, 233. 10.1038/s41419-022-04677-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Lishan W., Peng X., Zhengqing L., Yang Y., Fangfang H., et al. (2022a). Hsa_circ_0074298 promotes pancreatic cancer progression and resistance to gemcitabine by sponging miR-519 to target SMOC. J. Cancer 13, 34–50. 10.7150/jca.62927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Xu L., Jin L., Xu K., Tang W., Zhu Y., et al. (2022b). Exosomal circular RNA hsa_circ_0006220, and hsa_circ_0001666 as biomarkers in the diagnosis of pancreatic cancer. J. Clin. Lab. Anal. 36, e24447. 10.1002/jcla.24447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J.-P., Men X.-B., Yang L.-Y., Han E.-K., Han C.-Q., Liu L.-B. (2021). CircCCT3 acts as a sponge of miR-613 to promote tumor growth of pancreatic cancer through regulating VEGFA/VEGFR2 signaling. Balk. Med. J. 38, 229–238. 10.5152/balkanmedj.2021.21145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Xia R., Zhang X., Li T., Ye Y., Li G., et al. (2022). circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol. Cancer 21, 24. 10.1186/s12943-022-01501-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Gao J., Li T., Wang M., You L., Chen G., et al. (2021). The promoting effects of hsa_circ_0050102 in pancreatic cancer and the molecular mechanism by targeting miR-1182/NPSR1. Carcinogenesis 42, 471–480. 10.1093/carcin/bgaa130 [DOI] [PubMed] [Google Scholar]
- Huang L., Han J., Yu H., Liu J., Gui L., Wu Z., et al. (2020). CircRNA_000864 upregulates B-cell translocation gene 2 expression and represses migration and invasion in pancreatic cancer cells by binding to miR-361-3p. Front. Oncol. 10, 547942. 10.3389/fonc.2020.547942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Li Y., Luo Y., Zhu J., Zheng H., Gao B., et al. (2020). circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol. Cancer 19, 82. 10.1186/s12943-020-01205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A., Dijkema R., Arnberg A. C., Van Der Meide P. H., Schellekens H. (1986). The hepatitis delta (delta) virus possesses a circular RNA. Nature 323, 558–560. 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- Li J., Li Z., Jiang P., Peng M., Zhang X., Chen K., et al. (2018a). Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 37, 177. 10.1186/s13046-018-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yanfang W., Li J., Jiang P., Peng T., Chen K., et al. (2018b). Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 432, 237–250. 10.1016/j.canlet.2018.04.035 [DOI] [PubMed] [Google Scholar]
- Ling S., He Y., Li X., Hu M., Ma Y., Li Y., et al. (2020). CircRHOT1 mediated cell proliferation, apoptosis and invasion of pancreatic cancer cells by sponging miR-125a-3p. J. Cell. Mol. Med. 24, 9881–9889. 10.1111/jcmm.15572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Xu J. (2021). Circ_03955 promotes pancreatic cancer tumorigenesis and Warburg effect by targeting the miR-3662/HIF-1α axis. Clin. Transl. Oncol. 23, 1905–1914. 10.1007/s12094-021-02599-5 [DOI] [PubMed] [Google Scholar]
- Liu C. X., Chen L. L. (2022). Circular RNAs: Characterization, cellular roles, and applications. Cell 185, 2016–2034. 10.1016/J.CELL.2022.04.021 [DOI] [PubMed] [Google Scholar]
- Liu J., Gao J., Du Y., Li Z., Ren Y., Gu J., et al. (2012). Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. cancer 131, 683–691. 10.1002/IJC.26422 [DOI] [PubMed] [Google Scholar]
- Liu L., Liu F.-B., Huang M., Xie K., Xie Q.-S., Liu C.-H., et al. (2019). Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat. Dis. Int. 18, 580–586. 10.1016/j.hbpd.2019.03.003 [DOI] [PubMed] [Google Scholar]
- Liu T., Zhou L., He Z., Chen Y., Jiang X., Xu J., et al. (2021a). Circular RNA hsa_circ_0006117 facilitates pancreatic cancer progression by regulating the miR-96-5p/KRAS/MAPK signaling pathway. J. Oncol. 2021, 9213205. 10.1155/2021/9213205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Deng L., Xu A., Xiong X., Tao J., Chang J., et al. (2022). Identifying a novel IRF3/circUHRF1/miR-1306-5p/ARL4C axis in pancreatic ductal adenocarcinoma progression. Cell Cycle 21, 392–405. 10.1080/15384101.2021.2020450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhong L., Jiang W., Wen D. (2021b). Repression of circRNA_000684 inhibits malignant phenotypes of pancreatic ductal adenocarcinoma cells via miR-145-mediated KLF5. Pancreatology 21, 406–417. 10.1016/j.pan.2020.12.023 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhou L., Chen Y., Jiang X., Jiang J. (2021c). CircRNF13 promotes the malignant progression of pancreatic cancer through targeting miR-139-5p/igf1r Axis. J. Oncol. 2021, 6945046. 10.1155/2021/6945046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xia L., Dong L., Wang J., Xiao Q., Yu X., et al. (2020). CircHIPK3 promotes gemcitabine (GEM) resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag. Res. 12, 921–929. 10.2147/CMAR.S239326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Li G., Fan W., Xu Y., Song S., Guo K., et al. (2021). Circ-0005105 activates COL11A1 by targeting miR-20a-3p to promote pancreatic ductal adenocarcinoma progression. Cell Death Dis. 12, 656. 10.1038/s41419-021-03938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Modell A. E., Lim D., Nguyen T. M., Sreekanth V., Choudhary A. (2022). CRISPR-Based therapeutics: Current challenges and future applications. Trends Pharmacol. Sci. 43, 151–161. 10.1016/J.TIPS.2021.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., et al. (1991). Scrambled exons. Cell 64, 607–613. 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- Ou Z.-L., Luo Z., Wei W., Liang S., Gao T.-L., Lu Y.-B. (2019). Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: Role of circ_0000977/miR-153 axis. RNA Biol. 16, 1592–1603. 10.1080/15476286.2019.1649585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Hao X., Song W., Niu K., Yang X., Zhang X., et al. (2019). Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics 11, 53–63. 10.2217/epi-2018-0051 [DOI] [PubMed] [Google Scholar]
- Roberts T. C., Langer R., Wood M. J. A. (2020). Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694. 10.1038/S41573-020-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z., Shi S., Tan Z., Xu J., Meng Q., Hua J., et al. (2021a). Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol. Cancer 20, 106. 10.1186/s12943-021-01400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z., Xu J., Shi S., Tan Z., Meng Q., Hua J., et al. (2021b). Circular RNA in pancreatic cancer: A novel avenue for the roles of diagnosis and treatment. Theranostics 11, 2755–2769. 10.7150/THNO.56174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., et al. (2014). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Salzman J., Chen R. E., Olsen M. N., Wang P. L., Brown P. O. (2013). Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777. 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. (1976). Viroids are single stranded covalently closed circular RNA molecules existing as highly base paired rod like structures. Proc. Natl. Acad. Sci. U. S. A. 73, 3852–3856. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya T., Otsuka M., Iwata T., Shibata C., Tanaka E., Suzuki T., et al. (2020). Emerging roles of exosomal circular RNAs in cancer. Front. Cell Dev. Biol. 8, 568366. 10.3389/fcell.2020.568366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya T., Otsuka M., Iwata T., Tanaka E., Sekiba K., Shibata C., et al. (2021). Aberrant expression of a novel circular RNA in pancreatic cancer. J. Hum. Genet. 66, 181–191. 10.1038/S10038-020-00826-5 [DOI] [PubMed] [Google Scholar]
- Shao F., Cai M., Fan F.-F., Huang M., Tao Y., Wang C., et al. (2020). Overexpression of circRNA chr7:154954255-154998784+ in cancer-associated pancreatic stellate cells promotes the growth and metastasis of pancreatic cancer by targeting the miR-4459/KIAA0513 axis. Am. J. Transl. Res. 12, 5048–5063. [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Eltawil K. M., Renfrew P. D., Walsh M. J., Molinari M. (2011). Advances in diagnosis, treatment and palliation of pancreatic carcinoma: 1990-2010. World J. Gastroenterol. 17, 867–897. 10.3748/WJG.V17.I7.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Yang T., Chen Q., Yuan H., Wu P., Cai B., et al. (2021a). CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol. Cancer 20, 51. 10.1186/s12943-021-01333-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Zheng G., Zhou Y., Tong J., Xu S., Gao H., et al. (2021b). CircRNA circ_0092314 induces epithelial-mesenchymal transition of pancreatic cancer cells via elevating the expression of S100P by sponging miR-671. Front. Oncol. 11, 675442. 10.3389/fonc.2021.675442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Chen Y., Li J., Huang H., Liu C., Zhou N. (2021c). Identification of Circ_001569 as a potential biomarker in the diagnosis and prognosis of pancreatic cancer. Technol. Cancer Res. Treat. 20, 1533033820983302. 10.1177/1533033820983302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Li H., Zhen T., Dong Y., Pei X., Zhang X. (2020). hsa_circ_001653 implicates in the development of pancreatic ductal adenocarcinoma by regulating MicroRNA-377-mediated HOXC6 Axis. Mol. Ther. Nucleic Acids 20, 252–264. 10.1016/j.omtn.2019.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shi X., Yang J., Liu M., Zhang Y., Zhou Z., Luo W., et al. (2022). Circular RNA ANAPC7 inhibits tumor growth and muscle wasting via PHLPP2-AKT-TGF-β signaling Axis in pancreatic cancer. Gastroenterology 162, 2004–2017.e2. 10.1053/j.gastro.2022.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2022). Cancer statistics, 2022. Ca. Cancer J. Clin. 72, 7–33. 10.3322/CAAC.21708 [DOI] [PubMed] [Google Scholar]
- Sun H., Liu F., Zhang H. (2022). Circ_0072008, an oncogene in pancreatic ductal adenocarcinoma, contributes to tumour cell malignant progression and glycolysis by regulating miR-545-3p/SLC7A11 axis. Autoimmunity 55, 203–213. 10.1080/08916934.2022.2027919 [DOI] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. 10.3322/CAAC.21660 [DOI] [PubMed] [Google Scholar]
- Vo J. N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176, 869–881. 10.1016/J.CELL.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang F., Wu D., Wang Q., Nie L., Yu J. (2021). A novel circ_0099999/miR-330-5p/FSCN1 ceRNA crosstalk in pancreatic cancer. Autoimmunity 54, 471–482. 10.1080/08916934.2021.1963958 [DOI] [PubMed] [Google Scholar]
- Wong C. H., Lou U. K., Fung F. K.-C., Tong J. H. M., Zhang C.-H., To K.-F., et al. (2022). CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol. Cancer 21, 10. 10.1186/s12943-021-01481-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. H., Lou U. K., Li Y., Chan S. L., Tong J. H., To K.-F., et al. (2020). CircFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with RNA-binding proteins and sponging MiR-942. Cancer Res. 80, 2138–2149. 10.1158/0008-5472.CAN-19-3268 [DOI] [PubMed] [Google Scholar]
- Xie H., Zhao Q., Yu L., Lu J., Peng K., Xie N., et al. (2022). Circular RNA circ_0047744 suppresses the metastasis of pancreatic ductal adenocarcinoma by regulating the miR-21/SOCS5 axis. Biochem. Biophys. Res. Commun. 605, 154–161. 10.1016/j.bbrc.2022.03.082 [DOI] [PubMed] [Google Scholar]
- Xing C., Ye H., Wang W., Sun M., Zhang J., Zhao Z., et al. (2019). Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif. Cells Nanomed. Biotechnol. 47, 3920–3928. 10.1080/21691401.2019.1671856 [DOI] [PubMed] [Google Scholar]
- Xiong X., Feng J., Yang X., Li H., Shi Q., Tao J., et al. (2021). Circular RNA CDR1as promotes tumor progression by regulating miR-432-5p/E2F3 axis in pancreatic cancer. Cancer Cell Int. 21, 112. 10.1186/s12935-021-01812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chen R., Shen Q., Yang D., Peng H., Tong J., et al. (2021a). Overexpression of circular RNA circ_0013587 reverses erlotinib resistance in pancreatic cancer cells through regulating the miR-1227/E-cadherin pathway. Front. Oncol. 11, 754146. 10.3389/fonc.2021.754146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Qiu Z., Xu L., Qiu X., Hong L., Wang J. (2021b). Increased levels of circulating circular RNA (hsa_circ_0013587) may serve as a novel biomarker for pancreatic cancer. Biomark. Med. 15, 977–985. 10.2217/bmm-2020-0750 [DOI] [PubMed] [Google Scholar]
- Xu S., Lei S.-L., Liu K.-J., Yi S.-G., Yang Z.-L., Yao H.-L. (2021c). circSFMBT1 promotes pancreatic cancer growth and metastasis via targeting miR-330-5p/PAK1 axis. Cancer Gene Ther. 28, 234–249. 10.1038/s41417-020-00215-2 [DOI] [PubMed] [Google Scholar]
- Xu Y., Yao Y., Gao P., Cui Y. (2019). Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem. Biophys. Res. Commun. 509, 138–142. 10.1016/j.bbrc.2018.12.088 [DOI] [PubMed] [Google Scholar]
- Yang F., Liu D. Y., Guo J. T., Ge N., Zhu P., Liu X., et al. (2017). Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J. Gastroenterol. 23, 8345–8354. 10.3748/wjg.v23.i47.8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Cong X., Ren M., Sun H., Liu T., Chen G., et al. (2019). Circular RNA hsa_circRNA_0007334 is predicted to promote MMP7 and COL1A1 expression by functioning as a miRNA sponge in pancreatic ductal adenocarcinoma. J. Oncol. 2019, 7630894. 10.1155/2019/7630894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Shen P., Chen Q., Wu P., Yuan H., Ge W., et al. (2021). FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 40, 261. 10.1186/s13046-021-02063-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Zhang C., Chen Y., Gao S. (2019). Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 239, 116871. 10.1016/j.lfs.2019.116871 [DOI] [PubMed] [Google Scholar]
- Yao X., Mao Y., Wu D., Zhu Y., Lu J., Huang Y., et al. (2021). Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/β-catenin axis. Cancer Lett. 512, 38–50. 10.1016/j.canlet.2021.04.030 [DOI] [PubMed] [Google Scholar]
- Yu S., Wang M., Zhang H., Guo X., Qin R. (2021). Circ_0092367 inhibits EMT and gemcitabine resistance in pancreatic cancer via regulating the miR-1206/ESRP1 Axis. Genes (Basel) 12, 1701. 10.3390/genes12111701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Zhao Y., Chen Q., Zhu S., Niu Y., Ye Z., et al. (2021). Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 40, 5505–5517. 10.1038/s41388-021-01960-w [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Z. (2021). Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression. Open Life Sci. 16, 495–510. 10.1515/biol-2021-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Zhu W., Ma C., Ai K. (2021a). Silencing of circRNA circ_0001666 represses EMT in pancreatic cancer through upregulating miR-1251 and downregulating SOX4. Front. Mol. Biosci. 8, 684866. 10.3389/fmolb.2021.684866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qiu M., Gao S., Tian T. (2021b). Circular RNA PCDH10 regulates the tumorigenesis of pancreatic cancer through the miR-338-3p/hTERT axis. Am. J. Transl. Res. 13, 2181–2197. [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Li M., Lu H., Peng T. (2021c). Up-regulation of circEIF6 contributes to pancreatic cancer development through targeting miR-557/slc7a11/PI3K/AKT signaling. Cancer Manag. Res. 13, 247–258. 10.2147/CMAR.S280307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tan P., Zhuang Y., Du L. (2020a). hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am. J. Physiol. Gastrointest. Liver Physiol. 319, G703–G717. 10.1152/ajpgi.00118.2020 [DOI] [PubMed] [Google Scholar]
- Zhang X., Xue C., Cui X., Zhou Z., Fu Y., Yin X., et al. (2020b). Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J. Cell. Mol. Med. 24, 14596–14607. 10.1111/jcmm.16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Hu C., Lin H., Li G., Xia R., Zhang X., et al. (2022). circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-κB signaling pathway. J. Exp. Clin. Cancer Res. 41, 71. 10.1186/s13046-021-02237-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liu K., Cui J., Xiong J., Wu H., Peng T., et al. (2021). Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J. Exp. Clin. Cancer Res. 40, 124. 10.1186/s13046-021-01894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhou Y., Zhu S., Li F., Xu J., Zhang L., et al. (2021). circRNA circ_102049 implicates in pancreatic ductal adenocarcinoma progression through activating CD80 by targeting miR-455-3p. Mediat. Inflamm. 2021, 8819990. 10.1155/2021/8819990 [DOI] [PMC free article] [PubMed] [Google Scholar]


