Abstract
CRISPR-based gene editing technology represents a promising approach to deliver therapies for inherited disorders, including amyotrophic lateral sclerosis (ALS). Toxic gain-of-function superoxide dismutase 1 (SOD1) mutations are responsible for ~20% of familial ALS cases. Thus, current clinical strategies to treat SOD1-ALS are designed to lower SOD1 levels. Here, we utilized AAV-PHP.B variants to deliver CRISPR-Cas9 guide RNAs designed to disrupt the human SOD1 (huSOD1) transgene in SOD1G93A mice. A one-time intracerebroventricular injection of AAV.PHP.B-huSOD1-sgRNA into neonatal H11Cas9 SOD1G93A mice caused robust and sustained mutant huSOD1 protein reduction in the cortex and spinal cord, and restored motor function. Neonatal treatment also reduced spinal motor neuron loss, denervation at neuromuscular junction (NMJ) and muscle atrophy, diminished axonal damage and preserved compound muscle action potential throughout the lifespan of treated mice. SOD1G93A treated mice achieved significant disease-free survival, extending lifespan by more than 110 days. Importantly, a one-time intrathecal or intravenous injection of AAV.PHP.eB-huSOD1-sgRNA in adult H11Cas9 SOD1G93A mice, immediately before symptom onset, also extended lifespan by at least 170 days. We observed substantial protection against disease progression, demonstrating the utility of our CRISPR editing preclinical approach for target evaluation. Our approach uncovered key parameters (e.g., AAV capsid, Cas9 expression) that resulted in improved efficacy compared to similar approaches and can also serve to accelerate drug target validation.
Subject terms: Gene therapy, Gene delivery
Introduction
Amyotrophic lateral sclerosis (ALS), commonly known as Lou Gehrig’s disease, is a devastating neurodegenerative disorder that results in the selective loss and dysfunction of motor neurons (MNs) in the brain and spinal cord, resulting in progressive muscle weakness and atrophy [1]. Though the precise etiology of ALS remains unknown, approximately 10% of cases present an inherited or familial form of the disease (fALS), while the remaining 90% are deemed sporadic (sALS). Amongst the 20 or more genes associated with ALS, mutations in the superoxide dismutase 1 (SOD1) gene account for approximately 15–20% of genetically defined ALS cases [2]. Although the exact mechanism of SOD1 mutations remains incompletely understood, there is a consensus that a toxic gain-of-function could disrupt several cellular functions thereby collectively contributing to MN degeneration [3–10]. Thus, lowering levels of SOD1 is predicted to be therapeutic.
The SOD1G93A transgenic mouse carries a high number of tandem repeats of a human SOD1 (huSOD1) transgene harboring the G93A mutation and recapitulates many aspects of disease, including progressive muscle atrophy, impaired motor functions and death due to paralysis [6]. It was used to support the preclinical development of Tofersen, an antisense oligonucleotide (ASO) [11] targeting huSOD1 mRNA for degradation through a RNase H1-dependent mechanism [12]. Delivery of SOD1-ASO to the cerebral spinal fluid (CSF) significantly reduced huSOD1 protein expression in the brain and spinal cord of SOD1G93A mice, extended survival and slowed the rate of neurodegeneration [12].
The translatability of these preclinical studies was supported by results from the Tofersen clinical trials [13]. In the Phase 3 VALOR study, SOD1 CSF concentrations decreased in patients receiving 100 mg of Tofersen over 28 weeks. Plasma neurofilament light chain (NfL), a potential marker of neuronal degeneration, also decreased. With the 12-month data from the open-label extension of Tofersen, signs of reduced disease progression across multiple secondary and exploratory endpoints (including motor function, respiratory function and quality of life) were also observed, though the primary endpoint as measured by the Revised Amyotrophic Lateral Sclerosis Functional Rating Scale did not reach statistical significance [14].
In recent years, gene therapy has emerged as a promising modality to treat diseases with defined genetic causes (e.g., monogenic liver, retinal and blood cell diseases). Adeno-associated virus (AAV) vectors allow for prolonged expression of engineered genetic information in diverse cell populations in the central nervous system (CNS) [15, 16] and are the basis of most gene therapy approaches for the treatment of neurodegenerative diseases (e.g., Alzheimer’s disease, spinal muscular atrophy, Parkinson’s disease, Huntington’s disease). If successfully developed, an AAV-based gene therapy could provide a one-time treatment for neurodegenerative diseases, such as SOD1 ALS. Several preclinical studies utilized AAV-delivered RNA interference (RNAi) to decrease huSOD1 mRNA levels in transgenic SOD1G93A mice and showed varying degrees of huSOD1 protein reduction and protection against disease progression [17–21]. Although clearly disease-modifying, the survival benefits reported were incomplete (<70 days extension of median survival) and most animals eventually succumbed to ALS-like symptoms, thus attempts to improve efficacy are warranted either through refinement of RNAi methods or through alternative strategies.
Alternative approaches to decrease huSOD1 protein levels include employing AAV9 with genome editing methods such as clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 systems [22–27]. The CRISPR-Cas9 system generates DNA double-strand breaks (DSBs) at genomic loci specifically targeted by single-guide RNAs (sgRNAs) and, following DNA repair, introduces insertions and/or deletions (indels) that disrupt gene expression [28]. Two studies utilized CRISPR to target the huSOD1 transgene in SOD1G93A mice using the AAV9 serotype and observed modest survival benefits (extension of median survival by 12 days and 25 days, respectively) [29, 30], perhaps due to insufficient distribution and/or expression of CRISPR-Cas9 components in target cell populations.
Here, we set out to assess key parameters affecting CRISPR-based gene therapy, with the goal of increasing huSOD1 protein reduction and assessing whether greater huSOD1 knockdown improved the reported therapeutic benefits in preclinical studies. We screened immortalized cells for sgRNAs directed against huSOD1 to identify sequences that markedly reduced human SOD1 protein levels. To deliver our SOD1-targeting sgRNAs to disease-relevant-cells in the SOD1G93A model, we utilized AAV-PHP.B and AAV-PHB.eB variants, which provide broader tropism and/or transduction rates in mice compared to other serotypes [31–34]. For our in vivo model, we crossed the SOD1G93A transgenic mice with the H11Cas9 knock-in mice [35], which are engineered to express constitutively spCas9 from a CAG promoter. This eliminates the need to package Cas9 into viral constructs and allows for broad distribution of Cas9 expression in all disease-relevant cells. We show that a single intracerebroventricular (ICV) injection of AAV-PHP.B-huSOD1-sgRNA into neonatal H11Cas9 SOD1G93A mice extended disease-free survival in the SOD1G93A model by >110 days and resulted in strong disease suppression through robust and sustained reduction of mutant SOD1 protein in the cortex and spinal cord. Furthermore, we observed improved protection (>170 days in survival extension) using the AAV-PHP.eB capsid to delivery SOD1-targeting sgRNA to pre-onset adult SOD1G93A mice via one-time intrathecal (IT) or intravenous (IV) injections. These results support the notion of applying this approach to multiple genes of therapeutic potential for effects in other CNS disease areas and points to key parameters of preclinical work to support the development of CRISPR-based gene therapy.
Materials and methods
Animals and general procedures
H11Cas9 mice [B6J.129-Igs2tm1.1(CAG-cas9*) Mmw/J; stock #028239; laboratory of M. Winslow, Stanford University, Stanford, CA] and transgenic SOD1.G93A mice (B6.Cg-Tg(huSOD1*G93A)1Gur/J; stock #: 004435) were purchased from the Jackson Laboratory. All mice were backcrossed for at least eight generations to C57BL/6 mice. Homozygous H11Cas9 female mice and heterozygous SOD1-G93A male mice were crossed to generate H11Cas9−/+; SOD1G93A−/+ mice and age-matched H11Cas9−/+; huSOD1.G93A−/− littermates. Mice were housed in a 12/12 h light/dark cycle in a temperature-controlled room (22–24 °C) with access to food pellets and water provided ad libitum. Behavioral testing occurred between 8:00 A.M. and 5:00 P.M. For survival studies, SOD1-G93A mice were observed daily for paralysis once they reached 130 days old. The animals were sacrificed at the humane endpoint, when they were unable to right themselves within 15 s of being placed on their side, showed 20% weight loss of peak body weight or exhibited hunched posture, as directed by a veterinarian. For biochemical and histological studies, mice were euthanized at the indicated age prior to paralysis, and organs were removed and immediately frozen in liquid nitrogen or fixed in 10% neutral buffered formalin (NBF). Experimenters were blind to treatment. All animal use and treatments were approved by the Biogen Institutional Animal Care and Use Committee (IACUC) and followed the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Single guide RNA (sgRNA) design, vector design and production
The eight SOD1-targeting spacer sequences of ~20 nucleotides [36] were selected based on Benchling (San Francisco, CA, USA) bioinformatic output [37, 38] against the human SOD1 gene (Table S1). Each sgRNA spacer sequence was used in combination with sgRNA scaffolds, structurally optimized for sp.Cas9 binding [39]. The sgRNAs were synthesized and cloned into the expression constructs by PackGene (Worcester, MA, USA). Each AAV construct contains a U6 promoter that drives expression of a sgRNA, and either a CBA promoter that drives expression of a green fluorescence protein (eGFP) fused with a KASH domain or a CAG promoter that drives expression of a mCherry fluorescence protein. All AAV vectors, including AAV-PHP.B and AAV-PHP.eB, were purchased from PackGene (Worcester, MA, USA).
Neonatal intracerebroventricular (ICV) injection, intrathecal injection (IT) and intravenous injection (IV) of AAV
Postnatal day 0 (P0) pups were anesthetized by hypothermia for 2–4 minutes until movement ceased. Cryo-anesthetized pups were injected with 4 μl of AAV buffered with PBS containing 0.25% of FastGreen dye into the lateral ventricle(s) (2E11 vg per mouse). Injection sites were located halfway between lambda and bregma, 1 mm lateral to the superior sagittal sinus, to a depth of 2 mm. Injections were performed with a 33-gauge, 10 µL, 45o bevel Hamilton syringe (Hamilton Company, Reno, NV, USA) inserted perpendicular to the surface of the skull. Injection efficiency was monitored by the spread of the dye throughout the lateral and the third ventricles. Mice were euthanized for tissue collection at 20 weeks of age for histological analysis, and at 5, 10 or 20 weeks of age for biochemical and genomic analysis. Animal injection, group allocation and takedown were pseudorandomized.
For intrathecal injection (IT), 5 weeks of age mice were anesthetized with isoflurane first in the chamber and then continuously anesthetized with isoflurane to maintain deep anesthesia via a nose cone throughout the procedure pad. The fur on the back of the anesthetized mice were shaved from the tail to the caudal thoracic spine and placed on a sterile sheet laid on top of a heating pad. 100 μl of AAV buffered with PBS (1E12 vg per mouse) was injected into the intrathecal space between the L3 an L4 vertebrae with a 31-gauge, 0.5 mL insulin syringe [40]. A subset of injected mice was euthanized for tissue collection at 15 weeks of age for biochemical analysis.
For IV injection, 5 weeks of age mice were placed in a restraint that positioned the mouse tail in a lighted, heated groove (Braintree Scientific, Inc). The tail was swabbed with alcohol and then injected intravenously with 100 μl of AAV buffered with PBS (1E12 vg per mouse). The sample size for all mouse experiments was an n > 6/treatment was determined from prior unpublished data.
Open field
To assess locomotor activity, mice were placed within an open field apparatus and locomotor activity was captured by video tracking software. After, mice were acclimated to the testing room for 30 min they were individually placed in a cylindrical arena (43 cm × 36 cm: d x h). An overhead camera was used to track the animals using Noldus Ethovision (version 13) for 15 min. Mice were then returned to their home cage, and the arena was thoroughly cleaned prior to the next session. Locomotor activity was assessed on a monthly basis, starting at 7 weeks of age and concluding at 43 weeks of age.
Rotarod
For the Rotarod testing, mice were placed in one of five lanes on a slowly rotating rod (Ugo Basile) 16 cm above a stainless-steel trip box. Mice were acclimated to the testing room for a minimum of 30 min prior to testing. Once the mice were placed on the rotating rod, it gradually accelerated from 5 to 40 RPM (revolutions per minute) over 5 min. When the mice fell onto the trip box, a magnetic switch was activated and the latency to fall was recorded. For each mouse there were three test trials each day with a minimum 5-min interval between test trials. After each trial the mice were returned to their home cage. Observations were recorded at 11, 15, 19, 21, 23, 25, 27, 29, 31, 35, 39, and 43 weeks of age.
Inverted grid
Mice were placed on a small, inverted metal grid 25 cm above standard shaving bedding and latency to fall was recorded. Mice were acclimated to the test room for a minimum of 30 min prior to testing. The height of the grid allowed a soft landing when the mice fell but was high enough to prevent the mice from voluntarily jumping down. The time to fall was recorded by an experimenter with a stopwatch. The trial ended when the mouse fell or remained on the grid for 240 s. Up to two trials were done each test day with a rest interval of at least 2 min between trials. The second trial was only performed if the mouse did not reach 240 s on the first trial. After each trial and upon test completion, mice were returned to their home cage. Observations were recorded at 25, 27, 31, 35, 39, and 43 weeks of age.
Clasping
Mice were observed for limb clasping each day just prior to the rotarod assessment. Mice were lifted by the base of the tail and were observed for 10 sec and then placed back into their home cage. A score was assigned based on the following 5-point scale: limbs splayed outward (away from abdomen) = 0, one limb retracted toward abdomen = 1, two limbs retracted toward abdomen = 2, three limbs retracted toward abdomen = 3, four limbs retracted toward abdomen = 4, and all limbs retracted toward abdomen and body forming a ball =5. Observations were recorded at 11, 15, 19, 21, 23, 25, 27, 29, 31, 35, 39, and 43 weeks of age.
Compound Muscle Action Potential (CMAP)
Recordings were conducted by an experimenter blinded to genotype and treatment. All measurements were performed with animals maintained under isoflurane anesthesia (1.5–2.5%) and with body temperature maintained around 37 °C. Electrophysiological responses were obtained separately from both the left and right hind limbs of each mouse; the left hind limb was always recorded before the right. The period of testing (and time during which the animal was anesthetized) lasted approximately 10 minutes. Responses were recorded at 5, 8 and 10 weeks of age, and at monthly intervals subsequently until age week 46. Disposable monopolar needle electrodes (Teca 25 mm, 28G electrodes, Natus Medical Inc., San Carlos, CA) were used for both stimulation and recording. The sciatic nerve was stimulated with constant-current monophasic square-wave pulses (0.1 ms duration) produced by a WPI (model 365A) stimulator, with timing controlled by a data acquisition interface (National Instruments USB-6343). Current was delivered via a stimulating cathode placed sub-dermally near the sciatic notch to stimulate the sciatic nerve. The recording electrode was placed 1 mm intramuscularly into the belly of the tibialis anterior muscle and the reference electrode was placed at the ankle. For each recording, stimulation was applied every 2 s with incremental current levels (beginning at 1.0 mA and increasing in 0.5 mA steps) until the CMAP amplitude stopped increasing; recordings were performed using a current level 0.5 mA above this level. This supra-maximal stimulus intensity (typically found with a current between 1.5 mA and 3.5 mA) was used to record four CMAP responses which were then averaged for analysis. The minimum and maximal values of the response waveform were measured beginning 0.8 ms after stimulation to exclude the stimulus artifact. The absolute value of these numbers was taken and then summed for the final CMAP value of a response. The final CMAP value for a given animal is the average of the left and right leg peak-to-peak amplitudes. Each animal is considered one data point for statistical purposes. CMAP data was analyzed with two-way ANOVA followed by Sidak’s multiple comparison test, P < 0.05 is considered significant.
NMJ staining and counting
Tibialis anterior muscles were dissected and fixed in 10% NBF at 4 °C for 8 h, followed by 2% paraformaldehyde [11] buffered in 0.1 M sodium phosphate at 4 °C for 16 h, and then transferred to 20% sucrose in PBS at 4 °C for 24 h for cryoprotection. Samples were shipped to Jackson Laboratory for cryo-embedding in OCT, staining and analysis. Sections (20 μm) were collected by a cryostat and mounted directed onto glass slides. Sections were incubated overnight in primary antibodies containing a cocktail of mouse monoclonal anti-neurofilament 2H3 and anti-SV2 antibodies (Developmental Studies Hybridoma Bank), washed the following day and incubated overnight with Alexa-Fluor-488 conjugated anti-mouse IgG1 and Alexa-Fluor-594 conjugated α-bungarotoxin (BTX, Invitrogen). Sections were mounted to slides in fluorescence mounting media (DAKO, S3023) and imaged on a Leica SP5 laser confocal microscope. Manual counting of NMJ was performed by blinded individuals under a microscope as innervated, partially innervated (i.e., 50% or less anti-SV2 and anti-neurofilament positive signals overlapping with anti-bungarotoxin positive signals) and denervated NMJ. A total of 970 NMJs were counted in SOD1; sgLacZ group, 1531 NMJs were counted in SOD1; sgSOD1#5 group, and 1949 NMJs were counted in WT; sgLacZ group.
Spinal motor neuron staining and quantification
Lumbar spinal cord samples were fixed in 10% NBF for 48 h, processed on the Leica Peloris tissue processor and embedded into paraffin in a transverse orientation. Samples were bisected prior to embedding, such that every block contained two segments. Sectioning was performed on a Leica rotary microtome at a 5-micron thickness. Sections were collected at 6 levels, >50 µm apart. 6 sections per animals were used for IHC staining for choline acetyltransferase (ChAT), for a total of 12 technical replicates for quantification of motor neuron counts. Immunohistochemistry was performed for ChAT to assess MN numbers. The automated Ventana Discovery Ultra staining platform was used. Briefly, slides were deparaffinized and rehydrated. All reagents were from Roche Ventana, unless stated otherwise. Epitope retrieval was performed in the CC2 buffer (pH 6.0) for 64 min, at 93 °C. Primary antibody (rabbit monoclonal anti-choline acetyltransferase, clone EPR16590, Abcam, Cat. No. ab178850) was applied at the final concentration of 2 µg/mL for 60 min at ambient temperature. Goat anti-rabbit polyclonal antibody conjugated to nitropyrazole (NP) was applied for 12 min, followed by incubation with anti-NP reagent conjugated to alkaline phosphatase for 12 min. Positive staining was visualized with Discovery Red chromogen applied for 16 min. Slides were counterstained with hematoxylin. Slides were digitized on a 3D-Histech Pannoramic-250 whole slide scanner at 200x magnification. Custom made VisioPharm algorithms were used to quantify motor neurons in the ventral horns of the spinal cord.
Tibialis muscle staining and analysis
Tibialis Anterior muscle samples were fixed in 10% NBF for 7 days, processed on the Leica Peloris tissue processor and embedded into paraffin in a transverse orientation. Samples were bisected prior to embedding, such that every block contained two segments of the muscle. Sectioning was performed on a Leica rotary microtome at a 5-micron thickness. Gomori method for staining reticulin fibers was applied. Briefly, slides were deparaffinized in a series of xylene and graded ethanol solutions, and rehydrated. Slides were incubated with the following solutions (all from Poly Scientific R&D Corp unless stated otherwise): 0.5% aqueous potassium permanganate (Cat. No. S263), 2% aqueous potassium meta-bisulfite (Cat. No. S2005), 2% aqueous ferric ammonium sulfate (Cat. No. S179), Gomori’s ammonical silver nitrate (Cat. No. S114), 10% buffered formalin (Cat. No. S185), 0.2% aqueous gold chloride (Cat. No. S202), 2% aqueous potassium meta-bisulfite (Cat. No. S2005), and 2% aqueous sodium thiosulfate (Cat. No. S280). Slides were thoroughly washed between each incubation step. All subsequent reagents were from Roche Ventana, unless stated otherwise. Epitope retrieval was performed in the CC2 buffer (pH 6.0) for 64 min at 93 °C. To block the endogenous mouse IgG and non-specific background, slides were incubated with Rodent Block M (Biocare Medical, Cat. No. RBM961) for 16 min. Slides were counterstained with hematoxylin.
Slides were digitized on a 3D-Histech Pannoramic-250 whole slide scanner at a 200x magnification. Custom made VisioPharm algorithms were used to assess the average diameter of myosin fibers.
Blood neurofilament quantification
Roughly 30 μl blood was collected by facial vein puncture at the indicated time points. Serum samples were prepared by centrifugation through BD Microtainer SST Clog Activator/Gel tubes (Becton Dickinson) and stored at −80 °C until used. Levels of pNFH in serum were measured by the ELLA microfluidic ELISA platform according to the manufacturer’s instructions (Protein Simple).
Human SOD1 ELISA analysis
Cortex or spinal cord tissues were homogenized in tissue lysis buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X, 1% Na-deoxycholate, 0.1% SDS, 8 M Urea, 5 mM EDTA, supplemented with 1 mM dithiothreitol, 1 mM phenylmethanesulfonylfluoride fluoride (Sigma, 93482), complete protease inhibitor (Roche, 04693124001), PhosSTOP phosphatase inhibitor (Roche, 4906845001), 10 mM sodium fluoride, and 1 mM sodium orthovanadate (New England Biosciences, P07585)], by using TissueLyser II (QIAGEN). Homogenates were subsequently cleared via centrifugation at 20,000 x g for 30 min. Total protein concentration of the supernatant was measured by using BCA Protein Assay Kit (Pierce). Human SOD1 protein levels were measured from lysates with equal amount of total proteins, using Human Cu/ZnSOD1 Platinum ELISA Kit according to manufacturer’s instructions (BMS222, Invitrogen).
Nuclei isolation, human SOD1 indel analysis and copy number analysis
For Cohort #1, at 5-, 10-, or 20-weeks post-injection of AAV-U6-sgRNA-CBA-eGFP-KASH, cortices and half spinal cords were dissected, snap frozen and stored at −80 °C. Frozen tissues were thawed and homogenized in 1.5 mL ice-cold homogenization buffer (HBSS, 25 mM HEPES). The homogenate was passed through a 250 µm filter and spun at 600 x g for 5 min at 4 °C. The cell pellet was gently resuspended in 1 mL FBS and subsequently in 9 mL of 33% Percoll solution (GE Healthcare, Chicago, IL, USA) containing HBSS and 16.7 mM HEPES. An additional 1 mL of 10% FBS solution containing HBSS and 22.5 mM HEPES was carefully layered onto the top of cell suspension layer containing 30% Percoll. Density gradient centrifugation was performed at 800 x g for 15 min at 4 °C (1 acceleration and 1 brake). The supernatant was removed, and the nuclei pellet was resuspended and washed in FACS buffer (HBSS, 1% BSA, 2 mM EDTA, 25 mM HEPES, 0.09% sodium azide). Half a million intact EGFP positive nuclei labeled with Vybrant DyeCycle Violet Stain (Thermo Fisher Scientific) were isolated by FACS using MoFlo Astrios EQ (Beckman Coulter, Brea, CA, USA). Genomic DNA was isolated from the sorted nuclei or total nuclei by DNeasy Blood & Tissue Kit (catalog #: 69504) according to manufacturer’s instruction (QIAGEN).
For Cohort #4, at the end of study (~45 weeks post-injection of AAV-U6-sgRNA-CBA-eGFP-KASH), cortices and half spinal cords were dissected, snap frozen and stored at −80 °C. Total genomic DNA were isolated from frozen cortices and spinal cords using DNeasy Blood & Tissue Kit (catalog #: 69504) according to manufacturer’s instruction (QIAGEN).
For human SOD1 indel analysis, primers (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGagcttgctggaggttcactg-3′; 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGcacaacacccacctgctgta-3′) with Illumina adaptor (underlined) were used to amplify region of human SOD1 surrounding the sgRNA-targeted locus, using KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA). The library was constructed by indexing individual samples with Illumina’s Nextera XT indices (Illumina, San Deigo, CA, USA) with limited PCR cycles. The library was pooled by volume and purified using AMPure XP beads (Beckman Coulter). The final library pool was quantified by KAPA Library Quantification Kit (Kapa Biosystems) and loaded on to Illumina’s MiSeq at 10 pM for 2 × 150 bp cycle run.
For human SOD1 transgene copy number analysis, the following oligonucleotides were used for human SOD1: 5′-GGGAAGCTGTTGTCCCAAG - 3′ (primer #1); 5′- CAAGGGGAGGTAAAAGAGAGC - 3′ (primer #2); 5′-CTGCATCTGGTTCTTGCAAAACACCA - 3′ (probe conjugated with FAM). The follow oligonucleotides were used for ApoB as a reference gene with two copies per cell: 5′-CACGTGGGCTCCAGCATT - 3′ (primer #1); 5′- TCACCAGTCATTTCTGCCTTTG - 3′ (primer #2); 5′-CCAATGGTCGGGCACTGCTCAA - 3′ (probe conjugated with HEX). Oligonucleotides were purchased from IDT (Newark, NJ, USA). The ddPCR 2x Super Mix reagent (186–3010, Bio-Rad) was used for the real-time amplification, along with 5–10 ng of genomic DNA, 900 nM of primers and 250 nM of probe. Prior to thermal cycling, the reaction mixtures were further processed into droplets by QX100 Droplet Generator according to manufacturer’s instructions (1863002, Bio-Rad). After initial activation at 95 °C for 10 min, 40 PCR cycles of 96 °C for 30 s and 59 °C for 60 s were performed, followed by 98 °C for 10 min and then held at 4 °C. Amplified products in the droplets were subsequently analyzed on QX100 Droplet Reader (1863003, Bio-Rad).
Tissue culture, transfection, AAV transduction and RNA sequencing (RNAseq)
COS1 cells were purchased from ATCC (CRL-1650) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 1% Penicillin-Streptomycin (Pen-Strep) and 2 mM L-glutamine and incubated at 37 °C and 5% CO2. FUGENE HD (Promega, San Luis Obispo, CA, USA) was used for transfection according to the manufacturer’s instructions. The expression plasmid CMV-HA-human SOD1, CMV-FLAG-Cas9 and U6-sgSOD1-CAG-mCherry were mixed in a 1:4:5 ratio in Opti-MEM media (Thermo Fisher Scientific, Waltham, MA, USA) for transfection. The transfected COS1 cells were harvested 24 h after transfection.
The inducible Neuro-2a cells expressing sp.Cas9 were purchased from GeneCopoeia and maintained in DMEM supplemented with 10% HI-FBS, 1% L-glutamine and hygromycin (100 µg/mL final) and incubated at 37 °C and 5% CO2. Neuro-2a cells were treated with doxycycline inducer at a concentration of 0.01, 0.1, or 1 µg/mL to induce expression of sp.Cas9. The expression plasmid CMV-HA-human SOD1 and U6-sgSOD1-CAG-mCherry were mixed in a 1:9 ratio in Opti-MEM media (Thermo Fisher Scientific, Waltham, MA, USA) for transfection by FUGENE HD. The transfected Neuro-2a cells were harvested 24 h after transfection.
HeLa cells stably expressing Cas9 (GeneCopoeia SL503) were cultured in DMEM, 10% FBS, 1% Pen-Strep, 1x final GlutaMAX (Thermofisher Scientific 35050061), and Hygromycin (250 µg/mL final) at 37 °C and 5% CO2. After 24 h of seeding into 24-well PDL-coated tissue culture plates (Corning 354470), cells were transduced with AAV-PHP.B-sgLacZ and AAV-PHP.B-sgSOD1#5 at 1000k MOI (multiplicity of infection) for 48 h or 72 h. The cells were then harvested in QIAzol (Qiagen 79306) for RNA extraction using miRNeasy mini kit (Qiagen 217004). For bulk RNAseq, 250 ng of extracted RNA was used to prepare RNAseq libraries using the Kapa mRNA hyperprep kit (Kapa biosystems KK8581), following the manufactures instructions. Fragmentation of mRNA was carried out for 6 min at 94 °C, followed by RT, A-tailing, adaptor ligation, and 12 cycles of PCR, to produce Illumina sequencing libraries. Tagged libraries were pooled and sequenced on an Illumina NovaSeq6000 sequencer with a run parameter of 2 × 51 bp paired end reads. Quality control was performed using Illumina’s BaseSpace run summary tool, which showed a %Q30 of 94.14%. The fastq files were generated from the bcl files using bcl2fastq v2.20 (Illumina). The RNA-seq analysis pipeline consisted of alignment with STAR version 2.5.2a against human genome version GRCh38 and Gencode gene model release 27 [41, 42]. After alignment, quantification of gene expression was carried out with RSEM v1.2.26 [43]. Data quality metrics and plots for visualization were generated with Quickomics [44]. Differential expression analysis was performed with DESeq2 version 1.30.0 [45]. The function apeglm was used for LFC shrinkage to reduce noise and preserve large differences [46]. Cutoff values used for significant differential expression were fold change >2 and adjusted p-value < 0.05.
Immunoblotting and antibodies
For the detection of protein expression in total cell lysates, cells were lysed in Novex Tris-Glycine SDS sample buffer containing NuPAGE sample reducing agent (Invitrogen). Lysates were electrophoresed through Novex Tris-Glycine gels or NuPAGE Bis-Tris gels, transferred to nitrocellulose membranes, and subjected to immunoblot analysis. The blocking of membranes and subsequent antibody incubations were performed using Odyssey blocking buffer (LI-COR Biosciences) according to the manufacturer’s instructions. Primary antibodies against β-Tubulin (926–42211, LI-COR Biosciences), β-actin (926–42210, LI-COR Biosciences), FLAG (F1804, Millipore Sigma, Burlington, MA), mCherry (ab167453, Abcam, Cambridge, United Kingdom), GAPDH (ab8245, Abcam), and human SOD1 (ADI-SOD1–100, Enzo) were purchased from commercial sources. The IRDye 800CW-conjugated and IRDye 680-conjugated secondary antibodies were obtained from LI-COR Biosciences. Immunoblot signals were visualized by the Odyssey CLx infrared imaging system and quantified by ODYSSEY application software (LI-COR Biosciences).
Results
Neonatal ICV injection of AAV-sgSOD1 effectively reduces huSOD1 protein in the CNS and generates a high indel rate in the huSOD1 gene
To identify CRISPR/sgRNAs that disrupt huSOD1 expression, eight huSOD1-targeting sgRNAs (termed sgSOD1s) were assessed for their CRISPR activity in vitro in two different cell lines. COS-1 cells were co-transfected with expression vectors for huSOD1, Streptococcus pyogenes Cas9 nuclease (spCas9) and eight different sgSOD1s. These eight sgRNAs were compared for their ability to facilitate reduction of exogenously expressed huSOD1 protein by immunoblotting analysis (Fig. S1). Additionally, engineered Neuro-2a cells were induced to express spCas9, and were subsequently co-transfected with expression vectors for huSOD1 and sgSOD1s. Two of the huSOD1-targeting sgRNAs, sgSOD1#1 and sgSOD1#5, consistently resulted in marked reduction of exogenous huSOD1 protein levels in the COS-1 and Neuro-2a cell types and were used in subsequent in vivo studies (Fig. S1).
CRISPR-Cas can sometimes cause off-target genome editing which could lead to unintended changes in gene expression. To assess for potential off-target effects in an unbiased manner, we performed bulk RNA sequencing to detect differentially expressed genes (DEGs) in human HeLa cell lines stably expressing Cas9 transduced with AAV9-sgSOD1#5 at an MOI of 1000 K for 48 h or 72 h. The average sequencing depth was 100 million reads per sample, with mapping rates of 97% and 85% of the reads mapping to exons. As expected, AAV-sgSOD1#5 caused a pronounced reduction of 69% and 79% in SOD1 expression at 48 h and 72 h. When compared to the control samples transduced with AAV9-sgLacZ, two genes (SERPINE2, MYEOV) were slightly downregulated by AAV-sgSOD1#5 at 48 h post-transduction (Fig. S2). However, downregulation of these genes was not observed at 72 h post-transduction, suggesting that these genes are unlikely to be targeted directly by sgSOD1#5. Consistent with this notion, neither gene shares homology with the spacer sequence of sgSOD1#5 and neither gene was a predicted off-target based on Benchling’s “CRISPR Guide RNA Design” tool [38]. Taken together, these data indicate that sgSOD1#5 does not display any off-target effects in human HeLa cells.
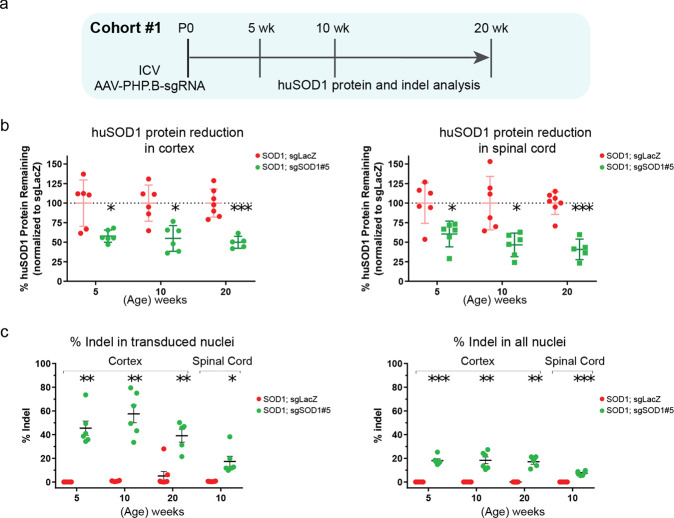
After confirming the ability of sgRNAs to reduce huSOD1 protein in vitro, we evaluated whether a huSOD1-targeting CRISPR system delivered by AAV (AAV-CRISPRSOD1) could reduce huSOD1 protein and alleviate disease progression in SOD1G93A mice crossed to H11Cas9 knock-in mice. sgSOD1#1 and sgSOD1#5 were packaged into AAV-PHP.B capsids, selected for their broad CNS transduction rates [31–34]. sgRNA targeting the non-mammalian LacZ gene (termed sgLacZ) was used as the control. In H11Cas9 SOD1G93A mice, AAV-sgRNAs (2E11 viral genome (vg) per animal) were delivered to the CSF via intracerebroventricular (ICV) injection on post-natal day 0 (P0) (Fig. 1a, Cohort #1). We first analyzed the degree of huSOD1 protein reduction by AAV-sgSOD1 using an ELISA assay. At 5 weeks after P0 ICV injection of AAV-PHP.B-sgRNA directed against huSOD1, we observed ~50–60% huSOD1 protein reduction in the cortex and spinal cord of sgSOD1-treated mice – this level of reduction was sustained at later time points (10 and 20 weeks of age, Fig. 1b). As a direct indicator of CRISPR-mediated gene editing efficiency, we then assessed the indel rates in the huSOD1 transgene by amplicon sequencing. In the cortex, we detected indels in ~20% of the huSOD1 transgenes in all nuclei, which included nuclei of transduced and non-transduced cells. Using FACS to enrich for GFP-positive nuclei transduced by AAV-PHP.B, the indel rate in the huSOD1 transgene doubled to ~40% (Fig. 1c). Consistent with stable reduction of huSOD1 protein over time, the indel rates in the cortex remained stable at 5-, 10- and 20-weeks post-injection. In the spinal cord, huSOD1 transgene indels were detected in ~7% of the total nuclei, as compared to an indel rate of ~17% in the AAV-transduced nuclei after enrichment by FACS (Fig. 1c).
Fig. 1. Neonatal ICV injection of AAV-sgSOD1 effectively reduces huSOD1 protein in the CNS and generates high indel rates in the huSOD1 gene.
a Study design of histological and biochemical analysis on H11Cas9 SOD1G93A mice treated with AAV-sgSOD1 (Cohort #1). b Percentage of remaining huSOD1 protein in cortex (left) and spinal cord (right) of H11Cas9 SOD1G93A mice treated with AAV-sgSOD1 (Cohort #1) as determined by ELISA. c Indel percentage in cortex and spinal cord of H11Cas9 SOD1G93A mice treated with AAV-sgSOD1 (Cohort #1) as determined by amplicon-Seq of transduced nuclei (left) and all nuclei (right). Data are presented as mean with standard deviation (SD) unless otherwise stated. Welch’s t-test with Bonferroni correction is performed unless otherwise stated. *p < 0.05, **p < 0.01, and ***p < 0.001, n.s. not significant. See also Fig. S3.
To assess the frequency of large excisions of SOD1 transgenes, we used droplet digital PCR (ddPCR) to directly assess huSOD1 transgene copy number remaining in the transduced nuclei from the cortex and spinal cord of 10-week-old animals. We found no difference in huSOD1 transgene copy numbers between sgSOD1-treated and sgLacZ-treated mice (Fig. S3), indicating that a large excision could not be detected following CRISPR editing in vivo using ddPCR.
Thus AAV-sgSOD1 delivered to neonates generates high indel rates in the huSOD1 gene and effectively reduces huSOD1 protein in the CNS in SOD1G93A mice. While we did not measure indel rates at the endogenous mouse SOD1 gene, sgSOD1#1 targets a region in the SOD1 gene that is 100% conserved between human and mouse. Unlike sgSOD1#1, sgSOD1#5 targets a region in the SOD1 gene that is poorly conserved between human and mouse and is unlikely to induce murine SOD1 suppression. As describe above, sgSOD1#5 did not result in any off-target gene expression changes in human HeLa cells. For these reasons, sgSOD1#5 was selected as the lead sgRNA used in all in vivo study cohorts while sgSOD1#1 was used in a subset of in vivo study cohorts.
We previously reported broad neuronal distribution and high rates of transduction in the hippocampus (~80%), cortex (~95%) and in the dorsal (~60%) and ventral (~80%) spinal cord upon neonatal ICV injection of AAV-PHP.B targeting NeuN [33]. As our current analysis used the same AAV-PHP.B serotypes as in the NeuN study, we elected not to perform histological experiments to assess cell type distribution and transduction efficiency. We expect similar cellular distribution and transduction efficiencies as those observed in Hana et al.
Neonatal ICV injection of AAV-sgSOD1 prevents motor function decline and extends survival in H11Cas9 SOD1G93A mice
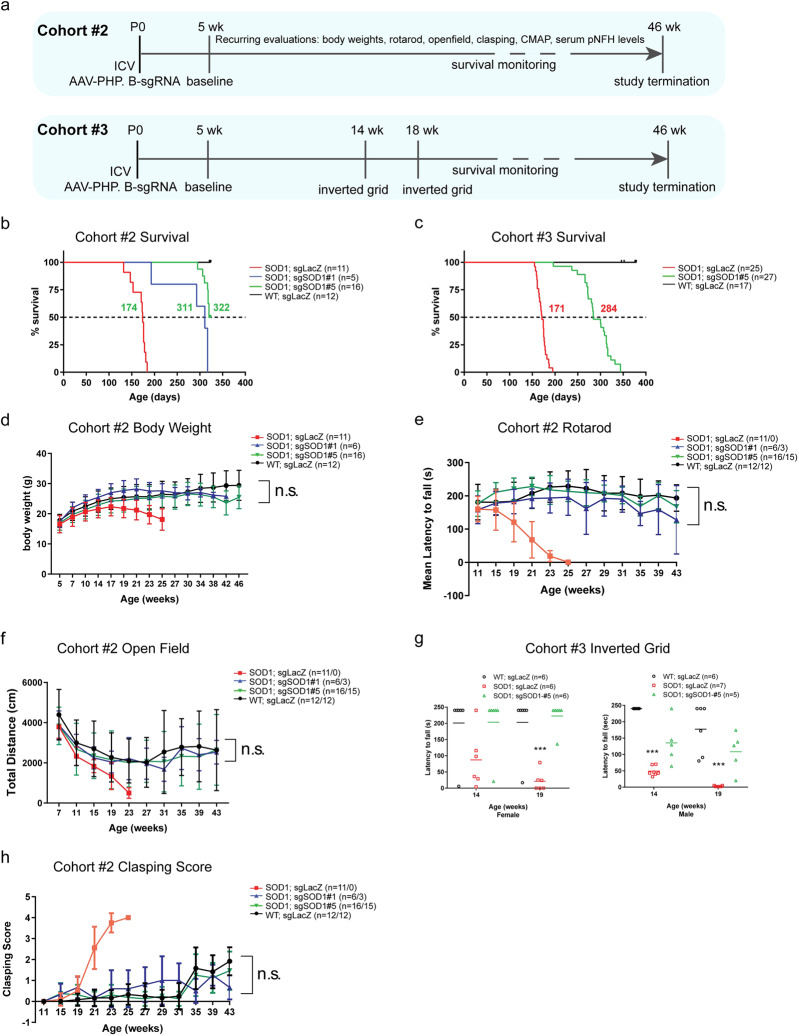
We next evaluated whether AAV-sgSOD1 treatment could alleviate disease progression in the H11Cas9 SOD1G93A mouse model of ALS. The effect of AAV-sgSOD1 on survival and motor functions in mice was evaluated in two independent study cohorts (Cohort #2 and Cohort #3, Fig. 2a). Again, AAV-sgRNAs (2E11 vg per animal) were delivered to the CSF via ICV injection on P0.
Fig. 2. Neonatal ICV injection of AAV-sgSOD1 prevents motor function decline and significantly extends survival in H11Cas9 SOD1G93A mice.
a Study design of two independent cohorts (Cohorts #1 and #2) of H11Cas9 SOD1G93A mice treated with AAV-sgSOD1. b, c Kaplan-Meier survival curves of Cohorts #1 and #2. Log-Rank test, P < 0.0001. d Body weight of mice over time (Cohort #1). Behavioral assessments of SOD1G93A mice treated with AAV-sgSOD1: rotarod performance measured as latency to fall over age (e), total distance travelled in open field over time (f), performance in inverted grid test measured as latency to fall at week 14 and 19 (g), and clasping score over age (h). Data are presented as mean with standard deviation (SD) unless otherwise stated. Two-way ANOVA was performed with all groups compared to WT group in d–f. One-way ANOVA Dunnett’s multiple comparisons test was performed with all groups compared to WT group in g (n.s. is not labeled). *p < 0.05, **p < 0.01, and ***p < 0.001, and n.s. not significant (p > 0.05).
Strikingly, both huSOD1-targeting sgRNAs extended the life span of H11Cas9 huSOD1G93A mice by >110 days: 137 days (~20 weeks) in Cohort #2 and 113 days (~16 weeks) in Cohort #3 (Fig. 2b, c). As further evidence of efficacy, out of a total of 48 treated animals (21 in Cohort #2 and 27 in Cohort #3), only one exhibited paralysis and succumbed to ALS-like symptoms at 193 days (~27 weeks) whereas 47 died or required euthanasia for humane reasons at ~300 days (~43 weeks) without paralysis or any other ALS-like symptoms. Euthanasia was required in this group because of hunched body posture and overall poor body condition, malocclusion or prolapse, or a combination of the two. These symptoms in our treated SOD1G93A mice could result from CNS-independent SOD1G93A toxicity and warrant further investigation [11, 47–49]. Furthermore, over time, sgSOD1#1- and sgSOD1#5-treated animals maintained body weights that were similar to those of wild type mice (Fig. 2d), indicating effective protection against disease progression. We further performed a battery of motor behavioral tests, including rotarod, open field, and inverted grid (Fig. 2e–g). sgLacZ-treated control mice exhibited motor deficits beginning at 14–15 weeks of age which became more severe by week 19. In contrast, the performance of sgSOD1-treated animals was not significantly different from that of wild type mice, even at 43 weeks of age (Fig. 2e–g). We also used the “clasping” reflex phenotype to monitor motor function (Fig. 2h). This reflex assesses motor control whereby wild type animals lifted by the tail extend their hindlimbs outward. In contrast, mice with motor deficits when handled in this manner exhibit abnormal “clasping” characterized by retraction of the hindlimbs toward the abdomen, an effect observed in SOD1G93A mice as well as other ALS models [50, 51]. While sgLacZ-treated H11Cas9 SOD1G93A mice showed abnormal clasping behavior starting at ~21 weeks of age, animals receiving the huSOD1-targeting sgRNAs showed minimal clasping, resembling wild type mice (Fig. 1h). Together, these data demonstrate that CRISPR-mediated in vivo disruption of the huSOD1 transgenes in SOD1G93A mice prevented the development of motor deficits and markedly extended survival.
Neonatal ICV injection of AAV-sgSOD1 prevents neuromuscular junction loss, muscle atrophy and neurodegeneration
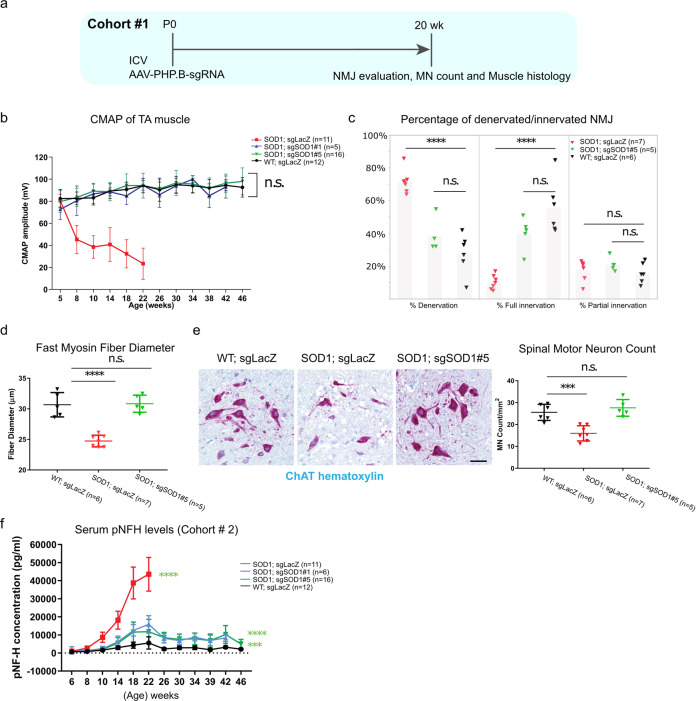
The first sign of disease pathophysiology in SOD1G93A mice is characterized by denervation of the neuromuscular junction (NMJ) in the hindlimbs [52, 53]. Denervation can be detected at the electrophysiological level as a decline over time in compound muscle action potential (CMAP), followed by MN loss [12]. To test whether CRISPR-mediated huSOD1G93A reduction could affect this phenotype, H11Cas9 SOD1G93A mice were injected with AAV-PHP.B-sgRNAs intracerebroventricularly at P0 and evaluated for CMAP (Cohort #2) and innervation at NMJ (Cohort #1) in the tibialis anterior (TA) muscles in the hind limbs (Figs. 2a, 3a). Remarkably, CMAP measurements of mice treated with either sgSOD1#1 or sgSOD1#5 were indistinguishable from their wild type littermates over the course of 46 weeks or until the end stage, whereas CMAP for sgLacZ-treated SOD1G93A animals was reduced by greater than 75% by 22 weeks of age (Fig. 3b). We double-labeled MN axon presynaptic terminals with anti-neurofilament and anti-SV2, and muscle post-synaptic membranes with α-bungarotoxin to evaluate the integrity of NMJ in the TA muscle. Consistent with electrophysiological preservation of CMAP, this analysis revealed that sgSOD1-treated mice-maintained innervation of TA muscles, whereas sgLacZ-treated SOD1G93A animals showed evidence of full denervation of ~70% of NMJ in muscle endplates by 20 weeks of age (Fig. 3c).
Fig. 3. Neonatal ICV injection of AAV-sgSOD1 prevents NMJ loss, muscle atrophy and neurodegeneration in H11Cas9 SOD1G93A mice.
a Study design of histological and biochemical analysis on H11Cas9 SOD1G93A mice treated with AAV-sgSOD1 (Cohort #1). b CMAP amplitude over disease progression (time). Two-way ANOVA is performed with all groups compared to WT group. c Percentage of fully innervated, denervated and partially innervated NMJs in male mice at 20 weeks of age. d Diameter of fast myosin muscle fiber of tibialis muscles at 20 weeks of age. e Left, representative images of spinal motor neurons stained by ChAT at 20 weeks of age. (scale bar, 50 µm); Right, spinal motor neuron numbers at 20 weeks of age. c–e One-way ANOVA Dunnett’s multiple comparisons test was performed with all groups compared to WT group. f Serum pNFH levels over disease progression (time). Two-way ANOVA was performed, all groups compared to WT group. b–f Data are presented as mean with standard deviation (SD) unless otherwise stated. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and n.s. not significant. Data was from collected from Cohort #3 in B-E and from Cohort #1 in F.
A myosin classification system based on ATPase reactivity distinguishes skeletal muscle fibers as either fast-twitch or slow-twitch types [54]. In SOD1G93A mice, fast-twitch muscle fibers are more susceptible to atrophy than slow-twitch fiber types [55, 56]. To determine if AAV-sgSOD1 protects against these changes in the TA muscles, we compared sgSOD1- and sgLacZ-treated H11Cas9 SOD1G93A mice for fast-myosin fiber diameter at 20 weeks of age. As expected, sgLacZ-treated SOD1G93A mice showed significant decrease in fast-myosin fiber diameter, indicating atrophy of fast-twitch muscle fibers (Fig. 3d and Fig. S4). The TA muscle of sgSOD1-treated H11Cas9 SOD1G93A mice did not show any of these phenotypes and were indistinguishable from wild type littermates.
Death of spinal MNs represents a hallmark of ALS, a phenotype that is recapitulated in the SOD1G93A mice [6]. To determine if CRISPR-mediated huSOD1G93A reduction can protect against MN loss, we counted MNs stained positive for choline acetyltransferase (ChAT) [18] in the spinal cords of sgLacZ- and sgSOD1-treated animals. We observed a 37% decrease in the number of ChAT+ MNs in sgLacZ-treated H11Cas9 SOD1G93A mice at 20 weeks of age (Fig. 3e), whereas no decrease in MN number was detected in sgSOD1-treated animals, (Fig. 3e).
Elevated levels of phospho-neurofilament heavy chain (pNFH), one of the main by-products of axonal damage [57], are detected in CSF and serum from patients with neurodegenerative diseases, including ALS [58]. As similar increases are observed in ALS rodent models [59], sgSOD1-treated H11Cas9 SOD1G93A mice (Cohort #2) showed lower levels of serum pNFH compared to controls (Fig. 3f). This reduction occurred in two phases: an initial rise that never exceeded 40% of the levels detected in SOD1 mice treated with control AAV and a subsequent plateau at <20% of the maximal levels observed in SOD1 controls. Therefore, AAV-sgSOD1 protects against neurodegeneration. Despite full correction of the morphological and functional readouts, AAV-sgSOD1 treatment did not completely extinguish the pNFH biomarker signal. This could not be explained by degeneration caused by the AAV vector alone, since wild type mice injected with control AAV did not show significant pNFH increases. It therefore seems likely that a set of neurons not required for normal function were not transduced and/or changes in pNFH are more sensitive indicators of ongoing low rates of MN degeneration than the functional or morphological assays.
Collectively, these data demonstrate that sgRNA-guided in vivo editing of toxic huSOD1 prevented NMJ loss and muscle atrophy, and strongly reduced neurodegeneration in the SOD1G93A mouse model.
Intrathecal and intravenous injection of AAV.PHP.eB-CRISPRSOD1 in adult mice fully preserves neuromuscular function and extends survival beyond one year
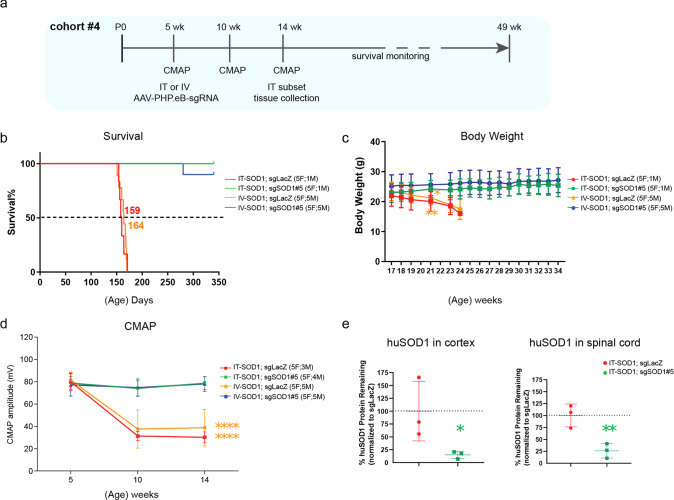
Next, we set out to evaluate whether the therapeutic effects of CRISPRSOD1 could be further enhanced by another novel AAV capsid, AAV.PHP.eB. AAV.PHP.eB was derived from AAV-PHP.B via AAV-targeted evolution for more efficient transduction of neurons and contains two different amino acids at positions 587 and 588 [31]. We treated a small cohort of adult H11Cas9 SOD1G93A mice (Cohort #4) with a single intrathecal (IT) or intravenous (IV) injection of AAV.PHP.eB-sgSOD1#5 (or sgLacZ; n = 10 per group; 1E12 vg per animal) at 5 weeks of age, immediately prior to the onset of CMAP decline (Fig. 4a). Strikingly, sgSOD1 extended survival of treated mice by at least 170 days (~24 weeks) independent of delivery method (Fig. 4b). This is a minimum estimate since we euthanized all sgSOD1-treated animals at postnatal day 340 (~49 weeks) due to COVID-19-related emergency shutdown of the animal facility. None of the treated mice showed any abnormal phenotypes at the time of euthanasia. sgSOD1 treatment prevented body weight loss observed in the sgLacZ-treated mice (Fig. 4c) and CMAP decline in the TA muscle, indicating protection against NMJ denervation (Fig. 4d). Notably, there was no difference in the degree of therapeutic benefit between IT and IV delivery methods, consistent with the report that AAV-PHP.eB crosses the blood-brain barrier efficiently in C57BL/6 mice [31]. When huSOD1 protein levels in the CNS were evaluated by ELISA in IT-injected mice at 10 weeks post injection, we observed a ~85% reduction of huSOD1 protein in the cortex and ~80% reduction of huSOD1 protein in the spinal cord of sgSOD1-treated mice (Fig. 4e). The higher degree of huSOD1 protein reduction (80% in Cohort #4 vs 50–60% in Cohort #1) and stronger therapeutic benefits (>170 days survival extension in Cohort #4 vs 113–137 days in Cohort #2 and #3) may reflect superior transduction efficiency in the CNS by AAV.PHP.eB versus AAV.PHP.B [33, 36] and/or the route of administration, highlighting two key considerations to potentially boost the efficacy of gene therapy. Amplicon sequencing performed on genomic DNA of a subset of sgSOD1-treated mice at the end of the study (~49 weeks) showed indel rates of ~39% in the cortex and ~24% in the spinal cord (Fig. S5). Together, these results demonstrated that AAV-CRISPR-mediated in vivo gene editing achieved robust reduction of huSOD1 transgene expression in the SOD1G93A mice, leading to pronounced survival extension.
Fig. 4. IT and IV injection of AAV-sgSOD1 in adult H11Cas9 SOD1G93A mice preserves neuromuscular function and significantly extends survival.
a Study design of IT and IV injection of AAV-sgSOD1 in adult H11Cas9 SOD1G93A mice (Cohort #4). b Kaplan-Meier survival curves of Cohorts #1 and #2. Log-Rank test, P < 0.0001. c Body weight of mice over age (Cohort #4). t-test was performed at week 21 before any mock-treated mice reach euthanasia endpoint: IT-sgLacZ vs. IT-sgSOD1; IV-sgLacZ vs. IV-sgSOD1. d CMAP amplitude of TA muscle at week 5, 10 and 14. Two-way ANOVA with Dunnett’s multiple comparisons test was performed. e Percentage of remaining huSOD1 protein at week 14 in cortex (left) and spinal cord (right) of H11Cas9 SOD1G93A mice IT injected with AAV-sgSOD1 (Cohort #4) as determined by ELISA. Unpaired t-test. c–e Data are presented as mean with standard deviation (SD). *p < 0.05, **p < 0.01, and ***p < 0.001, ****p < 0.0001, n.s, not significant. Color of * indicates the group on which statistics was performed.
Discussion
In recent years, AAVs have been extensively used as vehicles for gene transfer to the nervous system to modulate gene expression, via overexpression, knockdown or genome editing. As a result, gene therapy approaches have greatly facilitated the ability to evaluate potential therapeutic candidates in preclinical proof of concept studies for the treatment of neurological diseases, as performed here. Gene therapy approaches that employ CRISPR-associated genome editing systems hold enormous potential to treat several genetic forms of ALS. Here, we report that one-time delivery of SOD1-targeting sgRNAs using AAV-PHP.B and AAV-PHP.eB to the CNS of H11Cas9 SOD1G93A mice significantly extended disease-free survival through robust and sustained reduction of mutant huSOD1 protein in the cortex and spinal cord and resulted in strong disease suppression corroborated by comprehensive morphological and functional analyses (Summarized in Table 1).
Table 1.
Summary of all phenotypes assessed in Cohort #1–4.
| Study Summary | Study Cohort | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 4 | ||
| Serotype | AAV-PHP.B | AAV-PHP.B | AAV-PHP.B | AAV-PHP.eB | AAV-PHP.eB | |
| Delivery | ICV | ICV | ICV | IT | IV | |
| sgSOD1 | #1/#5 | #1/#5 | #1/#5 | #5 | #5 | |
| Survival | – | +137 days (20 weeks) | +113 days (16 weeks) | >170 days (24 weeks) | >170 days (24 weeks) | |
| Body weight | – | Resemble WT | resemble WT | Resemble WT | Resemble WT | |
| Motor functions | Rotarod | – | Resemble WT | – | – | – |
| Open field | – | Resemble WT | – | – | – | |
| Inverted grip | – | – | resemble WT | – | – | |
| “clasping” | – | Resemble WT | – | – | – | |
| CMAP (Tibialis anterior) | – | No decline | – | No decline | No decline | |
| Neuromuscular junction | Resemble WT | – | – | resemble WT | Resemble WT | |
| Motor neuron count | Resemble WT | – | – | – | – | |
| Serum pNFH | – | Lower than controls | – | – | – | |
| Fast-myosin muscle diameter | Resemble WT | – | – | – | – | |
| huSOD1 levels | Cortex | 50–60% reduction | – | – | 85% reduction | – |
| Spinal cord | 50–60% reduction | – | – | 80% reduction | – | |
| Indel rate bulk | Cortex | 20% | – | – | 39% | – |
| Spinal cord | 7% | – | – | 24% | – | |
| Indel rate FACS sorted | Cortex | 40% | – | – | – | – |
| Spinal cord | 17% | – | – | – | – | |
We found a larger degree of suppression than two previously published studies employing AAV-mediated delivery of CRISPR to treat SOD1G93A mice [29, 30]. We observed a median survival extension of >110 days vs 25 days and 12 days, respectively. Gaj and colleagues employed a single AAV9 vector delivery of saCas9 and sgRNA to introduce indels in the huSOD1 transgene. Lim and colleagues used two AAV9 vectors to deliver a split intein-mediated protein trans-splicing system to introduce early stop codons in the huSOD1 transgene. Several factors could contribute to the improved efficacy in our study. First, in our experiments spCas9 was constitutively expressed in target cell populations rather than being packaged into AAV and delivered exogenously. Second, we used newer AAV variants with reportedly broader distribution and/or higher transduction rates in neurons and astrocytes than AAV9 [33, 34, 36]. Consistent with this, we observed much higher indel rates (~7–17%) in the spinal cord of treated animals as compared to earlier studies (<1.2%). Remarkably, the 80% huSOD1 protein reduction achieved in Cohort #4 (vs. 50–60% in Cohort #1) translated to greater survival extension (>170 days vs >137 days), highlighting that phenotypic efficacy correlates with the degree of huSOD1 protein reduction. Lastly, we cannot exclude the possibility that the nuclease activity of spCas9 generates higher editing efficiency than saCas9 or spCas9-fused CBEs. Future studies to directly compare the nuclease activity of these different Cas proteins are warranted. Taken together, our approach provided a higher degree of therapeutic benefit in SOD1G93A mice, highlighting several key considerations to increase genome editing efficiency, including distribution and expression level of Cas9, and transduction efficiency of AAV capsids on target cell populations.
Though successful, our approach has some inherent limitations that preclude it from directly translating into clinical development. First, the introduction of the H11Cas9 transgene into SOD1G93A mice is highly artificial and does not translate as a general strategy for patients. In patients, there is a need to package a complete CRISPR/Cas9 system, which might limit the effectiveness of target modulation. Second, our study did not assess the immunological impact of constitutive Cas9 expression in the CNS, another factor highly relevant to gene therapy patients. Patients undergoing gene therapy treatment would need to be monitored constantly for an immunological response and potentially receive immuno-suppressants to compensate for such a response. A third limitation is the preventive nature of our treatment paradigm—animals were treated immediately prior to symptom onset. This paradigm does not parallel the treatment of patients as they would be treated after symptom onset. Despite this specific concern, other studies also administered treatments prior to symptom onset with less impactful effects on survival than in our study [29, 30, 60]. Finally, though the enhanced CNS tropism of AAV-PHP.B and AAV-PHP.eB is useful for evaluating and validating targets in pre-clinical models, it is not yet translatable to development of therapeutics in humans as their strong transduction in CNS appears to be limited to specific mouse strains, such as C57BL/6 and FVB/NCrl [30, 61–64]. The goal of the current study is not to develop therapeutics for clinical trials, but instead to demonstrate the therapeutic potential of CRISPR editing in preclinical models and identify key parameters (e.g., AAV capsid, Cas9 expression) critical to greater efficacy.
Like other work using CRISPR to target huSOD1 [29, 30], we observed a discrepancy between indel rate (~10–20%) and huSOD1 protein reduction (~50–60%) in target tissues. Several reasons could account for this discrepancy. First, while huSOD1 transgene is present in all cells in the target tissue, its expression level varies between different cell types (https://www.brainrnaseq.org/). As the AAV-PHP.B variant has strong tropism towards neurons and astrocytes where huSOD1 expression is also high (https://www.brainrnaseq.org/) [32, 34], it is conceivable that cells with high huSOD1 expression such as neurons and astrocytes also have high indel rates. Other cell types (e.g., microglia, endothelial cells) with low huSOD1 expression might have low indel rates. Single-cell or single-nucleus RNA-Seq could be employed in future studies to confirm this possibility. Secondly, larger deletions may also occur in the vicinity of the Cas9 cleavage sites in addition to small insertions and deletions. These deletions may not be detected by amplicon sequencing due the smaller size (~500 bp) of the amplicons. Indeed, a recent publication detected deletions, ranging in size from a few hundred base pairs (bp) to over 4000 bp by long-range PCR, cloning, and Sanger-sequencing in the vicinity of the Cas9 cleavage site [65]. While we have ruled out large deletions that affect huSOD1 transgene copy number by ddPCR, we cannot exclude the possibility AAV-sgSOD1 introduced intermediate-sized deletions (a few hundred to a few thousand bp), which led to huSOD1 protein reduction. Long-range PCR, cloning, and Sanger-sequencing approaches are needed in future studies to carefully examine this possibility.
In summary, our approach provided remarkable protection against multiple ALS-related phenotypes in the SOD1G93A mouse model and, importantly, resulted in substantial disease-free survival extension, highlighting the utility of combining CRISPR-based gene editing with robust novel AAV capsids in evaluating potential therapeutic targets in pre-clinical studies. The positive results from our proof-of-concept study in SOD1G93A mice, supports the notion of introducing the spCas9 gene into other genetic animal models of ALS for CRISPR-based gene editing. This would afford an opportunity to assess nearly any gene in the genome for their potential effects in multiple ALS mouse models. Genes found to be protective across multiple ALS models are more likely to identify potential disease-modifying therapeutic targets for sporadic ALS, which represents ~90% of all ALS cases. Such an approach would ultimately accelerate drug discovery while also aiding in elucidating the etiology and biology of ALS. Moreover, as this strategy is agnostic to disease, it can easily be applied more broadly to other CNS disease areas, potentially benefiting those patient communities as well.
Supplementary information
Supplementary Material Chen et al SOD1 CRISPR-2022
Acknowledgements
We thank Jianxin Hu, Brigitte Pettmann and Giulio Srubek Tomassy for helpful discussion of this research; Chia-yen Wu (Jackson Laboratory), Martin Lamb, and Linhong Sun for assistance with experiments; and Dr. Monte M. Winslow for providing constitutive H11Cas9 knock-in mice.
Author contributions
SCL, YAC, AM, and CEH conceived the project and designed the experiments. OM, NM, SL and SKL performed in vitro screening of sgRNAs. DF, DK. KK, YAC, YL, and KS, performed in vivo studies including injections, survival monitoring, body weight measurement, tissue collection, etc. JD and BW performed CMAP recordings. AF, HM, ML, EM, CS, PC and TC performed indel analysis and RNA sequencing. GM, SH, MC, and JS performed histology analysis. HMA, RD, and AS performed behavioral testing. SM and SL performed huSOD1 protein measurement. MIZ performed sequencing analysis. SCL, YAC, MWK., SH, and CEH wrote the paper.
Funding
This research was provided by Biogen Inc.
Data availability
The RNA-seq data generated from HeLa cell lines has been deposited in the Gene Expression Omnibus (GEO tracking number GSE213125).
Competing interests
The authors would like to declare they are/were employees of Biogen Inc.
Ethical approval
All animal use and treatments were approved by the Biogen Institutional Animal Care and Use Committee (IACUC) and followed the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41434-022-00375-w.
References
- 1.Brown RH, Al-Chalabi A. Amyotrophic lateral Sclerosis. N Engl J Med. 2017;377:162–72. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 3.Andersen PM, Nilsson P, Keranen ML, Forsgren L, Hagglund J, Karlsborg M, et al. Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain. 1997;120:1723–37. doi: 10.1093/brain/120.10.1723. [DOI] [PubMed] [Google Scholar]
- 4.Bruijn LI, Cleveland DW. Mechanisms of selective motor neuron death in ALS: insights from transgenic mouse models of motor neuron disease. Neuropathol Appl Neurobiol. 1996;22:373–87. doi: 10.1111/j.1365-2990.1996.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 5.Fischer LR, Glass JD. Oxidative stress induced by loss of Cu,Zn-superoxide dismutase (SOD1) or superoxide-generating herbicides causes axonal degeneration in mouse DRG cultures. Acta Neuropathol. 2010;119:249–59. doi: 10.1007/s00401-009-0631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurney ME, Pu H, Chiu AY, Canto MCD, Polchow CY, Alexander DD, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–5. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 7.Ratovitski T, Corson LB, Strain J, Wong P, Cleveland DW, Culotta VC, et al. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum Mol Genet. 1999;8:1451–60. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 8.Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–7. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 9.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65:S3–9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 10.Saccon RA, Bunton-Stasyshyn RK, Fisher EM, Fratta P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain. 2013;136:2342–58. doi: 10.1093/brain/awt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bravo-Hernandez M, Tadokoro T, Navarro MR, Platoshyn O, Kobayashi Y, Marsala S, et al. Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat Med. 2020;26:118–30. doi: 10.1038/s41591-019-0674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCampbell A, Cole T, Wegener AJ, Tomassy GS, Setnicka A, Farley BJ, et al. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J Clin Invest. 2018;128:3558–67. doi: 10.1172/JCI99081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller T, Cudkowicz M, Shaw PJ, Andersen PM, Atassi N, Bucelli RC, et al. Phase 1–2 trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N Engl J Med. 2020;383:109–19. doi: 10.1056/NEJMoa2003715. [DOI] [PubMed] [Google Scholar]
- 14.Miller T, Cudkowicz M on behalf of the VWG. Results from the Phase 3 VALOR study and its open-label extension: evaluating the clinical efficacy and safety of Tofersen in adults with ALS and confirmed SOD1 mutation. Am Neurol Assoc Annu Meet. 2021.
- 15.Broekman ML, Comer LA, Hyman BT, Sena-Esteves M. Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or -2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience. 2006;138:501–10. doi: 10.1016/j.neuroscience.2005.11.057. [DOI] [PubMed] [Google Scholar]
- 16.McLean JR, Smith GA, Rocha EM, Hayes MA, Beagan JA, Hallett PJ, et al. Widespread neuron-specific transgene expression in brain and spinal cord following synapsin promoter-driven AAV9 neonatal intracerebroventricular injection. Neurosci Lett. 2014;576:73–8. doi: 10.1016/j.neulet.2014.05.044. [DOI] [PubMed] [Google Scholar]
- 17.Borel F, Gernoux G, Sun H, Stock R, Blackwood M, Brown RH, et al. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci Transl Med. 2018;10:1–14. [DOI] [PubMed]
- 18.Dirren E, Aebischer J, Rochat C, Towne C, Schneider BL, Aebischer P. SOD1 silencing in motoneurons or glia rescues neuromuscular function in ALS mice. Ann Clin Transl Neurol. 2015;2:167–84. doi: 10.1002/acn3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iannitti T, Scarrott JM, Likhite S, Coldicott IRP, Lewis KE, Heath PR, et al. Translating SOD1 Gene Silencing toward the Clinic: A Highly Efficacious, Off-Target-free, and Biomarker-Supported Strategy for fALS. Mol Ther - Nucleic Acids. 2018;12:75–88. doi: 10.1016/j.omtn.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoica L, Todeasa SH, Cabrera GT, Salameh JS, Elmallah MK, Mueller C, et al. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang B, Qiu L, Yang C, Kramer J, Su Q, et al. Widespread spinal cord transduction by intrathecal injection of rAAV delivers efficacious RNAi therapy for amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23:668–81. doi: 10.1093/hmg/ddt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 23.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–7. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt RJJ, Chen S, Zhou Y, Yim MJJ, Swiech L, Kempton HRR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–50. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–14. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaj T, Ojala DS, Ekman FK, Byrne LC, Limsirichai P, Schaffer DV. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci Adv. 2017;3:1–11. doi: 10.1126/sciadv.aar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim CKW, Gapinske M, Brooks AK, Woods WS, Powell JE, Zeballos CM, et al. Treatment of a mouse model of ALS by in vivo base editing. Mol Ther. 2020;28:1177–89. doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WLL, et al. Engineered AAVs for Efficient Noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci. 2017;20:1172–9. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34:204–9. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hana S, Peterson M, McLaughlin H, Marshall E, Fabian AJ, McKissick O, et al. Highly efficient neuronal gene knockout in vivo by CRISPR-Cas9 via neonatal intracerebroventricular injection of AAV in mice. Gene Ther. 2021;28:646–58. [DOI] [PMC free article] [PubMed]
- 34.Torregrosa T, Lehman S, Hana S, Marsh G, Xu S, Koszka K, et al. Use of CRISPR/Cas9-mediated disruption of CNS cell type genes to profile transduction of AAV by neonatal intracerebroventricular delivery in mice. Gene Ther. 2021;28:456–68. doi: 10.1038/s41434-021-00223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–85. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowland LP. How amyotrophic lateral sclerosis got its name the clinical-pathologic genius of Jean-Martin Charcot. Arch Neurol. 2001;58:512–5. doi: 10.1001/archneur.58.3.512. [DOI] [PubMed] [Google Scholar]
- 37.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–7. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–73. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, Gingeras TR Optimizing RNA-Seq Mapping with STAR. In: Carugo O, Eisenhaber F, editors. Data Mining Techniques for the Life Sciences. New York, NY: Springer; 2016 [cited 2022 Aug 23]. 245–62. (Methods in Molecular Biology). 10.1007/978-1-4939-3572-7_13. [DOI] [PubMed]
- 43.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao S, Gordon W, Du S, Zhang C, He W, Xi L, et al. QuickMIRSeq: a pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinform. 2017;18:180. doi: 10.1186/s12859-017-1601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–92. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vecchia LD, Maria BD, Marinou K, Sideri R, Lucini A, Porta A, et al. Cardiovascular neural regulation is impaired in amyotrophic lateral sclerosis patients. A study by spectral and complexity analysis of cardiovascular oscillations. Physiol Meas. 2015;36:659–70. doi: 10.1088/0967-3334/36/4/659. [DOI] [PubMed] [Google Scholar]
- 48.Linden D, Diehl RR, Berlit P. Reduced baroreflex sensitivity and cardiorespiratory transfer in amyotrophic lateral sclerosis. Electroencephalogr Clin Neurophysiol Mot. Control. 1998;109:387–90. doi: 10.1016/s0924-980x(98)00035-6. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka Y, Yamada M, Koumura A, Sakurai T, Hayashi Y, Kimura A, et al. Cardiac sympathetic function in the patients with amyotrophic lateral sclerosis: analysis using cardiac [123I] MIBG scintigraphy. J Neurol. 2013;260:2380–6. doi: 10.1007/s00415-013-7005-0. [DOI] [PubMed] [Google Scholar]
- 50.Duchen LW, Strich SJ, Falconer DS. Clinical and pathological studies of an hereditary neuropathy in mice (Dystonia musculorum) Brain. 1964;87:367–78. doi: 10.1093/brain/87.2.367. [DOI] [PubMed] [Google Scholar]
- 51.Hatzipetros T, Kidd JD, Moreno AJ, Thompson K, Gill A, Vieira FG. A quick phenotypic neurological scoring system for evaluating disease progression in the SOD1-G93A Mouse Model of ALS. J Vis Exp. 2015;104:1–6. [DOI] [PMC free article] [PubMed]
- 52.Mancuso R, Osta R, Navarro X. Presymptomatic electrophysiological tests predict clinical onset and survival in SOD1G93A ALS mice. Muscle Nerve. 2014;50:943–9. doi: 10.1002/mus.24237. [DOI] [PubMed] [Google Scholar]
- 53.Mancuso R, Santos-Nogueira E, Osta R, Navarro X. Electrophysiological analysis of a murine model of motoneuron disease. Clin Neurophysiol. 2011;122:1660–70. doi: 10.1016/j.clinph.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 54.Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 55.Hegedus J, Putman CT, Tyreman N, Gordon T. Preferential motor unit loss in the SOD1G93A transgenic mouse model of amyotrophic lateral sclerosis. J Physiol. 2008;586:3337–51. doi: 10.1113/jphysiol.2007.149286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–40. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 57.McCombe PA, Pfluger C, Singh P, Lim CYH, Airey C, Henderson RD. Serial measurements of phosphorylated neurofilament-heavy in the serum of subjects with amyotrophic lateral sclerosis. J Neurol Sci. 2015;353:122–9. doi: 10.1016/j.jns.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 58.Steinacker P, Feneberg E, Weishaupt J, Brettschneider J, Tumani H, Andersen PM, et al. Neurofilaments in the diagnosis of motoneuron diseases: a prospective study on 455 patients. J Neurol Neurosurg Psychiatry. 2016;87:12–20. doi: 10.1136/jnnp-2015-311387. [DOI] [PubMed] [Google Scholar]
- 59.Lu CH, Petzold A, Kalmar B, Dick J, Malaspina A, Greensmith L. Plasma neurofilament heavy chain levels correlate to markers of late stage disease progression and treatment response in SOD1G93A Mice that Model ALS. PLoS ONE. 2012;7:e40998. doi: 10.1371/journal.pone.0040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foust KD, Salazar DL, Likhite S, Ferraiuolo L, Ditsworth D, Ilieva H, et al. Therapeutic AAV9-mediated Suppression of Mutant SOD1 Slows Disease Progression and Extends Survival in Models of Inherited ALS. Mol Ther. 2013;21:2148–59. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hordeaux J, Wang Q, Katz N, Buza EL, Bell P, Wilson JM. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice [Internet]. Vol. 26, Molecular Therapy. Cell Press; 2018. 664–8. www.moleculartherapy.org. [DOI] [PMC free article] [PubMed]
- 62.Hordeaux J, Yuan Y, Clark PM, Wang Q, Martino RA, Sims JJ, et al. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol Ther. 2019;27:912–21. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Q, Chan KY, Tobey IG, Chan YA, Poterba T, Boutros CL, et al. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. 2019 10.1371/journal.pone.0225206. [DOI] [PMC free article] [PubMed]
- 64.Challis RC, Kumar SR, Chan KY, Challis C, Beadle K, Jang MJ, et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat Protoc. 2019;14:379–414. doi: 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- 65.Deng HX, Zhai H, Shi Y, Liu G, Lowry J, Liu B, et al. Efficacy and long-term safety of CRISPR/Cas9 genome editing in the SOD1-linked mouse models of ALS. Commun Biol. 2021;4:1–11. doi: 10.1038/s42003-021-01942-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Chen et al SOD1 CRISPR-2022
Data Availability Statement
The RNA-seq data generated from HeLa cell lines has been deposited in the Gene Expression Omnibus (GEO tracking number GSE213125).