Abstract
In the design and development of therapeutic agents, macromolecules with restricted structures have stronger competitive edges than linear biological entities since cyclization can overcome the limitations of linear structures. The common issues of linear peptides include susceptibility to degradation of the peptidase enzyme, off-target effects, and necessity of routine dosing, leading to instability and ineffectiveness. The unique conformational constraint of cyclic peptides provides a larger surface area to interact with the target at the same time, improving the membrane permeability and in vivo stability compared to their linear counterparts. Currently, cyclic peptides have been reported to possess various activities, such as antifungal, antiviral and antimicrobial activities. To date, there is emerging interest in cyclic peptide therapeutics, and increasing numbers of clinically approved cyclic peptide drugs are available on the market. In this review, the medical significance of cyclic peptides in the defence against viral infections will be highlighted. Except for chikungunya virus, which lacks specific antiviral treatment, all the viral diseases targeted in this review are those with effective treatments yet with certain limitations to date. Thus, strategies and approaches to optimise the antiviral effect of cyclic peptides will be discussed along with their respective outcomes. Apart from isolated naturally occurring cyclic peptides, chemically synthesized or modified cyclic peptides with antiviral activities targeting coronavirus, herpes simplex viruses, human immunodeficiency virus, Ebola virus, influenza virus, dengue virus, five main hepatitis viruses, termed as type A, B, C, D and E and chikungunya virus will be reviewed herein.
Graphical Abstract
Keywords: Cyclic peptides, Antiviral activity, Coronavirus, Dengue virus
Introduction
In drug discovery, macromolecules, such as antibiotics, proteins, peptides and nucleic acids, are currently attracting increasing interest as therapeutic candidates, although small biological entities remain central to the market (Siang Ong et al. 2017). Peptides have relatively straightforward production, low toxicity, high structural diversity, good selectivity and strong binding affinity towards the desired biological targets (Ayoub and Scheidegger 2006; Passioura et al. 2014; Siang Ong et al. 2017). However, the existing barriers of linear peptide stability in the human body, including low oral bioavailability, high risk of proteolytic degradation and high excretion rate, greatly influence their development in therapeutic fields (Ayoub and Scheidegger 2006; Gongora-Benitez et al. 2014; Wang and Craik 2016; Siang Ong et al. 2017). Although polypeptides are used to treat various ailments and diseases, major drawbacks are their in vitro and in vivo stability. Currently, researchers overcome this problem by developing various novel drug delivery systems (Kumar et al. 2018; Kumar et al. 2019; Chellathurai et al. 2021; Tee et al. 2021). One of the smartest approaches that is predicted is converting these polypeptides into cyclic peptides, but their pharmacological properties and stability need to be proven once again after conversion. The findings from the researchers supported that cyclic peptides, polypeptide chains with a closed structure (Joo 2012), are the more favourable alternatives that are free from the majority of these challenges. Cyclic peptides demonstrate their resistance to exopeptidases and even endopeptidases, showing improved in vivo stability (Joo 2012; Siang Ong et al. 2017). With a bioactive conformation and decreased entropic penalty for binding, target selectivity can be improved and ensures stronger binding interactions (Siang Ong et al. 2017). In addition, cyclization of peptides has the advantage of cell penetration under some conditions (Siang Ong et al. 2017). Thus, drug discovery and development with cyclic peptides has recently become a significant direction of study based on their unique structural features.
Due to their wide spectrum of biological activities and the formation of unique folded structures, cyclic peptides have been employed in applications ranging from drug design to nanotechnology (Tapeinou et al. 2015). Recently, the generation of peptide nanostructures, such as nanospheres, nanofibers and nanotubes, has been exploited for the purpose of drug delivery, medical imaging tools, and tissue engineering. Ghadiri et al. demonstrated a pioneering design of a novel class of heterochiral cyclic peptides in nanotubular structures, and this transmembrane channel structure was proposed to be a potential delivery agent into living cells in gene therapy (Mandal et al. 2014). In the pharmaceutical industry, great success in the bioengineering of linear peptide cyclization from the N- to C-terminus has specifically contributed to peptide toxin progression (Thapa et al. 2014).
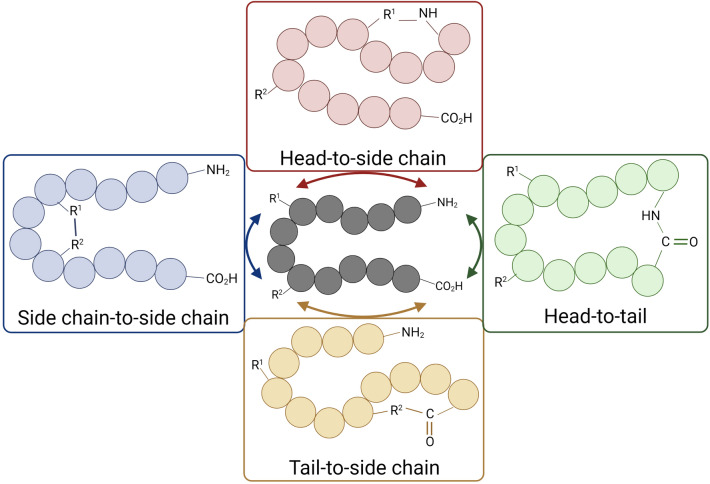
Current therapeutic cyclic peptides permitted by the Food and Drug Administration (FDA) consist of natural derivatives or artificial analogues resembling the natural substrate (Table 1) (Zorzi et al. 2017; Al Shaer et al. 2020; Choi and Joo 2020, Buckton et al. 2021, Zhang and Chen 2021). They are able to exhibit various biological activities. Examples of natural cyclic peptides are gramicidin S with antimicrobial activity, vancomycin with antibacterial effect and cyclosporine A with immunosuppressive activity (Ayoub and Scheidegger 2006; Joo 2012; Jin 2020). In discovering and designing cyclic peptide drugs, various sound approaches have been practised to optimise the desired properties for drug delivery. In addition to the well-established biological methods, such as split intein circular ligation of peptides and proteins system (SICLOPPS), phage display, and mRNA display (Siang Ong et al. 2017; Liras and Mcclure 2019), cyclization of peptides can be achieved chemically via techniques such as head-to-side chain, side chain-to-side chain, head-to-tail and side chain-to-tail, which are determined by the functional groups (Fig. 1) (White and Yudin 2011; Vitali and Fasan 2017; Hayes et al. 2021).
Table 1.
Cyclopeptides approved in the last 10 years (2011–2021) approved by the EMA and/or FDA
| Name | Activity or indication | Route of administration | First clinical use |
|---|---|---|---|
| [177Lu]-DOTATATE Al Shaer et al. (2020) | Gastroenteropancreatic neuroendocrine tumors | Intravenous | 2018 |
| [64Cu]-DOTATATE Al Shaer et al. (2020) | Positron emission tomography (PET) imaging | Intravenous | 2020 |
| [68 Ga]-DOTATATE Buckton et al. (2021) | PET imaging | Intravenous | 2016 |
| [68 Ga]-DOTATOC Al Shaer et al. (2020) | PET imaging | Intravenous | 2019 |
| [68 Ga]-PSMA-11 Al Shaer et al. (2020) | Diagnosis of recurrent prostate carcinoma by PET | Intravenous | 2020 |
| Bremelanotide Al Shaer et al. (2020) | Women hypoactive sexual desire disorder | Subcutaneous | 2019 |
| Dalbavancin Zorzi et al. (2017) | Complicated skin and skin structure infections (CSSSIs) | Intravenous infusion | 2014 |
| Linaclotide Choi and Joo (2020) | Irritable bowel syndrome (IBS-C), chronic idiopathic constipation (CIC) | Oral | 2012 |
| Oritavancin Zorzi et al. (2017) | CSSSIs | Intravenous infusion | 2014 |
| Pasireotide Zorzi et al. 2017) | Cushing’s disease, acromegaly, neuroendocrine tumors | Subcutaneous, intramuscular | 2012 |
| Peginesatide Zorzi et al. (2017) | Anemia associated with chronic kidney disease | Intravenous, subcutaneous | 2012 |
| Plecanatide Choi and Joo (2020) | CIC | Oral | 2017 |
| Setmelanotide Al Shaer et al. (2020) | Obesity | Subcutaneous | 2020 |
| Voclosporin Zhang and Chen (2021) | Lupus nephritis | Oral | 2021 |
Fig. 1.
Schematic illustration of peptide cyclization modes. Created with BioRender.com
Current and emerging viral diseases have evolved as public health threats, causing morbidity and death worldwide over the human development timeline (De Clercq and Li 2016; Hoffmann et al. 2020). However, there are still inadequate effective therapeutics for most viruses to date (Hoffmann et al. 2020). In fact, the current treatment of some viral infections faces a dilemma, as the approved antiviral treatments are less than satisfactory and presented with certain limitations (Balasubramanian et al. 2019; Farooq and Ngaini 2021; O’Donnell and Marzi 2021). Hence, to minimize the impact of virus outbreaks, novel antiviral drug discovery is of utmost importance. In conjunction with the fast-evolving biomedical and biotechnology developments, antiviral cyclic peptides can be synthesized not only through the fermentation of natural sources but also via synthetic modifications from linear peptides to achieve the anticipated biological activities. With high feasibility and possibility, substantial attention is given to cyclic peptides with antiviral activity to occupy this void space to combat viral infections (Hoffmann et al. 2020). In this review, antiviral cyclic peptides targeting different viral strains of infectious diseases will be discussed.
Natural and Synthetic Cyclic Peptides
With the advancement in the biotechnology, the cyclic peptides can be synthesised or modified accordingly in order to obtain the desired biological activities based on rational design despite the fact that the majority of the clinically approved cyclic peptides in the market are the natural product derivatives (Abdalla and McGaw 2018). Naturally occurring cyclopeptides have been extracted from various sources including algae, fungi, plants, mammals, and bacteria, be it on the land or in the ocean (Agarwal and Gabrani 2021). They have a wide range of size composing of the 20 proteinogenic amino acids with one or more disulphide bonds. The naturally synthesized cyclic peptides share common characteristics of well-defined structures and exceptional stability even in unfavourable biological environment (Bockus et al. 2013). In addition, they typically possess the ability on host defence of their producing organisms (Thorstholm and Craik 2012).
On the other hands, the natural linear peptides with significant drawback of instability towards proteolysis require structural modifications to maintain their feasibility for the pharmaceutical industry (Katsara et al. 2006; Tapeinou et al. 2015). To date, diverse strategies have been introduced and concentrated on the remodelling of the cyclic peptidomimetics. There are three principal technologies being introduced chronologically, which are direct isolation from host organism, chemical synthesis via solid phase peptide synthesis (SPPS) or chemo-enzymatic synthesis, and biological synthesis employing recombinant expression in plants or bacteria (Thorstholm and Craik 2012). For example, cyclization to increase stability, incorporate unnatural amino acids to improve specificity and introduce assorted structural restrictions. The novel modality forces the ordered secondary arrangement of a peptide, enhancing its metabolic stability and oral bioavailability. The cyclic mimetics are more preferred over the larger proteins in terms of lower immunogenicity while preserving the selectivity and potency (Tapeinou et al. 2015). However, it is difficult to obtain the native fold when the original sequence of the peptide is altered (Thorstholm and Craik 2012).
The Antiviral Activities of Synthetic Cyclic Peptides
Cyclic Peptides in Coronavirus Therapy
Coronaviruses (CoVs) are grouped under the Coronaviridae family (Ali and Alharbi 2020). The outbreak of CoV disease in December 2019 (COVID-19) is a viral disease caused by severe acute respiratory syndrome CoV 2 (SARS-CoV-2) (Ali and Alharbi 2020; Brodin 2021). In December 2021, the WHO reported that SARS-CoV-2 variants of concern (VOCs) variants are Alpha, Beta, Gamma, Delta, and Submicron, whereas the variants of interest (VOIs) includes the Epsilon, Theta, Lambda and Mu (Konings et al. 2021, Parums 2021, WHO 2021).
The mutation in SARS-CoV-2 variants mainly enhances viral attachment to the host cell and therefore facilitates its entry, thus having various degrees of spreadability (Davies et al. 2021). Researchers revealed that in the Omicron variant, there are 15 RBD region mutations among the variations concealed in its spike protein. Unlike Mu, Beta, and Gamma variants having E484K substitutions, E484A and Y505H substitutions are reported in the Omicron variant (Woo and Shah 2021; Kannan et al. 2022). From the investigations by Woo and his colleagues, the E484A substitution in the Omicron variant facilitates evasion of the immune response by significantly reducing the binding energy between its RBD and the approved therapeutic monoclonal antibodies, together with other replacements of T478K, Q493K, and Q498R, which strengthen ACE2 binding (Woo and Shah 2021). With greater binding affinity and antibody neutralization ability, the Omicron variant is estimated to have an infectability that is three to 6 times greater than that of the Delta variant, given the same period of time (Callaway and Ledford 2021).
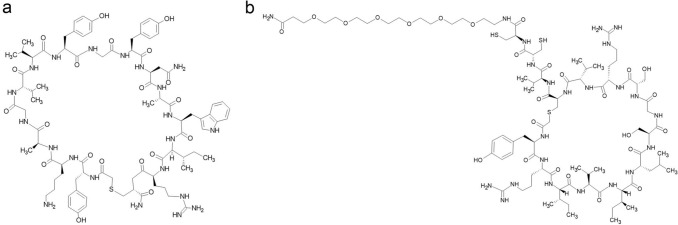
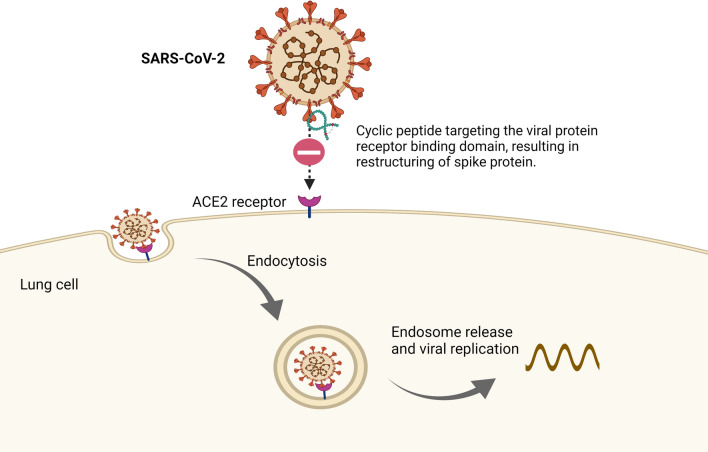
SARS-CoV-2 facilitates its entry into human cells through the binding of the spike protein RBD to cell-surface ACE2 receptors in the lung (Ali and Alharbi 2020; Shang et al. 2020; Norman et al. 2021). Thus, to develop new antiviral agents against SARS-CoV-2, targeting the RBD-ACE2 interaction is potentially an effective strategy (Norman et al. 2021). Norman et al. have utilized mRNA display to discover cyclic peptides with high glycoprotein binding affinity ligands towards SARS-CoV-2 spike protein (Norman et al. 2021). Among the nine target cyclic peptides being evaluated, peptide 4 (Fig. 2a) has the highest affinity to RBD with a detected nanomolar dissociation constant, KD with a value of 15 nM and a noticeably low dissociation rate of 1.4 × 10–3 s−1. Peptide 5 (Fig. 2b) recorded a KD value of 76 nM (Norman et al. 2021). However, useful detection of the spike protein was identified, although the tested peptides did not counteract the cells infected with SARS-CoV-2, which might be due to the distal ligand binding of RBD to the ACE2 binding site (Norman et al. 2021) (Fig. 3). The complexation of peptide 4 with the SARS-CoV-2 spike protein provided a unique binding mechanism of cyclic peptides neighbouring the C- and N-end domains. This result suggested that spike protein restructuring is vital for ligand accommodation. In addition, the possibility of hybridization by designing lipid-linked variants of peptide 4 is another interesting direction of study to improve spike RBD affinity and antiviral activity, as the binding site was found to be located next to a newly revealed SARS-CoV-2 RBD fatty acid binding pocket (Toelzer et al. 2020; Norman et al. 2021).
Fig. 2.
Cyclic peptide structure of a peptide 4 and b peptide 5 (Norman et al. 2021)
Fig. 3.
Mechanism of SARS-CoV-2 binding to the human ACE2 receptor and entering the host cell. Cyclic peptide is able to restructure the spike protein, influencing the nature of the interaction between the RBD and the binding site of the ACE2 surface receptor. (Norman et al. 2021) Created with BioRender.com
In addition, the main protease (Mpro) of SARS-CoV-2 is utilized as a specific intracellular target for the development of new antiviral agents (Zhang et al. 2021). With the number of cleavage sites of the two polyproteins pp1a and pp1ab not less than eleven sites, together with papain-like protease (PLpro), Mpro has an important role in viral replication and viability (V’kovski et al. 2021). Notably, the highly specific proteolytic activity of the Leu–Gln↓Xaa sequence (as ↓ indicates the cleavage site and Xaa could be Asn, Ala or Ser) has not been reported for human proteases (Zhang et al. 2020, Johansen-Leete et al. 2022). Hence, this unique finding leads to the potential search for peptidomimetic-based or peptide inhibitors of Mpro. Despite the currently available peptidomimetic Mpro inhibitors either in the market or under clinical trials (Boras et al. 2021; Owen et al. 2021; Yang et al. 2021), macrocyclic peptides are attractive candidates because of their high binding selectivity and greater proteolytic stability in comparison with their linear counterparts (Vinogradov et al. 2019). Johansen et al. utilized random nonstandard peptide integrated discovery technology coupling mRNA display with flexizyme-mediated genetic code reprogramming and successfully identified cyclic peptides with potential antiviral activity as inhibitors of SARS-CoV-2 Mpro (Johansen-Leete et al. 2022). The importance of peptide cyclization has been proven to impose strain on the peptide, avoiding the adoption of a wholly relaxed configuration (Johansen-Leete et al. 2022).
Cyclic Peptides in Dengue Therapy
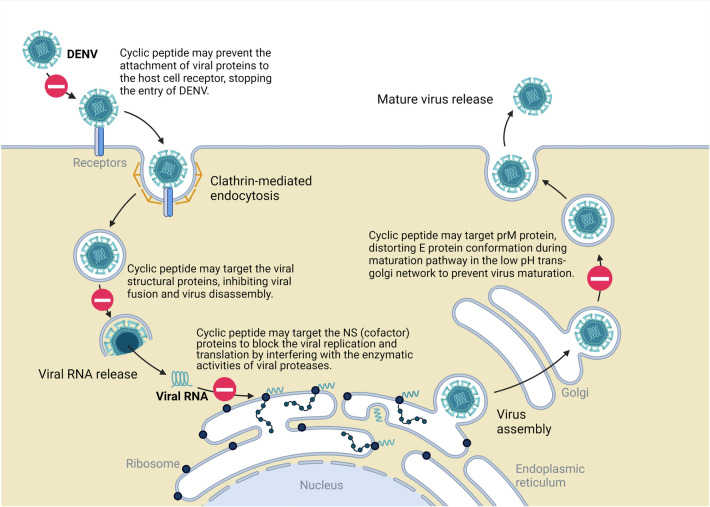
Dengue virus (DENV) is an enveloped vector-borne Flaviviridae family virus with positive single-stranded RNA (Muller et al. 2017). In the viral infection cycle, structural proteins, especially the envelope (E) protein, facilitate viral entry through interactions with host cell receptors (Fig. 4) (Chew et al. 2017). The E protein can be divided into envelope domains (ED) I, II and III (Rodenhuis-Zybert et al. 2010; Alhoot et al. 2013). The important contribution of EDIII in receptor recognition for viral attachment has been reported by several studies; therefore, targeting DENV E structural protein could be considered an effective strategy to inhibit the entry of the virus into the host cell (Hung et al. 2004; Swaminathan and Khanna 2009). Researchers have gained valuable information and a better understanding of the well-determined E structural protein by utilizing advanced techniques such as cryo-electron microscopy, nuclear magnetic resonance spectroscopy and X-ray crystallography (Modis et al. 2004; Volk et al. 2007; Fibriansah et al. 2015). Thus, various strategies have been applied in the development of antiviral drugs against the E protein. Among them, a novel peptide DN59 at < 25 µM with > 99% inhibition rate of DENV-2 was reported by Hrobowski et al. in 2005 (Hrobowski et al. 2005). However, a sequence of certain mechanisms is required for the peptide to otherwise lose its function in inhibiting DENV infection due to possible unnecessary intramolecular interactions. A constrained structure of the peptide can help overcome this issue. In 2013, a study was conducted to screen marketed cyclic peptides through in silico studies (Parikesit and Tambunan 2013). Among all ligands, brain natriuretic peptide (BNP) (7–32) porcine, amylin human and big endothelin (1–38) human are the three ligands able to enter the envelope protein cavity as a whole. From the molecular dynamic simulation, BNP (7–32) porcine is stable without conformational changes at different temperatures. Despite the promising results, further in vitro and in vivo studies are required to evaluate the practicability of cyclic peptides as effective ligands against DENV. In fact, with the property of a highly conserved E stem region among flaviviruses, it is likely for the antiviral cyclic peptide inhibiting one DENV serotype to exhibit the same action on other serotypes and even closely related flaviviruses, including Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBE), West Nile virus (WNV) and yellow fever virus (YFV) (Hrobowski et al. 2005; Schmidt et al. 2010; Chew et al. 2017).
Fig. 4.
DENV infection cycle. Virus binding and membrane fusion are organized by the envelope protein in a pH-dependent environment. (Chew et al. 2017) Adapted from “ZIKV Infection Cycle”, by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates
Among the nonstructural (NS) components, NS3 consists of adenosine triphosphate (ATP)-dependent helicase and serine protease at its C-terminus and N-terminal domain, respectively (Moreland et al. 2010; Ross 2010). It is responsible for viral processing. NS2B is an NS3 protease cofactor (Ross 2010). NS2B-NS3 protease is a two-component protease with a serine protease catalytic triad in a combination of Ser135, His51, and Asp75 that has a polybasic substrate recognition profile (Noble et al. 2012; Takagi et al. 2017). This lipophobic catalytic site obstructs the design of potent micromolecule inhibitors that mimic DENV NS2B-NS3 protease. With the incorporation of an electrophilic warhead into substrate mimetic inhibitors, the inhibitory activities can be improved but not much impact on the specificity to DENV protease and the cell penetrability to give an antiviral effect (Takagi et al. 2017).
Cyclic peptides target the regulation of protein–protein interfaces and other plane binding surfaces, which is difficult for the characteristic “rule of 5” small entities to accomplish (Matsson et al. 2016). Takagi et al. adopted the cyclization strategy by on-resin head-to-tail for high-throughput cyclic peptide synthesis (Takagi et al. 2017). From their findings, cyclic peptide 22 (Fig. 4, Table 2) was the strongest blocker (IC50 = 0.95 µM, where IC50 is the 50% of the maximal inhibitory concentration in the evaluation of the in vitro enzymatic inhibition). Using molecular modelling, the cyclic peptide consisting of a stable, active β-turn-like conformation with DDLL (D-Phe-D-Pro-L-Lys-L-Arg) sequencing at the P4’—P4—P3—P2 position was docked with the protease. Cyclic peptides with appropriate placement of the hydrophobic aromatic residue and an extra Arg revealed anti-DENV efficacy. Peptide 32 (Fig. 5, Table 2) had a low cytotoxic concentration with a remarkable antiviral effect (EC50 = 2 µM, where EC50 is the half maximal effective concentration to induce antiviral effect by 50% compared to an infected control culture without treatment). It is believed that peptide 32 (Fig. 5, Table 2) is the first cyclic peptide showing a hindering effect against NS2B-NS3 protease and possesses an antiviral effect towards DENV. Unlike peptide 22, peptide 33 (Fig. 5, Table 2), with the replacement of an amino methylene group as a substitute of an amide bond at site P1–P1’, was found to be stable against hydrolysis by the protease enzyme while maintaining enzyme inhibition and antiviral activity (IC50 = 2.1 µM, EC50 = 11.4 µM) (Takagi et al. 2017).
Table 2.
The moieties at different positions of the cyclic peptides. The amino acids are stereoisomers designated based on the optical direction of the plane-polarized light, with D-indicating dextro-rotatory (to the right) and L-indicating levo-rotatory (to the left). (Takagi et al. 2017)
| Cyclic peptide | Sequence | |||||||
|---|---|---|---|---|---|---|---|---|
| P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | |
| 22 | D-Pro | L-Lys | L-Arg | L-Lys | L-Ser | L-hPhe | L-Ser | D-Phe |
| 32 | D-Arg | L-Arg | L-Arg | L-Lys | D-2Nal | L-hPhe | L-hPhe | D-2Nal |
| 33 | D-Arg | L-Arg | L-Arg | L-Lys-Ψ[CH2NH]-L-Ala | L-hPhe | L-1Nal | D-Phe | |
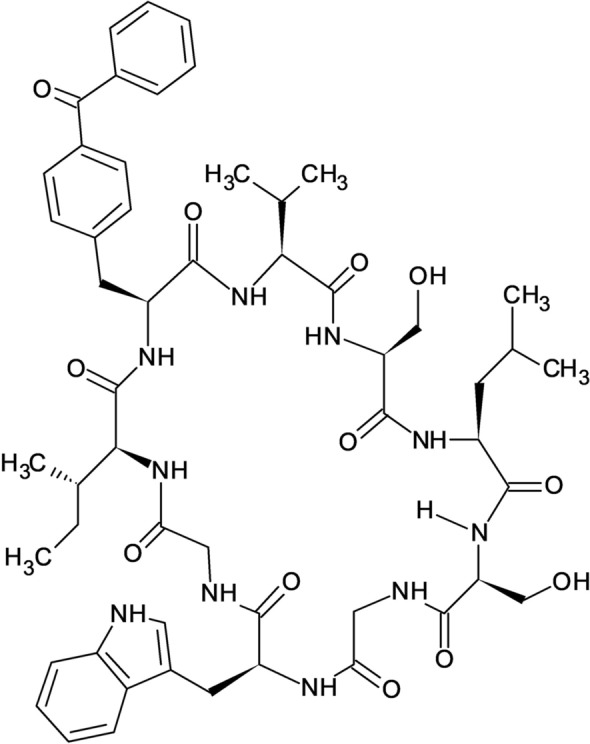
Fig. 5.
Cyclic peptide modelled by Takagi et al. (Takagi et al. 2017)
Other than the extremely hydrophilic and general nonprime side (P), the prime side (P’) of the active site of DENV protease (Saleem et al. 2019) was also reported to be highly effective against DENV protease (Ki = 26 nM) (Lin et al. 2017). Lin’s group studied the interaction between both sides of the active site of DENV protease and cyclic peptides as potential inhibitors. With an optimized amino acid sequence and length in addition to an appropriate cyclization linkage, the findings showed that the tightest cyclic peptide targeting both sides had the potential to reach specificity and lower hydrophilicity as an inhibitor (Ki = 2.9 µM) against wild-type (WT) DENV3 protease (Lin et al. 2017). However, even with the inclusion of a second loop to support the structure, its flexibility is greater than the equivalent loops in aprotinin, potentially leading to the decreased binding affinity and weaker interactions than those in aprotinin, as evidenced by the fact that aprotinin has a comparatively lower Ki value against DENV3 WT protease (Lin et al. 2017).
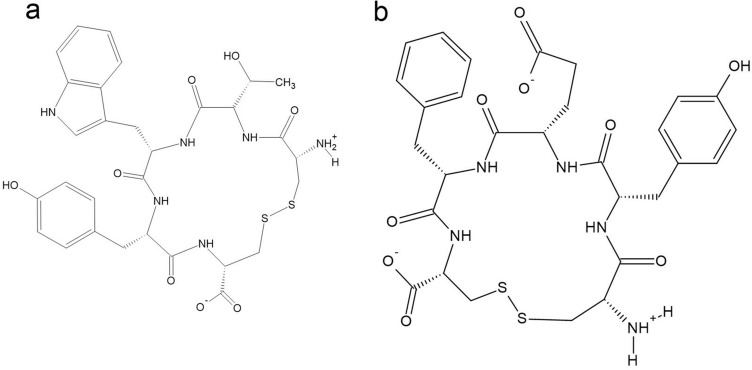
In addition, the enzymes NS3 and NS5 with known activity in DENV duplication are the supreme targets in the manufacturing of antiviral therapeutic agents (Noble et al. 2010; Lim et al. 2015). The Y-shaped slit of NS5 methyltransferase (NS5 Mtase) allows it to connect to two active sites (the RNA-cap binding site and S-adenosyl-l-methionine (SAM) binding site), where two-step methylation takes place from the binding site of SAM to the guanine base of RNA (Lim et al. 2011; Lim et al. 2013; Brecher et al. 2015). Idrus et al. reported the design of cyclic peptide inhibitors targeting both NS5 methyltransferase binding pockets (Idrus et al. 2012). The cyclic pentapeptide inhibitors consisted of terminal cysteines at both ends with a combination of three other amino acids in the middle, along with an S–S bridge to increase the stability of the ligand. The findings reported that the cyclic peptide CTWYC (Fig. 6a) has a more stable structure to inhibit the SAM active site, whereas the CYEFC structure (Fig. 6b) has better strength to bind with the RNA-cap pocket, with binding free energies (∆Gbinding) of − 30.72 and − 22.89 kcal.mol−1, respectively (Idrus et al. 2012). The lower ∆Gbinding indicated more stable binding of ligands to the target protein compared to the standards (Tambunan et al. 2017).
Fig. 6.
Cyclic peptides a CTWYC and b CYEFC illustrated by Indrus et al. (Idrus et al. 2012)
Cyclic Peptides in Human Immunodeficiency Virus Therapy
Human immunodeficiency virus (HIV) is a lentivirus that belongs to the Retroviridae family and causes acquired immunodeficiency syndrome (AIDS) (Girard et al. 2011). CD4+ helper T-cells are the desired target where depletion will cause immunocompromised, increasing the risk of opportunistic infections (Fauci 2003; Girard et al. 2011).
The interaction of the HIV Gag p6 region (a domain containing a P(T/S)AP tetrapeptide motif) with the domain ubiquitin E2 variant (UEV) of host protein tumour susceptibility gene 101 (TSG101) is vital to complete the vital life cycle (Garrus et al. 2001, Liu et al. 2008; Tavassoli et al. 2008). Tavassoli et al. reported the practice of the reverse two-hybrid system (RTHS) of bacteria to recognize the cyclic peptides that restrict the interaction between Gag and TSG101. It was observed that cyclic peptides with five amino acids consisting of threonine at the first position and tryptophan at the third position can interrupt and inhibit HIV egress (Tavassoli et al. 2008). Reducing virus-like particle (VLP) release has been suggested as a potential HIV infection treatment in practical use. The selected inhibiting molecule has a primary sequence different from the sites of interaction of Gag and TSG101. The HIV Tat-tagged cyclic peptide inhibited VLP production in addition to enhancing protein translocation across mammalian cell membranes (Tavassoli et al. 2008). The inhibitory effect on PTAP-dependent Gag budding is dose-dependent (IC50 = 7 µM) without any cytotoxic effects (Tavassoli et al. 2008). Therefore, cyclic peptides can interfere with TSG101-Gag protein interactions with the addition of translocation-promoting amino acid sequences, but this has no effect on the Gag protein mutation (Tavassoli et al. 2008). Therefore, conclusions can be made on small biological therapeutics that inhibit specific virus-host protein interactions and are believed to have wide-ranging applicability in antiviral therapy, but existing countermeasures may be readily circumvented by viral mutations, resulting in ineffective targeting of the viral budding process (Tavassoli et al. 2008).
In viral replication, Tat and Rev are both basic regulatory proteins of HIV-1 (Sherman and Greene 2002), having a similar arginine rich motif (ARM) that plays a role in the mediation of host-cell nuclear import, translocation over biological membranes, and binding to targeted viral RNA sequences (Futaki et al. 2001; Seelamgari et al. 2004; Hariton-Gazal et al. 2005). Hariton-Gazal et al. used karyophilic backbone cyclic (BC) ARM peptides to target the ARM domain to develop a therapeutic agent against HIV-1 and as a delivery carrier to living cells (Hariton-Gazal et al. 2005). The findings revealed that the BC Rev-ARM analog (Rev-13) (Fig. 7) effectively blocks Rev-GFP from binding to the importin β receptor. Moreover, it influences the penetration of bovine serum albumin (BSA). An ELISA-based study revealed that the majority of backbone cyclic peptides (BCPs) from the library possess both cell-penetrating characteristics and binding ability by transporting BSA into intact Colo-205 cells (Hariton-Gazal et al. 2005). In addition, the BC Rev-13 (Fig. 7) analog showed the ability to inhibit the expression of Rev-induced genes in the low micromolar range (2–5 µM) without showing any cytotoxic effect (Hariton-Gazal et al. 2005).
Fig. 7.
Peptide Rev-13 (Hariton-Gazal et al. 2005)
As mentioned previously, the life cycle of HIV-1 is dependent on Rev protein for the regulation of viral mRNA expression by enhancing the production of viral structural protein precursors (Chaloin et al. 2007). Chaloin et al. investigated in vitro the ability of a series of BCP analogues having a conformational-controlled ARM of Rev for HIV-1 replication suppression. Rev-BCPs showed remarkable inhibitory activity against HIV-1 in persistently infected T lymphocytic cells. In addition, the nuclear import process was affected by Rev-BCPs, and there was significant efficiency in blocking the action controlling polyprotein precursor protein Pr55Gag and envelope glycoprotein precursor gp160env production (Chaloin et al. 2007). Remarkably, Rev binding is necessary for the nuclear export of these mRNA-encoded protein precursors (Smagulova et al. 2005). By utilizing an HIV-1 Gag oligonucleotide probe conjugated with Cy-3, in situ hybridization revealed that Rev-BCPs are able to inhibit the intracellular accumulation of unspliced viral RNA (Chaloin et al. 2007). In silico studies revealed that recombinant simulation proposes that upon binding to the Rev-response element, Rev-BCP 14 (Fig. 8) is the most promising analogue with an observed IC50 = 6.2 µM and was able to broaden the distorted viral RNA major groove (Chaloin et al. 2007). In addition to the strong electrostatic interactions present between the positive charges of the peptides and the phosphate backbone, various contacts between RNA and peptide were discovered within the complex, and some are important for the interactions, for example, the hydrogen bonds and the hydrophobic contacts (van der Waals) between the carboxy-amidated peptide end and the RNA backbone phosphate (Chaloin et al. 2007; Young et al. 2011). Altogether, the use of structurally restricted Rev-BCPs is a promising tactic to develop novel cyclic peptide-based therapeutics against HIV-1, with further improvement on the reduction in the total inhibition concentration (Chaloin et al. 2007).
Fig. 8.
Peptide REV-14 (Hariton-Gazal et al. 2005)
To date, various affinity-based methods, such as encoded synthetic peptide libraries, DNA directed synthesis, and phage and ribosomal display, are commonly enriched for the synthetic and in vitro generation of large libraries of peptides from synthetic and natural amino acids (Sandman et al. 2000; Sattely et al. 2008; Young et al. 2011). Young et al. reported a microbial system for cyclic peptide evolution that utilizes a series of lengthened amino acid building blocks (Young et al. 2011). The combination of SICLOPPS with orthogonal aminoacyl-tRNA synthetase∕tRNACUA pairs was used to biosynthesize a library of ribosomal peptides encompassing amino acids with exclusive reactivity and structure (Young et al. 2011). According to cellular viability, the selection was performed in the peptide library, and the HIV protease inhibitor in Escherichia coli (E. coli) was evolved. After selection and isolation, two cyclic peptides containing p-benzoylphenylalanine (pBzF) amino acids were reported to have a moiety of electrophilic aryl ketone and a fairly large hydrophobic surface area (Young et al. 2011). With observed cyclic IC50 = 0.96 µM and linear IC50 of 6.67 µM, the encoded GIXVSL clone, in which X is pBzF, was the most potent HIV protease blocker (Fig. 9). Therefore, the pBzF amino acid residue is believed to covalently form a Schiff base adduct with the protease by binding to the ϵ-amino group of Lys-14, suggesting that in response to selective pressure, the expansion of the genetic code could have evolutionary benefits. Therefore, further studies could combine chemical building blocks with natural evolutionary processes to develop novel biologically active peptides (Young et al. 2011).
Fig. 9.
Cyclic peptide HIV protease inhibitor G12 (Young et al. 2011)
Cyclic Peptides in Ebola Virus Disease Treatment
In 1976, Ebola virus (EBOV) was first identified in the Democratic Republic of the Congo (Jacob et al. 2020). It is an enveloped virus with negative-stranded RNA belonging to the family Filoviridae (Geisbert et al. 2003) and has been discovered to be the causative agent of Ebola virus disease (EVD) (Baize et al. 2014). Studies have been conducted and they found out that by blocking the construction of the six-helix bundle structure of the glycoprotein 2 (GP2) ectodomain can restrict the entry of the genetic material of EBOV into the cytoplasm (Miller et al. 2011; Tambunan et al. 2017).
In silico modelling was utilized to investigate the interaction between the modified commercial cyclopeptide ligands and the EBOV N-terminal heptad repeat (NHR) GP2 ectodomain. The ligand that interacts with the amino acid residues Arg B580, Glu C578, Phe C572, and Thr B576 was placed in the active site of the target protein and was found to form a stable protein–ligand complex with the best molecular interaction under both 310 and 312 K (Tambunan et al. 2017). The temperatures mentioned above indicate the normal body condition and the infected human body with theEBOV. As evidenced, its root mean square deviation is 3.0–3.5 Å at 310 and 312 K, whereas ΔGbinding is − 53.6622 kcal/mol. Moreover, the pharmacokinetics evaluation revealed that this particular ligand has good pharmacological characteristics, and no toxic effect was detected. (Garabedian et al. 2018). The conjugation of HIV-1 Tat ligand with commercial cyclic peptides has been proven to be effective in exhibiting inhibitory effects against EBOV (Tambunan et al. 2017). Therefore, further in vitro and in vivo investigations of its activity against EBOV could be conducted for its future therapeutic value.
As previously mentioned, Tsg101 UEV domains are potential targets for HIV infection treatment. Studies have shown that the same strategy could also be utilized against EBOV. The goal is to protect the emerging virions from infected cells as immature particles. The resultant virus budding process failure can be achieved by interruption of the binding interaction between the PT/SAP motifs and UEV domain of Tsg101 proteins on EBOV Vp40 proteins (Pornillos et al. 2002; Lin et al. 2020). Due to the limited experimental 3D structure information available, a study conducted by Lin et al. utilized a beneficial methodology that involves various techniques, including Gaussian accelerated molecular dynamics (GaMD), normal molecular dynamics (cMD), solvated interaction energy (SIE), two-dimensional (2D), and potential of mean force (PMF) techniques, with the purpose of examining the binding conformations of cyclic peptides with UEV domain proteins (Lin et al. 2020). Their findings described Trp145, Tyr147, and Trp148 as the hot-spot residues of the cyclo-(Ser-Gly-Trp-Ile-Tyr-Trp-Asn-Val) inhibitor, which play a crucial role in the interaction with the UEV domain proteins via hydrogen bond formation and nonbonding interactions (Lin et al. 2020). Since the single point substitution might influence the 3D structures of the peptide and the protein receptor binding affinity (Iwazaki et al. 2005; Garabedian et al. 2018), Lin et al. revealed that the mutation of Tyr 147 reduces UEV domain protein binding abilities. From this study, the developments in computational approaches and calculating power (Wang and Cheng 2021) might escalate the prediction precision of cyclic peptide-protein conformation, which is able to expedite in tackling the blocking blocks towards cyclic peptide inhibitor design (Lin et al. 2020).
Promising Progress of Cyclic Peptides in Chikungunya Therapy
Chikungunya, an arthropod-borne virus (CHIKV), causes deleterious health effects globally for more than a half-century. This is distinguished as an alphavirus of the Togaviridae family, mainly spreading by mosquito-borne vectors Aedes albopictus and Aedes aegypti (Sourisseau et al. 2007, for Chikungunya 2021, Jagadesh et al. 2021).
To date, there is no approved therapeutic medicine or vaccine available for CHIKV (August et al. 2021). Multiapproach research exploring potential anti-CHIKV drugs is in progress (Kumar et al. 2020; Bappy et al. 2021). Peptides are now receiving attention as potential alternatives to many conventional antiviral drugs available on the market. Since several of these peptides have anionic characteristics, they are easily transported into tumor cells and execute antiproliferative functions (Hoskin and Ramamoorthy 2008). Previously, antibacterial peptides have been widely documented for countering antibiotic resistance (Parachin and Franco 2014). In this milieu, researchers have started experimenting with cyclic peptides to combat multidrug resistive pandemic viruses, as they will have a huge impact on clinical research and medical practice. It is natural and synthetic cyclic peptide products (as pure and formulated) that have been designed to treat CHIKV (Boas et al. 2019). Similarly, attempts to discover cyclic peptide vaccines have also been recently initiated because vaccines can prevent and provide permanent protection against CHIKV infection. In addition to exploring active peptides, subsequently, it is also imperative to develop strategies to construct an appropriate design and predict an appropriate target to have a potential outcome.
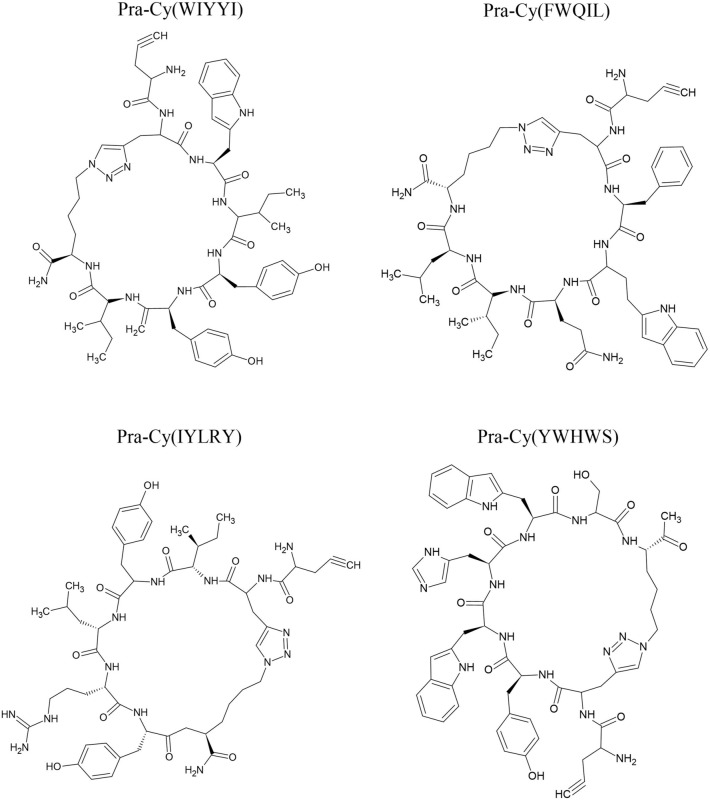
CHIKV uniquely contains approximately 240 copies of immunogenic E2 protein on its surface, which renders an attractive structure for finding suitable targets for detection as well as designing drugs or vaccines (Voss et al. 2010; Weger-Lucarelli et al. 2015). With the help of a library of one-bead-one-compound cyclic peptides, researchers have developed a multivalent maturation strategy to construct peptide-based capture receptors (peptide format: H2N-Pra-Cy(XXXXX)-TG) with satisfactory thermal stability to target unglycosylated E2 proteins of CHIKV. They synthesized macrocyclic peptides such as Pra-Cy(WIYYI), Pra-Cy(FWQIL), Pra-Cy(IYLRY), and Pra-Cy(YWHWS) (Fig. 10). Using high-performance computing algorithms, Pra-Cy(WIYYI) was found to be the best interacting macrocyclic peptide whose appropriate conformation for the effective E2 protein-peptide receptor affinity was identified (Coppock et al. 2019).
Fig. 10.
Structure of macrocyclic peptides synthesized by Coppock et al. (2019)
The analysis displayed the binding of macrocyclic peptides with lysine residues in the hydrophobic surface of domain B of the E2 protein, K107, K200, and K280. Each of the peptides found interacted with more than one residue. WIYYI, YWHWS, and FWQIL peptides established affinity at all three residues, while YWHWS and IYLRY preferred targeted K200 locations with high enthalpy (Coppock et al. 2019). These predictions are in agreement with previous studies that targeted domain B of the E2 protein (Asnet Mary et al. 2013, Lam et al. 2015, Deeba et al. 2017). After multivalent enhancement, WIYYI macrocyclic peptide top performed with up to ninefold better selectivity and displayed affinity in 50% human serum, which was comparatively better than the commercial anti-E2 antibody.
The multivalent enhancement of macrocyclic peptides was primarily attributed to their efficient binding with CHIKV E2. These peptides bind at multiple sites to viral surfaces and exert structural analogues and compatible physical features, such as hydrophobicity. This could limit their specificity to target a potential domain, as multivalency improves macrocyclic peptides to interact with several copies of the same target (Coppock et al. 2019). In terms of thermal stability, macrocyclic peptides are efficient over linear peptides since they have characteristic structural rigidity with stringent intramolecular arrangements, unlike their linear counterparts. Indeed, from the study, the best protein-peptide complex, (Pra-Cy(WIYYI)–CHIKV-E2), has also been successfully demonstrated with commercially available polyclonal antibodies, suggesting that cyclic peptides through this strategy can also be employed for detecting viruses other than CHIKV in sensor-based devices used in clinical diagnosis and environmental monitoring (Coppock et al. 2019).
On the other hand, researchers have tried and successfully designed antiviral drugs to mitigate multiplication in the host by blocking the NS proteins that interact with enzymes involved in the replication process. However, unlike other viruses, it is challenging to target CHIKV, as it possesses a complex or intricate NS protein design (Wang et al. 2011; Brandler et al. 2013). Although recombinant CHIKV vaccines have been used to invoke an immune response in the host, they still have not been subjected to clinical trials despite protecting against arthritis in experimental animals. In search for alternatives, researchers are attempting antigenic peptide candidates to explore conserved B cell and T cell epitopes of CHIKV structural polyprotein to discover vaccines that would hopefully evoke a defensive immunogenic response (Tahir ul Qamar et al. 2018). Using respective computational tools, B cell epitopes (PPFGAGRPGQFGDI, IPTGAGKPGDSGRP, NYPASHTTLGVQDI, PFHHDPPVIGREKF, TSAPCTITGTMGHF, TPYELTPGATVPFL, PEGYYNWHHGAVQY and WLKERGASLQHTAP) and T cell epitopes (MHC-I alleles: IFDNKGRVVAIVL, PLVPRNAEL, VMHKKEVVL, LSVTLEPTL, TAECKDKNL, GTLKIQVSL & LQISFSTAL and MHC-II alleles: FKRSSKYDL, LLANTTFPC, LKIQVSLQI, FVRTSAPCTI, VGFTDSRKI, VRYKCNCGG, WVMHKKEVV & LVVAVAALILIVVLCVSFSR) with higher antigenicity, flexibility, accessibility of surface, linearity, and hydrophilicity were predicted and docked with the solved 3D structure of the human HLA-B7 allele. The analysis showed VMHKKEVVL, LSVTLEPTL, GTLKIQVSL, and LQISFSTAL as potentially interacting peptides with minimized binding energy (− 20.2086, − 19.6688, − 17.3569, & − 14.5594) (Tahir ul Qamar et al. 2018).
The analysis showed that the best-interacted peptide, VMHKKEVVL, formed stable hydrogen bonds with Lys-243 and Glu-232 of chain A and Tyr-26 and Tyr63 of chain B of HLA-B7. In addition, the peptide also extended a water-mediated interaction with Thr-233 of the A-chain and Thr-10 of the B-chain (Tahir ul Qamar et al. 2018).
The above peptides showed no toxicity compliance to the FAO/WHO standards for nonallergenicity. Since these peptides were characterized only through in silico models, they may show unstable physiochemical characteristics if tested in vitro or in vivo. There are some physiochemical modification strategies (multi-valent enhancement, analogue formations, hydrophobic ion pairing, macromolecular conjugation, substitution-based delivery) through which peptides can be improvised as potential targets to construct vaccines (Mahajan et al. 2014).
Taking into account all these factors, it seems that cyclic peptides can provide a clear hope to successfully develop novel antiviral agents. In the near future, they may possibly emerge as suitable candidates for designing effective vaccines or medicines to combat the deadliest pandemic viruses.
Cyclic Peptides in Influenza Virus Therapy
The Orthomyxoviridae is a family of segmented RNA viruses, including influenza viruses (IVs), which are enveloped RNA viruses. Generally, the IVs that affect humans are three types: A, B and C. Type B and C viruses have no known subtypes, while Type A has many subtypes based on antigens (Kumar 2017, Suchitra Rao 2019). Polymerase (PB1, PB2, and PA), hemagglutinin (HA)and neuraminidase (NA)-glycoproteins, nucleoprotein (NP) and member protein (-M1 and -M2) are RNA-coded proteins (7 or 8 segments) of the influenza virus genome (Machała and Brydak 2006). These segments consist of one or more open-reading frames, segment-specific untranslated regions (UTRs) and partially complementary 5′ and 3′ termini. The double-helical rod-like structure is formed by the assembly of viral ribonucleoprotein complexes, as the viral RNA-dependent RNA polymerase bound the 5′ and 3′ termini and the oligomeric viral NP bound the rest of the RNA (Fodor and Te Velthuis 2020).
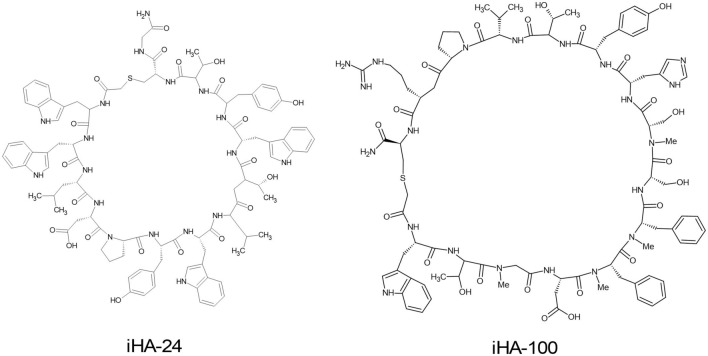
Saito et al. developed a class of macrocyclic peptides (iHAs) targeting the viral envelope HA protein of IV. The in vitro examination of the developed iHAs revealed that iHA-24 and iHA-100 (Fig. 11) are capable of inhibiting the replication of IVs. iHA-100 inhibits HA-mediated adsorption and membrane fusion through binding to the stalk domain of HA. This demonstrates the potential development of mid-sized multifunctional cyclic peptides as next-generation molecules against IVs (Saito et al. 2021).
Fig. 11.
The chemical structures of iHA-24 and iHA-100 synthesized by Saito et al. 2021 (Saito et al. 2021)
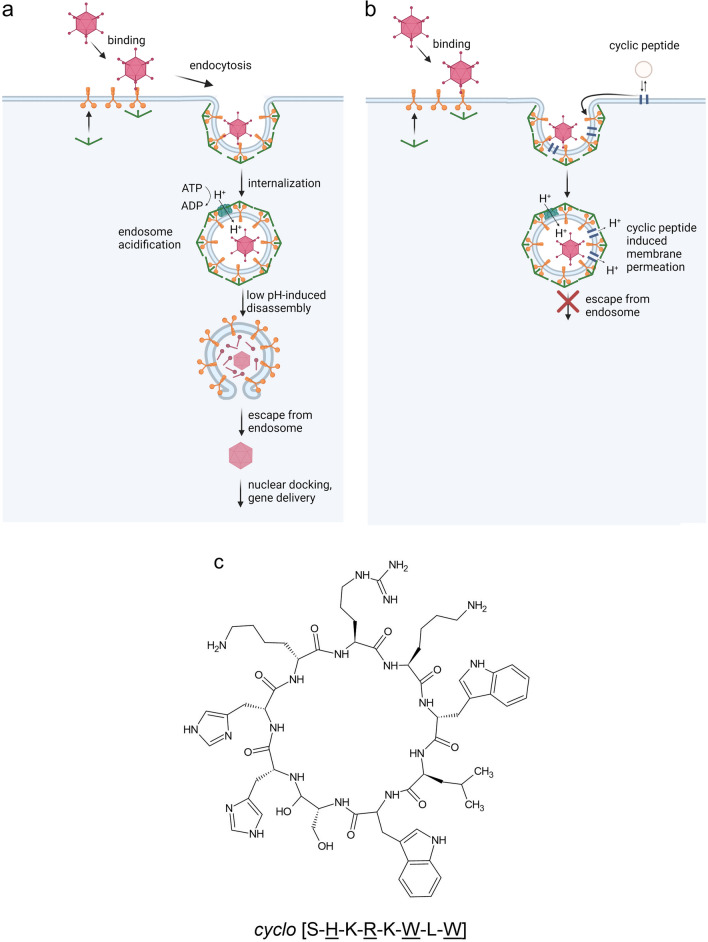
Horne et al. reported the selection of eight-residue cyclic d, l-peptide from a directed combinatorial library as a potential molecule to develop a low pH environment at endocytic vesicles, which could stop the emergence of virions from the endosome to prevent adenovirus infections without affecting cell viability. The inhibition of the emergence of IV type A from endosomes by the action of these peptides supports the potential utilization of this approach against other pH-dependent viral infections. Therefore, the discovery/design of broad-spectrum antiviral agents could be achieved by the self-assembling cyclic d, l-peptides as a new rational supramolecular approach. From the 20 hits found, peptide 1 (Fig. 12) was synthesized and used in the study because it significantly inhibited Ad5-mediated gene delivery. α-sarcin uptake has previously been found to correlate with enveloped viral entry. Interestingly, peptide 1 significantly reduced the delivery of influenza virus-mediated α-sarcin (Horne et al. 2005).
Fig. 12.
Schematic illustration of the antiviral action of membranes associating cyclic d, l-a-peptides to the endosomal membranes during virus internalization by offsetting the formation of a low pH environment inside endosomes via the membrane permeating supramolecular assemblies and the primary stages of the adenovirus infection cycle. a Schematic illustration of the primary stages of the adenovirus infection cycle. b The antiviral action of membranes associating cyclic d, l-a-peptides to the endosomal membranes during virus internalization by offsetting the formation of a low pH environment inside endosomes via the membrane permeating supramolecular assemblies. c The structure and sequence of the peptide identified by Horne et al. (Horne et al. 2005). (a and b are created with BioRender.com)
Kadam et al. introduced a strategy to design macrocyclic peptides with the ability to inhibit the fusion of group 1 influenza A viruses (Fig. 13). These peptides are capable of binding to the conserved HA stem with a similar approach of broad neutralizing antibodies, which might inhibit the process of generating escape mutants (Kadam et al. 2017).
Fig. 13.
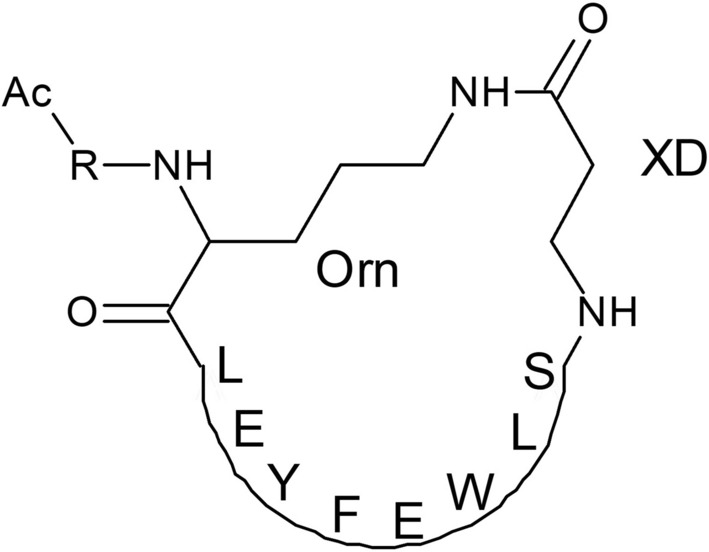
Amino acid sequences of cyclic peptides designed by Kadam et al. (Kadam et al. 2017)
Cyclic Peptides in Hepatitis Virus Therapy
The majority of the global burden of hepatitis is caused by hepatitis A virus (HAV), HBV, HCV, HDV, and HEV. HBV and HCV are responsible for the majority of chronic hepatitis morbidity cases and death (Lanini et al. 2019; Wang and Tsai 2021). Human HBV is characterized by an asymmetric replication mechanism and an unusual organization of its genome. It is circular in shape, with 3200 base pairs and four overlapping ORF regions: PreC/C, X, P, and Pré-S1/Pré-S2/S. The three envelope proteins of HBV, nuclear protein, core protein, surface antigen (HBeAg, HBcAg and HBsAg), viral polymerase (P), and X protein (affect viral proliferation and replication), are all encoded by these genes (Wen et al. 2008; Zhu et al. 2008, Souza et al. 2014). In a preferred but not obligated order, this polyprotein is processed into ten mature proteins; p7 separates the structural proteins (the envelope glycoproteins E1 and E2 and the core) from the NS3, NS4A, NS4B, NS5A and NS5B, which are essential for viral RNA replication (Lohmann 2013; Moradpour and Penin 2013; Niepmann 2013; Simmonds 2013; Madan and Bartenschlager 2015).
A study utilizing a binding assay in solution revealed that peptides bearing the amino acid sequences C-WPFWGPW-C and C-WSFFSNI-C tightly bind to HBcAg and inhibit L-HBsAg-HBcAg association with KD rel < 25 nM, which is at least 10 orders of magnitude higher than the peptide (LLGRMK) identified from a linear peptide library (Ho et al. 2003). The cyclic peptide CP11 (CGWIYWNV) inhibits HEV viral protein interaction with the host cell, preventing the release of the virus (Anang et al. 2018). In addition, HCV15R (CR-Nal-RV-(D)-P-Cha-HRYRC-CONH2) cyclic peptide inhibits the entry of HCV by targeting cell receptors (Khachatoorian et al. 2015). Through interactions with HCV RNA, human La protein stimulates the translation of HCV. A cyclopeptide (LaR2C-N7-cy) synthesized by Manna et al. mimics the β-turn of the human La-protein and can interact with HCV-RNA. It significantly inhibits the translation of HCV-RNA better than the corresponding linear peptide (Fig. 14). This cyclic peptide was also found to inhibit the replication of HCV by measuring replicon RNA levels using RT-PCR (Manna et al. 2013).
Fig. 14.
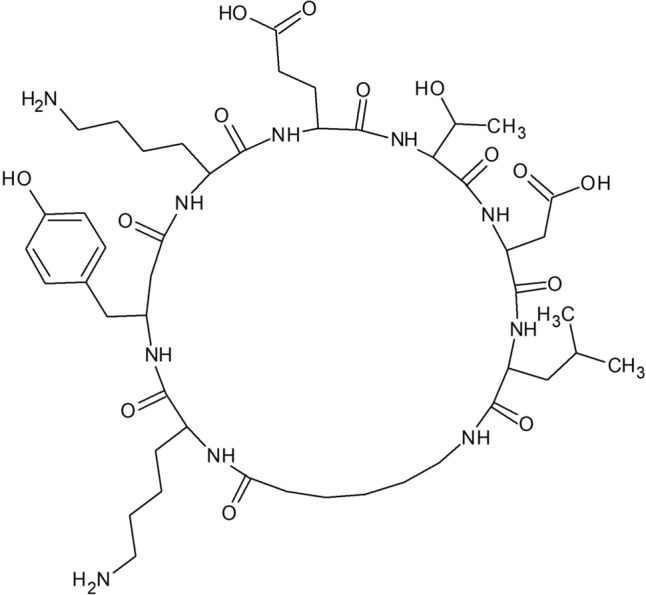
Chemical structure of LaR2C-N7-cy synthesized by Manna et al. (Manna et al. 2013)
The Antiviral Activities of Natural Isolated Cyclic Peptides
Naturally isolated cyclic peptides possess medical significance as therapeutic agents with a wide range of biological activities comprising antiviral, anticancer, antibacterial, antifungal effects and so on (Abdalla and McGaw 2018). The cyclic peptides from various natural sources are documented (Table 3) with their respective antiviral activities studied in the previous literature.
Table 3.
Cyclic peptides from various natural sources with respective enzyme inhibition (IC50) and antiviral activities (EC50)
| Compound name | Class | Source | IC50/EC50 | References |
|---|---|---|---|---|
| Animal (leukocytes) | ||||
| Retrocyclin-2 (RC-2, θ-defensin) | Cyclic octadecapeptides | Non-human ‘Old World’ primates |
HIV-1 -IC50: 2.33 µg/mL |
Wang et al. (2004) |
| Cyclosporine | Cyclic oligopeptide | Tolypocladium inflatum fungi |
IV -IC50: 1.45 µM |
Hamamoto et al. (2013); Abdalla and McGaw (2018) |
| Fungi | ||||
| Marine natural product | ||||
| Aspergillipeptide D | Cyclic pentapeptides | Marine gorgonian-derived fungal strain Aspergillus sp. SCSIO 41, 501 |
Herpes simplex virus type 1 (HSV-1) -IC50: 9.5 µM |
Ma et al. (2017) |
| Asperterrestide A | Cyclic tetrapeptide | Marine-derived fungus Aspergillus terreus SCSG AF0162 |
IAV/WSN/33 (H1N1) -IC50: 15.0 µM IAV/Hong Kong/8/68 (H3N2) -IC50: 8.1 µM |
He et al. (2013) |
|
Celebeside A Celebeside C |
Cyclic depsipeptide |
Indonesian marine sponge Siliquariaspongia mirabilis |
HIV-1 -IC50: 2.1 μM -IC50: > 62 μM |
Kang et al. (2015) |
| Didemnin A | Cyclic depsipeptides | Marine tunicate Trididemnum solidum |
Coxsackie virus, equine rhinovirus (both RNA), HSV-2 (DNA) -IC50: 1.5 µg/mL HSV-22 -IC50: 3 µg/mL |
Mata et al. (2017) |
|
Homophymine A-E Homophymine A1-E1 |
Cyclodepsipeptides | Sponge Homophymia sp. |
III B strain of HIV-1 -IC50: 75 nM |
Kang et al. (2015) |
|
Koshikmaide B Koshikmaide F–H |
Cyclic peptide lactone | Theonella sp. |
HIV-1 -IC50: 2.3 & 5.5 µM |
Kang et al. (2015) |
|
Mirabamide A Mirabamide C Mirabamide D |
Cyclic depsipeptides | Sponge Siliquariaspongia mirabilis |
HIV -IC50: 40–140 nM -IC50: 140 nM-1.3 µM -IC50: 190 nM-3.9 µM |
Plaza et al. (2007) |
|
Mirabamide E Mirabamide F Mirabamide G Mirabamide H |
Cyclic depsipeptides | Sponge Stelletta clavosa |
HIV-1 viral strain YU2-V3 -IC50: 121 nM -IC50: 62 nM -IC50: 68 nM -IC50: 41 nM |
Lu et al. (2011) |
| Mollamides B | Cyclic hexapeptide | Indonesian tunicate Didemnum molle |
HIV-1 -EC50: 48.7 µM |
Donia et al. (2008) |
|
Papuamide A Papuamide B |
Cyclic depsipeptide | Sponge Theonella sp. |
HIV -EC50: 3.6 ng/mL -IC50: 710 nM |
Ford et al. (1999); Sagar et al. (2010) |
| Sansalvamide A | Cyclodepsipeptide | Marine fungus Fusarium sp. |
Topoisomerase of Molluscum Contagiosum Virus (MCV) -IC50: 124 µM |
Moghadamtousi et al. (2015) |
| Simplicilliumtide J | Cyclolipodepsipeptide | Deep see-derived fungal strain Simplicillium obclavatum EIODSF 020 |
HSV-1 -EC50: 14.0 µM |
Liang et al. (2017) |
|
Theopapuamide A Theopapuamide B |
Cyclic undecapeptide |
Sponge Theonella swinhoei Siliquariaspongia mirabilis |
CEM-TART (T-cells that express both HIV-1 tat and rev) -IC50: 0.5 µM -IC50: 0.8 µg/mL |
Ngo et al. (2012) |
|
Verlamelin A Verlamelin B |
Cyclic hexadepsipeptide | Deep see-derived fungal strain Simplicillium obclavatum EIODSF 020 |
HSV-1 -EC50: 16.7 µM -EC50: 15.6 µM |
Liang et al. (2017) |
| Plant | ||||
|
Circulin A Circulin B |
Cyclotide | Chassalia parvifolia |
HIV -EC50: 70 nM |
Wani et al. (2020) |
|
Cycloviolacin VY1 Cycloviolacin Y5 |
Cyclotide | Viola yedoensis |
IAV (H1N1) -IC50: 2.27 µg/mL HIV-EC50: 40 nM |
Wang et al. (2008, Liu et al. (2014) |
|
Kalata B1 Kalata B8 |
Cyclotide | Oldenlandia affinis |
HIV-1-EC50: 140 Nm -EC50: 2.5 µM |
Gerlach and Mondal (2012) |
|
MCoT-I MCoT-II |
Cyclotide (Palicourein) | Momordica cochinchinensis |
HIV -EC50: 100 nM |
Gerlach and Mondal (2012) |
| Secondary metabolites | ||||
| Antimycin A | Cyclic lactone | Streptomyces kaviengensis |
Western equine encephalitis viruses (WEEV) -IC50: 3 nM |
Yi et al. (2020), Lin et al. (2020) |
| Daptomycin | Cyclic lipopeptide | Streptomyces spp. |
ZIKV -IC50: 1 µM |
Jakubiec-Krzesniak et al. (2018) |
Aspergillipeptide D
Aspergillipeptide D (Fig. 15) is a cyclic pentapeptide collected from the broth culture of the sea gorgonian-derived fungal strain Aspergillus sp. SCSIO 41, 501 (Ma et al. 2017; Abdalla and McGaw 2018; Jing and Jin 2020). It appears as a white crystal with the known molecular formula of C40H49N5O8 (Wang et al. 2020) From Ma et al. aspergillipeptide D (Fig. 15) showed a potent antiviral effect on HSV-1 (Ma et al. 2017). With their noncytotoxic concentrations (TC0) against the Vero cell line, aspergillipeptide D showed an IC50 of 9.5 µM. It also exhibited good antiviral effects towards two isolates, HSV-1-106 and HSV-V-153, which were clinically resistant to acyclovir at a concentration of 12.5 µM with approximately 50% inhibition rates (Ma et al. 2017).
Fig. 15.

Aspergillipeptide D (Ma et al. 2017)
Simplicilliumtide
Among the cyclic peptides discovered from the marine-derived fungal strain Simplicillium obclavatum EIODSF 020, Liang et al. showed that the novel cyclolipodepsipeptide simplicilliumtide J (Fig. 16a) has the molecular formula of C46H73N7O11 with the presence of a fatty acid chain moiety and lactone linkage, exhibiting an anti-HSV-1 effect with an EC50 value of 14.0 µM (Liang et al. 2017; Jing and Jin 2020). In addition, its noncytotoxic concentrations TC0 and TC50 values against Vero cells were observed to be 25.1 and 204 μM, respectively (Liang et al. 2017, Yi et al. 2020, Lin et al. 2020). The other simplicilliumtide cyclopeptides, for example simplicilliumtide K (Fig. 16b) and L (Fig. 16c), have less remarkable antiviral activity towards HSV-1 (El Maddah et al. 2020). It is believed that the activity reduction is due to the carbonyl substituent at C-13 or C-14 of (S)-5-hydroxy-13-ketotetradecanoic acid (HKTA) or (S)-5-hydroxy-12-ketotetradecanoic acid (HKTA), respectively (Liang et al. 2017; El Maddah et al. 2020).
Fig. 16.
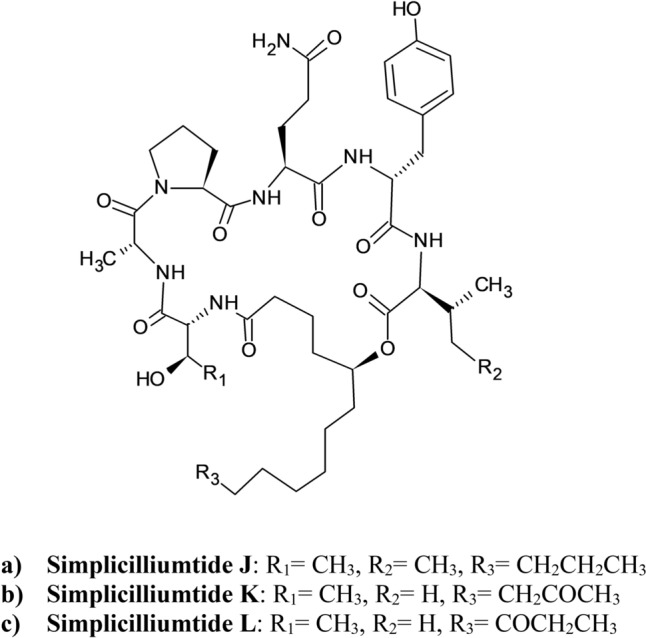
Structure of Simplicilliumtides (Liang et al. 2017; El Maddah et al. 2020)
Verlamelins
Verlamelins A (Fig. 17a) and B (Fig. 17b) are known analogs of the cyclic peptide simplicilliumtide J–M. Recently, they were discovered to be isolated from the same crude fungal strain as simplicilliumtide J–M (Liang et al. 2017). Formerly, verlamelin A (Fig. 17a) was isolated from Verticillium lamellicola because of its antifungal activity (El Maddah et al. 2020). Later, together with verlamelin B (Fig. 17b), both were reported from Lecanicillium sp. HF627 in 2014 for the absolute configuration determination (Liang et al. 2017). Liang et al. determined their anti-HSV-1 activity through plaque reduction assays. Their corresponding EC50 values were 16.7 and 15.6 µM under TC0 against the Vero cell line. Verlamelin A (Fig. 17a) displayed TC0 and TC50 values of 57.2 and 137.0 μM, respectively, whereas verlamelin B (Fig. 17b) reported TC0 and TC50 values of 49.4 and 101.1 μM, respectively (Liang et al. 2017).
Fig. 17.
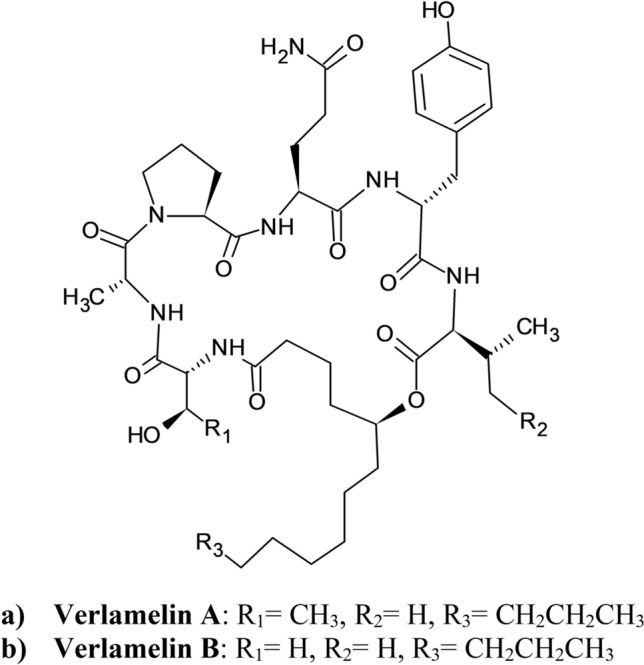
Structure of Verlamelins (Liang et al. 2017)
Asperterrestide A
With the investigation of the chemical constituents in the fermentation broth of Aspergillus terreus SCSGAF0162 isolated from gorgonian Echinogorgia aurantiaca tissue from Sanya, Hainan Province, China (Moghadamtousi et al. 2015), a new cyclic tetrapeptide asperterrestide A (Fig. 18) was isolated along with other compounds (He et al. 2013). From He et al. asperterrestide A (Fig. 18) was allocated with a molecular formula of C26H32N4O5. Asperterrestide A (Fig. 18) consists of a structure with an intermittent 3–OH–N–CH3-Phe moiety (He et al. 2013). From their research, the antiviral activity of asperterrestide A (Fig. 18) was tested on two strains of IV, A/WSN/33 (H1N1) and A/Hong Kong/8/68 (H3N2), by cytopathic effect (CPE) assay. These virus strains are the M2-resistant strain and the M2-sensitive strain, respectively. The results showed its inhibitory ability with corresponding IC50 values of 15 and 8.1 µM (He et al. 2013; Moghadamtousi et al. 2015).
Fig. 18.

Asperterrestide A (He et al. 2013)
θ-Defensins and Retrocyclins
θ-Defensins are the sole family of mammalian cyclic peptides found by scientists, in which they are isolated from baboons and rhesus macaque leukocytes (Lehrer et al. 2012). With a premature stop codon, the translation apparatus in human θ-defensin genes is unable to form peptides in human leucocytes, although the precursors are needed (Venkataraman et al. 2009). Thus, retrocyclin (RC) was synthesized with the corresponding sequences with those expressed in the human pseudogenes (Lehrer et al. 2012).
Activity Against Human Immunodeficiency Virus
Gp120 is a viral surface glycoprotein that engages two target receptors, CD4 and CXCR4 or CCR5. Following this initial binding of HIV, viral entry is mediated by six-helix bundle structure formation due to conformational changes in gp41 (Noah et al. 2008; Regula et al. 2013). Neither cytotoxic nor virotoxic effects of RC-1 inhibit the formation of proviral DNA, preventing viral entry (Lehrer et al. 2012). Several studies revealed that RC-1 can defend human target cells against HIV-1 infection and that HIV-1 strains employ either CXCR4 or CCR5 coreceptors; however, there is not much effect against HIV-2 viral strains and related retroviruses infecting blood cells that do not rely on the co-receptors CXCR4 or CCR5, such as simian immunodeficiency virus type 1 (Simmons et al. 2000; Lehrer et al. 2012). With the cyclic backbone and intact disulphide bonds, RC-1 was found to have high binding affinity to CD4, gp120, and a surface glycolipid that is galactosylceramide with KD values of 31 nM, 35.4 nM, and 24.1 nM, respectively (Lehrer et al. 2012; Xiong et al. 2021). From the fusion assays targeting the envelope glycoproteins of HIV-1, RC-1 showed its ability to prevent the viral entry of HIV-1 with neither the involvement of cross-linking membrane proteins nor impeding the interactions of gp120-CD4 (Cole et al. 2006; Lehrer et al. 2012).
RC, which is only 18 residues in length, is of interest for its lack of complexity in designing congeners with greater anti-HIV-1 activity (Penberthy et al. 2011). A congener, RC-101, formed from a charge-conservative substitution with lysine on one of the arginines (Owen et al. 2004), was found to have increased anti-HIV-1 potency (low micromolar EC50 activity of 1–5 µg/mL) compared with the parent molecule (Owen et al. 2004; Wang et al. 2004; Penberthy et al. 2011). Compared to RC1, the RC-101 congener has a much greater binding affinity to gp120 that is approximately 25-fold, while it has no significant superiority in binding to galactosylceramide with identical binding, as proven by the observed values of 20–30 nM (Wang et al. 2004).
To date, new HIV-1 viral strains have been found to commonly develop resistance to drugs targeting coreceptors gp41, CXCR5, or CCR4 within weeks after exposure. Therefore, the ability to obstinately block HIV-1 by affecting the key amino acid residues for infection is one of the criteria of anti-HIV-1 therapeutic interventions (Pang et al. 2009; Penberthy et al. 2011) To better understand the latent resistance of HIV-1 to RCs, a study revealed that most HIV-1 strains were expected to be vulnerable to inhibition by RC-101 treatment (Fuhrman et al. 2007). Further support by data reporting RC-101 induced minimal HIV-1 resistance of only 5–10-fold. It is significantly different from other HIV entry inhibitors showing 10,000–20,000-fold HIV resistance in similar passaging tests (Trkola et al. 2002; Nameki et al. 2005). Unpredictably, even with HIV-1 mutations, RC-101 is still able to reduce HIV-1 infectivity, although HIV-1 protein binding is prohibited (Cole et al. 2006; Fuhrman et al. 2007), therefore conveying viral fitness significance to undermining RC action.
Activity Against IAV
Aside from HIV-1, RC has potent antiviral activity on several viruses as well. For example, RC-1 showedan antiviral effect on IAV. In addition to the impairment of HA-mediated viral entry (Leikina et al. 2005), RC-1 causes the induction of viral aggregation, enhancing IAV ingestion by neutrophils (Doss et al. 2009). RC-2, with an additional arginine in structure, acts against IAV by preventing the membrane fusion mediated byHA (Doss et al. 2009; Lehrer et al. 2012). Similar to RC-1, RC-2, as a multivalent lectin, can form an immobilized surface glycoprotein cross-linkage network to block IAV entry (Leikina et al. 2005; Lehrer et al. 2012).
Activity Against HSV
From the in vitro testing on the effectiveness of antiviral activity against HSV-1- and HSV-2-mediated cervix epithelial cells infection (Yasin et al. 2004), RC-1, RC-2, and rhesus θ-defensin-3 (RTD-3) showed protective ability, but RC-2 stood out without viral preincubation. With a Kd value of 13.3 nM, RC-2 attached to HSV-2 glycoprotein B, but no binding was observed after deglycosylating glycoprotein B (Lehrer et al. 2012).
Diverse Peptides that Resemble RCs
Tachyplesin/polyphemusins were isolated from horseshoe crab haemocytes. They are peptides comprising 17, 18 amino acids with an alpha-amide group at the C-terminus that build a rigid double-stranded anti-parallel beta-pleated sheet structure in the connection by a beta-turn (Bulet et al. 2004). Although these natural host-defense peptides (HDPs) exhibit modest antiviral activity against HIV-1 X4 tropic viruses, in which viral entry exploits the CXCR4 coreceptor, they show undesirable haemolytic action against human erythrocytes, and more prominent activity is being observed in high salt environments mimicking the typical marine living settings of horseshoe crabs (Bulet et al. 2004; Penberthy et al. 2011). To reduce haemolytic effects, the backbone of synthetic tachyplesins is manipulated via peptide cyclization. By imitating the HIV-1 protein gp41 portions, synthetic tachyplesins (e.g., T22) bind gp120 and effectively minimize X4-tropic HIV-1 infection of T lymphocytes (Penberthy et al. 2011).
Cyclotides, a subset of HDPs that are cyclic in nature, were first identified and isolated from plant sources such as Rubiaceae (coffee), Violaceae (violet), and Cucurbitaceae (cucurbit) families (Gruber et al. 2008; de Veer et al. 2019). Structurally, they are small (approximately 30 amino acids) proteins having a cysteine knot motif from three interlocking disulphide bonds, which is significant for protein stability. In addition to being thermally stable and resistant to proteases, cyclotides are capable of being modified with amino acid sequence replacement to optimize their inherent antiviral activity (Penberthy et al. 2011).
Cycloviolins
In a study by Hallock and colleagues, an emerging class of cyclic peptides consisting of four new anti-HIV macromolecules with 28–31 amino acid residues, known as cycloviolins A–D, were quarantined from the genuine tropical plant Leonia cymosa from South American collections (Hallock et al. 2000). A combination of methods was employed to determine and analyse the amino acid configuration and sequence in the cyclic primary structure of the discovered entities (Hallock et al. 2000). Six cysteines, presented as three internal disulphide bonds, are commonly found in all cycloviolin structures, contributing to chemical stability and possibly reducing proteolysis resistance (Hallock et al. 2000; Northfield et al. 2014). Cycloviolin B is the smallest member among this peptide class, and a proline residue is absent in its structure, but it has a distinctive TSSQ sequence from residues 17–20 (Hallock et al. 2000). Captivatingly, cycloviolins A–D performed a high level of homology in sequence to the identified circulins A and B as well as cyclopsychotride A from the Rubiaceae family but much less matching with the Viola varv peptides isolated from a member of the Violaceae family (Hallock et al. 2000).
Apart from their potential chemotaxonomic interest, an anti-HIV-1 study on cycloviolins A–D reported that these peptides possessed similar antiviral activities with observed EC50 values at approximately 130 nM (Hallock et al. 2000). Compared to circulins A and B, no significant deviations of their direct cytotoxicity to the host cells with the reported EC50 values of approximately 560 nM, concluding that the variations in the amino acid sequence variations have no assessable influence on the general activity profile (Hallock et al. 2000).
Prospective
Most of the viral infections discovered currently still face the challenges of a lack of proper treatment with desired therapeutic outcomes. Worldwide, populations are being exposed to the public health threats of viral pandemics due to the emergence or mutation of some viral strains, or even worse, the development of viral resistance from the available antiviral therapies. Such alarming conditions should be avoided, as viral diseases can spread uncontrollably all over the world in a highly globalised world. For example, the COVID-19 pandemic has already reported 620,301,709 infected people, including 6,540,487 deaths globally by WHO as of 13 October 2022 since the outbreak was discovered in January 2020 (Ciotti et al. 2020; WHO 2020).
Although there are antiviral molecules undergoing clinical trials, the development of novel antiviral cyclic peptides with satisfactory clinical outcomes has been the unique direction of effort by research scientists because of their significant advantageous properties when compared to linear peptides and other biological entities. With the current available naturally isolated cyclopeptides, optimization of cyclic peptide analogues has been the drug discovery trend to achieve better potency and pharmacokinetic properties. Modern genetic recombinant technologies are combined with peptide chemistry to develop more diverse cyclic peptide libraries. Rapid screening with display systems and the introduction of cyclization approaches further promotes its evolution in cyclic peptide discovery.
The conjugation of cyclic peptides with nanocarriers plays an important role in improving tissue targetability. Cyclic peptide nanoparticles have excellent capability for a broad array of biomedical applications, especially in drug delivery, due to their great tissue permeation and selectivity to the target site with specific receptor interactions (Abdalla and McGaw 2018, Zhang and Chen 2021).
In the future, cyclic peptides will be one of the most important highlights in the advancement of therapeutic agents due to their outstanding characteristics. Significantly, the reduction in production cost will encourage growth and fasten the pave of cyclic peptide drug discovery as well as commercialization.
Conclusion
In conclusion, polypeptides with restricted structures are potential candidates for antiviral drug development to produce favourable therapeutic outcomes with outstanding properties compared to their linear counterparts. The progress of novel drug discovery towards antiviral therapy is receiving increasing attention and is being supported with sophisticated biotechnologies and useful approaches, from the fermentation of natural products to biological methods and then chemically structural modification techniques, to overcome the limitations and to optimise the antiviral activities of cyclic peptides. In designing cyclic peptides as effective treatment agents for viral infections such as CoVs, DENV, and HIV, a well-established understanding of the virus structures, their mechanism of action, and their interactions with the target site of host cells are vital to ensure the effectiveness and targetability of therapeutic drugs. In fact, the increasing mutation on the viral strains has placed obstacles in the path of the drug investigation in addition to the challenges of the existing antiviral cyclic peptides yet to handle. Nonetheless, because of the desired characteristics of cyclic peptides in medical and pharmaceutical areas, such blocking stones never demotivate researchers to investigate the possibilities of cyclic peptides for antiviral treatment. With continuous effort in optimising and utilising the distinctive properties of cyclic peptides, it will be the cross-century advancement of therapeutic agents.
Acknowledgements
This Project is supported by Fundamental Research Grant Scheme (FRGS) (Reference code: FRGS/1/2021/SKK0/UCSI/02/5), Ministry of Higher Education (MOHE), and Research excellence and innovation grant (Project Code: REIG-FPS-2020-052) Under Centre of Excellence in Research, Value Innovation and Entrepreneurship (CERVIE), UCSI University, Malaysia.
Abbreviations
- ∆Gbinding
Gibbs free binding energy
- 2D
Two-dimensional
- ACE2
Angiotensin converting enzyme 2
- ADMET
Absorption, distribution, metabolism, excretion and toxicity
- AIDS
Acquired immunodeficiency syndrome
- ARM
Arginine rich motif
- ATP
Adenosine triphosphate
- BCP
Backbone cyclic peptide
- bnAbs
Broad neutralizing antibodies
- BNP
Brain natriuretic peptide
- BSA
Bovine serum albumin
- CHIKV
Chikungunya virus
- CIC
Chronic idiopathic constipation
- cMD
Classical molecular dynamics
- CoV
Coronavirus
- CPE
Cytopathic effect
- CSSSIs
Complicated skin and skin structure infections
- DENV
Dengue virus
- E. coli
Escherichia coli
- EBOV
Ebolavirus
- EC50
Median effective concentration
- ED
Envelope domain
- ELISA
Enzyme-linked immunosorbent assay
- EVD
Ebola virus disease
- FAO
Food and agriculture organization
- FDA
Food and drug administration
- GaMD
Gaussian accelerated molecular dynamics
- Gp
Glycoprotein
- HA
Hemagglutinin
- HDPs
Host-defence peptides
- HIV
Human immunodeficiency viruses
- HKTA
Hydroxy ketotetradecanoic acid
- HSV
Herpes simples virus
- HV
Hepatitis virus
- IAV
Influenza A virus
- IBS-C
Irritable bowel syndrome
- IC50
Median inhibitory concentration
- JEV
Japanese encephalitis virus
- KD
Dissociation constant
- Ki
Inhibitory constant
- Koff
Rate of dissociation
- MCV
Molluscum contagiosum virus
- MHC-I
Major histocompatibility complex class I
- Mpro
Main protease
- mRNA
Messenger ribonucleic acid
- Mtase
Methyltransferase
- NA
Neuraminidase
- NHR
N-terminal heptad repeat
- NP
Nucleoprotein
- NS
Nonstructural
- ORF
Open-reading frames
- pBzP
P-benzoylphenylalanine
- PET
Positron emission tomography
- PLpro
Papain-like protease
- PMF
Potential of mean force
- RBD
Receptor binding domain
- RC
Retrocyclin
- RdRp
RNA-dependent RNA polymerase
- RTD
Rhesus Theta defensin
- RTHS
Reverse two-hybrid system
- RT-PCR
Reverse transcription–polymerase chain reaction
- SAM
S-adenosyl-l-methionine
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SICLOPPS
Split intein-catalysed ligation of proteins and peptides system
- SIE
Solvated interaction energy
- SPPS
Solid phase peptide synthesis
- S–S
Disulphide
- Tat
Transactivator of transcription
- TBE
Tick-borne encephalitis virus
- TC0
Noncytotoxic concentration
- TNFSF10
TNF ligand superfamily member 10
- tRNA
Transfer ribonucleic acid
- TSG
Tumour susceptibility gene
- UAA
Unnatural amino acid
- UEV
Ubiquitin E2 variant
- UTR
Untranslated regions
- VLP
Virus-like particle
- VOC
Variant of concern
- VOI
Variant of interest
- Vp
Viral matrix protein
- vRNP
Viral ribonucleoprotein
- WEEV
Western equine encephalitis viruses
- WHO
World health organisation
- WNV
West Nile virus
- WT
Wild type
- YFV
Yellow fever virus
- ZIKV
Zika virus
Author Contributions
The conceptualization of the review article was by PVK. CLY, MAAM, and GR searched the related articles and prepared the figures. CLY collected the literature data and prepared the tables. CLY, MAAM, and GR wrote the manuscript. MAAM, PVK and ST revised the manuscript. All authors reviewed the final version of the manuscript for submission.
Declarations
Conflict of interest
The authors have no relevant financial or nonfinancial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdalla MA, McGaw LJ. Natural cyclic peptides as an attractive modality for therapeutics: a mini review. Molecules. 2018;23(8):2080. doi: 10.3390/molecules23082080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal G, Gabrani R. Antiviral peptides: identification and validation. Int J Pept Res Ther. 2021;27:149–168. doi: 10.1007/s10989-020-10072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Shaer D, Al Musaimi O, Albericio F, de la Torre BG. 2019 FDA TIDES (peptides and oligonucleotides) harvest. Pharmaceuticals. 2020;13(3):40. doi: 10.3390/ph13030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhoot MA, Rathinam AK, Wang SM, Manikam R, Sekaran SD. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. Int J Med Sci. 2013;10(6):719. doi: 10.7150/ijms.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I, Alharbi OM. COVID-19: disease, management, treatment, and social impact. Sci Total Environ. 2020;728:138861. doi: 10.1016/j.scitotenv.2020.138861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anang S, Kaushik N, Hingane S, Kumari A, Gupta J, Asthana S, Shalimar B, Nayak C-K, Surjit M. Potent inhibition of hepatitis E virus release by a cyclic peptide inhibitor of the interaction between viral open reading frame 3 protein and host tumor susceptibility gene 101. J Virol. 2018;92(20):e00684. doi: 10.1128/JVI.00684-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnet Mary J, Paramasivan R, Tyagi B, Surender M, Shenbagarathai R. Identification of structural motifs in the E2 glycoprotein of Chikungunya involved in virus–host interaction. J Biomol Struct Dyn. 2013;31(10):1077–1085. doi: 10.1080/07391102.2012.721496. [DOI] [PubMed] [Google Scholar]
- August A, Attarwala HZ, Himansu S, Kalidindi S, Lu S, Pajon R, Han S, Lecerf J-M, Tomassini JE, Hard M. A phase 1 trial of lipid-encapsulated mRNA encoding a monoclonal antibody with neutralizing activity against Chikungunya virus. Nat Med. 2021;27(12):2224–2233. doi: 10.1038/s41591-021-01573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub M, Scheidegger D. Peptide drugs, overcoming the challenges, a growing business. Chim Oggi. 2006;24(4):46. [Google Scholar]
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba NF, Soropogui B, Sow MS, Keïta S, De Clerck H. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371(15):1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- Balasubramanian A, Pilankatta R, Teramoto T, Sajith AM, Nwulia E, Kulkarni A, Padmanabhan R. Inhibition of dengue virus by curcuminoids. Antiviral Res. 2019;162:71–78. doi: 10.1016/j.antiviral.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bappy SS, Sultana S, Adhikari J, Mahmud S, Khan MA, Kibria KK, Rahman MM, Shibly AZ. Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against envelope protein of Chikungunya virus: a computational biology approach. J Biomol Struct Dyn. 2021;39(4):1139–1154. doi: 10.1080/07391102.2020.1726815. [DOI] [PubMed] [Google Scholar]
- Boas LCPV, Campos ML, Berlanda RLA, de Carvalho Neves N, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci. 2019;76(18):3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockus AT, McEwen CM, Lokey RS. Form and function in cyclic peptide natural products: a pharmacokinetic perspective. Curr Top Med Chem. 2013;13(7):821–836. doi: 10.2174/1568026611313070005. [DOI] [PubMed] [Google Scholar]
- Boras B, Jones RM, Anson BJ, Arenson D, Aschenbrenner L, Bakowski MA, Beutler N, Binder J, Chen E, Eng H. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat Commun. 2021;12(1):1–17. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler S, Ruffié C, Combredet C, Brault J-B, Najburg V, Prevost M-C, Habel A, Tauber E, Desprès P, Tangy F. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine. 2013;31(36):3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- Brecher M, Chen H, Liu B, Banavali NK, Jones SA, Zhang J, Li Z, Kramer LD, Li H. Novel broad spectrum inhibitors targeting the flavivirus methyltransferase. PLoS ONE. 2015;10(6):e0130062. doi: 10.1371/journal.pone.0130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27(1):28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- Buckton LK, Rahimi MN, McAlpine SR. Cyclic peptides as drugs for intracellular targets: the next frontier in peptide therapeutic development. Chem Eur J. 2021;27(5):1487–1513. doi: 10.1002/chem.201905385. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198(1):169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Callaway E, Ledford H. How bad is omicron? What scientists know so far. Nature. 2021;600(7888):197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- Chaloin L, Smagulova F, Hariton-Gazal E, Briant L, Loyter A, Devaux C. Potent inhibition of HIV-1 replication by backbone cyclic peptides bearing the rev arginine rich motif. J Biomed Sci. 2007;14(5):565–584. doi: 10.1007/s11373-007-9180-4. [DOI] [PubMed] [Google Scholar]
- Chellathurai MS, Vivien WangTing L, Palanirajan V. Fabrication and evaluation of transdermal microneedles for a recombinant human keratinocyte growth factor. Tur J Pharm Sci. 2021;18(1):96. doi: 10.4274/tjps.galenos.2020.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew M-F, Poh K-S, Poh C-L. Peptides as therapeutic agents for dengue virus. Int J Med Sci. 2017;14(13):1342. doi: 10.7150/ijms.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikungunya TTRCT. Current status of chikungunya in India. Front Microbiol. 2021;12:695173. doi: 10.3389/fmicb.2021.695173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-S, Joo SH. Recent trends in cyclic peptides as therapeutic agents and biochemical tools. Biomol Ther. 2020;28(1):18. doi: 10.4062/biomolther.2019.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciotti M, Ciccozzi M, Terrinoni A, Jiang W-C, Wang C-B, Bernardini S. The COVID-19 pandemic. Crit Rev Clin Lab Sci. 2020;57(6):365–388. doi: 10.1080/10408363.2020.1783198. [DOI] [PubMed] [Google Scholar]
- Cole AL, Yang OO, Warren AD, Waring AJ, Lehrer RI, Cole AM. HIV-1 adapts to a retrocyclin with cationic amino acid substitutions that reduce fusion efficiency of gp41. J Immunol. 2006;176(11):6900–6905. doi: 10.4049/jimmunol.176.11.6900. [DOI] [PubMed] [Google Scholar]
- Coppock MB, Hurley M, Jones C, Erickson D, Stratis-Cullum DN. A novel discovery, maturation, and assay integration approach for the development of ruggedized multi-valent capture receptors exemplified against the chikungunya virus E2 protein. Sensing Bio-Sensing Res. 2019;22:100248. doi: 10.1016/j.sbsr.2018.100248. [DOI] [Google Scholar]
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CA, Russell TW, Tully DC, Washburne AD. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science. 2021;372:6538. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29(3):695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer SJ, Kan M-W, Craik DJ. Cyclotides: from structure to function. Chem Rev. 2019;119(24):12375–12421. doi: 10.1021/acs.chemrev.9b00402. [DOI] [PubMed] [Google Scholar]
- Deeba F, Malik MZ, Naqvi IH, Haider MSH, Shafat Z, Sinha P, Ishrat R, Ahmed A, Parveen S. Potential entry inhibitors of the envelope protein (E2) of Chikungunya virus: in silico structural modeling, docking and molecular dynamic studies. VirusDisease. 2017;28(1):39–49. doi: 10.1007/s13337-016-0356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Wang B, Dunbar DC, Desai PV, Patny A, Avery M, Hamann MT. Mollamides B and C, cyclic hexapeptides from the Indonesian tunicate Didemnum molle. J Nat Prod. 2008;71(6):941–945. doi: 10.1021/np700718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss M, White MR, Tecle T, Gantz D, Crouch EC, Jung G, Ruchala P, Waring AJ, Lehrer RI, Hartshorn KL. Interactions of α-, β-, and θ-defensins with influenza A virus and surfactant protein D. J Immunol. 2009;182(12):7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- El Maddah F, König GM, Fisch KM. Non-ribosomal peptides from marine-derived fungi: chemistry, bioactivity, and biosynthesis. Encycl Marine Biotechnol. 2020;4:2261–2322. doi: 10.1002/9781119143802.ch103. [DOI] [Google Scholar]
- Farooq S, Ngaini Z. Natural and synthetic drugs as potential treatment for coronavirus disease 2019 (COVID-2019) Chem Afr. 2021;4(1):1–13. doi: 10.1007/s42250-020-00203-x. [DOI] [Google Scholar]
- Fauci AS. HIV and AIDS: 20 years of science. Nat Med. 2003;9(7):839–843. doi: 10.1038/nm0703-839. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng T-S, Smith SA, Tan JL, Lim X-N, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349(6243):88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Te Velthuis AJ. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb Perspect Med. 2020;10(9):a038398. doi: 10.1101/cshperspect.a038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, Dilip de Silva E, Lassota P, Allen TM. Papuamides A−D, HIV-inhibitory and cytotoxic depsipeptides from the Sponges Theonella m irabilis and Theonella s winhoei collected in Papua New Guinea. J Am Chem Soc. 1999;121(25):5899–5909. doi: 10.1021/ja990582o. [DOI] [Google Scholar]
- Fuhrman CA, Warren AD, Waring AJ, Dutz SM, Sharma S, Lehrer RI, Cole AL, Cole AM. Retrocyclin RC-101 overcomes cationic mutations on the heptad repeat 2 region of HIV-1 gp41. FEBS J. 2007;274(24):6477–6487. doi: 10.1111/j.1742-4658.2007.06165.x. [DOI] [PubMed] [Google Scholar]
- Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides: an abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276(8):5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- Garabedian A, Bolufer A, Leng F, Fernandez-Lima F. Peptide sequence influence on the conformational dynamics and DNA binding of the intrinsically disordered AT-hook 3 peptide. Sci Rep. 2018;8(1):1–8. doi: 10.1038/s41598-018-28956-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163(6):2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach SL, Mondal D. Naturally occurring cyclic peptides and their potential application in HIV therapeutics. J Biol Active Prod Nat. 2012;2(1):1–29. [Google Scholar]
- Girard MP, Osmanov S, Assossou OM, Kieny M-P. Human immunodeficiency virus (HIV) immunopathogenesis and vaccine development: a review. Vaccine. 2011;29(37):6191–6218. doi: 10.1016/j.vaccine.2011.06.085. [DOI] [PubMed] [Google Scholar]
- Gongora-Benitez M, Tulla-Puche J, Albericio F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chem Rev. 2014;114(2):901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20(9):2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallock YF, Sowder RC, Pannell LK, Hughes CB, Johnson DG, Gulakowski R, Cardellina JH, Boyd MR. Cycloviolins A−D, anti-HIV macrocyclic peptides from Leonia c ymosa. J Org Chem. 2000;65(1):124–128. doi: 10.1021/jo990952r. [DOI] [PubMed] [Google Scholar]
- Hamamoto I, Harazaki K, Inase N, Takaku H, Tashiro M, Yamamoto N. Cyclosporin A inhibits the propagation of influenza virus by interfering with a late event in the virus life cycle. Jpn J Infect Dis. 2013;66(4):276–283. doi: 10.7883/yoken.66.276. [DOI] [PubMed] [Google Scholar]
- Hariton-Gazal E, Rosenbluh J, Zakai N, Fridkin G, Brack-Werner R, Wolff H, Devaux C, Gilon C, Loyter A. Functional analysis of backbone cyclic peptides bearing the arm domain of the HIV-1 Rev protein: characterization of the karyophilic properties and inhibition of Rev-induced gene expression. Biochemistry. 2005;44(34):11555–11566. doi: 10.1021/bi050752b. [DOI] [PubMed] [Google Scholar]
- Hayes HC, Luk LY, Tsai Y-H. Approaches for peptide and protein cyclisation. Org Biomol Chem. 2021;19(18):3983–4001. doi: 10.1039/D1OB00411E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Bao J, Zhang X-Y, Tu Z-C, Shi Y-M, Qi S-H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J Nat Prod. 2013;76(6):1182–1186. doi: 10.1021/np300897v. [DOI] [PubMed] [Google Scholar]
- Ho KL, Yusoff K, Seow HF, Tan WS. Selection of high affinity ligands to hepatitis B core antigen from a phage-displayed cyclic peptide library. J Med Virol. 2003;69(1):27–32. doi: 10.1002/jmv.10266. [DOI] [PubMed] [Google Scholar]
- Hoffmann AR, Guha S, Wu E, Ghimire J, Wang Y, He J, Garry RF, Wimley WC. Broad-spectrum antiviral entry inhibition by interfacially active peptides. J Virol. 2020;94(23):e01682. doi: 10.1128/JVI.01682-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne WS, Wiethoff CM, Cui C, Wilcoxen KM, Amorin M, Ghadiri MR, Nemerow GR. Antiviral cyclic d, l-α-peptides: targeting a general biochemical pathway in virus infections. Bioorg Med Chem. 2005;13(17):5145–5153. doi: 10.1016/j.bmc.2005.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim et Biophys Acta. 2008;1778(2):357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J. 2005;2(1):1–10. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung J-J, Hsieh M-T, Young M-J, Kao C-L, King C-C, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J Virol. 2004;78(1):378–388. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrus S, Tambunan USF, Zubaidi AA. Designing cyclopentapeptide inhibitor as potential antiviral drug for dengue virus ns5 methyltransferase. Bioinformation. 2012;8(8):348. doi: 10.6026/97320630008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwazaki T, Li X, Harada K. Evolvability of the mode of peptide binding by an RNA. RNA. 2005;11(9):1364–1373. doi: 10.1261/rna.2560905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob ST, Crozier I, Fischer WA, Hewlett A, Kraft CS, de La Vega M-A, Soka MJ, Wahl V, Griffiths A, Bollinger L. Ebola virus disease. Nat Rev Dis Primers. 2020;6(1):1–31. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadesh A, Jayaram A, Babu N, Mudgal PP, Sudandiradas R, Sheik S, Shetty U, Verma DK, Mahilkar S, Sunil S. Current status of chikungunya in India. Front Microbiol. 2021;12:1. doi: 10.3389/fmicb.2021.695173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec-Krzesniak K, Rajnisz-Mateusiak A, Guspiel A, Ziemska J, Solecka J. Secondary metabolites of actinomycetes and their antibacterial, antifungal and antiviral properties. Pol J Microbiol. 2018;67(3):259. doi: 10.21307/pjm-2018-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K. Developing cyclic peptide-based drug candidates: an overview. Future Med Chem. 2020;12(19):1687–1690. doi: 10.4155/fmc-2020-0171. [DOI] [PubMed] [Google Scholar]
- Jing X, Jin K. A gold mine for drug discovery: strategies to develop cyclic peptides into therapies. Med Res Rev. 2020;40(2):753–810. doi: 10.1002/med.21639. [DOI] [PubMed] [Google Scholar]
- Johansen-Leete J, Ullrich S, Fry S, Frkic R, Bedding M, Aggarwal A, Ashhurst A, Ekanayake K, Mahawaththa M, Sasi V. Antiviral cyclic peptides targeting the main protease of SARS-CoV-2. Chem Sci. 2022;13:3826. doi: 10.1039/D1SC06750H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo SH. Cyclic peptides as therapeutic agents and biochemical tools. Biomol Ther. 2012;20(1):19. doi: 10.4062/biomolther.2012.20.1.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WB, Kesteleyn B, Stoops B, Vreeken RJ, Vermond J, Goutier W. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358(6362):496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HK, Seo CH, Park Y. Marine peptides and their anti-infective activities. Mar Drugs. 2015;13(1):618–654. doi: 10.3390/md13010618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J Autoimmun. 2022;126:102779. doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsara M, Tselios T, Deraos S, Deraos G, Matsoukas M-T, Lazoura E, Matsoukas J, Apostolopoulos V. Round and round we go: cyclic peptides in disease. Curr Med Chem. 2006;13(19):2221–2232. doi: 10.2174/092986706777935113. [DOI] [PubMed] [Google Scholar]
- Khachatoorian R, Ruchala P, Waring A, Jung C-L, Ganapathy E, Wheatley N, Sundberg C, Arumugaswami V, Dasgupta A, French SW. Structural characterization of the HSP70 interaction domain of the hepatitis C viral protein NS5A. Virology. 2015;475:46–55. doi: 10.1016/j.virol.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings F, Perkins MD, Kuhn JH, Pallen MJ, Alm EJ, Archer BN, Barakat A, Bedford T, Bhiman JN, Caly L. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6:821. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- Kumar V. Influenza in children. The Indian J Pediatr. 2017;84(2):139–143. doi: 10.1007/s12098-016-2232-x. [DOI] [PubMed] [Google Scholar]
- Kumar PV, Abdelkarim Maki MA, Takahje ML, Wei YS, Tatt LM. Detection of formation of recombinant human keratinocyte growth factor loaded chitosan nanoparticles based on its optical properties. Curr Nanosci. 2018;14(2):127–135. doi: 10.2174/1573413713666171016150707. [DOI] [Google Scholar]
- Kumar PV, Maki MAA, Wei YS, Tatt LM, Elumalai M, Cheah S-C, Raghavan B, Majeed ABBA. Rabbit as an animal model for pharmacokinetics studies of enteric capsule contains recombinant human keratinocyte growth factor loaded chitosan nanoparticles. Curr Clin Pharmacol. 2019;14(2):132–140. doi: 10.2174/1574884714666181120103907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Shrivastava T, Samal S, Ahmed S, Parray HA. Antibody-based therapeutic interventions: possible strategy to counter chikungunya viral infection. Appl Microbiol Biotechnol. 2020;104(8):3209–3228. doi: 10.1007/s00253-020-10437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S., M. Nyo, P. Phuektes, C. W. Yew, Y. J. Tan and J. J. H. Chu (2015). A potent neutralizing IgM mAb targeting the N218 epitope on E2 protein protects against Chikungunya virus pathogenesis. MAbs, Taylor & Francis [DOI] [PMC free article] [PubMed]
- Lanini S, Ustianowski A, Pisapia R, Zumla A, Ippolito G. Viral hepatitis: etiology, epidemiology, transmission, diagnostics, treatment, and prevention. Infect Dis Clin. 2019;33(4):1045–1062. doi: 10.1016/j.idc.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Cole AM, Selsted ME. θ-Defensins: cyclic peptides with endless potential. J Biol Chem. 2012;287(32):27014–27019. doi: 10.1074/jbc.R112.346098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E, Delanoe-Ayari H, Melikov K, Cho M-S, Chen A, Waring AJ, Wang W, Xie Y, Loo JA, Lehrer RI. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nat Immunol. 2005;6(10):995–1001. doi: 10.1038/ni1248. [DOI] [PubMed] [Google Scholar]
- Liang X, Nong X-H, Huang Z-H, Qi S-H. Antifungal and antiviral cyclic peptides from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. J Agric Food Chem. 2017;65(25):5114–5121. doi: 10.1021/acs.jafc.7b01238. [DOI] [PubMed] [Google Scholar]
- Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286(8):6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Wang Q-Y, Noble CG, Chen Y-L, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D. Ten years of dengue drug discovery: progress and prospects. Antiviral Res. 2013;100(2):500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Lim SP, Noble CG, Shi P-Y. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Lin K-H, Ali A, Rusere L, Soumana DI, Kurt Yilmaz N, Schiffer CA. Dengue virus NS2B/NS3 protease inhibitors exploiting the prime side. J Virol. 2017;91(10):e00045. doi: 10.1128/JVI.00045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-W, Wang Y-J, Ko C-W, Cheng T-L, Wang Y-T. Cyclic peptide inhibitors of the Tsg101 UEV protein interactions refined through global docking and Gaussian accelerated molecular dynamics simulations. Polymers. 2020;12(10):2235. doi: 10.3390/polym12102235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liras S, Mcclure KF. Permeability of cyclic peptide macrocycles and cyclotides and their potential as therapeutics. ACS Med Chem Lett. 2019;10(7):1026–1032. doi: 10.1021/acsmedchemlett.9b00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Stephen AG, Fisher RJ, Burke TR., Jr Protected aminooxyprolines for expedited library synthesis: application to Tsg101-directed proline–oxime containing peptides. Bioorg Med Chem Lett. 2008;18(3):1096–1101. doi: 10.1016/j.bmcl.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-Z, Yang Y, Zhang S-X, Tang L, Wang H-M, Chen C-J, Shen Z-F, Cheng K-D, Kong J-Q, Wang W. A cyclotide against influenza A H1N1 virus from Viola yedoensis. Yao xue xue bao Acta Pharm Sinica. 2014;49(6):905–912. [PubMed] [Google Scholar]
- Lohmann V. Hepatitis C virus RNA replication. Hepat C Virus: Mol Virol Antiviral Ther. 2013 doi: 10.1007/978-3-642-27340-7_7. [DOI] [Google Scholar]
- Lu Z, Van Wagoner RM, Harper MK, Baker HL, Hooper JN, Bewley CA, Ireland CM. Mirabamides E−H, HIV-inhibitory depsipeptides from the sponge Stelletta clavosa. J Nat Prod. 2011;74(2):185–193. doi: 10.1021/np100613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Nong X-H, Ren Z, Wang J, Liang X, Wang L, Qi S-H. Antiviral peptides from marine gorgonian-derived fungus Aspergillus sp. SCSIO 41501. Tetrahedron Lett. 2017;58(12):1151–1155. doi: 10.1016/j.tetlet.2017.02.005. [DOI] [Google Scholar]
- Machała MK, Brydak LB. Various sides of influenza, part I–structure, replication, changeability of influenza viruses, clinical course of the disease, immunological response and laboratory diagnostics. Polski Merkuriusz Lekarski: Organ Polskiego Towarzystwa Lekarskiego. 2006;21(123):270–276. [PubMed] [Google Scholar]
- Madan V, Bartenschlager R. Structural and functional properties of the hepatitis C virus p7 viroporin. Viruses. 2015;7(8):4461–4481. doi: 10.3390/v7082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Rawat AS, Bhatt N, Chauhan MK. Structural modification of proteins and peptides. Indian J Pharm Educ Res. 2014;48(3):34–47. doi: 10.5530/ijper.48.3.6. [DOI] [Google Scholar]
- Mandal D, Shirazi AN, Parang K. Self-assembly of peptides to nanostructures. Org Biomol Chem. 2014;12(22):3544–3561. doi: 10.1039/C4OB00447G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna AK, Kumar A, Ray U, Das S, Basu G, Roy S. A cyclic peptide mimic of an RNA recognition motif of human La protein is a potent inhibitor of hepatitis C virus. Antiviral Res. 2013;97(3):223–226. doi: 10.1016/j.antiviral.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Mata ÉCGD, Mourão CBF, Rangel M, Schwartz EF. Antiviral activity of animal venom peptides and related compounds. J Venom Anim Toxins Incl Trop Dis. 2017 doi: 10.1186/s40409-016-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson P, Doak BC, Over B, Kihlberg J. Cell permeability beyond the rule of 5. Adv Drug Deliv Rev. 2016;101:42–61. doi: 10.1016/j.addr.2016.03.013. [DOI] [PubMed] [Google Scholar]
- Miller EH, Harrison JS, Radoshitzky SR, Higgins CD, Chi X, Dong L, Kuhn JH, Bavari S, Lai JR, Chandran K. Inhibition of Ebola virus entry by a C-peptide targeted to endosomes. J Biol Chem. 2011;286(18):15854–15861. doi: 10.1074/jbc.M110.207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Moghadamtousi SZ, Nikzad S, Kadir HA, Abubakar S, Zandi K. Potential antiviral agents from marine fungi: an overview. Mar Drugs. 2015;13(7):4520–4538. doi: 10.3390/md13074520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Hepat C Virus: Mol Virol Antiviral Ther. 2013 doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- Moreland NJ, Tay MY, Lim E, Paradkar PN, Doan DN, Yau YH, Geifman Shochat S, Vasudevan SG. High affinity human antibody fragments to dengue virus non-structural protein 3. PLoS Negl Trop Dis. 2010;4(11):e881. doi: 10.1371/journal.pntd.0000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DA, Depelsenaire AC, Young PR. Clinical and laboratory diagnosis of dengue virus infection. J Infect Dis. 2017;215:S89–S95. doi: 10.1093/infdis/jiw649. [DOI] [PubMed] [Google Scholar]
- Nameki D, Kodama E, Ikeuchi M, Mabuchi N, Otaka A, Tamamura H, Ohno M, Fujii N, Matsuoka M. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J Virol. 2005;79(2):764–770. doi: 10.1128/JVI.79.2.764-770.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo D-H, Vo T-S, Ngo D-N, Wijesekara I, Kim S-K. Biological activities and potential health benefits of bioactive peptides derived from marine organisms. Int J Biol Macromol. 2012;51(4):378–383. doi: 10.1016/j.ijbiomac.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Niepmann M. Hepatitis C virus RNA translation. Hepat C Virus: Mol Virol Antiviral Ther. 2013 doi: 10.1007/978-3-642-27340-7_6. [DOI] [Google Scholar]
- Noah E, Biron Z, Naider F, Arshava B, Anglister J. The membrane proximal external region of the HIV-1 envelope glycoprotein gp41 contributes to the stabilization of the six-helix bundle formed with a matching N′ peptide. Biochemistry. 2008;47(26):6782–6792. doi: 10.1021/bi7023139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Chen Y-L, Dong H, Gu F, Lim SP, Schul W, Wang Q-Y, Shi P-Y. Strategies for development of dengue virus inhibitors. Antiviral Res. 2010;85(3):450–462. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Noble CG, Seh CC, Chao AT, Shi PY. Ligand-bound structures of the dengue virus protease reveal the active conformation. J Virol. 2012;86(1):438–446. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman A, Franck C, Christie M, Hawkins PM, Patel K, Ashhurst AS, Aggarwal A, Low JK, Siddiquee R, Ashley CL. Discovery of cyclic peptide ligands to the SARS-CoV-2 spike protein using mRNA display. ACS Cent Sci. 2021;7(6):1001–1008. doi: 10.1021/acscentsci.0c01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northfield SE, Wang CK, Schroeder CI, Durek T, Kan M-W, Swedberg JE, Craik DJ. Disulfide-rich macrocyclic peptides as templates in drug design. Eur J Med Chem. 2014;77:248–257. doi: 10.1016/j.ejmech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- O’Donnell KL, Marzi A. Immunotherapeutics for ebola virus disease: hope on the horizon. Biol: Targets Ther. 2021;15:79. doi: 10.2147/BTT.S259069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen SM, Rudolph DL, Wang W, Cole AM, Waring AJ, Lal RB, Lehrer RI. RC-101, a retrocyclin-1 analogue with enhanced activity against primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 2004;20(11):1157–1165. doi: 10.1089/aid.2004.20.1157. [DOI] [PubMed] [Google Scholar]
- Owen DR, Allerton CM, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Pang W, Tam S-C, Zheng Y-T. Current peptide HIV type-1 fusion inhibitors. Antiviral Chem Chemother. 2009;20(1):1–18. doi: 10.3851/IMP1369. [DOI] [PubMed] [Google Scholar]
- Parachin NS, Franco OL. New edge of antibiotic development: antimicrobial peptides and corresponding resistance. Front Microbiol. 2014;5:147. doi: 10.3389/fmicb.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikesit A, Tambunan U. Screening of commercial cyclic peptides as inhibitor envelope protein dengue virus (DENV) through molecular docking and molecular dynamics. Pak J Biol Sci: PJBS. 2013;16(24):1836–1848. doi: 10.3923/pjbs.2013.1836.1848. [DOI] [PubMed] [Google Scholar]
- Parums DV. Revised world health organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med Sci Monitor: Int Med J Exp Clin Res. 2021;27:e933622. doi: 10.12659/MSM.933622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T, Katoh T, Goto Y, Suga H. Selection-based discovery of druglike macrocyclic peptides. Annu Rev Biochem. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Chari S, Cole AL, Cole AM. Retrocyclins and their activity against HIV-1. Cell Mol Life Sci. 2011;68(13):2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza A, Gustchina E, Baker HL, Kelly M, Bewley CA. Mirabamides A–D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J Nat Prod. 2007;70(11):1753–1760. doi: 10.1021/np070306k. [DOI] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21(10):2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regula LK, Harris R, Wang F, Higgins CD, Koellhoffer JF, Zhao Y, Chandran K, Gao J, Girvin ME, Lai JR. Conformational properties of peptides corresponding to the ebolavirus GP2 membrane-proximal external region in the presence of micelle-forming surfactants and lipids. Biochemistry. 2013;52(20):3393–3404. doi: 10.1021/bi400040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67(16):2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM. Dengue virus. Clin Lab Med. 2010;30(1):149–160. doi: 10.1016/j.cll.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar S, Kaur M, Minneman KP. Antiviral lead compounds from marine sponges. Mar Drugs. 2010;8(10):2619–2638. doi: 10.3390/md8102619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Itoh Y, Yasui F, Munakata T, Yamane D, Ozawa M, Ito R, Katoh T, Ishigaki H, Nakayama M. Macrocyclic peptides exhibit antiviral effects against influenza virus HA and prevent pneumonia in animal models. Nat Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-22964-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem HN, Batool F, Mansoor HJ, Shahzad-ul-Hussan S, Saeed M. Inhibition of dengue virus protease by eugeniin, isobiflorin, and biflorin isolated from the flower buds of Syzygium aromaticum (Cloves) ACS Omega. 2019;4(1):1525–1533. doi: 10.1021/acsomega.8b02861. [DOI] [Google Scholar]
- Sandman KE, Benner JS, Noren CJ. Phage display of selenopeptides. J Am Chem Soc. 2000;122(5):960–961. doi: 10.1021/ja992462m. [DOI] [Google Scholar]
- Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25(4):757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of flavivirus entry derived from the E protein stem. J Virol. 2010;84(24):12549–12554. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelamgari A, Maddukuri A, Berro R, de la Fuente C, Kehn K, Deng L, Dadgar S, Bottazzi ME, Ghedin E, Pumfery A. Role of viral regulatory and accessory proteins in HIV-1 replication. Front Biosci. 2004;9(9):2388–2413. doi: 10.2741/1403. [DOI] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MP, Greene WC. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 2002;4(1):67–73. doi: 10.1016/S1286-4579(01)01511-8. [DOI] [PubMed] [Google Scholar]
- Siang Ong Y, Gao L, Kalesh KA, Yu Z, Wang J, Liu C, Li Y, Sun H, Seong Lee S. Recent advances in synthesis and identification of cyclic peptides for bioapplications. Curr Top Med Chem. 2017;17(20):2302–2318. doi: 10.2174/1568026617666170224121658. [DOI] [PubMed] [Google Scholar]
- Simmonds P. The origin of hepatitis C virus. Hepat C Virus: Mol Virol Antiviral Ther. 2013 doi: 10.1007/978-3-642-27340-7_1. [DOI] [Google Scholar]
- Simmons G, Reeves JD, Hibbitts S, Stine JT, Gray PW, Proudfoot A, Clapham PR. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol Rev. 2000;177:112–126. doi: 10.1034/j.1600-065X.2000.17719.x. [DOI] [PubMed] [Google Scholar]
- Smagulova F, Maurel S, Morichaud Z, Devaux C, Mougel M, Houzet L. The highly structured encapsidation signal of MuLV RNA is involved in the nuclear export of its unspliced RNA. J Mol Biol. 2005;354(5):1118–1128. doi: 10.1016/j.jmb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Roux KL, Prevost M-C, Fsihi H. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007;3(6):e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BFDCD, Drexler JF, Lima RSD, Rosário MDOHVD, Netto EM. Theories about evolutionary origins of human hepatitis B virus in primates and humans. Braz J Infect Dis. 2014;18:535–543. doi: 10.1016/j.bjid.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchitra Rao A-CN, Stillwell Paul C. 27—Influenza kendig’s disorders of the respiratory tract in children. Amsterdam: Elsevier; 2019. pp. 460–465. [Google Scholar]
- Swaminathan S, Khanna N. Dengue: recent advances in biology and current status of translational research. Curr Mol Med. 2009;9(2):152–173. doi: 10.2174/156652409787581592. [DOI] [PubMed] [Google Scholar]
- Tahir ul Qamar M, Bari A, Adeel MM, Maryam A, Ashfaq UA, Du X, Muneer I, Ahmad HI, Wang J. Peptide vaccine against chikungunya virus: immuno-informatics combined with molecular docking approach. J Transl Med. 2018;16(1):1–14. doi: 10.1186/s12967-018-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Matsui K, Nobori H, Maeda H, Sato A, Kurosu T, Orba Y, Sawa H, Hattori K, Higashino K. Discovery of novel cyclic peptide inhibitors of dengue virus NS2B-NS3 protease with antiviral activity. Bioorg Med Chem Lett. 2017;27(15):3586–3590. doi: 10.1016/j.bmcl.2017.05.027. [DOI] [PubMed] [Google Scholar]
- Tambunan USF, Alkaff AH, Nasution MAF, Parikesit AA, Kerami D. Screening of commercial cyclic peptide conjugated to HIV-1 Tat peptide as inhibitor of N-terminal heptad repeat glycoprotein-2 ectodomain Ebola virus through in silico analysis. J Mol Graph Model. 2017;74:366–378. doi: 10.1016/j.jmgm.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Tapeinou A, Matsoukas MT, Simal C, Tselios T. Review cyclic peptides on a merry-go-round; towards drug design. Pept Sci. 2015;104(5):453–461. doi: 10.1002/bip.22669. [DOI] [PubMed] [Google Scholar]
- Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag−TSG101 interaction. ACS Chem Biol. 2008;3(12):757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- Tee YN, Kumar PV, Maki MA, Elumalai M, Rahman SA, Cheah S-C. Mucoadhesive low molecular chitosan complexes to protect rHuKGF from proteolysis: in-vitro characterization and FHs 74 int cell proliferation studies. Curr Pharm Biotechnol. 2021;22(7):969–982. doi: 10.2174/1389201021666201218124450. [DOI] [PubMed] [Google Scholar]
- Thapa P, Espiritu MJ, Cabalteja C, Bingham J-P. The emergence of cyclic peptides: The potential of bioengineered peptide drugs. Int J Pept Res Ther. 2014;20(4):545–551. doi: 10.1007/s10989-014-9421-0. [DOI] [Google Scholar]
- Thorstholm L, Craik D. Discovery and applications of naturally occurring cyclic peptides. Drug Discov Today Technol. 2012;9(1):e13–e21. doi: 10.1016/j.ddtec.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Toelzer C, Gupta K, Yadav SK, Borucu U, Davidson AD, Williamson MK, Shoemark DK, Garzoni F, Staufer O, Milligan R. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370(6517):725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman N, Cole AL, Ruchala P, Waring AJ, Lehrer RI, Stuchlik O, Pohl J, Cole AM. Reawakening retrocyclins: ancestral human defensins active against HIV-1. PLoS Biol. 2009;7(4):e1000095. doi: 10.1371/journal.pbio.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AA, Yin Y, Suga H. Macrocyclic peptides as drug candidates: recent progress and remaining challenges. J Am Chem Soc. 2019;141(10):4167–4181. doi: 10.1021/jacs.8b13178. [DOI] [PubMed] [Google Scholar]
- Vitali F, Fasan R. Biological and hybrid biological/chemical strategies in diversity generation of peptidic macrocycles. Pract Med Chem Macrocycles: Des Synth Case Stud. 2017 doi: 10.1002/9781119092599.ch7. [DOI] [Google Scholar]
- Volk DE, Lee Y-C, Li X, Thiviyanathan V, Gromowski GD, Li L, Lamb AR, Beasley DW, Barrett AD, Gorenstein DG. Solution structure of the envelope protein domain III of dengue-4 virus. Virology. 2007;364(1):147–154. doi: 10.1016/j.virol.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JE, Vaney M-C, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468(7324):709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Wang Y-T, Cheng T-L. Computational modeling of cyclic peptide inhibitor–MDM2/MDMX binding through global docking and Gaussian accelerated molecular dynamics simulations. J Biomol Struct Dyn. 2021;39(11):4005–4014. doi: 10.1080/07391102.2020.1773317. [DOI] [PubMed] [Google Scholar]
- Wang CK, Craik DJ. Cyclic peptide oral bioavailability: lessons from the past. Pept Sci. 2016;106(6):901–909. doi: 10.1002/bip.22878. [DOI] [PubMed] [Google Scholar]
- Wang C-R, Tsai H-W. Human hepatitis viruses-associated cutaneous and systemic vasculitis. World J Gastroenterol. 2021;27(1):19. doi: 10.3748/wjg.v27.i1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Owen SM, Rudolph DL, Cole AM, Hong T, Waring AJ, Lal RB, Lehrer RI. Activity of α-and θ-defensins against primary isolates of HIV-1. J Immunol. 2004;173(1):515–520. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- Wang CK, Colgrave ML, Gustafson KR, Ireland DC, Goransson U, Craik DJ. Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J Nat Prod. 2008;71(1):47–52. doi: 10.1021/np070393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Suhrbier A, Penn-Nicholson A, Woraratanadharm J, Gardner J, Luo M, Le TT, Anraku I, Sakalian M, Einfeld D. A complex adenovirus vaccine against chikungunya virus provides complete protection against viraemia and arthritis. Vaccine. 2011;29(15):2803–2809. doi: 10.1016/j.vaccine.2011.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jia J, Wang L, Li F, Wang Y, Jiang Y, Song X, Qin S, Zheng K, Ye J. Anti-HSV-1 activity of Aspergillipeptide D, a cyclic pentapeptide isolated from fungus Aspergillus sp. SCSIO 41501. Virol J. 2020;17(1):1–9. doi: 10.1186/s12985-020-01315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani SS, Dar PA, Zargar SM, Dar TA. Therapeutic potential of medicinal plant proteins: present status and future perspectives. Curr Protein Pept Sci. 2020;21(5):443–487. doi: 10.2174/1389203720666191119095624. [DOI] [PubMed] [Google Scholar]
- Weger-Lucarelli J, Aliota MT, Wlodarchak N, Kamlangdee A, Swanson R, Osorio JE. Dissecting the role of E2 protein domains in alphavirus pathogenicity. J Virol. 2015;90(5):2418–2433. doi: 10.1128/JVI.02792-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J Biol Chem. 2008;283(5):2793–2803. doi: 10.1074/jbc.M708419200. [DOI] [PubMed] [Google Scholar]
- White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat Chem. 2011;3(7):509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- WHO, W. H. O. (2021, 13 December 2021). Tracking SARS-CoV-2 variants. from https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- WHO. (2020). WHO COVID-19 Dashboard. Retrieved 13 October 2022, from https://covid19.who.int/
- Woo, H. G. and M. Shah (2021). Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. bioRxiv [DOI] [PMC free article] [PubMed]
- Xiong W, Zhou C, Yin S, Chai J, Zeng B, Wu J, Li Y, Li L, Xu X. Fejerlectin, a lectin-like peptide from the skin of Fejervarya limnocharis, inhibits HIV-1 entry by targeting Gp41. ACS Omega. 2021;6(9):6414–6423. doi: 10.1021/acsomega.1c00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KS, Ma XR, Ma Y, Alugubelli YR, Scott DA, Vatansever EC, Drelich AK, Sankaran B, Geng ZZ, Blankenship LR. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem. 2021;16(6):942–948. doi: 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring AJ, Herold BC, Wagar EA, Lehrer RI. θ defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J Virol. 2004;78(10):5147–5156. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lin S, Zhang B, Jin H, Ding L. Antiviral potential of natural products from marine microbes. Eur J Med Chem. 2020;207:112790. doi: 10.1016/j.ejmech.2020.112790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108(27):11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chen S. Cyclic peptide drugs approved in the last two decades (2001–2021) RSC Chem Biol. 2021;3:18–31. doi: 10.1039/D1CB00154J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Krumberger M, Morris MA, Parrocha CMT, Kreutzer AG, Nowick JS. Structure-based drug design of an inhibitor of the SARS-CoV-2 (COVID-19) main protease using free software: a tutorial for students and scientists. Eur J Med Chem. 2021;218:113390. doi: 10.1016/j.ejmech.2021.113390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Zhang H-P, Yu H, Li H, Ling Y-Q, Hu X-Q, Zhu H-G. Hepatitis B virus mutations associated with in situ expression of hepatitis B core antigen, viral load and prognosis in chronic hepatitis B patients. Pathol Res Pract. 2008;204(10):731–742. doi: 10.1016/j.prp.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Zorzi A, Deyle K, Heinis C. Cyclic peptide therapeutics: past, present and future. Curr Opin Chem Biol. 2017;38:24–29. doi: 10.1016/j.cbpa.2017.02.006. [DOI] [PubMed] [Google Scholar]