Summary
Background
Previous work on predicting type 2 diabetes by integrating clinical and genetic factors has mostly focused on the Western population. In this study, we use genome-wide polygenic risk score (gPRS) and serum metabolite data for type 2 diabetes risk prediction in the Asian population.
Methods
Data of 1425 participants from the Korean Genome and Epidemiology Study (KoGES) Ansan-Ansung cohort were used in this study. For gPRS analysis, genotypic and clinical information from KoGES health examinee (n = 58,701) and KoGES cardiovascular disease association (n = 8105) sub-cohorts were included. Linkage disequilibrium analysis identified 239,062 genetic variants that were used to determine the gPRS, while the metabolites were selected using the Boruta algorithm. We used bootstrapped cross-validation to evaluate logistic regression and random forest (RF)-based machine learning models. Finally, associations of gPRS and selected metabolites with the values of homeostatic model assessment of beta-cell function (HOMA-B) and insulin resistance (HOMA-IR) were further estimated.
Findings
During the follow-up period (8.3 ± 2.8 years), 331 participants (23.2%) were diagnosed with type 2 diabetes. The areas under the curves of the RF-based models were 0.844, 0.876, and 0.883 for the model using only demographic and clinical factors, model including the gPRS, and model with both gPRS and metabolites, respectively. Incorporation of additional parameters in the latter two models improved the classification by 11.7% and 4.2% respectively. While gPRS was significantly associated with HOMA-B value, most metabolites had a significant association with HOMA-IR value.
Interpretation
Incorporating both gPRS and metabolite data led to enhanced type 2 diabetes risk prediction by capturing distinct etiologies of type 2 diabetes development. An RF-based model using clinical factors, gPRS, and metabolites predicted type 2 diabetes risk more accurately than the logistic regression-based model.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2019M3E5D1A02070863 and 2022R1C1C1005458). This work was also supported by the 2020 Research Fund (1.200098.01) of UNIST (Ulsan National Institute of Science & Technology)
Keywords: Type 2 diabetes, Genome-wide polygenic risk score, Machine learning, Serum metabolites, KoGES, East Asian
Abbreviations: gPRS, genome-wide polygenic risk score; KoGES, Korean genome and epidemiology study; RF, random forest; LR, logistic regression; HOMA-B, homeostatic model assessment of beta-cell function; HOMA-IR, homeostatic model assessment of insulin resistance; BCAA, branched-chain amino acids; FOS, Framingham offspring study; GLU0, fasting glucose; HbA1c, glycated hemoglobin; BMI, body mass index; HTN, hypertension; TAC, total alcohol consumption; HDL, high-density lipoprotein; TG, triacylglycerol; TC, total cholesterol; HEXA, KoGES health examinee study; CAVAS, KoGES cardiovascular disease association study; GWAS, genome-wide association study; OR, odds ratio; LD, linkage disequilibrium; SNP, single nucleotide polymorphism; LoD, limit of detection; MDI, mean decrease impurity; AUC, area under receiver operating characteristic curve
Research in context.
Evidence before this study
Polygenic risk score (PRS) can boost up the predictive performance of type 2 diabetes risk in European ancestries (Framingham Offspring, and Finnish studies). In addition, incorporating metabolites can enhance the predictive ability of incident type 2 diabetes. However, no studies are conducted on combining both information for predicting type 2 diabetes risk in non-white ethnicity.
Added value of this study
In the current study, we proposed genome-wide polygenic risk score (gPRS), which has added value in discriminating the risk of type 2 diabetes in the Asian population as a genetic factor beyond clinical information. We also found that serum metabolites can also modestly improve the risk prediction accuracy for type 2 diabetes on top of clinical and genetic factors. We compared a RF-based machine learning model to a widely-used logistic regression model and the machine learning model is more effective in terms of gain in predictive powers such as discrimination performances and reclassification improvements and also comparable in terms of the interpretability.
Implications of all the available evidence
Our findings support that exploiting machine learning algorithms with gPRS derived from non-European ethnicity can enhance the predictive performance of type 2 diabetes risk. In addition, it can be implied that serum metabolites can also add values to capture information helpful for predicting incident type 2 diabetes risk combined with genetic factors in complementary manners.
Introduction
Type 2 diabetes is a chronic disease characterized by insulin resistance and beta-cell dysfunction1 that contributes to the development of cardiovascular disease and increased mortality.2 Globally, 416 million people were estimated to be affected by type 2 diabetes in 2019, and the number is expected to reach 630 million by 2040.3 Owing to the large-scale impact of type 2 diabetes on public health, several reliable risk prediction models have been developed for early screening and identification of individuals at high risk for type 2 diabetes using medical history, anthropometric measurements, and laboratory data.4, 5, 6 Such predictive models facilitate timely adoption of appropriate preventive measures such as incorporating lifestyle modifications or strategies to defer the onset of type 2 diabetes with medications such as metformin. However, the conventional type 2 diabetes risk models do not account for genetic predisposition or subclinical metabolic changes that precede metabolic impairments; hence, integrative approaches using polygenic risk score (PRS) or serum metabolites have been proposed to predict the risk for type 2 diabetes. Therefore, a new risk-prediction model using PRS and serum metabolites could improve the identification of high-risk individuals and reduce the burden of type 2 diabetes.7,8
PRS, generated from genetic variant data associated with type 2 diabetes risk, not only predicts the future risk for type 2 diabetes, but also improves the conventional type 2 diabetes risk models. The Framingham Offspring and Finnish studies reported that incorporating PRS, calculated from dozens to hundreds of genetic variants with genome-wide significance level, to a clinical type 2 diabetes risk prediction model modestly improved its performance9,10; however, this combination of PRS with clinical data did not enhance the performance of models built based on data from Asian populations.11, 12, 13, 14 Recently, genome-wide PRS (gPRS), which includes genetic variants with a lower threshold than the genome-wide association signal proposed by Purcell,15 has gained popularity in predicting type 2 diabetes risk due to its enhanced performance compared to that of PRS.
Data of several metabolites, including branched-chain amino acids (BCAA),16, 17, 18 aromatic amino acids,18,19 glutamine-to-glutamate ratio,20 and lipid species such as acylcarnitines21 and phosphatidylcholines,22, 23, 24 are also associated with type 2 diabetes risk-prediction along with gPRS and clinical factors. The incorporation of metabolite data as attributes improved type 2 diabetes risk prediction models in previous studies.16, 17, 18, 19,21 The utility of incorporating genomic or metabolomic data for type 2 diabetes risk prediction has been demonstrated.25 Further, the Framingham Offspring Study (FOS) reported slightly enhanced predictive performance when both types of information were used concurrently, thus capturing the information on insulin secretion and resistance in a complementary manner.25 The theoretical basis of this pioneering study stemmed from the hypothesis that genetic information is primarily associated with beta-cell function,25,26 whereas metabolite measurements were more closely related to insulin resistance.18,20 However, due to the limited availability of cohort studies including genetic and metabolite data, the cumulative effect of genomic and metabolomic information with clinical variables in type 2 diabetes risk prediction has not been validated. Therefore, our study tries to validate the theoretical hypothesis presented by the Caucasian ancestry in the Asian population by using enriched data containing both genetic and metabolite components collected from a considerable Korean population over a long time period. Furthermore, to confirm the predictive ability of gPRS rigorously, we designed gPRS from another representative cohort. Finally, we estimated the predictive ability of gPRS and metabolites on top of clinical factors for discriminating incident type 2 diabetes. A machine learning technique is adopted to enhance the predictive performance of risk prediction models and we compared their performances with that of the existing statistical model.
Methods
Study design and participants
The Ansan and Ansung study of the Korean Genome and Epidemiology Study (KoGES) is a population-based prospective cohort study that included urban (Ansan) and rural (Ansung) Korean participants aged between 40 and 69 years.23 The study recorded demographic variables, health status, medical history, and information on biochemical variables, genotype, and metabolites of the participants. The baseline survey was conducted in 2001–2002, and follow-up surveys were conducted biennially for 14 years. However, as metabolites were measured only from 2005 to 2006, we filtered the data to include only 7515 individuals who participated in the KoGES from 2005 to 2006 as the baseline data; the follow-up was conducted until 2015–2016. Participants with self-reported type 2 diabetes, on type 2 diabetes medications, or meeting the American Diabetes Association diagnostic criteria (fasting glucose (GLU0) ≥7.0 mmol/L, 2-h glucose ≥11.1 mmol/L, or glycated hemoglobin (HbA1c) ≥48 mmol/mol (6.5%))27 were defined as patients with type 2 diabetes. Among these participants, we selected 1905 participants with information on type 2 diabetes diagnosis, serum glucose, HbA1c, genotype, and metabolites for baseline examinations. We excluded 480 participants for the following reasons: diagnosed with type 2 diabetes at baseline (n = 415) and missing information on serum glucose, HbA1c, or other covariates (n = 65). Eventually, 1425 participants were included in the analysis. The flowchart for type 2 diabetes cohort selection is illustrated in Supplementary Fig. S1.
Data statement
Due to the regulations on exploiting private information of participants of KoGES study, the data used in this study is only accessible with consent from Korea Biobank.
Measurements
We selected 12 significant risk factors (P < 0.10) that had been included in type 2 diabetes risk prediction models in previous studies for the present correlation or prediction analysis of type 2 diabetes risk: age,6,9, 10, 11, 12, 13,25 body mass index (BMI),6,9, 10, 11, 12, 13,25 sex,6,9, 10, 11, 12, 13,25 hypertension (HTN),6,9, 10, 11, 12,25 family history of type 2 diabetes,6,9, 10, 11, 12,25 smoking status,6,9, 10, 11, 12, 13, 14, 15,25 total alcohol consumption (TAC),13,25 and biochemical variables such as HbA1c, GLU0, high-density lipoprotein (HDL) cholesterol, triacylglycerol (TG), and total cholesterol (TC).6,9, 10, 11, 12, 13, 14, 15,25 Individuals with systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg, or currently on HTN medication were defined as having HTN. A family history of type 2 diabetes was defined as having a first-degree relative who had been previously diagnosed with type 2 diabetes. Individuals based on smoking status were classified into three groups: never-, former-, and current-smoker. TAC was calculated by multiplying alcohol consumption frequency with amounts consumed per occasion.
Construction of ethnic-specific gPRS
We combined two population-based prospective cohorts, the KoGES health examinee (HEXA) study and the KoGES cardiovascular disease association study (CAVAS), consisting of South Korean adults aged ≥40 years at baseline to construct the ethnic-specific gPRS.23 KoGES HEXA study and KoGES CAVAS started in 2001 and 2004, comprising 58,701 and 8105 individuals with genotypic and clinical information, respectively. Among the 66,806 participants, we excluded 13 participants due to missing information on type 2 diabetes diagnosis, GLU0, HbA1c, or other covariates. Therefore, 66,793 participants were included in the analysis.
Genotyping was performed using the Korea Biobank Array (KoreanChip). The array comprised ≥833,300 markers, including ≥247,000 rare-frequency or functional variant markers.28 We phased the genotype data using ShapeITv2 and imputed phased genotype data using IMPUTEv2 with the 1000 Genomes Project Phase 3 East Asians (1KG EAS) data as a reference panel.29 SNPs were excluded based on the following criteria: call rates <95%, minor allele frequency <1%, and imputation quality score <0.8. In addition, we excluded SNPs that were below the P-value threshold (P < 1 × 10−6) from the Hardy–Weinberg equilibrium in the data. In total, 7,104,359 variants passed the quality criteria and were used in the analysis. Genome-wide type 2 diabetes-susceptible loci were identified using logistic regression (LR) analysis assuming the additive mode of inheritance in both KoGES HEXA and KoGES CAVAS studies. Age, sex, area of residence, and BMI were adjusted as covariates for the analysis. Based on the Bonferroni correction, the significance threshold of genome-wide association with type 2 diabetes was defined as P < 5 × 10−8, and the threshold of false-positive error was set at 0.05. The effect sizes and standard errors calculated by LR of both genome-wide association studies (GWASs) were combined using the fixed-effect inverse-variance weighted average method in a meta-analysis using PLINK (https://www.cog-genomics.org/plink/) (Supplementary Fig. S2). The results of genome-wide meta-analysis of type 2 diabetes is presented in Supplementary Fig. S3.
The risk alleles weighted by taking logarithm of the odds ratio (OR) from the LR analysis were added to calculate gPRS. Linkage disequilibrium (LD)-based clumping was used to obtain the strongest signals and to prune out the weaker ones in the LD block based on the following criteria: GWAS meta-analysis P-value threshold for single nucleotide polymorphism (SNP) is 0.2, LD threshold for clumping (r2) is 0.1, and the genomic distance for clumping is 250 kilobase (kb). Finally, a total of 239,062 SNPs were scored to construct gPRS. The circus plot to estimate the density of tagged SNPs around the known T2D-related genes was generated using R package (version 4.1.0.). We investigated the effects of gPRS on type 2 diabetes prediction by analyzing the gPRS distribution of the study participants and the change in the incidence ratio of type 2 diabetes.
Serum metabolite measurements and selection of variables
We measured 186 serum metabolites (40 acylcarnitines, 21 amino acids, 19 biogenic amines, 1 hexose, 90 glycerophospholipids, and 15 sphingolipids) using the AbsoluteIDQ® p180 kit. To ensure accurate measurement of the metabolites, the following quality control criteria were adopted: (a) variation coefficient for each metabolite in the reference range <25%, (b) half of the analyzed metabolite levels in the reference range >limit of detection (LoD), and (c) half of the analyzed metabolite levels in the experimental samples >LoD.22 Finally, 135 metabolites (13 acylcarnitines, 21 amino acids, 10 biogenic amines, 1 hexose, 78 glycerophospholipids, and 12 sphingolipids) were used to predict type 2 diabetes risk.
Considering high dimensionality of metabolites, Boruta algorithm,30 where the mean decrease impurity (MDI) of each variable is iteratively estimated and a subset of variables is selected based on the statistically significant MDI values, was used to select essential variables, and elicit improved prediction in the models. The Boruta selection method was incorporated in the 100-times repeated 10-fold cross-validation (CV) to reduce possible bias when estimating variable importance.31 Thus, 15 serum metabolites (spermine, hexose, isoleucine, valine, phosphatidylcholine acyl-alkyl (PC ae) C34:3, PC ae C36:3, PC ae C42:0, PC ae C42:1, PC ae C42:4, PC ae C44:6, PC aa C40:5, lysoPC a C18:2, glycine, alanine, and leucine) were consistently selected as accepted or tentative variables by the Boruta algorithm.32 The importance of the corresponding metabolite variable extracted from the variable selection procedure is depicted in Supplementary Fig. S4. Meanwhile, we conducted additional sensitivity analysis by adding tyrosine, phenylalanine, and glutamine to the metabolite list. These metabolites were selected based on a previous meta-analysis of prospective studies33 and used to evaluate if their inclusion enhanced prediction of type 2 diabetes.
Statistical and machine learning analyses
Baseline characteristics of patients with type 2 diabetes were compared with those of patients with non-type 2 diabetes using the Mann–Whitney U-test for quantitative variables and the χ2-test for qualitative variables. We also looked for any differences in the characteristics of the uncensored and censored participants. During pre-processing before model building, all qualitative variables (gender, HTN, family history, smoking status) were one-hot encoded, and remaining numerical variables were standardized using mean and standard deviation values calculated from training samples to be zero-centered with standard deviation 1 (i.e., z-scoring).
We applied LR with four different sets of independent variables for predicting incident type 2 diabetes, the dependent variable. Based on the risk factors involved, the four primary type 2 diabetes risk prediction models were denoted as model 1, 2, 3, and 4. Model 1 contained demographic variables and medical history such as sex, age, HTN, BMI, family history of type 2 diabetes, smoking, and TAC. Model 2 additionally included clinical variables: HbA1c, GLU0, TC, HDL, and TG. Model 3 consisted of demographic, clinical, and genetic variables, plus gPRS. Lastly, model 4 incorporated the data of selected metabolites in addition to the variables in model 3. To learn more complex, non-linear patterns inherent in the data, we conducted an additional analysis using a machine learning algorithm, RF. RF is a prediction algorithm based on aggregation of a set of multiple decision trees generated by bootstrap sampling.34 Note that all the listed variables were input equally to both LR and RF for a fair comparison. To accurately measure the bias and variance of each model, we conducted an internal validation via 10-fold CV with 100 bootstrap replicates.35 The tree-structured Parzen estimator-based Bayesian optimization technique was adopted for finding optimal hyperparameters for each type of LR and RF model using nested 10-fold stratified cross-validation scheme.36,37 Resulting hyperparameters were found to maximize the area under receiver operating characteristic curve (AUC). We used three metrics to estimate risk prediction performance of the models after the inclusion of gPRS and metabolites: AUC,38 Brier score, and log-loss. Model reclassification performance for estimating the improvement between the prediction models was also evaluated using net reclassification improvement (NRI), category-free NRI (cNRI), and integrated discrimination improvement (IDI).39,40 Unlike other model metrics that are interpreted in terms of probability, cNRI is used to define a model as weak (0.2), intermediate (0.4), or strong (0.8 or more).41 For additional estimation of the model consistency in measuring type 2 diabetes risk, calibration curve, net benefit-based decision curve, receiver operating characteristic curve, and precision–recall curve analyses were performed for all models.42
To evaluate the hypothesis that genetic information is closely associated with beta-cell function and that metabolic profiles are associated with insulin resistance, LR was used to test the association of gPRS and metabolites with homeostatic model assessment of beta-cell function (HOMA-B) and homeostatic model assessment of insulin resistance (HOMA-IR) values. HOMA-B and HOMA-IR values were calculated as follows: HOMA-B = 20 × fasting plasma insulin (FPI) (pmol/l)/[GLU0 (mmol/l)-3.5] and HOMA-IR = [FPI(pmol/l) × GLU0 (mmol/l)]/22.5. All analyses were performed using Python 3.6. Supplementary Figs. S5 and S6 demonstrate the overall workflow of type 2 diabetes risk prediction model building and its nested cross-validation scheme with feature importance calculation. Code is publicly available at https://github.com/vaseline555/T2D-Predictive-Modeling-for-Korean-with-gPRS-and-Metabolites.
Ethics
The present study was approved by the Institutional Review Board of Kosin University Gospel Hospital (IRB File No. KUGH 2019-08-042), and written informed consent was provided by all participants.
Role of funders
The funders of this study had no role in study design, data collection, data analyses, interpretation, and writing of the report and the submission of this study for publication.
Results
Baseline characteristics
Out of the 1425 participants followed up for 8.3 ± 2.8 years, 331 participants (23.2%) were eventually diagnosed with type 2 diabetes. Participants diagnosed with type 2 diabetes were likely to be older (P = 0.01, χ2-test), obese (P < 0.001, Mann–Whitney U-test), smokers (P = 0.002, χ2-test), having HTN (P < 0.001, χ2-test), and with a family history of type 2 diabetes (P = 0.02, χ2-test), compared to non-type 2 diabetes participants. Moreover, the type 2 diabetes participants consumed more alcohol (P = 0.002, Mann–Whitney U-test), had higher TG levels (P < 0.001, Mann–Whitney U-test), and lower HDL levels (P < 0.001, Mann–Whitney U-test) compared to non-type 2 diabetes individuals (Table 1). Additionally, there was no significant difference in the basic characteristics between the censored and uncensored participants (Supplementary Tables S1 and S2).
Table 1.
Baseline characteristics of participants for type 2 diabetes prediction analyses.
| Variable | Non-type 2 diabetes | Type 2 diabetes | P |
|---|---|---|---|
| Incident type 2 diabetes, n (%) | 1094 (76.8%) | 331 (23.2%) | |
| Gender (Female, n (%)) | 613 (56.0%) | 168 (50.8%) | 0.040 |
| Age (years) | 55.6 ± 8.9 | 57.0 ± 8.5 | 0.010 |
| HTN, n (%) | 273 (25.0%) | 112 (33.8%) | <0.001 |
| BMI (kg/m2) | 24.0 ± 3.1 | 25.2 ± 3.2 | <0.001 |
| Family history, n (%) | 22 (2%) | 8 (2.4%) | 0.020 |
| Smoking, n (%) | 0.002 | ||
| Never | 714 (65.3%) | 197 (59.5%) | |
| Ever | 185 (16.9%) | 64 (19.3%) | |
| Current | 195 (17.8%) | 70 (21.1%) | |
| TAC (g/week) | 8.0 ± 18.0 | 11.8 ± 25.4 | 0.002 |
| HbA1c (mmol/mol) | 36.0 ± 4.0 | 40.0 ± 4.0 | <0.001 |
| HbA1c (%) | 5.4 ± 0.3 | 5.8 ± 0.3 | <0.001 |
| GLU0 (mmol/L) | 4.9 ± 0.5 | 5.4 ± 0.6 | <0001 |
| TC (mmol/L) | 5.0 ± 0.9 | 5.1 ± 1.0 | 0.031 |
| HDL-cholesterol (mmol/L) | 1.2 ± 0.3 | 1.1 ± 0.2 | <0.001 |
| TG (mmol/L) | 1.4 ± 1.1 | 2.0 ± 1.8 | <0.001 |
| gPRS | 0.002845 ± 0.00003 | 0.002877 ± 0.00003 | <0.001 |
BMI, body mass index; GLU0, fasting glucose; HTN, hypertension; TAC, total alcohol consumption; TC, total cholesterol; TG, triglyceride.
Data are represented as mean ± SD for quantitative traits and n (%) (the number of participants) for qualitative traits. P represents the P-value at a significance level within the 5% (Mann–Whitney U test for each quantitative variable and the χ2-test for each qualitative variable between samples with and without type 2 diabetes).
Density of tagged SNPs around type 2 diabetes-associated genes and distribution of gPRS in relation to type 2 diabetes incidence
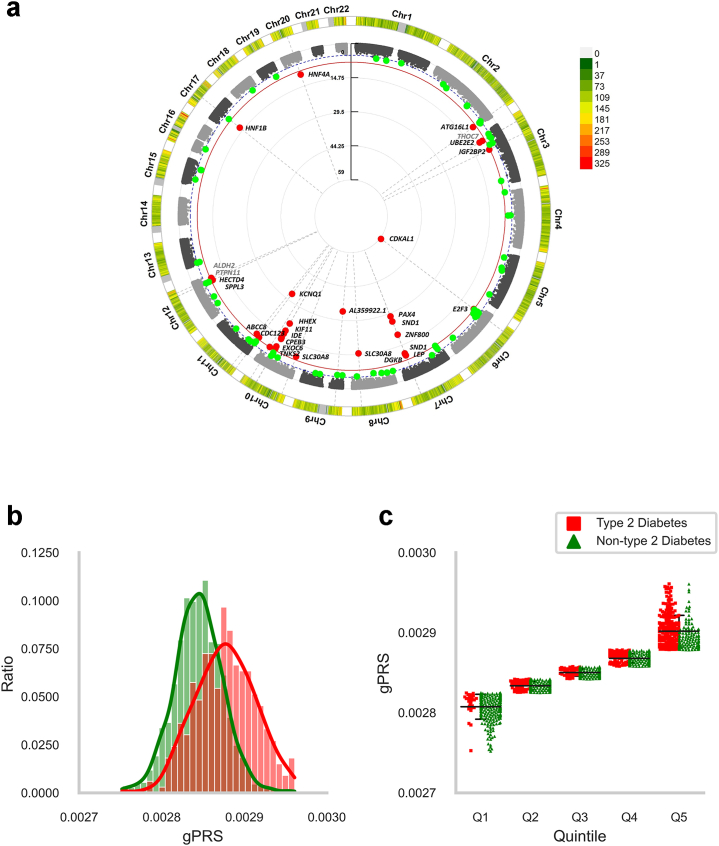
The density of tagged SNPs around type 2 diabetes-associated genes is presented using a circus plot (see Fig. 1a). As shown in Fig. 1a, a total of 34 tagged SNPs in 28 genes had genome-wide association with type 2 diabetes. Among these genes, 24 have been previously reported to be associated with type 2 diabetes (CDKAL1, AL359922.1, KCNQ1, PAX4, SND1, HHEX, ZNF800, KIF11, IDE, SLC30A8, CPEB3, EXOC6, TNKS2, HNF1B, UBE2E2, LEP, HNF4A, ABCC8, CDC123, ATG16L1, HECTD4, SPPL3, IGF2BP2, and DGKB), and the remaining genes were reported to be associated with waist-hip ratio (PTPN11 and ALDH2), BMI (E2F3), or neutrophil count (THOC7) (Supplementary Data 1).
Fig. 1.
(a) Circus plot represents the density of tagged SNPs around type 2 diabetes-associated genes. The outer track shows density of tagged SNPs for gPRS, and the inner track shows P-values of corresponding SNPs. The red line indicates genome-wide significance (P = 5.0 × 10–8, P-value was calculated using logistic regression to fit an additive model, and fixed-effect inverse-variance weighted average method) Red dots represent type 2 diabetes associated SNPs with genome-wide significance and green dots represent suggestive SNPs. Genes associated with type 2 diabetes were presented as black letters, and the others were presented as grey letters. (b) Depicts a histogram with the data density for type 2 diabetes status based on the standardized gPRS value. (c) Describes individual data points of both type 2 diabetes and non-type 2 diabetes by gPRS quintile groups (Q1: gPRS quintile < 20%, Q2: 20% ≤ gPRS quintile < 40%, Q3: 40% ≤ gPRS quintile < 60%, Q4: 60% ≤ gPRS quintile < 80%, and Q5: 80% ≤ gPRS quintile) along with individual average (longest horizontal line) and one standard deviation above and below each average (shorter horizontal lines).
In addition, the gPRS distribution of participants and the associated gPRS of the quintile groups with the ratios of incident type 2 diabetes are shown in Fig. 1b and c. The distribution of gPRS in patients with type 2 diabetes was skewed to the left more than that of the non-type 2 diabetes participants, indicating that the gPRS of patients with type 2 diabetes was larger than that of the non-type 2 diabetes participants. Furthermore, the type 2 diabetes incidence ratio in the fifth quintile (high gPRS group) was significantly higher than that in the first quintile (low gPRS group) (55.8% vs 8.8%).
Performance of type 2 diabetes prediction model
The predictive performance of model 1 in terms of AUC was 0.608 (95% confidence interval [CI]: 0.601, 0.615) for LR and 0.613 (95% CI: 0.606, 0.620) for RF. The AUC values for both LR (0.835 [95% CI: 0.830, 0.840], P < 0.001, bootstrapped t-test) and RF (0.844 [95% CI: 0.838, 0.850], P < 0.001, bootstrapped t-test) increased as clinical information was added to model 2. When gPRS was added to the demographic and clinical variables in model 3, the performance of the model increased by up to 3.6% (AUC = LR: 0.871 [95% CI: 0.866, 0.879], P < 0.001, bootstrapped t-test; RF: 0.876 [95% CI: 0.871, 0.881], P < 0.001, bootstrapped t-test) compared to that of model 2. Furthermore, the AUC of model 4, which contained both gPRS and selected metabolite data, was higher than that of model 3 without metabolite data (AUC = LR: 0.875 [95% CI: 0.871, 0.879], P = 0.015, bootstrapped t-test; RF: 0.883 [95% CI: 0.879, 0.887], P = 0.005, bootstrapped t-test) (Table 2 and Supplementary Fig. S7). Other performance metrics (e.g., accuracy, precision, recall, and F1-score) and the adjusted classification threshold value are shown in Table 3. The figures of four curves (i.e., calibration, net benefit-based decision, receiver operating characteristic, and precision–recall curves) are presented to show the additional estimation of the model consistency in measuring type 2 diabetes risk for both LR and RF models (Supplementary Figs. S8–S16). Though the sample ratio of non-type 2 diabetes and type 2 diabetes is imbalanced as 3.3:1, the risk prediction ability of both LR and RF were not much affected by the imbalanced setting. It is based on the results in Table 2 and Supplementary Figs. S8–S16, which supports the fact that no additional add-ons need to be considered for the imbalance issue. Further analyses such as the sensitivity analysis which additionally included tyrosine, phenylalanine, and glutamine on top of selected metabolites variables (Supplementary Table S6), or the comparison experiments adopting other machine learning techniques such as extreme gradient boosting and multi-layer perceptron (Supplementary Table S7), did not substantially improve the predictive performance in this cohort data.
Table 2.
Risk prediction performance and reclassification results of logistic regression and random forest for type 2 diabetes prediction analysis.
| Method | Performance | Model 1 | Model 2 | Model 3 | Model 4 |
|---|---|---|---|---|---|
| LR | 1) Discrimination | ||||
| Brier score | 0.175 (0.006) | 0.131 (0.013) | 0.112 (0.018) | 0.110 (0.013) | |
| Log-loss | 0.531 (0.016) | 0.407 (0.043) | 0.360 (0.055) | 0.348 (0.036) | |
| AUC | 0.608 (0.043) | 0.835 (0.032) | 0.871 (0.028) | 0.875 (0.023) | |
| P for AUC | <0.001 (vs Model 1) | <0.001 (vs Model 2) | 0.015 (vs Model 3) | ||
| 2) Reclassification | vs Model 1 | vs Model 2 | vs Model 3 | ||
| NRI | 0.321 (0.092) | 0.035 (0.026) | 0.002 (0.004) | ||
| cNRI | 1.041 (0.161) | 0.361 (0.174) | 0.325 (0.205) | ||
| IDI | 0.263 (0.039) | 0.135 (0.059) | 0.005 (0.002) | ||
| RF | 1) Discrimination | ||||
| Brier score | 0.172 (0.007) | 0.130 (0.008) | 0.109 (0.011) | 0.106 (0.015) | |
| Log-loss | 0.555 (0.016) | 0.390 (0.032) | 0.353 (0.034) | 0.332 (0.027) | |
| AUC | 0.613 (0.041) | 0.844 (0.037) | 0.876 (0.029) | 0.883 (0.023) | |
| P for AUC | <0.001 (vs Model 1) | <0.001 (vs Model 2) | 0.005 (vs Model 3) | ||
| 2) Reclassification | vs Model 1 | vs Model 2 | vs Model 3 | ||
| NRI | 0.393 (0.011) | 0.117 (0.024) | 0.042 (0.008) | ||
| cNRI | 1.003 (0.144) | 0.511 (0.109) | 0.336 (0.188) | ||
| IDI | 0.266 (0.033) | 0.039 (0.032) | 0.022 (0.015) | ||
| LR vs RF | P for AUC | 0.038 | 0.003 | 0.012 | 0.006 |
AUC, area under curve; cNRI, category-free NRI; IDI, integrated discrimination improvement; LR, logistic regression; NRI, net reclassification index; RF, random forest.
Model 1 contained sex, age, hypertension, body mass index, family history, smoking, and total alcohol consumption; Model 2 additionally included HbA1c, GLU0, total cholesterol, HDL-cholesterol, and triglyceride; Model 3 additionally included gPRS; Model 4 additionally included 15 selected serum metabolites (P represents the P-value at a significance level within the bootstrapped t-test for AUC between prediction models).
Table 3.
Results on discrimination performance metrics and adjusted classification thresholds.
| Method | Model | Accuracy | Equilibrium Accuracy | Sensitivity | Specificity | PPV | NPV | F1 | Youden's J statistic |
|---|---|---|---|---|---|---|---|---|---|
| LR | Model 1 (Demo.) | 0.565 (0.004) | 0.583 (0.004) | 0.615 (0.006) | 0.55 (0.005) | 0.295 (0.003) | 0.824 (0.003) | 0.398 (0.004) | 0.226 (0.001) |
| Model 2 (Demo. + Clin.) | 0.779 (0.003) | 0.756 (0.003) | 0.713 (0.005) | 0.799 (0.004) | 0.523 (0.005) | 0.902 (0.001) | 0.601 (0.004) | 0.289 (0.001) | |
| Model 3 (Demo. + Clin. + gPRS) | 0.806 (0.002) | 0.789 (0.003) | 0.758 (0.006) | 0.820 (0.003) | 0.564 (0.004) | 0.909 (0.002) | 0.645 (0.004) | 0.261 (0.003) | |
| Model 4 (Demo. + Clin. + gPRS + Metabo.) | 0.812 (0.003) | 0.797 (0.003) | 0.793 (0.006) | 0.816 (0.004) | 0.545 (0.005) | 0.930 (0.002) | 0.646 (0.004) | 0.218 (0.003) | |
| RF | Model 1 (Demo.) | 0.570 (0.004) | 0.583 (0.003) | 0.646 (0.005) | 0.561 (0.006) | 0.292 (0.003) | 0.829 (0.002) | 0.445 (0.003) | 0.483 (0.001) |
| Model 2 (Demo. + Clin.) | 0.784 (0.002) | 0.754 (0.004) | 0.743 (0.009) | 0.809 (0.003) | 0.549 (0.003) | 0.909 (0.003) | 0.637 (0.005) | 0.487 (0.002) | |
| Model 3 (Demo. + Clin. + gPRS) | 0.851 (0.002) | 0.849 (0.004) | 0.810 (0.008) | 0.864 (0.002) | 0.582 (0.008) | 0.910 (0.002) | 0.723 (0.007) | 0.545 (0.003) | |
| Model 4 (Demo. + Clin. + gPRS + Metabo.) | 0.854 (0.003) | 0.851 (0.003) | 0.832 (0.009) | 0.857 (0.004) | 0.608 (0.006) | 0.911 (0.002) | 0.729 (0.004) | 0.490 (0.002) |
Model 1 contained sex, age, hypertension, body mass index, family history, smoking, and total alcohol consumption; Model 2 additionally included HbA1c, GLU0, total cholesterol, HDL-cholesterol, and triglyceride; Model 3 additionally included genome-wide polygenic risk score, Model 4 additionally included selected metabolites. PPV and NPV are positive and negative predictive values.
Reclassification performance of type 2 diabetes prediction models
Inclusion of gPRS in the prediction model in addition to demographic and clinical variables led to NRI and IDI improvement by 3.5% and 13.5% for LR and 11.7% and 3.9% for RF, respectively. For cNRI, the gPRS-adjusted model (model 3) showed improvement in both LR (0.361) and RF (0.511) compared to those of model 2. Inclusion of metabolic information in model 4 further enhanced the reclassification results for RF compared to those in model 3 (NRI = 0.042, cNRI = 0.336, IDI = 0.022). However, only limited improvement was reported for LR (NRI = 0.002, cNRI = 0.325, IDI = 0.005) (Table 2).
Association of gPRS and serum metabolites with beta-cell function and insulin resistance
Although gPRS was negatively associated with HOMA-B values (P < 0.001, correlation test), no significant association was observed between gPRS and HOMA-IR values (Table 4). A total of 11 metabolites among the 15 top-ranked metabolites were significantly associated with HOMA-IR values, although in varied directions. Moreover, hexose (P < 0.001, correlation test), glycine (P = 0.012, correlation test), PC ae C36:3 (P < 0.001, correlation test), and lysoPC a C18:2 (P < 0.001, correlation test) were associated with HOMA-B values.
Table 4.
Correlation coefficients of gPRS and 15 serum metabolites (top-ranked ≤ 2) for HOMA-B and HOMA-IR.
| Variable | Correlation with HOMA-B (95% CI) | P | Correlation with HOMA-IR (95% CI) | P |
|---|---|---|---|---|
| gPRS | −0.09 (−0.14, −0.03) | <0.0010 | 0.01 (−0.04, 0.05) | 0.73 |
| Spermine | 0.02 (−0.04, 0.07) | 0.56 | 0.03 (−0.02, 0.08) | 0.18 |
| Hexose | −0.23 (−0.29, −0.17) | <0.0010 | 0.28 (0.22, 0.33) | <0.001 |
| Isoleucine | 0.07 (−0.06, 0.20) | 0.31 | 0.25 (0.13, 0.37) | <0.001 |
| Valine | 0.01 (−0.10, 0.12) | 0.85 | 0.20 (0.11, 0.30) | <0.001 |
| Glycine | 0.07 (0.02, 0.13) | 0.012 | −0.12 (−0.17, −0.07) | <0.001 |
| PC ae C42:4 | −0.04 (−0.15, 0.07) | 0.51 | −0.14 (−0.20, −0.12) | <0.001 |
| PC ae C42:0 | −0.04 (−0.11, 0.02) | 0.20 | −0.01 (−0.06, 0.05) | 0.85 |
| PC ae C36:3 | 0.20 (0.09, 0.30) | <0.0010 | 0.20 (0.11, 0.30) | <0.001 |
| PC ae C34:3 | −0.06 (−0.16, 0.04) | 0.26 | −0.10 (−0.20, −0.01) | 0.027 |
| PC ae C42:1 | 0.02 (−0.06, 0.11) | 0.56 | −0.14 (−0.22, −0.07) | <0.001 |
| PC aa C40:5 | 0.04 (−0.02, 0.10) | 0.20 | −0.02 (−0.07, 0.04) | 0.49 |
| lysoPC a C18:2 | −0.11 (−0.18, −0.05) | <0.0010 | −0.20 (−0.26, −0.14) | <0.001 |
| Alanine | 0.02 (−0.05, 0.08) | 0.62 | 0.23 (0.17, 0.28) | <0.001 |
| PC ae C44:6 | 0.03 (−0.07, 0.13) | 0.53 | −0.02 (−0.11, 0.07) | 0.62 |
| Leucine | −0.03 (−0.17, 0.10) | 0.62 | −0.37 (−0.49, −0.24) | <0.001 |
HOMA-B, homeostasis model assessment of beta-cell function; HOMA-IR, homeostatic model assessment for insulin resistance, PC, phosphatidylcholine.
P represents the P-value at a significance level within the Pearson correlation test.
Odds ratio and feature importance
The OR during LR and averaged MDI calculated internally using the Boruta algorithm for each model are shown in Supplementary Tables S3 and S4, respectively. The tendency of importance values in terms of the OR magnitude and averaged MDI of each variable was consistent between both models. The importance values of gPRS, HbA1c, GLU0, and spermine were higher than that of the other variables.
Discussion
Our population-based prospective cohort study assessed the value of novel approaches integrating gPRS and metabolite profiles with clinical information for type 2 diabetes risk prediction among the Korean population. The addition of gPRS and metabolite profiles to clinical risk factors resulted in better model performance for prediction of type 2 diabetes risk compared to that of conventional risk factor-based models. Furthermore, compared to conventional LR-based models, RF-based machine learning analysis was modestly better in predicting type 2 diabetes incidence. Previous research on predicting type 2 diabetes or related clinical parameters incorporated either genetic information43, 44, 45 or metabolomic information45, 46, 47 to enhance model prediction. Various types of models have been developed for type 2 diabetes risk prediction, from statistical models such as logistic regression-based ones48,49 to machine learning models such as those using random forests,48, 49, 50, 51 gradient boosting,52, 53, 54 and (deep) neural networks.51,55,56 However, few models exist that incorporate both factors, and no model exists specifically for the non-white ethnic population. To the best of our knowledge, this is the first study to evaluate the effects of genetic information combined with metabolic measurements on type 2 diabetes risk prediction using RF-based machine learning analysis.
This study showed that gPRS modestly enhanced the accuracy of type 2 diabetes risk prediction, and our findings substantiate the value of PRS in the prediction of type 2 diabetes risk.9,10 Although the performance of the previous type 2 diabetes risk prediction models that added PRS to clinical risk factors varied from null to only modest improvements, better prediction ability was achieved when PRS was combined with conventional type 2 diabetes risk factors in our study. Notably, the majority of the previous PRS-based type 2 diabetes risk prediction models from Asian countries did not outperform the conventional risk models, especially when GLU0 or HbA1c were included as predictors.11, 12, 13, 14 However, gPRS, which is developed by aggregating clumped SNPs at the sub-threshold genome-wide association with incident type 2 diabetes, captures an individual's comprehensive genetic predisposition for type 2 diabetes. Thus, its inclusion enhanced the model performance even in the presence of glucose and HbA1c as clinical factors. This finding was consistent with that of a previous study,57 which showed that including gPRS consisting of 6.9 million type 2 diabetes-associated variants significantly improved the type 2 diabetes risk prediction ability in terms of AUC.
The predictive power of PRS derived from European ancestry was compromised in the Asian population, due to differences in allele frequency, LD, and effect-size of the risk allele.58,59 Thus, we calculated the effect-size of type 2 diabetes variants based on GWAS meta-analysis of KoreanChip to develop gPRS optimized exclusively for the South Korean population.28 Although direct comparison may be limited due to the heterogeneity in study design, population, and analytical methods, the current gPRS-based model performed better compared to the models from previous PRS-based studies in European ancestry,9,10 South Korea,11,12 and Japan,13,14 which either combined PRS with clinical risk factors or were based solely on the PRS. In addition, greater reclassification and prediction ability was observed when gPRS was added along with the clinical risk factors for type 2 diabetes.
There was a modest improvement in the predictive performance when metabolites were sequentially incorporated into the model with gPRS and clinical predictors. Although Walford et al.25 suggested that the use of metabolic information improved type 2 diabetes risk prediction in the FOS study, it was uncertain whether the improvement in performance of the type 2 diabetes risk prediction model that uses concurrent genetic and metabolite information would be applicable to an independent cohort of non-European ethnicity. Despite the modest improvement in the predictive ability of metabolite data in predicting type 2 diabetes, the concurrent use of gPRS and metabolite data enhanced prediction of type 2 diabetes risk in the Asian population. The reason for the limited contribution of metabolite data to model performance is unclear; however, overlap of information between metabolites, genetic factors, and clinical predictors may explain this limited improvement. Most metabolites were closely linked to HOMA-IR values, while several metabolites, such as glycine, PC ae C36:3, and lysoPC a C18:2, were significantly associated with both HOMA-IR and HOMA-B values, suggesting that metabolic information may, at least in part, contribute to the performance of the prediction model, with gPRS. Furthermore, the attenuated strength of association between TG and type 2 diabetes in the analysis accounting for metabolites suggested that TG might account for some aspect of metabolic information in the prediction model as a surrogate marker for insulin resistance (Supplementary Table S5). However, since metabolites capture early detrimental changes in type 2 diabetes21 and could serve as an independent predictor in previous prediction models accounting for clinical variables,16, 17, 18 larger prospective studies are necessary to evaluate the precise predictive value of metabolite data for type 2 diabetes risk, along with gPRS and clinical predictors.
We selected metabolites associated with type 2 diabetes using the Boruta algorithm. Frequently selected metabolites comprised three BCAAs (isoleucine, valine, and leucine), alanine, spermine, 6 PC ae (34:3, 36:3, 42:0, 42:1, 42:4, 44:6), 1 PC aa (40:5), lysoPC a C18:2, glycine, and hexose. Among the metabolites that passed through the variable selection algorithm, BCAA and alanine have been consistently associated with type 2 diabetes in previous studies.16, 17, 18,22 In addition, an inverse association of PC ae (34:3, 42:1, 42:4) with insulin resistance has been previously reported.24
In the current study, gPRS was closely associated with estimation of HOMA-B values, whereas most metabolites were significantly linked to HOMA-IR values, suggesting that genetic variants associated with type 2 diabetes primarily influenced beta-cell function, and the type 2 diabetes-linked metabolites were principally associated with insulin resistance. This result is consistent with that of the previous FOS.25 However, another case–control study suggested that five amino acids (isoleucine, phenylalanine, tyrosine, leucine, and valine) were modestly associated with both beta-cell function and insulin resistance.18 Although the reason for this discrepancy is not clear, distinctive features of patients with type 2 diabetes among the Asian population, such as greater proportion of body fat, visceral adiposity, and leaner BMI, compared to that of people from European ethnicity could explain the association of metabolites with beta-cell function.60 Heterogeneity in study design could also be a reason for this inconsistency.
Accurate prediction of type 2 diabetes risk allows physicians to identify individuals at high risk of type 2 diabetes, thereby providing a window to apply preventive measures such as advising nutritional modification and regular exercise, or implementing strategies to defer the onset of type 2 diabetes with medications such as metformin. Although there was only a modest performance improvement after incorporating genetic information (3.2%), a significant proportion of participants (11.7%) were correctly reclassified after the gPRS was included in the prediction model (e.g., from type 2 diabetes to non-type 2 diabetes or vice versa).
The major strengths of our study are as follows: 1) integrating genetic information and metabolite profiles to predict type 2 diabetes risk, 2) use of a machine-learning approach to predict the type 2 diabetes risk beyond the scope of a conventional risk model, 3) extending the utility of gPRS using genetic information on non-European ancestry, 4) use of a long-term prospective cohort data in which phenotypic information was gathered on a biannual basis, and 5) use of ethnic-specific gPRS to estimate the performance of the type 2 diabetes risk prediction model.
The present study also has several limitations. First, the study participants were from the South Korean population, thus extrapolation of the utility of gPRS and metabolite data to predict type 2 diabetes risk in non-Korean populations could be limited. However, this approach was adopted from a previous study among European ancestral populations that was subsequently validated among our non-European population with an improved predictive performance. Secondly, although we conducted CV of the type 2 diabetes risk prediction model, the predictive performance might be ranged for the non-Asian population. However, an improved method to generate gPRS might mitigate transferability issues between the ancestries for PRSs to some degree. Nonetheless, it would be necessary to test and replicate the predictive ability of the current model in independent prospective cohorts. Third, SNPs used for gPRS generation were mainly derived from common genetic variants associated with type 2 diabetes, and genetic information on rare variants was not captured in the analyses. In addition, as we used targeted metabolite analysis, other unknown metabolites that might additionally explain the etiology of type 2 diabetes risk were not included. Fourth, gPRS was calculated based on the genetic information of the South Korean population, which substantially differs in effect-size, LD, and allele frequency from other ethnicities. Thus, some degree of attenuation in the predictive performance may occur when this model is applied to other datasets without ethnicity-specific modifications. Fifth, the incidence of type 2 diabetes in the present study was slightly higher than that in other cohort studies at the given BMI levels61,62 indicating that a higher proportion of individuals at high risk for type 2 diabetes might have been included in the analysis. Nevertheless, the incidence of type 2 diabetes in our study is comparable to that of the Korean National Health Insurance claim and census data (Supplementary Tables S8 and S9).63 Considering the higher visceral adiposity shown by the Asian population compared to that in people of European descent at any BMI level, the issue of under-representation of current data is minimal.
In conclusion, this work shows the possibility to improve the discriminability of type 2 diabetes incidence by harmonizing genetic and metabolomic factors in the east Asian population, where national-scale clinical studies are actively nourished. gPRS and metabolites reflected distinctive etiologies of type 2 diabetes, and genetic and metabolomic information enhanced the predictability of type 2 diabetes, in addition to the clinical risk factors in non-European ethnicity with the aid of a machine learning algorithm. Ethnic-specific gPRS could be a viable option for the earlier identification of individuals at high risk for type 2 diabetes. This novel approach should be validated in different prospective cohorts of diverse ethnicities.
Contributors
J.L., and J.K. conceived and designed the study. Y.S.C. and J.K. verified the underlying data used in this study. S.-J.H., S.K., and J.K. performed data processing and statistical analysis. All authors took part in data interpretation. S.-J.H., S.K., J.L., and J.K. wrote the manuscript. All authors contributed to analyses of experimental results. All authors verified the integrity and the reproducibility of results. All authors critically edited the manuscript and approved the final version. J.K. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data sharing statement
Due to the privacy and sensitive information contained in the data, the data used for this study will not be made publicly available. Data is only accessible after screening of a proposal with relevant ethics consent and an approval of Korea Biobank (http://koreabiobank.re.kr; +82-1661-9070) maintained by National Institute of Health of Republic of Korea.
Declaration of interests
None exists.
Acknowledgements
This study was conducted with bioresources from the National Biobank of Korea, the Disease Control and Prevention Agency, Republic of Korea (KBN-2019-050). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. 2019M3E5D1A02070863 and 2022R1C1C1005458). This work was also supported by the 2020 Research Fund (1.200098.01) of UNIST (Ulsan National Institute of Science & Technology).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104383.
Contributor Information
Junghye Lee, Email: junghyelee@unist.ac.kr.
Jihun Kang, Email: josua85@naver.com.
Appendix A. Supplementary data
References
- 1.Kahn S.E. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46(1):3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 2.Rawshani A., Rawshani A., Franzén S., et al. Mortality and cardiovascular disease in type 1 and type 2 diabetes. N Engl J Med. 2017;376(15):1407–1418. doi: 10.1056/NEJMoa1608664. [DOI] [PubMed] [Google Scholar]
- 3.Ogurtsova K., da Rocha Fernandes J.D., Huang Y., et al. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract. 2017;128:40–50. doi: 10.1016/j.diabres.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Lindström J., Tuomilehto J. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 2003;26(3):725–731. doi: 10.2337/diacare.26.3.725. [DOI] [PubMed] [Google Scholar]
- 5.Wilson P.W.F., Meigs J.B., Sullivan L., Fox C.S., Nathan D.M., D'Agostino R.B., Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham offspring study. Arch Intern Med. 2007;167(10):1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 6.Lim N.-K., Park S.-H., Choi S.-J., Lee K.-S., Park H.-Y. A risk score for predicting the incidence of type 2 diabetes in a middle-aged Korean cohort. Circ J. 2012;76(8):1904–1910. doi: 10.1253/circj.cj-11-1236. [DOI] [PubMed] [Google Scholar]
- 7.Ramachandran A., Snehalatha C., Mary S., Mukesh B., Bhaskar A.D., Vijay V. The Indian Diabetes Prevention Programme shows that lifestyle modification and metformin prevent type 2 diabetes in Asian Indian subjects with impaired glucose tolerance (IDPP-1) Diabetologia. 2006;49(2):289–297. doi: 10.1007/s00125-005-0097-z. [DOI] [PubMed] [Google Scholar]
- 8.Herman W.H., Hoerger T.J., Brandle M., et al. The cost-effectiveness of lifestyle modification or metformin in preventing type 2 diabetes in adults with impaired glucose tolerance. Ann Intern Med. 2005;142(5):323–332. doi: 10.7326/0003-4819-142-5-200503010-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talmud P.J., Hingorani A.D., Cooper J.A., et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meigs J., Shrader P., Sullivan L., et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park H., Choi H., Hong Y. Utilizing genetic predisposition score in predicting risk of type 2 diabetes mellitus incidence: a community-based cohort study on middle-aged Koreans. J Korean Med Sci. 2015;30(8):1101. doi: 10.3346/jkms.2015.30.8.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Go M., Lee Y., Park S., Kwak S., Kim B., Lee J. Genetic-risk assessment of GWAS-derived susceptibility loci for type 2 diabetes in a 10 year follow-up of a population-based cohort study. J Hum Genet. 2016;61(12):1009–1012. doi: 10.1038/jhg.2016.93. [DOI] [PubMed] [Google Scholar]
- 13.Inaishi J., Hirakawa Y., Horikoshi M., et al. Association between genetic risk and development of type 2 diabetes in a general Japanese population: the Hisayama study. J Clin Endocrinol Metab. 2019;104(8):3213–3222. doi: 10.1210/jc.2018-01782. [DOI] [PubMed] [Google Scholar]
- 14.Goto A., Noda M., Goto M., et al. Predictive performance of a genetic risk score using 11 susceptibility alleles for the incidence of type 2 diabetes in a general Japanese population: a nested case-control study. Diabet Med. 2018;35(5):602–611. doi: 10.1111/dme.13602. [DOI] [PubMed] [Google Scholar]
- 15.The International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahola-Olli A., Mustelin L., Kalimeri M., et al. Circulating metabolites and the risk of type 2 diabetes: a prospective study of 11,896 young adults from four Finnish cohorts. Diabetologia. 2019;62(12):2298–2309. doi: 10.1007/s00125-019-05001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peddinti G., Cobb J., Yengo L., et al. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia. 2017;60(9):1740–1750. doi: 10.1007/s00125-017-4325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T., Larson M., Vasan R., et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu G., Zheng Y., Wang H., et al. Plasma metabolomics identified novel metabolites associated with risk of type 2 diabetes in two prospective cohorts of Chinese adults. Int J Epidemiol. 2016;45(5):1507–1516. doi: 10.1093/ije/dyw221. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S., Rhee E., Larson M., et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L., Liang L., Gao X., et al. Early prediction of developing type 2 diabetes by plasma acylcarnitines: a population-based study. Diabetes Care. 2016;39(9):1563–1570. doi: 10.2337/dc16-0232. [DOI] [PubMed] [Google Scholar]
- 22.Yang S., Kwak S., Jo G., Song T., Shin M. Serum metabolite profile associated with incident type 2 diabetes in Koreans: findings from the Korean Genome and Epidemiology Study. Sci Rep. 2018;8(1):8207. doi: 10.1038/s41598-018-26320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y., Han B. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) Consortium. Int J Epidemiol. 2017;46(4):1350. doi: 10.1093/ije/dyx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietiläinen K., Sysi-Aho M., Rissanen A., et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects – a monozygotic twin study. PLoS One. 2007;2(2) doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walford G., Porneala B., Dauriz M., et al. Metabolite traits and genetic risk provide complementary information for the prediction of future type 2 diabetes. Diabetes Care. 2014;37(9):2508–2514. doi: 10.2337/dc14-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupuis J., DIAGRAM Consortium. Langenberg C., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Kim J., Kwak M., Bajaj M. Genetic prediction of type 2 diabetes using deep neural network. Clin Genet. 2018;93(4):822–829. doi: 10.1111/cge.13175. [DOI] [PubMed] [Google Scholar]
- 28.Moon S., Kim Y., Han S., et al. The Korea Biobank array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019;9(1):1382. doi: 10.1038/s41598-018-37832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 30.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J Stat Softw. 2010;36:1–13. [Google Scholar]
- 31.Hastie T., Tibshirani R., Friedman J. 2nd ed. 2017. The elements of statistical learning. [Google Scholar]
- 32.Kursa M., Jankowski A., Rudnicki W. Boruta – a system for feature selection. Fundam Inf. 2010;101(4):271–285. [Google Scholar]
- 33.Guasch-Ferré M., Hruby A., Toledo E., et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39(5):833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 35.Steyerberg E.W. 2nd ed. Springer Nature; Cham, Switzerland: 2020. Clinical prediction models: a practical approach to development, validation, and updating. [Google Scholar]
- 36.Bergstra J., Bardenet R., Bengio Y., Kégl B. Advances in neural information processing systems. 2011. Algorithms for hyper-parameter optimization; p. 24. [Google Scholar]
- 37.Cawley G.C., Talbot N.L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. [Google Scholar]
- 38.Pencina M.J., D'Agostino Sr R.B., D'Agostino R.B., Jr., Vasan R.S. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 39.Leening M.J., Vedder M.M., Witteman J.C., Pencina M.J., Steyerberg E.W. Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician's guide. Ann Intern Med. 2014;160(2):122–131. doi: 10.7326/M13-1522. [DOI] [PubMed] [Google Scholar]
- 40.Pencina M.J., D'Agostino R.B., Sr., Demler O.V. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med. 2012;31(2):101–113. doi: 10.1002/sim.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen J. Routledge; 2013. Statistical power analysis for the behavioral sciences. [Google Scholar]
- 42.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Läll K., Mägi R., Morris A., Metspalu A., Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19(3):322–329. doi: 10.1038/gim.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weedon M.N., McCarthy M.I., Hitman G., et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med. 2006;3(10):e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lango H., UK Type 2 Diabetes Genetics Consortium. Palmer C.N., et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57(11):3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Floegel A., Stefan N., Yu Z., et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vangipurapu J., Fernandes Silva L., Kuulasmaa T., Smith U., Laakso M. Microbiota-related metabolites and the risk of type 2 diabetes. Diabetes Care. 2020;43(6):1319–1325. doi: 10.2337/dc19-2533. [DOI] [PubMed] [Google Scholar]
- 48.Carter T.C., Rein D., Padberg I., et al. Validation of a metabolite panel for early diagnosis of type 2 diabetes. Metabolism. 2016;65(9):1399–1408. doi: 10.1016/j.metabol.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Battineni G., Sagaro G.G., Nalini C., Amenta F., Tayebati S.K. Comparative machine-learning approach: a follow-up study on type 2 diabetes predictions by cross-validation methods. Machines. 2019;7(4):74. [Google Scholar]
- 50.Zheng T., Xie W., Xu L., et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int J Med Inf. 2017;97:120–127. doi: 10.1016/j.ijmedinf.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou Q., Qu K., Luo Y., Yin D., Ju Y., Tang H. Predicting diabetes mellitus with machine learning techniques. Front Genet. 2018;9:515. doi: 10.3389/fgene.2018.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinh A., Miertschin S., Young A., Mohanty S.D. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med Inform Decis Mak. 2019;19(1):1–5. doi: 10.1186/s12911-019-0918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guang P., Huang W., Guo L., et al. Blood-based FTIR-ATR spectroscopy coupled with extreme gradient boosting for the diagnosis of type 2 diabetes: a STARD compliant diagnosis research. Medicine (Baltimore) 2020;99(15) doi: 10.1097/MD.0000000000019657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahamed B.S., Arya S. LGBM classifier based technique for predicting type-2 diabetes. EJMCM. 2021;8(3):454–467. [Google Scholar]
- 55.Ayon S.I., Islam M.M. Diabetes prediction: a deep learning approach. Int J Inf Eng Electron Bus. 2019;12(2):21. [Google Scholar]
- 56.Montaez C.A., Fergus P., Montaez A.C., Hussain A., Al-Jumeily D., Chalmers C. 2018 International joint conference on neural networks (IJCNN) IEEE; 2018. Deep learning classification of polygenic obesity using genome wide association study SNPs; pp. 1–8. [Google Scholar]
- 57.Khera A.V., Chaffin M., Aragam K.G., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin A.R., Gignoux C.R., Walters R.K., et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma R.C., Chan J.C. Type 2 diabetes in East Asians: similarities and differences with populations in Europe and the United States. Ann N Y Acad Sci. 2013;1281(1):64–91. doi: 10.1111/nyas.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abraham T.M., Pencina K.M., Pencina M.J., Fox C.S. Trends in diabetes incidence: the Framingham heart study. Diabetes Care. 2015;38(3):482–487. doi: 10.2337/dc14-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.González E.M., Johansson S., Wallander M.A., Rodríguez L.G. Trends in the prevalence and incidence of diabetes in the UK: 1996–2005. J Epidemiol Community Health. 2009;63(4):332–336. doi: 10.1136/jech.2008.080382. [DOI] [PubMed] [Google Scholar]
- 63.Song S.O., Lee Y.H., Kim D.W., et al. Trends in diabetes incidence in the last decade based on Korean National Health Insurance claims data. Endocrinol Metab. 2016;31(2):292–299. doi: 10.3803/EnM.2016.31.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.