Summary
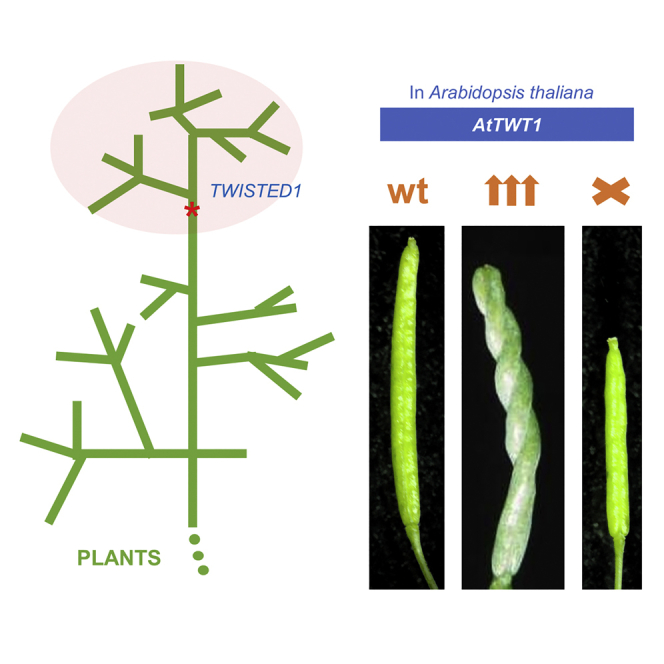
Evolution has long been considered to be a conservative process in which new genes arise from pre-existing genes through gene duplication, domain shuffling, horizontal transfer, overprinting, retrotransposition, etc. However, this view is changing as new genes originating from non-genic sequences are discovered in different organisms. Still, rather limited functional information is available. Here, we have identified TWISTED1 (TWT1), a possible de novo-originated protein-coding gene that modifies microtubule arrangement and causes helicoidal growth in Arabidopsis thaliana when its expression is increased. Interestingly, even though TWT1 is a likely recent gene, the lack of TWT1 function affects A. thaliana development. TWT1 seems to have originated from a non-genic sequence. If so, it would be one of the few examples to date of how during evolution de novo genes are integrated into developmental cellular and organismal processes.
Subject areas: Molecular biology, Evolutionary biology, Plant development
Graphical abstract
Highlights
-
•
The identification of a possible de novo-originated gene TWISTED1 in Brassicaceae
-
•
High expression of TWISTED1 causes right-handed helical growth of organs
-
•
Not the plant hormone auxin, but altered microtubule orientation causes helical growth
-
•
TWISTED1 CRISPR-Cas9 mutations affect plant development
Molecular biology; Evolutionary biology; Plant development.
Introduction
Genes have long been thought to arise almost exclusively from pre-existing genes through gene duplication, domain shuffling, horizontal transfer, overprinting, retrotransposition, etc.1,2,3 The genesis of new genes from “scratch,” non-coding, or non-genic sequences was considered extremely unlikely.1,4 However, it is becoming increasingly clear that the de novo birth of genes is more prevalent than was previously thought.5 “De novo” genes are present in a restricted number of related species, and have been found in different organisms including yeast, animals, and plants.6,7,8,9,10,11,12,13 For some such genes, functions have been identified, e.g., modulation of starch metabolism in plants, and adaptation to freezing temperatures in fish.14,15 However, for most de novo genes, their biological functions and the effects of their integration into the cellular machinery or networks remain unknown. Moreover, while the effects of new genes in the development of some animal species are starting to be clarified,16 the effects of these genes in the development of plants have not been explored.
Here, we describe the characterization of the seemingly taxonomically restricted TWISTED1 (TWT1) gene, which likely arose de novo from a non-genic sequence and is involved in plant development. TWT1 conspicuously modifies the growth and development of the plant Arabidopsis thaliana.
Results and discussion
Identification and characterization of the TWISTED1 gene in Arabidopsis
TWISTED1 (TWT1; At2g32275) was identified in a mutant (twt1-D) with peculiar, twisted siliques in Arabidopsis thaliana (Figures 1A, 1B, 1G, and 1H). The mutant was discovered in a forward genetic screening of a transposon-based activation-tag population.17 General plant development and reproduction were not severely affected in the mutant, but twt1-D presented a well-organized, prominent, right-handed twisted or helical growth pattern in the petioles, stem, petals, gynoecium, and inflorescences (Figures 1 and S1). Moreover, the epidermal cell files of twt1-D hypocotyls and roots were also arranged as right-handed helices (Figures 1K and 1M). At early stages of development, twt1-D exhibited shortened primary roots, with the early development of lateral roots (Figures 1I and 1J). The size of the root meristematic region and columella were reduced, though quiescent center (QC) markers such as WOX5 and QC46 were expressed in young seedlings (Figure S1). Furthermore, the vasculature of twt1-D cotyledons and leaves was less complex than their wild-type counterparts (Figure S1). Trichomes were also affected, most of them had only two branches, while most wild-type trichomes had three (Figure S1). Interestingly, the epidermal cells of twt1-D fruits (Figure 1G) appeared to have a different shape, less elongated and with wavy edges, compared to the epidermal cells of wild-type fruits (Figure 1H).
Figure 1.
twt1-D confers a twisted growth pattern in different organs
(A–J) See also Figures S1 and S2. Wild-type (WT) and twt1-D siliques (A, B), stamens and pistils (C, D), and rosettes (E, F). In (F) yellow arrows indicate right-handed twisting. Electron microscopy image of WT and twt1-D siliques (G, H). Seedlings presenting differences in root architecture (I, J).
(K and L) Confocal microscopy image of the upper hypocotyl region of WT (K), and twt1-D (L).
(M) Confocal microscopy image of a twt1-D root.
(N) Subcellular localization of TWT:GFP protein in the plasma membrane region of root cells (in a plant bearing the gtwt1-D:GFP construct).
(O) Schematic representation of the twt1-D genomic region, where the activating I element (AIE) was inserted. Blue lines represent the different regions used for constructs that recapitulated the phenotypes. The four arrows in the AIE represent the enhancer tetramer in the transposon. Dark green boxes represent exons, light green boxes represent UTRs, and triangles indicate the orientation of the gene. The black line represents non-coding genomic regions and introns.
(P–S) Phenotypes of stable transgenic lines bearing the constructs depicted in the scheme in (O). Each construct recovers the twt1-D phenotype (right-handed twists indicated by yellow arrows).
(T–V) Three-weeks-old WT (T), twt1-D (U), and twt1-D transformed with a construct causing the constitutive overexpression of an antisense version of cTWT1 (35S::antiTWT1) (V). The twt1-D phenotype can be rescued by the 35S::antiTWT1 construct.
(W and X) twt1-D (W) and twt1-D 35S::antiTWT1 (X) inflorescences. Scale bars: 2 mm (A, B, P-S, W-X), 1 mm (C, D), 1 cm (E-F, I-J, T-V), 100 μm (G), 200 μm (H), 50 μm (K-M), 5 μm (N).
The activation tagging population from where the mutant was isolated contains an engineered maize non-autonomous transposable element, Inhibitor (I, or defective Suppressor-Mutator, dSpm), that harbors a tetramer of the CaMV 35S enhancer. This element was named Activating I Element (AIE).17 The twt1-D phenotype was dominant, resulting from a single activation transposon insertion. The Activating I Element (AIE) was found to be inserted in chromosome 2 between a microRNA (MIR417; At2g32273) and a small peptide-coding region (At2g32275) 334 bp downstream of the transposon enhancer sequences (Figure 1O). miRNA417 has been reported to be involved in the salt stress response, but no twisted phenotype was described.18 When we tested two recapitulation constructs, including either 6.5- or 2.8-kb-long genomic fragments amplified from the original mutant, comprising the enhancer tetramer and its closest adjacent genomic sequence (Figure 1O), they produced phenotypes similar to those of twt1-D when introduced into wild-type plants (Figures 1P and 1Q). Both contained At2g32275 but not MIR417. Furthermore, At2g32275 expression was also found to be strongly increased in twt1-D compared to wild-type plants (Figure S2). After the increase in expression was confirmed, fragments of either the coding region together with the 3′-UTR of At2g32275 or the coding region alone were overexpressed using the CaMV 35S promoter in wild-type plants (Figure 1O). The increased expression of At2g32275 in these plants also recapitulated the twt1-D twisted leaves and siliques phenotype (Figures 1R, 1S, and S2). Furthermore, when the At2g32275-coding region placed in antisense orientation under the control of the CaMV 35S promoter (35S::antiTWT1) was introduced into twt1-D plants, the expression of At2g32275 was markedly reduced in comparison to the twt1-D mutant (Figure S2) and the characteristic twisting of rosette leaves and inflorescences was suppressed in these plants (Figures 1T–1X). Therefore, we concluded that the twt1-D phenotype was the result of the overexpression of the At2g32275 gene, which was renamed TWISTED1 (TWT1).
The annotation for the At2g32275 gene was “Expressed protein.” It has a relatively short CDS of 279 nucleotides and a single intron in its 3′-UTR (TAIR; arabidopsis.org). Surprisingly, homology searches in the NCBI database using BLASTx and tBLASTn revealed no similar genes or proteins in plants or other organisms besides close relatives of the Brassicaceae family (Arabidopsis lyrata, Capsella rubella, and lower similarity to Arabis alpina). This lack of gene homology with other organisms suggested that TWT1 could be a “new” gene, only present in A. thaliana and closely related species.
To evaluate whether TWT1 is indeed a gene and that its expression was not an artifact created by the nearby enhancer in twt1-D, we searched for transcripts in expression databases. TWT1 transcripts were found at low levels in the expression database ARS and by RT-PCR analysis using different Arabidopsis organs (Figure S2). A fusion of the TWT1 promoter with the β-glucuronidase (GUS) reporter gene was used to explore the pattern of TWT1 expression in more detail. In young plants, GUS activity was mainly observed in the vasculature of cotyledons and leaves, the root meristematic region, and inner tissues of the mature root (Figure S2). Thus, the TWT1 promoter and gene are active in specific tissues. We further explored whether the putative protein coded by TWT1 was stable and whether it had a specific localization in the cell. To visualize this protein, we used a translational fusion to GFP. To obtain enough signal, a genomic fragment cloned from the twt1-D mutant comprising the enhancer tetramer in the transposon and the adjacent region containing the TWT1 gene ending just before its stop codon was fused to GFP. This construct was named gtwt1-D:GFP. The roots of stable Arabidopsis transformants were analyzed, and fluorescence was observed at the edges of the cells, at the membrane (Figures 1N and S2). The construct was also used to perform transient expression assays by agroinfiltration in tobacco leaves, where a similar localization was observed (Figure S2). These findings corroborate previous proteomics studies that have identified TWT1 among plasma membrane proteins of Arabidopsis cells.19,20
Phenotypes due to increased TWISTED1 expression are related to microtubules
We then sought to ascertain the physiological processes affected by the increased expression of TWT1 in the mutant plant that caused the peculiar twisted or helicoidal morphological phenotype and root alterations. Auxin is a key plant hormone involved in many plant developmental processes. Alterations in the auxin pathway can cause organ torsions, twisted growth in non-fixed orientations, and root development defects.21,22 We analyzed reporter lines for the transcriptional auxin response and auxin transport in the roots of the twt1-D mutant (Figure S3). Furthermore, we evaluated the effects of IAA and NPA treatments (Figure S3). The results suggested that reduced growth of the root in twt1-D is not due to reduced auxin content or obvious differences in auxin transport. Moreover, they could also not explain the observed twisted phenotype.
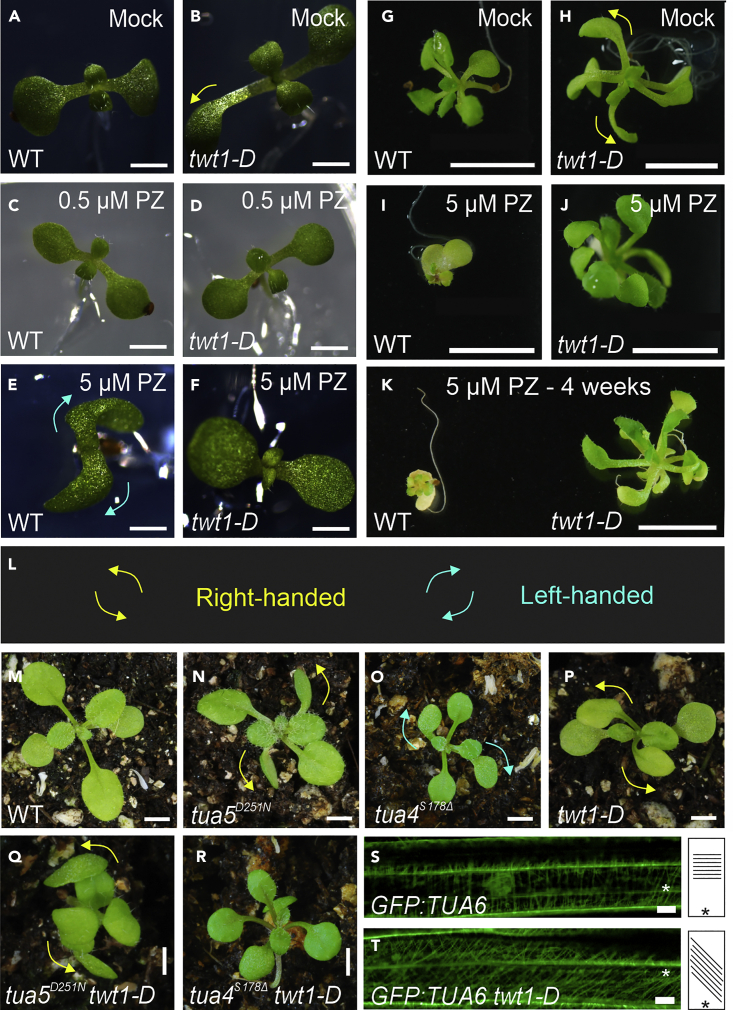
Helical growth phenotypes can also be caused by alterations in microtubules or microtubule-associated proteins and drugs that affect microtubule polymerization or depolymerization. When the properties of cortical microtubules are compromised, elongated organs can exhibit helical or twisted growth in a fixed orientation.23,24 Therefore, we examined the effects of propyzamide, a microtubule-disrupting drug, on twt1-D growth. Propyzamide is a benzamide that binds to β-tubulin, alters microtubule dynamics, and at low doses causes left-handed helical growth.25,26 Wild-type seedlings grown for a week on medium supplemented with 1, 3, or 5 μM propyzamide displayed definite left-handed helical growth (Figures 2A, 2E, and S3), while untreated twt1-D plants displayed a right-handed twisted phenotype (Figures 2B, 2H, and S3). At 0.5 μM propyzamide, the rightward twisting of twt1-D cotyledons decreased (Figures 2C and 2D) and was even reversed to leftward twisting at 1 and 3 μM (Figure S3). Moreover, twt1-D mutants grown for 3 or 4 weeks at 5 μM propyzamide were more resistant to this long-term treatment than wild-type plants (Figures 2G–2K). The growth of wild-type plants was severely affected by this treatment. In contrast, twt1-D plants appeared to be less altered, as evidenced by the larger size of the leaves and rosette, and the greener color (Figures 2G–2K). Therefore, it seems that the increased expression of TWT1 in the mutant counteracts the growth inhibition caused by long exposure to 5 μM propyzamide.
Figure 2.
Microtubule disruption, mutation, and orientation analyses in twt1-D
See also Figure S3.
(A–F) Effects of propyzamide on helical growth phenotypes in wild-type (WT) (A, C, E) and twt1-D mutants (B, D, F). Seedlings grown on control medium (A, WT; B, twt1-D) or medium containing 0.5 μM (C, WT; D, twt1-D) and 5 μM (E, WT; F, twt1-D) propyzamide for 7 days.
(G–J) WT (G, I) and twt1-D mutant (H, J) plants grown on control agar medium (G, H) or medium containing 5 μM propyzamide (I, J) for three weeks.
(K) WT (left) and twt1-D plants grown in medium containing 5 μM propyzamide for four weeks.
(L) Scheme indicating right- (yellow arrows) and left- (cyan arrows) handed twist.
(M–R) twt1-D interaction with tubulin mutants; (M) WT, (N) right-handed twisted TUA5D251N mutant, (O) left-handed twisted TUA4S178Δ mutant, (P) twt1-D, (Q) twt1-D TUA5D251N double mutant exhibiting increased right-handed twisting, and (R) twt1-D TUA4S178Δ double mutant displaying reduced twisting. Cortical microtubules in seedling hypocotyls of GFP:TUA6 (S) and GFP:TUA6 twt1-D (T), where microtubules are oriented to the left with respect to the top of the long axis of the cell. Asterisk indicates the basal part of the cell. Cartoons on the right illustrate the microtubule orientation in the cell (S, T). Scale bars: 2 mm (A–F), 5 mm (G-R), 10 μm (S, T).
Next, we tested the genetic interaction with tubulin mutants. twt1-D was crossed with tua5D251N and tua4S178Δ, which present right-handed and left-handed growth, respectively24 (Figures 2M−2O). The tua5D251N twt1-D double mutant displayed a prominent, enhanced, right-handed helical growth phenotype in the leaves and cotyledons (Figures 2M, 2N, 2P, and 2Q). Conversely, the tua4S178Δ twt1-D double mutant demonstrated a marked reduction or suppression of the left- and right-handed twisting displayed by the respective single mutants (Figures 2M, 2O, 2P, and 2R). The orientation of cortical microtubules is crucial for normal plant cell elongation, and alterations frequently result in helical or twisted growth.24 To analyze the orientation of cortical microtubules in twt1-D, we crossed it with the GFP:TUA6 marker line.27 In wild-type plants, cortical microtubules were aligned almost perpendicularly to the long axis of the cell (Figure 2S), whereas, in twt1-D, the microtubules did not show this orientation and appeared to produce left-handed oblique cortical arrays (Figures 2S and 2T). Therefore, the increased expression of TWT1 in the twt1-D mutant appears to affect microtubule orientation, and thereby the cytoskeleton. This was surprising, considering that TWT1 is a recent gene we did not expect it to influence basic cellular structures.
TWISTED1 likely originated from a non-genic sequence
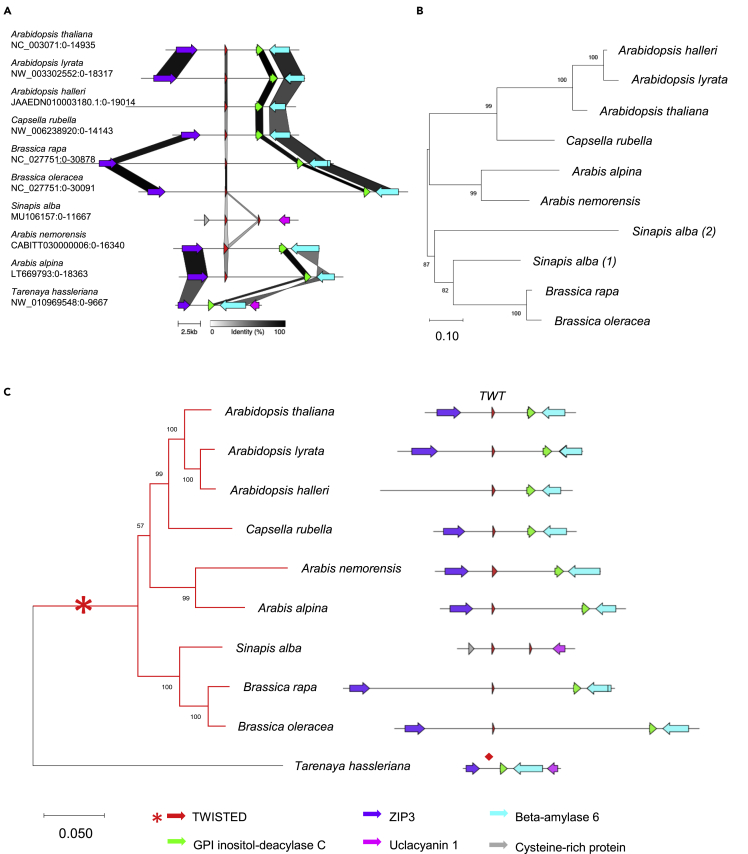
Next, we investigated the evolutionary origin of TWT1 by syntenic and phylogenetic analyses (Figures 3 and S4), which suggested a recent de novo origin of TWT1 in species related to A. thaliana. The syntenic regions of the genomes of these species were analyzed and searched for sequences, using tblastn, that would produce proteins with homology to TWT1. In nine Brassicaceae genomes analyzed, open reading frames (putative in some cases) with homology to TWT1 were identified. Though the identity of these putative TWTs was low (Table S1), at least three regions in the protein sequence appeared to be conserved, to some extent, as revealed by conservation analyses based on the multiple protein sequence alignments (Figure S4). We identified one of these regions as a “conserved TWT1 motif” (WxPxLxxIxExxE). The synteny of the genomic regions where these putative orthologues are is also well conserved in most of the analyzed related genomes (Figures 3A, 3C, and S4).
Figure 3.
Synteny and phylogenetic analyses of TWT1
See also Figure S4 and Table S1.
(A) Visualization of genomic regions surrounding the TWT1 gene based on GenBank files and the software clinker and clustermap.js. Syntenic genes are indicated in different colors. Gene percentage identity is presented in gray color scale.
(B) Phylogenetic tree inferred by maximum likelihood (ML) of putative TWT1 genes in Brassicaceae species.
(C) Left: Phylogenetic tree inferred by ML of Brassicaceae species coding for TWT1. Tarenaya hassleriana from the Brassicaceae sister family Cleomaceae was used as the outgroup. The origin of the TWT1 gene in the phylogeny is indicated with a red asterisk coinciding with the origin of Brassicaceae around 38 MYA. Right: Synteny of TWT1 (in red) and neighboring genes (in other colors). The diamond (red) indicates the non-genic region in T. hassleriana corresponding to the region where TWT is located in the genome of the other species. The tree was reconstructed from a concatenated alignment of the genes coding for GPI inositol-deacylase C (green) and beta-amylase 6 (cyan). In Sinapis alba, these two genes are located in a different contig. Bootstrap values are shown in tree nodes. The bars indicate the number of substitutions per site.
Tarenaya hassleriana is a plant that belongs to Cleomaceae, a sister family of Brassicaceae. The Cleomaceae and Brassicaceae lineages diverged only ∼38 million years ago.28 Interestingly, though the T. hassleriana genome has a very conserved syntenic region, the region that would correspond to TWT1 is an intergenic non-coding region (indicated by a red diamond in Figure 3C). After a careful inspection of putative ORFs present in this region between ZIP3 and GPI inositol-deacylase C, using tblastx we found a sequence that when translated in different translation frames showed significant similarity (at a low level, E-value 0.002) to different translation frames of TWT1 from A. thaliana (Figure S4D). There were three matches between different reading frames from A. thaliana TWT1 and the translated genomic sequence from T. hassleriana (Figure S4). A close inspection of one of the frames match, reveals the presence of five (WxPxLxxIxExxx) out of six (WxPxLxxIxExxE) amino acids of the conserved TWT1 motif (Figures S4 and S5). The DNA alignment between the A. thaliana TWT1 and the T. hassleriana putative 'TWT1' ORF is shown in Figure S5. Interestingly, two regions of the sequence, when translated in different open reading frames can give rise to one possibly conserved Cysteine at the beginning in one frame, and to an almost conserved TWT1 motif in the middle, in another frame (Figure S5). This sequence has the potential to become a TWT1 coding gene if various mutations would occur, meaning that this sequence could be ancestral to the Brassicaceae TWT1 coding genes.
These results suggest that coding TWT genes could be originated de novo in the Brassicaceae family. This finding coincides with a previous genome-scale study that sought lineage-specific genes in Arabidopsis that were restricted to the Brassicaceae family.11 Phylogenetic analyses of TWT1 (Figure 3B) and concatenated genes (Figure 3C) in the same syntenic region suggested that the origin of the TWT1 may coincide with the origin of Brassicaceae around 38 MYA (Figures 3B and 3C, indicated as an asterisk in Figure 3C).
Therefore, TWT1 likely originated recently in the Brassicaceae family. Different possible mechanisms of de novo protein genesis have been suggested, one of which proposes that these proteins arise when reading frames initially encoding disordered structures become expressed;29 young genes have indeed been found to encode highly disordered proteins.30,31 This appears to be the case for TWT1. The 92-amino acid (aa) TWT1 protein contains a predicted disordered domain that covers a large portion of the complete protein, from aa 19 to 76 (Figures 4 and S4). Despite the low sequence identity percentages of the AtTWT1 with TWTs from the very close species Arabidopsis lyrata, Capsella rubella, and Arabis nemorensis, these proteins also contain predicted disordered domains,32 though of different lengths and with variable sequences (Figure S4). Contrary to proteins that give rise to specific structures when they fold, an Intrinsically Disordered Protein or Domain does not fold spontaneously into a stable three-dimensional structure. These proteins or domains are disordered, and their structures are dynamic, changing their conformation continuously.33,34,35 Disordered domains were recognized some years ago and are gaining increasing attention due to their roles in different processes,35 including microtubule organization in animals36 and plants.37 It is thus plausible that the disordered domain of TWT1 could interact with microtubules, most likely cortical microtubules, considering the membrane localization of the protein.
Figure 4.
Phenotype and structure of edited TWT1 alleles
See also Figure S5.
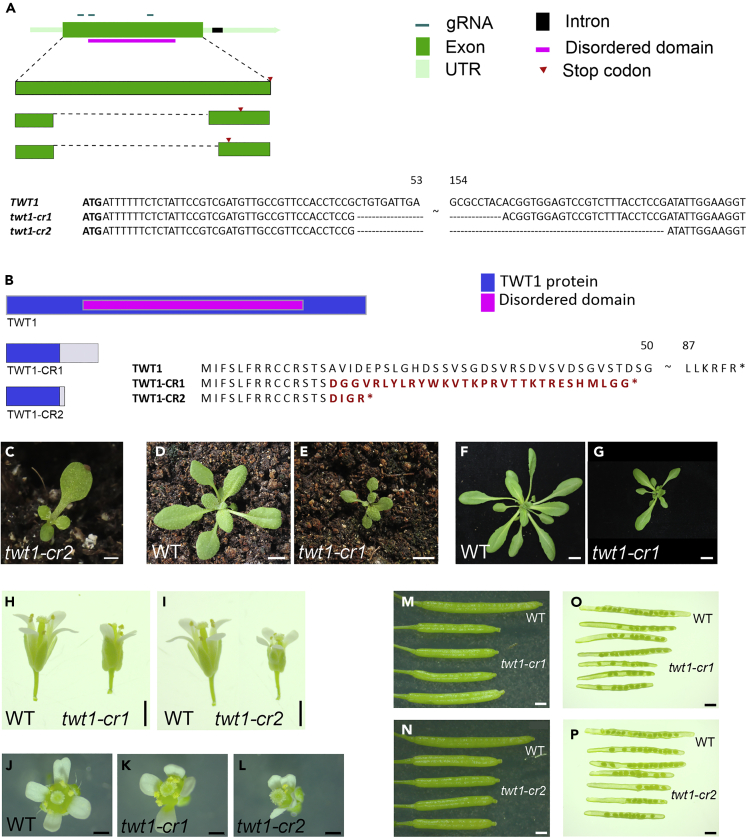
(A) Genomic structure of two twt1 CRISPR-Cas9 edited alleles.
(B) TWT1 protein structure and comparison to the proteins produced by the edited alleles. In both cases, a deletion results in a frameshift and premature stop codons that produce smaller proteins than TWT1. The 30 and 4 letters in red represent the amino acids that are different from the original TWT1 amino acids in each allele, due to frameshifts. Asterisks represent stop codons.
(C) Ten-day-old twt1-cr2 seedling with dissimilar cotyledons.
(D and E) Seventeen-day-old wild-type (WT) (D) and twt1-cr1 (E) plant.
(F and G) Twenty-six-day-old WT (F) and twt1-cr1 (G) plants.
(H–L) Flower phenotypes of WT and twt1-cr mutants.
(M and N) Comparison between WT and mutant siliques.
(O and P) Comparison between WT and mutant open siliques with exposed seeds. Scale bars: 1 mm (C, H, I, M-P), 5 mm (D, E), 1 cm (F, G), 0.5 mm (J-L).
TWISTED1 loss-of-function in Arabidopsis affects plant development
Finally, to know whether the loss of TWT1 function would affect plant development, the A. thaliana genome was edited using the CRISPR-Cas9 system.38 Three RNA guides were used to delete a large part of the coding region of the gene. Different deletion alleles were obtained. In two edited lines, in which 118 bp and 143 bp regions were deleted (Figures 4A and 4B), only the first 14 aa of the protein were maintained, whereas the remaining amino acids are changed due to frameshifts and premature stop codons in the coding region, eliminating the disordered region of the protein. No obvious differences were observed in microtubule organization in the epidermal cells of homozygous edited plants (Figure S6). However, the gene does not seem to be expressed in these cells, so more detailed analyses of inner tissues would be required to discard that microtubules are not affected. Nevertheless, homozygous edited plants displayed irregularities in their cotyledons such as elongated shape, dented edges, or size differences in the cotyledons of a seedling when grown on soil (Figure 4C). Older plants also exhibited delayed development, resulting in a reduced size phenotype (Figures 4D–4G). Furthermore, during reproductive development, edited plants presented smaller flowers (Figures 4H–4L) and reduced fruit length and seed number (Figures 4M−4P). Therefore, the loss of TWT1 function affects plant development. Other recent genes in plants have been reported to be involved in other kinds of processes such as starch accumulation14 or pathogen-induced defense,39 but there are seldom examples of recent genes involved in plant development.
In summary, TWT1 is likely a de novo-originated lineage-specific gene able to produce a strong, yet exquisite, morphological phenotype in Arabidopsis. As occurs with most de novo genes,8 it is a small gene that is expressed in many tissues at the low level. It encodes a 92 aa protein consisting mostly of a disordered domain that can be found at the plasma membrane. It appears to have integrated into processes involving basic cellular structures such as microtubules. Determining whether TWT1 binds to cortical microtubules, possibly affecting their dynamics and cellulose deposition and therefore affecting plant morphology, will help further clarify the role of this possibly “new” Brassicaceae protein. Moreover, since it seems to have originated recently from a non-genic sequence, it would be very interesting to reconstruct the ancient gene or putative transitory proto-gene(s), if they existed in more genomes.8
Limitations of the study
The identification of de novo-originated genes is not trivial. The erroneous conclusion of the identification of a lineage-specific gene that originated de novo could be the result of a homology detection failure.40 Furthermore, disordered domain protein-coding genes may evolve rapidly, and identification of homologs may be difficult. The putative ORF identified in the outgroup T. hassleriana may be ancestral to TWT1. Another scenario would be that the sequence in T. hassleriana was originally a TWT gene that was lost due to mutations. That would mean that the gene originated earlier. In the same line, we cannot exclude the possibility of horizontal gene transfer, though we did not find any evidence of similar proteins with the complete conserved TWT1 motif outside plant species. Only one hit to a bacterial sequence was found (MPZ85508.1), where a match between one amino acid (proline) matched the proline of the conserved TWT1 motif and other some less conserved residues. Furthermore, the bacterial sequence lacks the conserved Cysteine pair in the N-terminal part of TWT1, therefore we favor the hypothesis that the found hit to a bacterial sequence is not due to common ancestry but convergent evolution. Related to cortical microtubules, we cannot exclude that the natural function of TWT1 is not related to them. The affected microtubule arrangement, and other striking specific phenotypes such as helical growth of the fruit, were observed in an activation tag mutant and in overexpression lines, and therefore, care should be taken with drawing conclusions on the natural function of the gene.
Data and materials availability
All data and materials used in this study are available.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-α-tubulin (mouse, monoclonal) | Sigma-Aldrich | Cat#T5168 |
| Alexa Fluor 488 (donkey anti-mouse IgG) | Invitrogen | Cat#A-21202, RRID: AB_141607 |
| Chemicals, peptides, and recombinant proteins | ||
| Indole-3-Acetic Acid (IAA) | Duchefa Biochemie | Cat#I0901, CAS 87-51-4 |
| Naphthylphthalamic Acid (NPA) | Chemical Service | Cat#N-12507, CAS 132-66-1 |
| Propyzamide | Sigma-Aldrich | Cat#45645, CAS 23950-58-5 |
| X-gluc | Gold Biotechnology Inc | Cat#G1281C, CAS 114162-64-0 |
| Propidium iodide | FLUKA/Sigma-Aldrich | Cat#81845, CAS 25535-16-4 |
| Cellulase Onozuka R-10 | Duchefa Biochemie | Cat#C8001, CAS 9012-54-8 |
| Driselase | Sigma-Aldrich | Cat#D8037, CAS 85186-71-6 |
| Mazerozyme R-10 | Yakult | N/A |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana: twt1-D | This study17 | N/A |
| Arabidopsis thaliana: DR5rev::GFP | ABRC41,42 | CS9361 |
| Arabidopsis thaliana: PIN1::PIN1:GFP | ABRC41 | CS9362 |
| Arabidopsis thaliana: GFP:TUA6 | ABRC27 | CS6551 |
| Arabidopsis thaliana: PIN3::PIN3:GFP | Lars Østergaard43 | N/A |
| Arabidopsis thaliana: DR5::GUS | Luis Herrera-Estrella44,45 | N/A |
| Arabidopsis thaliana: PIN2::PIN2:GFP | Luis Herrera-Estrella46 | N/A |
| Arabidopsis thaliana: PIN7::PIN7:GFP | Luis Herrera-Estrella47 | N/A |
| Arabidopsis thaliana: WOX5::GFP | Luis Herrera-Estrella48 | N/A |
| Arabidopsis thaliana: QC46::GUS | Luis Herrera-Estrella49 | N/A |
| Arabidopsis thaliana: tua4S178Δ | Takashi Hashimoto24 | N/A |
| Arabidopsis thaliana: tua5D251N | Takashi Hashimoto24 | N/A |
| Arabidopsis thaliana: pARC170 (twt1-D genomic long) | This study | N/A |
| Arabidopsis thaliana: pGD625TR (twt1-D genomic short) | This study | N/A |
| Arabidopsis thaliana: pGD625GE5 (35S::TWT1) | This study | N/A |
| Arabidopsis thaliana: pGD121GE (35S::cTWT1) | This study | N/A |
| Arabidopsis thaliana: pTWT1::GUS | This study | N/A |
| Arabidopsis thaliana: gtwt1-D:GFP | This study | N/A |
| Arabidopsis thaliana: 35S::antiTWT1 (pGD121ANT) | This study | N/A |
| Oligonucleotides | ||
| See Table S2 | N/A | N/A |
| Recombinant DNA | ||
| pCR2.1 | Invitrogen | Cat#K202020 |
| pGEM-T easy | Promega | Cat#A1360 |
| pDONR207 | Invitrogen | N/A |
| pENTR/D-TOPO | Invitrogen | Cat#K240020 |
| pGD121 | de Folter et al.50 | N/A |
| pGD625 | de Folter et al.50 | N/A |
| pGD625-35S | This study | N/A |
| pBI121 | Jefferson et al.51 | N/A |
| pKGW | Karimi et al.52 | N/A |
| pMDC204 | Curti and Grossniklaus53 | N/A |
| pICSL01009::AtU6-26p (level 0) | Addgene54,55 | Cat#46968 |
| pICH47751 (level 1) | Addgene54,55 | Cat#48002 |
| pICH47761 (level 1) | Addgene54,55 | Cat#48003 |
| pICH47742 (level 1) | Addgene54,55 | Cat#48001 |
| pICH47742::pRPS5a-aCas9 | Renze Heidstra (Wageningen UR) | N/A |
| pICH47732::pOle-TagRFP | Renze Heidstra (Wageningen UR) | N/A |
| pICSL70008 (level 1) | Addgene55 | Cat#50336 |
| pICH47732 (level 1) | Addgene54,55 | Cat#48000 |
| pICSL4723 (level 2) | Addgene, Mark Youle55 | Cat#86173 |
| Software and algorithms | ||
| BLAST Arabidopsis | https://www.arabidopsis.org/Blast/index.jsp | N/A |
| ARS database, RNA-seq libraries | http://ipf.sustech.edu.cn/pub/athrna/ | N/A |
| BLAST NCBI | https://blast.ncbi.nlm.nih.gov/Blast.cgi | N/A |
| MUSCLE software | Edgar56 | N/A |
| MEGA for macOS software | Stecher et al.57 | N/A |
| Jalview | Waterhouse et al.58 | N/A |
| clinker & clustermap.js | Gilchrist and Chooi59 | N/A |
| MAUVE | http://darlinglab.org/mauve/mauve.html60,61 | N/A |
| genoPlotR | Guy et al.62 | N/A |
| MobiDB | https://mobidb.bio.unipd.it/32 | N/A |
| CRISPOR | http://crispor.org63 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stefan de Folter (stefan.defolter@cinvestav.mx).
Materials availability
Plant lines generated in this study will be made available on request, but we may require a completed Materials Transfer Agreement if there is potential for commercial application.
Experimental model and subject details
Plant materials and growth in soil
Seeds of the DR5rev::GFP (CS9361),41,42 PIN1::PIN1:GFP (CS9362),41 GFP:TUA6 (CS6551)27 lines were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/; ABRC, Ohio State University, Columbus). The PIN3::PIN3:GFP43 was obtained from Lars Østergaard. The DR5::GUS,44,45 PIN2::PIN2:GFP,46 PIN7::PIN7:GFP,47 WOX5::GFP48 and QC46::GUS49 were donated by Luis Herrera-Estrella. The tua5D251N and tua4 S178Δ mutants were supplied by Takashi Hashimoto.24 Arabidopsis plants in soil were grown under normal greenhouse or growth chamber conditions (∼22°C, long day light regime).
Method details
Activation tagging mutant screen
The stable I transposon (based on the En-I or Spm-dSpm transposon system) population of Arabidopsis thaliana plants, ecotype Wassilewskija (Ws-3)17 was visually screened for pistil/silique aberrations under normal greenhouse conditions. From the selected twt1-D mutant, the F1 segregating generation was grown to examine the dominance of the mutation and copy number of the I element.
Southern blot analysis
Genomic DNA from 24 segregating plants and the parental mutant was isolated64 and approximately 300 ng of DNA was digested with the restriction EcoRI enzyme. Equal loading of the DNA was checked by Ethidium bromide staining. DNA was electrophoresed in a 1.0% (w/v) agarose gel in 1x TBE (1.0 M Tris, 0.9 M boric Acid, 0.01 M EDTA) blotted onto a Hybond N+ membrane (Amersham Pharmacy Biotech) following the normal manufacturer’s instructions. A 1.3 kb PCR fragment amplified from the 5′ end of the BAR gene and from the 3′ end of the right transposon junction, was labelled by random oligonucleotide priming (Gibco BRL®) and used as a probe.17
Localization of the activating I element in the Arabidopsis genome
To identify the putative activated gene, genomic DNA was used to isolate flanking fragments of the Activating I element (AIE) by modified thermal asymmetric interlaced–PCR (TAIL-PCR) as described previously.17 Flanking sequences were compared to the Arabidopsis database using BLASTN65 in the TAIR database.66,67 The insertion was located in a genomic region between two genes, At2g32273 and At2g32275 (TWT1). The promoter of gene At2g32275 was found to be the closest to the 4 × 35S enhancer cluster located at the left border of the I transposable element.
Gene expression analysis
The expression of gene At2g32275 (TWT1) was analyzed by semiquantitative RT-PCR. RNA was isolated from rosette leaves, stem, and flower buds using the using TRIzol method (Invitrogen). After DNAse I treatment, cDNA was prepared using M-MLV (Invitrogen) according to the manufacturer’s instructions. Primers corresponding to the single coding exon (TWT1-exon), and the different splicing versions of the transcript (TWT1-splicing) were used.
Expression analysis of TWT1 was also performed using transcriptome data different tissues of Arabidopsis thaliana. Data was obtained from the ARS database; a collection of 20,000 RNA-seq libraries (http://ipf.sustech.edu.cn/pub/athrna/).68
Recapitulation and reporter constructs
For the genomic recapitulation construct pARC170, a two-step cloning strategy was used to clone a 6.5 kb genomic fragment of the twt1-D mutant, starting from the NOS terminator within the AIE element to 446 bp downstream of the predicted stop codon. This 6.5 kb twt1-D genomic fragment was divided into two fragments to facilitate cloning. In all PCR amplification steps, TAQ plus precision polymerase enzyme (Stratagene) was used. Fragment “A” was amplified using the primers PRO142 and PRO143, resulting in a 4.5 kb fragment spanning the NOS terminator to 1.2 kb upstream of the putative ATG. The “B” fragment was amplified with the primers PRO097 and PRO176. This 3.2 kb fragment contains the region 2.0 kb upstream of the ATG to 446 bp upstream of the predicted stop codon. Each fragment was cloned independently into pGEM®-T Easy (Promega), digested with the restriction enzymes SstII and BsmBI and ligated to each other resulting in the full 6.5 kb twt1-D genomic sequence in pGEM®-T Easy. Primers PRO142 and PRO176 contain additional GATEWAYTM sequences allowing recombination into the pDONR207 vector (Gibco BRL®), creating pARC169; followed by LR recombination step into the vector pKGW,52 establishing pARC170 (twt1-D genomic long). The primer sequences with GATEWAYTM sites underlined are included in Table S2.
For the genomic recapitulation construct pGD625TR (twt1-D genomic short), a 2.8 kb genomic fragment was amplified from genomic DNA isolated from the twt1-D mutant. The fragment was cloned from the NOS terminator within the AIE element to 500 bp downstream the predicted stop codon of At2g32275, using Pfx polymerase (Invitrogen). It was introduced into pENTR/D-TOPO (Invitrogen). For the overexpression recapitulation construct pGD625GE5 (35S::TWT1), a 795 bp genomic fragment was amplified from genomic DNA isolated from wild-type Ws-3 ecotype plants, the amplified region started from the ATG start codon to 500 bp downstream of the predicted stop codon. It was cloned in pENTR/D–TOPO. For the overexpression recapitulation construct pGD121GE (35S::cTWT1), the coding region of the At2g32275 gene was amplified with HiFi taq-polymerase (Invitrogen) using DNA from the same wild-type ecotype Ws-3 as the previous fragment and cloned into pCR2.1 (Invitrogen). All amplified fragments were verified by sequencing. The fragment used for pGD625TR was recombined with the binary Gateway vector pGD625-35S (the pGD625 vector without the 35S promoter). The fragments cloned for pGD625GE5 and pGD121GE were recombined or cloned into the binary overexpression vector pGD625 or pGD121 50, respectively.
To obtain a GUS reporter line, an 824 bp TWT1 (At2g32275) promoter fragment was amplified from genomic DNA isolated from wild-type Ws-3 plants using the HiFi taq-polymerase (Invitrogen). The fragment corresponds to the sequence directly upstream of the ATG start codon till the MIR417 gene and was cloned in pCR2.1 (Invitrogen). The amplified fragment was verified by sequencing. Finally, the fragment was cloned into the binary vector pBI121 51, generating a transcriptional fusion with the GUS reporter gene (pTWT1::GUS).
A translational fusion with the Green Fluorescent Protein (GFP) was made by cloning a 2.3 kb genomic fragment amplified from twt1-D mutant genomic DNA using Pfx polymerase (Invitrogen) in pENTR/D-TOPO (Invitrogen). This fragment includes the enhancer of the AIE element and the coding region minus the stop codon. This construct was recombined with the binary Gateway vector pMDC204 (gtwt1-D:GFP).53 To perform transient expression analysis to visualize protein localization, the gtwt1-D:GFP construct was agroinfiltrated in young Nicotiana tabacum leaves as previously described in69 and stably transformed in Arabidopsis plants.
The antisense construct 35S::antiTWT1 (pGD121ANT) was made by introducing the reverse sequence of the At2g32275 gene (cTWT1) into the pGD121 binary vector,50 and was stably transformed in twt1-D plants.
Plant transformations and transformant selection
The constructs were used to transform Agrobacterium tumefaciens strain GV3101 and introduced into wild-type Arabidopsis thaliana, ecotype Col-0, Ws-3, or the twt1-D mutant using a modified floral dip method.70 For selection, the harvested seeds were vapor phase surface-sterilized (http://plantpath.wisc.edu/%afb/vapster.html) or sterilized by rinsing with 70% ethanol, leaving them for 10 to 15 min in a 20% bleach solution, and rinsing at least 3 times with sterile water. Depending on the construct, the selection of first generation (T1) seedlings was done by germinating seed on medium containing Arabidopsis 0.5X MS nutrient solution (PhytoTechnology labs), 1% sucrose, and 0.6% or 0.8% agar. The medium was supplemented with the corresponding selective agent. They were stratified for 2 days at 4°C, and grown at 22°C, under a long day light regime. Resistant plants were transferred to soil for further analysis. Alternatively, seeds were sown directly on soil, and resistant plants were selected after spraying twice with 100 mg L−1 Finale®, 7 days after germination.
in vitro phenotypic analyses
Seeds were handled and sown on plates containing medium as for transformant selection, with some differences. The medium contained 0.5 or 2% sugar, depending on the experiment, and 1.5% agar. Plates were placed either horizontally or in a near vertical position. Day 0 of growth was defined as the time when plates were transferred to the growth chamber, after stratification. Indole-3-Acetic Acid (IAA) (Duchefa Biochemie, Amsterdam, The Netherlands) and Naphthylphthalamic Acid (NPA) (Chem Service, Westchester, PA) stock solutions were prepared in ethanol. The final concentration used for the experiments was 5 μM. IAA treatment experiments were repeated twice, with a total n of at least 25 plants per treatment. The data is presented as a graph of the average length and the standard deviation.
The Propyzamide (PZ; Sigma-Aldrich) stock solution was prepared by dissolving it first in dimethyl sulfoxide (DMSO). Propyzamide was then used at a final concentration of 0.5–5 μM. The final concentration of DMSO in the medium was lower than 0.3% and did not appear to affect the growth of seedlings. The experiment was repeated twice with an n of at least 10 plants per treatment.
Leaf morphology and vasculature analyses
For the analysis of leaf morphology and vasculature, the tissue was first fixed in 90% acetone at −20°C for one hour, followed by ethanol treatments at 80% and 40% (v/v) ethanol for a 24 h period each and fixed in 50% (v/v) glycerol. Cleared plants were imaged using an Olympus stereoscope SZH10.
GUS reporter staining
3- to 7-day-old seedlings were collected and placed in tubes containing GUS substrate solution composed of 50 mM sodium phosphate pH 7, 5 mM K3/K4 FeCN, 0.1% (w/v) Triton X-100, and 2 mM 5-bromo-4-chloro-3-indolyl-beta-GlcUA (Gold BioTechnology Inc). They were left in the solution for 2 to 6 days. After application of vacuum for 5 min, samples were incubated at 37°C for 24 h, processed to eliminate chlorophyll, and imaged.
Tissue preparation and confocal analysis
To observe the GFP fluorescence signal, hypocotyls and roots were collected and observed using a Zeiss LSM 510 META inverted confocal microscope (Carl Zeiss, Germany) with either a 20X or 40X air objective. 20 μM Propidium iodide (Fluka), was used as counterstain. GFP was excited with the 488 nm line of an Argon laser, and propidium iodide (PI) with a 514 nm laser line. GFP emission was filtered with a BP 500–520 nm filter and PI emission was filtered with a LP 575 nm filter.
Scanning electron microscope analysis
Fresh leaf and silique samples were visualized using a scanning electron microscope EVO40 (Carl Zeiss) using the VPSE G3 or the BSD detector with a 15–20 kV beam.
Immunolocalization of microtubules
Alpha-tubulin immunolabeling experiments were conducted following the protocol by71 with some modifications. Hypocotyls were dissected from six-days old Arabidopsis plants and transferred to 24-well plates to perform fixation, permeabilization, antibody incubation and washing steps. Cell permeabilization was done using 0.2% cellulase, 0.2% driselase, 0.2% mazerozyme, and 1% triton in 2 mM MES pH 5.0, during 1 h at room temperature. For primary antibody incubation an anti-α-tubulin primary antibody (T5168; mouse, monoclonal, Sigma) was diluted 1:800 and incubated for 16 h at 4°C in the dark; the secondary antibody was Alexa Fluor 488 (A-21202; donkey anti-mouse IgG, Invitrogen) diluted 1:1000 and incubated for 3 h at 37°C in the dark. Samples were mounted in 50% glycerol; images were acquired with a LSM800 confocal microscope using a 63X oil objective (Carl Zeiss).
Phylogenetic and sequence analyses
The protein coding sequence of TWT1 (AT2G32275.1) was used as a query in tblastn and blastp65 to search the nucleotide collection (nr/nt) and non-redundant (nr) databases at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). To improve sensibility, the search was initially restricted to Brassicales (taxid:3699). Subsequent blast searches excluding the Brassicales were performed to look for TWT1 homologs outside this clade (none were found). Homolog sequences in Fasta and Genbank formats were downloaded from NCBI. Genbank files included neighboring genes for synteny analyses.
Multiple sequence alignments were performed with MUSCLE software56 as implemented in MEGA for macOS software.57 MEGA software was also used for phylogenetic inference based on Maximum Likelihood (ML) principle. Multiple sequence alignment was visualized with Jalview.58 Synteny analysis was done with clinker & clustermap.js59 and with MAUVE (http://darlinglab.org/mauve/mauve.html)60,61 and genoPlotR.62
Generation of edited loss-of-function mutant alleles
The CRISPR-Cas9 system was used to edit TWT1.72,73 The CRISPR construct was generated using GoldenGate cloning and the MoClo toolkit.54,74,75 The plasmids were obtained from Addgene (https://www.addgene.org/). Possible off-targets were analyzed at http://crispor.org.63 Three guide RNAs were designed inside the coding region of the gene, and ordered as the primers indicated in Table S2, fused to the U6-26 promoter (pICSL01009::AtU6-26p) and ligated into level 1 vector pICH47751 or pICH47761. The RPS5a promoter76 was cloned into pICH47742 and used to drive an Arabidopsis-optimized Cas9 (aCas9). The FAST seed marker pOle::TagRFP77 obtained in the MoClo kit as pICSL70008 55, was cloned into pICH47732 and used as selection marker. These level 1 modules were subsequently combined into the binary level 2 vector pICSL4723. The resulting construct was transformed into Agrobacterium strain C58C1 and introduced in Arabidopsis Ws-3 and Col-0 using floral dip. Transformed seed was selected by visual inspection for the RFP marker. First transformants were evaluated by PCR (twt1-cr primers in Table S2) and seed was obtained from those that presented clear deletions.
To evaluate segregation of the deletion and the morphological phenotype, around sixty seeds (sown as fifteen seeds per tray, in 4 trays) of 4 Ws-3 and 5 Col-0 twt1 CRISPR mutant alleles (twt1-cr) were sown in soil (peat moss, perlite and vermiculite 3:1:1). They were stratified at 4°C for 3 nights and moved to a growth chamber at 22°C. Two weeks after germination, the plants were transferred to a greenhouse with a temperature range from 22 to 28°C, and natural light conditions. Day length varied in different seasons. Genomic DNA from plants that were homozygous for the deletion was used as template to obtain a PCR fragment that was subsequently sequenced (using the same twt1-cr primers indicated in Table S2). The fragment was sequenced from both sides and provided the same, but complementary, sequence.
Quantification and statistical analysis
For all the experiments, details of statistical tests used and error-bars in graphs are indicated in the relevant figure legends.
Acknowledgments
We thank Sarah Hake and David Smyth for critical reading of the article, the Arabidopsis Biological Resource Center (ABRC), Takashi Hashimoto, and Lars Ostergaard for seeds, Renze Heidstra for the RPS5a:aCas9 and pOle:Tag-RFP plasmids, and Karla L. González-Aguilera for lab support and logistics. J.I.R.O. was supported by the Mexican National Council of Science and Technology (CONACYT) with a PhD fellowship (210085), A.C.J. was financially supported by CAPES (Brazilian Agency for Higher Education Improvement from Ministry of Education) – Grant BEX 1519/98-0. CONACYT financed work in the N.M.M. laboratory (grant CB-2015-255069) and the S.d.F. laboratory (grants CB-2007-82826, CB-2012-177739, FC-2015-2/1061, CB-2017-2018-A1-S-10126).
Author contributions
J.I.R.O., A.C.J., A.C., R.E.G. investigation; M.B., Y.D.M., J.C.O. CRISPR-Cas9 and analyses; L.D. Analyses of evolutionary history; N.M.M., J.M., A.P., L.H.E., G.A., S.d.F. conceptualization and methodology; N.M.M., J.I.R.O., S.d.F. writing-original draft; N.M.M., S.d.F. writing-review and editing.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105627.
Contributor Information
Nayelli Marsch-Martínez, Email: nayelli.marsch@cinvestav.mx.
Stefan de Folter, Email: stefan.defolter@cinvestav.mx.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate new code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact.
References
- 1.Schlötterer C. Genes from scratch - the evolutionary fate of de novo genes. Trends Genet. 2015;31:215–219. doi: 10.1016/j.tig.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S., Krinsky B.H., Long M. New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 2013;14:645–660. doi: 10.1038/NRG3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long M., Vankuren N.W., Chen S., Vibranovski M.D. New gene evolution: little did we know. Annu. Rev. Genet. 2013;47:307–333. doi: 10.1146/ANNUREV-GENET-111212-133301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S. Springer-Verlag; 1970. Evolution by Gene Duplication. [Google Scholar]
- 5.McLysaght A., Hurst L.D. Open questions in the study of de novo genes: what, how and why. Nat. Rev. Genet. 2016;17:567–578. doi: 10.1038/nrg.2016.78. [DOI] [PubMed] [Google Scholar]
- 6.Begun D.J., Lindfors H.A., Thompson M.E., Holloway A.K. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics. 2006;172:1675–1681. doi: 10.1534/genetics.105.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai J., Zhao R., Jiang H., Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179:487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvunis A.R., Rolland T., Wapinski I., Calderwood M.A., Yildirim M.A., Simonis N., Charloteaux B., Hidalgo C.A., Barbette J., Santhanam B., et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles D.G., McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Ren Y., Yang T., Li G., Chen J., Gschwend A.R., Yu Y., Hou G., Zi J., Zhou R., et al. Rapid evolution of protein diversity by de novo origination in Oryza. Nat. Ecol. Evol. 2019;3:679–690. doi: 10.1038/s41559-019-0822-5. [DOI] [PubMed] [Google Scholar]
- 11.Donoghue M.T., Keshavaiah C., Swamidatta S.H., Spillane C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 2011;11:47. doi: 10.1186/1471-2148-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine M.T., Jones C.D., Kern A.D., Lindfors H.A., Begun D.J. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc. Natl. Acad. Sci. USA. 2006;103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin G., Ma P.F., Wu X., Gu L., Long M., Zhang C., Li D.Z. New genes interacted with recent whole-genome duplicates in the fast stem growth of bamboos. Mol. Biol. Evol. 2021;38:5752–5768. doi: 10.1093/MOLBEV/MSAB288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Foster C.M., Gan Q., Nettleton D., James M.G., Myers A.M., Wurtele E.S. Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 2009;58:485–498. doi: 10.1111/j.1365-313X.2009.03793.x. [DOI] [PubMed] [Google Scholar]
- 15.Baalsrud H.T., Tørresen O.K., Solbakken M.H., Salzburger W., Hanel R., Jakobsen K.S., Jentoft S. De novo gene evolution of antifreeze glycoproteins in codfishes revealed by whole genome sequence data. Mol. Biol. Evol. 2018;35:593–606. doi: 10.6084/m9.figshare.5509465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascual-Carreras E., Marin-Barba M., Herrera-Úbeda C., Font-Martín D., Eckelt K., de Sousa N., García-Fern'ndez J., Saló E., Adell T. Planarian cell number depends on blitzschnell, a novel gene family that balances cell proliferation and cell death. Development. 2020;147:dev184044. doi: 10.1242/dev.184044. [DOI] [PubMed] [Google Scholar]
- 17.Marsch-Martinez N., Greco R., van Arkel G., Herrera-Estrella L., Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol. 2002;129:1544–1556. doi: 10.1104/pp.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung H.J., Kang H. Expression and functional analyses of microRNA417 in Arabidopsis thaliana under stress conditions. Plant Physiol. Biochem. 2007;45:805–811. doi: 10.1016/j.plaphy.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J.R., Slijper M., Menke F.L.H. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Castellana N.E., Payne S.H., Shen Z., Stanke M., Bafna V., Briggs S.P. Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. USA. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida T., Thitamadee S., Hashimoto T. Twisted growth and organization of cortical microtubules. J. Plant Res. 2007;120:61–70. doi: 10.1007/s10265-006-0039-y. [DOI] [PubMed] [Google Scholar]
- 22.Weijers D., Wagner D. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 2016;67:539–574. doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- 23.Smyth D.R. Helical growth in plant organs: mechanisms and significance. Development (Camb.) 2016;143:3272–3282. doi: 10.1242/dev.134064. [DOI] [PubMed] [Google Scholar]
- 24.Ishida T., Kaneko Y., Iwano M., Hashimoto T. Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:8544–8549. doi: 10.1073/pnas.0701224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furutani I., Watanabe Y., Prieto R., Masukawa M., Suzuki K., Naoi K., Thitamadee S., Shikanai T., Hashimoto T. The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development. 2000;127:4443–4453. doi: 10.1242/dev.127.20.4443. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura M., Naoi K., Shoji T., Hashimoto T. Low concentrations of propyzamide and oryzalin alter microtubule dynamics in arabidopsis epidermal cells. Plant Cell Physiol. 2004;45:1330–1334. doi: 10.1093/pcp/pch300. [DOI] [PubMed] [Google Scholar]
- 27.Ueda K., Matsuyama T., Hashimoto T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma. 1999;206:201–206. doi: 10.1007/bf01279267. [DOI] [Google Scholar]
- 28.Cheng S., van den Bergh E., Zeng P., Zhong X., Xu J., Liu X., Hofberger J., de Bruijn S., Bhide A.S., Kuelahoglu C., et al. The tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell. 2013;25:2813–2830. doi: 10.1105/tpc.113.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornberg-Bauer E., Schmitz J., Heberlein M. Emergence of de novo proteins from “dark genomic matter” by “grow slow and moult. Biochem. Soc. Trans. 2015;43:867–873. doi: 10.1042/BST20150089. [DOI] [PubMed] [Google Scholar]
- 30.Wilson B.A., Foy S.G., Neme R., Masel J. Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth. Nat. Ecol. Evol. 2017;1:0146. doi: 10.1038/s41559-017-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James J.E., Willis S.M., Nelson P.G., Weibel C., Kosinski L.J., Masel J. Universal and taxon-specific trends in protein sequences as a function of age. Elife. 2021;10:573477–e57423. doi: 10.7554/eLife.57347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piovesan D., Necci M., Escobedo N., Monzon A.M., Hatos A., Mičetić I., Quaglia F., Paladin L., Ramasamy P., Doszt'nyi Z., et al. MobiDB: Intrinsically disordered proteins in 2021. Nucleic Acids Res. 2021;49:D361–D367. doi: 10.1093/NAR/GKAA1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uversky V.N. Intrinsically disordered proteins and their “mysterious” (Meta)Physics. Front. Phys. 2019;7:10. [Google Scholar]
- 34.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 36.Kadavath H., Hofele R.V., Biernat J., Kumar S., Tepper K., Urlaub H., Mandelkow E., Zweckstetter M. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. USA. 2015;112:7501–7506. doi: 10.1073/pnas.1504081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesten C., Wallmann A., Schneider R., McFarlane H.E., Diehl A., Khan G.A., van Rossum B.J., Lampugnani E.R., Szymanski W.G., Cremer N., et al. The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nat. Commun. 2019;10:857. doi: 10.1038/s41467-019-08780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao W., Liu H., Li Y., Li X., Xu C., Long M., Wang S. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One. 2009;4:e4603. doi: 10.1371/journal.pone.0004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisman C.M., Murray A.W., Eddy S.R. Many, but not all, lineage-specific genes can be explained by homology detection failure. PLoS Biol. 2020;18:e3000862. doi: 10.1371/JOURNAL.PBIO.3000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benkov' E., Michniewicz M., Sauer M., Teichmann T., Seifertov' D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 42.Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 43.Z'dníkov' P., Petr'sek J., Marhavý P., Raz V., Vandenbussche F., Ding Z., Schwarzerov' K., Morita M.T., Tasaka M., Hej'tko J., et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- 44.Ulmasov T., Hagen G., Guilfoyle T.J. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 45.Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J., Scheres B. Dissection of arabidopsis ADP-ribosylation factor 1 function in epidermal cell polarity. Plant Cell. 2005;17:525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 48.Pi L., Aichinger E., van der Graaff E., Llavata-Peris C.I., Weijers D., Hennig L., Groot E., Laux T. Organizer-Derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell. 2015;33:576–588. doi: 10.1016/j.devcel.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 49.Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/S0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- 50.de Folter S., Shchennikova A.v., Franken J., Busscher M., Baskar R., Grossniklaus U., Angenent G.C., Immink R.G.H. A B sister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006;47:934–946. doi: 10.1111/j.1365-313X.2006.02846.x. [DOI] [PubMed] [Google Scholar]
- 51.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/J.1460-2075.1987.TB02730.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karimi M., Inzé D., Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 53.Curtis M.D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engler C., Youles M., Gruetzner R., Ehnert T.M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 2014;3:839–843. doi: 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- 56.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilchrist C.L.M., Chooi Y.-H. <tt>Clinker & clustermap.js</tt> : automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 60.Darling A.E., Mau B., Perna N.T. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guy L., Kultima J.R., Andersson S.G.E. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/NAR/GKY354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pereira A., Aarts M.G. Transposon tagging with the En-I system. Methods Mol. Biol. 1998;82:329–338. doi: 10.1385/0-89603-391-0:329. [DOI] [PubMed] [Google Scholar]
- 65.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reiser L., Subramaniam S., Li D., Huala E. Using the arabidopsis information resource (TAIR) to find information about arabidopsis genes. Curr. Protoc. Bioinf. 2017;60:1.11.1–1.11.45. doi: 10.1002/cpbi.36. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H., Zhang F., Yu Y., Feng L., Jia J., Liu B., Li B., Guo H., Zhai J. A comprehensive online database for exploring ∼20, 000 public arabidopsis RNA-seq libraries. Mol. Plant. 2020;13:1231–1233. doi: 10.1016/j.molp.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 69.Marsch-Martínez N., Zúñiga-Mayo V.M., Herrera-Ubaldo H., Ouwerkerk P.B.F., Pablo-Villa J., Lozano-Sotomayor P., Greco R., Ballester P., Balanz' V., Kuijt S.J.H., et al. The NTT transcription factor promotes replum development in Arabidopsis fruits. Plant J. 2014;80:69–81. doi: 10.1111/tpj.12617. [DOI] [PubMed] [Google Scholar]
- 70.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313X.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 71.Du F., Zhao F., Traas J., Jiao Y. Visualization of cortical microtubule networks in plant cells by live imaging and immunostaining. STAR Protoc. 2021;2:100301. doi: 10.1016/J.XPRO.2021.100301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang W., Yang B., Weeks D.P. Efficient CRISPR/Cas9-Mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One. 2014;9:e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brooks C., Nekrasov V., Lippman Z.B., Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Werner S., Engler C., Weber E., Gruetzner R., Marillonnet S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs. 2012;3:38–43. doi: 10.4161/bbug.3.1.18223. [DOI] [PubMed] [Google Scholar]
- 76.Weijers D., Franke-van Dijk M., Vencken R.-J., Quint A., Hooykaas P., Offringa R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 77.Shimada T.L., Shimada T., Hara-Nishimura I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010;61:519–528. doi: 10.1111/j.1365-313X.2009.04060.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This study did not generate new code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact.