Abstract
Candida albicans is an opportunistic fungal pathogen that causes superficial infections in immunocompetent individuals, as well as life-threatening systemic disease in immunocompromised patients. A key virulence trait of this pathogen is its ability to transition between yeast and filamentous morphologies. A functional genomic screen to identify novel regulators of filamentation previously revealed VPS53 as being important for morphogenesis. Vps53 belongs to the Golgi-associated retrograde protein (GARP) complex, which mediates retrograde trafficking from the endosome to the trans-Golgi network. Here, we explored the role of the entire GARP complex in regulating morphogenesis. Deletion of any of the four genes encoding GARP complex subunits severely impaired filamentation in response to diverse filament-inducing cues, including upon internalization by macrophages. Genetic pathway analysis revealed that while hyperactivation of protein kinase A (PKA) signaling is insufficient to drive filamentation in GARP complex mutants, these strains are capable of filamentation upon overexpression of transcriptional activators or upon deletion of transcriptional repressors of hyphal morphogenesis. Finally, compromise of the GARP complex induced lipotoxicity, and pharmacological inhibition of sphingolipid biosynthesis phenocopied genetic compromise of the GARP complex by impairing filamentation. Together, this work identifies the GARP complex as an important mediator of filamentation in response to multiple inducing cues, maps genetic circuitry important for filamentation upon compromise of GARP function, and supports a model whereby GARP deficiency impairs lipid homeostasis, which is important for supporting filamentous growth in C. albicans.
Keywords: Candida albicans, fungal pathogen, morphogenesis, GARP complex
Plain language summary
Candida albicans causes serious infections in immunocompromised patients. A key virulence trait is its ability to transition between yeast and filamentous morphologies. Here, we identified a key role for the GARP complex in regulating hyphal morphogenesis in response to multiple inducing cues. Genetic analysis revealed that GARP complex mutants were capable of forming filaments and the defects observed were likely due to alterations in lipid homeostasis. Together, this work characterizes the GARP complex as important for C. albicans filamentation.
Introduction
The incredibly diverse fungal kingdom imposes profound impacts on agriculture, biodiversity, ecology, manufacturing, biomedical research, and global human health (Fisher et al. 2020). In the context of human disease, fungi cause serious infections in over 300 million individuals annually, with ∼1.5 million of these people succumbing to these infections (Bongomin et al. 2017). Candida albicans is an opportunistic pathogen that is a leading cause of invasive mycoses and the fourth leading cause of hospital-acquired infections (Pfaller and Diekema 2010). Those most susceptible include patients with indwelling medical devices or those with compromised immune systems from transplant surgeries or cancer treatments (Brown et al. 2012). The success of C. albicans as a pathogen is largely attributable to its arsenal of virulence traits, including its ability to transition between yeast and filamentous forms (Noble et al. 2017). Both morphotypes play distinct roles in the infection process, as the yeast form is crucial for colonization and dissemination through the bloodstream, while the filamentous form enables tissue invasion, deep-seated infection, and escape from macrophages (Saville et al. 2003). Morphogenesis is also fundamental to biofilm formation, which has a major clinical impact as C. albicans biofilms that form on medical implants are significant risk factors for infection and disease.
As an important virulence trait, filamentation is tightly regulated by multiple complex signaling pathways, with several morphogenetic regulators remaining to be identified and characterized. As a seminal example, the small GTPase Ras1 acts with adenylyl cyclase (Cyr1) to activate the cAMP–protein kinase A (PKA) pathway, a core signaling cascade of central importance for C. albicans morphogenesis, which culminates in the activation of the terminal transcription factor Efg1 (Shapiro et al. 2011). In addition, recent studies have highlighted the potential contribution of cAMP-independent pathways that mediate hyphal morphogenesis in vitro and in vivo (Parrino et al. 2017; Min et al. 2021; Wakade et al. 2022). The transition from yeast to filamentous growth in C. albicans is triggered by diverse host-relevant cues, including serum, nutrient limitation, alkaline pH, and elevated temperature (Shapiro et al. 2011). Most filament-inducing cues require a concurrent rise in temperature to induce filamentation to relieve repression of morphogenetic signaling mediated by the molecular chaperone Hsp90 (Shapiro et al. 2009; Kim, Iyer, et al. 2019). Specifically, pharmacological or genetic compromise of Hsp90 function induces the yeast-to-hyphal transition in the absence of any other inducing cue in both C. albicans and the related fungal pathogen Candida auris (Shapiro et al. 2009; Kim, Iyer, et al. 2019). Hence, Hsp90 has a central role in tuning cellular outputs important for morphogenesis and virulence in response to thermal input.
Functional genomic screens provide unprecedented power in identifying novel regulators of filamentation. For instance, an assessment of C. albicans genes important for filamentation using the gene replacement and conditional expression (GRACE) library identified 102 negative morphogenetic regulators and 872 positive regulators, revealing key roles for sterol biosynthesis and N-linked glycosylation in regulating filamentation in response to serum (O’Meara et al. 2015). Additional profiling of the GRACE collection to identify genes important for filamentation in response to Hsp90 inhibition unveiled several regulators of morphogenesis, many of which were previously unidentified, reinforcing the observation that genes required for hyphal morphogenesis are often specific to a particular environmental stimulus (Hossain et al. 2021). Through this analysis, it was observed that transcriptional repression of the largely uncharacterized gene VPS53 blocked filamentation in response to the Hsp90 inhibitor geldanamycin. This piqued our interest as VPS53 has not been previously reported to have any roles in filamentation in C. albicans.
Vps53 is a member of the Golgi-associated retrograde protein (GARP) complex that is comprised of subunits Vps51, Vps52, Vps53, and Vps54. This complex mediates retrograde trafficking of proteins and lipids from the endosomes to the trans-Golgi network (Conibear and Stevens 2000; Siniossoglou and Pelham 2002), such that GARP complex mutants display fragmented vacuoles, mis-sorting of vacuolar proteins, and aberrant lipid homeostasis (Conibear and Stevens 2000; Reggiori et al. 2003; Fröhlich et al. 2015). Previous studies in C. albicans have found that genetic impairment of VPS51 and VPS52 reduces filamentation in response to tissue culture-treated plastic, serum, or Spider medium (Park et al. 2009; Epp et al. 2010), however, a comprehensive assessment into the role of the GARP complex in mediating hyphal morphogenesis remains unexplored.
Here, we established that the entire GARP complex is important for the yeast-to-filament transition in C. albicans in response to diverse filament-inducing cues. Investigations into the genetic circuitry through which the GARP complex regulates hyphal morphogenesis revealed that hyperactivation of the cAMP–PKA signaling cascade in strains harboring homozygous deletion of GARP complex subunits blocked filamentation. However, overexpression of the filament-specific transcription factor UME6 or deletion of the transcriptional repressor NRG1, bypassed the requirement for the GARP complex in filamentation. Finally, as normal sphingolipid homeostasis is important for C. albicans filamentation, we propose that the GARP complex regulates this important virulence trait through its regulation of lipid homeostasis. Overall, this work establishes the GARP complex as a critical regulator of filamentation, furthering our understanding of this key virulence trait in a major human fungal pathogen.
Materials and methods
Strains and culture conditions
All strains and plasmids used in this study are listed in Supplementary Tables 1 and 2, respectively. All oligonucleotide sequences used in this study are listed in Supplementary Table 3. Strains were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) at 30°C. All strains were stored in 25% glycerol in YPD medium at −80°C. For solid media, 2% agar was added.
Stock solutions of geldanamycin (Toronto Research Chemicals Inc), aureobasidin A (Takara Bio), myriocin (Cayman Chemical Company), and nocodazole (Sigma-Aldrich) were prepared in DMSO. Doxycycline (BioBasic), nourseothricin (NAT) (Jena Bioscience), hygromycin (Bioshop), and caspofungin (provided by Merck) stocks were made in sterile water.
Strain construction
CaLC6642: The tetO-BCY1/tetO-BCY1 strain was made using a transient CRISPR approach adapted from Min et al. (2016). The promoter replacement cassette was PCR amplified from pLC1031 using oLC8586 and oLC8587. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA fusion cassette was made by PCR amplifying from pLC1081 with oLC6926 and oLC8588 (fragment A), as well as oLC6927 and oLC8589 (fragment B), at which point fusion PCR was performed on the fragments using the nested primers oLC6928 and oLC6929. The TAR-tetO cassette, sgRNA, and Cas9 DNA were transformed into CaLC2302. Transformants were selected by plating onto YPD plates supplemented with 600 µg/ml hygromycin B. Downstream integration was confirmed by PCR using oLC812 and oLC4714. Lack of the wild-type promoter was verified using oLC812 and oLC8433.
CaLC7507: The vps53Δ/vps53Δ strain was made using a transient CRISPR approach adapted from Min et al. (2016). The vps53Δ::NAT cassette with homology to the VPS53 locus was PCR amplified from pLC49 using oLC9804 and oLC9805. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA fusion cassette was made by PCR amplification from pLC963 with oLC5978 and oLC9807 (fragment A), as well as oLC5980 and oLC9806 (fragment B), at which point fusion PCR was performed on the fragments using the nested primers oLC5979 and oLC5981. The vps53Δ::NAT cassette, sgRNA, and Cas9 DNA were transformed into CaLC239. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration of the cassette was confirmed by PCR using oLC275 and oLC9808. Lack of a wild-type allele was verified by PCR using oLC9808 and oLC9810. The NAT marker was excised by culturing cells in YNB–BSA (0.17% yeast nitrogen base, 0.4% bovine serum albumin, 0.2% yeast extract, and 2% maltose) to induce the FLP recombinase.
CaLC7965: The vps51Δ/vps51Δ strain was made using a transient CRISPR approach adapted from Min et al. (2016). The vps51Δ::NAT cassette with homology to the VPS51 locus was PCR amplified from pLC49 using oLC10k167 and oLC10k168. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA fusion cassette was made by PCR amplification from pLC963 with oLC5978 and oLC10k170 (fragment A), as well as oLC5980 and oLC10k169 (fragment B), at which point fusion PCR was performed on the fragments using the nested primers oLC5979 and oLC5981. The vps51Δ::NAT cassette, sgRNA, and Cas9 DNA were transformed into CaLC239. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration of the cassette was confirmed by PCR using oLC275 and oLC10k171 and downstream integration was confirmed using oLC10k172 and oLC274. Lack of a wild-type allele was verified by PCR using oLC10k171 and oLC10k173. The NAT marker was excised by culturing cells in YNB–BSA (0.17% yeast nitrogen base, 0.4% bovine serum albumin, 0.2% yeast extract, and 2% maltose) to induce the FLP recombinase.
CaLC7967: The vps52Δ/vps52Δ strain was made using a transient CRISPR approach adapted from Min et al. (2016). The vps52Δ::NAT cassette with homology to the VPS52 locus was PCR amplified from pLC49 using oLC10k174 and oLC10k175. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA fusion cassette was made by PCR amplification from pLC963 with oLC5978 and oLC10k177 (fragment A), as well as oLC5980 and oLC10k176 (fragment B), at which point fusion PCR was performed on the fragments using the nested primers oLC5979 and oLC5981. The vps52Δ::NAT cassette, sgRNA, and Cas9 DNA were transformed into CaLC239. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration of the cassette was confirmed by PCR using oLC275 and oLC10k178 and downstream integration was confirmed using oLC10k179 and oLC274. Lack of a wild-type allele was verified by PCR using oLC10k178 and oLC10k180. The NAT marker was excised by culturing cells in YNB–BSA (0.17% yeast nitrogen base, 0.4% bovine serum albumin, 0.2% yeast extract, and 2% maltose) to induce the FLP recombinase.
CaLC7969: The vps54Δ/vps54Δ strain was made using a transient CRISPR approach adapted from Min et al. (2016). The vps54Δ::NAT cassette with homology to the VPS54 locus was PCR amplified from pLC49 using oLC10k181 and oLC10k182. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA fusion cassette was made by PCR amplification from pLC963 with oLC5978 and oLC10k184 (fragment A), as well as oLC5980 and oLC10k183 (fragment B), at which point fusion PCR was performed on the fragments using the nested primers oLC5979 and oLC5981. The vps54Δ::NAT cassette, sgRNA, and Cas9 DNA were transformed into CaLC239. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration of the cassette was confirmed by PCR using oLC275 and oLC10k185 and downstream integration was confirmed using oLC10k186 and oLC274. Lack of a wild-type allele verified by PCR using oLC10k185 and oLC10k187. The NAT marker was excised by culturing cells in YNB–BSA (0.17% yeast nitrogen base, 0.4% bovine serum albumin, 0.2% yeast extract, and 2% maltose) to induce the FLP recombinase.
CaLC8152: The tetO-BCY1/tetO-BCY1 vps51Δ/vps51Δ strain was constructed as described for CaLC7965, except the vps51Δ::NAT cassette was transformed in CaLC6642 (tetO-BCY1/tetO-BCY1).
CaLC8154: The tetO-BCY1/tetO-BCY1 vps52Δ/vps52Δ strain was constructed as described for CaLC7967, except the vps52Δ::NAT cassette was transformed in CaLC6642 (tetO-BCY1/tetO-BCY1).
CaLC8156: The tetO-BCY1/tetO-BCY1 vps53Δ/vps53Δ strain was constructed as described for CaLC7507, except the vps53Δ::NAT cassette was transformed in CaLC6642 (tetO-BCY1/tetO-BCY1).
CaLC8158: The tetO-BCY1/tetO-BCY1 vps54Δ/vps54Δ strain was constructed as described for CaLC7969, except the vps54Δ::NAT cassette was transformed in CaLC6642 (tetO-BCY1/tetO-BCY1).
CaLC8254: The tetO-UME6/UME6 strain was made by amplifying the tetO promoter construct from pLC605 using oLC10k356 and oLC10k357 and transforming into CaLC239. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration was confirmed using oLC2400 and oLC5034 and downstream integration was verified using oLC4714 and oLC2402.
CaLC8256: The tetO-UME6/UME6 vps51Δ/vps51Δ strain was constructed as described for CaLC8254, except the tetO cassette was transformed in CaLC7965 (vps51Δ/vps51Δ).
CaLC8258: The tetO-UME6/UME6 vps52Δ/vps52Δ strain was constructed as described for CaLC8254, except the tetO cassette was transformed in CaLC7967 (vps52Δ/vps52Δ).
CaLC8260: The tetO-UME6/UME6 vps53Δ/vps53Δ strain was constructed as described for CaLC8254, except the tetO cassette was transformed in CaLC7507 (vps53Δ/vps53Δ).
CaLC8262: The tetO-UME6/UME6 vps54Δ/vps54Δ strain was constructed as described for CaLC8254, except the tetO cassette was transformed in CaLC7969 (vps54Δ/vps54Δ).
CaLC8314: The nrg1Δ/nrg1Δ vps51Δ/vps51Δ strain was strain was made using a transient CRISPR approach adapted from Min et al. (2016). The nrg1Δ::NAT cassette with homology to the NRG1 locus was PCR amplified from pLC49 using oLC6663 and oLC6664. The CaCAS9 cassette was amplified from pLC963 using oLC6924 and oLC6925. The sgRNA guide to target NRG1 was amplified from pLC1075 using oLC5979 and oLC5981. The nrg1Δ::NAT cassette, sgRNA, and Cas9 DNA were transformed into CaLC7965. Transformants were selected by plating onto YPD plates supplemented with 150 µg/ml NAT. Upstream integration of the cassette was confirmed by PCR using oLC275 and oLC6669 and downstream integration was confirmed using oLC6670 and oLC274. Lack of a wild-type allele was verified by PCR using oLC3080 and oLC4107.
CaLC8316: The nrg1Δ/nrg1Δ vps52Δ/vps52Δ was constructed as described for CaLC8314, except the nrg1Δ::NAT cassette was transformed in CaLC7967.
CaLC8318: The nrg1Δ/nrg1Δ vps53Δ/vps53Δ was constructed as described for CaLC8314, except the nrg1Δ::NAT cassette was transformed in CaLC7507.
CaLC8320: The nrg1Δ/nrg1Δ vps54Δ/vps54Δ was constructed as described for CaLC8314, except the nrg1Δ::NAT cassette was transformed in CaLC7969.
Microscopy
To monitor filamentation in GARP complex mutants, overnight cultures were diluted to an OD600 of 0.1–0.2 in YPD medium with or without 10 µM geldanamycin and grown at 30°C with shaking for 7 h before imaging. For tetO-BCY1/tetO-BCY1 strains, overnights were diluted to an OD600 of 0.1 in YPD in the presence or absence of 20 μg/ml DOX and grown overnight at 30°C with shaking. The next day strains were diluted to an OD600 of 0.1 and grown in the same conditions for 6 h prior to imaging. For tetO-UME6/UME6 strains, strains were cultured overnight in the presence or absence of 20 µg/ml DOX prior to imaging. For nrg1Δ/nrg1Δ strains, overnight cultures were imaged. All imaging was performed using differential interference contrast microscopy with a Zeiss Axio Imager.MI (Carl Zeiss) using 40× magnification.
For treatment with inhibitors, an overnight culture of wild-type or vps53Δ/vps53Δ C. albicans was subcultured in the absence or presence of 0.025 µg/ml aureobasidin A, 0.4 µg/ml myriocin, or 8 ng/ml caspofungin and grown for 5 h at 39°C under static growth conditions prior to imaging with a Zeiss Axio Imager.MI (Carl Zeiss) using 40× magnification.
Liquid arrayed filamentation assay
To monitor C. albicans filamentation in GARP complex mutants, overnight cultures were transferred via a 96-well pin replicator into specific filament-inducing cues and incubated under the following static growth conditions: 6 h at 30°C for Hsp90 inhibition with 10 μM geldanamycin, 6 h at 30°C for cell cycle arrest with 40 μM nocodazole, 5 h at 42°C for high temperature, 5 h at 37°C for Spider medium (1% mannitol, 1% nutrient broth, and 0.2% K2HPO4), 5 h at 37°C for RPMI medium, 5 h at 37°C for GlcNAc (0.17% yeast nitrogen base without ammonium sulfate and without amino acids, 2% casamino acids, 0.008% uridine, 0.2% glucose, and 5 mM N-acetylglucosamine), 6 h at 37°C for anaerobic growth, 5 h at 37°C for serum (10% fetal bovine serum; Gibco#16000044), 5 h at 37°C for Lee’s medium (Lee et al. 1975). Images were captured using the IncuCyte S3 Live-Cell Analysis System (Sartorius) using 20× magnification.
BODIPY staining
Overnight cultures were diluted to an OD600 of 0.2 in YPD medium and grown at 30°C for 2–3 h with shaking at 200 rpm. After 2 h cells were treated with 5 µg/ml myriocin and grown at 30°C shaking for 3 h. Cells were centrifuged, media was removed, and cells were washed once in 1× PBS. To visualize lipid droplets, cells were stained with 1 µg/ml BODIPY (Cayman Chemical, 25892) for 10 min at 30°C and washed twice in 1× PBS before microscopy. To visualize fluorescence, an X-Cite series 120 light source with ET green fluorescent protein (GFP), was used.
FM4-64 staining
Overnight cultures were diluted to an OD600 of 0.2 in YPD medium and grown at 30°C for 3 h with shaking at 200 rpm. Cells were stained with 8 µM FM4-64 (Thermo Fisher Scientific, #T3166) in the dark for 30 min at 30°C with shaking. After 30 min, cells were resuspended in 1 ml YPD medium to remove free FM4-64 and added to 4 ml of fresh YPD medium. Cells were incubated in FM4-64 free medium for 90 min at 30°C, spun down, and resuspended in 1× PBS. Cells were imaged using a Zeiss Axio Observer.Z1 at 100× magnification. FM4-64 staining was observed using an X-Cite series 120 light source and an ET HQ tetramethylrhodamine isothiocyanate (TRITC)/DsRED filter set.
C. albicans filamentation in macrophages
J774A.1 (ATCC) macrophages were diluted to 1 × 105 cells/ml in RPMI-1640 medium (Gibco) containing 3% heat-inactivated fetal bovine serum (HI-FBS; Gibco). One hundred microliters were added to each well of a 96-well plate and incubated at 37°C with 5% CO2 for 18 h to activate the macrophages. Overnights of GARP complex mutants were diluted to an OD600 of 0.02 in RPMI-3% HI-FBS, and 50 µl of the diluted C. albicans cells were added to the macrophages. Cells were coincubated for 4 h at 37°C with 5% CO2. Cells were then fixed in 4% formaldehyde for 30 min at room temperature. Cells were washed 3 times in PBS. The macrophages were subsequently permeabilized by PBS containing 0.1% v/v Triton X-100 for 5 min. The cells were washed 3 times in PBS, blocked for 30 min in 3% bovine serum albumin in PBS, and stained with FITC-conjugated anti-Candida antibody (Abcam, ab21164) (1:2,000 dilution) for 1 h at room temperature. Cells were washed 3 times in PBS and incubated with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) to stain the macrophage nuclei prior to imaging on a Zeiss Axio Observer.Z1 at 40× magnification. Fluorescence imagining was performed using an X-Cite series 120 light source and ET GFP, and DAPI hybrid filter.
Wrinkly colony assays
For wrinkly colony formation on agar, overnights were washed once with sterile water and diluted to an OD600 of 10. Five microliters of the dilutions were spotted on indicated plates containing 2% agar, except for Spider medium where 1.35% agar was used. For serum plates, overnights were washed once with sterile water and diluted to an OD600 of 0.1. Five microliters of the dilutions were spotted. Plates were incubated at 30°C (control), 37°C (serum, Spider, and GlcNAc), or 42°C (high temperature) for 3 days and imaged using ChemiDoc.
Quantitative real-time PCR
qRT-PCR was utilized to monitor target gene repression upon treatment with DOX. Strains were grown in liquid YPD overnight at 30°C with shaking. Overnights were subcultured in YPD in absence or presence of 20 μg/ml DOX overnight. The next day, strains were subcultured into the same conditions and grown to mid-log phase. Cells were pelleted at 3,000 rpm at 4°C, washed with cold, distilled water, and then flash frozen in liquid nitrogen before storing pellets at −80°C. Cells were lysed by bead beating with acid washed glass beads (Sigma G8772-500g) with the MiniBeadBeater-16 (BioSpec Products) 4 times for 45 s, with 1 min on ice in between. RNA was extracted from the lysed cells using the QIAGEN RNeasy kit and DNase treated using the Invitrogen DNA-free DNA Removal Kit. Complementary DNA synthesis was performed using the iScript cDNA Synthesis Kit, utilizing both oligo (dT) and random primers for amplification. qRT-PCR was performed using the BioRad CFX-384 Real Time System in a 384-well plate, utilizing the Applied Biosystems Fast SYBR Green Master Mix with a 10-μl reaction volume. Amplification was performed with the following cycle conditions: 5°C for 3 min, then 95°C for 10 s and 60°C for 30 s, for 40 cycles. The melt curve was completed with the following cycle conditions: 95°C for 10 s and 65°C for 10 s with an increase of 0.5°C per cycle up to 95°C. Reactions were performed with technical triplicates and data were analyzed using the BioRad CFX Maestro software. Statistical significance was determined using unpaired t-test with GraphPad Prism. The primer sequences used for monitoring expression of ACT1 (oLC2285/oLC2286), BCY1 (oLC4258/oLC4259), and UME6 (oLC10k541/oLC10k542) are available in Supplementary Table 3.
Results
The GARP complex is important for filamentation in response to diverse inducing cues in C. albicans
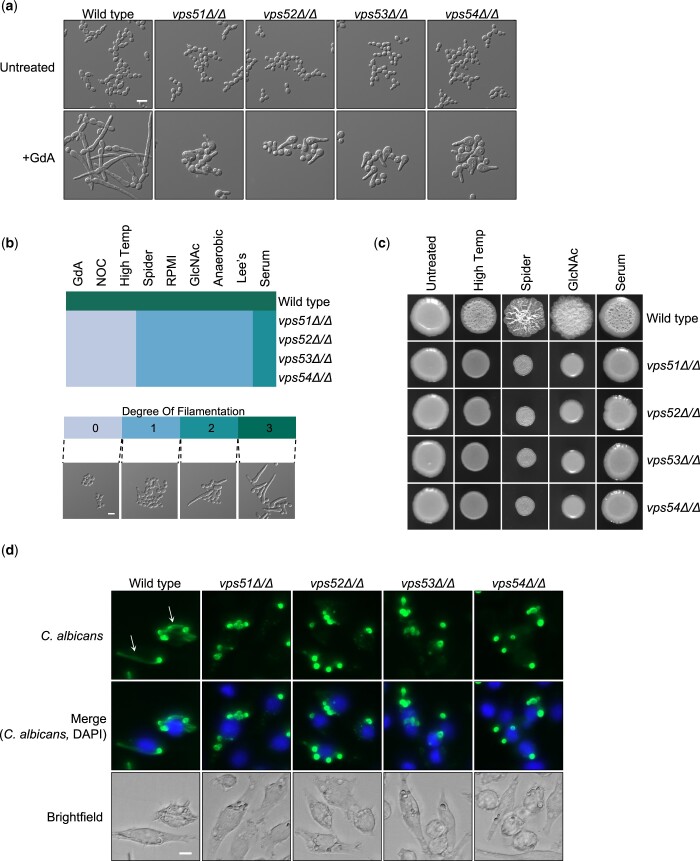
A previous functional genomic screen identified a novel role for Vps53 in filamentation in response to Hsp90 inhibition in C. albicans (Hossain et al. 2021). Thus, we wanted to explore the importance of this regulator, along with the other subunits in the GARP complex, in hyphal morphogenesis. First, we tested whether only Vps53 or the entire GARP complex is important for filamentation in response to Hsp90 inhibition. To do so we profiled the morphology of GARP complex homozygous deletion mutants in the presence of the Hsp90 inhibitor geldanamycin. Deletion of VPS51, VPS52, VPS53, or VPS54 impaired filamentation in response to Hsp90 inhibition (Fig. 1a).
Fig. 1.
The GARP complex is important for filamentation in response to diverse cues in C. albicans. a) Strains were grown in the presence or absence of 10 µM geldanamycin (GdA) for 7 h under shaking conditions prior to imaging. Scale bar represents 10 µm. b) Phenotype of GARP mutants grown in different filament-inducing media under static growth conditions. Strains were grown in the presence of the following inducing cues and conditions: Hsp90 inhibition with 10 μM geldanamycin (GdA) at 30°C for 6 h, cell cycle arrest with 40 μM nocodazole (NOC) at 30°C for 6 h, high temperature at 42°C for 5 h, Spider medium at 37°C for 5 h, RPMI medium at 37°C for 5 h, 5 mM N-acetyl-glucosamine (GlcNAc) at 37°C for 5 h, anaerobic growth at 37°C for 6 h, Lee’s medium at 37°C for 5 h, and 10% heat inactivated new born calf serum at 37°C for 5 h. Numbers correspond to degree of filamentation (DOF; see scale bar). Scale bar on microscopy image represents 10 µm. Heatmap was generated using the heatmap.2 function in R. c) Cells from overnight cultures were spotted onto filament inducing medium. Agar plates were incubated at 30°C (control), 37°C (serum, Spider, and GlcNAc) or 42°C (high temperature) for 3 days and imaged. d) J774A.1 macrophages were coincubated for 4 h with C. albicans strains, fixed with 4% formaldehyde, immunostained with a FITC anti-Candida antibody (green), and stained with DAPI to mark the macrophage nucleus (blue). Representative filaments are indicated with a white arrow. Scale bar represents 10 µm.
Next, we assessed whether the GARP complex was specifically required for morphogenesis in response to Hsp90 inhibition or if it was more broadly required in response to multiple inducing cues, including elevated temperature, serum, anaerobic conditions, amino acid-deficient Lee’s medium, Spider medium, RPMI medium, alternative carbon source with N-acetyl-glucosamine (GlcNAc), and cell cycle arrest with the microtubule disrupting agent nocodazole. The strains were qualitatively scored for their degree of filamentation on a scale from 0 to 3 in each filament-inducing condition, where 0 indicates a complete block in filamentation, 1 indicates minimal polarized growth, 2 indicates intermediate polarized growth, and 3 indicates a level of filamentation similar to a wild-type control (Fig. 1b) (Hossain et al. 2021). We observed genetic impairment of any GARP complex subunit largely blocked hyphal morphogenesis across all filament-inducing cues (Fig. 1b). Notably, only a partial defect was observed in response to serum (degree of filamentation score of 2). Additionally, we monitored the ability of GARP complex mutants to form wrinkly colonies, a hallmark of filamentous growth, on solid filament-inducing media. By spotting inoculum onto agar plates and allowing strains to grow for several days, we observed GARP complex mutants were defective in forming wrinkly colonies on all cues tested, including elevated temperature, Spider medium, GlcNAc, and serum (Fig. 1c).
C. albicans filamentation is also induced upon internalization by macrophages, promoting macrophage death and fungal escape (Miramón et al. 2013). To assess C. albicans morphology during infection, we inoculated murine macrophages with a wild-type strain or with the GARP complex mutants. Unlike the wild-type control, vps51, vps52, vps53, and vps54 homozygous deletion mutants were unable to filament upon internalization by macrophages, further highlighting the importance of the GARP complex in filamentation in response to diverse inducing cues (Fig. 1d).
Overexpression of PKA signaling is insufficient to drive filamentation in GARP complex mutants, while modulating transcriptional regulators can rescue the filamentation defect
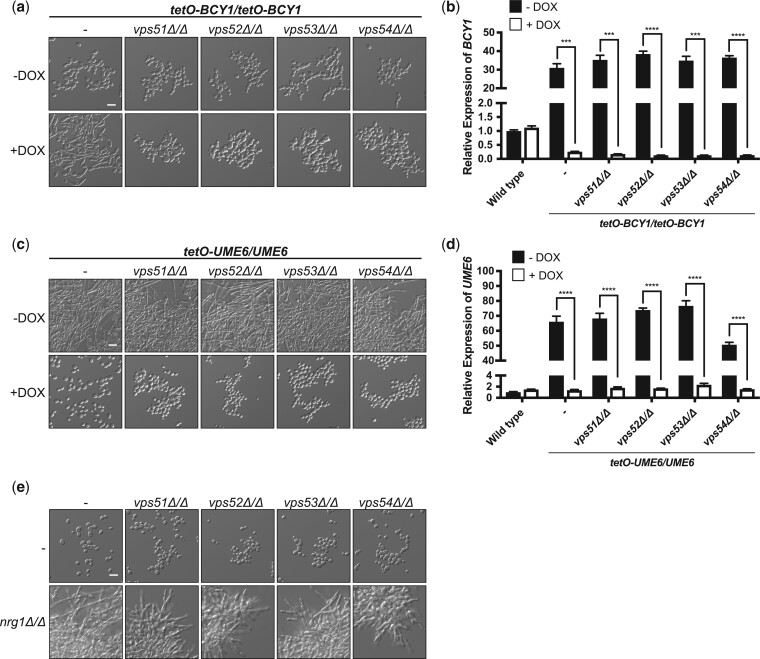
Morphogenesis in C. albicans is tightly regulated by interconnected and complex cellular signaling cascades. One of the core signaling pathways that governs morphogenesis in response to multiple inducing cues is the cAMP–PKA signaling pathway (Shapiro et al. 2011; Sudbery 2011). The protein kinase PKA is comprised of 2 catalytic subunits, Tpk1 and Tpk2, and a regulatory subunit, Bcy1. Elevated cAMP levels that ensue in the presence of a filament-inducing cue result in the binding of cAMP to Bcy1, leading to its release from the complex and activation of the catalytic subunits. Thus, impairment of BCY1 causes constitutive activation of PKA and in turn induces filamentation in the absence of an inducing cue (Ding et al. 2016). To determine whether hyperactivation of PKA could rescue the filamentation defect observed with GARP complex mutants, we generated a strain where both alleles of BCY1 were under the control of a tetracycline-repressible promoter. In the absence of doxycycline this strong promoter results in overexpression of its target gene. However, in the presence of tetracycline, or its analog doxycycline, expression of BCY1 is repressed, which leads to activation of the PKA and the induction of filamentation in C. albicans in the absence of any other inducing cue (Fig. 2a and b). However, depletion of BCY1 did not induce filamentation in any GARP complex mutant, suggesting that activation of PKA is not sufficient to induce hyphal morphogenesis in cells lacking Vps51, Vps52, Vps53, or Vps54 (Fig. 2a and b).
Fig. 2.
Identifying genes important for filamentation for GARP complex mutants. a) To repress BCY1 expression in the tetO-BCY1/tetO-BCY1 strains, cells were grown overnight at 30°C in YPD medium in the absence or presence of 20 µg/ml doxycycline (DOX) with shaking and subsequently grown in the absence or presence of 20 µg/ml DOX for 6 h at 30°C with shaking prior to imaging. Scale bar represents 10 µm. b) Cells were grown overnight in the absence or presence of 20 µg/ml DOX. Cells were subcultured into YPD with the same DOX conditions for 5 h before pelleting, RNA extraction, cDNA synthesis, and qRT-PCR. Transcript levels of BCY1 were normalized to ACT1 and are relative to the wild-type no DOX control. Data are presented as mean ± SEM of technical triplicates. Significance was determined by unpaired t-test. c) To overexpress UME6 in the tetO-UME6/UME6 strains, cells were grown overnight at 30°C in YPD medium in the absence or presence of 20 µg/ml DOX with shaking and subsequently imaged. Scale bar represents 10 µm. d) Cells were grown as in (b). Transcript levels of UME6 were normalized to ACT1 and are relative to the wild-type no DOX control. Data are presented as mean ± SEM of technical triplicates. Significance was determined by unpaired t-test. e) Strains were grown overnight at 30°C in YPD medium with shaking and subsequently imaged. Scale bar represents 10 µm. ***P < 0.001 and ****P < 0.0001.
Next, we wanted to assess whether GARP mutants are physically unable to filament or if distinct genetic perturbations could induce morphogenesis in these mutant backgrounds. We focused our efforts on two transcription factors, Ume6 and Nrg1. Ume6 is a transcription factor that upregulates hyphal-specific gene expression in response to filament-inducing cues, and overexpression is sufficient to drive hyphal formation in an otherwise wild-type background (Banerjee et al. 2008; Carlisle et al. 2009). We generated a strain in which a single allele of UME6 was under the control of a tetracycline-repressible promoter, given that in the absence of doxycycline this strong promoter results in overexpression of its target gene. Indeed, in the absence of an additional inducing cue, the tetO-UME6/UME6 strain was robustly filamentous due to UME6 overexpression (Fig. 2d). Interestingly, overexpression of UME6 also induced filamentation in the GARP complex mutant backgrounds (Fig. 2c and d). Next, we turned to Nrg1, a transcription factor that represses filamentous growth, such that homozygous deletion of NRG1 induces robust filamentation in the absence of an inducing cue (Braun et al. 2001; Murad et al. 2001). Deletion of NRG1 induced filamentous growth in the GARP complex mutants, albeit not to the same extent as overexpression of UME6 (Fig. 2e). Overall, these results highlight that while overexpression of PKA signaling is insufficient to drive filamentation in GARP complex mutants, these strains are capable of filamentation and do so upon overexpression of transcriptional activators or upon deletion of transcriptional repressors of hyphal morphogenesis.
GARP complex is required for lipid homeostasis, which is important for filamentation
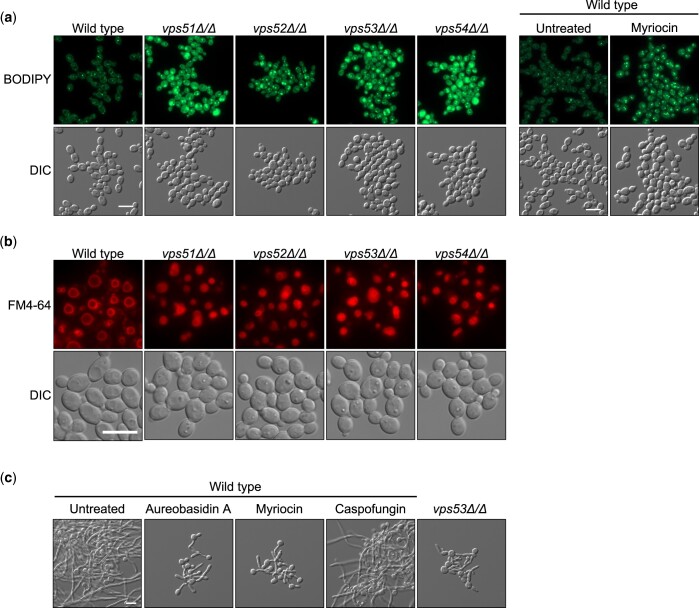
The GARP complex is an important regulator of sphingolipid homeostasis in the model yeast Saccharomyces cerevisiae, as disruption of any Vps subunit results in the accumulation sphingolipid intermediates, alters sterol distribution in the cell, and leads to vacuolar dysfunction (Fröhlich et al. 2015). To confirm the GARP genes were important for sphingolipid homeostasis in C. albicans, we monitored lipid droplet formation in cells. Lipid droplets are cytosolic fat storage organelles that are important for buffering excess lipids in the cell, providing immediate protection from lipotoxicity (Jarc and Petan 2019). GARP complex deficiency resulted in an increase in lipid droplet number and size compared with the wild-type control, similar to what was observed upon pharmacological inhibition of sphingolipid biosynthesis using myriocin (Fig. 3a). In order to visualize cellular membranes, including vacuolar membranes, we stained the GARP mutants with FM4-64. Compared with the wild-type strain, the vacuoles of GARP mutants were small and highly fragmented, indicative of vacuolar dysfunction (Fig. 3b), similar to what was previously observed in both S. cerevisiae and C. albicans (Conibear and Stevens 2000; Park et al. 2009).
Fig. 3.
GARP complex mutants have altered lipid homeostasis, which impairs filamentation. a) Strains were grown to exponential phase prior to staining with BODIPY to visualize lipid droplets. For drug treatment, exponential phase cells were exposed to 5 µg/ml myriocin for 3 h prior to staining with BODIPY. Scale bar represents 10 µm. b) Strains of C. albicans were grown to exponential phase prior to staining with membrane dye FM4-64, which fluoresces red. Scale bar represents 10 µm. c) Wild-type or vps53Δ/Δ C. albicans was grown in the absence or presence 0.025 µg/ml aureobasidin A, 0.4 µg/ml myriocin, or 8 ng/ml caspofungin and grown for 5 h under filament-inducing conditions of elevated temperature at 39°C and imaged. Scale bar represents 10 µm.
Based on these observations we hypothesized that genetic compromise of the GARP complex leads to aberrant sphingolipid homeostasis, resulting in a block in filamentous growth. To test this model, we treated wild-type C. albicans with myriocin or aureobasidin A, inhibitors of sphingolipid biosynthesis, in the presence of elevated temperature as the filament-inducing cue. Both myriocin and aureobasidin A impaired filamentation but did not completely abolish filamentous growth, similar to a GARP complex mutant, suggesting that sphingolipid biosynthesis plays an intermediate role in filamentation (Fig. 3c). Overall, this suggests disruption of sphingolipid homeostasis at least partially contributes to impairment of filamentous growth in GARP complex mutants.
Discussion
The ability to transition between yeast and filamentous forms is a key virulence trait in diverse fungal pathogens. Here, we established the GARP complex as an important regulator of morphogenesis in C. albicans in response to diverse filament-inducing cues, including engulfment by macrophages. Genetic pathway analysis revealed that activation of PKA signaling is not sufficient to induce filamentation in the absence of proper GARP function. Interestingly, overexpression of the filament-specific transcriptional activator UME6 or deletion of the transcriptional repressor NRG1 was sufficient to induce polarized growth in GARP-deficient cells, highlighting complex genetic connections between the GARP complex and core filament-inducing signaling cascades. Moreover, GARP deficiency led to the accumulation of lipid droplets, which is indicative of altered lipid homeostasis and lipotoxicity, leading us to postulate that GARP deficiency alters cellular sphingolipid homeostasis, thereby blocking filamentation in C. albicans.
Lipid homeostasis is crucial for polarized growth of C. albicans (Martin and Konopka 2004; O’Meara et al. 2015). Previous functional genomic analyses revealed that targeting the ergosterol biosynthetic pathway, either genetically or pharmacologically, impairs filamentation in response to serum due to the accumulation of specific sterol intermediates (O’Meara et al. 2015). Other work has shown polarization of lipid rafts, membrane microdomains highly enriched in sterols and sphingolipids, to the hyphal tip is important for hyphal growth in C. albicans (Martin and Konopka 2004). Notably, the manner by which sphingolipid levels regulate the yeast-to-filament transition in C. albicans remains poorly defined. Here, we confirm the GARP complex is required for maintaining lipid homeostasis in C. albicans such that mutations in the GARP complex result in an accumulation of lipid droplets, similar to what is observed with the sphingolipid biosynthesis inhibitor myriocin (Fröhlich et al. 2015). Further, we show that treatment of C. albicans with myriocin blocks filamentation, phenocopying what is observed with GARP complex mutants. Based on these observations, we propose that disruption of sphingolipid homeostasis in response to GARP complex deficiency, leads to impairment of filamentous growth.
The vacuole is a dynamic storage organelle that plays significant roles in many processes ranging from cellular trafficking, ion homeostasis, stress response signaling, adaptation to new environments, and cellular differentiation (Armstrong 2010), and the GARP complex is important for maintaining proper vacuolar integrity in both S. cerevisiae and C. albicans (Conibear and Stevens 2000; Park et al. 2009). In C. albicans, vacuolar function has been implicated in the yeast-to-filament transition as disruption of vacuolar proton-translocating ATPases (V-ATPases) impair vacuolar acidification, filamentous growth, and virulence in C. albicans (Lv et al. 2021). Specifically, deletion of genes encoding subunits of V-ATPases, including TFP1, VMA2, VMA5, VMA4, VMA7, VMA10, VPH1, STV1, VMA3, or VMA6, results in defects in filamentation to variable degrees (Poltermann et al. 2005; Kane 2006; Jia et al. 2014; Rane, Bernardo, et al. 2014; Rane, Hardison, et al. 2014; Zhang et al. 2017; Jia et al. 2018; Kim, Park, et al. 2019). Similarly, molecules that perturb vacuolar pH also block morphogenesis, highlighting the importance of V-ATPases in filamentous growth (Patenaude et al. 2013). Hyphal formation is also coupled to proper vacuolar trafficking (Lv et al. 2021). It has been postulated that disruption of vacuolar trafficking compromises the regulation of turgor pressure required to provide a force for directional hyphal elongation, and prevents necessary factors like V-ATPase subunits from localizing in the vacuole (Lv et al. 2021). Interestingly, overexpression of the hyphal-specific transcription factor UME6 induced filamentous growth even in the absence of a functional GARP complex, highlighting that these mutants are physically capable of undergoing filamentation, and suggesting the GARP complex regulates morphogenesis through alternate mechanisms that are at least in part independent of mechanical forces. Additionally, overexpression of UME6 is not sufficient to induce hyphal growth in V-ATPase mutants, including vma4Δ/vma4Δ or vma10Δ/vma10Δ (Kim, Park, et al. 2019), suggesting that the GARP complex and V-ATPase regulate filamentation through different mechanisms. UME6 expression is induced in response to diverse filament-inducing cues and Ume6 is critical for hyphal extension, as well as maintenance of filamentation (Banerjee et al. 2008). Given that overexpression of UME6 was sufficient to induce filamentation in the GARP complex mutants, it is conceivable that the GARP complex mutants are defective in morphogenesis because they are unable to maintain filamentation. This hypothesis is consistent with our findings as GARP complex mutants can initiate the filamentation program, as evidenced by the formation of short germ tubes in liquid filament-inducing conditions, but these mutants are defective in maintenance of filamentation (Fig. 1). Thus, our findings suggest a model whereby the GARP complex and sphingolipid homeostasis could be important for maintenance of filamentation. Further mechanistic insights into the connection between the GARP complex, sphingolipid biosynthesis, and the yeast-to-hyphal transition will undoubtedly further our understanding of the complex signaling that regulates this important virulence trait.
There is a rising demand for alternative strategies to combat fungal infections with antimicrobial resistance becoming ever more rampant. Antivirulence strategies are promising alternative approaches of antifungal drug development that remain largely unexplored (Vila et al. 2017). Targeting the GARP complex may serve as a viable strategy to attenuate C. albicans virulence and render the pathogen more susceptible to diverse environmental stressors. In fact, previous studies have demonstrated that vps51 mutants display increased susceptibility to environmental stressors, including cell wall, cell membrane, osmotic, and oxidative assaults. Additionally, VPS51 is required for virulence in a mouse model of oropharyngeal candidiasis (Park et al. 2009; Liu et al. 2014). Given that the GARP complex is conserved from yeasts to humans (Fröhlich et al. 2015), one important challenge for exploiting the GARP complex as an antifungal target will be the development of fungal-selective inhibitors. Recent efforts for the development of fungal-selective inhibitors of Hsp90, calcineurin, and mitochondrial bc1 complex have shown great promise (Vincent et al. 2016; Juvvadi et al. 2019; Whitesell et al. 2019). Overall, this work furthers our understanding of the complex cellular circuitry that regulates filamentation in this important human fungal pathogen and highlights a great opportunity in exploiting the GARP complex to combat C. albicans infections.
Supplementary Material
Acknowledgments
We thank all members of the Cowen lab for helpful discussions. We thank Dr Zhongle Liu for constructing the CaLC6642 strain used in this study.
Funding
SH is supported by Ontario Graduate Scholarship. LEC is supported by the Canadian Institutes of Health Research (CIHR) Foundation grant (FDN-154288) and a National Institutes of Health (NIH) R01 grant (R01AI127375). LEC is a Canada Research Chair (Tier 1) in Microbial Genomics & Infectious Disease and co-Director of the CIFAR Fungal Kingdom: Threats & Opportunities program.
Conflicts of interest
LEC is a cofounder and shareholder in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics, and a Science Advisor for Kapoose Creek, a company that harnesses the therapeutic potential of fungi.
Contributor Information
Saif Hossain, Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada.
Nicole Robbins, Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada.
Leah E Cowen, Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada.
Data Availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.
Literature cited
- Armstrong J. Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 2010;20(10):580–585. doi: 10.1016/j.tcb.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, López-Ribot JL, Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19(4):1354–1365. doi: 10.1091/mbc.e07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. JoF. 2017;3(4):57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20(17):4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, López-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci USA. 2009;106(2):599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11(1):305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao C, Zheng Q, Huang G. The regulatory subunit of protein kinase A (Bcy1) in Candida albicans plays critical roles in filamentation and white-opaque switching but is not essential for cell growth. Front Microbiol. 2016;7:2127. doi: 10.3389/fmicb.2016.02127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp E, Walther A, Lépine G, Leon Z, Mullick A, Raymond M, Wendland J, Whiteway M. Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol Microbiol. 2010;75(5):1182–1198. doi: 10.1111/j.1365-2958.2009.07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Gurr SJ, Cuomo CA, Blehert DS, Jin H, Stukenbrock EH, Stajich JE, Kahmann R, Boone C, Denning DW, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio. 2020;11(3):e00449-20. doi: 10.1128/mBio.00449-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich F, Petit C, Kory N, Christiano R, Hannibal-Bach H-K, Graham M, Liu X, Ejsing CS, Farese RV, Walther TC. The GARP complex is required for cellular sphingolipid homeostasis. eLife. 2015;4:e08712. doi: 10.7554/eLife.08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain S, Veri AO, Liu Z, Iyer KR, O’Meara TR, Robbins N, Cowen LE. Mitochondrial perturbation reduces susceptibility to xenobiotics through altered efflux in Candida albicans. Genetics. 2021;219(2):iyab095. doi: 10.1093/genetics/iyab095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarc E, Petan T. Lipid droplets and the management of cellular stress. Yale J Biol Med. 2019;92(3):435–452. [PMC free article] [PubMed] [Google Scholar]
- Jia C, Yu Q, Xu N, Zhang B, Dong Y, Ding X, Chen Y, Zhang B, Xing L, Li M. Role of TFP1 in vacuolar acidification, oxidative stress and filamentous development in Candida albicans. Fungal Genet Biol. 2014;71:58–67. doi: 10.1016/j.fgb.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Jia C, Zhang K, Zhang D, Yu Q, Zhao Q, Xiao C, Dong Y, Chu M, Li M. Roles of VPH2 and VMA6 in localization of V-ATPase subunits, cell wall functions and filamentous development in Candida albicans. Fungal Genet Biol. 2018;114:1–11. doi: 10.1016/j.fgb.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Juvvadi PR, Fox D, Bobay BG, Hoy MJ, Gobeil SMC, Venters RA, Chang Z, Lin JJ, Averette AF, Cole DC, et al. Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents. Nat Commun. 2019;10(1):1–18. doi: 10.1038/s41467-019-12199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70(1):177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Iyer KR, Pardeshi L, Muñoz JF, Robbins N, Cuomo CA, Wong KH, Cowen LE. Genetic analysis of Candida auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio. 2019;10(1):e02529-18. doi: 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Park YK, Joo YJ, Chun YJ, Hwang JY, Baek J-H, Kim J. Subunits of the vacuolar H+-ATPase complex, Vma4 and Vma10, are essential for virulence and represent potential drug targets in Candida albicans. Fungal Biol. 2019;123(10):709–722. doi: 10.1016/j.funbio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13(2):148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Liu Y, Solis NV, Heilmann CJ, Phan QT, Mitchell AP, Klis FM, Filler SG. Role of retrograde trafficking in stress response, host cell interactions, and virulence of Candida albicans. Eukaryot Cell. 2014;13(2):279–287. doi: 10.1128/EC.00295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Yan L, Jiang Y. The importance of vacuolar ion homeostasis and trafficking in hyphal development and virulence in Candida albicans. Front Microbiol. 2021;12:779176. doi: 10.3389/fmicb.2021.779176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SW, Konopka JB. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell. 2004;3(3):675–684. doi: 10.1128/EC.3.3.675-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere. 2016;1(3):e00130-16. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K, Jannace TF, Si H, Veeramah KR, Haley JD, Konopka JB. Integrative multi-omics profiling reveals cAMP-independent mechanisms regulating hyphal morphogenesis in Candida albicans. PLoS Pathog. 2021;17(8):e1009861. doi: 10.1371/journal.ppat.1009861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón P, Kasper L, Hube B. Thriving within the host: Candida spp. interactions with phagocytic cells. Med Microbiol Immunol. 2013;202(3):183–195. doi: 10.1007/s00430-013-0288-z. [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20(17):4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 2017;15(2):96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun. 2015;6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Liu Y, Solis N, Spotkov J, Hamaker J, Blankenship JR, Yeaman MR, Mitchell AP, Liu H, Filler SG. Transcriptional responses of Candida albicans to epithelial and endothelial cells. Eukaryot Cell. 2009;8(10):1498–1510. doi: 10.1128/EC.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrino SM, Si H, Naseem S, Groudan K, Gardin J, Konopka JB. cAMP-independent signal pathways stimulate hyphal morphogenesis in Candida albicans. Mol Microbiol. 2017;103(5):764–779. doi: 10.1111/mmi.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude C, Zhang Y, Cormack B, Köhler J, Rao R. Essential role for vacuolar acidification in Candida albicans virulence. J Biol Chem. 2013;288(36):26256–26264. doi: 10.1074/jbc.M113.494815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36(1):1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- Poltermann S, Nguyen M, Günther J, Wendland J, Härtl A, Künkel W, Zipfel PF, Eck R. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology (Reading). 2005;151(Pt 5):1645–1655. doi: 10.1099/mic.0.27505-0. [DOI] [PubMed] [Google Scholar]
- Rane HS, Bernardo SM, Hayek SR, Binder JL, Parra KJ, Lee SA. The contribution of Candida albicans vacuolar ATPase subunit V1B, encoded by VMA2, to stress response, autophagy, and virulence is independent of environmental pH. Eukaryot Cell. 2014;13(9):1207–1221. doi: 10.1128/EC.00135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane HS, Hardison S, Botelho C, Bernardo SM, Wormley F, Lee SA. Candida albicans VPS4 contributes differentially to epithelial and mucosal pathogenesis. Virulence. 2014;5(8):810–818. doi: 10.4161/21505594.2014.956648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Wang C-W, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem. 2003;278(7):5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2(5):1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75(2):213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA Signaling. Curr Biol. 2009;19(8):621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HRB. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277(50):48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9(10):737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, Lopez-Ribot JL. Targeting Candida albicans filamentation for antifungal drug development. Virulence. 2017;8(2):150–158. doi: 10.1080/21505594.2016.1197444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BM, Langlois J-B, Srinivas R, Lancaster AK, Scherz-Shouval R, Whitesell L, Tidor B, Buchwald SL, Lindquist S. A fungal-selective cytochrome bc1 inhibitor impairs virulence and prevents the evolution of drug resistance. Cell Chem Biol. 2016;23(8):978–991. doi: 10.1016/j.chembiol.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakade RS, Kramara J, Wellington M, Krysan DJ. Candida albicans filamentation does not require the cAMP-PKA pathway in vivo. mBio. 2022;13(3):e0085122. doi: 10.1128/mbio.00851-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Robbins N, Huang DS, McLellan CA, Shekhar-Guturja T, LeBlanc EV, Nation CS, Hui R, Hutchinson A, Collins C, et al. Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus. Nat Commun. 2019;10(1):1–17. doi: 10.1038/s41467-018-08248-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Jia C, Yu Q, Xiao C, Dong Y, Zhang M, Zhang D, Zhao Q, Zhang B, Li M. Contribution of VMA5 to vacuolar function, stress response, ion homeostasis and autophagy in Candida albicans. Future Microbiol. 2017;12:1147–1166. doi: 10.2217/fmb-2017-0029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.