Abstract
Chaperones facilitate the folding of other (“client”) proteins and can thus affect the adaptive evolution of these clients. Specifically, chaperones affect the phenotype of proteins via two opposing mechanisms. On the one hand, they can buffer the effects of mutations in proteins and thus help preserve an ancestral, premutation phenotype. On the other hand, they can potentiate the effects of mutations and thus enhance the phenotypic changes caused by a mutation. We study that how the bacterial Hsp90 chaperone (HtpG) affects the evolution of green fluorescent protein. To this end, we performed directed evolution of green fluorescent protein under low and high cellular concentrations of Hsp90. Specifically, we evolved green fluorescent protein under both stabilizing selection for its ancestral (green) phenotype and directional selection toward a new (cyan) phenotype. While Hsp90 did only affect the rate of adaptive evolution transiently, it did affect the phenotypic effects of mutations that occurred during adaptive evolution. Specifically, Hsp90 allowed strongly deleterious mutations to accumulate in evolving populations by buffering their effects. Our observations show that the role of a chaperone for adaptive evolution depends on the organism and the trait being studied.
Keywords: chaperone, Hsp90, protein evolution
Introduction
Mutations are essential for the evolution of new traits and of proteins with new functions. However, the vast majority of mutations negatively affect a protein’s function (Bershtein et al. 2006; Eyre-Walker and Keightley 2007), thereby constraining its evolution (DePristo et al. 2005; Zeldovich et al. 2007). Many such deleterious mutations can impair a protein’s ability to spontaneously fold into a structure that is necessary for its function (Covalt et al. 2001; Chakraborty et al. 2010). In addition, they can make a protein misfold and form nonfunctional and potentially toxic structures (Fersht 1997; Winklhofer et al. 2008; Hartl 2017). Mutations that alter the biochemical activity of a protein (neofunctionalizing mutations) may destabilize the protein (Tokuriki et al. 2008; Fromer and Shifman 2009; Studer et al. 2014). Conversely, high thermodynamic stability and fast folding of a protein, may favor its adaptive evolution (Bloom et al. 2006; Zheng et al. 2020).
A cell is a crowded environment brimming with proteins that occur at high concentrations. To ensure protein quality in this environment, organisms have evolved various mechanisms embodied in various chaperone proteins that help other proteins fold. These mechanisms include the stabilization of nascent unfolded polypeptides, the acceleration of protein folding, the prevention of misfolding, and the refolding of misfolded proteins (Kim et al. 2013; Saibil 2013; Ries et al. 2017; Imamoglu et al. 2020). Chaperones are especially active when cells are subject to stressors like heat, which destabilize proteins (Richter et al. 2010). Many chaperones are thus also known as heat shock proteins (Hsp). Like environmental stressors, mutations can also destabilize proteins, and chaperones can mitigate the destabilizing effects of such mutations by helping proteins to fold correctly (Tokuriki and Tawfik 2009; Wyganowski et al. 2013; Sadat et al. 2020).
By mitigating or “buffering” the effect of deleterious mutations, chaperones can help organisms that accumulate such mutations, maintain high fitness (Fares et al. 2002; Sabater-Muñoz et al. 2015; Aguilar-Rodríguez et al. 2016; Karras et al. 2017; Phillips et al. 2018). For example, Escherichia coli populations with multiple random genomic mutations grow faster and are less likely to go extinct when the two chaperones DnaK and GroEL are overexpressed (Fares et al. 2002; Sabater-Muñoz et al. 2015; Aguilar-Rodríguez et al. 2016). By buffering the effects of mutations, chaperones can also facilitate the evolution of new phenotypes (Rutherford and Lindquist 1998; Cowen and Lindquist 2005; Tokuriki and Tawfik 2009; Wyganowski et al. 2013; Agozzino and Dill 2018; Alvarez-Ponce et al. 2019). For example, they can buffer the destabilizing effects of neofunctionalizing mutations (Tokuriki and Tawfik 2009; Wyganowski et al. 2013). In sum, chaperones can play an important role in adaptive protein evolution.
In this study, we explore how the chaperone Hsp90 modifies the phenotypic effects of genetic variation that arises during experimental protein evolution. Like other chaperones, Hsp90 can accelerate protein folding (Kim et al. 2013). It does so by forming a dimer that provides an interface for unfolded proteins to bind and fold properly. More specifically, it takes over a partially folded protein molecule from the chaperone Hsp70, and facilitates its further folding (Genest et al. 2011; Morán Luengo et al. 2018; Wang et al. 2022).
Hsp90 is especially interesting for protein evolution, because it seems to affect protein mutations in two opposite ways. First, like other chaperones, it can buffer the effect of mutations (Rutherford and Lindquist 1998; Queitsch et al. 2002; Karras et al. 2017). Second, it can also enhance or exaggerate the effect of mutations. This phenomenon is also called potentiation (Xu et al. 1999; Cowen and Lindquist 2005; Whitesell et al. 2014). One pertinent study investigated the effects of Hsp90 on mutations that affect the cellular morphology of the yeast Saccharomyces cerevisiae (Geiler-Samerotte et al. 2016). It found that Hsp90 potentiated the effects of most mutations, but buffered the effects of some mutations. These buffered mutations preferentially survive selection, and gradually accumulate in populations during many generations of evolution. Although these two phenomena—buffering and potentiation—seem opposite to each other in their effects, we note that the underlying molecular mechanisms may be identical. Specifically, a chaperone may promote folding of its protein clients irrespective of the phenotypes they manifest. This enhancement in protein folding can minimize the effect of destabilizing mutations (buffering), and it can enhance (potentiate) the effect of mutations that change the protein’s activity. Many mutations that change a protein’s activity simultaneously reduce its stability and original activity (Tokuriki et al. 2008; Fromer and Shifman 2009; Studer et al. 2014). Thus by enhancing protein folding, a chaperone can preserve an ancestral phenotype but can also enhance a novel phenotype. Therefore, a chaperone’s effect on a protein phenotype can be called buffering or potentiation, depending on the point of view of the observer.
Hsp90 occurs in both eukaryotes and prokaryotes, and several key amino acids in the functional domains of the chaperone are conserved (Chen et al. 2006; Schopf et al. 2017). The vast majority of studies on Hsp90 and its role on adaptive evolution have been performed on eukaryotic Hsp90, in model systems such as yeast (S. cerevisiae) and animal cells (Rutherford and Lindquist 1998; Xu et al. 1999; Queitsch et al. 2002; Whitesell et al. 2014; Geiler-Samerotte et al. 2016; Karras et al. 2017; Schopf et al. 2017; Dorrity et al. 2018). Because we know comparatively little about prokaryotic Hsp90 and its role (if any) in protein evolution, we focus here on E. coli Hsp90. We note that despite the substantial sequence conservation of Hsp90 orthologs, different orthologs can affect protein folding differently (Jahn et al. 2018). Different orthologs may thus also differ in their effect on genetic mutations.
To characterize the phenotypic effects of Hsp90 on mutations in greater molecular detail than would be possible for phenotypes that are complex and affected by multiple genes (Geiler-Samerotte et al. 2016), we studied the effect of Hsp90 on a single evolving protein. Pertinent previous work exists, but it has focused on a different chaperone (GroEL/S; Tokuriki and Tawfik 2009; Wyganowski et al. 2013). Our work also overcomes a limitation of previous work, which studied only a modest number of ∼200 evolving protein genotypes (Tokuriki and Tawfik 2009; Wyganowski et al. 2013). Instead, we evolved populations of more than 105 proteins, which can explore a larger region of genotype space. More specifically, we studied the effect of bacterial Hsp90 (HtpG) on the directed evolution of green fluorescent protein (GFP). Our choice of GFP was motivated by three main considerations. First, the fluorescent phenotype of GFP can be easily quantified in a high-throughput manner and at single cell resolution using flow cytometry. Second, this phenotype also allowed us to perform selection in a highly controlled manner using fluorescence activated cell sorting (FACS). Third, as a widely used biomarker, GFP is presumably nontoxic to E. coli. It is also not endogenous to E. coli and thus interferes minimally with the bacterium’s proteome and physiology.
Another pertinent study focuses on the yeast Hsp90, and uses a high-throughput selection experiment to show that the chaperone can differently influence the phenotypic outcomes of different mutations on a single protein (Dorrity et al. 2018). Although our work is similar to this study in some aspects, such as in the use of random mutagenesis and high-throughput sequencing, it fundamentally differs in the main question we ask. Instead of focusing on the chaperone’s effect on specific mutations, we aim to understand how the chaperone influences protein evolution. To this end we analyze the phenotypes and the genotypes of evolving populations of GFP variants at two different expression levels of Hsp90, during multiple rounds of directed evolution by (PCR-mediated) mutation and (FACS-mediated) selection. Specifically, we analyzed the phenotypic evolution of these populations through flow cytometry and studied their genotypic evolution through high-throughput single-molecule real-time (SMRT) sequencing. Furthermore, we engineered selected mutants into ancestral GFP to study their phenotype in detail. We found that Hsp90 generally increases the activity of GFP variants. That is, it buffers the phenotypic effect of deleterious variants and potentiates the phenotypic effects of beneficial variants. However, in our evolving populations, Hsp90-mediated buffering far outweighs potentiation.
Materials and methods
Mutagenesis
We used nucleotide analog mediated error-prone PCR for mutagenesis of GFP with the following reaction mixture composition: 150 nM each of the nucleotide analogs 8-oxodeoxyguanosine triphosphate (8-oxo-dGTP, Trilink Biotechnologies) and 6-(2-deoxy-beta-d-ribofuranosyl)-3,4-dihydro-8H-pyrimido-[4,5-C] [1,2]oxazin-7-one triphosphate (dPTP, Trilink Biotechnologies), 200 nM each of forward and reverse primers (pHtpG-Mut-F and pHtpG-Mut-R; Supplementary Table 1), 400 µM of each dNTP (Thermo Scientific), 1× ThermoPol buffer (NEB) and 25 units/ml of Taq polymerase (NEB). We prepared 100 µl of the reaction mixture with 5 ng of plasmid DNA as the template (∼6 × 108 molecules), and split the reaction mixture into two 50 µl aliquots for efficient heat transfer during PCR. We performed the PCR with the following program: initial denaturation at 95°C for 5 min, 25 cycles of amplification with 95°C for 30 s, 56°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min.
We column-purified the PCR products (QIAquick PCR purification kit, QIAGEN), digested the purified amplicons using SalI-HF, SacI-HF, and DpnI, and column-purified the digested DNA again. Using T4 DNA ligase (NEB), we ligated the purified digested amplicons with digested (SalI, SacI) pHtpG7-rplN-GFP, replacing the ancestral GFP gene with the mutagenized variants. We desalted the ligated plasmid library using ethanol purification, and introduced this library into electrocompetent BW27784 E. coli cells with the glycerol–mannitol method using electroporation, as previously described (Iyengar and Wagner 2022). We maintained the transformed library in a liquid medium (LB-chl), plating a 1:500 dilution on LB-chl agar plate to estimate library size. Throughout our evolution experiments, we maintained a minimum library size of 105 transformants.
With this method, we obtained ∼1–2 nucleotide mutations per amplicon corresponding to ∼0.95 amino acid changes per GFP protein. We performed mutagenesis on ancestral GFP at every round (generation) of directed evolution (phases 1 and 2) to ensure that the mutation rate stays in this range. We also estimated the frequencies of different nucleotide substitutions, which was in accordance with what we had previously observed (Iyengar and Wagner 2022) AT → GC: 76%, GC → AT: 14.5%, AT → TA: 7%, AT → CG: 2%, GC → CG: 0.5%, and GC → TA: 0%.
Selection of transformed cells using FACS
We performed directed evolution in four replicate populations in which Hsp90 was expressed from the plasmid (H+), along with four control populations in which it was not expressed from the plasmid (H−). To prepare the cells for selection, we inoculated 4 ml of LB-chl in a 20-ml glass tube with 80 µl of the transformed library. We incubated the cells at 37°C with shaking at 220 rpm. After 60 min of incubation, we induced Hsp90 expression in H+ populations by adding l-arabinose to a final concentration of 0.1 µg/ml and continued the incubation. After 11 h of growth postinoculation, we harvested the cells from 700 µl of the suspension. We washed the cells once in cold PBS to remove traces of LB and resuspended them again in 1 ml cold PBS. We transferred 100 µl of this suspension to 1 ml cold PBS in a 5-ml polystyrene tube (Falcon), and performed cell sorting on a BD FACSAriaIII cell sorter with the following photomultiplier tube (PMT) voltages for different channels—FSC: 478 V, SSC: 282 V, FITC: 480 V, and AmCyan: 493 V. We excluded debris and other small particles by setting a threshold of 1,000 (arbitrary units) for FSC-H and SSC-H.
We used the FITC channel (488 nm excitation and 530 ± 15 nm emission) for measuring green fluorescence, and the AmCyan channel (405 nm excitation and 510 ± 25 nm emission) for measuring cyan fluorescence. We quantified the autofluorescence of cells by measuring the fluorescence of untransformed (nonplasmid bearing) cells in each channel. To select variants with green fluorescence, we sorted cells with an FITC-H value higher than the maximum FITC-H value of the untransformed cells (Supplementary Fig. 3). During phase 2 evolution we selected for the new phenotype of cyan fluorescence. Because green and cyan fluorescence are correlated, we selected variants that showed a higher cyan fluorescence relative to green fluorescence than the ancestral GFP (Supplementary Fig. 8a). This procedure ensures that selected variants show cyan fluorescence that cannot be merely explained by enhanced green fluorescence.
We sorted 105 cells and expanded their population by an overnight incubation at 37°C with shaking at 220 rpm. We then performed a second round of sorting to minimize contamination from cells that did not meet the selection criteria. To this end, we repeated the above procedure, treating the cell populations from the first round of sorting as our new library of variants. After overnight growth following the second round of sorting, we prepared glycerol stocks and isolated variant plasmids from each cell population. The plasmid libraries thus obtained served as templates for the next round of directed evolution.
Analysis of flow cytometry data
We used flow cytometry to analyze the phenotype (green and cyan fluorescence) of evolving populations. We prepared samples for flow cytometry using the same procedure as we described in the previous section on FACS based selection. We analyzed the cells on a BD LSR FortessaII flow cytometer with PMT voltages of different channels set to the same values as described above for FACS analysis. We recorded 100,000 events and analyzed the data using the R package flowCore (Ellis et al. 2019).
We measured the fluorescence of evolved populations after every round of directed evolution. To analyze the change in fluorescence over multiple rounds we fitted linear models using the R stats package (v3.4.4; R Core Team 2018) with median fluorescence as the response variable. To this end, we first fitted a linear model for each population separately (H+ and H−) using only time (round of evolution) as the independent variable, which allowed us to determine if fluorescence changes significantly over time. Specifically, we used the expression, Fluorescence ∼ time, in this fitting procedure.
Next, we fitted a model to include the state of Hsp90 overexpression (+ or –) as an additional predictor that may interact with time. This allowed us to determine if chaperone overexpression significantly affects the rate of fluorescence change. Specifically, we used the expression, Fluorescence ∼ time * condition, for this fitting procedure. For all models, we analyzed the significance of the fit using the anova function from the R stats package (v3.4.4; R Core Team 2018). The null hypotheses in these models is that the factors (time and Hsp90 overexpression) do not improve the fit significantly, i.e. that these factors do not affect fluorescence.
For analyzing the difference in fluorescence between the two populations (H+ and H−) at any one generation, we compared the median fluorescence (4 × 4) values using a Mann–Whitney U test.
Preparation of sequencing libraries
We sequenced the GFP coding sequence from plasmids isolated after every round (generation) of evolution using SMRT sequencing (Pacific Biosciences, PacBio). We used a multiplex amplicon sequencing procedure, as described previously (Iyengar and Wagner 2022). This procedure requires two rounds of PCR. In the first round, we amplified GFP coding regions using primers containing a “universal” sequence provided by PacBio (HtpG-ORF-PacBio-F and HtpG-ORF-PacBio-R; Supplementary Table 1). Using the products of this PCR as a template, we performed a second round of PCR using barcoded primers (Pacific Biosciences 2019a).
We determined the purity of the PCR products through agarose gel electrophoresis and found that most of the products were clean, without nonspecific bands or primer dimers. We purified these products using a QIAquick PCR purification kit (QIAGEN). For the few samples that contained high amounts of primer dimers, we purified the products using gel extraction (QIAGEN). We measured the concentration of the purified products on a Qubit 3.0 fluorometer (Life Technologies). For GFP libraries from each phase of evolution (1 and 2), we pooled 130 ng of every barcoded product and purified the two resulting pools using gel extraction.
For subsequent DNA sequencing we used two PacBio Sequel SMRT cells for the two pooled samples (phases 1 and 2). Sequencing was performed on the PacBio Sequel system (3.0 Chemistry) by the Functional Genomics Center Zurich.
Processing of raw data
We obtained ∼600,000 raw zero mode waveguide (ZMW) reads from both SMRT cells. We determined the consensus of circular sequences (CCS) from the raw data (subreads) using the ccs (v3.4.1) application from the PacBio SMRTlink package (Pacific Biosciences 2019b), with the following parameters: minimum length = 750, maximum length = 1,500, minimum passes = 3, minimum predicted accuracy = 99%. We kept the other application parameters at their default value. This procedure allowed us to use ∼55% of the ZMW reads for further analysis. We demultiplexed the post-CCS reads using lima (v1.9.0, SMRTlink), and aligned them to the reference sequence (GFPmut2) using minimap2 (v2.15-r905, SMRTLink). We analyzed the alignments in SAM format using a custom awk script. PacBio sequencing is known to produce a large number of artifactual indels (Goodwin et al. 2016; Giordano et al. 2017; Watson and Warr 2019). Our sequencing data also contained many indels. We confirmed that these were sequencing artifacts by Sanger-sequencing 20 random single clones, and excluded any indels from further analysis. We obtained lists of single mutations as well as genotypes with multiple mutations, along with their raw counts and frequencies, from the data thus filtered.
Calculation of mutation enrichment
For each round of evolution, we compared the enrichment of mutations under chaperone overexpression relative to the nonoverexpression controls using generalized linear models (GLM; R stats package v3.4.4; R Core Team 2018). Specifically, we fitted a GLM with a logit link function (binomial model) using mutation counts as the response variable, and the state of Hsp90 overexpression (+ or –) as the predictor variable. We analyzed the goodness of fit of the full model (slope + intercept) with respect to a reduced (intercept only) model, using the anova function from the R stats package v3.4.4. This function performs an analysis of deviance on the models with a likelihood ratio test (LRT) and determines if the additional parameters (slope in our case) significantly improve the fit. Here, a positive value of the slope denotes enrichment of the mutation in the chaperone overexpression condition. We adjusted the P-values thus obtained for multiple testing using a Bonferroni correction. We used the mutations with a corrected P-value of <0.05 for subsequent analyses.
Construction and analysis of specific mutants
We used PCR-based site-directed mutagenesis (Liu and Naismith 2008) to engineer specific mutations into the GFP gene, using a technique described previously (Iyengar and Wagner 2022).
We analyzed the fluorescence of the engineered mutants using flow cytometry, preparing samples as described in earlier sections. Specifically, we performed the fluorescence measurements in three biological replicate samples each for every mutant, with and without Hsp90 overexpression. To determine the effect of a mutation on the fluorescence of ancestral GFP, we compared the fluorescence distribution of the mutant with that of ancestral GFP using a Mann–Whitney U test. We considered a mutation as significantly deleterious to fluorescence only if the median fluorescence of the mutant was consistently and statistically significantly (Mann–Whitney U test, P < 0.05) lower than that of ancestral GFP in all the three replicate measurements. We identified beneficial mutations analogously.
To understand the effect of Hsp90 on fluorescence, we calculated the fold change in fluorescence of a mutant when Hsp90 was overexpressed relative to when it was not. Since we had three unpaired replicate measurements in each condition (overexpression and no overexpression), we calculated the fluorescence fold change for all the nine possible permutations of the sample pairs. We determined whether an Hsp90 mediated change in fluorescence was statistically significant by performing a Mann–Whitney U test for each pair (overexpression and no overexpression), and using a Bonferroni correction for multiple testing. In addition, we considered a fluorescence change to be significant only when for each pair the nature of the difference (reduction or increase in fluorescence) was the same in all pairs.
Results
To find out whether (and how) the chaperone Hsp90 affects the directed evolution of GFP, we first constructed a plasmid that expresses Hsp90 from the E. coli htpG gene downstream of an arabinose-inducible promoter. The same plasmid expresses GFP constitutively (Supplementary Fig. 1). This plasmid allowed us to study the evolution of GFP at different expression levels of Hsp90. Although Hsp90 is nonessential for E. coli under laboratory growth conditions (Bardwell and Craig 1988; Chen et al. 2006), we reasoned that more chaperone may be needed to cope with the additional burden of misfolded GFP variants. Specifically, if GFP is in excess and Hsp90 is present in limiting amounts, then the chaperone may not be available to bind to and fold all GFP molecules. Therefore, we performed our experiments for two expression levels of Hsp90. The first was the amount of chaperone expressed from the endogenous chromosomal htpG gene. The second was a higher amount, which we achieved by arabinose-induced expression of plasmid-borne htpG. We use the term Hsp90 overexpression to refer to this plasmid-borne expression throughout. We note that our ancestral GFP is a likely client of Hsp90, because its fluorescence increases upon chaperone overexpression (Supplementary Fig. 2).
Experimental design
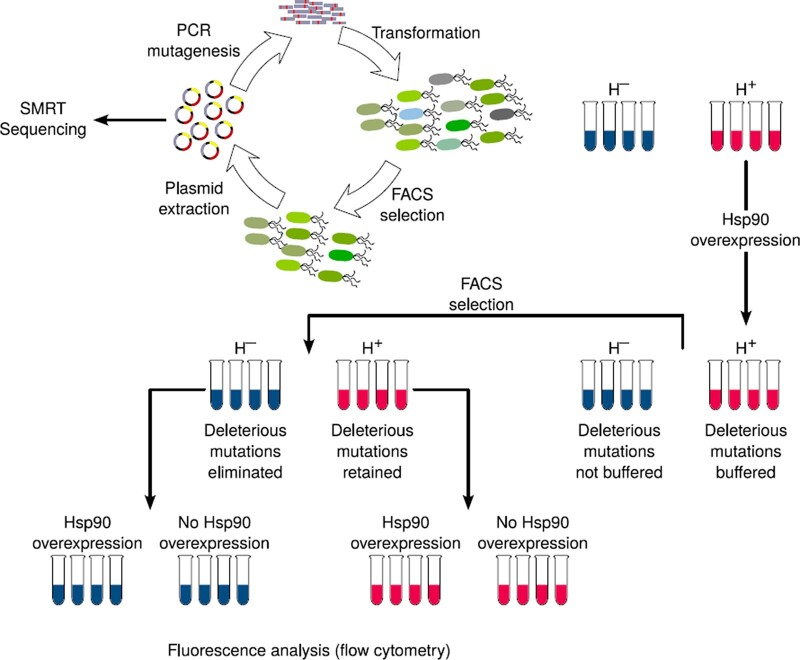
We performed multiple rounds (“generations”) of directed GFP evolution in four replicate populations with (H+) and four populations without (H−) overexpression of Hsp90 (Fig. 1). In each generation, we used error-prone PCR to introduce ∼1–2 random mutations per GFP coding region, corresponding to ∼0.95 amino acid changes per GFP protein. We used FACS to select GFP-expressing cells to survive into the next generation. Our evolving populations were large (≥105 individuals) which implies that genetic drift was negligible on the time scale of our experiment (Messer 2016).
Fig. 1.
Directed evolution of GFP with stabilizing selection for ancestral (green) fluorescence. We selected cells for green fluorescence in phase 1 (and for cyan fluorescence in phase 2). Each phase consisted of five rounds (generations) of directed evolution. We transiently overexpressed Hsp90 in H+ populations prior to selection, whereas we never overexpressed it in H- populations at any stage of evolution. We sequenced surviving GFP gene variants after each generation using SMRT sequencing and analyzed their phenotype using flow cytometry.
A chaperone might affect evolution differently during stabilizing or purifying selection for an ancestral phenotype than during directional selection on a new phenotype. To find out whether this is the case, we performed the evolution experiment in two phases. In phase 1, we evolved GFP under weak stabilizing selection for the ancestral phenotype of green fluorescence. In phase 2, we evolved GFP under directional selection for the novel phenotype of cyan fluorescence. Our analysis focuses mostly on phase 1, because phase 2 yielded similar observations (Supplementary Sections 8–12 and Supplementary Figs. 8–13). To determine a suitable strength of selection for both phases, we reasoned that stringent selection for high fluorescence may enrich mutants that fold well on their own, and that may thus not require chaperone assistance. Therefore, we employed weak selection in both phases. Specifically, we required that surviving cells fluoresce at an intensity that is higher than the background (autofluorescence) of cells that do not express GFP (Supplementary Fig. 3). In both phases, we performed five rounds of directed evolution.
Hsp90 buffers the effect of fluorescence-reducing mutations during weak selection
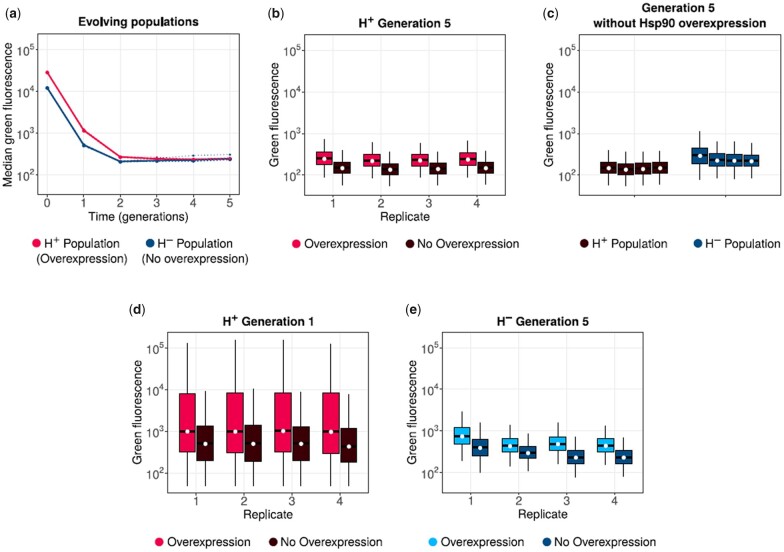
Most mutations destabilize a protein and are deleterious to its phenotype (Supplementary Fig. 3; Bershtein et al. 2006; Eyre-Walker and Keightley 2007). Since we used weak selection in our evolution experiments, many such destabilizing mutations are likely to accumulate in our evolving populations. This accumulation is expected to cause a steady decay of fluorescence. To test this hypothesis, we measured the distribution of green fluorescence for 105 single cells from each replicate H+ and H− populations after every generation of phase 1 evolution. Specifically, we measured the fluorescence of H+ populations with Hsp90 overexpression, and that of H− populations without Hsp90 overexpression, in order to match the conditions in which these populations evolved. Green fluorescence indeed decreased in all populations over time (Fig. 2a). However, this decrease in fluorescence initially differed between the H+ and H− populations. More precisely, Hsp90 overexpression caused a significantly higher median fluorescence in H+ population relative to H− populations in the first two generations (one-tailed Mann–Whitney U test, P = 0.014). However, this difference disappeared thereafter. By generation 3, both H+ and H− populations had reached statistically indistinguishable median green fluorescence (two-tailed Mann–Whitney U test, P > 0.9).
Fig. 2.
Hsp90 buffers the effect of fluorescence-reducing mutations. a) Green fluorescence (arbitrary units) of H+ (red) and H- (blue) populations at different generations (horizontal axis) of phase 1 directed evolution (post selection), in their respective selection environments. Dotted lines denote the median fluorescence of individual replicates, and the solid line denotes the median fluorescence value when data from all replicate populations are pooled. Generation 0 refers to the fluorescence of ancestral GFP. Box plots in (b)–(e) show the green fluorescence distribution of different populations (postselection): the boxes extend from the first quartile to the third quartile, and the whiskers have a length equal to the interquartile range (1 × IQR). The black horizontal line and the white circle in the center of each box denote the median of the corresponding distribution. b, d) The fluorescence distribution of H+ populations at generation 5 and generation 1, respectively. In both panels, the paired red and brown boxes denote the fluorescence distributions of the same population with and without Hsp90 overexpression, respectively. c) The fluorescence distribution of 5th generation H+ (brown) and H- (dark blue) populations (all replicates) in the absence of Hsp90 overexpression. e) shows the fluorescence distribution of 5th generation H- populations with (light blue) and without (dark blue) Hsp90 overexpression.
Although Hsp90 overexpression did not help retain higher fluorescence in H+ populations relative to H− populations, the chaperone might still affect the phenotype of protein variants in H+ populations. On the one hand, Hsp90 might help buffer the effect of destabilizing mutations and thus reduce this detrimental effect on fluorescence. In consequence, such mutations might accumulate in a population. On the other hand, the chaperone might potentiate (enhance) the effects of fluorescence-reducing mutations and lead to their faster elimination from a population. We note that in both scenarios, the combination of chaperone effects and evolutionary dynamics make a prediction of the rate of fluorescence decay difficult. For example, a faster accumulation of fluorescence-reducing mutations for a buffering chaperone in a H+ population may reduce the net beneficial effect of buffering and may cause fluorescence to decay at a similar rate as in an H− population.
To find out if and how Hsp90 affects the phenotypes of segregating variants, and whether it predominantly buffers or potentiates their effects, we asked whether the fluorescence of H+ populations is contingent on Hsp90 overexpression. To this end, we measured the fluorescence distribution of H+ populations at the end of phase 1 evolution, both with and without Hsp90 overexpression (Fig. 2b). We found that the fluorescence of all replicate populations was significantly higher (Mann–Whitney U test under Hsp90 overexpression, suggesting that the chaperone indeed buffers the effect of fluorescence-reducing mutations. Specifically, the median population fluorescence was between 41% and 49% higher when the chaperone was overexpressed, depending on the population.
We also found that H− populations had 38–57% higher fluorescence than H+ populations in the absence of chaperone overexpression (Mann–Whitney U test ; Fig. 2c). This finding suggests that H+ and H− populations accumulate a different spectrum of genetic variants, otherwise the fluorescence of H+ and H− should be similar at similar expression levels of the chaperone. This observation also indicates that H+ populations accumulate mutations whose fluorescence-reducing effect in the aggregate is greater than in H− populations (Fig. 2c). A principal component analysis of GFP genotypes supports this hypothesis (Supplementary Fig. 4), as does our more detailed analysis of individual genetic variants in the next section.
A previous study (Geiler-Samerotte et al. 2016) has shown that yeast Hsp90 potentiates the effects of newly acquired mutations, whereas it buffers the effects of mutations that have evolved under Hsp90 expression for many generations. To determine if an analogous phenomenon exists in our experiments, we measured the fluorescence distribution of first generation H+ populations with and without Hsp90 overexpression. Because all genetic variants in these populations have arisen recently, we hypothesized that Hsp90 would potentiate their effects. If so, Hsp90 overexpression should decrease the fluorescence of these populations. In contrast, we found that Hsp90 overexpression significantly increased the fluorescence of these populations (17–40% increase; Mann–Whitney U test, ; Fig. 2d). We concluded that Hsp90 also buffers the effects of newly acquired mutations in our study system.
To further validate the hypothesis that Hsp90 can buffer the phenotype of variants that have not evolved under high Hsp90 expression for multiple generations, we overexpressed Hsp90 in fifth generation H− populations (Fig. 2e). Recall that H− populations also accumulate fluorescence-reducing mutations during evolution (Fig. 2a). If Hsp90 potentiates the effect of these mutations, we would expect that chaperone expression causes a reduction in fluorescence. In contrast, we found that Hsp90 expression significantly increased fluorescence, and it did so in every replicate population (Mann–Whitney U test, ; Fig. 2e). Specifically, it increased fluorescence by 32–52%, depending on the population. This shows that Hsp90 can buffer the effects of fluorescence-reducing mutations even if such mutations had accumulated while the chaperone was not highly expressed.
During phase 2 evolution toward the new phenotype of cyan fluorescence, Hsp90 affected phenotypic evolution similarly. Specifically, whereas H+ and H− populations showed no significant difference in their cyan fluorescence intensities at the end of phase 2 evolution, fluorescence of H+ populations decreased when the chaperone was no longer overexpressed (see Supplementary Section 8).
Hsp90 mediated buffering leads to the accumulation of GFP variants with very low fluorescence
We next focused more closely on individual amino acid variants whose spreading was significantly influenced by Hsp90 expression. To this end, we sequenced the GFP coding regions from each H+ and H− population in every generation of evolution, to a coverage of 1,307–3,933 (average 2,490) single-molecule reads, depending on the population. From the sequencing reads, we calculated the frequencies of point mutations and multimutant genotypes in the corresponding protein sequences. While synonymous mutations can affect cotranslational folding by altering translation elongation rate (Buhr et al. 2016), they may not affect posttranslational folding, which depends only on the protein sequence. Because Hsp90 binds to completely translated polypeptides (Schopf et al. 2017; Wang et al. 2022), we focused our analyses on nonsynonymous mutations.
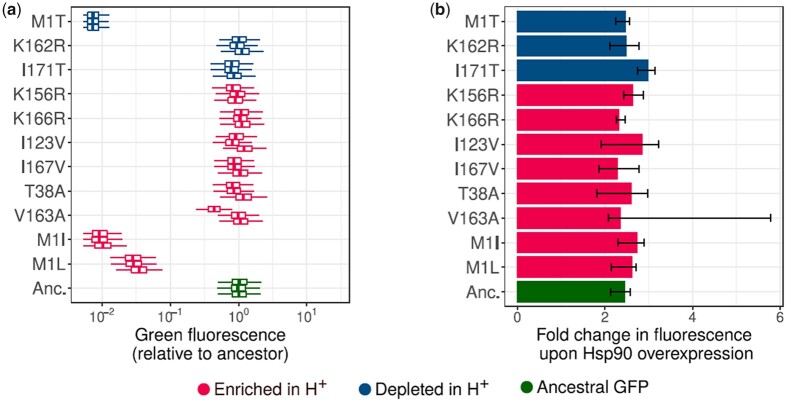
To focus on candidate variants with the most pronounced effect on population fluorescence, and to render this analysis manageable, we restricted ourselves to mutations with a frequency exceeding 3.5% at the end of phase 1 in at least one of the replicate populations. This frequency threshold is significantly higher than the expected frequency of any mutation caused by mutation pressure alone (Monte–Carlo simulations, ; Materials and methods). We used GLM to identify variants whose frequencies are significantly different between H+ and H− populations (analysis of deviance of GLM using LRT, P < 0.05). We identified eleven such differentially enriched variants: M1T, K162R, I171T, K156R, K166R, I123V, I167V, T38A, V163A, M1I, and M1L. Three of these variants (M1T, K162R, and I171T) were significantly more abundant (Supplementary Fig. 5a) in H+ populations, while the rest were significantly less abundant (Supplementary Fig. 5b).
To understand how Hsp90 influences the phenotype of the eleven variants, we engineered each of them into the ancestral GFP gene using site-directed mutagenesis. We then determined how they affect the fluorescence of ancestral GFP, in the absence of Hsp90 overexpression. Variants M1T, M1I, M1L, I171T, and K156R significantly reduced the fluorescence of ancestral GFP, and consistently did so in three independent experiments (Mann–Whitney U test, ; Fig. 3a). Among these fluorescence-reducing variants, the start codon variants M1T, M1I, and M1L had a strongly detrimental effect on fluorescence, leading to at least 16-fold reduction in fluorescence relative to ancestral GFP. In contrast, the mutations I171T and K156R reduced the fluorescence modestly (5–28% reduction in fluorescence; Fig. 3a). Among the three start codon mutations, M1T was the most detrimental to fluorescence, reducing the fluorescence of ancestral GFP by more than 75-fold. Of the remaining six differentially enriched variants, only one (K166R) caused a significant but modest increase in fluorescence by 5–11% (Mann–Whitney U test, ). The remaining variants did not significantly affect fluorescence.
Fig. 3.
Hsp90 enhances fluorescence of individual GFP variants. Enriched and depleted mutations in H+ populations (Supplementary Figs. 5, a and b) are color coded as red and blue, respectively. Ancestral GFP is denoted by green color. a) Box plots show fluorescence relative to median fluorescence of ancestral GFP (horizontal axis), for different GFP variants (vertical axis), in the absence of Hsp90 overexpression. In each boxplot, the boxes extend from the first quartile to the third quartile, and the whiskers have a length equal to the interquartile range (1 × IQR). Groups of three boxes corresponding to each mutation denote the three biological replicate measurements. b) Bar plots denoting the median fluorescence with Hsp90 overexpression relative to that without Hsp90 overexpression (fold change; horizontal axis), for different variants (vertical axis). We calculated the fold change for every pair (overexpression vs no-overexpression) of biological replicate measurements (3 × 3 = 9 pairs). The length of each solid bar corresponds to the median fold change in fluorescence (based on the 9 pairs). Error bars range from the minimum value to the maximum value of the fold change. For all mutations, and for every pair of biological replicates, Hsp90 overexpression significantly increased fluorescence (Mann–Whitney U test, ). Fluorescence fold change values of M1I, K156R, and I171T were modestly higher than that of ancestral GFP (one-tailed Mann–Whitney U test, P < 0.05). For absolute fluorescence values please see Supplementary Fig. 5c.
Next, we determined how Hsp90 overexpression affects the fluorescence of the differentially enriched variants. To this end, we measured the fluorescence of these mutants in the presence and absence of Hsp90 overexpression. We found that Hsp90 overexpression increased the fluorescence of all mutations, causing a 1.7- to 5.8-fold increase in fluorescence (Fig. 3b). This suggests that Hsp90 is a generic chaperone that enhances the folding of all the protein variants we studied (including the ancestral GFP). Such enhancement also exists for the modestly fluorescence-beneficial variant K166R, implying that Hsp90 potentiates the effect of this mutation. The same holds for beneficial (color shifting) variants we observed in phase 2 (Supplementary Fig. 13).
H+ populations fluoresce less intensely than H− populations in the absence of Hsp90 overexpression, suggesting that H+ populations accumulate mutations whose net detrimental effect on fluorescence is greater than that of the mutations accumulated in H− populations (Fig. 2c). In other words, Hsp90-mediated buffering leads to persistence of strongly fluorescence-reducing mutations in population. A good example that illustrates this phenomenon is the start codon mutation M1T. While all start codon mutations reduced fluorescence, M1T was the most detrimental to fluorescence (Fig. 3a). As a consequence of Hsp90 overexpression, this mutation accumulated to high frequencies in H+ populations (44–68%). In contrast, it was infrequent (<4.5%) in H− populations.
The weak selection we imposed allows fluorescence-reducing mutations to accumulate, but it does not explain why some of these mutations can attain very high frequencies that exceed not only the cutoff of 3.5% but reach values up to 75% (Supplementary Fig. 5). The explanation is that these mutants have a higher population growth rate, as we have also shown previously (Iyengar and Wagner 2022). Specifically, the start codon mutations M1I, M1L, and M1T reduce the rate of protein synthesis but not completely abolish it (Supplementary Fig. 6a; also see Hecht et al. 2017), which can lead to a reduced metabolic burden and a higher cell growth rate (Supplementary Fig. 6b; also see Kafri et al. 2016).
We did not investigate genotypic variants with more than one mutation because no such variant reached a high frequency in any population (frequencies <2.5%). Moreover, the most abundant mutations in different population (M1I, M1L, and M1T) were also abundantly present as single mutation variants (1.5–20% of all variants). These observations suggest that the fluorescence-reducing mutations we observed did not hitchhike to high frequency with other beneficial mutations.
If a chaperone buffers the effect of most deleterious mutations, chaperone overexpressing populations might accumulate higher genetic diversity than populations that do not overexpress the chaperone. However, in our experiments this is not the case. Hsp90 overexpression did not significantly affect genetic diversity in phase 1 (Supplementary Section 6 and Supplementary Fig. 7), and led to slower accumulation of such diversity in phase 2 (Supplementary Section 9 and Supplementary Fig. 10). A possible reason could be that fast accumulation of deleterious mutations in H+ populations due to Hsp90 mediated buffering, prevents any further accumulation of more such mutations. This may in turn prevent genetic diversification of the population.
Discussion
We aimed to answer the long-standing question whether chaperones predominantly buffer or potentiate the effects of mutations during adaptive evolution (Rutherford and Lindquist 1998; Queitsch et al. 2002; Cowen and Lindquist 2005; Whitesell et al. 2014; Geiler-Samerotte et al. 2016). To this end, we studied the effect of the E. coli chaperone Hsp90 on the directed evolution of GFP. Buffering means that a chaperone tends to mask the effect of a mutation, thereby preserving the original phenotype or reducing the mutation’s effect on the phenotype. At the organismal level, buffering may preserve cellular growth or morphology (Rutherford and Lindquist 1998; Queitsch et al. 2002). At the molecular level, it may preserve the activity of a biomolecule such as a protein (Tokuriki and Tawfik 2009; Wyganowski et al. 2013; Sadat et al. 2020). Potentiation is the opposite of buffering. In potentiation, a chaperone exposes or enhances the effect of a mutation (deleterious or beneficial) (Cowen and Lindquist 2005; Whitesell et al. 2014; Geiler-Samerotte et al. 2016). More importantly, a chaperone can simultaneously enhance one phenotype while suppressing another (Dorrity et al. 2018). Because natural selection acts on mutations with phenotypic effects, both buffering and potentiation can affect adaptive evolution. Conversely, how a chaperone affects a phenotype may depend on how natural selection acts on the phenotype.
We deliberately performed our evolution experiments under weak selection. This allowed our populations to accumulate mutations that are detrimental to protein activity, which are the most important class of mutations affecting protein evolution (Bershtein et al. 2006; Eyre-Walker and Keightley 2007). We found that Hsp90 not only enhanced the median fluorescence of all evolving populations (Fig. 2; Supplementary Figs. 3 and 9), it also enhanced the fluorescence of most genetic variants we engineered (Fig. 3b and Supplementary Fig. 12). Thus, Hsp90 unequivocally buffered the effects of fluorescence-reducing mutations. As a consequence, H+ populations, which evolved under Hsp90 overexpression also accumulated mutations that reduced fluorescence to a greater extent in the aggregate, than H− populations.
An important earlier study on eukaryotic Hsp90 focused on the chaperone’s effect on random genomic mutations that alter cellular morphology in a population of S. cerevisiae (Geiler-Samerotte et al. 2016). This study defined buffering as a chaperone-mediated reduction in morphological variation caused by mutations. Conversely, it defined potentiation as an increase in this variation. The study found that Hsp90 predominantly potentiated the effect of mutations. Furthermore, it showed that Hsp90 potentiated the effects of new mutations, but buffered the effects of older mutations that had segregated in the population for multiple generations.
In contrast to this earlier work, our experiments showed that bacterial Hsp90 chaperone enhanced the fluorescence of all our engineered GFP variants. It thus buffered the deleterious effects of fluorescence-reducing mutations, including new mutations (Fig. 2). This contrast may have at least three explanations. First, if bacterial Hsp90 functions differently from eukaryotic Hsp90 (Jahn et al. 2018), it may also have different effects on protein variants. Second, we allowed genetic variation to accumulate in only one protein, not in a whole genome. Complex epistatic (nonadditive) interactions between mutations in multiple genes may also affect how chaperones affect polygenic phenotypes. Finally, a mutation that is deleterious to protein activity, can be beneficial in other ways, for example, by reducing protein expression cost or toxicity. A chaperone can allow these mutants to survive selection and become fixed in the population after many generations of evolution. As a consequence, these mutants are always dependent on the chaperone for their function. Our results are consistent with this hypothesis. Although GFP is not essential for cell growth, it is rendered essential in our experiments due to the selection regime we impose. Thus, our results may also extend to genes essential for growth. Such genes, when mutagenized, are more likely to become chaperone dependent, because they are constantly under selection. For example, many oncoproteins that are crucial for cancer growth become Hsp90 dependent. They can render cancer cells “addicted” to the chaperone (Trepel et al. 2010; Park et al. 2020).
Even though Hsp90 buffered the effects of fluorescence-reducing mutations, it did not facilitate adaptive evolution in our experiments. One possible reason is that chaperone-mediated buffering allowed fluorescence-reducing mutations to attain greater frequency in our populations, which may have neutralized the benefit that the chaperone provided during evolution. Relatedly, the chaperone also did not lead to a greater accumulation of genetic diversity (Supplementary Figs. 7 and 10).
We speculate that strongly fluorescence-reducing mutations that originated early in populations evolving under Hsp90 overexpression prevented the accumulation of other fluorescence-reducing mutations, because their combined effect would have completely abolished fluorescence. Any such effect might be especially pronounced for start codon mutations, which strongly reduce fluorescence (Fig. 3a), while simultaneously providing a growth advantage to the cells harboring them (Supplementary Fig. 7). As a consequence, start codon mutations can rise to high frequency, and prevent further deleterious mutations from accumulating, because their combined effect on fluorescence would lead to their elimination under purifying selection. In this regard, it is relevant that most deleterious protein mutations show negative epistasis, that is, their combined deleterious effect is stronger than their individual effects (Bank et al. 2015, 2016; Sarkisyan et al. 2016).
Although Hsp90 increased the fluorescence of GFP variants with fluorescence-reducing mutations, it also did so for ancestral GFP and variants with high fluorescence. A possible explanation is that Hsp90 is a generic chaperone that does not discriminate between different client protein variants, possibly because Hsp90 is not involved in the initial stages of protein folding and binds only to partially folded proteins (Genest et al. 2011; Morán Luengo et al. 2018). Another possible explanation is that Hsp90 affects GFP synthesis or folding indirectly. For example, it may promote the folding of proteins that drive transcription or translation of GFP. Also, Hsp90 overexpression may cause metabolic changes that promote GFP maturation. It is difficult to distinguish such indirect effects from direct effects of chaperones on protein folding and evolution in vivo. In vitro evolution experiments may help resolve these two different effects. However, such experiments would entail their own limitations.
Regardless of the mechanism by which Hsp90 acts on GFP, the chaperone’s nearly uniform effect on different GFP variants raises another important concern regarding the definition of buffering. Although Hsp90 minimizes the harmful effect of fluorescence-reducing mutations and protects GFP variants from extinction, it does not completely restore wild-type activity. Additionally, Hsp90 does not only increase the activity of fluorescence reducing (deleterious) mutations, it also seems to enhance the effect of beneficial mutations. In consequence, the seemingly opposite phenomena of “buffering” and “potentiation” depend on the point of view of the observer. Therefore, the definition of these terms requires careful reevaluation. An alternative definition of chaperone action could be based on the effect of the chaperone on a variant relative to that on the ancestral protein.
Taken together, these considerations show that knowing the effect of a chaperone on a phenotype does not suffice to understand its consequences for adaptive evolution. The reason is that chaperone effects interact in complicated ways with the evolutionary dynamics of evolving populations.
Our study provides several directions for future work. One of them is to better understand the interaction between chaperone effects and evolutionary dynamics, for example, through population genetic modeling. This challenge is augmented by the observation that chaperones may have different effects on different variants of the same protein (Tokuriki and Tawfik 2009; Wyganowski et al. 2013), just like some proteins depend to a greater extent on chaperones than others (Kerner et al. 2005; Calloni et al. 2012). Along the same lines, a chaperone may possibly even buffer the effect of some mutations, while potentiating the effect of other mutations on the same protein. Another direction is to understand mechanistically how some chaperones can buffer while others potentiate the phenotypic effects of a mutation. This is not only the case for Hsp90 from E. coli and yeast. We have shown in previous work that the bacterial chaperone GroEL/S, unlike bacterial Hsp90, predominantly potentiates mutational effects (Iyengar and Wagner 2022). A third challenge is to understand how the multiple chaperones typically expressed in any one cell interact to affect adaptive evolution. What little we know suggests that few if any universal rules will apply. The organism, the kind of chaperone, and the evolving trait may all affect how chaperones affect adaptive evolution.
Supplementary Material
Acknowledgments
We acknowledge support by the flow cytometry facility and the functional genomics center at the University of Zurich. We thank Andrei Papkou for his suggestions on statistical analyses and Miriam Olombrada Sacristan, Jia Zheng, and Shraddha Karve for their assistance during the experiments.
Funding
This project has received funding from the European Research Council under Grant Agreement No. 739874 and Swiss National Science Foundation grant 31003A_172887, by the University Priority Research Program in Evolutionary Biology.
Conflicts of interest
None declared.
Contributor Information
Bharat Ravi Iyengar, Department of Evolutionary Biology and Environmental Studies, University of Zurich, 8057 Zurich, Switzerland; Swiss Institute of Bioinformatics, Quartier Sorge-Batiment Genopode, 1015 Lausanne, Switzerland; Institute for Evolution and Biodiversity, Westfalian Wilhelms—University of Münster, 48149 Münster, Germany.
Andreas Wagner, Department of Evolutionary Biology and Environmental Studies, University of Zurich, 8057 Zurich, Switzerland; Swiss Institute of Bioinformatics, Quartier Sorge-Batiment Genopode, 1015 Lausanne, Switzerland; The Santa Fe Institute, Santa Fe, NM 87501, USA; Stellenbosch Institute for Advanced Study (STIAS), Wallenberg Research Centre at Stellenbosch University, 7600 Stellenbosch, South Africa.
Data Availability
All data are available in the manuscript or supplementary materials. SMRT sequencing data are available at NCBI Sequence Read Archive (SRA) under the BioProject ID, PRJNA718260. Codes used for sequencing data analysis are available on GitHub: https://github.com/BharatRaviIyengar/pacbioanalysis.
Supplemental material is available at GENETICS online.
Literature cited
- Agozzino L, Dill KA.. Protein evolution speed depends on its stability and abundance and on chaperone concentrations. Proc Natl Acad Sci USA. 2018;115(37):9092–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Rodríguez J, Sabater-Muñoz B, Montagud-Martínez R, Berlanga V, Alvarez-Ponce D, Wagner A, Fares MA.. The molecular chaperone DnaK is a source of mutational robustness. Genome Biol Evol. 2016;8(9):2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Aguilar-Rodríguez J, Fares MA.. Molecular chaperones accelerate the evolution of their protein clients in yeast. Genome Biol Evol. 2019;11(8):2360–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C, Hietpas RT, Jensen JD, Bolon DN.. A systematic survey of an intragenic epistatic landscape. Mol Biol Evol. 2015;32(1):229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C, Matuszewski S, Hietpas RT, Jensen JD.. On the (un)predictability of a large intragenic fitness landscape. Proc Natl Acad Sci USA. 2016;113(49):14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JC, Craig EA.. Ancient heat shock gene is dispensable. J Bacteriol. 1988;170(7):2977–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS.. Robustness–epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444(7121):929–932. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH.. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103(15):5869–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA.. Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell. 2016;61(3):341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, chun Chang H, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU.. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1(3):251–264. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M.. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142(1):112–122. [DOI] [PubMed] [Google Scholar]
- Chen B, Zhong D, Monteiro A.. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 2006;7:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covalt JC, Roy M, Jennings PA.. Core and surface mutations affect folding kinetics, stability and cooperativity in IL-1β: does alteration in buried water play a role? J Mol Biol. 2001;307(2):657–669. [DOI] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S.. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309(5744):2185–2189. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL.. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6(9):678–687. [DOI] [PubMed] [Google Scholar]
- Dorrity MW, Cuperus JT, Carlisle JA, Fields S, Queitsch C.. Preferences in a trait decision determined by transcription factor variants. Proc Natl Acad Sci USA. 2018;115(34):E7997–E8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B, Haaland P, Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J, Jiang M, Finak G.. flowCore: flowCore: Basic Structures for Flow Cytometry Data, R package version 1.52.1; 2019.
- Eyre-Walker A, Keightley PD.. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610–618. [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E.. GroEL buffers against deleterious mutations. Nature. 2002;417(6887):398–398. [DOI] [PubMed] [Google Scholar]
- Fersht AR. Nucleation mechanisms in protein folding. Curr Opin Struct Biol. 1997;7(1):3–9. [DOI] [PubMed] [Google Scholar]
- Fromer M, Shifman JM.. Tradeoff between stability and multispecificity in the design of promiscuous proteins. PLoS Comput Biol. 2009;5(12):e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML.. Selection transforms the landscape of genetic variation interacting with Hsp90. PLoS Biol. 2016;14(10):e2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S.. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc Natl Acad Sci USA. 2011;108(20):8206–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Aigrain L, Quail MA, Coupland P, Bonfield JK, Davies RM, Tischler G, Jackson DK, Keane TM, Li J, et al. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci Rep. 2017;7(1). 10.1038/s41598-017-03996-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR.. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Protein misfolding diseases. Annu Rev Biochem. 2017;86:21–26. [DOI] [PubMed] [Google Scholar]
- Hecht A, Glasgow J, Jaschke PR, Bawazer LA, Munson MS, Cochran JR, Endy D, Salit M.. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017;45(7):3615–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoglu R, Balchin D, Hayer-Hartl M, Hartl FU.. Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat Commun. 2020;11(1). 10.1038/s41467-019-14245-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar BR, Wagner A.. GroEL/S overexpression helps to purge deleterious mutations and reduce genetic diversity during adaptive protein evolution. Mol Biol Evol. 2022;39(6):msac047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M, Tych K, Girstmair H, Steinmaßl M, Hugel T, Buchner J, Rief M.. Folding and domain interactions of three orthologs of Hsp90 studied by single-molecule force spectroscopy. Structure. 2018;26(1):96–105.e4. [DOI] [PubMed] [Google Scholar]
- Kafri M, Metzl-Raz E, Jona G, Barkai N.. The cost of protein production. Cell Rep. 2016;14(1):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Yi S, Sahni N, Fischer M, Xie J, Vidal M, D'Andrea AD, Whitesell L, Lindquist S.. Hsp90 shapes the consequences of human genetic variation. Cell. 2017;168(5):856–866.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang H-C, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122(2):209–220. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F.. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. [DOI] [PubMed] [Google Scholar]
- Liu H, Naismith JH.. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. Neutral models of genetic drift and mutation. In: RM Kliman, editor. Encyclopedia of Evolutionary Biology. Oxford: Academic Press, 2016. p. 119–123. [Google Scholar]
- Morán Luengo T, Kityk R, Mayer MP, Rüdiger SGD.. Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol Cell. 2018;70(3):545–552.e9. [DOI] [PubMed] [Google Scholar]
- Pacific Biosciences. Products and Services: Multiplexing; 2019a. https://www.pacb.com/products-and-services/analytical-software/multiplexing/.
- Pacific Biosciences. SMRT® Tools Reference Guide; 2019b. https://www.pacb.com/wp-content/uploads/SMRT-Tools-Reference-Guide-v8.0.pdf.
- Park HK, Yoon NG, Lee JE, Hu S, Yoon S, Kim SY, Hong JH, Nam D, Chae YC, Park JB, et al. Unleashing the full potential of Hsp90 inhibitors as cancer therapeutics through simultaneous inactivation of Hsp90, Grp94, and TRAP1. Exp Mol Med. 2020;52(1):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AM, Ponomarenko AI, Chen K, Ashenberg O, Miao J, McHugh SM, Butty VL, Whittaker CA, Moore CL, Bloom JD, et al. Destabilized adaptive influenza variants critical for innate immune system escape are potentiated by host chaperones. PLoS Biol. 2018;16(9):e3000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S.. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618–624. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna (Austria: ): R Foundation for Statistical Computing, 2018. [Google Scholar]
- Richter K, Haslbeck M, Buchner J.. The heat shock response: life on the verge of death. Mol Cell. 2010;40(2):253–266. [DOI] [PubMed] [Google Scholar]
- Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F.. Structural and molecular comparison of bacterial and eukaryotic trigger factors. Sci Rep. 2017;7(1):10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S.. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–342. [DOI] [PubMed] [Google Scholar]
- Sabater-Muñoz B, Prats-Escriche M, Montagud-Martínez R, López-Cerdán A, Toft C, Aguilar-Rodríguez J, Wagner A, Fares MA.. Fitness trade-offs determine the role of the molecular chaperonin GroEL in buffering mutations. Mol Biol Evol. 2015;32(10):2681–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadat A, Tiwari S, Verma K, Ray A, Ali M, Upadhyay V, Singh A, Chaphalkar A, Ghosh A, Chakraborty R, et al. GROEL/ES buffers entropic traps in folding pathway during evolution of a model substrate. J Mol Biol. 2020;432(20):5649–5664. [DOI] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14(10):630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, et al. Local fitness landscape of the green fluorescent protein. Nature. 2016;533(7603):397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J.. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18(6):345–360. [DOI] [PubMed] [Google Scholar]
- Studer RA, Christin PA, Williams MA, Orengo CA.. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci USA. 2014;111(6):2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Serrano L, Tawfik DS.. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4(2):e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS.. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459(7247):668–673. [DOI] [PubMed] [Google Scholar]
- Trepel J, Mollapour M, Giaccone G, Neckers L.. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RYR, Noddings CM, Kirschke E, Myasnikov AG, Johnson JL, Agard DA.. Structure of Hsp90–Hsp70–Hop–GR reveals the Hsp90 client-loading mechanism. Nature. 2022;601(7893):460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Warr A.. Errors in long-read assemblies can critically affect protein prediction. Nat Biotechnol. 2019;37(2):124–126. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Santagata S, Mendillo ML, Lin NU, Proia DA, Lindquist S.. Hsp90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proc Natl Acad Sci USA. 2014;111(51):18297–18302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklhofer KF, Tatzelt J, Haass C.. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27(2):336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyganowski KT, Kaltenbach M, Tokuriki N.. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 2013;425(18):3403–3414. [DOI] [PubMed] [Google Scholar]
- Xu Y, Singer MA, Lindquist S.. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc Natl Acad Sci USA. 1999;96(1):109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Chen P, Shakhnovich EI.. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104(41):16152–16157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Guo N, Wagner A.. Selection enhances protein evolvability by increasing mutational robustness and foldability. Science. 2020;370(6521):eabb5962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript or supplementary materials. SMRT sequencing data are available at NCBI Sequence Read Archive (SRA) under the BioProject ID, PRJNA718260. Codes used for sequencing data analysis are available on GitHub: https://github.com/BharatRaviIyengar/pacbioanalysis.
Supplemental material is available at GENETICS online.