Abstract
Lactobacillaceae are an important family of lactic acid bacteria that play key roles in the gut microbiome of many animal species. In the honey bee (Apis mellifera) gut microbiome, many species of Lactobacillaceae are found, and there is functionally important strain-level variation in the bacteria. In this study, we completed whole-genome sequencing of 3 unique Lactobacillaceae isolates collected from hives in Virginia, USA. Using 107 genomes of known bee-associated Lactobacillaceae and Limosilactobacillus reuteri as an outgroup, the phylogenetics of the 3 isolates was assessed, and these isolates were identified as novel strains of Apilactobacillus kunkeei, Lactobacillus kullabergensis, and Bombilactobacillus mellis. Genome rearrangements, conserved orthologous genes (COG) categories and potential prophage regions were identified across the 3 novel strains. The new A. kunkeei strain was enriched in genes related to replication, recombination and repair, the L. kullabergensis strain was enriched for carbohydrate transport, and the B. mellis strain was enriched in transcription or transcriptional regulation and in some genes with unknown functions. Prophage regions were identified in the A. kunkeei and L. kullabergensis isolates. These new bee-associated strains add to our growing knowledge of the honey bee gut microbiome, and to Lactobacillaceae genomics more broadly.
Keywords: Lactobacillus, Apilactobacillus, Bombilactobacillus, whole genome, honey bee microbiome, gut microbiome, prophage, bacteriophage
Introduction
In recent years, honey bees have emerged as an important system for understanding the functional roles of bacteria in the gut microbiome. Relative to vertebrates, the gut microbiome of honey bees is simplified, with 8–10 dominant bacterial phylotypes (Moran 2015), and important strain-level variation (Engel et al. 2014; Ellegaard and Engel 2016; Ellegaard et al. 2019). Lactobacillaceae (phylum: Firmicutes, class: Bacilli, order: Lactobacillales) is a family of lactic acid bacteria found within the gut of both vertebrates and invertebrates (Makarova et al. 2006; Zheng et al. 2020). Lactobacillaceae taxonomy was recently updated, with all genera in the family Leuconostoceae incorporated into Lactobacilliceae, and the predominant genus, Lactobacillus, split into 25 genera representing distinct clades (Zheng et al. 2020). Genome sequences of new Lactobacillaceae species and strain variants are regularly being published (Kwong et al. 2014; Olofsson et al. 2014), and these species can vary widely in genome size (from 1,700,000 to ≥3,000,000 base pairs) and GC content (from 31% to 56%) (Felis and Dellaglio 2007; Kant et al. 2011). The recent taxonomic changes (Zheng et al. 2020) resulted in some honey bee Lactobacillus species reclassified as Apilactobacillus (with subsequent changes to their specific epithets), and some bumble bee species reclassified as Bombilactobacillus. As for many bacterial species, Lactobacillaceae genomes are composed of both species specific genes and additional genetic elements, including bacteriophages (Medini et al. 2005), that can result in larger genomes than expected. Temperate bacteriophages have stable relationships with their bacterial hosts and are incorporated into the host genome as prophages (Casjens 2003). A recent study of 1,472 Lactobacillaceae genomes found that 99.8% of the genomes contained predicted prophage regions (Pei et al. 2021).
In the honey bee microbiome, Lactobacillaceae phylotypes previously comprised 2 clades: Firm-4 and Firm-5. Firm-5 was reclassified to the Lactobacillus melliventris clade (Zheng et al. 2020), and are the most common of the honey bee bacterial phylotypes, although other Lactobacillus species are also found in high abundance in honey bees (Corby-Harris et al. 2014; Anderson et al. 2016; Ellegaard and Engel 2019). There is variation in microbiome composition along the honey bee gut, from crop to midgut to hindgut (Powell et al. 2014). The crop may be dominated by species that favor acidic, sugar-rich environments, such as Apilactobacillus kunkeei (Corby-Harris et al. 2014). The midgut contains a small bacterial community, dominated by the Gamma-1 phylotype (Martinson et al. 2012). Most bacteria reside in the hindgut, which can be divided into the ileum and the rectum. The ileum contains mainly Gram-negative species, such as Frischella perrara, Snodgrassella alvi, Gilliamella apicola, and some species of the L. melliventris clade, while the rectum contains large populations of Gram-positive species, including species in the L. melliventris clade, other Lactobacillus species, and Bifidobacterium species (Martinson et al. 2012; Powell et al. 2014; Kwong and Moran 2016).
Here, we present 3 new Lactobacillaceae genomes from isolates obtained from honey bees in the Eastern US, and compare them to existing honey bee Lactobacillaceae genomes, which have predominantly been collected in Europe.
Materials and methods
Honey bee collection and bacterial isolation
Honey bees were collected from hives at the Virginia Tech apiary in Montgomery County, VA, USA in August (N = 1 hive) and September (N = 6 hives), 2016. Bees were collected from inside the hive in sterile 50 ml centrifuge tubes and placed on ice until they were returned to the laboratory, where they were frozen at −80°C until they could be dissected.
In the laboratory, bees were briefly thawed and then surface-sterilized using 5% bleach followed by 3 rinses in sterile water (Engel et al. 2013). For each individual bee (N = 10), the whole gut was dissected using sterile technique, the mid- and hind-guts were separated, and the separate gut regions were placed into 1.5 ml microcentrifuge tubes, each containing 500 μl of sterile 10 mM MgSO4 (Kwong and Moran 2013). Sterile pestles were used to homogenize the gut samples, which were serially diluted in 10 mM MgSO4 (10−1 to 10−5), and 200 μl of each dilution was plated onto de Man Rogosa Sharpe (MRS) agar plates. Plates were incubated at 37°C for 2–3 days under low oxygen culture conditions using incubation chambers (BD GasPak™ EZ Gas Generating Systems Incubation Containers) and GasPak™ sachets (BD Bioscience’s GasPak™ Sachets). Morphologically distinct bacterial colonies were further isolated to obtain pure cultures; this was done by visual inspection of colony morphology to ensure pure colonies from between 1 and 3 subcultures.
For each of these isolates, we extracted DNA using the MoBio Laboratories UltraClean Microbial DNA Isolation kit or the Qiagen DNeasy Blood & Tissue kit (Qiagen Inc., Valencia, CA), then amplified and sequenced the full-length 16S rRNA region using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), as described in Lauer et al. (2008). Isolate PCR products were cleaned using the Qiagen QIAquick PCR Purification Kit and eluted in 50 μl of molecular water prior to Sanger sequencing for assignment to genus. Sequences were determined to be different species based on taxonomic differences from BLAST analysis. Within the set of 27 sequenced isolates, we obtained 17 Lactobacillus isolates, and of these, we chose 3 that appeared to be separate species based on the full-length 16S rRNA gene sequence for whole-genome sequencing. To extract DNA for whole-genome sequencing, isolates were regrown in 750 μl of MRS broth, shaking at 150 rpm at room temperature for 24 h. The cultures were then centrifuged at 7,500 rpm for 10 min, and the resulting pellet was resuspended in 180 μl of lysis buffer containing 20 mg/ml lysozyme and processed through the Gram-positive bacteria protocol of the Qiagen DNeasy Blood and Tissue Kit, with final elution in 150 μl molecular water.
Genome assembly
Extracted DNA was sent to the Duke Center for Genomic and Computational Biology for library construction and sequencing on the Illumina Hi-Seq 4000 platform, 2 × 150 bp. This generated an average of 4 Gbp of reads per sample library with an average Q score of 34% and 96% of reads >Q30. Raw reads were adapter trimmed using Trimmomatic v.0.35 (Bolger et al. 2014) with default settings, including standard Illumina adapters, and visually checked for quality using FastQC (Andrews 2010). Processed raw reads were de novo assembled using Minia v.2.0 (Chikhi and Rizk 2013) with the command line arguments, -kmer-size 121. K-mer size was optimized by iteratively assembling the genomes -kmer-size = 41–141 and the optimal k-mer size was selected based on assembly statistics (Chikhi and Rizk 2013). Comparative assembly statistics were compiled using Quast v 5.0.0 (Gurevich et al. 2013). For the average assembly, the total length was 1.75 Mbp (range 1.55:2.05 Mbp), with 25 total contigs (range 19:38). The average N50 was 327 kb (range 72.6:491 kb). The average L50 was 4 (2:8), and GC content was 36% (range 35.7%:36.8%) (Supplementary Table 1). Minia generated contig files were used for downstream analyses.
To assess assembly completeness (accuracy of assembled orthologs), we analyzed the genome assemblies for Benchmarking Universal Single-Copy Orthologs (BUSCOs), which are genes that are expected to be present in closely related bacteria. To do this, we used BUSCO v.4.1.3 (Simão et al. 2015) with the Lactobacillales database v.10. All 3 genomes contained at least 98% of expected BUSCOs, suggesting they were relatively complete.
Phylogenetic analysis
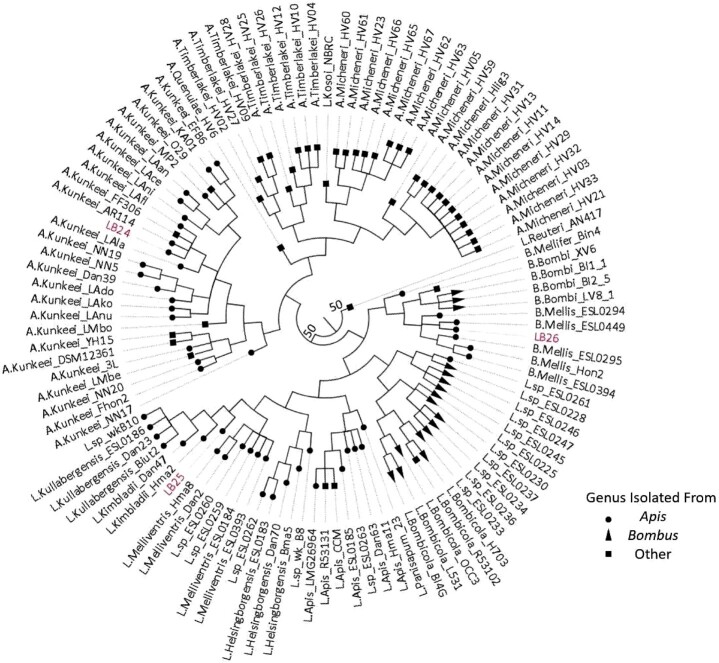
In addition to our 3 Lactobacillus isolates, 107 additional whole-genome sequences of all bee-associated members of the Lactobacillaceae family, as well as the genome of Limosilactobacillus reuteri AN417 [=Lactobacillus reuteri (Zheng et al. 2020)], were downloaded from Genbank for use in phylogenetic analysis (Supplementary Table 2). Limosilactobacillus reuteri AN417 was used as an outgroup for our bee-associated Lactobacillaceae tree (N = 111 genomes). All genomes were annotated using Prokka v.1.14.6 using the default databases (Seemann 2014). To generate a core gene alignment, PIRATE (Bayliss et al. 2019) was run using the general feature format files generated by Prokka. The resulting core alignment contained 359 genes. A Lactobacillaceae phylogenetic tree was reconstructed using this core genome alignment in RAxML HPC v.8.2.12 (Stamatakis 2014). RAxML was run using a random number seed for parsimony inferences, rapid bootstrapping with 1,000 replicates, and the GTRCAT nucleotide substitution model. The resulting consensus tree was rooted and converted to nexus format using Geneious prime v.2020.2.1 (Biomatters Ltd). The tip labels were aligned, and tip colors were changed to highlight the placement of our isolates. Bootstrap values below 100 were visualized using FigTree v.1.4.4 (Rambaut 2010) (Fig. 1).
Fig. 1.
Phylogenetic tree of 110 bee-associated members of the Lactobacilliaceae family plus L. reuteri AN417 (outgroup). Tip shape is based on the host genus the bacteria was isolated from; circle (Apis = honey bee), triangle (Bombus = bumble bee), and square (other). Only bootstrap values below 100 are shown.
Comparative genomic analysis
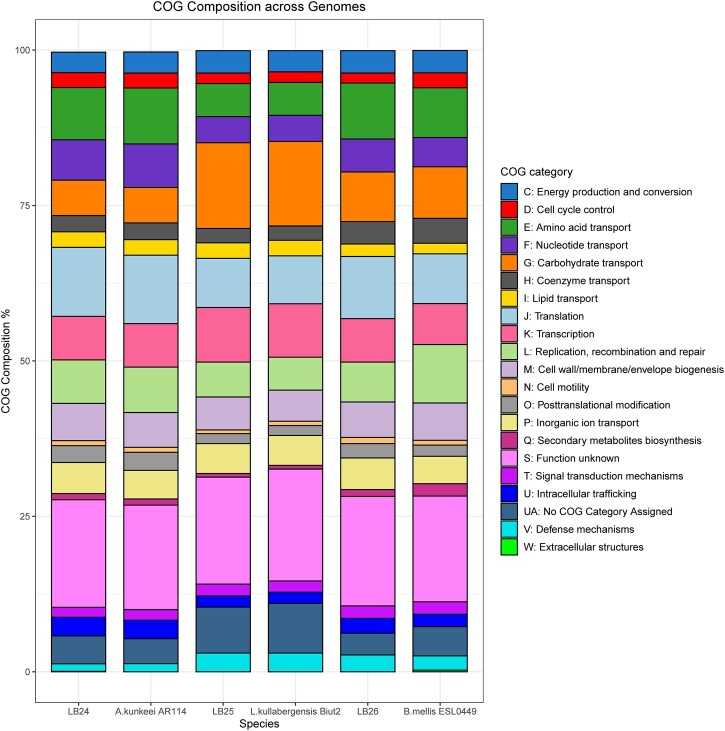
To visualize rearrangements within the genomes, Mauve build v.2.4.0 (Darling et al. 2004, 2010) was used. Our isolates, as well as a few of their closest relatives selected from the phylogenetic tree produced from all 111 genomes, were aligned using the default settings of progressive mauve. A phylogenetic tree was then added to these alignments in R v.4.0.5 (R Core Team 2021) using the genoPlotR package v.0.8.11 (Guy et al. 2010) (Fig. 2). To compare COG functional categories across genomes, our isolates, as well as the genomes of their closest relatives (Apilatobacillus kunkeei AR114, Lactobacillus kullabergensis Biut2 and Bombilactobacillus mellis ESL0449; Genbank accession numbers GCF_000830375.1, GCF_000967195.1, and GCF_013346905.1, respectively), were annotated using eggNOG-mapper v.2.0 with eggNOG database v.5.0 and the DIAMOND algorithm (Buchfink et al. 2015; Huerta-Cepas et al. 2017, 2019). The total number of genes per COG category were divided by the total number of genes within the genome. The resulting percentages were then plotted in R v.4.0.5 using ggplot2 v.3.3.5 (Wickham 2016) (Fig. 3). For the sake of brevity, the COG descriptions used in the legend of Fig. 3 have been truncated. The full COG descriptions are included in Supplementary Table 3.
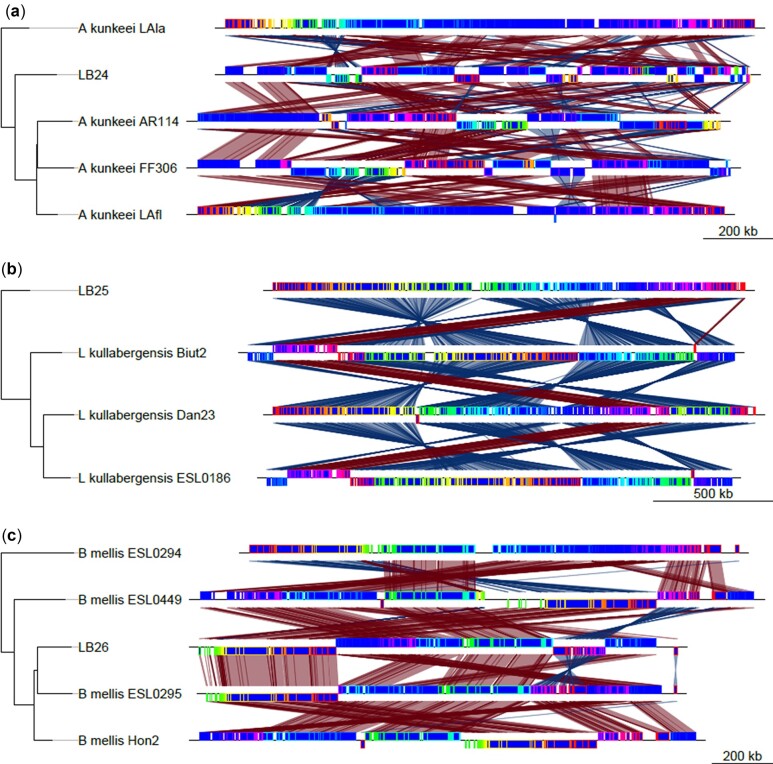
Fig. 2.
Mauve alignments of the 3 Lactobacillaceae isolates with closely related known species. Isolate LB24 aligned with 4 closely related Apilactobacillus kunkeei isolates (a). Isolate LB25 aligned with 3 closely related Lactobacillus kullabergensis isolates (b). Isolate LB26 aligned with 4 closely related Bombilactobacillus mellis isolates. Rearrangement of syntenic blocks are shown in red, and inversion of syntenic blocks are shown in blue.
Fig. 3.
Stacked bar chart comparing the COG category composition between LB24, LB25, LB26, and their most closely related isolate: Apilactobacillus kunkeei AR114, Lactobacillus kullabergensis Biut2, and Bombilactobacillus mellis ESL0449, respectively. LB24 and B. mellis ESL0449 both contained genes that were assigned to COG category “W”; however, in both species that category represented <0.5% of the genes which makes it difficult to see on the figure. A more detailed description of each COG category can be found in Supplementary Table 3.
Prophage investigation
To check for the presence of prophage, the 3 genomes were analyzed with VirSorter2 v.2.2.2 (Guo et al. 2021). To reduce false positives and trim bacterial ends from potential prophage regions, potential regions scored above 0.9 by VirSorter2 were then analyzed with VIBRANT v.1.2.1, and those classified as viral were considered prophages (Kieft et al. 2020).
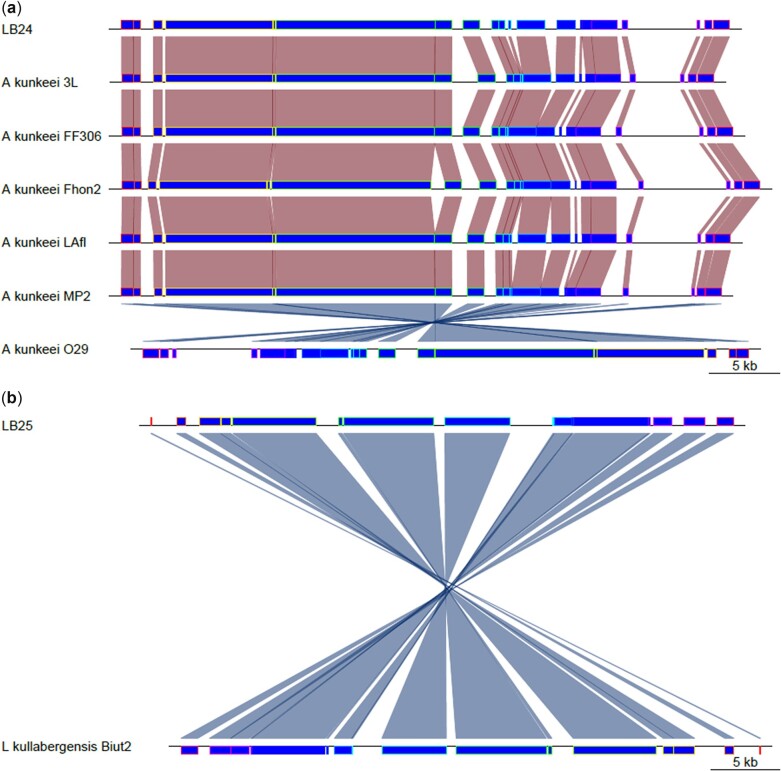
Additionally, a BLAST database was created using the 2 phages found in LB24 and LB25 and run against all 107 bee-associated members of the Lactobacillaceae family used in this study. Bacterial genomes with E scores of 0 were then analyzed with VirSorter2 and VIBRANT. The score threshold for VirsSorter2 was relaxed to 0.5 to include putative prophage regions in the reference genomes that may be partially degraded or cryptic but were still confirmed and trimmed with VIBRANT. The open reading frames (ORFs) of each putative prophage were identified with Prodigal (Hyatt et al. 2010) and average nucleic acid (ANI) similarity between phages was calculated using BLAST. Prophage from reference genomes were also aligned to the LB24 and LB25 prophage regions using progressive Mauve and visualized in R v.4.0.5 (R Core Team 2021) using the genoPlotR package v.0.8.11 (Guy et al. 2010) (Fig. 4).
Fig. 4.
Mauve alignments of the most similar prophage(s) to those in LB24 and LB25 found in related species. Alignments shown share more than 80% of ANI to at least half of either LB24 or LB25’s prophage ORFs. Isolate LB24 with 6 Apilactobacillus kunkeei isolates (a). Isolate LB25 with one other Lactobacillus kullabergensis isolate (b). Prophage regions were predicted bioinformatically and therefore, this does not necessarily confirm the presence of active phages.
Results and discussion
Using the nucleotide sequence of the 16S rRNA gene extracted from the whole-genome sequences, we identified LB24 as a novel strain of Apilactobacillus kunkeei, LB25 as Lactobacillus kullabergensis and LB26 as Bombilactobacillus mellis (Zheng et al. 2020). The core genome of the 110 bee-associated members of the Lactobacillaceae family contained 359 genes, as did the Limosilactobacillus reuteri strain AN417 isolated from domestic pigs that served as an outgroup in our phylogeny. The phylogenetic tree created using the core genome alignment confirmed the initial identifications of LB24 (A. kunkeei) and LB26 (B. mellis) (Fig. 1). However, in this tree, LB25 was placed at the root of L. kullabergenis and L. kimbladii, suggesting it was likely to be closely related to both of these, but could not be unambiguously placed as one of those 2 species (Fig. 1). Because of this unclear phylogenetic placement, we used sequence homology to determine the most closely related species to LB25 for subsequent comparative analyses. In this case, 30 full-length genes were aligned to the NCBI nr database using BLAST. Of these genes, 100% retrieved a strain of Lactobacillus kullabergensis as the best hit, so we used that species for comparative analyses for LB25.
Apilactobacillus kunkeei [=Lactobacillus kunkeei (Zheng et al. 2020)] is a fructophilic lactic acid bacterium, with previous isolates found within wine, flowers, honey, and honey bees (Endo et al. 2012; Neveling et al. 2012). The assembled A. kunkeei isolate (LB24) has a similar genome size of 1,558,246 bp (1.407–1.634 Mb) and GC content, 37% (35.47–36.57%) compared with the 59 published A. kunkeei genomes. We identified a lower number of predicted coding sequences (1,294 compared with 1,338–1,384) and similar number of tRNA genes (62 compared with 31–65), as 3 other newly described species isolated from honey bee guts (Crovadore et al. 2021), which may result from assembly level differences. Unlike Crovadore et al. (2021), we identified a prophage region in the LB24 assembly, consistent with other A. kunkeei MP2 assemblies (Asenjo et al. 2016). Additionally, the prophage identified in LB24 shares approximately half its genes, with high nucleotide similarity, to prophage regions found in MP2 and 5 other strain assemblies of A. kunkeei. Functional analysis of the LB24 assembly revealed that 1,229 of the 1,291 predicted coding sequences were assigned COG categories, which along with BUSCO scores supports the integrity of this assembly. Categorization was largely similar between the LB24 assembly and a reference strain (Fig. 3). Investigating the genes unique to LB24 we identified S (Function unknown; 23% of genes) as the largest, with L (Replication, recombination and repair; 12% of genes) as the second most abundant of the assigned categories. Many of the genes in these 2 groups (18% and 22%, respectively) included some with annotations related to phage (e.g. ybl78: conserved phage C-terminus, and sip: phage integrase family). While the functional importance of this categorical expansion, and prophage presence in LB24 needs to be tested, in studies of human commensal Lactobacillus johnsonii, these genes have been associated with genome scale rearrangements that have facilitated host specific adaptation (Guinane et al. 2011). While a direct connection remains to be established, consistent with this concept, we identified rearrangements between this assembly and reference strains (Fig. 2a).
Lactobacillus kullabergensis has been isolated from both the honey stomach and gut of honey bees (Olofsson et al. 2014). The assembly of LB25, putatively, L. kullabergensis, had a genome size of 2,052,702 bp (2.019–2.118 Mb) and the same GC content 36%, as seen in the 6 published genomes on NCBI. The LB25 assembly has marginally lower predicted coding sequences (1,833 compared with 1,844) as well as number of tRNA genes (53 compared with 50) compared with the sequence used by Ellegaard et al. (2015) in their investigation of intraphylotype diversity in Lactobacilli. As with our A. kunkeei isolate (LB24), a prophage region was identified within our L. kullabergensis isolate. The prophage in our isolate shared some similarity to a prophage identified in the L. kullabergensis Biut2 isolate, and similarity to putative prophage in L. melliventris isolates, albeit with much weaker support. Functional analysis revealed that 1,686 of the 1,827 predicted coding sequences from the LB25 assembly were assigned COG categories. As with the LB24 assembly, categorization was largely similar to that of the reference strain (Fig. 3). Investigating genes that were unique to the LB25 assembly, we found an increased number of unique genes associated with carbohydrate transport, with 34% of the uniquely identified genes falling in this category. This is consistent with other studies of honey bee isolated L. kullabergensis strains (Ellegaard et al. 2015) and it has been associated with increased metabolic flexibility among host-associated Lactobacillus spp. (Barrangou et al. 2006). When LB25 is aligned with the closely related L. kullabergensis Biut2, the inversion of several syntenic blocks is observed, and these are also seen when L. kullabergensis Biut2 is aligned with the 2 other known L. kullabergensis strains (Fig. 2b). Together these suggest potential, testable, mechanisms through which LB25 and other L. kullabergensis strains may adapt to diverse hosts.
Bombilactobacillus mellis [=Lactobacillus mellis (Zheng et al. 2020)] has been isolated from honey, bee bread, pollen, the honey stomach, gut, and digestive tract of A. mellifera and other Apis species (Olofsson et al. 2014). The LB26 B. mellis assembled with a genome size of 1,644,951 bp (1.684–1.811 MB) and a GC content of 36%, similar to the 8 published genomes on NCBI. This assembly had a lower number of predicted coding sequences (1,415 compared with 1,572), and fewer tRNA genes (38 compared with 53) when compared with the strain sequenced by Ellegaard et al. (2015) in their investigation of intraphylotype diversity in Lactobacilli. Functional analysis of the LB26 assembly assigned 1,359 of the 1,411 predicted coding sequences to COG categories. Once again, categorization was similar to that of the reference strain (Fig. 3). Analysis of the unique genes revealed a dominance of genes with unknown function, similar to the A. kunkeei isolate LB24. In contrast, the number of annotations associated with differences in transcription or transcriptional regulation were markedly greater (17% vs 7%) than those found in common between LB26 and the reference strain. While beyond the scope of the present study, in other lactobacilli, changes in the regulation of transcription have been associated with increased host-specificity (Goh et al. 2021). Unlike the A. kunkeei and L. kullabergensis isolates in our study, no prophage regions were identified within the B. mellis isolate. While the lack of phage in this isolate is rare, as Pei et al. (2021) found 99.8% of Lactobacillaceae genomes investigated had prophage regions, no B. mellis strains were included in that study. For LB26 and the 4 most closely related strains of B. mellis (ESL0294, ESL0449, ESL0295, and Hon2), the Mauve alignment suggested many of the syntenic blocks had undergone rearrangements among the genomes (Fig. 2c).
Intriguingly, we identified predicted prophage regions in 2, LB24 and LB25, of the 3 assembled genomes. No prophage was predicted in LB26. In LB24, a single predicted prophage region was identified with a probability >0.9 using VirSorter2 (dsDNA phage, 89,958 bp), which was trimmed to 56,656 bp by VIBRANT. In LB25, there was also 1 predicted prophage region identified with a probability >0.9 (dsDNA phage, 43,795 bp), which was not trimmed by VIBRANT. It is possible that the predicted prophage in LB24 prophage may be partially degraded or possess novel genes, disrupting the analysis software. Conversely, the prophage predicted in the LB25 prophage may be more intact and/or have genes that are more recognizably phage-like. The comparative alignments of prophage regions among closely related strains are consistent with general genomic rearrangements (Fig. 4).
While it remains to be determined that these predicted prophage regions are inducible, we identified shared homology with these regions and prophage identified in other bee-associated Lactobacillaceae. The putative prophage of LB24 shared high ANI across aligned ORFs with 12 other predicted phage regions, appearing most similar to prophages found in 6 isolates of A. kunkeei (LAfl, FF306, 3L, Fhon2, O29, MP2), with 92.23–96.80% ANI across 55.3–81.6% of the LB24 assembled phage ORFs (Fig. 4a and Supplementary Table 4). Phage from other isolates of A. kunkeei (Laan, Dan39) and other species like Apilactobacillus quenuiae (HV6) and Apilactobacillus micheneri (HV05, HV61, HV60) shared between 1 and 3 ORFs (1.3–4.0% of LB24’s phage ORFs) with ANI ranging from 73.9 to 93.86% (Supplementary Table 4). This may indicate that a high amount of horizontal gene transfer occurred between these prophages, resulting in mosaic genomes (Vale et al. 2017; Moura de Sousa et al. 2021). The most similar prophage region to that found in LB25 prophage was identified in L. kullabergensis Biut2 with an ANI of 88.0% across 53.3% of the LB25 assembled phage ORFs (Fig. 4b and Supplementary Table 4). Additional less confident matches to the phage occurred in 4 isolates of L. melliventris (ESL0184, ESL0393, Hma8, Dan2), 2 isolates of L. helsingborgensis (ESL0183, Dan70) and in 1 Lactobacillus apis isolate (Dan63), with a range of 84.8–90.5% ANI across 20.0–28.3% of the assembled phage ORFs (Supplementary Table 4). Finally, LB25’s phage also matched slightly with phage found in 3 isolates of Lactobacillus bombicola (BI4G, L531, OCC3), each sharing 3 ORFs (5.0% of LB25’s ORFs) with ANI of 76.7–76.9% (Supplementary Table 4). Additionally, the LB25 prophage region shared similarities with prophage regions found in 11 unidentified species of Lactobacillaceae (wkB10, ESL0263, ESL0262, ESL0261, ESL0237, ESL236, ESL0234, ESL0233, ESL0230, ESL0228, and ESL0225) with 76.5–90.5% ANI across 1.7–21.7% of LB25’s ORFs (Supplementary Table 4).
This study presents the assembly and annotation of 3 novel strains of Lactobacillaceae isolated from the honey bee gut. Phylogenetic placement of the strains was well supported, with additional support coming from syntenic and functional analyses. While we have a number of insights on the probiotic nature of this family in the human gut (Zhang et al. 2018), their role in the honey bee gut is just emerging. These complete genome assemblies can serve as references for assembling additional strains, and gleaning further genomic and mechanistic insights among members of this group that form close associations with honey bees.
Supplementary Material
Acknowledgments
The authors thank Chenming Cui for drafting an initial assembly pipeline and Alaina Weinheimer for the script to calculate average nucleotide identity between prophages.
Funding
This project was funded by the Virginia Agricultural Council (project no. 677 to JBW, LKB, and RF) and National Science Foundation grants (MCB-1817736 to LKB, RF, and DCH and MCB-1817717 to JBW).
Conflicts of interest
None declared.
Contributor Information
Emma L Bradford, Department of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
Noah Wax, Department of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
Emma K Bueren, Department of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
Jenifer B Walke, Department of Biology, Eastern Washington University, Cheney, WA 99004, USA.
Richard Fell, Department of Entomology, Virginia Tech, Blacksburg, VA 24061, USA.
Lisa K Belden, Department of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
David C Haak, School of Plant and Environmental Sciences, Virginia Tech, Blacksburg, VA 24061, USA.
Data Availability
Genome assemblies and raw data can be found here: http://www.ncbi.nlm.nih.gov/bioproject/866146, but note that these assemblies have been cleaned of any contigs <200 bp by NCBI requirements. Full assemblies, processing scripts, and associated metadata can be obtained from, https://doi.org/10.7294/19524946. File LB24_final_contigs.fasta contains the complete final contig list of LB24. File LB25_final_contigs.fasta contains the complete final contig list of LB25. File LB26_final_contigs.fasta contains the complete final contig list of LB26. File lactobacillus_MS_metadata_scripts_readme.docx contains all scripts used in the metadata analysis in a word document. File 09_scripts.zip contains all scripts used in the metadata analysis.
Supplemental material is available at G3 online.
Literature cited
- Anderson KE, Rodrigues PAP, Mott BM, Maes P, Corby-Harris V.. Ecological succession in the honey bee gut: shift in Lactobacillus strain dominance during early adult development. Microb Ecol. 2016;71(4):1008–1019. . [DOI] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data [Online]; 2010. [accessed 2021]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Asenjo F, Olmos A, Henríquez-Piskulich P, Polanco V, Aldea P, Ugalde JA, Trombert AN.. Genome sequencing and analysis of the first complete genome of Lactobacillus kunkeei strain MP2, an Apis mellifera gut isolate. PeerJ. 2016;4(4):e1950. 10.7717/peerj.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Azcarate-Peril MA, Duong T, Conners SB, Kelly RM, Klaenhammer TR.. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci USA. 2006;103(10):3816–3821. 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ.. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience. 2019;8(10):giz119. 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49(2):277–300. . [DOI] [PubMed] [Google Scholar]
- Chikhi R, Rizk G.. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithms Mol Biol. 2013;8(1):22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V, Maes P, Anderson KE.. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS One. 2014;9(4):e95056. 10.1371/journal.pone.0095056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crovadore J, Chablais R, Raffini F, Cochard B, Hänzi M, Gérard F, Jensen KK, Lefort F.. Draft genome sequences of 3 strains of Apilactobacillus kunkeei isolated from the bee gut microbial community. Microbiol Resour Announc. 2021;10(13):e00088-21. 10.1128/MRA.00088-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT.. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT.. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, Brochet S, Bonilla-Rosso G, Emery O, Glover N, Hadadi N, Jaron KS, van der Meer JR, Robinson-Rechavi M, Sentchilo V, et al. ; SAGE Class 2016-17. Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol Ecol. 2019;28(9):2224–2237. 10.1111/mec.15075. [DOI] [PubMed] [Google Scholar]
- Ellegaard KM, Engel P.. Beyond 16S rRNA community profiling: Intra-species diversity in the gut microbiota. Front Microbiol. 2016;7:1475. 10.3389/fmicb.2016.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, Engel P.. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun. 2019;10(1):1–13. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SGE, Vásquez A.. Extensive intra-phylotype diversity in Lactobacilli and Bifidobacteria from the honeybee gut. BMC Genomics. 2015;16(1):1–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Irisawa T, Futagawa-Endo Y, Takano K, Toit MD, Okada S, Dicks LMT.. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int J Syst Evol Microbiol. 2012;62(Pt 3):500–504. 10.1099/ijs.0.031054-0. [DOI] [PubMed] [Google Scholar]
- Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA.. Standard methods for research on Apis mellifera gut symbionts. J Apicultural Res. 2013;52(4):1–24. 10.3896/IBRA.1.52.4.07. [DOI] [Google Scholar]
- Engel P, Stepanauskas R, Moran NA.. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 2014;10(9):e1004596. 10.1371/journal.pgen.1004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felis GE, Dellaglio F.. Taxonomy of Lactobacilli and Bifidobacteria. Curr Issues Intest Microbiol. 2007;8(2):44–61. [PubMed] [Google Scholar]
- Goh YJ, Barrangou R, Klaenhammer TR.. In vivo transcriptome of Lactobacillus acidophilus and colonization impact on murine host intestinal gene expression. mBio. 2021;12(1):1–19. 10.1128/mBio.03399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinane CM, Kent RM, Norberg S, Hill C, Fitzgerald GF, Stanton C, Ross RP.. Host specific diversity in Lactobacillus johnsonii as evidenced by a major chromosomal inversion and phage resistance mechanisms. PLoS One. 2011;6(4):e18740. 10.1371/journal.pone.0018740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitúa MC, Vik D, Sullivan MB, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9(1):1–13. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SGE.. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26(18):2334–2335. 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P.. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–2122. 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R, Blom J, Palva A, Siezen RJ, de Vos WM.. Comparative genomics of Lactobacillus. Microb Biotechnol. 2011;4(3):323–332. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft K, Zhou Z, Anantharaman K.. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8(1):855387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, Mancenido AL, Moran NA.. Genome sequences of Lactobacillus sp. strains wkB8 and wkB10, members of the Firm-5 clade, from honey bee guts. Genome Announc. 2014;2(6):e01176-14. 10.1128/genomeA.01176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, Moran NA.. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63(Pt 6):2008–2018. 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- Kwong WK, Moran NA.. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14(6):374–384. 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer A, Simon MA, Banning JL, Lam BA, Harris RN.. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J. 2008;2(2):145–157. 10.1038/ismej.2007.110. [DOI] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine M, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103(42):15611–15616. 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, Moy J, Moran NA.. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol. 2012;78(8):2830–2840. 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R.. The microbial pan-genome. Curr Opin Genet Dev. 2005;15(6):589–594. 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Moran NA. Genomics of the honey bee microbiome. Curr Opin Insect Sci. 2015;10:22–28. 10.1016/j.cois.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura de Sousa JA, Pfeifer E, Touchon M, Rocha EPC.. Causes and consequences of bacteriophage diversification via genetic exchanges across lifestyles and bacterial taxa. Mol Biol Evol. 2021;38(6):2497–2512. 10.1093/molbev/msab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling DP, Endo A, Dicks LMT.. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and bee-hives. Curr Microbiol. 2012;65(5):507–515. . [DOI] [PubMed] [Google Scholar]
- Olofsson TC, Alsterfjord M, Nilson B, Butler È, Vásquez A.. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int J Syst Evol Microbiol. 2014;64(Pt 9):3109–3119. 10.1099/ijs.0.059600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Sadiq FA, Han X, Zhao J, Zhang H, Paul Ross R, Lu W, Chen W.. Comprehensive scanning of prophages in Lactobacillus: distribution, diversity, antibiotic resistance genes, and linkages with CRISPR-Cas systems. mSystems. 2021;6(3):e0121120. 10.1128/mSystems.01211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JE, Martinson VG, Urban-Mead K, Moran NA.. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol. 2014;80(23):7378–7387. 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. FigTree v1.3.1 [Online]; 2010. https://github.com/rambaut/figtree [accessed 2021].
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna (Austria: ): R Foundation for Statistical Computing; 2021. https://www.R-project.org/. [Google Scholar]
- Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale FF, Nunes A, Oleastro M, Gomes JP, Sampaio DA, Rocha R, Vítor JMB, Engstrand L, Pascoe B, Berthenet E, et al. Genomic structure and insertion sites of Helicobacter pylori prophages from various geographical origins. Sci Rep. 2017;7:42471. 10.1038/srep42471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: legant Graphics for Data Analysis. New York (NY: ): Springer-Verlag; 2016. [Google Scholar]
- Zhang Z, Lv J, Pan L, Zhang Y.. Roles and applications of probiotic Lactobacillus strains. Appl Microbiol Biotechnol. 2018;102(19):8135–8143. . [DOI] [PubMed] [Google Scholar]
- Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. 2020;70(4):2782–2858. 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies and raw data can be found here: http://www.ncbi.nlm.nih.gov/bioproject/866146, but note that these assemblies have been cleaned of any contigs <200 bp by NCBI requirements. Full assemblies, processing scripts, and associated metadata can be obtained from, https://doi.org/10.7294/19524946. File LB24_final_contigs.fasta contains the complete final contig list of LB24. File LB25_final_contigs.fasta contains the complete final contig list of LB25. File LB26_final_contigs.fasta contains the complete final contig list of LB26. File lactobacillus_MS_metadata_scripts_readme.docx contains all scripts used in the metadata analysis in a word document. File 09_scripts.zip contains all scripts used in the metadata analysis.
Supplemental material is available at G3 online.