Abstract
Transcription factors play important roles in the development of the intestinal epithelium and its ability to respond to endocrine, nutritional, and microbial signals. Hepatocyte nuclear factor 4 family nuclear receptors are liganded transcription factors that are critical for the development and function of multiple digestive organs in vertebrates, including the intestinal epithelium. Zebrafish have 3 hepatocyte nuclear factor 4 homologs, of which, hnf4a was previously shown to mediate intestinal responses to microbiota in zebrafish larvae. To discern the functions of other hepatocyte nuclear factor 4 family members in zebrafish development and intestinal function, we created and characterized mutations in hnf4g and hnf4b. We addressed the possibility of genetic redundancy amongst these factors by creating double and triple mutants which showed different rates of survival, including apparent early lethality in hnf4a; hnf4b double mutants and triple mutants. RNA sequencing performed on digestive tracts from single and double mutant larvae revealed extensive changes in intestinal gene expression in hnf4a mutants that were amplified in hnf4a; hnf4g mutants, but limited in hnf4g mutants. Changes in hnf4a and hnf4a; hnf4g mutants were reminiscent of those seen in mice including decreased expression of genes involved in intestinal function and increased expression of cell proliferation genes, and were validated using transgenic reporters and EdU labeling in the intestinal epithelium. Gnotobiotics combined with RNA sequencing also showed hnf4g has subtler roles than hnf4a in host responses to microbiota. Overall, phenotypic changes in hnf4a single mutants were strongly enhanced in hnf4a; hnf4g double mutants, suggesting a conserved partial genetic redundancy between hnf4a and hnf4g in the vertebrate intestine.
Keywords: nuclear receptor, transcription factor, HNF4, NR2A, intestine, microbiota, zebrafish development, host–microbe interaction
Introduction
The intestinal epithelium is a highly proliferative tissue that integrates and responds to diverse signals from the diet and microbiome within the intestinal lumen and to signals from the body’s endocrine and nervous systems to perform its primary function of nutrient absorption. Transcription factors establish intestinal cell identities during development, and direct tissue renewal, differentiation, and response to external stimuli in the developed intestinal epithelium. To carry out these activities, transcription factors primarily function by binding to specific DNA sequences and regulating the expression of neighboring genes. This allows them to transmit information, generate cellular identity, and respond to the environment in a tissue-specific manner. As multiple transcription factors contribute to the identity of a tissue, understanding how they work both independently and together is challenging. This issue is further complicated when considering how transcription factors from the same family function in the same tissues and cells, as they often bind to similar or identical DNA sequences and have related functions. Multiple transcription factor families, including hepatocyte nuclear factor 4 (Hnf4 or NR2A) family transcription factors, have been implicated in intestinal development and function (Heppert et al. 2021).
Hnf4 family transcription factors are members of the nuclear receptor superfamily of liganded transcription factors which are essential for the development and function of multiple digestive tissues in mammals, including the intestinal epithelium (Sladek et al. 1990). Hnf4 family transcription factors have been linked to human diseases such as hepatic and GI cancers (Chellappa et al. 2016; Wang et al. 2020; Lv et al. 2021), inflammatory bowel diseases (UK IBD Genetics Consortium et al. 2009; Jostins et al. 2012; Marcil et al. 2012; Yang et al. 2014), obesity (Berndt et al. 2013), and maturity-onset diabetes of the young 1 (Yamagata et al. 1996). Mammals encode 2 Hnf4 family members Hnf4a and Hnf4g, and many other vertebrates outside of the mammalian lineage also encode a third factor, Hnf4b. Hnf4a has historically been the most studied Hnf4 family member due to its roles in multiple digestive tissues and strong association with many of the human conditions mentioned above. In mice, whole animal knockouts of Hnf4a are lethal because it is essential in extra embryonic tissues prior to gastrulation (Chen et al. 1994; Duncan et al. 1997; Li et al. 2000). Conditional deletions of Hnf4a in the mouse intestinal epithelium have demonstrated that it regulates intestinal functions including lipid metabolism and absorption, barrier function, suppressing inflammation, hormone signaling, intestinal epithelial cell (IEC) proliferation, and enterocyte differentiation (Garrison et al. 2006; Ahn et al. 2008; Babeu et al. 2009; Cattin et al. 2009; Darsigny et al. 2009; Verzi et al. 2010; Frochot et al. 2012; Girard et al. 2019). Further, microbiota suppress Hnf4a activity in the intestinal epithelium by reducing Hnf4a DNA occupancy and target gene expression (Davison et al. 2017; Qin et al. 2018; Lickwar et al. 2022).
Although Hnf4a has received the most attention, it is not clear how it functions together with other Hnf4 family members, and whether their independent and cooperative functions are conserved. Compared with Hnf4a conditional deletions, mouse Hnf4g knockouts display only minor intestinal phenotypes suggesting their roles in the intestinal epithelium are not identical (Gerdin et al. 2006; Baraille et al. 2015). However, Hnf4a and Hnf4g are both broadly expressed in the mouse intestinal epithelium, have a common predicted DNA binding motif, and have been shown to bind the same gene regulatory regions in vivo (Fang et al. 2012; Davison et al. 2017; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019; Chen et al. 2020). Further, both factors respond similarly to microbial colonization, with reduced occupancy at most of their (shared) binding sites compared to germ-free conditions (Davison et al. 2017), suggesting that they may play overlapping roles in the intestinal epithelium. A series of recent studies created double Hnf4 mutants by conditionally eliminating Hnf4a from the intestine in Hnf4g mutant mice at different developmental stages. These studies revealed that loss of both Hnf4 orthologs leads to defects in fetal intestinal maturation resulting in early lethality, in enterocyte differentiation leading to an increased relative number of secretory goblet cells in the double knockout epithelium, in promoting expression of genes involved in lipid metabolism and fatty acid oxidation in intestinal stem cells resulting in stem cell loss from the intestinal epithelium, and in brush border formation (Chen, Toke, Luo, Vasoya, Aita, et al. 2019; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019; Chen et al. 2020, 2021). The increased severity of the double mutant phenotypes compared to that of the single mutants suggests that there is at least partial genetic redundancy between the 2 Hnf4 factors in the intestinal epithelium of mice. However, how Hnf4 family members are coordinated to simultaneously function across multiple intestinal epithelial cell types and how metabolic functions are integrated with cell differentiation and proliferation is not fully understood.
Hnf4 is thought to be one of the earliest branching transcription factor families in animals and is prevalent across extant animal lineages (Bridgham et al. 2010). In organisms as diverse as Caenorhabditiselegans, Drosophila, Xenopus, zebrafish, mice, and humans, Hnf4 family transcription factors regulate gene expression and processes related to lipid mobilization and transport, metabolism, and fatty acid oxidation (Hayhurst et al. 2001; Palanker et al. 2009; Frochot et al. 2012; Barry and Thummel 2016; Davison et al. 2017; Goh et al. 2018; Lee et al. 2021) and also function in diverse developmental processes and stem cell maintenance (Weber et al. 1996; Hahn-Windgassen and Van Gilst 2009; Gracida and Eckmann 2013; Chen, Toke, Luo, Vasoya, Aita, et al. 2019; Storelli et al. 2019; Chen et al. 2020; Dubois et al. 2020; Cheng et al. 2020), suggesting that these basic functions of the Hnf4 family are highly conserved. A comparison of the transcription factor binding motifs near the transcription start sites of genes expressed in the intestinal epithelium across multiple vertebrate species revealed a core set of signature genes involved in intestinal epithelial biology, including transcription factors in the Hnf4 family (Lickwar et al. 2017). Together, these findings suggest that Hnf4 family transcription factors may play conserved roles in intestinal function across the vertebrate lineage.
Zebrafish (Danio rerio) have emerged as a useful system for studying the role of transcription factors in intestinal biology. Multiple aspects of intestinal function, development, gene expression, anterior–posterior patterning, cell types, and microbial response in zebrafish are conserved with those in mice and humans (Ng et al. 2005; Wallace et al. 2005; Bates et al. 2007; Muncan et al. 2007; Chen et al. 2009; Cheesman et al. 2011; Davison et al. 2017; Lickwar et al. 2017; Park et al. 2019; Ye et al. 2019, 2021; Li et al. 2020; Wen et al. 2021; Tavakoli et al. 2022). Like in other fishes and vertebrates outside of the mammalian lineage, the zebrafish genome encodes 3 Hnf4 family transcription factors hnf4a, hnf4g, and hnf4b. hnf4a and hnf4g are thought to be direct orthologs of the mammalian Hnf4a and Hnf4g, suggesting loss of hnf4b in the mammalian lineage (Bertrand et al. 2004). Previous work from our lab utilized zebrafish gnotobiotics to identify Hnf4 family members as microbially responsive transcription factors (Camp et al. 2012; Davison et al. 2017), and further comparisons with mouse and human datasets demonstrated that the Hnf4a-microbiota signaling axis plays a conserved role in suppressing inflammation in the intestinal epithelium (Davison et al. 2017).
Here we sought to expand our knowledge of transcription factor function in the zebrafish intestine, explore the potential role of other Hnf4 family transcription factors in microbial response, and address the potential redundancy of Hnf4 family member function in zebrafish. To do so, we created and characterized mutants in the remaining Hnf4 family members, hnf4g and hnf4b. By crossing these mutants and the existing hnf4a mutants, we identified phenotypes of double and triple mutants for the Hnf4 transcription factor family in zebrafish. Finally, we compared gene expression in the digestive tracts of hnf4a and hnf4g single mutants to hnf4a; hnf4g double mutants to assess their individual roles and potential genetic redundancy between these factors in zebrafish larvae. The changes in gene expression and phenotypes we observed in hnf4a and hnf4g single mutants and the hnf4a; hnf4g double mutants in larval zebrafish are strikingly similar to those observed in mice. We conclude that hnf4a and hnf4g show partial genetic redundancy in the intestinal epithelium in zebrafish and play highly conserved roles in intestinal function in vertebrates.
Materials and methods
Zebrafish husbandry
Zebrafish were reared and all experiments were performed following protocols established and approved by the Duke University Medical Center Institutional Animal Care and Use Committee (IACUC Protocol Number A096-19-04). Zebrafish stocks were raised on a recirculating system with a 14/10 h light/dark cycle at 28.5°C, and adults were fed Artemia (Brine Shrimp Direct, BSEACASE) twice a day and a 1:1 mixture of Gemma Micro 500 and Wean 0.5 (Bio-Oregon, B1473 and B2818) once a day. New zebrafish lines were generated on a mixed TL/Tü background and were subsequently ingressed into an EK background. Embryos for experiments were collected from natural matings. Conventionally raised larvae were maintained on a 14/10 h light/dark cycle at 28.5°C in petri dishes in Egg Water (0.3% [w/v] crystal sea salt, 0.75 mM CaCO3, 0.45 mM NaHCO3, methylene blue). We used the transgenic lines Tg(−4.5fabp2: DsRed)pd1000 (Kanther et al. 2011), Tg(fabp6: GFP)rdu22 (Lickwar et al. 2017), Tg(in3.4: cfos: GFP)nc2b (Camp et al. 2012), which were previously reported by us. All zebrafish lines used in this study are included in the Reagent Table.
Wholemount in situ hybridization
Wholemount in situ hybridization for hnf4a, hnf4b, and hnf4g were performed as described previously (Park et al. 2019). To make riboprobes, primers were designed to amplify the 3′ end of the transcripts, targeting all characterized isoforms of each gene (Fig. 1a and Supplementary Table 2). A T7 promoter sequence was included in the reverse primer of the antisense probes and the forward primer of the sense probes. Products were amplified from zebrafish larval cDNA using the primers specified in Supplementary Table 2 and Q5 DNA polymerase (NEB) and visualized on an agarose gel. DNA from six 25 μl PCR reactions was pooled and ethanol precipitated, and 1 μg of DNA was used in an in vitro transcription reaction with T7 RNA polymerase (NEB) and 10× DIG RNA Labeling Mix (Roche). Probes were visualized on an agarose gel, the concentration of each was measured via Nanodrop, and 150 ng of labeled probe was used in 200 μl hybridization mix for each sample. Unfed conventionally raised 6 dpf zebrafish larvae were fixed overnight in 4% paraformaldehyde at 4°C. Chromogenic reaction was stopped at various intervals, larvae were washed, fixed, and mounted in glycerol for imaging.
Fig. 1.
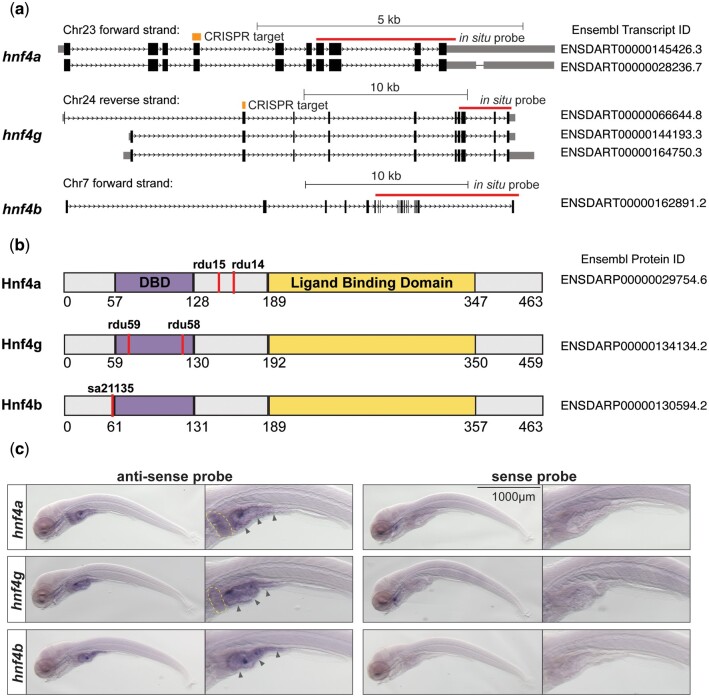
Members of the hnf4 gene family are expressed in the zebrafish intestinal epithelium and liver at 6 dpf. a) Gene models of the major known isoforms for the 3 zebrafish Hnf4 homologs are shown. Splice forms depicted are those predicted to be protein encoding and complete by Ensembl (Supplementary Table 1). Gene models were derived from UCSC genome browser tracks for GRCz11/danRer11 and are shown in 5′-3′ orientation; black bars are exons and gray bars are noncoding regions; thin red bars indicate exons covered by in situ hybridization riboprobes; thick orange bars indicate regions targeted by CRISPR gRNAs. Where characterized, 3′ UTRs are shown in gray at the 3′ end of each transcript. b) Protein models of Hnf4 homologs are shown with the DBD (purple) and ligand binding domain (yellow) defined by SMART protein domain prediction (Letunic and Bork 2018). Alleles encoding nonsense mutations are indicated by vertical red lines where stop codons occur. c) Wholemount in situ hybridization against hnf4 transcripts in 6 dpf wild-type zebrafish larvae. Antisense probe results are shown at left, and control sense probes are shown at right for each gene. Representative whole animals are shown, plus an inset focused on the digestive tract. For all 3 genes in the antisense condition, purple staining indicates the chromogenic reaction in the intestinal epithelium (arrow heads). A yellow dotted line outlines the liver in hnf4a larva and in hnf4g larva where some purple staining in the region is observed in the antisense condition.
Generating hnf4g and hnf4b mutant lines
Targeted mutagenesis of the hnf4g locus was performed using CRISPR/Cas-based genome editing as described in Davison et al. (2017). Briefly, 2 guide RNAs were designed using the MIT CRISPR Design Tool (http://cripsr.mit.edu; last accessed 2017), cloned (see Supplementary Table 2 for primers), and ligated into plasmid pT7-gRNA (http://www.addgene.org/46759/). Guide RNAs were produced via an in vitro transcription reaction using the MEGAshortscript T7 kit (Ambion/Invitrogen AM1354). Cas9 RNA was produced from plasmid pT3TS-nls-zCas9-nls (http://www.addgene.org/46757/) using the mMESSAGE mMACHINE T3 kit (Ambion/Invitrogen 1348) for in vitro transcription. One- to 2-cell stage zebrafish embryos (Tü background) were injected with 150 ng/µl nls-zCas9-mls, 34 ng/µl of each guide RNA (both individually and in combination), 0.05% phenol red, 120 mM KCl, and 20 mM Hepes Buffer (pH 7.0). The resulting zebrafish were screened for mutagenesis events using the Melt Doctor High Resolution Melting Assay (Thermo Fisher Scientific 4409535) and then confirmed by cloning and screening PCR products by gel electrophoresis and sequencing. Two separate alleles within the CRISPR-targeted region were isolated: the first, hnf4grdu58, was a single base-pair deletion which was predicted to result in a nonsense mutation beginning at amino acid 62 and a premature stop codon after amino acid 126. The second, hnf4grdu59, was an 8 bp insertion which was predicted to result in a nonsense mutation also beginning at amino acid 62 and ending after amino acid 73 in a premature stop codon (Fig. 1b and Supplementary Fig. 1a and b). hnf4grdu58 is the allele utilized for the experiments throughout this manuscript. The hnf4bsa21135 allele was generated and acquired from the Zebrafish Mutation Project (Kettleborough et al. 2013). hnf4bsa21135 allele is a point mutation which results in a stop codon being introduced after amino acid 55 (Fig. 1b and Supplementary Fig. 2, a and b).
qRT-PCR and sequencing of Hnf4 transcripts from hnf4g and hnf4b mutants
hnf4g+/+ and hnf4g−/− siblings were in-crossed, and 5 groups of 25 larvae from each genotype were euthanized and collected into Trizol (Invitrogen) at 6 dpf for RNA extraction. For hnf4b mutants, hnf4b+/+ and hnf4b−/− siblings were in-crossed, and groups of 10 larvae from each genotype were collected at 6 dpf for RNA extraction (4-5 groups per genotype). RNA extraction and cDNA synthesis were performed as in Davison et al. (2017), and as described elsewhere in Materials and Methods for RNA sequencing experiments. Primer sequences can be found in Supplementary Table 2. 18S primers were used at 2 μM, and all other primers were used at 10 μM concentration. Quantitative PCR was performed using 2× Sybr Green Master Mix (Applied Biosystems) in 25 μl reactions per the manufacturer’s instructions and run on a QuantStudio 3 Real-Time PCR System (Thermo Fisher). Transcript abundance for Hnf4 genes were normalized to 18S transcript levels and plotted using Prism Software (GraphPad). Sanger sequencing of both the hnf4g−/− and hnf4b−/− transcripts was performed using cDNA extracted for qPCR. The transcripts were amplified from the cDNA using gene-specific primers (hnf4g F: 5′-CTCCAACTCCTCTCCCTAAATCC, R: 5′-GTGAAAGTGCTTCAGCGTGAG; hnf4b F: 5′-ATGAAGATCTCTGGGTCGTC, R: 5′-TTAAAGGCACGTGTGGACG) and Q5 High-Fidelity 2X-Master Mix (NEB), and the products were column purified and sequenced via Sanger sequencing.
Genotyping for hnf4a, hnf4g, and hnf4b
For genotyping larvae older than 6 dpf and adults, zebrafish were anesthetized in 0.75 mM Tricaine solution and a few millimeters of tissue was clipped from the tail fin and placed in 50 μl of 50 mM sodium hydroxide using sterilized scissors and forceps. For genotyping larvae, whole 6 dpf larva were euthanized and transferred into 50 μl of 50 mM sodium hydroxide in PCR tubes or 96 well PCR plates using sterilized forceps. DNA was extracted for genotyping of adult and larval zebrafish by lysing tissue at 98°C for 25 min followed by vortexing, the addition of 5 μl 1 M Tris Buffer (pH 8), vortexing to mix, and spinning down the samples at max speed for 1 min. The resulting supernatant was used as the template in PCR reactions.
Standard genotyping PCR reactions used 2× GoTaq Master Mix (Promega), primers at 10 μM concentration, and 1 μl lysis product for DNA template. The following primers were used to amplify regions containing genetic lesions for each gene respectively: hnf4a (F: 5′-TGATTCACACTACTTACTTGTCTAG, R: 5′-GATTAAAAGTAGTTATCTCATCCTCAG); hnf4g (F: 5′-GAGTGTGTCTGTTGTTGTTGACAG, R′-GTAGACATGGCTCTTCCTGATACTG); hnf4b (F: 5′-CCATCTAAGATCCACTAATGCC, R: 5′-CCTGCATGTGTAGGCATGGTTCTTG) (also available in Supplementary Table 2). The 43-bp deletion in hnf4ardu14 was detectable by size on a 1% agarose gel run at 100 V for 1 h. For hnf4grdu58 and hnf4grdu59, 3 μl of PCR products (∼200 bp) were digested with the restriction enzyme BstUI (NEB) in a 20-μl reaction volume, per manufacturer’s instructions, for a minimum of 3 h to ensure complete product cleavage. Wild-type products are cleaved completely by BstUI whereas hnf4grdu58 and hnf4grdu59 mutations destroy the BstUI restriction site and PCR products remain uncut. Therefore, wild-type lanes result in a 2-band doublet (∼100 bp), heterozygote reactions result in lanes with a mutant band (∼200 bp) and a 2-band doublet (∼100 bp), and mutants have only a single band (∼200 bp). Bands were best resolved on 2–2.5% agarose gels. Unpurified PCR reactions for hnf4bsa21135 genotyping were Sanger sequenced using the reverse primer. Sequences were compared to a wild-type reference sequence using ApE (A Plasmid Editor) software and determined to be wild type, heterozygous, or mutant based on chromatogram peaks.
Brightfield imaging
Zebrafish embryos were harvested from breeding tanks and conventionally raised in 10 cm dishes in egg water (see Zebrafish Husbandry). On the indicated developmental day, larvae were anesthetized with Tricaine (0.75 mM), mounted in 3% methylcellulose, and imaged using a Leica M205 FA with a Leica DFC 365FX camera. Animals were then euthanized and genotyped for the alleles indicated by the methods described earlier.
RNA sequencing
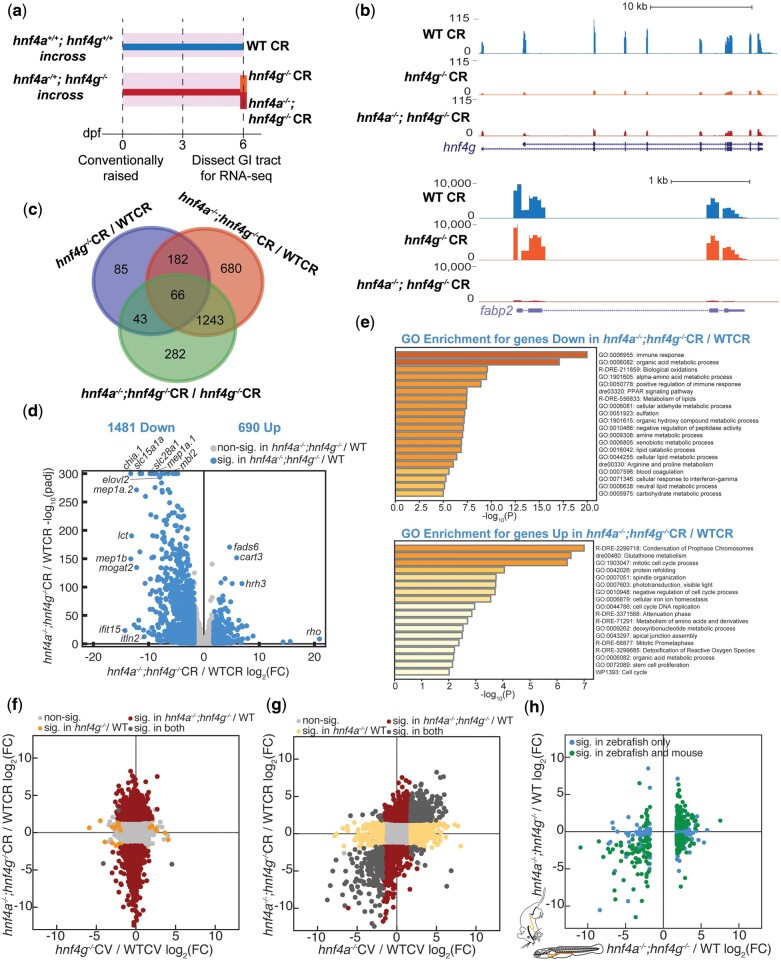
Larvae for RNA sequencing experiments were raised as described previously (see Zebrafish Husbandry). For hnf4g CV/GF RNA-seq, embryos were harvested separately from in-crosses of hnf4g+/+ and hnf4g−/− siblings. The embryos were made axenic by antibiotic treatment and surface sterilization (Pham et al. 2008). Half of the embryos from each genotype were left germ-free and half were conventionalized (colonized with a microbiota collected from zebrafish system water) at 3 dpf. At 6 dpf, digestive tracts from 12 to 15 larvae per flask were dissected by hand using fine forceps under a dissecting microscope and placed in Trizol. Conditions and genotypes of the groups being dissected were rotated, and all dissections for a given experiment occurred within a 5 h window of time. For hnf4g CV/GF RNA-seq, 4 samples of each genotype and condition were submitted for sequencing (16 total samples), which were the combined result of 2 separate gnotobiotic derivations. For the hnf4a; hnf4g CR RNA-seq experiment, hnf4a+/+; hnf4g+/+ and hnf4a−/+; hnf4g−/− adult siblings were in-crossed and larvae were raised conventionally. At 6 dpf, the digestive tracts of 4 groups of 10 wild-type larvae resulting from the hnf4a+/+; hnf4g+/+ cross were dissected and placed in Trizol. From the hnf4a−/+; hnf4g−/− in-cross progeny, 192 larval digestive tracts were dissected and placed in 50 μl of Trizol in individual wells in two 96 well plates and frozen at −80°C, the remainder of the carcass was placed in the same well of a corresponding plate with genotyping buffer. The larvae were then genotyped for hnf4a as described previously, and approximately Mendelian ratios of hnf4a alleles were recovered (hnf4a+/+-54; hnf4a−/+-91 hnf4a−/−-43; failed PCR-4). Wells with hnf4a+/+; hnf4g−/−, and hnf4a−/−; hnf4g−/− were then pooled into 4 groups of 10–11 digestive tracts per genotype for RNA extraction and sequencing.
The same RNA extraction, library preparation, and sequencing protocols were used for both hnf4g and hnf4a; hnf4g RNA sequencing experiments. Larval digestive tracts in Trizol were kept on ice and homogenized with a 27-Ga needle on a 1-ml syringe for 12 passages/sample, and then incubated at room temperature for 5 min. To extract RNA, the PureLink RNA Mini Kit (Invitrogen) was used according to the manufacturer’s instructions for using the kit with Trizol reagent. Briefly, 200 μl of chloroform was added to each sample, and the tubes were shaken vigorously by hand for 30 seconds. After centrifugation at 4°C, the aqueous phase was taken to a new tube and 1 ml of 70% ethanol was added. Samples were then transferred to a PureLink RNA Mini Kit column, the kit protocol was followed. On-column DNase (New England Biolabs M0303L) treatment was performed, and samples were eluted in 30 μl nuclease-free water. The purified RNA was stored at −80°C before submission to the Sequencing and Genomic Technologies Shared Resource at the Duke Center for Genomic and Computational Biology (GCB) for sequencing. RNA-sequencing libraries for stranded mRNA-sequencing were made by the GCB Core and 50 bp paired-end sequencing was performed using one NovaSeq 6000 S-Prime flow cell (Illumina) for each experiment. The average number of reads for each sample in the hnf4g dataset was ∼52 million reads and in the hnf4a; hnf4g dataset was ∼85 million reads.
RNA-sequencing analysis
For the hnf4g CV vs GF RNA-sequencing experiment, the raw sequencing read files were loaded into Galaxy, and paired read files were concatenated and then trimmed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The trimmed reads were then mapped to the Danio rerio genome (danRer11, Ensembl) using STAR RNA-seq (Dobin et al. 2013). FeatureCounts was used to quantify the number of reads mapped per gene from the BAM file (Liao et al. 2014). DESeq2 was run using the featureCounts output to obtain differential gene expression (log2(FC)) and statistics (adjusted P-values) at each loci in the different conditions (Supplementary Table 3) (Love et al. 2014). The BAM files were converted to Bigwig files using bamCoverage in Galaxy with RPKM normalization and uploaded to the UCSC Genome Browser for visualization (http://genome.ucsc.edu).
To map the reads from our hnf4a; hnf4g CR RNA-sequencing experiment and re-map the previously collected hnf4a CV vs GF RNA-sequencing experiment (PRJNA354631) (Davison et al. 2017), raw reads were processed using the Trim Galore toolkit (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), which employs Cutadapt (Martin 2011) to remove low quality bases and adapter sequences from the 3′ ends of the reads. Only reads that were 20 nt or longer after trimming were kept for further analysis. Reads were mapped to the Danio rerio genome (danRer11, Ensembl) using the STAR RNA-Seq alignment tool (Dobin et al. 2013). Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool (Putri et al. 2021).
To facilitate comparison of all 3 datasets and correct for variation amongst the datasets that was due to the technical limitation that they were performed and sequenced separately, we performed a batch effect adjustment. The read counts from all 3 datasets were compiled and genes that weren’t present with at least 10 reads in any one sample were excluded. The data were normalized using the Anders and Huber method (Love et al. 2014) and then batch corrected in Limma (Ritchie et al. 2015) using the “removeBatchEffect” algorithm. Differential expression analysis of the merged datasets was performed using a linear model framework in Limma using an empirical Bayes’ test statistic. The false discovery rate was used to correct for multiple hypothesis testing. To determine which genes within a given comparison were differentially expressed, we applied a log2 fold change (log2(FC)) cutoff of −/+1.75 and an adjusted P-value threshold of less than 0.05 to all the datasets analyzed in this study. This cutoff was selected because (1) functional enrichment analysis becomes relatively uninformative if a differential gene set is too large, (2) we wanted to select a single cutoff to apply to all comparisons across individual datasets and the batch corrected dataset, and (3) comparison of hnf4a−/−; hnf4g−/− double mutants to wild-type controls revealed differential expression of a very large number of genes (3,006 using the chosen cutoff, 8,888 genes if we had used a typical log2(FC) cutoff of −/+1.0 with an adjusted P-value threshold of less than 0.05, which is approximately a third of the genes in the genome). When possible, the data in our analyses were represented in such a way as to include all genes with their log2(FC) values and not just those that passed the significance threshold. Log2(FC) values and adjusted P-values for all genes are available in the Supplementary Table 3 Excel file. Venn Diagrams were created using a web tool (https://bioinformatics.psb.ugent.be/webtools/Venn/). Scatter plots were generated using Excel and Plot2 graphing software. Metascape was used to analyze gene sets for Gene Ontology (GO) Terms (Zhou et al. 2019).
Fluorescence imaging and analysis of transgenic larvae
Adult hnf4g−/+ zebrafish were crossed with hnf4g−/+; Tg(in3.4: cfos: GFP), embryos were collected, and beginning at 3 dpf larvae were housed in egg water at a density of 1 larva per ml. At 6 dpf, larvae were mounted in 3% methyl cellulose and imaged using a Leica M205 FA stereo fluorescence microscope with GFP bandpass filter set. After imaging, larvae were euthanized and genotyped for both hnf4g and GFP (for genotyping primer sequences, see Supplementary Table 2). The fluorescence channel images of animals determined to be hnf4g+/+ or hnf4g−/− and GFP positive, were quantified using FIJI image analysis software. An ROI was defined for each larva encompassing both epithelial monolayers of the anterior intestine and a background ROI for each image was measured just below each larva. The total fluorescence of the tissue was measured by subtracting the average background fluorescence from the integrated density of the fluorescence in the intestinal ROI. The fluorescence intensities measured for the individual larva were plotted in PRISM and the average fluorescence intensity for wild type and mutants was compared via Student’s t-test. The data presented are 3 separate biological replicates with a total of n = 24 hnf4g+/+ and n = 23 fish hnf4g−/−, respectively.
Fluorescence was quantified for Tg(fabp2: DsRed) and Tg(fabp6: GFP) transgenic reporter lines by euthanizing 6 dpf zebrafish larvae, embedding larvae in 3% methylcellulose, and imaging via a Leica M205 FA stereomicroscope with identical exposure time and magnification in the same experiment. Imaged larvae were placed in 50 mM NaOH in preparation for the appropriate genotyping protocols. Image analysis was performed using FIJI software. We analyzed the changes in fluorescence for the 2 transgenes by different methods because in one case we observed only a change in intensity of fluorescence (Tg(fabp2: DsRed)) and in the other, we observed changes in the expression pattern (Tg(fabp6: GFP)) (Fig. 5c). To quantify fabp2 reporter expression, the shape tool in FIJI was used to outline the intestine as the region of interest (ROI) with attention paid to avoid capturing yolk expression. The mean fluorescence values within this ROI were calculated and then plotted using GraphPad Prism statistical software. To quantify fabp6 reporter, fluorescence intensity along the length of the intestine, starting from the cloaca, was measured using the line and plot profile functions in FIJI software. Mean fluorescence plots were graphed in GraphPad Prism software.
Fig. 5.
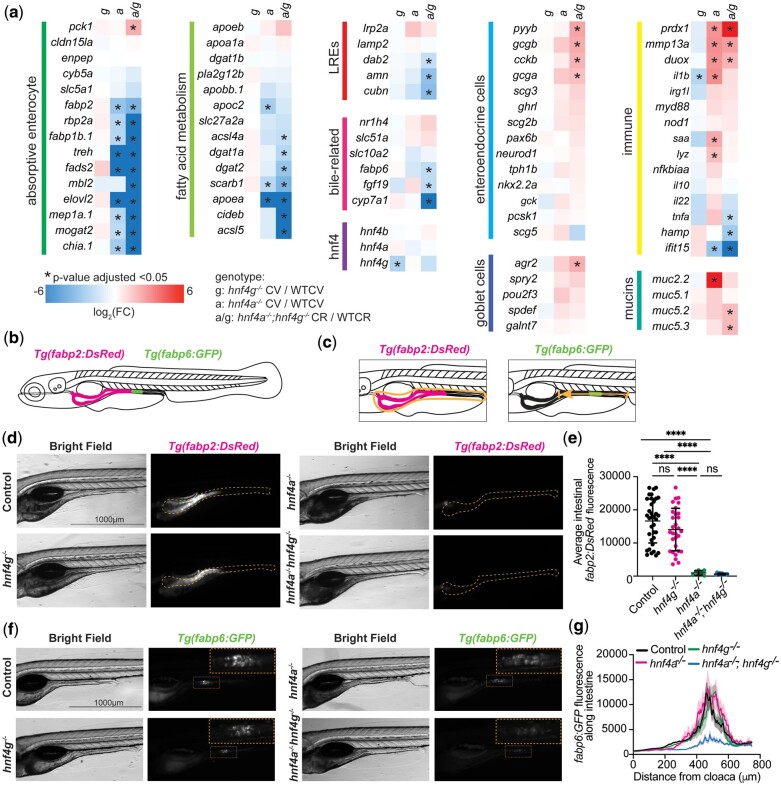
Multiple cell types in the intestinal epithelium are significantly altered in hnf4a−/− and hnf4a−/−; hnf4g−/− larvae. a) Heatmaps showing changes in gene expression in hnf4g−/− CV/WTCV, hnf4a−/− CV/WTCV, hnf4a−/−; hnf4g−/− CR/WTCR for groups of genes involved in specific intestinal cell types or functions. Red indicates an increase in gene expression and blue, a decrease. Genes that are significantly different between genotypes are marked with an asterisk. b) A schematic depicting the localization of Tg(fabp2: DsRed) and Tg(fabp6: GFP) in a wild-type zebrafish larva at 6 dpf. c) Shows the method of quantification for changes in fluorescence for each of the transgenes to produce the graphs in panels (e) and (g). d) Representative images of 6 dpf larvae expressing Tg(fapb2: DsRed) grouped by genetic mutant background. e) The average fluorescence intensity of the intestinal area is plotted for each mutant group. Error bars represent the mean and 95% confidence interval for each condition, and statistical differences determined by a 1-way ANOVA with multiple comparisons (**** P < 0.0001) are shown above the plot (control n = 35, hnf4g−/−n = 30, hnf4a−/−n = 7, hnf4a−/−; hnf4g−/−n = 10). f) Representative images of 6 dpf larvae expressing Tg(fabp6: GFP) grouped by genetic mutant background. g) Quantification of fluorescence intensity along the intestine from the cloaca opening to the swim bladder. Heavy lines represent mean intensity values and shading around the line is the standard error (SEM) (control n = 19, hnf4g−/−n = 20, hnf4a−/−n = 9, hnf4a−/−; hnf4g−/−n = 7). Area under the curve measurements show overlapping 95% confidence intervals for the control and hnf4g−/− groups, but a significantly larger area for hnf4a−/− and smaller area for the hnf4a−/−; hnf4g−/− groups respectively, and comparing the curves by Tukey’s 1-way ANOVA over multiple comparisons reveals control vs hnf4g−/− (ns; P = 0.9836), hnf4a−/− (P = 0.0004), and hnf4a−/−; hnf4g−/− (P < 0.0001); hnf4a−/− vs hnf4g−/− (P = 0.0002) and hnf4a−/−; hnf4g−/− (P < 0.0001); and hnf4g−/− vs hnf4a−/−; hnf4g−/− (P < 0.0001).
Quantifying proliferative cells
Adult hnf4a−/+; hnf4g−/+ zebrafish were crossed with hnf4a+/−; hnf4g−/− and embryos were collected and housed in egg water until 5 dpf. At 5 dpf, larvae were incubated in 200 µM EdU (Cayman Chemical, 20518) diluted in 2% DMSO in egg water for 24 h. Following EdU incubation, larvae were euthanized in tricaine, tails were removed for genotyping, and the remaining carcasses were fixed in 4% paraformaldehyde overnight. After fixation, larvae were grouped in their respective genotypes, rinsed with wash buffer (PBS + 3% BSA), incubated with ice cold acetone for 7 min, rinsed with wash buffer, and permeabilized in PBS, 1% DMSO, and 1% Triton X-100 for 1 h. Following permeabilization, larvae were rinsed in a wash buffer, then incubated with Click-iT EdU Alexa Fluor 647 reagent (Invitrogen) for 1 h. After Click-iT EdU reaction, larvae were rinsed in the wash buffer, co-stained with DAPI, and mounted onto slides with coverslip. Slides were imaged using the Zeiss 780 upright confocal microscope detecting DAPI with the 405 nm Diode laser and EdU stained cells with the HeNe 633 nm laser. Zebrafish were visualized on the midsagittal plane, containing the length of the intestinal tract posterior to the intestinal bulb, which is the region containing the most intestinal proliferation at 6 dpf. Total intestinal epithelial cells (detected by DAPI) and EdU positive cells (intensity above a set threshold) along the length of the intestine were counted from 1 representative image for each fish. Data were analyzed while blinded to genotypes. Anterior and posterior regions were defined by the midway point between the intestinal bulb and cloaca.
Results
Hnf4 family gene expression and mutant generation
To better understand the role of Hnf4 family transcription factors in regulating gene expression in the zebrafish intestinal epithelium, we characterized single and compound mutants for each of the 3 zebrafish Hnf4 family members: hnf4a, hnf4g, and hnf4b. The protein encoding isoforms predicted for the 3 Hnf4 family transcription factors result in peptides with a conserved domain structure, including the DNA binding domain (DBD) and ligand binding domains, which are hallmarks of Hnf4 family members and other nuclear receptor transcription factors (Fig. 1, a and b, Ensembl, GRCz11) (Howe et al. 2021). In mammals, alternative splicing results in multiple isoforms of Hnf4a that have been shown to have distinct functions and change target gene expression (Dean et al. 2010; Chellappa et al. 2016; Ko et al. 2019; Lambert, Babeu, et al. 2020). Although the role of the multiple predicted hnf4a and hnf4g isoforms in zebrafish has not yet been explored, the exonic regions of both genes targeted for CRISPR-based mutagenesis are predicted to be present in all protein-encoding isoforms (Fig. 1a). For the hnf4a locus, a deletion mutant hnf4ardu14 was previously created and mutant phenotypes including changes in gene expression were characterized (Fig. 1, a and b) (Davison et al. 2017). We refer to the genotype of the hnf4ardu14 allele as hnf4a+/+, hnf4a−/+, or hnf4a−/−. To understand the role of each Hnf4 gene in development and intestinal homeostasis, we generated additional mutations in the hnf4g and hnf4b genes.
To create mutations in the hnf4g locus, we utilized CRISPR-Cas9 to create lesions in an exon encoding the DBD and isolated 2 alleles. We isolated the allele hnf4grdu58, a single base-pair deletion which was predicted to result in a nonsense mutation beginning at amino acid 62 and a premature stop codon after amino acid 126 (Fig. 1b and Supplementary Fig. 1, a and b). Here, we refer to the genotype of the hnf4grdu58 allele as hnf4g+/+, hnf4g−/+, or hnf4g−/−, where appropriate. Homozygous hnf4g−/− larva isolated from heterozygote hnf4g−/+ in-crosses were found to be viable at 6 dpf and displayed no obvious phenotypic differences with their wild-type and heterozygous siblings, in contrast to hnf4a−/− 6 dpf larvae which showed a narrowed intestinal lumen (Supplementary Fig. 1, c and d) (Davison et al. 2017). Fish resulting from hnf4g−/+ heterozygote in-crosses and raised in standard conditions survived to adulthood in ratios which were not different from the expected Mendelian ratios, as determined by a Chi-square statistical test (Supplementary Fig. 1e). To test for transcriptional adaptation for loss of hnf4g function by the other Hnf4 family genes, we performed qRT-PCR to measure gene expression using RNA extracted from 6 dpf hnf4g+/+ and hnf4g−/− larvae derived from separate, sibling in-crosses of hnf4g+/+ or hnf4g−/− adults (El-Brolosy et al. 2019). We found no significant differences in the levels of hnf4a and hnf4b transcripts between hnf4g+/+ and hnf4g−/− larvae, however, we did confirm a significant decrease in hnf4g transcript quantity (Supplementary Fig. 1f). The decrease in mutant transcript and lack of significant compensatory increase in the other Hnf4 alleles mirrored what was observed for the Hnf4 transcripts in hnf4a mutant alleles (Davison et al. 2017). To check for possible compensatory changes in mRNA processing (Anderson et al. 2017), we sequenced the hnf4g cDNA isolated from the hnf4g−/− larvae and found that it matched the expected transcript sequence and contained the deletion.
For the hnf4b locus, we acquired an ENU-induced mutant allele isolated as part of the Zebrafish Mutation Project (Kettleborough et al. 2013). hnf4bsa21135 is a point mutation which results in a stop codon being introduced after amino acid 55 (Fig. 1b and Supplementary Fig. 2, a and b). We refer to the genotype of hnf4bsa21135 as hnf4b+/+, hnf4b−/+, or hnf4b−/− throughout this manuscript. hnf4b−/− fish which were the result of hnf4b heterozygote adult in-crosses appeared phenotypically normal, were viable both as 6 dpf larvae and as adults and survived in approximately Mendelian ratios (Supplementary Fig. 2, c–e). We used qRT-PCR to check for transcriptional adaptation amongst Hnf4 genes in hnf4b−/− larvae and found none (Supplementary Fig. 2f). However, in contrast to the reduction in transcript observed in the hnf4a (Davison et al. 2017) and the hnf4g mutants, we found no difference in the amount of hnf4b transcript between the hnf4b+/+ and hnf4b−/− larvae. We went on to sequence the mRNA isolated from the hnf4b−/− larvae and found that the sequence matched the predicted sequence for the mutant, suggesting a lack of compensatory changes in mRNA processing that can occur with such alleles (Anderson et al. 2017).
We were specifically interested in the functions of Hnf4 family transcription factors in the intestinal epithelium and their dual role in maintaining proper tissue and cell specification, while simultaneously responding to environmental signals, including nutrition and intestinal microbes. Therefore, we focused on 6 dpf larvae for our studies, because maternal yolk has been completely utilized, larva are feeding and absorbing nutrients from ingested food, and we can successfully rear larvae germ-free through 6 dpf (Pham et al. 2008). Transcriptomic data from the dissected digestive tracts of wild-type larvae from a previous study suggests that all 3 Hnf4 family transcription factors are expressed in the digestive tract at 6 dpf (hnf4a 73.03, hnf4g 36.19, hnf4b 42.21 average FPKM; Davison et al. 2017). Using wholemount in situ hybridization in wild-type larvae, we confirmed that all 3 transcription factors appear to be expressed in the anterior intestinal epithelium, in the intestinal bulb and into the postbulb anterior epithelium at 6 dpf (Fig. 1c). This expression pattern closely mirrors that of the reporter transgene Tg(in3.4: fos: gfp), a cis-regulatory element which harbors multiple Hnf4 binding sites and is responsive to loss of hnf4a activity (Davison et al. 2017). We also observed hnf4a and hnf4g expression in the liver (Fig. 1c). These data are consistent with the expression observed for hnf4a, hnf4g, and hnf4b in 2 single-cell sequencing experiments which profiled gene expression in intestinal epithelia and the digestive tracts (Supplementary Fig. 3) (Wen et al. 2021; Willms et al. 2022).
Variable lethality dependent on Hnf4 mutant combinations
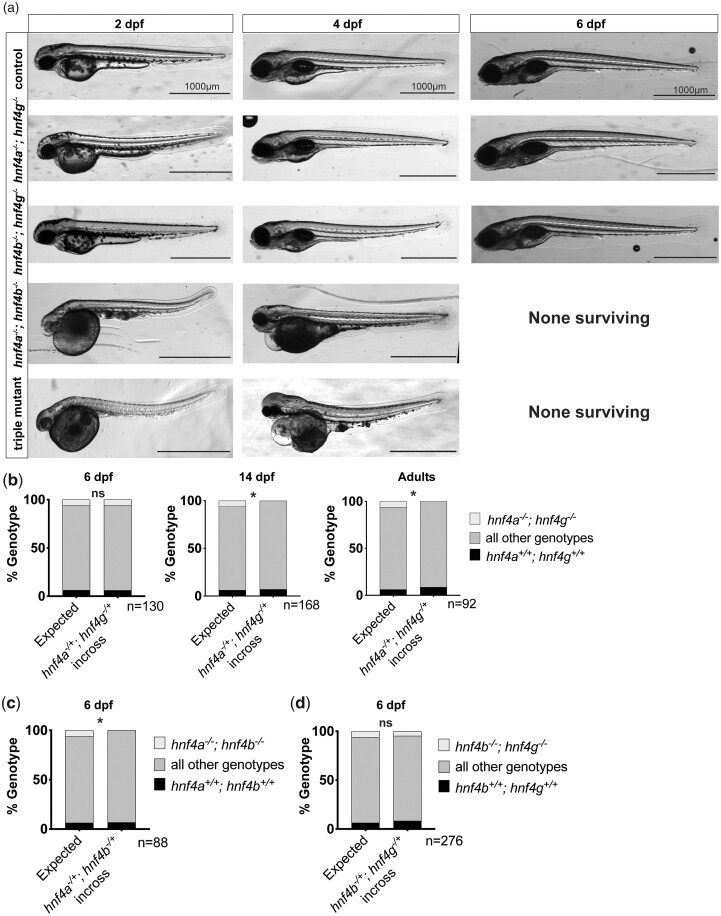
To better understand the genetic contributions of each Hnf4 family transcription factor, we sought to make double and triple mutants of hnf4a, hnf4g, and hnf4b. We first crossed our mutant alleles to generate adults which were heterozygous for multiple Hnf4 mutant alleles (e.g. hnf4a−/+; hnf4g−/+ or hnf4a−/+; hnf4b−/+). Triple heterozygous adults were in-crossed and their progeny were examined and imaged with a dissecting microscope, and subsequently genotyped at multiple stages of larval development (Fig. 2a). Both hnf4a−/−; hnf4g−/− and hnf4b−/−; hnf4g−/− larvae survived to 6 dpf and were phenotypically similar to control animals (controls were defined as larvae which were not homozygous mutant for any other hnf4 alleles), with the exception that hnf4a−/−; hnf4g−/− larvae displayed a more slender anterior intestinal lumen than controls, similar to what was previously observed in hnf4a−/− single mutants at 6 dpf (Davison et al. 2017) (Fig. 2a). Genotyping 6 dpf larvae from double heterozygous in-crosses of hnf4a−/+; hnf4g−/+ or hnf4b−/+; hnf4g−/+ revealed offspring, including double mutants, surviving in approximately Mendelian ratios (Fig. 2b;n = 130, and Fig. 2d;n = 276). However, for hnf4a−/−; hnf4b−/− and hnf4a−/−; hnf4b−/−; hnf4g−/− larvae, significant defects including developmental delay, cloudy yolk, edema, and death were observed as early as 2 dpf. Consistent with these results, when surviving 6 dpf larvae were genotyped from hnf4a−/+; hnf4b−/+ double mutant in-crosses, no viable hnf4a−/−; hnf4b−/− double mutant larva were recovered (Fig. 2c;n = 88).
Fig. 2.
Hnf4 compound mutants display variable rates of survival. a) Brightfield images of wild type, double, and triple mutant zebrafish larva recovered at 2, 4, and 6 dpf. Each row shows fish of the specified genotype and controls that are either wild type or heterozygous for each hnf4 allele. b–d) Allelic distribution of progeny genotyped at the indicated developmental stage following an in-cross of (b) hnf4a−/+; hnf4g−/+ (6 dpf, n = 130; P > 0.9999; 14 dpf, n = 168; P = 0.0447; adults n = 92; P = 0.0306), (c) hnf4a−/+; hnf4b−/+ (6 dpf, n = 88; P = 0.0447), and (d) hnf4b−/+; hnf4g−/+ (6 dpf, n = 276; P = 0.4864) individuals. The expected distribution is shown on the left of each graph, while the observed percentage of animals of each genotypic group is depicted by the bar on the right. The total number of individuals genotyped in each experiment is indicated by the “n” value below the graphs. Genotypic ratios were determined to be significantly different from expected by chi-square statistical test, * P < 0.05.
Though hnf4b−/− had no immediately observable phenotypes on its own, the fact that when combined with hnf4a−/−, we recovered no 6 dpf double mutant larvae suggests that the hnf4bsa21135 mutation results in a loss-of-function allele. The phenotypes and apparent early larval lethality observed for the hnf4a−/−; hnf4b−/− double mutant and hnf4a−/−; hnf4b−/−; hnf4g−/− triple mutants, combined with previous in situ hybridization observations that hnf4b is prominently expressed in the developing zebrafish embryo (Bertrand et al. 2007), suggest that hnf4a and hnf4b together may be required in zebrafish development prior to digestive tract formation and function at 6 dpf. Because of the early larval phenotypes we observed, we did not explore the role of hnf4b further and instead focused our investigations on hnf4a and hnf4g. Interestingly, despite surviving in Mendelian ratios at 6 dpf, we never recovered any hnf4a−/−; hnf4g−/− double mutant adults from hnf4a−/+; hnf4g−/+ in-crosses (Fig. 2b;n = 92) nor from triple heterozygous in-crosses. To better understand when hnf4a−/−; hnf4g−/− fish were dying, we genotyped hnf4a−/+; hnf4g−/+ in-cross progeny at 14 dpf (Fig. 2b;n = 168), and failed to recover any double mutant larvae, revealing that hnf4a−/−; hnf4g−/− double mutant larvae likely die sometime between 6 and 14 dpf. We focused our future analyses on the 6 dpf time point to uncover the essential roles of hnf4a and hnf4g at this developmental stage.
Deletion of hnf4g alone has a relatively minor impact on gene expression in the digestive tract
To better understand the role of hnf4g in zebrafish digestive tract physiology and response to microbiota, we analyzed changes in gene expression from the dissected digestive tracts of 6 dpf wild-type and hnf4g−/− larvae raised either germ-free (WTGF, hnf4g−/− GF) or ex-GF, colonized with a conventional microbiome from 3 dpf (WTCV, hnf4g−/− CV) (Fig. 3a). We identified transcripts with differential expression in WT and hnf4g−/− including a prominent reduction in hnf4g transcript levels in hnf4g−/− mutants. In addition, for previously characterized microbially responsive genes, such as saa, we observed an increase in CV compared to GF larvae (Fig. 3b). Using a log2(FC) cutoff of −/+1.75 and adjusted an P-value threshold of less than 0.05 (see Materials and Methods), we found 326 differentially expressed genes across the 4 experimental groups (Fig. 3c), with 42 transcripts were found to be significantly increased (12) or decreased (30) in hnf4g−/− CV compared to WTCV (Fig. 3d). We did not observe significant changes in any signature hnf4a target genes in hnf4g−/− CV compared to WTCV larvae (e.g. those involved in lipid and fatty acid metabolism), and the small number of significantly differential genes precluded systematic GO Term analysis. Amongst the downregulated genes, meltf, hpxb, and frrs1a are involved in iron metabolism, slc13a4 and slc30a8 are solute transporters, and itln3, irak3, and il1b function in immunity. In the upregulated gene set, we found nr0b2b, another nuclear receptor superfamily member and homolog of SHP/NR0B2, which is associated with metabolic disease and hepatocarcinoma (Zhang et al. 2011).
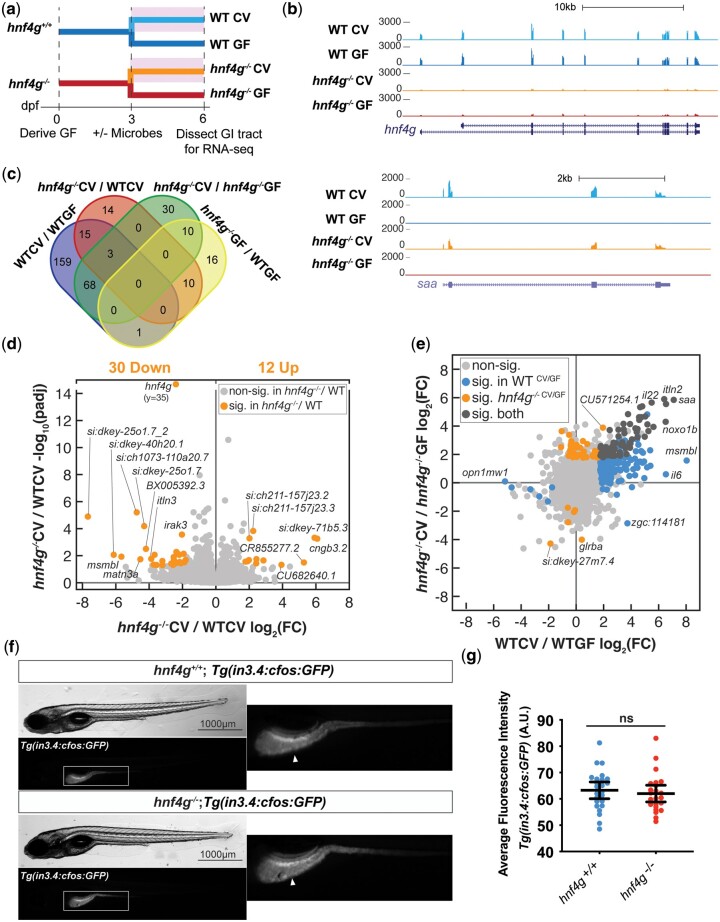
Fig. 3.
Deletion of hnf4g alone has a minimal impact on gene expression in the digestive tract. a) Schematic representation of zebrafish rearing and treatment across the experiment. Separate clutches of hnf4g+/+ and hnf4g−/− embryos were derived germ-free, and then half of each group was exposed to microbes at 3 dpf (shaded rectangle). Larvae were sacrificed at 6 dpf for RNA sequencing. b) Representative RNA-seq tracks for genes showing differential expression in WT vs hnf4g−/− (hnf4g) conditions and CV vs GF conditions (saa). c) Four-way Venn diagram showing the overlap of differential genes between the conditions analyzed. d) Volcano plot depicting the fold change and significance of all genes, with genes significantly differentially expressed in hnf4g−/−CV/WTCV (log2(FC)>1.75; adj P < 0.05) colored in orange. e) For each gene, the log2(FC) in WTCV/WTGF and hnf4g−/− CV/hnf4g−/− GF were plotted and colored according to significance (see legend) with a strong correlation between the datasets suggesting that loss of hnf4g−/− alone has minimal impact on intestinal gene expression. f) Expression of the fluorescent transgene Tg(in3.4: cfos: GFP) was found to be unchanged in hnf4g−/− 6 dpf larvae. Brightfield and fluorescence images of whole larvae are shown for both hnf4g+/+ and hnf4g−/−, with the boxed inset in the fluorescence channel depicting the intestinal epithelial transgene expression (arrowheads) enlarged to the right. g) The average fluorescence intensity across the anterior intestine was quantified for each larva and graphed (hnf4g+/+,n = 24; hnf4g−/−, n = 23; P = 0.5709, unpaired t-test) with no significant differences found between wild-type and hnf4g−/− larvae.
Because we had previously shown that hnf4a positively regulates many microbially responsive genes, we were interested in how the response to microbes might differ in wild-type and hnf4g−/− larvae (Davison et al. 2017). Therefore, we compared differentially expressed genes in hnf4g−/− CV/hnf4g−/− GF and WTCV/WTGF. We found a strong general trend amongst genes with increased expression in response to microbial colonization in wild type and hnf4g−/−, including amongst those genes with the highest fold-changes in both conditions: itln2, il22, and saa (Fig. 3e—dark gray). There were a number of microbially responsive genes that were significantly increased or decreased only in wild type (175) or hnf4g−/− (40) suggesting the potential for modest differences in hnf4g−/− in response to microbiota (Fig. 3e—blue and orange). To better understand this phenomenon, and to test whether hnf4g is functionally similar to hnf4a, we utilized the transgene Tg(in3.4: cfos: GFP) which had previously been shown to have decreased expression in the intestinal epithelium in the absence of hnf4a (Davison et al. 2017). We did not observe a reduction of fluorescence intensity in the intestinal epithelium of hnf4g−/− larvae compared with wild-type larvae (Fig. 3, f and g), suggesting differences in the role of hnf4a and hnf4g in regulating gene expression. Taken together these data suggest that hnf4g appears to play more subtle and distinct roles in the intestinal epithelium and microbial response compared to hnf4a (Davison et al. 2017).
Changes in hnf4a−/−hnf4g−/− double mutant gene expression suggests defects in intestinal function and proliferation
To further investigate the unique and redundant functions of hnf4a and hnf4g in larval zebrafish, we added to our existing datasets for hnf4a and hnf4g single mutants by performing RNA-seq on dissected GI tracts from wild type, hnf4g−/− single mutants, and hnf4a−/−; hnf4g−/− double mutants, which were all conventionally raised with a microbiota (Fig. 4a). Because we never recovered any hnf4a−/−; hnf4g−/− double mutant adults, we relied on postdissection genotyping to pool digestive tracts for sequencing (see Materials and Methods). Differential expression analysis confirmed a modest number of transcripts (376) that were differentially expressed in conventionally raised hnf4g−/− single mutants compared to wild type, but many more genes (2,171) that were influenced by loss of hnf4a and hnf4g in the double mutant condition (Fig. 4, b–d), including genes involved in intestinal function such as fabp2 (Fig. 4b). About half as many genes were significantly differentially upregulated (690) compared to downregulated (1,481) in hnf4a−/−; hnf4g−/− CR/WTCR. GO analysis of the genes significantly down regulated revealed many processes associated with intestinal function including multiple metabolic pathways (Fig. 4e). We also observed signatures of lipid metabolism which are a hallmark of Hnf4 function identified previously in multiple other systems (Hayhurst et al. 2001; Palanker et al. 2009; Frochot et al. 2012; Barry and Thummel 2016; Davison et al. 2017; Goh et al. 2018; Lee et al. 2021). These data suggest that together hnf4a and hnf4g positively regulate many genes involved in intestinal function in the larval zebrafish digestive tract, consistent with previous findings for Hnf4 factors in mice (Verzi et al. 2010; Davison et al. 2017; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Immune response was the top GO category amongst the downregulated genes in hnf4a−/−; hnf4g−/− CR/WTCR, suggesting that loss of these factors may prevent the intestine from properly responding to colonizing microbes in conventionally raised larvae. The genes upregulated in the absences of both hnf4a and hnf4g were primarily associated with GO categories pertaining to cell division processes, suggesting that cell proliferation may be increased in hnf4a−/−; hnf4g−/− larvae compared to wild-type larvae.
Fig. 4.
Deletion of both hnf4a−/− and hnf4g−/− results in large-scale changes in intestinal gene expression. a) Wild-type, hnf4g−/−, and hnf4a−/−; hnf4g−/− larva were raised conventionally, digestive tracts were dissected at 6 dpf, and gene expression changes were measured by RNA-sequencing. All animals in these experiments were exposed to microbes (shaded rectangles). b) Representative RNA-seq tracks for genes showing decreased gene expression hnf4g−/− and hnf4a−/−; hnf4g−/−(hnf4g) or just hnf4a−/−; hnf4g−/−(fabp2) compared to wild type. c) Venn diagram depicting the number of differentially expressed genes across the experimental conditions. d) Volcano plot showing the substantial number of downregulated and upregulated genes when wild type and hnf4a−/−; hnf4g−/− gene expression are compared. e) GO term enrichment analysis for the downregulated genes revealed immune response and many metabolic processes involved in normal intestinal function as the major pathways affected by the loss of hnf4a−/−; hnf4g−/−, while the upregulated genes revealed cell division processes to be among the pathways most affected by loss of hnf4a−/−; hnf4g−/−. f) log2(FC) for hnf4g−/− CV/WTCV and g) hnf4a−/− CV/WTCV compared to hnf4a−/−; hnf4g−/− CV/WTCV reveal that unlike gene expression changes in hnf4a−/− CV/WTCV which largely mirror those found in hnf4a−/−; hnf4g−/− CV/WTCV, very few genes change in hnf4g−/− CV/WTCV by comparison. h) Comparison of the changes in gene expression for the one-to-one orthologs of mouse and zebrafish genes which were significantly differentially expressed in hnf4a−/−; hnf4g−/− larval zebrafish or mouse (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019) compared to wild type.
We next set out to compare the results of the 3 RNA-seq experiments characterizing gene expression changes in dissected GI tracts from 6 dpf hnf4a−/− (Davison et al. 2017), hnf4g−/− (Fig. 3), and hnf4a−/−; hnf4g−/− (Fig. 4) (Supplementary Fig. 4a). To assess what variables might be driving differences in the datasets, we conducted a principal components analysis (PCA) on all of the replicates from each dataset. The replicates from each experimental dataset clustered together on the first 2 components, with only very small separation within those groups based on genotype (Supplementary Fig. 3b), suggesting that differences in experimental conditions was the primary source of variation in the datasets. Therefore, to facilitate comparison between the 3 RNA-seq experiments, we performed a batch correction on the combined 3 datasets (see Materials and Methods). After batch correction, PCA showed that the samples no longer grouped according to experiment on the first 2 components, but instead stratified according to the major independent variables in our experiments: genotype (X-axis; PC1 38% variance) and microbial colonization (Y-axis; PC2: 12% variance) (Supplementary Fig. 4, c and d).
Utilizing the batch-corrected data (Supplementary Table 3), we compared hnf4a−/−; hnf4g−/− CR/WTCR to hnf4a−/− CV/WTCV, and to hnf4g−/− CV/WTCV to understand the shared differences between the double mutants and each of the single mutants. The comparison of hnf4g−/−CV/WTCV and hnf4a−/−; hnf4g−/− CR/WTCR, showed not only very few genes are significantly differentially expressed in both data sets, but that there was no apparent correlation across all genes (Fig. 4f). In contrast, there was a large amount of concordance between hnf4a−/− CV/WTCV and hnf4a−/−; hnf4g−/− CR/WTCR, with many common differentially expressed genes changing in the same direction (Fig. 4g, dark gray). To better understand how the changes in gene expression we observed in the zebrafish digestive tract in hnf4a−/−; hnf4g−/− double mutants compare with mouse intestinal hnf4a−/−; hnf4g−/− double mutants, we identified the genes in our dataset with one-to-one orthologs in mouse (GSE112946) (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Of the genes with one-to-one orthologs, 356 were significantly differentially regulated in both datasets, and many of these genes (278; 78%) showed the same directional change in mouse and zebrafish (Fig. 4h). This trend was most noticeable in the genes that had decreased expression in the double mutants, consistent with the previous findings from the mouse intestinal epithelium, that Hnf4a and Hnf4g are primarily activating gene expression (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019).
Hnf4 factors influence cell type-specific gene expression in the intestinal epithelium
We next sought to first, confirm whether the changes in gene expression we observed were due to a nonspecific developmental delay, and second, to understand the contribution of nonintestinal tissues (included in larval zebrafish gastrointestinal tract dissection, including the liver and pancreas) to our observed gene expression changes (San et al. 2018; Willms et al. 2022). We found limited expression differences for developmentally regulated intestinal genes (Supplementary Fig. 5). In analyzing genes associated the pancreas and liver, we found that over half of the hepatocyte-defining genes as determined an scRNA-sequencing experiment of dissected zebrafish larval digestive tissues (Willms et al. 2022) are significantly downregulated in the double mutants compared to wild type controls (Supplementary Fig. 6), which suggests hnf4a and hnf4g are important for the positive regulation of hepatocyte-defining transcripts.
To better understand what cell types in the intestinal epithelium are impacted by the loss of Hnf4 transcription factors, we leveraged an intestinal scRNA-seq dataset produced in our lab that identified sets of genes which define distinct intestinal cell clusters or types in 6 dpf larval zebrafish (Wen et al. 2021) (Supplementary Table 4 and Fig. 7). Consistent with our previous analyses, we observed that none of the genes that define the cell types were observed to be differentially expressed in hnf4g−/− CV/WTCV, and the mean log2(FC) for the groups of genes rarely deviated from zero (Supplementary Fig. 7). Loss of hnf4a−/− or hnf4a−/−; hnf4g−/− showed similar trends in the changes in gene expression within a given cell type, with the double mutant appearing to exacerbate the changes observed in hnf4a−/− CV/WTCV (Supplementary Fig. 7).
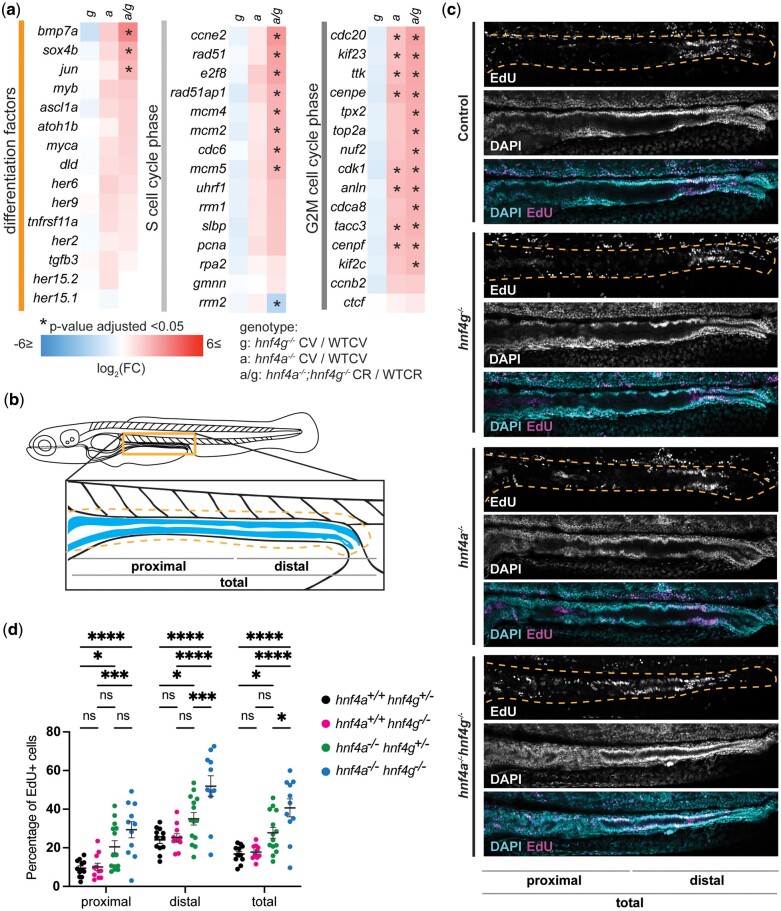
Two groups of genes, those defining anterior enterocytes (Ent1) and those defining ileal and mid-intestinal cells (Ent2) were strongly downregulated, while groups of genes that define most other intestinal epithelial cells types, were upregulated on average in hnf4a−/− and hnf4a−/−; hnf4g−/− mutants compared to wild type. Of the genes which define anterior enterocytes and those which define ileal and mid-intestinal cells (Wen et al. 2021), 50% and 35% of genes respectively, were significantly reduced in hnf4a−/−; hnf4g−/− double mutants (Supplementary Fig. 7). Consistent with this finding, in hnf4a−/− and hnf4a−/−; hnf4g−/− mutants we observed decreased expression of genes involved in enterocyte function and fatty acid metabolism, ileal bile-metabolism related genes, and genes which define the lysosome-rich enterocyte cell population (Fig. 5a), suggesting that these Hnf4s positively regulate genes involved in processes central to the intestinal functions of nutrient absorption and metabolism. Another salient pattern in the hnf4a−/−; hnf4g−/− double mutants was the significant upregulation of several genes encoding hormones secreted by enteroendocrine cells (EECs) such as pyyb, gcga, gcgb, and cckb along with goblet cell marker agr2 (Fig. 5a). This may suggest a proportional increase in secretory cell populations or their activities, or increased expression of these genes within other cell types in the intestinal epithelium in the absence of hnf4a or hnf4a; hnf4g function in zebrafish, as has been reported in mice (Babeu et al. 2009; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Overall, goblet cells (GC1–GC3) and EEC (EEC1–EEC5) populations showed weaker and mixed trends than enterocytes in our cell-type analysis (Supplementary Table 4).
To confirm that the changes in gene expression observed in our RNA-seq data (Fig. 4) and findings from our comparisons to IEC-specific scRNA-seq data (Supplementary Fig. 7) translate to in vivo phenotypes in the larval intestinal epithelium, we utilized 2 existing transgenic lines: Tg(fabp2: DsRed), which drives expression of DsRed under the control of 4.5 kb region of the zebrafish fabp2 promoter and labels anterior enterocytes in the intestinal epithelium (Her et al. 2004; Kanther et al. 2011) and Tg(fabp6: GFP), which drives expression of GFP the under control of 250 bp region of the zebrafish fabp6 promoter and labels cells in the mid-intestine which make up the zebrafish ileum (Lickwar et al. 2017) (Fig. 5b). We crossed these transgenes into our hnf4a; hnf4g compound mutant background to obtain groups of 6 dpf larvae that were heterozygous or wild type at both loci (control), mutant for hnf4g, but wild type or heterozygous for hnf4a (hnf4g−/−), mutant for hnf4a, but wild type or heterozygous for hnf4g (hnf4a−/−), or double mutant for both hnf4a and hnf4g (hnf4a−/−; hnf4g−/−) (Fig. 5, d–g).
We found that there was no significant difference in Tg(fabp2: DsRed) average intestinal fluorescence in hnf4g−/− and wild-type larvae (Fig. 5, d and e). However, in both hnf4a−/− and hnf4a−/−; hnf4g−/−, we observed very little Tg(fabp2: DsRed) expression in the intestinal epithelium compared to wild-type controls (Fig. 5, d and e). These results are consistent with our RNA-seq results for the fabp2 transcript and other genes known to be expressed in absorptive enterocytes (Fig. 5a), and further are consistent with our cell-type analysis which indicated that anterior enterocytes were negatively impacted by the loss of hnf4a−/− and hnf4a−/−; hnf4g−/− (Supplementary Fig. 7—group Ent1).
We next examined the expression of Tg(fabp6: GFP) in the same genotypic groups and timepoints. Similar to our results for Tg(fabp2: DsRed), we found that the Tg(fabp6: GFP) expression pattern in hnf4g−/− larva was indistinguishable from wild type (Fig. 5, f and g). In hnf4a−/− larva, we consistently observed a broadening of the domain of Tg(fabp6: GFP) expressing cells in the intestinal epithelium (Fig. 5, f and g). However, in hnf4a−/−; hnf4g−/− larva, we observed a significant decrease in the overall fluorescence of Tg(fabp6: GFP) that was not observed in any other mutant group (Fig. 5, f and g). Our RNA-seq results for fabp6 had led us to predict that Tg(fabp6: GFP) expression would be decreased in both hnf4a−/− and hnf4a−/−; hnf4g−/− (Fig. 5a). Although this was not the case on average for hnf4a−/−, the hnf4a−/−; hnf4g−/− larva displayed significant loss of Tg(fabp6: GFP) fluorescence as predicted (Supplementary Fig. 7—group Ent2).
hnf4a; hnf4g double mutants cause an increase in proliferation in the distal intestinal epithelium
Another category of genes that was strongly impacted by the loss of Hnf4 factors were cell proliferation factors (Figs. 4e and 6a). To test whether this was in fact due to an increase in dividing cells in the intestinal epithelium, we treated larval zebrafish with the thymidine analog EdU for 24 h from 5 to 6 dpf, and then sacrificed larvae for EdU visualization. We observed the most EdU+ cells in the intestinal region posterior to the intestinal bulb (Fig. 6b), and therefore, focused on comparing the number of EdU+ cells in control (hnf4a+/+; hnf4g+/−), hnf4g−/− (hnf4a+/+; hnf4g−/−), hnf4a−/− (hnf4a−/−; hnf4g+/−), and hnf4a−/−; hnf4g−/− double mutant larvae in that region (Fig. 6, b and c). We quantified the number of EdU+ cells across the total length of the posterior intestinal epithelium and further divided this region into proximal and distal segments. In hnf4a−/−, we observed a significant increase only in the distal segment (Fig. 6, c and d). In hnf4a−/−; hnf4g−/− double mutants we observed a notable increase in the percentage of EdU+ in both the proximal and distal segments of our imaged region (Fig. 6, c and d). This raises the possibilities that either cells are aberrantly entering the cell cycle in the double mutants or that cells are entering the cell cycle at the correct rate but are not exiting the cell cycle and properly differentiating.
Fig. 6.
Cell proliferation across the intestinal epithelium increases in hnf4a−/−; hnf4g−/− double mutant zebrafish larvae. a) Heatmaps showing changes in gene expression in hnf4g−/− CV/WTCV, hnf4a−/− CV/WTCV, hnf4a−/−; hnf4g−/− CR/WTCR for groups of genes involved in differentiation and the proliferative phases of the cell cycle (S and G2M). b) The number of EdU+ cells were counted within the posterior region of the larval zebrafish intestinal epithelium, the blue tissue in the diagram. c) Representative images of the posterior two-thirds of the intestinal epithelium (from the swim bladder to the cloaca) showing cells stained with EdU and DAPI for larva grouped by genetic mutant background. The intestinal epithelium is circled by the orange dotted line, as shown in panel (b). d) The number of EdU+ cells in the intestinal epithelium for larvae of each genotype as a percentage of the total number of intestinal epithelial cells (hnf4a+/+; hnf4g+/−n = 12, hnf4a+/+; hnf4g−/−n = 11, hnf4a−/−; hnf4g+/−n = 14, hnf4a−/−; hnf4g−/−n = 11). A 2-way ANOVA with multiple comparisons was performed and the significant differences within groups are shown on the graph (*P > 0.05, **P > 0.01, ***P > 0.001, ****P > 0.0001).
Discussion
The Hnf4 family of transcription factors regulates gene expression and cellular function in the digestive tract of mammals, including playing protective roles against human GI and metabolic diseases. To better understand the roles of Hnf4 family transcription factors across the vertebrate lineage, we compared and analyzed the roles of the 3 Hnf4 family homologs in larval zebrafish. We found that the individual hnf4a, hnf4g, and hnf4b mutant zebrafish were fully viable as homozygous mutant adults. In contrast, reduced viability in the double and triple mutants revealed genetic redundancies and potential stage specific functions for the homologs.
We did not recover any triple mutants or hnf4a−/−; hnf4b−/− double mutant larva at 6 dpf, suggesting that hnf4a and hnf4b may be synthetically lethal and play important roles during early development. Previous wholemount in situ hybridization studies showed hnf4b localization in the zebrafish yolk syncytial layer of embryos and larvae (Bertrand et al. 2007). In this study, we observed that the yolk of hnf4a−/−; hnf4b−/− double mutants and triple mutants showed decreased opacity, which is a hallmark of defects in lipid metabolism in that embryonic tissue (Wilson et al. 2020). These findings are also consistent with data from Xenopus, another vertebrate encoding an hnf4b homolog, in which HNF4A and HNF4B proteins were found to be maternally provisioned in the developing oocytes, and Hnf4b transcripts, specifically, were found to be present in fertilized eggs (Holewa et al. 1997). Taken together, these results suggest that hnf4b and hnf4a may redundantly regulate genes involved in yolk lipid mobilization and utilization during early zebrafish development and that this function might be conserved in other egg-laying vertebrates. We did not observe any phenotype in the hnf4b single mutants, but it is possible that the allele we analyzed results only in a partial loss of function due to either genetic or transcriptional compensation. It therefore remains possible that a true null allele of hnf4b may show more severe developmental phenotypes both alone and in combination with Hnf4 family members.
We focused our analysis of Hnf4 transcription factor function in the larval intestine on hnf4a and hnf4g. We set out to define the relationship between hnf4a−/−, hnf4g−/−, and hnf4a−/−; hnf4g−/− phenotypes in terms of redundancy and function. Our comparison of the RNA-seq from the dissected digestive tracts of these mutants (Fig. 4), analysis of intestinal cell types (by Tg(fabp2: DsRed) and Tg(fabp6: GFP), Fig. 5) and proliferation (by EdU staining, Fig. 6) revealed that loss of hnf4g had a weaker phenotypic impact than loss of hnf4a. Further, in the case of these phenotypes, loss of hnf4a and hnf4g simultaneously had a stronger phenotypic effect than the intermediate phenotype seen in hnf4a mutants, and ultimately resulted in larval lethality (Fig. 2). A similar unequal contribution of Hnf4a and Hnf4g to enterocyte gene expression and specification and proliferation phenotypes has been documented in mouse intestinal epithelial knockouts, in addition to lethality caused by simultaneous deletion of Hnf4a and Hnf4g in both fetal development and adult stages (Chen, Toke, Luo, Vasoya, Aita, et al. 2019; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019; Cheng et al. 2020). Importantly, the identification of this conserved relationship suggests that this unequal contribution of Hnf4 family members to these phenotypes is indeed selected for and not a synthetic effect that could occur spuriously or by chance. Taken together, our results provide evidence that the specific roles of Hnf4 transcription factors and their interactions in transcriptional regulation within the intestinal epithelium are conserved between zebrafish and mice.
One possible explanation for partial or unequal redundancy of 2 genes in a tissue could be overlapping but nonidentical expression patterns or subtle but significant differences in expression levels (Briggs et al. 2006). Examining hnf4a and hnf4g expression from 2 zebrafish intestinal scRNA-seq experiments revealed the potential for subtle differences in expression of the 2 factors, especially across the secretory cell types (Supplementary Fig. 3) (Wen et al. 2021; Willms et al. 2022). One consistent finding across the 2 datasets is that hnf4g is expressed in a higher percentage of secretory cells than hnf4a (i.e. goblet cells and EECs). However, this type of analysis could be somewhat limited by the fact that transcription factors, including hnf4 family members, are expressed at relatively low levels (Fig. 1c and Supplementary Table 3), and therefore, analyzing their presence in a scRNA-seq experiment where the sequencing depth could hinder the detection of lowly expressed genes.
The cell-type specific protein localization of Hnf4a and Hnf4g in the zebrafish intestinal epithelium are not known, and in mouse they are broadly present in overlapping cell types, but may show differences across the crypt-villus axis with Hnf4a showing increased crypt localization and Hnf4g more restricted to villus cells (Sauvaget et al. 2002; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). There is also evidence that the different Hnf4a isoforms have differential localization, which could impact the interactions between these factors (Chellappa et al. 2016). Another possibility is that these factors bind to different sites in the genome or have different affinity for the same sites. This seems unlikely considering ChIP-seq in the mouse intestinal epithelium for both factors found <80% of sites were shared and they are predicted to have the same DNA binding motif (Fang et al. 2012; Davison et al. 2017; Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Alternatively, they could regulate each other's activity or form heterodimers or have differential affinity for fatty-acid ligands or protein cofactors (Wisely et al. 2002; Daigo et al. 2011). These various potential explanations for partial or unequal redundancy between Hnf4a and Hnf4g could also help explain our observation that expression of some genes was significantly affected in hnf4a or hnf4g single mutants but not in the hnf4a; hnf4g double mutants compared to their respective wild-type controls (Figs. 4, f and g and 5a). However, it remains unclear if those differences are due to biological interactions between hnf4a and hnf4g or a result of RNA-seq batch correction, variation between biological replicates, or husbandry differences between these independent experiments (Supplementary Fig. 4a).
Our evidence suggests that the phenotypic homology between the zebrafish and mouse hnf4a−/−; hnf4g−/− double mutants appears to extend to the loss of specific cell types in the intestinal epithelium. In hnf4a−/−; hnf4g−/− zebrafish we observed the loss of Tg(fabp2: DsRed) expressing cells and decrease in genes involved in absorptive enterocyte functions (Figs. 4 and 5 and Supplementary Fig. 7), and the coincident increase in proliferation associated genes and EdU+ cells (Fig. 6). Loss of absorptive enterocytes and an increase in proliferation were also observed in adult double knockouts in the mouse intestinal epithelium (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). In the mouse double knockouts, the authors also observed a conversion of the enterocytes to enterocyte-like cells that express goblet cell markers. Although, we did not look directly at goblet cell numbers, our cell type analysis did show that the groups of genes associated with most secretory lineages, including goblet cells, increased in the zebrafish hnf4a−/−; hnf4g−/− double mutants (Supplementary Fig. 7). This effect did not appear to be as specific to the goblet cell lineage in 6 dpf larval zebrafish as it was in the mouse (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Regardless, the level of phenotypic conservation between double mutant phenotypes in the larval zebrafish and mouse intestinal epithelium was striking, especially considering the differences in developmental stage and experimental conditions.
As part of our study we did not determine the cause of hnf4a−/−; hnf4g−/− double mutant larvae lethality between 6 and 14 dpf. One possibility is that because of the changes we observed in genes and cell types involved in intestinal function and specifically, loss of absorptive enterocytes (Figs. 4 and 5), that the larvae simply cannot absorb enough nutrients once they begin fully relying on food from their environment instead of maternally supplied yolk. This would be similar to phenotypes observed in adult mouse double knockouts which do not survive long after conditional double deletion (Chen, Toke, Luo, Vasoya, Fullem, et al. 2019). Another possibility is that double mutant larvae fail in subsequent steps of intestinal development such as the significant length increase, development of intestinal rugae, or restriction of cell division to the base of the intestinal folds that occurs during the 6 and 14 dpf window (Li et al. 2020). If this is the case, these phenotypes could be similar to the defects and lethality observed in mouse fetal double mutants where the tissue fails to properly form villi (Chen, Toke, Luo, Vasoya, Aita, et al. 2019). It would be interesting for future studies to explore this critical developmental time window in these double mutant zebrafish larvae.
Unknown impacts on other tissues in these zebrafish larvae could also be a cause of premature lethality. In mammals, Hnf4 family transcription factors are critical for the development and function of digestive tract tissues outside of the intestinal epithelium. Hnf4a functions in hepatocyte differentiation, liver development, homeostasis, and gene expression (Li et al. 2000; Hayhurst et al. 2001; Parviz et al. 2003; Battle et al. 2006) and in pancreatic development and beta-cell function in humans (Yamagata 2014; Legøy et al. 2020). Our analysis of liver and pancreatic-specific genes revealed that hnf4a and hnf4g together likely have a combined role in regulating expression of many hepatocyte-defining genes (Supplementary Fig. 6b) (Willms et al. 2022). The genetic reagents described here will allow for future examination of these factors in liver and pancreas development and function. Further, conditional and tissue-specific knockouts for these genes would also be helpful in dissecting tissue-specific phenotypes, and even intestinal phenotypes in adult zebrafish for genotypes which proved lethal prior to reaching adulthood.
A few other factors complicate our understanding of how multiple Hnf4 transcription factors function both specifically and redundantly. In many vertebrates, including in zebrafish, Hnf4 factors have multiple isoforms. Both homodimeric and heterodimeric complexes can form between the isoforms, and in some cases, these complexes have been shown to have differential activity (Dean et al. 2010; Chellappa et al. 2016; Ko et al. 2019; Lambert, Depernet, et al. 2020). However, the relative contribution of these different complexes to the intestinal activity of Hnf4 transcription factors is currently unknown and fully unexplored in zebrafish. In addition, the activity of nuclear receptor superfamily members is commonly modulated by ligand binding. While the ligands of Hnf4 transcription factors are believed to be fatty acids (Hertz et al. 1998; Yuan et al. 2009), the impact of ligand binding on the activity of Hnf4 transcription factors in vivo remains poorly understood both in zebrafish and across other vertebrate and invertebrate models.
Together, this work expands our understanding of the roles of the Hnf4 family transcription factors in zebrafish, and further demonstrates the utility of the larval zebrafish as a model for exploring intestinal physiology and the genetic programs that underlie intestinal function. Despite differences between the digestive systems of mouse and zebrafish larva (e.g. a lack of true intestinal crypts, a more aerobic environment, and a lack of stomach in the zebrafish), our study highlights that many intestinal transcriptional programs, their functions, and even their interactions are likely to be conserved across these organisms. It seems likely that the intestinal functions of Hnf4 family transcription factors we find to be conserved between zebrafish and mice were present the last common ancestor of mammals and fishes, and that Hnf4 factors are involved in the function of the intestine in earlier branching vertebrates or even invertebrates (Martín-Durán and Hejnol 2015; Miglioli et al. 2021). Our work establishes the zebrafish as a model for investigating the conserved and novel functions of Hnf4 family transcription factors.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Susan Henning for guidance throughout the project and for providing valuable input on the manuscript. Members of the Center for Gastrointestinal Biology and Disease and Gastroenterology Research Training Program (NIH T32-DK007737) at the University of North Carolina at Chapel Hill for providing support, training, and feedback throughout the project. Dr. Jieun Park for assistance with wholemount in situ hybridization. The Duke Sequencing and Genomic Technologies Shared Resource for assistance with RNA-sequencing and analysis. The Duke University School of Medicine Zebrafish Core Facilities (Z-Core) including Jim Burris and Eileen Gu for their help in maintaining the zebrafish lines and facility which housed the zebrafish used in this study.
Funding
This work was supported by grants from the National Institutes of Health (R01-DK093399, P01-DK094779, R01-DK111857, R01-DK131742) and the Gordon and Betty Moore Foundation to JFR, and by a Gastroenterology Research Training Fellowship from the University of North Carolina at Chapel Hill (T32-DK007737) to JKH.
Conflicts of interest
None declared.
Contributor Information
Jennifer K Heppert, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA; Department of Medicine, Division of Gastroenterology and Hepatology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Colin R Lickwar, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
Matthew C Tillman, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
Briana R Davis, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
James M Davison, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
Hsiu-Yi Lu, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
Wei Chen, Center for Genomics and Computational Biology, Duke University School of Medicine, Durham, NC 27710, USA.
Elisabeth M Busch-Nentwich, School of Biological and Behavioural Sciences, Queen Mary University of London, E1 4NS London, UK.
David L Corcoran, Center for Genomics and Computational Biology, Duke University School of Medicine, Durham, NC 27710, USA.
John F Rawls, Department of Molecular Genetics and Microbiology, Duke Microbiome Center, Duke University School of Medicine, Durham, NC 27710, USA.
Data availability
All new sequencing data produced in this work can be found on NCBI Genome Expression Omnibus (GSE183799). Supplemental information includes Supplementary Figs. 1–7 and Supplementary Tables 1–4. Supplementary Tables 3–4 and Reagents Table are available at figshare: https://doi.org/10.25386/genetics.19775128.
Supplemental material is available at GENETICS online.
Literature cited
- Ahn S-H, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, et al. Hepatocyte nuclear factor 4α in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14(7):908–920. doi:10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Mulligan TS, Shen M-C, Wang H, Scahill CM, Tan FJ, Du SJ, Busch-Nentwich EM, Farber SA.. mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 2017;13(11):e1007105. doi:10.1371/journal.pgen.1007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babeu J-P, Darsigny M, Lussier CR, Boudreau F.. Hepatocyte nuclear factor 4α contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol. 2009;297(1):G124–G134. doi:10.1152/ajpgi.90690.2008. [DOI] [PubMed] [Google Scholar]
- Baraille F, Ayari S, Carrière V, Osinski C, Garbin K, Blondeau B, Guillemain G, Serradas P, Rousset M, Lacasa M, et al. Glucose tolerance is improved in mice invalidated for the nuclear receptor HNF-4γ: a critical role for enteroendocrine cell lineage. Diabetes. 2015;64(8):2744–2756. doi:10.2337/db14-0993. [DOI] [PubMed] [Google Scholar]
- Barry WE, Thummel CS.. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. eLife. 2016;5:e11183. doi:10.7554/eLife.11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K.. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2(6):371–382. doi:10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA.. Hepatocyte nuclear factor 4α orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA. 2006;103(22):8419–8424. doi:10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–512. doi:10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M.. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol. 2004;21(10):1923–1937. doi:10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet P-L, Escrivà H, Duffraisse M, Marchand O, Safi R, et al. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3(11):e188. doi:10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier MEA, Ortlund EA, Degnan BM, Thornton JW.. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010;8(10):e1000497. doi:10.1371/journal.pbio.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS.. Unequal genetic redundancies in Arabidopsis–a neglected phenomenon? Trends Plant Sci. 2006;11(10):492–498. doi:10.1016/j.tplants.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Camp JG, Jazwa AL, Trent CM, Rawls JF.. Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription. PLoS Genetics. 2012;8(3):e1002585. doi:10.1371/journal.pgen.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A-L, Le Beyec J, Barreau F, Saint-Just S, Houllier A, Gonzalez FJ, Robine S, Pinçon-Raymond M, Cardot P, Lacasa M, et al. Hepatocyte nuclear factor 4α, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29(23):6294–6308. doi:10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K.. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4570–4577. doi:10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briançon N, Bolotin E, Lytle C, Nair MG, Sladek FM, et al. Opposing roles of nuclear receptor HNF4α isoforms in colitis and colitis-associated colon cancer. eLife. 2016;5:e10903. doi:10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Luo S, Dupre A, Vasoya RP, Parthasarathy A, Aita R, Malhotra R, Hur J, Toke NH, Chiles E, et al. The nuclear receptor HNF4 drives a brush border gene program conserved across murine intestine, kidney, and embryonic yolk sac. Nat Commun. 2021;12(1):2886. doi:10.1038/s41467-021–22761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Toke NH, Luo S, Vasoya RP, Aita R, Parthasarathy A, Tsai Y-H, Spence JR, Verzi MP.. HNF4 factors control chromatin accessibility and are redundantly required for maturation of the fetal intestine. Development. 2019;146(19). doi:10.1242/dev.179432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Toke NH, Luo S, Vasoya RP, Fullem RL, Parthasarathy A, Perekatt AO, Verzi MP.. A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat Genet. 2019;51(5):777–785. doi:10.1038/s41588-019–0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Vasoya RP, Toke NH, Parthasarathy A, Luo S, Chiles E, Flores J, Gao N, Bonder EM, Su X, et al. HNF4 regulates fatty acid oxidation and is required for renewal of intestinal stem cells in mice. Gastroenterology. 2020;158(4):985–999.e9. doi:10.1053/j.gastro.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE.. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8(20):2466–2477. doi:10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Li Y, Li W, Song Y, Zeng R, Lu K.. Effect of hepatocyte nuclear factor 4 on the fecundity of Nilaparvata lugens: insights from RNA interference combined with transcriptomic analysis. Genomics. 2020;112(6):4585–4594. doi:10.1016/j.ygeno.2020.08.002. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, Lu Y-F, Ko T-Y, Tsai M-Y, Lin CY, Lin C-C, Hwang S-PL.. Zebrafish cdx1b regulates differentiation of various intestinal cell lineages. Dev Dyn. 2009;238(5):1021–1032. doi:10.1002/dvdy.21908. [DOI] [PubMed] [Google Scholar]
- Daigo K, Kawamura T, Ohta Y, Ohashi R, Katayose S, Tanaka T, Aburatani H, Naito M, Kodama T, Ihara S, et al. Proteomic analysis of native hepatocyte nuclear factor-4α (HNF4α) isoforms, phosphorylation status, and interactive cofactors. J Biol Chem. 2011;286(1):674–686. doi:10.1074/jbc.M110.154732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsigny M, Babeu J-P, Dupuis A-A, Furth EE, Seidman EG, Lévy E, Verdu EF, Gendron F-P, Boudreau F.. Loss of hepatocyte-nuclear-factor-4α Affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One. 2009;4(10):e7609. doi:10.1371/journal.pone.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF.. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor hepatocyte nuclear factor 4 alpha. Genome Res. 2017;27(7):1195–1206. doi:10.1101/gr.220111.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S, Tang JI, Seckl JR, Nyirenda MJ.. Developmental and tissue-specific regulation of hepatocyte nuclear factor 4-alpha (HNF4-alpha) isoforms in rodents. Gene Expr. 2010;14(6):337–344. doi:10.3727/105221610x12717040569901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi:10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V, Staels B, Lefebvre P, Verzi MP, Eeckhoute J.. Control of cell identity by the nuclear receptor HNF4 in organ pathophysiology. Cells. 2020;9(10):2185. doi:10.3390/cells9102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W.. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(−/−) embryos. Development. 1997;124(2):279–287. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai S-L, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568(7751):193–197. doi:10.1038/s41586-019–1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM.. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res. 2012;40(12):5343–5356. doi:10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frochot V, Alqub M, Cattin A-L, Carrière V, Houllier A, Baraille F, Barbot L, Saint-Just S, Ribeiro A, Lacasa M, et al. The transcription factor HNF-4α: a key factor of the intestinal uptake of fatty acids in mouse. Am J Physiol Gastrointest Liver Physiol. 2012;302(11):G1253–G1263. doi:10.1152/ajpgi.00329.2011. [DOI] [PubMed] [Google Scholar]
- Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA.. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130(4):1207–1220. doi:10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdin AK, Surve VV, Jönsson M, Bjursell M, Björkman M, Edenro A, Schuelke M, Saad A, Bjurström S, Lundgren EJ, et al. Phenotypic screening of hepatocyte nuclear factor (HNF) 4-gamma receptor knockout mice. Biochem Biophys Res Commun. 2006;349(2):825–832. doi:10.1016/j.bbrc.2006.08.103. [DOI] [PubMed] [Google Scholar]
- Girard R, Darsigny M, Jones C, Maloum-Rami F, Gélinas Y, Carpentier AC, Laplante M, Perreault N, Boudreau F.. HNF4α is a novel regulator of intestinal glucose-dependent insulinotropic polypeptide. Sci Rep. 2019;9(1):4200. doi:10.1038/s41598-019–41061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh GYS, Winter JJ, Bhanshali F, Doering KRS, Lai R, Lee K, Veal EA, Taubert S.. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell. 2018;17(3):e12743. doi:10.1111/acel.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X, Eckmann CR.. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr Biol. 2013;23(7):607–613. doi:10.1016/j.cub.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Hahn-Windgassen A, Van Gilst MR.. The Caenorhabditis elegans HNF4α Homolog, NHR-31, mediates excretory tube growth and function through coordinate regulation of the vacuolar ATPase. PLoS Genet. 2009;5(7):e1000553. doi:10.1371/journal.pgen.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ.. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–1403. doi:10.1128/MCB.21.4.1393–1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppert JK, Davison JM, Kelly C, Mercado GP, Lickwar CR, Rawls JF.. Transcriptional programmes underlying cellular identity and microbial responsiveness in the intestinal epithelium. Nat Rev Gastroenterol Hepatol. 2021;18(1):7–23. doi:10.1038/s41575-020–00357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her GM, Chiang C-C, Wu J-L.. Zebrafish intestinal fatty acid binding protein (I-FABP) gene promoter drives gut-specific expression in stable transgenic fish. Genesis. 2004;38(1):26–31. doi:10.1002/gene.10248. [DOI] [PubMed] [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar-Tana J.. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392(6675):512–516. doi:10.1038/33185. [DOI] [PubMed] [Google Scholar]
- Holewa B, Zapp D, Drewes T, Senkel S, Ryffel GU.. HNF4beta, a new gene of the HNF4 family with distinct activation and expression profiles in oogenesis and embryogenesis of Xenopus laevis. Mol Cell Biol. 1997;17(2):687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi:10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. International IBD Genetics Consortium (IIBDGC). Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi:10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Sun X, Mühlbauer M, Mackey LC, Flynn EJ, Bagnat M, Jobin C, Rawls JF.. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141(1):197–207. doi:10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496(7446):494–497. doi:10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HL, Zhuo Z, Ren EC.. HNF4α combinatorial isoform heterodimers activate distinct gene targets that differ from their corresponding homodimers. Cell Rep. 2019;26(10):2549–2557.e3. doi:10.1016/j.celrep.2019.02.033. [DOI] [PubMed] [Google Scholar]
- Lambert É, Babeu J-P, Simoneau J, Raisch J, Lavergne L, Lévesque D, Jolibois É, Avino M, Scott MS, Boudreau F, et al. Human hepatocyte nuclear factor 4-α encodes isoforms with distinct transcriptional functions. Mol Cell Proteomics. 2020;19(5):808–827. doi:10.1074/mcp.RA119.001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert GG, Depernet H, Gotthard G, Schultz DT, Navizet I, Lambert T, Adams SR, Torreblanca-Zanca A, Chu M, Bindels DS, et al. Aequorea’s secrets revealed: new fluorescent proteins with unique properties for bioimaging and biosensing. PLoS Biol. 2020;18(11):e3000936. doi:10.1371/journal.pbio.3000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Veeriah V, Levine F.. Liver fat storage is controlled by HNF4α through induction of lipophagy and is reversed by a potent HNF4α agonist. Cell Death Dis. 2021;12(6):1–18. doi:10.1038/s41419-021–03862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legøy TA, Mathisen AF, Salim Z, Vethe H, Bjørlykke Y, Abadpour S, Paulo JA, Scholz H, Ræder H, Ghila L, et al. In vivo environment swiftly restricts human pancreatic progenitors toward mono-hormonal identity via a HNF1A/HNF4A mechanism. Front Cell Devel Biol. 2020;8:109. doi: 10.3389/fcell.2020.00109. eCollection 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D496. doi:10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA.. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000;14(4):464–474. doi:10.1101/gad.14.4.464. [PMC free article] [PubMed] [Google Scholar]
- Li J, Prochaska M, Maney L, Wallace KN.. Development and organization of the zebrafish intestinal epithelial stem cell niche. Dev Dyn. 2020;249(1):76–87. doi:10.1002/dvdy.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi:10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Camp JG, Weiser M, Cocchiaro JL, Kingsley DM, Furey TS, Sheikh SZ, Rawls JF.. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017;15(8):e2002054. doi:10.1371/journal.pbio.2002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Davison JM, Kelly C, Mercado GP, Wen J, Davis BR, Tillman MC, Semova I, Andres SF, Vale G, et al. Transcriptional integration of distinct microbial and nutritional signals by the small intestinal epithelium. Cell Mol Gastroenterol Hepatol. 2022;14(2):465–493. doi:10.1016/j.jcmgh.2022.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014–0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D-D, Zhou L-Y, Tang H.. Hepatocyte nuclear factor 4α and cancer-related cell signaling pathways: a promising insight into cancer treatment. Exp Mol Med. 2021;53(1):8–18. doi:10.1038/s12276-020–00551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcil V, Sinnett D, Seidman E, Boudreau F, Gendron F-P, Beaulieu J-F, Menard D, Lambert M, Bitton A, Sanchez R, et al. Association between genetic variants in the HNF4A gene and childhood-onset Crohn’s disease. Genes Immun. 2012;13(7):556–565. doi:10.1038/gene.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi:10.14806/ej.17.1.200. [Google Scholar]
- Martín-Durán JM, Hejnol A.. The study of Priapulus caudatus reveals conserved molecular patterning underlying different gut morphogenesis in the Ecdysozoa. BMC Biol. 2015;13:29. doi:10.1186/s12915-015–0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglioli A, Canesi L, Gomes IDL, Schubert M, Dumollard R.. Nuclear receptors and development of marine invertebrates. Genes (Basel). 2021;12(1):83. doi:10.3390/genes12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muncan V, Faro A, Haramis A-PG, Hurlstone AFL, Wienholds E, van Es J, Korving J, Begthel H, Zivkovic D, Clevers H, et al. T-cell factor 4 (Tcf7l2) maintains proliferative compartments in zebrafish intestine. EMBO Rep. 2007;8(10):966–973. doi:10.1038/sj.embor.7401071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng ANY, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PDS, Stainier DYR, Heath JK.. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286(1):114–135. doi:10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS.. Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metab. 2009;9(3):228–239. doi:10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Levic DS, Sumigray KD, Bagwell J, Eroglu O, Block CL, Eroglu C, Barry R, Lickwar CR, Rawls JF, et al. Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev Cell. 2019;51(1):7–20.e6. doi:10.1016/j.devcel.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34(3):292–296. doi:10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Pham LN, Kanther M, Semova I, Rawls JF.. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3(12):1862–1875. doi:10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F.. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. 2022;38(10):2943–2945. doi: 10.1093/bioinformatics/btac166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA.. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):7. doi:10.1186/s13059-018–1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San B, Aben M, Elurbe DM, Voeltzke K, den Broeder MJ, Rougeot J, Legler J, Kamminga LM.. Genetic and epigenetic regulation of zebrafish intestinal development. Epigenomes. 2018;2(4):19. doi:10.3390/epigenomes2040019. [Google Scholar]
- Sauvaget D, Chauffeton V, Citadelle D, Chatelet F-P, Cywiner-Golenzer C, Chambaz J, Pinçon-Raymond M, Cardot P, Le Beyec J, Ribeiro A, et al. Restriction of apolipoprotein A-IV gene expression to the intestine villus depends on a hormone-responsive element and parallels differential expression of the hepatic nuclear factor 4alpha and gamma isoforms. J Biol Chem. 2002;277(37):34540–34548. doi:10.1074/jbc.M206074200. [DOI] [PubMed] [Google Scholar]
- Sladek FM, Zhong WM, Lai E, Darnell JE.. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4(12B):2353–2365. doi:10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Storelli G, Nam H-J, Simcox J, Villanueva CJ, Thummel CS.. Drosophila HNF4 directs a switch in lipid metabolism that supports the transition to adulthood. Dev Cell. 2019;48(2):200–214.e6. doi:10.1016/j.devcel.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli S, Shiwen Z, Matsudaira P.. Cell clusters containing intestinal stem cells line the zebrafish intestine intervillus pocket. iScience. 2022;25(5):104280. doi:10.1016/j.isci.2022.104280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK IBD Genetics Consortium, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi:10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA, et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19(5):713–726. doi:10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M.. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122(2):157–173. doi:10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Wu D, Yu S, Wang Y, Leung Chan F.. Nuclear receptor HNF4α performs a tumor suppressor function in prostate cancer via its induction of p21-driven cellular senescence. Oncogene. 2020;39(7):1572–1589. doi:10.1038/s41388-019–1080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Holewa B, Jones EA, Ryffel GU.. Mesoderm and endoderm differentiation in animal cap explants: identification of the HNF4-binding site as an activin A responsive element in the Xenopus HNF1alpha promoter. Development. 1996;122(6):1975–1984. [DOI] [PubMed] [Google Scholar]
- Wen J, Mercado GP, Volland A, Doden HL, Lickwar CR, et al. Fxr signaling and microbial metabolism of bile salts in the zebrafish intestine. Sci Adv. 2021;7:eabg1371. doi:10.1126/sciadv.abg1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms RJ, Jones LO, Hocking JC, Foley E.. A cell atlas of microbe-responsive processes in the zebrafish intestine. Cell Rep. 2022;38(5):110311. doi:10.1016/j.celrep.2022.110311. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Rajan S, Danoff A, White RJ, Hensley MR, Quinlivan VH, Recacha R, Thierer JH, Tan FJ, Busch-Nentwich EM, et al. A point mutation decouples the lipid transfer activities of microsomal triglyceride transfer protein. PLoS Genet. 2020;16(8):e1008941. doi:10.1371/journal.pgen.1008941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10(9):1225–1234. doi:10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- Yamagata K. Roles of HNF1α and HNF4α in pancreatic β-cells: lessons from a monogenic form of diabetes (MODY). Vitam Horm. 2014;95:407–423. doi:10.1016/B978-0–12-800174-5.00016-8. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI, et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature. 1996;384(6608):458–460. doi:10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- Yang S-K, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, Kim KM, Ye BD, Kim K-J, Park SH, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63(1):80–87. doi:10.1136/gutjnl-2013–305193. [DOI] [PubMed] [Google Scholar]
- Ye L, Bae M, Cassilly CD, Jabba SV, Thorpe DW, Martin AM, Lu H-Y, Wang J, Thompson JD, Lickwar CR, et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe. 2021;29(2):179–196.e9. doi:10.1016/j.chom.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Mueller O, Bagwell J, Bagnat M, Liddle RA, Rawls JF.. High fat diet induces microbiota-dependent silencing of enteroendocrine cells. eLife. 2019;8:e48479. doi:10.7554/eLife.48479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM.. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4(5):e5609. doi:10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hagedorn CH, Wang L.. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812(8):893–908. doi:10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019–09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new sequencing data produced in this work can be found on NCBI Genome Expression Omnibus (GSE183799). Supplemental information includes Supplementary Figs. 1–7 and Supplementary Tables 1–4. Supplementary Tables 3–4 and Reagents Table are available at figshare: https://doi.org/10.25386/genetics.19775128.
Supplemental material is available at GENETICS online.