Abstract
Background
Heterozygous TERT (telomerase reverse transcriptase) promoter mutations (TPMs) facilitate TERT expression and are the most frequent mutation in glioblastoma (GBM). A recent analysis revealed this mutation is one of the earliest events in gliomagenesis. However, no appropriate human models have been engineered to study the role of this mutation in the initiation of these tumors.
Method
We established GBM models by introducing the heterozygous TPM in human induced pluripotent stem cells (hiPSCs) using a two-step targeting approach in the context of GBM genetic alterations, CDKN2A/B and PTEN deletion, and EGFRvIII overexpression. The impact of the mutation was evaluated through the in vivo passage and in vitro experiment and analysis.
Results
Orthotopic injection of neuronal precursor cells (NPCs) derived from hiPSCs with the TPM into immunodeficient mice did not enhance tumorigenesis compared to TERT promoter wild type NPCs at initial in vivo passage presumably due to relatively long telomeres. However, the mutation recruited GA-Binding Protein and engendered low-level TERT expression resulting in enhanced tumorigenesis and maintenance of short telomeres upon secondary passage as observed in human GBM. These results provide the first insights regarding increased tumorigenesis upon introducing a TPM compared to isogenic controls without TPMs.
Conclusion
Our novel GBM models presented the growth advantage of heterozygous TPMs for the first time in the context of GBM driver mutations relative to isogenic controls, thereby allowing for the identification and validation of TERT promoter-specific vulnerabilities in a genetically accurate background.
Keywords: genome editing, glioma, neural progenitor cell, telomerase, TERT promoter
Key Points.
TERT promoter mutation is not required for tumor initiation in cells with long telomeres.
TERT promoter mutation facilitates tumor growth upon reduced telomere length.
Importance of the Study.
We have established the first gene-edited glioblastoma models with the heterozygous TERT promoter mutation, the most frequent mutation in glioblastoma, that recapitulate shortened telomere length seen in clinical samples. Our models are suitable for examining the function of the TERT promoter mutation throughout gliomagenesis and will be an essential tool to test novel precision therapies specifically for TERT promoter mutant tumors.
Telomerase reverse transcriptase (TERT) promoter mutations, known to be the most recurrent mutation in glioblastoma (GBM) and the most common noncoding mutation across human cancer, increase TERT expression and are prominently associated with EGFR amplified cases.1,2 Studies have produced conflicting results regarding the clonality of the TERT promoter mutation, but there is agreement that TERT promoter mutations are an early event in gliomagenesis that is required for clonal expansion.3,4 Possibly due to relatively high TERT expression in somatic cells of mice, their long telomere length, and promoter sequence that diverges from humans, genetically engineered mouse models of GBM are not able to address the function of this mutation, and thus an ideal heterozygous TERT promoter mutant human model with isogenic controls has yet to be established.5,6 Recently, we established strategies for generating brain tumor models using neural progenitor cells (NPCs) derived from human induced pluripotent stem cells (hiPSCs) that have been CRISPR/Cas9-engineered with different combinations of authentic GBM-related genetic drivers.7 Here we used this isogenic human system for the introduction of the TERT promoter mutation and examined its functions in gliomagenesis in the context of knockout of PTEN and CDKN2A/B tumor suppressor genes, which are commonly deleted in TERT promoter mutant GBM, combined with overexpression of EGFRvIII, a constitutively active mutated form of EGFR frequently found in these tumors.3,8
Materials and Methods
Cell Culture
Experiments using hiPSCs were conducted under regulations of the UCSD Human Research Protections Program, project number 151330ZX. The hiPSCs were propagated as previously reported.7 Briefly, an hiPSC line, iPS12 (female, StemoniX, San Diego, CA), was cultured on plates coated with Matrigel hESC-Qualified Matrix (Corning) in mTeSR1 media (Stemcell Technologies). NPCs were cultured on Matrigel-coated plates in NPC maintenance media consisting of DMEM/F12 with GlutaMAX, 1 × N-2 supplement, 1 × B-27 supplement (Thermo Fisher Scientific), 50 mM ascorbic acid, 3 µM CHIR99021, and 0.5 µM purmorphamine (Tocris). Sphere cells were cultured in suspension in DMEM/F12 with GlutaMAX (Thermo Fisher Scientific) with 1 × B-27 supplement, 20 ng/mL EGF, and 20 ng/mL bFGF (Stemcell Technologies) either in floating sphere condition or in adherent condition on matrigel-coated plates. The period of in vitro culture of the sphere lines before re-injection was matched between cell lines and limited to a few week. Astrocyte were cultured in Astrocyte Medium (Sciencell Research Laboratories, Carlsbad, CA). U87MG vIII was developed and maintained as previously described.9
Generation and Validation of Genetically Engineered hiPSC Clones
Gene-edited hiPSC clones were generated and validated as previously reported.7 For CRISPR/Cas9 genome engineering of the TERT promoter, the sgRNA sequences to delete the region including the TERT promoter, and the template plasmids for reconstitution of the deleted region with either wild type or mutant sequence, were obtained from Dr. Dirk Hockemeyer (UC Berkeley).10 The sgRNA target sequences for each of the targeted genes were cloned into px458 plasmid (Addgene Plasmid #48138) using combinations of the top and bottom oligonucleotides listed in Supplementary Table 1. Dissociated hiPSCs were electroporated with the plasmids in Human Stem Cell Nucleofector Kit 1 (Lonza) using B-016 program of Nucleofector 2b (Lonza). 1–2 × 104 sorted GFP-positive cells were plated on a 10-cm Matrigel-coated plate in mTeSR1 and isolated colonies were picked manually. For candidate clones, DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen), and genotyping PCR was performed using Platinum Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific). For TERT promoter lesion-specific PCR reactions, we used 94°C for 2 min, 40 cycles of 94°C for 15 s, 63.7°C for 30 s, and 68°C for 90 s. Primers used for genotyping PCR are listed in Supplementary Table 1.
Differentiation of hiPSCs to NPCs and Astrocytes
Generation of small molecule neural progenitor cells (smNPCs) from hiPSCs was adapted from a previous study11 and described in detail as previously reported.7 Briefly, embryoid body (EB) formation was achieved by suspending hiPSCs in NPC maintenance media with 1 µM Dorsomorphin, 10 µM SB431542 (Tocris) (NPC differentiation media), and 5 mM Y-26732 (Stemcell Technologies, day 1 only). Cells were transferred to an uncoated 6-well tissue culture plate and incubated at 37°C, 5% CO2 on a shaker at 90 rpm with full or half media change occurring every day. On day 6, media was changed to NPC maintenance media and on day 8, EBs were triturated by pipetting and plated onto matrigel-coated 10-cm plates. After 3–4 days, attached EB and outgrown cells were split at a 1:6 to 1:8 ratio onto matrigel-coated plates. After the first passage, cells were passaged at a 1:10 to 1:15 ratio every 3–6 days. Astrocyte differentiation from NPCs was performed using STEMdiff Astrocyte Differentiation Kit and STEMdiff™ Astrocyte Maturation Kit (STEMCELL Technologies, Canada) following the manufacturer’s instruction.
Cell Growth Assay
3–4 replicates were plated in each well of black-walled, clear-bottom 96-well plates. Cell growth was analyzed using ATPlite 1step assay kit (PerkinElmer 6016731) following the manufacturer’s instruction.
Lentivirus Production and Infection
Lentivirus for EGFRvIII overexpression was produced as previously reported.9 Briefly, 293T cells were transfected with the pLV-EF1a-Hyg plasmid (Biosettia) cloned with EGFRvIII cDNA, VSVg, and Δ8.9 packaging constructs using Lipofectamine 2000 reagent (Life Technologies). Supernatants-containing virus were collected at 48 and 72 h after transfection, filtered through a 0.45 µm cellulose acetate filter, and concentrated by ultracentrifugation. NPCs were infected and viral supernatant was replaced by fresh culture media after 24 h of incubation at 37°C. Infected cells were selected with hygromycin.
RT-qPCR
Total RNA was purified using RNeasy Mini or Micro Kit (Qiagen) and was reverse transcribed using RNA to cDNA EcoDry Premix (Clontech) according to the manufacturer’s instruction. Triplicate RT-qPCR reactions containing cDNA obtained from 10 ng RNA were run with 3 replicates per cell line on CFX96 Real Time System (Bio-Rad) to confirm designated targeting of the genes with the following reaction conditions: 95°C for 5 min, 40 cycles of 95°C for 15 s, 56°C for 30 s. Primers used for RT-qPCR reactions are listed in Supplementary Table 1. All RT-qPCR expression data shown were compared to internal control genes using 2−∆Ct formula. A CT value of 40 was used to indicate an undetected gene for the calculation.
Intracranial Tumor Formation
Animal research experiments were conducted under the regulations of the UCSD Animal Care Program, protocol number S00192M. Cells were inoculated into the striatum of 4–7 week-old female Nod-scid mice (Charles River Laboratory) by stereotactic injections (1.0 mm anterior and 2.0 mm right to the bregma, and 3 mm deep from the inner plate of the skull).7 The image of brain tumor injection was created at BioRender.com.
Immunohistochemistry
Staining of paraffin-embedded tissue sections with H&E and NM95 (Abcam Cat # ab190710) was done at UCSD Moore’s Cancer Center Pathology Core and the Center for Advanced Laboratory Medicine (UCSD). Cryopreserved brain tissues were embedded in Optimal Cutting Temperature (OCT) matrix compound (Tissue-Tek, Sakura Finetek), frozen with dry ice, and mounted on the cryostat. Three sections (30-µm thickness) were taken from each brain and brain-stem level (2.4 mm, 4.8 mm, and 7.2 mm caudal from bregma) for hNuMA-positive cell counting in each animal (ie, total 9 sections used per animal). Free-floating sections were washed three times in PBS with 0.3% Triton-X100 followed by a blocking step with 4% serum in ×1 PBS with 0.3% Triton-X100 for 1-hour. Sections were then incubated with the primary antibodies (in the blocking solution) overnight at 4°C. The following day, the sections were washed three times with ×1 PBS with 0.3% Triton-X100, and incubated with a secondary antibody in PBS, 0.3% Triton-X100 for 1 h at room temperature. Sections were mounted on slides, dried at room temperature, and cover-slipped in ProLong Gold antifade mounting medium, with DAPI (Invitrogen).12 The following primary antibodies and dilutions were used; hNuMA (Mouse, EMD Millipore, MAB1281, 1:100), Vimentin (Chicken, EMD Millipore, AB5733, 1:500), Ki-67 (Rabbit, Abcam, AB6667, 1:500).
RNA Sequencing
RNA sequencing was performed by Novogene Corporation Inc, Sacramento, CA (read length: paired-end 150). Analysis was performed as previously described.13 Briefly, RNA-seq reads were aligned to the human genome (hg19) with STAR aligner. Reads were counted using featureCounts with default settings, and differential expression analysis conducted using DESeq2.14 Significant genes were those with a Benjamini–Hochberg adjusted P-value of greater than .05 and a fold-change of greater than 2. For gene ontology networks, gProfiler15 was used for the analysis and Cytoscape16 software with EnrichmentMap plugin17 was used for output file visualization.
Telomerase Activity Quantification
Telomerase activity quantification qPCR assay kit (#8928 ScienCell Research Laboratories, Carlsbad, CA) was used for quantification of the telomerase activity. Three replicate PCR reactions were performed for each sample. Positive and negative control samples were provided by the kit.
Telomere Length Quantification
Absolute human telomere length quantification qPCR assay kit (#8918 ScienCell Research Laboratories, Carlsbad, CA) was used for quantification of the telomere length. Two replicate PCR reactions were performed for each sample.
Chromatin Immunoprecipitation (ChIP)
ChIP for GA-binding protein, alpha subunit (GABPA) was performed as described previously using the ActiveMotif High Sensitivity kit.18 Cells were grown in 15-cm plates and fixed with 4% formaldehyde. Chromatin was sonicated by the Diagenode Biorupter. 14–20 ug of chromatin was used for GABPA (Santa Cruz Biotechnology: sc-22810) and IgG control (Cell Signaling: 2729) immunoprecipitations for each clone. Enrichment at the TERT promoter was determined by qPCR with the SsoAdvanded Universal SYBR Green Supermix (Bio-Rad). Primer sets used are listed in Supplementary Table 1. 1M of Resolution Solution (Roche GC-Rich kit) was added to each standard SsoAdvanced qPCR reaction. PCR was performed on an Applied Biosystems 7900HT Fast Real-Time System. Two technical replicate PCR reactions were performed for each biological replicate of the TERT promoter wild type and TPM clone. Primers used for ChIP-PCR are listed in Supplementary Table 1.
Scratch Assay
Scratches were made with 200 ul pipette tips after cells reached 100% confluence on matrigel-coated 48-well plates. Images were taken using a phase-contrast microscope on 4× magnification every 24H (0, 24H, 48H). The wound area was calculated by imageJ19 manually.
Western Blotting
Cells were harvested in RIPA buffer lysis (50 mM Tris–HCl, 1 mM EDTA, 20% SDS, 5 mM DTT 10 mM phenylmethylsulfonyl fluoride). Equal amounts of protein lysates (30 μg each) were separated by SDS-PAGE and then transferred to PVDF membranes. The membranes were blocked with a buffer containing 5% nonfat dry milk in phosphate-buffered saline with 0.1% Tween-20 for 1 h and incubated overnight with antibodies in 5% BSA at 4°C. After a second wash with phosphate-buffered saline with 0.1% Tween-20, the membranes were incubated with peroxidase-conjugated secondary antibodies (Sigma–Aldrich), and developed with an enhanced chemiluminescence detection kit (Pierce). Western blot analysis was performed with antibodies specific to GAPDH (CST#3683), E2F1a (CST#3742), and CyclinA2 (CST#67955).
Digital Karyotyping
Digital karyotyping was performed at the UC San Diego Institute for Genomic Medicine. Briefly, 200 ng of DNA was hybridized to Infinium CoreExome-24 arrays (Illumina), and stained per Illumina’s standard protocol. Copy Number Variation (CNV) calling was carried out in Nexus CN (version 7.5) and manually inspected, visualizing the B-allele frequencies (proportion of A and B alleles at each genotype) and log R ratios (ratio of observed to expected intensities) for each sample, as described.20
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 8 software. Unpaired t-test was used to assess significance in samples sets of two. One-way ANOVA was used to assess for the multiple sample sets comparisons (P < .05). Two-way ANOVA (alfa = 0.05) was performed for cell growth assay and scratch assay. Kaplan–Meier curves and comparison of survival were analyzed using Log-rank (Mantel–Cox) test.
Data Accessibility
Data have been deposited at Gene Expression Omnibus. Bulk RNA-seq data are available under accession number GSE186289.
Results
Engineering a Heterozygous TERT Promoter Mutation in Human hiPSC
Of the two hot spot TERT promoter mutations found in GBM, C228T, and C250T, we focused on the C228T mutation based on its greater frequency in patient samples (approximately 70% C228T and 30% C250T),1 and its association with worse prognosis when compared to C250T.21 A previously reported two-step CRISPR/Cas9 method for TERT promoter genome engineering was modified to obtain the heterozygous TERT promoter mutation in hiPSC.10 The first round of CRISPR/Cas9 mediated editing used two sgRNAs to delete a large portion of the promoter followed by a second round of CRISPR/Cas9 editing targeting a novel sequence created by the first round of editing to restore the promoter region using C228T mutant and wild type template plasmids in an hiPSC clone with homozygous deletions of PTEN, CDKN2A/B (Figure 1a, Supplementary Fig. 1a, Supplementary Table 1). The homozygous TERT promoter wild type clone was also created to ensure a true isogenic comparison and to control for effects of editing. Gene-edited clones were genotyped by PCR (Figure 1b) and sequenced to confirm the introduction of heterozygous C228T mutation, or restoration of wild type sequence (Figure 1b). Subsequently, the recovery of TERT expression was confirmed by RT-qPCR (Figure 1c). We then differentiated the engineered hiPSCs to NPCs, which were confirmed with elevated expression of Pax6 and decreased expression of pluripotent markers, Oct4 and Nanog (Supplementary Fig. 1b), and further to astrocytes with decreased Pax6 and increased GFAP (Supplementary Fig. 1c). TERT expression was retained only in the TERT promoter mutant (TPM) astrocytes while it was lost in the TERT promoter wild type (TPW) astrocytes (Supplementary Fig. 1d) Furthermore, TPM astrocytes continued to proliferate while the TPW cells became quiescent at later passages after more than 3 months (Supplementary Fig. 1e), in agreement with data by Chiba et al. examining terminally differentiated neurons and fibroblasts10,22 with mutant TERT promoters.
Fig. 1.
Engineering a heterozygous TERT promoter mutation in hiPSC. a) Schema of CRISPR editing strategy to introduce heterozygous C228T TERT promoter mutation. b) Genotyping PCR and Sanger sequencing confirmation of the TERT promoter mutation. c) RT-qPCR of TERT in engineered hiPSC clones with a TPM or TPW reconstituted promoter region. Data represents mean ± SD.
Impact of TERT Promoter Mutation on Tumor Initiation
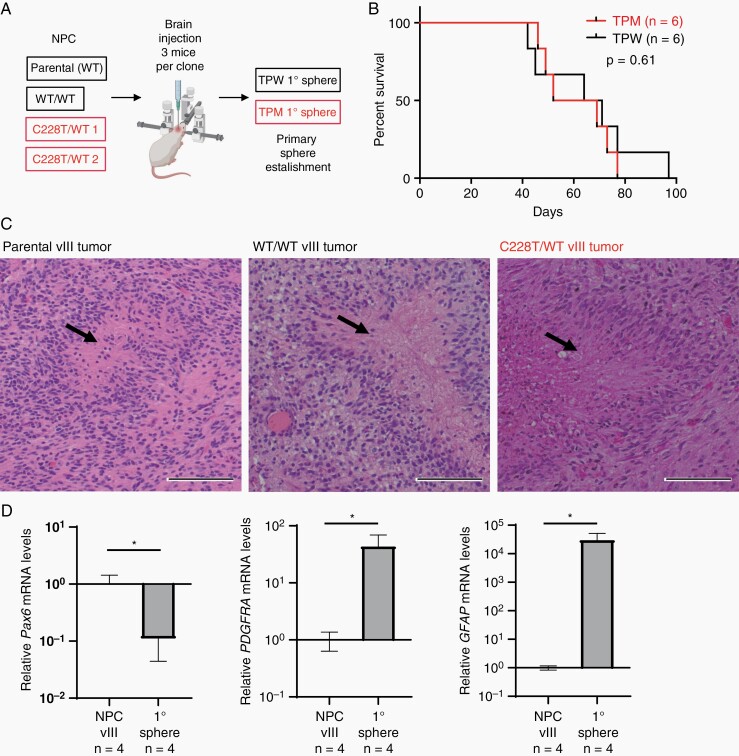
To test the effect of TERT promoter mutation on glioma formation, PTEN and CDKN2A/2B null, EGFRvIII-expressing NPC clones without editing of TERT promoter (parental), and the ones engineered either with TPW (WT/WT) or TPM (C228T/WT) were implanted into the right cerebral hemisphere of NOD-SCID mice (Figure 2a). The survival analysis of the mice implanted with the engineered NPCs showed that tumors formed with similar latency periods between TPM and TPW clones (Figure 2b). Tumors with and without TPM similarly presented extensive growth with psuedopalisading necrosis, thus recapitulating human GBM morphological features (Figure 2c, Supplementary Fig. 2a). Additionally, analysis of in vitro cell growth of TPM (n = 4) and TPW (n = 4) spheres obtained from these primary tumors (2 from each NPC clone) showed identical proliferation capacity (Supplementary Fig. 2b) and expressed markers of oligodendrocyte precursor cells (PDGFRA) and astrocytes (GFAP), while Pax6, a NPC marker, was downregulated (Figure 2d). RNA sequencing of these 8 spheres also showed a very similar expression pattern between the two groups (Supplementary Fig. 2c). We confirmed the similar expression level of EGFRvIII in these groups as well (Supplementary Fig. 2c). These results indicate the TERT promoter mutation does not enhance tumor initiation from these NPCs with the gliomagenic mutations.
Fig. 2.
Impact of the TERT promoter mutation on tumorigenesis. a) Workflow of the in vivo experiments. b) Survival analysis of the mice implanted with PTEN−/−; CKDKN2A/2B−/−; EGFRvIIIOE; TPM or TPW NPCs. 3 mice for each NPC clone were injected for the analysis. c) H&E staining of the parental, TPW, and TPM primary tumors illustrating pseudopalisading necrosis (arrow). Scale bars indicate 100 µm. d) RT-qPCR of Pax6, PDGFRA, and GFAP in 4 injected NPC lines and 4 primary tumor sphere lines (2 TPW and 2 TPM). Data represents mean ± SD (*P < .05, n.s = not significant).
Functional Analysis of the TERT Promoter Mutation in Primary Tumor Spheres
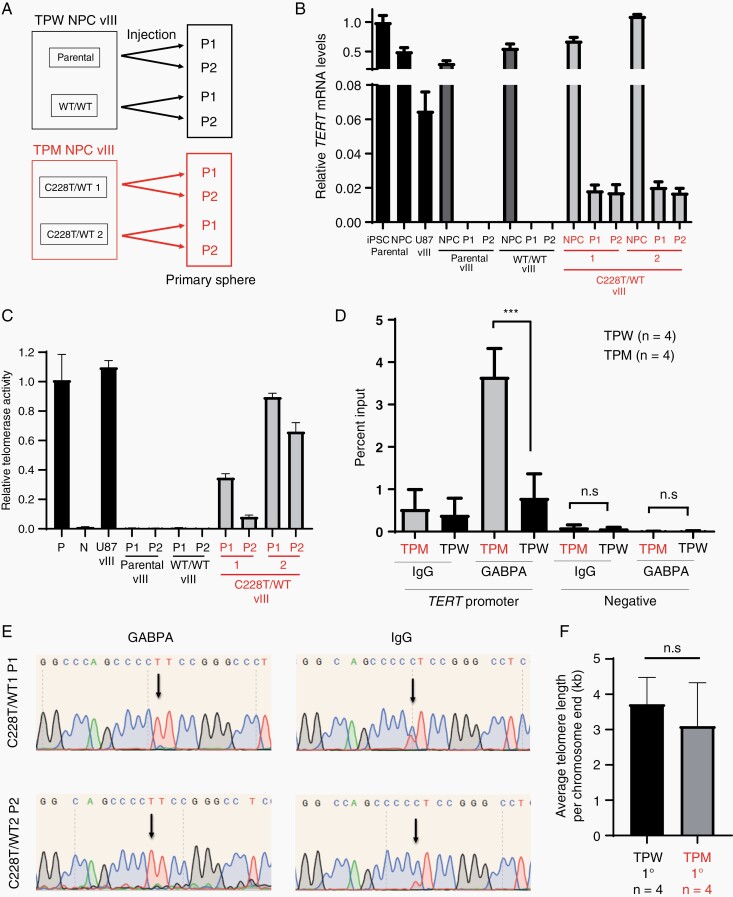
To elucidate the functions of TPM in these cellular models, we first evaluated the expression of TERT and found that it was silenced both in the parental and WT/WT TPW primary spheres, while the TPM spheres maintained a low level of expression comparable to that detected in U87vIII cells, which is known to have TERT promoter C228T mutation (Figure 3a and b). Correspondingly, telomerase activity was present in the TPM but not in the TPW primary spheres (Figure 3a and c). Consistent with the mechanism of TERT upregulation by the selective binding of GA-binding protein transcription factor,18 ChIP-qPCR and ChIP-PCR showed GABPA was selectively enriched on the promoter of the TPM allele (Figure 3d and e). These results indicate the function of the TPM in our models recapitulates that seen in patient-derived GBM cell lines. We further investigated the telomere length in these tumor sphere cells and found that the telomeres in both TPW and TPM cells were similarly long (Figure 3f).
Fig. 3.
Functional analysis of the TPW and TPW primary tumor spheres. a) Schema explaining the cell lines used. b) TERT RT-qPCR of the 4 biological replicates of the TPM and TPW primary tumor spheres. c) Telomerase activity of the 4 biological replicates of the TPM and TPW primary tumor spheres. (P: positive control, N: negative control). d) GABPA ChIP-qPCR of the 2 TPM and 2 TPW primary sphere cell lines. The quantitative results of qPCR from IgG ChIP and GABPA ChIP DNA using TERT promoter lesion primers (left half) and negative control primers (right half) are shown. e) Sanger sequencing results of ChIP-PCR products. f) Average telomere length in the TPM and TPW primary tumor spheres. Data represent mean ± SD (***P < .0001, n.s = not significant).
The Function of TERT Promoter Mutation Upon Serial In Vivo Passage
To investigate potential delayed effects of the TPM in our models of gliomagenesis, tumors were harvested two months after reimplantation of one million of the TPM and TPW primary sphere cells and the number of invading tumor cells away from the implantation site, were quantified in multiple sections (Figure 4a). The number of invading tumor cells in the brain, derived from implanted sphere cells, presented a striking difference between the isogenic TPM and TPW groups in the shared microenvironment, although invasiveness was similar in vitro setting (Figure 4b and c, Supplementary Fig. 3a,b). The survival analysis after implantation of two independent sphere cell lines from each group of TPW and TPM showed worse survival for the TPM tumor-bearing mice with a similar pathological finding at the time of morbidity. Correspondingly, the TPM maintained cell growth when cultured long term (Supplementary Fig. 4a-e). Next, the secondary sphere cell lines (TPM (n = 3) and TPW (n = 4) sphere lines) were established by recovering the tumors obtained by reimplantation of the primary tumor sphere cells and were assessed for TERT promoter mutation status and TERT expression by genomic sequencing and RT-qPCR, respectively, confirming persistence of the mutation status (data not shown), TERT silencing in the TPW, and sustained TERT expression in the TPM spheres (Figure 5a). RNA sequencing of the TPM (n = 3) and TPW (n = 3) secondary spheres presented more prominent difference in gene expression profiles compared to the primary spheres (Figure 5b, Supplementary Fig. 2c). GO analysis of the differentially expressed genes between the TPM and TPW secondary spheres showed cell proliferation and mitosis related pathways upregulated in the TPM secondary spheres, despite similar EGFR expression between the two groups (Figure 5c, Supplementary Fig. 5a). As confirmation of this result, upregulation of the cell cycle genes, E2F1a and CyclinA2, was confirmed by immunoblot analysis (Supplementary Fig. 5b). In parallel, the TPM secondary spheres presented in vitro growth advantage compared with the TPW secondary spheres (Supplementary Fig. 5c). Surprisingly, the telomere length was significantly longer in the TPW secondary spheres (Figure 5d) even though mice harbored the secondary TPW tumors for a longer duration than the secondary TPM tumors, possibly indicating slower growing TPW cells maintaining their telomere length were selected in vivo. However, when the TPW secondary spheres were cultured in vitro for one month, telomere length was shortened compared to the length at the initiation of the culture (Supplementary Fig. 5d), indicating a lack of telomere maintenance function in TPW cells. When the TPW secondary spheres were reinjected orthotopically (3 clones), none of the injected mice (0/5) formed tumors up to 10 months of observation. In contrast, half of the TPM secondary sphere-injected mice (1/2 from each clone) formed tumors during the same observation period (Figure 6a and b). The tertiary TPM tumor sphere cell lines maintained TERT expression, short telomeres, and proliferation capacity comparable to the secondary TPM spheres (Figure 6c–e). Lastly, we assessed chromosomal abnormalities in our models by digital karyotyping. We found several chromosomal aberrations both in the TPW and TPM primary sphere cells. Since these cells were re-adapted to culture prior to being karyotyped, it was not possible to distinguish if these aberrations were acquired in vivo or upon adaption to in vitro culture. Nonetheless, we did detect additional abnormalities in the TPM models (Supplementary Fig. 6). Taken together, these results highlight the advantage of TERT promoter mutation in maintaining proliferation in cells with shortened telomere length after extensive cell divisions in vivo.
Fig. 4.
Impact of the TERT promoter mutation upon serial in vivo passage. a) Workflow schema of in vivo experiments. b) Graph represents the number of hNuMA-positive cells at each level of sectioned brains in the mice implanted with the 4 TPM and 4 TPW primary sphere cell lines. Data represents mean ± SEM (*P < .05). c) Representative images of hNuMA, vimentin, and Ki-67 staining of the TPM and TPW secondary tumors in the sections 2.4 mm posterior to the bregma.
Fig. 5.
Function of the TERT promoter mutation in serial passaged tumors. a) TERT RT-qPCR of the primary/secondary TPM and TPW tumor spheres. b) Differential gene expression analysis of the TPM (n = 3) and TPW (n = 3) secondary tumor spheres. Volcano plot showing gene expression comparison between the TPM and TPW tumor spheres, with significant genes colored in teal. c) Gene ontology networks of the genes differentially expressed between the TPM and TPW secondary tumor spheres are shown. d) Average telomere length in the TPM and TPW secondary spheres. All data represents mean ± SD (*P < .05, n.s = not significant).
Fig. 6.
TERT promoter mutant tumors maintain growth capacity and telomere length during serial passage. a) Survival curve for tumor associated death of TPM and TPW secondary sphere-injected mice. b) Representative histological staining of the TPM tertiary tumor (left H&E, right human nuclei staining. Bar 5 mm). c) Relative TERT expression in the TPM secondary spheres compared to the tertiary tumor spheres. d) Relative telomere lengths in the TPM secondary and tertiary spheres. e) Growth rate comparison between the TPM tertiary (red) and secondary (black) spheres. All Data represents mean ± SD (*P < .05, n.s = not significant).
Discussion
Recent studies support the idea that cells in an undifferentiated state can serve as a glioma cell of origin.23–25 Our current study indicates the TERT promoter mutation has little impact on tumor initiation from the NPC state, where cells have longer telomeres and can escape from TERT expression silencing during the tumorigenic process. Furthermore, our results showed this mutation conveys a proliferation advantage upon secondary passaging in vivo, where the telomere length of the tumor cells shortens. The relatively shorter telomere length in TPM GBM is well known, and maintenance of this shorter length telomere plays a role in promoting genomic instability.22,26 A recent study showed that short telomeres are susceptible to break fusion breaks and correlate with moderate TERT expression as found in TPM cells.27 Although the digital karyotyping data cannot distinguish clonal selection and gain of chromosomal abnormalities, our results also support a dual effect of this mutation, which are maintenance of minimal telomere length and increase in chromosome instability. Importantly, our findings show that a heterozygous mutation is sufficient to illustrate the advantage of this mutation in tumor formation, thus explaining the predominance of heterozygous mutations in patients.21
On the contrary, no difference in our TPM and TPW system during tumor initiation could be explained by sufficient telomere length in the hiPSC-derived NPCs used for our experiments, and similarly, long telomeres in patient tumors could explain the lack of necessity for TPMs for tumor initiation proposed by some models.6,26 Single nucleotide polymorphisms associated with longer telomeres detected in leukocytes are known to increase glioma risks, and longer telomeres could allow for more cell divisions before cells experience a selective pressure to acquire a TERT promoter mutation followed by clonal expansion.28,29 Indeed, we demonstrated that human hiPSC-derived NPCs with three key GBM genetic alterations in the context of the TPW are sufficient for tumor initiation. The recent patient-derived trajectory model by Körber et al. suggests TPMs occur soon after chromosomal abnormalities and are predicted to be necessary for clonal expansion once the telomeric crisis is reached, which our results do not contradict.3 However, our study has the limitation of the use of a single female hiPSC line and only female mice in the scope of sex differences in GBM biology. Further, although we were not able to obtain a single clone with TPM without other genetic alterations, the function of this mutation in tumor initiation should ideally be studied using cells with this mutation by itself. Lastly, our results would also be consistent with a model for TPMs for tumor initiation from a more differentiated cell of origin with shorter telomeres rather than NPC.30
Interestingly, our in vivo results indicate TERT expression by the TERT promoter mutation may increase invasiveness not only enabling enhancement of proliferation in vivo, however in the in vitro setting, increased invasion was not detected (Figure 4, Supplementary Fig. 3a,b). Although our present study focused more on the growth advantage of TPM tumors, rather than migration, the involvement of TERT in increasing invasive capacity has been reported in different cancer and hTERT overexpressed model31–33 and would be an intriguing subject for future studies using our models. In general, the overexpression is supra-physiologic especially compared to the very low expression from the endogenous mutant TERT promoter.
The timing of the generation of this mutation might also be related to the status of chromatin in the TERT locus, which is open and accessible in undifferentiated cells but compacted in differentiated cells.34,35 Findings from recent longitudinal analyses of patient samples suggest gliomagenesis is a process occurring over a decade, and the cells with TPMs in stem-like states could be a reservoir for the processes of gliomagenesis.3,36 In our study, we could not confirm the activation of telomere maintenance function in the TPW cell lines through our in vivo and in vitro observations. This may be due to the limited period of observation and may also be due to the combination of the gene alterations we engineered in our models.3,8
It is known that the gain of chromosome 7, one of the most common chromosomal abnormalities in all glioblastoma subtypes, enhances the expression of not only EGFR but also PDGFA,37,38 which induces tumorigenesis of subventricular zone progenitor cells in vivo in mice and leads to radical chromosomal abnormalities in vitro.37,39–41 In fact, we previously reported double minute amplification observed in tumor models with constitutively active PDGFRA.7 In our current study, increased expression of PDGFRA was observed when cells were transformed from NPCs, perhaps indicating the need for PDGFA/PDGFRA autocrine signaling to further recapitulate patient tumors (Figure 2d). The necessity of PDGFRA signaling during tumorigenesis in our models will be evaluated in future studies.
Diverse mutations among patients and heterogeneity within each patient’s tumor are well-known factors that relate to treatment resistance in GBM.3,36,42,43 Not only it is the most common abnormality in GBM but the status of the TERT promoter mutation is also stable throughout tumor progression from onset to recurrence as shown in several studies,3,44,45 thus, mutant TERT promoter is an ideal target to overcome these issues of heterogeneity as indicated by recently attempted therapeutic strategies.46,47 The fact that the cells with this mutation develop a growth advantage in the later in vivo growth phase of our models but have shorter telomeres encourages the reconsideration of precision therapy targeting TERT promoter mutant brain tumors, coupled with a TERT promoter molecular diagnosis.30,48,49 The failure of Imetelstat, an oligonucleotide targeting the RNA component of the telomerase, in a phase II study directed at pediatric brain tumors might be due to patient selection without TERT promoter molecular diagnosis, as it is now known that most pediatric brain tumors are TPW.50,51 Our novel model of the TPM in the context of other GBM oncogenic drivers, therefore, allows testing of this hypothesis and for identification of TERT promoter-specific vulnerabilities.
Supplementary Material
Acknowledgments
We thank Drs. Chiba and Hockemeyer for sharing their plasmids for the TERT promoter targeting by CRISPR/Cas9. We thank the Center for Advanced Laboratory Medicine, University of California San Diego for assistance with immunohistochemistry.
Contributor Information
Shunichiro Miki, Department of Medicine, University of California San Diego, La Jolla, California, USA.
Tomoyuki Koga, Department of Neurosurgery, University of Minnesota, Minneapolis, Minnesota, USA.
Andrew M Mckinney, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA.
Alison D Parisian, Department of Medicine, University of California San Diego, La Jolla, California, USA; Biomedical Sciences Graduate Program, University of California, San Diego, La Jolla, California, USA .
Takahiro Tadokoro, Neuroregeneration Laboratory, Department of Anesthesiology, University of California San Diego, La Jolla, California, USA.
Raghavendra Vadla, Department of Medicine, University of California San Diego, La Jolla, California, USA.
Martin Marsala, Neuroregeneration Laboratory, Department of Anesthesiology, University of California San Diego, La Jolla, California, USA.
Robert F Hevner, Department of Pathology, University of California San Diego, La Jolla, California, USA.
Joseph F Costello, Department of Neurological Surgery, University of California, San Francisco, San Francisco, California, USA.
Frank Furnari, Department of Medicine, University of California San Diego, La Jolla, California, USA; Laboratory of Tumor Biology, Ludwig Cancer Research, San Diego Branch, La Jolla, California, USA.
Funding
This work was supported by National Institutes of Health R01NS080939 (F.F.), National cancer institute P50CA097257 (J.F.C.), a gift from the Dabbiere family (J.F.C.), and a gift from the Hana Jabsheh Research Initiative (J.F.C.), National Cancer Institute fellowship F31 CA243187 (A.M.M.), the National Institute of General Medical Sciences T32GM008666 (A.P.), the Defeat GBM Research Collaborative, a subsidiary of the National Brain Tumor Society (F.F.). S.M. was supported by Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowships.
Conflict of interest statement. J.F.C. is co-founder of Telo Therapeutics and has ownership interests.
Authorship statement. S.M., T.K., J.F.C., and F.F. conceived and designed the study. S.M., T.K., A.M., and R.V. conducted the experiments. A.P. performed bioinformatics analyses. R.H., T.T., and M.M. conducted histological analyses. J.F.C., and F.F. supervised all aspects of the study. S.M. and F.F. wrote the manuscript with input from other authors.
References
- 1. Arita H, Narita Y, Fukushima S, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013; 126(2):267–276. [DOI] [PubMed] [Google Scholar]
- 2. Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013; 110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Körber V, Yang J, Barah P, et al. Evolutionary trajectories of IDHWT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell. 2019; 35(4):692–704.e12. [DOI] [PubMed] [Google Scholar]
- 4. Mahlokozera T, Vellimana AK, Li T, et al. Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro-Oncology. 2018; 20(4):472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang F, Cheng D, Wang S, Zhu J. Human specific regulation of the telomerase reverse transcriptase gene. Genes. 2016; 7(7):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyai M, Tomita H, Soeda A, et al. Current trends in mouse models of glioblastoma. J Neurooncol. 2017; 135(3):423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koga T, Chaim IA, Benitez JA, et al. Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat Commun. 2020; 11(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015; 372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zanca C, Villa GR, Benitez JA, et al. Glioblastoma cellular cross-talk converges on NF-κB to attenuate EGFR inhibitor sensitivity. Genes Dev. 2017; 31(12):1212–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiba K, Johnson JZ, Vogan JM, et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015; 4:e07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reinhardt P, Glatza M, Hemmer K, et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013; 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bravo-Hernandez M, Tadokoro T, Navarro MR, et al. Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat Med. 2020; 26(1):118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parisian AD, Koga T, Miki S, et al. SMARCB1 loss interacts with neuronal differentiation state to block maturation and impact cell stability. Genes Dev. 2020; 34(19-20):1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raudvere U, Kolberg L, Kuzmin I, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47(W1):W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010; 5(11):e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bell RJ, Rube HT, Kreig A, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015; 348(6238):1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis . Nat Methods. 2012; 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panopoulos AD, D’Antonio M, Benaglio P, et al. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Rep. 2017; 8(4):1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nonoguchi N, Ohta T, Oh J-E, et al. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013; 126(6):931–937. [DOI] [PubMed] [Google Scholar]
- 22. Chiba K, Lorbeer FK, Shain AH, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017; 357(6358):1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weng Q, Wang J, Wang J, et al. Single-Cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell. 2019; 24(5):707–723.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Llaguno SA, Sun D, Pedraza AM, et al. Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat Neurosci. 2019; 22(4):545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee JH, Lee JE, Kahng JY, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018; 560(7717):243–247. [DOI] [PubMed] [Google Scholar]
- 26. Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016; 164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dewhurst SM, Yao X, Rosiene J, et al. Structural variant evolution after telomere crisis. Nat Commun. 2021; 12(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walsh KM, Codd V, Rice T, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015; 6(40):42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saunders CN, Kinnersley B, Culliford R, et al. Relationship between genetically determined telomere length and glioma risk. Neuro-Oncology. 2022; 24(2):171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lorbeer FK, Hockemeyer D. TERT promoter mutations and telomeres during tumorigenesis. Curr Opin Genet Dev. 2020; 60:56–62. [DOI] [PubMed] [Google Scholar]
- 31. Liu Z, Li Q, Li K, et al. Telomerase reverse transcriptase promotes epithelial–mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013; 32(36):4203–4213. [DOI] [PubMed] [Google Scholar]
- 32. Tang B, Xie R, Qin Y, et al. Human telomerase reverse transcriptase (hTERT) promotes gastric cancer invasion through cooperating with c-Myc to upregulate heparanase expression. Oncotarget. 2016; 7(10):11364–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park Y-J, Kim EK, Bae JY, Moon S, Kim J. Human telomerase reverse transcriptase (hTERT) promotes cancer invasion by modulating cathepsin D via early growth response (EGR)-1. Cancer Lett. 2016; 370(2):222–231. [DOI] [PubMed] [Google Scholar]
- 34. Wang S, Zhu J. The hTERT gene is embedded in a nuclease-resistant chromatin domain. J Biol Chem. 2004;279(53):55401–55410. [DOI] [PubMed] [Google Scholar]
- 35. Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol Biol Cell. 2007; 18(2):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016; 48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ozawa T, Riester M, Cheng Y-K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer cell. 2014; 26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Westermark B. Platelet-derived growth factor in glioblastoma—driver or biomarker? Ups J Med Sci. 2014; 119(4):298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, et al. PDGFRα-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006; 51(2):187–199. [DOI] [PubMed] [Google Scholar]
- 40. Assanah M, Bruce J, Suzuki S, et al. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. 2009; 57(16):1835–1847. [DOI] [PubMed] [Google Scholar]
- 41. Bohm AK, DePetro J, Binding CE, et al. In vitro modeling of glioblastoma initiation using PDGF-AA and p53-null neural progenitors. Neuro-oncology. 2020; 22(8):1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014; 344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Z, Chan AK-Y, Ding X, et al. Glioma groups classified by IDH and TERT promoter mutations remain stable among primary and recurrent gliomas. Neuro-Oncology. 2017; 19(7):1008–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miki S, Satomi K, Ohno M, et al. Highly sensitive detection of TERT promoter mutations in recurrent glioblastomas using digital PCR. Brain Tumor Pathol. 2020; 37(4):154–158. [DOI] [PubMed] [Google Scholar]
- 46. Li X, Qian X, Wang B, et al. Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nat Cell Biol. 2020; 22(3):282–288. [DOI] [PubMed] [Google Scholar]
- 47. Mancini A, Xavier-Magalhães A, Woods WS, et al. Disruption of the β1L isoform of GABP reverses glioblastoma replicative immortality in a TERT promoter mutation-dependent manner. Cancer Cell. 2018; 34(3):513–528.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guterres AN, Villanueva J. Targeting telomerase for cancer therapy. Oncogene. 2020; 39(36):5811–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aquilanti E, Kageler L, Wen PY, Meyerson M. Telomerase as a therapeutic target in glioblastoma. Neuro-Oncology. 2021; 23(12):2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Salloum R, Hummel TR, Kumar SS, et al. A molecular biology and phase II study of imetelstat (GRN163L) in children with recurrent or refractory central nervous system malignancies: a pediatric brain tumor consortium study. J Neurooncol. 2016; 129(3):443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013; 126(6):907–915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited at Gene Expression Omnibus. Bulk RNA-seq data are available under accession number GSE186289.