Abstract
Background
Glioblastoma (GBM) is the most aggressive primary brain tumor. Its cellular composition is very heterogeneous, with cells exhibiting stem-cell characteristics (GSCs) that co-determine therapy resistance and tumor recurrence. Bone Morphogenetic Protein (BMP)-4 promotes astroglial and suppresses oligodendrocyte differentiation in GSCs, processes associated with superior patient prognosis. We characterized variability in cell viability of patient-derived GBM cultures in response to BMP4 and, based on single-cell transcriptome profiling, propose predictive positive and early-response markers for sensitivity to BMP4.
Methods
Cell viability was assessed in 17 BMP4-treated patient-derived GBM cultures. In two cultures, one highly-sensitive to BMP4 (high therapeutic efficacy) and one with low-sensitivity, response to treatment with BMP4 was characterized. We applied single-cell RNA-sequencing, analyzed the relative abundance of cell clusters, searched for and identified the aforementioned two marker types, and validated these results in all 17 cultures.
Results
High variation in cell viability was observed after treatment with BMP4. In three cultures with highest sensitivity for BMP4, a substantial new cell subpopulation formed. These cells displayed decreased cell proliferation and increased apoptosis. Neuronal differentiation was reduced most in cultures with little sensitivity for BMP4. OLIG1/2 levels were found predictive for high sensitivity to BMP4. Activation of ribosomal translation (RPL27A, RPS27) was up-regulated within one day in cultures that were very sensitive to BMP4.
Conclusion
The changes in composition of patient-derived GBM cultures obtained after treatment with BMP4 correlate with treatment efficacy. OLIG1/2 expression can predict this efficacy, and upregulation of RPL27A and RPS27 are useful early-response markers.
Keywords: BMP, drug therapy, glioblastoma, single-cell RNA-sequencing, tumor heterogeneity
Key Points.
OLIG1/2 presence appears associated with sensitivity to therapeutic BMP4.
Neurodevelopmental lineage choices in GBM correlate with response to BMP4.
Upregulation of RPL27A and RPS27 marks early efficacy of treatment with BMP4.
Importance of the Study.
Failure of making progress in the treatment of glioblastoma is mainly due to resistance of tumor cells to current therapies. Since almost two decades no new agent against glioblastoma has been approved in Europe. This is predominantly due to insufficient understanding of glioma tumor biology and lack of patient stratification. BMP4 recently entered the clinic in a Phase-I trial and, if deemed safe, will likely be tested in a Phase-II trial. More biological knowledge on inter- and intra-tumoral differences in therapy sensitivity and potential markers for patient stratification then become relevant. Understanding of the heterogeneous response to differentiation cues also helps development of other pro-differentiation therapies. For this, we performed a high-resolution analysis of 17 patient-derived GBM cultures. We found a specific cluster of cells that, upon treatment with BMP4, was formed only in cultures with high sensitivity, and identified OLIG1 and OLIG2 as potential predictive markers for therapeutic efficacy of BMP4.
Patients diagnosed with glioblastoma (GBM) and submitted to standard therapy, a combination of surgical resection and chemoradiation with temozolomide (TMZ), have a median life expectancy of 18 months. High intra-tumoral cell hete-rogeneity and plasticity downsize therapeutic effectiveness, together with resistance to therapy, which results in tumor recurrence and lethal progression.1,2 The glioblastoma stem cell-like cells (GSCs) are a major factor in both acquisition of chemoradiation resistance and tumor recurrence.3 GSCs are a self-renewing, small subpopulation of GBM cells, express stemness marker genes and can differentiate in vitro to several neural lineages.4,5 Their progeny constitutes novel subpopulations of tumor cells able to resist treatment or survive environmental changes.6,7 Hence, GSCs are considered as important targets for therapy.8,9
During embryogenesis, BMP actions must be antagonized first for enabling central nervous system (CNS) formation. However, at later stages BMPs co-stimulate cell differentiation along the astroglial lineage.10,11 In vitro and in vivo studies have shown that BMP4 promotes astroglial differentiation of GSCs, which depletes the GSC pool.12 Furthermore, an orthotopic GBM model in mice, upon administration of BMP4, had a significantly better survival rate than with TMZ.13
Novel therapeutic anti-glioma agents are often assessed first in cell cultures.14–16 The readout of such assays is generally done at cell-population, not single-cell level. Incomplete understanding of tumor cell responses to targeted treatments at such high-resolution contributes to translational gaps for glioma.17 Effective application of novel therapies, such as administration of BMP4 (which recently entered a first-in-human Phase-I trial for GBM18) requires thorough understanding of its intra-tumoral biological effects. Here we provide an analysis at single-cell resolution of differently responding cultures to differentiation-inducing cues and highlight novel resistance mechanisms to directed differentiation therapies. As GBM tumors also have high inter-tumoral heterogeneity, patient stratification will be essential in defining effectiveness of any other new therapeutic strategy. Hence, we wanted to gain novel insight into inter-tumoral and intra-tumoral variability, starting from cell viability monitoring after treatment with BMP4 and applying single-cell RNA-sequencing (scRNA-seq). This combinatorial strategy enabled us to identify both predictive markers and markers of early-therapeutic efficacy.
Materials and Methods
Cell Culture
Tumors were collected by the Department of Neurosurgery, Erasmus University Medical Center (Rotterdam) after receiving patient’s informed consent and in accordance with protocols approved by the institutional review board. All cultures were routinely tested for mycoplasma and were authenticated with either SNP-array or whole-exome sequencing in the used passages. Further details are mentioned in Supplementary Methods.
Cell Viability Assay
Seven days after treatment with 0.75-180 ng BMP4/ml BMP4, using three-fold serial dilutions, cell viability was measured (CellTiter-Glo kit, Promega). See details in Supplementary Methods .
Apoptosis Assay
The AnnexinV-iFluor 555 Apoptosis Staining/Detection Kit (Abcam) was used to assess apoptosis after 4 days of treatment with 60 ng BMP4/ml. See details in Supplementary Methods .
Proliferation Assay
5-Ethynyl-2′-deoxyuridine (EdU) incorporation was conducted using the EdU Staining Proliferation Kit (iFluor 647), according to the manufacturer’s protocol (Abcam). See details in Supplementary Methods .
Indirect Immunofluorescence
Indirect immunofluorescence was performed using antibodies and concentrations as described in Supplementary Methods.
Flow Cytometry
Dissociated cells were incubated for 30 min at 24°C with conjugated antibodies (Supplementary Table 3). Cells were then washed with and diluted in PBS. Samples were analyzed using a BD LSRFortessa, raw data was analyzed with FlowJoTM v10.6.2.
Self-Renewal Assay
The self-renewal assay was performed in soft-agar.19 For further details see Supplementary Methods.
Quantitative PCR
Total RNA was isolated with TRIzol reagent20; cDNA was synthesized using RevertAid (both Thermo Scientific). For primers, see Supplementary Table 4. Real-time PCR was done using the Bio-Rad CFX96 system. All qPCR experiments were done as biological and technical triplicates.
Western Blots
Cell lysates supplemented with phosphatase/protease inhibitors were separated by gel electrophoreses. Specifics are described in Supplementary Methods .
Single-Molecule RNA FISH
Cells seeded on coverslips were treated with BMP4 and fixed according to the manufacturer’s protocol. The RNAscope® Multiplex Fluorescent Reagent Kit v2 Assay was used for hybridization and visualization of probes of interest. See Supplementary Methods for further details.
Sample Preparation for scRNA-seq
At passage 7, cells were seeded at 200,000 cells/well in six-well plates, one day before treatment with BMP4 (60 ng BMP4/ml, 24 h). Cells were harvested after detachment with Accutase (Invitrogen) and dissolved in PBS-1% Dispase (ThermoFisher). The Controller and v3 Library and Gel Bead kit (all 10xGenomics) were used to obtain single-cell emulsions. The Chromium 3′ v3 protocol was used to generate libraries for RNA-seq, which were sequenced using a NovaSeq6000 (Illumina). Information with regard to data analysis and classification is described in Supplementary Methods.
Integration Public Dataset
The Seurat anchorpoints method compared the GBM cells cultured as serum-free monolayer with freshly resected tumor.21,22 Cells were annotated using unsupervised annotation (scCATCH).23
Availability of Data and Materials
The scRNA-seq files were deposited with NCBI-GEO (accession number GSE148196).
Statistical Analysis
Descriptive statistics were calculated in Graphpad Prism 8.2.1. Means of two sets of data were compared by student’s t-test.
Results
Patient-Derived Monolayer Cultures Self-Renew and Activate BMP-SMAD Signaling
Seventeen patient-derived cultures were assessed for their cell self-renewal, and all formed neurospheres, albeit with high variation between tumors (Supplementary Fig. 1A,B). After 24 h of treatment with 60 ng BMP4/ml, activation of BMP-SMADs [indicated as pSMAD1/5] was detected in 16 cultures (and was very weak in GS630) (Supplementary Fig. 2A,B). In addition, SMAD1 steady-state mRNA levels did not change drastically upon addition of BMP4 (Supplementary Fig. 2C). SMAD1/5 activation resulted in upregulation of ID family genes and SMAD7, acknowledged direct targets of BMP-pSMADs, in all 17 patient-derived cultures (Supplementary Fig. 2D).
Cell Viability After Treatment with BMP4 is Highly Variable in Patient-Derived GBM Cultures
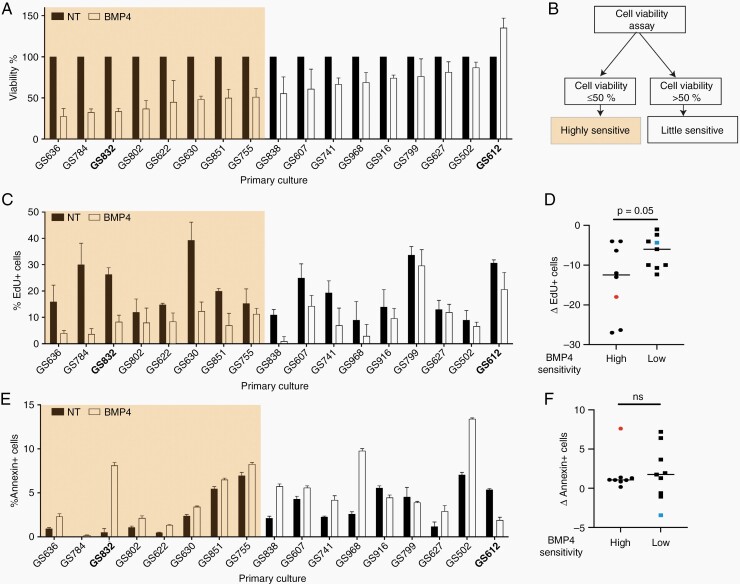
The effect of BMP4 on GBM cell viability has been a matter of controversy.24–27 We first assessed the variation in sensitivity to BMP4 in the 17 GBM cultures (their characteristics are summarized in Supplementary Table 1). We used three-fold dilutions (from 180 ng down to 0.74 ng BMP4/ml) to determine the dose that best allowed discrimination for therapeutic efficacy (Supplementary Figure 3A). This was 60 ng/ml, a classic dose of BMPs for stimulation of multiple cell types in culture. Treatment for 7 days at 60 ng/ml resulted in decreased cell viability for 16 cultures (Figure 1A). The remaining cell viability after BMP4 ranged from 28% (GS636) up to 87% (GS502) compared to the respective untreated cultures, whereas in GS612 the cell viability increased by 35%. Cell viability reduction after BMP treatment of monolayers correlated significantly with its effects in neurosphere cultures (Supplementary Figure 3B). We interpret this reduction in cell viability as a guide for in vitro therapeutic efficacy and accordingly stratified our culture panel into two groups. We define therapeutic efficacy as high-sensitive (i.e. viability in the presence of BMP4 decreased to ≤50% of non-treated cells) or low-sensitive (>50% remaining viability) (Figure 1B).
Fig. 1.
Cell viability after treatment with BMP4. (A) Average cell viability with standard deviation 7 days post-treatment with 60 ng BMP4/ml normalized to untreated (NT) cells (n = 3). Cultures used for single-cell analysis are in bold. (B) Flow-chart of stratification into the ‘highly sensitive or the “little sensitive” cultures. (C) Mean percentage with standard deviation of EdU-positive (EdU+) cells after 2 days of 60 ng BMP/ml (n = 3). (D) Mean difference in EdU+ cells after treatment with BMP4. (F) Mean percentage with standard deviation of Annexin-V+ cells after 4 days of 60 ng BMP4/ml (n = 3). (G) Mean difference of Annexin+ cells after treatment. ns = Not-significant. In E and F, the orange and blue square indicate single-cell sequenced tumors GS832 and GS612.
The differences in post-BMP4 cell viability can result from changes in survival versus apoptosis, and proliferation versus differentiation. In all cultures the cell proliferation decreased by adding BMP4 (Figure 1C). However, the median decrease in EdU+ (i.e. proliferating) cells was 12.5% in the highly and 6% in the low-sensitive cultures (Figure 1D) (P = 0.05). Measurement of the apoptotic (Annexin+) cells in each culture revealed that BMP4 generally promoted apoptosis, independent of its in vitro therapeutic efficacy. Upon additional analysis BMP4 induced limited necrosis and therefore we disregarded it in further analyses (Supplementary Figure 3C–E). Nonetheless, in 3 cultures with low-sensitivity (GS916, GS799 and GS612), apoptosis was reduced by BMP4 (Figure 1E). The variability in apoptosis resulting from BMP4 was larger in cultures with low-sensitivity to BMP4 (ranging from -3.4 to +6.4%) compared to those with high-sensitivity (+0.2 to +7.6%) (Figure 1F). Taken together, therapeutic efficacy of BMP4 in patient-derived GBM cultures has a stronger association with the proliferation rate than level of apoptosis.
Single-Cell Transcriptomic Profiling Reveals Cell Clusters and Cell Types Commonly Found in Primary GBM
To further investigate the effects of BMP4 we proceeded to scRNA-seq, with focus on the comparison between a highly (i.e. GS832) and a low-sensitive culture (i.e. GS612). GS612 was selected because it is the only culture in which BMP4 increased the viability (see Figure 1A). GS636, GS784 and GS832 were each highly-sensitive; we selected GS832 based on three assumed favorable features: its high sphere-forming capacity, comparable to GS612; a clear difference in proliferation after adding BMP4; and limited apoptosis in untreated cells (see Figure 1E).
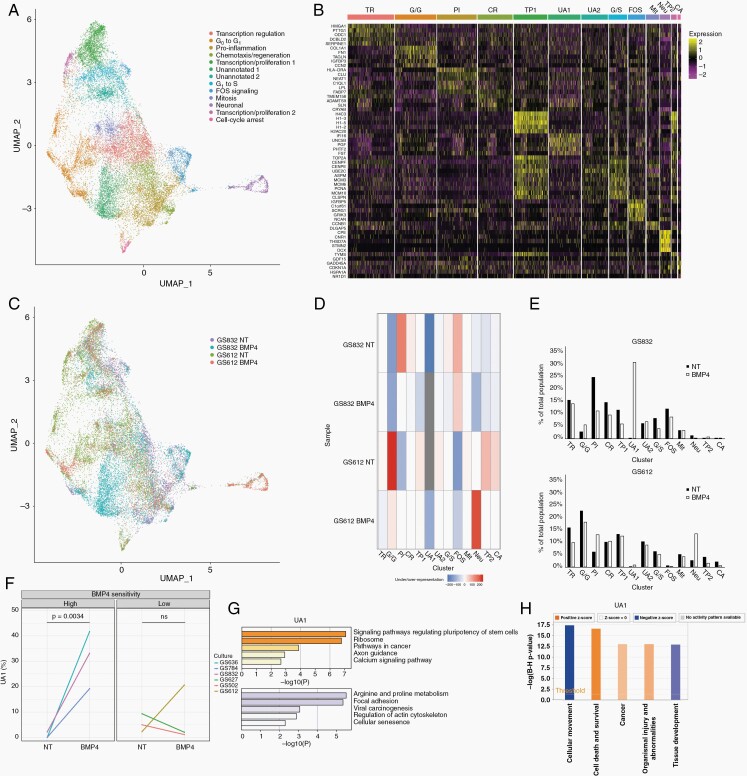
We clustered our datasets to focus on transcriptional dynamics in the cultures and assessed transcriptomic responses to BMP4 (Figure 2A). Based on known marker genes we annotated 13 cell clusters (Figure 2A, B). Two of these (i.e. unannotated (UA) 1/2; Figure 2A) did not have signatures indicative for a given cell state or type, a previously observed common characteristic of GBM.28,29 Gene transcription and cell proliferation (TP1/2) signatures were marked by upregulated TOP2A and H1-2, 3 and 5 (Figure 2B),30 and split in two clusters based on genes relating to extracellular matrix, COL3A1 and COL1A2 (for TP2, Figure 2B).31 Both clusters were enriched for proliferative/progenitor cells as classified according to Garofano et al. (Supplementary Figure 4B).32 The FOS cluster was marked by upregulated expression of FOS and C1orf61 (Figure 2B),33 while MCM3 and MCM6 marked G1 to S phase transition (G/S, Figure 2B).33
Fig. 2.
Single-cell transcriptomes of BMP4 treated patient-derived cultures. (A) UMAP embedding of single cells from two (un)treated GBM cultures. Clusters of transcriptionally similar cells are colored and labeled by ontology. (B) Heatmap of top-5 uniquely expressed genes per cluster. TR = Transcription regulation, G/G = G0 to G1, PI = Pro-inflammation, CR = Chemotaxis/regeneration, TP = Transcription/proliferation, UA = Unannotated, G/S = G1 to S, FOS = FOS signaling, Mit = Mitosis, Neu = Neuronal, CA = Cell-cycle arrest. (C) UMAP colored by sample. (D) Heatmap of the representation of each sample per cluster as determined by hypergeometric testing. (E) Proportion of each subpopulation per sample before and after treatment with BMP4 in GS832 (top panel) and GS612 (bottom panel). (F) Percentage of cells belonging to UA1 in the RNA-scope analysis before (NT) and after treatment with BMP4. ns = Not-significant (G) Enriched KEGG pathways of upregulated (orange) and downregulated genes (purple) in UA1. (H) Ingenuity Pathway Analysis of enriched biological functions ordered by adjusted P-value and filled according to z-score (activation) of UA1.
Gene-set enrichment analysis (GSEA) annotated one cluster (CR) as chemotaxis and regeneration (ANGPT2 and LGAL) (Figure 2B). The FOS, G1/S and CR clusters display upregulated PTPRZ1 and FABP7, markers of radial glial cells (Supplementary Figure 4A; Supplementary Table 2).28 We identified DCX, MAP2 and NHLH1 positive (+) neuronal cells (Figure 2A, B; Supplementary Figure 4B).34 These same cells were also annotated as neuronal cells after classification according to Neftel et al. (Supplementary Figure 4A).29 The proportion of DCX+ cells per sample followed the same trend in scRNA-seq and RNAscope (Supplementary Figure 5A, B). CDKN1A+ marked those cells in cell-cycle arrest (CA) (Figure 2B)35; these were found enriched in untreated GS612 in scRNA-seq and RNAscope (Supplementary Figure 5C, D).
We also examined to what extent the RNA-seq profiled cultures resemble freshly resected GBM, so cross-referenced our scRNA-seq datasets to those of freshly resected glioma tumors published by Wang et al.22 Unsupervised annotation of this merged dataset revealed an array of expected cell types (Supplementary Figure 6A). All neural subtypes found previously in freshly resected tissue could also be identified in our model. However, our cultures were enriched for cells with GSC-profile compared to previously published freshly resected tumors.22 Cell types that were not present in our dataset were macrophages and microglia (Supplementary Figure 6B, C).
Treatment with BMP4 Depletes Neuronal and Pro-inflammatory Subpopulations of Cells in a Highly-Sensitive Culture
Exposure to BMP4 may influence the distribution of the RNA-seq-defined cell clusters. Prior to treatment, this distribution differed between GS832 and GS612 (Figure 2C, D). Cells in G0 to G1 transition (the G/G cluster) were more frequent in GS612, while pro-inflammatory (PI) cells (with upregulated HLA-DRA, CLU and NEAT1) were over-represented in the highly-sensitive culture (GS832). COL1A1 was prominently expressed in the G/G cluster. RNAscope validated the over-representation of these cells in untreated GS612 (Supplementary Figure 5E, F). The TP1 cluster (H4C3+) in GS832 showed a clear decrease upon exposure to BMP4 both in scRNA-seq and RNAscope (see Figure 2C, D; Supplementary Figure 5E, G).
In GS832, a profound change was noted in the distribution of two specific clusters: the PI cluster decreased by ~50% of its original size, while UA1 emerged and eventually constituted almost 33% of the culture (Figure 2E, top graph; Supplementary Table 2). Validation by RNAscope of the six most and least BMP4-sensitive cultures revealed that the UA1 (PGF_hi, UNC5b_hi, IGFBP5_low and MCM3_low) cluster always arose in the high-sensitive, and sporadically in low-sensitive cultures (Figure 2F; Supplementary Figure 8). The cells in UA1 displayed, in relative terms, upregulated pathways regulating pluripotency states of stem cells (i.e. BMP, Hippo) (Figure 2G). Furthermore, ingenuity pathway analysis marked decreased migration and increased cell-death associated genes (Figure 2H). Although the PI cluster shrank with BMP4 in GS832, it is one of the two clusters, together with the population of neuronal cells (Neu), that expanded most in the low-sensitive GS612. This trend is also present after classification as described by Neftel et al. and Garofano et al. (Supplementary Figure 4C–F).29,32 Thus, BMP4 causes a shift in cell state/type composition in both cultured tumors, but most significantly for the highly-sensitive GS832 (Figure 2C–E).
Treatment with BMP4 Reduces Plasticity Between Cell States in Highly-Sensitive Cultures
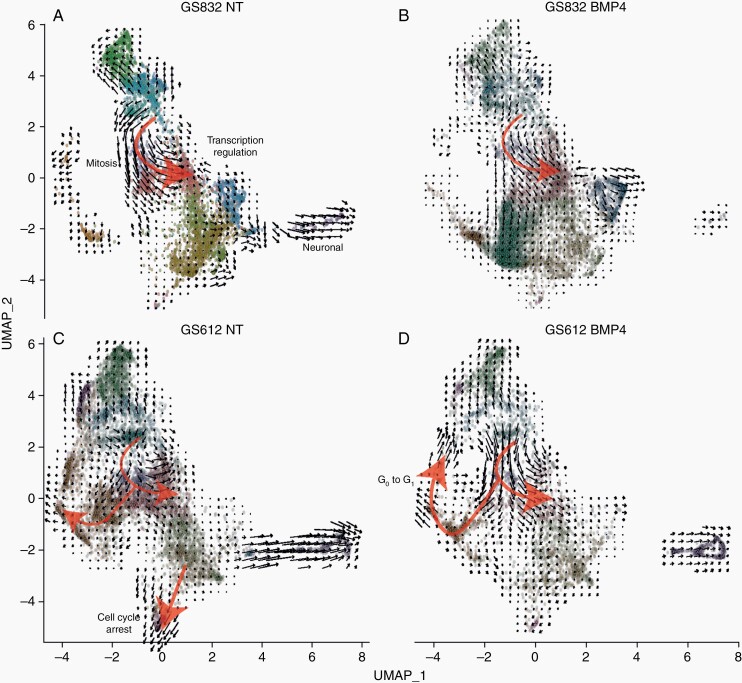
We used RNA-velocity analysis to assess the transcriptional dynamics upon BMP4 addition. Untreated GS832 cells followed the trajectory from unannotated to mitotic cells (CCNB1+ and DLGAP5+) towards cells involved in transcription regulation (HMGA1+) (red arrow, Figure 3A). Upon treatment with BMP4, the RNA-velocity generally decreased in all GS832 cells (Figure 3B). GS612 cells were moving at high velocity towards cell-cycle arrest (Figure 3C), which is halted after addition of BMP4 (Figure 3D). The cells followed the same trajectory as in GS832, but ended not only with annotated transcription regulation, but also with G0 to G1 cell-cycle transition, especially when exposed to BMP4.
Fig. 3.
UMAP embedding of (un)treated GBM cells with the black arrows showing RNA velocities and red arrows highlighting the main trajectories in untreated (A) treated (B) BMP4 highly-sensitive GS832 and of untreated (C) and treated (D) little-sensitive GS612.
Post-BMP4 MAP2 Expression is Discriminative for in Vitro Therapy Efficacy
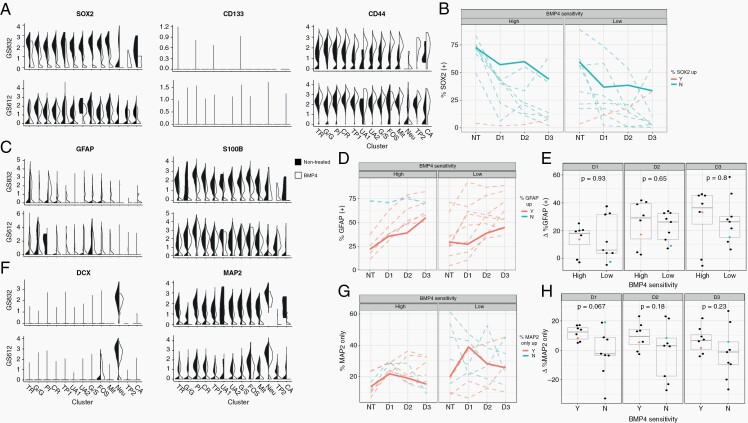
BMP4 as therapeutic agent in GBM became of interest following the report of its ability to induce astroglial differentiation in GSCs.26 Therefore, we documented upregulation of astroglial and downregulation of stemness marker genes (SOX2, CD44, CD133) after BMP4 addition (Figure 4). SOX2 and CD133 were downregulated after stimulation with BMP4, irrespective of therapeutic efficacy (Figure 4A, B, Supplementary Figures 9, 10A). After 24-h treatment, the low-sensitive cultures had enhanced downregulation of CD133 compared to high-sensitive cultures (Supplementary Figure 10B). BMP4 induced the upregulation of GSC marker CD44 (Figure 4A, right panels, Supplementary Figure 10C, D).
Fig. 4.
Lineages of GSC differentiation. (A) Violin plots depicting normalized mean and variance of glioma stemness-associated genes SOX2, CD133 and CD44 in the culture with high (GS832, left) and low (GS612, right) BMP4-sensitivity. The black side of the violin plots represents the non-treated cultures, and the white side the BMP4-treated cultures. The subsequent panels display the mean percentage of SOX2+ (B), GFAP+ (D) and MAP2+ (G) cells, as determined by indirect immunofluorescence staining (n = 5) of untreated cultures (NT), or after 1 day (D1), 2 days (D2) or 3 days (D3) of treatment with BMP4. The bold unbroken lines indicate the highly BMP4-sensitive GS832 and the little-sensitive GS612. Upregulation after 3 days of treatment is shown in red, while downregulation is depicted in blue. (C) Violin plots depicting normalized mean and variance of astrocyte markers GFAP and S100B. Boxplots of the difference in percentage GFAP+ (E) and MAP2+ cells (H). The orange and blue dot represent GS832 and GS612. (F) Violin plots depicting normalized mean and variance of neuronal lineage markers DCX and MAP2.
The astrocyte marker GFAP was expressed in few subpopulations, although more of these gained its expression after BMP4 was added (left panels, Figure 4C). Upon treatment with BMP4 the percentage GFAP+ cells increased in all cultures, regardless BMP4-sensitivity (Figure 4D, E). The other astrocyte marker gene, S100B, was downregulated after treatment, especially in GS832.
Neuronal marker genes DCX and MAP2 were expressed by GS612 and GS832. BMP4 only caused their downregulation in GS832 (Figure 4F). In a previous study, in which scRNA-seq was done for one BMP4-treated GBM culture, diminished proliferation of treated GBM cells was associated with neuronal cell-type genes.36 In our set of 17 cultures there was a trend (P = 0.067) of reduced expression of MAP2 in low-sensitive cultures compared to high-sensitive ones over the first 24 h (Figure 4G, H).
BMP4 has a Higher Therapeutic Efficacy in Cultures with an Oligodendrocyte Signature
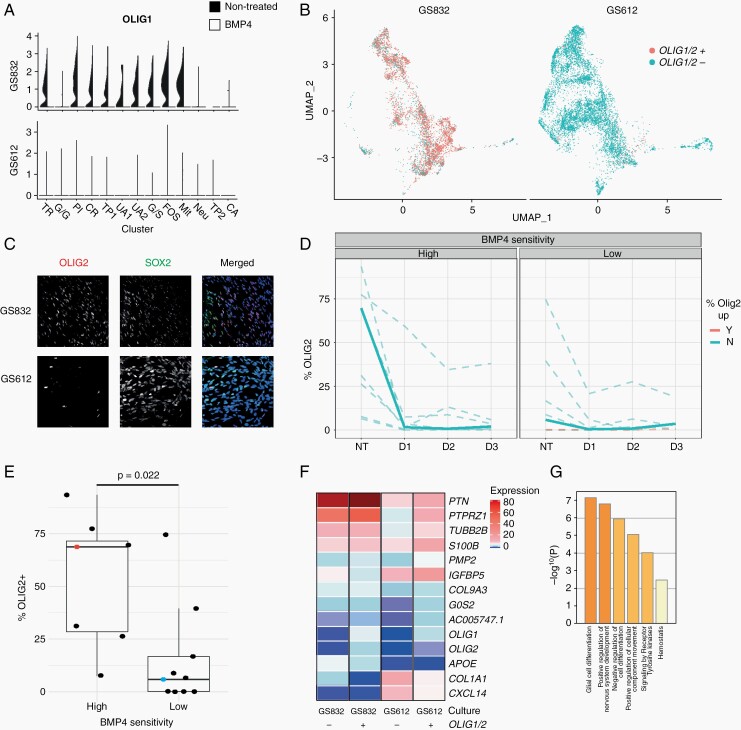
Most cells in GS832 were OLIG1+ and OLIG2+ before treatment, unlike those in GS612 (Figure 5A, B) and adding BMP4 caused loss of both. Validation by immunofluorescence in our 17-tumor panel confirmed that upon BMP4 addition almost no OLIG2+ cells were observed after 24 h (Figure 5C, D; Supplementary Figure 11). A low percentage of OLIG2+ cells within a cell culture was significantly (P = 0.022) associated with little sensitivity to BMP4 (Figure 5E).
Fig. 5.
OLIG1/2 predicts in vitro therapeutic efficacy of BMP4. (A) Violin plots depicting mean and variance of oligodendroglial lineage markers in highly BMP4-sensitive GS832 (top) and little-sensitive GS612 (bottom). The black side represents the non-treated, and the white side the BMP4-treated cultures. (B) UMAP of untreated cells discriminated based on scaled OLIG1 or OLIG2 (≥0.75 red; <0.75 blue) expression of cultures GS832 (left) and GS612 (right). (C) Representative images of untreated cultures stained for OLIG2 (red), SOX2 (green) and DAPI (blue). (D) Percentage of SOX2+ cells. NT (untreated), D1 (day 1), D2 (day 2), D3 (day 3). The bold lines represent GS832 (high-sensitivity) and GS612 (low-sensitivity). Loss of OLIG2+ cells after 3 days of BMP4 is colored blue, and gain red. (E) Percentage of OLIG2+ cells expressing related to dichotomized (viability <50% after 7 days of BMP4) sensitivity. (F) Scaled expression of differentially expressed genes between OLIG1/2+ and OLIG1/2- cells. (G) Overrepresented gene ontologies in differentially expressed genes mentioned in (F).
As OLIG1/2 positivity relates to oligodendrocyte (precursor) cells we hypothesized that it is the cellular state, rather than these specific genes, that is associated with sensitivity to BMP4. Therefore, we performed a differential gene expression (DEG) analysis and Metascape GSEA on OLIG1/2-positive and OLIG1/2-negative cells. Indeed, all cells identified as OLIG1/2+ expressed the oligodendrocyte signature (Figure 5F, G).
Activation of Ribosomal Translation Signaling is an Early Marker for BMP4-Sensitivity
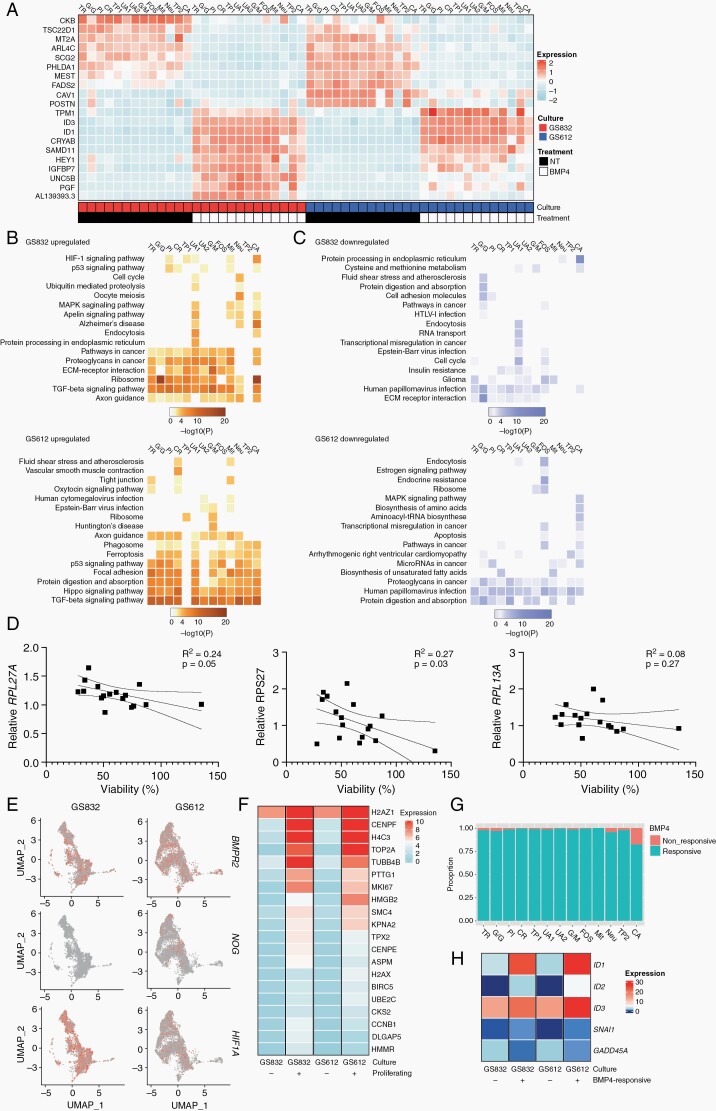
We also investigated the effect of treatment with BMP4 within each individual cell cluster. Most DEGs after treatment were similar within both cultures and most clusters therein (Figure 6A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping revealed that the TGFβ/BMP family signaling pathway was enriched in the upregulated gene sets (Figure 6B, left panels). In GS832, also axon guidance and interaction of ECM and ECM receptors were enriched, while in GS612 Hippo signaling and focal adhesion were. In GS612, the response between clusters was more heterogeneous than in GS832, although the clusters generally showed similar trends in pathway mapping. In the highly-sensitive GS832 culture ribosomal translation was one of the prominent pathways, unlike in GS612 where we found it was not (Figure 6B, left panels).
Fig. 6.
Subpopulation-specific responses to treatment with BMP4. (A) Expression of the 10 most differentially expressed genes after treatment with BMP4 in GS832 (red) and GS612 (blue). NT = untreated. Heatmap of enriched KEGG pathways for upregulated (B) and downregulated (C) genes after treatment with BMP4 in GS832 (top panel) and GS612 (bottom panel). (D) Correlation with standard deviation of mean relative expression of ribosomal translation genes after 24 h treatment with BMP4 normalized to GAPDH and relative to untreated cells, compared to viability after 7 days of treatment with BMP4 (n = 3). (E) Top-20 differentially expressed genes in proliferating vs non-proliferating cells after BMP4. (F) Proportion of cells per cluster that do not upregulate ID1 and ID2 (non-responsive) vs those that do (responsive) after BMP4. (G) Heatmap illustrating the anti-correlation of GADD45A with ID1 and ID2. (H) Expression of BMPR2, NOG and HIG1A in untreated cultures GS832 and GS612.
To validate the latter finding we performed RT-qPCR on the three most common DEGs in this pathway (RPL17A, RPS27 and RPL13A). We correlated the relative expression of these genes after 24 h of BMP4 treatment with the cell viability after 7 days of treatment (Figure 6D). Only the first two genes had a significantly higher expression in cultures with high compared to those with low-sensitivity to BMP4 (Figure 6D; for RPL17A P = 0.05, RPS27 P = 0.03). Interestingly, the glioma signature decreased in the culture with high therapeutic efficacy, i.e. GS832 (Figure 6C, top panel). Also, several clusters of GS612 had decreased apoptosis after treatment with BMP4, which can explain the increased viability measured after treatment (Figure 6C, bottom panel).
Several other genes that could influence therapeutic efficacy of BMP4 are those encoding the BMP-antagonists NOG, GREM1 and GREM2, the TGFβ-family receptors or the hypoxia associated gene HIF1A.37BMPR2, NOG and HIF1A were expressed in the untreated samples of GS832 and GS612, but not enriched in the latter (Figure 6E, Supplementary Figure 12).
Since decreased proliferation rate was significantly associated with low viability after treatment with BMP4 we further assessed the gene signature associated with this phenotype. We defined proliferating cells as MKI67+ and TOP2A+. The cells that continue proliferating with BMP4 expressed marker genes for epigenetic control (H2AZ1, H4C3) and DNA-configuration (HMGB2, SMC4) (Figure 6F). We hypothesized that those cells are likely near-non-responsive to BMP4 (i.e. do not upregulate ID1/2). Those “non-responsive” cells were rare, but still enriched in the CA cluster of cells (Figure 6G). The only gene that showed a negative correlation to upregulation of ID1/2 with BMP4 was the DNA-repair gene GADD45A (Figure 6H).
Discussion
In the search for transcriptional profiles that serve prediction of response to BMP4 in patient-derived cultures of GBM we found that scRNA-seq was the approach that yielded most useful results. This technique might not (yet) be the ideal candidate for near-routine clinical implementation, but it is very informative for further deciphering the underlying biology and future prediction of experimental BMP administration.
We show large inter-tumoral differences in sensitivity to in vitro treatment with BMP4 in line with published differential effects of BMP4 on glioma tumors.8,25 BMP4 effects on cell viability were found to be predominantly attributed to the rate of proliferation and not by degree of apoptosis. Cells with unaffected proliferation after exposure to BMP4 had upregulated genes involved in epigenetic regulation. Those undergoing cell-cycle arrest do not upregulate ID1/2, but GADD45A. This gene can confer pluripotency, while BMP4 induces differentiation.38 Consistently, upregulation of CDKN1A was found associated with a cytoprotective and quiescence-inducing effect of BMP4 on glioma cells.39 In our RNA-velocity analysis (Figure 3) we find decreased velocity after treatment with BMP4, which suggests induced quiescence as described by Sachdeva et al.39 However, as CDKN1A is only expressed in a limited number of cells in our population, it is unlikely that this gene can solely explain this response. Our least-sensitive culture GS612 had the most heterogeneous response to BMP4, which can partially explain the differences in sensitivity.3,40 As some differences in gene expression were relatively subtle, one could argue that these are caused by stochasticity of gene expression in cell cultures over time. Therefore, we have repeated each experiment in multiple passages and different cultures, which allowed us to discriminate unambiguously the BMP-driven effects.
Contrary to our expectation, the stemness marker CD44 is upregulated after stimulation with BMP4. CD44 is produced by astrocyte-progenitor neural stem cells,41 so CD44 upregulation is likely one of the first changes in astroglial differentiation. The astrocyte marker S100B is downregulated in GS832 after treatment with BMP4. As S100B marks both astrocytes and CSPG4+ oligodendrocytes,42 this downregulation is likely caused by depletion of oligodendrocytes by BMP4. In agreement with Videla et al., we also confirm that with BMP443 GSCs either undergo astroglial or neuronal differentiation, as revealed by GFAP vs. MAP2 expression, although we also observed that these were not mutually exclusive. Dalmo et al.44 recently reported that, in a panel of 40 primary GBM cultures, decreased expression of SOX2 would be predictive for decreased proliferation after treatment with BMP4. Indeed, in our panel we do see this trend as well. However, our least BMP4-sensitive culture still has a relatively high expression of SOX2. Therefore, we decided to address other, novel predictive markers for BMP4-therapy response.
We found that OLIG1/2 are potential predictive markers for therapeutic efficacy, irrespective of the genetic background. IDH-mutant gliomas with high OLIG1/2 expression are highly dependent on GSCs and specifically downregulate BMP4 expression.45,46 This dependency on GSCc might also explain the sensitivity to BMP4 we observed in OLIG1/2+ GBM cells. We also discovered that genes involved in ribosomal translation (RPL27A and RPS27) can be used as markers for early-response to BMP4. Ribosome translation signaling is known to become upregulated concomitant with stress response in GBM.47,48 As discussed above, GBM is characterized by a large inter-tumoral heterogeneity. Therefore, neither predictive nor response marker identified in this study will be solely able to predict or explain the differential response, with regard to cell viability in the presence of BMP4. Thus, we recommend to strive at creating further a multi-factorial model, with the least possible variables, to aid clinicians in stratifying their patients instead of expecting to perform one-marker based stratification.
We find that scRNA-seq of patient-derived cultures can improve our understanding of the mechanism of action of novel therapeutic agents in general, as well as provide a start for predictive models at an early stage of therapy selection. It would also help to clarify possible pitfalls and resistance mechanisms, such as the induction of novel cell entities by such treatment, as we demonstrate here with cluster UA1. Furthermore, based on found markers, patients could be stratified alternatively in novel clinical trials, contributing to precision medicine in the long run. For BMP4, markers as OLIG1/2 could be further investigated in clinical setting. Early-response markers can then be used for monitoring effectiveness.
Supplementary Material
Acknowledgement
We thank Bert Eussen, Annelies de Klein, Annick Francis for genetic validation of cultures, Eric Bindels for assistance with 10X-Genomics, Remco Hoogenboezem for pre-processing scRNA-seq data, Lisette Vogelezang and Maurice van der Gaag for 3D-cell-viability assays and Eelke Bos and Clemens Dirven for helpful discussions.
Contributor Information
Iris S C Verploegh, Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands; Department of Neurosurgery, Erasmus University Medical Center, Rotterdam, The Netherlands.
Andrea Conidi, Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Rutger W W Brouwer, Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands; Center for Biomics, Erasmus University Medical Center, Rotterdam, The Netherlands.
Hayri E Balcioglu, Department of Medical Oncology, Erasmus Medical Center Cancer Institute, Rotterdam, The Netherlands.
Samira Makhzami, Laboratory for Molecular Cancer Biology, Center for Cancer Biology, VIB, Leuven, Belgium; Laboratory for Molecular Cancer Biology, Department of Oncology, KU Leuven, Leuven, Belgium.
Anne Korporaal, Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Jean-Christophe Marine, Laboratory for Molecular Cancer Biology, Center for Cancer Biology, VIB, Leuven, Belgium; Laboratory for Molecular Cancer Biology, Department of Oncology, KU Leuven, Leuven, Belgium.
Martine Lamfers, Department of Neurosurgery, Erasmus University Medical Center, Rotterdam, The Netherlands.
Wilfred F J Van IJcken, Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Sieger Leenstra, Department of Neurosurgery, Erasmus University Medical Center, Rotterdam, The Netherlands.
Danny Huylebroeck, Department of Development and Regeneration, KU Leuven, Leuven, Belgium; Department of Cell Biology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Funding
This work was supported by Erasmus MC (extra investment funds of the Cell Biology Dept., to DH) and Fund for Scientific Research-Flanders (FWO-V G.0A3116N; DH).
Competing interests
The authors declare no competing interests.
Authorship Statement
Conception and design: ISCV and AC; data analysis and interpretation: all authors; writing, and/or revision of the manuscript: all authors; study supervision: AC, SL, DH.
References
- 1. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-oncology. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Carlo DT, Cagnazzo F, Benedetto N, Morganti R, Perrini P. Multiple high-grade gliomas: epidemiology, management, and outcome. A systematic review and meta-analysis. Neurosurg Rev. 2019;42(2):263–275. [DOI] [PubMed] [Google Scholar]
- 3. Dirkse A, Golebiewska A, Buder T, et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10(1):1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. [DOI] [PubMed] [Google Scholar]
- 6. Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. [DOI] [PubMed] [Google Scholar]
- 7. Venteicher AS, Tirosh I, Hebert C, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355(6332):eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. [DOI] [PubMed] [Google Scholar]
- 9. Taylor JT, Ellison S, Pandele A, et al. Actinomycin D downregulates Sox2 and improves survival in preclinical models of recurrent glioblastoma. Neuro-oncology. 2020;22(9):1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gamez B, Rodriguez-Carballo E, Ventura F. BMP signaling in telencephalic neural cell specification and maturation. Front Cell Neurosci. 2013;7:87. Published Jun 4, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chalazonitis A, D’Autreaux F, Pham TD, Kessler JA, Gershon MD. Bone morphogenetic proteins regulate enteric gliogenesis by modulating ErbB3 signaling. Dev Biol. 2011;350(1):64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campos B, Wan F, Farhadi M, Ernst A. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer. 2010;16(10):2715–2728. [DOI] [PubMed] [Google Scholar]
- 13. Piccirillo SGM, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. [DOI] [PubMed] [Google Scholar]
- 14. Li Y, Wei X, Wang Q, Li W, Yang T. Inverse screening of Simvastatin kinase targets from glioblastoma druggable kinome. Comput Biol Chem. 2020;86:107243. [DOI] [PubMed] [Google Scholar]
- 15. Mallm JP, Windisch P, Biran A, et al. Glioblastoma initiating cells are sensitive to histone demethylase inhibition due to epigenetic deregulation. Int J Cancer. 2020;146(5):1281–1292. [DOI] [PubMed] [Google Scholar]
- 16. Medarova Z, Pantazopoulos P, Yoo B. Screening of potential miRNA therapeutics for the prevention of multi-drug resistance in cancer cells. Sci Rep. 2020;10(1):1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guishard AF, Yakisich JS, Azad N, Iyer AKV. Translational gap in ongoing clinical trials for glioma. J Clin Neurosci. 2018;47:28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. A Dose Escalation Phase I Study Of Human-Recombinant Bone Morphogenetic Protein 4 Administrated Via CED In GBM Patients. Accessed May 19, 2020. https://ClinicalTrials.gov/show/NCT02869243.
- 19. Seyfrid M, Bobrowski D, Bakhshinyan D, et al. In vitro self-renewal assays for brain tumor stem cells. Methods Mol Biol. 2019;1869:79–84. [DOI] [PubMed] [Google Scholar]
- 20. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. [DOI] [PubMed] [Google Scholar]
- 21. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L, Babikir H, Muller S, et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019;9(12):1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shao X, Liao J, Lu X, et al. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23(3):100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caren H, Stricker SH, Bulstrode H, et al. Glioblastoma stem cells respond to differentiation cues but fail to undergo commitment and terminal cell-cycle arrest. Stem Cell Rep. 2015;5(5):829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hover LD, Owens P, Munden AL, et al. Bone morphogenetic protein signaling promotes tumorigenesis in a murine model of high-grade glioma. Neuro-oncology. 2016;18(7):928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu S, Yin F, Zhao M, et al. The homing and inhibiting effects of hNSCs-BMP4 on human glioma stem cells. Oncotarget. 2016;7(14):17920–17931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Z, Sun L, Wang Y, et al. Bone morphogenetic protein 4 inhibits cell proliferation and induces apoptosis in glioma stem cells. Cancer Biother Radiopharm. 2011;26(1):77–83. [DOI] [PubMed] [Google Scholar]
- 28. Bhaduri A, Di Lullo E, Jung D, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26(1):48–63.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neubauer E, Wirtz RM, Kaemmerer D, et al. Comparative evaluation of three proliferation markers, Ki-67, TOP2A, and RacGAP1, in bronchopulmonary neuroendocrine neoplasms: Issues and prospects. Oncotarget. 2016;7(27):41959–41973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christner PJ, Ayitey S. Extracellular matrix containing mutated fibrillin-1 (Fbn1) down regulates Col1a1, Col1a2, Col3a1, Col5a1, and Col5a2 mRNA levels in Tsk/+ and Tsk/Tsk embryonic fibroblasts. Amino Acids. 2006;30(4):445–451. [DOI] [PubMed] [Google Scholar]
- 32. Garofano L, Migliozzi S, Oh YT, et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat Cancer. 2021;2(2):141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeffrey PL, Capes-Davis A, Dunn JM, et al. CROC-4: a novel brain specific transcriptional activator of c-fos expressed from proliferation through to maturation of multiple neuronal cell types. Mol Cell Neurosci. 2000;16(3):185–196. [DOI] [PubMed] [Google Scholar]
- 34. Yao Z, Mich JK, Ku S, et al. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell. 2017;20(1):120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704(1-3):12–20. [DOI] [PubMed] [Google Scholar]
- 36. Darmanis S, Gallant CJ, Marinescu VD, et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 2016;14(2):380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pistollato F, Chen HL, Rood BR, et al. Hypoxia and HIF1alpha repress the differentiative effects of BMPs in high-grade glioma. Stem Cells. 2009;27(1):7–17. [DOI] [PubMed] [Google Scholar]
- 38. Li L, Chen K, Wu Y, et al. Gadd45a opens up the promoter regions of miR-295 facilitating pluripotency induction. Cell Death Dis. 2017;8(10):e3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sachdeva R, Wu M, Johnson K, et al. BMP signaling mediates glioma stem cell quiescence and confers treatment resistance in glioblastoma. Sci Rep. 2019;9(1):14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis JB, Krishna SS, Abi Jomaa R, et al. A new model isolates glioblastoma clonal interactions and reveals unexpected modes for regulating motility, proliferation, and drug resistance. Sci Rep. 2019;9(1):17380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Han SS, Wu Y, et al. CD44 expression identifies astrocyte-restricted precursor cells. Dev Biol. 2004;276(1):31–46. [DOI] [PubMed] [Google Scholar]
- 42. Steiner J, Bernstein HG, Bielau H, et al. Evidence for a wide extra-astrocytic distribution of S100B in human brain. BMC Neurosci. 2007;8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Videla Richardson GA, Garcia CP, Roisman A, et al. Specific preferences in lineage choice and phenotypic plasticity of glioma stem cells under BMP4 and noggin influence. Brain Pathol. 2016;26(1):43– 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dalmo E, Johansson P, Niklasson M, et al. Growth inhibitory activity of bone morphogenetic protein 4 in human glioblastoma cell lines is heterogeneous and dependent on reduced SOX2 expression. Mol Cancer Res. 2020;18(7):981–991. [DOI] [PubMed] [Google Scholar]
- 45. Tirosh I, Venteicher AS, Hebert C, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Augustus M, Pineau D, Aimond F, et al. Identification of CRYAB(+) KCNN3(+) SOX9(+) astrocyte-like and EGFR(+) PDGFRA(+) OLIG1(+) Oligodendrocyte-like tumoral cells in diffuse IDH1-mutant gliomas and implication of NOTCH1 signalling in their genesis. Cancers. 2021;13(9):2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen HY, Lin LT, Wang ML, et al. Musashi-1 promotes chemoresistant granule formation by PKR/eIF2alpha signalling cascade in refractory glioblastoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt A):1850–1861. [DOI] [PubMed] [Google Scholar]
- 48. Klacanova K, Pilchova I, Klikova K, Racay P. Short chemical ischemia triggers phosphorylation of eIF2alpha and death of SH-SY5Y cells but not proteasome stress and heat shock protein response in both SH-SY5Y and T98G cells. J Mol Neurosci. 2016;58(4):497–506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq files were deposited with NCBI-GEO (accession number GSE148196).