Abstract
Background
IDH-mutant gliomas are separate based on the codeletion of the chromosomal arms 1p and 19q into oligodendrogliomas IDH-mutant 1p/19q-codeleted and astrocytomas IDH-mutant. While nuclear loss of ATRX expression excludes 1p/19q codeletion, its limited sensitivity prohibits to conclude on 1p/19q status in tumors with retained nuclear ATRX expression.
Methods
Employing mass spectrometry based proteomic analysis in a discovery series containing 35 fresh frozen and 72 formalin fixed and paraffin embedded tumors with established IDH and 1p/19q status, potential biomarkers were discovered. Subsequent validation immunohistochemistry was conducted on two independent series (together 77 oligodendrogliomas IDH-mutant 1p/19q-codeleted and 92 astrocytomas IDH-mutant).
Results
We detected highly specific protein patterns distinguishing oligodendroglioma and astrocytoma. In these patterns, high HIP1R and low vimentin levels were observed in oligodendroglioma while low HIP1R and high vimentin levels occurred in astrocytoma. Immunohistochemistry for HIP1R and vimentin expression in 35 cases from the FFPE discovery series confirmed these findings. Blinded evaluation of the validation cohorts predicted the 1p/19q status with a positive and negative predictive value as well as an accuracy of 100% in the first cohort and with a positive predictive value of 83%; negative predictive value of 100% and an accuracy of 92% in the second cohort. Nuclear ATRX loss as marker for astrocytoma increased the sensitivity to 96% and the specificity to 100%.
Conclusions
We demonstrate that immunohistochemistry for HIP1R, vimentin, and ATRX predict 1p/19q status with 100% specificity and 95% sensitivity and therefore, constitutes a simple and inexpensive approach to the classification of IDH-mutant glioma.
Keywords: ATRX, HIP1R, IDH, proteomics, Vimentin, 1p/19q
Key Points.
Inverse proteomic abundances of HIP1R and vimentin strongly associate with 1p/19q status.
HIP1R/vimentin staining strongly associates with 1p/19q status of IDH mutant gliomas.
HIP1R/vimentin/ATRX immunohistochemistry minimizes the need for 1p/19q analyses.
Importance of the Study.
Assessment of the chromosomal 1p/19q status is mandatory to distinguish between IDH mutant astrocytoma and oligodendroglioma. Genetic analyses need to be carried out, which are expensive, time consuming, and not readily available everywhere. Immunohistochemical loss of ATRX is currently the only established surrogate marker for a non-1p/19q-codeleted genotype. We identified inverse abundances of HIP1R and vimentin as candidate biomarkers using mass spectrometry. Translation of this pattern to immunohistochemistry was feasible and in an independent validation cohort 1p/19 status could be correctly assigned in more than 90% of all cases. Including immunohistochemistry for ATRX increased the sensitivity to 95%. Performance of the HIP1R/vimentin/ATRX approach could be confirmed in a second independent validation cohort analyzed in another laboratory. Therefore, immunohistochemistry for HIP1R, VIM, and ATRX has a great potential to minimize the “not otherwise specified” diagnoses in IDH-mutant gliomas for which genetic analyses are not available.
Since 2016 WHO requires molecular testing for the diagnoses of oligodendroglioma IDH-mutant and 1p/19q-codeleted and astrocytoma IDH-mutant.1 IDH and 1p/19q status is diagnostic, prognostic, and predictive and, therefore, essential for postsurgical treatment.2
In the overwhelming majority of cases IDH status can be determined by mutation specific antibodies due to the very high prevalence of the IDH1-R132H mutation. Only approximately 10% of tumors require sequencing due to other rare IDH1 mutations or due to infrequent IDH2 mutations.3,4 The 1p/19q status requires more laborious approaches and relies either on FISH analyses, MLPA analysis, or copy number profiling based on array data.5 Therefore, surrogate markers for determining 1p/19q are highly requested. To this end, two genes associated with telomere modelling have been employed so far. Loss of nuclear ATRX expression is highly associated with astrocytoma, IDH-mutant.6 While this can be readily determined by immunohistochemistry, many of these tumors do not exhibit this feature resulting in a reduced sensitivity of this highly specific marker for astrocytoma, IDH-mutant. On the other hand, TERT exhibits quite specific promoter mutations in IDH-mutant oligodendroglioma but its determination requires a DNA sequencing approach not ubiquitously available.7
With this project we first set out to determine mass spectrometry (MS)-based protein patterns for oligodendroglioma IDH-mutant and 1p/19q-codeleted and for astrocytoma IDH-mutant. In a second step, we explored the possibility of re-translating differences in these expression patterns to simple immunohistochemical tests for the determination of 1p/19q status.
Materials and Methods
Tumor Series
Tumor tissues were obtained from the archives of the Departments of Neuropathology and Neurosurgery at the Heidelberg University Hospital and the Biobank of the University Cancer Center (UCT), Institute of Neurology, (Edinger Institute) at the University Hospital Frankfurt Am Main. Tissue and data collection were performed in consideration of local ethics regulations and approval. Inclusion criterium was availability of an Illumina Infinium HumanMethylation450 (450k) or Infinium Methylation EPIC (850k) BeadChip-based copy number profile determined in previous studies.6,8 An overview of the cohorts is provided in Supplementary Table S1 (sample overview). All tumors of the discovery cohort were evaluated and marked for tissue extraction by a neuropathologist (DER) using HE-stained sections. The tumors from 12 patients were represented in FF and FFPE discovery cohorts. Tumors from all cohorts were reviewed by neuropathologists and integrated diagnoses were given according to the WHO 2016 criteria.
Sample Preparation of FF and FFPE Tissue for Mass Spectrometry
Starting from FF tissue we employed protocols including pressure cycling technology for efficient tissue lysis and rapid protein digestion.9,10 Minimum tumor cell content of 70% was ensured on frozen sections by microscopy followed by processing 2 mm3 tumor tissue. Details are provided in Supplementary Methods.
Starting with FFPE tissue suitable regions on the tissue blocks where punched with 1.5 mm disposable biopsy punches. The tumor core was transferred into a bead tube (Bertin Technologies SAS, Montigny Le Bretonneux, France) and further processed. Details are provided in Supplementary Methods.
LC-MS/MS Acquisition by DDA-PASEF Mode
An Easy nLC 1200 (Thermo Fischer Scientific) system was coupled online to timsTOF pro (Bruker Daltonics, Bremen, Germany). Peptides (equivalent to 500 ng) were loaded onto a 50 cm reversed-phase column with 75 μm inner diameter. Chromatography was performed at 60°C with a flow of 400 nL/min on a binary buffer system. Mobile phases A (0.1% formic acid (v/v)) and B (80/20/0.1% ACN/water/formic acid (v/v/vol)) were used for low pH peptide separation. Peptides were separated in a linear gradient from 7.5% to 55% solvent B within 100 min.
The timsTOF pro was operated in data dependent parallel accumulation-serial fragmentation (DDA-PASEF) mode. For a single TIMS-MS survey scan, 10 PASEF MS/MS scans were acquired per topN acquisition cycle. Ion accumulation and ramp time was set to 50 ms each and ion mobility range from 1/K0 = 1.6 to 0.6 Vs cm−2 was analyzed. Mass range was set from m/z 100 to 1700. Details are provided in Supplementary Methods.
Peptide Identification and Data Analysis
The raw data were analyzed in the MaxQuant environment (version 1.6.17.0) using default settings.11 The data were searched against the human Uniprot database (August 2018 release). Match between runs was enabled with default settings. Label-free quantification was performed with a minimum ratio count of one. Postprocessing of MaxQuant data output and statistical analysis were carried out by R (version 4.0.4). Proteins identified in more than 90% of all oligodendroglioma or astrocytoma in the fresh frozen or FFPE cohort were selected for further analysis. The R-package “DEP” was used for normalization (variance stabilizing normalization), missing value imputation (downshift imputation, shift = 1.5, scale = 0.5) and for downstream differential expression analysis. Differential expression was examined by the R-package “limma” and Benjamini–Hochberg was used for FDR controlling of the resulting p-values. TCGA data was obtained through the R-Package curatedTCGAdata.12 mRNA expression values of VIM and HIP1R were log2 transformed and visualized in boxplots according to 1p/19q codeletion status.
Immunohistochemistry Heidelberg
Immunohistochemistry was conducted on 5-µm-thick formalin-fixed, paraffin-embedded (FFPE) tissue sections mounted on StarFrost Advanced Adhesive slides (Engelbrecht, Kassel, Germany). Immunohistochemistry was performed on a BenchMark Ultra immunostainer (Ventana Medical Systems, Tucson, AZ, USA). Sections were stained with anti-IDH1-R132H antibody H09 (Dianova, Hamburg, Germany) as previously described.4 For ATRX (Clone BSB-108, Medac, Wedel, Germany, 1:2000), HIP1R (ab140608, Abcam, Cambridge, UK, 1:200), and VIM (GA63061-2, Dako, Jena, Germany, 1:300) staining detailed protocols are provided in Supplementary Methods.
Immunohistochemistry Frankfurt
By use of a microtome (Leica SM 2000R, Wetzlar, Germany) 4 µm thin tissue sections were cut and mounted on slides (Superfrost Plus, Thermo Scientific, Braunschweig, Germany) for immunohistochemical stainings. Immunohistochemical stainings against the antigens Vimentin (dilution 1:300) and HIP1R (dilution 1:200) were performed using established protocols for the LEICA BOND-III automated stainer (Leica, Wetzlar, Germany). Tissue sections were evaluated by two board certified neuropathologists (P.N.H., K.F.).
Results
Proteomes from FF and FFPE Samples are Comparable
In FF tissue, label free proteome quantification using DDA-PASEF mode13 with a 2 h gradient (single shot) was performed. This allowed peptide identifications averaging 26 830 (±2586) unique peptides corresponding to 5171 (±322) proteins per sample by MS/MS. In total we identified 8220 proteins and 82 885 unique peptides in the FF cohort at an FDR of 1% on the peptide and protein level.
In FFPE tissue, we identified 16 681 (±3549) peptides per sample allotted to 3905 (±493) proteins resulting in a total of 72 867 unique peptides and 7912 proteins for the cohort. Pearson-correlations from the 12 matched pairs of FF and FFPE tissues from the same tumor ranged from 0.7 to 0.87 (Figure 1B, C).
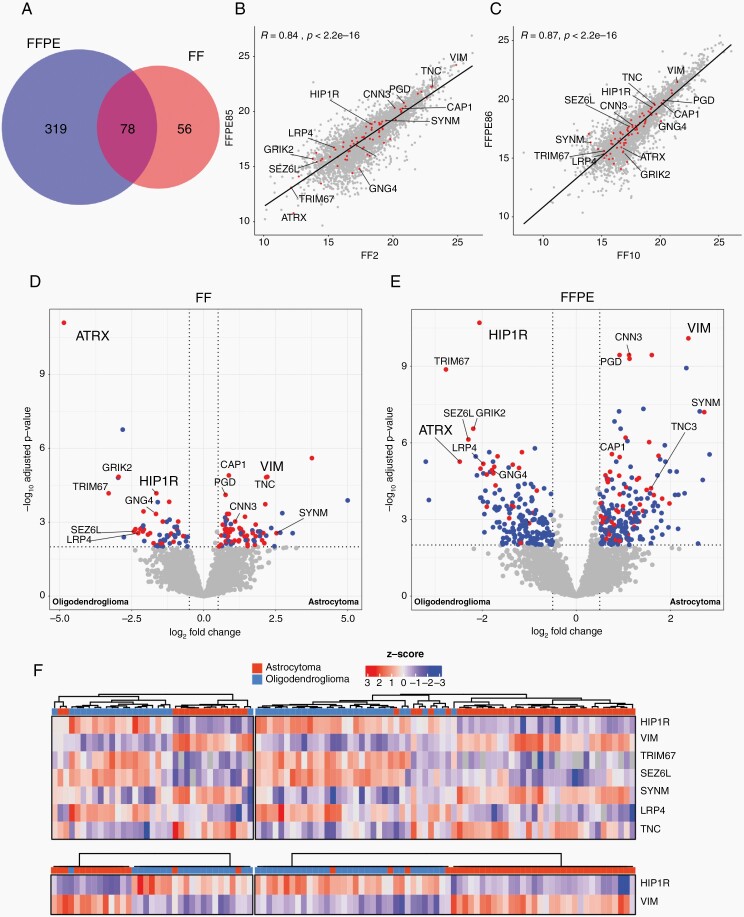
Fig. 1.
Differential protein abundance analysis of astrocytoma and oligodendroglioma. (A) Venn diagramm depicting number of significantly DAP in FF and FFPE tissue. Overlap shows the number of proteins which were significantly different in both cohorts. Pearson correlation analyses between a matched pair of FF and FFPE of an astrocytoma (B) and an oligodendroglioma (C). Volcano plot of significantly differentially expressed proteins (DAP) between astrocytoma and oligodendroglioma in FF (D) and FFPE (E) tissue. Blue dots represent proteins which were found either in the FF or FFPE tissues exclusively, red dots represent proteins which were found consistently in both cohorts. Dashed lines represent threshold for significance (P-value <.01) and log2 fold change (1,5). (F): Hierarchical clustering of the FF (left) and FFPE (right) cohort using a subset of DAP (top) and HIP1R and VIM (bottom) between astrocytoma and oligodendroglioma.
Differentially Abundant Proteins for Astrocytoma and Oligodendroglioma
To identify significantly differentially abundant proteins (DAP), astrocytoma and oligodendroglioma MS data were compared for each tissue cohort. From patients with paired FF and FFPE samples only FF samples were analysed to prevent statistical inflation. Overall, we found 134 DAPs in the FF cohort and 397 in the FFPE cohort (Supplementary Table S1, online resource). 78 DAPs were found in both cohorts (Figure 1A and Supplementary Table S1).
ATRX was one of the most significantly differentially abundant proteins between astrocytoma and oligodendroglioma (Figure 1D, E). Additionally, highly significantly upregulated in oligodendroglioma in both the FF and the FFPE cohorts were “Huntingtin Interacting Protein 1 Related” (HIP1R), “Tripartite Motif Containing 67” (TRIM67), “Seizure Related 6 Homolog Like” (SEZ6L), and “low density lipoprotein receptor-related protein 4” (LRP4). Most significantly upregulated proteins in astrocytoma not associated with 1p/19q localisation of their coding genes were vimentin (VIM), synemin (SYNM), and tenascin C (TNC).
HIP1R and VIM have an Inverse Abundance Pattern in Astrocytoma and Oligodendroglioma
Unsupervised hierarchical clustering of all samples using the protein abundances of just these top differentially abundant proteins (HIP1R, TRIM67, SEZ6L, LRP4, VIM, SYNM, TNC) excluding ATRX as already established marker, resulted in a good separation of oligodendrogliomas and astrocytomas (Figure 1F, top). The abundance patterns of HIP1R, TRIM67, SEZ6L, and LRP4 on the one side and VIM, SYNM, and TNC on the other side were largely overlapping. We evaluated antibodies for immunohistochemical detection of these proteins. However, for several proteins no suitable antibodies were available (TRIM67), no satisfactory immunohistochemical stains could be obtained or several proteins provided largely redundant information (e.g. VIM and TNC). HIP1R and VIM were chosen for detailed analyses because these provided the most consistent stains and clustering analyses using only abundances of HIP1R and VIM showed a good separation of oligodendrogliomas and astrocytomas (Figure 1F, bottom). Remarkably, samples displaying inverse abundances of HIP1R and VIM showed a strong tumor type association. In fact, 37/38 tumors with a high abundance of HIP1R and a low abundance of VIM were oligodendrogliomas and 38/39 tumors with high abundance of VIM and a low abundance of HIP1R were astrocytomas. A small group of tumors without reverse abundances of VIM and HIP1R consisted of both astrocytomas and oligodendrogliomas. An investigation of mRNA levels using independent data from the TCGA database12 confirmed an inverse expression pattern of HIP1R and VIM in IDH-mutant glioma with and without a 1p/19q codeletion (Figure 2). This suggested that the presence of an inverse abundance pattern of these proteins could be useful as a biomarker.
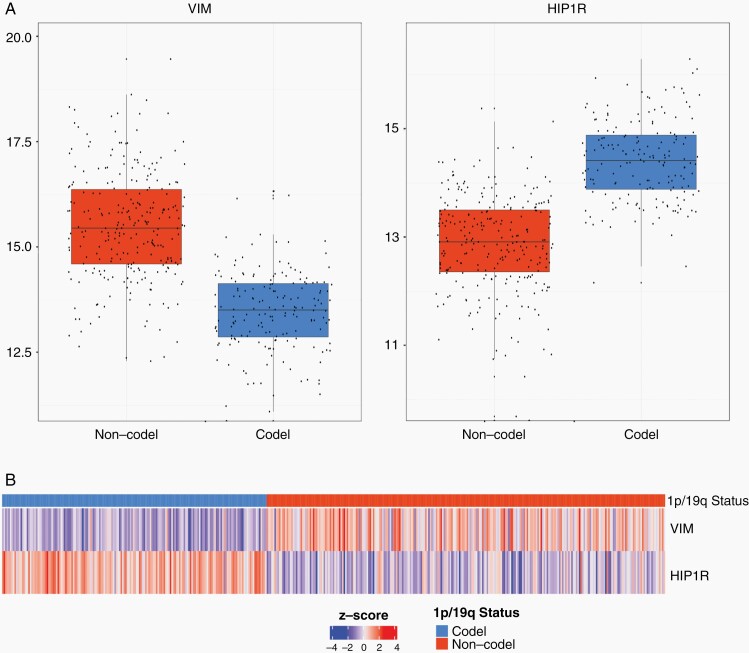
Fig. 2.
mRNA expression levels of IDH-mutant glioma. Top: Boxplots log2 normalized expression values of VIM (left) and HIP1R (right) originating from the TCGA database. High levels of VIM coding mRNA in IDH-mutant glioma without 1p/19 codeletion and low levels of VIM in glioma with 1p/19q codeletion are observed. Vice versa high levels of HIP1R coding mR NA are found in 1p/19q codeleted glioma and low levels of HIP1R in glioma without 1p/19 codeletion. Bottom: Heatmap showing Z-scored expression values, samples were ordered by their 1p/19q status. Pairwise comparison of z-scored expression values demonstrates that 1p/19q codeleted glioma exhibiting low levels of VIM and high levels of HIP1R. In contrast glioma without 1p/19q codeletion exhibit high levels of VIM and lower levels of HIP1R.
Immunohistochemical Expression Patterns of HIP1R and VIM
Based on staining tumors of the discovery cohort we defined four immunohistochemical expression levels for each, HIP1R (H0-3) and VIM (V0-3). HIP1R showed cytoplasmic as well as membranous positivity. Score H0 was given if tumor cells did not bind antibody or only a very few cells were weakly labelled (H0, <10% are weakly positive). Score H1 represents cases where many tumor cells were weakly positive and/or few cells were moderately positive (H1, 10%–75% are positive with less than 30% of tumor cells being strongly positive). Tumors of score H2 showed >50% positive tumor cells with >30% being strongly positive which however were not closely packed and showed space in between. Alternatively, H2 score was also given if just moderately positive tumor cells were closely packed forming diffusely positive areas (~100% are moderately positive). Score H3 tumors showed closely packed, strong, and diffusely positive areas of tumor cells with only vessels being negative (~100% are strongly positive). Representative examples and corresponding scores for HIP1R are given in Supplementary Figure S1. IDH1-R132H-mutant oligodendrogliomas exhibited binding of HIP1R and IDH1-R132H antibodies in identical distribution patterns, suggesting that HIP1R-expression was restricted to tumor cells only (Supplementary Figure S2). Of note, the tumor cell specificity of HIP1R clearly depended on the antibody titration, since higher antibody concentrations resulted in staining of nonneoplastic cells like endothelial cells (Supplementary Figure S3).
In contrast, VIM was present in a broad range of cell types. VIM antibodies invariably bound to vessels making them suitable internal controls. VIM was also positive in reactive astrocytes (IDH1-R132H negative) and leptomeningeal structures. For evaluation of VIM expression immunoreactivity within the tumor was scored, however, vascular signals were excluded. Separation of tumor derived or reaction derived VIM expression appeared not to be feasible.
Score V0 was given if <10% of tumor cells were labelled and only vessels or other nonneoplastic cells were positive. Score V1 represents tumors where 10%–20% of (tumor) cells and several processes were labelled. In score V2 about 20%–70% of (tumor) cells were labelled including many processes but a fraction of ≥30% of tumor cells is negative. Sometimes, a prominent star-like pattern of positive cells was present. Score V3 tumors showed moderate or strong expression in >70% of the tumor cells resulting in diffusely positive tumor tissue. Typical examples for VIM expression are provided in Supplementary Figure S4. In oligodendrogliomas with typical morphology, VIM associated with mini-gemistocytes and sparse fibrillary processes which could not be assigned to a specific cell body.
Tumor Type-specific HIP1R/VIM Expression Patterns in the Discovery Series
Two independent observers evaluated the expression of HIP1R and VIM expression patterns in the discovery series. In general, oligodendrogliomas showed moderate or high abundance levels of HIP1R (H2, H3) and absent (V0) or low (V1) levels of VIM expression. In contrast astrocytomas commonly showed absent (H0) or low (H1) levels of HIP1R but moderate (V2) to high (V3) levels of VIM (Figure 3). Staining patterns for HIP1R and VIM in most cases were homogenous, we also encountered intratumoral heterogeneity: In some samples we observed focal high HIP1R coupled with low VIM expression contrasted by low HIP1R and high VIM expression in other regions (Supplementary Figure S5). Restricting to tumors with heterogeneous pattern and reaching an H2 score in any area, this phenomenon was observed only in oligodendroglioma 1p/19q-codeleted. Frequently high VIM expression could be attributed to infiltrated CNS tissue demonstrated by lack of IDH1-R132H binding in infiltrated and reactive cells (Supplementary Figure S6).
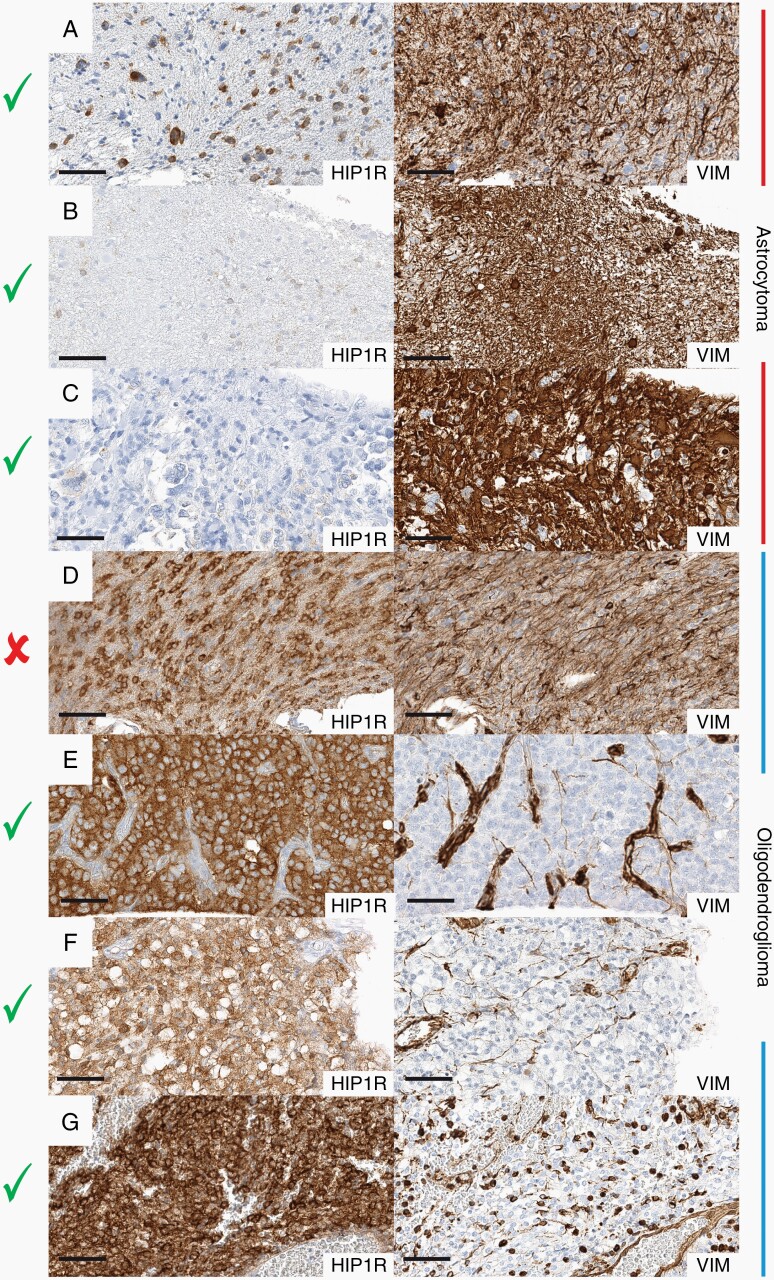
Fig. 3.
Examples of HIP1R/VIM immunohistochemistry for the classification of IDH-mutant glioma. Pairwise comparison of HIP1R (left side) and VIM (right side) in astrocytomas (top 3) and oligodendrogliomas (bottom 3). (A–C) Astrocytomas with low-level HIP1R expression and high-level VIM expression. (D) A nondeterminable oligodendroglioma with similar HIP1R and VIM staining intensities. (E–G) Oligodendrogliomas with high-level HIP1R expression and low-level VIM expression. Check marks indicate determinable cases, cross indicates not-determinability. Black line indicates a length of 60 µm.
Following a two-tiered approach, all cases could be systematically evaluated: First, identifying cases with homogenous staining pattern and clear dominance of either HIP1R or VIM expression. These represent the majority of all cases which could be immediately attributed to oligodendroglioma and astrocytoma accordingly. Second, identifying cases with heterogeneous staining pattern or ambiguous expression of both markers. In cases with heterogeneous staining patterns special attention was given to areas with strongest HIP1R expression. If HIP1R expression in these areas was stronger than VIM expression, these tumors were scored oligodendroglioma. This means that focal presence of a typical oligodendroglioma HIP1R-high/VIM-low expression pattern was sufficient for classification as oligodendroglioma irrespective of other tumor areas. For example, even tumors with numerous VIM-positive mini-gemistocytes being present focally, could be assigned as oligodendrogliomas because other areas of the same tumor presented with much less VIM expression.
In the minority of cases where the scoring results were more ambiguous, direct comparison of the HIP1R and VIM immunoreactivity at low magnification (10–20 fold) often still revealed a predominant stain and the tumor was classified accordingly (Supplementary Figure S7). By convention in those cases the VIM score was adjusted to match the impression of direct comparison, for example an initial H2/V2 score was adjusted to H2/V1 if HIP1R predominated and to H2/V3 if VIM predominated in direct comparison. Predominance of VIM intensity prompted an astrocytoma score, as long as no low-level VIM expression was present in any area of the tumor. In cases with equal intensities no decision was made (Figure 3, middle row). Due to the reciprocal behaviour of both markers, the combination of very low or very high expression in both markers, both of which are nondiagnostic, was not or only very rarely observed. A graphical algorithm is provided in Figure 4.
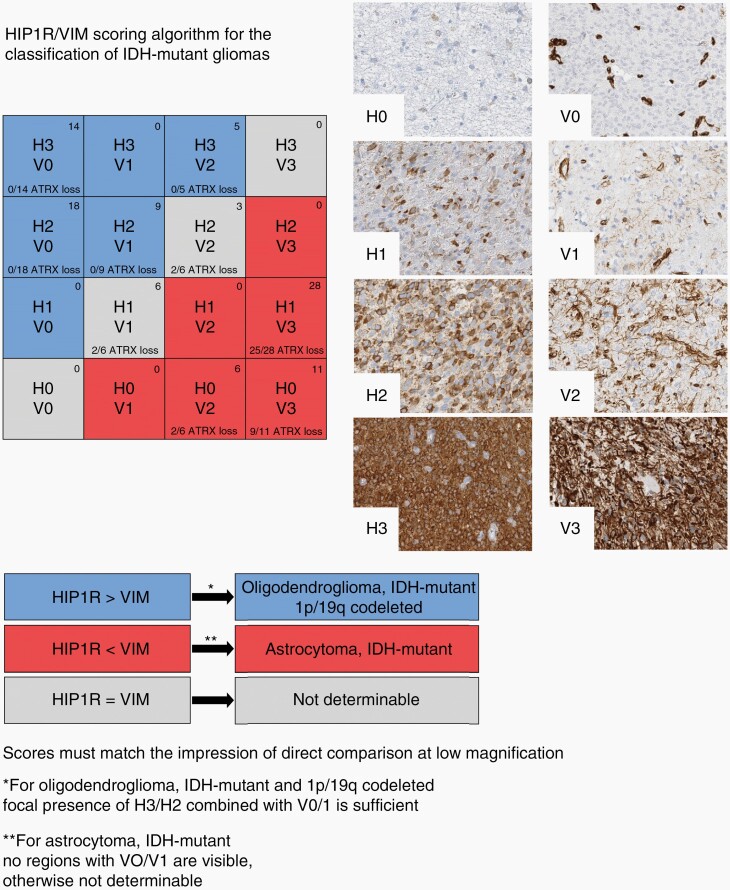
Fig. 4.
Graphical algorithm for the classification of IDH-mutant glioma according to HIP1R/VIM immunohistochemistry. The HIP1R (H) level should be evaluated first. If the H score is higher than the VIM (V) score the tumor is identified as oligodendroglioma. Note that focal presence of a moderate to strong HIP1R level (H3/H2) in combination with low-level VIM (V0/V1) in the same area is sufficient for making this conclusion. If the VIM score is higher than the HIP1R score the tumor is identified as astrocytoma, as long as there are no areas in the tumor with sparse VIM expression (V0, V1) as well. If the HIP1R and the VIM levels are ambigous, the stains should be compared directly at low magnification. If HIP1R is clearly stronger, the tumor is identified as oligodendroglioma. If VIM is clearly stronger, the tumor is identified as astrocytoma. Similar expression levels of HIP1R and VIM in the tumor are interpreted as not determinable. The distribution of HIP1R and VIM expression from the HD validation cohort (n = 100; observer 1) is shown in the top right corner and the fraction of samples with ATRX loss is noted on the bottom of every combination box.
HIP1R Expression Patterns in Oligodendroglioma-mimics
In the light of the strong diffuse HIP1R positivity in oligodendroglioma including those with a classic “honeycomb”-morphology we wondered whether histological mimics of IDH-wt tumors show a similar staining pattern. Remarkably, none of 10 IDH-wt tumors with oligoid features (4 GBM-O, 2 clear cell ependymomas, 2 DLGNT, 1 PLYNT) analysed, showed the typical oligodendroglioma HIP1R expression pattern. IDH-wt tumors were either negative for HIP1R or showed a positivity in which the perinuclear halos of tumor cells were completely or partly spared from HIP1R immunopositivity contrasting oligodendrogliomas in which the perinuclear halos displayed a strong diffuse cytoplasmic staining of HIP1R (Supplementary Figure S8).
Validation of HIP1R/VIM/ATRX Immunohistochemistry as Surrogate for 1p/19q Status in IDH-Mutant Glioma
Two independent neuropathologists scored HIP1R and VIM immunohistochemistry in an independent series of 50 oligodendrogliomas and astrocytomas (HD validation cohort) each blinded for the histological diagnosis, the 1p/19q- and ATRX-status. Based on the established algorithm (Figure 4) observer 1 called 46/50 (sensitivity 92%) oligodendrogliomas and 45/50 (90%) astrocytomas with 100% specificity. 9 cases remained undetermined. Observer 2 identified 47/50 (sensitivity 94%) as oligodendroglioma and 47/50 (sensitivity 94%) tumors as astrocytoma with 100% specificity while 6 cases remained undetermined (positive and negative predictive value as well as accuracy 100%). The interobserver concordance was high with 91/100 of the calls being identical. Observer 1 called 3 tumors correctly which were undeterminable for observer 2 and observer 2 called 6 tumors correctly which were undeterminable for observer 1. As all calls were correct the interobserver variance occurred only between correct entity calls and calls as nondeterminable (Supplementary Table S2, online resource). Excluding ATRX immunohistochemistry from the analysis proved the HIP1R/VIM based algorithm to be very powerful on its own. By including loss of ATRX as marker for astrocytomas observer 1 identified 4 further cases as astrocytoma and observer 2 identified 2 further cases as astrocytoma. The sensitivity of a HIP1R/VIM/ATRX triple-marker approach was 98% for astrocytomas and 95%–96% for the whole series, all with 100% specificity.
Similar to the discovery set, the validation set demonstrated the reciprocal intensity of staining patterns of HIP1R and VIM in oligodendroglioma and astrocytoma. Combinations with low scores on both sides exhibiting H0/V0, H0/V1, H1/V0, and H1/V1 scores occurred in only 3 patients and the combination with high scores H3/V3 was not encountered at all. The distribution of HIP1R and VIM expression in the HD validation series is given in Supplemental Table S2.
Finally, to evaluate the reproducibility of our approach in a different laboratory, a second independent cohort was analysed by the neuropathology Frankfurt (Frankfurt validation cohort). Immunohistochemistry for HIP1R and VIM was established according to the Heidelberg protocol using the identical antibodies at the Frankfurt site. Two neuropathologists not involved in the analyses of HD cohorts, scored an independent series of 69 IDH-mutant gliomas (42 astrocytomas, 27 oligodendrogliomas). Correctly called were 26/27 (sensitivity 96%) oligodendrogliomas and 32/42 (sensitivity 76%) astrocytomas. 5 astrocytomas were falsely called as oligodendrogliomas and 6 cases remained undetermined (positive predictive value for 1p/19q codeletion 83%; negative predictive value 100%; accuracy: 92%). Loss of ATRX correctly revealed 35 cases as astrocytomas (83% sensitivity; 100% specificity) including 3 previously nondeterminable cases and the 5 falsely called tumors. Considering loss of ATRX as the superior astrocytoma marker the triple ATRX/HIP1R/VIM approach reached a sensitivity of 96% and a specificity of 100% in the Frankfurt cohort (Supplementary Table 2).
Discussion
Improvement of diagnostic approaches aims at high sensitivity, high specificity, and simplicity. The latter may require complex intermediate steps ultimately leading to straight forward procedures.
Proteomic Profiling of IDH-mutant Gliomas
LC-MS/MS identified protein patterns highly specific for astrocytoma, IDH-mutant, and oligodendroglioma, IDH-mutant, and 1p/19q-codeleted. Pearson correlation showed high concordance of results from FF and FFPE tissues demonstrating reproducibility and suitability of FFPE material for LC-MS/MS analyses. Higher DAP count in the FFPE material was attributed to the larger size of the FFPE cohort. High expression differences and ability to separate the entities by unsupervised clustering prompted focus on three of the 78 DAP.
DAP as Candidates for Immunohistochemistry
ATRX is well established for its loss of expression in IDH-mutant astrocytoma. In both FFPE and FF based analyses, ATRX indeed emerged as highly differentially abundant with low protein abundances in astrocytomas (Figure 1D, E).
VIM is an intermediate filament commonly expressed in astrocytic brain tumors.14,15 VIM has already been found in earlier proteomic studies to be downregulated in oligodendroglioma.16 Of note, most of these studies fall in the pre-IDH era. A recent study revisited VIM expression patterns with molecularly defined tumors and suggested it as a biomarker for distinguishing oligodendroglioma from astrocytoma.17
HIP1R was identified as a relative of huntingtin-interacting protein 1 (HIP1).18 HIP1 is abundantly present in several different tumor types and has transforming activity in vitro.19 While this is not reported for HIP1R there seems to be at least some functional overlap with HIP1, both being involved in receptor tyrosine kinase stabilisation and endocytosis.20 Consistent with our results, HIP1R was recently found downregulated in astrocytomas compared to oligodendrogliomas on the mRNA level in a gene expression meta-analysis of histology-defined low-grade gliomas.21
Potential of ATRX, HIP1R, and VIM Immunohistochemistry for Predicting 1p/19q Status
Nuclear loss of ATRX expression is observed in 87%–95% of supratentorial IDH mutant astrocytomas and is a strong indicator for 1p/19q wildtype status.22,23 Thus, for WHO conforming diagnosis determination of 1p/19q status is required for about 10% of astrocytomas and for all oligodendrogliomas.
Combined ATRX, HIP1R, and VIM immunohistochemistry in the discovery and two validation series allowed identification of astrocytomas IDH-mutant and oligodendrogliomas IDH-mutant and 1p/19-codeleted with high sensitivity and perfect specificity. Codeletion of 1p/19q was not seen in the astrocytomas but present in all oligodendrogliomas. Thus, the combination of ATRX, HIP1R, and VIM immunohistochemistry could clearly predict 1p/19q status.
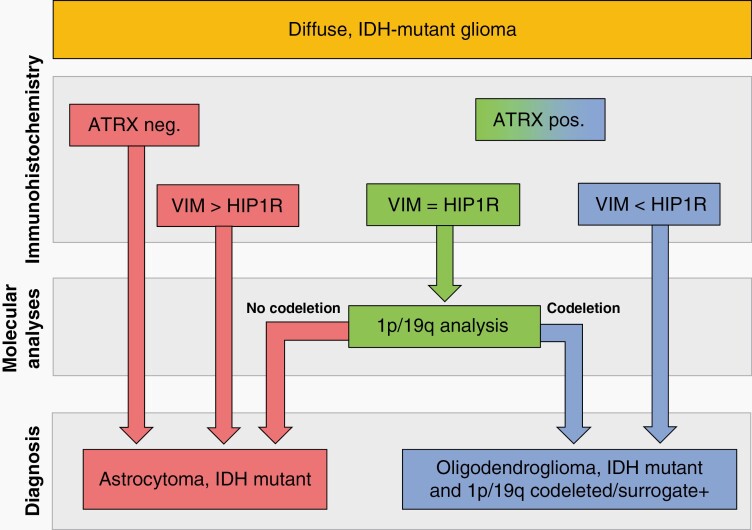
HIP1R/VIM and ATRX complemented each other. From collectively 94 cases with retained or undeterminable ATRX analyzed in this study, 86 (91%) could be correctly classified by HIP1R/VIM. The Frankfurt cohort revealed a small risk of misclassifying astrocytomas as oligodendrogliomas by HIP1R/VIM alone. All of these were high-grade tumors, for which the Frankfurt cohort was strongly enriched. Importantly however, combined evaluation with ATRX minimizes the risk for misinterpretation. A rational approach for the usage of these markers for the classification of IDH-mutant gliomas will therefore start with ATRX. In case of a nuclear ATRX loss the case can be classified as astrocytoma. In case of a retained of unevaluable ATRX status, HIP1R/VIM will allow classification of 90% of the remaining cases either as oligodendroglioma or astrocytoma (Figure 5).
Fig. 5.
Flowchart for the efficient use of ATRX and HIP1R/VIM for the classification of IDH-mutant gliomas. Tumors with nuclear loss of ATRX can be classified as astrocytoma. Tumors with retained or nondeterminable ATRX status are evaluated with HIP1R/VIM immunohistochemistry. VIM > HIP1R tumors are classified as astrocytomas, VIM < HIP1R are classified as oligodendrogliomas. VIM = HIP1R tumors need molecular 1p/19q testing for classification.
Current standard approaches to 1p/19q status determination include FISH, MLPA, NGS-sequencing, and array-based analyses. While both, NGS sequencing and array approaches are highly suitable, this technology is not widely available for diagnostic routine. Most frequently employed is FISH analysis limited for its inability to differentiate between segmental deletions and whole arm deletions.5 The estimated false-positive FISH 1p/19q codeletion rate is reported to be 3.6%.24
Other Surrogate Markers for 1p/19q Codeletion Status
Strong and diffuse staining of p53 has been suggested by cIMPACT -NOW 2 to be used in a similar fashion as ATRX loss and as sufficient to exclude 1p/19q deletion.25 According to WHO 2021, p53 accumulation “supports” the diagnoses of astrocytoma and is listed as a desirable feature. However, only the loss of ATRX is listed as an essential criterion being sufficient on its own to substitute 1p/19q exclusion in IDH-mutant glioma.
Only a few surrogate markers have been previously suggested for the identification of oligodendroglioma, none of which is accepted by the WHO. INA has been suggested as a positive marker for 1p/19q-codeleted oligodendroglioma26,27 but results of different studies showed limited specificity.26,28–30 Of note, the LC-MS/MS results of the present study also showed an increased abundance of INA in oligodendrogliomas which however did not reach statistical significance.
Reduced amounts of H3K27me3 frequently associated with an immunohistochemical lack of H3K27me3 (using monoclonal antibody C36B11) has been demonstrated to occur in the vast majority of oligodendrogliomas.31,32 Its usefulness as a surrogate marker for 1p/19q codeletion is limited by a notable fraction of astrocytomas similarly displaying lack of H3K27me3, a reported selectivity for IDH1-R132H mutant oligodendrogliomas and an intra-laboratory variability of results.33,34
However, in a resource limited set-up and for cases which remain undeterminable by HIP1R/VIM/ATRX immunohistochemistry, p53 and H3K27me3 may represent valuable additional markers.
Limitations and Caveats
Molecular analyses are the gold standard for 1p/19q determination and are mandatory in ambiguous cases. In principle, interpretation of immunohistochemical results always has to consider the possibility of artificial results. To this end we strongly recommend the use of on slide controls with known HIP1R/VIM expression levels. Careful establishing the immunohistochemical stains including possibly necessary adaptions to the local technique will be critical for an optimal dynamic range and successful application of the method. We do not recommend the usage of HIP1R or VIM as single markers as only the combined evaluation of HIP1R/VIM reaches a high degree of specificity which can be further enhanced by ATRX. The immunohistochemical patterns identified here base on solid tumor tissues only. While infiltration zones with a high percentage of tumor cells may still provide informative results in many cases, the performance of the approach is expected to decline significantly with a decreasing tumor cell content. Representative tumor tissues such as those obtained during a tumor resection is another prerequisite for accurate interpretation of HIP1R/VIM stains. The approach is intended for conventional IDH-mutant gliomas only because the recently characterised rare variant oligosarcoma, IDH-mutant does not show the same HIP1R/VIM expression patterns as conventional oligodendrogliomas despite harbouring 1p/19q codeletion.35
Conclusions
LC-MS/MS is a powerful approach for identifying and quantifying protein prevalence in FFPE brain tumors and can serve as a tool for recognizing diagnostic protein patterns. We demonstrate that the combination of three immunohistochemical tests predicts 1p/19q status with high sensitivity and specificity. Our approach reduces the technical complexity for assessing this parameter relevant for WHO diagnosis of IDH-mutant glioma.
Supplementary Material
Acknowledgement
We thank Madelaine Stoll, Marlene Hoffarth, Viktoria Zeller and Ulrike Vogel for excellent technical assistance.
Contributor Information
Marius Felix, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Dennis Friedel, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Ashok Kumar Jayavelu, Clinical Cooperation Unit Pediatric Leukemia, German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Pediatric Oncology, Hematology and Immunology, University of Heidelberg, Heidelberg, Germany; Hopp Children’s Cancer Center Heidelberg - KiTZ, Heidelberg, Germany; Molecular Medicine Partnership Unit, EMBL, Heidelberg, Germany.
Katharina Filipski, Institute of Neurology, (Edinger Institute), University Hospital, Frankfurt Am Main, Germany; German Cancer Consortium (DKTK), Partner Site Frankfurt/Mainz, Heidelberg, Germany; German Cancer Research Center (DKFZ), Heidelberg, Germany; University Cancer Center (UCT), Frankfurt, Germany.
Annekathrin Reinhardt, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Uwe Warnken, Clinical Cooperation Unit Neurooncology, German Consortium for Translational Cancer Research (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Damian Stichel, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Daniel Schrimpf, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Andrey Korshunov, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Yueting Wang, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Tobias Kessler, Clinical Cooperation Unit Neurooncology, German Consortium for Translational Cancer Research (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany; Department of Neurology and Neurooncology Program, National Center for Tumor Diseases, Heidelberg University Hospital, Heidelberg, Germany.
Nima Etminan, Department of Neurosurgery, University Hospital Mannheim, University of Heidelberg.
Andreas Unterberg, Department of Neurosurgery, Heidelberg University Hospital, Heidelberg, Germany.
Christel Herold-Mende, Department of Neurosurgery, Heidelberg University Hospital, Heidelberg, Germany.
Laura Heikaus, Bruker Daltonics GmbH & Co. KG, Bremen, Germany.
Felix Sahm, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Wolfgang Wick, Clinical Cooperation Unit Neurooncology, German Consortium for Translational Cancer Research (DKTK), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Patrick N Harter, Institute of Neurology, (Edinger Institute), University Hospital, Frankfurt Am Main, Germany; German Cancer Consortium (DKTK), Partner Site Frankfurt/Mainz, Heidelberg, Germany; German Cancer Research Center (DKFZ), Heidelberg, Germany; Frankfurt Cancer Institute (FCI), Frankfurt Am Main, Germany.
Andreas von Deimling, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
David E Reuss, Department of Neuropathology, Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ) German Consortium for Translational Cancer Research (DKTK), Heidelberg, Germany.
Funding
A.v.D. and D.E.R. are funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 404521405, SFB 1389 - UNITE Glioblastoma, Work Package A05. A.v.D. is funded by the Bundesministerium für Bildung und Forschung (BMBF) program MSCorSys, SMART-CARE, 031L0212A. K.F. is funded by the Deutsche Krebshilfe (Mildred Scheel Career Center Frankfurt) and the Frankfurt Research Funding (FFF) program ‘Nachwuchswissenschaftler’. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interest statement. A.v.D. and D.E.R. were non-financially supported by Bruker Daltonics. L.H. is employee of Bruker Daltonics. All terms are being managed by the University of Heidelberg in accordance with its conflict of interest policies.
Authorship statement.
MF, AvD and DER conceived the study. MF performed sample preparation. AKJ performed MS measurements. AKR, KF, PNH and DER performed immunohistochemical analysis. MF, DF, DS and DSc analysed proteomic data and TCGA data. MF and DER interpreted data and drafted the manuscript. The other authors contributed with tissue samples and/or by consultancy in data analysis and interpretation and/or by proposals regarding form and content of the manuscript. All authors read and approved the final manuscript.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification and Grading of Tumours of the Central Nervous System. Lyon: IARC Press; International Agency for Research on Cancer; 2016. [Google Scholar]
- 2. Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021; 18(3):170–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balss J, Meyer J, Mueller W, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008; 116(6):597–602. [DOI] [PubMed] [Google Scholar]
- 4. Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009; 118(5):599–601. [DOI] [PubMed] [Google Scholar]
- 5. Woehrer A, Hainfellner JA. Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncol. 2015; 4(5):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015; 129(1):133–146. [DOI] [PubMed] [Google Scholar]
- 7. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015; 372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018; 555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo T, Kouvonen P, Koh CC, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat Med. 2015; 21(4):407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu Y, Guo T. High-throughput proteomic analysis of fresh-frozen biopsy tissue samples using pressure cycling technology coupled with SWATH mass spectrometry. Methods Mol Biol. 2018; 1788:279–287. [DOI] [PubMed] [Google Scholar]
- 11. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008; 26(12):1367–1372. [DOI] [PubMed] [Google Scholar]
- 12. Ramos M, Geistlinger L, Oh S, et al. Multiomic integration of public oncology databases in bioconductor. JCO Clin Cancer Inform. 2020; 4:958–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meier F, Brunner AD, Koch S, et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol Cell Proteomics. 2018; 17(12):2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dehghani F, Schachenmayr W, Laun A, Korf HW. Prognostic implication of histopathological, immunohistochemical and clinical features of oligodendrogliomas: a study of 89 cases. Acta Neuropathol. 1998; 95(5):493–504. [DOI] [PubMed] [Google Scholar]
- 15. Ikota H, Kinjo S, Yokoo H, Nakazato Y. Systematic immunohistochemical profiling of 378 brain tumors with 37 antibodies using tissue microarray technology. Acta Neuropathol. 2006; 111(5):475–482. [DOI] [PubMed] [Google Scholar]
- 16. Grzendowski M, Wolter M, Riemenschneider MJ, et al. Differential proteome analysis of human gliomas stratified for loss of heterozygosity on chromosomal arms 1p and 19q. Neuro Oncol. 2010; 12(3):243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim SI, Lee K, Bae J, et al. Revisiting vimentin: a negative surrogate marker of molecularly defined oligodendroglioma in adult type diffuse glioma. Brain Tumor Pathol. doi: 10.1007/s10014-021-00411-4. [DOI] [PubMed] [Google Scholar]
- 18. Engqvist-Goldstein AE, Kessels MM, Chopra VS, Hayden MR, Drubin DG. An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J Cell Biol. 1999; 147(7):1503–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao DS, Bradley SV, Kumar PD, et al. Altered receptor trafficking in Huntingtin Interacting Protein 1-transformed cells. Cancer Cell. 2003; 3(5):471–482. [DOI] [PubMed] [Google Scholar]
- 20. Hyun TS, Rao DS, Saint-Dic D, et al. HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J Biol Chem. 2004; 279(14):14294–14306. [DOI] [PubMed] [Google Scholar]
- 21. Wang S, Jin F, Fan W, et al. Gene expression meta-analysis in diffuse low-grade glioma and the corresponding histological subtypes. Sci Rep. 2017; 7(1):11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banan R, Stichel D, Bleck A, et al. Infratentorial IDH-mutant astrocytoma is a distinct subtype. Acta Neuropathol. 2020; 140(30):569–581. [DOI] [PubMed] [Google Scholar]
- 23. Leeper HE, Caron AA, Decker PA, et al. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015; 6(4):30295–30305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ball MK, Kollmeyer TM, Praska CE, et al. Frequency of false-positive FISH 1p/19q codeletion in adult diffuse astrocytic gliomas. Neurooncol Adv. 2020; 2(1):vdaa109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018; 135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 26. Ducray F, Criniere E, Idbaih A, et al. alpha-Internexin expression identifies 1p19q codeleted gliomas. Neurology. 2009; 72(2):156–161. [DOI] [PubMed] [Google Scholar]
- 27. Ducray F, Idbaih A, de Reynies A, et al. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer. 2008; 7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ducray F, Mokhtari K, Criniere E, et al. Diagnostic and prognostic value of alpha internexin expression in a series of 409 gliomas. Eur J Cancer. 2011; 47(5):802–808. [DOI] [PubMed] [Google Scholar]
- 29. Durand K, Guillaudeau A, Pommepuy I, et al. Alpha-internexin expression in gliomas: relationship with histological type and 1p, 19q, 10p and 10q status. J Clin Pathol. 2011; 64:793–801. [DOI] [PubMed] [Google Scholar]
- 30. Eigenbrod S, Roeber S, Thon N, et al. alpha-Internexin in the diagnosis of oligodendroglial tumors and association with 1p/19q status. J Neuropathol Exp Neurol. 2011; 70:970–978. [DOI] [PubMed] [Google Scholar]
- 31. Feller C, Felix M, Weiss T, et al. Histone epiproteomic profiling distinguishes oligodendroglioma, IDH-mutant and 1p/19q co-deleted from IDH-mutant astrocytoma and reveals less tri-methylation of H3K27 in oligodendrogliomas. Acta Neuropathol. 2020; 139:211–213. [DOI] [PubMed] [Google Scholar]
- 32. Filipski K, Braun Y, Zinke J, et al. Lack of H3K27 trimethylation is associated with 1p/19q codeletion in diffuse gliomas. Acta Neuropathol. 2019; 138:331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habiba U, Sugino H, Yordanova R, et al. Loss of H3K27 trimethylation is frequent in IDH1-R132H but not in non-canonical IDH1/2 mutated and 1p/19q codeleted oligodendroglioma: a Japanese cohort study. Acta Neuropathol Commun. 2021; 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pekmezci M, Phillips JJ, Dirilenoglu F, et al. Loss of H3K27 trimethylation by immunohistochemistry is frequent in oligodendroglioma, IDH-mutant and 1p/19q-codeleted, but is neither a sensitive nor a specific marker. Acta Neuropathol. 2020; 139:597–600. [DOI] [PubMed] [Google Scholar]
- 35. Suwala AK, Felix M, Friedel D, et al. Oligosarcomas, IDH-mutant are distinct and aggressive. Acta Neuropathol. 2022; 143:263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.