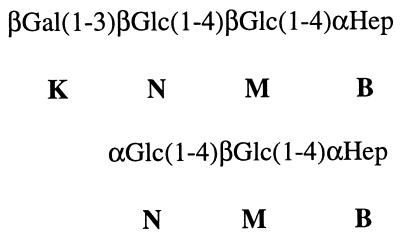Abstract
Haemophilus somnus undergoes antigenic and structural phase variation in its lipooligosaccharide (LOS). A gene (lob-1) containing repetitive 5′-CAAT-3′ sequences that may, in part, contribute to phase variation was cloned and sequenced (T. J. Inzana et al., Infect. Immun. 65:4675–4681, 1997). We have now identified another putative gene (lob-2A) immediately upstream from lob-1. Lob-2A contained homology to several LOS biosynthesis proteins of the family Pasteurellaceae and the LgtB and LgtE galactosyltransferases of Neisseria meningitidis and N. gonorrhoeae. Unlike lob-1, lob-2A contained 18 to 20 5′-GA-3′ repeats 141 bp upstream of the termination codon as determined by PCR amplification of DNA from individual colonies. Twenty repeats were most common, but when 19 5′-GA-3′ repeats were present a stop codon would occur 1 bp after the last 5′-GA-3′ repeat. A 630-bp SalI-BsgI fragment within lob-2A was deleted, and a kanamycin resistance (Kmr) gene was inserted into this site to create pCAATΔlob2A. Following electroporation of pCAATΔlob2A into H. somnus 738, several allelic exchange mutants were isolated. The LOS electrophoretic profile of one mutant, strain 738-lob2A1::Km, was altered, and the phase variation rate was reduced but phase variation was not eliminated. A variant with 19 5′-GA-3′ repeats in lob-2A had an LOS profile similar to that of 738-lob2A1::Km, suggesting that lob-2A was turned off in this phase. Nanoelectrospray mass spectrometry (nES-MS) and nuclear magnetic resonance spectroscopy showed that 738-lob2A1::Km was deficient in the terminal βGal(1-3)βGlcNAc residue present in parent strain 738. Mutant 738-lob2A1::Km was significantly more sensitive to the bactericidal action of normal bovine serum and was less virulent in mice than was parent strain 738. When H. somnus 129Pt was electrotransformed with shuttle vector pLS88 containing lob-2A, its LOS electrophoretic profile was modified and additional N-acetylhexosamine residues were present, as determined by nES-MS analysis. These results indicated that lob-2A may be an N-acetylglucosamine transferase involved in LOS biosynthesis and phase variation and that LOS structure is important to H. somnus virulence.
Haemophilus somnus is the etiologic agent of various bovine diseases, including pneumonia, septicemia, abortion, thrombotic meningoencephalitis, arthritis, myocarditis, and others (7, 20). Proposed virulence factors for H. somnus include adherence (7), immunoglobulin-binding proteins (42–44), survival in and toxicity to phagocytic cells (7, 9, 29), serum resistance (6), and lipooligosaccharide (LOS) (24, 25). Several H. somnus genes have been cloned (7, 25, 32), and recently a gene that encodes a 76-kDa immunoglobulin-binding surface protein associated with serum resistance was electrotransformed on shuttle vector pLS88 into a serum-sensitive strain, converting the strain to serum resistance (36). However, little else has been done to identify and characterize potential virulence factors of H. somnus at the molecular level.
In addition to being an endotoxin (26), the oligosaccharide region of H. somnus LOS can undergo antigenic phase variation (24, 25) similar to that reported for H. influenzae and Neisseria species (34). Some epitopes phase vary at a rate of 12% or greater (25). Isolates of H. somnus recovered from calves following intratracheal challenge randomly change their LOS electrophoretic profile. Immunoblots of LOS from these isolates with sera obtained from the calves at different postchallenge times showed that the calves made an immune response to LOS epitopes of previous isolates but not to those of current isolates. Calves that had cleared the infection had antibodies that recognized the LOS epitopes of all of the isolates recovered from them (24). Therefore, LOS phase variation may enable H. somnus to avoid the host immune response. Recent characterization of the LOS structure of H. somnus 738 (8) has provided us with a template for assessment of the structural consequences of phase variation in the LOS biosynthesis genes. In H. influenzae, some genes involved in LOS biosynthesis contain repetitive DNA sequences of 5′-CAAT-3′ or other sequences immediately downstream of potential start codons. These repeats can change in number through slipped-strand mispairing during DNA replication, resulting in phase-variable expression of LOS genes (13, 31, 41).
A putative H. somnus LOS gene that may be involved in phase variation has been cloned and sequenced and also contains variable repeats of the tetranucleotide sequence 5′-CAAT-3′ immediately downstream of potential start codons. This gene, which has been named lob-1, contains 59% DNA homology to the H. influenzae type b LOS biosynthesis gene lex-2B (25). We now report the cloning and mutagenesis of another putative LOS biosynthesis gene, designated lob-2A, which appears to be an N-acetylglucosamine transferase, is required for full bacterial virulence, and has homology to several LOS biosynthesis genes (5, 11, 13, 33).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Bacterial strains were maintained in 10% skim milk at −80°C. H. somnus strains were grown on agar plates or in broth as previously described (23). For electroporation, bacteria were grown in brain heart infusion broth with 10% Levinthal base and 0.1% thiamine monophosphate. Antibiotics were used in growth media for maintenance of plasmids in H. somnus and Escherichia coli at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 85 μg/ml; streptomycin, 85 μg/ml. Kanamycin was used at 145 μg/ml for selection of double-crossover mutants of H. somnus 738.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| H. somnus strains | ||
| 738 | LOS phase variant of pneumonia isolate | 24 |
| 129pt | Preputial isolate; does not phase vary | 6 |
| 738-lob2A1–5::Km | LOS knockout mutants containing deletion in lob-2A; Kmr | This work |
| E. coli XL1-Blue | Commercial E. coli strain (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac) | Stratagene |
| Plasmids | ||
| pGEM-3Z | Cloning vector, 2.74 kb; Ampr | Promega |
| pUC4-kixx | Commercial plasmid containing Tn5 Kmr gene | Pharmacia Biotech |
| pLS88 | Broad-host-range shuttle vector from H. ducreyi; Strr Kmr | 10 |
| pCAAT | 5′-CAAT-3′-rich 3.9-kb EcoRI fragment of H. somnus 738 cloned into pGEM-3Z; Ampr | 25 |
| pCAAT-SalI | 3.9-kb EcoRI fragment of pCAAT cloned into HincII site of pGEM-3Z | This work |
| pCAATΔlob2A | pCAAT-SalI with 630-bp SalI-BsgI fragment deleted and 1.2-kb SmaII fragment of pUC4-kixx inserted | This work |
| pLSGA | 3.9-kb EcoRI fragment of pCAAT in SmaII site of pLS88; lob-2A in same orientation as Kmr gene of pLS88 | This work |
| pLSlob2A | pLSGA with 1,960 bp XbaI-HpaI fragment deleted | This work |
Recombinant DNA methods and reagents.
Genomic DNA was isolated as previously described (40). Plasmid DNA was isolated by a rapid alkaline lysis method (27) and purified by using a Qiagen Midi column (Qiagen Inc., Santa Clarita, Calif.). Restriction fragments required for cloning and probe synthesis were eluted from gels as previously described (45). Restriction digests, agarose electrophoresis, DNA ligations, and other recombinant DNA procedures were performed by standard methods (35). Restriction endonucleases were purchased from Promega Corp. (Madison, Wis.). BsgI and HhaI methylase were purchased from New England Biolabs (Beverly, Mass.).
DNA sequencing and analysis.
The nucleotide sequence of both strands of the 3.9-kb EcoRI DNA fragment of pCAAT was determined by the dideoxy-chain termination method (37) by using the Sequenase version 2.0 DNA sequencing kit. The nucleotide sequence of pCAAT was analyzed by using DNASTAR analysis software (DNASTAR, Inc., Madison, Wis.). Sequence similarity searches of the EMBL, GenBank, and DDBJ databases at the National Center for Biotechnology Information were performed by using BLAST software (1). Amino acid alignments were analyzed by the Jotun-Hein method with MegAlign (DNASTAR). PCR products were sequenced by dideoxy-chain termination using the Autoload solid-phase sequencing kit (Amersham-Pharmacia, Picataway, N.J.) and a Cy5-labeled sequencing primer.
DNA hybridization analysis.
Southern blotting was conducted by downward capillary action to MagnaGraph nylon membranes (Micron Separations Inc., Westboro, Mass.) using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) (35, 38). DNA was covalently linked to nylon membranes by UV irradiation using a UV Stratalinker (Stratagene, La Jolla, Calif.). Digoxigenin-labeled probes for DNA hybridizations were synthesized by the random primer method using the Genius System nonradioactive labeling and detection kit (Boehringer Mannheim Corp., Indianapolis, Ind.). A probe was made to the kanamycin resistance (Kmr) gene from Tn5 by labeling a 1.2-kb SmaI fragment from pUC4-kixx. Probes were also made to a 630-bp SalI-BsgI fragment within lob-2A and to the vector pGEM-3Z (Promega Corp.) by using the entire 2.7-kb plasmid as a template. DNA hybridizations were performed at 68°C in 5× SSC containing 0.1% N-lauroylsarcosine, 0.025% sodium dodecyl sulfate (SDS), and 1% Genius blocking reagent.
Creation of H. somnus suicide vector pCAATΔlob2A.
The vector pCAAT was created by cloning a 3.9-kb EcoRI genomic DNA fragment containing a (5′-CAAT-3′)n region from H. somnus 738 into pGEM-3Z as previously described (25). The suicide vector pCAATΔlob2A was created by cloning the 3.9-kb EcoRI fragment from pCAAT into the HincII site of pGEM-3Z, thus destroying the overlapping SalI site within pGEM-3Z and allowing the SalI site within lob-2A to be a unique restriction site (Fig. 1). A 630-bp region within lob-2A was deleted with SalI and BsgI, and a 1.2-kb SmaI fragment from pUC4-kixx containing the Tn5 Kmr gene was cloned into this site, creating the suicide vector pCAATΔlob2A (Fig. 1), which could not replicate within H. somnus.
FIG. 1.
Restriction endonuclease map and construction of plasmids used to make allelic exchange mutants and to complement LOS genes in H. somnus. Plasmid pCAAT has been previously described (25). pCAAT was digested with EcoRI to obtain the 3.9-kb fragment, and pGEM-3Z was digested with HincII to obtain a 2.7-kb fragment. These fragments were ligated together to obtain 6.6-kb plasmid pCAAT-SalI, which was further digested with SalI-BsgI to obtain a 6.1-kb fragment with a 630-bp deletion in lob-2A. The 1.2-kb SmaI fragment from pUC4-kixx containing the Tn5 Kmr cassette was then ligated into the SalI-BsgI site of pCAAT-SalI to obtain 7.3-kb plasmid pCAATΔlob2A. The 8.8-kb plasmid pLSGA was constructed by digesting pCAAT with EcoRI and cloning the 3.9-kb fragment into the SmaI site of pLS88. pLSGA was then cut with XbaI and HpaI to remove part of lob-1 and the non-LOS afu-like genes, and the 6.84-kb plasmid was ligated to itself. All ligations were done following blunt ending of the plasmids.
Electrotransformation.
H. somnus was electrotransformed by using a BTX ECM 600 electroporator (BTX, Inc., San Diego, Calif.) with 0.5 to 2.0 μg of plasmid DNA methylated with HhaI methylase by modification of a previously described method (36). Briefly, a 50-ml volume of bacteria was grown to about 109 CFU/ml (150 Klett units), washed twice in 272 mM sucrose buffer, and resuspended in 100 μl of sucrose buffer. A 39-μl volume of bacteria was mixed with 1 μl of DNA and electroporated under the conditions previously described (36). For transformations involving the broad-host-range shuttle vector pLS88 and its derivatives, cells were recovered by being spun for 1 h at 37°C at 180 rpm. For suicide plasmid transformations, cells were recovered by being spun for 3 to 4 h at 37°C at 60 rpm. After recovery, recombinant cells were cultured on blood agar plates containing streptomycin at 85 μg/ml or, to select for knockout mutants, on kanamycin at 145 μg/ml at 37°C in a candle extinction jar.
Complementation of H. somnus 129Pt.
The 3.9-kb EcoRI fragment of pCAAT containing lob-1, lob-2A, and part of lob-2B was cloned into the SmaI site of pLS88 to obtain plasmid pLSGA (Fig. 1). A 1.9-kb XbaI-HpaI fragment of pLSGA was deleted upstream of lob-2A, leaving lob-2A as the only intact gene on the plasmid. The new vector, designated pLSlob2A, was introduced into non-phase-variable H. somnus 129Pt by electroporation as described above to obtain strain 129Pt(pLSlob2A).
Colony immunoblotting.
H. somnus 738 colonies were transferred to nitrocellulose (Nitrobind; Micron Separations, Westboro, Mass.), the membrane was dried at 68°C for 5 min and gently washed in Tris-buffered saline (TBS), pH 7.5. The membrane was incubated for 1 h at room temperature in TBS containing 1% skim milk, washed five times in TBS, and incubated in TBS containing monoclonal antibody (MAb) 5F5.9, which is specific for phosphorylcholine (PC) (17), for 1 h. Bound antibodies were detected as previously described (25).
Screening of recombinant colonies by PCR.
All primers were designed by DNASTAR software. For analysis of recombinant strains, primers YWA (5′-AGCAATCTCGCAAATAAA-3′) and YWB (5′-GAATAATAATGGCAGGCT-3′) were used to amplify by PCR a 451-bp region of lob-2A within the 630-bp SalI-BsgI fragment that would be deleted in the event of a double-crossover mutation. PCRs were carried out by using Ready-to-Go PCR beads (Amersham-Pharmacia, Piscataway, N.J.). Thirty cycles of PCR were used, with each cycle consisting of denaturation at 96°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min. A probe of this fragment was used to screen Kmr mutants following electrotransformation with the suicide plasmid pCAATΔlob2A.
For analysis of 5′-GA-3′ repeats in lob-2A, primers YWC (5′-TATCCGGTTTATCAATGTG-3′) and YWD (5′-biotin-ATCTTTCAAATATCCTTCTTCTAA-3′) were used to amplify a 218-bp region of lob-2A containing the 5′-GA-3′ repeats. Some colonies were selected for analysis by PCR based on reactivity to MAb 5F5.9. The area of nitrocellulose containing a colony of interest was excised and boiled in water for 10 min in order to extract the DNA (30). Ten microliters of supernatant was used as the template for Ready-to-Go PCR beads (Amersham-Pharmacia). Forty cycles of PCR were used as described above, but annealing was done at 50°C for 1 min. An additional primer, YWE (5′-Cy5-GAGCCTGCCATTATTATTCA-3′), was used to sequence the resulting PCR product by using the Autoload sequencing kit.
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was used to determine if inactivation of lob-2A through variation of the number of 5′-GA-3′ repeats or mutagenesis would affect transcription of the putative downstream open reading frame (ORF) lob-2B. H. somnus 738 was grown to 109 CFU/ml, and RNA was isolated by using the RNeasy mini kit (Qiagen, Inc.) in accordance with the manufacturer's instructions. Contaminating DNA was removed by incubating 5 μl (about 5 μg) of RNA with 1 μl of 10× PCR buffer (Gibco Bethesda Research Laboratories, Gaithersburg, Md.), making the mixture 2 mM in MgCl2, adding 1 μl of RQ1 DNase I (Promega), and adding sterile distilled water to 10 μl. The mixture was incubated at 37°C for 1 h, and then 1 μl of 25 mM EDTA was added and the mixture was incubated for 15 min at 65°C. The sample was considered free of DNA when primers to lob-2B failed to generate any PCR product in the absence of RT. RNA was converted into first-strand cDNA by using the 3′ reverse primer to lob-2B (see below) with the SuperScript Preamplification System (Gibco Bethesda Research Laboratories) in accordance with the manufacturer's instructions. Primers YWC and YWD (see above) were used for amplification of lob-2A from the cDNA strand. The primers for amplification of lob-2B from cDNA were YWF 5′-TTCCTTTTTTAATGCGTTTCAACC-3′; forward) and YWG (5′-CCGCTCTTTTAGCCAATCATATT-3′; reverse). The predicted DNA product for lob-2A was 218 bp, and that for lob-2B was 240 bp.
LOS microextraction and electrophoresis.
LOS was extracted from H. somnus cells grown in broth using a hot phenol-water microextraction method (19). For analysis of LOS from individual colonies, a single colony that had been identified with MAbs as having the desired phenotype was transferred to a separate plate and grown overnight, and the cells were removed from the plate with a swab and phosphate-buffered saline (PBS). A dilution of this suspension was replated to confirm that the majority of the cells were of the desired antigenic phenotype. For analysis of LOS from immunoblots following colony transfer, the original plate was reincubated to promote regrowth of the colonies and selected colonies were subcultured for LOS microextraction. Purified LOS was subjected to polyacrylamide gel electrophoresis (PAGE) through a 14% polyacrylamide separating gel containing urea and a bilayer stacking gel as previously described (22). LOS profiles were visualized by silver staining (39). Band intensity was adjusted by washing the gel two or three times in 5% acetic acid (5 min per wash) and then storing it in cold water.
Preparation of LOS for chemical analysis.
LOS was purified in bulk as previously described (8). O-deacylated LOS was prepared by treatment of the LOS with anhydrous hydrazine with stirring at 37°C for 30 min. The reaction mixture was cooled (0°C), cold (−70°C) acetone was added gradually to destroy excess hydrazine, and precipitated O-deacylated LOS was obtained by centrifugation. LOS (100 mg) was hydrolyzed at 100°C for 2 h in 1% acetic acid (20 ml). Insoluble material was removed by centrifugation, the supernatant solution was lyophilized, and the resulting oligosaccharide was purified by gel filtration chromatography on a Bio-Gel P2 column eluted with pyridinium acetate (0.05 M, pH 4.5). Column eluates were monitored for refractive index changes, and collected fractions (4.5 ml) were assayed colorimetrically for neutral glycoses (8).
Mass spectrometry analysis.
For nanoelectrospray-mass spectrometry (nES-MS) analysis, samples were analyzed on a VG Quattro triple quadrupole mass spectrometer (Fisons Instruments) with an electrospray ion source as previously described (8). Deacylated samples (8) were dissolved in an aqueous solvent containing 50% acetonitrile–0.1% formic acid. The electrospray tip voltage was 2.5 kV, and the mass spectrometer was scanned from m/z 150 to m/z 2,500 with a scan time of 10 s.
For capillary electrophoresis (CE)–nES-MS, a Crystal model 310 CE instrument (ATI Unicam, Boston, Mass.) was coupled to the mass spectrometer via a coaxial sheath flow interface. Mass spectral analyses were conducted on an API 300 triple quadrupole mass spectrometer (Perkin-Elmer/Sciex, Concord, Ontario, Canada) for analyses involving orifice stepping functions. Separations were obtained on 90-cm-long, bare fused silica capillary using 30 mM aqueous morpholine–acetate, pH 9, containing 5% methanol for negative-ion detection and 30 mM aqueous ammonium acetate, pH 8.5, containing 5% methanol for positive-ion detection. A voltage of 25 kV was typically applied at the injection end of the capillary. The outlet of the fused silica capillary (185 μm [outer diameter] by 50 μm [inner diameter]) was tapered to a 75-μm outer diameter and a 20-μm inner diameter.
NMR spectroscopy.
Nuclear magnetic resonance (NMR) spectra were obtained on a Varian Inova 500 spectrometer by using standard Varian software. Spectral measurements of oligosaccharides (10 mg/ml) in D2O were made at 300 K subsequent to repeated lyophilization in D2O.
Serum bactericidal assay.
The bactericidal assay was done as previously described (21, 26) in 100-μl reaction volumes containing 10, 20, 30, 40, and 50% normal bovine serum. The percent viability of H. somnus allelic exchange mutant 738-lob2A1::Km or parent strain 738 was evaluated after 0 and 60 min of incubation at 37°C.
Murine virulence studies.
The abilities of H. somnus 738-lob2A1::Km and 738 to cause mortality and bacteremia in 4- to 5-week-old Swiss-Webster mice were compared. Bacteria were grown in Columbia-yeast extract-TMP broth at 37°C to mid-logarithmic phase and resuspended to approximately 109 CFU/ml in PBS. Each mouse was inoculated intraperitoneally with 0.5 ml of a dilution of this suspension in 2% mucin. At 6 and 30 h postinoculation, blood was collected from the surviving mice, diluted in PBS, and inoculated onto Columbia blood agar plates to quantitate bacteremia.
Statistical analysis.
The two-sided P value for mouse mortality was determined by Fisher's exact test. The two-sided P value for bacteremia and serum resistance was determined by t test for unpaired samples. Calculations were made by using InStat software (GraphPad, San Diego, Calif.).
Nucleotide sequence accession number.
The nucleotide sequence of lob-2A was submitted to the EMBL and GenBank databases and assigned accession no. AF096997.
RESULTS
Cloning and characterization of lob-2A.
We previously cloned a 3.9-kb EcoRI genomic DNA fragment from H. somnus 738 that contained repeating sequences of the tetrameric oligonucleotide 5′-CAAT-3′ (25). This fragment was cloned into the commercial vector pGEM-3Z to create pCAAT (Fig. 1). The 5′-CAAT-3′ repeat tracts were located at the 5′ end of an ORF that contained 59% DNA base pair homology to lex-2B from H. influenzae (25). Based on homology, this putative gene has been designated lob-1 (LOS biosynthesis gene). In addition to lob-1, sequence analysis of the entire 3.9-kb EcoRI fragment from pCAAT identified an additional ORF and part of a second DNA region that contained homology to known LOS biosynthesis genes from H. influenzae. These regions were designated lob-2A and lob-2B, respectively, and were located adjacent to lob-1 and transcribed in the opposite direction. Two additional putative genes with homology to genes encoding iron uptake proteins in Actinobacillus pleuropneumoniae (afuBC) (3) were also identified but were not further examined (Fig. 1).
The putative AUG initiation codon for lob-2A was 121 nucleotides upstream from the AUG initiation codon of lob-1, and the initiation codon of lob-2B was 30 bases downstream from the UAG termination codon of lob-2A. Putative promoter sequences similar to the E. coli ς70 −10 (TATAAT) and −35 (TTGACA) consensus sequence promoter (12) were identified upstream of lob-2A and overlapped the −35 consensus sequence of lob-1. Repetitive sequences of 5′-CAAT-3′ were not present in lob-2A. However, 20 consecutive sequences of 5′-GA-3′ were present 141 bp upstream of the UAG termination codon. As cloned, lob-2A encoded a deduced protein of 33,691 kDa (282 amino acids), which showed 40.6, 39.6, and 38.1% overall amino acid identity to the lipopolysaccharide biosynthesis proteins LpsA of Pasteurella haemolytica A1 and Lex-1/Lic-2A and Lic-2B of H. influenzae type b, respectively (DNASTAR Inc.). Overall homologies of 37 and 35% were also found between Lob-2A and the LgtE and LgtB galactosyltransferase proteins, respectively, of Neisseria gonorrhoeae and N. meningitidis. However, 58.2% of the Lob2A amino acids were identical to the corresponding amino acids of one or more of the other proteins and 68.4% were similar (within 2 distance units) to amino acids in at least one of the other proteins. The least similarity was at the C terminus of Lob-2A, which contained 13 alternating pairs of the amino acids glutamic acid and arginine, which corresponded to the 20 5′-GA-3′ tracts present in the nucleotide sequence (data not shown). If one 5′-GA-3′ dinucleotide repeat was lost through slipped-strand mispairing, a UAA termination codon would be placed in frame 1 bp from the last 5′-GA-3′ sequence, encoding a truncated protein of 235 amino acids. If two 5′-GA-3′ sequences were lost, a UAG termination sequence would be in frame 55 bp downstream of the last 5′-GA-3′ sequence, encoding a deduced protein of 252 amino acids.
Production of Kmr double-crossover mutants.
In order to assess the role of lob-2A in LOS biosynthesis, allelic exchange mutants were created as described in Materials and Methods. A suicide plasmid derived from pCAAT and designated pCAATΔlob2A was constructed (Fig. 1). This plasmid contained a 630-bp SalI-BsgI deletion in lob-2A and the Kmr gene from Tn5 cloned into the deletion site. This vector was introduced into H. somnus 738 by electroporation, and Kmr transformants were selected. The Kmr-encoding gene within the chromosome was identified in 20 isolates by Southern hybridization of genomic DNA to a probe made to the Tn5 Kmr-encoding gene. Double-crossover mutations were differentiated from single-crossover mutations by PCR using primers YWA and YWB (see Materials and Methods), which are specific for a region within the 630-bp deletion of lob-2A. Of the 20 Kmr colonies, only 1 failed to yield a PCR product. This strain was designated 738-lob2A1::Km. In addition, probes to the SalI-BsgI 630-bp deletion from lob-2A and to vector pGEM-3Z failed to hybridize to 738-lob2A1::Km (data not shown).
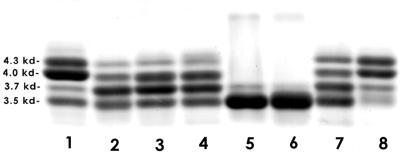
Electrophoretic analysis of the LOS from H. somnus 738-lob2A1::Km indicated that the profile had changed in comparison to that of parent strain 738 (Fig. 2, lanes 2 and 1, respectively). Specifically, there was a dramatic decrease in the 4.3- and 4.0-kDa bands, with a concomitant increase in the 3.7- and 3.5-kDa bands. Unlike strain 738, when the LOS electrophoretic profiles of 37 randomly selected 738-lob2A1::Km colonies grown in broth were analyzed, cells derived from only 1 colony changed their profile. Unlike that of the parent or mutant, this LOS contained a major band at 4.1 to 4.2 kDa and no bands at 4.0 or 4.3 kDa; the 3.7- and 3.5-kDa bands were unchanged (data not shown). Thus, the phase variation rate of strain 738-lob2A1::Km was greatly reduced but not eliminated. To confirm that the LOS profile alteration was due to insertion of the Kmr-encoding gene into lob-2A, additional allelic exchange mutants were generated in a separate experiment. Sixteen additional Kmr colonies were screened for a double-crossover mutation as described above. Four of these transformants were confirmed to contain the Kmr-encoding gene in place of the 630-bp SalI-BsgI deletion. Electrophoretic analysis showed that the LOSs of two of these mutants were similar or identical to that of 738-lob2A1::Km, while the other two mutants had predominately a single LOS band of 3.5 kDa (Fig. 2, lanes 3 to 6, respectively). Analysis of LOS extracts of 24 colonies of 738-lob2A1::Km immunoblotted with anti-PC MAb 5F5.9 showed that the 5F5.9-nonreactive colonies contained an LOS with the same profile as that of the original isolate shown in Fig. 2, lane 2. However, colonies that were reactive with MAb 5F5.9 contained an LOS expressing only the 3.5-kDa band. A 5F5.9-reactive colony was subcultured in broth and recultured on plates, and a 5F5.9-nonreactive colony was selected. The 5F5.9-nonreactive colony had only a slightly lower-molecular-mass LOS, which would be consistent with the loss of PC (data not shown). This result suggested that the PC component of the strain 738-lob2A1::Km LOS was phase variable.
FIG. 2.
Electrophoretic profile of LOSs extracted from H. somnus 738, lob-2A allelic exchange mutants of strain 738 and strain 738 isolates containing 18 or 19 5′-GA-3′ repeats in lob-2A. Lanes and strains: 1, strain 738 containing 20 5′-GA-3′ repeats in lob-2A; 2, strain 738-lob2A1::Km; 3, strain lob2A2::Km; 4, strain lob2A3::Km; 5, strain lob2A4::Km; 6, strain lob2A5::Km; 7, strain 738 containing 19 5′-GA-3′ repeats in lob-2A; 8, strain 738 containing 18 5′-GA-3′ repeats in lob-2A. Molecular masses of the major bands are indicated on the left.
Variation in the number of lob-2A 5′-GA-3′ sequences.
To determine if the number of 5′-GA-3′ sequences varied, DNAs were extracted from 13 individual colonies of strain 738 previously blotted with MAb 5F5.9, the 5′-GA-3′ region was amplified by PCR using primers YWC and YWD, and the LOSs from these isolates were analyzed by SDS-PAGE. More than one-third of the strain 738 colonies contained an LOS with a profile different from that of the parent, but there was no clear relationship between 5F5.9 reactivity and the number of 5′-GA-3′ sequences. However, 1 of the 13 colonies contained 19 5′-GA-3′ dimeric repeats in lob-2A and 1 colony contained 18 5′-GA-3′ repeats, confirming that the number of 5′-GA-3′ sequences could change in vitro. When 19 5′-GA-3′ repeats were present, a UAA termination codon would be in frame 1 bp from the last 5′-GA-3′, resulting in a premature stop to transcription. It was of interest, however, that the LOS derived from the colony with 19 5′-GA-3′ repeats was similar in profile to the LOS from 738-lob2A1-3::Km knockout mutants (Fig. 2, lanes 7 and 2 to 4, respectively). The LOS of the colony containing 18 5′-GA-3′ repeats, however, was similar to the LOS of the parent (Fig. 2, lanes 8 and 1, respectively). The LOSs from 5 of the 11 remaining colonies containing 20 repeats of 5′-GA-3′ also changed in profile, although none of the profiles were similar to that of the colony with 19 5′-GA-3′ repeats (data not shown). Therefore, the biosynthetic enzyme encoded by lob-2A appeared to be active if it contained 282 or 252 amino acids (20 or 18 5′-GA-3′ repeats, respectively) but was inactive if composed of 235 amino acids (19 5′-GA-3′ repeats).
Transcription of lob-2A and lob-2B.
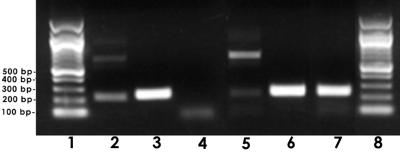
Sequence analysis indicated that the initiation codon of lob-2B was within 30 bp of the UAG termination codon of lob-2A when 20 repeats of 5′-GA-3′ were present, and the only promoter consensus sequence identified was upstream of lob-2A. The presence of a variable number of 5′-GA-3′ sequences near potential stop codons in lob-2A could therefore affect the transcription of downstream lob-2B. To investigate this possibility, cDNA was synthesized by using RT and the downstream primer of lob-2B. DNA fragments of lob-2A and lob-2B were amplified from the cDNA by PCR using primers YWC and YWD (lob-2A) or primers YWF and YWG (lob-2B) with strain 738 containing 20 sequences of 5′-GA-3′ in lob-2A (Fig. 3, lanes 2 and 3, respectively) or 19 sequences of 5′-GA-3" (Fig. 3, lanes 5 and 6, respectively) or with knockout mutant 738-lob2A1::Km using primers YWF and YWG (lob-2B) (Fig. 3, lane 7). Amplification of lob-2B resulted in a single band of 218 bp whether 19 or 20 dinucleotide repeats of 5′-GA-3′ were present or when there was a deletion in lob-2A. These results indicated that an alteration in the number of 5′-GA-3′ repeats or inactivation of lob-2A by insertion with a Kmr cassette did not affect the transcription of lob-2B.
FIG. 3.
Cotranscription of lob-2A and lob-2B from H. somnus 738 and 738-lob2A1::Km. Primers to lob-2B were used in an RT-PCR assay to determine if 19 or 20 5′-GA-3′ sequences or a deletion in lob-2A would affect the transcription of lob-2A or lob-2B. Lanes: 1 and 8, 100-bp DNA ladder; 2, cDNA 218-bp product of strain 738 lob-2A containing 20 repetitions of 5′-GA-3′ (bands above 700 bp are nonspecific amplification products); 3, cDNA 240-bp product of strain 738 lob-2B containing 20 sequences of 5′-GA-3′ in lob-2A; 4, control well (same components as in lane 2 except lacking RT; the faint small-molecular-size band represents primer dimers); 5, cDNA products of strain 738 lob-2A containing 19 sequences of 5′-GA-3′ (bands above 700 bp are nonspecific amplification products); 6, cDNA product of strain 738 lob-2B containing 19 sequences of 5′-GA-3′ in lob-2A; 7, cDNA product of strain 738-lob2A1::Km lob-2B. Amplification of lob-2A from 738-lob2A1::Km was not done because the primers were within the deleted region.
Chemical analysis of H. somnus 738-lob2A1::Km.
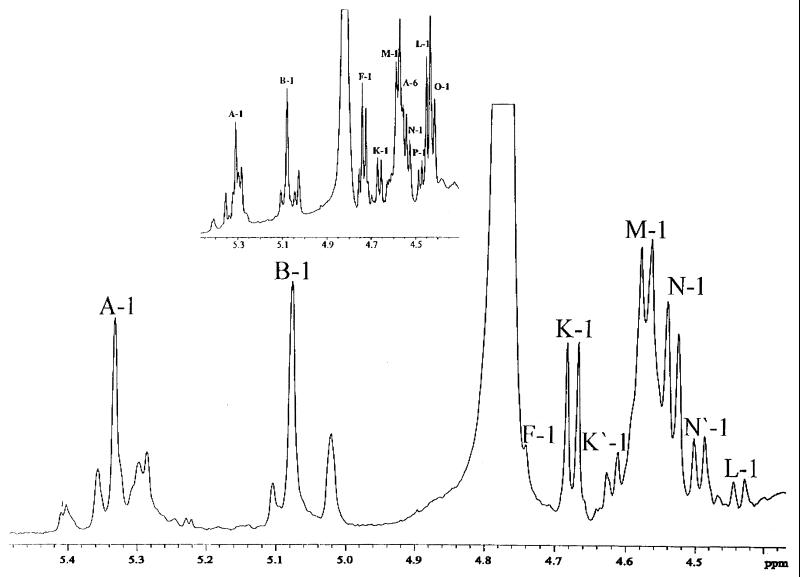
nES-MS of the O-deacylated 738 and 738-lob2A1::Km LOSs showed that the terminal βGal(1-3)βGlcNAc component, which is present in the two major glycoforms in the 738 LOS (8), was deficient in 738-lob2A::Km (see Table 2). Two doubly charged ions at m/z 1,111.1 and m/z 1,192.0 and a smaller amount of a third doubly charged ion at m/z 1,273.0 were present at low levels in the parent but were major ions in the mutant (Table 2). nES-MS of the purified core oligosaccharide from the mutant produced similar results (not shown). There were comparable amounts of the ions corresponding to the 2-hexose (2-Hex)- and 3-Hex-containing glycoforms and very little of the 4-Hex-containing glycoform in 738 LOS. CE–nES-MS analysis (not shown) indicated that very small amounts of the strain 738 parental (fully extended) glycoforms were present in the mutant LOS, which was consistent with the presence of minor bands at 4.3 and 4.0 kDa determined by gel electrophoresis (Fig. 2). NMR spectroscopy confirmed the inferences made from MS analyses. Examination of the anomeric region of the 1H-NMR spectrum of the 738-lob2A1::Km core oligosaccharide revealed a spectrum very similar to that of the core oligosaccharide derived from parent 738 but lacking the terminal βGal(1-3)βGlcNAc disaccharide anomeric resonances (Fig. 4). Minor resonances (labeled F-1 and L-1 in Fig. 4) were observed that correspond to the anomeric resonances of the terminal disaccharide in the fullest extended parental glycoforms corroborating the CE–nES-MS evidence. Detailed two-dimensional NMR analyses and selective one-dimensional excitation experiments confirmed the outer core regions of the mutant's 2-Hex- and 3-Hex-containing glycoforms, as shown in Fig. 5 (coding corresponds to the designations in reference 8).
TABLE 2.
Negative-ion nES-MS data and proposed compositions of O-deacylated LOSs from H. somnus 738, knockout mutant 738-lob2A1::Km, strain 129Pt, and recombinant strain 129Pt(pLSlob2A)
| Strain | Observed ions (m/z)
|
Molecular mass (Da)a
|
Relative intensity (fold)b | Proposed compositionc | ||
|---|---|---|---|---|---|---|
| (M-2H)2− | (M-H)3− | Observed | Calculated | |||
| 738 | 1,111.1 | 740.3 | 2,224.0 | 2,224.1 | 0.2 | 2Hex, 2Hep, PEtn, 2Kdo, lipidA |
| 1,192.0 | 794.4 | 2,386.0 | 2,386.2 | 0.2 | 3Hex, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,194.0 | 795.2 | 2,389.0 | 2,389.1 | 0.2 | 2Hex, 2Hep, PEtn, PCho, 2Kdo, lipid A | |
| 1,294.0 | 862.8 | 2,590.0 | 2,589.4 | 1.0 | HexNAc, 3Hex, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,375.0 | 916.1 | 2,752.0 | 2,751.6 | 0.5 | HexNAc, 4Hex, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,458.0 | 971.3 | 2,917.0 | 2,916.7 | 0.3 | HexNAc, 4Hex, 2Hep, PEtn, PCho, 2Kdo, lipid A | |
| 738-lob2A1::Km | 1,111.0 | 740.3 | 2,224.0 | 2,224.1 | 0.7 | 2Hex, 2Hep, PEtn, 2 Kdo, lipid A |
| 1,192.0 | 794.3 | 2,386.0 | 2,386.2 | 1.0 | 3Hex, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,273.0 | 848.3 | 2,548.0 | 2,548.4 | 0.3 | 4Hex, 2Hep, PEtn, 2Kdo, lipidA | |
| 129Pt | 1,131.6 | 2,265.0 | 2,265.1 | 0.75 | Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,212.8 | 2,427.0 | 2,427.2 | 0.20 | 2Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,293.7 | 2,589.0 | 2,589.4 | 1.00 | 3Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 129Pt(pLSlob2A) | ||||||
| Analysis 1 | 1,131.7 | 2,265.0 | 2,265.1 | 0.40 | Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,172.1 | 2,345.0 | 2,345.1 | 0.40 | Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A, P | ||
| 1,212.5 | 2,427.0 | 2,427.2 | 0.10 | 2Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,293.6 | 2,589.0 | 2,589.4 | 1.00 | 3Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,314.1 | 2,630.0 | 2,630.4 | 1.10 | 2Hex, 2HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,333.4 | 2,669.0 | 2,669.4 | 0.90 | 3Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,354.2 | 2,710.0 | 2,710.4 | 1.25 | 2Hex, 2HexNAc, 2Hep, PEtn, 2Kdo, lipid A, P | ||
| Analysis 2 | 1,131.7 | 2,265.0 | 2,265.1 | 0.50 | Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | |
| 1,172.1 | 2,345.0 | 2,345.1 | 0.05 | Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A, P | ||
| 1,212.5 | 2,427.0 | 2,427.2 | 0.15 | 2Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,293.8 | 2,589.0 | 2,589.4 | 1.00 | 3Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,314.1 | 2,630.0 | 2,630.4 | 0.70 | 2Hex, 2HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,334.0 | 2,669.0 | 2,669.4 | 0.20 | 3Hex, HexNAc, 2Hep, PEtn, 2Kdo, lipid A | ||
| 1,353.6 | 2,710.0 | 2,710.4 | 0.20 | 2Hex, 2HexNAc, 2Hep, PEtn, 2Kdo, lipid A, P | ||
Average mass units were used for calculation of molecular weight based on the proposed composition as follows: Hex, 162.15; Hep, 192.17; HexNAc, 203.19; Kdo, 220.18; PEtn, 123.05. PCho, 165.05, lipid A, 953.01.
Relative intensity is expressed with respect to the most intense peak of the doubly charged ions in the spectrum.
Abbreviations: Hep, heptose; PEtn, phosphoethanolamine; PCho, phosphorylcholine; Kdo, 3-deoxy-d-manno-2-octulosonic acid; P, phosphate.
FIG. 4.
Anomeric region of the 1H-NMR spectrum of the oligosaccharide from knockout mutant H. somnus 738-lob2A1::Km. The spectrum was recorded in D2O at pH 7.0 and 300 K. The inset is the anomeric region of the 1H-NMR spectrum of the fraction 3 oligosaccharide (described in reference 8) from H. somnus 738. The spectrum was recorded in D2O at pH 7.0 and 295 K. Both spectra were referenced to the methyl resonance of acetone (δH, 2.225 ppm). Resonances are labeled in accordance with the designations assigned previously (8).
FIG. 5.
Outer core regions of 2-Hex and 3-Hex glycoforms of mutant strain 738-lob2A1::Km LOS.
Modification of H. somnus 129Pt LOS by lob-2A.
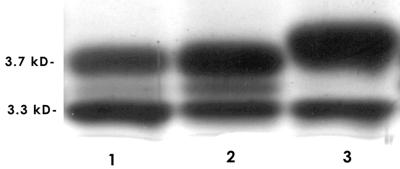
The entire 3.9-kb H. somnus fragment from pCAAT was cloned into the broad-host-range shuttle vector pLS88 to obtain pLSGA. From pLSGA, an HpaI-XbaI region that included part of lob-1 through both afu-like ORFs was deleted, leaving lob-2A and its promoter region as the only complete ORF. This construct was named pLSlob2A (Fig. 1). However, attempts to electrotransform pLSlob2A into strain 738-lob2A1::Km were unsuccessful. We also could not obtain transformants when parent strain 738 was electrotransformed with shuttle vector pLS88. Mobilization of plasmids into H. somnus by conjugation has also not been reported. In contrast, we obtained 106 transformants of strain 129Pt/μg of pLS88. Strain 129Pt is an isolate from a normal bovine prepuce that has not been observed to phase vary following daily serial passage of individual colonies. In contrast, all of the isolates examined from animals with H. somnus disease have phase varied during in vitro passage (7, 25). Therefore, additional evidence that lob-2A is involved in LOS biosynthesis was obtained by electrotransforming pLSlob2A into strain 129Pt to obtain strain 129Pt(pLSlob2A). LOS extracts from strain 129Pt(pLSlob2A) exhibited a LOS profile different from that of parent strain 129Pt or strain 129Pt containing pLS88 (Fig. 6, lanes 3, 1, and 2, respectively). Strain 129Pt contains two major bands of 3.7 and 3.3 kDa and a minor band of 3.5 kDa. The 3.3-kDa band was unaffected in 129pt(pLSlob2A), whereas the minor 3.5-kDa band was deficient and the 3.7-kDa band increased in molecular size to about 3.8 kDa. Thus, lob-2A was capable of modifying the LOS of strain 129Pt in trans.
FIG. 6.
Electrophoretic profile of LOSs extracted from the parent strain and recombinant H. somnus preputial isolate 129Pt. Lanes and strains: 1, 129Pt; 2, 129Pt(pLS88) (contains shuttle vector pLS88 only); 3, 129Pt(pLSlob2A) (contains lob-2A expressed in shuttle vector pLS88).
To obtain additional evidence that the LOS of 129Pt(pLSlob2A) was modified, O-deacylated LOSs from strains 129Pt and 129Pt(pLSlob2A) were analyzed by nES-MS (Table 2). Three major glycoforms were identified in 129Pt LOS, which contained a single N-acetylhexosamine (HexNAc) moiety and one to three Hex moieties. In contrast, two separate analyses of O-deacylated 129Pt(pLSlob2A) LOS indicated that some glycoforms contained two HexNAc moieties. When two HexNAc components were present, there was never a 3-Hex glycoform. An additional HexNAc in place of a Hex would account for the increased size of the largest LOS band on SDS-PAGE. In strain 738, these HexNAc residues have been determined to be N-acetylglucosamine (8). These results, in combination with homology data on the Neisseria lgt genes (11), suggested that lob-2A may encode an N-acetylglucosamine transferase.
Presence of lob-2A in other H. somnus strains.
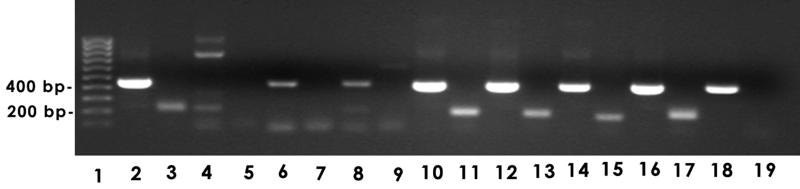
Primers YWA and YWB and primers YWC and YWD were used to amplify a central region within lob-2A and the region containing the 5′-GA-3′ repeats from strains 738 and 129Pt and seven additional preputial and disease isolates (Fig. 7). As expected, neither the 451-bp central region nor the 218-bp 5′-GA-3′ region was amplified from strain 129Pt (lanes 4 and 5, respectively), although some nonspecific products were amplified with primers YWA and YWB (lane 4). A lesser amount of the 451-bp product was amplified from preputial isolates 127P and 1P but not the 218-bp 5′-GA-3′ region (lanes 6 to 9, respectively). Both regions were well amplified from disease isolates 738, 649, TI93, TI5, and 8025 (lanes 2 and 3 and 10 to 17, respectively), but the 218 bp region was not amplified from disease isolate TI25 (lane 19).
FIG. 7.
Amplification of lob-2A from H. somnus disease isolates and preputial isolates using primers YWA and YWB (451-bp central region) and primers YWC and YWD (218-bp 5′-GA-3′ region), respectively. Lanes and strains: 1, 100-kb ladder; 2 and 3, pneumonia isolate 738; 4 and 5, preputial isolate 129Pt; 6 and 7, preputial isolate 127P; 8 and 9, preputial isolate 1P; 10 and 11, abortion isolate 649; 12 and 13, pneumonia isolate TI93; 14 and 15, pneumonia isolate TI5; 16 and 17, meningoencephalitis isolate 8025; 18 and 19, pneumonia isolate TI25.
Contribution of lob-2A to H. somnus serum resistance and virulence.
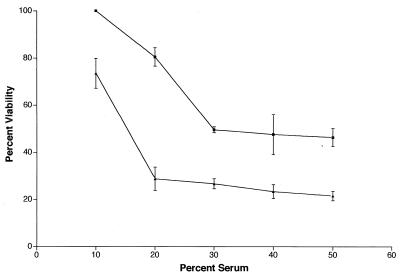
Strain 738-lob2A1::Km was more susceptible than parent strain 738 to the bactericidal action of 10 to 50% normal bovine serum at all of the concentrations tested, but the difference was most significant in 10 to 20% serum (P = 0.0012 at a serum concentration of 20%) (Fig. 8). Thus, inactivation of lob-2A diminished serum resistance.
FIG. 8.
Bactericidal activity of normal bovine serum against parent strain 738 (■) and knockout mutant strain 738-lob2A1::Km (▴). Points and error bars represent the averages ± the standard deviations of at least three separate experiments. In each experiment, percent viability was determined with cultures plated in duplicate.
Although H. somnus is a bovine-specific pathogen, disease isolates of H. somnus cause greater mortality and bacteremia than preputial isolates when inoculated into the peritoneums of mice in the presence of 2% porcine gastric mucin (18). This model was used to compare the virulence of isogenic strains 738 and 738-lob2A1::Km (Table 3). At challenge doses of about 2.5 × 107 CFU of strain 738 and 4.8 × 107 CFU of strain 738-lob2A1::Km, both strains killed 100% of the challenged mice by 24 h and the bacteria per milliliter of blood were too numerous to count at a 1:10 dilution at 6 h postchallenge (data not shown). At the lower challenge dose of about 107 CFU, strain 738 caused significantly greater mortality than strain 738-lob2A1::Km (P = 0.0152) (Table 3). All mice challenged with strain 738 that died (six of eight) did so between 6 and 28 h postchallenge. However, the two mice from this group that survived may have received a smaller challenge dose, as some leakage of the inoculum from the injection site was observed. Only one of nine mice challenged with a similar dose of strain 738-lob2A1::Km died at 32 h postchallenge. The mean level of bacteremia at 6 h postchallenge, when all of the mice were alive, was also significantly greater in mice challenged with strain 738 than in mice challenged with strain 738-lob2A1::Km (P = 0.0014). At 30 h postchallenge, both of the surviving mice challenged with strain 738 were still bacteremic (500 and 1,100 CFU/ml) whereas only one moribund mouse of nine challenged with strain 738-lob2A1::Km was bacteremic. These results indicated that the mutation in lob-2A reduced the virulence of strain 738 in mice.
TABLE 3.
Virulence of H. somnus parent and recombinant strains in mice
| Strain | Challenge dose (CFU/mouse [107]) | No. of mice dead/total (% mortality)a | Bacteremiab (CFU/ml of blood [105]) |
|---|---|---|---|
| 738 | 1.1 | 6/8 (75)c | 3.7 |
| 738-lob2A1::Km | 1.0 | 1/9 (11)d | 1.7e |
By 48 h postchallenge.
Ten microliters of blood was obtained from the tail vein of each surviving mouse 6 h after challenge, diluted 1:10 in PBS, and inoculated onto blood agar.
In two mice, approximately 20% of the inoculum leaked out of the injection site. All mice that died did so within 30 h postchallenge.
This mouse died at 32 h postchallenge. The difference in mortality is significant (P = 0.0152).
The difference in bacteremia is significant (P = 0.0014). Both surviving mice challenged with strain 738 remained bacteremic at 30 h postchallenge, whereas only one of eight surviving mice challenged with strain 738-lob2A1::Km remained bacteremic at this time.
DISCUSSION
The primary mechanism controlling phase variation in H. influenzae appears to be the presence of repetitive base pair sequences, such as (5′-CAAT-3′)n, downstream of potential start codons. A change in the number of these tetranucleotide repeats due to slipped-strand mispairing can cause the downstream ORF to go in or out of frame with the start codon, resulting in phase-variable expression of the gene (41). Repeating tracts of other nucleotide sequences have also been identified in various genes (16). Similar LOS phase-variable mechanisms exist in Neisseria spp. but are controlled by tracts of dinucleotide repeats or a homopolymeric tract of guanine residues (11, 28). Using the oligonucleotide (5′-CAAT-3′)7 as a probe, we previously cloned a 3.9-kb fragment from the H. somnus genome that contained repetitive sequences of 5′-CAAT-3′ downstream of potential start codons within a putative gene now designated lob-1 (25). Within this same fragment, but transcribed in the opposite direction, we have now sequenced another putative gene (lob-2A) and the 5′ end of a second region (lob-2B) which contained homology to lic2B of H. influenzae. Sequence analysis indicated that Lob-2A had homology to several LOS or lipopolysaccharide biosynthesis proteins, including Lex-1/Lic-2A and Lic-2B of H. influenzae (5, 13, 14), the Neisseria spp. LgtB and LgtE proteins (11), and LpsA of P. haemolytica (33). The LgtB and LgtE proteins of N. gonorrhoeae have been proposed to be galactosyltransferases involved in transfer of β1-4 galactose in the assembly of the lacto-N-neotetraose terminal structure (11, 28). All of these proteins have been shown to have homology to each other and may therefore be part of a family of glycosyltransferases.
Although this family of genes is conserved in regions, they vary in regard to the presence or absence of repetitive DNA sequences that act as a genetic on-off switch. The H. influenzae lex-1 and lic-2A genes have repeating tracts of 5′-CAAT-3′, but the lgtB and lgtE genes of Neisseria spp. lack the polymeric G tracts of the lgtA and lgtD genes (11, 28). Likewise, the P. haemolytica lpsA gene also lacks repetitive DNA sequences. Uniquely, lob-2A contained 18 to 20 repetitive sequences of 5′-GA-3′ near the 3′ end of the gene, which has not been previously reported. However, a homopolymeric tract of cytidine residues were recently identified in the lgtG gene encoding a glucosyltransferase involved in biosynthesis of the LOS β chain in N. gonorrhoeae (2). The poly(C) tracts are located near the middle of the gene, and 11 C residues are required in this gene to enable the strain to react with MAb 3G9. If 10 or 12 C residues are present, premature termination of the gene occurs, resulting in loss of binding by MAb 3G9. A similar premature termination would occur in lob-2A if 19 5′-GA-3′ repeats were present, which would place a stop codon 1 bp after the dinucleotide repeats. Unfortunately, we did not have a MAb to the lob-2A epitope to confirm that the 5′-GA-3′ repeats act as a genetic on-off switch for a particular epitope. However, LOS from a clonal isolate containing 19 5′-GA-3′ repeats in lob-2A had an electrophoretic LOS profile similar to that of three recombinant strains with a knockout mutation in lob-2A. The deficiency in the 4.3- and 4.0-kDa bands in the isolate with 19 5′-GA-3′ repeats was not as great as in the mutants, probably because some cells from the subculture had switched to the phase containing 20 5′-GA-3′ repeats, which would make more of the higher-molecular-weight glycoforms. Of interest, however, was the fact that when 18 5′-GA-3′ repeats were present in lob-2A, the 4.3-, 4.0-, and 3.7-kDa bands were normal but there was a deficiency in the 3.5-kDa band. The reason for this is unknown. The LOS electrophoretic profile of 738-lob2A1::Km was much more stable than that of parent strain 738, suggesting that lob-2A was involved in LOS phase variation. However, colonies that were strongly reactive with MAb 5F5.9 (indicating expression and accessibility of PC) contained only the 3.5-kDa band. Subculture of these colonies yielded some nonreactive colonies. Therefore, mutagenesis of lob-2A reduced the rate of phase variation but did not eliminate it.
Analysis of deacylated oligosaccharide LOS by nES-MS indicated that the terminal βGal(1-3)βGlcNAc present in 738 LOS was deficient in 738-lob2A1::Km. The presence but deficiency of the βGal(1-3)βGlcNAc component was confirmed by more sensitive CE–nES-MS and NMR. Therefore, lob-2A may act in conjunction with another gene to fully express this epitope. For example, if only lob-2A is turned off, the βGal(1-3)βGlcNAc component would be present in only small amounts, but if another gene acting in conjunction with lob-2A is turned off, then the disaccharide may not be expressed.
We were unable to complement the mutation in 738-lob2A1::Km due to inefficient electrotransformation of the H. ducreyi pLS88 shuttle vector into the mutant or parent strain. However, pLS88 was efficiently transformed into strain 129Pt, which is a non-phase varying isolate from a normal prepuce (6, 36). When 129Pt was transformed with pLSlob2A, the largest LOS band increased in molecular size. Furthermore, the LOS of recombinant strain 129Pt(pLSlob2A) contained an additional HexNAc not present in the parent strain 129Pt LOS. These results, combined with homology to the Neisseria LgtB and -E galactosyltransferases (11), suggested that Lob-2A is an N-acetylglucosamine transferase.
The presumed absence of lob-2A in strain 129Pt was confirmed by PCR with two sets of primers within lob-2A, including one set of primers that spanned the 5′-GA-3′ region. Both primers amplified lob-2A products from disease isolates 738, 649, TI93, TI5, and 8025. However, the primers that spanned the 5′-GA-3′ region did not amplify a product from preputial isolate 1P or 127P or pneumonia isolate TI25. We did not determine if a functional lob-2A gene was present in each of these isolates. However, the lack of the 5′-GA-3′ region in the preputial isolates and its presence in most of the disease isolates were consistent with our observation that none of the preputial isolates thus far examined undergo LOS phase variation, whereas most disease isolates do.
The distal location of the 5′-GA-3′ repeats in lob-2A suggested that a change in the number of 5′-GA-3′ repeats may result in the use of different translational stop sites, thereby affecting the length and terminal amino acid sequence of the proposed protein. It is possible that one or more of these variable-size proteins is nonfunctional or modified in function. This hypothesis is supported by the similarity in LOS profile between the strain 738 isolate with 19 5′-GA-3′ repeats and isolates with knockout mutations in lob-2A. Alternatively, these repeats could affect the expression of lob-2B. However, RT-PCR analysis indicated that lob-2B was transcribed when 19 or 20 5′-GA-3′ repeats were present in lob-2A or if lob-2A was knocked out by allelic exchange. This also suggested that the insertion of the Kmr-encoding gene in lob-2A did not have a downstream polar effect on lob-2B. The complete cloning of lob-2B and characterization of its potential relationship with lob-2A are in progress.
The LOS of H. influenzae has been shown to play an important role in virulence through mutagenesis of specific LOS genes (4, 5, 15, 16). In fact, a correlation between LOS structure and virulence has been proposed (15). Mutagenesis of lob-2A significantly increased the susceptibility of strain 738-lob2A1::Km to the bactericidal activity of normal bovine serum. Furthermore, strain 738-lob2A1::Km was significantly less virulent in mice than was parent strain 738 in regard to both mortality and ability to cause bacteremia when the bacteria were suspended in 2% mucin. Although this mouse model is not ideal for investigation of the natural pathogenesis of H. somnus, it does demonstrate that alteration of LOS structure can attenuate H. somnus virulence.
In summary, lob-2A was required for full expression of the terminal βGlcNAc residue of the strain 738 LOS. This gene is novel in having varying repeat sequences of 5′-GA-3′ near its 3′ end. Mutagenesis of lob-2A reduced but did not eliminate phase variation, indicating that multiple genes are involved in this molecular switching. Normal expression of lob-2A enhanced serum resistance and virulence and may, in part, account for the difference in virulence between disease and preputial isolates of H. somnus. The LOS of H. somnus therefore appears to be an important component in the pathogenesis of H. somnus disease.
ACKNOWLEDGMENTS
This work was supported by grant 94-37204-0406 from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program to T.J.I. and HATCH formula funds to the Virginia State Agricultural Experiment Station.
We thank Eric Hansen, Michael Apicella, and Alan Lesse for contributing monoclonal antibodies; Jerry Sanders and Lynette Corbeil for strains and plasmids; and Don Krajcarski, Pierre Thibault, and Jianjun Li for mass spectrometry. We thank Abey Bandara, Michael Howard, Wendy Maduff, John McQuiston, Mark Lawrence, and Gretchen Glindemann for excellent technical assistance and advice and Ramesh Vermulapali and Stephen Boyle for valuable discussions.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Wang R, Uljon S N, Rice P A, Gotschlich E C, Stein D C. Identification of the gene (lgtG) encoding the lipooligosaccharide b chain synthesizing glucosyl transferase from Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:10872–10877. doi: 10.1073/pnas.95.18.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin N, Frey J, Chang C F, Chang Y F. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1996;143:1–6. doi: 10.1111/j.1574-6968.1996.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 4.Cope L D, Yogev R, Mertsola J, Argyle J C, McCracken G H, Jr, Hansen E J. Effect of mutations in lipooligosaccharide biosynthesis genes on virulence of Haemophilus influenzae type b. Infect Immun. 1990;58:2343–2351. doi: 10.1128/iai.58.7.2343-2351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cope L D, Yogev R, Mertsola J, Latimer J L, Hanson M S, McCracken G H, Jr, Hansen E J. Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type b. Mol Microbiol. 1991;5:1113–1124. doi: 10.1111/j.1365-2958.1991.tb01884.x. [DOI] [PubMed] [Google Scholar]
- 6.Corbeil L B, Blau K, Prieur D J, Ward A C S. Serum susceptibility of Haemophilus somnus from bovine clinical cases and carriers. J Clin Microbiol. 1985;22:192–198. doi: 10.1128/jcm.22.2.192-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbeil L B, Gogolewski R P, Stephens L R, Inzana T J. Haemophilus somnus: antigen analysis and immune responses. In: Donachie W, Lainson F A, Hodgson J C, editors. Haemophilus, Actinobacillus, and Pasteurella. New York, N.Y: Plenum Press; 1995. pp. 63–73. [Google Scholar]
- 8.Cox A D, Howard M D, Brisson J-R, Van der Zwan M, Thibault P, Perry M B, Inzana T J. Structural analysis of the phase-variable lipooligosaccharide from Haemophilus somnus. Eur J Biochem. 1998;253:507–516. doi: 10.1046/j.1432-1327.1998.2530507.x. [DOI] [PubMed] [Google Scholar]
- 9.Czuprynski C J, Hamillton H L. Bovine neutrophils ingest but do not kill Haemophilus somnus in vitro. Infect Immun. 1985;50:431–436. doi: 10.1128/iai.50.2.431-436.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon L G, Albritton W L, Willson P J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pSL88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- 11.Gotschlich E C. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.High N J, Deadman M E, Moxon E R. The role of repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope aGal(1-4)bGal. Mol Microbiol. 1993;9:1275–1282. doi: 10.1111/j.1365-2958.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 14.High N J, Jennings M P, Moxon E R. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol Microbiol. 1996;20:165–174. doi: 10.1111/j.1365-2958.1996.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 15.Hood D W, Deadman M E, Allen T, Masoud H, Martin A, Brisson J R, Fleischmann R, Venter J C, Richards J C, Moxon E R. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol. 1996;22:951–965. doi: 10.1046/j.1365-2958.1996.01545.x. [DOI] [PubMed] [Google Scholar]
- 16.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard M D. M.S. thesis. Blacksburg: Virginia Polytechnic Institute and State University; 1999. [Google Scholar]
- 18.Inzana, T. J. 1998. Unpublished data.
- 19.Inzana T J. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis. 1983;148:492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- 20.Inzana T J. The Haemophilus somnus complex. In: Howard J L, Smith R A, editors. Current veterinary therapy: food animal practice 4. Vol. 4. Philadelphia, Pa: The W. B. Saunders Co.; 1999. pp. 358–361. [Google Scholar]
- 21.Inzana T J, Anderson P. Serum factor-dependent resistance of Haemophilus influenzae type b to antibody to lipopolysaccharide. J Infect Dis. 1985;151:869–877. doi: 10.1093/infdis/151.5.869. [DOI] [PubMed] [Google Scholar]
- 22.Inzana T J, Apicella M A. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis. 1999;20:462–465. doi: 10.1002/(SICI)1522-2683(19990301)20:3<462::AID-ELPS462>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Inzana T J, Corbeil L B. Development of a defined medium for Haemophilus somnus from cattle. Am J Vet Res. 1987;48:366–369. [PubMed] [Google Scholar]
- 24.Inzana T J, Gogolewski R P, Corbeil L B. Phenotypic phase variation in Haemophilus somnus lipooligosaccharide during bovine pneumonia and after in vitro passage. Infect Immun. 1992;60:2943–2951. doi: 10.1128/iai.60.7.2943-2951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inzana T J, Hensley J, McQuiston J, Lesse A J, Campagnari A A, Boyle S M, Apicella M A. Phase variation and conservation of lipooligosaccharide epitopes in Haemophilus somnus. Infect Immun. 1997;65:4675–4681. doi: 10.1128/iai.65.11.4675-4681.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inzana T J, Iritani B, Gogolewski R P, Kania S A, Corbeil L B. Purification and characterization of lipooligosaccharides from four strains of “Haemophilus somnus.”. Infect Immun. 1988;56:2830–2837. doi: 10.1128/iai.56.11.2830-2837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings M P, Hood D W, Peak I R A, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 29.Lederer J A, Brown J F, Czuprynski C J. “Haemophilus somnus,” a facultative intracellular pathogen of bovine mononuclear phagocytes. Infect Immun. 1987;55:381–387. doi: 10.1128/iai.55.2.381-387.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maskell D, Szabo M, High N. PCR amplification of DNA sequences from nitrocellulose-bound, immunostained bacterial colonies. Nucleic Acids Res. 1993;21:171–172. doi: 10.1093/nar/21.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maskell D J, Szabo M J, Butler P D, Williams A E, Moxon E R. Molecular analysis of a complex locus from Haemophilus influenzae involved in phase-variable lipopolysaccharide biosynthesis. Mol Microbiol. 1991;5:1013–1022. doi: 10.1111/j.1365-2958.1991.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 32.Pontarollo R A, Rioux C R, Potter A A. Cloning and characterization of bacteriophage-like DNA from Haemophilus somnus homologous to phages P2 and HP1. J Bacteriol. 1997;179:1872–1879. doi: 10.1128/jb.179.6.1872-1879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potter M D, Lo R Y. Cloning and characterization of a gene from Pasteurella haemolytica A1 involved in lipopolysaccharide biosynthesis. FEMS Microbiol Lett. 1995;129:75–81. doi: 10.1016/0378-1097(95)00140-Z. [DOI] [PubMed] [Google Scholar]
- 34.Preston A, Mandrell R E, Gibson B W, Apicella M A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit Rev Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanders J D, Tagawa Y, Briggs R E, Corbeil L B. Transformation of a virulence associated gene of Haemophilus somnus into a strain lacking the gene. FEMS Microbiol Lett. 1997;154:251–258. doi: 10.1111/j.1574-6968.1997.tb12652.x. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 39.Tsai C-M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 40.Ward C K, Inzana T J. Identification and characterization of a DNA region involved in the export of capsular polysaccharide by Actinobacillus pleuropneumoniae serotype 5a. Infect Immun. 1997;65:2491–2496. doi: 10.1128/iai.65.6.2491-2496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of Haemophilus influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 42.Widders P R, Smith J W, Yarnall M, McGuire T C, Corbeil L B. Non-immune immunoglobulin binding of Haemophilus somnus. J Med Microbiol. 1988;26:307–311. doi: 10.1099/00222615-26-4-307. [DOI] [PubMed] [Google Scholar]
- 43.Yarnall M, Gogolewski R P, Corbeil L B. Characterization of two Haemophilus somnus Fc receptors. J Gen Microbiol. 1988;134:1993–1999. doi: 10.1099/00221287-134-7-1993. [DOI] [PubMed] [Google Scholar]
- 44.Yarnall M, Widders P R, Corbeil L B. Isolation and characterization of Fc receptors from Haemophilus somnus. Scand J Immunol. 1988;28:129–137. doi: 10.1111/j.1365-3083.1988.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhen L, Swank R T. A simple and high yield method for recovering DNA from agarose gels. BioTechniques. 1993;14:894–898. [PubMed] [Google Scholar]