Abstract
Each vertebrate species appears to have a unique timing mechanism for forming somites along the vertebral column, and the process in human remains poorly understood at the molecular level due to technical and ethical limitations. Here, we report the reconstitution of human segmentation clock by direct reprogramming. We first reprogrammed human urine epithelial cells to a presomitic mesoderm (PSM) state capable of long‐term self‐renewal and formation of somitoids with an anterior‐to‐posterior axis. By inserting the RNA reporter Pepper into HES7 and MESP2 loci of these iPSM cells, we show that both transcripts oscillate in the resulting somitoids at ~5 h/cycle. GFP‐tagged endogenous HES7 protein moves along the anterior‐to‐posterior axis during somitoid formation. The geo‐sequencing analysis further confirmed anterior‐to‐posterior polarity and revealed the localized expression of WNT, BMP, FGF, and RA signaling molecules and HOXA‐D family members. Our study demonstrates the direct reconstitution of human segmentation clock from somatic cells, which may allow future dissection of the mechanism and components of such a clock and aid regenerative medicine.
Keywords: anteroposterior axis, oscillation, presomitic mesoderm, somitogenesis, UiSomitoid
Subject Categories: Chromatin, Transcription & Genomics; Development; Stem Cells & Regenerative Medicine
Direct reprogramming of urine epithelial cells into presomitic mesoderm establishes long‐term cultures mimicking spatio‐temporal dynamics of human somitogenesis and body axis formation.
Introduction
Vertebrate embryos have a highly conserved developmental plan that leads to characteristic spatial patterning of tissues and organs. The most striking characteristic is the conserved skeleton pattern formation during early development when somites bud off the presomitic mesoderm or PSM along the anteroposterior axis (Wilson et al, 2009; Henrique et al, 2015; Saito & Suzuki, 2020). Analysis in model organisms such as chicken, zebrafish, and mouse provided much of our understanding of somitogenesis, e.g., somites emerge anteriorly from presomitic mesoderm or PSM, which is derived from the caudal lateral epiblast (CLE) and adjacent node streak border (NSB) of E8.5 mouse embryos, then migrate from E10.5 chordoneural hinge (CNH) of tail bud (Henrique et al, 2015). Thus, PSM stem cells or PSMsc may serve as a model for analyzing human somite development.
There have been reports that human ESCs can be differentiated into PSM in vitro (van den Brink et al, 2014; Henrique et al, 2015). The recent generation of mouse and human gastruloids from both ESCs and iPSCs not only recapitulates part of early embryogenesis but also uncovers previously unexpected properties such as self‐organizing or assembling of differentiating cells into structures very similar to those found in vivo (Beccari et al, 2018; Moris et al, 2020; van den Brink et al, 2020). Remarkably, in vitro regeneration of a segmentation clock that controls rhythmical production of somites using human ESCs and iPSCs suggests that human somitogenesis can be now analyzed in greater detail (Diaz‐Cuadros et al, 2020; Moris et al, 2020; Matsuda et al, 2020a). Indeed, the human segmentation clock runs at ~5 h oscillation period versus ~2.5 h for mouse, and the clock appears to be cell autonomous and unique to each species (van den Brink et al, 2014; Matsuda et al, 2020a, 2020b). However, human gastruloids generated from pluripotent cells still possess endoderm development and express neuroectoderm cell‐associated genes (OTX2, SOX1, and SOX9). They may complicate the further analysis of somitogenesis (Moris et al, 2020). It would be desirable to generate human PSM capable of self‐renewal and generating somitoids with a functional segmentation clock.
We have previously shown that epithelial cells from human urine samples can be reprogrammed to iPSCs and NPCs by an episomal system (Wang et al, 2013; Li et al, 2016). In this report, we further hypothesize that self‐renewing PSM stem cells can be regenerated from these somatic cells and showed that indeed iPSMsc cell lines can be generated efficiently, resulting in somitoids with both a functional segmentation clock and an anteroposterior axis.
Results
Generation of UiPSM by reprogramming
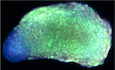
Somites emerge anteriorly from presomitic mesoderm or PSM, which migrate from the caudal lateral epiblast (CLE) and adjacent node streak border (NSB) where dual‐fated neuromesodermal progenitor (NMP) cells reside (Henrique et al, 2015; Fig 1A). NMPs require T/Bra and the Cdx gene co‐expression as well as FGF and Wnt signaling, to form posterior vertebral regions, presomitic mesoderm (PSM; Chawengsaksophak et al, 2004; Boulet & Capecchi, 2012). We hypothesize that human somatic cells can be reprogrammed directly to the PSM state. If true, the segmentation clock should be able to be regenerated from somatic cells, thus, allowing molecular dissection. To test this hypothesis, we collected urine cells (UC) from urine samples and cultured them with REGM to expand both the epithelial and mesenchymal cells but picked the epithelial cells for subsequent experiments (Zhou et al, 2012; Schutgens et al, 2019; Data not show). We then formulated induction conditions based on previous reports, i.e., activation of Wnt ß‐catenin signaling pathway (Xi et al, 2017; Chu et al, 2019; Moris et al, 2020), inhibition of histone H3K79 methyltransferase DOT1L (Halstead & Wright, 2015; Wang et al, 2019), and the epidermal growth factor (EGF) signaling for self‐renewal (Date & Sato, 2015). As illustrated in Fig 1B, UCs were electroporated with pEP4E02SET2K and pCEP4‐miR‐302‐367 and then cultured with WNT agonist CHIR99021, bFGF, Dot1L inhibitor EPZ5676, and EGF. We observed cells beginning to come together around day 3 and forming tightly clustered ones on day 9 (Fig 1B and C). Next, we evaluated the induction efficiency with T (also known as Brachyury and TBXT), a key gene involved in paraxial mesoderm specification (Yamaguchi et al, 1999), and showed that T‐positive cells reached up to 44% on day 9 (Fig 1C), with co‐expression of MIXL1, TBX6, and CDX2, SOX2 and SALL4, SOX2, and CDX2 (Appendix Fig S1A), all known to be involved in the maintenance of PSM at the posterior region and vertebral patterning (Hart et al, 2002; Zhao & Duester, 2009; Takemoto et al, 2011; Koch et al, 2017; Tahara et al, 2019). We also monitored the expression of PSM and UC‐specific genes and showed that T, MIXL1, TBX6, and CDX2 are unregulated on day 3 reaching peak level around day 9 while PAX8 and CD13 downregulated rapidly (Fig 1D). These observations support our hypothesis about PSM reprogramming.
Figure 1. Generation and characterization of UiPSM reprogramming.
- Schematic representation of the vertebrate embryonic central nervous system (CNS) indicating cell populations that give rise to the CNS. The posterior spinal cord arises from neuromesodermal progenitors (NMPs; red/green), which are located in the anterior primitive streak (PS; brown) and in the adjacent caudal lateral epiblast (CLE; light gray). NMPs give rise to new neural progenitors (NP; green), which contribute to the CLE (light gray) and then the preneural tube (PNT; dark gray), and to new mesoderm progenitors (MP; red), which contribute to presomitic mesoderm (PSM; brown), then give rise to somites.
- Schematic overview of UiPSM reprogramming from UC. UC, human‐derived urine cells; UiPSM, presomitic mesoderm progenitor cells induced from human‐derived urine cells.
- Representative images showing morphologic changes during UiPSM reprogramming (n = 3 independent experiments). Scale bars, 100 μm. Representative flow cytometry of T protein expression during UiPSM reprogramming, with hES‐derived HiPSM as a positive control (n = 3 independent experiments).
- qRT–PCR analysis of PSM‐specific genes T, MIXL1, TBX6, and CDX2; and UC‐specific genes CD13 and PAX8 during the extended reprogramming time points, including days 0, 3, 6, 9, 12, 15, and 18. Data are mean ± s.d., n = 3 independent experiments.
- UMAP analysis of 32,798 individual cells from UiPSM reprogramming on days 0, 3, 6, 9, and human iPSC differentiated PSM at d0 iPSC, d1 NMP, d2 MPC, and d3‐4 aPSM based on Margarete Diaz‐Cuadros et al (2020).
- UMAP analysis of UiPSM on day 9 and published data (d1 NMP, d2 MPC, and d3‐4 aPSM) as in (E).
- The cells in (F) are clustered and colored by the Louvain algorithm with four clusters.
- Bar chart showing the proportion of UiPSM on day 9, d1 NMP, d2 MPC, and d3‐4 aPSM in each cluster.
- Dot plot showing the expression level and ratio of PSM marker genes in four clusters.
- Expression level of PSM marker genes in UMAP projection.
- Gene ontology analyses of differentially expressed genes in four clusters with significantly enriched terms ranked by P‐value.
We then performed single‐cell sequencing (scRNA‐seq) of 7,246 individual cells on day 9 (Appendix Fig S1B and C) and identified four clusters (C0–3) with C0 (4,184 cells) expressing PSM genes involved in segmentation, somitogenesis, limb development, and PSM stemness genes expressed in C1 (CDX2, SOX2, SALL4, and GDF3) (Bisgrove et al, 2017; Saito & Suzuki, 2020). Minor clusters C2 and C3 are intermediates or residual UC expressing SOX2 or PAX8 (Appendix Fig S1C and D). As SOX2, T and TBX6 appear to regulate NMP fate in a complex way (Takemoto et al, 2011; Koch et al, 2017), we performed further analysis pairwise with CDX2 and SALL4 and showed that the co‐expression of each pair is more than 50% with T and MIXL1 exceeding 70% (Appendix Fig S1E). While PSM characteristics were quite apparent, no neural characteristics were observed (Takemoto et al, 2011; Tahara et al, 2019). This reprogramming process was highly reproducible. So far, we have established UiPSM colonies from 18 urine cell samples from 10 healthy volunteers aged 20–40 years (Appendix Fig S1F–H), and the induction efficiency varied from 15 to 60% due to individual differences independent of gender. We named them temporally UiPSM for human‐derived urine cell‐induced presomitic mesoderm progenitor cells.
We also analyzed the cell state during reprogramming by comparing to similar cells from human iPSC (Diaz‐Cuadros et al, 2020) and showed that cells on day 9 correspond to d1 NMP, d2 MPC, and d3‐4 aPSM but not with D0 iPSC (Fig 1E). Further comparing this similarity, we found day 9 UiPSM cells in cluster 0/1 almost covered a majority of d3‐4 aPSM, showing expression of HOXB2, LEF1, DLL3, HES7, TBX6, WNT5A, FGF8, and CDX2, which were enriched for somitogenesis, somite development, and pattern specification process. Those cells in cluster 3 highly resembled d1 NMP, showing expression of T, NODAL, SOX2, MIXL1, and SALL4. Besides, day 9 UiPSM cells in cluster 2 had a partial overlap with d2 MPC and, related to the Wnt signaling pathway (Fig 1F–K). These results revealed our UiPSM possessed features of previously reported d1 NMP and d3‐4 aPSM.
Characterization of UiPSM reprogramming by scRNA‐seq
To analyze the reprogramming process further, we performed qPCR during reprogramming and showed that pluripotency markers (endoPOU5F1, NANOG), nor endoderm markers (SOX17, FOXA2), or ectoderm markers (SOX1, PAX6) were induced during the 9‐day reprogramming process (Appendix Fig S2A–C). Consistent with the results shown above, we can only detect the expression of endoSOX2, SALL4, and LIN28A, not endoPOUF1, endoKLF4, NANOG, ESRRB, DPPA3, DPPA5 even during extended reprogramming time (Appendix Fig S2D). Since both Lin28a and sall4 are involved in mesoderm and axial stem cell proliferation (Robinton et al, 2019; Tahara et al, 2019), these results support the direct route of reprogramming to the PSM state.
To better understand the reprogramming process, we collected 32,798 individual cells from days 0, 3, 6, and 9 and identified five stages along the reprogramming process by trajectory analysis (Appendix Fig S2E and F). Stage 5 marked the successful branch that includes cells mostly from day 9, enriched for somitogenesis, segmentation, somite development, and pattern specification. Cells on day 0 and part of cells on days 3 and 6 are in stages 1 and 2. These two stages were marked as prebranch and mostly with renal system development and Wnt signaling pathway. The rest of the cells on days 3 and 6 are at stages 3 and 4, which were failed branches and/or intermediates enriched with genes involved in ribosome biogenesis and oxidative phosphorylation (Appendix Fig S2E–G). These results once again suggested that UiPSM reprogramming did not pass through a pluripotent phase, but a potential novel intermediate phase with enhanced metabolic activities.
To compare our dataset with previously published ones, we reconstructed trajectory from all data throughout the whole UiPSM reprogramming process and the above published human iPSC‐differentiated PSM data (d1 NMP, d2 MPC, and d3‐4 aPSM) (Diaz‐Cuadros et al, 2020). Similarly, all of these cells can be divided into three states (P1, successful branch; P2, failed branch; P3, prebranch). Only one failed branch could match the larger failed branch in the UiPSM reprogramming, mainly those cells on days 3 and 6 during UiPSM, some from d3‐4 aPSM, a few from d2 MPC, and day 9 UiPSM (Appendix Fig S2H–J). Further analysis indicated that cells in this branch not only express prebranch‐specific genes, PAX2, PAX8, and SAA1, associated with nephron tubule development but also express some of the genes enriched in the successful branch, CDX2, DLL3, HES7, TBX6, SOX2, SALL4, LEF1, MIXL1, T, related to somite development (Appendix Fig S2K and L). Trajectory analysis suggested cells in this branch include features from starting urine cells and PSM. GO analysis showed that these cells have no cell type specificity but with cell adhesion and apoptosis (Appendix Fig S2L). Nevertheless, these comparisons suggest that our day 9 UiPSM as well as those from NMP, MPC, and aPSM are quite similar in having the majority of cells in P1, enriched with segmentation or related features (Appendix Fig S2L).
UiPSM undergo long‐term expansion in vitro and differentiate into mesoderm cell types in vivo
The embryonic neuromesodermal progenitor (NMP) cells of mammalian embryos contributing to the spinal cord and the paraxial mesoderm are capable of only limited self‐renewal (Cambray & Wilson, 2002; McGrew et al, 2008; Edri et al, 2019). Based on the results obtained so far, we hypothesized that UiPSM clones obtained through reprogramming may be expanded in vitro under optimized conditions. Considering that UiPSM colonies were not able to survive in long‐term culture under induction conditions, we then embarked on an optimization process by showing that removing EPZ5676 could improve survival. Moreover, the TGF‐ß signal inhibitor (LDN193189) has been shown to be compatible with PSM cells (Diaz‐Cuadros et al, 2020). Taken these considerations together, we developed a defined medium (hereafter, DM medium) including CHIR99021, TGF‐ß inhibitor A8301, FGF2, and EGF and showed that UiPSM colonies can be expanded in DM medium without morphological changes over 30 passages in 120 days while maintaining presomitic mesodermal characteristics (Fig 2A–D). For instance, UiPSM colonies at passage 1, 5, 9, and 18 all expressed presomitic markers T, MIXL1, TBX6, and CDX2 (Fig 2A and B, and Appendix Fig S3A and B). As expected, we did not detect any pluripotent genes such as POU5F1 and NANOG, endodermal genes SOX17 and FOXA2, and ectodermal genes SOX1 and PAX6 within 20 passages (Appendix Fig S3C). In addition, UiPSM colonies passaged for over 24 generations can maintain a normal karyotype (Appendix Fig S3D). These results suggested that UiPSM is capable of long‐term self‐renewal.
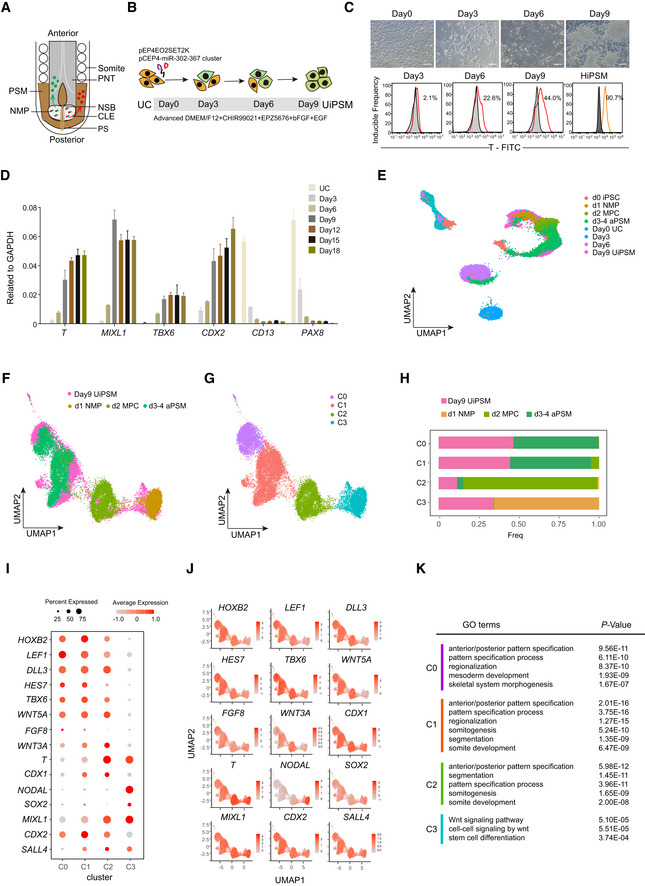
Figure 2. UiPSM self‐renewal and differential potentials.
- Representative images of UiPSM maintained in a defined medium (DM) at different passages. n = 4 biological experiments. Scale bars, 100 μm.
- Immunofluorescence of DAPI, T, MIXL1, TBX6, and CDX2 in UiPSM colonies in (A). n = 4 biological experiments. Scale bars, 200 μm.
- Growth curve of UCs and UiPSMs. Cells were seeded at 1 × 105 cells/well (24‐well plate) and passaged every 5 days. Data were mean, n = 3 wells from three biological experiments.
- Correlation analysis showing the similarity among UiPSM and the difference between UC and UiPSM. UiPSM1/2/3 derived from donor 1/2/3.
- Representative images of UiPSM‐ and UiPSC‐derived teratomas at 1 month after transplantation. n = 3 biological experiments.
- Representative images of HES‐stained UiPSM (d–f) and UiPSC (a–c)‐derived teratomas. (a) gland epithelium (endoderm); (b) osteoblast (mesoderm); (c) neural epithelium (ectoderm); (d) chondrocyte (mesoderm); (e) osteoblasts, osteoclasts, osteocytes (mesoderm); (f) muscle cell (ectoderm); n = 3 biological experiments. Scale bars, 100 μm.
- t‐SNE projection of 12,456 individual cells from UiPSM and UiPSC teratomas.
- t‐SNE projection in (G) colored by clusters identified with Louvain algorithm.
- Representative marker expression shown in heatmap corresponding to each cluster in (G) and (H).
- Gene ontology analyses for each cluster in (G) and (H). P‐value was < 0.05.
To further determine the differentiation potential and tumorigenicity of UiPSM with the self‐renewal ability in vivo, we transplanted UiPSM into an immunodeficient mouse (MITRG mice; Rongvaux et al, 2014) subcutaneously for 1 month and showed that UiPSM could grow into solid tissues with a much smaller size than UiPSC (Fig 2E). Then, we performed paraffin section and hematoxylin–eosin staining for UiPSC‐ and UiPSM‐derived teratoma/tissues, respectively, and showed that UiPSCs gave rise to teratoma with relevant tissue structures of three germ layers (e.g., gland epithelium, chondrocyte, and neural epithelium) whereas UiPSM differentiated into only mesoderm tissues (e.g., osteoblasts, chondrocytes, muscle cells; Fig 2F). To resolve the cellular diversity generated by UiPSM in vivo, we performed single‐cell RNA‐seq of UiPSC‐ and UiPSM‐derived teratoma or tissues, respectively. We selected UiPSC‐ and UiPSM‐transplanted tissues at one and 2 months for a better comparison and collected a total of 12,456 individual cells to show that cells derived from UiPSM in vivo overlap very little with that from UiPSCs (Fig 2G and H). UiPSCs gave rise to four clusters that could be categorized into mesoendoderm, axonogenesis, cartilage development, and endoderm development (Fig 2I and J), while UiPSM gave rise to six clusters among which cluster M5 and M6 are significantly consistent with muscle development (Fig 2I and J). Further analysis revealed that cells from these two clusters express muscle‐associated genes like MYL4, TNNT2, MYH6, TNNI1, ACTC1, MYL7, TPM1, and TPM2 (Appendix Fig S3E). To further analyze features of cluster M1 in UiPSM‐derived tissues, we classified them into seven subclusters and showed that these subclusters are mainly involved in mesodermal lineage fate like muscle, cartilage, bone, cardiac, and limb morphogenesis (Appendix Fig S3F and G). Thus, we conclude that UiPSM possesses the potential to give rise to cells of the mesoderm lineage, distinct from those from UiPSCs.
We then compared the scRNA‐seq data of the UiPSM clone at P10 with day 9 UiPSM and the published data (d3‐4 aPSM), and showed that all these data could be aligned nicely and clustered into six groups (Appendix Fig S4A and B). Cells in clusters 0 and 2 are from day 9 UiPSM, UiPSM_P10, and d3‐4 aPSM and are highly enriched with PSM‐specific genes, TBX6, CDX2, HOXB1, DLL3, HES7, TBX6, T, MIXL1, and WNT5A (Appendix Fig S4D and E), related to somitogenesis and somite development (Appendix Fig S4F). Furthermore, UiPSM_P10 had a higher distribution in clusters 1 and 4, showed a higher proportion of cells expressing NMP‐specific genes, SOX2 and SALL4, and a lower proportion of cells expressing PSM‐specific genes (Appendix Fig S4C–F). This analysis suggests that cells of UiPSM_P10 clone have better enrichment of PSM features (Appendix Fig S4C). In addition, we counted the proportion of cells co‐expressing PSM‐specific genes in the sequenced cells of UiPSM_P10, SOX2 and TBX6, SOX2 and T, TBX6 and CDX2, T and SALL4, T, and MIXL1 and showed that the ratio varies between 26 and 52% (Appendix Fig S4G). Therefore, UiPSM_P10 clone in DM maintains the PSM characteristics nicely consistent with the self‐renewal of these cells.
Notably, we have previously reported that iPSCs and NPCs colonies induced from urine cells have no detectable pEP4E02SET2K and pCEP4‐miR‐302‐367 after 10 passages (Wang et al, 2013; Li et al, 2016). By contrast, no obvious loss of episomal vectors occurred as expected in the UiPSM colonies, until 18 passages. Indeed, EBAN1 as a key factor for episomal vectors is stably expressed (Malecka et al, 2019). We detected EBAN1 steadily expressed in UiPSM reprogramming process and UiPSM colonies, as well as episomal vector carrying factors POU5F1, KLF4, and SOX2 with exogenous expression at various UiPSM colonies. However, only SOX2 was endogenously activated in the induced PSM state. The miR302‐367 cluster has variable integration (Appendix Fig S5A–C). Furthermore, urine cells failed to enter the reprogramming process without the induction medium (Data not shown). We knocked out total POU5F1 and another activated factor (SALL4) in UiPSM reprogramming and showed that POU5F1 is dispensable for maintaining the PSM state, whereas SALL4 is required for PSM maintenance (Appendix Fig S5D and E). These results suggest that these vectors, even being integrated at various sites, play a minimal role in maintaining PSM.
Generation of somitoids from UiPSM
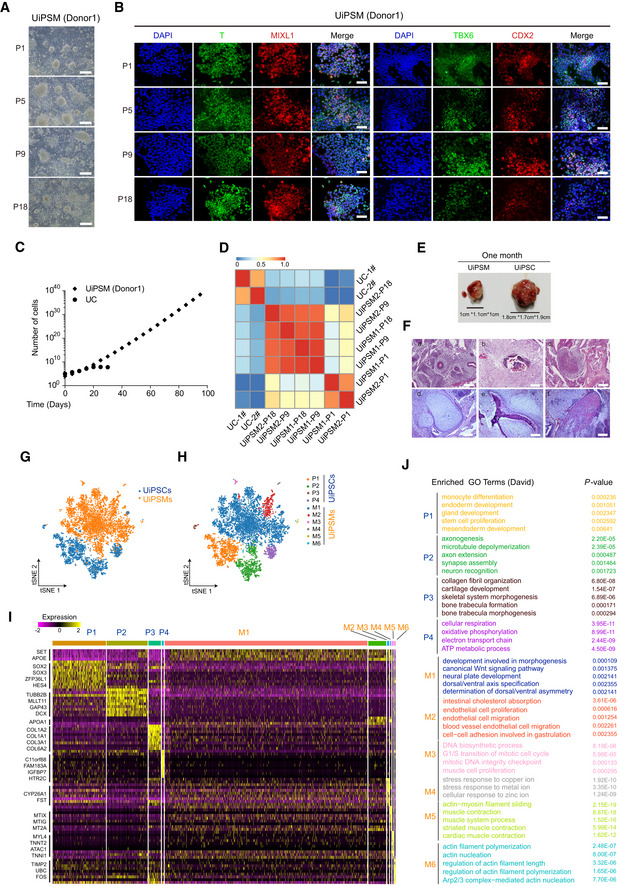
The caudal presomitic mesoderm (PSM) cells are known to form embryonic tissue by narrowing and elongating posteriorly through a process called convergence/extension during the early stages of primitive streak and axis formation (Sirbu & Duester, 2006; Benazeraf et al, 2010; Thomson et al, 2021). To test whether UiPSM self‐organizes into somite like structure, we followed the protocol established by Moris et al (2020) and seeded 100–1,000 UiPSM cells in wells of low‐adherence plates, and showed that 300–600 UiPSM cells are sufficient to aggregate into a sphere up to 150 μm in diameter with high frequency when treated with 3uM CHIR99021 (Appendix Fig S6A and C). But these spheres could not extend symmetrically. Recent studies have suggested that Wnt, Nodal, and BMP signaling appear to control the patterning of human self‐organized gastruloids (Chhabra et al, 2019; Moris et al, 2020). Therefore, we further optimized conditions for somitoid elongation by including the inhibitors for BMP, WNT, and Nodal signaling (Appendix Fig S6B) and showed that CHIR99021 (3 μM) and SB431542 (5 μM) appeared to be optimal for 300–600 seeding cells (Appendix Fig S6C–E). Thus, we developed a two‐step approach for UiSomitoids generation, including UiPSM cells aggregated in DM and the UiPSM aggregations in DM supplemented with SB431542 (hereafter, CS medium).
Under this optimized condition, we routinely seed 300–600 UiPSM cells in wells of low‐adherence plates, which allow them to form compact spherical aggregates within 48 h followed by elongation in a medium containing CHIR99021 and SB431542 for 9 days (Fig 3A). These aggregates progressively break symmetry and form elongated structures and continue to grow over time as measured by the length (L) over width (W) axis displaying a “rostral” cell‐dense region and a polar extension toward a “caudal” extremity, reaching up to 1,000 μm (L) on day 9, and resembling the elongating embryonic tailbud, tentatively termed UiSomitoids for urine‐derived induced somitoids (Matthews et al, 2021; Fig 3A–C), that maintain the expression of somitogenesis related genes, but no expression of other adjacent tissue‐associated genes (Appendix Fig S6F and G). By contrast, UiPSCs and UiNSCs generated previously could neither aggregate nor elongate under identical condition (Data not show; Wang et al, 2013; Li et al, 2016). We further show that UiSomitoids display polarized patterns of expression for SOX2/CDX2, SOX2/T, and CDX2/T in pairwise co‐staining with antibodies on day 9 (Fig 3D), indicative of anterior–posterior organization.
Figure 3. UiPSM‐derived somitoids with a functional segmentation clock.
-
ASchematic overview of UiSomitoid generation from UiPSM in a two‐stage process, CS (Advanced DMEM/F12 supplemented with 3 μM CHIR99021 and 5 μM SB431542) medium from day 2 to 9 after 2‐day aggregation in DM medium. Scale bars, 200 μm.
-
BRepresentative images of UiSomitoids (n = 10 biological experiments). Scale bars, 100 μm. Detected the ratio of length (L) over width (W) axis in (A). Top to bottom on the box chart represents maximum value, 75th percentile, median, 25th percentile, and minimum value. Twenty‐four aggregates calculated in one experiment. n = 3 independent experiments. Scale bars, 100 μm.
-
CScanning electron micrographs of UiSomitoids on day 9. Two presentative examples were shown in two independent experiments (n = 24 UiSomitoids). Scale bars, 200 μm.
-
DImmunofluorescence of SOX2 (green) co‐stained with T (red) or CDX2 (red), CDX2 (green) co‐stained with T (red) within UiSomitoids on day 9 (n = 4 biological experiments). Scale bars, 200 μm.
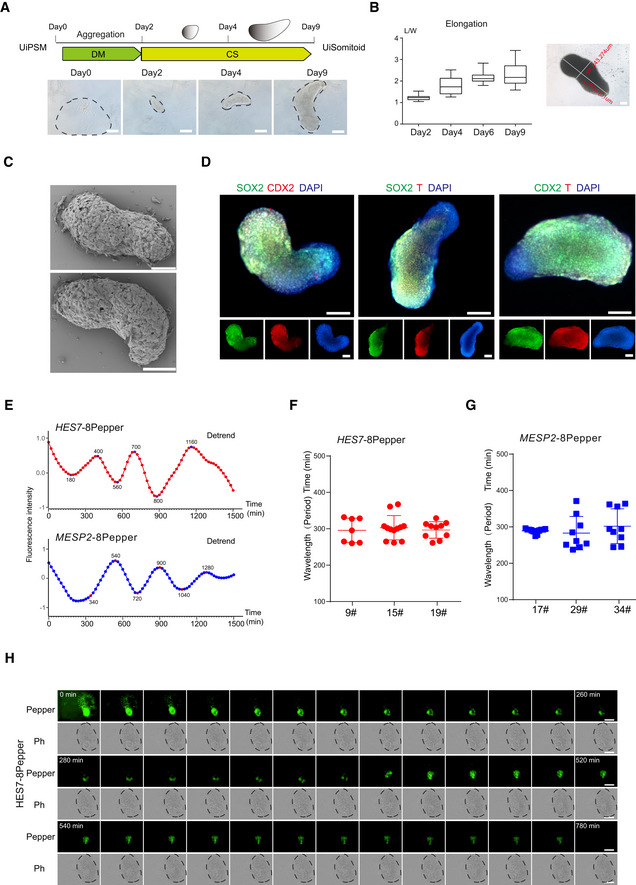
-
ERepresentative quantification of HES7 and MESP2 mRNA–8pepper fluorescence intensity in a Uisomitoid (red and blue lines, n = 29 independent experiments for each plot).
-
F, GOscillatory HES7 (F) and MESP2 (G) reporter activity measured with Incucyte® S3 Live‐Cell Analysis System and calculated oscillation periods. Data are mean ± s.d. n = 7, 12, 10 independent experiments for HES7 and n = 11, 9, 9 independent experiments for MESP2.
-
HRepresentative time‐lapse imaging of a single UiSomitoid (n = 10 biological experiments), pepper, HES7 mRNA–8pepper; Ph—phase. Scale bar, 400 μm.
HES7 and MESP2 oscillate in UiSomitoids
A defining feature of somitogenesis is a segmentation clock oscillating in a species‐specific manner (Bessho et al, 2003; Oginuma et al, 2008; Diaz‐Cuadros et al, 2020; Matsuda et al, 2020a, 2020b). We hypothesized that UiSomitiods should be able to recapitulate the human segmentation clock. To test this hypothesis, we engineered pepper aptamers, which bind and activate fluorescent dyes HBC (could image diverse RNA species in live cells; Chen et al, 2019) into UiPSM at two loci, HES7 and MESP2 (Appendix Fig S7A and B), thus, allowing us to monitor the oscillations of both RNAs. HES7 and MESP2 are expressed periodically in the presomitic mesoderm in a dynamic manner (Bessho et al, 2003; Oginuma et al, 2008). Indeed, we show that both HES7 and MESP2 RNAs oscillate ~300 min in UiSomitoids (Fig 3E–G, Appendix Fig S7C–F, Movies EV1 and EV2), with ectopically expressed GFP as a reference (Appendix Fig S7G and H). We observed a gradual decay of HES7 pepper oscillation fluorescence (Fig 3H), consistent with a delayed autoregulatory negative feedback mechanism (Bessho et al, 2003). These results suggest that the human segmentation clock can be regenerated in UiPSM‐derived Somitoids.
Anteroposterior organization of UiSomitoid
Studies from model organisms such as chick and mouse have demonstrated that PSM generates somites along the anterior–posterior axis with RA/BMP and opposing FGF/WNT signaling gradients by orchestrating the expression of key regulators such as HES7 (Niwa et al, 2007; Diez Del Corral & Morales, 2017; Ghyselinck & Duester, 2019; Anderson et al, 2020; Guzzetta et al, 2020). Indeed, our UiSomitoids recapitulate this feature well as HES7‐GFP regresses posteriorly along the extending axis (Fig 4A, Appendix Fig S8A–C, Movie EV3). To gain a more comprehensive view of this gradient and further confirm these UiPSM cells elongate into a structure with an anterior–posterior axis, we performed Geo‐seq (Chen et al, 2017) by freshly embedding and sectioning individual somitoids (using 10‐μm sections) along their anteroposterior axis into 110 slices and sequencing every other slice. The resulting dataset revealed tightly organized expression of key genes along the A‐P axis with the top 1,500 high variation genes in each slice (Fig 4B and C, Appendix Fig S9A). We identified four well‐organized clusters (U2–5): U2 enriched with genes involved in skeletal system morphogenesis and ossification, including COL3A1, COL6A3, and BMP4; U3 with genes regulating somitogenesis and segmentation, including MEOX1, TCF15, LFNG, HES1, HEY1; U4 with RIPPLY, FOXC2 MESP2 related to somite development and oxygen response; U5 with genes resembling those from mouse tailbud, including TBX6, CDX2, and FGF8 (Fig 4B–D). Selected genes from each cluster are expressed with similar patterns as expected (Appendix Fig S9B and C).
Figure 4. Characterization of UiSomitoid by Geo‐seq.

- Illustration of somitogenesis in mammalian embryos. Dark yellow, FGF/WNT signaling; light yellow, RA/BMP signaling; green, HES7. Lower panel: Diagram of representative images during the elongation of UiSomitoids along A‐P axis (green HES7‐GFP).
- Bar charts showing the anteroposterior distribution of tailbud‐associated genes T, SOX2, and CDX2 in UiSomitoid.
- Geo‐seq of an UiSomitoid with ~1,500 top highly variable genes selected by calculating variances along the anteroposterior axis. Selected genes are classified into six groups by the fuzzy clustering method.
- Gene trends along the anteroposterior axis for clusters U2–U5 with Gene Ontology (GO) terms on the right. Line means; shading, s.d.
- Localization distribution of HOX family along the anteroposterior axis. The level of expression is in scaled expression value.
- Expression distribution of Wnt, BMP, RA, and FGF signaling components along the A‐P axis. Red color highlights genes located along the posterior axis. Blue color for genes located along the anterior axis.
HOX genes should sequentially express along the anteroposterior axis, consistent with the locus distribution on the chromosome based on studies from model organisms and in vitro models (Mallo & Alonso, 2013; Gaunt, 2018; Mallo, 2018; Li et al, 2019). Indeed, paralogues from the four clusters of HOX genes appear to be expressed in a well‐organized manner (Fig 4E), exhibiting expression domains along the length of the anteroposterior axis according to their respective locations on chromosomes. For example, in HOXA cluster, a posterior‐biased distribution for groups 1–4, anterior‐biased for groups 5–10 is detected while 11 and 13 appear to be quite variable. Similarly, the HOXB members also follow the established pattern on their chromosome positions. For example, groups 1–4 are at the posterior while 5 and 6 are at the anterior with the exception that the anteriorly localized groups 8, 9 expressed in the opposite posterior area. For HOXC cluster, paralogues 4–10 distribute to the posterior end, while groups 11–13 exhibit variable expression. In HOXD cluster, groups 3–11 exhibit middle region expression similar to locus distribution on the chromosome, but HOXD1 shows a middle‐biased distribution different from posterior locus distribution on chromosome. Together, the expression patterns of the HOX genes are quite consistent with their chromosome location (Fig 4E, Appendix Fig S9D). Despite variations, the localized expression of HOX genes in UiSomitoids suggests that their roles in human development may be analyzed at the molecular level in this system in the near future.
Signaling patterns in UiSomitoid
We next focused on the signaling pathways critical for somitogenesis (Niwa et al, 2007; Diez Del Corral & Morales, 2017; Ghyselinck & Duester, 2019; Anderson et al, 2020; Guzzetta et al, 2020). We show that there is a regionalized distribution of the Wnt, BMP, RA, and FGF signaling pathways (Fig 4F, Appendix Fig S9E). First, WNT3A, 5A/B, and 8A are tightly packed at the posterior (Yamaguchi, 2001; Nakaya et al, 2005; Lin et al, 2010; Mallo & Alonso, 2013; Andre et al, 2015), while WNT4/11/16 at the anterior (Lee et al, 2000; Mallo & Alonso, 2013; Martinez‐Bartolome & Range, 2019). Second, BMP2/4/5 are localized at the anterior while 1/7/1A at the posterior (Zakin et al, 2005; Jasuja et al, 2006; Yoney et al, 2018; Li et al, 2021), confirming the key role of BMP signaling along the axial extension (Fig 4F). For RA signaling, while the cognate receptors for RA (RARA, RARB, RARG) have no obvious pattern along the axis, enzymes controlling RA synthesis and metabolism appear to be highly regionalized with three dehydrogenases (ALDH1A1, ALDH1A2, and ALDH1A3) at the anterior region, P450 enzymes (CYP26A1, CYP26B1, and CYP26C1) at the posterior‐most region, consistent with the axial distribution of RA during early embryonic development (Diaz‐Cuadros et al, 2020). The FGF signaling members also show remarkable concentration at the posterior‐ (FGF10, FGF4, FGF8, FGF3, FGF17) and anterior most localization of FGF20, FGFRL1 (Fig 4F; Trueb & Taeschler, 2006; Yoney et al, 2018; Ho et al, 2019). Based on these results, we conclude that UiSomitoids recapitulate signaling pathways quite faithfully along the anteroposterior axis.
Discussion
We present here, to the best of our knowledge, the first attempt to regenerate human PSM stem cells from somatic cells. By utilizing a well‐characterized system of urine epithelial cell reprogramming, we optimized conditions such that human PSM cells can be regenerated efficiently and reproducibly. Remarkably, these stem cells recapitulate PSM quite nicely by forming somitoids in vitro with a functional segmentation clock and patterning an anterior–posterior axis. We believe that this approach and the resulting cells can be used to understand the segmentation clock and anterior‐to‐posterior elongation as well as model diseases of the somite lineage, including dermis, tendon/ligament, skeletal muscle, cartilage, and bone. Unlike iPSC (Takahashi & Yamanaka, 2006; Takahashi et al, 2007), these UiPSM cells cannot generate teratoma, although mesoderm tissues can be formed when transplanted into immunodeficient mice, a feature consistent with a safer profile required for regenerative medicine.
Unlike hESC‐ or iPSC‐derived PSM reported recently, UiPSM reported here can undergo self‐renewal and can also be manipulated directly at the PSM stage, rather than at the hESC level. If a gene is involved in the differentiating process from hESCs or iPSCs to PSM, it would be difficult to obtain PSM and its role in PSM function could not be analyzed experimentally. But such a gene should be analyzed directly in UiPSM cells as we demonstrated by generating HES7 and MESP2 reporters (Fig 3E–G). In other words, genome‐wide screening can be carried out directly in UiPSM.
Developmentally, the starting urine epithelial cells come from the intermediate mesoderm while the resulting UiPSM are part of the paraxial mesoderm, suggesting that the reprogramming process occurs within the mesoderm lineage. Our limited dissection of this process should be further expanded to understand this process better. As the segmentation clock functions early during embryonic development, reprogramming has seemingly erased years of development epigenetically. As human embryos research faces ethical controversies (Wu & Izpisua Belmonte, 2015), UiPSM and the regeneration process may offer an alternative for embryo‐related investigation, especially somitogenesis, by extension, can be used to model diseases of paraxial mesoderm‐related organs and tissues.
Another technical point that awaits further clarification in the near future is the detection of residual reprogramming vectors. It would be highly desirable to generate integration‐free UiPSM. A few approaches may be attempted. First, the vectors may be further optimized such that integration is not possible. Secondly, mRNA‐based reprogramming may be utilized to avoid DNA‐based vectors. Lastly, the culture condition may be further optimized to lessen the length required for reprogramming, presumably minimizing integration.
Materials and Methods
MITRG mice
MITRG (Rongvaux et al, 2014) were Rag2‐deficient, Il2rg‐deficient mice with human versions of three cytokines important for innate immune cell development (M‐CSFh; IL‐3/GM‐CSFh; TPOh; Rag2−; γc−). MITRG mice were highly permissive to human hematopoiesis and enabled the full recapitulation of human myeloid development and function in the mouse, since the human cytokines supported the development and function of monocytes, macrophages, and NK cells derived from liver or adult CD34+ progenitor cells injected into the mice. This humanized mouse model may be used for human cell engraftment and patient‐derived tumor xenografts (PDX; https://www.jax.org/strain/017711).
Generation of UiPSM
Urine cell collection
Harvested urine samples were centrifuged in 500 g for 8 min for gathering cell precipitation, which was washed with DPBS twice and plated on gelatin‐coated 6‐cm dishes. Cells were cultured with REGM medium (Lonza, CC‐3190) supplemented with 50 ng/ml primocin (InvivoGen, ant‐pm‐05) in the first 4 days and cultured for 7–10 days until epithelial‐like appeared and clumped together. Next, digest these epithelial‐like urine cells with 0.25% trypsin–EDTA (Gibco, 25200072) and seed in a new 6 cm dish (Lonza, CC‐3190).
UiPSM induction
Preparing 1.5 × 106–2.0 × 106 urine cells at passage1 for electrical transduction with episomal vectors (pEP4EO2SET2K and pCEP4‐miR‐302‐367). The electric transfer kit was Amaxa™ Basic Nucleofector™ Kit (Lonza, VPI‐1005). The treated cells were plated on Matrigel (Corning, 354227) coated 12‐well plate at 1:6–1:8 and then cultured for 3–6 days in REGM medium until reaching 60% confluence. Next, induction medium or IM (advanced DMEM/F12 (Gibco, 12634‐010), 3 μM CHIR99021 (Synthesized in GIBH), 10 ng/μl bFGF (PeproTech P09038), 5 ng/μl EGF (R&D systems, 236‐EG), and 5 μM EPZ5676 (Selleck Chemicals S7062)) was added and changed every 2 days for 8–12 days.
UiPSM colonies culture
The UiPSM colonies can be digested with 0.25% Trypsin–EDTA (Gibco, 25200056), passaged at intervals of 5 days, and seeded at a density of 1 × 105 cells per well in Matrigel‐coated 12‐well plate. UiPSM colonies will be maintained in a defined medium of more than 40 passages.
Defined medium (DM): advanced DMEM/F12 supplemented with 3 μM CHIR99021, 10 ng/ml bFGF, 5 ng/ml EGF, and 1 μM A8301 (R&D systems, 2939).
pEP4EO2SET2K and pCEP4‐miR‐302‐367 cluster
The episomal vector pEP4EO2SET2K has been used in the derivation of human iPS cells and expresses OCT4, SOX2, SV40LT, and KLF4. The vector was first constructed in James Thomson's lab (Yu et al, 2009). We bought this vector from Addgene and used to generate both UiPSC and UiNPC from urine cells (Wang et al, 2013; Li et al, 2016). Our lab also developed pCEP4‐miR‐302‐367 cluster that promotes reprogramming (Wang et al, 2013; Li et al, 2016).
Generation of DEs (human definitive endoderm) and NPCs (human neural progenitor cells)
Generation of hDEs (human definitive endoderm induced from hESCs) or NPCs (human neural progenitor cells induced from human‐derived urine cells) was performed based on previously described methods (Wang et al, 2013; Loh et al, 2014).
Immunofluorescence staining
Adherent UiPSM cells were fixed with 4% formaldehyde for 30 min (TM) and washed three times (PBS). They were then permeabilized and blocked‐in perm/blocking buffer (PBS adding 0.1% Triton X‐100) (Sigma, 9002‐93‐1) and 3% BSA (Sigma, 9048‐46‐8) for 1 h (TM) and then stained overnight (4°C) with primary antibody diluted in blocking buffer (PBS adding 3% BSA). Subsequently, cells were washed three times (PBS) and stained with appropriate secondary antibodies (diluted in perm/blocking buffer) for 1 h (TM). For nuclear counterstaining, the cells were stained with DAPI (1:5,000, diluted in blocking buffer) for 3 min and then washed over twice prior to conducting fluorescent microscopy. Suspending UiSomitoids was operated in a droplet under the microscope, and then, the above operations were performed. Goat polyclonal anti‐Human/Mouse Brachyury (T) (R&D systems, AF2085), Rabbit polyclonal anti‐Human MIXL1 (Invitrogen, PA5‐64903), Goat polyclonal anti‐Human TBX6 (R & D systems, AF4744), Mouse monoclonal anti‐Human CDX2 (R&D systems, MAB3665), SOX2 (D6D9) XP® Rabbit mAb (Cell signaling Technology, 3579), Mouse monoclonal anti‐Human SALL4 (R&D systems, MAB6374). Mouse monoclonal anti‐Human MYOD (R&D systems, MAB5966), Mouse monoclonal anti‐Myosin Heavy Chain (MF20) (R&D systems, MAB4470), Mouse monoclonal anti‐Human Laminin a3 (R&D systems, MAB2144), Rabbit polyclonal anti‐Human Desmin (R&D systems, abs106139).
Quantitative RT–PCR
Total RNAs were isolated from cells with TRIzol and converted into cDNAs with HiScript II Q RT SuperMix (Vazyme, R222‐01) for qPCR, and then, qPCR was analyzed with specific qPCR primers (Appendix Table S1) and stained with ChamQ SYBR qPCR Master Mix (Vazyme, R311‐02).
Intracellular flow cytometry
To quantitatively analyze the expression of T and NAONG in urine cell‐derived UiPSM cells or human ESC, respectively, intracellular flow cytometry was conducted using the Accuri C6 Plus. Adherent cell populations were briefly washed with DPBS to remove dead or floating cells, dissociated with 0.25% Trypsin–EDTA, washed with DPBS twice, and centrifugated to prepare cell suspensions. Subsequently, cells were fixed in 100 μl 4% PFA (20 min, TM), and washed twice in DPBS; Cold methanol was slowly added to the precooled cells through mild vortex mixing to permeate the cells at least 10 min on the ice and washed twice in DPBS. The cells were resuspended in a 100 μl diluted primary antibody with a blocking buffer. For the flow cytometry antibody with a fluorescent label, T (1:100, Millipore, FCMAB302), incubated at room temperature for 30 min, was then washed twice in DPBS; cells were resuscitated with DPBS for detection on Accuri C6 Plus. For the unlabeled common antibody, NANOG (1:100, Cell Signaling Technology, 4903), incubated at room temperature for 1 h, and washed twice in DPBS. The cells were resuspended in a 100 μl diluted secondary antibody (1:500), with blocking buffer, incubated at room temperature for 30 min, and washed twice in DPBS; cells were resuscitated with DPBS for detection on Accuri C6 Plus. Data analysis was performed with FlowJo7.6.1.
Generation of UiSomitoids
300–600 UiPSM cells were seeded in low‐adherence 96‐well plates (Corning, 7007), which can form compact and spherical aggregations within 48 h in “DM” medium, and then elongated in CS medium (advanced DMEM/F12 supplemented with CHIR99021 and 5 μM ALK5 inhibitor SB431542 (PeproTech, 301836‐41‐9)), reaching up to 600–1,000 μm on day 9.
Elongation quantification
To quantify the elongation of UiSomitoids, bright‐field channel wide‐field images were collected. The length (L) and width (W) axis were measured by the line tool. We followed calculated the length (L) over width (W) axis ratio at virous timing points including days 2, 4, 6, and 9, and then drew bow plots with Prism8.
CRISPR‐Cas9 gene editing
For the human HES7 and MESP2 reporters, 3′ untranslated region (UTR) was fused to an 8pepper RNA loop with HDR. Oligonucleotides encoding sgRNA protospacer sequences were annealed in pX330. sgRNAs were verified by sequencing. Donor construction with 8pepper or GFP, 3′ untranslated region (UTR), and parts of the CDS region. Donor plasmid DNA (6 μg) and pX330‐sgRNA plasmid DNA (4 μg) were co‐transfected into UiPSM cells with Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher, L3000001) followed by selection with puromycin (1 μg/ml) and Hygromycin B (100 μg/ml) for 48 h. Surviving cells were allowed to recover and then replated at a low density before picking isolated colonies.
Oscillation assay
UiSomitoids were labeled with HES7‐8pepper reporter and MESP2‐8pepper reporter. The fluorescence of 8pepper was measured in the presence of HBC525 (0.5 μM) with Incucyte® S3 Live‐Cell Analysis System (Essen Bioscience).
The recorded picture signal was converted into a digital signal via ImageJ. The obtained 8pepper fluorescence signal of the UiSomitoids was calculated via ImageJ and then recorded the mean fluorescence intensity for subsequent analysis. Detrend was performed first to remove trends and enhance data quality. When appropriate, the moving average was subtracted with a window size of 10 units, and then, data were normalized between 0 and 1. To remove noise, the Sgolay filtering was applied. Finally, for smoothening data, we applied RLOESS. All operations were done in MATLAB. The plots were generated in R using ggplot2 package. We also applied the scale function to make it clearer.
In addition, the mean fluorescence intensity and corresponding time points fitted sine wave with nonzero baseline (Y = Amplitude*sin((2*pi*X/Wavelength) + PhaseShift) + Baseline) in prism 8.0. A phase change was defined as the time for the reporter cell line undergoing an oscillation, the oscillation transfer period. Moreover, we drew scatter plots of the calculated periods, mean ± s.d.
On this basis, we detected the oscillation signal value of three HES7‐8pepper reporter cell lines and three MESP2‐8pepper reporter cell lines, with a lenti‐GFP UiPSM cell line as a negative control for the waken GFP expression. We showed the latter did not oscillate.
Axial patterning quantification
UiSomitoids labeled with HES7‐GFP reporter were monitored by Incucyte® S3 Live‐Cell Analysis System. The expression of the HES7 moved toward the posterior area, following the elongation of UiSomitoids along the A‐P axis.
Teratoma assay
All of the animal experiments were performed according to the Animal Protection Guidelines of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China.
UiPSM (P20, 1.5 × 106, n = 3) cells and UiPSC (P20, 1.0 × 106, n = 3) cells were harvested, washed with PBS, and digested with 0.25% Trypsin–EDTA (Gibco) and accutase (Stem Cell, 07992). After centrifugation, cells were resuspended in a small volume (100–150 μl) with cold Matrigel (diluted with an equal volume of cold DMEM/F12). These UiPSM and UiPSC cell suspensions were both transplanted subcutaneously into immunodeficient MITRG mice (aged 6–8 weeks) for 1–2 months.
Notice: The Matrigel was precooled and defrosted in a 4‐degree refrigerator for 3 h.
Histological analysis of teratoma
The fresh graft tissues of UiPSM and UiPSC were isolated from mice after 2 months and a month. These tissues were fixed in 4% paraformaldehyde for more than 48 h and paraffin‐embedded. We transected each tissue into four segments of equal thickness. Then, we continuously cut tissues into 8‐μm sections along the cross‐section. Twenty‐four transverse sections corresponding to a 192‐μm length in each segment were used for quantitative analysis. Histological analysis was done on these HES‐stained sections.
High throughput data collection and analysis
Bulk‐population RNA‐seq libraries construction
For bulk‐population RNA‐seq, RNA samples with a high RNA integrity (RIN) value were used for RNA‐seq. Purified total RNA was captured with a magnetic bead with oligo‐polyA, interrupted, and reverse‐transcribed into cDNA using the VAHTSTM mRNA‐seq V3 Library Prep Kit for Illumina (Vazyme, NR611), purified using VAHTS DNA Clean Beads (Vazyme, N411). Next, cDNAs ligated adapters and amplified libraries using the amplification mix (Vazyme, NR611). Finally, we sequenced the cDNAs library to obtain 150 bp paired‐end reads.
Bulk RNA‐seq and expression analysis
RNA‐seq was preprocessed as described in a published paper (Hutchins et al, 2014). In brief, reads were mapped to the human reference genome hg38 (v81), using RSEM (Li & Dewey, 2011) and bowtie2 (very sensitive). The gene counts were normalized using EDAseq (Risso et al, 2011). Differentially expressed genes were filtered to select only those with log2 fold change > 2. RNA‐seq data were expressed in units of GC‐normalized tag counts.
Transcription factor motif discovery and gene ontology
Transcription factor motif analysis was performed by using HOMER2 with settings (‐p 4 ‐size given). Gene ontology enrichment analysis was performed using clusterProfiler.
Single‐cell RNA‐seq libraries construction
Cell suspension preparation
The cells in the UiPSM reprogramming process including days 0, 3, 6, and 9 samples were briefly washed (DPBS), dissociated (Accutase), strained with a 100‐μm filter (Solarbio, F8190) twice, pelleted, and resuspended in DPBS containing 0.04% BSA to give a final concentration of 800 cells/μl.
The fresh graft tissues of UiPSM and UiPSC isolated from mice after 2 months and 1 month were, respectively, immersed in DMEM with 0.1% penicillin–streptomycin on ice when isolated from the mice, and transferred into a 6 cm cell plate, washed with DPBS and mechanically dissociated using a pipette until a single‐cell suspension with 1 ml 0.25% Trypsin–EDTA (Gibco), then incubated with 3 ml 0.25% Trypsin–EDTA (Gibco) for 30 min at 37°C. The cell suspension was run through a 200‐mesh cell strainer (Miltenyi Biotec) to remove aggregates and debris. Cell precipitation obtained by centrifugation was resuspended in DPBS and then strained (100‐μm filter) twice. Finally, cells were resuspended with DPBS containing 0.04% BSA to give a final concentration of 800 cells/μl.
Libraries construction
For single‐cell capture, cells were diluted to a concentration of 800 cells/μl. Cells were then resuspended in the 0.04% BSA buffer, after cells, cell barcodes, and oil were loaded onto 10× Chromium chip A Single Cell Kit (10× GENOMICS, 120236). Single‐cell capture was verified on a 10× Chromium Controller, followed by cell lysis, reverse transcription, and cDNA synthesis using the Chromium chip A Single Cell Kit. Subsequently, the single‐cell cDNA libraries were preamplified. Then, preamplified cDNA libraries were fragmented and amplified with added adapter (Chromium Multiplex kit 10× GENOMICS, PN‐120262) using 10× Chromium chip A Single Cell Kit. Finally, diluted single‐cell cDNA libraries were sent to Novogene company for deep sequencing using Novo seq.
Analysis of single‐cell RNA‐seq
We mapped single‐cell sequencing data to human genome by using CellRanger 3.0.2 with setting “‐‐localcores=10 ‐‐localmem=3 ‐‐mempercore=3 ‐‐maxjobs=10 ‐‐r1‐length=26 ‐‐r2‐length=151.” We processed mapped data by using Seurat (Stuart et al, 2019). The count matrix was generated by using merge() function in Seurat and excluding low‐quality cells with < 200 genes and low‐quality genes expressing < 10 cells. The filtered count matrix was normalized by the method of SCTransform in Seurat website (https://satijalab.org/seurat/v3.1/sctransform_vignette.html). In brief, SCTransform() regressed out mitochondrial mapping percentage, normalized, scaled, and found highly variable genes (HVG). The top 5,000 HVG were used for principal component analysis (PCA), and we picked first 50 principal components (PC) to perform downstream analysis. t‐SNE (t‐distributed Stochastic Neighbor Embedding) was performed using RunTSNE() in Seurat package. UMAP (Uniform Manifold Approximation and Projection) was run using RunUMAP(). Louvain algorithm was applied to detect clusters.
Single cell trajectory analysis
The analysis of single‐cell trajectory used the expression matrix of SCTransform without HVG. The idea of constructing single‐cell trajectory was followed from Monocle2 website. In brief, we first filtered cells < 300 and > 7,500. Then, we removed cells whose counts of RNA (sum of SCT value) were > 50,000 and mitochondrial mapping percentage ≥ 25. Trajectory ordering genes were chosen on the basis of different clusters and reconstructing trajectories. Next, we reduced dimensions using reduceDimension (max_components = 2, num_dim = 100, reduction_method = “t‐SNE”) and clustered cells by clusterCells(). After the clustering, we can check the clustering results through plot_rho_delta (rho_threshold = 200, delta_threshold = 5). We then re‐ran clustering based on the threshold. After re‐clustering, we identified differential expression genes differentialGeneTest (fullModelFormulaStr = “~Cluster + Day,” cores = 10, verbose = T) and selected top 5,000 based on q value. Finally, we constructed trajectory using orderCells().
Comparison analysis of public scRNA‐seq data
Presomitic mesoderm data were obtained from (Diaz‐Cuadros et al, 2020) (GEO: GSE114186). The data were processed as described in the previous section. Integration was done following the standard pipeline with Seurat (Stuart et al, 2019). In brief, we first merged data into one object. Then, the object was split according to batch. Normalization and finding HVG were performed. Integration features were selected based on HVG. Finally, integrated data were generated by finding anchors and integrating data. The trajectory was reconstructed as mentioned above.
Geo‐seq libraries construction
Geo‐seq was performed using a protocol published method (Chen et al, 2017). In brief, rapidly cryogenic treatment was done on one UiSomitoid with liquid nitrogen and stored at −80°C overnight. Then, the pretreated UiSomitoid was cut into 110 slices along their anteroposterior axis with a Frozen slicing machine. The mRNA content of each interval section was extracted using Geo‐seq library building kit. Geo‐seq libraries' construction was profiled by Peng Guangdun's Group.
Geo‐seq analysis
The method of processing geo‐seq data were similar to bulk RNA‐seq. The data were first mapped using RSEM and bowtie2 (Langmead & Salzberg, 2012). Then, we normalized data using EDAseq. Geo‐seq data were expressed in units of GC‐normalized tag counts. After mapping and normalization, noncoding, mitochondrial, and ribosomal genes were removed. We selected top 1,500 highly variable genes by sorting their variances across sections for downstream analysis.
We used Mfuzz() to identify gene patterns across samples (Kumar & Futschik, 2007). First, filtering the genes was performed by “filter.std (min.std = 0).” Then, using “standardize ()” function to standardize the data and calculate the m1 coefficient with “mestimate ().” After that, genes were clustered into six groups.
Statistical analysis
Data were presented as mean ± s.e.m, as indicated in the figure legends. The unpaired two‐tailed Student's t‐test and two‐way ANOVA test were used to assess statistical significance. The P‐value and t‐ratio were calculated with the Prism 5 software. P‐value < 0.05 was considered as statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
RNA‐seq data qualification
All RNA‐seq samples were assessed by FastQC before downstream analysis. Reads less than Q30 were considered as the low‐quality reads and removed by Trimgalore. As for single‐cell RNA‐seq data, Q30 reads were more than 85%. The mapping rate was nearly 98%, where the average ratio of reads mapped onto the exonic region was over 75%. More than 1,200 genes were detected in each cell. As for bulk RNA‐seq samples, mapping rates were no < 80% in most samples. Furthermore, the number of detected genes ranged from 10,000 to 15,000. We do not display biological replicates in this project.
Author contributions
Duanqing Pei: Conceptualization; resources; formal analysis; supervision; funding acquisition; investigation; writing – original draft; project administration; writing – review and editing. Yue Qin: Resources; data curation; formal analysis; validation; investigation; methodology; writing – original draft. Xingnan Huang: Software; formal analysis. Zepo Cai: Data curation. Baomei Cai: Data curation; investigation. Jiangping He: Software; formal analysis; methodology. Yuxiang Yao: Data curation; software; formal analysis. Chunhua Zhou: Investigation. Junqi Kuang: Investigation. Yihang Yang: Data curation. Huan Chen: Investigation; methodology. Yating Chen: Investigation. Sihua Ou: Investigation. Lijun Chen: Investigation. Fang Wu: Investigation. Ning Guo: Investigation. Yapei Yuan: Investigation. Xiangyu Zhang: Investigation. Wei Pang: Investigation. Ziyu Feng: Investigation. Shengyong Yu: Formal analysis; investigation. Jing Liu: Formal analysis; investigation. Shangtao Cao: Conceptualization; formal analysis; supervision; investigation; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest. DP is EMBO Journal editorial advisory board member. This has no bearing on the editorial consideration of this article for publication.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (numbers 92068201, 31830060, 31970681, and 21907095); the “Strategic Priority Research Program” of the Chinese Academy of Sciences, Grant No: XDA16010505; XDA16010204; XDA16010401; the National Key Research and Development Program of China (2017YFA0504100, 2016YFA0101800, 2018YFE0204800, and 2020YFA0112400); the “Frontier Science Research Program” of the Chinese Academy of Sciences, Grant No: QYZDJ‐SSW‐SMC009; the Science and Technology Planning Project of Guangdong Province (numbers 2017B030314056 and 2019A1515011024); Guangdong Science and Technology Project (2020B1212060052). We thank G.D. Peng, J. Xu, and G.Z. Cui for Geo‐seq experimental (slice and library construction) assistance; X.J. Chen for providing RNA fluorescent reporter system; Z. Zhang for the operation of Incucyte® S3 Live‐Cell Analysis System; and the Guangzhou Branch of the Supercomputing Center of Chinese Academy of Sciences, and the Cloud Computing Center of Chinese Academy of Sciences for their support. The urine samples from 10 consent individuals were collected for this work. The donors have full knowledge of this work and signed the informed consent forms.
The EMBO Journal (2022) 41: e110928
Contributor Information
Shangtao Cao, Email: cao_shangtao@grmh-gdl.cn.
Duanqing Pei, Email: peiduanqing@westlake.edu.cn.
Data availability
The data supporting the conclusions of this article, the reprogramming process of UiPSM scRNA‐seq data including samples on days 0, 3, and 6 are available at GEO under accession GSE185136. The UiPSM on day 9 scRNA‐seq data is available at GEO under accession GSE181110. The UiPSM‐ and UiPSC‐derived tissue scRNA‐seq data are available at GEO under accession GSE189195. The bulk RNA‐seq data of the UiPSM on day 9 and UiPSM colonies including P1, P9, and P18 from three donors are available at GEO under accession GSE185039 and GSE186674. The UiSomitoid geo‐seq data and bulk RNA‐seq data are available at GEO under accession GSE181181.
References
- Anderson MJ, Magidson V, Kageyama R, Lewandoski M (2020) Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. Elife 9: e55608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre P, Song H, Kim W, Kispert A, Yang Y (2015) Wnt5a and Wnt11 regulate mammalian anterior‐posterior axis elongation. Development 142: 1516–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari L, Moris N, Girgin M, Turner DA, Baillie‐Johnson P, Cossy AC, Lutolf MP, Duboule D, Arias AM (2018) Multi‐axial self‐organization properties of mouse embryonic stem cells into gastruloids. Nature 562: 272–276 [DOI] [PubMed] [Google Scholar]
- Benazeraf B, Francois P, Baker RE, Denans N, Little CD, Pourquie O (2010) A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466: 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R (2003) Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev 17: 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Su YC, Yost HJ (2017) Maternal Gdf3 is an obligatory cofactor in nodal signaling for embryonic axis formation in zebrafish. Elife 6: e28534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR (2012) Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev Biol 371: 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray N, Wilson V (2002) Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development 129: 4855–4866 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F (2004) Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci USA 101: 7641–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Suo S, Tam PP, Han JJ, Peng G, Jing N (2017) Spatial transcriptomic analysis of cryosectioned tissue samples with Geo‐seq. Nat Protoc 12: 566–580 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang D, Su N, Bao B, Xie X, Zuo F, Yang L, Wang H, Jiang L, Lin Q et al (2019) Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol 37: 1287–1293 [DOI] [PubMed] [Google Scholar]
- Chhabra S, Liu L, Goh R, Kong X, Warmflash A (2019) Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self‐organized fate patterning in human gastruloids. PLoS Biol 17: e3000498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LF, Mamott D, Ni Z, Bacher R, Liu C, Swanson S, Kendziorski C, Stewart R, Thomson JA (2019) An in vitro human segmentation clock model derived from embryonic stem cells. Cell Rep 28: e2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date S, Sato T (2015) Mini‐gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol 31: 269–289 [DOI] [PubMed] [Google Scholar]
- Diaz‐Cuadros M, Wagner DE, Budjan C, Hubaud A, Tarazona OA, Donelly S, Michaut A, Al Tanoury Z, Yoshioka‐Kobayashi K, Niino Y et al (2020) In vitro characterization of the human segmentation clock. Nature 580: 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez Del Corral R, Morales AV (2017) The multiple roles of FGF signaling in the developing spinal cord. Front Cell Dev Biol 5: 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri S, Hayward P, Baillie‐Johnson P, Steventon BJ, Martinez Arias A (2019) An epiblast stem cell‐derived multipotent progenitor population for axial extension. Development 146: dev168187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt SJ (2018) Hox cluster genes and collinearities throughout the tree of animal life. Int J Dev Biol 62: 673–683 [DOI] [PubMed] [Google Scholar]
- Ghyselinck NB, Duester G (2019) Retinoic acid signaling pathways. Development 146: dev167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzetta A, Koska M, Rowton M, Sullivan KR, Jacobs‐Li J, Kweon J, Hidalgo H, Eckart H, Hoffmann AD, Back R et al (2020) Hedgehog‐FGF signaling axis patterns anterior mesoderm during gastrulation. Proc Natl Acad Sci USA 117: 15712–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead AM, Wright CV (2015) Disrupting Foxh1‐Groucho interaction reveals robustness of nodal‐based embryonic patterning. Mech Dev 136: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PPL, Elefanty AG, Robb L (2002) Mixl1is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development 129: 3597–3608 [DOI] [PubMed] [Google Scholar]
- Henrique D, Abranches E, Verrier L, Storey KG (2015) Neuromesodermal progenitors and the making of the spinal cord. Development 142: 2864–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WKW, Freem L, Zhao D, Painter KJ, Woolley TE, Gaffney EA, McGrew MJ, Tzika A, Milinkovitch MC, Schneider P et al (2019) Feather arrays are patterned by interacting signalling and cell density waves. PLoS Biol 17: e3000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins AP, Jauch R, Dyla M, Miranda‐Saavedra D (2014) glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high‐throughput sequencing data. Cell Regen 3: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasuja R, Voss N, Ge G, Hoffman GG, Lyman‐Gingerich J, Pelegri F, Greenspan DS (2006) bmp1 and mini fin are functionally redundant in regulating formation of the zebrafish dorsoventral axis. Mech Dev 123: 548–558 [DOI] [PubMed] [Google Scholar]
- Koch F, Scholze M, Wittler L, Schifferl D, Sudheer S, Grote P, Timmermann B, Macura K, Herrmann BG (2017) Antagonistic activities of Sox2 and brachyury control the fate choice of neuro‐mesodermal progenitors. Dev Cell 42: e517 [DOI] [PubMed] [Google Scholar]
- Kumar L, Futschik ME (2007) Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped‐read alignment with bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Buttitta LA, May NR, Kispert A, Fan CM (2000) SHH‐N upregulates Sfrp2 to mediate its competitive interaction with WNT1 and WNT4 in the somitic mesoderm. Development 127: 109–118 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA‐seq data with or without a reference genome. BMC Bioinformatics 12: 323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang L, Hou J, Shen Q, Chen Q, Wang X, Du J, Cai X, Shan Y, Zhang T et al (2016) Optimized approaches for generation of integration‐free iPSCs from human urine‐derived cells with small molecules and autologous feeder. Stem Cell Reports 6: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang Y, Song G, Zhang X, Gao S, Liu H (2019) HOX cluster‐embedded antisense long non‐coding RNAs in lung cancer. Cancer Lett 450: 14–21 [DOI] [PubMed] [Google Scholar]
- Li N, Liu J, Liu H, Wang S, Hu P, Zhou H, Xiao J, Liu C (2021) Altered BMP‐Smad4 signaling causes complete cleft palate by disturbing osteogenesis in palatal mesenchyme. J Mol Histol 52: 45–61 [DOI] [PubMed] [Google Scholar]
- Lin S, Baye LM, Westfall TA, Slusarski DC (2010) Wnt5b‐Ryk pathway provides directional signals to regulate gastrulation movement. J Cell Biol 190: 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CY, Nichane M et al (2014) Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14: 237–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecka KA, Dheekollu J, Deakyne JS, Wiedmer A, Ramirez UD, Lieberman PM, Messick TE (2019) Structural basis for cooperative binding of EBNA1 to the Epstein‐Barr virus dyad symmetry minimal origin of replication. J Virol 93: e00487‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M (2018) Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet 34: 209–217 [DOI] [PubMed] [Google Scholar]
- Mallo M, Alonso CR (2013) The regulation of Hox gene expression during animal development. Development 140: 3951–3963 [DOI] [PubMed] [Google Scholar]
- Martinez‐Bartolome M, Range RC (2019) A biphasic role of non‐canonical Wnt16 signaling during early anterior‐posterior patterning and morphogenesis of the sea urchin embryo. Development 146: dev168799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Hayashi H, Garcia‐Ojalvo J, Yoshioka‐Kobayashi K, Kageyama R, Yamanaka Y, Ikeya M, Toguchida J, Alev C, Ebisuya M (2020a) Species‐specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369: 1450–1455 [DOI] [PubMed] [Google Scholar]
- Matsuda M, Yamanaka Y, Uemura M, Osawa M, Saito MK, Nagahashi A, Nishio M, Guo L, Ikegawa S, Sakurai S et al (2020b) Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580: 124–129 [DOI] [PubMed] [Google Scholar]
- Matthews KRW, Wagner DS, Warmflash A (2021) Stem cell‐based models of embryos: the need for improved naming conventions. Stem Cell Reports 16: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Sherman A, Lillico SG, Ellard FM, Radcliffe PA, Gilhooley HJ, Mitrophanous KA, Cambray N, Wilson V, Sang H (2008) Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development 135: 2289–2299 [DOI] [PubMed] [Google Scholar]
- Moris N, Anlas K, van den Brink SC, Alemany A, Schroder J, Ghimire S, Balayo T, van Oudenaarden A, Martinez Arias A (2020) An in vitro model of early anteroposterior organization during human development. Nature 582: 410–415 [DOI] [PubMed] [Google Scholar]
- Nakaya MA, Biris K, Tsukiyama T, Jaime S, Rawls JA, Yamaguchi TP (2005) Wnt3a links left‐right determination with segmentation and anteroposterior axis elongation. Development 132: 5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Masamizu Y, Liu T, Nakayama R, Deng CX, Kageyama R (2007) The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell 13: 298–304 [DOI] [PubMed] [Google Scholar]
- Oginuma M, Niwa Y, Chapman DL, Saga Y (2008) Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development 135: 2555–2562 [DOI] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, Dudoit S (2011) GC‐content normalization for RNA‐seq data. BMC Bioinform 12. 10.1186/1471-2105-12-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Chal J, Lummertz da Rocha E, Han A, Yermalovich AV, Oginuma M, Schlaeger TM, Sousa P, Rodriguez A, Urbach A et al (2019) The lin28/let‐7 pathway regulates the mammalian caudal body axis elongation program. Dev Cell 48: 396–405.e3. 10.1016/j.devcel.2018.12.016 [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK et al (2014) Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 32: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Suzuki T (2020) How do signaling and transcription factors regulate both axis elongation and Hox gene expression along the anteroposterior axis? Dev Growth Differ 62: 363–375 [DOI] [PubMed] [Google Scholar]
- Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M et al (2019) Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol 37: 303–313 [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Duester G (2006) Retinoic‐acid signalling in node ectoderm and posterior neural plate directs left‐right patterning of somitic mesoderm. Nat Cell Biol 8: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single‐cell data. Cell 177: e1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara N, Kawakami H, Chen KQ, Anderson A, Yamashita Peterson M, Gong W, Shah P, Hayashi S, Nishinakamura R, Nakagawa Y et al (2019) Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos. Development 146: dev177659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell‐Badge R, Papaioannou VE, Kondoh H (2011) Tbx6‐dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470: 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson L, Muresan L, Steventon B (2021) The zebrafish presomitic mesoderm elongates through compaction‐extension. Cells Dev 168: 203748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueb B, Taeschler S (2006) Expression of FGFRL1, a novel fibroblast growth factor receptor, during embryonic development. Int J Mol Med 17: 617–620 [PubMed] [Google Scholar]
- van den Brink SC, Baillie‐Johnson P, Balayo T, Hadjantonakis AK, Nowotschin S, Turner DA, Martinez Arias A (2014) Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141: 4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Alemany A, van Batenburg V, Moris N, Blotenburg M, Vivie J, Baillie‐Johnson P, Nichols J, Sonnen KF, Martinez Arias A et al (2020) Single‐cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582: 405–409 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang L, Huang W, Su H, Xue Y, Su Z, Liao B, Wang H, Bao X, Qin D et al (2013) Generation of integration‐free neural progenitor cells from cells in human urine. Nat Methods 10: 84–89 [DOI] [PubMed] [Google Scholar]
- Wang L, Su Y, Huang C, Yin Y, Zhu J, Knupp A, Chu A, Tang Y (2019) FOXH1 is regulated by NANOG and LIN28 for early‐stage reprogramming. Sci Rep 9: 16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Olivera‐Martinez I, Storey KG (2009) Stem cells, signals and vertebrate body axis extension. Development 136: 1591–1604 [DOI] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC (2015) Dynamic pluripotent stem cell states and their applications. Cell Stem Cell 17: 509–525 [DOI] [PubMed] [Google Scholar]
- Xi H, Fujiwara W, Gonzalez K, Jan M, Liebscher S, Van Handel B, Schenke‐Layland K, Pyle AD (2017) In vivo human somitogenesis guides somite development from hPSCs. Cell Rep 18: 1573–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP (2001) Heads or tails: Wnts and anterior–posterior patterning. Curr Biol 11: R713–R724 [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP (1999) T (brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev 13: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoney A, Etoc F, Ruzo A, Carroll T, Metzger JJ, Martyn I, Li S, Kirst C, Siggia ED, Brivanlou AH (2018) WNT signaling memory is required for ACTIVIN to function as a morphogen in human gastruloids. Elife 7: e38279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga‐Otto K, Tian S, Stewart R, Slukvin II, Thomson JA (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, Reversade B, Kuroda H, Lyons KM, De Robertis EM (2005) Sirenomelia in Bmp7 and Tsg compound mutant mice: requirement for bmp signaling in the development of ventral posterior mesoderm. Development 132: 2489–2499 [DOI] [PubMed] [Google Scholar]
- Zhao X, Duester G (2009) Effect of retinoic acid signaling on Wnt/beta‐catenin and FGF signaling during body axis extension. Gene Expr Patterns 9: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, Wang Y, Zhang Y, Zhuang Q, Li Y et al (2012) Generation of human induced pluripotent stem cells from urine samples. Nat Protoc 7: 2080–2089 [DOI] [PubMed] [Google Scholar]


