Abstract
The unsuitable substitution ratio of fish meal by plant protein will reshape the intestinal microbial composition and intestine immunity. However, previous studies were mostly limited to investigating how different feed or probiotics characterized the microbial composition but ignored the biological interactions between bacteria and host physiology through secondary metabolites. Therefore, this study integrates the apparent indicators monitoring, 16S rDNA sequencing, and metabonomics to systematically investigate the effects of cottonseed protein concentrate (CPC) substitution of fish meal and Bacillus coagulans intervention on gut microbes, secondary metabolites, and intestinal immunity of Macrobrachium rosenbergii. Prawns were fed with three diets for 70 days: HF diets contained 25% fish meal, CPC in LF diets were replaced with 10% fish meal, and LF diets supplemented with 2 × 108 CFU/g diet B. coagulans were designated as BC diets. Results showed that CPC substitution induced a significant decrease in digestive enzyme activities (trypsin and lipase) and gut barrier protein PT-1 expression and a significant increase in γ-GT enzyme activity and inflammatory-related factors (Relish and Toll) expression. B. coagulans treatment mitigated the negative changes of the above indicators. Meanwhile, it significantly improved the expression levels of the barrier factor PT-1, the reparative cytokine IL-22, and Cu/Zn-SOD. CPC substitution resulted in a remarkable downregulated abundance of Firmicutes phyla, Flavobacterium spp., and Bacillus spp. B. coagulans treatment induced the callback of Firmicutes abundance and improved the relative abundance of Sphingomonas, Bacillus, and Ralstonia. Functional prediction indicated that CPC substitution resulted in elevated potential pathogenicity of microbial flora, and B. coagulans reduces the pathogenesis risk. Pearson’s correlation analysis established a significant positive correlation between differential genera (Sphingomonas, Bacillus, and Ralstonia) and secondary metabolites (including sphingosine, dehydrophytosphingosine, amino acid metabolites, etc.). Meanwhile, the latter were significantly associated with intestinal immunoregulation-related genes (Cu/Zn-SOD, IL-22, PT-1, Toll, and Relish). This study indicated that B. coagulans could mediate specific gut microbes and the combined action of multiple functional secondary metabolites to affect intestinal barrier function, digestion, and inflammation. Our study revealed the decisive role of gut microbes and derived secondary metabolites in the model of dietary composition-induced intestinal injury and probiotic treatment from a new perspective.
Keywords: Macrobrachium rosenbergii, cottonseed protein concentrate, bacillus coagulans, fish meal, intestinal microbial, secondary metabolites
Introduction
As a high-quality protein source, fish meal is widely used in aquafeeds, especially crustacean feeds, thanks to its good palatability and balanced nutritional composition (1). Due to the rapid development of aquaculture and the depletion of wild capture fisheries resources, the feed cost is gradually increasing, and the sustainable development of aquaculture is being seriously challenged (2). Therefore, one of the solutions is using more widely available and inexpensive plant proteins to partially or entirely replace fish meal in aquafeeds. In recent years, aquatic nutritionists have been conducting a series of studies on substituting fish meal with plant proteins (3).
However, with the in-depth study, plant protein substitution problems have gradually come to the fore. The excessive substitution of plant protein for fish meal leads to dysbacteriosis, impaired intestinal barrier function, reduced digestion and absorption of nutrients, and intestinal inflammation in aquatic animals. These side effects are attributed to the inherent anti-nutritional factors and unbalanced nutritional composition, which have received immense attention across aquaculture nutrition. For example, Miao et al. found that different dietary soybean meal substitutions for defatted fish meal resulted in a decrease in the relative abundance of beneficial intestinal bacteria and an increase in the abundance of conditionally pathogenic bacteria (4).
Persistent elevation of the substitutions proportion led to a significant upregulation of intestinal mucosal inflammatory factor expression (4). Zhang et al. reported that a 20% soybean meal substitution (basal fish meal level of 50%) induced intestinal mucosal barrier injury, downregulated immunization parameters, and enteritis of juvenile grouper (5). With the upgrading of the processing industry, the palatability and nutritional value of cottonseed protein concentrate obtained by low-temperature extraction, dephenolization, and moderate desugarization have significantly improved. Recent reports indicated that cottonseed protein concentrate could replace fish meal and soybean protein with a higher level, which is currently a preferable alternative (6). However, high levels of cottonseed protein concentrate (CPC) substitution will still cause oxidative damage and intestinal barrier damage in aquatic animals. Fu et al. (7) reported that substituting low-gossypol cottonseed meal for more than 20% fish meal gradually aggravated the intestinal barrier function damage of juvenile golden pompano (Trachinotus ovatus). The phenotypes appeared as a suppressed expression of tight junction and antioxidant-related genes and an intensified inflammation response (7).
Recently, probiotic bacteria alleviating the adverse effects of aquatic animals induced by plant protein substitution is arousing great interest. Studies on zebrafish (8) and bullfrog (Lithobates catesbeianus) (9) have shown that probiotics can improve the growth inhibition, metabolic disorder, intestinal microdysbiosis, and intestinal inflammation of aquatic animals caused by fish meal replacement. As such, increasingly comprehensive probiotic solutions for fish meal substitution to improve the host’s gut health and stress resistance are being proposed in the aquaculture industry. The probiotic Bacillus coagulans is a Gram-positive lactic acid-producing, spore-forming bacterial species. Recent studies have shown that B. coagulans GBI-30, 6086 could enhance plant protein digestion, absorption, and amino acid bioavailability (10, 11). Moreover, B. coagulans could promote the intestinal health of aquatic animals by strengthening the gut barriers, relieving oxidative stress, and modulating the intestinal microflora (12). Given this, this study chose B. coagulans as a regulator for mitigating the side effects of plant protein replacement fish meal.
Notably, recent medical studies have provided us with a new perspective: the regulation essence of host metabolic diseases and inflammatory diseases by probiotics, prebiotics, medicine, and other regulators is via the modulation of secondary metabolites, such as secondary bile acids (13), short-chain fatty acids (14), and branched-chain amino acids (15). These microbiota-derived metabolites signal to host immune and metabolism organs by ligand–receptor interaction, enabling the implementation of microbe–host communication (16). However, previous studies were mostly limited to investigating how different probiotics or feed types characterized intestinal microbial composition. Alternatively, it only evaluated the variations of growth performance and host physiological index but ignored the biological interactions between intestinal bacteria and host, i.e., the functions of bacterial exo-metabolites.
Therefore, our study evaluated the effects of B. coagulans on intestinal microbial composition and functional secondary metabolites related to the gut health of Macrobrachium rosenbergii based on dietary fish meal substitution treatment. Above all, we provided new complementary insights combining 16S flora sequencing profiling with metabolomics datasets of intestinal contents, aimed to reveal their interactions with host gut immune barrier function and interpret the mechanism of probiotics in alleviating the adverse effects of plant protein substitution.
Materials and methods
Ethics statement and prawn management
The use of prawns in this study was approved by the Animal Care and Use Committee of the Committee on the Ethics of Animal Experiments of Freshwater Fisheries Research Center. M. rosenbergii was obtained from the Freshwater Fisheries Research Center breeding base at the Chinese Academy of Fishery Science. After 15 days of domestication, 600 healthy prawns with a uniform initial weight (0.28 ± 0.02 g) were randomly assigned to three treatment groups with five replicates in each treatment and 40 individuals per replicate bucket (150-cm inner diameter, 45-cm water depth). The three experimental diets were fed at a rate of 5%–10% of body weight at 9:00, 13:00, and 18:00 daily. The feeding amount was observed and timely adjusted based on the amount of residual bait after 30 min, and the residual bait and feces were sucked out by siphoning prior to feeding. Prawns’ mortality was recorded daily. Daily continuous aeration was performed to ensure adequate dissolved oxygen (≥6 mg/ml). Water quality was measured once a week, and the water temperature was 28°C–32°C, nitrite ≤ 0.02 mg/L, ammonia ≤ 0.2 mg/L, pH = 7.0–8.5, and the trial period was 70 days.
Experimental design, diet preparation, and growth assessment
The group containing 25% fish meal as the positive control group was designated as the HF group. The negative control group replaced 10% fish meal with CPC and was designated the LF group. Subsequently, B. coagulans JSSW-LA-07-1 was added to the LF diet at 2 × 108 CFU/g diet under sterilized conditions and was designated as the BC group. The additional amount of B. coagulans JSSW-LA-07-1 was referred to in previous reports (17, 18). The strain was kept by the China General Microbiological Culture Collection Center (CGMCC No. 10182). The ingredients and proximal composition of the control groups are shown in Table 1 . All raw materials were finely ground and passed through a 60-mesh sieve. Premix additives and raw materials were weighed accurately according to the formula, mixed step by step, and then squeezed into strips by a twin-screw extruder with a 1-mm aperture.
Table 1.
Ingredients and proximate analysis of HF diets and LF diets.
| Ingredients | HF | LF |
|---|---|---|
| Concentration (% dry matter) | ||
| Fish meal | 25 | 15 |
| Cottonseed protein concentrate | 10 | |
| Chellocken meal | 3 | 3 |
| Poultry by-product powder | 2 | 2 |
| Squid paste | 3 | 3 |
| Shrimp meal | 4 | 4 |
| Soybean meal | 19 | 19 |
| Rapeseed meal | 15.5 | 15.5 |
| α-Starch | 20.5 | 19.9 |
| Fish oil:soybean oil | 2 | 2.6 |
| Lecithin powder | 1 | 1 |
| Cholesterol | 0.3 | 0.3 |
| Ecdysterone | 0.01 | 0.01 |
| Choline chloride | 1 | 1 |
| Premix a | 1 | 1 |
| Bentonite | 0.69 | 0.69 |
| Monocalcium phosphate | 2 | 2 |
| Proximate analysis | ||
| Moisture | 10.46 | 10.61 |
| Crude protein | 40.14 | 40.3 |
| Ether extracts | 5.98 | 5.42 |
| Ash | 11.19 | 12.1 |
Premix supplied the following vitamins (IU or mg/kg) and minerals (g/kg): vitamin A, 25,000 IU; vitamin D3, 20,000 IU; vitamin E, 200 mg; vitamin K3, 20 mg; thiamin, 40 mg; riboflavin, 50 mg; calcium pantothenate, 100 mg; pyridoxine HCl, 40 mg; cyanocobalamin, 0.2 mg; biotin, 6 mg; folic acid, 20 mg; niacin, 200 mg; inositol, 1,000 mg; vitamin C, 2,000 mg; choline, 2,000 mg; calcium biphosphate, 20 g; sodium chloride, 2.6 g; potassium chloride, 5 g; magnesium sulfate, 2 g; ferrous sulfate, 0.9 g; zinc sulfate, 0.06 g; cupric sulfate, 0.02 g; manganese sulfate, 0.03 g; sodium selenate, 0.02 g; cobalt chloride, 0.05 g; potassium iodide, 0.004 g.
After a 70-day feeding trial, the survival rate and the weight of prawns in each group were recorded. The weight gain rate, specific growth rate, and feed conversion ratio were calculated in the following formulas:
Sample collection, RNA extraction, and DNA extraction
Hemolymph was collected using sterile 1-ml syringes from the pericardial cavity, mixing 1:1 with precooling anticoagulant solution, and immediately centrifuged at 8,000 r/min, 4°C for 10 min. The supernatant was collected and stored at −20°C before measuring antioxidant indicators. Part of the intestine was dissected aseptically and stored at −20°C for subsequent biochemical analysis. The remaining intestine was dissected aseptically, quick-frozen in liquid nitrogen, and stored at −80°C for RNA isolation and PCR analysis. The total RNA of intestine tissues was isolated using TRIzol (TaKaRa Biomedical Technology Co., Ltd., Beijing, China). RNA integrity was tested using 1% agarose gel electrophoresis. RNA concentration and purity were determined based on OD 260/280 readings (ratio > 1.8) using a NanoDrop ND-1000 UV Spectrophotometer (NanoDrop Technologies Co., Ltd., Wilmington, DE, USA). In addition, the adequate intestinal contents were collected aseptically and stored in a −80°C freezer for subsequent 16S rDNA sequencing and metabolomics analysis. Microbial DNA was extracted from the samples following the manufacturer’s protocol using the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA). The integrity of the extracted DNA was measured using a NanoDrop ND2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and 1% agarose gel electrophoresis. Concentrations of isolated DNA were measured using a Quant-iT PicoGreen dsDNA Assay kit (Invitrogen, Carlsbad, CA, USA) and a fluorometer and diluted to 20 ng/μl. Samples were stored at −20°C until sequencing.
Detection of biochemical indicators in hemolymph and intestine
The content of malondialdehyde (MDA) (A003-1-2) and the antioxidant enzymatic activities of catalase (CAT) (A00-1-1) and total superoxide dismutase (T-SOD) (A001-3-2) in hemolymph were detected and quantitated by using the commercial kits purchased from Nanjing Jiancheng Institute of Bioengineering (Nanjing, China), according to the manufacturer’s instructions. Approximately 0.1 g of intestinal tissue was minced and homogenized in ice-cold 0.86% stroke-physiological saline solution (w/v, 1:9) using an Ultra-Turrax homogenizer (Tekmar Co., Cincinnati, OH, USA) and centrifuged at 4,000 r/min at 4°C for 10 min to obtain the supernatant for further analysis. The digestive enzymes and brush border enzyme indicators including trypsin, lipase, amylase, and γ-GT were determined using commercial assay kits (A080-2-2, A054-2-1, C016-1-1, and C017-2-1, Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s protocols.
Quantitative real-time PCR
The extracted total RNA from the intestine was synthesized using a Perfect Real Time SYBR Prime Script TM RT Reagent Kit (TaKaRa Biotechnology, Dalian, China) as per the manufacturer’s instructions. The reaction parameters of RTqPCR are as follows: pre-run at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 15 s, and a 60°C annealing step for 34 s. The reaction mixture comprised 2 µl of cDNA, 0.4 µl of forward primer, 0.4 µl of reverse primer, 10 µl of SYBR Premix ExTaqTM (TaKaRa, Dalian, Liaoning, China), 0.4 µl of ROX Reference Dye (TaKaRa, Dalian, Liaoning, China), and 6.8 µl of double-distilled water. β-Actin gene was selected as a reference gene (19), and each sample was tested in triplicate. Primers were designed online (NCBI, Bethesda, MA, USA) ( Table 2 ). Notably, the CDS sequences used in the primer design of some genes were obtained from our laboratory’s database of the hepatopancreas transcriptome sequencing of M. rosenbergii. The results were calculated by the 2−ΔΔCt method, and the gene-specific primers were synthesized by Shanghai Generay Biotech Co., Ltd. (Shanghai, China).
Table 2.
Primer sequences for real-time PCR.
| Gene | Primers | Sequence 5′–3′ | Product length (bp) | Reference |
|---|---|---|---|---|
| Toll | F | TTCGTGACTTGTCGGCTCTC | 145 | KX610955.1 |
| R | GCAGTTGTTGAAGGCATCGG | |||
| Relish | F | GATGAGCCTTCAGTGCCAGA | 149 | KR827675.1 |
| R | CCAGGTGACGCCATGTATCA | |||
| IL-22 | F | ACGAGCTGCGATCCAGTAAG | 101 | Cluster-21039.10593 |
| R | GCAACGCACTGCTCCTTAAC | |||
| Cu-Zn-SOD | F | AGAGCAGTTGTAGGCCGAAG | 116 | EU077527.1 |
| R | GTGCAGCAAGCCAATCTAGC | |||
| PT-1 | F | TTGCTTGGTCAGTCTCCTGC | 106 | Cluster-1871.0 |
| R | CTCTAAGGTCTGGGCCTGTC | |||
| β-Actin | F | TCCGTAAGGACCTGTATGCC | 101 | AY651918.2 |
| R | TCGGGAGGTGCGATGATTTT |
16S rRNA sequencing of intestinal microorganisms
The target fragment library was prepared by PCR amplification of the V4–V5 region of the 16S rRNA gene. The universal primers 515 F (5′-GTGCCAGCMGCCGCGG-3′) and 907 R (5′-CCGTCAATTCMTTTRAGTTT-3′) were used to amplify the V4–V5 region of the 16S rRNA gene. PCR products were detected by 2% agarose gel electrophoresis, and target fragments were collected using the AxyPrep DNA Gel Extraction Kit. Amplicons of every sample were combined, purified, and quantified. Library construction and double-ended sequencing were conducted using the Illumina HiSeq platform to produce 2 × 250-bp paired-end reads. Raw fastq files were de-multiplexed and quality-filtered by removing the reads’ barcode and linker sequences and merging with the paired-end reads into a longer fragment. Then, reads that were truncated at any site from the beginning of the window to the 3′ terminus and received an average quality score<20 over a 6-bp sliding window, and truncated reads<100 bp were discarded. Mismatching primer sequences or ambiguous bases (Ns) exceeding 5% were removed from the downstream analyses, and reads that could not be assembled were discarded. FLASH (fast length adjustment of short reads, v1.2.8, http://ccb.jhu.edu/software/FLASH) was used for spliced paired-end sequences, and chimeric sequences were removed using VSEARCH (https://github.com/torognes/vsearch, v2.3.4). Finally, the operational taxonomic units (OTUs) were generated after denoising. Quality-filtered reads were clustered into OTUs following the criterion of more than or equal to 97% similarity. For detailed steps and parameters of 16S rRNA, refer to our previous description (20).
Metabolomics analysis of intestinal contents
Intestinal contents samples were prepared from the prawns of the LF and BC groups. Each sample measuring 150 mg was accurately weighed and placed into the EP tube, added with 800 μl of pre-cooled 50% methanol–water (1:1, V:V), and grinded thoroughly in a tissue grinder (50 Hz, 5 min). After three centrifugations at 4°C, 2,500 g, 550 μl of supernatant was extracted and dried in the freeze dryer. Then, 600 μl methanol–water (1:9, V:V) was added to re-dissolve and vortex the residue and perform ultrasonic treatment, and finally, the supernatant was obtained. Meanwhile, 50 μl of supernatant from each sample was mixed to be the quality control (QC) sample, and all the specimens were stored at −80°C until liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. The ACQUITY UPLC BEH C18 column (100 mm * 2.1 mm, 1.7 μm, Waters, Cheshire, UK, column temperature 50°C) was used for chromatographic separation, and the injection volume was 10 μl. The high-resolution tandem mass spectrometry Xevo G2-XS QTOF (Waters, UK) was used to collect the small molecules eluted from the column, under the positive (ES+) and negative ion modes (ES−). The raw data were imported into commercial software Progenesis QI (version 3.0.3) for peak alignment, peak extraction, and peak identification and data correction, and the mass charge ratio, retention time, and ion area of metabolites were obtained. Then, the metabolomics R software package-Metax was used to remove low-quality ions. The univariate and multivariate analyses were used to identify the differential metabolites with different abundances under different treatment conditions. The variable importance in projection (VIP) was calculated to assist in the screening of metabolic markers. This study used multivariate analysis of VIP values of the first two principal components of the partial least squares discriminant analysis (PLS-DA) model, combined with univariate analysis of fold change and p-value to screen the differential ions. The differential ions should simultaneously satisfy the following three conditions: 1) VIP ≥ 1, (2) fold change ≥1.2 or ≤0.8333, and 3) p-value< 0.05. The identification databases of metabolites included the Kyoto Encyclopedia of Genes and Genomes (KEGG), Human Metabolome Database (HMDB), and LIPID MAPS commercial databases. The differential metabolites were identified and screened out based on the fragment score, isotope similarity, mass error, and the pathway annotation results of the differential ions. Finally, the KEGG pathway analysis of different metabolites was performed. For detailed steps and parameters of metabolomics, refer to our previous description (21).
Statistical analysis
The differences in growth performance parameters, biochemical indicators, and gene expression parameters were analyzed using one-way ANOVA and Tukey’s honestly significant difference (Tukey’s HSD) (SPSS 22.0 software, Chicago, IL, USA). Statistically significant was considered when p< 0.05. The above data were expressed as means ± SEMs, and before statistical analysis, all data were tested for the normality of distribution and homogeneity of variances with Levene’s test. The alpha diversity was calculated using QIIME software, including Chao1, observed species, Shannon index, and Simpson index. An independent-samples t-test was performed for all diversity indices and presented as mean ± standard deviation (SD). Statistically significant was considered when p< 0.05. The R package drew the principal coordinates analysis (PCoA) diagram. The microbial community and relative abundance values analysis were conducted using STAMP software. Mean differences were considered significant at p< 0.05. Furthermore, intestinal differential metabolite clusters were conducted using a Pearson’s correlation test with differential microbial genera abundance and intestinal gene richness. Tables were generated using Excel 2016, while figures were made using GraphPad Prism 8.
Results
Growth performance indicator determination
As shown in Table 3 , the levels of growth performance indicators were not significantly different among the three groups; however, WGR, SGR, and SR clearly presented a trend of decreasing followed by increasing, and FCR showed contrary trends.
Table 3.
Effect of cottonseed protein concentrate as a substitute for fish meal and Bacillus coagulans supplementation on the growth performance of Macrobrachium rosenbergii.
| Group | FW (g) | WGR (%) | SGR (%/day) | FCR | SR (%) |
|---|---|---|---|---|---|
| HF | 4.22 ± 0.28 | 1,521.85 ± 71.92 | 3.98 ± 0.06 | 1.63 ± 0.12 | 84.17 ± 3.63 |
| LF | 4.29 ± 0.16 | 1,489.40 ± 37.13 | 3.95 ± 0.03 | 1.69 ± 0.04 | 80.83 ± 1.67 |
| LF+BC | 4.93 ± 0.47 | 1,582.71 ± 134.00 | 4.02 ± 0.11 | 1.42 ± 0.18 | 86.67 ± 4.17 |
FW, final weight; WGR, weight gain rate; SGR, specific growth rate; FCR, feed conversion ratio; SR, survival rate.
Analysis of hemolymph antioxidant indicators
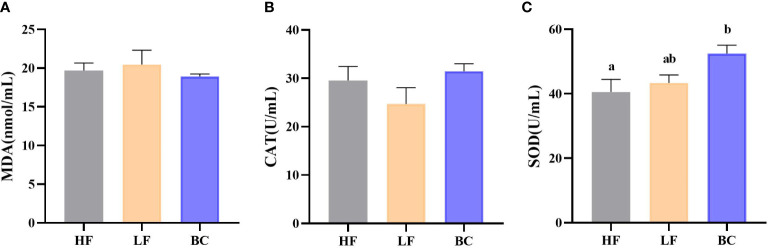
Figure 1 shows no significant difference in hemolymph MDA contents and CAT levels of prawns among these three groups. The hemolymph SOD activities of prawns were significantly increased in the BC group compared with the HF group.
Figure 1.
(A–C) Hemolymph antioxidant indicators. (A) Hemolymph MDA content; (B) Hemolymph CAT activity; (C) Hemolymph SOD activity.
Intestinal digestive enzymes and brush border enzyme activity determination
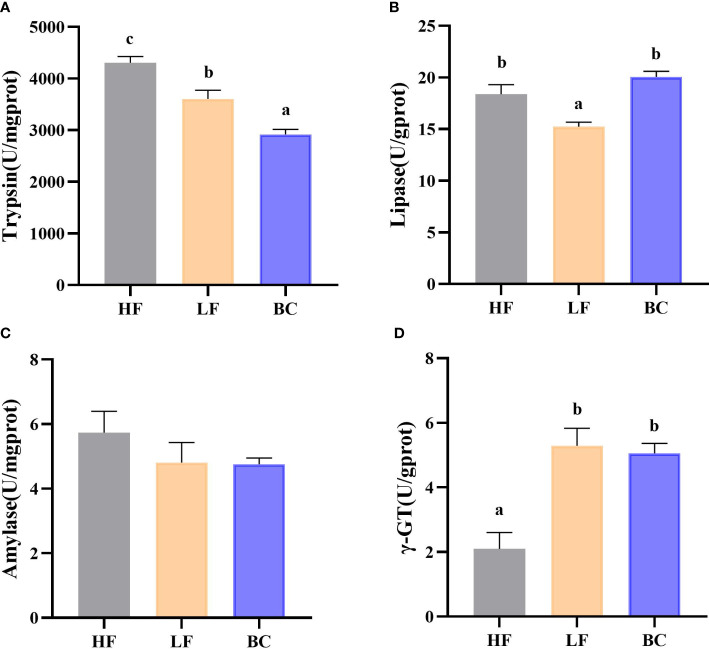
As shown in Figure 2 , the trypsin activities of prawns were significantly decreased in the LF group compared with the HF group, and the lowest level was found in the BC group. No significant difference in lipase activity was found between the HF and BC groups; the activity in LF was significantly decreased compared with these two groups. There was no significant difference in amylase activities among these three groups. The γ-GT activities of prawns were considerably increased in the LF and BC groups compared with the HF group.
Figure 2.
(A–D) Intestinal digestive enzymes and brush border enzyme activity. (A) Intestinal trypsin activities; (B) Intestinal lipase activities; (C) Intestinal amylase activities; (D) Intestinal γ-GT activities.
Analysis of intestinal gene expression profile
As shown in Figure 3 , the Toll and Relish expression levels of prawns were significantly upregulated in the LF group compared with the HF group. BC intervention downregulated the Toll and Relish expression levels compared to the LF group. BC intervention significantly downregulated the expression of Toll to a level close to the HF group; however, the Relish expression level in the BC group was significantly higher than in the HF group. The IL-22 and Cu/Zn-SOD expression levels of prawns were significantly upregulated in the BC group than the HF and LF groups, and no significant difference was found between the HF and LF groups. The PT-1 expression in the BC group was significantly upregulated compared with the LF group, and there was no significant difference between the HF and LF groups.
Figure 3.
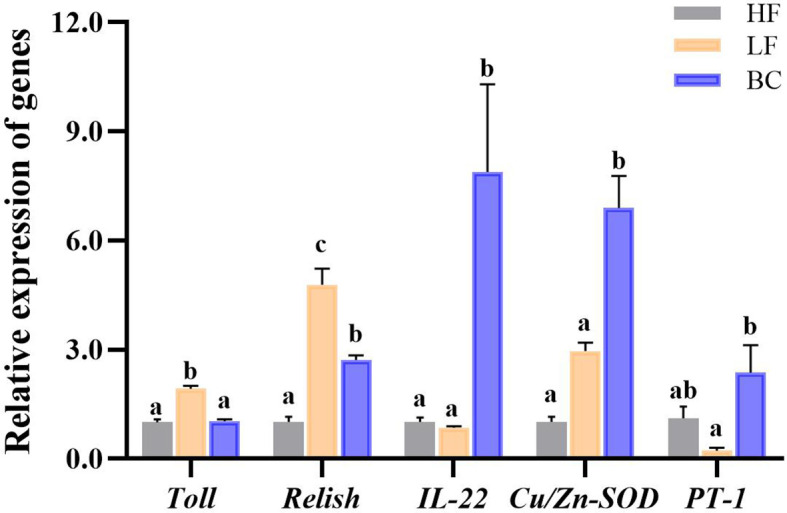
Intestinal gene expression profile. Different lowercase letters indicate significant differences (P < 0.05) between the three groups.
Community diversity analysis of prawns’ intestinal microflora
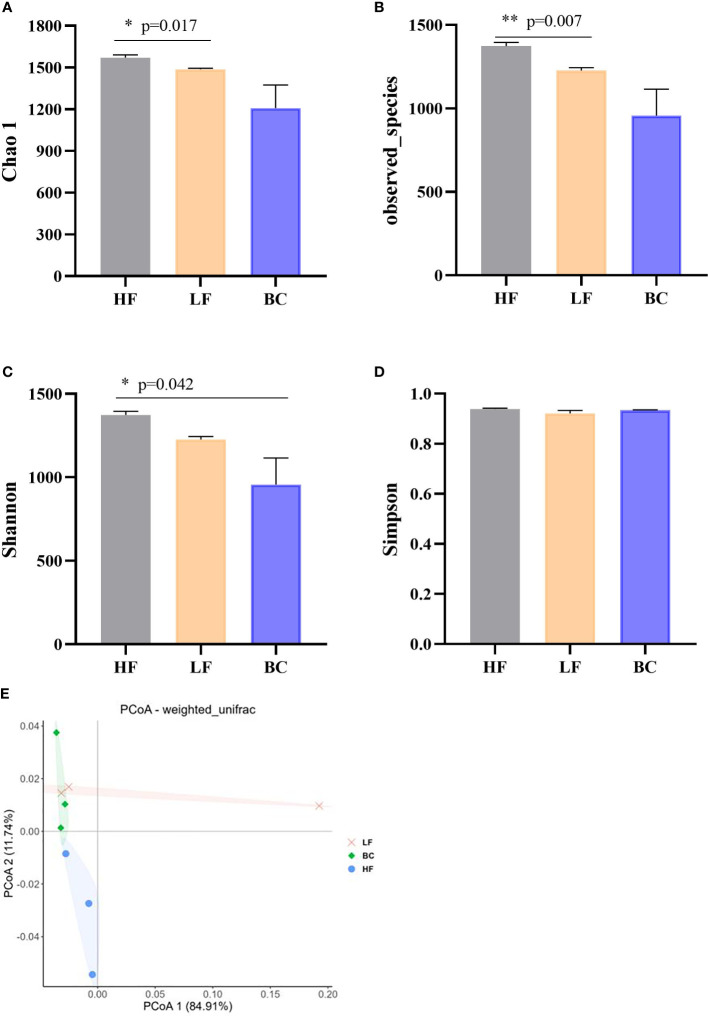
This study obtained 2,682 OTUs from all samples. The Chao1, observed species, Shannon, and Simpson indices were calculated to evaluate the microbial community richness and diversity of prawns’ intestines. As shown in Figure 4 , the Chao 1 and observed_species index of prawns’ intestinal microflora in HF were significantly higher than in LF prawns. There was no significant difference in Chao 1 and observed_species index between the BC group and the other two groups. The Shannon index of prawns’ intestinal microflora in the BC group was significantly lower than in the HF group. No significant difference was found in the Simpson index among these three groups. PCoA of bacterial communities of M. rosenbergii fed with three different diets showed that the LF group was distant from the HF group samples. In contrast, the BC group sample cluster intersected the LF and HF groups, shortening the distance from the HF group samples.
Figure 4.
(A–E) Community diversity analysis and principal coordinates analysis (PCoA) of bacterial communities of Macrobrachium rosenbergii fed with different diets. * P < 0.05, ** P < 0.01.
Gut microbiota function prediction
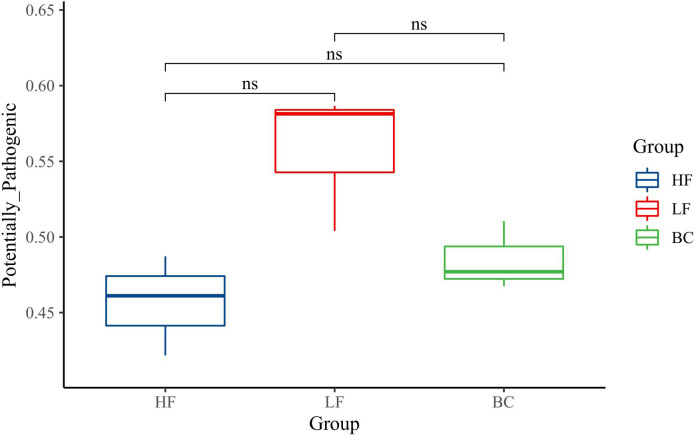
The predicted_phenotypes of potentially_pathogenic by BugBase software are shown in Figure 5 . The potentially_pathogenic of prawns in the LF group was higher than in the HF group, and the BC group’s predicted_phenotypes were decreased compared with the LF group. There was no significant difference among these three groups.
Figure 5.
Gut microbiota function prediction. ns, P > 0.05.
Microbial community structure of Macrobrachium rosenbergii treated with different diets
As shown in Figure 6 , four predominant phyla (mean relative abundance >1%) were detected in M. rosenbergii samples, including Proteobacteria (81.33%), Firmicutes (11.84%), Tenericutes (3.85%), and Bacteroidetes (1.11%). Compared with the HF group, the mean relative abundance of Proteobacteria, Firmicutes and Bacteroidetes decreased in the LF group, while the Tenericutes increased. B. coagulans treatment adjusted the relative abundance of these changed genera to a level similar to that of the HF group, except for Bacteroidetes. At the genus level, compared with the HF group, the main differential bacteria in the LF group were Flavobacterium, Bacillus, Muricauda, Nocardioides, Hyphomicrobium, Nitrospira, and Acinetobacter, all of which showed decreased abundance. Compared with the LF group, the different main bacteria in the BC group were Sphingomonas, Bacillus, Ralstonia, Demequina, and Sphingopyxis, of which the abundance of the former was significantly upregulated, and the latter two were significantly decreased. Compared with the BC group, a higher abundance of Nocardioides, Hyphomicrobium, Sphingopyxis, Nitrospira, and Acinetobacter was found in the HF group.
Figure 6.
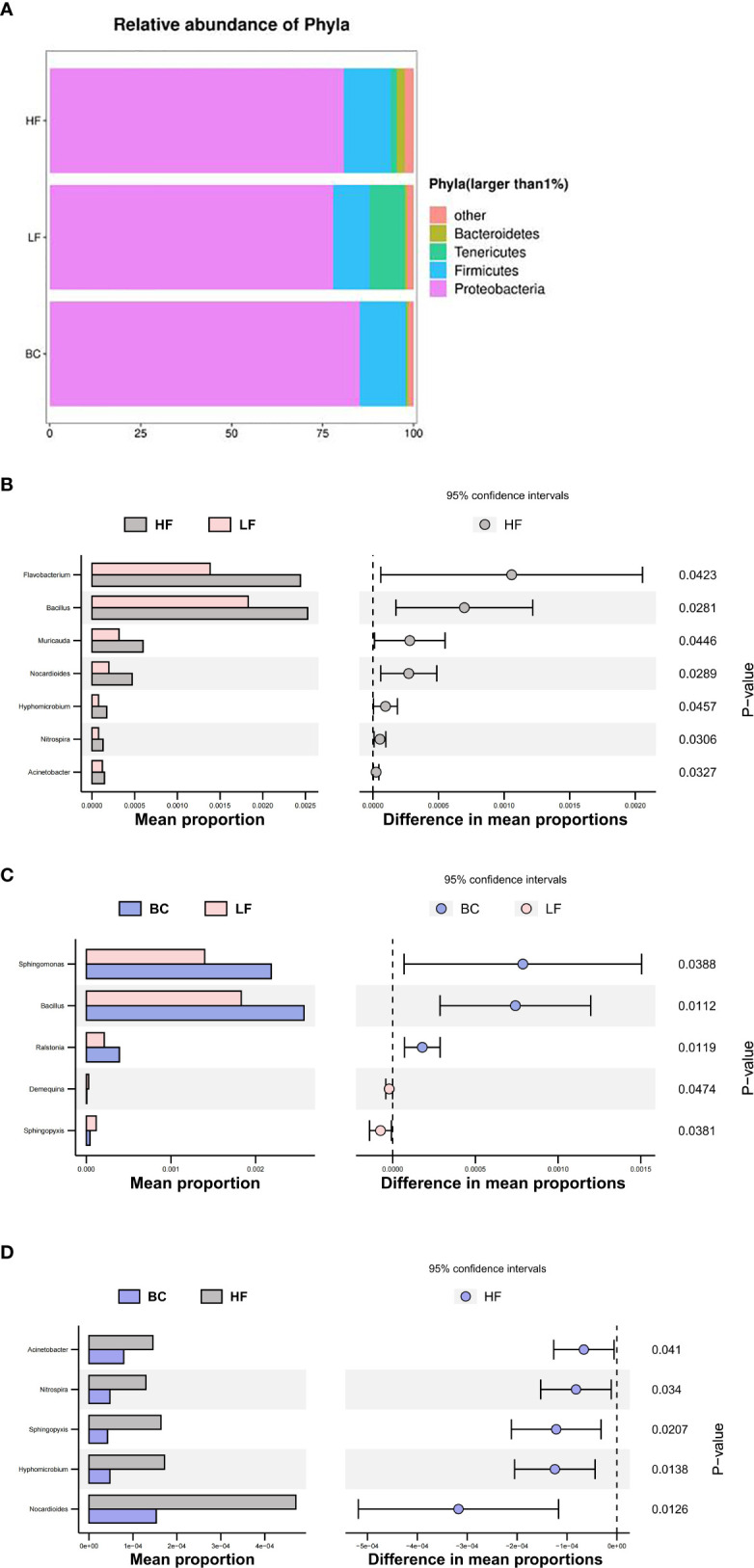
(A–D) Microbial community structure of Macrobrachium rosenbergii treated with different diets.
Analysis of the differential metabolites in fecal between LF and BC groups
The differential metabolites of intestinal flora between the LF and BC groups are listed in Table 4 . Among them, 89 metabolites were significantly upregulated, and one metabolite was significantly downregulated in the BC group. The differential metabolites mainly included amino acids and amino acid derivatives, sphingolipids and other lipids, aromatic compounds, energy metabolism-related metabolites, secondary metabolism of plants, nucleotides, and nucleotide derivatives.
Table 4.
The differential metabolites in fecal between LF and BC groups.
| Metabolites | rt | Mass | MeanLF | MeanBC | Trend | VIP | p-Value | FC |
|---|---|---|---|---|---|---|---|---|
| Amino acids and amino acid derivatives | ||||||||
| l-Serine | 389.699 | 106.050 | 1.437 | 3.838 | Up | 1.235 | 0.047 | 2.671 |
| l-Lysine | 532.865 | 147.112 | 3.324 | 19.025 | Up | 1.318 | 0.006 | 5.723 |
| beta-Alanine | 359.246 | 90.055 | 10.680 | 24.322 | Up | 1.243 | 0.024 | 2.277 |
| d-Ornithine | 547.123 | 133.097 | 0.386 | 1.065 | Up | 1.229 | 0.036 | 2.760 |
| d-Proline | 514.857 | 116.070 | 1.293 | 4.484 | Up | 1.316 | 0.010 | 3.467 |
| Tryptophan | 0.876 | 205.096 | 4.911 | 16.538 | Up | 1.242 | 0.018 | 1.717 |
| 5-Hydroxy-l-tryptophan | 297.080 | 221.091 | 0.051 | 0.190 | Up | 1.217 | 0.039 | 3.701 |
| l-Norleucine | 279.448 | 130.086 | 255.886 | 427.238 | Up | 1.283 | 0.045 | 1.670 |
| l-Valine | 313.992 | 118.086 | 24.588 | 58.664 | Up | 1.250 | 0.037 | 2.386 |
| l-Phenylalanine | 273.762 | 166.085 | 35.455 | 102.182 | Up | 1.275 | 0.029 | 2.882 |
| l-Tyrosine | 316.869 | 182.080 | 8.767 | 33.059 | Up | 1.225 | 0.027 | 3.771 |
| Prolylglycine | 311.634 | 173.091 | 0.045 | 0.160 | Up | 1.236 | 0.019 | 3.531 |
| l-Glutamic acid | 411.255 | 148.059 | 10.094 | 24.526 | Up | 1.204 | 0.035 | 2.430 |
| l-Asparagine | 389.770 | 133.060 | 3.387 | 9.566 | Up | 1.228 | 0.029 | 2.825 |
| Argininosuccinic acid | 484.113 | 291.127 | 0.052 | 0.179 | Up | 1.230 | 0.028 | 3.430 |
| Glutamyllysine | 506.193 | 276.153 | 0.014 | 0.089 | Up | 1.331 | 0.001 | 6.526 |
| Gamma-glutamyl ornithine | 465.060 | 262.138 | 0.116 | 0.419 | Up | 1.316 | 0.005 | 3.601 |
| Lysyl-Glycine | 470.018 | 204.133 | 0.543 | 1.578 | Up | 1.251 | 0.021 | 2.906 |
| Leucyl-Glycine | 120.022 | 189.122 | 1.042 | 2.057 | Up | 1.174 | 0.043 | 1.974 |
| Lysyl-Arginine | 572.540 | 303.211 | 0.080 | 0.462 | Up | 1.332 | 0.003 | 5.748 |
| Lysyl-Aspartate | 482.117 | 262.138 | 0.022 | 0.093 | Up | 1.276 | 0.028 | 4.164 |
| Aspartyl-Arginine | 456.445 | 290.143 | 0.254 | 0.816 | Up | 1.282 | 0.021 | 3.215 |
| Lysyl-Lysine | 574.436 | 275.206 | 0.043 | 0.237 | Up | 1.344 | 0.000 | 5.507 |
| Prolyl-Lysine | 485.670 | 244.164 | 0.412 | 1.618 | Up | 1.247 | 0.020 | 3.924 |
| Hydroxyprolyl-Arginine | 380.695 | 288.165 | 0.094 | 0.160 | Up | 1.171 | 0.048 | 1.706 |
| N 6-Acetyl-l-lysine | 403.155 | 189.122 | 0.108 | 0.230 | Up | 1.195 | 0.036 | 2.125 |
| Epsilon-(gamma-glutamyl)-lysine | 342.282 | 114.091 | 0.426 | 0.164 | down | 1.294 | 0.002 | 0.385 |
| Vinylacetylglycine | 445.764 | 144.065 | 0.948 | 1.564 | Up | 1.215 | 0.035 | 1.650 |
| N-Acetyldopamine | 389.971 | 196.096 | 0.052 | 0.139 | Up | 1.206 | 0.035 | 2.658 |
| 5-Aminopentanal | 268.421 | 102.091 | 0.855 | 1.795 | Up | 1.146 | 0.050 | 2.100 |
| 5-Aminopentanoic acid | 285.672 | 118.086 | 153.482 | 232.481 | Up | 1.193 | 0.037 | 1.515 |
| (3S,5S)-3,5-Diaminohexanoate | 358.914 | 147.112 | 0.082 | 0.127 | Up | 1.221 | 0.031 | 1.543 |
| Sphingolipids | ||||||||
| Sphinganine | 126.829 | 302.303 | 0.150 | 0.483 | Up | 1.245 | 0.030 | 3.213 |
| Dehydrophytosphingosine | 150.057 | 316.283 | 2.173 | 4.968 | Up | 1.266 | 0.018 | 2.286 |
| Phospholipids | ||||||||
| PC(15:0/15:0) | 165.938 | 706.534 | 0.876 | 2.051 | Up | 1.219 | 0.044 | 2.341 |
| PC(16:0/16:0) | 143.618 | 734.567 | 0.113 | 0.247 | Up | 1.166 | 0.049 | 2.182 |
| PC(16:1(9Z)/14:0) | 165.938 | 704.517 | 0.871 | 1.784 | Up | 1.224 | 0.027 | 2.048 |
| PC(16:1(9Z)/15:0) | 165.148 | 718.532 | 0.430 | 1.172 | Up | 1.282 | 0.006 | 2.726 |
| PC(18:1(11Z)/14:0) | 163.671 | 732.548 | 7.850 | 16.581 | Up | 1.262 | 0.017 | 2.112 |
| PC(16:1(9Z)/16:1(9Z)) | 164.080 | 730.535 | 2.463 | 5.134 | Up | 1.203 | 0.042 | 2.085 |
| PC(18:2(9Z,12Z)/18:0) | 158.697 | 786.596 | 11.337 | 23.771 | Up | 1.263 | 0.008 | 2.097 |
| PC(15:0/18:2(9Z,12Z)) | 163.281 | 744.549 | 0.774 | 1.979 | Up | 1.248 | 0.015 | 2.557 |
| PC(24:1(15Z)/14:1(9Z)) | 61.254 | 814.623 | 0.138 | 0.500 | Up | 1.257 | 0.012 | 3.630 |
| PC(20:2(11Z,14Z)/15:0) | 160.063 | 772.580 | 0.970 | 2.096 | Up | 1.263 | 0.013 | 2.162 |
| PC(16:1(9Z)/P-18:1(11Z)) | 157.795 | 742.570 | 0.117 | 0.261 | Up | 1.303 | 0.009 | 2.221 |
| PC(18:3(6Z,9Z,12Z)/15:0) | 163.278 | 742.532 | 0.383 | 0.964 | Up | 1.277 | 0.010 | 2.517 |
| PC(18:3(9Z,12Z,15Z)/14:0) | 164.912 | 728.519 | 0.237 | 0.600 | Up | 1.246 | 0.041 | 2.536 |
| PC(22:2(13Z,16Z)/16:1(9Z)) | 157.827 | 812.611 | 1.076 | 1.904 | Up | 1.194 | 0.042 | 1.769 |
| PC(18:3(6Z,9Z,12Z)/18:1(11Z)) | 59.786 | 782.561 | 4.189 | 14.056 | Up | 1.185 | 0.028 | 3.355 |
| PC(18:3(6Z,9Z,12Z)/P-18:1(11Z)) | 156.085 | 766.568 | 2.699 | 4.163 | Up | 1.169 | 0.046 | 1.542 |
| PC(18:4(6Z,9Z,12Z,15Z)/18:1(11Z)) | 60.817 | 780.549 | 5.307 | 23.397 | Up | 1.270 | 0.010 | 4.409 |
| PC(18:4(6Z,9Z,12Z,15Z)/P-18:1(11Z)) | 154.689 | 764.555 | 1.898 | 3.151 | Up | 1.242 | 0.013 | 1.661 |
| PC(20:2(11Z,14Z)/20:4(5Z,8Z,11Z,14Z)) | 153.256 | 834.593 | 4.658 | 9.579 | Up | 1.230 | 0.044 | 2.056 |
| PC(22:5(7Z,10Z,13Z,16Z,19Z)/20:1(11Z)) | 150.997 | 862.623 | 0.162 | 0.494 | Up | 1.290 | 0.009 | 3.048 |
| PC(P-18:1(11Z)/22:5(4Z,7Z,10Z,13Z,16Z)) | 149.122 | 818.601 | 0.802 | 1.068 | Up | 1.237 | 0.042 | 1.332 |
| PE(P-16:0/20:5(5Z,8Z,11Z,14Z,17Z)) | 155.152 | 722.508 | 1.542 | 2.651 | Up | 1.181 | 0.048 | 1.719 |
| LysoPE(0:0/14:0) | 224.850 | 426.258 | 0.013 | 0.048 | Up | 1.235 | 0.022 | 3.755 |
| LysoPC(16:0) | 212.374 | 496.336 | 6.094 | 25.253 | Up | 1.253 | 0.021 | 4.144 |
| LysoPC(P-16:0) | 205.967 | 480.341 | 0.194 | 0.479 | Up | 1.250 | 0.020 | 2.470 |
| LysoPC(18:1(9Z)) | 209.520 | 522.353 | 14.907 | 66.989 | Up | 1.226 | 0.034 | 4.494 |
| SM(d18:1/16:0) | 200.851 | 703.569 | 0.195 | 0.937 | Up | 1.332 | 0.001 | 4.798 |
| SM(d18:1/20:0) | 197.867 | 759.630 | 0.029 | 0.118 | Up | 1.267 | 0.018 | 4.072 |
| SM(d18:0/18:1(9Z)) | 198.900 | 731.601 | 0.113 | 0.611 | Up | 1.319 | 0.002 | 5.425 |
| Other lipids | ||||||||
| (2E)-Decenoyl-ACP | 532.921 | 130.085 | 1.731 | 8.939 | Up | 1.317 | 0.007 | 5.164 |
| Phosphodimethylethanolamine | 460.647 | 170.057 | 0.123 | 0.561 | Up | 1.312 | 0.003 | 4.561 |
| 5α-Cholesta-8,24-dien-3-one | 31.273 | 383.328 | 0.201 | 0.341 | Up | 1.225 | 0.026 | 1.702 |
| Aromatic compounds | ||||||||
| Quinone | 223.123 | 109.028 | 0.106 | 0.386 | Up | 1.220 | 0.026 | 3.653 |
| Phenol | 316.869 | 95.049 | 0.194 | 0.763 | Up | 1.212 | 0.037 | 3.929 |
| Parabanic acid | 16.343 | 112.998 | 0.825 | 0.976 | Up | 1.495 | 0.012 | 1.183 |
| Benzaldehyde | 273.701 | 107.049 | 0.358 | 1.088 | Up | 1.291 | 0.022 | 3.037 |
| 4-Hydroxybenzaldehyde | 316.993 | 123.044 | 1.072 | 4.121 | Up | 1.221 | 0.031 | 3.844 |
| 2-Phenylacetamide | 184.356 | 128.143 | 11.688 | 20.587 | Up | 1.280 | 0.018 | 1.761 |
| 1H-Indole-3-carboxaldehyde | 276.993 | 146.059 | 0.518 | 1.876 | Up | 1.190 | 0.028 | 3.619 |
| 1H-Indole-2,3-dione | 47.215 | 148.038 | 0.163 | 0.464 | Up | 1.209 | 0.029 | 2.855 |
| Energy metabolism-related | ||||||||
| O-Phosphotyrosine | 492.769 | 262.049 | 0.271 | 1.058 | Up | 1.308 | 0.002 | 3.902 |
| Isocitral | 183.881 | 153.127 | 0.087 | 0.177 | Up | 1.147 | 0.041 | 2.034 |
| 3-Dehydroxycarnitine | 390.707 | 146.117 | 31.835 | 78.692 | Up | 1.260 | 0.018 | 2.472 |
| Secondary metabolites of plants | ||||||||
| 7-Hydroxy-2-phenyl-4H-chromen-4-one | 202.998 | 237.054 | 0.326 | 0.640 | Up | 1.569 | 0.011 | 1.961 |
| Glycitein | 145.862 | 307.060 | 0.154 | 0.317 | Up | 1.327 | 0.001 | 2.061 |
| Luteolin 4′-sulfate | 26.207 | 367.009 | 0.032 | 0.100 | Up | 1.318 | 0.001 | 3.154 |
| Momordicinin | 30.867 | 439.355 | 0.026 | 0.045 | Up | 1.311 | 0.002 | 1.708 |
| Isoquinoline | 137.025 | 130.064 | 0.099 | 0.269 | Up | 1.279 | 0.018 | 2.713 |
| 2-Propylpiperidine | 184.356 | 128.143 | 11.688 | 20.587 | Up | 1.280 | 0.018 | 1.761 |
| Pipericine | 1.954 | 336.324 | 0.007 | 0.016 | Up | 1.139 | 0.044 | 2.296 |
| Nucleotides and nucleotide derivatives | ||||||||
| 8-Hydroxyguanine | 309.359 | 168.051 | 0.253 | 0.403 | Up | 1.189 | 0.045 | 1.593 |
| Pyrimidine | 172.823 | 81.045 | 0.134 | 0.585 | Up | 1.259 | 0.043 | 4.361 |
| 3-Aminoisobutanoic acid | 350.392 | 102.055 | 0.655 | 1.177 | Up | 1.500 | 0.027 | 1.796 |
| Cyclic AMP | 281.191 | 328.044 | 0.319 | 1.564 | Up | 1.461 | 0.041 | 4.903 |
| 3′-AMP | 420.136 | 346.055 | 0.424 | 0.863 | Up | 1.330 | 0.041 | 0.662 |
| Vitamin D3 | 31.910 | 385.343 | 0.164 | 0.326 | Up | 1.204 | 0.039 | 1.988 |
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differential metabolites between LF and BC groups
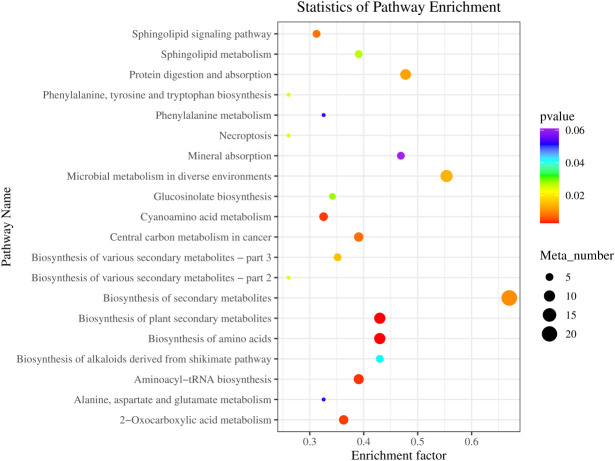
As shown in Figure 7 , the top 20 KEGG pathways with significant enrichment are as follows: Secondary metabolite synthesis-related (Biosynthesis of secondary metabolites, Biosynthesis of plant secondary metabolites, Glucosinolate biosynthesis, Biosynthesis of alkaloids derived from shikimate pathway, Biosynthesis of various secondary metabolites-part 3, Biosynthesis of various secondary metabolites-part, Sphingolipids metabolism-related, Sphingolipid signaling pathway, and Sphingolipid metabolism), Amino acid synthesis, metabolism and transfer-related (Biosynthesis of amino acids; Aminoacyl-tRNA biosynthesis; Cyanoamino acid metabolism; Phenylalanine metabolism; Alanine, aspartate, and glutamate metabolism; Phenylalanine, tyrosine, and tryptophan biosynthesis), Microbial metabolism in diverse environments, Protein digestion and absorption, Central carbon metabolism in cancer, 2-Oxocarboxylic acid metabolism, Necroptosis, and Mineral absorption.
Figure 7.
KEGG pathway enrichment of differential metabolites in fecal between LF and BC groups. Colors indicate enriched Q-value; the redder color represents a smaller Q-value. The size of the bubble indicates the number of metabolites mapped within a KEGG pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Correlation analysis of intestinal microbes and metabolites
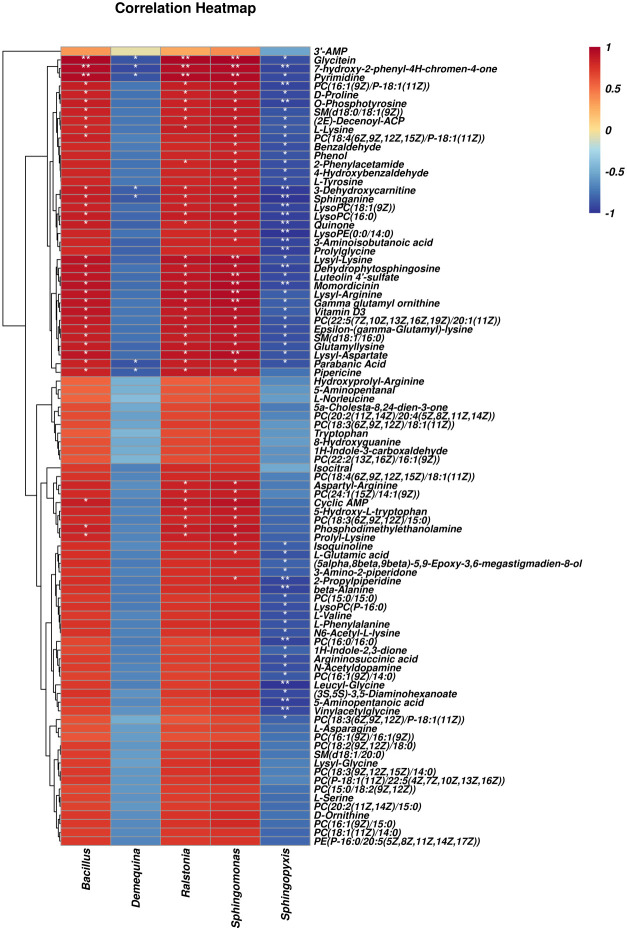
Figure 8 shows significant correlations between 63 differential metabolites and five differential microbial genera found through Pearson’s correlation analysis. The correlation analysis heat map showed that Bacillus, Ralstonia, and Sphingomonas were positively correlated with differential metabolites and that Demequina and Sphingopyxis negatively correlated with differential metabolites. Among them, 32 metabolites were significantly positively correlated with the Bacillus genus, 36 with the Ralstonia genus, 46 with the Sphingomonas genus, and 57 with the Sphingopyxis genus, and seven were negatively associated with the Demequina genus.
Figure 8.
Correlation analysis of intestinal microbes and metabolites between LF and BC groups. Red represents a positive correlation; blue represents a negative–positive correlation. The darker the color, the stronger the correlation. *p < 0.05, **p < 0.01.
Correlation analysis of selected differential metabolites and intestinal genes
As shown in Figure 9 , differential metabolites with significant microbial correlation were selected for further correlation analysis with intestinal genes. The results showed that immune and intestinal barrier-related genes (Cu/Zn-SOD, IL-22, and PT-1) were positively correlated with differential metabolites. Inflammation-related factor (Toll and Relish) expression was negatively correlated with differential metabolite levels. Pearson’s correlation analysis revealed that the primary differential metabolites (significant correlation with more than three genes) associated with intestinal immune genes were as follows: l-Lysine, Glycitein, Pipericine, Isoquinoline, 7-Hydroxy-2-phenyl-4H-chromen-4-one, PC(16:1(9Z)/P-18:1(11Z)), Quinone, PC(15:0/15:0), Vinylacetylglycine, Vitamin D3, N 6-Acetyl-l-lysine, Sphinganine, LysoPC(16:0), N-Acetyldopamine, Lysyl-Lysine, SM(d18:1/16:0), SM(d18:0/18:1(9Z)), 1H-Indole-2,3-dione, 5-Hydroxy-l-tryptophan, Glutamyllysine, Dehydrophytosphingosine, l-Glutamic acid, and PC(16:1(9Z)/14:0).
Figure 9.
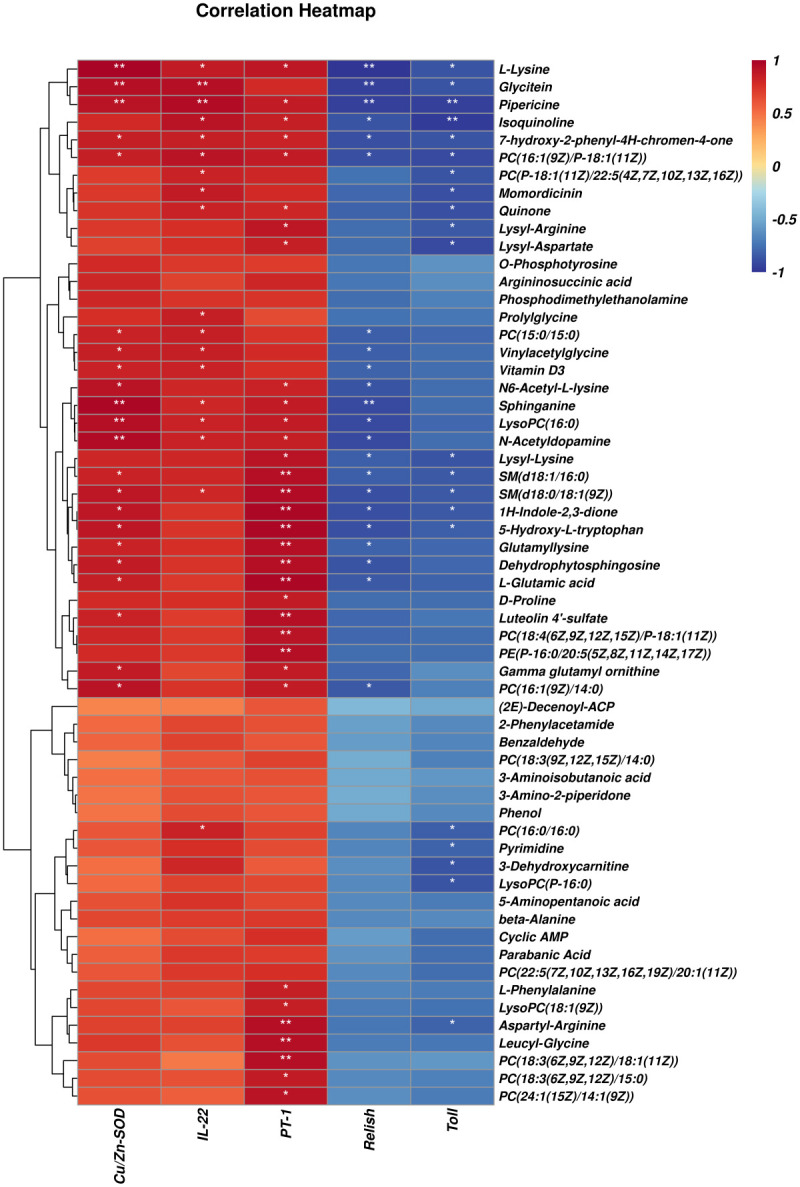
Correlation analysis of selected differential metabolites and intestinal genes between LF and BC groups. Red represents a positive correlation; blue represents a negative–positive correlation. The darker the color, the stronger the correlation. *p < 0.05, **p< 0.01.
Discussion
Aquaculture relies heavily on fish meal to produce aquafeeds, leading to its continually inflated price. As a cost-effective and sustainable alternative to fish meal, various plant proteins are used to partially or entirely replace fish meal. However, an inappropriate plant protein substitution ratio could adversely affect aquatic animals’ growth due to limiting amino acids and anti-nutritional factors (6, 22). Using probiotic bacteria to alleviate the adverse effects of growth suppression in aquatic animals induced by plant protein substitution is becoming an effective auxiliary strategy (23). Similarly, in our study, 10% fish meal substitution (basal fish meal level of 25%) by CPC depressed the growth to some extent, and B. coagulans supplementation alleviated the downregulated levels of WGR and SGR. However, there were no significant differences in growth and survivability among these three groups. Meanwhile, the phenotypic differences in hemolymph antioxidant indicators (MDA and CAT) coincided with the growth indicators variation and trend among the groups. The significantly upregulated SOD activity reflected the improvement of the overall antioxidant capacity of M. rosenbergii after treatment with B. coagulans, which echoed the upregulated growth performance following B. coagulans fortification, indicating the growth promotion function of B. coagulans in the model of fish meal replacement by plant protein. The growth performance of animals is closely related to intestinal physiological function. The activity of digestive enzymes, the integrity of barrier structure, and the immune function of the intestine directly determine the utilization efficiency of nutrients in the feeds and the body’s defense against environmental pathogens. Also, the primary mechanism by which dietary probiotics augment the growth of cultured species is by improving intestinal digestive and absorptive activity, and health (24, 25). Given this, we further explored the effects of fish meal replacement and probiotic supplementation on intestinal digestive enzyme activity and health status.
In this study, substituting 40% fish meal with CPC considerably decreased intestinal digestive enzyme activities (trypsin and lipase). This result is consistent with previous studies that fish meal replaced with excessive plant proteins elicited intestinal villus morphology destruction and suppressed the digestive and absorptive-related enzyme activities in aquatic animals (26). The inherent anti-nutritional factors or unbalanced amino acid composition in high-plant-protein diets may be intrinsically responsible for this (27). Gamma-glutamyl transferase (γ-GT) is located on the surface of the brush border epithelium in the intestine. It is essential in the epithelial cells’ amino acid uptake and dipeptide transport (28). In addition, gene expression of γ-GT is highly sensitive to oxidative stress as part of the cellular antioxidant defensive system (29), as γ-GT is involved in the degradation and resynthesis of glutathione in vivo and regulates GSH levels. There is a relationship between the intestine structural damage and the upregulated level of γ-GT in the intestine. In this study, the intestine γ-GT enzyme activity was significantly upregulated following the replacement of fish meal with CPC. On the one hand, the differentiation in amino acid composition between the two diets may result in the intestinal epithelial cells’ differential amino acid absorption strategy. On the other hand, reflecting that plant protein substitution may stimulate the onset of intestinal oxidative stress. Contrary to other research results, the dietary supplementation with B. coagulans significantly increased the activities of the digestive enzymes, including trypsin. In this study, B. coagulans supplementation with high-protein plant diets significantly downregulated or somewhat did not improve to a higher level of the trypsin activities, speculating for a reason related to the timing and tissue region of sampling. The spatial and temporal orders of the digestive enzymes involved in protein and peptide digestion in aquatic animals differ (30). Our sampling node may miss the active phase of trypsin action or B. coagulans altered the strategy for protein digestion and absorption in the intestine. However, the exact reasons need to be further explored.
In addition to digestive and absorptive capacity, the barrier and immune functions of the gut are also crucial to the health of aquatic animals. To investigate the effects on intestinal barrier function and inflammatory response in M. rosenbergii, we evaluated the variations in gene expression levels of the gut barrier protein PT-1, inflammatory-related factors (Relish, Toll, and IL-22), and antioxidant factor (Cu/Zn-SOD). In invertebrates, the peritrophic membrane (PM) protects the epithelium cells from mechanical abrasion and prevents pathogens from adhering to intestine epithelial cells (31). Moreover, the dominant proteins of PMs were identified mainly including digestion-related, immune-related, antioxidant proteins, and structural proteins of PM in Litopenaeus vannamei, highlighting its essential role in the intestinal immune system (32). Peritrophins (PTs) are essential for the generation of PM, which directly affect the permeability, elasticity, porosity, and strength of PM that lines the crustacean midgut. Therefore, PT plays a role in the immune response against pathogenic bacteria by protecting the intestine from colonization, intrusion, and secondary tissue distribution of pathogenic bacteria in crustaceans (33). The expression of intestinal inflammatory transcription factors (Toll and Relish) was significantly upregulated, while the expression of barrier factor PT-1 was significantly downregulated following the substitution of 40% fish meal by cottonseed protein concentrate, as determined. This indicates that plant protein substitution caused impairment of intestinal barrier function and secondary inflammatory responses. This may be attributed to anti-nutritional factors in plant proteins, which could impair the intestine peritrophic membrane’s morphological structure and thus induce gut inflammation (significantly upregulated inflammation factor) (34). Interleukin (IL)-22 is a homeostatic cytokine that promotes the expression of a series of genes that enforce intestinal epithelial cell defense function (35). For example, IL-22 signaling promoted the upregulation of intestine membrane mucin Muc17 in neonatal mice, suggesting its critical role in the epithelial barrier function of the intestine (36). Moreover, the reparative cytokine IL-22 was confirmed to drive repair in the intestinal epithelium through decreasing inflammation response in the colitis model (37). Additional supplementation with B. coagulans significantly alleviated the upregulated expression of inflammatory factors. Meanwhile, it significantly upregulated the expression levels of the barrier factor PT-1, the reparative cytokine IL-22, and the antioxidant factor Cu/Zn-SOD. The above results indicate that B. coagulans can alleviate the side effects of plant protein substitution and that the critical target regulatory molecules were PT-1 and IL-22. Similar studies have shown that probiotic Lactobacillus casei treatment strengthened the function of the gut mucosal epithelium by boosting IL-22 and tight junction protein (Zonulin-1 and Claudin-1) expression levels in the intestinal inflammatory model of chicks infected with Salmonella pullorum (38).
From the gut microbiota perspective, partial substitution of fish meal with plant protein sources affected the composition and diversity of gut microbes, and inappropriate substitution ratio induced host intestine mucosa lesions and hypoimmunity (26, 39). At the phylum level, consistent with the previous study, Proteobacteria, Firmicutes, Bacteroidetes, and Tenericutes were determined as the predominant phyla in M. rosenbergii intestine (40). We noted a remarkable downregulation of Firmicutes abundance after plant protein substitution. Speculating that the fiber content in the CPC substitution group diet is higher than the HF group, a recent report suggested that dietary fiber intake decreases the proportion of Firmicutes (41). The Firmicutes phylum includes the vast majority of lactic acid bacteria genera, such as Streptococcus, Lactobacillus, Leuconostoc, and Carnobacterium, which are generally considered to be probiotics for intestinal health (42). The phylum Firmicutes produces short-chain fatty acids (SCFAs) through collaboration with oligosaccharide fermentation bacteria such as Bifidobacteria (43). Thus, the callback of Firmicutes abundance after B. coagulans intervention suggests that B. coagulans has the potential to improve the structure of the intestinal flora positively. In addition, substituting 40% fish meal with CPC significantly decreased the richness and diversity of the intestinal flora, as well as downregulated the abundance of Flavobacterium and Bacillus, which were regarded as antagonistic probiotics in aquaculture (44). Furthermore, the potential pathogenicity in the CPC substitution group apparently increased through the phenotype prediction of potentially_pathogenic by BugBase software. This indicated that this substitution ratio led to the unbalance of a game between probiotics and conditionally pathogenic bacteria, disrupting intestinal health. This is consistent with previous studies showing that appropriate levels of CPC substitution can increase the richness and diversity of gut flora, while excessive substitution induces enteritis and disrupt the homeostasis and part of the functions of the gut microbiota (45, 46). In recent years, Bacillus spp. has been reported as a potential probiotic candidate in aquatic farming sectors due to its high antagonistic activities and extracellular enzyme synthesis (47). Dietary Bacillus supplementation benefits the host’s growth, intestinal microbial homeostasis, stress resistance, and immune and anti-inflammatory response (48–51). Sphingomonas is also considered to be a potential probiotic for aquaculture. The prebiotic effects mainly include the following ways: 1) regulates nutrient metabolism by secreting cellulase, protease, and amylase (52); 2) degrades toxic compounds, ammonia nitrogen, and nitrite (53); 3) produces oligosaccharide prebiotics (54); 4) inhibit Vibrio (53, 55)). In this study, we noted that B. coagulans treatment significantly improved the relative abundance of Sphingomonas, Bacillus, and Ralstonia while reducing the potential pathogenic flora after CPC substitution, echoing the trend in gene expression of intestinal inflammatory and barrier factors. This indicates that B. coagulans treatment is conducive to the colonization of Bacillus spp. in the gut and has a positive regulatory effect on intestinal immunity. This is similar to a recent study on turbot that different species of Bacillus treatment induced a significant positive correlation between the immune and antioxidant indexes and intestinal probiotics such as Bacillus, Ralstonia, and Bifidobacterium and a negative correlation with pathogenic bacteria (50).
Strikingly, a new research perspective has been condensed through integrating clinical monitoring, 16S rDNA and metagenomic sequencing, metabonomics analysis, and whole-genome sequencing. Accumulating medical reports have revealed the decisive role of specific gut microbes and gut microbes-derived secondary metabolites in metabolic and gastrointestinal disease (insulin resistance, non-alcoholic fatty liver, and chronic enteritis) (56–58). Studies likewise highlighted that the secondary metabolites of intestinal microorganisms, for instance, secondary bile acids, short-chain fatty acids, and branched-chain amino acids, play a substantial role in the process of medicine and diet treatment-induced remission (13, 59). These findings raised our focus on gut microbiota-derived metabolites. In this study, we identified several differential metabolites through metabonomics analysis, mainly sphingolipids, amino acids, amino acid derivatives, phospholipids, etc. Sphingolipids are a class of lipids with a long chain of amino alcohol sphingolipid backbones and amide-bound fatty acyl chains. As signaling molecules, sphingolipids mediated microbial-host interactions, such as immunity and inflammation. Sphingolipids have been identified as the most differential biomarker metabolites in the stool of inflammatory bowel disease patients (60). Known sphingolipid-producing bacteria include most of the Bacteroidetes phylum, a few members of the Chlorobi phylum, and a subset of α-Proteobacteria (Sphingomonas, Acetobacter, and Neospora). Sphingolipid metabolites are essential signaling molecules mediating host cell processes, such as ceramide, sphingosine, and sphingosine-1-phosphate (S1P) (61). Sphingosine and dehydrophytosphingosine are the precursors of S1P, which play critical immunological functions in intestinal tissue (62). Our study identified the increased sphingosine and dihydrosphingosine, and the upregulated abundance of sphingosine-producing bacteria Sphingomonas after B. coagulans intervention. Meanwhile, Sphingolipid metabolism-related pathways (Sphingolipid signaling pathway and Sphingolipid metabolism) were enriched. In addition, correlation analysis established a significant positive correlation between sphingosine, Sphingomonas, and immune-related genes (Cu/Zn-SOD, IL-22, and PT-1) and a significant negative correlation with inflammatory factor Relish. The above results indicated that the Sphingomonas–sphingosine axis is involved in the intestinal inflammatory repair and immune regulation of M. rosenbergii mediated by B. coagulans.
In terms of amino acids, studies have proved that intestinal microorganisms can synthesize, incorporate, and metabolize several available amino acids to maintain amino acid homeostasis through bidirectional switching with the host. In addition, the microbially derived metabolites mediate mucosal immunoregulation, epithelial cell barrier maintenance, and enteroendocrine regulation, such as serotonin, ammonia, hydrogen sulfide, branched-chain amino acids, polyamines, and phenolic and indolic compounds (63). In mammals, l-tryptophan, endogenous tryptophan metabolites, and bacterial tryptophan metabolites-indole derivatives could enhance intestinal barrier function by promoting the expression of tight junction protein of intestinal epithelial cells (63) and regulating the expression of inflammatory-related factors (such as anti-inflammatory cytokine IL-22) (64). In addition, sufficient evidence suggested that lysine (65), glycine (66), arginine (67), etc., can shape the intestinal microbial composition and improve the host intestinal mucosal immunity. In the present study, we noted that B. coagulans treatment considerably affected the amino acid metabolic profile of the M. rosenbergii gut microbes, such as l-Lysine, Lysyl-Lysine, Epsilon-(gamma-glutamyl)-lysine, Tryptophan, 5-Hydroxy-l-tryptophan, 1H-Indole-2,3-dione, Lysyl-Arginine, Aspartyl-Arginine, and Leucyl-Glycine. Moreover, the correlation analysis also established a significant positive correlation between the above amino acid metabolites and the intestinal factors PT-1 and Cu/Zn-SOD. These results showed that changes in the amino acid metabolic spectrum of gut microorganisms induced by B. coagulans treatment are correlated with the intestinal immunity and barrier function of M. rosenbergii. However, to date, studies mainly focused on exogenous amino acid’s contribution to hosting intestine immunity but rarely explored the effect and mechanism of bacterial amino acid intermediates themselves on the host physiology through the intestinal epithelial cells; their contribution degree and fate remain to be further explored. In addition, our study also established the correlation between other differential metabolites (glycitein, pipericine, isoquinoline, 7-hydroxy-2-phenyl-4H-chromen-4-one, PC(16:1(9Z)/P-18:1(11Z)), PC(15:0/15:0), PC(16:1(9Z)/14:0), LysoPC(16:0), SM(d18:1/16:0), SM(d18:0/18:1(9Z)), Vitamin D3, and N-acetyldopamine) and intestinal function genes (Cu/Zn-SOD, IL-22, PT-1, Toll, and Relish). Our results suggested that the effects of B. coagulans on gut microbe metabolism are multifaceted, and the positive regulation of intestinal immunity, barrier function, and inflammatory responses in M. rosenbergii depends on the combined action of multiple metabolites ( Figure 10 ). However, the precise mechanisms require further research.
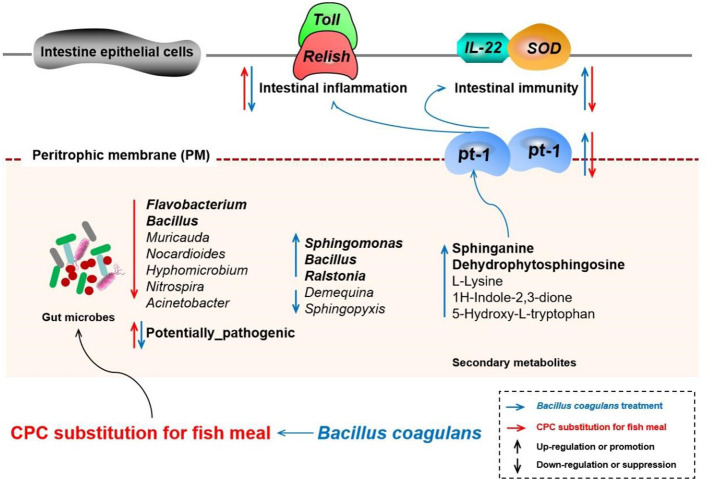
Figure 10.
Schematic diagram depicting the effects and possible mechanisms of the effects of CPC substitution and Bacillus coagulans intervention on gut microbes, derived secondary metabolites, and intestinal physiological functions of Macrobrachium rosenbergii. The red arrow represents the action path of CPC substitution, and the blue arrow represents the action path of Bacillus coagulans intervention. The upward arrow represents upregulation or promotion, and the downward arrow represents downregulation or suppression. CPC, cottonseed protein concentrate.
Data availability statement
The raw dataset for 16SrDNA sequencing have been deposited in the National Institutes of Health’s Short Read Archive database (SRA accession no. PRJNA894136; BioSample accessions: SAMN31438493, SAMN31438494, SAMN31438495, SAMN31438496, SAMN31438497, SAMN31438498, SAMN31438499, SAMN31438500, SAMN3138501).
Author contributions
XZ contributed in the areas of experimental design, sampling, data analysis, and write-up. BL, YZ, QZ and XZ contributed to the experimental design and manuscript review. JY and NW contributed to feeding and cultivating experimental prawns, sampling, and statistics. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Project of National Key R&D Program of China (2019YFD0900200), China Agriculture Research System of MOF and MARA (CARS-48), Central Public-interest Scientific Institu-tion Basal Research Fund, CAFS (2020TD59).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1074399/full#supplementary-material
References
- 1. Bian F, Zhou H, He G, Wang C, Peng H, Pu X, et al. Effects of replacing fishmeal with different cottonseed meals on growth, feed utilization, haematological indexes, intestinal and liver morphology of juvenile turbot (Scophthalmus maximus l.) . Aquacult Nutr (2017) 23(6):1429–39. doi: 10.1111/anu.12518 [DOI] [Google Scholar]
- 2. Olsen RL, Hasan MR. A limited supply of fishmeal: Impact on future increases in global aquaculture production. Trends Food Sci Technol (2012) 27(2):120–8. doi: 10.1016/j.tifs.2012.06.003 [DOI] [Google Scholar]
- 3. Cavrois-Rogacki T, Leeming D, Lopez PM, Davie A, Migaud H. Plant-based protein ingredients can successfully replace fish meal in the diet of ballan wrasse (LABRUS BERGYLTA) juveniles. Aquaculture (2022) 546:737419. doi: 10.1016/j.aquaculture.2021.737419 [DOI] [Google Scholar]
- 4. Miao SY, Zhao CZ, Zhu JY, Hu JT, Dong XJ, Sun LS. Dietary soybean meal affects intestinal homoeostasis by altering the microbiota, morphology and inflammatory cytokine gene expression in northern snakehead. Sci Rep (2018) 8(1):113. doi: 10.1038/s41598-017-18430-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang W, Tan BP, Ye GL, Wang JX, Dong XH, Yang QH, et al. Identification of potential biomarkers for soybean meal-induced enteritis in juvenile pearl gentian grouper, epinephelus lanceolatus♂ × epinephelus fuscoguttatus♀. Aquaculture (2019) 512:734337. doi: 10.1016/j.aquaculture.2019.734337 [DOI] [Google Scholar]
- 6. Xu XY, Yang H, Zhang CY, Bian YH, Yao WX, Xu Z, et al. Effects of replacing fishmeal with cottonseed protein concentrate on growth performance, flesh quality and gossypol deposition of largemouth bass (Micropterus salmoides) . Aquaculture (2022) 548:737551. doi: 10.1016/j.aquaculture.2021.737551 [DOI] [Google Scholar]
- 7. Fu SL, Qian K, Liu HS, Song F, Ye JM. Effects of fish meal replacement with low-gossypol cottonseed meal on the intestinal barrier of juvenile golden pompano (Trachinotus ovatus). Aquacult Res (2022) 53(1):285–99. doi: 10.1111/are.15576 [DOI] [Google Scholar]
- 8. Santos KO, Costa-Filho J, Riet J, Spagnol KL, Nornberg BF, Kütter MT, et al. Probiotic expressing heterologous phytase improves the immune system and attenuates inflammatory response in zebrafish fed with a diet rich in soybean meal. Fish Shellfish Immunol (2019) 93:652–8. doi: 10.1016/j.fsi.2019.08.030 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z, Yang MQ, Wang L, Lu KL, Song K, Zhang CX. Bacillus subtilis LCBS1 supplementation and replacement of fish meal with fermented soybean meal in bullfrog (Lithobates catesbeianus) diets: Effects on growth performance, feed digestibility and gut health. Aquaculture (2021) 545:737217. doi: 10.1016/j.aquaculture.2021.737217 [DOI] [Google Scholar]
- 10. Jäger R, Purpura M, Farmer S, Cash HA, Keller D. Probiotic bacillus coagulans GBI-30, 6086 improves protein absorption and utilization. Probiotics Antimicrob Proteins (2017) 10(1):1–5. doi: 10.1007/s12602-017-9354-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller D, Dinter RV, Cash H, Farmer S, Venema K. Bacillus coagulans GBI-30, 6086 increases plant protein digestion in a dynamic, computer-controlled in vitro model of the small intestine (TIM-1). Beneficial Microbes (2017) 8(3):1–6. doi: 10.3920/BM2016.0196 [DOI] [PubMed] [Google Scholar]
- 12. Chang XL, Kang M, Shen YH, Yun LL, Yang GK, Zhu L, et al. Bacillus coagulans SCC-19 maintains intestinal health in cadmium-exposed common carp (Cyprinus carpio l.) by strengthening the gut barriers, relieving oxidative stress and modulating the intestinal microflora. Ecotoxicol Environ Saf (2021) 228:112977. doi: 10.1016/j.ecoenv.2021.112977 [DOI] [PubMed] [Google Scholar]
- 13. Sun LL, Xie C, Wang G, Wu Y, Wu Q, Wang XM, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med (2018) 24(12):1919–29. doi: 10.1038/s41591-018-0222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab (2021) 33(5):988–1000.e1007. doi: 10.1016/j.cmet.2021.03.002 [DOI] [PubMed] [Google Scholar]
- 15. Li YY, Ji XY, Wu HN, Li X, Zhang HM, Tang DL. Mechanisms of traditional Chinese medicine in modulating gut microbiota metabolites-mediated lipid metabolism. J Ethnopharmacol (2021) 278:114207. doi: 10.1016/j.jep.2021.114207 [DOI] [PubMed] [Google Scholar]
- 16. Schroeder B, Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med (2013) 22(10):1079–89. doi: 10.1038/nm.4185 [DOI] [PubMed] [Google Scholar]
- 17. Amoah K, Huang QC, Tan BP, Zhang S, Chi SY, Yang QH, et al. Dietary supplementation of probiotic bacillus coagulans ATCC 7050, improves the growth performance, intestinal morphology, microflora, immune response, and disease confrontation of pacific white shrimp, litopenaeus vannamei. Fish Shellfish Immunol (2019) 87:796–808. doi: 10.1016/j.fsi.2019.02.029 [DOI] [PubMed] [Google Scholar]
- 18. Chang XL, Chen YY, Feng JC, Huang MY, Zhang JX. Amelioration of cd-induced bioaccumulation, oxidative stress and immune damage by probiotic bacillus coagulans in common carp (Cyprinus carpio l.) . Aquacult Rep (2021) 20:100678. doi: 10.1016/j.aqrep.2021.100678 [DOI] [Google Scholar]
- 19. Sun CX, Shan F, Liu MY, Liu B, Zhou QL, Zheng XC, et al. High-Fat-Diet-Induced oxidative stress in giant freshwater prawn (Macrobrachium rosenbergii) via NF-κB/NO signal pathway and the amelioration of vitamin e. Antioxidants (2022) 11:228. doi: 10.3390/antiox11020228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu B, Song CY, Gao Q, Liu B, Zhou QL, Sun CX, et al. Maternal and environmental microbes dominate offspring microbial colonization in the giant freshwater prawn macrobrachium rosenbergii . Sci Total Environ (2021) 790:148062. doi: 10.1016/j.scitotenv.2021.148062 [DOI] [PubMed] [Google Scholar]
- 21. Zheng XC, Jiang WB, Zhang L, Espe M, Abasubong KP, Zhang CN, et al. Investigation of the regulatory effect of icariin on the hepatopancreas glycolipid metabolism and energy metabolism of female eriocheir sinensis during exogenous vitellogenesis based on the transcriptomics and metabolomics. Aquacult Nutr (2021) 27(6):2066–83. doi: 10.1111/anu.13341 [DOI] [Google Scholar]
- 22. Magbanua TO, Ragaza JA. Selected dietary plant-based proteins for growth and health response of Nile tilapia oreochromis niloticus . Aquacult Fish (2022). doi: 10.1016/j.aaf.2022.04.001 [DOI] [Google Scholar]
- 23. Li WK, Pan L, Liu HY, Tan BP, Dong XH, Yang QH, et al. Effects of the clostridium butyricum on growth performance, antioxidant capacity, immunity and disease resistance of litopenaeus vannamei fed with cottonseed protein concentrate (CPC) replacement of fishmeal in diet. Fish Shellfish Immunol (2022) 126:283–91. doi: 10.1016/j.fsi.2022.05.030 [DOI] [PubMed] [Google Scholar]
- 24. Andani HRR, Tukmechi A, Meshkini S, Sheikhzadeh N. Antagonistic activity of two potential probiotic bacteria from fish intestines and investigation of their effects on growth performance and immune response in rainbow trout (Oncorhynchus mykiss) . J Appl Ichthyol (2012) 28(5):728–34. doi: 10.1111/j.1439-0426.2012.01974.x [DOI] [Google Scholar]
- 25. Han B, Long WQ, He JY, Liu YJ, Si YQ, Tian LX. Effects of dietary bacillus licheniformis on growth performance, immunological parameters, intestinal morphology and resistance of juvenile Nile tilapia (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol (2015) 46(2):225–31. doi: 10.1016/j.fsi.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 26. Zhu BP, Zhou JC, Wang ZQ, Hu YJ, Cai ML, Yang LL, et al. Interactions between intestinal morphology, digestion, inflammatory responses, and gut microbiota of juvenile channel catfish elicited by dietary enzymatic rice protein. Fish Shellfish Immunol (2022) 127:155–65. doi: 10.1016/j.fsi.2022.06.018 [DOI] [PubMed] [Google Scholar]
- 27. Dong YW, Jiang WD, Wu P, Liu Y, Kuang SY, Tang L, et al. Nutritional digestion and absorption, metabolism fates alteration was associated with intestinal function improvement by dietary threonine in juvenile grass carp (Ctenopharyngodon idella). Aquaculture (2022) 555:738194. doi: 10.1016/j.aquaculture.2022.738194 [DOI] [Google Scholar]
- 28. Kovacs-Nolan J, Rupa P, Matsui T, Tanaka M, Konishi T, Sauchi Y, et al. In vitro and ex vivo uptake of glutathione (GSH) across the intestinal epithelium and fate of oral GSH after in vivo supplementation. J Agric Food Chem (2014) 62(39):9499–506. doi: 10.1021/jf503257w [DOI] [PubMed] [Google Scholar]
- 29. Lim JS, Yang JH, Chun BY, Kam S, Jacobs DR, Lee DH. Is serum γ-glutamyltransferase inversely associated with serum antioxidants as a marker of oxidative stress? Free Radical Biol Med (2004) 37(7):1018–23. doi: 10.1016/j.freeradbiomed.2004.06.032 [DOI] [PubMed] [Google Scholar]
- 30. Hirji KN, Courtney WAM. Leucine aminopeptidase activity in the digestive tract of perch, perca fluviatilis l. J Fish Biol (1982) 21(6):615–22. doi: 10.1111/j.1095-8649.1982.tb02865.x [DOI] [Google Scholar]
- 31. Wang LY, Li FH, Wang B, Xiang JH. A new shrimp peritrophin-like gene from exopalaemon carinicauda involved in white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol (2013) 35(3):840–6. doi: 10.1016/j.fsi.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 32. Wang LY, Li FH, Wang B, Xiang JH. Structure and partial protein profiles of the peritrophic membrane (PM) from the gut of the shrimp litopenaeus vannamei . Fish Shellfish Immunol (2012) 33(6):1285–91. doi: 10.1016/j.fsi.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 33. Huang Y, Ma FT, Wang W, Ren Q. Identification and molecular characterization of a peritrophin-like gene, involved in the antibacterial response in Chinese mitten crab, eriocheir sinensis. Dev Comp Immunol (2015) 50(2):129–38. doi: 10.1016/j.dci.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 34. Han FL, Wang XD, Guo JL, Qi C, Xu CL, Luo Y, et al. Chen, Effects of glycinin and β-conglycinin on growth performance and intestinal health in juvenile Chinese mitten crabs (Eriocheir sinensis) . Fish Shellfish Immunol (2019) 84:269–79. doi: 10.1016/j.fsi.2018.10.013 [DOI] [PubMed] [Google Scholar]
- 35. Hanash AM, Dudakov JA, Hua G, O’Connor MH, Young LF, Singer NV, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immun (2012) (2012) 37(2):339–50. doi: 10.1016/j.immuni.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Layunta E, Jäverfelt S, Dolan B, Arike L, Pelaseyed T. IL-22 promotes the formation of a MUC17 glycocalyx barrier in the postnatal small intestine during weaning. Cell Rep (2021) 34(7):108757. doi: 10.1016/j.celrep.2021.108757 [DOI] [PubMed] [Google Scholar]
- 37. Ihekweazu FD, Engevik MA, Ruan W, Shi Z, Fultz R, Engevik KA, et al. Bacteroides ovatus promotes IL-22 production and reduces trinitrobenzene sulfonic acid–driven colonic inflammation. Am J Pathol (2021) 191(4):704–19. doi: 10.1016/j.ajpath.2021.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng ZT, Han DP, Wang YY, Wang QZ, Yan X, Wang SJ, et al. Lactobacillus casei protects intestinal mucosa from damage in chicks caused by salmonella pullorum via regulating immunity and the wnt signaling pathway and maintaining the abundance of gut microbiota. Poult Sci (2021) 100(8):101283. doi: 10.1016/j.psj.2021.101283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei HC, Xing SJ, Chen P, Wu XF, Gu X, Luo L, et al. Plant protein diet-induced hypoimmunity by affecting the spiral valve intestinal microbiota and bile acid enterohepatic circulation in amur sturgeon (Acipenser schrenckii) . Fish Shellfish Immunol (2020) 106:421–30. doi: 10.1016/j.fsi.2020.08.025 [DOI] [PubMed] [Google Scholar]
- 40. Xue HB, Wu XJ, Li ZH, Liu Y, Yin XL, Wang WN. Correlation and causation between the intestinal microbiome and male morphotypes in the giant freshwater prawn macrobrachium rosenbergii. Aquaculture (2021) 531:735936. doi: 10.1016/j.aquaculture.2020.735936 [DOI] [Google Scholar]
- 41. Zhang ZY, Shi L, Pang WH, Liu WW, Li JF, Wang HB, et al. Dietary fiber intake regulates intestinal microflora and inhibits ovalbumin-induced allergic airway inflammation in a mouse model. PloS One (2016) 11(2):e0147778. doi: 10.1371/journal.pone.0147778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinto EJ, Jiménez P, Caro I, Tejero J, Mateo J, Girbés T. Probiotic lactic acid bacteria: a review. Food Nutr Sci (2014) 5:1765–75. doi: 10.4236/fns.2014.518190 [DOI] [Google Scholar]
- 43. Huang Z, Boekhorst J, Fogliano V, Capuano E, Wells JM. Wells, distinct effects of fiber and colon segment on microbiota-derived indoles and short-chain fatty acids. Food Chem (2023) 398:133801. doi: 10.1016/j.foodchem.2022.133801 [DOI] [PubMed] [Google Scholar]
- 44. Qi Z, Zhang XH, Boon N, Bossier P. Probiotics in aquaculture of China-current state, problems and prospect. Aquaculture (2009) 290(1):15–21. doi: 10.1016/j.aquaculture.2009.02.012 [DOI] [Google Scholar]
- 45. Wang HM, Hu X, Zheng YD, Chen J, Tan BP, Shi LL, et al. Effects of replacing fish meal with cottonseed protein concentrate on the growth, immune responses, digestive ability and intestinal microbial flora in litopenaeus vannamei. Fish Shellfish Immunol (2022) 128:91–100. doi: 10.1016/j.fsi.2022.07.067 [DOI] [PubMed] [Google Scholar]
- 46. Yin B, Liu HY, Tan BP, Dong XH, Chi SY, Yang QH, et al. Cottonseed protein concentrate (CPC) suppresses immune function in different intestinal segments of hybrid grouper ♀Epinephelus fuscoguttatus×♂Epinephelus lanceolatu via TLR-2/MyD88 signaling pathways. Fish Shellfish Immunol (2018) 81:318–28. doi: 10.1016/j.fsi.2018.07.038 [DOI] [PubMed] [Google Scholar]
- 47. Goutam B, Ray AK. The advancement of probiotics research and its application in fish farming industries. Res Vet Sci (2017) 115:66–77. doi: 10.1016/j.rvsc.2017.01.016 [DOI] [PubMed] [Google Scholar]
- 48. Hassaan MS, Mohammady EY, Soaudy MR, Elashry MA, Moustafa MMA, Wassel MA, et al. Synergistic effects of bacillus pumilus and exogenous protease on Nile tilapia (Oreochromis niloticus) growth, gut microbes, immune response and gene expression fed plant protein diet. Anim Feed Sci Technol (2021) 275:114892. doi: 10.1016/j.anifeedsci.2021.114892 [DOI] [Google Scholar]
- 49. Kuebutornye FKA, Abarike ED, Lu Y. A review on the application of bacillus as probiotics in aquaculture. Fish Shellfish Immunol (2019) 87:820–8. doi: 10.1016/j.fsi.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 50. Ma SK, Yu DN, Liu Q, Zhao MJ, Xu C, Yu JC. Relationship between immune performance and the dominant intestinal microflora of turbot fed with different bacillus species. Aquaculture (2022) 549:737625. doi: 10.1016/j.aquaculture.2021.737625 [DOI] [Google Scholar]
- 51. Sutthi N, Doan HV. Saccharomyces crevices and bacillus spp. effectively enhance health tolerance of Nile tilapia under transportation stress. Aquaculture (2020) 528:735527. doi: 10.1016/j.aquaculture.2020.735527 [DOI] [Google Scholar]
- 52. Zhao Q, Yue SJ, Bilal M, Hu HB, Wang W, Zhang XH. Comparative genomic analysis of 26 sphingomonas and sphingobium strains: Dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci Total Environ (2017) 609:1238–47. doi: 10.1016/j.scitotenv.2017.07.249 [DOI] [PubMed] [Google Scholar]
- 53. Yun L, Yu ZH, Li YY, Luo P, Jiang X, Tian YS, et al. Ammonia nitrogen and nitrite removal by a heterotrophic sphingomonas sp. strain LPN080 and its potential application in aquaculture. Aquaculture (2019) 500:477–84. doi: 10.1016/j.aquaculture.2018.10.054 [DOI] [Google Scholar]
- 54. He MM, Guo M, Zhang X, Chen KK, Irbis CG. Purification and characterization of alginate lyase from sphingomonas sp. ZH0. J Biosci Bioeng (2018) 126(3):310–6. doi: 10.1016/j.jbiosc.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 55. Chaudhary A, Qazi JI. Probiotic antagonism of sphingomonas sp. against vibrio anguillarum exposed labeo rohita fingerlings. Adv Life Sci (2014) 4(3):156–65. doi: 10.5923/j.als.20140403.11 [DOI] [Google Scholar]
- 56. Friedman SL, Neuschwander-Tetri BA, Mary R, Sanyal AJ. Sanyal, mechanisms of NAFLD development and therapeutic strategies. Nat Med (2018) 24(7):908–22. doi: 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Geach T. GUT MICROBIOTA: Mucin-munching bacteria modulate glucose metabolism. Nat Rev Endocrinol (2017) 13(2):66. doi: 10.1038/nrendo.2016.201 [DOI] [PubMed] [Google Scholar]
- 58. Wang S, Martins R, Sullivan MC, Friedman ES, Misic AM, El-Fahmawi A, et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. MICROBIOME (2019) 7(1):126–45. doi: 10.1186/s40168-019-0740-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li CY, Liang YQ, Qiao Y. Messengers from the gut: Gut microbiota-derived metabolites on host regulation. Front Microbiol (2022) 13:863407. doi: 10.3389/fmicb.2022.863407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heaver SL, Johnson EL, Ley RE. Sphingolipids in host-microbial interactions. Curr Opin IN Microbiol (2018) 43:92–9. doi: 10.1016/j.mib.2017.12.011 [DOI] [PubMed] [Google Scholar]
- 61. Duan RD, Nilsson A. Metabolism of sphingolipids in the gut and its relation to inflammation and cancer development. Prog IN Lipid Res (2009) 48(1):62–72. doi: 10.1016/j.plipres.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 62. Kunisawa J, Kiyono H. Immunological function of sphingosine 1-phosphate in the intestine. NUTRIENTS (2012) 4(3):154–66. doi: 10.3390/nu4030154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu LT, Tang ZR, Chen HY, Ren ZX, Ding Q, Liang KY, et al. Mutual interaction between gut microbiota and protein/amino acid metabolism for host mucosal immunity and health. Anim Nutr (2021) 7(1):11–6. doi: 10.1016/j.aninu.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lanis JM, Alexeev EE, Curtis VF, Kitzenberg DA, Kao DJ, Battista KD, et al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol (2017) 10(5):1133–44. doi: 10.1038/mi.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang DY, Maulu S, Ren MC, Liang HL, Ge XP, Ji K, et al. Dietary lysine levels improved antioxidant capacity and immunity via the TOR and p38 MAPK signaling pathways in grass carp, ctenopharyngodon idellus fry. Front IN Immunol (2021) 12:635015. doi: 10.3389/fimmu.2021.635015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ji Y, Fan XX, Zhang YC, Li J, Dai ZL, Wu ZL. Glycine regulates mucosal immunity and the intestinal microbial composition in weaned piglets. Amino Acids (2022) 54(3):385–98. doi: 10.1007/s00726-021-02976-y [DOI] [PubMed] [Google Scholar]
- 67. Wu MM, Xiao H, Shao FY, Tan B, Hu SL. Arginine accelerates intestinal health through cytokines and intestinal microbiota. Int Immunopharmacol (2020) 81:106029. doi: 10.1016/j.intimp.2019.106029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw dataset for 16SrDNA sequencing have been deposited in the National Institutes of Health’s Short Read Archive database (SRA accession no. PRJNA894136; BioSample accessions: SAMN31438493, SAMN31438494, SAMN31438495, SAMN31438496, SAMN31438497, SAMN31438498, SAMN31438499, SAMN31438500, SAMN3138501).