Abstract
The intestinal microbiota of the preterm neonate has become a major research focus, with evidence emerging that the microbiota influences both short and long-term health outcomes, in the neonatal intensive care unit and beyond. Similar to the term microbiome, the preterm gut microbiome is highly influenced by diet, specifically formula and human milk use. This study aims to analyze next-generation products including preterm formula, human milk-oligosaccharide term formula, and preterm breastmilk. We used a culture-based model to differentially compare the growth patterns of individual bacterial strains found in the human intestine. This model probed 24 strains of commensal bacteria and 8 pathobiont species which have previously been found to cause sepsis in preterm neonates. Remarkable differences between strain growth and culture pH were noted after comparing models of formulas and between human milk and formula. Both formula and human milk supported the growth of commensal bacteria; however, the formula products but not human milk supported the growth of several specific pathogenic strains. Computational analysis revealed potential connections between long-chain fatty acid and iron uptake from formula in pathobiont organisms. These findings indicate that there is a unique profile of growth in response to human milk and formula and shed light into how the infant gut microbiota could be influenced.
Introduction
The infant gut microbiota undergoes significant fluxes in the domains of bacterial community structure and function during the first few years of life.1–4 Shortly after birth, the intestine is colonized with facultative anaerobes, like Streptococcus, Enterococcus, Enterobacter, and Staphylococcus, which reduce the intestinal oxygen content and pave the way for strict anaerobes, like Bifidobacteria, Bacteroides, and Clostridia.5–8 Several factors affect the infant microbiota composition, but the dominant influencers are gestational age and diet. The gut microbiota of preterm infants (born before 37 weeks of gestation) is characterized by limited microbial diversity and delayed colonization.5, 9 In general, there are decreased levels of commensal, obligate anaerobes and increased levels of facultative pathobionts.7, 10–12 Preterm infants are dominated by Enterococci, Staphylococci, and Enterbacteriacea (Enterobacter, Escherichia, and Klebsiella spp.) and exhibit a wider variation in microbial constituents when compared to term infants.5, 13–18 Diet is another major driver of the composition of the infant gut microbiota.1 The intake of human milk is associated with increased Bifidobacteria abundance, while the intake of formula correlates with increased Escherichia, Clostridia, Bacteroides, and Enterobacteriaceae.8, 19–25 Although multiple studies have examined the microbial communities of infant stool by 16S rRNA sequencing in relationship to diet, these complex communities obscure which microbes are responding to milk or formula.26–28
A healthy preterm gut microbiota is supported by maternal milk rather than formula or banked donor human milk.27 Human milk is associated with numerous benefits to the preterm neonate, including lower risks of necrotizing enterocolitis, less feeding intolerance and better lean body mass indices.29 Studies mapping the preterm microbiota development are lacking. Large neonatal cohorts characterizing the stool microbiome of infants in the neonatal intensive care unit provide evidence of the optimal microbiome diversity and make-up,3, 27, 30 but these analyses are unable to draw clear associations between infant diet and species growth. This is due, in part, to the frequently changing enteral diet that a preterm neonate might receive over a short course of time. For example, an infant might receive mother’s own milk, donor pasteurized milk, formula, bovine or human fortification, and other supplements all within the course of a few weeks.28, 31, 32 Additionally, the use of antibiotics is prevalent. More than 75% of very low birth weight infants admitted to neonatal intensive care units in the United States receive antibiotics in the first days of life.33 The goal of this study was to characterize the growth of select gut microbes present in the neonatal gut to determine mechanistic associations between human milk and formula. Since alterations in the infant gut microbiota have been linked to childhood health and development, understanding how individual bacteria differentially utilize whole human milk or formula may provide insights into which microbes are driven by diet-related changes.
Experimental
Consent and Human Milk Preparation
This study was approved by the Medical University of South Carolina Institutional Review Board (#103782). Consent was obtained from 3 mothers of preterm infants who were admitted to the Shawn Jenkin’s Hospital Neonatal Intensive Care Unit. Maternal details are provided in Supplemental Table 1. Mothers were educated about hygienic pumping techniques by hospital lactation specialists as part of standard practice. Expressed milk was frozen at −20°C and stored in the hospital’s nutrition management center before use, which strictly adheres to standard milk-handling protocols.34, 35 To eliminate the impact of natural human milk microbes on the experiments in the present study, milk was pasteurized directly before use at 72°C for 15 seconds, also known as high-temperature short-time (HTST) pasteurization, which also inactivates viruses and milk cells.36–39
Bacterial Culture Conditions
For culturing experiments, Bifidobacteria and Lactobacilli were selected based on their isolation in infant feces.40–50 Streptococci can be found in both human milk and feces51–53 and Enterococcus and Klebsiella have also been identified in infant stool, particularly those born preterm.43, 54–57 Bacteria in Table 1 were grown on agar plates (MRS agar for all Bifidobacteria and Lactobacillus strains and BHI agar for all Streptococcus, Enterococcus, and Klebsiella strains) and then single colonies were used to inoculate rich media (MRS for Bifidobacteria and Lactobacillus species and BHI supplemented with 2% yeast extract and 0.2% cysteine (BHIS) for all Streptococcus Enterococcus, and Klebsiella species). Bacteria were grown in rich media overnight at 37°C in an anaerobic chamber (Anaerobe Systems, AS-150, Morgan Hill, CA). After the incubation, all bacteria were examined for morphology by light microscopy at 40x (Motic AE 2000, Schertz, TX) and cultures were then examined at OD600nm on a Spectronic 200 Spectrophotometer (Carolina Biological Supply Company, Burlington, NC). Sub-cultures were generated by inoculating pre-reduced chemically defined media (LDM4 for Bifidobacteria and Lactobacillus species and ZMB1 Streptococcus, Enterococcus, and Klebsiella species) at an OD600nm of 0.1. To test bacterial growth in response to whole human milk, milk from 3 mothers of preterm infants was pooled, pasteurized at 72°C for 15 seconds, and introduced into bacterial cultures at a 1:100 dilution. Preterm formula, (Neosure Similac® 22 kcal/oz) and term formula with oligosaccharides (Similac® Pro-Advance with HMO, 2-Fl, 20 kcal/oz) was likewise introduced into bacterial cultures at a 1:100 dilution. This dilution was selected because it did not interfere with the optical density, yet significantly supported the growth of bacteria in media. Nutritional content of both preterm human milk and formula was generated from available references (Table 2).58, 59 Growth was monitored after anaerobic incubation at 20 hr by measuring OD600nm. OD600nm measurements were back-calculated to standard curves made from each type of bacteria using the Quantum Tx Bacterial Cell Counter (Logos Biosystems, Annandale, VA) (see Supplemental Table 2). Each experiment was performed two independent times in triplicate.
Table 1:
Bacterial strains and growth conditions used in this study.
| Bacteria | Designation | Rich Media | Defined Media |
|---|---|---|---|
| Bifidobacterium bifidum | MIMB | MRS | LDM4 |
| Bifidobacterium longum | ATCC 55813 | MRS | LDM4 |
| Bifidobacterium longum subsp. infantis | ATCC 15697 | MRS | LDM4 |
| Bifidobacterium dentium | ATCC 27678 | MRS | LDM4 |
| Bifidobacterium gallicum | ATCC 20093 | MRS | LDM4 |
| Bifidobacterium angulatum | ATCC 27535 | MRS | LDM4 |
| Bifidobacterium breve | ATCC 15698 | MRS | LDM4 |
| Bifidobacterium animalis | Bi-07 | MRS | LDM4 |
| Lactobacillus brevis | ATCC 27303 | MRS | LDM4 |
| Lactobacillus gasseri | ATCC 3323 | MRS | LDM4 |
| Lactobacillus acidophilus | ATCC 4796 | MRS | LDM4 |
| Lactobacillus plantarum | ATCC 14917 | MRS | LDM4 |
| Lactobacillus johnsonii | ATCC 33200 | MRS | LDM4 |
| Lactobacillus paracasei | ATCC 25302 | MRS | LDM4 |
| Lactobacillus fermentum | ATCC 14931 | MRS | LDM4 |
| Lactobacillus delbrueckii | ATCC 11842 | MRS | LDM4 |
| Streptococcus salvarius | ATCC 13419 | BHIS | ZMB1 |
| Streptococcus sanguinis | ATCC 49296 | BHIS | ZMB1 |
| Streptococcus parasanguinis | ATCC 15912 | BHIS | ZMB1 |
| Streptococcus thermophilus | ATCC 491 | BHIS | ZMB1 |
| Streptococcus mitis | ATCC 6249 | BHIS | ZMB1 |
| Streptococcus pasteurianus | ATCC 49133 | BHIS | ZMB1 |
| Streptococcus oralis | ATCC 35037 | BHIS | ZMB1 |
| Streptococcus pyogenes | CB1 | BHIS | ZMB1 |
| Enterococcus faecalis | NCTC 775 | BHIS | ZMB1 |
| Enterococcus faecium | ATCC 8459 | BHIS | ZMB1 |
| Enterococcus faecium | NCTC 12204 | BHIS | ZMB1 |
| Klebsiella aerogenes | CB1 | BHIS | ZMB1 |
| Klebsiella aerogenes | ATCC 35029 | BHIS | ZMB1 |
| Klebsiella pneumoniae | ATCC 700607 | BHIS | ZMB1 |
| Klebsiella pneumoniae | ATCC 13883 | BHIS | ZMB1 |
| Klebsiella pneumoniae | ATCC 35657 | BHIS | ZMB1 |
Table 2.
Nutrient comparison between milk and formula.
| Macronutrients & minerals (per 100 mL) | Preterm Human Milk* (Ref values51’ 84) | Preterm formula Similac® Neosure® | Similac® ProAdvance with HMO, 2-Fl |
| Calories, kcal | 70–80 | 74 | 68 |
| Protein, g/dL | 2.1–2.7 | 2.1 | 1.4 |
| Fat (%) | 3–4.1 | 4.1 | 3.8 |
| Carbohydrates, g | 6.7–8.5 | 7.5 | 7.1 |
| IgA, Lactoferrin, Lysozyme g | 1.3, 4–5, 3–4 | - | - |
| HMOs, g | 1.5–2 | - | Quantity Not reported |
| Sodium, meq | 0.6–1 | 1 | 0.7 |
| Potassium, meq | 1–1.7 | 2.7 | 1.8 |
| Calcium, mg | 25 | 78 | 53 |
| Magnesium, mg | 2–5 | 6.7 | 4.1 |
| Chloride, meq | 1–2 | 1.6 | 1.2 |
| Phosphate, mg | 9–18 | 46 | 28 |
| Iron, mg | 0.7–1.6 | 1.3 | 1.2 |
| Zinc, mg | 0.1–0.3 | 0.9 | 0.5 |
| Manganese, mcg | 3–6 | 7.4 | 3.4 |
| Copper, mcg | 20–40 | 89 | 61 |
| Selenium, mcg | 1.5–2.5 | 1.7 | 1.4 |
| Iodine, mcg | 3–17* | 11 | 10 |
| Vitamins (per 100mL) | Preterm Human Milk (Ref values51, 102¥) | Preterm formula Similac® Neosure® | Similac® ProAdvance with HMO, 2-Fl |
| Vitamin A, IU | 100–200 | 260 | 203 |
| Vitamin D, IU | 0.4–4 | 52 | 41 |
| Vitamin E, IU | 0.4–1 | 3 | 1 |
| Vitamin K, mcg | 0.2–0.3 | 8 | 5 |
| Thiamin (B1), mcg | 22–23 | 130 | 68 |
| Riboflavin (B2), mcg | 48–58 | 111 | 101 |
| Vitamin B6, mcg | 13–31 | 74 | 41 |
| Vitamin B12, mcg | 0.06–0.1 | 0.3 | 0.2 |
| Niacin, mcg | 180–230 | 1450 | 710 |
| Folic Acid (Folacin), m | :g 8–13 | 19 | 10 |
| Biotin, mcg | 0.5 | 6.7 | 3.0 |
| Vitamin C, mg | 3.8 | 1 1 | 6 |
| Choline, mg | <0.1 | 12 | 16 |
| Inositol, mg | 22 | 26 | 16 |
| Panthothenic Acid, mc | g 250 | 595 | 304 |
| Fatty Acids (Per 100ml) | Preterm Human Milk (Ref values50, 51) | Preterm formula Similac® Neosure® | Similac® ProAdvance with HMO, 2-Fl, |
| Linoleic Acid, mg | 400–500 | 750 | 676 |
Mature and transitional human milk is defined as 30–60 days in post-partum. Term-gestation infant milk reference values were substituted for premature gestation milk for reference values of contents (Fe, Zn, Mn, Cu, Se, vitamins, and linoleic) acid due to lack of available references ranges.
Highly geographically dependent.
Preterm milk ranges of water-soluble vitamins are highly dependent on stage of lactation, maternal intake and preterm delivery, and term-infant ranges were used for B1, B2, B6, B12, Niacin, Folic Acid, Biotin, Vitamin C, and Panthothenic Acid using populations from the Western countries and the U.S.A. Formula nutrition is listed on manufacture’s website.
pH assay
Bacterial supernatant pH was measured using a UV-Vis absorbance measurement as previously described.60 Briefly, a standard curve was generated of ZMB1 or LDM4 at the pH values 2–8 in 0.5 increments. These standards were mixed with Dulbecco’s Modified Eagle’s Medium (DMEM) at a ratio 1 volume bacterial media: 3 volumes DMEM. DMEM contains phenol red which has an isosbestic point of 470 nm (a wavelength where the light absorption of an indicator dye is independent of solution pH and a lambda max (λmax) of 560 nm.) To measure pH, bacterial cultures were centrifuged to pellet bacteria and 1 volume of bacterial culture was added to 3 volumes DMEM and the absorbance was measured at 470 nm and 560 nm. The ratio of these values was back-calculated to the standard curve to approximate pH for all cultures.
Genome Analysis
Glycosyl hydrolase families associated with human milk oligosaccharide consumption from human gut microbes were examined using the Carbohydrate-Active enZYmes (CAZy) database (www.cazy.org) as previously described.61–64 HMO-related GH’s include: GH33, GH2, GH20, GH95, GH29, GH42, GH101, GH89, GH110, GH112, and GH27. Glycosyl hydrolase families were examined in Bifidobacteria (179 genomes representing 14 different species), Lactobacilli (452 genomes representing 23 different species), Streptococci (526 genomes representing 19 different species), Enterococcus (228 genomes, representing 9 different species), and Klebsiella (884 genomes, representing 10 species). We also identified bacterial transporters involved in long-chain-fatty acid (LCFA) uptake: fadL (K06076); iron uptake: TonB (K03832), exbB (K03561), FeoB (K04759), FeoA (K04758), ECF (K16787) and FepA (K19611); as well as phosphate uptake: PiT (K16322) and pstS (K02040) using the KEGG database (KEGG: Kyoto Encyclopedia of Genes and Genomes, https://www.genome.jp). KEGG IDs were input into the gene search tool of the Integrated Microbial Genomes (IMG) database v5.0 (http://img.jgi.doe.gov)65 and genomes were binned for analysis as previously described.48 Bacterial transporters were identified in the following pathobionts: E. faecalis (416 genomes), E. faecium (488 genomes), Enterococcus spp. (16 genomes), K. aerogenes (102 genomes), K. pneumoniae (1,049 genomes), and Klebsiella spp. (71 genomes). Bacterial transporters were also identified in the following commensals: B. angulatum (4 genomes), B. animalis (37 genomes), B. bifidum (33 genomes), B. breve (66 genomes), B. dentium (7 genomes), B. longum (126 genomes), B. longum subsp infantis (36 genomes), Bifidobacterium spp. (10 genomes), L. acidophilus (23 genomes), L. johnsonii (18 genomes), L. paracasei (154 genomes), L. plantarum (215 genomes), L. fermentum (42 genomes), L. gasseri (15 genomes), L. delbrueckii (48 genomes), L. brevis (54 genomes), Lactobacillus spp. (44 genomes), S. salivarius (19 genomes), S. sanguinis (28 genomes), S. parasanguinis (26 genomes), S. pyogenes (351 genomes), S. thermophilus (34 genomes), S. mitis (41 genomes), S. pasteurianus (7 genomes), S. oralis (27 genomes) and Streptococcus spp. (92 genomes).
Statistics
Data are presented as mean ± standard deviation (stdev). Comparisons between groups were made with a One-way Analysis of Variance (ANOVA), using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons. To determine associations between factors, a Pearson correlation was performed and the coefficient (r) was recorded. Graphs and statistics were generated using GraphPad (GraphPad Software, Inc., La Jolla, CA). A *p < 0.05 value was considered significant as indicated by *, while n is the number of experiments performed.
Results
To determine which gastrointestinal microbes responded to human milk and formula, we systematically examined the growth of commensal and pathobiont gut and breast milk associated microorganisms. We inoculated a chemically defined media, LDM4, with B. longum subspecies infantis ATCC 15697, B. breve ATCC 15698, B. bifidum MIMB, B. angulatum ATCC 27535, B. animalis Bi-07, B. longum ATCC 55813, B. dentium ATCC 27678 and B. gallicum ATCC 20093 at an OD600nm of 0.1 and examined growth after 20 hr. We observed that all Bifidobacteria spp. had elevated growth in response to pasteurized preterm human milk compared to media alone controls (Figure 1A–H). As a comparison, we also examined the growth of Bifidobacteria spp. in LDM4 supplemented with preterm infant formula or term infant formula with the human milk oligosaccharide (HMO) 2’-Fucosyllactose (2-FL). We found that preterm formula enhanced the growth of 6 of the 8 strains: B. infantis, B. breve, B. bifidum B. angulatum, B. animalis, and B. longum (Figure 1A–F). Full-term formula containing HMOs only elevated the growth of 5 strains: B. infantis, B. breve, B. bifidum, B. angulatum, and B. animalis (Figure 1A–D).
Figure 1: Bifidobacteria species growth with human milk, preterm and term formula.
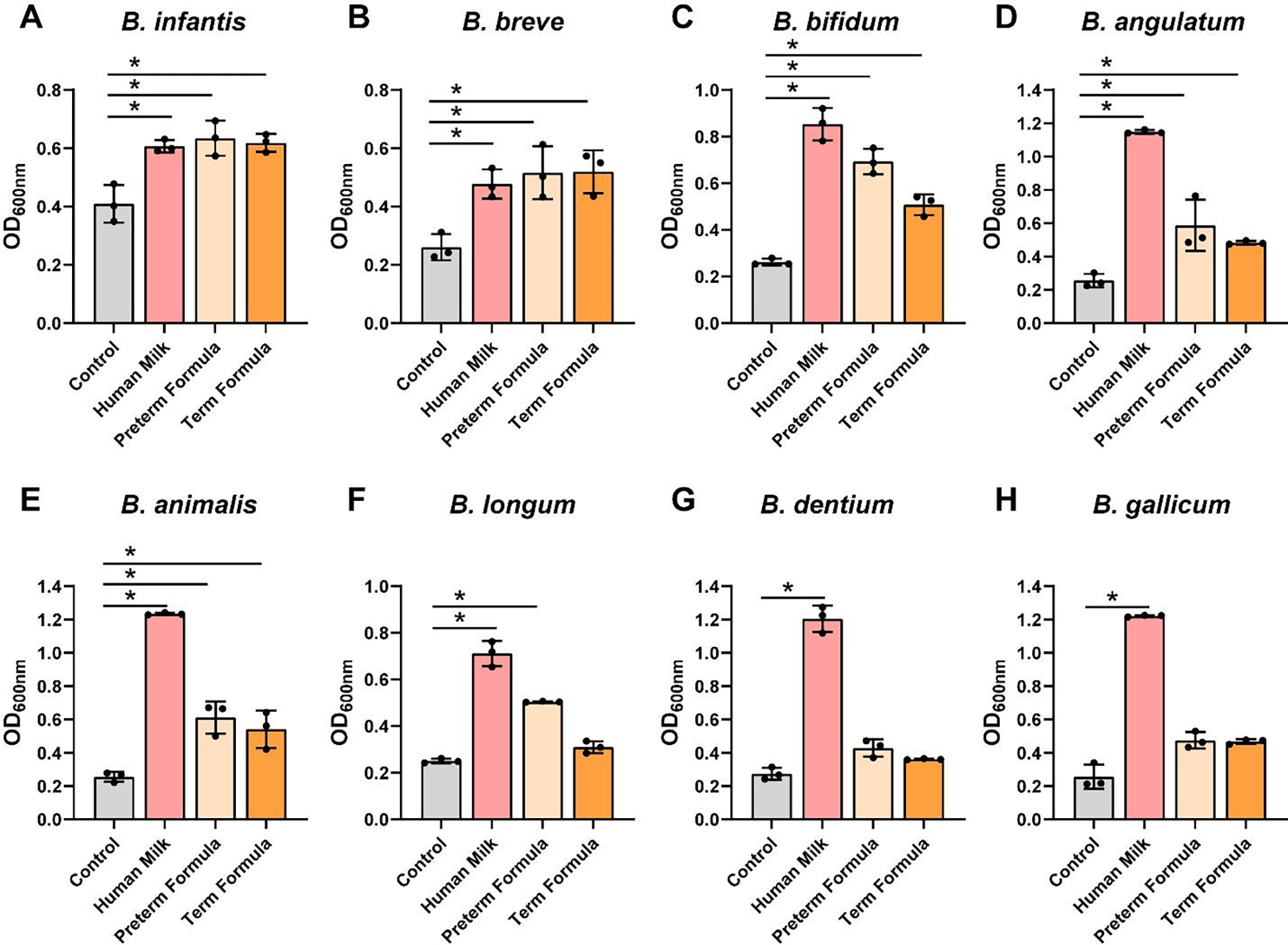
Bifidobacteria species A. B. longum subsp. infantis ATCC 15697, B. B. breve ATCC 15698, C. B. bifidum MIMB, D. B. angulatum ATCC 27535, E. B. animalis Bi-07, F. B. longum ATCC 55813, G. B. dentium ATCC 27678, and H. B. gallicum ATCC 20093 were grown anaerobically at 37°C in a chemically-defined media (LDM4) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. Growth was monitored at OD600nm after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA, using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p < 0.05.
Lactic acid bacteria such as Bifidobacteria are known for their ability to lower the pH and pH can serve as a functional output of microbial growth.66 Consistent with our growth analysis, we found that all Bifidobacteria spp. grown with preterm milk exhibited significant reductions in pH (Figure 2A–H). Similarly, preterm formula was associated with reduced pH in 6 of the Bifidobacteria strains and full-term formula with HMOs reduced pH in 3 of the strains. These data indicate that the majority of Bifidobacteria spp. can grow with preterm milk and formula.
Figure 2: Bifidobacteria species acidification in cultures with human milk, preterm and term formula.
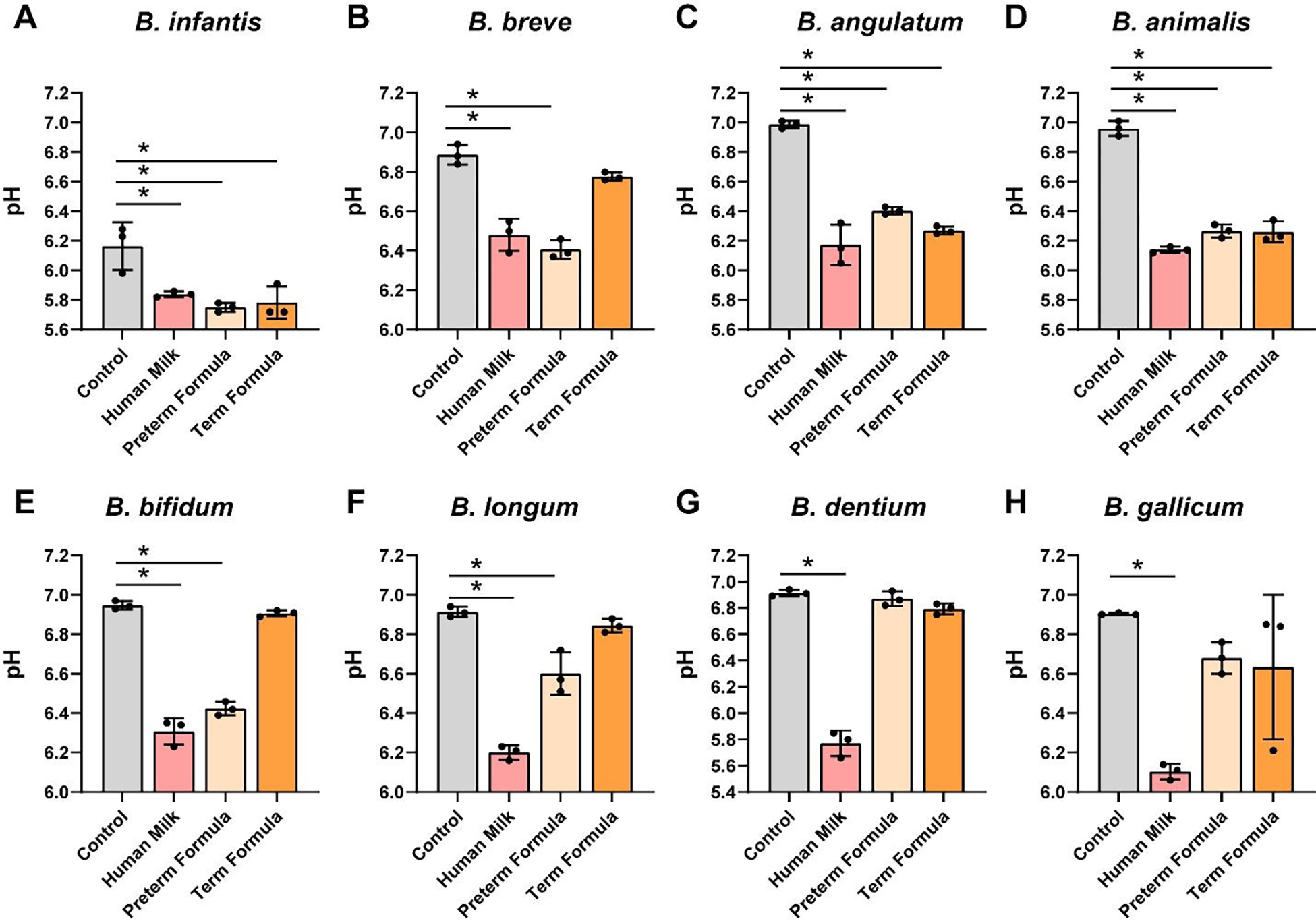
Bifidobacteria species A. B. longum subsp. infantis ATCC 15697, B. B. breve ATCC 15698, C. B. bifidum MIMB, D. B. angulatum ATCC 27535, E. B. animalis Bi-07, F. B. longum ATCC 55813, G. B. dentium ATCC 27678, and H. B. gallicum ATCC 20093 were grown anaerobically at 37°C in a chemically-defined media (LDM4) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. pH was monitored after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA using the Holm-Sidak post-hoc test to determine significance between pairwise comparison; *p < 0.05.
We observed that all Lactobacillus spp. (L. acidophilus ATCC 4796, L. johnsonii ATCC 33200, L. paracasei ATCC 25302, L. plantarum ATCC 14917, L. fermentum ATCC 14931, L. gasseri ATCC 3323, L. delbrueckii ATCC 11842, and L. brevis ATCC 27303) had increased growth in the presence of preterm human milk and preterm formula compared to media controls (Figure 3A–H). Six of the 8 Lactobacillus strains had elevated growth with full-term formula supplemented with HMOs. In the presence of preterm milk, L. acidophilus, L. paracasei, L. plantarum, L fermentum, L. gasseri and L. delbrueckii significantly reduced the pH compared to media controls (Figure 4A–H). Although we observed increased growth in the presence of preterm formula, we found that only 6 of the strains (L. paracasei, L. fermentum, L. gasseri and L. delbrueckii) lowered the pH in response to preterm milk. Moreover, only 3 strains reduced the pH in media supplemented with formula with HMOs. These findings suggest that lactic acid production by Lactobacilli is influenced by multiple factors and that formula supplementation does not necessarily promote pH reduction.
Figure 3: Lactobacillus species growth with human milk, preterm and term formula.
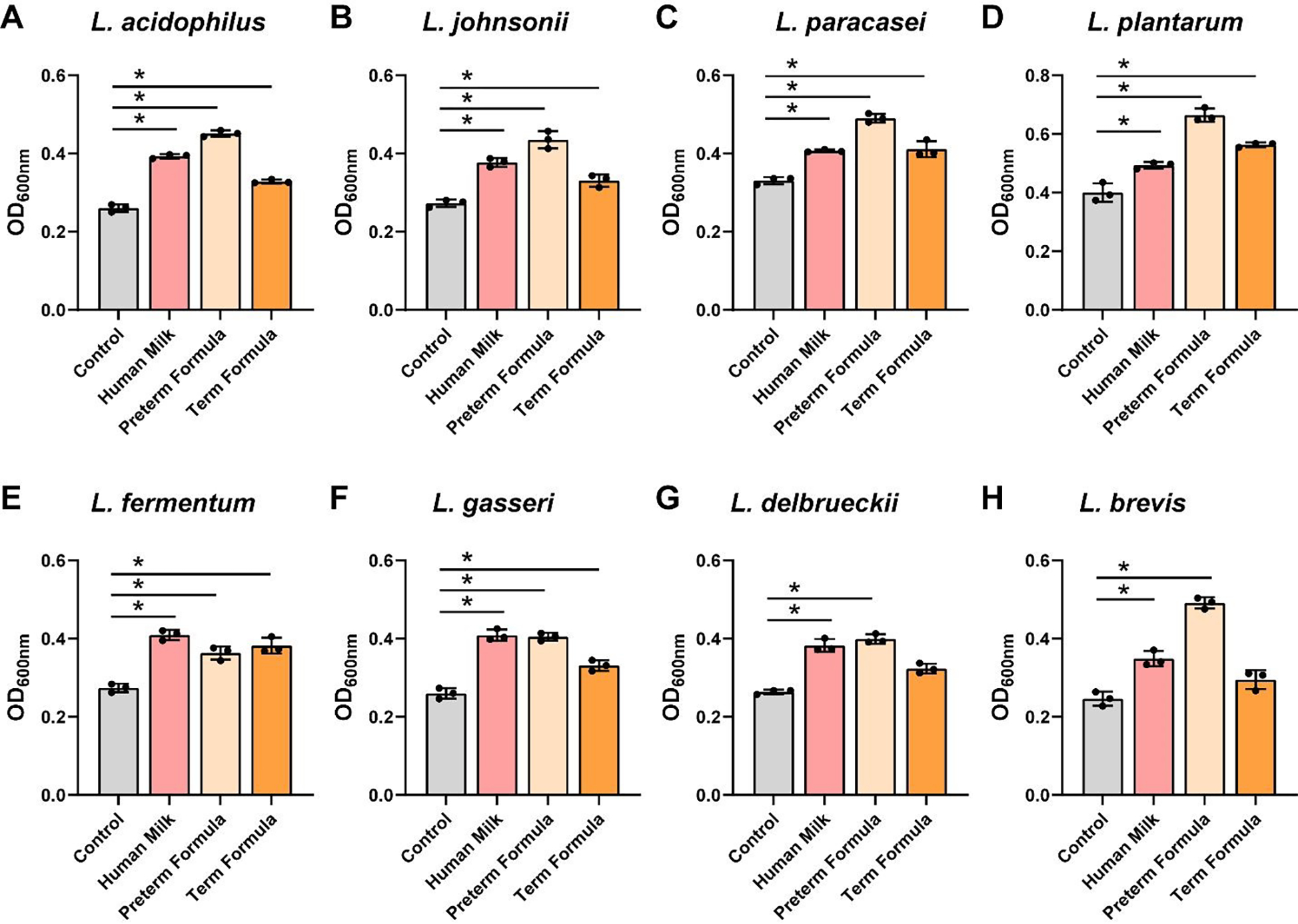
Lactobacillus species A. L. acidophilus ATCC 4796, B. L. johnsonii ATCC 33200, C. L. paracasei ATCC 25302, D. L. plantarum ATCC 14917, E. L. fermentum ATCC 14931, F. L. gasseri ATCC 3323, G. L. delbrueckii ATCC 11842, and H. L. brevis ATCC 27303 were grown anaerobically at 37°C in a chemically-defined media (LDM4) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. Growth was monitored at OD600nm after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA, using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p < 0.05.
Figure 4: Lactobacillus species acidification in cultures with human milk, preterm and term formula.
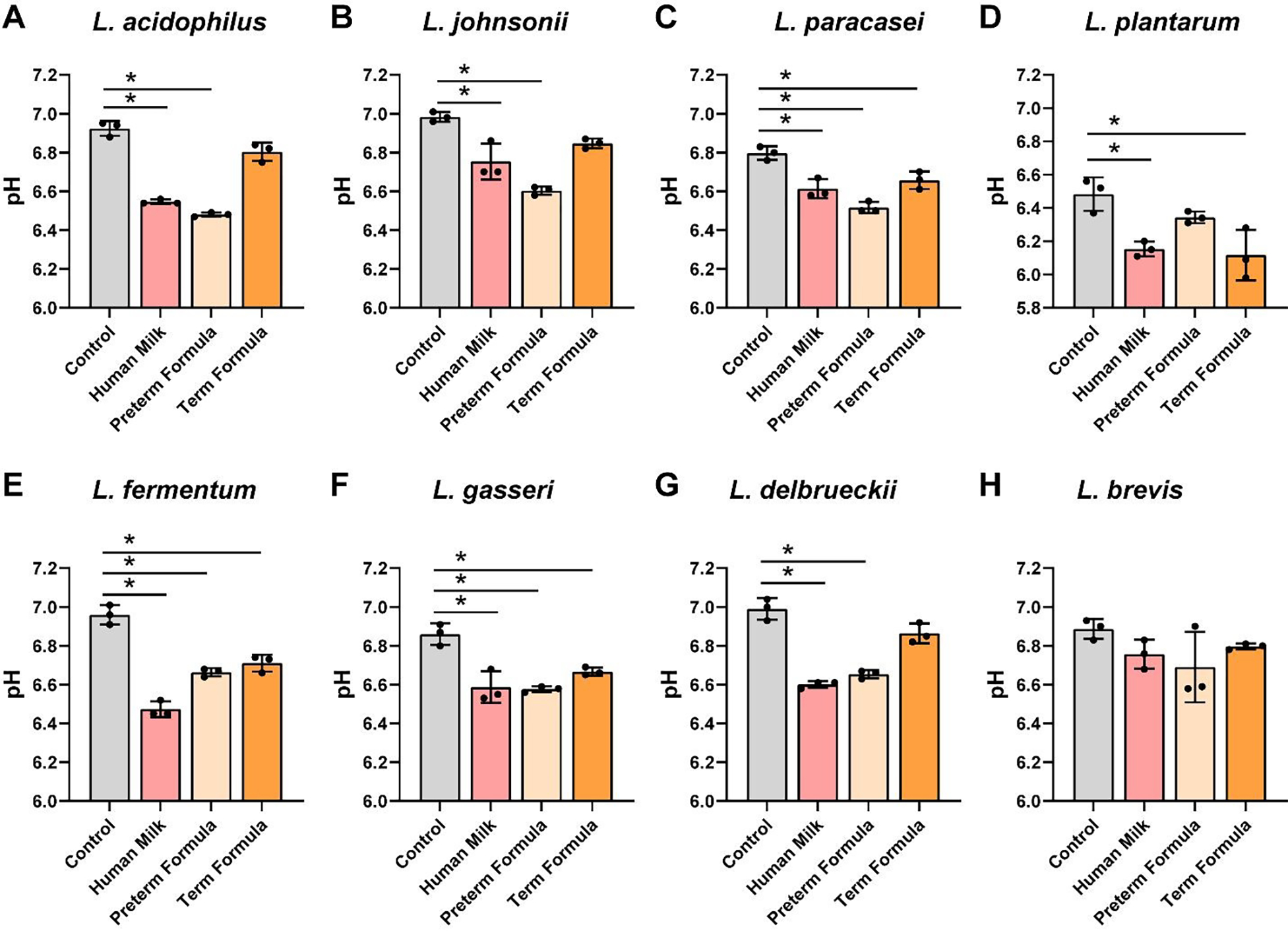
Lactobacillus species A. L. acidophilus ATCC 4796, B. L. johnsonii ATCC 33200, C. L. paracasei ATCC 25302, D. L. plantarum ATCC 14917, E. L. fermentum ATCC 14931, F. L. gasseri ATCC 3323, G. L. delbrueckii ATCC 11842, and H. L. brevis ATCC 27303 were grown anaerobically at 37°C in a chemically-defined media (LDM4) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. pH was monitored after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA, using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p< 0.05.
To assess the effects of preterm milk and formula on Streptococci growth, we grew S. salivarius ATCC 13419, S. sanguinis ATCC 49296, S. parasanguinis ATCC 15912, S. thermophilus ATCC 491, S. pyogenes CB1, S. mitis ATCC 6249, S. pasteurianus ATCC 49133, and S. oralis ATCC 35037 in another chemically defined media, ZMB1, and examined growth after 20 hr of incubation at OD600nm (Figure 5A–H). We found that only 2 Streptococcus spp. grew with preterm human milk compared to media controls: S. sanguinis and S. parasanguinis (Figure 5B,C). Moreover, 2 strains were suppressed by preterm milk: S. salivarius and S. thermophilus (Figure 5A,D). Also, 3 Streptococcus strains could use preterm and full-term formula with HMOs to support their growth. Of the Streptococci examined, we found that S. pyogenes, S. mitis, S. pasteurianus and S. oralis did not respond in terms of growth to any dietary interventions. As members of the lactic acid bacteria group, Streptococci can also produce lactic acid and lower the pH.67 When we assessed media pH (Figure 6A–H), we found that S. sanguinis and S. parasanguinis, which had improved growth with preterm milk, lowered the pH compared to media controls (Figure 6B,C). We also found that 3 Streptococcus spp. reduced the pH in the presence of preterm formula and 2 strains reduced the pH in presence of full-term formula with HMOs. No changes in pH were observed in S. thermophilus, S. mitis, S. pasteurianus or S. oralis (Figure 6D,F–H). These data indicate that not all Streptococci can use human milk or formula to support their growth.
Figure 5: Streptococcus species growth with human milk, preterm and term formula.
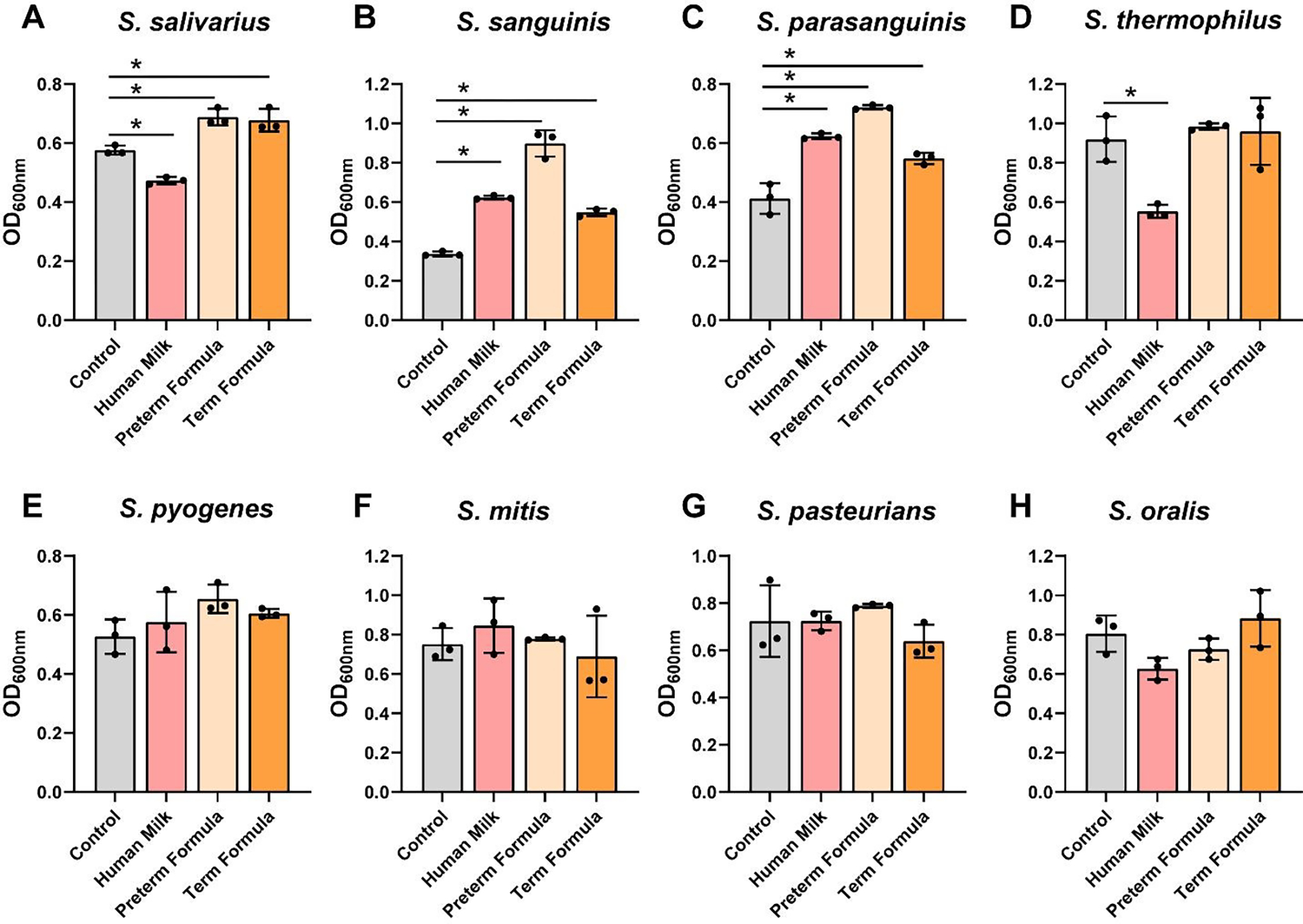
Streptococcus species A. S. salivarius ATCC 13419, B. S. sanguinis ATCC 49296, C. S. parasanguinis ATCC 15912, D. S. thermophilus ATCC 491, E. S. pyogenes CB1, F. S. mitis ATCC 6249, G. S. pasteurianus ATCC 49133, and H. S. oralis ATCC 35037 were grown anaerobically at 37°C in a chemically-defined media (ZMB1) with or pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. Growth was monitored at OD600nm after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA, using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p < 0.05.
Figure 6: Streptococcus species acidification in cultures with human milk, preterm and term formula.
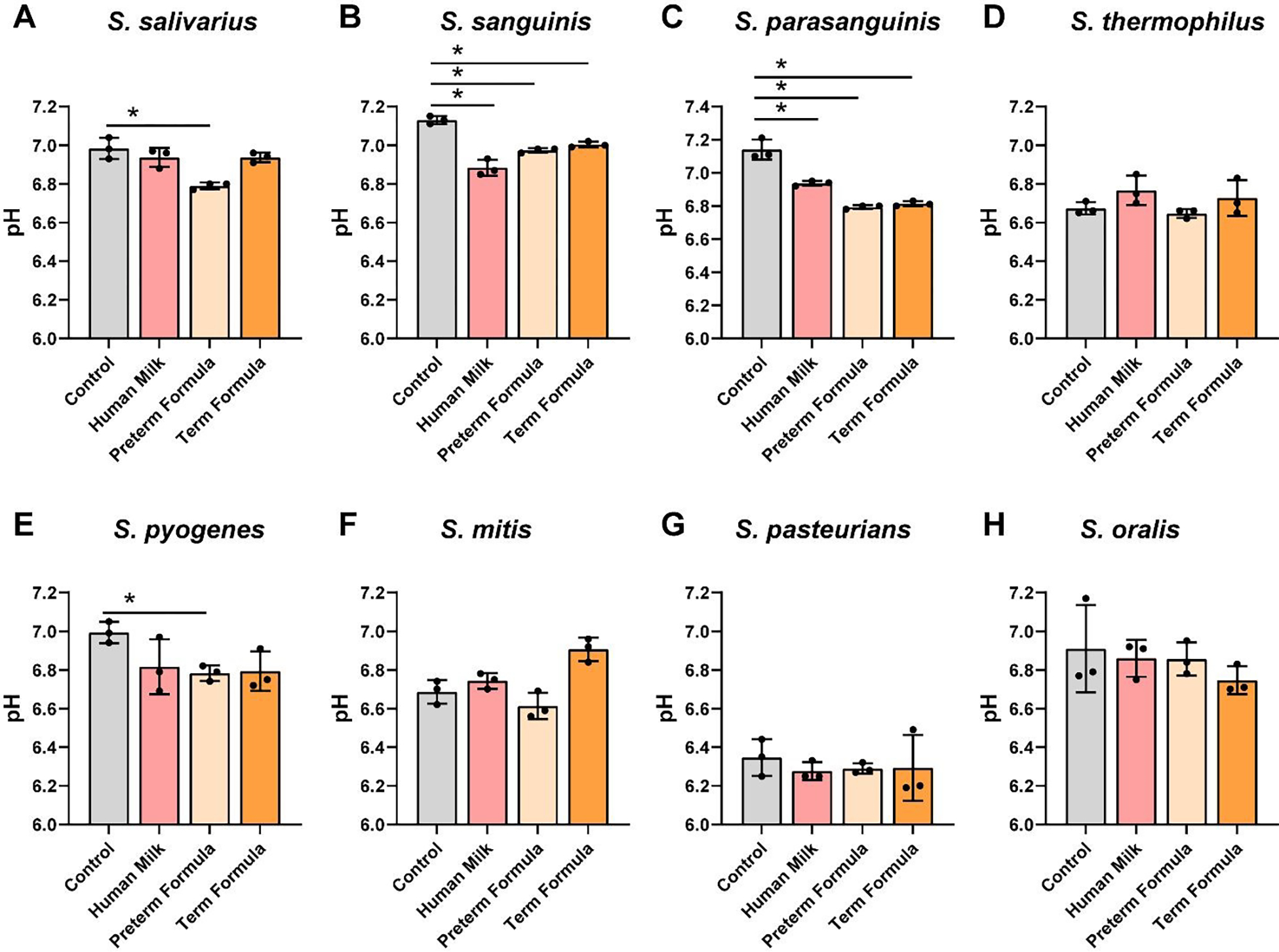
Streptococcus species A. S. salivarius ATCC 13419, B. S. sanguinis ATCC 49296, C. S. parasanguinis ATCC 15912, D. S. thermophilus ATCC 491, E. S. pyogenes CB1, F. S. mitis ATCC 6249, G. S. pasteurianus ATCC 49133, and H. S. oralis ATCC 35037 were grown anaerobically at 37°C in a chemically-defined media (ZMB1) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. Growth was monitored at OD600nm after 20 hr. pH was monitored after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p< 0.05.
To address pathobiont species, we grew E. faecalis NCTC 775, E. faecium ATCC 8459, E. faecium NCTC 12204, K. aerogenes CB1, K. aerogenes ATCC 35029, K. pneumoniae ATCC 700607, K. pneumoniae ATCC 13883, and K. pneumoniae ATCC 35657 in ZMB1 in the presence of preterm milk or formula (Figure 7A–H). Compared to commensal Bifidobacteria, Lactobacilli and Streptococci, the Enterococcus and Klebsiella spp. only nominally used preterm milk for growth. All the pathobiont strains exhibited robust growth with preterm formula and full-term formula harboring HMOs. The highest OD600nm values were largely obtained with preterm formula. Enterococcus spp. are part of the lactic acid bacteria group and although these species had only a minimal improvement in growth with preterm milk, all 3 Enterococcus spp. significantly reduced the pH in the presence of milk (Figure 8A–C). Reduced pH was also found in response to formula and HMO containing formula for all Enterococcus spp. In contrast to Enterococcus spp., Klebsiella strains had no effect on pH (Figure 8D–H).
Figure 7: Enterococcus and Klebsiella species growth with human milk, preterm and term formula.
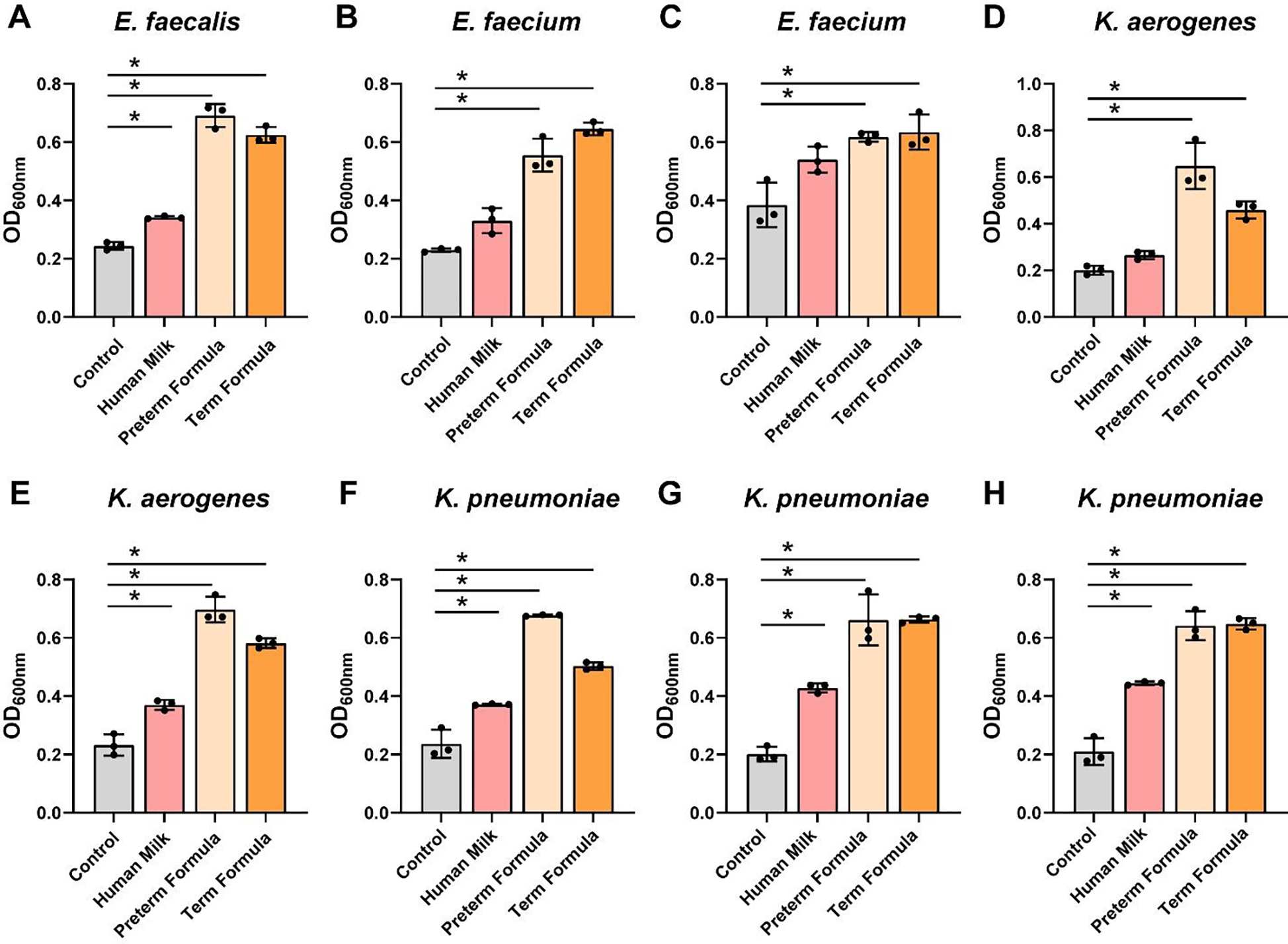
Pathobiont species A. Enterococcus faecalis NCTC 775, B. Enterococcus faecium ATCC 8459, C. Enterococcus faecium NCTC 12204, D. Klebsiella aerogenes CB1, E. Klebsiella aerogenes ATCC 35029, F. Klebsiella pneumoniae ATCC 700607, G. Klebsiella pneumoniae ATCC 13883, and H. Klebsiella pneumoniae ATCC 35657 were grown anaerobically at 37°C in a chemically-defined media (ZMB1) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. Growth was monitored at OD600nm after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA, using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; * p < 0.05.
Figure 8: Enterococcus and Klebsiella species acidification in cultures with human milk, preterm and term formula.
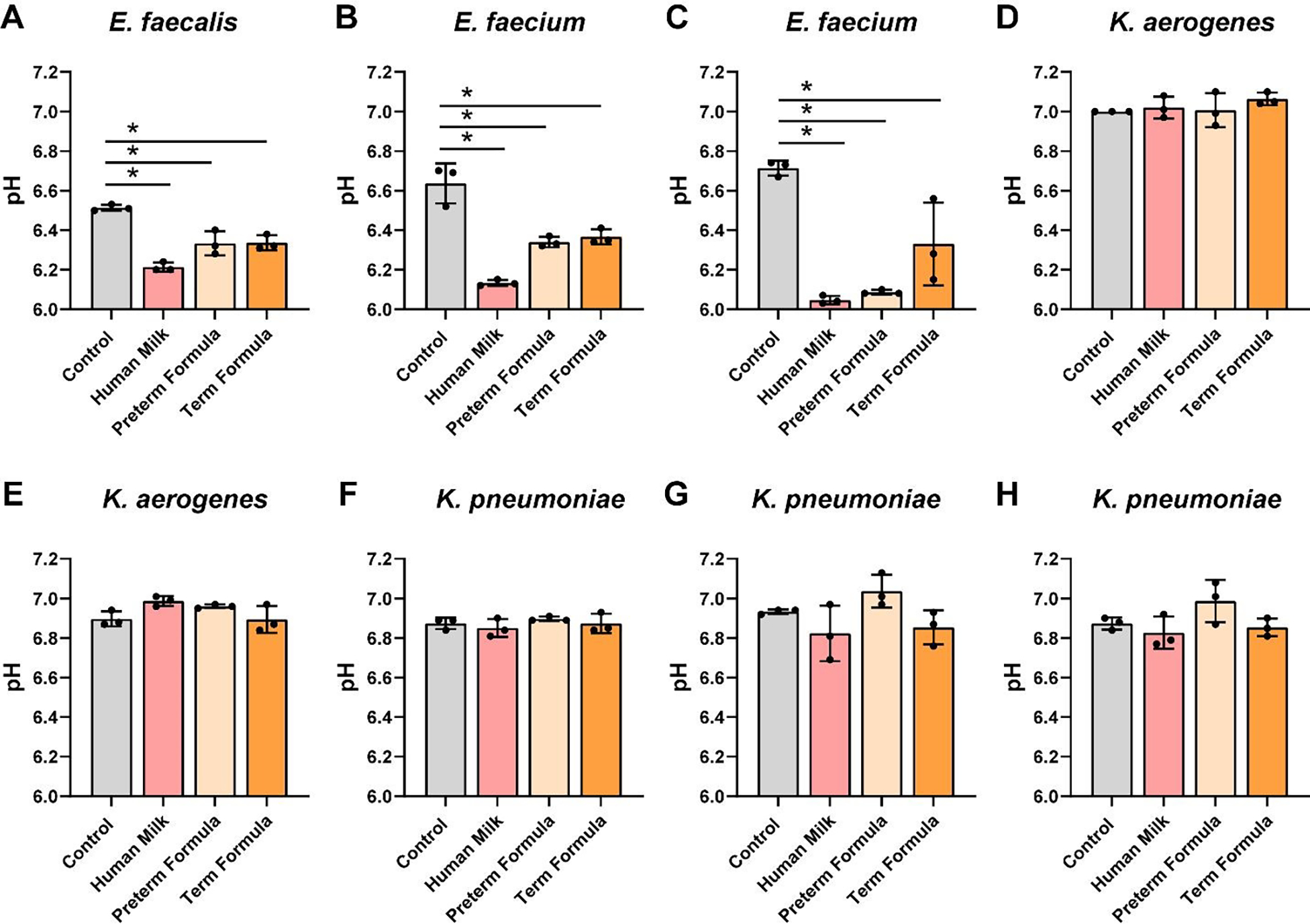
Pathobiont species A. Enterococcus faecalis NCTC 775, B. Enterococcus faecium ATCC 8459, C. Enterococcus faecium NCTC 12204, D. Klebsiella aerogenes CB1, E. Klebsiella aerogenes ATCC 35029, F. Klebsiella pneumoniae ATCC 700607, G. Klebsiella pneumoniae ATCC 13883, and H. Klebsiella pneumoniae ATCC 35657 were grown anaerobically at 37°C in a chemically-defined media (ZMB1) with or without pooled whole preterm breast milk (pooled from n=3), preterm formula or full-term formula supplemented with HMOs. pH was monitored after 20 hr. All data are presented as mean ± stdev, n=3 replicates, performed 2 independent times. One-way ANOVA using the Holm-Sidak post-hoc test to determine significance between pairwise comparisons; *p< 0.05.
Finally, we sought to determine if there was a correlation between diet (formula vs. milk) and microbial composition. One unique component of human milk is human milk HMOs (see Table 2). To degrade HMOs, bacteria must possess specific glycosyl hydrolases which enzymatically cleave HMOs. HMO degrading glycosyl hydrolase families include GH33, 2, 20, 95, 29, 42, 101, 89, 110, and 27. We profiled HMO-related glycosyl hydrolase families using the CAZy database and filtered human gut microbes using the Integrated Microbial Genome Database (IMG). We identified 179 Bifidobacteria genomes (14 different species), 452 Lactobacilli genomes (23 species), 526 Streptococci genomes (19 species), 228 Enterococcus genomes (9 species) and 884 Klebsiella genomes (10 species). Genome analysis revealed that the majority of Bifidobacteria spp. harbored multiple HMO-related glycosyl hydrolases, with B. bifidum and B. longum subspecies infantis exhibiting the largest repertoire (Figure 9A). Compared to Bifidobacteria, Lactobacilli had far fewer HMO-related glycosyl hydrolases (Figure 9B); although most species had GH2 and GH42. Streptococci had varying levels of HMO-related glycosyl hydrolases with species such as S. mitis, S. oralis, and S. sanguinis, which harbour the highest number of glycosyl hydrolase families (Figure 9C). Enterococcus and Klebsiella genomes had fewer HMO-related glycosyl hydrolases than Bifidobacteria; with most species possessing GH2 and GH42. To assess whether glycosyl hydrolase profiles correlated with growth in response to milk or formula, we calculated the total number of HMO-related glycosyl hydrolase families (1–11 GHs) and correlated these values with fold change growth compared to media alone controls (Figure 9E–J). Regression curves revealed a correlation between HMO-related glycosyl hydrolases and commensal growth in human milk (*p = 0.03) (Figure 9E). No correlations were identified between preterm or full-term formula and HMO-degrading glycosyl hydrolases expression in commensal organisms (Figure 9F,H). Additionally, no correlations were found between the HMO-related glycosyl hydrolases in pathobiont microbes and the consumption of human milk or formula (Figure 9H–J). These data indicate that HMOs contribute to the growth of commensal microbes in preterm milk, but that other components also are participating in the growth of certain commensal and pathobiont organisms.
Figure 9. Heat Maps and Correlation between Growth and HMO-related Glycosyl Hydrolase Families.
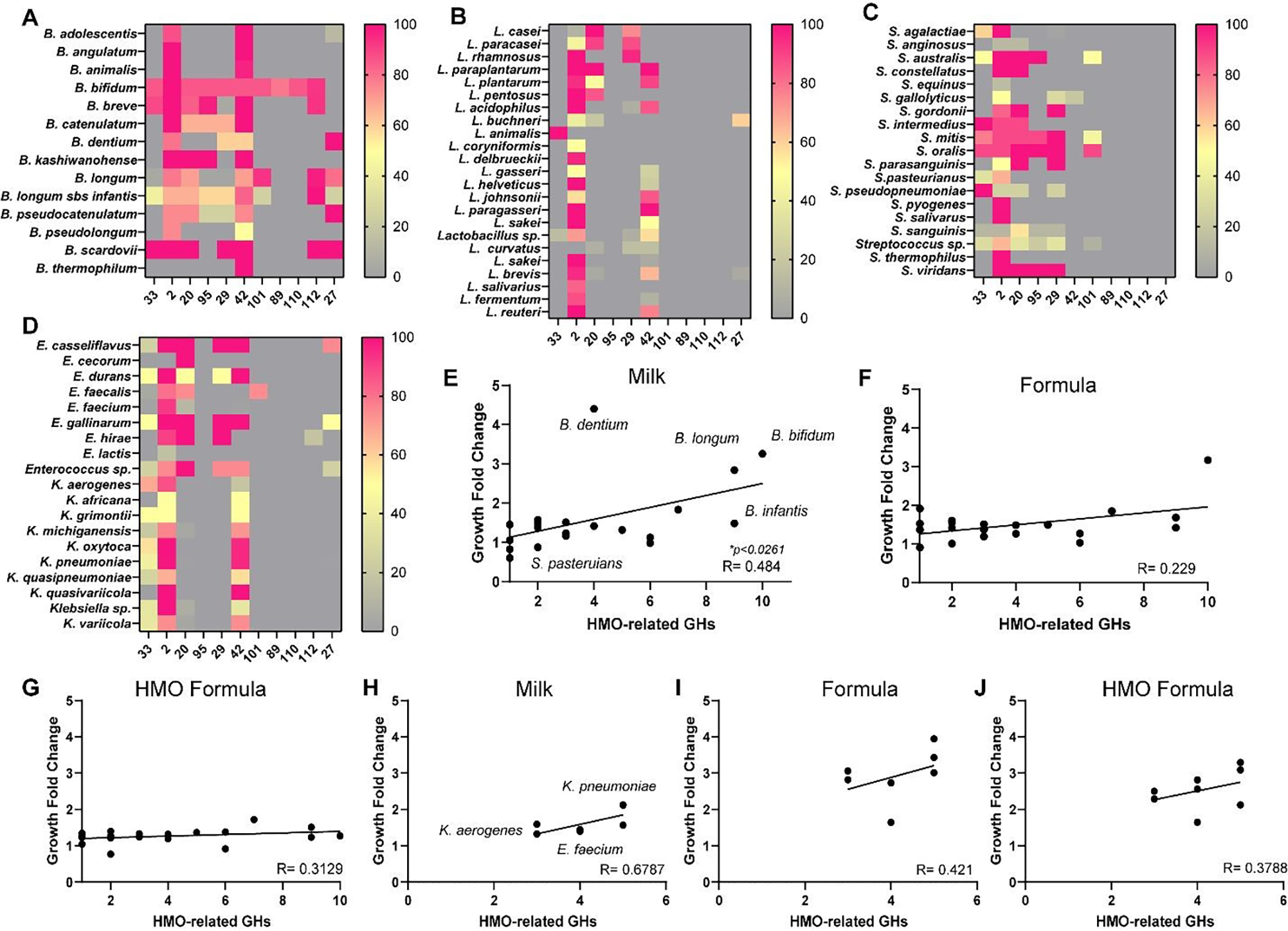
Heat map of the percentage genomes that have a least one gene copy of Human Milk Oligosaccharide (HMO)-related Glycosyl hydrolase families. GH profiles were examined in A. Bifidobacterium, B. Lactobacillus, C. Streptococcus, and D. Enterococcus and Klebsiella species. Correlation analysis of the number of HMO-related GH families with the average fold change of growth in commensal Bifidobacterium, Lactobacillus and Streptococcus species in the presence of E. pooled preterm human breast milk, F. preterm formula, or G. HMO formula. Correlation analysis of the number of HMO-related GH families with the fold change of growth in pathobiont Enterococcus and Klebsiella species in the presence of H. human breast milk, I. preterm formula, or J. HMO formula. P values are shown in the figure.
A comparison between breast milk and formula revealed that formula has significantly higher levels of linoleic acid (a long-chain fatty acid), iron and phosphate (Table 2). We speculated that these components may be driving the growth enhancement of pathobionts. To address this, we examined bacterial transporters involved in the transport of long-chain-fatty acid (fadL); iron (TonB, exbB, FeoB, FeoA, ECF and FepA) and phosphate (PiT, pstS) using the KEGG database and Integrated Microbial Genomes (IMG) database. We examined 319 Bifidobacteria genomes, 613 Lactobacilli genomes, 625 Streptococci genomes, 920 Enterococci genomes, and 1,222 Klebsiella genomes. We found that almost all Klebsiella possessed the long-chain fatty acid transporter fadL, while this gene was absent in Bifidobacteria, Lactobacilli, Streptococci and Enterococci (Figure 10A,B). We observed a similar trend for iron transporter components TonB and exbB. Some expression on iron transporters FeoB, FeoA and ECF were found in the commensal microbes, but in general, far more iron transporters were observed in pathobiont Enterococci and Klebsiella. No commensal microbes possessed the phosphate transporter PiT, while all K. aerogenes and K. pneumoiae harbored this gene. All bacteria were found to have the pstS gene. These data suggest that iron uptake in pathobionts may promote the growth of pathogens. Correlation analysis revealed a positive correlation between iron uptake related genes in pathobionts and growth in formula (*p = 0.03) (Figure 10C). We speculate that iron uptake may provide an essential nutrient for pathobiont organisms in the infant gut. Collectively, these data provide more targeted information on which microbes respond to human milk and formula.
Figure 10. Heat Maps and Correlations between Growth and Long-chain Fatty Acid, Iron and Phosphate Transporters.
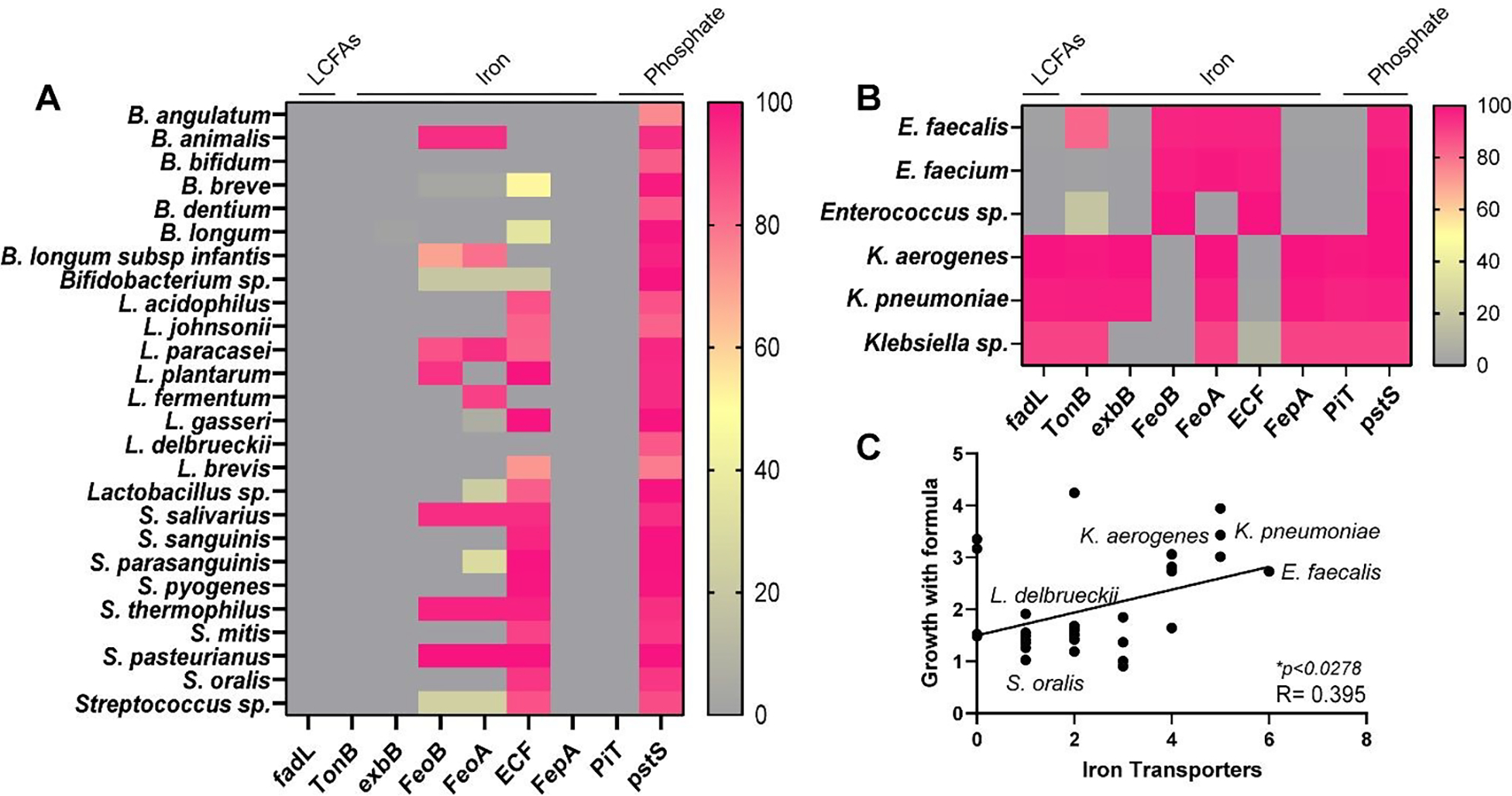
Heat map of the percentage genomes that have a least one gene copy of long-chain fatty acid, iron or phosphate transporters. Transporter profiles were examined in A. Bifidobacterium, Lactobacillus, and Streptococcus species, or B. Enterococcus and Klebsiella species. C. Correlation analysis of the number of iron transporters with the fold change of growth in the presence of formula. P values are shown in the figure.
Discussion
In this study, we sought to dissect how individual microbes respond to intact preterm milk or formula. We used intact diets because microbial communities do not just encounter single compounds in the intestinal milieu. We selected strains based on relevant clinical outcomes noted in the literature. We first examined Bifidobacteria; a group which dominates the gut of term breastfed infants and is associated with numerous health benefits including reducing feeding intolerance, lower rates of necrotizing enterocolitis and a lower the risk of asthma.24, 68 Next, we examined Lactobacilli, another lactic acid bacterial group commonly found in infants which is associated with decreased NEC in clinical trials.69 Streptococci are commonly found in human milk and can make up part of the preterm infant gut microbiota.51, 70, 71 This group has been associated with childhood wheezing72 and NEC.73 The gut microbiota of preterm infants can harbor varying levels of pathobionts linked to NEC and late-onset sepsis such Klebsiella and Enterococcus spp.13–18
Using this approach, we found representative infant gut microbes from the genus Bifidobacteria and Lactobacilli exhibit robust growth in the presence of whole preterm human milk. This phenomenon was not observed with other genera, such as Streptococci, Enterococci and Klebsiella, which had minimal growth with human milk. These studies support the dominance of Bifidobacteria and Lactobacilli in breast-fed infants and the high levels of Enterococcus and Klebsiella in formula-fed infants.26, 67, 73–75 This work can serve as a foundation for more complex interactions with microbial communities and diet.
The composition of breast milk is influenced by multiple factors, including the phase of nursing, lactation period, genetic background of the mother, maternal diet, and gestation length.51, 76–80 It has been shown that the breast milk from mothers who deliver full term infants significantly differs from the milk of mothers whose infants were delivered prematurely.81 Preterm human milk is higher in fat, protein, free amino acids, and sodium in the first few weeks, but these components decrease over subsequent weeks.81 Additionally, calcium is significantly lower in preterm milk than term milk and does not increase with time. HMO content is highly variable, but in general preterm human milk contains more lacto-N-tetraose and the HMOs are not consistently fucosylated.82, 83 Unfortunately, the HMOs of preterm milk do not become more mature over the course of lactation and it has been speculated that there is a failure to produce mature HMOs.83 Our data indicates that preterm milk is sufficient to increase the growth of all tested Bifidobacteria and Lactobacilli strains and does not readily promote the growth of pathobionts when strains were grown individually. It is difficult to decipher how these microbes behave as a community and future work will be needed to address how representative gut microbes respond to diet in the setting of defined communities. Despite these limitations, this work does support the notion that preterm human milk can still benefit preterm infants and supports the recommendation that preterm infants receive human milk.
It has been speculated that the development of the early gut microbiota is driven by specific dietary compounds present in human milk that support selective colonization of gut microbes.84 The beneficial role of breast milk on the composition of gut microbiota development has largely been attributed to the presence of HMOs.85 We demonstrate that HMO degrading glycosyl hydrolase profiles correlate with milk utilization in commensal microbes, but we did observe some outliers. For example, B. dentium, which only harbors 4 HMO-glycosyl hydrolases had robust growth in response to milk, while E. faecalis, which also has 4 HMO-glycosyl hydrolases had limited growth in response to milk. These findings suggest that other factors in milk influence certain commensal microbes. Human breast milk comprises both macronutrients (fat, proteins, and carbohydrates), micronutrients (vitamins, minerals, hormones, etc.), cellular compartments, and the milk fat globule membrane dispersed between aqueous and colloidal phases.86 Among the macronutrients, breast milk contains carbohydrates such as a fructose and maltose87, 88 and we have previously shown that B. dentium has robust growth in response to both these carbohydrates in LDM4.89 Therefore, it is possible that these components may contribute to the growth of B. dentium and other minimal HMO degrading microbes.
In this study we compared preterm formula and full-term formula supplemented with the HMO 2-FL with preterm human milk. The preterm formula resembled preterm human milk in terms of commensal growth. For example, B. infantis, B. breve, B. angulatum, B. animalis, B. bifidum, L. acidophilus, L. johnsonii, L. plantarum, L. fermentum, L. paracasei, L. gasseri, L. delbrueckii and L. brevis had the same or higher growth with preterm formula as they did with preterm human milk. Interestingly, the full-term formula containing 2-FL in our study supported the growth of many but not all Bifidobacteria and Lactobacilli. Preterm formula contains higher levels of protein, vitamins, minerals, specifically calcium and phosphorous, compared to term infant formula and these factors may be contributing to the more robust growth with many of our commensal microbes. A potential issue with both the preterm and full-term formula was that it also promoted the growth of Enterococcus and Klebsiella; microbes that are associated with a dysbiotic infant gut microbiota and NEC.
A nutrient that may enhance to the growth of pathobionts in this model is iron, which in 1.5x higher in formula than preterm human milk.90 Iron deficiency anaemia related to cow’s milk products and exclusive breastfeeding is prevalent, and this risk is increased due to low stores in the preterm infant.91 Iron in human milk is thought be highly bioavailable for absorption, and is tightly regulated in low concentrations, regardless of maternal iron status or maternal iron intake.91 Yet, iron can also selectively increase pathobiont growth and even augment the virulence of pathogenic bacteria such as Klebsiella pneumoniae.92 Interestingly, iron supplementation is related to slower weight gain and head growth but higher psychomotor developmental tasks in a recent meta-analysis of exclusively breastfed infants.93 The mechanistic influences of iron on the preterm gut and microbiome are still unclear. Deciphering which components of breast milk that promote robust growth of commensal microbes -while only nominally promoting the growth of pathobionts- is the next step to creating customized preterm diets.
One caveat of this study is that we did not determine the secretory status of these three preterm mothers. This may be important because non-secretors lack of Fucosyltransferase 2 (FUT2) and cannot produce oligosaccharides such as 2’-FL. The proportion of non-secretor of population is about 20%. As a result, the chances of having all non-secretary mothers representing our sample is <1%. We speculate that 2-Fl and other HMOs are likely present in our pooled preterm milk within the reference range values. We were unable to determine how much HMO has been added to term formula by the manufacture’s website, but likely is within the range of 0.2g/dL-1g/dL per a clinical trial.94‡ Future studies should include the secretor status of the mother’s to provide a more robust analysis of microbe-HMO interactions.
One of the observations of this study was that human milk resulted in bacterial fermentation and reductions in culture pH. Decreased bacterial pH in response to diet has been noted with non-structural carbohydrates95 and high concentrate diets.96 Human milk has multiple carbohydrates which could be used by bacteria to generate lactic acid, including lactose and glucose. Additionally, fermentation and lactic acid production has been demonstrated in response to HMOs present in milk and galacto-oligosaccharides that can be present in formula.97 Our data indicates that the majority of commensal Bifidobacteria and Lactobacillus strains could ferment preterm milk and preterm formula to effectively lower the pH. Reductions in pH in the gastrointestinal tract has been speculated to limit pathogen colonization98 and thus fermentation of milk or formula may represent an important factor in controlling the establishment of subsequent communities.
Literature suggests that breast milk can serve as a reservoir for gut microbes. For example, Pannaraj et al. found that ~30% of the infant gut microbiota could be traced to their mother’s milk.99 This is consistent with other studies which found certain bacteria such as Streptococcus, Veillonella, Bifidobacterium, Lactobacillus, and Staphylococcus spp. co-occurred in mothers’ milk and their infants’ stool.100–102 These studies indicate that breastmilk may transfer commensal microbes and influence the infant gut microbiota development. In contrast to full-term infants, preterm infants are often given donor milk that has been pasteurized and is therefore missing the contribution of milk-associated microbes. In this setting, gut microbes lack the competition of natural human milk microbes and this may play a role in limited microbial diversity observed in preterm infants. Co-delivery of probiotics and diet (milk or formula) may temper this phenomenon.
The method of pasteurization used in this study simulates commercial pasteurization of skim/1%/2% table milk in the dairy industry. High-temperature short time (HTST) pasteurization has been shown to better preserve the function of milk including protecting bioactive ingredients and protein integrity than Holder pasteurization (62.5°C for 30 minutes).103 By using the HTST pasteurization method, the approach may optimally have simulated the effect of unpasteurized maternal preterm milk on bacteria, but it may not necessarily reflect how banked donor milk may affect bacterial growth. Banked donor milk uses Holder pasteurization, generally is from mothers of term infants, and has been stored for much longer periods than mother’s own milk. Therefore, conclusions of these data should not be over-extended to banked donor milk or commercially sterilized human milk products.
Conclusion
The establishment of a complex intestinal microbiota during the first 1,000 days of life plays a crucial role in the long-term health and well-being of the individual.41, 104, 105 Premature infants are at a higher risk of psychomotor and metabolic disorders (obesity, type 2 diabetes, cardiovascular diseases) in adulthood compared to full-term infants99–101.106–108 Several health factors, including neurodevelopment, have been associated with infant growth rate; thereby raising the issue of how best to deliver proper nutrition to premature infants. Human milk remains the optimal enteral nutrition, and the many benefits of this “liquid gold” are yet to be discovered. The rapidly changing advances in preterm diets necessitate more sophisticated models that can simulate effects in the neonatal intestine, as new products are introduced. Using our in vitro model, with microbes grown individually and, in the future, grown together, we believe we have a tuneable model that has the potential to identify new strategies for manipulating bacterial communities to alter developmental trajectories. The approach here stands as an alternative to large ‘-omic perspectives, which are useful to reveal over-arching patterns of diet on systemic disease, but are unable to offer detailed information between specific dietary components and bacterial strains in preterm neonates. This study offers a basic model that crystallizes a species-specific mechanistic influence of infant diet on the preterm microbiome.
Supplementary Material
Acknowledgements
We would like to thank Allison Rohrer, R.D. for her help advising formula and human milk nutrient information.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to disclose.
This citation references a commercially-supported study.
Notes and references
- 1.Sugino KY, Ma T, Paneth N and Comstock SS, Effect of Environmental Exposures on the Gut Microbiota from Early Infancy to Two Years of Age, Microorganisms, 2021, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J and Wang J, Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life, Cell Host Microbe, 2015, 17, 852. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler AG, She JX, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP and Petrosino JF, Temporal development of the gut microbiome in early childhood from the TEDDY study, Nature, 2018, 562, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mesa MD, Loureiro B, Iglesia I, Fernandez Gonzalez S, Llurba Olive E, Garcia Algar O, Solana MJ, Cabero Perez MJ, Sainz T, Martinez L, Escuder-Vieco D, Parra-Llorca A, Sanchez-Campillo M, Rodriguez Martinez G, Gomez Roig D, Perez Gruz M, Andreu-Fernandez V, Clotet J, Sailer S, Iglesias-Platas I, Lopez-Herce J, Aras R, PallasAlonso C, de Pipaon MS, Vento M, Gormaz M, Larque Daza E, Calvo C and Cabanas F, The Evolving Microbiome from Pregnancy to Early Infancy: A Comprehensive Review, Nutrients, 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderickx JGE, Zwittink RD, van Lingen RA, Knol J and Belzer C, The Preterm Gut Microbiota: An Inconspicuous Challenge in Nutritional Neonatal Care, Frontiers in Cellular and Infection Microbiology, 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA and Stobberingh EE, Factors influencing the composition of the intestinal microbiota in early infancy, Pediatrics, 2006, 118, 511–521. [DOI] [PubMed] [Google Scholar]
- 7.Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E and Picaud JC, Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients, J Pediatr, 2011, 158, 390–396. [DOI] [PubMed] [Google Scholar]
- 8.Thompson-Chagoyan OC, Maldonado J and Gil A, Colonization and impact of disease and other factors on intestinal microbiota, Dig Dis Sci, 2007, 52, 2069–2077. [DOI] [PubMed] [Google Scholar]
- 9.Rougé C, Goldenberg O, Ferraris L, Berger B, Rochat F, Legrand A, Göbel UB, Vodovar M, Voyer M, Rozé JC, Darmaun D, Piloquet H, Butel MJ and de La Cochetière MF, Investigation of the intestinal microbiota in preterm infants using different methods, Anaerobe, 2010, 16, 362–370. [DOI] [PubMed] [Google Scholar]
- 10.Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, Hernandez-Barranco A, Margolles A, de Los Reyes-Gavilan CG and Gueimonde M, Establishment and development of intestinal microbiota in preterm neonates, FEMS Microbiol Ecol, 2012, 79, 763–772. [DOI] [PubMed] [Google Scholar]
- 11.Barrett E, Kerr C, Murphy K, O’Sullivan O, Ryan CA, Dempsey EM, Murphy BP, O’Toole PW, Cotter PD, Fitzgerald GF, Ross RP and Stanton C, The individualspecific and diverse nature of the preterm infant microbiota, Arch Dis Child Fetal Neonatal Ed, 2013, 98, F334–340. [DOI] [PubMed] [Google Scholar]
- 12.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernandez L, Rodriguez JM and Jimenez E, Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life, PLoS One, 2013, 8, e66986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aujoulat F, Roudière L, Picaud J-C, Jacquot A, Filleron A, Neveu D, Baum T-P, Marchandin H and Jumas-Bilak E, Temporal dynamics of the very premature infant gut dominant microbiota, BMC Microbiol, 2014, 14, 325–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiltunen H, Collado MC, Ollila H, Kolari T, Tölkkö S, Isolauri E, Salminen S and Rautava S, Spontaneous preterm delivery is reflected in both early neonatal and maternal gut microbiota, Pediatric Research, 2021, DOI: 10.1038/s41390-021-01663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwiertz A, Gruhl B, Lobnitz M, Michel P, Radke M and Blaut M, Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants, Pediatr Res, 2003, 54, 393–399. [DOI] [PubMed] [Google Scholar]
- 16.Arboleya S, Ang L, Margolles A, Yiyuan L, Dongya Z, Liang X, Solis G, Fernandez N, de Los Reyes-Gavilan CG and Gueimonde M, Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota, Anaerobe, 2012, 18, 378–380. [DOI] [PubMed] [Google Scholar]
- 17.Adlerberth I and Wold A, Establishment of the gut microbiota in Western infants, Acta paediatrica, 2009, 98, 229–238. [DOI] [PubMed] [Google Scholar]
- 18.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N and Mai V, Intestinal microbial ecology in premature infants assessed with non-culture-based techniques, J Pediatr, 2010, 156, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, Field CJ, Lefebvre D, Sears MR, Becker AB, Mandhane PJ, Turvey SE, Moraes TJ, Subbarao P, Scott JA, Kozyrskyj AL and Canadian I Healthy Infant Longitudinal Development Study, Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life, JAMA Pediatr, 2018, 172, e181161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB and Dantas G, Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes, Nat Med, 2018, 24, 1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA and Lebrilla CB, Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study, J Proteome Res, 2015, 14, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, Bender JM, Azad MB, Thompson AL, Weiss ST, Azcarate-Peril MA, Litonjua AA, Kozyrskyj AL, Jaspan HB, Aldrovandi GM and Kuhn L, Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations, Nat Commun, 2018, 9, 4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D and Holtz LR, Early life dynamics of the human gut virome and bacterial microbiome in infants, Nat Med, 2015, 21, 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR and Ventura M, Diversity of bifidobacteria within the infant gut microbiota, PLoS One, 2012, 7, e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J and Tanaka R, Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota, PLoS One, 2013, 8, e78331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozé JC, Ancel PY, Lepage P, Martin-Marchand L, Al Nabhani Z, Delannoy J, Picaud JC, Lapillonne A, Aires J, Durox M, Darmaun D, Neu J and Butel MJ, Nutritional strategies and gut microbiota composition as risk factors for necrotizing enterocolitis in very-preterm infants, Am J Clin Nutr, 2017, 106, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parra-Llorca A, Gormaz M, Alcantara C, Cernada M, Nunez-Ramiro A, Vento M and Collado MC, Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk, Front Microbiol, 2018, 9, 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford SL, Lohmann P, Preidis GA, Gordon PS, O’Donnell A, Hagan J, Venkatachalam A, Balderas M, Luna RA and Hair AB, Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother’s own milk compared with donor breast milk, The American Journal of Clinical Nutrition, 2019, 109, 1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chetta KE, Schulz EV and Wagner CL, Outcomes improved with human milk intake in preterm and full-term infants, Semin Perinatol, 2021, 45, 151384. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB and Claud EC, 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis, The ISME Journal, 2009, 3, 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belfort MB, Edwards EM, Greenberg LT, Parker MG, Ehret DY and Horbar JD, Diet, weight gain, and head growth in hospitalized US very preterm infants: a 10-year observational study, The American Journal of Clinical Nutrition, 2019, 109, 1373–1379. [DOI] [PubMed] [Google Scholar]
- 32.Ong ML and Belfort MB, Preterm infant nutrition and growth with a human milk diet, Semin Perinatol, 2021, 45, 151383. [DOI] [PubMed] [Google Scholar]
- 33.Flannery DD, Ross RK, Mukhopadhyay S, Tribble AC, Puopolo KM and Gerber JS, Temporal Trends and Center Variation in Early Antibiotic Use Among Premature Infants, JAMA Network Open, 2018, 1, e180164–e180164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele C, Best Practices for Handling and Administration of Expressed Human Milk and Donor Human Milk for Hospitalized Preterm Infants, Frontiers in Nutrition, 2018, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Group PNP, Infant and Pediatric Feedings: Guidelines for Preparation of Human Milk and Formula in Health Care Facilities, Academy of Nutrition and Dietetics; Chicago, IL, 3rd Ed edn., 2019. [Google Scholar]
- 36.Volk ML, Hanson CV, Israel-Ballard K and Chantry CJ, Inactivation of Cell-Associated and Cell-Free HIV-1 by Flash-Heat Treatment of Breast Milk, JAIDS Journal of Acquired Immune Deficiency Syndromes, 2010, 53, 665–666. [DOI] [PubMed] [Google Scholar]
- 37.Bapistella S, Hamprecht K, Thomas W, Speer CP, Dietz K, Maschmann J, Poets CF and Goelz R, Short-term pasteurization of breast milk to prevent postnatal cytomegalovirus transmission in very preterm infants, Clinical Infectious Diseases, 2019, 69, 438–444. [DOI] [PubMed] [Google Scholar]
- 38.Charm SE, Landau S, Williams B, Horowitz B, Prince AM and Pascual D, High-temperature short-time heat inactivation of HIV and other viruses in human blood plasma, Vox sanguinis, 1992, 62, 12–20. [DOI] [PubMed] [Google Scholar]
- 39.Terpstra FG, Rechtman DJ, Lee ML, Hoeij KV, Berg H, Engelenberg FAV and Wout A. B. V. t., Antimicrobial and antiviral effect of high-temperature short-time (HTST) pasteurization applied to human milk, Breastfeeding Medicine, 2007, 2, 27–33. [DOI] [PubMed] [Google Scholar]
- 40.Haarman M and Knol J, Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula, Appl Environ Microbiol, 2006, 72, 2359–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Mushajiang S, Luo B, Tian F, Ni Y and Yan W, The Composition and Concordance of Lactobacillus Populations of Infant Gut and the Corresponding Breast-Milk and Maternal Gut, Frontiers in Microbiology, 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh NS, Joung JY, Lee JY and Kim Y, Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces, PLoS One, 2018, 13, e0192021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albesharat R, Ehrmann MA, Korakli M, Yazaji S and Vogel RF, Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies, Syst Appl Microbiol, 2011, 34, 148–155. [DOI] [PubMed] [Google Scholar]
- 44.Makino H, Bifidobacterial strains in the intestines of newborns originate from their mothers, Biosci Microbiota Food Health, 2018, 37, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J-H and O’Sullivan DJ, Genomic insights into bifidobacteria, Microbiol Mol Biol Rev, 2010, 74, 378–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan W, Luo B, Zhang X, Ni Y and Tian F, Association and Occurrence of Bifidobacterial Phylotypes Between Breast Milk and Fecal Microbiomes in Mother-Infant Dyads During the First 2 Years of Life, Frontiers in microbiology, 2021, 12, 669442–669442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Junick J and Blaut M, Quantification of human fecal bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene, Appl Environ Microbiol, 2012, 78, 2613–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luck B, Horvath TD, Engevik KA, Ruan W, Haidacher SJ, Hoch KM, Oezguen N, Spinler JK, Haag AM, Versalovic J and Engevik MA, Neurotransmitter Profiles Are Altered in the Gut and Brain of Mice Mono-Associated with Bifidobacterium dentium, Biomolecules, 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinne MM, Gueimonde M, Kalliomaki M, Hoppu U, Salminen SJ and Isolauri E, Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota, FEMS Immunol Med Microbiol, 2005, 43, 59–65. [DOI] [PubMed] [Google Scholar]
- 50.Nagpal R, Kurakawa T, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K and Yamashiro Y, Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: a quantitative assessment, Scientific Reports, 2017, 7, 10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seferovic MD, Mohammad M, Pace RM, Engevik M, Versalovic J, Bode L, Haymond M and Aagaard KM, Maternal diet alters human milk oligosaccharide composition with implications for the milk metagenome, Sci Rep, 2020, 10, 22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, D. G. K., Kvist LJ, Otoo GE, García-Carral C, Jiménez E, Ruiz L, Rodríguez JM, Pareja RG, Bode L, McGuire MA and McGuire MK, What’s Normal? Microbiomes in Human Milk and Infant Feces Are Related to Each Other but Vary Geographically: The INSPIRE Study, Frontiers in Nutrition, 2019, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsunoda Y, Asahara T, Nomoto K, Yoshioka Y and Fukuma E, Bacterial profile of infant feces associated with lactation infectious breasts, Pediatric Health Med Ther, 2018, 9, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Brook TC, Soe CZ, O’Neill I, Alcon-Giner C, Leelastwattanagul O, Phillips S, Caim S, Clarke P, Hall LJ and Hoyles L, Preterm infants harbour diverse Klebsiella populations, including atypical species that encode and produce an array of antimicrobial resistance- and virulence-associated factors, Microb Genom, 2020, 6, e000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta A, Hospital-acquired infections in the neonatal intensive care unit--Klebsiella pneumoniae, Semin Perinatol, 2002, 26, 340–345. [DOI] [PubMed] [Google Scholar]
- 56.Seki D, Mayer M, Hausmann B, Pjevac P, Giordano V, Goeral K, Unterasinger L, Klebermass-Schrehof K, De Paepe K, Van de Wiele T, Spittler A, Kasprian G, Warth B, Berger A, Berry D and Wisgrill L, Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage, Cell Host Microbe, 2021, 29, 1558–1572 e1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paveglio S, Ledala N, Rezaul K, Lin Q, Zhou Y, Provatas AA, Bennett E, Lindberg T, Caimano M and Matson AP, Cytotoxin-producing Klebsiella oxytoca in the preterm gut and its association with necrotizing enterocolitis, Emerging Microbes & Infections, 2020, 9, 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen RG, Bitman J, Carlson SE, Couch SC, Hamosh M and Newburg DS, in Handbook of Milk Composition, ed. Jensen RG, Academic Press, San Diego, 1995, DOI: 10.1016/B978-012384430-9/50023-8, pp. 495–542. [DOI] [Google Scholar]
- 59.Zibadi S, RR W and Preedy V, Handbook of dietary and nutritional aspects of human breast milk, 2013 [Google Scholar]
- 60.Engevik K, Engevik MA, Oezuguen N, Haag A and Horavth TD, Development of a high-throughput method for examining bacterial supernatant pH using ratiometric UV-VIS spectrophotometry, The FASEB Journal, 2021, 35. [Google Scholar]
- 61.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V and Henrissat B, The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics, Nucleic Acids Res, 2009, 37, D233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.El Kaoutari A, Armougom F, Gordon JI, Raoult D and Henrissat B, The abundance and variety of carbohydrate-active enzymes in the human gut microbiota, Nat Rev Microbiol, 2013, 11, 497–504. [DOI] [PubMed] [Google Scholar]
- 63.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM and Henrissat B, The carbohydrate-active enzymes database (CAZy) in 2013, Nucleic Acids Res, 2014, 42, D490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park BH, Karpinets TV, Syed MH, Leuze MR and Uberbacher EC, CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database, Glycobiology, 2010, 20, 1574–1584. [DOI] [PubMed] [Google Scholar]
- 65.Chen IMA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN and Nikos C Kyrpides, The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities, Nucleic Acids Research, 2021, 49, D751–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang T and Savaiano DA, Modification of Colonic Fermentation by Bifidobacteria and pH In Vitro (Impact on Lactose Metabolism, Short-Chain Fatty Acid, and Lactate Production), Digestive Diseases and Sciences, 1997, 42, 2370–2377. [DOI] [PubMed] [Google Scholar]
- 67.Toit M. d., Huch M, Cho G-S and Franz CMAP, in Lactic Acid Bacteria, 2014, DOI: 10.1002/9781118655252.ch28, pp. 457–505. [DOI] [Google Scholar]
- 68.Akay HK, Bahar Tokman H, Hatipoglu N, Hatipoglu H, Siraneci R, Demirci M, Borsa BA, Yuksel P, Karakullukcu A, Kangaba AA, Sirekbasan S, Aka S, Mamal Torun M and Kocazeybek BS, The relationship between bifidobacteria and allergic asthma and/or allergic dermatitis: A prospective study of 0–3 years-old children in Turkey, Anaerobe, 2014, 28, 98–103. [DOI] [PubMed] [Google Scholar]
- 69.Liu D, Shao L, Zhang Y and Kang W, Safety and efficacy of Lactobacillus for preventing necrotizing enterocolitis in preterm infants, Int J Surg, 2020, 76, 79–87. [DOI] [PubMed] [Google Scholar]
- 70.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N and Knight R, Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns, Proc Natl Acad Sci U S A, 2010, 107, 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, Kim H, Zoratti EM, Lukacs NW, Boushey HA, Ownby DR, Lynch SV and Johnson CC, Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity, Sci Rep, 2016, 6, 31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang HH, Lang A, Teo SM, Judd LM, Gangnon R, Evans MD, Lee KE, Vrtis R, Holt PG and Lemanske RF Jr, Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk, Journal of Allergy and Clinical Immunology, 2021, 147, 1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roze JC, Ancel PY, Marchand-Martin L, Rousseau C, Montassier E, Monot C, Le Roux K, Butin M, Resche-Rigon M, Aires J, Neu J, Lepage P, Butel MJ and Group ES, Assessment of Neonatal Intensive Care Unit Practices and Preterm Newborn Gut Microbiota and 2-Year Neurodevelopmental Outcomes, JAMA Netw Open, 2020, 3, e2018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibson MK, Wang B, Ahmadi S, Burnham C-AD, Tarr PI, Warner BB and Dantas G, Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome, Nature Microbiology, 2016, 1, 16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tarracchini C, Milani C, Longhi G, Fontana F, Mancabelli L, Pintus R, Lugli GA, Alessandri G, Anzalone R, Viappiani A, Turroni F, Mussap M, Dessi A, Cesare Marincola F, Noto A, De Magistris A, Vincent M, Bernasconi S, Picaud JC, Fanos V and Ventura M, Unraveling the Microbiome of Necrotizing Enterocolitis: Insights in Novel Microbial and Metabolomic Biomarkers, Microbiol Spectr, 2021, 9, e0117621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michaelsen KF, Skafte L, Badsberg JH and Jorgensen M, Variation in macronutrients in human bank milk: influencing factors and implications for human milk banking, J Pediatr Gastroenterol Nutr, 1990, 11, 229–239. [DOI] [PubMed] [Google Scholar]
- 77.Ballard O and Morrow AL, Human milk composition: nutrients and bioactive factors, Pediatr Clin North Am, 2013, 60, 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andreas NJ, Kampmann B and Mehring Le-Doare K, Human breast milk: A review on its composition and bioactivity, Early Hum Dev, 2015, 91, 629–635. [DOI] [PubMed] [Google Scholar]
- 79.Bravi F, Wiens F, Decarli A, Dal Pont A, Agostoni C and Ferraroni M, Impact of maternal nutrition on breast-milk composition: a systematic review, Am J Clin Nutr, 2016, 104, 646–662. [DOI] [PubMed] [Google Scholar]
- 80.Samuel TM, Zhou Q, Giuffrida F, Munblit D, Verhasselt V and Thakkar SK, Nutritional and Non-nutritional Composition of Human Milk Is Modulated by Maternal, Infant, and Methodological Factors, Front Nutr, 2020, 7, 576133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Underwood MA, Human milk for the premature infant, Pediatric clinics of North America, 2013, 60, 189–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gabrielli O, Zampini L, Galeazzi T, Padella L, Santoro L, Peila C, Giuliani F, Bertino E, Fabris C and Coppa GV, Preterm milk oligosaccharides during the first month of lactation, Pediatrics, 2011, 128, e1520–1531. [DOI] [PubMed] [Google Scholar]
- 83.De Leoz ML, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, Tancredi DJ, Smilowitz JT, Kalanetra KM, Mills DA, German JB, Lebrilla CB and Underwood MA, Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm, J Proteome Res, 2012, 11, 4662–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, Di Pierro F, van Sinderen D and Ventura M, The infant gut microbiome as a microbial organ influencing host well-being, Italian Journal of Pediatrics, 2020, 46, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walsh C, Lane JA, van Sinderen D and Hickey RM, Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health, J Funct Foods, 2020, 72, 104074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boudry G, Charton E, Le Huerou-Luron I, Ferret-Bernard S, Le Gall S, Even S and Blat S, The Relationship Between Breast Milk Components and the Infant Gut Microbiota, Frontiers in nutrition, 2021, 8, 629740–629740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goran MI, Martin AA, Alderete TL, Fujiwara H and Fields DA, Fructose in Breast Milk Is Positively Associated with Infant Body Composition at 6 Months of Age, Nutrients, 2017, 9, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fanos V, Metabolomics, milk-oriented microbiota (MOM) and multipotent stem cells: the future of research on breast milk, Journal of Pediatric and Neonatal Individualized Medicine (JPNIM), 2015, 4, e040115. [Google Scholar]
- 89.Engevik MA, Danhof HA, Hall A, Engevik KA, Horvath TD, Haidacher SJ, Hoch KM, Endres BT, Bajaj M, Garey KW, Britton RA, Spinler JK, Haag AM and Versalovic J, The metabolic profile of Bifidobacterium dentium reflects its status as a human gut commensal, BMC Microbiol, 2021, 21, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernández-Sánchez ML, Rafael R, Iglesias HG, López-Sastre JB, Fernández-Colomer B, Pérez-Solís D and Sanz-Medel A, Iron content and its speciation in human milk from mothers of preterm and full-term infants at early stages of lactation: A comparison with commercial infant milk formulas, Microchemical Journal, 2012, 105, 108–114. [Google Scholar]
- 91.Neville MC, Zhang P and Allen JC, in Handbook of Milk Composition, ed. Jensen RG, Academic Press, San Diego, 1995, DOI: 10.1016/B978-012384430-9/50025-1, pp. 577–592. [DOI] [Google Scholar]
- 92.Chen T, Dong G, Zhang S, Zhang X, Zhao Y, Cao J, Zhou T and Wu Q, Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess, BMC Microbiology, 2020, 20, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai C, Granger M, Eck P and Friel J, Effect of daily iron supplementation in healthy exclusively breastfed infants: A systematic review with meta-analysis, Breastfeeding Medicine, 2017, 12, 597–603. [DOI] [PubMed] [Google Scholar]
- 94.Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG and Buck RH, Similar to Those Who Are Breastfed, Infants Fed a Formula Containing 2’-Fucosyllactose Have Lower Inflammatory Cytokines in a Randomized Controlled Trial, J Nutr, 2016, 146, 2559–2566. [DOI] [PubMed] [Google Scholar]
- 95.Russell JB and Diez-Gonzalez F, in Advances in Microbial Physiology, ed. Poole RK, Academic Press, 1997, vol. 39, pp. 205–234. [DOI] [PubMed] [Google Scholar]
- 96.Calsamiglia S, Ferret A and Devant M, Effects of pH and pH fluctuations on microbial fermentation and nutrient flow from a dual-flow continuous culture system, J Dairy Sci, 2002, 85, 574–579. [DOI] [PubMed] [Google Scholar]
- 97.Schwab C and Gänzle M, Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides, FEMS Microbiology Letters, 2011, 315, 141–148. [DOI] [PubMed] [Google Scholar]
- 98.Engevik MA and Versalovic J, Biochemical Features of Beneficial Microbes: Foundations for Therapeutic Microbiology, Microbiol Spectr, 2017, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, Bailey A, Bushman FD, Sleasman JW and Aldrovandi GM, Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome, JAMA Pediatr, 2017, 171, 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, Yonemitsu C, Field CJ, Becker AB, Mandhane PJ, Sears MR, Khafipour E, Moraes TJ, Subbarao P, Finlay BB, Turvey SE and Azad MB, Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: the CHILD Cohort Study, Cell Host Microbe, 2020, 28, 285–297 e284. [DOI] [PubMed] [Google Scholar]
- 101.Martín V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, Campo RD, Fernández L, Rodríguez JM and Jiménez E, Sharing of bacterial strains between breast milk and infant feces, J Hum Lact, 2012, 28, 36–44. [DOI] [PubMed] [Google Scholar]
- 102.Murphy K, Curley D, O’Callaghan TF, O’Shea C-A, Dempsey EM, O’Toole PW, Ross RP, Ryan CA and Stanton C, The Composition of Human Milk and Infant Faecal Microbiota Over the First Three Months of Life: A Pilot Study, Scientific Reports, 2017, 7, 40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Escuder-Vieco D, Espinosa-Martos I, Rodríguez JM, Fernández L and Pallás-Alonso CR, Effect of HTST and Holder Pasteurization on the Concentration of Immunoglobulins, Growth Factors, and Hormones in Donor Human Milk, Frontiers in Immunology, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G and de La Cochetiere MF, Development of intestinal microbiota in infants and its impact on health, Trends Microbiol, 2013, 21, 167–173. [DOI] [PubMed] [Google Scholar]
- 105.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG and Hart A, The gut microbiota and host health: a new clinical frontier, Gut, 2016, 65, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hofman PL, Regan F, Jackson WE, Jefferies C, Knight DB, Robinson EM and Cutfield WS, Premature birth and later insulin resistance, N Engl J Med, 2004, 351, 2179–2186. [DOI] [PubMed] [Google Scholar]
- 107.Boquien C-Y, Human Milk: An Ideal Food for Nutrition of Preterm Newborn, Frontiers in Pediatrics, 2018, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stettler N, Zemel BS, Kumanyika S and Stallings VA, Infant weight gain and childhood overweight status in a multicenter, cohort study, Pediatrics, 2002, 109, 194–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


