Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive and difficult to treat cancers with tumors typically exhibiting high levels of chronic hypoxia. Hypoxia activates hypoxia-inducible factors (HIFs) that mediate cellular responses to adapt to low oxygen environments. Hypoxia also causes endoplasmic reticulum (ER) stress, increasing activating transcription factor 4 (ATF4), a master regulator of the unfolded protein response (UPR) pathway that mediates cellular response to ER stress. ATF4 is overexpressed in PDAC and is associated with poor prognoses. While ATF4 promotes cell proliferation and tumorigenesis, most studies have been conducted under normoxia or acute hypoxia. The functions of ATF4 in chronic hypoxia remain largely unexplored. Using siRNA knockdown experiments of healthy skin fibroblast cells WS1 and PDAC cell lines PANC-1 and Mia-PaCa2 to analyze mRNA and protein expression levels, a novel ATF4 function was identified, in which it decreases HIF2α mRNA and increases HIF1α mRNA in chronic hypoxia while having no effect in acute hypoxia. A scratch assay was used to show that ATF4 decreases cell migration in chronic hypoxia as opposed to the increase in cell migration ATF4 imparts in acute hypoxia. Colony formation assay and cell viability assay showed that ATF4 promotes colony formation and cell viability in both chronic and acute hypoxia. In addition to the differential response of ATF4 in chronic hypoxia compared with acute hypoxia, this is the first time ATF4 has been implicated in regulation of response to hypoxia via interaction with HIF proteins in PDAC.
Keywords: ATF4, HIF1α, HIF2α, PDAC, chronic hypoxia, UPR pathway
Introduction
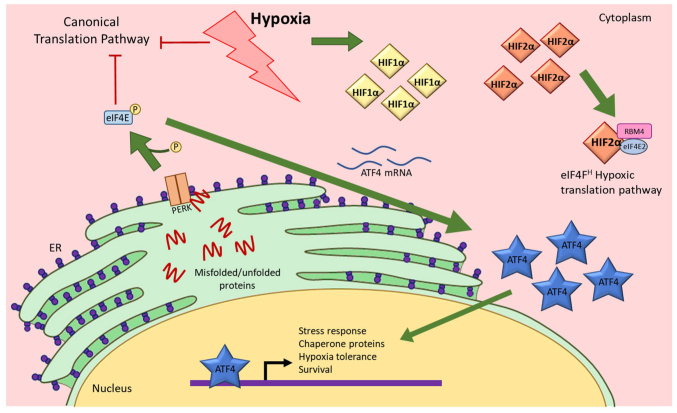
Pancreatic ductal adenocarcinoma (PDAC) is among the most aggressive cancers with a 5-year survival rate of 8–10% (1–3). Hypoxia, a stress condition in which oxygen levels are insufficient for typical cellular function, contributes to PDAC pathophysiology and treatment resistance (4). Protein synthesis in the endoplasmic reticulum (ER) demands more energy and oxygen compared with other cellular processes (5,6); as such hypoxia significantly hinders protein translation and folding (7). This results in accumulated misfolded or unfolded proteins activating ER stress sensors, thus triggering the unfolded protein response (UPR) (Fig. 1) (7,8). Within the UPR pathway, activating transcription factor 4 (ATF4) is one of the master regulators that activates transcription of genes required for amino acid metabolism, synthesis and transport (7). These genes, among others, generally alleviate hypoxia-induced ER stress to promote cell recovery and homeostasis (7,9–11).
Figure 1.
Hypoxia activates the UPR pathway and causes ATF4 expression to increase. Hypoxia causes inhibition of the canonical translation pathway, resulting in misfolded or unfolded proteins. These peptides activate PERK that consequently induces ATF4 mRNA translation, resulting in an increase in ATF4 protein expression. ATF4 then translocates to the nucleus to initiate gene expression changes. Hypoxia also causes increases in HIF1α and HIF2α. Both HIFs also translocate to the nucleus to function as transcription factors. HIF2α also functions as a translation factor that initiates translation when cells are under hypoxic stress (17). UPR, unfolded protein response; ATF4, activating transcription factor 4; eIF4E, eukaryotic initiation factor 4E; ER, endoplasmic reticulum; HIF1α, hypoxia-inducible factor 1α; HIF2α, hypoxia-inducible factor 2α; PERK, protein kinase R (PKR)-like endoplasmic reticulum kinase; P, phosphate.
PDAC exhibits enhanced cellular response to hypoxic stress and significantly higher ATF4 expression compared with healthy pancreatic cells (12–14). Numerous chemotherapeutic drugs are ineffective in PDAC due to ATF4 functions that promote drug resistance in hypoxia (14). ATF4 is unique in the UPR pathway because it is translated more efficiently in hypoxia compared with normoxia (7,15). This higher translation efficiency is orchestrated by upstream open reading frames (uORFs) in the mRNA transcript that promote translation of a truncated, inactive protein under normal conditions and initiate translation of functional ATF4 protein under cell stress conditions, such as hypoxia. Increased ATF4 protein expression in hypoxia is also promoted by post-translational modifications that stabilize the protein (16).
In addition to activating ATF4 in the UPR pathway, cells also respond to hypoxia by activating hypoxia-inducible factors (HIFs), a family of proteins that initiate hypoxia-response pathways (17). HIFs are well known for their roles as transcription factors in hypoxia because they activate the transcription of genes that promote cell adaptation during oxygen deprivation (17). HIF1α and HIF2α subunits are capable of transcribing genes that play important roles in cancer progression such as carbonic anhydrase IX (CA9) and octamer-binding transcription factor 4 (Oct4), among others (18,19). Independent of its transcriptional functions, HIF2α is also a translational factor that forms a complex with other proteins to activate an alternative cap-dependent translation mechanism to synthesize proteins from select mRNA that are necessary for response to hypoxia (17,20–22). In chronic hypoxia, the canonical translation initiation pathway is inhibited, resulting in significantly decreased global protein synthesis (20).
Previous studies on ATF4 and HIFs in pancreatic cancers have been conducted at atmospheric oxygen levels, but not in prolonged hypoxia of more than 24 h, a more pathophysiologically relevant condition in which the majority of PDAC cases exist. In vitro studies performed in normoxia demonstrated that ATF4 inhibition prevents cancer progression and treatment resistance. However, while ATF4 expression is higher in PDAC cells compared with healthy pancreatic cells in these studies, the cells used in normoxic experiments do not express HIFs at levels similar to those that are observed in patients, making it difficult to identify any relevant interactions between ATF4 and HIFs. Furthermore, other studies showed that more severe hypoxia of <1% oxygen levels are necessary for significant UPR activation (23,24). In the present study, ATF4 activity was explored in the hypoxia response pathway mediated by HIFs in conditions of severe, chronic hypoxia in pancreatic cancer cells.
Materials and methods
Cell culture
Pancreatic epithelial carcinoma cell lines PANC-1 (cat. no. CRL-1469) and MiaPaCa-2 [cat. no. CRL-1420; both from American Type Culture Collection (ATCC)] were cultured in Advanced DMEM media supplemented with 10% FBS, 1% GlutaMAX (cat. no. 35050061; Gibco; Thermo Fisher Scientific, Inc.), and 1% penicillin and streptomycin (cat. no. 15140-163; Gibco; Thermo Fisher Scientific, Inc.). Healthy skin fibroblasts WS1 cells (cat. no. CRL-1502; ATCC) were cultured in advanced DMEM supplemented with 15% FBS, penicillin and streptomycin, GlutaMAX, β-mercaptoethanol, and basic fibroblast growth factor. All cells were cultured under standard conditions of 37°C with 5% CO2, 21% O2 (normoxic) or as indicated in hypoxia. All cells were tested for mycoplasma ~every 3 months and confirmed to be free of contamination using a PCR mycoplasma detection kit (cat. no. 30-1012K; ATCC).
Hypoxia treatment
For hypoxia treatment, Ruskinn InvivO2 500 hypoxia workstation was used (Baker Ruskinn). Cells were seeded or plated and incubated under standard conditions in normoxia for 24 h prior to hypoxia exposure. Cells were then placed into a sterile hypoxia glove box pre-set to either 1% oxygen for healthy fibroblasts or 0.2% for cancer cell lines, 5% CO2, a balance of N2 and at 37°C. WS1 fibroblasts were exposed to 1% oxygen rather than 0.2% oxygen since cancer cells have a lower threshold for oxygen before hypoxia response pathways are activated, while normal fibroblasts are less tolerant of hypoxia. Cells were handled inside of the hypoxia glove box for the duration of the experiments until they were either fixed or lysed.
Transfection
To assess the effects of gene inhibition on hypoxic PDAC, PANC-1 cells were transiently transfected using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, Inc.) and pre-designed Silencer Select small interfering (si)RNAs (Thermo Fisher Scientific, Inc.) at 1 pmol per well: HIF1α (cat. no. s6539), HIF2α (cat. no. s4698) and ATF4 (cat. no. s1702). The siRNA-lipid complexes were constructed in reduced serum Opti-MEM media (cat. no. 31985070; Thermo Fisher Scientific, Inc.) according to manufacturer protocols, incubated at room temperature for at least 5 min and were added to each well in either six-, 12- or 96-well plates. The cells were then seeded into the plates in normal growth media. Lipofectamine RNAiMAX Transfection Reagent only requires reduced serum media when siRNA-lipid complexes are made and not when treated to cells in growth media containing serum. The cells were incubated at 37°C under normoxic conditions for 24 h before they were placed in the hypoxia chamber or remained in the normoxic incubator. Knockdown efficacy was determined by reverse transcription-quantitative (RT-q) PCR.
RNA extraction, quantification and RT-q PCR
Cells incubated in normoxia were washed with PBS while hypoxic cells were washed with de-oxygenated PBS. RNA was extracted using TRIzol® (Thermo Fisher Scientific, Inc.) and purified by RNeasy column centrifugation kit (Qiagen). Purified RNA was quantified using Nanodrop spectrophotometer at a wavelength of 260 nm (Thermo Fisher Scientific, Inc.). cDNA was synthesized using qScript cDNA synthesis kit (Quantabio) according to the manufacturer's protocol and qPCR was conducted using Taqman primer-probes (Thermo Fisher Scientific, Inc.) and Taqman Gene Expression Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer'σ protocol provided for the Taqman Master Mix. The Taqman probes used were Hs00153153_m1 (HIF1α), Hs01026149_m1 (HIF2α), Hs00154208_m1 (CA9), Hs00999632_g1 (Oct4) and Hs00909569_g1 (ATF4). The RT-qPCR data was quantified by calculating the 2−ΔΔCq values as defined by Livak and Schmittgen, 2001.
Protein extraction and western blot analysis
Normoxic and hypoxic cells were washed in either oxygenated PBS or de-oxygenated PBS, respectively. Mammalian Protein Extraction Reagent was supplemented with Halt Protease Inhibitor cocktail (Thermo Fisher Scientific, Inc.) and added to the cells for lysis. The cells were frozen for at least 6 h to promote lysis, and the samples were sonicated. The samples were centrifuged at 4°C for 10 min at 14,000 × g. The supernatant was transferred to new tubes and Bicinchoninic Acid (BCA) assay (Pierce; Thermo Fisher Scientific, Inc.) was conducted and measured using the EnVision plate reader (PerkinElmer, Inc.) to quantify protein.
For western blotting, the Criterion Blotter Western Blot system was used (Bio-Rad Laboratories, Inc.); 30 µg of total protein in Laemmli sample buffer was boiled for 10 min at 95–100°C. The samples were loaded into 10% gels (Bio-Rad Laboratories, Inc.) and electrophoresis commenced at 60–100 V. The protein in the gel was transferred to PVDF membranes by wet-transfer at 100 V for 30 min. The membranes were blocked in 5% w/v fat-free powdered milk in 1X Tris buffered solution with 0.1% Tween 20 detergent (TBS-T; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature, probed with primary antibodies overnight at 4°C in 5% BSA (Sigma-Aldrich; Merck KGaA) and with secondary antibodies for 1 h at room temperature in 5% BSA. The primary antibodies used were HIF1α (1:1,000; cat. no. MA1516; Thermo Fisher Scientific, Inc.), HIF2α (1:1,000; cat. no. ab8365; Abcam), ATF4 (1:1,000; cat. no. 11815S; Cell Signaling Technology, Inc.) and β-actin (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.). Horseradish peroxidase-conjugated mouse IgGκ light chain binding protein (1:1,000; cat. no. sc-516102; Santa Cruz Biotechnology, Inc.) and mouse anti-rabbit secondary antibodies (1:1,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) were used. The blots were visualized by enhanced chemiluminescence (ECL) incubation and C-DiGit Blot Scanner (LI-COR Biosciences). The images were quantified by ImageJ software version 1.53e (National Institutes of Health).
Cell viability assay
Cells (2,000/well) were seeded into 96-well plates and incubated overnight in normoxia under standard culturing conditions. The plates were placed into hypoxia or maintained at normoxia. After hypoxic exposure, the cells were removed from the chamber and reconstituted CellTiterGlo (Promega Corporation) reagents were added to the plate, following the manufacturer's protocol. The plate was incubated at room temperature for at least 10 min and the luminescence at 560 nm was measured on the PerkinElmer Envision plate reader.
Colony formation assay
Cells were seeded into each well (2,000 cells per well) in six-well plates. The cells were incubated at 37°C for 24 h and then placed in 0.2% oxygen or maintain in normoxia. The cells in hypoxia were incubated for 16 or 48 h and then placed in normoxia for 7–10 days of incubation for colony formation. Afterwards, the cells were washed with cold PBS, fixed with acetic acid and methanol fixing solution (1:4 mixture) at room temperature for at least 15 min until dry and stained at room temperature for at least 30 min with 0.5% w/v crystal violet (cat. no. C0775-25G; Sigma Aldrich; Merck KGaA) in 25% ethanol solution. The plates were then washed, and colonies were counted manually.
Cell migration (scratch) assay
Cells were transiently transfected following the previously outlined protocol. After siRNA-lipid complexes were added to 24-well plates, 1×105 PANC-1 cells in growth media were seeded into each well and incubated at 37°C for 24 h. Each well was then ‘scratched’ using a P1000 pipette tip down the middle of each well. Images of all wells were obtained. The plates were then placed in either 0.2% oxygen or maintained in normoxia and incubated for either 16 or 48 h. After incubation in normoxia or hypoxia, images of each well were captured using an inverted light microscope (Invertoskop 40C; Carl Zeiss AG) and analyzed using an ImageJ/Fiji plugin tool (25).
Statistical analysis
All experiments were conducted in at least three replicates. For all experiments consisting of two groups, unpaired Student's t-test was conducted for statistical significance. For all experiments consisting of more than two groups, tests for homogeneity of variance, such as Levene's test, were performed. One-way analyses of variance (ANOVA) tests were conducted, followed by the Bonferroni multiple variance post hoc tests. Data were graphed and analyzed using GraphPad Prism software version 9.3 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
ATF4 expression increases in acute and chronic hypoxia
ATF4 is over-expressed in PDAC cells compared with healthy pancreatic cells and is associated with poor prognosis (26). To first determine ATF4 expression in PDAC cells in hypoxia, PANC-1 and Mia-PaCa2 cells were cultured in a 0.2% oxygen environment or in a 21% oxygen environment for 24 h. Hypoxia was defined as 0.2% oxygen in PDAC cells because while different tissue types activate the hypoxia-responsive pathways at varying levels of oxygen, pancreatic cancers have been shown to be severely hypoxic (<1%) with ~0.4% oxygen in tumors (27–29). It was determined that 0.2% oxygen was necessary to ensure the PDAC cells adequately activate hypoxia-response pathways by confirming HIF and ATF4 activity at 0.2% oxygen (Figs. 2 and 3). Furthermore, the UPR pathway activates under severe hypoxic conditions, which is needed to investigate ATF4 (23,24).
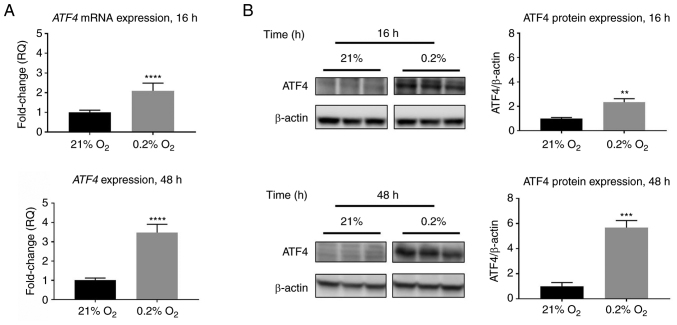
Figure 2.
ATF4 mRNA and protein expression increase in response to acute and chronic hypoxia in PANC-1 cells. PANC-1 cells were treated with hypoxia (0.2% oxygen) or normoxia (21% oxygen) for 16 or 48 h. (A) RT-qPCR was conducted to determine ATF4 mRNA levels. Fold changes for RT-qPCR are shown relative to normoxic cells. Error bars represent the mean ± SD for n=9. The fold changes were analyzed using unpaired Student's t-test. (B) Western blot analysis was conducted to determine ATF4 protein expression levels after 16 or 48 h of hypoxia or normoxia in PANC-1 cells. ATF4 protein levels were normalized to β-actin and are shown relative to normoxic levels. Error bars represent the mean ± SD for n=3. The fold changes were analyzed using unpaired Student's t-test. **P<0.01, ***P<0.001 and ****P<0.0001. ATF4, activating transcription factor 4; RT-qPCR, reverse transcription-quantitative PCR.
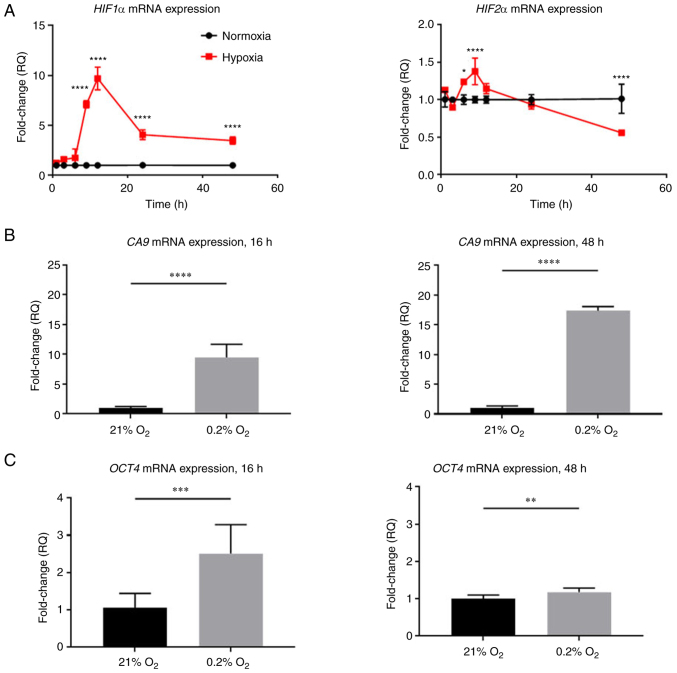
Figure 3.
HIF1α and HIF2α mRNA levels and HIF target genes change over time in hypoxia in PANC-1 cells. PANC-1 cells were treated with hypoxia (0.2% oxygen) or normoxia (21% oxygen) for 16 or 48 h. (A-C) RT-qPCR was conducted to determine (A) HIF1α, HIF2α, (B) CA9 and (C) Oct4 mRNA levels. Fold changes for RT-qPCR are shown relative to normoxic cells. Error bars represent the mean ± SD for n=9. The fold changes were analyzed using unpaired Student's t-test. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. HIF, hypoxia-inducible factor; RT-qPCR, reverse transcription-quantitative PCR; CA9, carbonic anhydrase IX; Oct4, octamer-binding transcription factor 4.
PANC-1 cells were used to assess ATF4 expression in acute and chronic hypoxia. Literature values for acute hypoxia generally do not surpass 24 h, while chronic hypoxia tends to be more than 24 h. In the present study, acute was defined as 16 h and chronic as 48 h (26,30,31). RT-qPCR data revealed that ATF4 mRNA expression increased by 2.1±0.1-fold in acute hypoxia compared with cells exposed to normoxia (P<0.0001), while ATF4 mRNA levels in chronic hypoxia increased 3.5±0.4-fold compared with normoxia levels (P<0.0001; Fig. 2A). Western blot analysis revealed similar expression patterns in which ATF4 protein levels increase in hypoxia compared with normoxic levels. Under normoxic conditions, little to no ATF4 protein expression was detected, but upon hypoxic exposure, ATF4 expression increased significantly. In acute hypoxia, the PANC-1 cells exhibited a 2.3±0.1-fold increase in ATF4 protein expression (P<0.01), while in chronic hypoxia, ATF4 levels increased by 5.7±0.3-fold compared with normoxia (P<0.001; Fig. 2B).
HIFs and expression of HIF target genes vary over time in hypoxia
In addition to ATF4 expression changes, hypoxia leads to increased HIF1α and HIF2α expression in PDAC (17). However, HIF expression profiles differ depending on cell type and duration in hypoxia (17). While all HIF levels oscillate over time, increased HIF1α expression is generally associated with acute hypoxic exposure whereas increased HIF2α expression is typically associated with chronic hypoxia (17,32). HIF expression profiles can vary greatly among different cell types, and, therefore, HIF expression and activity in PANC-1 cells were determined in acute and chronic hypoxia. To confirm HIF1α and HIF2α expression and activity, the cells were incubated in either normoxic conditions or hypoxia over time. As compared with normoxic control cells, HIF1α mRNA expression significantly increased up to 9.7±1.1-fold after 12 h of hypoxic exposure, decreased at 24 h compared with the 12-h timepoint and then ultimately plateaued at a level 3.5±0.4-fold change higher than cells in normoxia (P<0.0001). HIF2α mRNA expression levels, while lower than HIF1α expression levels, increased significantly up to 1.4±0.1-fold higher than normoxic levels at 9 h (P<0.0001), and then decreased to 0.6±0.02-fold lower than normoxic levels (P<0.0001; Fig. 3A).
HIFs are subject to post-translational regulation and degradation in normoxia, whereas in hypoxia, HIF proteins are stabilized and accumulate. While HIF mRNA levels may vary over time, known HIFα target genes were also analyzed to assess HIF activities in acute and chronic hypoxia. To confirm HIF1α activity, the mRNA levels of HIF1α-specific transcriptional target gene CA9 were measured (Fig. 3B) (33). CA9 expression increased 9.5±2.2-fold after 16 h of hypoxia (P<0.0001) and continued to increase up to 17.8±0.7-fold after 48 h of hypoxia compared with normoxic controls (P<0.0001). To measure HIF2α activity, the mRNA levels of HIF2α transcriptional target gene Oct were analyzed (Fig. 3C) (19). Oct4 expression increased 2.5±0.8-fold after 16 h of hypoxia (P<0.001), and in chronic hypoxia, Oct4 mRNA levels decreased close to normoxic levels with only a 1.2±0.1-fold decrease compared with normoxic levels (P<0.01). The increase in CA9 and Oct4 confirmed that HIF1α and HIF2α are active in acute and chronic hypoxia.
HIF1α and HIF2α knockdowns do not affect ATF4 mRNA or protein expression in chronic hypoxia
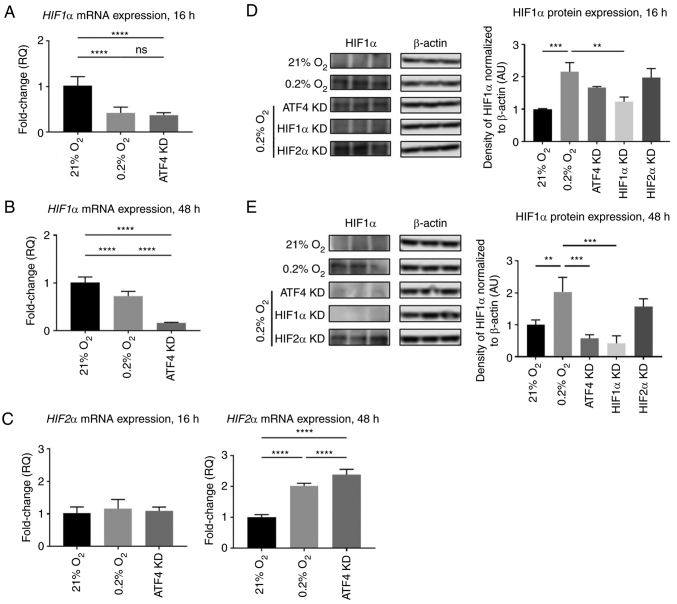
HIF transcriptional and translational functions make it possible that HIF1α or HIF2α affect ATF4 expression to mediate the UPR pathway in response to hypoxia. To understand the effects that HIF1α and HIF2α have on ATF4 in acute and chronic hypoxia in PDAC cells, siRNA was used to knockdown ATF4, HIF1α or HIF2α for loss-of-function experiments (Fig. S1). After 24 h of incubation in normal cell culture conditions, the cells were either placed in hypoxia or maintained in normoxia for 16 or 48 h. mRNA and protein expression levels of ATF4, HIF1α and HIF2α were determined. In acute hypoxia, knockdown of either HIF1α or HIF2α caused significant decreases in ATF4 mRNA levels compared with the hypoxia control (P<0.0001 and P<0.001, respectively; Fig. 4A). In chronic hypoxia, ATF4 mRNA expression significantly increased after HIF1α knockdown compared with the hypoxia (0.2% O2) control group (P<0.05) and was not affected after HIF2α knockdown compared with the hypoxia (0.2% O2) control group (Fig. 4B).
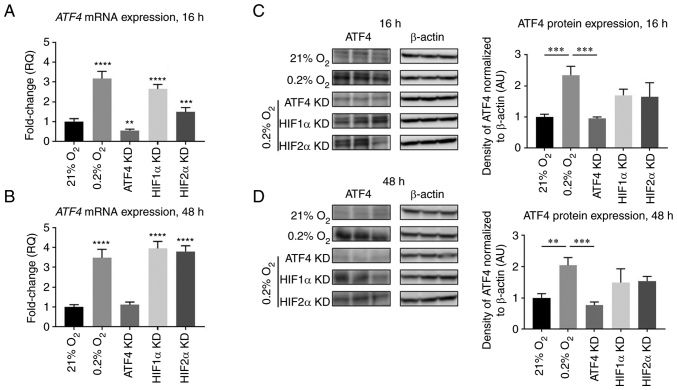
Figure 4.
ATF4 mRNA and protein expression do not change upon HIF1α or HIF2α KD in chronic hypoxia in PANC-1 cells. PANC-1 cells were treated with RNAiMAX Lipofectamine reagent and small interfering RNA to transiently KD ATF4, HIF1α or HIF2α. The cells were placed in hypoxia (0.2% oxygen) or normoxia (21% oxygen) for 16 or 48 h. All cells with KD were placed in hypoxia. (A and B) RT-qPCR was conducted to determine ATF4 mRNA levels after (A) 16 and (B) 48 h of hypoxia. Fold changes for RT-qPCR are shown relative to normoxic cells. Error bars represent mean ± SD for n=9. The fold changes were analyzed by one-way ANOVA followed by Bonferroni statistical hypothesis test. (C and D) Western blot analysis was conducted to determine the protein expression levels of ATF4 after (C) 16 or (D) 48 h of hypoxia. Fold changes are shown relative to normoxic cells. Error bars represent the mean ± SD for n=3. Fold changes were analyzed by one-way ANOVA followed by Bonferroni statistical hypothesis test. **P<0.01, ***P<0.001 and ****P<0.0001. ATF4, activating transcription factor 4; HIF, hypoxia-inducible factor; KD, knockdown; RT-qPCR, reverse transcription-quantitative PCR.
The protein expression levels of ATF4 were also determined after ATF4, HIF1α or HIF2α knockdowns. In normoxia, ATF4 protein expression levels were minimal, and with hypoxic exposure, ATF4 protein levels increased by 2.3±0.3-fold after 16 h (P<0.001) and 2.0±0.2-fold after 48 h compared with normoxic levels, respectively (P<0.05; Fig. 4C and D). ATF4 protein levels significantly decreased after ATF4 knockdown (P<0.001; Fig. 4C and D). HIF1α knockdown did not affect ATF4 protein expression levels in either acute (P>0.05) or chronic hypoxia (P>0.05). HIF2α knockdown had no effect on ATF4 protein expression in either acute (P>0.05) or chronic hypoxia (P>0.05).
ATF4 knockdown decreases HIF1α and increases HIF2α expression in chronic, but not acute, hypoxia
ATF4 is a master regulator within the UPR pathway when cells are under stress, such as in hypoxia. To understand the effects that ATF4 have on HIF1α and HIF2α in acute and chronic hypoxia in PDAC cells, ATF4, HIF1α or HIF2α were knocked down using siRNA, incubated in normoxia for 24 h and then placed in either hypoxia or maintained in normoxia for 16 or 48 h. HIF1α and HIF2α mRNA and protein levels were then determined.
Under acute hypoxia, HIF1α mRNA expression was 0.4±0.1-fold lower in the hypoxic vehicle-treated cells with no siRNA compared with normoxic cells (P<0.0001; Fig. 5A). There was no significant difference in HIF1α mRNA expression between the hypoxic vehicle-treated cells and the ATF4 knockdown cells (P>0.05; Fig. 5A). HIF1α protein expression in hypoxia alone was 2.2±0.3-fold higher than in normoxia, indicating that HIF protein levels do not appear to be solely determined by the corresponding hypoxic mRNA levels (P<0.001; Fig. 5D). Upon ATF4 knockdown in acute hypoxia, HIF1α protein expression was 1.7±0.1-fold higher compared with the normoxia-treated cells (P<0.05) but not significantly different compared with the hypoxia treatment cells (P>0.05; Fig. 5D). HIF1α knockdown in hypoxia significantly reduced HIF1α protein expression compared with the hypoxia treatment group with no knockdown (P<0.01). HIF2α knockdown did not affect HIF1α protein expression compared with hypoxia treatment alone (P>0.05; Fig. 5D). ATF4 knockdown had no effect on HIF2α mRNA expression in acute hypoxia with only a 1.1±0.1-fold increase compared with normoxia treated cells (P>0.05). However, in chronic hypoxia, HIF2α mRNA expression levels increased by 2.4±0.2-fold compared with normoxia-treated cells (P<0.0001; Fig. 5C).
Figure 5.
ATF4 KD affects HIF1α and HIF2α expression in acute and chronic hypoxia. PANC-1 cells were treated with RNAiMAX Lipofectamine reagent and small interfering RNA to transiently KD ATF4, HIF1α or HIF2α. The cells were placed in hypoxia (0.2% oxygen) or normoxia (21% oxygen) for 16 or 48 h. All cells with KD were placed in hypoxia. (A and B) RT-qPCR was conducted to determine HIF1α after (A) 16 and (B) 48 h of hypoxia. (C) HIF2α mRNA levels were measured using RT-qPCR after ATF4 KD in acute or chronic hypoxia. (D and E) Western blot analysis was conducted to determine the protein expression levels of HIF1α after (D) 16 or (E) 48 h of hypoxia. Fold changes are shown relative to normoxic cells. Error bars represent the mean ± SD for n=3. Fold changes were analyzed by one-way ANOVA followed by Bonferroni statistical hypothesis test. Fold changes for RT-qPCR are shown relative to normoxic cells. Error bars represent the mean ± SD for n=9. Fold changes were analyzed by one-way ANOVA followed by Bonferroni statistical hypothesis test. **P<0.01, ***P<0.001 and ****P<0.0001. ATF4, activating transcription factor 4; HIF, hypoxia-inducible factor; KD, knockdown; RT-qPCR, reverse transcription-quantitative PCR.
Furthermore, in chronic hypoxia, ATF4 knockdown significantly decreased HIF1α mRNA levels to 0.15±0.02-fold of the normoxia-treated and hypoxia-treated cells (P<0.05; Fig. 5B). This expression pattern was also identified in protein expression where ATF4 knockdown resulted in a 0.4±0.2-fold decrease in HIF1α protein expression levels (P<0.0001; Fig. 5E). While ATF4 did not affect HIF1α or HIF2α during acute hypoxia, it did affect HIF1α and HIF2α mRNA and protein expression levels during chronic hypoxia.
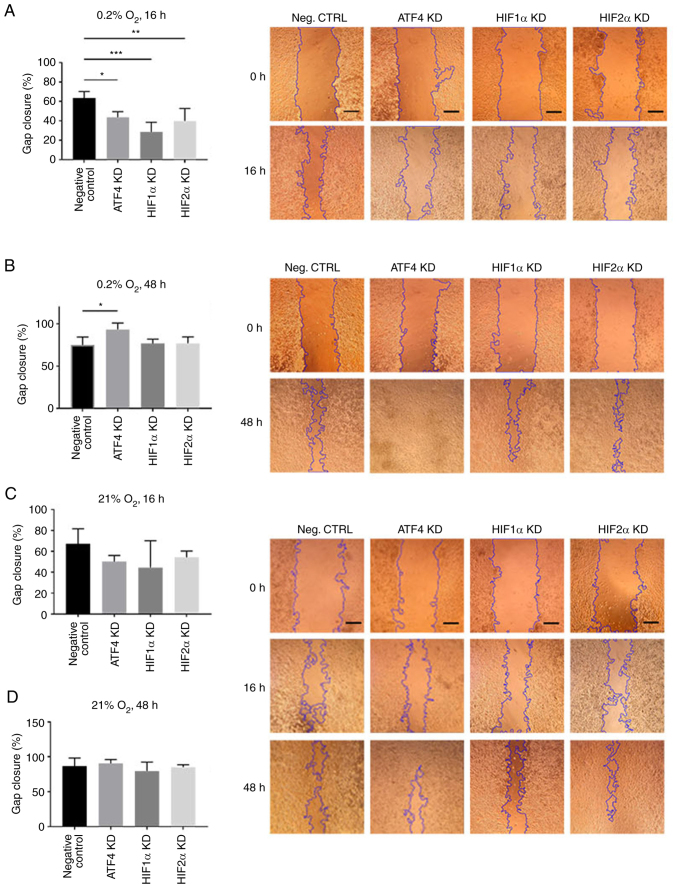
ATF4 knockdown increases cell migration in chronic but not acute hypoxia
To determine the effect of ATF4 inhibition on cell migration in acute and chronic hypoxia, a scratch assay was performed. PANC-1 cells transiently transfected with ATF4, HIF1α or HIF2α siRNA were seeded and scratched prior to 16 or 48 h of hypoxic exposure. After 16 h of hypoxic exposure, the negative control group, PANC-1 cells with no knockdown siRNA, exhibited the greatest gap closure among the treatment groups with 63.4±6.8% gap closure (Fig. 6A). In normoxia, there were no changes in gap closure rates among the treatment groups, with all treatments after 16 h of normoxic incubation resulted in at least 45% closure (Fig. 6C) and at least 80% closure after 48 h of incubation (Fig. 6D). Relative to the negative control group, hypoxia-treated cells without siRNA knockdowns, the ATF4 knockdown cells in acute hypoxia closed 43.7±5.7% of the gap measured at the initial scratch (P<0.05). The HIF1α and HIF2α knockdown cells also exhibited a significant decline in cell migration with 28.60±9.95% (P<0.001) and 39.8±13.0% (P<0.01) gap closure, respectively (Fig. 6A). In chronic hypoxia, the results demonstrated that HIF1α and HIF2α knockdown cell migration rates were comparable with those of the hypoxia-treated cells without siRNA knockdowns. The negative control cells closed 75.1±9.4% of the gap after 48 h of hypoxia (Fig. 6B). HIF1α knockdown cells showed 76.7±5.2% gap closure, and HIF2α knockdown cells exhibited 77.0±7.6% gap closure. The ATF4 knockdown cells, on the other hand, showed significantly more cell migration with 93.4±7.6% gap closure (P<0.05) compared with the negative control group and HIF1α or HIF2α knockdown cells (Fig. 6B). The scratch assay revealed that ATF4 knockdown inhibits cell migration in acute hypoxia.
Figure 6.
ATF4, HIF1α and HIF2α KD decrease cell migration in acute hypoxia, but ATF4 KD in chronic hypoxia increases cell migration. (A-D) PANC-1 cells were transiently transfected, scratched and incubated in hypoxia for (A) 16 or (B) 48 h or in normoxia for (C) 16 or (D) 48 h. The negative control were PANC-1 cells exposed to 0.2% oxygen with transfection reagent but no KD small interfering RNA. Images were captured by light microscope at ×50 magnification. Scale bar=500 µm. The gap closure was determined by using the ImageJ plugin tool to measure the gap area of each image and the gap closure of each treatment group was calculated after hypoxic incubation relative to images captured before hypoxic exposure. Error bars represent the mean ± SD for n=4. The fold changes were analyzed by two-way ANOVA followed by Bonferroni post hoc statistical hypothesis test. *P<0.05, **P<0.001 and ***P<0.0005. ATF4, activating transcription factor 4; HIF, hypoxia-inducible factor; KD, knockdown.
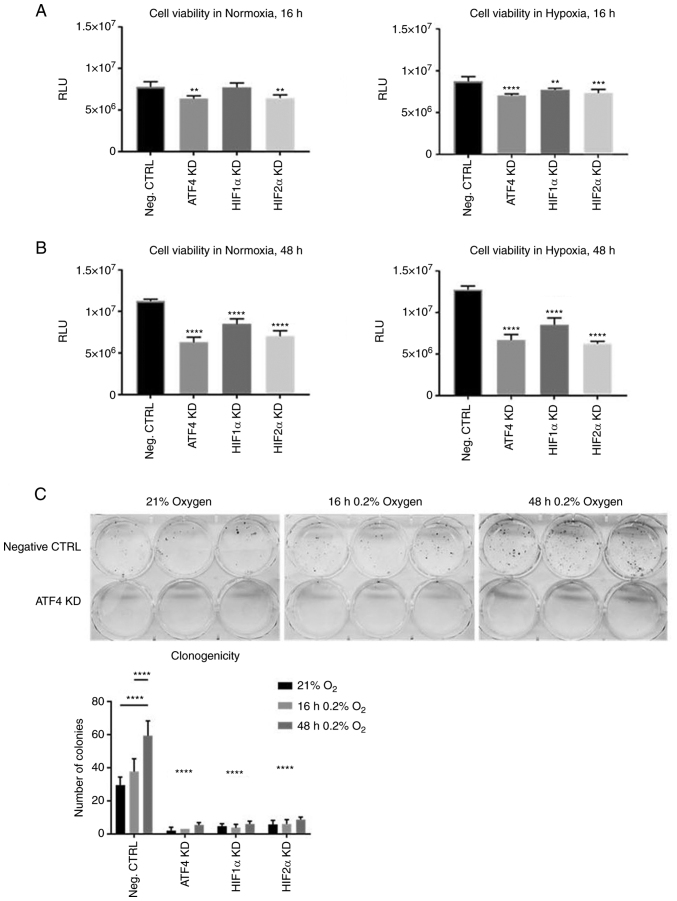
ATF4 inhibition decreases cell viability and colony formation
Considering that ATF4 knockdown promoted cell migration in chronic hypoxia, the effects of ATF4 knockdown on cell viability and clonogenicity were next determined by conducting cell viability assays and colony formation assays. PANC-1 cells were transiently transfected and seeded into 96-well plates. The cells were incubated in normoxia for 24 h, and then placed in either normoxia or hypoxia for 16 or 48 h. A CellTiterGlo Luminescent assay was used to measure ATP. After ATF4 knockdown, cells in normoxia and hypoxia for 16 and 48 h all showed significant decrease in cell viability compared with the PANC-1 cells with no knockdowns (P<0.05; Fig. 7A and B). HIF1α knockdown in normoxia did not affect cell viability but did in cells in acute hypoxia (Fig. 7A). HIF1α and HIF2α knockdowns significantly decreased cell viability (P<0.05) in both normoxia and chronic hypoxia (Fig. 7B) and there were no significant differences in cell viability between normoxic and chronically hypoxic cells in each treatment group (Fig. S2). The colony formation assays showed significantly less colonies after ATF4 knockdown (P<0.0001; Fig. 7C), as well as after HIF1α and HIF2α knockdown (P<0.0001; Figs. 7C and S3). The negative control group cells that were exposed to chronic hypoxia had significantly more colonies compared with the acute hypoxia cells and normoxic cells (P<0.05; Fig. 7C). Overall, ATF4 inhibition significantly decreases cell viability and colony formation abilities in PANC-1 cells in both normoxia and chronic hypoxia (26).
Figure 7.
ATF4, HIF1α and HIF2α KD decrease cell viability in chronic hypoxia. (A and B) PANC-1 cells were transiently transfected and seeded into 96-well plates and incubated in hypoxia or normoxia for (A) 16 or (B) 48 h. Cell viability was measured by CellTiter Glo luminescence. The negative control were PANC-1 cells exposed to 0.2% oxygen with transfection reagent but no KD small interfering RNA. Error bars represent the mean ± SD for n=5. The relative luminescence unit (RLU) were analyzed by one-way ANOVA followed by Bonferroni statistical hypothesis test. (C) Colony formation assays were performed by transiently transfecting PANC-1 cells and seeded into plates. The plates were placed in either normoxia or hypoxia for 16 or 48 h. The cells were fixed, stained and colonies were counted. Error bars represent the mean ± SD for n=3. **P<0.01, ***P<0.001 and ****P<0.0001. ATF4, activating transcription factor 4; HIF, hypoxia-inducible factor; KD, knockdown.
Transforming growth factor beta (TGF-β) has been shown to be prevalent in tumors and is secreted by cancer-associated fibroblasts to supplement cancer cells and promote growth. One proposed pathway that TGF-β promotes cancer survival and growth is by inducing ATF4 expression and activity (26). However, the effect that TGF-β may have on PDAC cells in hypoxia has not yet been studied. To understand if TGF-β has the same function in hypoxic PDAC, PANC-1 wells were treated with TGF-β in acute and chronic hypoxia. Western blot analysis revealed that TGF-β did not induce ATF4 expression in normoxic cells (Fig. S4A). ATF4 was upregulated in 16 h of hypoxic exposure, but there was little ATF4 expression detected in cells exposed to chronic hypoxia, despite TGF-β treatment (Fig. S4A). The colony formation assay used to assess clonogenicity of the cells revealed that TGF-β did improve clonogenicity in cells exposed to acute hypoxia, but not chronic hypoxia, compared with normoxic cells (Fig. S4B). There were minimal colonies formed in cells with ATF4 knockdown.
HIF1α, HIF2α and ATF4 expression increases in healthy WS1 skin fibroblast cells but not in Mia-PaCa2 PDAC cell line
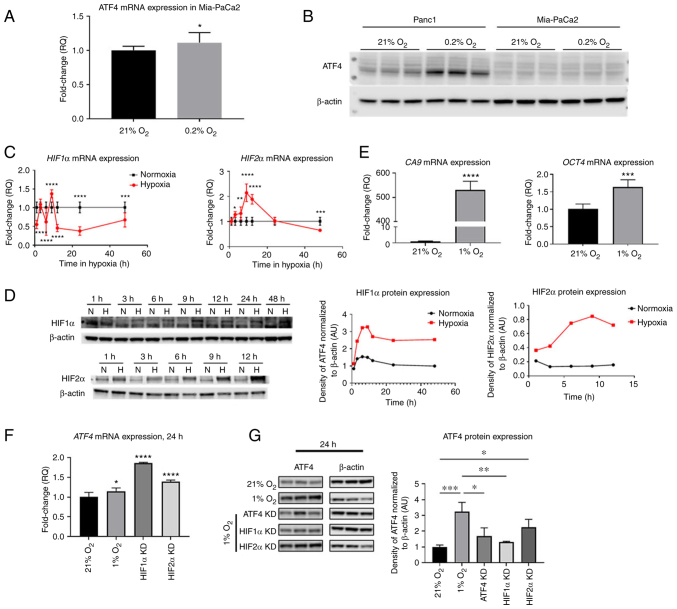
Next, it was investigated how ATF4 and HIFs regulation in PANC-1 PDAC cells compares to another PDAC, MiaPaCa2 and to non-cancerous WS1 fibroblasts. To determine whether ATF4 is expressed in Mia-PaCa2 cells in 21 and 0.2% oxygen conditions, cells were incubated in normoxia and hypoxia for 24 h. ATF4 mRNA levels in Mia-PaCa2 cells increased in hypoxia by 1.1±0.1-fold (P<0.1) and protein levels in Mia-PaCa2 were not detectable in either normoxia or hypoxia (Fig. 8A and B). In PANC-1 cells, however, ATF4 protein expression increased in hypoxia compared with normoxia (Fig. 8B).
Figure 8.
HIF1α, HIF2α and ATF4 mRNA levels and HIF target genes change over time in hypoxia in WS1 healthy cells but ATF4 was not detectable in hypoxic Mia-PaCa2 cells. PANC-1 and Mia-PaCa2 cells were placed in either normoxia (21% oxygen) or hypoxia (0.2% oxygen) for 24 h. (A) ATF4 mRNA levels in Mia-PaCa2 cells were determined by RT-qPCR, and (B) ATF4 protein levels were determined in Mia-PaCa2 cells using western blot analysis. WS1 fibroblast cells were treated with hypoxia (1% oxygen) or normoxia (21% oxygen) for durations from 1 to 48 h. (C) RT-qPCR and (D) western blot analyses were conducted to determine HIF1α and HIF2α mRNA and protein levels, respectively. (E) CA9 and Oct4 mRNA levels were also determined by RT-qPCR. (F and G) ATF4 mRNA and protein levels were also measured using (F) RT-qPCR and (G) western blot analysis, respectively. Fold changes for RT-qPCR are shown relative to normoxic cells. Error bars represent the mean ± SD for n=9. The fold changes were analyzed using unpaired Student's t-test. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. HIF, hypoxia-inducible factor; ATF4, activating transcription factor 4; RT-qPCR, reverse transcription-quantitative PCR; CA9, carbonic anhydrase IX; Oct4, octamer-binding transcription factor 4; KD, knockdown.
To determine HIF1α, HIF2α and ATF4 expression in hypoxia in healthy cells, WS1 skin fibroblast cells were incubated in normoxia (21% oxygen) or hypoxia (1% oxygen) from 1 to 48 h of treatment. After 1 h of hypoxic exposure, HIF1α mRNA significantly decreased 0.6±0.1-fold (P<0.0001). At 3 h, there was no significant difference of HIF1α mRNA levels in hypoxia compared with normoxia (1.1±0.1-fold). After 6 h of hypoxic exposure, mRNA levels decreased 0.6±0.4-fold, and they significantly increased after 9 h of exposure by 1.4±0.1-fold (P<0.0001). The HIF1α mRNA levels became significantly lower than normoxic levels from 12 h of hypoxia by 0.5±0.1-fold (P<0.0001) to 48 h of hypoxia (0.7±0.2-fold) (P<0.001; Fig. 8C). HIF2α mRNA expression steadily increased from 3 h of hypoxia and reached a peak at 9 h of hypoxia by 2.1±0.4-fold. The mRNA levels decreased continually from 12 h of exposure to 48 h, at which point, the mRNA levels were 0.7±0.1-fold lower than normoxic levels (Fig. 8C). It was confirmed that HIF1α and HIF2α protein levels were increased in hypoxia by western blotting, which demonstrated steady increases of HIF protein expression in all time points compared with normoxic levels (Fig. 8D).
The expression of HIF transcriptional target genes CA9 and Oct4 was also measured. CA9 is a HIF1α-specific target gene while Oct4 is a HIF2α-specific target gene. After 24 h of hypoxic exposure, results showed that CA9 mRNA levels significantly increased by 530.6±33.7-fold compared with normoxia while Oct4 mRNA levels were significantly higher in hypoxia by 1.6±0.2-fold compared with normoxic levels (Fig. 8E). ATF4 expression was also analyzed in normoxia, hypoxia and with HIF1α and HIF2α knocked down. ATF4 mRNA levels were significantly higher after 24 h in hypoxia than in normoxia by 1.1±0.08-fold (P<0.05; Fig. 8F). Upon HIF1α and HIF2α knockdowns, ATF4 mRNA expression significantly increased by 1.9±0.01 and 1.4±0.04-fold, respectively (Fig. 8F). Western blot analysis revealed that hypoxia significantly increased ATF4 protein expression in healthy cells by 3.2±0.6-fold compared with normoxia. While ATF4 mRNA increased upon HIF1α and HIF2α knockdown, ATF4 protein expression significantly decreased upon HIF1α knockdown in hypoxia (1.3±0.05-fold) compared with cells in hypoxia without knockdowns (P<0.01; Fig. 8G). There was no significant difference in cells with HIF2α knockdown compared with hypoxia-treated cells without knockdowns (Fig. 8G).
Discussion
Pancreatic cancer cells exist in a hypoxic environment, and yet, most research conducted is performed under normoxic conditions or in acute hypoxia, which is less pathophysiologically relevant. Despite the chronic hypoxic condition observed in PDAC in patients, the function of ATF4 during chronic hypoxia has remained largely unstudied in pancreatic cancers. In the present study, evidence was provided that ATF4 regulates HIF1α and HIF2α expression and significantly affects cell migration, cell viability and colony formation in chronically hypoxic pancreatic cancer cells.
Loss-of-function experiments were conducted to ascertain the relationship between ATF4 and HIFs in PANC-1 cells in chronic hypoxia. HIF1α and HIF2α are regulators of cellular hypoxic response (17). The two isoforms have unique roles in mediating cell survival in hypoxia (32). Generally, HIF1α activates genes that promote changes to cell metabolism while HIF2α activates genes that promote processes such as stemness and extracellular signaling and remodeling (31,34,35). HIF1α and HIF2α are also regulated in a type of ‘switch’ in which when HIF1α expression decreases or increases, HIF2α expression tends to change inversely. This ‘HIF switch’ is critical to regulate the different response pathways as the time in hypoxia increases (32). The switch is largely attributed to microRNAs that control HIF1α and HIF2α levels by preventing HIF mRNA translation, reducing the amount of protein synthesized (32). Hypoxia-associated factors (HAFs) also downregulate HIF1α by inducing ubiquitination and protein degradation while upregulating HIF2α activation, including in a variety of different cancers such as pancreatic cancer and renal clear cell carcinoma (31,36). The present data revealed, for the first time to the best of our knowledge, that ATF4 functions as a HIF regulator by decreasing HIF2α and increasing HIF1α expression in chronic hypoxia. ATF4 regulates the HIFs inversely to microRNAs and HAFs that increase HIF2α and decrease HIF1α in chronic hypoxia (31,37,38). This may possibly prevent a complete shutdown of HIF1α specific functions by counteracting the negative regulation of HIF1α by microRNAs and HAFs. While HIF2α is commonly associated with chronic hypoxia and HIF1α with acute hypoxia, it was demonstrated that HIF1α expression remains essential in chronic hypoxia for maintaining certain cell survival pathways, such as metabolic reprogramming (31,35,39). ATF4 may function to promote HIF1α expression to maintain critical these pathways that are activated by only HIF1α and prevent a complete inhibition of HIF1α functions and improve hypoxic tolerance in chronic hypoxia. Whether ATF4 affects HIFs in chronic hypoxia directly or indirectly remains to be understood. Further studies are planned to analyze downstream ATF4 transcriptional target genes to determine potential mechanisms used to affect HIF expression.
Our data also showed that ATF4 promotes colony formation and cell viability in chronically hypoxic PDAC cells (26,40). In chronic hypoxia, ATF4 knockdown significantly decreased colony formation and cell viability. Hence, ATF4 acts as a pro-survival factor for PDAC cells under conditions of chronic hypoxia and may be a viable target for PDAC therapies. ATF4 has been previously shown to promote colony formation and cell viability by regulating the Wnt/β-catenin pathway and the PERK/eIF2α/ATF4 axis in different cancers, including lung cancer and glioblastoma cells (41,42). It is plausible that ATF4 affects colony formation and viability of PDAC cells in chronic hypoxia the same way it does in normoxia. However, these functions of ATF4 may be more robust in chronic hypoxia compared with normoxia. Targeting ATF4 may be a therapeutic strategy for treating hypoxic PDAC which is otherwise difficult to treat.
ATF4 roles in cell migration in hypoxic PDAC cells were also determined. ATF4 stimulates cell migration and metastasis in breast cancer cells, as well as in PDAC (26,40). However, it was demonstrated that under chronic hypoxia, ATF4 inhibition increased cell migration in PDAC cells, evidence that rather than promoting cell migration as observed in normoxia, ATF4 negatively regulates cell migration in chronic hypoxia. While it has been previously revealed in breast cancer that ATF4 stimulates cell migration, these studies were conducted under normoxic conditions or in oxygen levels that are likely markedly higher than in patients (43,44). Previous studies showed that breast cancers typically exhibit oxygen levels of 0.2% oxygen or less, similar levels to that of PDAC (29,40,45,46). Furthermore, these experiments did not exceed 24 h of hypoxic exposure. There may also be differences in the physiology of the two cancers that can account for this discrepancy, which remain to be explored. Another study showed that ATF4 promotes cell migration in PDAC (26). However, this was not conducted in hypoxia, acute or chronic, and our data indicated that ATF4 functions differently in hypoxia than in normoxia. This suggested that ATF4 functions change or are different depending on the presence or absence of oxygen and the duration of hypoxia. The present results not only indicated a negative regulatory role of ATF4 on cell migration, but they also emphasized the importance of conducting pancreatic cancer research in pathophysiologically relevant conditions, particularly chronic hypoxia. The discovery of the ‘HIF switch’, particularly the dominance of HIF1α in acute hypoxia and HIF2α in chronic hypoxia, has illuminated the importance of distinguishing acute and chronic hypoxia in cancers (31,32,37,47). However, the same principles should be applied when investigating other stress response pathways in hypoxia, including ATF4 in the UPR pathway. As it was revealed, the role of ATF4 in acute and chronic hypoxia is not the same, indicative of a mechanism that facilitates this switch in function that is yet to be found.
It is unknown whether ATF4 directly interacts with HIF1α and HIF2α mRNA or protein to regulate expression or indirectly by way of another downstream factor that functions as a mediator between ATF4 and HIFs. It was demonstrated that HIF1α, HIF2α and ATF4 are all expressed at the protein level in hypoxia in healthy cells and in PANC-1 cells. Somewhat surprisingly, our data showed that hypoxia does not induce ATF4 expression in Mia-PaCa2 cells compared with PANC-1 cells which exhibit a robust increase in ATF4 protein expression. This can be due to several reasons, including the different phenotypes exhibited by Mia-PaCa2 cells and PANC-1 cells. For instance, low expression of neural cell adhesion molecule CD56, a protein found in PANC-1 cells but not Mia-PaCa2 cells, has been revealed to result in low levels of ER stress markers, including ATF4 (48,49). The lack of CD56 in Mia-PaCa2 cells may contribute to the low levels of ATF4, despite hypoxic exposure. Another protein that may contribute to the low ATF4 levels in Mia-PaCa2 but high induction in PANC-1 cells is E-cadherin, a cell-to-cell adhesion molecule. High E-cadherin levels, as observed in Mia-PaCa2 cells but not in PANC-1 cells, are associated with low expression of ATF4 (43). These types of differing expression profiles observed in the two cell lines may contribute to the low ATF4 expression identified in Mia-PaCa2 cells and high levels in PANC-1 cells. Notably, PANC-1 have been described as more aggressive and with greater metastasizing potential (48). The ATF4 related mechanism, found only in the PANC-1 cells and which is notably quite different than healthy WS-1 cells, is a candidate for mediating such a phenotype and therefore represents a potential biomarker for metastatic potential, though further studies are required to explore this hypothesis.
One major finding from the present study was that in chronically hypoxic PANC-1 cells, ATF4 was revealed to affect HIF1α expression while HIFs did not affect ATF4. This, however, was not observed in healthy cells. As observed in PANC-1 cells, ATF4 mRNA expression significantly increased when HIF1α was knocked down. However, in healthy cells, HIF1α knockdown caused ATF4 protein expression levels to be significantly lower than the hypoxic cells expressing HIF1α. This result is in contrast to what was observed in PANC-1 cells, in which HIF1α knockdown did not change ATF4. The present findings indicated that in healthy cells, HIF1α may regulate ATF4 in hypoxia, while in PDAC cells, this regulation pathway is either inhibited or is negated by other mechanisms that cause ATF4 overexpression (14,26,50). One such mechanism is the protein CD56 found in PANC-1 cells that causes high ATF4 expression (48,49). Since HIF1α has never been shown to affect ATF4 in hypoxia in healthy cells, the mechanism in which HIF1α may affect ATF4 protein expression requires further study. It will also be important to further validate these findings with other methods in in vitro PDAC models, such as the use of 3D spheroids, and in in vivo models which replicate in an improved way the heterogeneity of the tumor microenvironment observed in patients. This shall be the focus of our future efforts.
In conclusion, it was demonstrated that ATF4 is upregulated in chronically hypoxic PDAC cells and that HIF1α expression is dependent on ATF4 while ATF4 negatively regulates HIF2α in PDAC cells in chronic hypoxia. It was also demonstrated that while ATF4 may decrease cell migration in chronic hypoxia, its inhibition is detrimental to clonogenicity and cell viability in hypoxia, and therefore, is an attractive target for chronically hypoxic PDAC. The present study has emphasized the importance of conducting research in pathophysiologically relevant systems related to hypoxia. These findings elucidated a novel understanding of PDAC in which hypoxia causes the UPR pathway to regulate the hypoxia response pathways, possibly contributing to the aggressive qualities of PDAC.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ATF4
activating transcription factor 4
- CA9
carbonic anhydrase IX
- HAF
HIF activating factor
- HIF1α
hypoxia inducible factor 1α
- HIF2α
hypoxia inducible factor 2α
- Oct4
octamer-binding transcription factor 4
- PDAC
pancreatic ductal adenocarcinoma
- uORF
upstream open reading frame
- UPR
unfolded protein response
Funding Statement
The present study was supported by the National Institutes of Health (grant nos. R01NS092671 and R01MH110441), the University of Miami Sylvester Comprehensive Cancer Center Molecular Therapeutics Shared Resource (MTSR) and the Jay Weiss Institute for Health Equity.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NTC and SPB conceptualized the idea and designed experiments. NTC conducted experiments, procured and analyzed data and led manuscript writing and organization. ZM and CHC provided feedback on data interpretation and experimental design. SW was involved in planning and executing experiments. NTC and SPB confirm the authenticity of all raw data. All authors contributed to the analysis of results and writing of the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Hessmann E, Buchholz SM, Demir IE, Singh SK, Gress TM, Ellenrieder V, Neesse A. Microenvironmental determinants of pancreatic cancer. Physiol Rev. 2020;100:1707–1751. doi: 10.1152/physrev.00042.2019. [DOI] [PubMed] [Google Scholar]
- 3.Orth M, Metzger P, Gerum S, Mayerle J, Schneider G, Belka C, Schnurr M, Lauber K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat Oncol. 2019;14:141. doi: 10.1186/s13014-019-1345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuen A, Díaz B. The impact of hypoxia in pancreatic cancer invasion and metastasis. Hypoxia (Auckl) 2014;2:91–106. doi: 10.2147/HP.S52636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312:163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fähling M. Surviving hypoxia by modulation of mRNA translation rate. J Cell Mol Med. 2009;13:2770–2779. doi: 10.1111/j.1582-4934.2009.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving stress: Modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab. 2017;28:794–806. doi: 10.1016/j.tem.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B'chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579:427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pathria G, Scott DA, Feng Y, Sang Lee J, Fujita Y, Zhang G, Sahu AD, Ruppin E, Herlyn M, Osterman AL, Ronai ZA. Targeting the Warburg effect via LDHA inhibition engages ATF4 signaling for cancer cell survival. EMBO J. 2018;37:e99735. doi: 10.15252/embj.201899735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez MR, Cleveland JL. ATF4-amino acid circuits: A recipe for resistance in melanoma. EMBO J. 2018;37:e100600. doi: 10.15252/embj.2018100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou SH, Risca VI, Wang GX, Yang D, Grüner BM, Kathiria AS, Ma RK, Vaka D, Chu P, Kozak M, et al. BLIMP1 induces transient metastatic heterogeneity in pancreatic cancer. Cancer Discov. 2017;7:1184–1199. doi: 10.1158/2159-8290.CD-17-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mesclon F, Lambert-Langlais S, Carraro V, Parry L, Hainault I, Jousse C, Maurin AC, Bruhat A, Fafournoux P, Averous J. Decreased ATF4 expression as a mechanism of acquired resistance to long-term amino acid limitation in cancer cells. Oncotarget. 2017;8:27440–27453. doi: 10.18632/oncotarget.15828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palam LR, Gore J, Craven KE, Wilson JL, Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 2015;6:e1913. doi: 10.1038/cddis.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ait Ghezala H, Jolles B, Salhi S, Castrillo K, Carpentier W, Cagnard N, Bruhat A, Fafournoux P, Jean-Jean O. Translation termination efficiency modulates ATF4 response by regulating ATF4 mRNA translation at 5′ short ORFs. Nucleic Acids Res. 2012;40:9557–9570. doi: 10.1093/nar/gks762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chee NT, Lohse I, Brothers SP. mRNA-to-protein translation in hypoxia. Mol Cancer. 2019;18:49. doi: 10.1186/s12943-019-0968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logsdon DP, Shah F, Carta F, Supuran CT, Kamocka M, Jacobsen MH, Sandusky GE, Kelley MR, Fishel ML. Blocking HIF signaling via novel inhibitors of CA9 and APE1/Ref-1 dramatically affects pancreatic cancer cell survival. Sci Rep. 2018;8:13759. doi: 10.1038/s41598-018-32034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Zhao L, Wang J, Chen N, Yan J, Pan X. HIF-2α and Oct4 have synergistic effects on survival and myocardial repair of very small embryonic-like mesenchymal stem cells in infarcted hearts. Cell Death Dis. 2017;8:e2548. doi: 10.1038/cddis.2016.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho JJD, Wang M, Audas TE, Kwon D, Carlsson SK, Timpano S, Evagelou SL, Brothers S, Gonzalgo ML, Krieger JR, et al. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 2016;14:1293–1300. doi: 10.1016/j.celrep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uniacke J, Perera JK, Lachance G, Francisco CB, Lee S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014;74:1379–1389. doi: 10.1158/0008-5472.CAN-13-2278. [DOI] [PubMed] [Google Scholar]
- 23.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29:4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 24.Koumenis C, Wouters BG. ‘Translating’ tumor hypoxia: Unfolded protein response (UPR)-dependent and UPR-independent pathways. Mol Cancer Res. 2006;4:423–436. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- 25.Suarez-Arnedo A, Torres Figueroa F, Clavijo C, Arbeláez P, Cruz JC, Muñoz-Camargo C. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One. 2020;15:e0232565. doi: 10.1371/journal.pone.0232565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei L, Lin Q, Lu Y, Li G, Huang L, Fu Z, Chen R, Zhou Q. Cancer-associated fibroblasts-mediated ATF4 expression promotes malignancy and gemcitabine resistance in pancreatic cancer via the TGF-β1/SMAD2/3 pathway and ABCC1 transactivation. Cell Death Dis. 2021;12:334. doi: 10.1038/s41419-021-03420-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koong AC, Mehta VK, Le QT, Fisher GA, Terris DJ, Brown JM, Bastidas AJ, Vierra M. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48:919–922. doi: 10.1016/S0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 28.Graffman S, Björk P, Ederoth P, Ihse I. Polarographic pO2 measurements of intra-abdominal adenocarcinoma in connection with intraoperative radiotherapy before and after change of oxygen concentration of anaesthetic gases. Acta Oncol. 2001;40:105–107. doi: 10.1080/028418601750071163. [DOI] [PubMed] [Google Scholar]
- 29.McKeown SR. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radiol. 2014;87:20130676. doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena K, Jolly MK. Acute vs chronic vs cyclic hypoxia: Their differential dynamics, molecular mechanisms, and effects on tumor progression. Biomolecules. 2019;9:339. doi: 10.3390/biom9080339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh MY, Lemos R, Jr, Liu X, Powis G. The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 2011;71:4015–4027. doi: 10.1158/0008-5472.CAN-10-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh MY, Powis G. Passing the baton: The HIF switch. Trends Biochem Sci. 2012;37:364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Downes NL, Laham-Karam N, Kaikkonen MU, Ylä-Herttuala S. Differential but complementary HIF1α and HIF2α transcriptional regulation. Mol Ther. 2018;26:1735–1745. doi: 10.1016/j.ymthe.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmquist-Mengelbier L, Fredlund E, Löfstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1alpha, leading to its oxygen-independent degradation. Mol Cell Biol. 2008;28:7081–7095. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serocki M, Bartoszewska S, Janaszak-Jasiecka A, Ochocka RJ, Collawn JF, Bartoszewski R. miRNAs regulate the HIF switch during hypoxia: A novel therapeutic target. Angiogenesis. 2018;21:183–202. doi: 10.1007/s10456-018-9600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu ZJ, Lu LG, Tao KZ, Chen DF, Xia Q, Weng JJ, Zhu F, Wang XP, Zheng P. MicroRNA-185 suppresses growth and invasion of colon cancer cells through inhibition of the hypoxia-inducible factor-2α pathway in vitro and in vivo. Mol Med Rep. 2014;10:2401–2408. doi: 10.3892/mmr.2014.2562. [DOI] [PubMed] [Google Scholar]
- 39.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: Implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 40.Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du J, Liu H, Mao X, Qin Y, Fan C. ATF4 promotes lung cancer cell proliferation and invasion partially through regulating Wnt/β-catenin signaling. Int J Med Sci. 2021;18:1442–1448. doi: 10.7150/ijms.43167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dadey DYA, Kapoor V, Khudanyan A, Thotala D, Hallahan DE. PERK regulates glioblastoma sensitivity to ER stress although promoting radiation resistance. Mol Cancer Res. 2018;16:1447–1453. doi: 10.1158/1541-7786.MCR-18-0224. [DOI] [PubMed] [Google Scholar]
- 43.Zeng P, Sun S, Li R, Xiao ZX, Chen H. HER2 upregulates ATF4 to promote cell migration via activation of ZEB1 and downregulation of E-cadherin. Int J Mol Sci. 2019;20:2223. doi: 10.3390/ijms20092223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González-González A, Muñoz-Muela E, Marchal JA, Cara FE, Molina MP, Cruz-Lozano M, Jiménez G, Verma A, Ramírez A, Qian W, et al. Activating transcription factor 4 modulates TGFβ-induced aggressiveness in triple-negative breast cancer via SMAD2/3/4 and mTORC2 signaling. Clin Cancer Res. 2018;24:5697–5709. doi: 10.1158/1078-0432.CCR-17-3125. [DOI] [PubMed] [Google Scholar]
- 45.Rundqvist H, Johnson RS. Tumour oxygenation: Implications for breast cancer prognosis. J Intern Med. 2013;274:105–112. doi: 10.1111/joim.12091. [DOI] [PubMed] [Google Scholar]
- 46.Vaupel P, Schlenger K, Knoop C, Höckel M. Oxygenation of human tumors: Evaluation of tissue oxygen distribution in breast cancers by computerized O2 tension measurements. Cancer Res. 1991;51:3316–3322. [PubMed] [Google Scholar]
- 47.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 48.Gradiz R, Silva HC, Carvalho L, Botelho MF, Mota-Pinto A. MIA PaCa-2 and PANC-1-pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci Rep. 2016;6:21648. doi: 10.1038/srep21648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida T, Ri M, Kinoshita S, Narita T, Totani H, Ashour R, Ito A, Kusumoto S, Ishida T, Komatsu H, Iida S. Low expression of neural cell adhesion molecule, CD56, is associated with low efficacy of bortezomib plus dexamethasone therapy in multiple myeloma. PLoS One. 2018;13:e0196780. doi: 10.1371/journal.pone.0196780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Carbonero N, Li W, Cabeza-Morales M, Martinez-Useros J, Garcia-Foncillas J. New hope for pancreatic ductal adenocarcinoma treatment targeting endoplasmic reticulum stress response: A systematic review. Int J Mol Sci. 2018;19:2468. doi: 10.3390/ijms19092468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.