Abstract
The human microbiome represents a new frontier in understanding the biology of human health. While epidemiology in this area is still in its infancy, its scope will likely expand dramatically over the coming years. To rise to the challenge, we argue that epidemiology should capitalize on its “population perspective” as a critical complement to molecular microbiome research, allowing for the illumination of contextual mechanisms that may vary more across populations rather than individuals. We first briefly review current research on social context and the gut microbiome, focusing specifically on socioeconomic status (SES) and race/ethnicity. Next, we reflect on the current state of microbiome epidemiology through the lens of one specific area--the association of the gut microbiome and metabolic disorders. We identify key methodological shortcomings of current epidemiological research in this area, including extensive selection bias, the use of non-compositionally robust measures, and a lack of attention to social factors as confounders or effect modifiers.
Keywords: microbiome, microbiota, SES, race/ethnicity, social, metabolic, epidemiology
I. INTRODUCTION
Since the first publication of results from the Human Microbiome Project (HMP) in 2012 (60), research on the microbiome- the trillions of microorganisms that inhabit the human body and their genes-has grown exponentially1, generating new knowledge on everything from how diet affects the microbiome to how the microbiome may influence the brain, behavior, and mental illness (4; 116). Robust evidence points to a stronger role for the “environment” in shaping the human gut microbiome relative to genetics (110), compelling researchers to better define and measure the “environment” to understand the role of the microbiome in human health and disease (52). Use of model systems such as germ-free mice allows strong causal inference on isolated aspects of microbiome biology, but analysis of human populations in their full complexity is necessary to move beyond general principles towards actionable knowledge (82). Thus while it remains important to understand microbiome function at the molecular level with an eye towards novel prognostic and treatment breakthroughs, it is equally important to “zoom out” and consider a population perspective in microbiome research (108). A population perspective reminds us that individual-level determinants of the microbiome are not necessarily the same as those that explain differences across populations, especially those living within quite homogeneous environments with respect to geography, culture, and nutrition (108). Social and geographical context, for instance, are emerging as crucial determinants of both the microbiome itself as well as modifiers of existing microbiome-health associations (28; 49). Epidemiology is well-poised to make important contributions to the description of the microbiome across person, place, and time, as well as to improving population-based research design and causal inference to understand how the microbiome impacts health and disease across the life course (107).
This review briefly summarizes current research on social context and the gut microbiome, focusing specifically on socioeconomic status (SES) and race/ethnicity. We then reflect on the current state of microbiome epidemiology through the lens of one specific area--the association of the gut microbiome and metabolic traits. As in any new research area spurred forward by new technology, each step forward often illuminates an even longer path forward towards actionable knowledge. While epidemiological research on the microbiome is in its infancy, this review aims to raise awareness of key methodological challenges and the importance of the broader social and environmental contexts influencing both the microbiome and health.
Social Context and the Microbiome
While providing much needed novel description, the HMP project was designed to characterize the human microbiome for the first time only in a small group of healthy volunteers(60). An important next step has been to describe how the microbiome varies across more diverse populations, and especially across characteristics known to have large associations with overall health and longevity such as socioeconomic status (SES) and race/ethnicity.
Socioeconomic Status and the Microbiome
Through pathways such as living conditions, psychosocial stress, and nutrition, it is likely that social conditions across the life course act to significantly shape “environmental” drivers of the gut microbiome (29; 52). We define socioeconomic status (SES) as reflecting human capital and economic resources such as education, income and wealth, and occupation. Socioeconomic resources can also be conceptualized and measured at the neighborhood level. Though frequently treated as a unitary concept in biomedical research, we encourage consideration of separable influences of socioeconomic indicators where possible to better understand the pathways and potential targets for intervention (51).
To date, we identified two studies that specifically link varying socioeconomic conditions to alterations in the gut microbiome (10; 86). Miller, et al. related a composite indicator of neighborhood socioeconomic status to the microbiota composition of 44 healthy volunteers in Chicago, finding higher α-diversity and greater abundance of Bacteroides with increased neighborhood socioeconomic status (86). In a large sample of UK Twins, Bowyer, et al identified associations between individual and area-level income and diversity and relative abundance of OTUs in the gut microbiome (10). Community composition measured by Bray-Curtis dissimilarity was found to differ by education and area level income, and lower individual and area-level income was associated with lower gut α-diversity, with a weaker association for education (10). Of note, the identified SES differences were only slightly attenuated with controls for diet, BMI, and current health deficits, while the associations between these variables and the gut microbiome were significantly attenuated by adjustment for SES, suggesting that SES may be an important confounder that has not been previously accounted for in most microbiome-health research. These two papers are an important first step in describing associations of social factors and the microbiome, but data that can better test the mechanisms underlying these associations are needed. As we outline later in this review, not only are socioeconomic conditions a key driver of broader morbidity and mortality, they play an outsized role in patterning metabolic disorders, making socioeconomic factors an important confounder and/or effect measure modifier to consider in gut microbiome research in this area.
Race/ethnicity and the microbiome
Race/ethnicity reflects a range of influences on the microbiome including common genetic ancestry, shared residence, culture, migration history, socioeconomic resources, and exposure to racism (34). Even with a small number of non-whites in the HMP sample, investigators found that a “wide variety of taxa, gene families and metabolic pathways were differentially distributed with subject ethnicity at every body habitat, representing the phenotype with the greatest number…of total associations with the microbiome (60).” These incidental findings of strong associations with race/ethnicity suggest the need for further investigation in more diverse population-based studies, although these data are still limited. Brooks et al. analyzed data from the American Gut Project (AGP) and the HMP and identified 12 microbial genera and families varied in abundance by ethnicity and that the associations of ethnicity with the microbiota were stronger in effect size than BMI, age, and sex (11). It should be noted that the AGP sample is a highly selected (25) volunteer sample with a very limited number of non-whites (13 African-Americans, 37 Hispanics, 88 Asian Pacific Islanders, and 1237 Caucasians) included in the analysis, thus likely underestimating true differences in the gut microbiome by ethnicity. One interesting finding was that the typical association of Christensenellaceae with BMI was not consistent across ethnicities, suggesting the importance of sociodemographic factors in modifying microbiome-phenotype associations.
The Healthy Life in an Urban Setting (HELIUS) study, a population-based sample of residents of Amsterdam with an oversample of ethnic minorities, is one of few studies to capture large numbers of respondents across different ethnic groups (439 Dutch, 367 Ghanians, 280 Moroccons, 197 Turks, 443 African Surinamese and 359 South-Asian Surinamese). Ethnic Dutch respondents had the highest level of α-diversity, with South-Asian Surinamese having the smallest (27). Ethnicity was also significantly associated with dissimilarities gut microbiota composition as measured by Bray-Curtis index, suggesting that individuals of the same ethnicity shared more similar microbiomes. Ethnicity was also associated with relative abundance of 559 out of 744 OTUs. Intriguingly, in models adjusted for diet, age, sex, education, BMI, alcohol, smoking, physical activity, and area of residence, ethnicity remained the strongest predictor of both α-diversity and β-diversity, while no other factor reached the effect size of ethnicity in the model, and most associations of the other variables weakened or disappeared when adjusted for ethnicity. While the authors point to genetic factors underlying these associations, 94% of the non-Dutch participants were first generation migrants, suggesting that early-life exposures prior to migration may have contributed.
II. Case Study: Epidemiology of the gut microbiome and metabolic conditions
In order to better understand the general methodological challenges and implications of social contexts for microbiome research in epidemiology, we conducted a review of one focal area to assess the strengths and weaknesses of existing sample selection, research design, and inference. We first provide background on the association of the gut microbiome and metabolic disorders, then describe how the strong social patterning of metabolic conditions can serve as a model for thinking about social context and the gut microbiome. Next, we present findings from a mini-systematic review of existing literature on the gut microbiome and metabolic conditions, highlighting the types of samples used, the research design, adjustments for confounding, and use of compositionally robust measures of the microbiome. Implications for overall inference and future directions are discussed.
The gut microbiome and metabolic conditions
The largest existing area of research linking the gut microbiome and health outcomes is that focused on the gut microbiome and obesity/metabolic traits. Studies on mice and humans have shown definitive links between the composition of the microbiota and obesity (7; 104). For example, transplanting fecal samples from obese mice to lean mice (7) and from twins discordant for obesity into germ-free mice (104) can successfully transmit adiposity phenotype. In turn, host gut microbiotas in humans change significantly after bariatric surgeries for weight loss, though one cannot parse out the separable influences of obesity and nutrition (30; 47). More generally, however, while the overall body of research finds connections between the gut microbiome and obesity, there is a lack of consensus as to the size and mechanisms underlying those relationships(13). Some specific results have failed to replicate across human studies, most notably the association of the Firmicutes/Bacteroidetes ratio, initially discovered in mice (6) and in early human studies (124), but not replicated in later, larger studies (33; 128; 133).
SES, race/ethnicity, and metabolic conditions
Just as the gut microbiome is linked to obesity and metabolic disorders, so too are socioeconomic conditions and race/ethnicity. Obesity, for example is strongly patterned by educational attainment and income, and these patterns originate in early life. Children whose parents have higher incomes, and especially higher educational attainment, are significantly less likely to become overweight and obese through adolescence (93). The socioeconomic gradient in obesity remains among adults. In the U.S., the obesity rate for adult women living below 130 percent of the poverty level is 45 percent for women compared to 30 percent for those living above 350 percent of the poverty level (94). Studies generally find that educational attainment is a more robust predictor of obesity than is income (17). While white men are less likely to be obese compared to white men, the reverse pattern is true for women (97). Other metabolic disorders, specifically hypertension, blood lipid levels, glucose levels and insulin resistance are also patterned by socioeconomic conditions (66). Moreover, the disparity emerges early; children whose parents had lower educational attainment had higher glucose levels and insulin resistance (125). There is evidence that these patterns are also stronger for education compared to income, specifically for higher levels of cholesterol and hypertension (130). Race differences are especially robust for glucose and hypertension—and while socioeconomic conditions explain some of this difference, they do not explain all of these differences, with stress and discrimination pathways of significant interest as possible mediators(54).
Importantly in the context of microbiome-health research, it has been shown that the health risks associated with obesity, such as diabetes and hypertension, vary by socioeconomic status (12). For example, among those with similar BMI levels, those with lower educational attainment are at greater risk for both developing and dying from diabetes and cardiovascular disease—the possible explanations for which are linked to everything from occupational environments to variation in nutrition (42). A recent meta-analysis also found that obesity is a far greater risk for diabetes among those with low, compared to high, socioeconomic status (127).
Mini-review: The gut microbiome and metabolic conditions
In this section, we explore the extent to which existing human studies on the gut microbiome and metabolic disorders employ standard epidemiological methodological practices, and whether existing research on the gut microbiome and metabolic disorders accounts for the above social contextual factors. To this end, we conducted a mini-review of the current state of epidemiological research on the gut microbiome and metabolic conditions. We searched PubMed for human non-intervention studies analyzing associations between the fecal, colonic, or intestinal microbiome and metabolic phenotypes, including obesity, lipids, blood pressure, glucose, and metabolic syndrome, excluding major cardiovascular disease. We also mined references of existing systematic reviews in this area, passing relevant titles forward for abstract and full-text screening. We retained studies using untargeted 16S rRNA genomic sequencing survey methods, the most common measurement in recent studies. One author (AR) screened based on the abstract and full text, and extracted the following information from each article marked for inclusion:
(1). Sample selection (recruitment):
We identified samples as population representative or not based on whether a form of random sampling was used. We defined community-based recruitment as involving home visit, phone or mail invitation, or in-person recruitment at a public school. Volunteer recruitment was defined as workplace, university students, or an existing clinical trial population. We defined samples relying on patients in a hospital or outpatient clinic as clinical samples.
(2). Covariate control methods:
adjustment (inclusion of covariate(s) in a regression model or propensity score, or analysis of residuals of the outcome variable regressed on covariate(s)), restriction (participants excluded in certain levels of covariate(s)), matching (controls selected to have similar levels of a covariate to cases), stratification (analysis conducted separately according to levels of a covariate), and Mendelian randomization. We recorded the covariate set separately for each method.
(3). Study design:
We classified studies as longitudinal cohort (explicitly used measurements from more than one time point in the analysis), cross-sectional (microbiome and phenotype measured at approximately the same time point and no selection based on disease of interest), case-control (microbiome and phenotype measured at approximately the same time point with disease/phenotype of interest used to select the sample, or unclear (could not be determined whether (a) different time points were involved or (b) sample selection depended on disease of interest). Where interest was in the effect of the disease on the microbiome, what is typically classified as a case control study was classified instead as cross sectional (selection based on exposure).
(4). Temporal ordering:
If interest was in the effect of the microbiome on disease, we required cross-sectional studies to measure recent incidence rather than prevalence of discrete disease in order to be classified as having proper temporal ordering, and classified as improper all such studies measuring a continuous trait such as BMI or blood pressure. For longitudinal cohort studies, proper temporal ordering entailed microbiome measurement precededing diagnosis of a disease, and for continuous traits at least one measurement following the microbiome. If interest was in the effect of the disease on the microbiome, cross-sectional or longitudinal studies with prevalent cases of disease were classified as proper.
(6). Compositionally robust methods:
A major validity issue that has been often overlooked in microbiome science has been the compositional structure of 16S data, in which each sample is a vector of counts representing the number of reads detected for each taxon, and typically only the relative proportions of each taxon within a sample are of interest. This type of data is referred to as a composition, meaning it carries only information about relative percentages of a total. In a composition, the act of normalizing to a total, as is commonly performed, is termed closure, and creates spurious correlations between the elements, a feature noted by Karl Pearson in 1897 (99). These spurious correlations resulting from closure are a massive concern for microbiome science, resulting in roughly ¾ of detected correlations between taxa being false, and 60% of true correlations missed (35; 80). Importantly, they also induce spurious correlations between the relative abundances and exposures or outcomes of interest.(48) This problem likely also influences beta diversity findings, as common beta diversity metrics depend on relative abundances (41). Drawing on the work of Aitchison showing that only ratios between elements of a composition allow for the application of common statistical techniques (2), tools have recently been developed analyze correlational taxon networks (35; 71), differential abundance (31; 83), and phylogenetic beta diversity (115) in a compositionally-robust way.
For each study we recorded which compositionally robust and non-robust analytic methods were used. Non-robust methods included analysis of differentially abundance (DA) taxa using raw counts or closed compositions, including direct comparisons (e.g. t-tests) of taxa percentages, as well as newer methods such as LEfSe (113), DESeq2 (79) and metagenomeSeq (97), which also rely on normalization methods not directly accounting for compositionality. Non-robust methods also included rarefaction (equalizing the total number of reads across samples), beta diversity measures such as weighted and unweighted UniFrac (81) and Bray-Curtis distances (75), and correlational network methods using closure (e.g. Pearson’s correlation). Compositionally-robust methods included DA using a log-ratio transform such as ANCOM (83) and ALDEx2 (31) or a log-linear model with a log-ratio-based offset (15), Aitchison (2) or PhiLR (115) distance as beta diversity measures, and correlational network methods using log-ratio transforms such as SparCC (35) and SPEIC-EASI (71).
Results
Of 1899 studies meeting initial search criteria, 71 studies were selected based on abstract and full-text review, 90% of which were published since 2015 (Supplemental Table 1). The majority had small sample sizes (median=100), although several published in the past 2 years have over 1000 participants (9; 49; 62; 74; 132; 133). Clinical and volunteer-based sampling dominated, with community-based recruitment less common (n=13). Among studies using community-based recruitment, only four used a sampling strategy aimed at population-representativeness: He et al. (2018) used a multistage probability sample based in Guangdong, China (49); Org et al. (2017) used a random sample of a population register in Kuopio, Finland (95); Rampelli et al. (2018) aimed to recruit all children attending kindergarten and primary school in pre-selected municipalities across eight European countries (103); and Sun et al. (2019) used the CARDIA study, a representative sample of black and white adults in Minneapolis (120). Cross-sectional and case control studies were the most common, but a substantial number of longitudinal cohort studies have recently been published, primarily in the last year (Figure 1). Notably, in a surprising number of studies (n=16) it was impossible to determine the method of recruitment (22-24; 59; 65; 76; 101; 105; 111; 112; 118; 124; 135), study design was unclear in n=4 (either because measurement time points were not specified (74) or selection based on outcomes was possible but unclear (78; 88; 124)).
Figure 1.
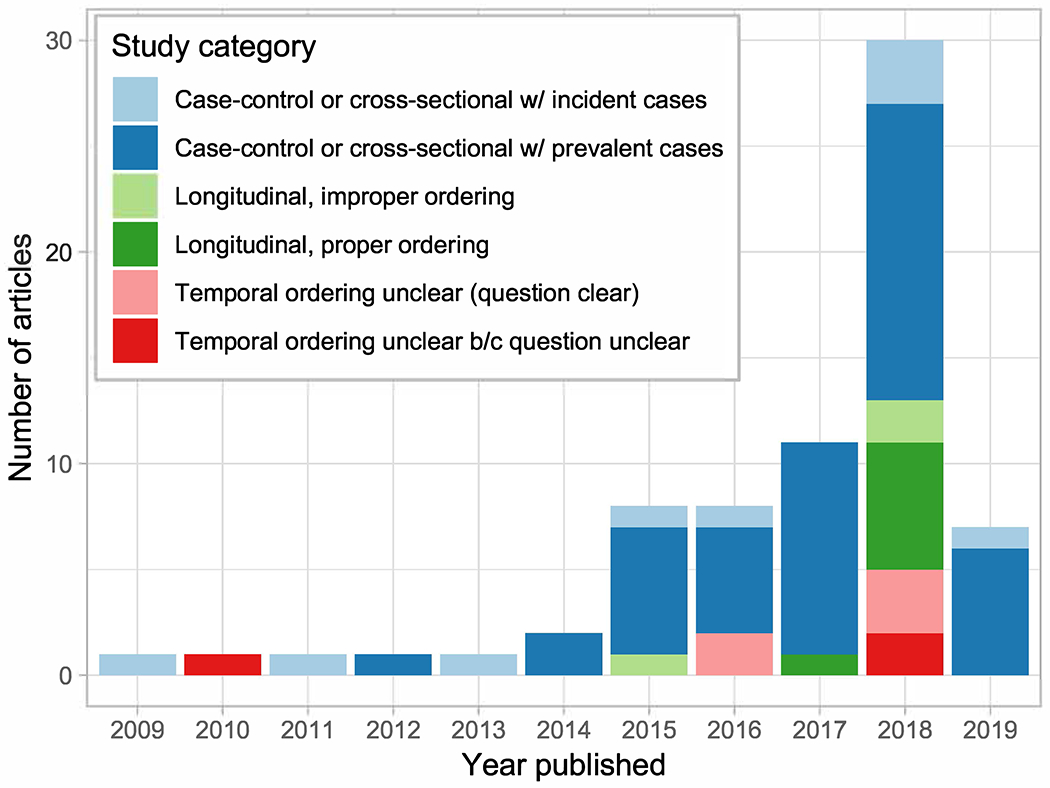
Study design and temporal ordering issues in the 16S microbiome literature on cardiometabolic phenotypes, 2009-2019 (n=71).
The prevalence of studies over time by study design and temporal ordering categorizations are shown in Figure 1. In the majority (n=39) of studies, we classified temporal ordering as improper in some way. Most typically (n=36), these were case control or cross sectional studies that relied on prevalent phenotypes, and in nearly half (n=7) of studies in which temporal ordering was proper, the interest was in the effect of phenotype on the microbiome, suggesting this is a question for which the available data are more informative. Longitudinal cohort studies achieved proper temporal ordering most frequently (n=7). Notably, temporal ordering was difficult or impossible to determine in 8 studies (See Supplemental Table 1 for classification of specific studies).
Supplemental Figure 1 catalogues the covariates controlled in some way by each study, according to the method used. Restriction and adjustment were used most frequently, although n=7 studies used no apparent method to control confounders. Nearly half (n=32) of studies excluded participants recently on antibiotics/antimicrobials (1; 3; 22–24; 32; 33; 39; 46; 59; 64; 67; 72; 76–78; 85; 88–91; 96; 100–102; 105; 111; 120; 124; 126; 131; 136) , and the majority (n=42) restricted on health in some way (1; 3; 8; 14; 16; 22–24; 32; 33; 39; 44; 56; 61; 64; 65; 72; 76; 78; 85; 88–92; 95; 96; 100–102; 105; 111; 112; 120; 123; 126; 131; 134–136). Such studies typically excluded patients with major chronic diseases or cardiometabolic phenotypes (such as major CVD) other than that being studied. Less than half (33) of studies used adjustment (1; 8; 9; 23; 24; 32; 37; 39; 45; 49; 58; 62; 63; 65; 67; 70; 74; 77; 78; 89; 90; 95; 96; 100; 103; 105; 118–120; 122; 123; 133; 136), typically for age and sex. Some studies (n=8) adjusted for diet in some way (9; 24; 32; 67; 70; 100; 120; 133). A few studies (n=11) used matching (26; 44; 69; 73; 74; 76; 77; 88; 102; 112; 124), typically for age and sex, and three studies (74; 77; 124) matched monozygotic twins, accounting for genetics and early life environment. One study (132) used Mendelian randomization, which leverages genetic instrumental variables to adjust for measured and unmeasured confounders. Of note, only three studies adjusted for education (63; 119; 120), two for race/ethnicity (119; 120), and four for geographic location (22; 24; 49; 70).
Finally, we examined compositionally robust and non-robust analytic methods used in each study. All 71 studies use at least one compositionally non-robust method, and none used compositionally robust methods for differential abundance or beta diversity. In contrast, correlation network methods, which calculate correlations between taxa in order to determine groups of co-occurring taxa, were the only compositionally robust method observed in n=5 studies (22; 23; 39; 45; 105), whereas 7 used non-robust correlation network methods (22; 23; 38; 95; 103; 122; 136).
Discussion
Over the past 5-10 years, there has been massive growth in both the number and sample sizes of human observational studies examining associations between various metabolic phenotypes and the gut microbiome. Despite significant advances, our review highlights several methodological areas in need of innovation and attention, specifically issues around sample selection, study design and confounding.
Sample Selection.
Of great importance for the generalizability of findings, only a small percentage of studies (4 of 71) had a population-based random sampling design. 20 studies used volunteer recruitment, typically based in a workplace (55; 78; 133), university (90), or existing clinical trials (5; 16; 43; 44; 56; 63; 92; 134). Several studies utilized data from the TwinsUK cohort (9; 45; 62; 74) recruited through media campaigns (87), and the American Gut project (8), in which participants were recruited online (84). Another 22 relied on in- or out-patient clinical samples (1; 3; 14; 18; 32; 37; 46; 57; 61; 64; 70; 72; 73; 88; 89; 100; 102; 119; 122; 123; 126; 131). Volunteer and convenience samples are known to be self-selected on both high SES and good health, with potentially serious implications for inference (36).
Figure 2 provides a stylized illustration of how non-random samples selected for high SES sample can lead to an underestimate of associations between gut microbiome diversity and obesity. As previously detailed, the health risks associated with obesity are often found to be less severe for higher SES individuals, likely reflecting a more favorable underlying health profiles on both observable (smoking) and unobservable factors (early life conditions, sleep, stress), all factors which likely affect the gut microbiome. Among the entire population of obese respondents therefore, we would expect a higher fraction of high SES individuals to have a “healthier”, or in this case more diverse, gut microbiome composition compared to lower SES individuals. Figure 2 shows how this selection would thus underestimate the association between diversity of the gut microbiome and obesity relative to the true association in the entire population by minimizing differences in the microbiome between obese and non-obese individuals. This may help explain lack of reproducibility of results from animal models to human populations thus far and emphasizes the need for population representative datasets with a full range of variability in SES (109), which is still very rare in this field.
Figure 2:
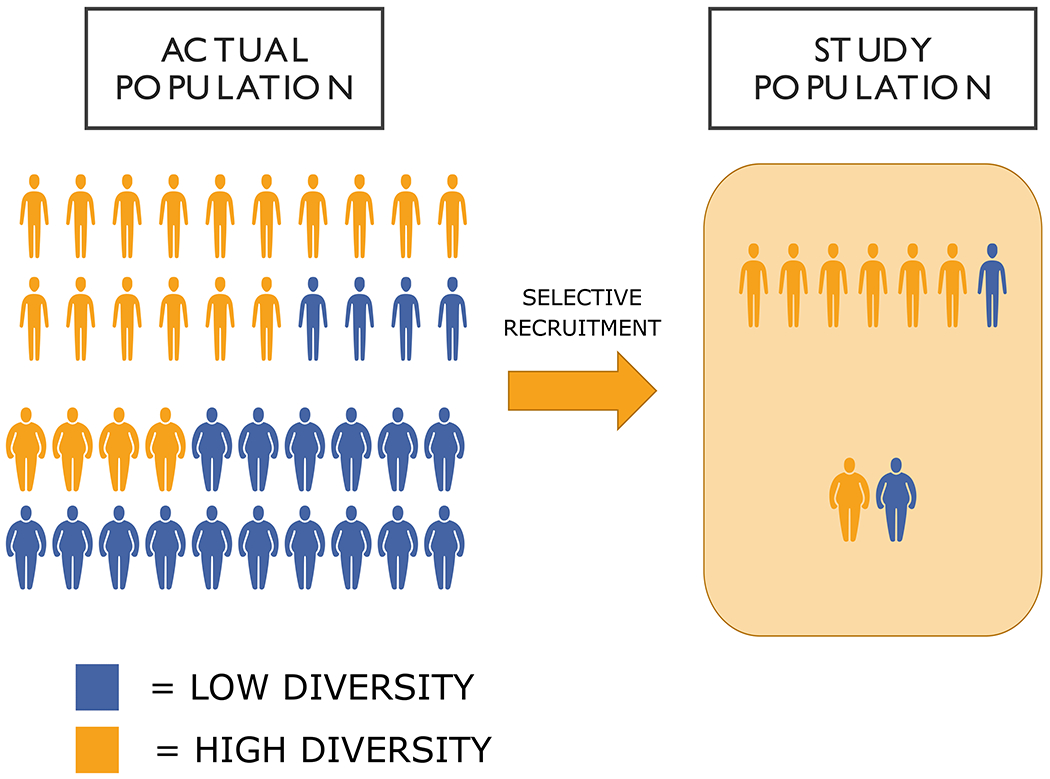
Implications of selection on SES for obesity-microbiome associations. On the left, the actual population is 30% obese. While the obese population is more likely to have low microbiome diversity, some fraction of obese, likely those of higher SES, will still will have relatively high diversity levels. In a self-selected study sample, which tends to be healthier and higher SES, this can minimize differences between lean and obese individuals on diversity measures. In the actual population, while 80% of lean individuals and 20% of obese individuals will have high diversity, the select study population would be 85% and 50% respectively, thus underestimating the association between obesity and diversity.
Confounder adjustment.
As in most observational research, causal inference on effects of the gut microbiota on host cardiometabolic phenotype is challenged by the fact that many exposures that impact the microbiome also affect health outcomes through different pathways. These include individual-level factors such as diet, smoking, physical activity, and prescription medications (especially antibiotics) (40), early life factors such as birth mode and gestational age at birth (19), as well as broader population-level context such as geography, SES, and race/ethnicity (49). Studies included in our review typically aimed to account in some way for age, sex, antibiotics, other medications, usually by restriction or regression adjustment (Supplemental Figure 1). Additionally, though nearly every study attempted to account for existing health status in some way, possible bidirectional relationship between nearly all organ systems and the gut microbiome (114) makes systemic health one of the thorniest validity issues in this field. This means that single-time point studies conditioning on health status in any way risk creating collider bias, as illustrated in Figure 3. Longitudinal studies with proper temporal ordering are better situated to measure and adjust for such time-varying confounding (106).
Figure 3.
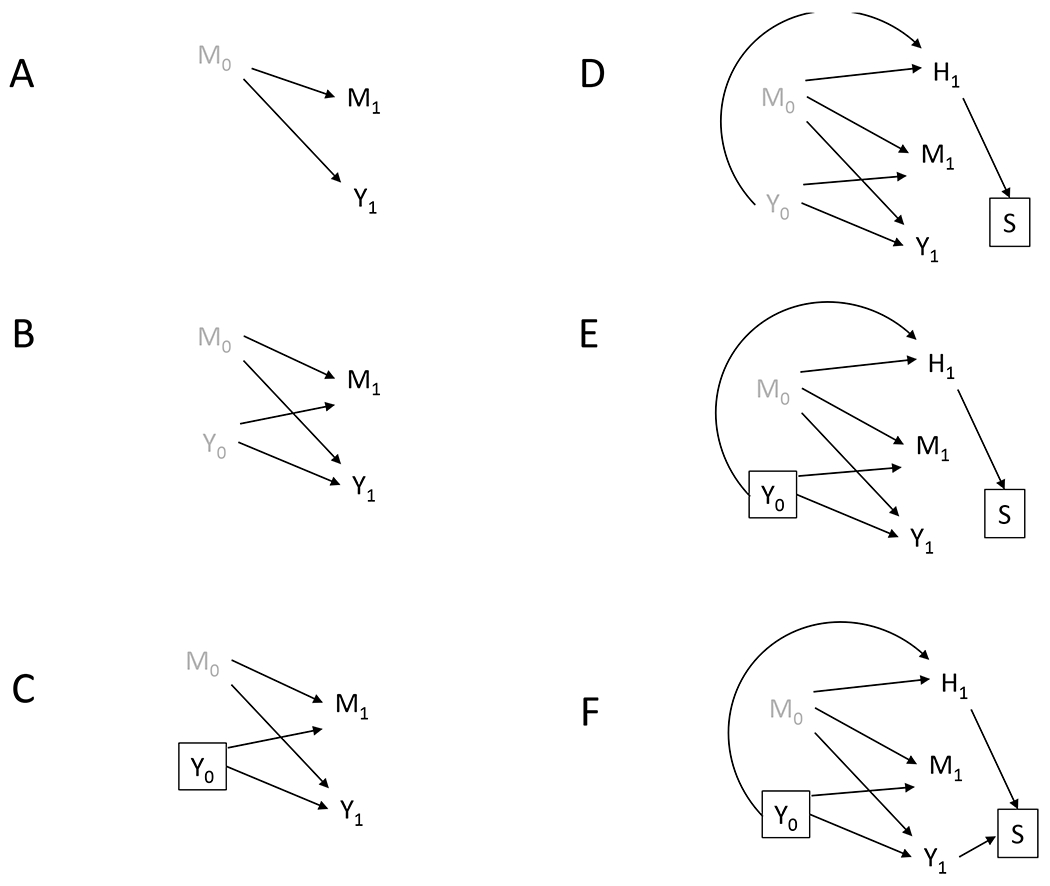
Directed acyclic graphs illustrating common study design issues present in cardiometabolic microbiome human subjects literature. Nodes represent variables (black=measured, grey=unmeasured) and the arrows represent causal relationships. A square around a node means the analysis is conditional on that variable in some way, whether by adjusting for it, restricting on it, or other means. Subscripts indicate time points. (A) A cross-sectional study where microbiome (M1) and disease outcome (Y1) are measured concurrenty. M1 is unlikely to affect simulatenous disease (Y1), but is meant as a proxy for previous microbiome, M0. (B) A cross-sectional study where prevalent cases are analyzed. Y1 can now be a marker for previous disease, Y0, which can affect M1. (C) A cross-sectional study where incident cases are anlyzed. Y1 is now no longer a marker for previous disease and confounding by Y0 is controlled. (D) A cross-sectional study where prevalent cases are analyzed and participants are selected (S) according to health variables (H1). Selection bias exists due to conditioning on a (descendent of a) collider, S. (E) A cross-sectional study where incident cases are analyzed and participants are selected (S) according to health variables (H1). Selection bias in (D) is alleviated because a non-collider (Y0) on the collider path present in (D) is controlled. (F) A case control study where incident cases are analyzed and participants are selected (S) according to health variables (H1). Selection bias exists due to conditioning on a collider, S.
One promising development is the use of Mendelian randomization (MR), employed in one study in our review (132). This technique capitalizes on findings from recent microbiome genome wide association studies (129) to strengthen causal inference, potentially bypassing temporal ordering concerns, unmeasured confounders, and other complex forms of endogeneity present in host-microbiome interactions (121), under some fairly strong assumptions (20). Although its use is likely to increase in coming years, MR in microbiome studies is currently challenging because of poor replicability in existing microbiome GWAS (129), limited functional understanding of observed genetic associations (117), and the need for very large samples (20) of which only a few currently exist owing to the expense of high-throughput sequencing. Despite the evidence reviewed above showing the strength of associations with race/ethnicity and SES and that adjustment for these factors can reduce or eliminate the strength of focal microbiome-health associations, these were rarely considered.
Study Design and Selection bias.
In Figure 3, we illustrate several issues concerning temporal ordering and selection in metabolic microbiome research, using causal diagrams, where M0 and Y0 and M1 and Y1 represent the microbiome at baseline and follow-up, respectively. (98). Many studies restrict eligibility to otherwise healthy individuals, illustrated in figure Supplemental Figure 1D, where H1 represents health at baseline, and S represents selection into the study. The rationale may be that health status can impact the microbiome and the disease of interest and is thus a confounder (58; 126). However, H1 could be affected by both prevalent metabolic syndrome (Y0) and previous microbiome (M0), so H1 is termed a collider, meaning a common effect of exposure and outcome (53; 98). Restricting participation, or otherwise conditioning, based on such a variable is known to induce selection bias, generating false associations that do not exist in the population (53). Heuristically, among otherwise healthy people, those who have metabolic syndrome are more likely to have a protective microbiome (53). Fortunately, we can control this bias by including only incident cases (i.e., controlling Y0) (Supplemental Figure 1E). Alternatively, Supplemental Figure 1F depicts a case-control study, where we would not be able to control selection bias by restricting to incident cases. This is because, in a case-control study, selection has already occurred based on case status, (hence the arrow from Y1 to S), and now S is a collider between M0 and Y1, inducing selection bias. (For a more complete explanation, see reference 56.)
Two major takeaways of Figure 3 are (a) for both cross-sectional and case-control studies, restricting analysis to incident cases limits confounding by previous disease, and (b) restricting a case-control study to individuals without other health conditions (other than the disease in question) may result in selection bias that could be avoided by eliminating such exclusion criteria. Therefore, both unmeasured confounding and selection bias likely affect a huge swath of the literature on gut microbiomes and cardiometabolic phenotypes.
Longitudinal cohort studies on this topic are frequently more informative as to the causal question of whether the microbiome is involved in the etiology of disease. For example, Rampelli et al. (2018) 104 conducted a longitudinal cohort study in which baseline fecal samples were collected for 70 children aged 2-9 years, all of whom were classified as normal weight on study entry, and about half of whom developed overweight or obesity throughout the 4-year study period. Authors explored associations between baseline microbiome and weight change over the study period, controlling for age. Such a design eliminates the specific biases discussed above because (a) there is no selection based on health or other variables affected by both previous microbiome and previous disease (i.e. no collider bias), and (b) since all participants were normal weight at baseline, only incidence of overweight is observed. Additionally, baseline microbiome (M0 in Figure 3) has been observed rather than inferred from later microbiome (M1 in Figure 3), a much less risky strategy as gut microbiomes have been observed to change rapidly, for example in response to dietary changes (21). A nearly equivalent approach is the use of stored fecal samples in case-control studies, as in the study of incident T1D conducted by Kostic et al. (2015) (70).
Analytic methods and compositional robustness.
Notably, none of the reviewed studies used compositionally-robust techniques, with the exception of correlation networks, employed by 5 studies (22; 23; 39; 45; 105). The compositionally non-robust methods used in the majority of the reviewed research call into question the truth of observed associations and suggest that findings from these studies are unlikely to replicate. However, this trend is expected, as the methods that have been most effectively disseminated and popularized, including UniFrac and Bray-Curtis distances, LEfSe and DESeq2, do not explicitly consider the compositional structure. Though developed in the 1980s, Aitchison’s work establishing the mathematical properties of compositions has only recently been considered in expert recommendations for microbiome data analysis (68), and compositionally-robust differential abundance (31; 83), and phylogenetic distance (115) methods emerged several years later than other highly popular methods (81; 97; 113).
III. Conclusion
Methodologically, as we move forward from the early days of microbiome research, it is clearly time for epidemiology to work towards best practices such as the use of compositionally robust measures and as proper attention to basic issues as temporal order, avoiding selection bias, and adjustment for confounding. In an alarming number of the studies we reviewed (Supplemental Table 1), the sample recruitment and study design could not be ascertained from the paper’s methods, a serious problem which we can surely collectively overcome in our own writing and reviewing in this emerging field.
Overall, descriptive epidemiology of the microbiome by social context remains limited, especially with the current lack of population representative, randomly selected samples. Thus far, it is clear that variables measuring important aspects of the social environment -- race/ethnicity and socioeconomic status- show strong associations with the gut microbiome, often explaining more variation than most other salient individual-level determinants such as diet. Given the strong social patterning of obesity and metabolic conditions, existing studies on the gut microbiome and metabolic conditions that do not take account for SES and race/ethnicity are at strong risk serious bias. At the same time, the strong associations between social context, the gut microbiome, and metabolic conditions across the life course provides an opportunity to explore the gut microbiome as a mechanism underlying social disparities in metabolic disease.
Moreover, microbiome-health associations themselves are increasingly found to vary across these social contexts, compelling researchers to carefully consider the inferences they can make from homogenous and volunteer samples. A recent notable paper highlighted the importance of geography in the generalizability of microbiome-disease associations using data from 14 districts within one large province in China (50). The effect sizes of geographic variation dominated in predicting gut microbiome composition, exceeding those of metabolic diseases, type 2 diabetes, obesity, and fatty liver. The authors applied a disease model for Type 2 Diabetes trained in one location to another location and found predictive power was reduced to no better than random guessing, suggesting that “healthy” reference baselines for gut microbiota can be heavily dependent on location. Taken together with similar results for SES and race/ethnicity, this strongly suggests that predictive models will need to seriously consider social and geographic context before they can be broadly applied, and that this population-level perspective may be key to ultimately understanding and intervening on the microbiome. As Geoffrey Rose might say, ‘Why do some individuals have sick microbiomes’ is a different question from ‘Why do some populations have sick microbiomes?’
Supplementary Material
Acknowledgements
The authors acknowledge support from the National Institute of Allergy and Infectious Diseases (R21AI121784-01) and National Institute of Child Health and Development (5T32HD091058-02)
Footnotes
From 43 papers in 2000 to 5032 in 2018, via PubMed search for “microbiota OR microbiome AND human,” 5/24/19.
Literature cited:
- 1.Aatsinki A-K, Uusitupa H-M, Munukka E, Pesonen H, Rintala A, et al. 2018. Gut Microbiota Composition in Mid-Pregnancy Is Associated with Gestational Weight Gain but Not Prepregnancy Body Mass Index. Journal of Women’s Health 27:1293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitchison J 1986. The Statistical of Analysis of Compositional Data. Chapman Hall, London [Google Scholar]
- 3.Al-Obaide M, Singh R, Datta P, Rewers-Felkins K, Salguero M, et al. 2017. Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD. Journal of Clinical Medicine 6:86- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen AP, Dinan TG, Clarke G, Cryan JF. 2017. A psychology of the human brain–gut–microbiome axis. Social and personality psychology compass 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelakis E, Armougom F, Carrière F, Bachar D, Laugier R, et al. 2015. A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLoS ONE 10:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, et al. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences 101:15718–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. 2007. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences 104:979–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai J, Hu Y, Bruner DW. 2019. Composition of gut microbiota and its association with body mass index and lifestyle factors in a cohort of 7–18 years old children from the American Gut Project. Pediatric Obesity 14. [DOI] [PubMed] [Google Scholar]
- 9.Beaumont M, Le Roy CI, Jackson MA, Steves CJ, Spector TD, Bell JT. 2017. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biology:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowyer RC, Jackson MA, Le Roy CI, Ni Lochlainn M, Spector TD, et al. 2019. Socioeconomic Status and the Gut Microbiome: A TwinsUK Cohort Study. Microorganisms 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks AW, Priya S, Blekhman R, Bordenstein SR. 2018. Gut microbiota diversity across ethnicities in the United States. PLoS biology 16:e2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkert NT, Rásky E, Großschädl F, Muckenhuber J, Freidl W. 2013. The influence of socioeconomic factors on health parameters in overweight and obese adults. PloS one 8:e65407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castaner O, Goday A, Park Y-M, Lee S-H, Magkos F, et al. 2018. The gut microbiome profile in obesity: a systematic review. International journal of endocrinology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chávez-Carbajal A, Nirmalkar K, Pérez-Lizaur A, Hernández-Quiroz F, Ramírez-del-Alto S, et al. 2019. Gut Microbiota and Predicted Metabolic Pathways in a Sample of Mexican Women Affected by Obesity and Obesity Plus Metabolic Syndrome. International Journal of Molecular Sciences 20:438- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Reeve J, Zhang L, Huang S, Wang X, Chen J. 2018. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ 6:e4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciubotary I, Green SJ, Kukreja S, Barengolts E. 2015. Significant differences in fecal microbiota are associated with various stages of glucose tolerance in African American male veterans. 25:289–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen AK, Rehkopf DH, Deardorff J, Abrams B. 2013. Education and obesity at age 40 among American adults. Social science & medicine 78:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crusell MKW, Franke A, Pedersen O, Nielsen T, Heinsen F-A, et al. 2018. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome 6:89- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahl C, Stigum H, Valeur J, Iszatt N, Lenters V, et al. 2018. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. International journal of epidemiology 47:1658–69 [DOI] [PubMed] [Google Scholar]
- 20.Davey Smith G, Ebrahim S. 2003. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? International journal of epidemiology 32:1–22 [DOI] [PubMed] [Google Scholar]
- 21.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De La Cuesta-Zuluaga J, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. 2018. Body size phenotypes comprehensively assess cardiometabolic risk and refine the association between obesity and gut microbiota. International Journal of Obesity 42:424–32 [DOI] [PubMed] [Google Scholar]
- 23.de la Cuesta-Zuluaga J, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, Abad JM, Escobar JS. 2018. Gut microbiota is associated with obesity and cardiometabolic disease in a population in the midst of Westernization. Scientific Reports 8:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, et al. 2019. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debelius JW, Vázquez-Baeza Y, McDonald D, Xu Z, Wolfe E, Knight R. 2016. Turning participatory microbiome research into usable data: lessons from the American Gut Project. Journal of microbiology & biology education 17:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Chierico F, Abbatini F, Russo A, Quagliariello A, Reddel S, et al. 2018. Gut microbiota markers in obese adolescent and adult patients: Age-dependent differential patterns. Frontiers in Microbiology 9:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, et al. 2018. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nature medicine 24:1526. [DOI] [PubMed] [Google Scholar]
- 28.Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, et al. 2019. Close social relationships correlate with human gut microbiota composition. Scientific reports 9:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowd JB, Renson A. 2018. “Under the Skin” and into the Gut: Social Epidemiology of the Microbiome. Current Epidemiology Reports [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ejtahed H-S, Angoorani P, Hasani-Ranjbar S, Siadat S-D, Ghasemi N, et al. 2018. Adaptation of human gut microbiota to bariatric surgeries in morbidly obese patients: a systematic review. Microbial pathogenesis 116:13–21 [DOI] [PubMed] [Google Scholar]
- 31.Fernandes AD, Reid JN, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB. 2014. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrocino I, Ponzo V, Gambino R, Zarovska A, Leone F, et al. 2018. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM). Scientific reports 8:12216- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. 2014. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE 9:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortenberry JD. 2013. The uses of race and ethnicity in human microbiome research. Trends in Microbiology 21:165–6 [DOI] [PubMed] [Google Scholar]
- 35.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS computational biology 8:e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, et al. 2017. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. American journal of epidemiology 186:1026–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, et al. 2015. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circulation Research 117:817–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao R, Zhu C, Li H, Yin M, Pan C, et al. 2018. Dysbiosis Signatures of Gut Microbiota Along the Sequence from Healthy, Young Patients to Those with Overweight and Obesity. Obesity 26:351–61 [DOI] [PubMed] [Google Scholar]
- 39.Gao X, Zhang M, Xue J, Huang J, Zhuang R, et al. 2018. Body mass index differences in the gut microbiota are gender specific. Frontiers in Microbiology 9:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nature medicine 24:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Frontiers in microbiology 8:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldman D, Smith JP. 2011. The increasing value of education to health. Social science & medicine 72:1728–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M. 2016. Increased Systolic and Diastolic Blood Pressure is Associated with Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension 68:974–81 [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, et al. 2016. Connections between the gut microbiome and metabolic hormones in early pregnancy in overweight and obese women. Diabetes 65:2214–23 [DOI] [PubMed] [Google Scholar]
- 45.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, et al. 2014. Human genetics shape the gut microbiome. Cell 159:789–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gózd-Barszczewska A, Koziol-Montewka M, Barszczewski P, Młodzińska A, Humińska K. 2017. Gut microbiome as a biomarker of cardiometabolic disorders. Annals of Agricultural and Environmental Medicine 24:416–22 [DOI] [PubMed] [Google Scholar]
- 47.Guo Y, Huang Z-P, Liu C-Q, Qi L, Sheng Y, Zou D-J. 2018. Modulation of the gut microbiome: a systematic review of the effect of bariatric surgery. European journal of endocrinology 178:43–56 [DOI] [PubMed] [Google Scholar]
- 48.Hawinkel S, Mattiello F, Bijnens L, Thas O. 2017. A broken promise: microbiome differential abundance methods do not control the false discovery rate. Briefings in bioinformatics 20:210–21 [DOI] [PubMed] [Google Scholar]
- 49.He Y, Wu W, Wu S, Zheng HM, Li P, et al. 2018. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome 6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y, Wu W, Zheng H-M, Li P, McDonald D, et al. 2018. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nature medicine 24:1532. [DOI] [PubMed] [Google Scholar]
- 51.Herd P, Goesling B, House JS. 2007. Socioeconomic position and health: the differential effects of education versus income on the onset versus progression of health problems. Journal of health and social behavior 48:223–38 [DOI] [PubMed] [Google Scholar]
- 52.Herd P, Palloni A, Rey F, Dowd JB. 2018. Social and population health science approaches to understand the human microbiome. Nature Human Behaviour [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernán MA, Hernández-Díaz S, Robins JM. 2004. A structural approach to selection bias. Epidemiology 15:615–25 [DOI] [PubMed] [Google Scholar]
- 54.Hicken MT, Lee H, Morenoff J, House JS, Williams DR. 2014. Racial/ethnic disparities in hypertension prevalence: reconsidering the role of chronic stress. American journal of public health 104:117–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoel H, Hove-Skovsgaard M, Hov JR, Gaardbo JC, Holm K, et al. 2018. Impact of HIV and Type 2 diabetes on Gut Microbiota Diversity, Tryptophan Catabolism and Endothelial Dysfunction. Scientific Reports 8:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houttu N, Mokkala K, Laitinen K. 2018. Overweight and obesity status in pregnant women are related to intestinal microbiota and serum metabolic and inflammatory profiles. Clinical Nutrition 37:1955–66 [DOI] [PubMed] [Google Scholar]
- 57.Hsu C-N, Lu P-C, Lo M-H, Lin IC, Chang-Chien G-P, et al. 2018. Gut Microbiota-Dependent Trimethylamine N-Oxide Pathway Associated with Cardiovascular Risk in Children with Early-Stage Chronic Kidney Disease. International Journal of Molecular Sciences 19:3699- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu HJ, Park SG, Jang HB, Choi MG, Park KH, et al. 2015. Obesity alters the microbial community profile in Korean Adolescents. PLoS ONE 10:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Y, Li SC, Hu J, Ruan HB, Guo HM, et al. 2018. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Research and Clinical Practice 141:256–63 [DOI] [PubMed] [Google Scholar]
- 60.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, et al. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoue R, Ohue-Kitano R, Tsukahara T, Tanaka M, Masuda S, et al. 2017. Prediction of functional profiles of gut microbiota the Society for Free Radical Research Japan 1880 50860912-0009 Original Article Kyj bn17-4 10.3164/j4cb .17-44 JJCBN c oto, Japan ournal of Clinical Biochemistry and Nutrition from 16S rRNA metagenomic d. Journal of Clinical Biochemistry and Nutrition 61:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, et al. 2018. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nature Communications 9:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karvonen AM, Sordillo JE, Gold DR, Bacharier LB, O’Connor GT, et al. 2019. Gut microbiota and overweight in 3-year old children. International Journal of Obesity 43:713–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, et al. 2015. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterology 15:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kashtanova D, Tkacheva O, Doudinskaya E, Strazhesko I, Kotovskaya Y, et al. 2018. Gut Microbiota in Patients with Different Metabolic Statuses: Moscow Study. Microorganisms 6:98- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kavanagh A, Bentley RJ, Turrell G, Shaw J, Dunstan D, Subramanian S. 2010. Socioeconomic position, gender, health behaviours and biomarkers of cardiovascular disease and diabetes. Social science & medicine 71:1150–60 [DOI] [PubMed] [Google Scholar]
- 67.Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, et al. 2016. Gut Microbiome Associates with Lifetime Cardiovascular Disease Risk Profile among Bogalusa Heart Study Participants. Circulation Research 119:956–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, et al. 2018. Best practices for analysing microbiomes. Nature Reviews Microbiology:1. [DOI] [PubMed] [Google Scholar]
- 69.Koren O, Spor A, Felin J, Fak F, Stombaugh J, et al. 2011. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences 108:4592–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, et al. 2015. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host and Microbe 17:260–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. 2015. Sparse and compositionally robust inference of microbial ecological networks. PLoS computational biology 11:e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lambeth S, Carson T, Lowe J, Ramaraj T, Leff J, et al. 2015. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J Diabetes Obes. 2:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsen N, Vogensen FK, Van Den Berg FWJ, Nielsen DS, Andreasen AS, et al. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le Roy CI, Beaumont M, Jackson MA, Steves CJ, Spector TD, Bell JT. 2018. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biology:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Legendre P, Legendre L. 1998. Numerical ecology: developments in environmental modelling. Elsevier, Amsterdam:63–75 [Google Scholar]
- 76.Leiva-Gea I, Sánchez-Alcoholado L, Martín-Tejedor B, Castellano-Castillo D, Moreno-Indias I, et al. 2018. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: A case-control study. Diabetes Care 41:2385–95 [DOI] [PubMed] [Google Scholar]
- 77.Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, et al. 2017. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66:1031–8 [DOI] [PubMed] [Google Scholar]
- 78.López-Contreras BE, Morán-Ramos S, Villarruel-Vázquez R, Macías-Kauffer L, Villamil-Ramírez H, et al. 2018. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatric Obesity 13:381–8 [DOI] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lovell D, Pawlowsky-Glahn V, Egozcue JJ, Marguerat S, Bähler J. 2015. Proportionality: a valid alternative to correlation for relative data. PLoS computational biology 11:e1004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. The ISME journal 5:169–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mai V, Prosperi M, Yaghjyan L. 2016. Moving microbiota research toward establishing causal associations that represent viable targets for effective public health interventions. Annals of epidemiology 26:306–10 [DOI] [PubMed] [Google Scholar]
- 83.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial ecology in health and disease 26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, et al. 2018. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems 3:e00031–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Méndez-Salazar EO, Ortiz-López MG, Granados-Silvestre MDLÁ, Palacios-González B, Menjivar M. 2018. Altered gut microbiota and compositional changes in firmicutes and proteobacteria in mexican undernourished and obese children. Frontiers in Microbiology 9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller GE, Engen PA, Gillevet PM, Shaikh M, Sikaroodi M, et al. 2016. Lower Neighborhood Socioeconomic Status Associated with Reduced Diversity of the Colonic Microbiota in Healthy Adults. PLoS One 11:e0148952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. 2013. Cohort Profile: TwinsUK and healthy ageing twin study. International journal of epidemiology 42:76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moreno-Indias I, Sánchez-Alcoholado L, García-Fuentes E, Cardona F, Queipo-Ortuño MI, Tinahones FJ. 2016. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. American journal of translational research 8:5672–84 [PMC free article] [PubMed] [Google Scholar]
- 89.Moreno-Navarrete JM, Serino M, Blasco-Baque V, Azalbert V, Barton R, et al. 2018. Gut microbiota interacts with markers of adipose tissue browning, insulin action and plasma acetate in morbid obesity. Molecular nutrition and food research [DOI] [PubMed] [Google Scholar]
- 90.Mörkl S, Lackner S, Müller W, Gorkiewicz G, Kashofer K, et al. 2017. Gut microbiota and body composition in anorexia nervosa inpatients in comparison to athletes, overweight, obese, and normal weight controls. International Journal of Eating Disorders 50:1421–31 [DOI] [PubMed] [Google Scholar]
- 91.Murugesan S, Ulloa-Martínez M, Martínez-Rojano H, Galvánez-Rodríguez FM, Miranda-Brito C, et al. 2015. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. European Journal of Clinical Microbiology and Infectious Diseases 34:1337–46 [DOI] [PubMed] [Google Scholar]
- 92.Naderpoor N, Mousa A, Gomez-Arango L, Barrett H, Dekker Nitert M, de Courten B. 2019. Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. Journal of Clinical Medicine 8:452- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogden CL, Carroll MD, Fakhouri TH, Hales CM, Fryar CD, et al. 2018. Prevalence of obesity among youths by household income and education level of head of household—United States 2011–2014. Morbidity and Mortality Weekly Report 67:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ogden CL, Fakhouri TH, Carroll MD, Hales CM, Fryar CD, et al. 2017. Prevalence of obesity among adults, by household income and education—United States, 2011–2014. MMWR. Morbidity and mortality weekly report 66:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, et al. 2017. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biology 18:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, et al. 2018. Connection between BMI-Related Plasma Metabolite Profile and Gut Microbiota. Journal of Clinical Endocrinology and Metabolism 103:1491–501 [DOI] [PubMed] [Google Scholar]
- 97.Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pearl J 1995. Causal diagrams for empirical research. Biometrika 82:669–88 [Google Scholar]
- 99.Pearson K On a form of spurious correlation which may arise when indices are useed in the measurement of organs. Proc. Royal Soc., London, Proc, 1897, 60:489–502: [Google Scholar]
- 100.Peters BA, Shapiro JA, Church TR, Miller G, Trinh-Shevrin C, et al. 2018. A taxonomic signature of obesity in a large study of American adults. Scientific Reports 8:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pushpanathan P, Srikanth P, Seshadri K, Selvarajan S, Pitani R, et al. 2016. Gut Microbiota in Type 2 Diabetes Individuals and Correlation with Monocyte Chemoattractant Protein1 and Interferon Gamma from Patients Attending a Tertiary Care Centre in Chennai, India. Indian Journal of Endocrinology and Metabolism 20:523- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qi CJ, Zhang Q, Yu M, Xu JP, Zheng J, et al. 2016. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chinese Medical Journal 129:1298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rampelli S, Guenther K, Turroni S, Wolters M, Veidebaum T, et al. 2018. Pre-obese children’s dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Communications Biology 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, et al. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Riva A, Borgo F, Lassandro C, Verduci E, Morace G, et al. 2017. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environmental Microbiology 19:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Robins JM, Hernan MA, Brumback B. 2000. Marginal structural models and causal inference in epidemiology. LWW [DOI] [PubMed] [Google Scholar]
- 107.Robinson CK, Brotman RM, Ravel J. 2016. Intricacies of assessing the human microbiome in epidemiologic studies. Annals of epidemiology 26:311–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rose G 1985. Sick Individuals and Sick Populations. Int. J. Epidemiol 14:32–8 [DOI] [PubMed] [Google Scholar]
- 109.Rothman KJ, Gallacher JE, Hatch EE. 2013. Why representativeness should be avoided. International journal of epidemiology 42:1012–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, et al. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210. [DOI] [PubMed] [Google Scholar]
- 111.Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozińska S, et al. 2018. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on next-generation sequencing of the 16S rRNA gene fragment. Polish Archives of Internal Medicine 128:336–43 [DOI] [PubMed] [Google Scholar]
- 112.Sanchez-Alcoholado L, Castellano-Castillo D, Jordán-Martínez L, Moreno-Indias I, Cardila-Cruz P, et al. 2017. Role of gut microbiota on cardio-metabolic parameters and immunity in coronary artery disease patients with and without type-2 diabetes mellitus. Frontiers in Microbiology 8:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. 2011. Metagenomic biomarker discovery and explanation. Genome biology 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shreiner AB, Kao JY, Young VB. 2015. The gut microbiome in health and in disease. Current opinion in gastroenterology 31:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Silverman JD, Washburne AD, Mukherjee S, David LA. 2017. A phylogenetic transform enhances analysis of compositional microbiota data. Elife 6:e21887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, et al. 2017. Influence of diet on the gut microbiome and implications for human health. Journal of translational medicine 15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith GD, Ebrahim S. 2008. Mendelian randomization: genetic variants as instruments for strengthening causal inference in observational studies. In Biosocial Surveys: National Academies Press (US). Number of. [Google Scholar]
- 118.Stanislawski MA, Dabelea D, Wagner BD, Iszatt N, Dahl C, et al. 2018. Gut Microbiota in the First 2 Years of Life and the Association with Body Mass Index at Age 12 in a Norwegian Birth Cohort. mBio 9:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stanislawski MA, Dabelea D, Wagner BD, Sontag MK, Lozupone CA, Eggesbø M. 2017. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome 5:113- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun S, Lulla A, Sioda M, Winglee K, Wu MC, et al. 2019. Gut Microbiota Composition and Blood Pressure. Hypertension:998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thomas H. 2019. Mendelian randomization reveals causal effects of the gut microbiota. Nature Reviews Gastroenterology & Hepatology:1. [DOI] [PubMed] [Google Scholar]
- 122.Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, et al. 2018. Roles of birth mode and infant gut microbiota in intergenerational transmission of overweight and obesity from mother to offspring. JAMA Pediatrics 172:368–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tun MH, Tun HM, Mahoney JJ, Konya TB, Guttman DS, et al. 2018. Postnatal exposure to household disinfectants, infant gut microbiota and subsequent risk of overweight in children. Cmaj 190:E1097–E107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Ley RE, et al. 2012. A core gut microbiome in obese and lean twins. Nature 486:222–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van den Berg G, van Eijsden M, Vrijkotte TG, Gemke RJ. 2012. Socioeconomic inequalities in lipid and glucose metabolism in early childhood in a population-based cohort: the ABCD-Study. BMC public health 12:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Villanueva-Millán MJ, Pérez-Matute P, Recio-Fernández E, Rosales J-ML, Oteo J-A. 2019. Characterization of gut microbiota composition in HIV-infected patients with metabolic syndrome. Journal of Physiology and Biochemistry:1–11 [DOI] [PubMed] [Google Scholar]
- 127.Volaco A, Cavalcanti AM, Roberto Filho P, Précoma DB. 2018. Socioeconomic status: the missing link between obesity and diabetes mellitus? Current diabetes reviews 14:321–6 [DOI] [PubMed] [Google Scholar]
- 128.Walters WA, Xu Z, Knight R. 2014. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS letters 588:4223–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang J, Kurilshikov A, Radjabzadeh D, Turpin W, Croitoru K, et al. 2018. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. BioMed Central [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Winkleby MA, Jatulis DE, Frank E, Fortmann SP. 1992. Socioeconomic status and health: how education, income, and occupation contribute to risk factors for cardiovascular disease. American journal of public health 82:816–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamashita M, Okubo H, Kobuke K, Ohno H, Oki K, et al. 2019. Alteration of gut microbiota by a Westernized lifestyle and its correlation with insulin resistance in non-diabetic Japanese men. Journal of Diabetes Investigation:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang Q, Lin SL, Kwok MK, Leung GM, Schooling CM. 2018. The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a mendelian randomization study. American journal of epidemiology 187:1916–22 [DOI] [PubMed] [Google Scholar]
- 133.Yun Y, Kim HN, Kim SE, Heo SG, Chang Y, et al. 2017. Comparative analysis of gut microbiota associated with body mass index in a large Korean cohort. BMC Microbiology 17:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zacarías MF, Collado MC, Gómez-Gallego C, Flinck H, Aittoniemi J, et al. 2018. Pregestational overweight and obesity are associated with differences in gut microbiota composition and systemic inflammation in the third trimester. PLoS ONE 13:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, et al. 2013. Human gut microbiota changes reveal the progression of glucose intolerance. PloS one 8:e71108–e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, et al. 2012. Analysis of the gut microbiota in the old order amish and its relation to the metabolic syndrome. PLoS ONE 7:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


