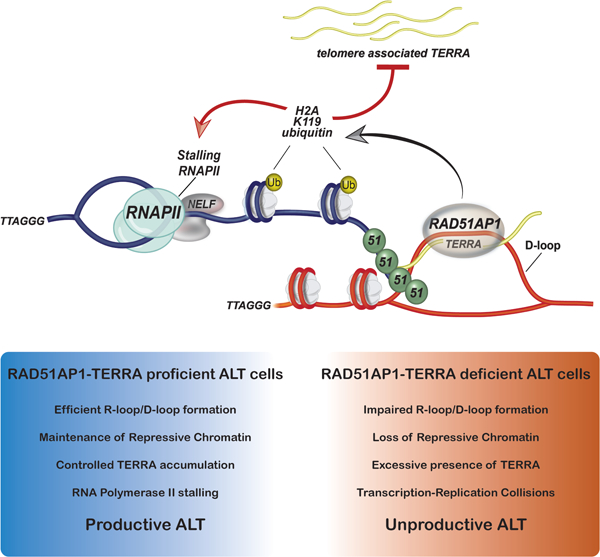
SUMMARY
Alternative lengthening of telomeres (ALT) is a homology-directed repair (HDR) mechanism of telomere elongation that controls proliferation in subsets of aggressive cancer. Recent studies have revealed that TERRA (telomere repeat-containing RNA) promotes ALT-associated HDR (ALT-HDR). Here we report that RAD51AP1, a crucial ALT factor, interacts with TERRA and utilizes it to generate D- and R- loop HR intermediates. We also show that RAD51AP1 binds to and may generate and potentially stabilize TERRA-containing R-loops as RAD51AP1 depletion reduces R-loop formation at telomere DNA breaks. Proteomic analyses uncover a role for RAD51AP1 mediated TERRA R-loop homeostasis in a mechanism of chromatin-directed suppression of TERRA and prevention of transcription-replication collisions (TRCs) during ALT-HDR. Intriguingly, we find that both TERRA binding and this non-canonical function of RAD51AP1 require its intrinsic SUMO-SIM regulatory axis. These findings provide insights into the multi-contextual functions of RAD51AP1 within the ALT mechanism and regulation of TERRA.
Keywords: ALT, TERRA, Telomere, Homology-Directed Repair, RAD51AP1, Chromatin, Transcription, Cancer
Graphical Abstract
eTOC Blurb
Kaminski et al., show the cooperative roles of RAD51AP1 and TERRA in generating HR intermediates during ALT-HDR. Proteomic analyses uncover that RAD51AP1 binding of R-loops might serve to maintain chromatin that suppresses TERRA and prevents transcription-replication collisions (TRCs) during ALT-HDR.
INTRODUCTION
Telomere length maintenance is a hallmark of cancer cells (Hanahan and Weinberg, 2011). Most cancers up-regulate the expression of hTERT, the catalytic subunit of telomerase. However, significant subsets of cancers use the Alternative Lengthening of Telomeres (ALT) pathway. In contrast to telomerase, which in addition to cancer cells is active in stem and germ cells, ALT is only used in cancer cells and thus represents a target for cancer therapy (Sieverling et al., 2020). Current evidence indicates that ALT arises as a latent response to the inactivation of chromatin modifiers including the ATRX (α-thalassemia/mental retardation, X-linked) and DAXX (Death domain associated protein) chromatin remodeling/histone deposition complex that regulates histone H3.3 deposition at telomeres (Heaphy et al., 2011; Sieverling et al., 2020). The ensuing alterations in telomeric chromatin render telomeres unstable and chronically prone to DNA breaks (Clynes et al., 2015; Li et al., 2019). Telomere DNA breaks generated during G2 cell cycle phase are repaired by homology-directed repair (HDR) mechanisms involving the RAD51 and RAD52 DNA recombinases. These recombinases facilitate the pairing of homologous telomere sequences and the establishment of homologous recombination (HR) intermediates that prime the synthesis of new telomere DNA sequences by the PCNA-RFC-Polδ replisome to extend telomere length (Dilley et al., 2016; Verma et al., 2019; Zhang et al., 2019).
Among the most potent stressors that stimulate ALT are increased levels and residency of TERRA (telomere repeat-containing RNA) lincRNA at telomeres (Silva et al., 2021). TERRA is transcribed from sub-telomeres and telomeres of several chromosome ends and has been implicated in heterochromatin formation, protein recruitment and telomere protection (Azzalin et al., 2007; Chu et al., 2017; Porro et al., 2014). Recent evidence implicated TERRA in fostering telomere instability and replicative stress that sets the stage for ALT-HDR and telomere extension (Silva et al., 2021). TERRA can exert ‘cis’ and ‘trans’ effects by binding to telomeres and other chromosomal regions (Chu et al., 2017). By hybridizing with telomere DNA sequences forming RNA-DNA hybrid structures, termed R-loops. R-loops created in cis (i.e., by TERRA transcription) can destabilize telomeres, generating DNA breaks that provide substrates for RAD51/RAD52-dependent HDR (Arora et al., 2012; 2014; Silva et al., 2021). Excessive TERRA and R-loop formation cause spontaneous telomere deletions that adversely affect cell viability. Conversely, depletion of TERRA significantly impedes or diminishes ALT initiation implying that TERRA homeostasis and minimal levels of R-loop-induced telomere instability are necessary for ALT (Arora et al., 2014; Silva et al., 2021).
Recent studies have uncovered direct associations between TERRA and HR factors like RAD51, BRCA1 and RTEL (Feretzaki et al., 2020; Vohhodina et al., 2021; Ghisays et al., 2021). TERRA was shown to initiate RAD51-dependent strand invasion and promote R-loop formation at telomeres (Feretzaki et al., 2020; Vohhodina et al., 2021). Similarly, RAD51–associated protein 1 (RAD51AP1), a factor that fulfills crucial roles in ALT, harbors RNA binding activity (Kovalenko et al., 1997). RAD51AP1 used RNA as part of a mechanism that generates R-loops and displacement (D)-Loops via specialized HR-intermediates termed DR-loops (Ouyang et al., 2021). TERRA and RAD51AP1 are essential for ALT. Therefore, understanding how TERRA and RAD51AP1 functionally co-operate to regulate ALT remains a crucial mechanistic question.
We show that RAD51AP1 directly binds TERRA in vitro and in ALT cancer cells. We provide biochemical evidence that TERRA stimulates RAD51AP1-dependent D-loop formation and binds to TERRA R-loops. Furthermore, we find that RAD51AP1 deficiency reduces telomere DSB-associated TERRA R-loops and that RAD51AP1 association with telomeres increases upon SETX depletion, reflecting its potential binding to TERRA containing HR intermediates or R-loops in cells. These data show that RAD51AP1 has a crucial role in TERRA R-loop homeostasis. By proximity-dependent biotinylation and proteomic analyses, we uncovered RAD51AP1 proximal associations with proteins involved in transcription-coupled double-strand break repair, chromatin-remodeling, and R-loop homeostasis. Functional studies revealed that RAD51AP1-dependent R-loop formation is required for chromatin-directed suppression of excessive TERRA accumulation at induced telomere DNA breaks, as well as at unchallenged telomeres, during ALT-HDR. This appears crucial to prevent transcription-replication collisions (TRCs) at telomeres. Intriguingly, RAD51AP1s role in suppressing TERRA requires not only its intrinsic nucleic acids binding but also its SUMO-SIM (SUMO interaction motif) module. These findings further illustrate the multi-functional roles of RAD51AP1 in maintaining productive ALT.
RESULTS
RAD51AP1 interacts with TERRA lincRNA.
The mounting evidence of transcription and R-loops stimulating homology-directed repair (Feretzaki et al., 2020; Hatchi et al., 2021; Vohhodina et al., 2021) and RAD51AP1’s role in this process (Ouyang et al., 2021) prompted us to assess if RAD51AP1 and TERRA interact in ALT cancer cells. To do so, we performed native RNA immunoprecipitations with antibodies to pulldown RNAs bound to endogenous RAD51AP1 in ALT U2OS cells. As a control, we also included RAD51 antibodies. Immunoprecipitation of RAD51AP1 specifically retrieved TERRA and not the 18S ribosomal RNA as determined by standard dot-blots with radio-labeled probes, and at levels comparable to RAD51 (Figure 1A). Notably, the TERRA signals of RAD51 and RAD51AP1 were comparable (Figure 1B). RAD51AP1 binding to TERRA was also confirmed in another ALT cell line, LM216J (Figure 1B). Through these RIP experiments, we also found that the interaction of these proteins with TERRA was more robust in these ALT cell lines than in telomerase expressing cells such as HeLa LT that have long telomeres (Figure 1B), which disfavor TERRA binding (Feretzaki et al., 2020).
Figure 1. RAD51AP1 binds to TERRA and TERRA R-loops.
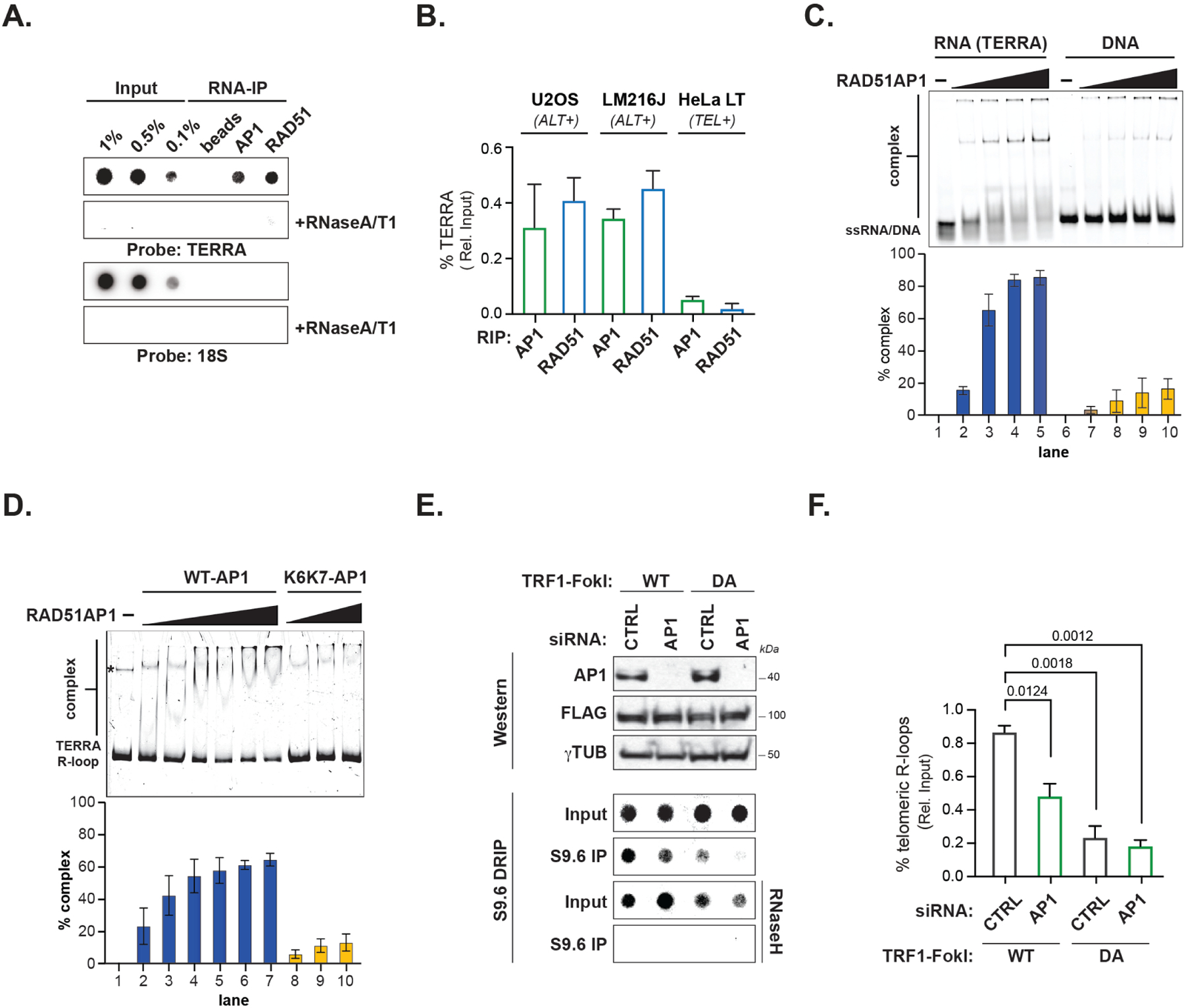
(A) Northern dot-blot of TERRA RNA immunoprecipitation (RIP) of endogenous RAD51AP1 and RAD51 in U2OS (n=4) and LM216J (n=2) ALT cells and HeLa LT (n=2) telomerase expressing (TEL+) cells. (B) Quantification of % TERRA detected in RAD51AP1 and RAD51 RIPs. Data represent mean ± SEM. (C) Electromobility Shift Assay (EMSA) of RAD51AP1 binding TERRA RNA (left) and ssDNA oligonucleotides (right) and quantification of shifted nucleic acid substrates. Data represent mean ± S.D, n=3. (D) EMSA of RAD51AP1 WT (left) and K6K7 (nucleic acid binding mutant, right) binding synthetic TERRA R-loops and quantification of shifted R-loop. * indicates a DNA contaminant in the substrate. Data represent mean ± S.D, n=3. (E) Top: Western blots showing RAD51AP1 and FLAG-TRF1-FokI protein levels after RAD51AP1 knockdown in (WT, left)- and (DA, right)- TRF1-FokI induced U2OS cells. Bottom: Southern dot-blot of DNA-RNA immunoprecipitation (DRIP) with S9.6 antibodies after RAD51AP1 knockdown in (WT, left)- and (DA, right)- TRF1-FokI induced U2OS cells. (F) Quantification of telomeric R-loops. Data represent mean ± SEM, n=3. p values are indicated and generated by One way ANOVA. See also Figure S1.
We determined that RAD51AP1’s interaction with TERRA was direct by using purified recombinant RAD51AP1 protein in an electrophoretic mobility shift assay (EMSA). By incubating increasing concentrations of recombinant RAD51AP1 with Cy-5 labeled TERRA containing (UUAGGG)6 or ssDNA oligonucleotides with (TTAGGG)6 we found that higher concentrations of RAD51AP1 binds TERRA RNA with five-to six-fold greater affinity when compared to ssDNA containing the TTAGGG sequence (Figure 1C). These results indicate that RAD51AP1 can directly associate with TERRA and this association is enhanced in ALT cancer cells.
Multi-contextual associations between RAD51AP1 and TERRA.
TERRA promotes RAD51-dependent strand invasion and formation of both D and R-loops (Feretzaki et al., 2020). RAD51AP1 was shown to act analogously by using RNA to generate unique HR intermediates referred to as DR-loops (Ouyang et al., 2021). First, we performed in vitro assays to examine whether the presence of RNA affects D-loop formation mediated by RAD51 and RAD51AP1. We tested various RNA species i.e., having sequences related to the invading ssDNA (RNA 1 or 2 in Figure S1A) or the sequences in the vicinity of the D-loop (RNA 4–7) or non-related RNA (RNA3). Notably, the RNA complementary to the displaced loop (RNA2) in conjunction with RAD51AP1 promotes D-loop formation (Figure S1A) in a RAD51AP1 concentration-dependent manner (Figure S1B). In addition, we tested strand invasion of TERRA-RAD51-RAD51AP1 and ssDNA-RAD51-RAD51AP1. TERRA D-loop formation activity of RAD51-RAD51AP1 was ~10fold less than when the equivalent DNA sequence was used as the invading strand (Figure S1C). These data suggest that RAD51AP1’s interaction with TERRA promotes D-loop formation, and potentially maintains an association with the products of strand invasion. Despite RAD51AP1’s weaker activity in generating TERRA R-loops, by performing EMSA we found that recombinant RAD51AP1 can bind to R-loops containing TERRA. This binding was mostly abrogated in a compound K6R/A-K7W/A nucleic acid binding defective RAD51AP1 mutant protein (K6/K7-RAD51AP1) (Dunlop et al., 2012) (Figure 1D).
To examine how RAD51AP1 deficiency affects TERRA R-loops in ALT cells we performed DNA-RNA Immunoprecipitations (DRIP) with the RNA: DNA hybrid-specific S9.6 antibody. First, we applied the TRF1-FokI telomere double-strand DNA break (t-DSB) system to stimulate ALT-HDR (Cho et al., 2014). The direct generation of t-DSBs could, in principle, bypass TERRA’s role in creating DNA damage and instead enable the formation of HR intermediates where RAD51AP1 and TERRA’s roles could be assessed. Indeed, by using DRIP, we found t-DSBs induced by wildtype (WT) TRF1-FokI caused a 4-fold increase in telomere R-loops in U2OS cells compared to the inactive (nuclease-dead, DA) TRF1-FokI (Figure 1E). Significantly, RNaseH treatment abolished these DRIP signals, confirming R-loops as their source. However, the increase of R-loops observed upon WT-TRF1-FokI was significantly reduced in RAD51AP1-depleted cells (Figure 1F). The reduction of telomere-associated R-loops in RAD51AP1 depleted cells was corroborated by visualization of the t-DSB localization of GFP-tagged RNaseH1 containing the D210N mutation that inactivates its catalytic activity but preserves RNaseH1s R-loop binding. We found that while GFP-RNaseH1-D210N robustly accumulated at t-DSBs following WT-TRF1-FokI induction, this frequency was significantly diminished after RAD51AP1 depletion (Figure S1D). Thus, RAD51AP1 appears to be required for R-loop generation or maintenance at telomeres in response to telomeres.
We then examined how modulating TERRA or R-loops might affect RAD51AP1 association at telomeres. We employed antisense oligonucleotides (ASOs) that trigger RNaseH1dependent degradation of TERRA in U2OS cells (Chu et al., 2017). Dot-blot analysis of bulk TERRA and RNA-FISH of TERRA in single cells confirmed the efficient degradation of TERRA in U2OS cells (Figure S1E-F). We observed that the localization to telomeres of GFPRAD51AP1 was drastically impaired in TERRA-depleted cells (Figure S1G). In contrast, inhibition of the elongating form of RNAPII, and thus nascent transcription, with DRB (5,6dichloro-1-β-D-ribofuranosyl-1H-benzimidazole) did not alter RAD51AP1 localization (Figure S1H). Interestingly, depletion of the SETX R-loop resolvase that provokes the persistence of unresolved R-loops enhanced the telomeric localization of RAD51AP1 (Figure S1I). Collectively, these studies provide evidence for RAD51AP1 binding to TERRA in vitro and in ALT cancer cells, where they form a complex that assists in the generation and subsequent stabilization of HR intermediates such as D or R-loops.
Interrogating the RAD51AP1 interactome in ALT cancer cells.
To decipher the RAD51AP1 interactome and possibly uncover factors that facilitate its TERRA or R-loop related activities during ALT-HDR we applied proximity-dependent biotinylation and protein identification (bioID) that detects direct and proximal protein associations (Roux et al., 2012). Full-length RAD51AP1 was N-terminally tagged with the BirA ligase and stably expressed in U2OS cells (Figure 2A). We conducted parallel bioID mass spectrometry experiments with BirA-TRF1 and BirA alone (Figure 2A). Asynchronously growing U2OS cells were pulsed with biotin for 24hrs, and biotinylated proteins were captured by streptavidin pulldown. Four independent bioIDs were subjected to liquid chromatography-mass spectrometry (LC-MS) and analyzed per condition (see methods for details). We then conducted a comparative analysis of the factors recovered only in BirA-RAD51AP1 and BirA-TRF1 BioIDs (García Expósito et al., 2016) (Table S1). Shared proteins that were detected in both BirA-TRF1 and Bir-RAD51AP1 were functionally annotated and classified according to biological processes and molecular functions in Uniprot and DAVID (Huang et al., 2009). Functional convergence between TRF1 and RAD51AP1 linked proteins was found in chromatin modification, RNA/DNA unwinding, topology, telomere maintenance and DSB repair by HDR (Figure 2B).
Figure 2. The RAD51AP1 proximal interactome contains RNA processing and transcription-associated complexes.
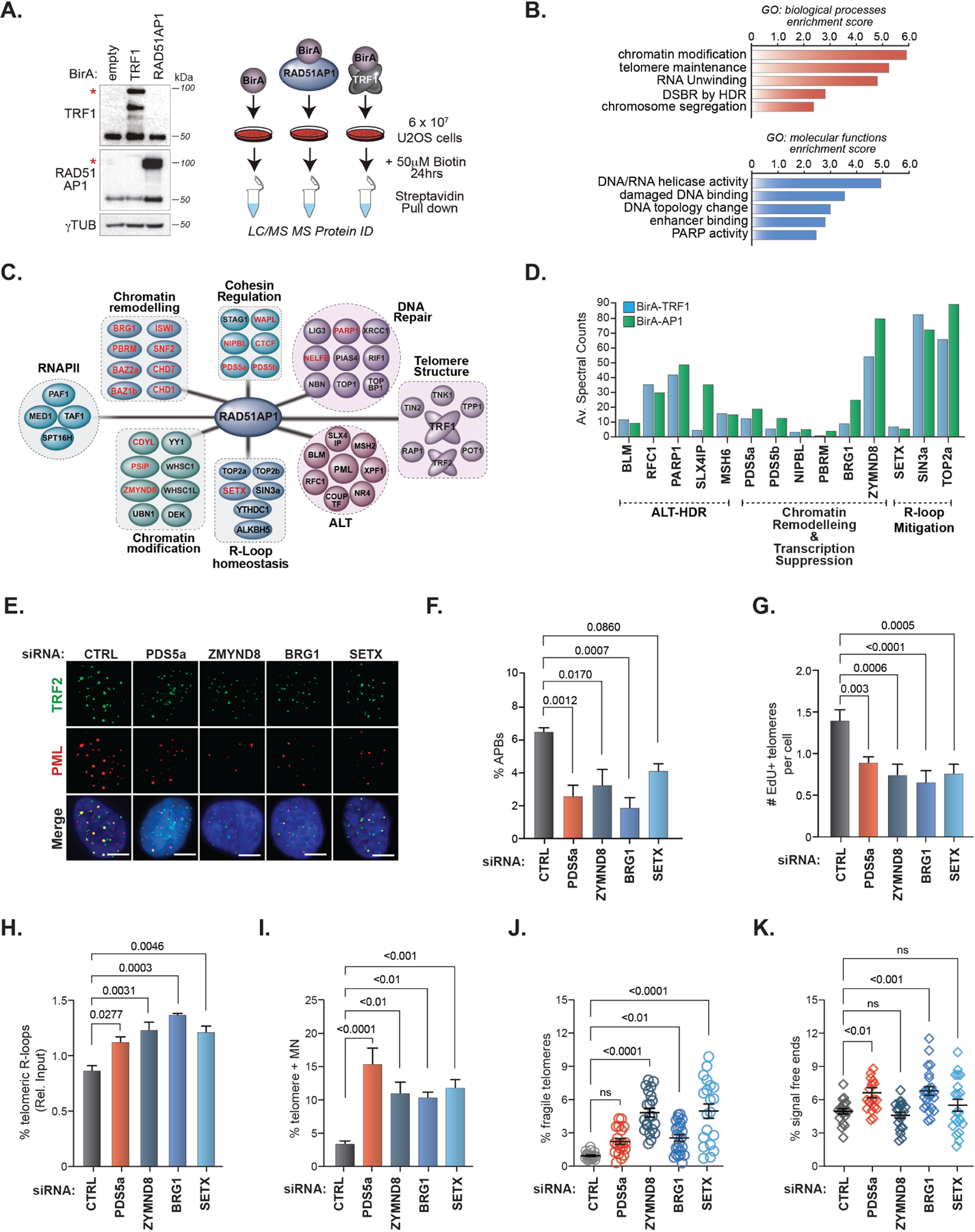
(A) Right. Schematic of TRF1 and RAD51AP1 BioID. Left. Western blots showing the expression of BirA-TRF1 and BirA-RAD51AP1 in U2OS cells. * indicates the band corresponding to the expected protein. (B) GO-term annotation and ranking of TRF1RAD51AP1 enriched proteins by biological processes and molecular functions using DAVID. (C) Clustering of distinct functional protein groups identified by TRF1-RAD51AP1 BioID. (D) Spectral counts of selected TRF1 (left bars) and RAD51AP1 (right bars) BioID hits from the indicated functional groups. (E) Representative images and (F) quantification of APBs (PML and TRF2 localization) after knockdown of the indicated proteins in U2OS cells. All data represent mean ± SEM, n=3. All scale bars, 5μm. (G) Quantification of telomeres staining positive for EdU incorporation after knockdown of the indicated proteins in U2OS cells synchronized in G2. All data represent mean ± SEM, n=4 (H) Box plot showing the quantification of telomere DRIP assays (n=3). Horizontal lines and boxes represent the mean ± SEM and min-max range of values from individual experiments. (I-K) Quantification of (I) micronuclei with TTAGGG FISH signals (n=2), (J) fragile telomeres (n=2) and (K) signal-free chromosome ends (i.e., telomere loss) (n=2) after knockdown of the indicated proteins in U2OS cells. Each circle in J and K represents a single data point, mean ± SEM are represented by black lines. p values are indicated and generated by One way ANOVA. See also Figures S2, S3 and Table S1.
Further grouping revealed networks of proteins that have been implicated in ALT like BLM (Sobinoff et al., 2017), RFC1 (Dilley et al., 2016), SLX4IP (Panier et al., 2019), MSH6 (Barroso-González et al., 2021), and PARP1 (Hoang et al., 2020) (Figure 2C-D). However, multiple proteins that facilitate RNA Polymerase II-dependent transcription (PAF1, MED1), RNA processing (YTHDC1, ALKBH5), R-loop mitigation (SETX, SIN3a, TOP2a/b), HIRA-mediated chromatin assembly (UBN1, DEK) as well as several chromatin remodeling and modification factors were represented as shared TRF1 and RAD51AP1 proximal proteins (Figure 2C). These included various subunits of the BAF (BRG1, PBRM, SNF2) (Brownlee et al., 2014; Chabanon et al., 2021; Kakarougkas et al., 2014; Meisenberg et al., 2019), SWI/SNF (ISWI, BAZ1B, BAZ2B) (Aydin et al., 2014; Lan et al., 2010), and NuRD (ZMYND8, CHD1, CHD7) (Gong et al., 2015; Luijsterburg et al., 2016; Rother et al., 2020) complexes, and mediators of Cohesin loading (PDS5A, PDS5B, NIPBL, WAPL, CTCF) (Meisenberg et al., 2019), which in addition to their canonical functions, are implicated in suppressing transcription at DNA breaks through establishing repressive chromatin as well as R-loop mitigation (Bayona-Feliu et al., 2021; Cohen et al., 2007; Kakarougkas et al., 2015; Kim et al., 2019; Meisenberg et al., 2019; Skourti-Stathaki et al., 2011). Other proteins such as CDYL (Abu-Zhayia et al., 2018), PSIP/LEDGF (Aymard et al., 2017), PARP1, KDM5A and NELF-E (Awwad et al., 2017) have been similarly implicated in chromatin-directed pausing of RNA Polymerase II at DNA breaks (Figure 2C).
The impact of RAD51AP1 proximal chromatin remodelers on ALT-HDR.
Consistent with the bioID analysis of unchallenged U2OS cells (Figure S2A), GFP-tagged PDS5a, ZMYND8, BRG1 and SETX were visualized at telomeres in cells expressing DA-TRF1-FokI (i.e., no t-DSB induction) (Figure S2B). Their localization was enhanced following the induction of t-DSBs with WT-TRF1-FokI indicating a DNA break dependency (Figure S2B). However, whereas depletion of RAD51AP1 had no effect on the localization of these chromatin remodelers to t-DSBs (Figure S2D), we observed that depleting PDS5a, BRG1, or ZMYND8 strongly impaired RAD51AP1 localization to t-DSBs (Figure 2D-E). This indicated that these proteins may act upstream to facilitate RAD51AP1 recruitment. The exception to this was the depletion of SETX, which as was seen with respect to telomeres in unchallenged U2OS cells (Figure S1I), enhanced the presence of GFP-RAD51AP1 at t-DSBs (Figure S2E). Interestingly, we observed that TERRA depletion using ASOs markedly diminished the localization of each of PDS5a, BRG1, ZMYND8 and SETX at t-DSBs (Figure S2F).
These data suggest a sequence of events whereby the chromatin remodeler factors accumulate at t-DSBs, likely by tracking with or being recruited to stalled RNAP II, where they modulate t-DSB proximal chromatin to facilitate ALT-HDR involving RAD51AP1. The retention of these proteins at DSBs may be dictated by TERRA, which acts as a scaffold to retain proteins on chromatin. RAD51AP1 on the other hand might be retained at telomeres following strand invasion and D-R loop formation through binding to R-loops, which are subsequently resolved by SETX.
We next assessed if these proteins contribute to ALT-HDR mechanism and telomere integrity in ALT cancer cells. Recent studies have underscored the importance of ALT-associated PML bodies (APBs) as crucial subnuclear compartments in which telomere DNA synthesis takes place (Zhang et al., 2019). We found that depleting each of these factors diminished the overall frequency of ALT-associated PML bodies (APBs) that could be detected in populations of U2OS and LM216J ALT cancer cells (Figure 2E-F and S3A). We visualized telomere DNA synthesis by monitoring the incorporation of the nucleoside analog of thymine, 5ethynyl-2'-deoxyuridine (EdU) directly at telomeres in U2OS cells. As with APBs, we observed a reduced frequency of EdU positive telomeres following depletion of PDS5a, ZYMND8, BRG1 or SETX (Figure 2G). These effects were not attributed to altered cell cycle progression or changes in PML protein levels (data not shown). We also assessed C-circles, an extrachromosomal partially single-stranded circular DNA species containing telomeric sequences, which are proposed to be generated via a break-induced replication (BIR) pathway at telomeres in ALT cancer cells. However, we did not detect significant alterations (Figure S3B). This could be due to redundancy in the mechanisms that regulate C-circle production.
We supplemented these analyses of unchallenged ALT cells by directly assessing the impact that depleting these factors has on RAD51-dependent telomere clustering (Cho et al., 2014), and break-induced telomere DNA synthesis (BITS) (Dilley et al., 2016) at DSBs acutely generated by TRF1-FokI (Figure S3C-E). Indeed, depletion of PDS5a, BRG1, ZMYND8, and SETX diminished the size of clustered telomeric foci (Figure S3D) and frequency of t-DSB foci containing EdU positive signals (Figure S3E). We noted in these EdU experiments that the depletion of the major R-loop resolvase SETX most strongly impaired DNA synthesis (Figure 2G and S3E). This indicates that SETX-dependent R-loop resolution may be required for efficient telomere DNA synthesis initiation at telomere DSBs, which is consistent with the studies linking impaired R-loop homeostasis and a dysfunctional non-productive ALT mechanism (Arora et al., 2014; 2012; Silva et al., 2021). Accordingly, by conducting DRIP with S9.6 we found that depletion of PDS5a, ZMYND8, BRG1 and SETX increased t-DSB generated TERRA R-loops in U2OS cells (Figure 2H). These data indicate that PDS5a, BRG1, ZMYND8, and SETX may promote RAD51-dependent HR that precedes telomere extension.
Lastly, we examined whether a deficiency of these factors affects telomere integrity. Prior studies showed that RAD51AP1 depletion elicited more frequent micronuclei containing fragments of telomeric DNA (Barroso-González et al., 2019). Similarly, a significant increase in micronuclei containing telomere DNA was observed by IF-FISH of PDS5a, BRG1, ZMYND8, and SETX depleted U2OS and LM216J cells (Figure 2I and S3F-G). An examination of telomeres on metaphase chromosomes revealed evidence of marked telomere fragility, which is linked with defects in telomere replication, particularly following depletion of ZMYND8, BRG1, SETX and, to a lesser extent, PDS5a (Figure 2J-K and S3H). Moreover, we found that signal-free ends (i.e., chromatids without detectable telomere FISH signals) were more prevalent in PDS5a and BRG1 depleted cells. Thus, as was previously reported for RAD51AP1 (Barroso-González et al., 2019), these RAD51AP1 vicinal factors appear to be required for maintaining an efficient ALT mechanism and the integrity of telomeres in ALT cancer cells.
Chromatin-directed suppression of TERRA during ALT-HDR.
At sites of DNA damage, RNA polymerase II (RNAPII) must be quickly stalled to prevent its collision with DNA replication machinery and DNA repair complexes (Marnef et al., 2017). This involves the recruitment of factors like NELF-E (Negative Elongation Factor-E) and KDM5A (Lysine Demethylase 5A) by PARP1 to pause RNAPII (Awwad et al., 2017; Gong et al., 2017). Chromatin-directed suppression of transcription and RNAPII at DSBs can occur through a canonical pathway that begins with ATM activation and culminates in the establishment of transcriptionally repressive chromatin characterized by ubiquitination of histone H2A at lysine 119 (Ub-H2A) by Polycomb Repressive Complex 1 (PRC1) and tri-methylation of histone H3 at lysine 27 (H3K27me3) by Polycomb Repressive Complex 2 (PRC2). Furthermore, although most chromatin-associated RNAs are evicted from the DSB site, trans-acting RNAs can reinforce transcriptionally inactive chromatin states (Marnef and Legube, 2021). Similarly, R-loops can promote repressive chromatin formation to terminate transcription (Skourti-Stathaki et al., 2014; 2019). PDS5a, BRG1, and ZMYND8 have been implicated in chromatin-directed suppression of RNAPII and transcription at DNA breaks (Gong et al., 2015; Meisenberg et al., 2019).
Thus, we first examined if TERRA is altered by telomere DSBs by RNA-FISH using fluorescently labeled (TAACCC)5 oligonucleotide probes complementary to TERRA. We observed that the generation of t-DSBs by WT-TRF1-FokI, but not DA-TRF1-FokI, elevated the levels of telomeric TERRA (Figure 3A-B). By combining immunofluorescence to visualize DSB-induced Ub-H2A foci with TERRA RNA-FISH, we observed that the increased presence of TERRA was accompanied by increased detection of Ub-H2A foci at t-DSBs generated by WT-TRF1-FokI, and not after DA-TRF1-FokI expression (Figure 3A-B). No alterations in TERRA levels were detected in Northern blot of bulk TERRA in these conditions (data not shown) indicating that it is only the abundance of telomere-bound TERRA that is altered. Yet, the addition of AZD0156, which inhibits ATMs kinase activity, further enhanced the presence of TERRA at t-DSBs but strongly diminished Ub-H2A foci (Figure 3A-B). Similarly, treating cells expressing WT or DA TRF1-FokI with an enzymatic inhibitor of EZH2 (EZH2i; GSK126), the catalytic histone lysine methyltransferase subunit of the PRC2 complex, elicited significantly more TERRA but strongly reduced UbH2A accumulation at t-DSBs (Figure 3A-B).
Figure 3. RAD51AP1 co-regulates TERRA suppression during ALT.
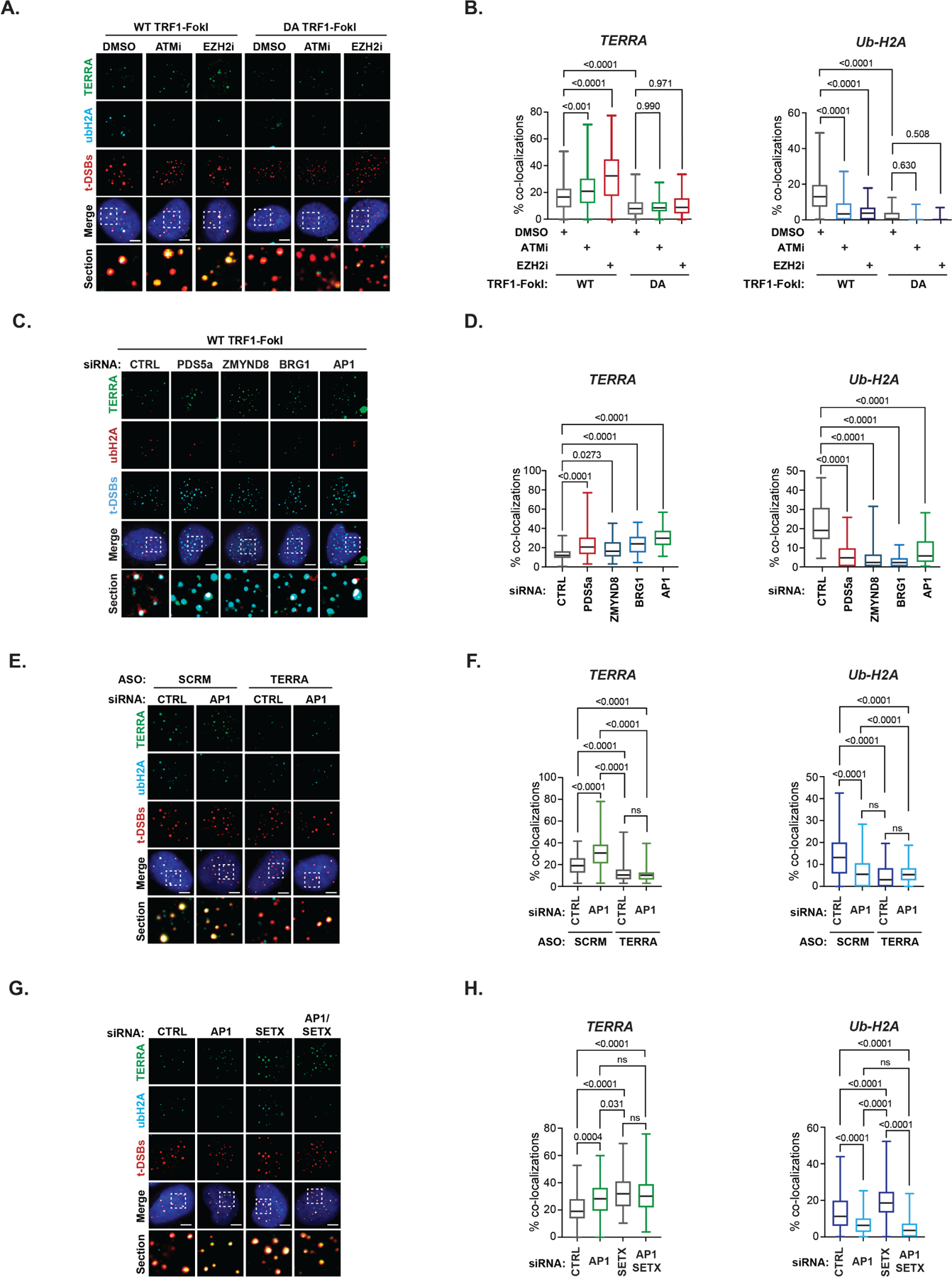
(A) Representative images and (B) quantification of TERRA (RNA FISH) and Ubiquitin-K119 of histone H2A (Ub-H2A) (IF) at t-DSBs after ATMi or EZH2i (10μM, 4hrs each). (C) Representative images and (D) quantification of TERRA (RNA-FISH) and ubiquitinated histone H2A-K119 (Ub-H2A) (IF) at telomeric double-strand breaks (t-DSBs generated by FLAG tagged WT-TRF1-FokI) after siRNA-mediated knockdown of indicated proteins. (E) Representative images and (F) quantification of TERRA (left) and Ub-H2A (right) at t-DSBs in scrambled (SCRM) and TERRA ASO depleted U2OS cells that were co-transfected with either control (CTRL) or RAD51AP1 siRNAs. (G) Representative images and (H) quantification of TERRA (left) and Ub-H2A (right) at t-DSBs in U2OS cells that were transfected with either control (CTRL), SETX or/and RAD51AP1 siRNAs. All box and whiskers plots show the interquartile and min-max ranges. The median is represented by the horizontal black line. Data from triplicate independent experiments are shown. p values are indicated and generated by One way ANOVA. All scale bars, 5μm. See also Figure S4.
While the increase in TERRA localization at t-DSBs seemed inconsistent with transcription suppression having a role at telomeres, we considered the evidence that ongoing TERRA transcription can initiate ALT by forming replicative barriers at stalled replication forks. TERRA also binds to many early-response DNA repair factors many of which are regulated by PARP1 (e.g., FUS, NONO, hnRNPs) that have roles in seeding the DDR (Altmeyer et al., 2015; Chu et al., 2017; Petti et al., 2019; Quinodoz et al., 2021), as well as factors like NELF-E/A and KDM5A that facilitate pausing of RNAPII and switching of active to repressive chromatin states. NELF-E is the smallest subunit of the Negative Elongation Complex that physically interacts with RNAPII and is required for RNAPII pausing (Awwad et al., 2017). NELFA and NELFE were present in the cohort of RAD51AP1 proximal proteins (Table S1). KDM5A is recruited to the damaged sites and demethylates the H3K4me3 to repress transcription (Gong et al., 2017). By the expression of GFP-tagged cDNAs, we observed that the induction of WT-TRF1-FokI led to the accumulation of NELF-E and KDM5A at t-DSBS (Figure S4A-B). These experiments provide evidence that TERRA is subject to chromatin-directed suppression via the canonical ATM-PRC1-PRC2 pathway.
RAD51AP1 mediated TERRA R-loop assembly promotes repressive chromatin assembly during ALT-HDR.
Next, we asked if the RAD51AP1-associated chromatin remodeling complexes, and potentially RAD51AP1 itself, might suppress TERRA association with telomere DSBs during ALT. As before, we depleted PDS5a, BRG1, ZMYND8 and RAD51AP1 by siRNA (Figure S2D). We confirmed by analysis of bulk TERRA that the transient depletion of these factors did not alter global TERRA levels (data not shown). However, by combining TERRA RNA-FISH and immunofluorescence for Ub-H2A, we found that the deficiency of PDS5a, BRG1 or ZMYND8 elevated the localization of TERRA but diminished Ub-H2A at t-DSBs (Figure 3C-D). Surprisingly, RAD51AP1 depletion elicited the same pattern, whereby the presence of TERRA was significantly elevated, and Ub-H2A was strongly reduced at t-DSBs (Figure 3C-D). The same alterations in TERRA and Ub-H2A were observed in factor-depleted U2OS, LM216J, GM847 ALT cells synchronized in the G2 cell-cycle phase but not in non-ALT HeLa LT cells that express telomerase (Figure S4C-F). These data consistently showed that RAD51AP1 and associated factors PDS5a, BRG1, ZYMND8 limit TERRA accumulation and residency at telomere DSBs and unchallenged telomeres in ALT cancer cells.
The relationship between TERRA and RAD51AP1 in the assembly of repressive chromatin remained unclear. We depleted TERRA using ASOs in U2OS cells expressing WT-TRF1-FokI. As before, we confirmed the efficient reduction in bulk TERRA by RNA blot (Figure S4G), as well as at the single cell level by TERRA FISH (Figure 3E-F). We also observed that TERRA depletion prevented the accumulation of Ub-H2A at t-DSBs, which was unchanged when TERRA and RAD51AP1 were co-depleted (Figure 3E-F). This indicated that TERRA is required for repressive chromatin. As these experiments were performed using WT-TRF1-FokI, TERRAs role in this respect is likely distinct from its role in destabilizing chromatin that initiates ALT and likely linked with its other functions in ALT-HDR. Interestingly, SETX deficiency, which causes R-loop accumulation at telomeres robustly increased Ub-H2A at t-DSBs (Figure 3G-H and S4C-E). This raised the intriguing scenario that chromatin repression is linked to the formation of TERRA R-loops that is mediated by RAD51AP1 and their resolution by SETX. Accordingly, we found that as with individual knockdown, combined depletion of RAD51AP1 and SETX caused TERRA to accumulate at t-DSBs (Figure 3G-H). Whereas elevated TERRA in RAD51AP1 depleted cells is due to defects in TERRA suppression at t-DSBs, the observed increase in TERRA at t-DSBs in SETX depleted cells is likely due to the elevated presence of RAD51AP1 and TERRA R-loops. Most importantly we found that co-depletion of RAD51AP1 in conjunction with SETX reversed the increased Ub-H2A that was observed following knockdown of SETX alone (Figure 3G-H). These data indicate that the chromatin-directed suppression of TERRA may depend on the formation of R-loops by RAD51AP1 and is coordinated with subsequent TERRA R-loop resolution by SETX.
RAD51AP1-directed TERRA suppression prevents transcription-replication collisions (TRCs).
Chromatin-directed suppression of transcription and faithful R-loop mitigation is important to prevent transcription-replication collisions (TRCs) at DSBs (Hamperl et al., 2017; Marnef and Legube, 2021). Given the defects in TERRA suppression and reduced R-loops in RAD51AP1 deficient cells, we tested if TRCs also ensue following the depletion of RAD51AP1, as well as its proximal chromatin remodelers. Due to the orientation of telomere replication, TRCs would occur co-directionally between RNAPII and the DNA replisome moving in the same direction (Hamperl et al., 2017). We performed proximity-ligation assays (PLA) to detect close associations between the elongating form of RNAPII (phospho-Ser2-RNAPII) and PCNA, a subunit of the ALT replisome in U2OS cells expressing WT-TRF1-FokI that were transfected with siRNAs to deplete RAD51AP1, PDS5a, BRG1, ZMYND8 or SETX (Figure 4A). Control PLA reactions in which primary antibodies were omitted and cells expressing DA-TRF1-FokI served as negative controls (Figure 4B). In comparing control cells expressing WT or DA-TRF1-FokI, we observed a modest increase in co-localization between PLA signals and t-DSBs generated by WT-TRF1-FokI (Figure 4B-C). A more robust increase in colocalization between PLA signals and t-DSBs was observed in RAD51AP1 deficient cells (Figure 4B-C). Likewise, the relative frequency of colocalizing PLA and t-DSB foci was significantly greater after depletion of PDS5a, BRG1, ZMYND8 or SETX (Figure 4B-C). This is consistent with the loss of RAD51AP1 and those factors compromising both ALT-HDR and telomere integrity shown here and in prior studies. Considered together these data implicate RAD51AP1 as acting within a network of factors that together coordinate the mechanism that limits the accumulation of TERRA remaining at telomere DSB sites and thus preventing TRCs.
Figure 4. RAD51AP1 prevents transcription-replication collisions (TRCs) during ALT.
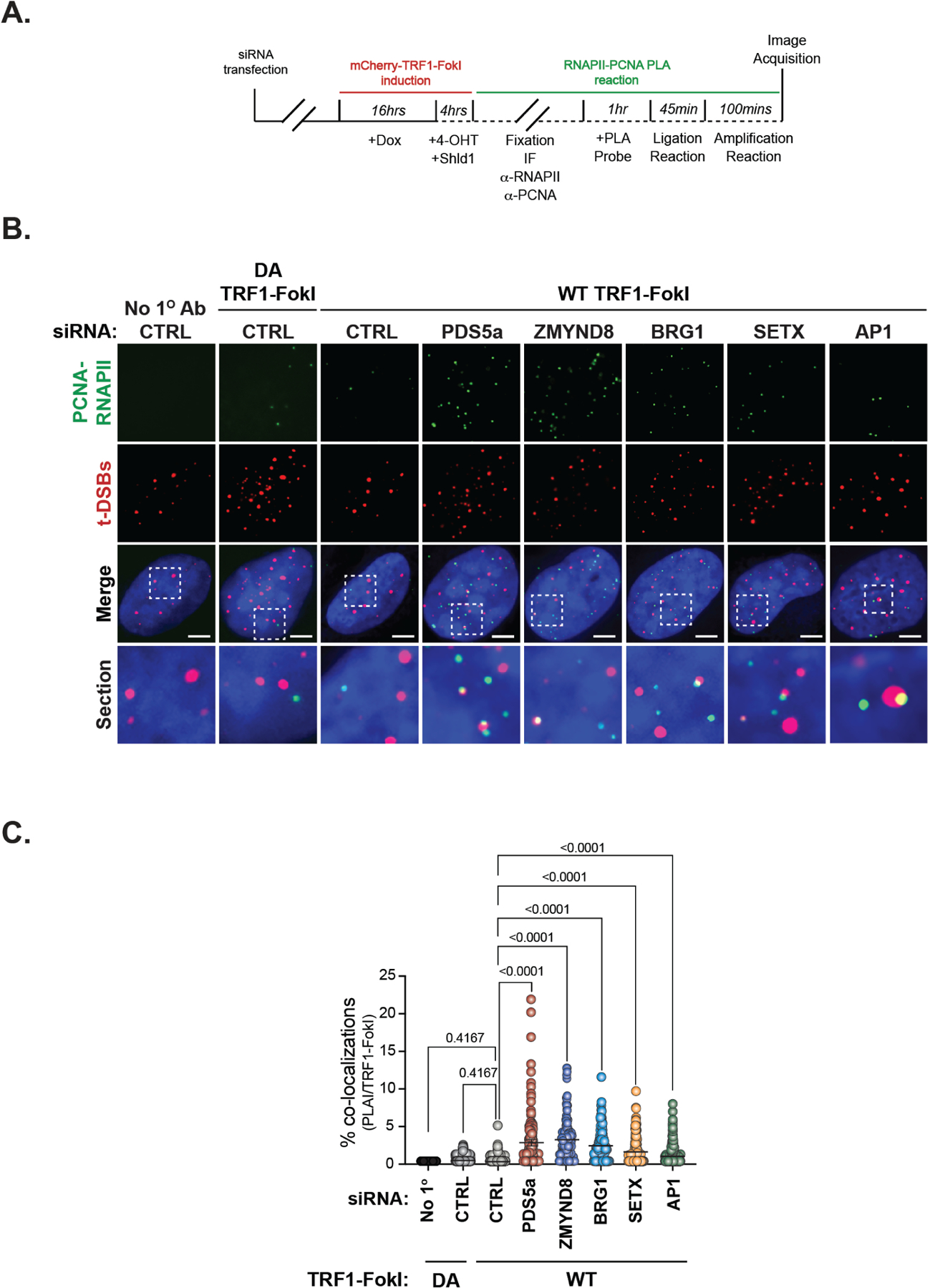
(A) Schematic of proximity ligation assay (PLA) of RNAPII (phospho-Serine 2) and PCNA at sites of t-DSB induction (mCherry-TRF1-FokI). (B) Representative IF images of PLA signals (green) and t-DSBs (red) in PDS5a, ZMYND8, BRG1 and RAD51AP1 depleted U2OS cells expressing WT-TRF1-FokI. Controls include PLA reactions where no primary antibody was added and expression of DA-TRF1-FokI. (C) Quantification of the % PLA signals (in PLA positive cells) that co-localize with t-DSBs from B. Each circle in C represents a single data point i.e., % PLA-TRF1-FokI co-localizations per cell. Mean ± SEM are represented by black lines. Data from at least 3 independent experiments per condition are shown. p values are indicated and generated by One way ANOVA. All scale bars, 5μm.
The SUMO-SIM regulatory axis of RAD51AP1 is required for TERRA suppression.
RAD51AP1 consists of an N-terminal PALB2 and C terminal RAD51/RAD52 interaction domain. The compound amino acid substitutions for lysines located within the N and C-termini in the K6-K7-RAD51AP1 mutant abolish RAD51AP1 nucleic acid binding in vitro (Dunlop et al., 2012). In addition to those HR and nucleic acid binding domains, RAD51AP1 contains an intrinsically disordered region (IDR) that extends from the N-terminal PALB2 domain (aa1–120) through the central portion to the C-terminal RAD51/RAD52 (aa283–352) interaction module (Barroso-González et al., 2019; Dray et al., 2010; Modesti et al., 2007; Wiese et al., 2007). RAD51AP1’s IDR contains a SUMO-interacting motif (SIM) domain (aa137–140) that is required to mediate synaptic complex and D-loop assembly through direct binding with UAF1 (Liang et al., 2016; 2020) (Figure 5A). Previous studies showed that SUMOylation of RAD51AP1 at lysine 269 (K269) was determined to stabilize RAD51AP1 in ALT cancer cells, prolonging its half-life (Barroso-González et al., 2019). Furthermore, SUMO defective RAD51AP1-K269R and SUMO-binding defective ΔSIM-RAD51AP1 (L137A, I140A, V142A) mutants failed to rescue defects in ALT-HDR, including APB formation, telomere clustering, and telomere length. This pointed to the SUMO-SIM axis's pivotal role in regulating RAD51AP1, but whether it contributes to TERRA suppression was unknown. We tested whether re-expression of SUMO-defective RAD51AP1 mutants could restore TERRA suppression and chromatin silencing observed when endogenous RAD51AP1 was depleted. FLAG-tagged full-length (FL), K269R, ΔSIM-RAD51AP1 as well as K6-K7 and ΔRAD51-RAD51AP1 mutants were transiently expressed in RAD51AP1 depleted U2OS cells (Figure 5B). Consistent with previous studies, we found that GFP-FL-RAD51AP1, GFP-tagged K269R, and ΔSIM-RAD51AP1 localized to telomeres, but not the K6-K7 or ΔRAD51 mutants (Figure 5C-E). By examining TERRA and Ub-H2A by RNA-FISH and IF respectively, we found that in contrast with FL-RAD51AP1, which reduced TERRA and increased Ub-H2A (i.e., restoring TERRA suppression), none of these mutants, including the K269R and ΔSIM-RAD51AP1 mutants, could re-establish TERRA suppression and Ub-H2A following t-DSBs (Figure 5F-G). This raised the possibility that the defects in TERRA suppression could be due to the altered binding of these mutants to TERRA in ALT cells. Therefore, we conducted RNA immunoprecipitations (RIP) in U2OS cells expressing myc-tagged WT, K269R, and ΔSIM RAD51AP1 (Figure 5H). Unlike the WT-RAD51AP1 protein, we did not detect TERRA association with either the K269R or ΔSIM-RAD51AP1 protein (Figure 5I). This finding implicates the stabilization of RAD51AP1 by the SUMO-SIM regulatory axis as a crucial factor in regulating its association with TERRA and chromatin-directed TERRA suppression during ALT.
Figure 5. The RAD51AP1 SUMO-SIM regulatory axis is required for TERRA suppression.
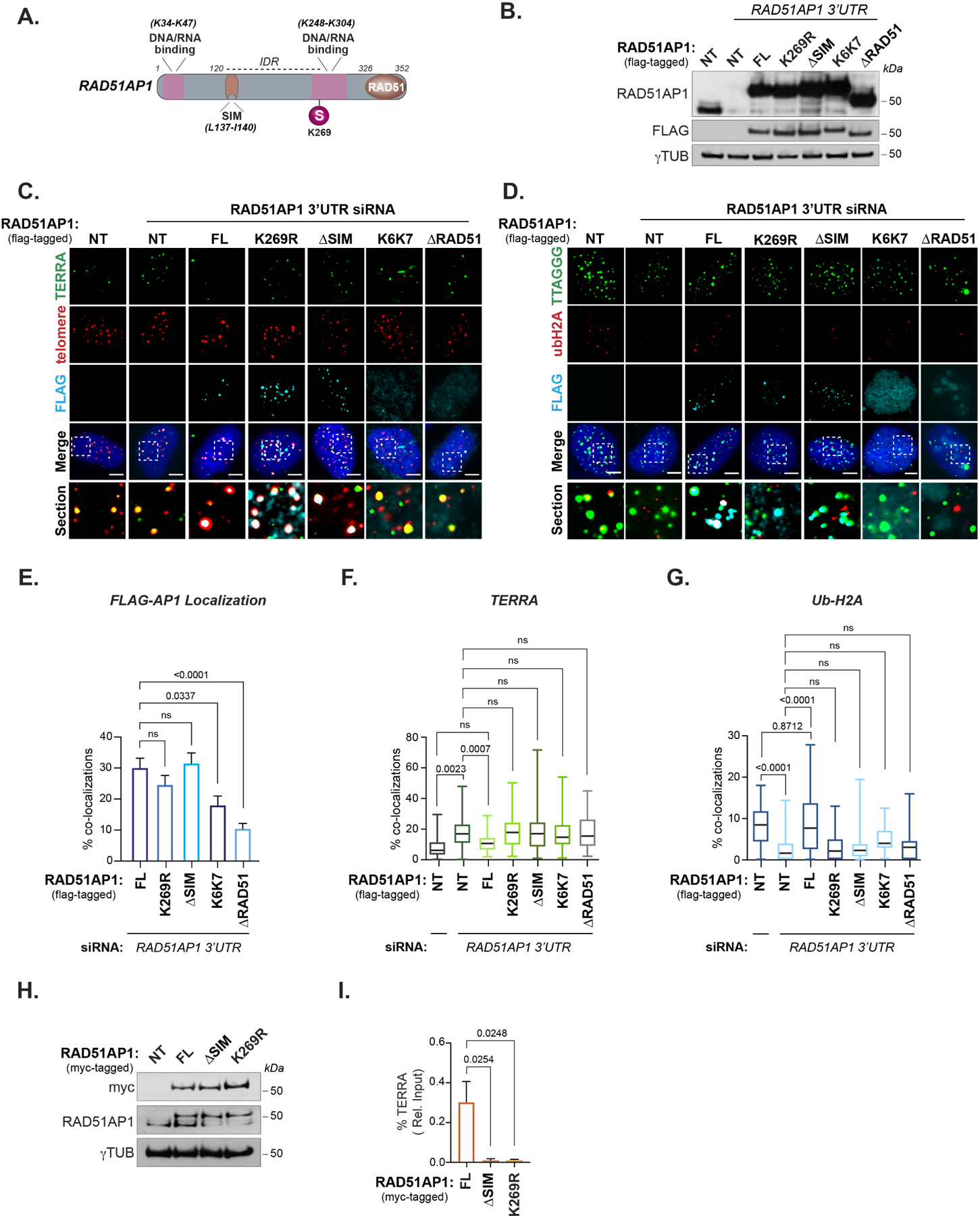
(A) Schematic showing the location of RAD51AP1 functional domains, SUMO modifications and known amino acid substitutions that alter DNA/RNA binding. (B) Western blot of RAD51AP1 knockdown and complementation with wildtype and mutant FLAG-tagged RAD51AP1 proteins. (C) Representative images of TERRA (RNA-FISH) at telomeres (TRF2) after knockdown of RAD51AP1 and complementation with FLAG-tagged RAD51AP1 mutants in U2OS cells synchronized in G2-phase. (D) Representative image of Ub-H2A at telomeres (TRF2) after knockdown of RAD51AP1 and complementation with FLAG-tagged RAD51AP1 mutants in U2OS cells synchronized in G2. (E) Quantification of full-length (FL) and mutant FLAG-tagged RAD51AP1 localization at telomeres in U2OS cells depleted of endogenous RAD51AP1. (F) Quantification of TERRA localization and (G) Ub-H2A accumulation at telomeres. Box and whiskers plots in F and G show the interquartile and min-max ranges. The median is represented by the horizontal black line. Data from triplicate independent experiments are shown. (H) Western blot of U2OS cells stably expressing myc-tagged RAD51AP1. (I) Quantification of TERRA recovered from U2OS cells expressing myc-tagged wildtype, K269R and ΔSIM RAD51AP1. Data represent mean ± SEM, n=3 biological replicates. p values are indicated and generated by One way ANOVA. All scale bars, 5μm.
DISCUSSION
RAD51AP1 and TERRA interactions in ALT cancer cells.
While TERRA can initially be a destabilizing force that induces replication complications and telomere damage (Silva et al., 2021) (Figure 6), it is also involved in generating HR intermediates and R-loops (Feretzaki et al., 2020). We uncovered a pronounced association between TERRA and RAD51AP1 in ALT cancer cells. Together RAD51AP1 and TERRA stimulate RAD51-dependent D-loop formation. RAD51AP1 also binds to TERRA R-loops in vitro, and its depletion reduces TERRA R-loops generated at telomere DSBs in ALT cells. These findings reveal RAD51AP1 as a major factor that assembles R-loops as well as TERRA-containing D-loops which are essential for downstream telomere DNA synthesis. Consistent with these activities, an accompanying paper from Yadav et al., (Yadav et al., 2022) shows that the TERRA-RAD51AP1 mediated generation of R-loops subsequently promotes G4-quadruplexes conferring a dynamic conversion of R-loops to D-loops. As it does at D-loops (Wiese et al., 2007), RAD51AP1 could stabilize TERRA R-loops or bind TERRA within the D-loop during such a switch or prevent their premature resolution by SETX or other helicases (Figure 6). Accordingly, we found that interfering with TERRA R-loop resolution and increasing R-loops by depleting SETX increased RAD51AP1s localization at telomere DSBs. Our study also uncovered that RAD51AP1’s SUMO-SIM module is necessary for binding to TERRA, suggesting some TERRA-associated role for this domain that remains unclear. Prior studies showed that RAD51AP1’s SUMO-SIM regulatory module is required for ALT and SUMOylation may extend the half-life of RAD51AP1 in ALT cancer cells (Barroso-González et al., 2019). RAD51AP1’s SIM domain interacts with the SUMO-like domains (SLD) of the UAF1-USP1 complex. RAD51AP1 and UAF1 can synergize to enhance synaptic complex formation during HR and coordinate the ubiquitination of Fanconi Anemia proteins (Liang et al., 2016; 2019). UAF1 was shown to stimulate RAD51AP1s RNA-directed R-loop formation activity and is necessary for RAD51AP1 accumulation at DNA breaks (Ouyang et al., 2021). Both UAF1 and USP1 bind DNA (Liang et al., 2016). It remains unclear whether they can engage with RNA during HR. Conceivably, UAF1 binding to RAD51AP1 via its SIM domain could stabilize RAD51AP1-TERRA associations with D or R -loops to facilitate its actions during ALT-HDR (Figure 6). RAD51AP1’s SIM domain could also foster interactions with itself or other SUMO-regulated HR or RNA modifying proteins (Psakhye and Jentsch, 2012). In agreement with Yadav et al., (Yadav et al., 2022), we propose that the delineation of factors and interactions that help RAD51AP1 binding or stabilization of R-Loops and assist in orchestrating the R to D switch ALT is of high importance.
Figure 6. Proposed model of RAD51AP1 dependent TERRA regulation during ALT-HDR.
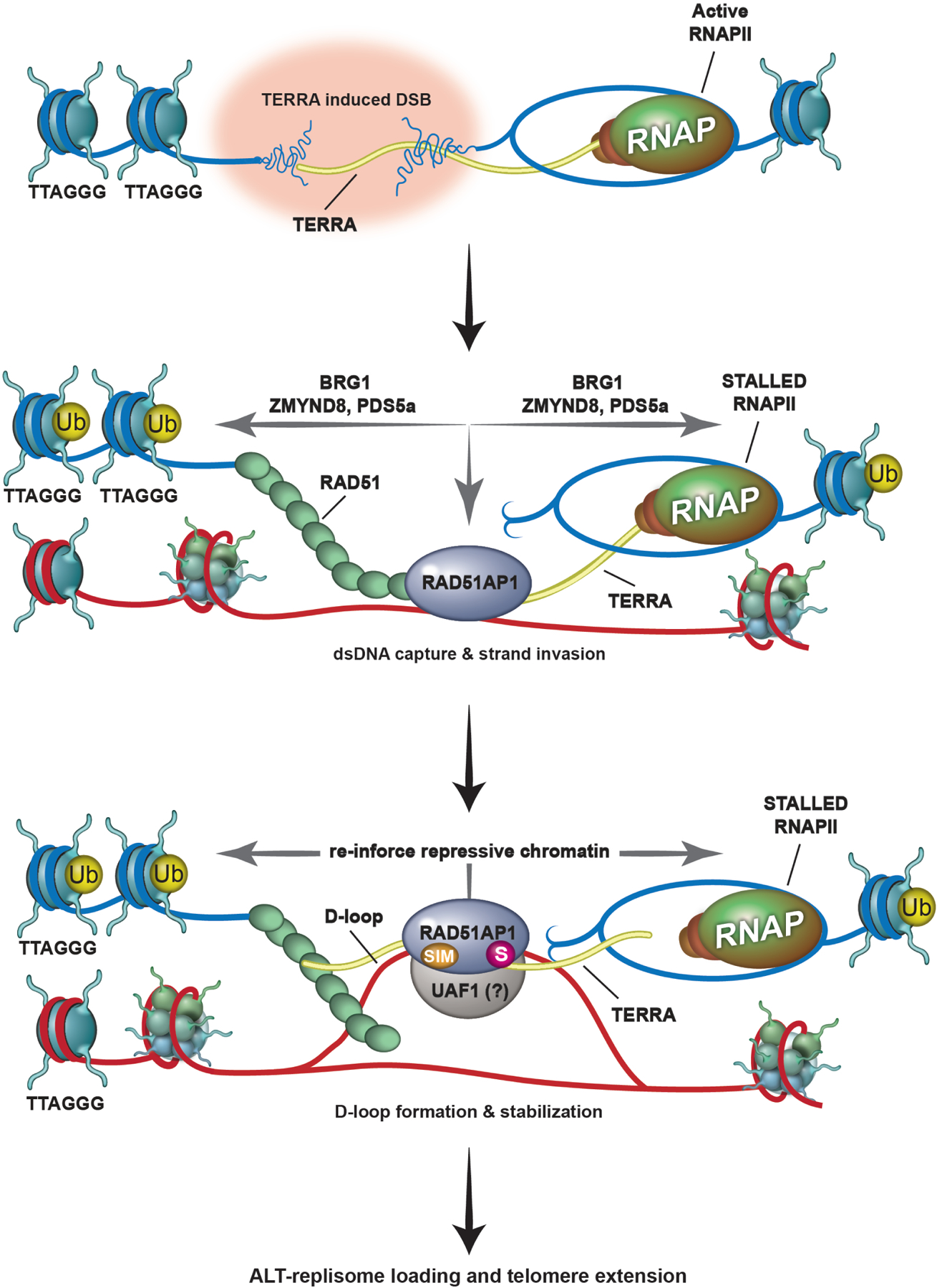
Top. TERRA (in yellow) transcription destabilizes telomeric chromatin, causing telomere replicative stress and DNA breaks. Middle. RNAPII and TERRA transcription are paused at the telomere DSB (t-DSB) by chromatin modulation involving BRG1, PDS5a and ZMYND8, and ubiquitination of chromatin (yellow circle; Ub). In addition, by sliding nucleosomes aside, chromatin remodelers like BRG1 make the break site accessible for repair. RAD51-RAD51AP1 binding to TERRA promotes the capture of homologous telomeric DNA sequences and strand invasion. Bottom. Together, TERRA and RAD51AP1 promote D-loop formation by RAD51. It is possible that TERRA located proximal to or annealed with the displaced strand of the D-loop allows RAD51AP1 binding to the D-loop. This D/R loop structure might then be stabilized by SUMO-SIM mediated interactions between RAD51AP1 and the UAF1 complex that further reinforces TERRA suppression by maintaining repressive chromatin at the t-DSB and possibly shielding these DNA: RNA hybrids from their premature resolution by factors that include SETX. Once telomere DNA synthesis and extension (blue dashed arrow) has successfully initiated and completed, these structures can be dismantled and RNAPII mediated transcription across telomeres reinstated.
A role for RAD51AP1 in maintaining repressive chromatin during ALT-HDR.
Stemming from unbiased proteomics, we unexpectedly uncovered that RAD51AP1 participates in an ATM/EZH2-dependent mechanism that limits RNAPII progression and the presence of excessive TERRA at telomere DSBs. We observed that the depletion of RAD51AP1 (and BRG1, ZMYND8 or PDS5a) caused the accumulation of TERRA, loss of histone H2A lysine (K) 119 ubiquitination (Ub-H2A) and increased transcription-replication collisions at telomere breaks. However, in contrast with RAD51AP1 knockdown that yielded fewer TERRA R-loops, knockdown of PDS5a, BRG1 (or ZMYND8) produced more TERRA R-loops. Why this distinction? BRG1, PDS5a and ZMYND8 are linked with RNAPII pausing at DSBs and modulating chromatin to establish repressive chromatin (Figure 6). Thus, these factors could prevent the excessive formation of TERRA R-loops generated co-transcriptionally in cis. While ZMYND8 evidently acts in pausing RNAPII in response to DNA breaks (Gong et al., 2015), BRG1 and PDS5a could slide or evict nucleosomes (Bayona-Feliu et al., 2021; Phelan et al., 2000) or establish chromatin domains, respectively, which are conducive for ALT-HDR initiation, RAD51AP1-TERRA dependent strand invasion and D- or R-loop formation (Figure 6). Accordingly, we observed that BRG1, ZMYND8 and PDS5a each facilitate RAD51AP1 association with telomere DNA breaks.
As shown in this study, and in Yadav et al., RAD51AP1 has a pivotal role in manipulating TERRA during strand invasion and generating both R-loops and D-loops. We identified that SETX has a key role in ALT-HDR, by dismantling TERRA-intermediates that are generated by RAD51AP1. Intriguingly, we observed that RAD51AP1 deficiency abrogated repressive chromatin while, in contrast, SETX depletion enhanced the appearance of repressive chromatin at t-DSBS, which coincided with elevated RAD51AP1occupancy at those sites. These observations implicate TERRA R-loops, which are generated by and possibly stabilized via RAD51AP1 binding, as being intimately involved in maintaining repressive chromatin during the later steps of ALT-HDR (Figure 6). Elegant studies that made the connection between R-loops and reinforcing established repressive chromatin to maintain RNAPII pausing at transcription termination sites support this premise (Skourti-Stathaki et al., 2019; 2014). Analogously, we envision that RAD51AP1 binding to R-loops serves as a benchmark that “locks-in” and maintains repressive chromatin and paused RNAPII until key intermediates are established so that PCNA-RFC-Polδ mediated telomere DNA synthesis can efficiently begin without the interference of oncoming RNAPII or free TERRA (Figure 6). R-loops can later be safely resolved, and RNA removed from HR intermediates without provoking adverse effects like TRCs or premature restart of productive transcription by RNAPII. This is consistent with those crucial studies that established the paradigm of tight regulation of TERRA and TERRA R-loops being essential for efficient ALT-HDR (Arora et al., 2014; Silva et al., 2021).
The evidence presented here, and in Yadav et al., shows that RAD51AP1 binding to TERRA is crucial to generate HR intermediates and R-loops necessary for ALT. That RAD51AP1 acts with a network of factors and in a mechanism that modulates telomeric chromatin during ALT-HDR further exemplifies the crucial functions that are directed through this important protein. Further dissection of these activities and advances in protein structure determination could be harnessed to design molecules that target key interfaces of RAD51AP1 or disrupt its cognate interactions could be immensely useful parts of the toolkit being developed to kill ALT cancer cells.
Limitations of the study
RAD52 is a crucial factor in ALT, involved in D-loop formation and telomere DNA synthesis (Min et al., 2017; Verma et al., 2019; Zhang et al., 2019). RAD52 and RAD51AP1 can interact in ALT cells and RAD52 localization to telomeres is diminished in RAD51AP1 deficient ALT cells (Barroso-González et al., 2019). The precise functional relationship between these two proteins is unclear. Several studies have implicated RAD52 in RNA-templated HR and the processing of R-loops to facilitate HR (Keskin et al., 2014; Storici et al., 2007). RAD52 localizes to telomere breaks induced by reactive oxygen species (ROS) (Tan et al., 2020) and generates R-loops in vitro, although this activity appears to be extremely weak (Ouyang et al., 2021). The study by Yadav et al., shows that this interaction is not required to generate TERRA R-loops and that RAD51AP1 acts with TERRA predominantly in a RAD52-independent HDR mechanism during ALT. But RAD52 can catalyze RNA-DNA annealing and inverse strand exchange between ssRNA and dsDNA molecules (Mazina et al., 2017). Thus, it remains possible that RAD51AP1 and RAD52 might act in alternative transactions during ALT-HDR, that also involve TERRA. Those possibilities were not explored in this study and will require further investigation.
Lastly, this study examined the effects of telomere DNA breaks and endogenous telomere instability on TERRA and chromatin across entire chromosomes. Thus, it is unclear whether the TERRA that accumulates at telomere breaks emanates from the same chromosome (in cis) or from other chromosomes (in trans). Using systems such as those that permit overexpression of TERRA from individual sub-telomeric promoters or chimeric reporters (Feretzaki et al., 2020) might be useful in this regard.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Roderick J. O’Sullivan (osullivanr@upmc.edu)
Materials availability
Most reagents generated in this study are available upon request to the Lead Contact and upon signature of the corresponding Material Transfer Agreement, if necessary. RAD51AP1 antibodies were generated in the lab of Dr. Mauro Modesti and can be provided upon request.
Data and code availability.
Mass spectrometry data is deposited at proteomeXchange database and made available at the following accession number (PXD035943).
Original western blot and data files are available on Mendeley Data as of the date of publication. The DOI is listed in the Key resources table.
This study does not report any original code
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| RAD51AP1 | Mauro Modesti | #805 (Barroso-González et al., 2019; Modesti et al., 2007) |
| RAD51AP1 | Mauro Modesti | #806 (Barroso-González et al., 2019; Modesti et al., 2007) |
| RAD51 | Abcam | Cat. # ab133534; RRID:AB_272261 |
| TRF2 | Novus | Cat. #NB110–57130; RRID:AB_844199 |
| TRF1 | Jan Karlseder | N/A |
| FLAG (D6W5B) | Cell Signaling | Cat. # 14793S; RRID:AB_2572291 |
| FLAG (M2) | Millipore | Cat. # F1804; RRID:AB_262044 |
| γ-TUB | Sigma-Aldrich | Cat. # T6557; RRID:AB_477584 |
| PML (PG-M3) | Santa Cruz | Cat. # sc966; RRID:AB_628162 |
| Ubiquitin histone H2A (Lys119) | Millipore | Cat. # AB10029; RRID:AB_10617517 |
| c-Myc (9E10) | Abcam | Cat. # ab32; RRID:AB_303599 |
| RNAPII | Millipore | Cat. # 8WG16; RRID:AB_10013665 |
| PCNA | Abcam | Cat. # ab92552; RRID:AB_10561973 |
| S9.6 | Kerafast | Cat. # ENH001; RRID:AB_2687463 |
| BRG1 | Abcam | Cat. # ab4081; RRID:AB_304271 |
| PDS5a | ThermoFisher | Cat. # A300–089; RRID:AB_2162213 |
| ZMYND8 | ThermoFisher | Cat. # A302–090; RRID:AB_1604208 |
| SETX | ThermoFisher | Cat. # A301–104; RRID:AB_873128 |
| Critical commercial assays | ||
| ATMi (AZD0156) | Sellekchem | Cat. # S8375 |
| EZH2i (GSK126) | Sellekchem | Cat. # S7061 |
| 5,6-dichloro-1-β-D-ribofuranosyl-1H-benzimidazole (DRB) | Sigma | Cat. # D1916 |
| RO-3306 | Sigma-Aldrich | Cat. # SML0569 |
| Benzonase | Merck Millipore | Cat. # 70746 |
| 4-hydroxytamoxifen | Sigma | Cat. # H7904 |
| Doxycycline | Clontech | Cat. # 631311 |
| Shield Ligand | Clontech | Cat. # 632189 |
| Lipofectamine 2000 | Invitrogen | Cat. # 11668030 |
| Lipofectamine 3000 | Invitrogen | Cat. # L3000015 |
| Dharmafect Reagent 1 | Dharmacon | Cat. # T-2001 |
| Tel C PNA probe | PNA Bio | Cat. # F1004 |
| TERRA PNA Probe | IdT | (TAACCC)5-Alexa488 |
| ULTRA-hyb | ThermoFisher | Cat. # AM8663 |
| Duolink In Situ Detection Reagents | Sigma | Cat. # DUO-9214 |
| PDS5A targeting siRNA pool | Dharmacon | Cat. # L-014071 |
| BRG1 targeting siRNA pool | Dharmacon | Cat. # L-010431 |
| ZMYND8 targeting siRNA pool | Dharmacon | Cat. # L-017354 |
| Control non-targeting siRNA pool | Dharmacon | Cat. # D-001810 |
| RAD51AP1 targeting siRNA pool | Dharmacon | Cat. # L-017166 |
| SETX targeting siRNA pool | Dharmacon | Cat. # L-021420 |
| Deposited data | ||
| Proteomics Data | This study | ProteomeXchange Accession # PXD035943 |
| Raw data files | This study | Mendeley doi:10.17632/mbhnd2b9c7.1 |
| Experimental models: Cell lines | ||
| U2OS | ATCC | Cat# HTB-96; RRID:CVCL_0042 |
| HeLa LT | This study | N/A |
| GM847 | This study | N/A |
| LM216J | This study | N/A |
| U2OS TRF1-FokI WT/D450A | Roger Greenberg | (Dilley et al., 2016) |
| Oligonucleotides | ||
| Scramble antisense oligo (ASO) | IdT | cacgtctatacaccac (Chu et al., 2017) |
| TERRA ASO | IdT | taaccctaaccctaac (Chu et al., 2017) |
| RAD51AP15primeR | This study | ttcttatgtctcacaggccg |
| RAD51AP15primeF | This study | Gtgagacataagaaaccagtcaattactcaca gtt |
| RAD51AP13primerR | This study | caaaggtttaactcgtgcta |
| RAD51AP13primeF | This study | Gagttaaacctttgcatccaaatgccactagca cc |
| RAD51AP1 Isoform1Insert F | This study | tagtattccagctagtgcagtgccttgtacaaagatggctttagatgacaagctct |
| RAD51AP1 Isoform1Insert R | This study | ctagctggaatactaaaagtaccttcagggagcctttttttaggggttttctcttgt |
| Oligo-T | This study | Cy5-ttagggttagggttagggttagggttagggttagggttagg |
| Oligo-R-T | This study | cattgcatatttaaaacatgttggatctcaatttctattttatttttattttttatttctttattttaccatgaattcaaatgacctcttatcaagtgac |
| Oligo-R-B | This study | gtcacttgataagaggtcatttgaattcatcctaaccctaaccctaaccctaaccctaaccctaaccctaagagatccaacatgttttaaatatgcaatg |
| Oligo-D | This study | Cy5aaatcaatctaaagtatatatgagtaaacttggtctgacagttaccaatgcttaatcagtgaggcactatctcagcgatctgtctattt |
| TERRA RNA | This study | Cy5uuaggguuaggguuaggguuaggguuaggguuaggguuag |
| RNA-1 | This study | cugacaguuaccaaugcuuaaucagugagg |
| RNA-2 | This study | ccucacugauuaagcauugguaacugucag |
| RNA-3 | This study | uuauggcagcacugcauaauucucuuacug |
| RNA-4 | This study | gacggggagucaggcaacuauggaugaacg |
| RNA-5 | This study | cguucauccauaguugccugacuccccguc |
| RNA-6 | This study | aaaacuucauuuuuaauuuaaaaggaucua |
| RNA-7 | This study | uagauccuuuuaaauuaaaaaugaaguuuu |
| Recombinant DNA | ||
| BirA | O’Sullivan Lab | (García-Expósito et al., 2016) |
| BirA-TRF1 | O’Sullivan Lab | (García-Expósito et al., 2016) |
| BirA-RAD51AP1 | O’Sullivan Lab | This study |
| FLAG-RAD51AP1 K6K7 | O’Sullivan Lab | This study |
| GFP-RNaseH1-D210N | Lee Zou | (Nguyen et al., 2017) |
| GFP-RAD51AP1 | Mauro Modesti | (Modesti et al., 2007) |
| GFP-PDS5A | Jan Michael Peters | (Ladurner et al., 2016) |
| GFP-PDS5B | Jan Michael Peters | (Ladurner et al., 2016) |
| GFP-STAG1 | Jan Michael Peters | (Holzmann et al., 2019) |
| GFP-ZMYND8 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65401; RRID: Addgene_65401) |
| GFP-BRG1 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 6539; RRID: Addgene_65391 |
| GFP-PBRM1 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65387; RRID: Addgene_65387 |
| GFP-KDM5A | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65387; RRID: Addgene_65387 |
| GFP-NELF-E | Nabieh Ayoub | (Awwad et al., 2017) |
| GFP-SETX | Stephen West | (Yüce and West, 2013) |
| FLAG-RAD51AP1 | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP1 SIM | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP1 K269R | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP11 ΔRAD51 | O’Sullivan Lab | (Barroso-González et al.,2019) |
| MBP-RAD51AP1-His | Sung lab | (Dunlop et al., 2012) |
| MBP-RAD51AP1 K6K7-His | Sung lab | (Dunlop et al., 2012) |
| RAD51 | Eric Greene Lab | (Xue et al., 2021) |
Experimental Model and Subject Details
U2OS cell lines were obtained, authenticated by STR profiling, and confirmed mycoplasma free by ATCC cell line authentication services. WT/DA TRF1-FokI U2OS cell lines and the LM216J cell line were generously provided by Roger Greenberg (University of Pennsylvania). Myc-RAD51AP1 U2OS cell lines were described previously (Barroso-González et al., 2019). Each cell line was cultured in Glutamax-DMEM supplemented with 10% bovine growth serum. Cells were cultured at normal oxygen conditions of 20% O2 and 7.5% CO2 at 37°C.
METHOD DETAILS
Purification of recombinant proteins.
MBP-WT RAD51AP1-His6 or RAD51AP1 K7/K6 mutant protein was expressed in the E. coli BL21 cells (Novagen) transformed with pMAL-C5-RAD51AP1-His6 or pMAL-C5-RAD51AP1-K7/K6 with 0.1mM IPTG for 1hr at 16°C. The cells were harvested by centrifugation, and stored at −80°C. All purification steps were carried out at 0°C to 4°C. 20g of cells from 2L culture was resuspended in 150mL of CBB-1 buffer (50 mM Tris-HCl, pH 7.5, 10% sucrose, 10mM EDTA, 600mM NaCl, 1mM TCEP, 0.01% Igepal CA-630 (Sigma), Protease inhibitor mixture (aprotinin, chymostatin, leupeptin, and pepstatin A, 3μg/ml each, 1 mM PMSF) and lysed by sonication. The cell lysate was clarified by ultracentrifugation at 100K x g for 45min at 4°C. Clarified cell lysate was incubated at for 1h with 2ml of amylose resin (NEB). The affinity resin was washed with 100ml TN buffer (25mM Tris-HCl pH7.5, 10% glycerol, 1mM TCEP) + 1M NaCl, 20ml of TN buffer + 1M NaCl and 2mM ATP/MgCl2, followed by 10ml of TN buffer + 150mM NaCl. Bound proteins were eluted with 3ml fraction each of TN buffer + 15mM NaCl and 20mM maltose. Fractions containing RAD51AP1 protein were pooled and further fractionated with a 7ml Source 15S column (Cytiva) using a 140ml gradient of 100 –1000mM NaCl in TN buffer. Peak fractions were pooled and concentrated using an Amicon Ultra-15 centrifugal filter (EMD Millipore). The protein was resolved with Superdex 200 (10/30) (Cytiva) equilibrated with K-150 buffer (20mM KH2PO4, pH7.4, 150mM KCl, 0.5mM EDTA, 1mM TCEP, 10% glycerol, 0.01% Igepal). Peak fractions were pooled, concentrated, aliquoted, and stored at −80°C.
His6-Smt3-RAD51 protein was expressed in E coli Rosetta cells transformed with pET-Duethis-smt3-Rad51 (Xue et al., 2021) with 0.5 mM IPTG at 16°C for 16hr. The cells were harvested by centrifugation, and stored at −80°C. All purification steps were performed at 4°C. 2g of cells from 2L culture were resuspended in 150mL of CBB-2 buffer (50 mM Tris -HCl, pH 7.5, 10% sucrose, 10mM EDTA, 600mM NaCl, 1mM DTT, 0.01% Igepal CA-630, Protease inhibitor mixture), and lysed by sonication. The cell lysate was clarified by ultracentrifugation at 100K x g for 45min at 4°C and treated with 45% (final concentration) ammonium sulfate over 1hr. The precipitate was collected by centrifugation at 18,00g for 20min and stored at −80°C. The precipitate was dissolved in 50ml of T buffer (25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5mM EDTA, 1mM DTT, 0.01%, Igepal CA-630) + 500mM KCl and incubated with 2ml Ni-NTA agarose (Qiagen) for 14hr. The mixture was poured into a column and then washed with 20ml T buffer + 500mM KCl followed by 30ml wash with T buffer + 300mM KCl. The Ni-NTA resin was incubated with 100 ng/ml Ulp1 for 3hr to cleave off His6-Smt3 and release untagged form of RAD51. The eluate was applied to an 8ml Macro hydroxyapatite column (Bio-Rad) with a 90ml gradient of 0–300 mM KH2PO4 in T buffer. The fractions containing RAD51 were identified by SDS-PAGE and Coomassie blue staining. The pooled RAD51 fractions were diluted with an equal volume of T buffer and further fractionated in 1mL Mono Q (Cytiva) with a 30mL gradient of 100–400 mM KCl in T buffer. RAD51 fractions were pooled, concentrated with an Amicon Ultra-15 centrifugal filter, aliquoted, and stored at −80°C.
DNA substrates.
All RNA or DNA oligonucleotides (see Key Resource Table) were obtained from IDT. The Rloop was prepared by annealing of three oligonucleotides (TERRA-RNA, Oligo-R-T, and OligoR-B) at an equal molar ratio in 50mM Tris-HCl, pH 7.5, 10mM MgCl2, 100mM NaCl by heating at 90°C and slow cooling to 4°C.
DNA/RNA Mobility Shift Assay.
In a 10μl reaction, the indicated nucleic acid substrate (10nM TERRA or Oligo T (Figure 1C), 2nM TERRA R-loop (Figure 1D) was incubated for 5min at 37 °C with RAD51AP1 or the mutant in RB buffer (35mM Tris-HCl, pH7.5, 1mM MgCl2, 6% glycerol, 100ng/μl BSA, 60mM KCl). Figure 1C: RAD51AP1 25, 50, 75, 100nM; Figure 1D: RAD51AP1 WT (20, 40, 60, 80, 120, 240nM) or RAD51AP1 K6K7 (80, 120, 240nM) were tested. The reaction mixtures were run in 6% polyacrylamide gels in TB buffer (90mM Tris-borate, pH 8.3). The gels were imaged with ChemiDoc imaging system (Biorad) and analyzed by Image Lab software.
D-loop & R-loop assays.
In a 12.5μl reaction, 2nM Cy5-labeled 90-mer ssDNA (Oligo-D, 2.4 μM nucleotides) was incubated with 0.8μM RAD51 in RD buffer (35mM Tris-HCl, pH7.5, 1mM MgCl2, 2mM ATP, 100ng/μl BSA) for 10min at 37°C. 400nM RAD51AP1 was added, with or without 27nM RNA (RNA 1 to RNA7) followed by a 5min incubation at 37°C, and then an addition of 12nM pBluescript (3μM base pairs) to initiate strand invasion reaction. Figure S1B: 112, 160, 240, 320nM RAD51AP1 were tested. For R-loop or D-loop reactions in Figure S1C, 10nM Cy5TERRA (or Oligo-T) was incubated with 200nM RAD51 in RD2 buffer (35mM Tris-HCl, pH7.5, 1mM MgCl2, 1mM CaCl2, 1mM AMP-PNP, 100ng/μl BSA) and RAD51AP1 (100, 200, 400nM) was tested. R-loop or D-loop reaction was initiated by adding 4.8nM pBluescript derived plasmid harboring 103bp TTAGGG repeats (Bower and Griffith, 2014). The reaction mixtures were incubated for 10 min at 37°C and the reaction was terminated by adding 1μl each of 5% SDS, 1 mg/ml proteinase K, and a 5min incubation at 37°C. The reaction products were resolved by electrophoresis in 0.9% agarose gels in TAE buffer (40mM Tris–acetate, pH 7.5 0.5mM EDTA). The gels were imaged with ChemiDoc imaging system (Biorad) and analyzed with Image Lab software.
See Key Resource Table for the sequences of oligonucleotides used in these experiments.
Direct Immunofluorescence (IF).
Cells on glass coverslips were washed twice in PBS and fixed with 2% paraformaldehyde (PFA) for 10mins. Cells were permeabilized with 0.1% (w/v) sodium citrate and 0.1 % (v/v) Triton X100 for 5mins and incubated with fresh blocking solution (1mg/mL BSA, 10% normal goat serum, 0.1% Tween) for 30mins. Primary antibodies were diluted in blocking solution and added to cells for 1hr at RT or overnight in refrigerated conditions. Next, cells were washed three times with PBS for 5mins and incubated with Alexa coupled secondary antibodies (488nm, 555nm, 647nm) (Life Technologies) for 1hr at RT. Then, cells were washed three times with PBS and mounted on slides with Prolong Gold Anti-fade reagent with DAPI (Life Technologies). Once the Prolong Anti-fade polymerized and cured, cells were visualized by conventional fluorescence with 40X and 63X Plan λ objective (1.4 oil) using a Nikon 90I.
IF-FISH.
After secondary antibody, cells were washed, and then the IF staining was fixed with 2% paraformaldehyde (PFA) for 10mins. PFA was washed off with PBS and coverslips dehydrated with successive washes in 70%, 95%, and 100% EtOH for 3mins, allowed to air-dry completely. Next, the coverslips were mounted on glass slides with 15ml per coverslip of hybridization mix (70% deionized Formamide, 1mg/ml of Blocking Reagent [Roche], 10mM Tris-HCl pH 7.4) containing Alexa 488-(CCCTAA)4 PNA probe. DNA was denatured by setting the slides on a heating block set to 72°C for 10mins and then incubating overnight at RT in the dark. The coverslips were then washed twice for 15mins with Wash Solution A (70% deionized formamide and 10mM Tris-HCl pH7.2) and three-time with Solution B (0.1M Tris-HCl pH7.2, 0.15M NaCl, and 0.08% Tween) for 5mins at RT. EtOH dehydration was repeated as above, and finally, the samples were mounted and analyzed as mentioned above.
Proximity Ligation Assay
PLA was performed as previously described (Hamperl et al., 2017). Briefly, cells on glass coverslips were washed twice in PBS and fixed with 2% paraformaldehyde (PFA) for 10mins. Cells were permeabilized with 0.1% (w/v) sodium citrate and 0.1 % (v/v) Triton X-100 for 5mins and blocked for 1hr at 37°C with blocking solution (Sigma). The coverslips were incubated in primary antibody overnight at 4°C (1:1000 mouse RNAPII, 1:8000 rabbit PCNA). Cells were incubated in a pre-mixed solution of PLA probe anti-mouse minus and PLA probe anti-rabbit plus (Sigma) for 1hr at 37°C. According to the manufacturer's instructions, the Duolink In Situ Detection Reagents (Sigma) was used to perform the PLA reaction. Then, cells were washed three times with PBS, and the samples were mounted and analyzed as mentioned above.
RNA-FISH.
RNA-FISH was conducted as described (Azzalin et al., 2007). After secondary antibody, cells were washed, and then the IF staining was fixed with 2% paraformaldehyde (PFA) for 10mins. Coverslips were dehydrated in 70%, 95%, and 100% EtOH for 3mins, allowed to air dry completely. Next, the coverslips were mounted on glass slides with 40μl per coverslip of hybridization mix (50% deionized Formamide, 2X SSC, 2mg/mL BSA, 10% Dextran Sulfate) containing Alexa 488-(TAACCC)7 PNA probe. Glass slides were incubated in a hybridization chamber at 40°C overnight. The coverslips were then washed three times for 5mins with Wash Buffer 1 (2X SSC, 50% deionized formamide) and three times for 5mins with Wash Buffer 2 (2X SSC) at 40°C. EtOH dehydration was repeated as above, and finally, the samples were mounted and analyzed as mentioned above.
Telomere DNA Synthesis detection by EdU.
Following siRNA knockdown, TRF1-FokI U2OS cells were induced with doxycycline (40ng/μL1) and synchronized in G2 with RO-3306 (Sigma) (10μM) 24hrs before harvest, followed by tamoxifen (4-OHT, Sigma) (1μM) and Shield1 Ligand (Takara Clontech) (1μM) 3hrs before harvest. Cells were pulsed with EdU (10μM) 1hr before harvest. Cells on glass coverslips were washed twice in PBS and fixed with 2% paraformaldehyde (PFA) for 10mins. Cells were permeabilized with 0.1% (w/v) sodium citrate and 0.1 % (v/v) Triton X-100 for 5mins. The Click-IT Plus EdU Cell Proliferation Kit with Alexa Flour 488 (Invitrogen) was used to detect EdU.
DNA-RNA Immunoprecipitation (DRIP).
DRIP was conducted as described (Feretzaki et al., 2020). RAD51AP1 siRNAs were transfected twice (0 and 3 days) in (WT)- or (D450A)- TRF1-FokI cells. Cells were induced with doxycycline (40ng μL-1) 24hrs before harvest at day 6, followed by tamoxifen (1μM) and SHIELD (1μM) 3hrs before harvest. Approximately 107 cells per condition were harvested by trypsinization and placed on ice. Cells were washed with 1X PBS and centrifuged (500G, 5mins, 4°C). Cells were dissolved in 200μL cold RLN buffer (50mM Tris-HCl pH 8.0, 140mM NaCl, 1.5mM MgCl2, 0.5% NP-40, 1mM dithiothreitol (DTT), and 100 U ml−1 RNasIN PLUS) on ice for 5mins. Cells were centrifuged (500G, 5mins, 4°C), and the liquid was carefully discarded. Cell pellets were lysed with 500μL RLT lysis buffer (RNAeasy Plus, Qiagen) and homogenized through a 20G × 11/2 syringe (0.9 mm × 40 mm). The homogenized extracts were mixed with 250μL H2O and 750μL ultra-pure phenol:chloroform: isoamyl alcohol (25:24:1). The samples were transferred to a heavy phase-lock tube and centrifuged (13,000G, 5mins). The top aqueous phase was poured into a tube containing 750μL ice-cold propanol and 50mM NaCl. The samples were inverted until nucleic acids precipitated. The samples were centrifuged (13,000G, 30mins, 4°C) to form nucleic acid pellets. The pellets were washed twice with 70% ice-cold EtOH and air-dried. The pellets were dissolved in 130μL of H2O and sonicated on a Covaris system (Peak power- 50, Duty Factor- 20, Cycles/Burst- 200, 150sec) to obtain fragments of approximately 200bp. 30μg of nucleic acids was incubated with RNaseH or H2O in RNaseH buffer (New England Biosystems) at 37°C overnight. Protein G beads (Dynabeads) were washed three times in DIP1 Buffer (10mM HEPES-KOH pH 7.5, 275 mM NaCl, 0.1% Na-deoxycholate, 0.1% SDS, 1% Triton X-100). Samples were diluted by ten in DIP1 Buffer, and 40μL of Protein G beads were added for 1 hour of pre-clearing rotating at 4°C. One percent of the sample was saved as input. Samples were transferred to a new tube, and 40μL of Protein G beads with 3μg of S9.6 antibody were added to each sample and incubated overnight, rotating at 4°C. Next, beads were washed with DIP2 Buffer (50mM HEPES-KOH pH 7.5, 140mM NaCl, 1mM EDTA pH 8.0, 1% Triton X-100, 0.1% Na-deoxycholate), DIP3 Buffer (50mM HEPES-KOH pH 7.5, 500mM NaCl, 1mM EDTA pH 8.0, 1% Triton-X100, 0.1% Na-deoxycholate), and DIP4 Buffer (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0, 250mM LiCl, 1% NP-40, 1% Na-deoxycholate). The sample was eluted in 100μL of Elution Buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 0.5% SDS, Proteinase K) shaking at 55°C for 30mins twice. DNA was purified (PCR cleanup, Machery-Nagel) and eluted in 100μL H2O. Input (0.5%) and IP samples were dot-blotted onto a positively charged nylon membrane for detection. Samples were denatured (0.5M NaOH and 1.5M NaCl) for 15mins and neutralized (0.5M Tris-HCL pH7.5 and 1M NaCl) for 10mins. Samples were cross-linked to the membrane (UV Stratalinker 2400, Stratagene) The membrane was prehybridized in a hybridization buffer (Ultrahyb Ultra-sensitive Hybridization Buffer, Invitrogen) rotating at 55°C for 1hr. The membrane was hybridized with 32P-labeled (AATCCC)4 oligonucleotides at 55°C overnight. The next day, the membrane was washed three times in 2 × SSC buffer and once in 2x SSC 0.5% SDS, exposed onto a storage phosphor screen, and scanned using Typhoon 9400 PhosphoImager (GE Healthcare). Sample intensity was measured with Fiji.
RNA Immunoprecipitation (RIP).
RIP was performed as described (Feretzaki et al., 2020). Approximately 5 x 107 U2OS cells per condition were harvested by trypsinization and placed on ice. Cells were washed with 1X PBS and centrifuged (500G, 5mins, 4°C). Cells were dissolved in 500μL cold RLN buffer (50mM Tris-HCl pH 8.0, 140mM NaCl, 1.5mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol (DTT), and 100 U ml−1 RNasIN PLUS), mixed vigorously, and left to rotate at 4°C for 20mins. Cells were centrifuged (13,000G, 10mins, 4°C). The liquid was transferred to a new tube, and the cell pellet was discarded. Inputs (10%) were taken from each sample and stored overnight at 4°C. The samples were then incubated with 6μg of antibody overnight, rotating at 4°C. The beads (Protein A/G, Dynabeads) were washed three times. Beads were precleared with yeast tRNA (Invitrogen) rotating for 1hr at 4°C. The beads were washed with RLN buffer then 35μL of beads was added to the samples containing antibodies. The samples were incubated with the beads for 2hrs rotating at 4°C. The beads were washed 4 times with RLN wash buffer (50mM Tris-HCl pH 8.0, 140mM NaCl, 1.5mM MgCl2, 1% NP-40, 1mM dithiothreitol (DTT), 100 U ml−1 RNasIN PLUS, and 6mM EDTA). Samples were eluted twice with 100μL of elution buffer (5mM EDTA, 1% SDS, and 10% β-mercaptoethanol) at 42°C shaking at 1500RPM for 30min. Elutions were pooled (200μL total), the RNA was isolated (RNeasy Plus, Qiagen), and eluted in 100μL H2O. Half of each sample (50μL) was digested with an RNase cocktail (Invitrogen) at 37oC for 3hrs. TERRA immunoprecipitation was detected using the dot-blot method (mentioned above) and 32P-labeled (TAACCC)7 oligonucleotides. For RNA-IP of myc-tagged RAD51AP1, ~5 x 107 U2OS cells stably expressing myc-tagged RAD51AP1 were harvested by trypsinization and placed on ice. The protocol above was followed with 35μL of myc-TRAP beads (Chromotek) added to each sample and incubated overnight, rotating at 4°C. Samples were not precleared with myc beads.
TERRA Northern Blot.
Approximately 5 x 107 U2OS cells per condition were harvested by trypsinization and placed on ice. Cells were washed with 1X PBS and centrifuged (1500 G, 5mins, 4°C). RNA from samples were isolated including an on column DNaseI digest (RNeasy Plus, Qiagen). 0.5μg of RNA was incubated with an RNase cocktail (Invitrogen) or H2O at 37°C for 3hrs. Samples were denatured with denaturation solution (50% Formamide, 15% Formaldehyde, and 0.5M MOPs Buffer) at 65 for 15mins. TERRA RNA was detected using the dot-blot method (mentioned above) and 32P-labeled (TAACCC)7 oligonucleotides.
Western Blotting.
Cells were harvested with trypsin, quickly washed in PBS, counted with Cellometer Auto T4 (Nexcelom Bioscience), and directly lysed in 4X NuPage LDS sample buffer at 10000 cells per μl. Proteins were gently homogenized using a Benzonase (ThermoFisher), denatured for 10mins at 70°C, and resolved by SDS-Page electrophoresis, transferred to nitrocellulose membranes, blocked in 5% milk in TBST for 30mins and probed. HRP-linked anti-rabbit or mouse (Amersham) was used for secondary antibodies, and the HRP signal was visualized with SuperSignal ECL substrate (Pierce) as per the manufacturer's instructions.
siRNA Transfections.
The following siRNA Smartpools from Dharmacon (GE) were used (see Key Resource Table). For complementation studies of RAD51AP1, the siRNA targeting the 3’UTR of RAD51AP1 was used. Briefly, 200,000 and 700,000 cells were seeded per well of a 6-well plate and 10cm dish containing growth medium without antibiotics. ~6hrs later, cells were transfected. siRNAs and Dharmafect were diluted in OptiMEM (Life Technologies). A working siRNA concentration of 50nM was used. We used 2.5μL and 10μL Dharmafect transfection reagent per well and 10cm plate. Transfection medium was replaced with complete culture media 24hrs later, or cells were split for the desired application and harvested at 72hrs post-transfection.
Anti-sense oligo (ASO) transfection.
TERRA knockdown by ASO was conducted as described (Chu et al., 2017). Approximately 150,000 cells were seeded per well of a 6-well plate containing 2mL of growth medium without antibiotics. ~6hrs later, cells were transfected with 6pmol of scrambled or anti-TERRA ASO, and 4μL of RNAiMax (ThermoFisher) diluted in OptiMEM (Life Technologies). Transfection medium was replaced with complete culture media 24hrs later, and cells were harvested at 72hrs post-transfection.
Bio-ID and mass spectrometry.
U2OS stably expressing myc-BirA alone, myc-BirA-TRF1 or myc-BirA-RAD51AP1 were generated by retroviral infection with particles generated from amphotropic 293T viral packaging cell lines. Infected cells were selected using puromycin (2μg/mL) for 5–7 days, and stable protein expression was validated by western blot and immunofluorescence. Proximitydependent biotinylation and streptavidin capture of biotinylated proteins was performed as described. Immunoprecipitated samples were separated ~1.5cm on a 10% Bis-Tris Novex minigel (Invitrogen) using the MES buffer system. The gel was stained with Coomassie, and each lane was excised into ten equally sized segments. Gel pieces were processed using a robot (ProGest, DigiLab) as follows: First washes were with 25mM ammonium bicarbonate followed by acetonitrile. Then, reduced with 10mM dithiothreitol at 60°C followed by alkylation with 50mM iodoacetamide at RT. Samples were digested with trypsin (Promega) at 37°C for 4hrs and then quenched with formic acid. Samples supernatants were analyzed directly without further processing using a nano-LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Q Exactive. Peptides were loaded on a trapping column and eluted over a 75mm analytical column at 350nL/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with MS and MS/MS performed in the Orbitrap at 70,000 FWHM resolution and 17,500 FWHM resolution, respectively. The fifteen most abundant ions were selected for MS/MS.
Proteomics Analysis.
Raw spectral files were converted to mzML format using Proteowizard. The mzML files were searched using X!Tandem search engine (Craig and Beavis, 2004). The searches were performed using the following parameters: trypsin digestion, allowing up to two missed tryptic cleavages, parent ion tolerance window of 10 ppm, and fragment ion tolerance of 0.02 Da. Oxidation of Methionine (+15.9949@M) was specified as variable modification and carbamidomethylation of Cysteine (+57.0215@C) as fixed. All protein searches were performed using the Human UniProt protein database appended with common contaminants and reversed protein sequences to serve as decoys. The X!Tandem results were then post-processed with PeptideProphet (Keller et al., 2002)and ProteinProphet (Nesvizhskii et al., 2003). The data was filtered to 1% protein-level FDR using the target decoy strategy (PMID: 20816881). Protein abundance in each sample was estimated using spectral counts. Spectral counts were extracted from the search results for all reported proteins across all 12 experiments (4 replicates for each of TRF1 and RAD51AP1, and the negative controls experiments) using Abacus (Fermin et al., 2011). The resulting table of spectral counts was processed using SAINT (run using reprint-apms.org resource) to calculate fold change (FC) scores and probabilities of true interactions using default settings (Choi et al., 2011; Mellacheruvu et al., 2013). Using spectral counts as measures of protein abundance in each sample, SAINT calculates the probability for each identified protein that it is a true interactor of the bait protein. In doing so, IP replicates of cells expressing only the empty vector were used as negative controls. Proteins with a SAINT probability of >=0.7 for either or both bait proteins were kept as potential interacting proteins. Data are available via ProteomeXchange with identifier PXD035943.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data in this study were analyzed in GraphPad Prism, ImageJ, and Microsoft Excel. Detection, colocalization, and quantification of cells were performed using the ComDet v.0.5.3 plugin for ImageJ. Statistical tests used are indicated in the figure legend accompanying each figure. In most cases, one-way analysis of variance (ANOVA) was used. Typically, unless otherwise stated n refers to the # of independent experiments and SEM refers to the standard error of means. The sample size was not pre-determined.
Supplementary Material
TABLE S1. List of proteins found in Bio ID of BirA, BirA-TRFl and BirA-RAD51AP1. Related to Figure 2.
Table lists the protein ID, fold change average (FCA) over BirA (CTRL) alone, SAINT probability (SP) and average spectral counts (SpC). Uniprot information is also provided.
Highlights.
RAD51AP1 and TERRA interact and stimulate the formation of D and R Loops
TERRA R-loops generated by RAD51AP1 regulate repressive chromatin at telomeres
RAD51AP1 protects telomeric regions from transcription-replication collisions
SUMMARY SENTENCE.
RAD51AP1 binds to TERRA to generate R-loops and HR intermediates while simultaneously using R-loops as platforms to maintain chromatin suppression of TERRA that prevents transcription-replication collisions during ALT-HDR.
ACKNOWLEDGMENTS
We are indebted to Joachim Lingner (EPFL, Lausanne) for sharing detailed RNA/RNA-DNA immunoprecipitations protocols. We thank members of the Bakkenist lab for sharing reagents and equipment. We thank Nicolas Paquet (UTHSA) for RAD51AP1 preparation. We thank Stephen West (Crick Institute, London), Ana Losada (CNIO, Madrid), Nabieh Ayoub (Technion, Israel), Eric Greene (Columbia University) and Jan-Michael Peters (IMP, Vienna) for sharing plasmids. We thank Lee Zou (Harvard) for sharing reagents and communicating data before publication. We thank Dr. Richard Jones at MS Bioworks (Michigan) for guidance on proteomics. N.K. is a recipient of the Ruth Kirschstein Predoctoral Individual National Research Service Award F31/#CA264885. M.L.L and A.R.W. are recipients of the John S. Lazo Cancer Pharmacology Fellowship from the Department of Pharmacology & Chemical Biology, University of Pittsburgh School of Medicine. R.B is a recipient of the Hillman Postdoctoral Fellowship for Innovative Cancer Research. The UPMC Hillman Cancer Center facilities were supported by Comprehensive Cancer Center Support Grant NCI/#P30CA047904. Research funding was provided to investigators from the following agencies: R.J.O.; NCI/# R01CA207209, R01CA262316, R37CA263622, and American Cancer Society #RSG-18–038-01-DMC; S.C.W.; S10OD019973, P.S.; R35CA241801 and RO1ES007061, A.I.N.: GM094231. H.Z; U01CA260851. M.M. is a recipient of funding from the French National League against Cancer. K.M.M is a recipient of funding from Cancer Prevention and Research Institute of Texas #RP220330.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATIONS OF INTERESTS
The authors have no conflicts of interest to declare.
REFERENCES
- Abu-Zhayia ER, Awwad SW, Ben-Oz BM, Khoury-Haddad H, Ayoub N, 2018. CDYL1 fosters double-strand break-induced transcription silencing and promotes homology-directed repair. J Mol Cell Biol 10, 341–357. doi: 10.1093/jmcb/mjx050 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gaillard H, 2014. Transcription and recombination: when RNA meets DNA. Cold Spring Harb Perspect Biol 6, a016543. doi: 10.1101/cshperspect.a016543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask MBD, Streicher W, Jungmichel S, Nielsen ML, Lukas J, 2015. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6, 8088–12. doi: 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Brun CM, Azzalin CM, 2012. Transcription regulates telomere dynamics in human cancer cells. RNA 18, 684–693. doi: 10.1261/rna.029587.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM, 2014. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5, 5220–11. doi: 10.1038/ncomms6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awwad SW, Abu-Zhayia ER, Guttmann-Raviv N, Ayoub N, 2017. NELF-E is recruited to DNA double-strand break sites to promote transcriptional repression and repair. EMBO Rep 18, 745–764. doi: 10.15252/embr.201643191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin ÖZ, Marteijn JA, Ribeiro-Silva C, Rodríguez López A, Wijgers N, Smeenk G, van Attikum H, Poot RA, Vermeulen W, Lans H, 2014. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res 42, 8473–8485. doi: 10.1093/nar/gku565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, Ginalski K, Rowicka M, Fraser P, Legube G, 2017. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol 24, 353–361. doi: 10.1038/nsmb.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J, 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. doi: 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Barroso-González J, García-Expósito L, Galaviz P, Lynskey ML, Allen JAM, Hoang S, Watkins SC, Pickett HA, O'Sullivan RJ, 2021. Anti-recombination function of MutSα restricts telomere extension by ALT-associated homology-directed repair. Cell Rep 37, 110088. doi: 10.1016/j.celrep.2021.110088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-González J, García-Expósito L, Hoang SM, Lynskey ML, Roncaioli JL, Ghosh A, Wallace CT, Modesti M, Bernstein KA, Sarkar SN, Watkins SC, O'Sullivan RJ, 2019. RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol. Cell 76, 11–26.e7. doi: 10.1016/j.molcel.2019.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Feliu A, Barroso S, Muñoz S, Aguilera A, 2021. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet 1–14. doi: 10.1038/s41588-021-00867-2 [DOI] [PubMed]
- Bower BD, Griffith JD, 2014. TRF1 and TRF2 differentially modulate Rad51-mediated telomeric and nontelomeric displacement loop formation in vitro. Biochemistry 53, 5485–5495. doi: 10.1021/bi5006249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee PM, Chambers AL, Cloney R, Bianchi A, Downs JA, 2014. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell Rep 6, 973–981. doi: 10.1016/j.celrep.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon RM, Morel D, Eychenne T, Colmet-Daage L, Bajrami I, Dorvault N, Garrido M, Meisenberg C, Lamb A, Ngo C, Hopkins SR, Roumeliotis TI, Jouny S, Hénon C, Kawai-Kawachi A, Astier C, Konde A, Del Nery E, Massard C, Pettitt SJ, Margueron R, Choudhary JS, Almouzni G, Soria J-C, Deutsch E, Downs JA, Lord CJ, Postel-Vinay S, 2021. PBRM1 Deficiency Confers Synthetic Lethality to DNA Repair Inhibitors in Cancer. Cancer Res 81, 2888–2902. doi: 10.1158/0008-5472.CAN-21-0628 [DOI] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA, 2014. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 159, 108–121. doi: 10.1016/j.cell.2014.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, Nesvizhskii AI, 2011. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73. doi: 10.1038/nmeth.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H-P, Cifuentes-Rojas C, Kesner B, Aeby E, Lee H-G, Wei C, Oh HJ, Boukhali M, Haas W, Lee JT, 2017. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 170, 86–101.e16. doi: 10.1016/j.cell.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, Mitson M, Taylor S, Higgs DR, Gibbons RJ, 2015. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun 6, 7538–11. doi: 10.1038/ncomms8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR, 2007. Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853. doi: 10.1126/science.1138596 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC, 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. doi: 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA, 2016. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58. doi: 10.1038/nature20099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai M-S, Sy SM-H, Schild D, Egelman E, Chen J, Sung P, 2010. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol 17, 1255–1259. doi: 10.1038/nsmb.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai M-S, Leung SG, Schild D, Wiese C, Sung P, 2012. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J. Biol. Chem 287, 12343–12347. doi: 10.1074/jbc.C112.352161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J, 2020. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587, 303–308. doi: 10.1038/s41586-020-2815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermin D, Basrur V, Yocum AK, Nesvizhskii AI, 2011. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics 11, 1340–1345. doi: 10.1002/pmic.201000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Expósito L, Bournique E, Bergoglio V, Bose A, Barroso-González J, Zhang S, Roncaioli JL, Lee M, Wallace CT, Watkins SC, Opresko PL, Hoffmann J-S, O'Sullivan RJ, 2016. Proteomic Profiling Reveals a Specific Role for Translesion DNA Polymerase η in the Alternative Lengthening of Telomeres. Cell Rep 17, 1858–1871. doi: 10.1016/j.celrep.2016.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD, 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol 11, 861–871. doi: 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisays F, Garzia A, Wang H, Canasto-Chibuque C, Hohl M, Savage SA, Tuschl T, Petrini JHJ. RTEL1 influences the abundance and localization of TERRA RNA. Nat Commun 2021. May 21;12(1):3016. doi: 10.1038/s41467-021-23299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Chiu L-Y, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM, 2015. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev 29, 197–211. doi: 10.1101/gad.252189.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM, 2017. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol 216, 1959–1974. doi: 10.1083/jcb.201611135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA, 2017. TranscriptionReplication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786.e19. doi: 10.1016/j.cell.2017.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2011. Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hatchi E, Goehring L, Landini S, Skourti-Stathaki K, DeConti DK, Abderazzaq FO, Banerjee P, Demers TM, Wang YE, Quackenbush J, Livingston DM, 2021. BRCA1 and RNAi factors promote repair mediated by small RNAs and PALB2-RAD52. Nature 591, 665–670. doi: 10.1038/s41586-020-03150-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SKN, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK, 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425–425. doi: 10.1126/science.1207313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang SM, Kaminski N, Bhargava R, Barroso-González J, Lynskey ML, GarcíaExpósito L, Roncaioli JL, Wondisford AR, Wallace CT, Watkins SC, James DI, Waddell ID, Ogilvie D, Smith KM, da Veiga Leprevost F, Mellacharevu D, Nesvizhskii AI, Li J, Ray-Gallet D, Sobol RW, Almouzni G, O'Sullivan RJ, 2020. Regulation of ALT-associated homology-directed repair by polyADP-ribosylation. Nat. Struct. Mol. Biol 27, 1152–1164. doi: 10.1038/s41594-020-0512-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Politi AZ, Nagasaka K, Hantsche-Grininger M, Walther N, Koch B, Fuchs J, Dürnberger G, Tang W, Ladurner R, Stocsits RR, Busslinger GA, Novák B, Mechtler K, Davidson IF, Ellenberg J, Peters J-M, 2019. Absolute quantification of cohesin, CTCF and their regulators in human cells. Elife 8. doi: 10.7554/eLife.46269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kakarougkas A, Downs JA, Jeggo PA, 2015. The PBAF chromatin remodeling complex represses transcription and promotes rapid repair at DNA double-strand breaks. Mol Cell Oncol 2, e970072. doi: 10.4161/23723548.2014.970072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, Löbrich M, Jeggo PA, Downs JA, 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 55, 723–732. doi: 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R, 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74, 5383–5392. doi: 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F, 2014. Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439. doi: 10.1038/nature13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang C-M, Xhemalce B, Paull TT, Brodbelt JS, Marcotte EM, Miller KM, 2019. Systematic bromodomain protein screens identify homologous recombination and Rloop suppression pathways involved in genome integrity. Genes Dev 33, 1751–1774. doi: 10.1101/gad.331231.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM, 1997. A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res 25, 4946–4953. doi: 10.1093/nar/25.24.4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner R, Kreidl E, Ivanov MP, Ekker H, Idarraga-Amado MH, Busslinger GA, Wutz G, Cisneros DA, Peters J-M, 2016. Sororin actively maintains sister chromatid cohesion. EMBO J 35, 635–653. doi: 10.15252/embj.201592532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S-I, Yasui A, 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 40, 976–987. doi: 10.1016/j.molcel.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Li F, Deng Z, Zhang L, Wu C, Jin Y, Hwang I, Vladimirova O, Xu L, Yang L, Lu B, Dheekollu J, Li J-Y, Feng H, Hu J, Vakoc CR, Ying H, Paik J, Lieberman PM, Zheng H, 2019. ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalization. EMBO J 38, e96659. doi: 10.15252/embj.201796659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Longerich S, Miller AS, Tang C, Buzovetsky O, Xiong Y, Maranon DG, Wiese C, Kupfer GM, Sung P, 2016. Promotion of RAD51-Mediated Homologous DNA Pairing by the RAD51AP1-UAF1 Complex. Cell Rep 15, 2118–2126. doi: 10.1016/j.celrep.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Miller AS, Longerich S, Tang C, Maranon D, Williamson EA, Hromas R, Wiese C, Kupfer GM, Sung P, 2019. DNA requirement in FANCD2 deubiquitination by USP1-UAF1-RAD51AP1 in the Fanconi anemia DNA damage response. Nat Commun 10, 2849–8. doi: 10.1038/s41467-019-10408-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Miller AS, Tang C, Maranon D, Williamson EA, Hromas R, Wiese C, Zhao W, Sung P, Kupfer GM, 2020. The DNA-binding activity of USP1-associated factor 1 is required for efficient RAD51-mediated homologous DNA pairing and homology-directed DNA repair. J. Biol. Chem 295, 8186–8194. doi: 10.1074/jbc.RA120.013714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, van Attikum H, 2016. PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Nonhomologous End-Joining. Mol. Cell 61, 547–562. doi: 10.1016/j.molcel.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Cohen S, Legube G, 2017. Transcription-Coupled DNA Double-Strand Break Repair: Active Genes Need Special Care. J. Mol. Biol 429, 1277–1288. doi: 10.1016/j.jmb.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Marnef A, Legube G, 2021. R-loops as Janus-faced modulators of DNA repair. Nat. Cell Biol 23, 305–313. doi: 10.1038/s41556-021-00663-4 [DOI] [PubMed] [Google Scholar]
- Mazina OM, Keskin H, Hanamshet K, Storici F, Mazin AV, 2017. Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair. Mol. Cell 67, 19–29.e3. doi: 10.1016/j.molcel.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberg C, Pinder SI, Hopkins SR, Wooller SK, Benstead-Hume G, Pearl FMG, Jeggo PA, Downs JA, 2019. Repression of Transcription at DNA Breaks Requires Cohesin throughout Interphase and Prevents Genome Instability. Mol. Cell 73, 212–223.e7. doi: 10.1016/j.molcel.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, AlHakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJR, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras A-C, Nesvizhskii AI, 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736. doi: 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, Shay JW, 2017. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol. Cell. Biol 37, 405. doi: 10.1128/MCB.00226-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M, Budzowska M, Baldeyron C, Demmers JAA, Ghirlando R, Kanaar R, 2007. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell 28, 468–481. doi: 10.1016/j.molcel.2007.08.025 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R, 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75, 4646–4658. doi: 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L, 2017. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 65, 832–847.e4. doi: 10.1016/j.molcel.2017.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Yadav T, Zhang J-M, Yang H, Rheinbay E, Guo H, Haber DA, Lan L, Zou L, 2021. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 594, 283–288. doi: 10.1038/s41586-021-03538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Maric M, Hewitt G, Mason-Osann E, Gali H, Dai A, Labadorf A, Guervilly J-H, Ruis P, Segura-Bayona S, Belan O, Marzec P, Gaillard P-HL, Flynn RL, Boulton SJ, 2019. SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol. Cell 76, 27–43.e11. doi: 10.1016/j.molcel.2019.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti E, Buemi V, Zappone A, Schillaci O, Broccia PV, Dinami R, Matteoni S, Benetti R, Schoeftner S, 2019. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat Commun 10, 1001–14. doi: 10.1038/s41467-019-08863-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE, 2000. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol 20, 6380–6389. doi: 10.1128/MCB.20.17.6380-6389.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires E, Sharma N, Selemenakis P, Wu B, Huang Y, Alimbetov DS, Zhao W, Wiese C, 2021. RAD51AP1 mediates RAD51 activity through nucleosome interaction. J. Biol. Chem 297, 100844. doi: 10.1016/j.jbc.2021.100844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J, 2014. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 5, 5379–13. doi: 10.1038/ncomms6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S, 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. doi: 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Quinodoz SA, Jachowicz JW, Bhat P, Ollikainen N, Banerjee AK, Goronzy IN, Blanco MR, Chovanec P, Chow A, Markaki Y, Thai J, Plath K, Guttman M, 2021. RNA promotes the formation of spatial compartments in the nucleus. Cell 184, 5775–5790.e30. doi: 10.1016/j.cell.2021.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother MB, Pellegrino S, Smith R, Gatti M, Meisenberg C, Wiegant WW, Luijsterburg MS, Imhof R, Downs JA, Vertegaal ACO, Huet S, Altmeyer M, van Attikum H, 2020. CHD7 and 53BP1 regulate distinct pathways for the re-ligation of DNA double-strand breaks. Nat Commun 11, 5775–19. doi: 10.1038/s41467-020-19502-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B, 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol 196, 801–810. doi: 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieverling L, Hong C, Koser SD, Ginsbach P, Kleinheinz K, Hutter B, Braun DM, Cortés-Ciriano I, Xi R, Kabbe R, Park PJ, Eils R, Schlesner M, PCAWG Structural Variation Working Group, Brors B, Rippe K, Jones DTW, Feuerbach L, PCAWG Consortium, 2020. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat Commun 11, 733–13. doi: 10.1038/s41467-019-13824-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Arora R, Bione S, Azzalin CM, 2021. TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat Commun 12, 3760–12. doi: 10.1038/s41467-021-24097-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ, 2014. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439. doi: 10.1038/nature13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N, 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805. doi: 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Torlai Triglia E, Warburton M, Voigt P, Bird A, Pombo A, 2019. RLoops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Mol. Cell 73, 930–945.e4. doi: 10.1016/j.molcel.2018.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Allen JA, Neumann AA, Yang SF, Walsh ME, Henson JD, Reddel RR, Pickett HA, 2017. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36, 2907–2919. doi: 10.15252/embj.201796889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA, 2007. RNA-templated DNA repair. Nature 447, 338–341. doi: 10.1038/nature05720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Duan M, Yadav T, Phoon L, Wang X, Zhang J-M, Zou L, Lan L, 2020. An Rloop-initiated CSB-RAD52-POLD3 pathway suppresses ROS-induced telomeric DNA breaks. Nucleic Acids Res 48, 1285–1300. doi: 10.1093/nar/gkz1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, Greenberg RA, 2019. RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev 33, 221–235. doi: 10.1101/gad.319723.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohhodina J, Goehring LJ, Liu B, Kong Q, Botchkarev VV, Huynh M, Liu Z, Abderazzaq FO, Clark AP, Ficarro SB, Marto JA, Hatchi E, Livingston DM, 2021. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat Commun 12, 3542–16. doi: 10.1038/s41467-021-23716-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai M-S, Williams GJ, Rydberg B, Sung P, Schild D, 2007. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell 28, 482–490. doi: 10.1016/j.molcel.2007.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Molnarova L, Steinfeld JB, Zhao W, Ma C, Spirek M, Kaniecki K, Kwon Y, Belan O, Krejci K, Boulton SJ, Sung P, Greene EC, Krejci L, 2021. Singlemolecule visualization of human RECQ5 interactions with single-stranded DNA recombination intermediates. Nucleic Acids Res 49, 285–305. doi: 10.1093/nar/gkaa1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce Ö, West SC, 2013. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol 33, 406–417. doi: 10.1128/MCB.01195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-M, Yadav T, Ouyang J, Lan L, Zou L, 2019. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep 26, 955–968.e3. doi: 10.1016/j.celrep.2018.12.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1. List of proteins found in Bio ID of BirA, BirA-TRFl and BirA-RAD51AP1. Related to Figure 2.
Table lists the protein ID, fold change average (FCA) over BirA (CTRL) alone, SAINT probability (SP) and average spectral counts (SpC). Uniprot information is also provided.
Data Availability Statement
Mass spectrometry data is deposited at proteomeXchange database and made available at the following accession number (PXD035943).
Original western blot and data files are available on Mendeley Data as of the date of publication. The DOI is listed in the Key resources table.
This study does not report any original code
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| RAD51AP1 | Mauro Modesti | #805 (Barroso-González et al., 2019; Modesti et al., 2007) |
| RAD51AP1 | Mauro Modesti | #806 (Barroso-González et al., 2019; Modesti et al., 2007) |
| RAD51 | Abcam | Cat. # ab133534; RRID:AB_272261 |
| TRF2 | Novus | Cat. #NB110–57130; RRID:AB_844199 |
| TRF1 | Jan Karlseder | N/A |
| FLAG (D6W5B) | Cell Signaling | Cat. # 14793S; RRID:AB_2572291 |
| FLAG (M2) | Millipore | Cat. # F1804; RRID:AB_262044 |
| γ-TUB | Sigma-Aldrich | Cat. # T6557; RRID:AB_477584 |
| PML (PG-M3) | Santa Cruz | Cat. # sc966; RRID:AB_628162 |
| Ubiquitin histone H2A (Lys119) | Millipore | Cat. # AB10029; RRID:AB_10617517 |
| c-Myc (9E10) | Abcam | Cat. # ab32; RRID:AB_303599 |
| RNAPII | Millipore | Cat. # 8WG16; RRID:AB_10013665 |
| PCNA | Abcam | Cat. # ab92552; RRID:AB_10561973 |
| S9.6 | Kerafast | Cat. # ENH001; RRID:AB_2687463 |
| BRG1 | Abcam | Cat. # ab4081; RRID:AB_304271 |
| PDS5a | ThermoFisher | Cat. # A300–089; RRID:AB_2162213 |
| ZMYND8 | ThermoFisher | Cat. # A302–090; RRID:AB_1604208 |
| SETX | ThermoFisher | Cat. # A301–104; RRID:AB_873128 |
| Critical commercial assays | ||
| ATMi (AZD0156) | Sellekchem | Cat. # S8375 |
| EZH2i (GSK126) | Sellekchem | Cat. # S7061 |
| 5,6-dichloro-1-β-D-ribofuranosyl-1H-benzimidazole (DRB) | Sigma | Cat. # D1916 |
| RO-3306 | Sigma-Aldrich | Cat. # SML0569 |
| Benzonase | Merck Millipore | Cat. # 70746 |
| 4-hydroxytamoxifen | Sigma | Cat. # H7904 |
| Doxycycline | Clontech | Cat. # 631311 |
| Shield Ligand | Clontech | Cat. # 632189 |
| Lipofectamine 2000 | Invitrogen | Cat. # 11668030 |
| Lipofectamine 3000 | Invitrogen | Cat. # L3000015 |
| Dharmafect Reagent 1 | Dharmacon | Cat. # T-2001 |
| Tel C PNA probe | PNA Bio | Cat. # F1004 |
| TERRA PNA Probe | IdT | (TAACCC)5-Alexa488 |
| ULTRA-hyb | ThermoFisher | Cat. # AM8663 |
| Duolink In Situ Detection Reagents | Sigma | Cat. # DUO-9214 |
| PDS5A targeting siRNA pool | Dharmacon | Cat. # L-014071 |
| BRG1 targeting siRNA pool | Dharmacon | Cat. # L-010431 |
| ZMYND8 targeting siRNA pool | Dharmacon | Cat. # L-017354 |
| Control non-targeting siRNA pool | Dharmacon | Cat. # D-001810 |
| RAD51AP1 targeting siRNA pool | Dharmacon | Cat. # L-017166 |
| SETX targeting siRNA pool | Dharmacon | Cat. # L-021420 |
| Deposited data | ||
| Proteomics Data | This study | ProteomeXchange Accession # PXD035943 |
| Raw data files | This study | Mendeley doi:10.17632/mbhnd2b9c7.1 |
| Experimental models: Cell lines | ||
| U2OS | ATCC | Cat# HTB-96; RRID:CVCL_0042 |
| HeLa LT | This study | N/A |
| GM847 | This study | N/A |
| LM216J | This study | N/A |
| U2OS TRF1-FokI WT/D450A | Roger Greenberg | (Dilley et al., 2016) |
| Oligonucleotides | ||
| Scramble antisense oligo (ASO) | IdT | cacgtctatacaccac (Chu et al., 2017) |
| TERRA ASO | IdT | taaccctaaccctaac (Chu et al., 2017) |
| RAD51AP15primeR | This study | ttcttatgtctcacaggccg |
| RAD51AP15primeF | This study | Gtgagacataagaaaccagtcaattactcaca gtt |
| RAD51AP13primerR | This study | caaaggtttaactcgtgcta |
| RAD51AP13primeF | This study | Gagttaaacctttgcatccaaatgccactagca cc |
| RAD51AP1 Isoform1Insert F | This study | tagtattccagctagtgcagtgccttgtacaaagatggctttagatgacaagctct |
| RAD51AP1 Isoform1Insert R | This study | ctagctggaatactaaaagtaccttcagggagcctttttttaggggttttctcttgt |
| Oligo-T | This study | Cy5-ttagggttagggttagggttagggttagggttagggttagg |
| Oligo-R-T | This study | cattgcatatttaaaacatgttggatctcaatttctattttatttttattttttatttctttattttaccatgaattcaaatgacctcttatcaagtgac |
| Oligo-R-B | This study | gtcacttgataagaggtcatttgaattcatcctaaccctaaccctaaccctaaccctaaccctaaccctaagagatccaacatgttttaaatatgcaatg |
| Oligo-D | This study | Cy5aaatcaatctaaagtatatatgagtaaacttggtctgacagttaccaatgcttaatcagtgaggcactatctcagcgatctgtctattt |
| TERRA RNA | This study | Cy5uuaggguuaggguuaggguuaggguuaggguuaggguuag |
| RNA-1 | This study | cugacaguuaccaaugcuuaaucagugagg |
| RNA-2 | This study | ccucacugauuaagcauugguaacugucag |
| RNA-3 | This study | uuauggcagcacugcauaauucucuuacug |
| RNA-4 | This study | gacggggagucaggcaacuauggaugaacg |
| RNA-5 | This study | cguucauccauaguugccugacuccccguc |
| RNA-6 | This study | aaaacuucauuuuuaauuuaaaaggaucua |
| RNA-7 | This study | uagauccuuuuaaauuaaaaaugaaguuuu |
| Recombinant DNA | ||
| BirA | O’Sullivan Lab | (García-Expósito et al., 2016) |
| BirA-TRF1 | O’Sullivan Lab | (García-Expósito et al., 2016) |
| BirA-RAD51AP1 | O’Sullivan Lab | This study |
| FLAG-RAD51AP1 K6K7 | O’Sullivan Lab | This study |
| GFP-RNaseH1-D210N | Lee Zou | (Nguyen et al., 2017) |
| GFP-RAD51AP1 | Mauro Modesti | (Modesti et al., 2007) |
| GFP-PDS5A | Jan Michael Peters | (Ladurner et al., 2016) |
| GFP-PDS5B | Jan Michael Peters | (Ladurner et al., 2016) |
| GFP-STAG1 | Jan Michael Peters | (Holzmann et al., 2019) |
| GFP-ZMYND8 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65401; RRID: Addgene_65401) |
| GFP-BRG1 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 6539; RRID: Addgene_65391 |
| GFP-PBRM1 | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65387; RRID: Addgene_65387 |
| GFP-KDM5A | Kyle Miller | (Gong et al., 2015); (Addgene plasmid # 65387; RRID: Addgene_65387 |
| GFP-NELF-E | Nabieh Ayoub | (Awwad et al., 2017) |
| GFP-SETX | Stephen West | (Yüce and West, 2013) |
| FLAG-RAD51AP1 | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP1 SIM | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP1 K269R | O’Sullivan Lab | (Barroso-González et al.,2019) |
| FLAG-RAD51AP11 ΔRAD51 | O’Sullivan Lab | (Barroso-González et al.,2019) |
| MBP-RAD51AP1-His | Sung lab | (Dunlop et al., 2012) |
| MBP-RAD51AP1 K6K7-His | Sung lab | (Dunlop et al., 2012) |
| RAD51 | Eric Greene Lab | (Xue et al., 2021) |


