Abstract
Chemically diverse in compounds, urine can give us an insight into metabolic breakdown products from foods, drinks, drugs, environmental contaminants, endogenous waste metabolites, and bacterial by-products. Hundreds of them are volatile compounds; however, their composition has never been provided in detail, nor has the methodology used for urine volatilome untargeted analysis. Here, we summarize key elements for the untargeted analysis of urine volatilome from a comprehensive compilation of literature, including the latest reports published. Current achievements and limitations on each process step are discussed and compared. 34 studies were found retrieving all information from the urine treatment to the final results obtained. In this report, we provide the first specific urine volatilome database, consisting of 841 compounds from 80 different chemical classes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12575-022-00184-w.
Keywords: Urine, Volatilome, Metabolomics, Untargeted analysis, Biomedical, Methodology, Database
Introduction
Volatilomics is a branch of metabolomics that comprehensively analyses the volatile compounds released from biological samples. These compounds are products of metabolic processes in organisms, which are of great interest in clinical research [1, 2]. The volatilome constituents are endogenous when they are naturally produced by human metabolism or exogenous when they are produced by the interaction with an external exposure via inhalation, ingestion, or dermal absorption. A recent review has summarized the discriminant effect of the volatilome between different diseases and matrices [3]. Currently, up to 2,746 volatile compounds have been identified across 7 biofluids -breath, blood, faeces, milk, saliva, semen, skin, and urine- in healthy humans [4]. However, the accurate and complete composition of all volatiles that form the human volatilome is still unknown.
Urine is a complex matrix in terms of compounds as it contains metabolic breakdown products from foods, drinks, drugs, environmental contaminants, endogenous waste metabolites, and bacterial by-products. The benefits of using urine as a diagnostic biofluid are numerous, from the ease and non-invasive collection, easy storage and richness of compounds. Recently, over 400 volatiles have been identified in urine, belonging to more than 15 chemical classes, including hydrocarbons, carbonyl, carboxylic acids, and alcohols, among others [4]. Moreover, the urinary volatilome has been used to detect several pathologies and diseases, such as cancer [2], and tuberculosis [5], among others [1].
Measurement of the urinary volatilome usually requires a pre-concentration step to enhance sensitivity, traditionally based on the sample’s headspace (HS) fraction. Some of the techniques used are needle-trap [6], e-nose technologies [7], headspace sorptive extraction [8], thermal desorption sorbent tubes (TD) [9], and solid-phase microextraction methods (SPME) [10]. Microextraction techniques are the evolution of traditional liquid–liquid or solid–liquid extraction techniques, which require less sample volume and solvent [11]. The miniaturization of the pre-concentration techniques has opened new possibilities in medical diagnosis and improved the analytical limits. Among them, SPME has gained high popularity for its simplicity, sensitivity, and cost. Introduced by Pawliszyn et al. in the 90 s, SPME allows the concentration and extraction of sample compounds in a single solvent-free step [12]. Since then, SPME has become a popular method, and the methodology has been applied to multiple matrices with a wide range of purposes, from disease’s biomarkers identification to forensics studies [13–15]. Another emerging technique is the so-called needle-trap device (NTD), an evolution of the purge-and-trap method designed to detect trace organic compounds. In this case the sample is drawn inside the needle [16]. Despite the similar characteristics, NTDs devices have an exhaustive character, being able to deal with larger volumes. The initial NTDs were based in gas samples, although now it is used in a wide range of matrices like water, breath, or urine [14, 17]. Thermal desorption tubes are sampling methods based on diffusion; the thermal tubes sorbents have high hydrophobic properties reducing the interference of water—especially suitable for humid samples—but capturing a wide range of volatile compounds [18].
Microextraction techniques are based on two steps; first, the sorption of volatile headspace compounds to a stationary phase or sorbent; then, thermal desorption of the retained compounds. For its automatic and convenient sample introduction, microextraction methods achieve the highest potential when linked with gas chromatography-mass spectrometry (GC–MS) [19]. Although microextraction methods can also be linked to liquid chromatography-mass spectrometry, this is not easily accomplished in a single step [20]. Thus, high-throughput technologies, such as gas chromatography-mass spectrometry (GC–MS), have the advantages of robustness, high separation capability, sensitivity, and reproducibility [21]. Resolving power between peaks depends on gas chromatography equipment and methods, which can be improved using hyphenated techniques like comprehensive gas chromatography coupled to mass spectrometry (GCxGC-MS). The GCxGC-MS consists of two columns connected in a serial configuration where a modulator transfers the sample portions on the first column to the second one. All these advantages are highly desirable for the simultaneous detection and identification of compounds in complex matrices [22].
Analysis in volatilomics studies can follow targeted or untargeted strategies. Targeted analysis focuses on the extraction, determination, and quantification of specific volatiles of interest using a methodology optimized for that purpose. In contrast, untargeted strategies (fingerprinting) aim to identify the maximum number of volatiles in a sample. These untargeted strategies rely on the use of extraction and determination methods suitable for a broad range of volatility and polarity of volatiles. Then compounds are identified by matching each peak data with existing spectral libraries, followed by data analysis for biological relevance based on statistical methods [23]. Finally, driving the discovery of metabolomic patterns related to diseases.
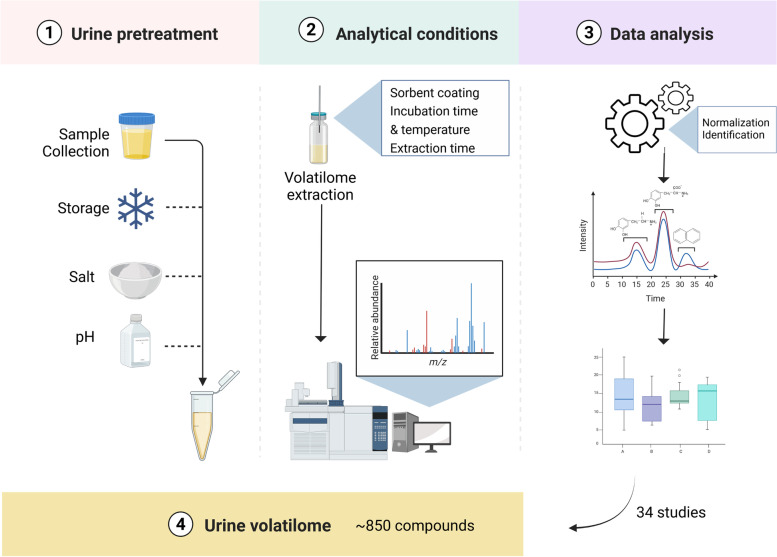
This review focuses on untargeted volatilomics analysis of urine for biomarker discovery in diseases. The key elements of the untargeted workflow methodology are reviewed in detail (see Fig. 1), including: inherent urine matrix needs (collection, storage, and enhancement of compounds volatility); analytical procedures (extraction technique, optimal GC column); and data analysis (normalization, compound ID, etc.). Finally, the applications derived from the urinary volatilome are summarized, and a urinary volatilome database is provided (Table S1).
Fig. 1.
Schematic workflow for untargeted urinary volatilome methodology. The workflow steps include: 1) Urine pretreatment, including collection and storage conditions, and matrix modifications to enhance the extraction of the volatile compounds; 2) Analytical conditions, including the selection and tuning of several parameters from extraction parameters, sample incubation, and analytical instrumentation; 3) Data analysis including normalization and identification sources. 4) Urine volatilome obtained after combining results of the 34 studies analysed. Created with BioRender.com
Studies inclusion
The selection of the reported studies was done via PubMed, Web of Science, and Scopus search using the keywords “VOCS OR ‘volatile organic compound’”, “urine OR urinary”, and “human” and “GC–MS OR ‘proton transfer’”. Until May 2022 we gathered 230 studies (after duplicates exclusion). From those, after the title and the abstract screening, only the studies that conducted an untargeted analysis of human urine were considered. We discarded studies not involving urine (106 studies), no humans matrix (10 studies), targeted approach (41 studies), reviews or book chapters (24 studies), new materials (6 studies), or not disease related (9 studies). Therefore, a total of 34 articles with biomedical applications and have been assessed and included in the biomedical urinary volatilome database (see Table S1). All the 34 studies are GC–MS based, even though we considered proton transfer reaction mass spectrometry, but the studies in this detector kind were not about biomedical application in urine volatiles.
Urine pretreatment
Collection and storage of urine are of paramount importance to preserve the volatilome as urine composition varies between day times. There are many approaches to urine collection, being the most used one spot urine or timed (many times during 24 h or shorter periods). The use of 24-h urine has the lowest day-time variability, but it is inconvenient for the volunteers; in such cases, the variability can be minimized using shorter periods for timed or one-spot collection with pre-analytical normalization methods [24]. Morning urine collection prevents variation due to external factors like physical activity [25]. Liu et al. tested the differences between morning sampling times, being the second-morning urine preferred for biomarker studies as it contains lower levels of dietary metabolites than first-morning urine [26]. In that sense, for volatilome analysis with clinical applications, the best option for sampling is second-morning urine, which includes overnight fasting by-products, but the dietary compounds are minimized. In Table S2 are summarized the different conditions of urine pretreatment by the 34 studies analysed.
Once collected, sample storage should maintain the volatilome. Urine can contain cells or bacteria that break upon freezing, thus pretreatment steps are used to remove them by centrifugation, filtering, and/or protein precipitation [24]. But centrifugation at high speed can cause cell breakage, provoking alterations in its volatilome [25]. Hence, direct aliquoting of samples is the preferred option to preserve the volatilome integrity, as it minimizes sample manipulation and the loss of volatile compounds. For measurements on the same day of collection, urine should be stored at 4ºC. For longer storage times, urine must be freeze. Some studies found that storage of samples at -80ºC was the condition that best preserves the compounds compared to fresh samples as no statistically significant differences were found between both conditions [27]. Storage for long times at -20ºC causes a considerable reduction in the amount of volatiles compared to the fresh condition [24, 27]. Another concern for the volatilome is the freeze–thaw cycles, Semren et al. showed that more than two cycles of freeze–thaw in samples stored at -80ºC influence significally the number of compounds detected [27].
There is a limited quantity of compounds in the headspace of urine. Therefore, to improve its extraction, it is necessary to enhance their affinity for the gaseous phase over the liquid phase. The most used method to enhance compounds to headspace in urine is based on salt addition, which produces a variation in the equilibrium where neutral organic compounds move to the gas phase [28]. Studies that tested the salt addition found an increase in the number of compounds extracted in the headspace[27, 29–32]. Nevertheless, salt saturation can be contra-productive due to opposite effects: a decrease in the number of compounds [30, 33], and an increase in the degraded compounds [34]. There is homogeneity between the analysed studies in choosing the same salt, sodium chloride (NaCl), in the range of 0.1 mg to 0.6 g NaCl/mL. Another alternative to increase the volatility is pH modification. The average normal urine pH is around 6, and its acidification (pH around 2) will increase the number of extracted compounds, mainly acids and sulfurs [34, 35]. On the other hand, the basification of urine (pH around 12) will show a limited increase in compound extraction, favouring mainly alcohols and heterocyclic compounds [30, 36]. The processing of samples with pH modification should consider the possibility of new compound formation due to side reactions of acidic media at high temperatures [30]. Considering the number of identified compounds as the figure of merit, the best performance is obtained (> 227 compounds identified) when combining all strategies (salt addition, acidic and alkaline pH) [36]. Even though it will be necessary to analyse the sample twice, one for acidic and another for alkaline conditions (see Table 1). Nevertheless, to maximize the results of a single measurement the urine pretreatment should include salt addition and acidification, only if the user needs to favour acidic compounds.
Table 1.
Summary of included studies
| Study design | Sampling | Analytical conditions | Post-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref | Application | Sample size/ study design | Storage temperature | Volume sample/ vial | Pretreatment | Sorbent coating Temperature/ Incubation time/ Extraction time | Desorption Temperature/ Time | Instrumentation / Column phase | Analysis software/ Identification (match factor)/ RI | Normalization | Metabolites ID (statistically significant) |
| Cancer | |||||||||||
| [37] | leukemia, colorectal and lymphoma | 33 cancer vs. 21 controls | -80ºC | 4 mL/ 8 mL | Salt + Acid |
CAR/PDMS 50ºC/-/60 min |
250ºC/6 min | SPME-GC-qMS/ PG |
Agilent MS ChemStation/ NIST05 (> 0.8)/ Yes* |
82 (6) | |
| [31] | Breast cancer (BC) | 30 BC patients vs. 41 controls | -80ºC | 4 mL/ 8 mL | Salt + Acid + IS |
CAR/PDMS 50ºC/-/75 min |
250ºC/10 min | SPME-GC-qMS/ PG |
Agilent MS ChemStation/ NIST05 (> 0.8)/ No* |
116 (10) | |
| [38] | Breast invasive ductal carcinoma (IDC) | 65 IDC vs. 70 controls | -80ºC | 4 mL/ 8 mL | Salt + Acid |
CAR/PDMS 50ºC/-/60 min |
250ºC/6 min | SPME-GC-qMD/ PG |
Agilent MS ChemStation/ NIST11 (> 0.80)/ Yes |
Quantile | 94 (14) |
| [6] | Breast cancer (BC) + Colorectal cancer CCRC) | 30 BC + 30 CRC vs. 60 controls | -80ºC | 4 mL/ 20 mL | Salt + Acid |
DVB/CAR1000/CARX 50ºC/40 min/4 min |
250ºC/ 0.5 min | NTD-GC-qMS / OG |
Agilent MS ChemStation/ NIST05 (> 0.80)/ Yes |
Median | 130 |
| [39] | Prostate cancer (Pca) | 58 Pca vs. 60 controls | -80ºC | 1 mL/ 10 mL | IS |
DVB/CAR/PDMS 44ºC/11 min/30 min |
250ºC/5 min | SPME-GC-qMS/ BDPDP |
Mzmine 2 software/ NIST14 / Yes* |
Total area | 122 (31) |
| [32] | Prostate cancer (Pca) | 20 Pca vs. 30 controls | -21ºC | 3 mL/ 10 mL | Salt |
PDMS 50ºC/20 min/20 min |
250ºC/4 min | SPME-GC-qMS/ 5DPDP |
R dtw package/ -/- |
n.a. (15) | |
| [40] | Prostate cancer (Pca) | 59 Pca vs. 43 controls | -20ºC | 0.75 mL/n.d | Base |
CAR/PDMS 60ºC/30 min/20 min |
220ºC/5 min | SPME-GC-qMS/ 6CPDP |
AMDIS & R metab/ NIST02 (> 0.8)/ No |
n.a. (4) | |
| [41] | Clear cell renal cell carcinoma (ccRCC) | 75 ccRCC vs. 75 controls | -80ºC | 2 mL/ 10 mL | Salt |
DVB/CAR/PDMS 44ºC/11 min/30 min |
250ºC/4 min | SPME-GC-qMS/ BDPDP |
Mzmine 2.52/ NIST14/ Yes* |
Total area | 101 (11) |
| [42] | Renal cell carcinoma (RCC) | 22 RCC vs. 25 controls | n.a | 2 mL / 20 mL |
CAR/PDMS 40ºC/-/40 min |
200ºC/2 min | SPME-GC-qMS/ 5DPDP |
Shimadzu GC–MS Post Run & XCMS/ NIST11/ No |
Total area | n.a. (14) | |
| [43] | Renal cell carcinoma (RCC) | 30 RCC vs. 30 controls | -80ºC | 2 mL/ 10 mL | Salt + Acid |
DVB/PDMS 68ºC/9 min/24 min |
n.d |
SPME-GC-qMS/ 5DPDP |
Saturn Workstation/ NIST14/Yes* | PQN + scaled to unit | 181 (21) |
| [44] | Head and neck squamous cell carcinoma (HNSCC) | 53 HNSCC vs. 82 controls | -20ºC | 10 mL/ 20 mL | Salt + Acid |
CAR/PDMS 50ºC/-/30 min |
250ºC/6 min | SPME-GC-MSD/ 6CPDP |
Agilent MS ChemStation/ NIST08 (> 0.8)/ No |
Mean peak area | 81 (35) |
| [45] | Head and Neck cancer (HCN) | 29 HNC vs. 31 controls | -80ºC | 4 mL/ 8 mL | Salt + Acid |
CAR/PDMS 50ºC/-/60 min |
250ºC/6 min | SPME-GC-MSD/ PG |
Agilent MS ChemStation/ NIST11 (> 0.75)/ No |
Median & cube root | 110 (28) |
| [36] | B-cell non-Hodgkin's lymphoma | 101 lymphatic vs. 30 controls | n.a | 10 mL/ 20 mL | Salt + Acid / Base |
CAR/PDMS 40ºC/30 min/30 min |
250ºC/n.d | SPME-GC-qMS/ 5DPDP | Shimadzu GC–MS Post Run/ NIST05/ No* | 227 (3) | |
| [46] | Non-Hodgkin lymphoma | 18 Non-Hodgkin lymphoma vs. 40 controls | -80ºC | 1.47 mL/ n.d | Salt |
DVB/CAR/PDMS 90ºC/15 min/15 min |
250ºC/5 min | SPME-GC-qMS/ 55DPDP |
Mass Hunter Quantitative Analysis & MS-DIAL/ Fiehn8 & NIST14 (> 0.8)/ No |
LOWESS | n.a. (28) |
| [47] | Lung Cancer (LC) | 20 LC vs. 20 controls | -80ºC | 0.1 mL/ 2 mL |
DVB/CAR/PDMS 45ºC/10 min/50 min |
240ºC/10 min | SPME-GC-TOF/ 5DPDP |
MassLynx & XCMS/ NIST08/ No* |
19 (9) | ||
| [48] | Lung Cancer (LC) | 17 LC vs. 30 controls | -80ºC | 4 mL/ 20 mL | Salt + Acid |
DVB/CAR1000/CARX 50ºC/40 min/4 min |
250ºC/ 0.5 min | NTD-GC-qMS / OG |
Agilent MS ChemStation/ NIST05 (> 0.80)/ Yes |
Median | |
| [49] | Bladder Cancer (BdC) | 9 BdC vs 7 controls | -80ºC | 20 mL / n.d | DB-1 25ºC/15 min/- | n.d | NeedleEx®-GC-qMS/DMPS |
Matlab/ NIST/ Yes |
12 (5) | ||
| [50] | Bladder Cancer (UBC) | 96 UBC vs. 209 controls | -20ºC | 0.9 mL / 10 mL | Acid |
DVB/CAR/PDMS 60ºC/30 min/20 min |
n.d |
SPME-GC-qMS/ 6CPDP |
AMDIS + R Metab/ NIST11 (> 0.8)/ No |
Median, log-transform and auto-scaled | n.a.(10) |
| [51] | Colorectal Cancer (CRC) | 58 CRC vs 38 controls | -80ºC | 5 mL / 20 mL | C2-AXXX-5149 40ºC/10 min/10 min | 250ºC/10 min | TD-GC–MS/n.d | TOF-DS software/NIST 11 (n.d.)/No | 23 | ||
| [52] | Colorectal Cancer (CRC) | 18 CRC vs. 31 polyps vs. 34 controls | -80ºC | 4 mL |
TD w/Tenax® + Sulficarb TA 60ºC/30 min/ 2 min |
n.d |
TD-GC–MS/ 6CPDP |
XCMS/ NIST(> 0.8)/No |
Square root | 64 (31) | |
| [53] | Cervical Cancer (CC) | 12 CC + 5 CIN vs. 10 controls | -80ºC | 2 mL / 20 ml |
PDMS 60ºC/30 min/20 min |
270ºC/ 5 min |
SPME-GC-qMS/ 5DPDP |
Agilent MS ChemStation/ NIST14 | Autoscaling | 220 | |
| Exposure | |||||||||||
| [54] | Presence of exogenous factors (tobacco and air pollutants) | 47 smokers vs. 68 non-smokers | -80ºC | 3 mL/ 20 mL | Acid / Base |
CAR/PDMS 37ºC/-/45 min |
290ºC/1 min | SPME-GC-MSD/ PP | Agilent MS ChemStation / NIST08/ No* | Creatinine | 108 (27) |
| [55] | Health impact of environmental pollutants in high contaminated areas | 36 Land of Fires vs. 14 Valley of Sacco River | -80ºC | 1 mL/ 5 mL | IS |
CAR/PDMS 60ºC/-/15 min |
250ºC/n.d | SPME-GC-qMS/ 6CPDP |
Enhanced Data Analysis/ NIST14 (> 0.6)/ No* |
164 (4) | |
| [56] | Human Exposures to Pyrethroids using an Exposure Reconstruction Approach | 28 exposed to Pyrethroids | -80ºC | 1.5 mL / 75 mL | TD w/Tenax® TA 25ºC/-/24 h | 260ºC/10 min |
TD-GC–MS/ 5DPDP |
n.d | 28 (n.d.) | ||
| [57] | Smoking | 4 smokers vs. 3 non-smokers | -80ºC | 0.5 mL/ 5 mL | Salt + Acid |
DVB/CAR/PDMS 40ºC/-/30 min |
250ºC/n.d | SPME-GCxGC-TOF / 5DPDP |
ChromaTOF LECO/ Wiley 275 (> 0.9)/Yes* |
294 | |
| Nephrotic diseases | |||||||||||
| [58] | Mesangial Proliferative Glomerulonephritis (MsPGN) and IgA Nephropathy |
A) 15 MsPGN + 21 IgAN vs. 15 controls B) 15 MsPGN vs. 21 IgAN |
n.a | n.d |
CAR/PDMS 40ºC/-/30 min |
200ºC/2 min | SPME-GC-qMS/ 5DPDP |
XCMS toolbox / NIST11 (> 0.72)/ No |
Total area | 15 (14) | |
| [59] | Minimal Change type Nephrotic Syndrome (MCNS) | 38 MCNS vs. 15 controls | n.a | n.d |
CAR/PDMS 40ºC/-/20 min |
200ºC/2 min | SPME-GC-qMS/ 5DPDP |
XCMS toolbox/ NIST11 (> 0.75)/ No |
n.a. (6) | ||
| [60] | Chronic kidney disease (CDK) | 27 CDK vs. 20 controls | -20ºC | 10 mL/ 20 mL | Salt |
DVB/PDMS 40ºC/30 min/45 min |
230ºC/1 min | SPME-GCxGC-TOF/ PG |
ChromaTOF LECO/ NIST11 (> 0.9)/No |
Specific gravity | 282 (15) |
| [61] | Idiophatic membranous nephropathy (iMN) | 63 iMN vs. 15 controls | -80ºC | 5 mL/ 10 mL |
PDMS/ 60ºC/5 min/10 min |
250ºC/n.d |
SPME-GC-qMS/ 6CPDP |
Shimadzu GCMS + XCMS/ NIST11/No |
n.a.(6) | ||
| Other diseases | |||||||||||
| [35] | Autism spectrum disorders (ASDs) | 24 ASD children vs 21 controls | -80ºC | 4 mL/ 20 mL | Salt + Acid / Base + IS + 1 mL H20 |
DVB/CAR/PDMS 40ºC/-/30 min |
230ºC/25 min | SPME-GC-MSD/ PG |
n.d./ NIST05 and Wiley07/ No* |
75 (9) | |
| [62] | Overweight in children (OW/OB) | 21 OW/OB vs. 28 normal weight | -80ºC | 4 mL/ 20 mL | Salt + Acid / Base + IS + 1 mL H20 |
DVB/CAR/PDMS 40ºC/30 min/10 min |
240ºC/25 min | SPME-GC-MSD/ PG |
n.d./ NIST05 and in-house library/ Yes |
110 acid / 83 alkaline (14) | |
| [63] | Psychological disorders | 30 stress patients vs. 30 controls | -80ºC | 5 mL/ 10 mL |
DVB/CAR/PDMS 50ºC/5 min/45 min |
250ºC/n.d | SPME- GCxGC-TOF/ 6CPDP | ChromaTOF LECO/ NIST11 (> 0.6)/Yes | PQN | 512(4) | |
| [5] | Tuberculosis | 117 tuberculosis + 56 non-TB controls | -80ºC | 2 mL/ 20 mL | Acid | HS 100ºC/50 min/1 min | 105ºC/1 min |
HS-GC-MSD/ Propietary |
AMDIS/ NIST /No |
Total area | n.d. (5) |
| [64] | Coeliac Disease (CD) / Irritable Bowel Syndrome (IBD) | 27 CD vs. 20 IBD | -80ºC | 5 mL/ 10 mL |
PDMS 60ºC/5 min/10 min |
250ºC/n.d |
SPME-GC-qMS/ 6CPDP |
n.d./NIST/ No |
70 | ||
*Confirmation by standards when available
n.a.: not applicable; IS Internal standard, GC-MSD Gas chromatography-Mass selective detector, GCxGC-TOF Comprehensive gas chromatography – time of flight detector, SPME Solid phase micro-extraction, NTD Needle trap device, LOWESS Locally weighted scatterplot smoothing, MSTUS MS total useful signal, PQN Probabilistic quotient normalization, PG Polyethylene glycol, BDPDP 1,4-bis(dimethylsiloxy)phenylene dimethyl polysiloxane, 5DPDP 5% Diphenyl—95% Dimethylpolysiloxane, 6CPDP 6% Cyanopropylphenyl – 94% Dimethylpolysiloxane, PP Porus polymer, 35PMP (35%-phenyl)-methylpolysiloxane
Analytical conditions
Needle-based microextraction is based on the time of contact and the affinity between the sample and the extraction phase of the device. The selection of the extraction mode depends on the sample matrix, analyte volatility, and the affinity of the analyte to the coating [19]. For the measurement of the untargeted urine volatilome, headspace SPME extraction is the preferred mode based on the studies found. The urine HS-SPME extraction opens the possibility to obtain cleaner extracts, higher selectivity, and guarantee longer fibre coating life-time. Nevertheless, Needle-trap techniques or Thermal desorption are also selected to have a broad picture of the urinary volatilome. Table 1 summarizes other parameters involved in the extraction, such as partitioning, sorbent coating, extraction time and temperature, analytes transfer, and instrumental conditions. In Table S3 are summarized other relevant extraction parameters, like coating type, the volume of urine used, incubation time and temperature, extraction time and temperature, and if stirring was used.
The SPME fibre sorbent coating materials will determine the affinity of the volatile compounds [12]. Nowadays, manufacturers offer a broad range of coating chemistries with different selectivity (affinity to different compounds), being the most used for volatilome applications polydimethylsiloxane (PDMS), Carboxen/PDMS (CAR/PDMS), Divinylbenzene/CAR/PDMS (DVB/CAR/PDMS), and polyacrylate (PA). We compared the outcome from each clinical study, despite the differences between urine pretreatment conditions (like volume, salt, or pH), where the highest number of compounds extracted—227- were obtained with CAR/PDMS, followed by DVB/CAR/PDMS with 176 compounds. Other coatings, like PDMS or PA, extracted less than 60 compounds. Five of the studies included in this review have compared the performance of different fibres for urinary volatiles determination, being the CAR/PDMS fibre selected when samples were under acidic conditions [27, 29, 37]. In contrast, under acidic/alkaline conditions or with non-pH modifications, the choice is a DVB/CAR/PDMS fiber [30, 35]. When used a GCxGC-MS, the selected coating is also DVB/CAR/PDMS despite whether urine has acidification or not. Thus, combination of matrix modification – salt and pH – with CAR/PDMS coating disclose the best results for urinary volatilome determination.
The time and the temperature the fibre spends exposed to the sample headspace (the so-called “extraction time”) also affect the efficiency of SPMEs. In the studies, the extraction time ranges from 15 to 90 min, and the temperature ranges from 37ºC to 90ºC. The best conditions for urinary volatilome are obtained with an extraction of 30–45 min at 40-60ºC (see Table 1). Cozolino et al. reported the smallest temperature after their optimization method, detecting 75 analytes with an extraction condition of 30 min at 40ºC [35]. Nevertheless, when the same extraction parameters are analysed with GCxGC-MS, the number of identified compounds increases to 294. Silva et al. 2011, and Drabinska et al. obtained their best performance at 60ºC with 60 min and 45 min extraction times, respectively [30, 37]. Nonetheless, Silva et al. 2019 found an optimal temperature of 70ºC [31], but this high temperature caused fibre damage and sample degradation, in accordance with previous studies [33, 37].
Thermal desorption tube coatings have different affinities for the volatiles based on the coating’s combination, the most famous is the Tenax TA coating – porous polymer –. Tenax was used in all studied evaluated for TD, except for one study where Tenax TA coating was combined with another sorbent, Sulficarb TA – carbonised molecular sieve – [52]. Despite the differences in pretreatment conditions, the best conditions for TD tubes was when Tenax was combined with Sulficarb with 30 min extraction at 60ºC identifying 64 compounds [52]. Comparable results are observed using a 24 h incubation at 25ºC and 20 min incubation at 40ºC with 28 and 23 compounds, respectively [51, 56]. Another type of extraction is needle trap microextraction, two different coatings are evaluated. Divinylbenzene/Carboxen 1000/ Carbopack X(DVB/CAR1000/CARX) extracted 130 and 98 compounds. However, when Dimethylpolysiloxane (DB-1) was used only 12 compounds are identified [49]. Table S4 summarizes the relevant GC and MS conditions of the analysed studies.
In the final step, the analytes are transferred from the devices to the GC, known as the desorption phase. The device is placed in the GC injector at a high temperature for the complete transfer of analytes by desorption [65]. Studies about best desorption parameters range from 1 to 25 min and 200ºC to 290ºC. Only Song et al. compared different desorption times for SPME and found after optimization that the optimal desorption time was 5 min [29]. Similar ranges are used in all devices except for direct headspace, where the temperature was 105ºC and 1 min for desorption.
Data analysis
The basis of all untargeted analysis is to identify as many compounds in a sample as possible to obtain a profile, in this case, the volatilome. For that, the strategy followed is based on the use of software, commercial or open-source, which includes all the analysis workflow from raw data to the list of identified compounds suitable for statistical analysis. More than half of the studies used commercial software for data processing, whereas the others used free software or open-source solutions, such as MS-DIAL [66]. The pre-processing pipeline includes peak detection, noise removal, deconvolution, alignment, and compound identification. Deconvolution separates overlapped peak signals, and then they are identified based on their spectra and elution time [67]. One strength of GC is the electron impact ionization (EI) source, which is generally used at 70 eV, considered a hard ionization since it completely breaks the compounds and produces reliable and reproducible patterns of their fragments. Thus, the independence of the patterns to the instrument allows an accurate peak identification by matching with open or commercial spectral libraries [68], especially if the Retention Index is also included.
Compound identification is a complex process, as with current tools is not possible to identify all the compounds detected in a sample. The reason lies in the identification process which is based on libraries or databases that are not yet completed because not all known metabolites can be purchased or even synthesized [69]. Moreover, not all libraries include RI, key information that allows a more secure and specific way to ensure proper identification. Here starts the user interaction. Once the peak table is obtained, the user has to select the minimum similarity factor between experimental and library spectra (usually cosine similarity) for the identification query, with a minimum acceptable value between 0.6 to 0.8 (see Table 1), based on different instrument and library conditions. The highest level of identification, metabolite standard level 1 (MS1), is achieved using two independent and orthogonal datasets, which is usually a confirmation using reference standard compounds [70], nominal mass spectra and retention index. But on a routinely untargeted analysis, the number of putative compounds identified is in the range of hundreds, and the confirmation by reference standard compounds of all of them becomes tedious and expensive (if not right away impossible). Nevertheless, in GC–MS the retention index (RI) achieves a MS1 level. RI is an orthogonal confirmation based on retention time (RT), a measure independent of experimental conditions and unique for each compound [71]. To obtain RI values, a set of compounds (aliphatic alkanes or FAMEs) are used as indicators [69]. Despite the increase in confidence that offers this measure, only 7 authors used the retention index (see Table 1). The applicability of RI in volatilome analysis has some limitations, as RI library values depend on the column polarity selected, and some of the volatile-specific columns used (e.g. ZB-624) have non-specific RI libraries. Moreover, in available commercial libraries, like NIST, only 11% of the compounds include the RI [72], which makes identification a limiting step.
The GC–MS profiles obtained after the data processing are tables containing the relative abundance of each peak detected: intensity, area, or both; its RI or RT and their identification. Comparative analysis uses this information to evaluate the differences between group samples and find compounds of interest [73]. Although this approach is used widely in metabolomics, it has considerable unwanted experimental and biological variation [74]. Experimental variation due to human error and instrument bias is corrected with the use of internal standards (IS). IS are compounds added in constant amounts to all samples, usually deuterated forms like 1,4-Dichlorobenzene-d4; or not biological compounds, like 4-Fluorobenzaldehyde [39, 55]. Notwithstanding their usefulness, only 25% of the studies reported the use of IS. Moreover, the biological variation in urine concentration is high, as it is a biofluid that is not homeostatically regulated, which can mask the variations due to internal factors. Compounds concentration in urine will depend on the hydration status of the individual. Thus, normalization becomes a fundamental step in metabolomics that is poorly implemented. However, only in 40% of the studies analysed it was somehow accounted for (see Table 1). Among the strategies used for normalization, there are several approaches like normalization by total area, creatinine, quantile, and median. The strategy most used is to normalize by total area: briefly, it is the division of the area of each peak by the sum of all the peaks’ areas. However, this strategy can mask variations due to differences in peak number as new peaks in samples are diffused across all the samples [73]. Similarly, in creatinine normalization, the peaks are normalized by the creatinine concentration of the sample. Creatinine concentration has been widely used in clinical applications as a urine normalization method. In the Human Metabolome Data Base (HMDB) [75], the urine compounds concentrations are reported normalized to the creatinine concentration. However, recent reports have proven that other factors such as diet, exercise, or gender, influence the excretion of creatinine [72]. Similarly, MS total useful signal (MSTUS) has been proposed as a normalization method, where the signal is divided by the sum of features common in all samples [76]. Quantile normalization refers to an intensity-dependent scaling factor and transformation of peaks [74]. Finally, median normalization is the division of the profiles by the median of all study profiles [73]. Other more statistically-oriented normalization methods exist [77], like the locally weighted scatterplot smoothing (LOWESS) algorithm (based on a local regression) or the probabilistic quotient normalization (PQN). Mack et al. compared 5 normalization methods in urine volatilome (creatinine, osmolarity, urine volume, MSTUS and PQN) [78]. All methods showed comparable results, but none of them could deal with problems in renal function. Urinary volatilomics for clinical research is still emerging in part because there is not a generally agreed standard normalization method yet, so the researcher must choose a determined strategy. Nevertheless, normalization methods for urine are highly established for other purposes. It is the case of epidemiological studies where is used the normalization by specific gravity [79, 80]. Specific gravity is used as pre-processing normalization method where the samples are diluted to the same concentration. But, there is only study using specific gravity applied as a sample selecting parameter [60].
Applications
Urine GC–MS untargeted analysis has been applied to several clinical topics: cancer, harmful chemicals, and nephrotic diseases, among others (see Table 1 and Table S5). Cancer is the most studied disease as twenty-one studies evaluated biomarker discovery for a range of cancer types such as prostate, breast, or renal. Another topic of relevance is the study of harmful chemicals in humans, where four studies evaluated the effects of polluted environments and tobacco. By the direct relationship with the urinary system, nephrotic diseases are an interesting topic, where the urinary volatilome is used to improve the diagnosis of some nephropathies. Finally, diseases not included in the previous groups are classified as other diseases, including autism, overweight children, psychological disorders, tuberculosis and coeliac disease.
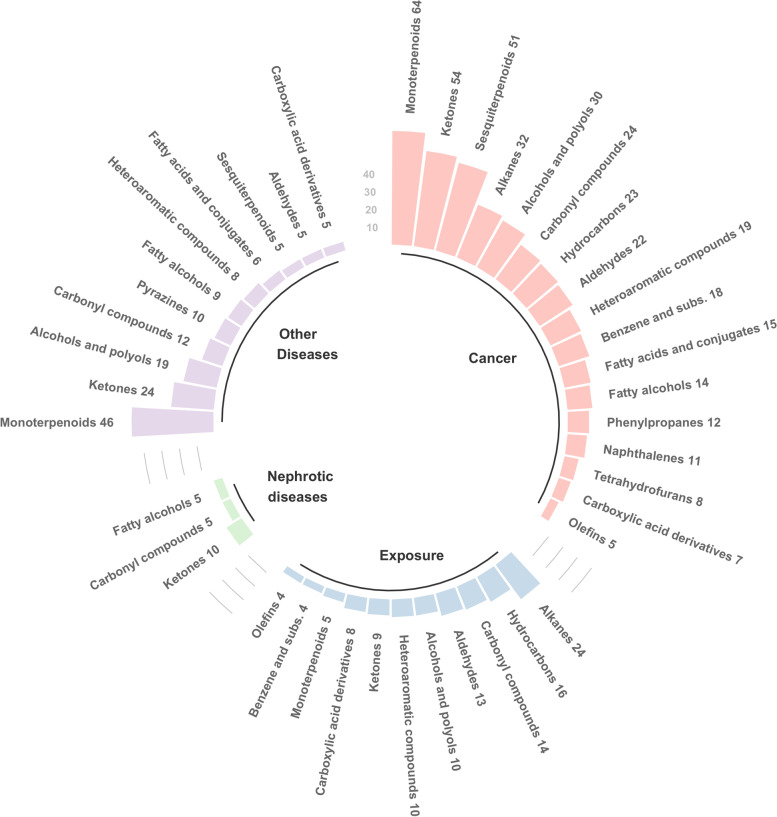
The biomedical untargeted urinary volatilome database includes the compounds identified in studies of biomedical applications using the urine volatilome; in total, we retrieved the information from 34 studies. One study on the cancer group was not included in the database creation, as they did not provide the compounds identification [32]. We retrieved and harmonised all detected compounds for each of the 33 included studies using the same InChIKey identifier [81]. The included studies reported 841 different volatiles (Table S1), of which 2-pentanone, and 4-heptanone were found in at least half of the studies (Table 2). From the urinary volatilome list, only 267 compounds were retrieved in two or more studies. The smallest compound detected is acetonitrile (C2H3N), whereas the biggest is Allyl octadecyl oxalate (C23H42O4). Given the complexity of comparing specific compounds per group, we performed the comparison by the chemical classes found within each group, which were retrieved with the ClassyFire tool [81]. The number of chemical classes found spans from 63 in cancer to 23 in nephrotic diseases. The chemical classes more present in all groups are organooxygen compounds, organic disulfides, and phenols. But with different abundance across groups. Focusing on subclasses, ketones are highly represented in all groups, being the more abundant in cancer. In contrast, the exposure groups have a higher abundance of alkanes, whereas the other diseases group is characterized by monoterpenoids (see Fig. 2). For each group, we performed an enrichment analysis. However, only for the cancer group, the enrichment analysis returned significant pathways (with p-values < 0.05 and false discovery rates FDR < 0.05).
Table 2.
Compounds of the biomedical untargeted urinary volatilome database found at least ten times in the included studies (n = 33)
| Nº studies | Compound | Chemical Subclass | HMDB ID |
|---|---|---|---|
| 21 | 2-Pentanone | Ketones | HMDB0034235 |
| 18 | 4-Heptanone | Ketones | HMDB0004814 |
| 15 | Hexanal | Aldehydes | HMDB0005994 |
| 14 | Dimethyl disulfide | Dialkyldisulfides | HMDB0005879 |
| 13 | 2-Butanone | Ketones | HMDB0000474 |
| 13 | 2-Propanone | Ketones | HMDB0001659 |
| 12 | 2-Heptanone | Ketones | HMDB0003671 |
| 12 | Nonanal | Aldehydes | HMDB0059835 |
| 12 | Phenol | Phenols | HMDB0000228 |
| 11 | Acetic acid | Carboxylic acids | HMDB0000042 |
| 10 | 3-Hexanone | Ketones | HMDB0000753 |
| 10 | Carvone | Monoterpenoids | HMDB0035824 |
| 10 | Furan | Heteroaromatic compounds | HMDB0013785 |
Fig. 2.
Circle bar plot for the chemical classes identified in human urine volatilome in biomedical conditions classified by application group, for the studies reviewed with a compound list disclosed. Studies included are divided into 4 groups by the application. The number of compounds within each class corresponds to the number of unique species identified for that chemical subclass
Cancer
The main application in clinical research is cancer, as several authors have proven the usefulness of volatilome analysis in urine samples for biomarker discovery in this disease. B-cell non-Hodgkin’s lymphoma disclosed the higher number of compounds identified among the cancer types tested (227 compounds, as seen in Table 1). Mesquita et al. also evaluate Non-Hodgkins lymphoma finding 28 volatiles statistically significant [46]. Prostate cancer is one of the most studied, as it is evaluated in three studies using different approaches of SPME conditions and GC column phases. Khalid et al. identified 197 compounds with CAR/PDMS at 60ºC, Lima et al. identified 122 compounds with DVB/CAR/PDMS at 44ºC, and Deev et al. used PDMS at 50ºC but none of them performed compound identifications [32, 39, 40]. Similarly, 3 authors evaluate renal cell carcinoma, Monteiro et al., Pinto et al., and Wang et al. found 21, 11 and 14 compounds statistically significant, respectively [41–43]. All used SPME extraction but with different conditions, being the combination of DVB/PDMS at 68ºC the one with more compounds identified [43]. Head and neck carcinoma was evaluated by two authors using the same SPME fibre coating and time; Taware et al. identified 110 compounds, whereas Opitz et al. found 81 compounds [44, 45]. Conditions for breast cancer only differ in the SPME exposure time used: Silva et al. 2019 applied 15 min longer extraction time with 116 compounds identified; and Taunk et al., identified 94 compounds but they get higher number of compounds statistically significant [31, 38]. Similar conditions were used to evaluate a set of samples from leukaemia, colorectal cancer, and lymphoma. In this case, 6 compounds allow to differentiate between cancer and healthy patients. Lung cancer is usually studied through breath, but in two studies it was also evaluated using the urine volatilomics profile. The authors selected different methodologies; Hanai et al. used SPME extraction identifying 19 compounds whereas Porto-Figueira et al. used NTD to identify 98 compounds [47, 48]. Bladder cancer has a direct relation with urine, Jobu et al. evaluate it by NeedleEx whereas Lett et al. used SPME [49, 50]. Comparison of techniques is not possible as results are not completely disclosed. Colorectal cancer was analysed by thermal desorption by two authors differing in time and temperature of extraction, studies showed 12 and 23 compounds identified [51, 52]. Díaz de León-Martinez et al. evaluated the urine volatilome for cervical cancer with SPME obtaining one of the highest number of compounds identified -220- using only 2 ml of urine [53]. Discrimination between cancer patients and controls is possible with the use of urine volatilomics [82]. Together, with the elevated number of studies, the cancer studies’ group shows the highest number of volatiles classes detected. The chemical classes found in higher proportion are terpenoids and carbonyl compounds (including ketones and aldehydes). There are 31 chemical classes unique for the cancer group, the most abundant being tetralins, cinnamaldehydes, and lactones. Three compounds are found in at least half of the cancer studies analysed (see Table S6): 2-pentanone, phenol, and dimethyl disulfide.
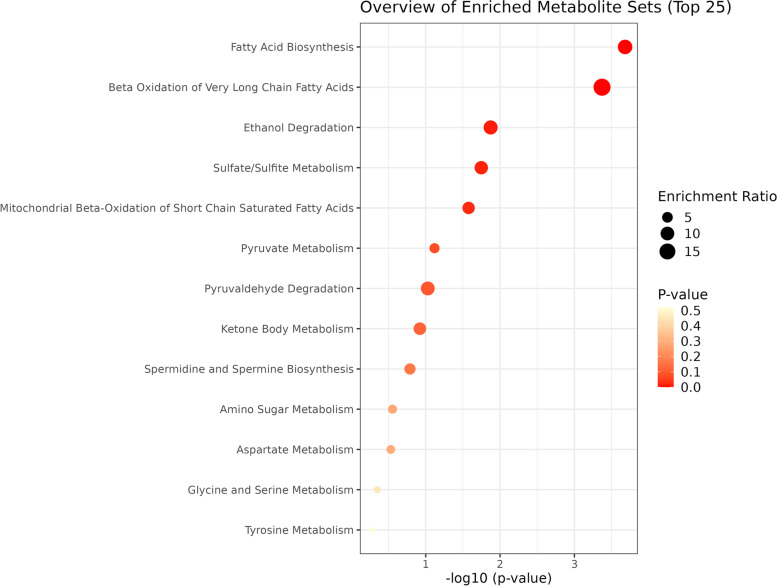
An enrichment metabolites analysis was performed to identify classes of metabolites that are over-represented in the large set of metabolites that conform the cancer group (641) and may have an association with cancer. Up to thirteen pathways are found to be over-represented (see Fig. 3), however, only the fatty acids biosynthesis and the beta oxidation of very long chain fatty acids are significant (p-value is 0.0002 and 0.0004, respectively and the FDR is 0.02 for both pathways).
Fig. 3.
Dot plot of the enrichment analysis results for the cancer application. The size of the circles per metabolite set represents the Enrichment Ratio and the colour represents the p-value. Analysis performed with MetaboAnalyst Enrichment Analysis [83] module using identifiers from HMDB and KEGG
Chemical exposure
Urine is the biofluid most used to assess the exposure of some chemical compounds that may be harmful to humans, including third hand tobacco [84]. Longo et al. assess the fingerprint in urine for areas with high air pollution in Italy [55]. The comparison between two areas with different pollution identified 164 volatiles, where only 4 of them were statistically significant. Previously, Filipiak et al. assessed the potential of smoking and the environmental exposure in two biofluids, breath and urine, finding 108 volatiles in urine [54]. SPME conditions were different in both studies, but they used the same fibre coating (CAR/PDMS). When SPME is coupled to GCxGC-MS the number of compounds detected increase, as reported by Rocha et al. for smoking comparison with 294 compounds identified [57]. One author selected another extraction methods for exposure analysis, O’Lenick et al. studied the exposure to pyrethroids using thermal desorption with the identification of 28 compounds [56]. Exposure related compounds belong to 52 chemical classes. The chemical classes found in higher proportion are carbonyl compounds (ketones and aldehydes), alkanes and hydrocarbons, where 10 are unique for exposure applications, such as benzofurans, oxazinanes and azolines. One compound is found in all the chemical exposure studies (see Table S6): hexanal.
Nephrotic disease
As part of the urinary system, some nephrotic diseases have been evaluated through the volatilome of urine. Biomarker discovery was successfully used in minimal change type nephrotic disease (MCNS), a disease with an invasive diagnosis that affects mostly children. Liu et al. identified 6 volatiles as possible biomarkers of MCNS [59]. In the same line, 5 volatiles were identified as possible biomarkers for mesangial proliferative glomerulonephritis [58]. Diseases may lead to chronic kidney disease (CDK), which was investigated by Ligor et al. using GCxGC-MS. The CDK patients showed a panel of 4 volatiles upregulated (methyl hexadecanoate, 9-hexadecen-1-ol, 6,10-dimethyl-5,9-undecadien-2-one, and 2-pentanone) [60]. Wang et al. evaluate the idiopathic membranous nephropathy with 6 compounds found as significant [61]. Despite the small number, 23 chemical classes are included, where 3 are unique for this groups such as the dihydrofurans. One compound is found in all the nephrotic related studies (see Table S6): 4-heptanone.
Other diseases
Applications with one or two studies are included in this group, such as children’s diseases, tuberculosis or coeliac disease. Cozzolino et al. used HS-SPME on urine samples to investigate children’s urine related to diseases [35, 62]. In one study, they aimed to find perturbations in the volatilome caused by overweight. The results showed 14 volatiles that distinguish between over-weight and normal children, from the more than 150 volatiles identified [62]. In a previous study, they compared the urinary profile of autistic children, finding that 3 volatiles under acidic conditions and 3 volatiles under alkaline conditions discriminated between groups [35]. Eshima et al. studied complex disorders, such as psychological disorders, due to the lack of quantitative diagnosis tools [63]. A multiple regression model led to 2 volatiles influenced by glucocorticoid signalling mechanism. SPME was used by Arasaradnam et al. to distinguish between coeliac patients and irritable bowel disease patients, 70 compounds were identified but with non-statistical significance [85]. Only one author used direct HS sampling, Banday et al. applied it to tuberculosis with 5 compounds found statistically significant [5]. Combination of the distinct studies increases the number of chemical classes found to 29 within this group. Where 2 chemical classes were not present in any other group,—cancer chemical exposure or nephrotic disease—(phenol ethers and pyrans). One compound is found in four of the five studies from other diseases (see Table S6): carvone.
Discussion
In our report, we review the untargeted analysis of the urinary volatilome, in response to the demand for alternatives for more comprehensive and environmentally friendly compound extraction methods than the traditional ones. Among the different techniques available, solid phase microextraction (SPME) is simple, sensitive, robust, and an easily automated technique. The optimum SPME configuration depends on various factors, such as urine pre-analytical conditions, the fibre coating, time, and temperature of extraction. Data analysis also includes some crucial steps. For instance, normalization of urine, to which there is no clear consensus up to now, and compound identification, which depends on the available libraries of standard compounds and the information they provide (mass spectra alone or accompanied by RI). In summary, the HS–SPME–GC–MS technique to measure urine has been used in 26 studies with clinical applications finding/identifying/reporting utmost 227 volatiles. Other technologies like NTD-GC–MS showed similar results as SPME, reporting differences in the chemical classes found. Although the similarity in the technique, TD showed poor results, capturing a number of compounds distant of SPME or NTD results The use of hyphenated techniques, such as GCxGC-MS showed an increase in the urinary compounds detected with the identification of utmost 512 compounds [63].
Further developments are still needed in untargeted HS–SPME–GC–MS urinary analysis. Each SPME fibre covers only a narrow spectrum compared to the broad chemical spectrum of urine volatile compounds. In that sense, new tools like Arrow SPMEs, which have higher capacity and are more robust (are thicker and do not blend as easily as regular SPMEs), still have not been used in urinary samples. Another advantage of the Arrow SPME is that it also reduces the exposure time of the fibre [21, 86]. First studies with SPME Arrow-GC–MS in water samples have detected trace levels at the ng L−1 [87] concentrations. Concerning the newer fibres, newer developments have not yet been tested in urine samples like new coating materials which will be useful in all evaluated extraction techniques. As an example, we can mention new non-toxic coatings such as Carbon Nanotubes (CNTs), which have been applied to PAHs analysis from water samples, or the Sol–gel extraction phase, which has been proposed for polar compounds extractions in urine samples [86]. However, these techniques will be always dependent on the coatings affinity for the compounds of interest. Some authors recommend the use of more traditional extraction methods such as liquid–liquid extraction to broader the chemical classes obtained in a single analysis [88].
Resolving power between peaks depends on gas chromatography. Use of hyphenated techniques, like GCxGC-MS, have proven a remarkable increase in peak capacity (selectivity) and peak resolution. Coupling PAL-SPME Arrow extraction with GCxGC-MS promises an improvement on the number of compounds extracted and overall resolving power for urine samples.
Technology advances do not go hand in hand with sample processing advances. Compound identification is a bottleneck in metabolomics, the process is limited by the commercial availability of standard compounds. Confident identification of compounds should be done by standard comparison, however, not all compounds are commercially available or even their structure is not known properly. Some stablished methods, like specific gravity is not being used in urinary volatilomics, but are used in several longitudinal studies and by the World Anti-Doping Agency [79, 89, 90]. This method has the advantage of fewer cofounding effects and ease of measurement [91]. Although it is not an automatic method, it allows to correct sample concentration based on the hydration status of the individual. However, in urine volatilomics for biomedical applications, it is only used by one author which used it to select individuals between a specific range [60].
The major drawback when comparing results from different studies is the different nomenclature among the studies. The use of several libraries returns several options of name for the same chemical compound. To overcome this problem we used unique identifiers (IDs) like InChIKey instead the chemical name. The final biomedical untargeted urinary volatilome database (uBIOVOC DB) consists of 841 compounds all of them with a unique InChIKey and PubChem CID (Table S1). For re-usability of the UVDDB a part from chemical information (molecular formula and molecular weight), we provided several IDs. Up to 721 compounds have CAS number, but sixteen of them are used by more than one compounds. Databases IDs include KEGG (211 compounds), ChEBI (266 compounds), HMDB (387 compounds), LIPIDMAPS (152 compounds) and Drugbank ID (62 compounds). Moreover only 19 compounds had an entry in all the public databases retrieved, whereas 114 do not have any identifier associated.
Cancer is one of the pathologies most evaluated in the bibliography (n = 20), however the compound found more times was reported only 13 times (65% of the studies). Also, cancer is a very broad term for a wide range of tumours in different body localizations with different behaviours. Even though this heterogeneity, the enrichment analysis returned two significant pathways, both related to fatty acids metabolism: the fatty acids biosynthesis and the beta oxidation of very long chain fatty acids. None of the compounds found in at least half of the cancer studies analysed (2-pentanone, phenol, and dimethyl disulfide) was found relevant in the enrichment analysis. In the fatty acids metabolism are involved four volatiles, which are the lowest fatty acids: acetic acid (FA 2:0), hexanoic acid or caproic acid (FA 6:0), octanoic acid or caprylic acid (FA 8:0) and decanoic acid or capric acid (FA 10:0). Fatty acid metabolism supports tumorigenesis and disease progression through a range of processes including energy production (β-oxidation), membrane biosynthesis, energy storage and production, and generation of signalling intermediates [92]. Worth to mention the fact that most VOCs do not have HMDB or KEGG identifiers. From the 641 cancer-related compounds 422 do not have either of HMDB nor KEGG identifiers. Also, enrichment analysis is based upon a comparison to a set library. In our case, we used the small molecule pathway database (SMPDB) based on metabolites pathways. The improvement of SMPDB or even KEGG pathways databases with the incorporation of volatiles will have a very highly and positive impact in further studies.
For the exposure and nephrotic disease groups where all studies evaluated share one compound, hexanal, and 4-heptanone, respectively. The other diseases group also has a compound found in 80% of the articles, but it is a terpenoid related to food (carvone). The nephrotic disease group showed the lowest number of compounds (56 compounds), despite only half of the studies disclosed the complete list of compounds identified. In contrast, cancer group with the 80% of studies disclosing the complete list gathered 640 compounds, almost three times the number of compounds gathered in the other applications. Up to ten compounds are found in all the applications considered: 2-pentanone, 4-heptanone, hexanal, 2-heptanone, nonanal, 3-hexanone, benzaldehyde, Pyrroline, Dimethyl trisulfide, and Phenylmethylketone. However only three are found in more than 40% of all studies analysed: 2-pentanone, 4-heptanone, and hexanal. The 2-Pentanone has been associated to several diseases such as ulcerative colitis, non-alcoholic fatty liver disease, crohn's disease; and coeliac disease [75] [REF]. The 4-Heptanone has been also associated to several diseases such as kidney disease, perillyl alcohol administration for cancer treatment, and coeliac disease [75] [REF]. Hexanal is one of the most common aldehydes found in urine, as it is a major breakdown product of linoleic acid [75] [REF].
Among the high number of studies evaluating VOCs related to cancer there are initiatives of database creation for cancer [93, 94], however the websites are not maintained or the results are not specified by matrix. To overcome similar situations, we have included the biomedical untargeted urinary volatilome database as a supplementary material and is also available at Zenodo so that further reuse will be possible. Also, due the increase of studies evaluating urine and volatilome, the author’s intention is to update the biomedical untargeted urinary volatilome database every two years. The increase of evidence of urinary volatilome as a source of non-invasive and reliable testing, will promote its use in more biomedical applications. Therefore, this database will be of interest to a broad audience, ranging from basic researchers doing biomarkers discovery, to personalized medicine applications as it opens the floor for the development of predictive medicine devices, such as point-of-care or home testing devices [95].
Conclusions
Our analysis compiled the largest database generated to date of urinary volatilomics data, with 841 compounds. Despite the high number of compounds reported, we have not restricted the inclusion of compounds by the level of identification or extraction technique. This is because a high level of confidence, comparison to reference standard or use of RI, is still limited in the bibliography. To overcome naming differences all compounds have been compared using a unique identificatory (the InChIKey in our case). However, the few compounds shared between studies show discrepancies in the results caused by different study designs or device coatings. The vast possibilities on the analysis technique contributes to the range of compounds obtained. In fact, less than 1% of all compounds is found in at least half of the studies evaluated, and no compound is reported in all the studies. Nonetheless, three compound are reported in all clinical groups: 2-pentanone, 4-heptanone and hexanal. Despite the different range of clinical applications, the comparison is usually done with healthy individuals (controls). The few compounds reported commonly reveals a need for standardization procedures, standardized analysis and reporting for the urinary volatilome. Nevertheless, the observed pattern of chemical classes found in the urinary volatilome will be helpful in deciding targeted compounds and methodology for further studies.
Supplementary Information
Additional file 1: Table S1. The biomedical untargeted urinary volatilome database (uBIOVOC DB). Table S2. Summary of the sample collection conditions used by the 34 analysed studies. Table S3. Summary of the analytical conditions used by the 34 studies using them. Table S4. Summary of the GC-MS conditions used by the 34 analysed studies. Table S5. Summary of the applications analysed by the 34 analysed studies. Table S6. The urinary volatilome database by biomedical applications.
Acknowledgements
We want to acknowledge Dr. Noelia Ramírez for her insightful comments on the shaping of this review.
Abbreviations
- HS
Headspace
- TD
Thermal desorption
- SPME
Solid-phase microextraction
- NTD
Needle trap device
- GC–MS
Gas chromatography – mass spectrometry
- GCxGC-MS
Comprehensive gas chromatography – mass spectrometry
- RI
Retention index
- ID
Identified
- IS
Internal standard
- CAR
Carboxen
- PDMS
Polydimethylsiloxane
- DVB
Divinylbenzene
- PA
Polyacrylate
- CARX
Carbopack
- LOWESS
Locally weighted scatterplot smoothing
- PQN
Probabilistic quotient normalization
- MSTUS
MS total useful signal
- PG
Polyethylene glycol
- BDPDP
1,4-Bis(dimethylsiloxy)phenylene dimethyl polysiloxane
- 5DPDP
5% Diphenyl—95% Dimethylpolysiloxane
- 6CPDP
6% Cyanopropylphenyl – 94% Dimethylpolysiloxane
- PP
Porus polymer
- DMPS
Dimethylpolysiloxane
- 35PMP
(35%-Phenyl)-methylpolysiloxane
- BC
Breast cancer
- IDC
Beast invasive ductal carcinoma
- CRC
Colorectal cancer
- Pca
Prostate cancer
- ccRCC
Clear cell renal cell carcinoma
- RCC
Renal cell carcinoma
- HNSCC
Head and Neck squamous cell carcinoma
- HCN
Head and neck cancer
- LC
Lung cancer
- BdC
Bladder cancer
- MsPGN
Mesangial proliferative glomerulonephritis
- MCNS
Minimal change type nephrotic syndrome (MCNS)
- iMN
Idiopathic membranous nephropathy
- n.d.
Not disclosed
- n.a.
Not applicable
- CDK
Chronic kidney disease
- ASDs
Autism spectrum disorders
- OW/OB
Overweight/obesity
- MS1
Metabolite standard level 1
- RT
Retention time
- HMDB
The human metabolome database
- EI
Electron impact ionization
- CNTs
Carbon nanotubes
- PAHs
Polycyclic aromatic hydrocarbons
Authors’ contributions
MLL and RC conceptualized the work and curated the database. MLL wrote the manuscript. RC and JB corrected the manuscript. The author(s) read and approved the final manuscript.
Funding
This research was supported by the Spanish MINECO (ref. RTI2018-098577-B-C21 and PID2021-126543OB-C21). This project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No (798038). MLL is thankful for her pre-doctoral fellowship from the URV PMF-PIPF program (ref. 2019PMF-PIPF-37). We would like to acknowledge the Departament d’Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya (expedient 2017 SGR 1119). IISPV is member of the CERCA Programme/Generalitat de Catalunya.
Availability of data and materials
The datasets generated during the current study are available in the Zenodo repository, link 10.5281/zenodo.6883341
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jesús Brezmes, Email: jesus.brezmes@urv.cat.
Raquel Cumeras, Email: raquel.cumeras@iispv.cat.
References
- 1.Djago F, Lange J, Poinot P. Induced volatolomics of pathologies. Nat Rev Chem. 2021;5:183–196. doi: 10.1038/s41570-020-00248-z. [DOI] [PubMed] [Google Scholar]
- 2.da Costa BRB, De Martinis BS. Analysis of urinary VOCs using mass spectrometric methods to diagnose cancer: A review. Clin Mass Spectrom. 2020;18:27–37. doi: 10.1016/j.clinms.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roszkowska A, Miękus N, Bączek T. Application of solid-phase microextraction in current biomedical research. J Sep Sci. 2019;42:285–302. doi: 10.1002/jssc.201800785. [DOI] [PubMed] [Google Scholar]
- 4.Drabińska N, Flynn C, Ratcliffe N, Belluomo I, Myridakis A, Gould O, et al. A literature survey of all volatiles from healthy human breath and bodily fluids: The human volatilome. J Breath Res 2021;15. 10.1088/1752-7163/abf1d0. [DOI] [PubMed]
- 5.Banday KM, Pasikanti KK, Chan ECY, Singla R, Rao KVS, Chauhan VS, et al. Use of Urine Volatile Organic Compounds To Discriminate Tuberculosis Patients from Healthy Subjects. Anal Chem. 2011;83:5526–5534. doi: 10.1021/ac200265g. [DOI] [PubMed] [Google Scholar]
- 6.Porto-Figueira P, Pereira JAM, Câmara JS. Exploring the potential of needle trap microextraction combined with chromatographic and statistical data to discriminate different types of cancer based on urinary volatomic biosignature. Anal Chim Acta. 2018;1023:53–63. doi: 10.1016/j.aca.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 7.Westenbrink E, Arasaradnam RP, O’Connell N, Bailey C, Nwokolo C, Bardhan KD, et al. Development and application of a new electronic nose instrument for the detection of colorectal cancer. Biosens Bioelectron. 2015;67:733–738. doi: 10.1016/j.bios.2014.10.044. [DOI] [PubMed] [Google Scholar]
- 8.Gao Q, Su X, Annabi MH, Schreiter BR, Prince T, Ackerman A, et al. Application of Urinary Volatile Organic Compounds (VOCs) for the Diagnosis of Prostate Cancer. Clin Genitourin Cancer. 2019;17:183–190. doi: 10.1016/j.clgc.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Yu Q, Xu S, Shi W, Tian Y, Wang X. Mass spectrometry coupled with vacuum thermal desorption for enhanced volatile organic sample analysis. Anal Methods. 2020;12:1852–1857. doi: 10.1039/d0ay00175a. [DOI] [Google Scholar]
- 10.Souza-Silva ÉA, Reyes-Garcés N, Gómez-Ríos GA, Boyacı E, Bojko B, Pawliszyn J. A critical review of the state of the art of solid-phase microextraction of complex matrices III. Bioanalytical and clinical applications TrAC Trends. Anal Chem. 2015;71:249–64. doi: 10.1016/j.trac.2015.04.017. [DOI] [Google Scholar]
- 11.Pereira J, Silva CL, Perestrelo R, Gonçalves J, Alves V, Câmara JS. Re-exploring the high-throughput potential of microextraction techniques, SPME and MEPS, as powerful strategies for medical diagnostic purposes Innovative approaches, recent applications and future trends Microextraction Techniques. Anal Bioanal Chem. 2014;406:2101–22. doi: 10.1007/s00216-013-7527-4. [DOI] [PubMed] [Google Scholar]
- 12.Górecki T, Yu X, Pawliszyn J. Theory of analyte extraction by selected porous polymer SPME fibres. Analyst. 1999;124:643–649. doi: 10.1039/a808487d. [DOI] [Google Scholar]
- 13.Huang S, Chen G, Ye N, Kou X, Zhu F, Shen J, et al. Solid-phase microextraction: An appealing alternative for the determination of endogenous substances - A review. Anal Chim Acta. 2019;1077:67–86. doi: 10.1016/j.aca.2019.05.054. [DOI] [PubMed] [Google Scholar]
- 14.Paiva AC, Crucello J, de Aguiar PN, Hantao LW. Fundamentals of and recent advances in sorbent-based headspace extractions. TrAC Trends Anal Chem. 2021;139:116252. doi: 10.1016/J.TRAC.2021.116252. [DOI] [Google Scholar]
- 15.Bojko B, Reyes-Garcés N, Bessonneau V, Goryński K, Mousavi F, Souza Silva EA, et al. Solid-phase microextraction in metabolomics. TrAC Trends Anal Chem. 2014;61:168–180. doi: 10.1016/J.TRAC.2014.07.005. [DOI] [Google Scholar]
- 16.Laaks J, Jochmann MA, Schmidt TC. Solvent-free microextraction techniques in gas chromatography. Anal Bioanal Chem. 2012;402:565–571. doi: 10.1007/s00216-011-5511-4. [DOI] [PubMed] [Google Scholar]
- 17.Kędziora-Koch K, Wasiak W. Needle-based extraction techniques with protected sorbent as powerful sample preparation tools to gas chromatographic analysis: Trends in application. J Chromatogr A. 2018;1565:1–18. doi: 10.1016/j.chroma.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 18.Grabowska-Polanowska B, Faber J, Skowron M, Miarka P, Pietrzycka A, Śliwka I, et al. Detection of potential chronic kidney disease markers in breath using gas chromatography with mass-spectral detection coupled with thermal desorption method. J Chromatogr A. 2013;1301:179–189. doi: 10.1016/j.chroma.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Theodoridis G, Koster EHM, de Jong GJ. Solid-phase microextraction for the analysis of biological samples. J Chromatogr B Biomed Sci Appl. 2000;745:49–82. doi: 10.1016/S0378-4347(00)00203-6. [DOI] [PubMed] [Google Scholar]
- 20.Pragst F. Application of solid-phase microextraction in analytical toxicology. Anal Bioanal Chem. 2007;388:1393–1414. doi: 10.1007/s00216-007-1289-9. [DOI] [PubMed] [Google Scholar]
- 21.Beale DJ, Pinu FR, Kouremenos KA, Poojary MM, Narayana VK, Boughton BA, et al. Review of recent developments in GC–MS approaches to metabolomics-based research. vol. 14Beale, D. Springer US; 2018. 10.1007/s11306-018-1449-2. [DOI] [PubMed]
- 22.Vazquez-Roig P, Pico Y. Gas chromatography and mass spectroscopy techniques for the detection of chemical contaminants and residues in foods. Chem. Contam. Residues Food, Elsevier Inc.; 2012, p. 17–61. 10.1533/9780857095794.1.17.
- 23.Misra BB. Advances in high resolution GC-MS technology: A focus on the application of GC-Orbitrap-MS in metabolomics and exposomics for FAIR practices. Anal Methods. 2021;13:2265–2282. doi: 10.1039/d1ay00173f. [DOI] [PubMed] [Google Scholar]
- 24.Smith L, Villaret-Cazadamont J, Claus SP, Canlet C, Guillou H, Cabaton NJ, et al. Important considerations for sample collection in metabolomics studies with a special focus on applications to liver functions. Metabolites 2020;10. 10.3390/metabo10030104. [DOI] [PMC free article] [PubMed]
- 25.González-Domínguez R, González-Domínguez Á, Sayago A, Fernández-Recamales Á. Recommendations and best practices for standardizing the pre-analytical processing of blood and urine samples in metabolomics. Metabolites. 2020;10:1–18. doi: 10.3390/metabo10060229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Yin P, Shao Y, Wang Z, Wang B, Lehmann R, et al. Which is the urine sample material of choice for metabolomics-driven biomarker studies? Anal Chim Acta. 2020;1105:120–127. doi: 10.1016/j.aca.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 27.Živković Semren T, Brčić Karačonji I, Safner T, Brajenović N, Tariba Lovaković B, Pizent A. Gas chromatographic-mass spectrometric analysis of urinary volatile organic metabolites: Optimization of the HS-SPME procedure and sample storage conditions. Talanta. 2018;176:537–543. doi: 10.1016/j.talanta.2017.08.064. [DOI] [PubMed] [Google Scholar]
- 28.Endo S, Pfennigsdorff A, Goss KU. Salting-out effect in aqueous NaCl solutions: Trends with size and polarity of solute molecules. Environ Sci Technol. 2012;46:1496–1503. doi: 10.1021/es203183z. [DOI] [PubMed] [Google Scholar]
- 29.Song H-N, Kim CH, Lee W-Y, Cho S-H. Simultaneous determination of volatile organic compounds with a wide range of polarities in urine by headspace solid-phase microextraction coupled to gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2017;31:613–622. doi: 10.1002/rcm.7827. [DOI] [PubMed] [Google Scholar]
- 30.Drabińska N, Małgorzata S, Krupa-Kozak U. Headspace Solid-Phase Microextraction Coupled with Gas Chromatography-Mass Spectrometry for the Determination of Volatile Organic Compounds in Urine. J Anal Chem. 2020;75:792–801. doi: 10.1134/S1061934820060088. [DOI] [Google Scholar]
- 31.Silva CL, Perestrelo R, Silva P, Tomás H, Câmara JS. Implementing a central composite design for the optimization of solid phase microextraction to establish the urinary volatomic expression: a first approach for breast cancer. Metabolomics. 2019;15:64. doi: 10.1007/s11306-019-1525-2. [DOI] [PubMed] [Google Scholar]
- 32.Deev V, Solovieva S, Andreev E, Protoshchak V, Karpushchenko E, Sleptsov A, et al. Prostate cancer screening using chemometric processing of GC–MS profiles obtained in the headspace above urine samples. J Chromatogr B Anal Technol Biomed Life Sci. 2020;1155:122298. doi: 10.1016/j.jchromb.2020.122298. [DOI] [PubMed] [Google Scholar]
- 33.Aggio RBM, Mayor A, Coyle S, Reade S, Khalid T, Ratcliffe NM, et al. Freeze-drying: An alternative method for the analysis of volatile organic compounds in the headspace of urine samples using solid phase micro-extraction coupled to gas chromatography - mass spectrometry. Chem Cent J. 2016;10:1–12. doi: 10.1186/s13065-016-0155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aggarwal P, Baker J, Boyd MT, Coyle S, Probert C, Chapman EA. Optimisation of urine sample preparation for headspace-solid phase microextraction gas chromatography-mass spectrometry: Altering sample ph, sulphuric acid concentration and phase ratio. Metabolites. 2020;10:1–17. doi: 10.3390/metabo10120482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cozzolino R, De Magistris L, Saggese P, Stocchero M, Martignetti A, Di Stasio M, et al. Use of solid-phase microextraction coupled to gas chromatography-mass spectrometry for determination of urinary volatile organic compounds in autistic children compared with healthy controls. Anal Bioanal Chem. 2014;406:4649–4662. doi: 10.1007/s00216-014-7855-z. [DOI] [PubMed] [Google Scholar]
- 36.Hua Q, Wang L, Liu C, Han L, Zhang Y, Liu H. Volatile metabonomic profiling in urine to detect novel biomarkers for B-cell non-Hodgkin’s lymphoma. Oncol Lett. 2018;15:7806–7816. doi: 10.3892/ol.2018.8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva CL, Passos M, Câmara JS. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br J Cancer. 2011;105:1894–1904. doi: 10.1038/bjc.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taunk K, Taware R, More TH, Porto-Figueira P, Pereira JAM, Mohapatra R, et al. A non-invasive approach to explore the discriminatory potential of the urinary volatilome of invasive ductal carcinoma of the breast. RSC Adv. 2018;8:25040–25050. doi: 10.1039/c8ra02083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lima AR, Pinto J, Azevedo AI, Barros-Silva D, Jerónimo C, Henrique R, et al. Identification of a biomarker panel for improvement of prostate cancer diagnosis by volatile metabolic profiling of urine. Br J Cancer. 2019;121:857–868. doi: 10.1038/s41416-019-0585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalid T, Aggio R, White P, De Lacy CB, Persad R, Al-Kateb H, et al. Urinary volatile organic compounds for the detection of prostate cancer. PLoS ONE. 2015;10:1–15. doi: 10.1371/journal.pone.0143283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto J, Amaro F, Lima AR, Carvalho-Maia C, Jerónimo C, Henrique R, et al. Urinary Volatilomics Unveils a Candidate Biomarker Panel for Noninvasive Detection of Clear Cell Renal Cell Carcinoma. J Proteome Res. 2021;20:3068–3077. doi: 10.1021/acs.jproteome.0c00936. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Wang C, Pi X, Guo L, Wang Y, Li M, et al. Urinary volatile organic compounds as potential biomarkers for renal cell carcinoma. Biomed Reports. 2016;5:68–72. doi: 10.3892/br.2016.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monteiro M, Moreira N, Pinto J, Pires-Luís AS, Henrique R, Jerónimo C, et al. GC-MS metabolomics-based approach for the identification of a potential VOC-biomarker panel in the urine of renal cell carcinoma patients. J Cell Mol Med. 2017;21:2092–2105. doi: 10.1111/jcmm.13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opitz P, Herbarth O. The volatilome - Investigation of volatile organic metabolites (VOM) as potential tumor markers in patients with head and neck squamous cell carcinoma (HNSCC) J Otolaryngol - Head Neck Surg. 2018;47:1–13. doi: 10.1186/s40463-018-0288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taware R, Taunk K, Pereira JAM, Dhakne R, Kannan N, Soneji D, et al. Investigation of urinary volatomic alterations in head and neck cancer: a non-invasive approach towards diagnosis and prognosis. Metabolomics. 2017;13:111. doi: 10.1007/s11306-017-1251-6. [DOI] [Google Scholar]
- 46.de Sousa Mesquita A, Zamora-Obando HR, Neves dos Santos F, Schmidt-Filho J, Cordeiro de Lima V, D’Almeida Costa F, et al. Volatile organic compounds analysis optimization and biomarker discovery in urine of Non-Hodgkin lymphoma patients before and during chemotherapy. Microchem J 2020;159:105479. 10.1016/j.microc.2020.105479.
- 47.Hanai Y, Shimono K, Matsumura K, Vachani A, Albelda S, Yamazaki K, et al. Urinary volatile compounds as biomarkers for lung cancer. Biosci Biotechnol Biochem. 2012;76:679–684. doi: 10.1271/bbb.110760. [DOI] [PubMed] [Google Scholar]
- 48.Porto-Figueira P, Pereira J, Miekisch W, Câmara JS. Exploring the potential of NTME/GC-MS, in the establishment of urinary volatomic profiles. Lung cancer patients as case study. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-31380-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jobu K, Sun C, Yoshioka S, Yokota J, Onogawa M, Kawada C, et al. Metabolomics study on the biochemical profiles of odor elements in urine of human with bladder cancer. Biol Pharm Bull. 2012;35:639–642. doi: 10.1248/bpb.35.639. [DOI] [PubMed] [Google Scholar]
- 50.Lett L, George M, Slater R, De Lacy CB, Ratcliffe N, García-Fiñana M, et al. Investigation of urinary volatile organic compounds as novel diagnostic and surveillance biomarkers of bladder cancer. Br J Cancer. 2022;127:329–336. doi: 10.1038/s41416-022-01785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyagi H, Daulton E, Bannaga AS, Arasaradnam RP, Covington JA. Non-invasive detection and staging of colorectal cancer using a portable electronic nose. Sensors. 2021;21:1–17. doi: 10.3390/s21165440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boulind CE, Gould O, Costello B de L, Allison J, White P, Ewings P, et al. Urinary Volatile Organic Compound Testing in Fast-Track Patients with Suspected Colorectal Cancer. Cancers (Basel) 2022;14. 10.3390/cancers14092127. [DOI] [PMC free article] [PubMed]
- 53.Díaz de León-Martínez L, Flores-Ramírez R, López-Mendoza CM, Rodríguez-Aguilar M, Metha G, Zúñiga-Martínez L, et al. Identification of volatile organic compounds in the urine of patients with cervical cancer. Test concept for timely screening. Clin Chim Acta 2021;522:132–40. 10.1016/j.cca.2021.08.014. [DOI] [PubMed]
- 54.Filipiak W, Ruzsanyi V, Mochalski P, Filipiak A, Bajtarevic A, Ager C, et al. Dependence of exhaled breath composition on exogenous factors, smoking habits and exposure to air pollutants. J Breath Res 2012;6. 10.1088/1752-7155/6/3/036008. [DOI] [PMC free article] [PubMed]
- 55.Longo V, Forleo A, Ferramosca A, Notari T, Pappalardo S, Siciliano P, et al. Blood, urine and semen Volatile Organic Compound (VOC) pattern analysis for assessing health environmental impact in highly polluted areas in Italy. Environ Pollut. 2021;286:117410. doi: 10.1016/j.envpol.2021.117410. [DOI] [PubMed] [Google Scholar]
- 56.O’Lenick CR, Pleil JD, Stiegel MA, Sobus JR, Wallace MAG. Detection and analysis of endogenous polar volatile organic compounds (PVOCs) in urine for human exposome research. Biomarkers. 2019;24:240–248. doi: 10.1080/1354750X.2018.1548031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocha SM, Caldeira M, Carrola J, Santos M, Cruz N, Duarte IF. Exploring the human urine metabolomic potentialities by comprehensive two-dimensional gas chromatography coupled to time of flight mass spectrometry. J Chromatogr A. 2012;1252:155–163. doi: 10.1016/j.chroma.2012.06.067. [DOI] [PubMed] [Google Scholar]
- 58.Wang C, Feng Y, Wang M, Pi X, Tong H, Wang Y, et al. Volatile Organic Metabolites Identify Patients with Mesangial Proliferative Glomerulonephritis, IgA Nephropathy and Normal Controls. Sci Rep. 2015;5:2–11. doi: 10.1038/srep14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu D, Zhao N, Wang M, Pi X, Feng Y, Wang Y, et al. Urine volatile organic compounds as biomarkers for minimal change type nephrotic syndrome. Biochem Biophys Res Commun. 2018;496:58–63. doi: 10.1016/j.bbrc.2017.12.164. [DOI] [PubMed] [Google Scholar]
- 60.Ligor T, Zawadzka J, Straczyński G, González Paredes RM, Wenda-Piesik A, Ratiu IA, et al. Searching for potential markers of glomerulopathy in urine by HS-SPME-GC×GC TOFMS. Molecules 2021;26. 10.3390/molecules26071817. [DOI] [PMC free article] [PubMed]
- 61.Wang M, Xie R, Jia X, Liu R. Urinary Volatile Organic Compounds as Potential Biomarkers in Idiopathic Membranous Nephropathy. Med Princ Pract. 2017;26:375–380. doi: 10.1159/000478782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cozzolino R, De Giulio B, Marena P, Martignetti A, Günther K, Lauria F, et al. Urinary volatile organic compounds in overweight compared to normal-weight children: Results from the Italian I.Family cohort. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-15957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eshima J, Davis TJ, Bean HD, Fricks J, Smith BS. A metabolomic approach for predicting diurnal changes in cortisol. Metabolites 2020;10. 10.3390/metabo10050194. [DOI] [PMC free article] [PubMed]
- 64.Arasaradnam RP, Westenbrink E, McFarlane MJ, Harbord R, Chambers S, O’Connell N, et al. Differentiating coeliac disease from irritable bowel syndrome by urinary volatile organic compound analysis - A pilot study. PLoS ONE. 2014;9:1–9. doi: 10.1371/journal.pone.0107312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mills GA, Walker V. Headspace solid-phase microextraction procedures for gas chromatographic analysis of biological fluids and materials. J Chromatogr A. 2000;902:267–287. doi: 10.1016/S0021-9673(00)00767-6. [DOI] [PubMed] [Google Scholar]
- 66.Stanstrup J, Broeckling CD, Helmus R, Hoffmann N, Mathé E, Naake T, et al. The metaRbolomics toolbox in bioconductor and beyond. Metabolites 2019;9. 10.3390/metabo9100200. [DOI] [PMC free article] [PubMed]
- 67.Baccolo G, Quintanilla-Casas B, Vichi S, Augustijn D, Bro R. From untargeted chemical profiling to peak tables – A fully automated AI driven approach to untargeted GC-MS. TrAC - Trends Anal Chem. 2021;145:116451. doi: 10.1016/j.trac.2021.116451. [DOI] [Google Scholar]
- 68.Mastrangelo A, Ferrarini A, Rey-Stolle F, García A, Barbas C. From sample treatment to biomarker discovery: A tutorial for untargeted metabolomics based on GC-(EI)-Q-MS. Anal Chim Acta. 2015;900:21–35. doi: 10.1016/j.aca.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-Mcintyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 70.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kovats E. Gas-Chromatographische Charakterisierung Organischer Verbindungen .1. Retentionsindices Aliphatischer Halogenide, Alkohole, Aldehyde Und Ketone. Helv Chim Acta 1958;41:1915–32. 10.1002/hlca.19580410703.
- 72.Khodadadi M, Pourfarzam M. A review of strategies for untargeted urinary metabolomic analysis using gas chromatography–mass spectrometry. Metabolomics. 2020;16:1–14. doi: 10.1007/s11306-020-01687-x. [DOI] [PubMed] [Google Scholar]
- 73.Noonan MJ, Tinnesand H V., Buesching CD. Normalizing Gas-Chromatography–Mass Spectrometry Data: Method Choice can Alter Biological Inference. BioEssays 2018;40. 10.1002/bies.201700210. [DOI] [PubMed]
- 74.Cuevas-Delgado P, Dudzik D, Miguel V, Lamas S, Barbas C. Data-dependent normalization strategies for untargeted metabolomics—a case study. Anal Bioanal Chem 2020:6391–405. 10.1007/s00216-020-02594-9. [DOI] [PubMed]
- 75.Wishart DS, Guo A, Oler E, Wang F, Anjum A, Peters H, et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–31. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gagnebin Y, Tonoli D, Lescuyer P, Ponte B, de Seigneux S, Martin PY, et al. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal Chim Acta. 2017;955:27–35. doi: 10.1016/j.aca.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 77.Han TL, Yang Y, Zhang H, Law KP. Analytical challenges of untargeted GC-MS-based metabolomics and the critical issues in selecting the data processing strategy. F1000Research 2017;6:1–17. 10.12688/f1000research.11823.1. [DOI] [PMC free article] [PubMed]
- 78.Mack CI, Egert B, Liberto E, Weinert CH, Bub A, Hoffmann I, et al. Robust Markers of Coffee Consumption Identified Among the Volatile Organic Compounds in Human Urine. Mol Nutr Food Res. 2019;63:1–12. doi: 10.1002/mnfr.201801060. [DOI] [PubMed] [Google Scholar]
- 79.Edmands WMB, Ferrari P, Scalbert A. Normalization to specific gravity prior to analysis improves information recovery from high resolution mass spectrometry metabolomic profiles of human urine. Anal Chem. 2014;86:10925–10931. doi: 10.1021/ac503190m. [DOI] [PubMed] [Google Scholar]
- 80.Vollmar AKR, Rattray NJW, Cai Y, Santos-Neto ÁJ, Deziel NC, Jukic AMZ, et al. Normalizing untargeted periconceptional urinary metabolomics data: A comparison of approaches. Metabolites 2019;9. 10.3390/metabo9100198. [DOI] [PMC free article] [PubMed]
- 81.Djoumbou Feunang Y, Eisner R, Knox C, Chepelev L, Hastings J, Owen G, et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform. 2016;8:61. doi: 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lange J, Eddhif B, Tarighi M, Garandeau T, Péraudeau E, Clarhaut J, et al. Volatile Organic Compound Based Probe for Induced Volatolomics of Cancers. Angew Chemie - Int Ed. 2019;58:17563–17566. doi: 10.1002/anie.201906261. [DOI] [PubMed] [Google Scholar]
- 83.Pang Z, Chong J, Zhou G, De Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:388–96. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torres S, Merino C, Paton B, Correig X, Ramírez N. Biomarkers of exposure to secondhand and thirdhand Tobacco smoke: Recent advances and future perspectives. Int J Environ Res Public Health. 2018;15:1–25. doi: 10.3390/ijerph15122693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arasaradnam RP, Mcfarlane MJ, Ryan-Fisher C, Westenbrink E, Hodges P, Thomas MG, et al. Detection of colorectal cancer (CRC) by urinary volatile organic compound analysis. PLoS One 2014;9. 10.1371/journal.pone.0108750. [DOI] [PMC free article] [PubMed]
- 86.Reyes-Garcés N, Gionfriddo E, Gómez-Ríos GA, Alam MN, Boyacl E, Bojko B, et al. Advances in Solid Phase Microextraction and Perspective on Future Directions. Anal Chem. 2018;90:302–360. doi: 10.1021/acs.analchem.7b04502. [DOI] [PubMed] [Google Scholar]
- 87.Kaziur-Cegla W, Salemi A, Jochmann MA, Schmidt TC. Optimization and validation of automated solid-phase microextraction arrow technique for determination of phosphorus flame retardants in water. J Chromatogr A. 2020;1626:461349. doi: 10.1016/j.chroma.2020.461349. [DOI] [PubMed] [Google Scholar]
- 88.Drabińska N, Młynarz P, De Lacy Costello B, Jones P, Mielko K, Mielnik J, et al. An optimization of liquid-liquid extraction of urinary volatile and semi-volatile compounds and its application for gas chromatography-mass spectrometry and proton nuclear magnetic resonance spectroscopy. Molecules 2020;25. 10.3390/molecules25163651. [DOI] [PMC free article] [PubMed]
- 89.Reinke SN, Naz S, Chaleckis R, Gallart-Ayala H, Kolmert J, Kermani NZ, et al. Urinary metabotype of severe asthma evidences decreased carnitine metabolism independent of oral corticosteroid treatment in the U-BIOPRED study. Eur Respir J. 2021 doi: 10.1183/13993003.01733-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.World Anti-Doping Agency. Urine Sample Collection Guidelines. Int Stand Test Investig 2014:1–45.
- 91.Meister I, Zhang P, Sinha A, Sköld CM, Wheelock ÅM, Izumi T, et al. High-Precision Automated Workflow for Urinary Untargeted Metabolomic Epidemiology. Anal Chem. 2021;93:5248–5258. doi: 10.1021/acs.analchem.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagarajan SR, Butler LM, Hoy AJ. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metab. 2021;9:1–28. doi: 10.1186/s40170-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agarwal SM, Sharma M, Fatima S. VOCC: a database of volatile organic compounds in cancer. RSC Adv. 2016;6:114783–114789. doi: 10.1039/C6RA24414A. [DOI] [Google Scholar]
- 94.Janfaza S, Khorsand B, Nikkhah M, Zahiri J. Digging deeper into volatile organic compounds associated with cancer. Biol Methods Protoc. 2019;4:1–11. doi: 10.1093/biomethods/bpz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giró Benet J, Seo M, Khine M, Gumà Padró J, Pardo Martnez A, Kurdahi F. Breast cancer detection by analyzing the volatile organic compound (VOC) signature in human urine. Sci Rep. 2022;12:1–13. doi: 10.1038/s41598-022-17795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The biomedical untargeted urinary volatilome database (uBIOVOC DB). Table S2. Summary of the sample collection conditions used by the 34 analysed studies. Table S3. Summary of the analytical conditions used by the 34 studies using them. Table S4. Summary of the GC-MS conditions used by the 34 analysed studies. Table S5. Summary of the applications analysed by the 34 analysed studies. Table S6. The urinary volatilome database by biomedical applications.
Data Availability Statement
The datasets generated during the current study are available in the Zenodo repository, link 10.5281/zenodo.6883341