Abstract
Membrane vesicles having a diameter of 30–150 nm are known as exosomes. Several cancer types secrete exosomes, which may contain proteins, circular RNAs (circRNAs), microRNAs, or DNA. CircRNAs are endogenous RNAs that do not code for proteins and can create continuous and covalently closed loops. In cancer pathogenesis, especially metastasis, exosomal circRNAs (exo-circRNAs) have a crucial role mainly due to the frequently aberrant expression levels within tumors. However, neither the activities nor the regulatory mechanisms of exo-circRNAs in advancing lung cancer (LC) are obvious. A better understanding of the regulation and network connections of exo-circRNAs will lead to better treatment for LCs. The main objective of the current review is to highlight the functions and mechanisms of exo-circRNAs in LC and assess the relationships between exo-circRNA dysregulation and LC progression. In addition, underline the possible therapeutic targets based on exo-circRNA modulating.
Keyword: Lung Cancer (LC), Circular RNA (circRNA), Exosomal circular RNA (exo-circRNA)
Introduction
Lung cancer (LC) is the most frequent type of cancer worldwide and the leading cause of cancer mortality [1]. An essential factor in LC deaths is the invasion and metastasis of cancer cells through the circulation or lymphatic systems, Which is a significant cause of mortality in patients [2]. Tumor-derived exosomes (TDEs) play a vital function in the tumor microenvironment by facilitating the development of a pre-metastatic niche [3]. Exosomes are small membrane vesicles with a diameter of 30–150 nm that are made in the endosomal part of a cell. They are involved in the intercellular regulation of pathophysiologic processes and serve as intercellular messengers that transport a variety of substances in a phospholipid bilayer membrane [4]. Exosomal circRNAs (exo-circRNAs) refer to the circRNAs discovered in exosomes [5]. When exosomes are released from cells, they are taken up by distant cells. Exosomes containing circRNAs regulate the TME to promote tumor cell proliferation, invasion, and metastasis [6, 7].
CircRNAs are closed, single-stranded RNA molecules without poly (A) tails and 5′-3′ ends, and compared to linear transcripts, they are more stable as they resist exonuclease-mediated destruction [8]. In 1979, endogenous circRNAs were discovered to be a byproduct of eukaryotic RNA splicing [9]. In 1986, the hepatitis delta virus caused circRNAs to be found in humans [10]. Almost 10,000 circRNAs have been identified, occurring naturally in many different organisms, from fungi to plants to vertebrates [11]. Currently, circRNAs are categorized into four classes: intergenic circRNAs, ecircRNAs, EIciRNAs, and exon–intron circRNAs [12]. Several studies have indicated that circRNAs are associated with various human disorders, including malignancies [13–17]. However, the mechanism and function of circRNAs have not been completely understood.
Exosomes are vesicles released from cancer cells; they carry circRNAs, which play an important role in cancer progression at multiple stages, including the proliferation of malignant tumors, formation of premetastatic niches, and metastasis of cancer cells to distant places [18, 19]. Li and his colleagues published the first study to know the expression levels of circRNAs in extracellular vesicles using the RNA-seq technique. They found that circRNAs are abundant at least twofold in exosomes than in cells and more stable [20]. In humans, around 60% of genes can express circRNA [21]. However, the tissue expression of these genes is still low, making up just 5–10% of the average mRNA expression in a specific tissue [22, 23].
Nevertheless, the relationship between exo-circRNAs and the promotion or inhibition of LC is still not well understood. Hence, this study provides recent studies on the functions and mechanisms of exo-circRNAs in LC and explains the connections between the dysregulation of exo-circRNAs and lung cancer progression. We also focused on possible therapeutic targets based on circRNA modulation and their potential function in promoting or inhibiting LC progression.
Biogenesis of exosomes
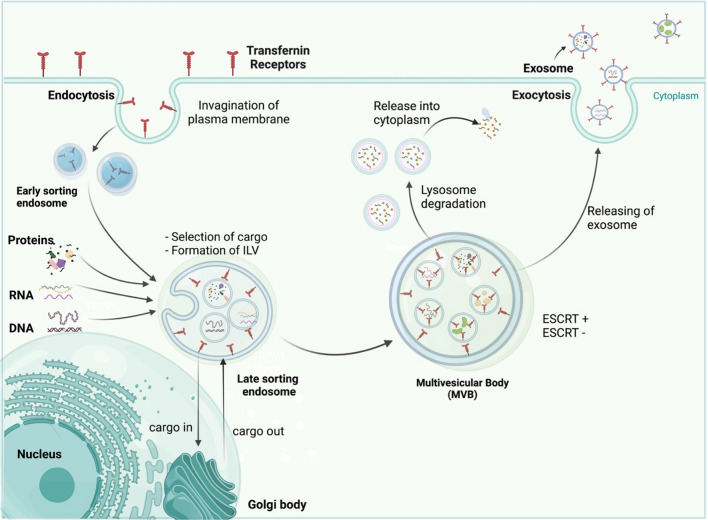
Exosomes originate from late endosomes, formed by the inward budding of the limited multivesicular body (MVB) membrane. The invagination of late endosomal membranes leads to the release of intraluminal vesicles (ILVs) inside massive MVBs [24]. Several proteins are taken to the invaginating membrane during this process. Meanwhile, the cytosolic components are taken up by the ILVs. Following fusion with the plasma membrane, most ILVs are discharged into the extracellular space, called exosomes, and move into body fluids [25, 26]. Eventually, these elements are taken by lysosomes, where they are broken down or released into the extracellular space after fusion with the plasma membrane [27] (Fig. 1). Endosomal-sorting complex that is required for transport (ESCRT) is necessary for both exosome biosynthesis and secretion [28]. Proteins such as ALIX, Tsg101, VPS4, and the four subunits of ESCRT (ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III), make up ESCRT. ESCRT-0 carries out the sorting of cargo proteins into the lipid domain. Membrane deformation is carried out by the other ESCRTs I and II; the VPS4 complex is recruited to ESCRT-III, responsible for the vesicle neck scission and the dissociation or recycling of the ESCRT-III complex [29–31]. Through its interaction with the syndecan receptor, the exosomal protein Alix has been demonstrated to play a role in endosomal membrane budding and abscission and the selection of specific exosomal cargo [32]. In light of these findings, it was hypothesized that the ESCRT has a vital role in exosomal biogenesis.
Fig. 1.
This illustration shows how exosomes are formed in the body and then released. Three processes contribute to exosome secretion: exosome biosynthesis, MVB transport to the cell membrane, and MVB fusion with the cell membrane
After exosomes are released, they can send signals to target cells through endocytosis, a fusion of membranes, and interactions between receptors and ligands. Clathrin, caveolin, and lipid raft-mediated endocytosis can engulf exosomes into specific cells [33]. Endocytosed exosomes can either combine with nearby endosomes or be transported to lysosomes, where they are degraded [34]. The exosomal membrane also can bind to particular receptors on the plasma membrane of the recipient cell to initiate signaling pathways or to fuse with the plasma membrane of the recipient cell to distribute its contents [35–37].
Biogenesis of circRNAs
Synthesis of circRNAs from segments of pre-messenger RNAs can occur by back-splicing, a process in which the 5' splice donor joins with the 3' splice receiver through a phosphodiester bond. This biological process can create a circular structure with one or more exonic/intronic regions [38]. Numerous nuclear back-splicing and linear splicing processes have been described, including exon skipping, intron pairing, and RNA-binding proteins (RBPs) [39] (Fig. 2). The first is an RBP-assisted circularization process that generally involves the association of two neighboring exons and skipping the intronic region, producing an exonic-circRNA. Numerous RBPs regulate this process, including RNA helicase DHX9 [40], FUS [41], ADAR1 [42], NF90/NF110 [43], MBL [44], QKI [45], and heterogeneous nuclear ribonucleoprotein L [46].
Fig. 2.
The process of biogenesis that occurs during lung cancer, in addition to the roles that exosomal circRNAs play in the disease
Exon–intron circRNAs are made when two or more exons and their correlating introns circle. Intron pairing back-splicing is a popular approach in the conserved RNAs with many Alu repetitions in the sequences on either side. These Alu components work well together, promoting the configuration of hairpins and more back-splicing, leading to mono-EcircRNAs [47]. Another type of this category is the intronic circRNAs, but it is still unknown how these molecules are produced.
CircRNAs are exported into the cytoplasm after being synthesized in the nucleus. According to recent studies, the UAP56/URH49 helicases are actively involved in this size-mediated mechanism. Transferring molecules larger than 1300 nucleotides requires UAP55, whereas URH49 only interferes with short transcript exports [48]. Following their entry into the cytoplasm, circRNAs accumulate and regulate transcription by sponging certain types of miRNAs, as seems to be usual for most cells. Although the process by which circRNA degrades is still unknown, recent research has provided insights into this issue and shown some exciting pathways that explain circRNA disintegration. For example, Hansen et al. revealed a mechanism whereby Ago2 and miR-671 degrade circRNA-CDR1as [49]. Likewise, Park and his colleagues showed that a circRNA cleavage process is mediated by RNase P/MRP and outlined in N6-methyladenosine (m6A)-enriched circRNAs [50]. In recent work, Liu et al. [51] showed that certain circRNAs tend to form complicated duplexes, which renders them vulnerable to destruction by RNase L during viral infection.
Biological functions of circRNAs
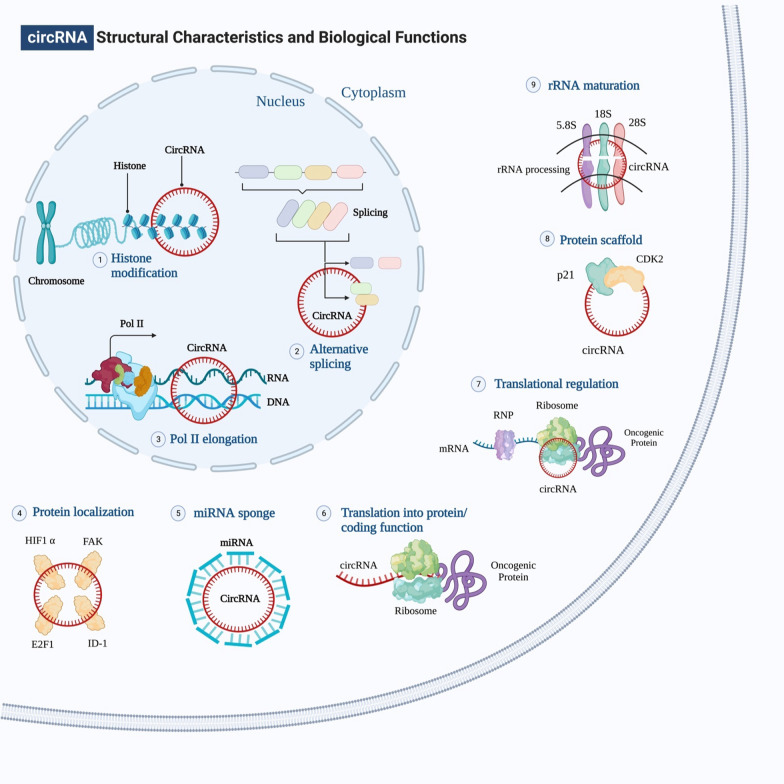
Many studies have highlighted that circRNAs may control gene expression either directly or indirectly by binding to miRNAs, RBPs, and other regulators of gene expression and managing various biological processes (Fig. 3). The mechanisms of circRNAs that are used in regulating gene expression are as follows.
Fig. 3.
Structural characteristics and biological functions of circRNAs. Inside the cell, circular RNAs have many multiple roles to do. In the nucleus, circRNAs can silence a specific locus by interacting with the histone methylation pattern. They can also control the transcription of their own gene by interacting with RNA polymerase II. Finally, alternative splicing can be blocked by their competition with mRNA for splice sites. CircRNAs are found in the cytoplasm, where they can function as miRNA sponges, decoy certain transcription factors, be translated into proteins, bind with RNA-binding proteins to regulate translation of particular mRNAs, and serve as protein scaffolds. In addition to promoting cell death by interfering with the processing of pre-rRNA components, circRNAs can bind to some proteins and inhibit their signal transduction activity
As miRNA sponge
The most critical function of circRNAs is to act as a miRNA sponge to regulate the expression of a target gene by inhibiting the activity of miRNA [52]. A single circRNA can bind to one or more miRNAs at one or more locations by perfect or near-perfect binding [53]. ‘‘Super sponges’’ like circRNAs are selectively attracted to miRNAs rather than other ceRNAs, such as lncRNAs and pseudogenes. The first example of a circRNA that functions as a miRNA sponge is CDR1as [54]. It has 74 miR-7 binding sites and is closely attached to AGO proteins. [55]. Gao and Ye et al. found that circ-SOX4 stimulated the growth of LUAD and activated the WNT axis by sponging miRNA-1270 and altering PLAGL2, providing a relevant conceptual framework for studying the therapeutic LUAD targets [56]. Additionally, circHIPK3 is derived from Exon2 of the HIPK3 gene, a key player in cell proliferation in human cancer, by sponging nine miRNAs with 18 binding sites into cells [57].
Despite the above, according to Militello et al. [58], some types of circRNAs, such as (circ_0005939 and circ_0013647) are unable to act as miRNA sponges. Therefore, additional work is needed to determine how circRNAs, miRNAs, and mRNAs work together.
Alternative splicing and transcriptional regulation
One of the most prevalent methods of controlling gene expression is alternative splicing, which is essential for enhancing functional proteins' complexity. Recently, it’s been shown that some circRNAs are highly concentrated in the nucleus, where they could potentially inhibit transcription. For instance, circURI1 may influence alternative splicing to promote cancer development and metastasis [59]. Likewise, EIciRNAs are circRNAs that have introns and exons [60]. Therefore, EIciRNAs are found in the nucleus and act as transcriptional regulators [61]. Besides, EIciRNAs regulate RNA polymerase II (Pol II) activity and trigger the transcription of parental genes [62]. EIciRNAs and Pol II work together to promote transcriptional initiation by making it easier for Pol II to bind with the core promoter of EIciRNA parent genes [63]. Similarly, the EIciRNAs and the U1 snRNA (small nuclear ribonucleoprotein) attach in an RNA-RNA manner, which makes it possible for the EIciRNAs and pol II to interact with one another [64]. Additionally, circRNAs, such as exon–intron circular RNAs (circPAIP2, circEIF3J), could attach to Pol II and control their host gene expression [65]. Accordingly, these studies suggest that intron-derived circRNAs are responsible for regulating the transcription process in the nucleus.
Translation
Endogenous circRNAs have been shown recently to be capable of protein translation. The protein-coding capacity of circRNAs was previously thought to be low, but it has been proven that circRNAs with IRES or N6-methyladenosine modifications can often be translated into peptides [66, 67]. In eukaryotic cells, untranslated regions (UTR) are necessary for the beginning of the translation process, specifically 5' and 3' positions. Due to the absence of 5' and 3' ends, circRNAs were previously categorized as ncRNAs. Growing data proved that circRNAs might be able to code for proteins since they can be coupled to polysomes, and some of them have AUG start codon in addition to putative ORFs with favorable lengths [68, 69]. According to the Legnini et al. study, the back splicing result of ZNF609 exon 2, known as circ-ZNF609, can be translated into a protein in both a splicing-dependent and a splicing-independent manner throughout the process of myogenesis [70]. However, it's not clear how standard circRNA translation occurs yet, and it's also not clear what the translated proteins might perform or what components are involved in the process. Despite its novelty and significance, the study of how circRNAs are translated into peptides or proteins has been published in only a few studies due to limitations in analysis and validation methodologies.
CircRNAs and RNA-binding proteins
Recent research has shown that circRNAs work like miRNA sponges, inhibiting miRNA function while also taking part in splicing target genes, translating genes into proteins, and interacting with RNA-binding proteins (RBPs). Interaction with RBPs is a crucial component in the actions of circRNAs, which include biogenesis, translation, control of target genes, and extracellular transport [71]. For instance, circBIRC6 is highly represented in the Ago2 binding complex and mediates pluripotency in hESCs by inhibiting differentiation through direct interactions with miR34a and miR145 [72]. Similarly, stat3 binding circAmotl1 and increasing nuclear translocation enhanced cell activity. Nuclear Stat3 would bind to Dnmt3a's promoter, increasing transcription and translation. Then, the miR-17 promoter is demethylated by Dnmt3a, which reduces the production of miR-17-5p [73]. These show a feedback loop in which circRNA-based RBPs bind together and perform different regulatory functions.
Implication of exosomal circRNA in lung cancer progression
According to several studies, exosomes contain a variety of non-coding RNAs (ncRNAs), including miRNA, lncRNA, circRNA, and rRNA [74–76]. In contrast to cells that release circRNA, also circRNAs are highly concentrated and persistent in exosomes, particularly in those generated from tumors.
Exosomal circRNAs are involved in several critical biological processes that promote or inhibit cancer [77, 78]. More evidence suggests that exo-circRNAs play a crucial role in several malignancies, including lung cancer, through different mechanisms (Table 1). Exosomal circRNAs have a similar physiological role in malignancies via the miRNA sponge [79]. For instance, circ 0013958, a molecular sponge for miR-134 in LC, was connected with lymphatic metastasis and the TNM stage [80]. Likewise, circFARSA promotes the progression of LC through sponging miR-326 and miR-330-5p, thereby allowing these miRNAs to lose their control of the FASN oncogene, which is the gene that causes cancer [65, 81]. Moreover, exosomes containing exo-hsa_circRNA_0056616 were highly expressed in tissues from lung adenocarcinomas that had lymph node metastases [82]. Similarly, overexpression of circCCDC66 by STAT3 increases the growth of NSCLC by affecting the miR-33a-5p/KPNA4 pathway [83]. Furthermore, circABCB10 altered the miR-584-5p/E2F5 axis to accelerate the development of NSCLC [84]. On the other hand, exosomal circPVT1, which is produced by LC cells, activates the axis of miR-124-3p/EZH2 to polarize macrophages and increase lung tumor cell invasion and migration [85]. Exo-circRNAs, taken as a whole, might be an important factor in the advancement of LC. Table 1 lists the patterns of oncogenic exo-circRNA expression, along with the genes they target and the mechanisms of actions with their functions.
Table 1.
Oncogenic exo-circRNAs and their role in lung cancer
| Exo-circRNA | Number of clinical samples | Types of samples | Animal model | Expression | Target genes | Mechanisms | Functions | Refs |
|---|---|---|---|---|---|---|---|---|
| CDR1-AS | 104 LUAD patients | PAEC, LUAD PC9, A549 | – | Up | PTX, CDDP | EGFR/PI3K pathway | Independent prognostic biomarker for LUAD patients | [165] |
| hsa_circ_0014235 | Tumor tissues 35 samples and adjacent 35 samples | A549, H1299, 16HBE | Nude mice | Up | miR-520a-5p, DDP, CDK4 | miR-520a-5p/CDK4 regulatory axis | An increase in DDP resistance and promotion of cancerous cell activity | [100] |
| hsa_circ_0056616 | 42 lung adenocarcinomas with lymph node metastases, 48 without | PC9, PC14 | – | Up | CXCR4 | – | CXCR4 knockdown inhibits colony formation, cell proliferation, migration, and invasiveness | [82] |
| Circ-MAN2B2 | – | BESA-2B, A549, H226, H1299, H446 | – | Up | miR-1275, FOXK1 | CircMAN2B2/miR-1275/FOXK1 signaling | Act as an oncogene, which promotes lung cancer cell proliferation and invasion | [166] |
| hsa_circ_0013958 | 49 pairs of LAC samples | A549, H1299, BEAS-2B | – | Up | miR-34, CCND1 | – | Encouraging cell growth and invasion while discouraging cell death | [167] |
| hsa-circRNA-002178 | 105 paired LUAD and noncancerous tissue samples | 95D, PC9, A549, BEAS-2B | – | Up | miR-34a, miR-28-5p, PDL1, PD1 | – | Increase PDL1 and PD1 expression in tumor cells | [168] |
| Circ-CPA4 | NSCLC patients (N = 50) | cA549, H1299, SK-MES-1, Calu-3, HBE | Nude mice | Up | miR-134, let-7 miRNA, PD-L1 | Let-7 miRNA/PDL-1 axis | Immunity evasion | [169] |
| CircFARSA | 10 pairs of tumor and adjacent normal tissues | A549 | – | Up | miR-330-5p, miR-1270, miR-1178-3p, miR-620, miR-326 | – | A novel biomarker for NSCLC | [170] |
| Circ_0014130(circPIP5K1A) | – | H1299, PC9, H1975, A549, H1650, BEAS‐2B | Nude mice | Up | miR-600, HIF‐1α gene 3′‐UTR | CircPIP5K1A/miR‐600/HIF‐1α axis | MiR600 reduced HIF1-mediated metastasis and cancer growth | [171] |
| Circ_RAD23B | 40 NSCLC samples and paired adjacent normal tissue specimens | H1299, H1581, H358, A549, 16HBE | – | Up | miR593-3p, miR-653-5p, CCND2, TIAM1 | MiR-593e3p/CCND2 axis, and miR-653e5p/TIAM1 pathway | Function as oncogene by miRNA sponge | [172] |
| CircPVT1 | – | – | – | Up | miR-124-3p, EZH2 | MiR-124-3p/EZH2 axis | Exosomal circPVT1 enhances proliferation and metastasis by polarizing macrophages through miR-124-3p/EZH2 | [85] |
| Circ-MEMO1 | Tissue samples 52 tumor and adjacent normal | H1650, A549, H1299, PC9, HBE | Nude mice | Up | miR-101-3p, 3′’ UTR of KRAS | MiR-101-3p/KRAS Axis | MiR-101-3p targeting KRAS increased NSCLC progression and glycolysis | [173] |
| Serum samples; 30 patients and 25 healthy | ||||||||
| Circ-PRMT5 | 90 pairs of cancer and adjacent normal tissues | A549, 95-D, HCC827, H1299, SK-MES-1, HBE | Nude mice | Up | miR-377/382/498 | MiR-377/382/498-EZH2 | Circ-PRMT5 promotes NSCLC growth by miR-377/382/498 sponging and upregulating EZH2 | [174] |
| pathway | ||||||||
| CircHIPK3 | 3 different primary lung cancer | A549, BEAS-2B | – | Up | miR124 | CircHIPK3-miR-124 pathway | Promotes lung cancer cell progression via miRNA sponging | [175] |
| patients | ||||||||
| CircRNA CCDC66 | 628 patients with newly diagnosed NSCLC | H125, H23, H226, H838, H1437, H2009, H2087, A549, H125, H23, H838, H1437, H2009, H2087, A549 | – | Up | ATAD3A, SAE2, CCDC66, EGFR | HGF/c-Met axis | Enhance LADC cell EMT and drug resistance | [176] |
| Circ-STXBP5L | – | Up | miR-224-3p and miR-512-3p | – | Circ-STXBP5L target miRNAs, causing LC progression | [177] | ||
| hsa_circ_0000064 | – | A549, H1299 | – | Up | Caspase-3, 9, BAX, p21, cyclin D1, CDK6, MMP-2, 9 | – | Induces cancer cell proliferation, apoptosis and metastasis | [178] |
| Circ-FOXM1 | 80 NSCLC patients | H1299, A549, SK-MES-1, Calu-3HBE | – | Up | miR-1304-5p, PPDPF, MACC1 | Circ-FOXM1/miR-1304-5p/PPDPF/MACC1 axis | Increases cellular growth and proliferation by sponging miR-1304-5p to target PPDPF and MACC1 | [179] |
| Circ_0047921 | patients (n = 60) | H1299, A549, H1650, Calu3, SK-MES1, BEAS-2B | Nude mice | Up | miR-1287-5p, LARP1 | Circ_0047921/miR-1287-5p/LARP1 axis | Circ0047,921 serves as miR-1287-sponge, controlling LC cell proliferation, migration, and glycolysis | [180] |
| Circ-0006006 | – | A549, H1299 | Nude mice | – | miR-924, SRSF7 | MiR-924/SRSF7 axis | Accelerated NSCLC development by regulation of SRSF7 expression via miR-924 sponging | [181] |
| Circ_0008717 | 48 NSCLC patients and 48 control samples | A549, H1299, BEAS-2B | Nude mice | High expression | miR-1287-5p, PAK2 | miR-1287-5p/P21-mediated kinase 2 (PAK2) pathway | Promotes carcinogenesis in NSCLC by increasing expression of PAK2 via miR-1287-5p sponging | [182] |
| CircMAGI3 | 30 NSCLC patients | H322, H460, A549, H1299, NHBE | Nude mice | up | HDGF, miR-515-5p | CircMAGI3/miR-515-5p/HDGF pathway | Stimulates cell glycolysis and NSCLC cell proliferation | [183] |
| Circ-ABCB10 | 40 NSCLC patient samples | SPC-A1, HCC827, H1975, H1650, PC9, A549 | Nude mice | up | MiR-584-5p, E2F5 | MiR-584-5p/ E2F5 pathway | Participate in the upregulation of E2F5 expression by sponging miR-584-5p | [84] |
| hsa_circ_0062389 | 33 paired of NSCLC samples | H1650, H23, H522, A549, H1703, H460, BEAS-2B | – | up | MiR-103a-3p, CCNE1 | MiR-103a-3p/CCNE1 axis | Utilize miR-103a-3p as a sponge to control CCNE1 expression in LC | [184] |
| Circ_0072088 | 20 patients with LUAD | H1299, H1975, H520, H827 | – | Up | MiR-1261, PIK3CA | Circ_0072088/miR-1261/PIK3CA regulatory pathway | Tumorigenesis and progression of LUAD | [185] |
| Circ_0007385 | – | – | Nude mice | Up | MiR-1253, FAM83A | MiR-1253/FAM83A axis | Promoted NSCLC cell proliferation and stemness | [186] |
| CircTUBA1C | 30 pairs of LC tissue samples | Calu-3, A549 | Nude mice | Up | MiR-143-3p, Cyclin B1, PCNA, BAX, caspase-3 | CircTUBA1C/miR-143-3p axis | CircTUBA1C sponges miR-143-3p to promote NSCLC | [187] |
| Circ-PITX1 | 40 patients with primary NSCLC | H1975, A549, BEAS | Nude mice | Up | MiR-30e-5p, ITGA6 | MiR-30e-5p/ITGA6 axis and ITGA6/PI3K/Akt pathway | MiRNA sponge | [188] |
| CircFECR1 | 35 moderate and 26 extensive SCLC patients | NCI-H460,NCI-H446, NCI-H2170,NCI-H1688, NCI-H1299, HCC-827 | Nude mice | Up | MiR584-3p, ROCK1 | MiR584–ROCK1 pathway | miR584-3p ensnared and deactivated by FECRs, which triggered the ROCK1 pathway | (189) |
| CircRNA-102481 | 58 NSCLC patients | PC9 | – | Up | MiR-30a-5p sponge, ROR1 | CircRNA_1024810/miR-30a-5p/ROR1 axis | promotes EGFR-TKI resistance through the miR-30a-5p/ROR1 pathway | [190] |
| CircSATB2 | 59 NSCLC and normal tissue samples | BEAS-2B, A549, H460, H1299, H226, MES-1 | – | Up | MiR-326, FSCN1 | – | Encourages LC to grow, spread, and invade | [191] |
| Hsa_circ_0002130 | 28 osimertinib-resistant LC (non-response) and 32 sensitive (response) | HCC827, H1975 | Nude mice | Highly expressed | MiR-498, GLUT1, HK2, LDHA | – | Osimertinib-resistant NSCLC promotion | [192] |
| Circ_100876 | – | A549, NCI-H23 | – | Up | Targeting miR-636, RET | MiR-636/RET axis | CircRNA 100876 downregulation decreased NSCLC via the miR-636/RET pathway | [193] |
| Circ_0002346 | 45 NSCLC tissue specimens | HBE, A549, H1299 | Nude mice | Up | miR-582-3p, STXBP6 | miR-582-3p/STXBP6 pathway | Circ 0002346 sponges miR-582-3p to promote STXBP6 in NSCLC cells | [194] |
| Hsa_circ_0018818 | 30 pairs of LC and normal tissues | A549, NCI-H1650, PC-9, 293 T, NCI-H441, BEAS-2B | Nude mice | Up | miR-767-3p, NID1 | miR-767-3p/NID1 signaling pathway | Targeted shRNA decreased NSCLC cell growth, invasion and induced the apoptosis process | [195] |
Exosomal circRNAs and EMT
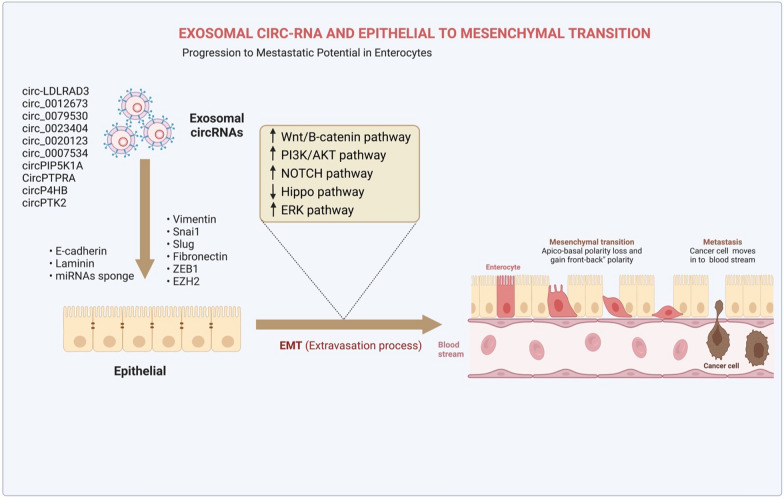
Once epithelial cells gain motility, a process known as the epithelial-mesenchymal transition (EMT) takes place and adopts a mesenchymal phenotype while retaining their invasive abilities [86]. Such an approach has been extensively seen in various biological phenomena, such as embryogenesis, fibrosis, cancer growth, and metastasis [87]. Like other malignant tumors, LC can spread and invade tissue due to the EMT process [88]. A high abundance of circRNAs is observed in LC, and some of them play oncogenic functions by promoting EMT processes in vitro (Fig. 4). For example, Inhibition of microRNA-137 by circ-LDLRAD3 led to an increase in glutamine transporter, a member of the SLC1A5in NSCLC cells, hence promoting proliferation and EMT [89]. Specifically, SLC1A5 was crucial for developing and controlling LC, and its inactivation was found to reduce the viability of LC cells [90]. Additionally, circ 0012673 enhances the proliferation and invasion of LUADs [91]. Reducing circ 0012673 levels inhibited cell growth, motility, and EMT via upregulation of LIM domain kinase 1in LUAD cell lines while simultaneously triggering apoptosis via miR-320a targeting [91]. According to Li et al., overexpression of hsa circ 0079530 stimulated cancer cells to migrate and invade through controlling EMT processes [92]. Similarly, EMT-related protein expression is regulated by hsa circ 0023404 through modulation of the miR-217/zinc finger E-box-binding homeobox 1 (ZEB1) axis and promoting LC cell growth [93].
Fig. 4.
This illustration highlights the key roles of exosomal circRNAs in the EMT process in lung cancer. Exosomal circRNAs which are overexpressed and play oncogenic functions by promoting EMT processes through promoting or/and inhibiting different pathways in lung cancer
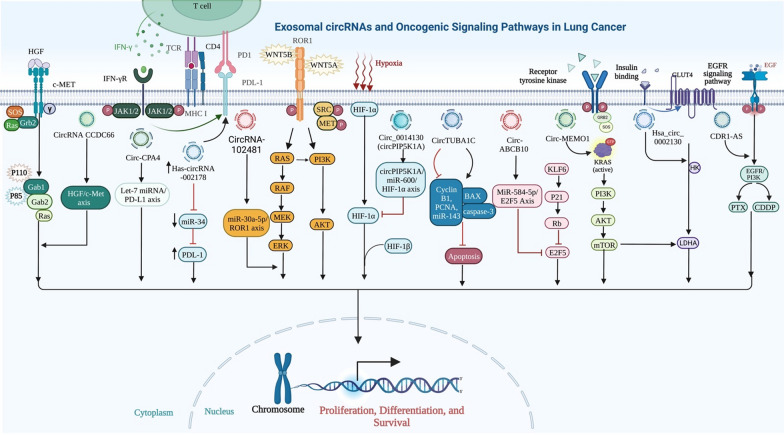
Furthermore, EMT plays a crucial role in LC, and numerous in vivo and in vitro studies have demonstrated that oncogenic circRNAs speed up this process through a number of pathways (Fig. 5). For instance, Qu and colleagues revealed that hsa circ 0020123 inhibited LC apoptosis by decreasing miRNA-144 and increasing ZEB1 and EZH2 expression [94]. Their results demonstrated that knocking down hsa_circ_0020123 slowed the growth and spread of LC cells. According to a recent study, circPIP5K1A functioned as a miR-600 sponge to increase LC development by increasing HIF-1α and inhibiting miR-600's effect on EMT-related proteins [95]. Similarly, in vitro experiments showed that circP4HB stimulated EMT processes in LC via sponging miR-133a-5p, as demonstrated by an increase in vimentin expression [96].
Fig. 5.
Illustration shows the connection between oncogenic signaling pathways and exosomal circRNAs in LC. CircRNA can promote tumor cell proliferation, invasion, migration, and survival by targeting particular genes and sponging various types of microRNAs, such as miR-101-3p, miR-498, miR-584-5p, miR-143-3p, and miR-600. CircRNA can act as an oncogene and promote the proliferation of cancer cells by involving in several essential signaling pathways in lung cancer, including EGFR/PI3K, miR-101-3p/KRAS, and miR-584-5p/E2F5 pathways
Despite this, a number of circRNAs are significantly suppressed in vitro and in vivo in LC, and, through positively regulating the EMT process, they prevent cancer progression (Table 2). For example, circPTK2, a miRNA sponge, was positively correlated with TIF1-y expression in human NSCLC tissue. [97]. Furthermore, overexpression of circPTK2 was found to elevate TIF1-y levels and suppress the TGF-β signaling pathway (Fig. 6). Additionally, by entrapping miR-96-5p and increasing the expression of RASSF8, circPTPRA inhibited EMT processes in LC cells and decreased cancer cell metastasis in a mouse xenograft model [98]. These results have given new insights into the EMT-mediated perspectives of the function of circRNAs within LC.
Table 2.
Exo-circRNAs which are functioned as tumor suppressors in lung cancer
| Circ-RNA | Number of clinical samples | Types of specimens | Animal model | Expression | Target Genes | Mechanisms | Functions | Refs |
|---|---|---|---|---|---|---|---|---|
| CircNOL10 | 61 pairs of cancerous and paracancerous lung tissue samples | A549, H1299, H226, H460, H661, SK-MES-1, BEAS-2B | Nude mice | Down | ESRP1, SCML, HN | – | Promotes SCLM1-mediated transcriptional regulation, hence suppressing LC development | [196] |
| Circ0006916 | 49 patients | 16HBE-T, A549, H460, 16HBE | – | Down | miR-522-3p, TNRC6A, PHLPP1 | – | Upregulating PHLPP1 with miR-522-3p inhibits cellular proliferation and tumor growth | [197] |
| hsa_circ_100395 | 69 pairs of LC tissues and normal tissues | A549, H460 | Nude mice | Down | miR-1228, TCF21 | miR‐1228/TCF21 axis | miRNA sponge | [198] |
| CircPTK2 | 73 pairs of LC tissues and normal tissues | A549,H1299, H1650, SPC-A1, Calu3, H226, H520, SK-MES-1, BEAS-2B | Nude mice | Down | miR-429/miR-200b-3p, 3’-UTR of TIF1γ, TGF-β | – | Targeting TIF1 to reduce TGF- γ induced EMT as miR-429/miR-200b-3p sponges | [199] |
| Circ_0001649 | 53 paired of tissue specimens | A549, H358, H1299, H1581, 16HBE | Nude mice | Down | miR-331-3p and miR-338-5p | Circ_0001649 miR-331-3p/miR-338-5p regulatory pathway | Represses LC development by sponging miR-331-3p and miR-338-5p | [200] |
| Circ_103820 | 20 paracarcinoma and lung cancer pairs | SPCA1, A549, HEK-293 T | – | Down | miRNA-200b-3p, LATS2, SOCS6 | Circ_103820/miRNA-200b-3p axis | Lung cancer miR-200b-3p sponge modulates LATS2 and SOCS6 expression | [201] |
| Circ-SLC7A6 | 110 pairs of NSCLC and precancerous normal tissues | A549, H460 | Nude mice | Down | miR-21 | Circ-SLC7A6/miR-21 axis | circ-SLC7A6 suppressed LC proliferation through sponging miR-21 | [202] |
| CircPTPRA | NSCLC patients (n = 34) | H522, H23, H1755, BEAS-2B | Nude mice | Down | miR-96-5p, RASSF8 | miR-96-5p/RASSF8/E-cadherin pathway | prevents LC cells from undergoing EMT and spreading by sponging miR-96-5p | [203] |
Fig. 6.
Illustration shows the relationship between tumor suppressor signaling pathways and exosomal circRNAs in lung cancer
Exosomal circRNAs and cell proliferation
Dysproliferation is a significant contributor to tumor progression, therefore the control of cell growth has attracted more attention [99]. Recently, exo-circRNAs have been shown to influence cell proliferation in a variety of malignancies, including lung cancer. Fig. 7 For instance, Xu et al. found that hsa_circ_0014235 promoted tumor development in non-small cell lung cancer through modulating the miR-520a-5p/CDK4 regulatory axis [100]. They revealed that hsa_circ_0014235 increased tumor growth by promoting cell proliferation, migration, and DDP resistance in vivo. In addition, Ying et al. demonstrated that the expression of circPVT1 was upregulated and stimulated cell proliferation in blood-derived exosomes isolated from lung cancer patients [85]. They found that exo-circPVT1 promotes LC proliferation through targeting the miR-124/EZH2 axis and induces macrophage polarization. Furthermore, circ-FOXM1 Table. 3 increases cell proliferation in NSCLC by targeting PPDPF and MACC1 with miR-1304-5p and is directly linked to lymph node invasion, a high TNM grade, and a poor prognosis [101]. Likewise, in NSCLC tissues and cells, Wei et al. proved that the levels of circ-FOXM1 and ATG5 were elevated, whereas the level of miR-149-5p was downregulated. Circ-FOXM1 knockdown reduced autophagy and cancer cell survival [102]. They observed that miR-149-5p functioned by inhibiting ATG5 expression, and circ-FOXM1 functioned by suppressing miR-149-5p expression. Similarly, exo-circaARHGAP10 expression level was increased in NSCLC tissues and serum samples. In vitro proliferation and glycolysis of NSCLC cells were suppressed by circARHGAP10 knockdown, while tumor growth was inhibited in vivo [103]. Recently, exosomes, according to Hongya et al., were responsible for transmitting circVMP1, which accelerated the proliferation of NSCLC and DDP resistance by targeting the miR-524-5p-METTL3/SOX2 axis [104].
Fig. 7.
Circular RNAs have been demonstrated to have therapeutic promise and have a potential to be applied in the treatment of a wide range of diseases including lung cancer
Table 3.
The potential role of exo-circRNAs as biomarkers in the diagnosis and treatment of lung cancer
| CircRNA | Role/Function | Regulation | Mechanism | Sample | Refs |
|---|---|---|---|---|---|
| Circ_0047921 | Biomarker | ↑ | – | Serum Exosome | [204] |
| Circ_005628 | Biomarker | ↑ | Circ_005628/miR-1244/TRIM44 | Serum Exosome | [205] |
| circ_0001492 circ_0001346 circ_0000690 | Biomarker | ↑ | circ_0001492/miR-93-5p | Plasma Exosome | [206] |
| Circ_0001439 | Biomarker | ↑ | – | Plasma Exosome | [207] |
| CircFARSA | Biomarker/ enhances NSCLC metastasis | ↑ | CircFARSA/PTEN/PI3K/AKT axis | Cell Line Exosome | [116] |
| hsa_circ_0069313 | Biomarker | ↑ | – | Serum Exosome | [208] |
| Circ_0043278 | Increased expression of ROCK1, CDKN1B, and AKT3 promotes proliferation, invasion, and migration | ↑ | miR-520f /ROCK1/CDKN1B/AKT3 axis | Cell Line Exosome | [209] |
| Circ CDYL | Sponges miR-185-5p and controls TNRC6A to suppress cell growth and trigger cell death | ↓ | Circ CDYL/miR-185-5p/TNRC6A axis | Cell Line Exosome | [210] |
| CircARHGAP10 | Boost cell division, migration, invasion, and glucose metabolism | ↑ | CircARHGAP10/miR-638/FAM83F axis | Serum Exosome | [211] |
| hsa_circ_0002130 | Involves facilitating resistance to osimertinib | ↑ | Hsa_circ_0002130/miR-498 axis | Serum Exosome | [192] |
| Circ_0008928 | Upregulation of miR-488 and HK2 in CDDP-resistant LC promotes cell proliferation, migration, and glycolysis metabolism | ↑ | Circ_0008928/miR-488/HK2 axis | Serum Exosome | [212] |
| CircSETDB1 | Promotes growth and metastasis | ↑ | CircSETDB1/miR-7/Sp1 axis | Cell Line Exosome | [213] |
| circRNA-002178 | Immune escape | ↑ | CircRNA-002178/miR-34/PDL1 | Plasma Exosome | [168] |
| Circ_0076305 | DDP resistance in NSCLC is controlled by upregulating ABCC1 expression via miR-186-5p sponging | ↑ | Circ_0076305/miR-186-5p/ABCC1 axis | Cell Line Exosome | [214] |
| CircVMP1 | miR-524-5p-METTL3/SOX2 axis targeting promotes NSCLC development and DDP resistance | ↑ | miR-524-5p-METTL3/SOX2 axis | Cell Line Exosome | [104] |
In contrast to the above, several circRNAs act as tumor suppressors, and they inhibit lung cancer cell proliferation. For example, the expression level of circ_0006677 was lower in LC cells and NSCLC tissues from patients compared to nearby healthy tissues. Poorer patient survival was considerably related to lower expression of circ 0006677 [105]. Circ_0006677’s overexpression drastically reduced NSCLC cells' capacity for proliferating, invading, and metabolizing glucose. By controlling the expression of the signal transducer inhibitor SOSC2 through sponging miR-578, circ_0006677 could prevent the growth of NSCLC and glycolysis [105]. Additionally, Shi et al. found that hsa_circ_0069244 also acts as a sponge for miR-346 to limit the proliferation of lung cancer via regulating XPC expression [106]. Recently, in both NSCLC tissues and cell lines, hsa_circ_0003176 had the typical characteristics of circRNAs, which were downregulated. Functionally, hsa_circ_0003176 was overexpressed, which prevented NSCLC cells from proliferating, invading other cells, and growing both in vitro and in vivo [107]. These findings might improve our understanding of the molecular processes behind the development of NSCLC into a malignant state.
Exosomal circRNAs mediated regulation of angiogenesis
Tumors are distinguished by their capacity for unrestricted reproduction, independent maintenance of their nutritional status, and aberrant regulation of their cellular energy metabolism [108]. Angiogenesis is essential to the microenvironment in which this severe and uncontrolled growth occurs [109]. When the tumor's "angiogenesis switch" is activated, the vascular system responds or becomes more dynamic and produces new blood vessels to supply the growing tumor [110]. Exo-circRNAs have recently been found to play a crucial role in tumor angiogenesis [111]. For example, Yang et al. showed that the abundance of circ_0006988 was increased in tissues and NCSLC cells. They proved that the angiogenesis process was slowed by silencing circ_0006988 [112]. Circ_0006988 can sponge miR-491-5p, which leads to overexpressing of MAP3K3. The growth of xenograft tumors was also inhibited when circ 0006988 was silenced or knocked down. This was accomplished by reducing tumor-promoting angiogenesis [112]. Moreover, the expression of circ_0016760 was significantly higher in NSCLC tissues and cells than in normal lung tissues. Because of its ability to behave as a miR-29b sponge, circ 0016760 was able to prevent miR-29b from binding to HIF1A. Furthermore, circ_0016760 silencing inhibited cell proliferation, invasion, and angiogenesis or tube formation [113].
There has only been a limited of research done on how circRNAs participate in the process of LC angiogenesis. However, circRNA-based molecular therapy may be an option for treating LC due to its advantages, such as its low molecular weight and high stability.
Exosomal circRNAs and metastasis
Tumor metastasis is the term for the spread of malignant tumor cells from their initial site and metastasis is the main factor that leads to cancer-related mortality [114]. Adhesion, disintegration, and migration are the three main steps of tumor cell metastatic progression. Through miRNA sponging, circRNAs regulate NSCLC invasion and metastasis. For instance, the serum exosomal FECR1 circRNA is a novel oncogenic driver that promotes tumor metastasis via the miR584-ROCK1 pathway; it is highly expressed in SCLC tissues and is positively correlated with lymph node metastasis [115]. Additionally, Chen et al. found that the PTEN/PI3K/AKT pathway is used by tumor-derived exosomal circFARSA to polarize M2 macrophages and promote NSCLC metastasis [116]. Moreover, using TGF-β as a model, Wang et al. demonstrated that circPTK2 suppresses TGF-β induced EMT and metastasis in NSCLC by regulating TIF1 [97]. Overexpression of circPTK2 may offer a treatment option for advanced non-small cell lung cancer and illuminate a novel approach by which circRNA regulates TGF-β induced EMT and tumor metastasis. Circ_0000519, another oncogenic circRNAs, overexpression of circ_0000519 promoted metastasis by targeting miR-1258 in NSCLC. Meanwhile, circ_0000519 inhibition decreased cell metastasis by reducing cyclin D1, vimentin, and MMP-9 expression levels. CircRNA hsa_circ_0020123 promotes metastasis via sponging miR-144 to relieve ZEB1 and EZH2 from inhibition [94]. In vitro and in vivo, suppressing hsa_circ_0020123 decreased NSCLC development and metastasis.
Further, circRNAs bind to RBPs in non-small cell lung cancer, which then allows them to influence EMT, invasion, and metastasis. CircLARP4 is a La-related RNA-binding protein and inhibits cell proliferation and metastasis by regulating SMAD7 expression [117]. A worse prognosis is related to reduced expression of circLARP4 in NSCLC. Moreover, the capacity for SPCA1 cells to metastasize is inhibited by overexpression of the circLARP4 gene [118]. Another circRNA down-regulated in NSCLC that may prevent lymphatic metastasis is hsa_circ_0033155. Inhibition of tumor growth, colony formation, and migration occur after ectopic expression of hsa_circ_0033155 [119].
Exosomal circRNAs and apoptosis
The development of LC is linked to circRNAs, which have been implicated in several cellular processes, including proliferation, growth, metastasis, aging, and apoptosis [120]. Exo-circRNAs that are increased in LC have been found in several studies to decrease the apoptotic process and enhance tumor growth by sponging miRNAs. Recently, Li Chuankui and his colleagues showed that exosomal circPLK1 upregulation enhances the proliferation of NSCLC via acting on the miRNA-1294/high mobility cluster protein A1 pathway and inhibits apoptotic cell death [121]. According to Yang et al., circRNA TUBA1C sponging miR-143-3p increased the progression of NSCLC [122]. Furthermore, they found that circTUBA1C silencing led to elevated levels of cleaved caspase-3 and Bax protein expression which makes increasing apoptosis. Additionally, hsa circ 0012673 circular RNA, through regulating the miR-320a/LIMK18521 pathway, promotes LC cell growth and invasion [91]. By targeting miR-320a and upregulating LIM domain kinase 1, circ 0012673 could reduce proliferation, motility, and EMT and increase apoptosis in LUAD cell lines upon knockdown [91]. Likewise, according to Ding et al., increased circ-MEMO1 levels boosted aerobic glycolysis, cell cycle progression, and proliferation while inhibiting LC cell death through the miR-101-3p/KRAS pathway and was associated with poor prognosis [123].
Several studies have revealed that circular RNAs that are overexpressed in LC make tumors grow by increasing the expression of Bcl-2 or decreasing the expression of Bax, which inhibits the process of apoptosis. For example, by sponging miR-195 and triggering Bcl-2, circVANGL1 overexpression was found to behave as an oncogene and suppress LC apoptosis [124]. Furthermore, inhibition of apoptosis in LC cells was achieved by has_circ_0109320's ability to upregulate Bcl-2, downregulate Bax, and cleave caspase 3 and by its ability to sponge miR-595, induce E2F transcription factor 7 expression [125]. According to Qin et al. work, circPVT1 facilitates the progression of NSCLC cells by suppressing apoptosis and modulating the miR-497/Bcl-2 pathway. They discovered that circPVT1 controls the miR-497/Bcl-2 pathway and inhibits cell death by sponging miR-497 [126].
Despite this, several circRNAs are downregulated in LC and appear to have an antagonistic role in LC growth by inhibiting the Wnt axis. For example, through downregulating Wnt/β-catenin signaling and elevating ITCH expression, circ-ITCH served as a sponge for the expression of oncogenic miR-7 and miR-214 [127]. Tian et al. also revealed that the hsa circ 0043256 serves as a miR-1252 sponge, allowing it to bind ITCH and interfere with the Wnt/β-catenin pathway. They found that cinnamaldehyde-treated LC cells increased circ 0043256, which decreased cell growth and triggered apoptosis through ITCH in LUAD cell lines [128].
In contrast, the expression of circNOL10 was shown to be suppressed in LC, and it was also shown to promote apoptosis, which reduced LC proliferation in both in vivo and in vitro studies [129]. The molecular mechanism by which circNOL10 influenced SCML1's regulation of the human polypeptide family was the inhibition of transcription factor ubiquitination. Ultimately, circNOL10 induced cell death by upregulating the expression of Bax and caspase-9 while downregulating Bcl-2 expression [129]. Through interactions with members of the Bcl-2 family, circRNAs were found to regulate apoptosis in lung cancer. This finding opens the new approach for the development of targeted therapies.
Exosomal circRNAs modulate drug resistance
Drug resistance is a significant concern in the management of cancer patients. Cancer cells can show resistance to treatment in a number of ways. Exosomes have gained universal attention as a novel therapeutic to treat cancer [130, 131]. Importantly, exosomes deliver non-coding RNAs (including circRNAs) and proteins linked with multi-drug resistance (MDR) to target cells [132]. Two MDR phenotypes exist. The first is the fundamental chemoresistance that predated medication exposure. However, the other is acquired resistance, which develops after extensive treatment [133]. Acquired MDR often develops during clinical cancer therapy and is a significant barrier to effectively inhibiting metastasis and cell proliferation, leading to a poor prognosis and short overall survival [134]. In addition, exosomes send functional P-glycoprotein to drug-sensitive recipient cells. This protein is a crucial part of the signaling pathways that help drug-sensitive recipient cells become resistant to drugs [135].
Numerous studies have found that circRNAs have a regulatory function in the resistance to cancers. For instance, lung adenocarcinoma (LAD) patients with high circPVT1 expression are less likely to respond to cisplatin and pemetrexed. CircPVT1 also leads to treatment resistance against these drugs by targeting the miR-145-5p/ABCC1 pathway [136]. Furthermore, Cao et al. found that inhibiting circ-PVT1 through the miR-429/FOXK1 signaling axis slowed LC growth and increased sensitivity to cisplatin [137]. Similarly, the lung cancer cell line circular RNA CDR1-AS promotes resistance to cisplatin and pemetrexed via activating the EGFR signaling pathway [138]. Additionally, the production of PD-L1 exosomes by NSCLC cells increased cell stemness, which in turn made tumor cells more resistant to cisplatin. By inhibiting PD-L1, chemoresistant tumor cells could be more sensitive to chemotherapy drugs such as cisplatin [139].
Recently, circRNAs that are increased in NSCLC have been identified to increase cisplatin resistance by promoting the expression of STAT3. For instance, circ 0076305 targeted miR-296-5p to actively modulate cisplatin resistance by overexpressing STAT3 in NSCLC [140]. Likewise, in LC cells, circAKT3 inhibited glycolysis and cisplatin resistance by controlling the miR-516b-5p/STAT3 pathway [141]. Meanwhile, Ma et al. observed that hsa_circRNA_0002130 had a high level of expression in the serum exosomes of osimertinib-resistant LC patients and osimertinib-resistant LC cells [142]. Accordingly, it has been hypothesized that circRNAs are critically involved in LC resistance pathways. Nevertheless, additional investigations will be needed to study those pathways that are triggered by exo-circRNAs in cancer patients.
Therapeutic potential of exo-circRNAs
Exosomes are a promising therapeutic tool for many diseases because of their practical ability to transport small molecules between cells [143]. They may also be useful as biomarkers in a variety of diseases via modulating cell communications [144]. Due to their unique properties, such as their nano size, double lipid membrane, ability to act as multiple carriers, strong histocompatibility [145], high bioavailability [146], low cytotoxicity, and immunogenicity [147], exosomes can be used to deliver therapeutics to cancer cells. Furthermore, surface receptors make it easier for exosomes to target tumor cells and have less of a negative effect on healthy tissue [148].
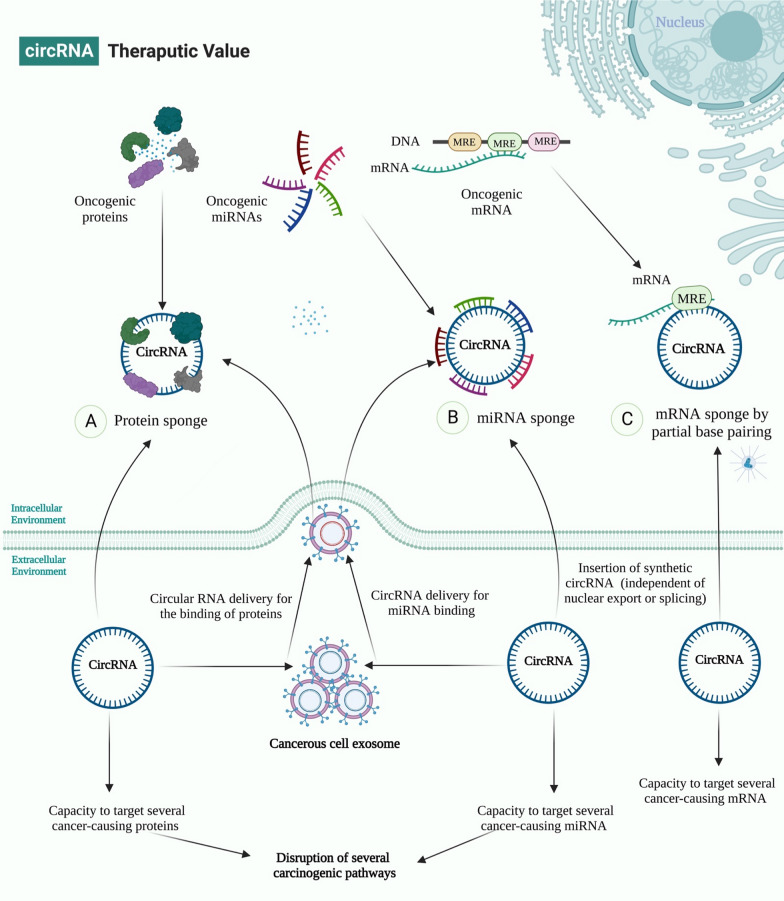
Recent advancements in RNA-based therapies and altered RNA expression in cancers offer promising therapeutic strategies [149–151]. A new method is to develop synthesized circRNAs with high-affinity domains for specific oncogenic proteins, mRNAs, lncRNAs, and miRNAs that might be delivered exogenously to restore the cell's normal signaling pathway and inhibit tumor progression [152, 153] (Fig. 7). Additionally, exosomes, which are thought to be circRNA transporters, may be able to increase the number of circRNAs in cancer cells [154]. This will probably make cancer less aggressive and may act as a biomarker.
The production of synthetic circRNA sequences that can inhibit oncogenic miRNAs has become a very effective way to treat cancer because it can reduce the effectiveness of cancer’s compensatory mechanisms. For example, Kristensen et al. found that hybrid circRNAs might target oncogenic miRNAs and oncoproteins of the same pathway [155].
Additionally, circRNAs can also be used as sponges for oncomiRs [156]. Their expression level is also considered a treatment approach, such as sponging miRNA-9 via circMTO1, which makes it possible for p21 expression and inhibits cancer progression [157]. Similarly, Liu et al. revealed that synthetic circRNA named scRNA21 acts as a miR-21 sponge to inhibit the proliferation of cancer cells [158].
Furthermore, circRNAs can target oncoproteins and leads to inhibit the proliferation of tumor cells. For instance, inhibiting the Wnt/β-catenin axis with circular RNA-ITCH could also be used to treat different types of cancer [15]. Molecular analysis showed that oncogenic miR-7 and miR-214 were found to behave as a sponge for circRNA-ITCH, which increased ITCH expression and consequently reduced Wnt/β-catenin signaling in LC [127]. Likewise, by attaching to cell cycle proteins CDK2 and p21, circ-Foxo3 suppressed cell cycle progression when it was overexpressed [159].
Other circRNAs sponge miRNAs and mRNAs also proposed as a therapeutic option. The relevant mRNA expression in physiologic processes and pathological mechanisms was controlled by cross-talk between circRNAs and miRNAs [160]. The relative processes of interaction between circRNAs, miRNAs, and mRNAs are still being argued. However, two types of strategies have been described: (1) circRNAs sponge microRNAs, such as circHMCU can sponge the let-7 family and lead to cancer development and metastasis [161]. (2) Circular RNA is mediated by miRNAs. For example, in an Ago2-slicer-dependent manner, miR-671 cleaves a circular antisense transcript of the Cerebellar Degeneration-Related protein 1 locus (CDR1) [49]. CDR1 mRNA levels decreased due to circular antisense downregulation, even if heterochromatin does not occur.
In another way, some circRNAs are upregulated in malignant cells and can sponge tumor suppressor miRNAs, such as circGFRA1 and miR-34a [162]; and circUBAP2 and miR-143 [163], could be subjected to inhibition as a strategic way in cancer therapeutics. Furthermore, circRNA-MYLK acts as a ceRNA by binding miR-29a and facilitating the production of VEGFA [164]. The treatment of cancer has also been proposed for the silencing of this circRNA. The above studies consider that exosomal cirRNA-modulating may have potential applications in cancer therapies.
Conclusion
Exosomes originating from LC cells, known as Lung cancer cell derived-exosomes (LCCDEs), play a role in the progression of LC. Exosomes, the smallest vesicle, deliver important cargo such as nucleic acids, lipids, and proteins. These molecules perform critical functions in cell-to-cell communication and are identified as promising markers for their diagnostic properties.
Exo-circRNAs are enriched in tumors and, with multiple configurations, have also recently received interest in their crucial function in LC carcinogenesis. It acts as sponge for microRNAs, binds to proteins, and interacts with the tumor microenvironment (TME). In addition, exo-circRNAs can be used in early diagnosis, therapeutic response, exosome drug-delivery design for target therapy, and prognosis.
Although the future holds great promise, various challenges should be overcome. Despite ongoing studies, several open concerns remain about the clinical use of mRNAs and exosomal circRNAs. From our perspective, exo-circRNAs will be one of the most hotly debated topics in the future, and further studies will be required to verify their clinical applications.
Acknowledgements
The authors would like to thank the clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Abbreviations
- LC
Lung Cancer
- CircRNA
Circular RNA
- TDEs
Tumor-derived exosomes
- exo-circRNAs
Exosomal circular RNA
- TME
Tumor microenvironment
- MVB
Multivesicular body
- ILVs
Intraluminal vesicles
- ESCRT
Endosomal-sorting complex that is required for transport
- VPS4
Vacuolar protein sorting-associated protein4
- TSG101
Tumor susceptibility gene 101
- ALIX
ALG-2-interacting protein X
- ESCRT
Endosomal sorting complex required for transport
- CHMP4
Charged multivesicular body protein 4a
- RBPs
RNA-binding proteins
- DHX9
DExH-box helicase 9
- FUS
Fused in sarcoma
- ADAR1
RNA-specific adenosine deaminase 1
- NF90/NF110
Nuclear factor 90/110
- MBL
Mannose‐binding lectin
- QKI
KH domain containing RNA Binding
- LUAD
Lung adenocarcinoma
- WNT
Wingless-related integration site
- PLAGL2
Pleomorphic adenoma gene like-2
- EIciRNAs
Exon–intron circRNAs
- Pol II
Polymerase II
- U1 snRNA
U1 spliceosomal RNA
- lncRNAs
Long non-coding RNAs
- IRES
Internal ribosome entry site
- UTR
Untranslated region
- ORFs
Open reading frames
- AUG
The codon for Methionine
- hESCs
Human embryonic stem cells
- Ago2
Argonaute 2
- rRNA
Ribosomal RNA
- TNM
Tumor, nodes, and metastases
- FASN
Fatty acid synthase
- STAT3
Signal transducers and activators of transcription 3
- NSCLC
Non-small cell lung cancer
- KPNA4
Karyopherin subunit alpha-4
- E2F5
E2F Transcription Factor 5
- EZH2
Enhancer of zeste 2 polycomb repressive complex 2 subunit
- SLC1A5
Solute carrier family 1 member 5
- ZEB1
Zinc finger E-box-binding homeobox 1
- TIF1-y
Transcription intermediary factor 1-gamma
- RASSF8
Ras association domain family nember 8
- TGF-β
Transforming growth factor beta
- KRAS
Kirsten rat sarcoma virus
- Bcl-2
B-cell lymphoma 2
- E2F
Family of transcription factors
- SCML1's
Scm polycomb group protein like 1
- MDR
Multiple drug resistance
- LAD
Lamina-associated domains
- ABCC1
ATP binding cassette subfamily C member 1
- FOXK1
Forkhead box K1
- EGFR
Epidermal growth factor receptor
- PD-L1
Programmed death-ligand 1
- FOXO3
Factor forkhead box O-3
- CDR1
Cerebellar degeneration related protein 1
- LCCDEs
Linear constant-coefficient difference equation
Author contributions
MT and SGF designed and supervised the study. BMH, SRA, GSHF, MFR, AS and MM wrote the draft and revised it. BMH, MM and AS collected the data and designed the figures and tables. All the authors read the submitted version and approved it. All authors read and approved the finalmanuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Taheri, Email: Mohammad.taheri@uni-jena.de.
Majid Mokhtari, Email: majimokh@gmail.com.
References
- 1.Thandra KC, Barsouk A, Saginala K, Aluru JS, Barsouk A. Epidemiology of lung cancer. Contemp Oncol. 2021;25(1):45–52. doi: 10.5114/wo.2021.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thawani R, Fakhoury K, Becker KD. Cause of mortality in patients with lung cancer and brain metastasis. J Clin Oncol. 2020;38:21743. doi: 10.1200/JCO.2020.38.15_suppl.e21743. [DOI] [Google Scholar]
- 3.Yin L, Liu X, Shao X, Feng T, Xu J, Wang Q, et al. The role of exosomes in lung cancer metastasis and clinical applications: an updated review. J Transl Med. 2021;19(1):312. doi: 10.1186/s12967-021-02985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, Peng X, et al. Exosomes, a new star for targeted delivery. Front Cell Develop Biol. 2021 doi: 10.3389/fcell.2021.751079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang C, Zhang N, Hu X, Wang H. Tumor-associated exosomes promote lung cancer metastasis through multiple mechanisms. Mol Cancer. 2021;20(1):117. doi: 10.1186/s12943-021-01411-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amicone L, Marchetti A, Cicchini C. Exosome-associated circRNAs as key regulators of EMT in cancer. Cells. 2022;11(10):1716. doi: 10.3390/cells11101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 10.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 12.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):1–8. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussen BM, Honarmand Tamizkar K, Hidayat HJ, Taheri M, Ghafouri-Fard S. The role of circular RNAs in the development of hepatocellular carcinoma. Pathol Res Pract. 2021;223:153495. doi: 10.1016/j.prp.2021.153495. [DOI] [PubMed] [Google Scholar]
- 14.Ghafouri-Fard S, Khoshbakht T, Hussen BM, Sarfaraz S, Taheri M, Ayatollahi SA. Circ_CDR1as: a circular RNA with roles in the carcinogenesis. Pathol Res Pract. 2022;236:153968. doi: 10.1016/j.prp.2022.153968. [DOI] [PubMed] [Google Scholar]
- 15.Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Samsami M. Emerging role of circular RNAs in the pathogenesis of ovarian cancer. Cancer Cell Int. 2022;22(1):172. doi: 10.1186/s12935-022-02602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghafouri-Fard S, Najafi S, Hussen BM, Basiri A, Hidayat HJ, Taheri M, et al. The role of circular RNAs in the carcinogenesis of bladder cancer. Front Oncol. 2022 doi: 10.3389/fonc.2022.801842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asadi MR, Moslehian MS, Sabaie H, Sharifi-Bonab M, Hakimi P, Hussen BM, et al. CircRNA-associated CeRNAs regulatory axes in retinoblastoma: a systematic scoping review. Front Oncol. 2022 doi: 10.3389/fonc.2022.910470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26(12):3444–60.e5. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 22.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44(1):11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 28.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juan T, Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. 2018;74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Frankel EB, Audhya A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol. 2018;74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9(1):19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Sorting it out regulation of exosome loading. Semin Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. 2021;19(1):47. doi: 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3(1):24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pironti G, Strachan RT, Abraham D, Mon-Wei YuS, Chen M, Chen W, et al. Circulating exosomes induced by cardiac pressure overload contain functional angiotensin II type 1 receptors. Circulation. 2015;131(24):2120–2130. doi: 10.1161/CIRCULATIONAHA.115.015687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cossetti C, Iraci N, Mercer TR, Leonardi T, Alpi E, Drago D, et al. Extracellular vesicles from neural stem cells transfer IFN-γ via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56(2):193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue B, Yang H, Wang J, Ru W, Wu J, Huang Y, et al. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020;53(7):e12857. doi: 10.1111/cpr.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dragomir M, Calin GA. Circular RNAs in cancer—lessons learned from microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 40.Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 41.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67(2):214–27.e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 44.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 45.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA. 2017;114(26):E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo j. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74(3):494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 51.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–80.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 52.Yu C-Y, Kuo H-C. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):1–12. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guria A, Sharma P, Natesan S, Pandi G. Circular RNAs—the road less traveled. Front Mol Biosci. 2020;6:146. doi: 10.3389/fmolb.2019.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Z, Cao Q, Zhao Z, Song C. Biogenesis features functions and disease relationships of a specific circular RNA CDR1 as. Aging Dis. 2020;11(4):1009–1020. doi: 10.14336/AD.2019.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panda AC. Circular RNAs act as miRNA sponges. In: Xiao Junjie., editor. Circular RNAs. Berlin: Springer; 2018. [DOI] [PubMed] [Google Scholar]
- 56.Gao N, Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 2020;20:2. doi: 10.1186/s12935-019-1065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Hua J, Li J, Zhang J, Dzakah EE, Cao G, et al. Mechanisms of non-coding RNA-modulated alternative splicing in cancer. RNA Biol. 2022;19(1):541–547. doi: 10.1080/15476286.2022.2062846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscipl Rev RNA. 2017;8(2):e1386. doi: 10.1002/wrna.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 63.Yu B, Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus. 2016;7(2):155–166. doi: 10.1080/19491034.2016.1179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eidem TM, Kugel JF, Goodrich JA. Noncoding RNAs: regulators of the mammalian transcription machinery. J Mol Biol. 2016;428(12):2652–2659. doi: 10.1016/j.jmb.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di X, Jin X, Li R, Zhao M, Wang K. CircRNAs and lung cancer: biomarkers and master regulators. Life Sci. 2019;220:177–185. doi: 10.1016/j.lfs.2019.01.055. [DOI] [PubMed] [Google Scholar]
- 66.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14(8):1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 72.Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25(9):2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 75.Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):1–18. doi: 10.1186/s13046-018-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):1–9. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang X-Y, Huang Z-L, Huang J, Xu B, Huang X-Y, Xu Y-H, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):1–16. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21(1):1–9. doi: 10.1186/s12885-021-08669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in cancer. Mol Ther-Nucl Acids. 2020;21:367–383. doi: 10.1016/j.omtn.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu W, Peng W, Sha H, Li J. Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326 in lung adenocarcinoma cells. Oncol Res. 2019;27(5):623. doi: 10.3727/096504018X15420734828058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang C, Wu Y, Lei Q, Jiang Y, Shao J, Liu D, et al. The emerging role of circular RNAs in non-small cell lung cancer. Int J Clin Exp Med. 2019;12(5):6049–6059. [Google Scholar]
- 82.He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. 2020;11(14):4037. doi: 10.7150/jca.30360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. doi: 10.1016/j.biopha.2020.110019. [DOI] [PubMed] [Google Scholar]
- 84.Ma D, Qin Y, Huang C, Chen Y, Han Z, Zhou X, et al. Circular RNA ABCB10 promotes non-small cell lung cancer progression by increasing E2F5 expression through sponging miR-584-5p. Cell Cycle. 2020;19(13):1611–1620. doi: 10.1080/15384101.2020.1761617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, Li L, Song X. Exosomal circPVT1 derived from lung cancer promotes the progression of lung cancer by targeting miR-124-3p/EZH2 axis and regulating macrophage polarization. Cell Cycle. 2022;21(5):514–530. doi: 10.1080/15384101.2021.2024997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prieto-García E, Díaz-García CV, García-Ruiz I, Agulló-Ortuño MT. Epithelial-to-mesenchymal transition in tumor progression. Med Oncol. 2017;34(7):122. doi: 10.1007/s12032-017-0980-8. [DOI] [PubMed] [Google Scholar]
- 88.Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020;30(10):764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xue M, Hong W, Jiang J, Zhao F, Gao X. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol. 2020;17(12):1811–1822. doi: 10.1080/15476286.2020.1789819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19(3):560–570. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin H, Liu J, Du ZH, Hu R, Yu YK, Wang QA. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1841–1852. doi: 10.26355/eurrev_202002_20362. [DOI] [PubMed] [Google Scholar]
- 92.Li J, Wang J, Chen Z, Chen Y, Jin M. Hsa_circ_0079530 promotes cell proliferation and invasion in non-small cell lung cancer. Gene. 2018;665:1–5. doi: 10.1016/j.gene.2018.04.059. [DOI] [PubMed] [Google Scholar]
- 93.Liu C, Zhang Z, Qi D. Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco Targets Ther. 2019;12:6181–6189. doi: 10.2147/OTT.S201834. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 95.Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120(11):19019–19030. doi: 10.1002/jcb.29225. [DOI] [PubMed] [Google Scholar]
- 96.Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513(4):904–911. doi: 10.1016/j.bbrc.2019.04.108. [DOI] [PubMed] [Google Scholar]
- 97.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu X, Pan Y, Ma H, Li W. Simvastatin inhibits proliferation and induces apoptosis in human lung cancer cells. Oncol Res. 2013;20(8):351–357. doi: 10.3727/096504013X13657689382897. [DOI] [PubMed] [Google Scholar]
- 100.Xu X, Tao R, Sun L, Ji X. Exosome-transferred hsa_circ_0014235 promotes DDP chemoresistance and deteriorates the development of non-small cell lung cancer by mediating the miR-520a-5p/CDK4 pathway. Cancer Cell Int. 2020;20(1):1–15. doi: 10.1186/s12935-020-01642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu G, Shi H, Deng L, Zheng H, Kong W, Wen X, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212. doi: 10.1016/j.bbrc.2019.03.213. [DOI] [PubMed] [Google Scholar]
- 102.Wei H, Li L, Zhang H, Xu F, Chen L, Che G, et al. Circ-FOXM1 knockdown suppresses non-small cell lung cancer development by regulating the miR-149-5p/ATG5 axis. Cell Cycle. 2021;20(2):166–178. doi: 10.1080/15384101.2020.1867780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fang K, Chen X, Qiu F, Xu J, Xiong H, Zhang Z. Serum-derived exosomes-mediated circular RNA arhgap10 modulates the progression of non-small cell lung cancer through the miR-638/FAM83F axis. Cancer Biother Radiopharm. 2022;37(2):96–110. doi: 10.1089/cbr.2019.3534. [DOI] [PubMed] [Google Scholar]
- 104.Xie H, Yao J, Wang Y, Ni B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Delivery. 2022;29(1):1257–1271. doi: 10.1080/10717544.2022.2057617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang B, Zhao F, Yao L, Zong Z, Xiao L. CircRNA circ_0006677 inhibits the progression and glycolysis in non-small-cell lung cancer by sponging miR-578 and regulating SOCS2 expression. Front Pharmacol. 2021;12:657053. doi: 10.3389/fphar.2021.657053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shi J, Wang H, Feng W, Huang S, An J, Wang L, et al. Hsa_circ_0069244 acts as the sponge of miR-346 to inhibit non-small cell lung cancer progression by regulating XPC expression. Hum Cell. 2021;34(5):1490–1503. doi: 10.1007/s13577-021-00573-5. [DOI] [PubMed] [Google Scholar]
- 107.Yang F, Pei Y, Xu W, Rong L. hsa_circ_0003176 suppresses the progression of non-small-cell lung cancer via regulating miR-182-5p/RBM5 Axis. Dis Markers. 2022;2022:8402116. doi: 10.1155/2022/8402116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baeriswyl V, Christofori G. The angiogenic switch in carcinogenesis Seminars in cancer biology. Amsterdam: Elsevier; 2009. [DOI] [PubMed] [Google Scholar]
- 109.Jiang S, Fu R, Shi J, Wu H, Mai J, Hua X, et al. CircRNA-mediated regulation of angiogenesis: a new chapter in cancer biology. Front Oncol. 2021;11:553706. doi: 10.3389/fonc.2021.553706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86(3):353–64. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 111.Bremnes RM, Camps C, Sirera R. Angiogenesis in non-small cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer. 2006;51(2):143–158. doi: 10.1016/j.lungcan.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 112.Yang C, Shi J, Wang J, Hao D, An J, Jiang J. Circ_0006988 promotes the proliferation, metastasis and angiogenesis of non-small cell lung cancer cells by modulating miR-491-5p/MAP3K3 axis. Cell Cycle. 2021;20(13):1334–1346. doi: 10.1080/15384101.2021.1941612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H, Zhang H, Zhu J, Liu H, Zhou Q. PESV represses non-small cell lung cancer cell malignancy through circ_0016760 under hypoxia. Cancer Cell Int. 2021;21(1):628. doi: 10.1186/s12935-021-02336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3(1):55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 115.Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res. 2019;25(4):1302–1317. doi: 10.1158/1078-0432.CCR-18-1447. [DOI] [PubMed] [Google Scholar]
- 116.Chen T, Liu Y, Li C, Xu C, Ding C, Chen J, et al. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun. 2021;28:100412. doi: 10.1016/j.ctarc.2021.100412. [DOI] [PubMed] [Google Scholar]
- 117.Shi JQ, Wang B, Cao XQ, Wang YX, Cheng X, Jia CL, et al. Circular RNA_LARP4 inhibits the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Eur Rev Med Pharmacol Sci. 2020;24(4):1863–1869. doi: 10.26355/eurrev_202002_20364. [DOI] [PubMed] [Google Scholar]
- 118.Wang R, Liu H, Dong M, Huang D, Yi J. Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis. Open Med. 2022;17(1):826–840. doi: 10.1515/med-2022-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghafouri-Fard S, Dinger ME, Maleki P, Taheri M, Hajiesmaeili M. Emerging role of circular RNAs in the pathobiology of lung cancer. Biomed Pharmacother. 2021;141:111805. doi: 10.1016/j.biopha.2021.111805. [DOI] [PubMed] [Google Scholar]
- 120.Cao YZ, Sun JY, Chen YX, Wen CC, Wei L. The roles of circRNAs in cancers: Perspectives from molecular functions. Gene. 2021;767:145182. doi: 10.1016/j.gene.2020.145182. [DOI] [PubMed] [Google Scholar]
- 121.Li C, Wang G, Ma X, Tao T, Li Q, Yang Y, et al. Upregulation of exosomal circPLK1 promotes the development of non-small cell lung cancer through the miR-1294/ high mobility group protein A1 axis. Bioengineered. 2022;13(2):4185–4200. doi: 10.1080/21655979.2022.2026727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J, Jia Y, Wang B, Yang S, Du K, Luo Y, et al. Circular RNA TUBA1C accelerates the progression of non-small-cell lung cancer by sponging miR-143-3p. Cell Signal. 2020;74:109693. doi: 10.1016/j.cellsig.2020.109693. [DOI] [PubMed] [Google Scholar]
- 123.Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal circ-memo1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS Axis. Front Genet. 2020;11:962. doi: 10.3389/fgene.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang L, Ma H, Kong W, Liu B, Zhang X. 2019. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. [DOI] [PMC free article] [PubMed] [Retracted]
- 125.Bai Q, Li L, Chen F, Zhu J, Cao L, Yang Y, et al. Suppression of circular RNA Hsa_circ_0109320 attenuates non-small cell lung cancer progression via MiR-595/E2F7 axis. Med Sci Monit. 2020;26:e921200. doi: 10.12659/MSM.921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qin S, Zhao Y, Lim G, Lin H, Zhang X, Zhang X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother. 2019;111:244–250. doi: 10.1016/j.biopha.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 127.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tian F, Yu CT, Ye WD, Wang Q. Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;493(3):1260–1266. doi: 10.1016/j.bbrc.2017.09.136. [DOI] [PubMed] [Google Scholar]
- 129.Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H, et al. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv Sci. 2019;6(2):1800654. doi: 10.1002/advs.201800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zafar S, Beg S, Panda SK, Rahman M, Alharbi KS, Jain GK, et al. Novel therapeutic interventions in cancer treatment using protein and peptide based targeted smart systems Seminars in cancer biology. Amsterdam: Elsevier; 2021. [DOI] [PubMed] [Google Scholar]
- 131.Bastos N, Ruivo CF, da Silva S, Melo SA. Exosomes in cancer: use them or target them? Seminars in cell & developmental biology. Amsterdam: Elsevier; 2018. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):1–10. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.FernandeseSilva E, Figueira FDS, Lettnin AP, Carrett-Dias M, Filgueira DMVB, Kalil S, et al. C-Phycocyanin: cellular targets mechanisms of action and multi drug resistance in cancer. Pharmacol Rep. 2018;70(1):75–80. doi: 10.1016/j.pharep.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 134.Algorashi I, Goldvaser H, Ribnikar D, Cescon DW, Amir E. Evolution in sites of recurrence over time in breast cancer patients treated with adjuvant endocrine therapy. Cancer Treat Rev. 2018;70:138–143. doi: 10.1016/j.ctrv.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 135.Mostafazadeh M, Samadi N, Kahroba H, Baradaran B, Haiaty S, Nouri M. Potential roles and prognostic significance of exosomes in cancer drug resistance. Cell Biosci. 2021;11(1):1–15. doi: 10.1186/s13578-020-00515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zheng F, Xu R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed Pharmacother. 2020;124:109828. doi: 10.1016/j.biopha.2020.109828. [DOI] [PubMed] [Google Scholar]
- 137.Cao L, Zhou X, Ding X, Gao D. Knockdown of circ-PVT1 inhibits the progression of lung adenocarcinoma and enhances the sensitivity to cisplatin via the miR-429/FOXK1 signaling axis. Mol Med Rep. 2021 doi: 10.3892/mmr.2021.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mao Y, Xu R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed Pharmacother. 2020;123:109771. doi: 10.1016/j.biopha.2019.109771. [DOI] [PubMed] [Google Scholar]
- 139.Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC) J Exp Clin Cancer Res. 2020;39(1):149. doi: 10.1186/s13046-020-01648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984. doi: 10.1016/j.lfs.2019.116984. [DOI] [PubMed] [Google Scholar]
- 141.Xu Y, Jiang T, Wu C, Zhang Y. CircAKT3 inhibits glycolysis balance in lung cancer cells by regulating miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol Lett. 2020;42(7):1123–1135. doi: 10.1007/s10529-020-02846-9. [DOI] [PubMed] [Google Scholar]
- 142.Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307. doi: 10.2147/OTT.S243214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hu G, Drescher KM, Chen X-M. Exosomal miRNAs: biological properties and therapeutic potential. Front Genet. 2012;3:56. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Molibeli KM, Hu R, Liu Y, Xiong D, Tang L. Potential clinical applications of exosomal circular RNAs: more than diagnosis. Front Mol Biosci. 2021 doi: 10.3389/fmolb.2021.769832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ke W, Afonin KA. Exosomes as natural delivery carriers for programmable therapeutic nucleic acid nanoparticles (NANPs) Adv Drug Deliv Rev. 2021;176:113835. doi: 10.1016/j.addr.2021.113835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kooijmans SA, Schiffelers RM, Zarovni N, Vago R. Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment. Pharmacol Res. 2016;111:487–500. doi: 10.1016/j.phrs.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 147.Nam GH, Choi Y, Kim GB, Kim S, Kim SA, Kim IS. Emerging prospects of exosomes for cancer treatment: from conventional therapy to immunotherapy. Adv Mater. 2020;32(51):2002440. doi: 10.1002/adma.202002440. [DOI] [PubMed] [Google Scholar]
- 148.Ye D, Gong M, Deng Y, Fang S, Cao Y, Xiang Y, et al. Roles and clinical application of exosomal circRNAs in the diagnosis and treatment of malignant tumors. J Transl Med. 2022;20(1):1–17. doi: 10.1186/s12967-022-03367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rasul MF, Hussen BM, Salihi A, Ismael BS, Jalal PJ, Zanichelli A, et al. Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Mol Cancer. 2022;21(1):64. doi: 10.1186/s12943-021-01487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghafouri-Fard S, Taheri M, Hussen BM, Vafaeimanesh J, Abak A, Vafaee R. Function of circular RNAs in the pathogenesis of colorectal cancer. Biomed Pharmacother. 2021;140:111721. doi: 10.1016/j.biopha.2021.111721. [DOI] [PubMed] [Google Scholar]
- 152.Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bai H, Lei K, Huang F, Jiang Z, Zhou X. Exo-circRNAs: a new paradigm for anticancer therapy. Mol Cancer. 2019;18(1):1–10. doi: 10.1186/s12943-019-0986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ghafouri-Fard S, Hussen BM, Taheri M, Ayatollahi SA. Emerging role of circular RNAs in breast cancer. Pathol Res Pract. 2021;223:153496. doi: 10.1016/j.prp.2021.153496. [DOI] [PubMed] [Google Scholar]
- 157.Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 158.Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucl Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucl Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song G, Yang Z, Guo J, Zheng Y, Su X, Wang X. Interactions Among lncRNAs/circRNAs, miRNAs, and mRNAs in neuropathic pain. Neurotherapeutics. 2020;17(3):917–931. doi: 10.1007/s13311-020-00881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Song X, Liang Y, Sang Y, Li Y, Zhang H, Chen B, et al. circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Mol Ther-Nucl Acids. 2020;20:518–533. doi: 10.1016/j.omtn.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33(6):2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 163.Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8(37):61687–61697. doi: 10.18632/oncotarget.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Corrigendum to "Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway" [Cancer Letter 403(2017), 305–317] Cancer Lett. 2022;534:215631. doi: 10.1016/j.canlet.2022.215631. [DOI] [PubMed] [Google Scholar]
- 165.Mao Y, Xu R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed Pharmacother. 2020;123:109771. doi: 10.1016/j.biopha.2019.109771. [DOI] [PubMed] [Google Scholar]
- 166.Ma X, Yang X, Bao W, Li S, Liang S, Sun Y, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498(4):1009–1015. doi: 10.1016/j.bbrc.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 167.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. Hoboken: Wiley Online Library; 2017. [DOI] [PubMed] [Google Scholar]
- 168.Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):1–11. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC) J Exp Clin Cancer Res. 2020;39(1):1–19. doi: 10.1186/s13046-020-01648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circ FARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791. doi: 10.1002/cam4.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120(11):19019–19030. doi: 10.1002/jcb.29225. [DOI] [PubMed] [Google Scholar]
- 172.Han W, Wang L, Zhang L, Wang Y, Li Y. Circular RNA circ-RAD23B promotes cell growth and invasion by miR-593–3p/CCND2 and miR-653–5p/TIAM1 pathways in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;510(3):462–466. doi: 10.1016/j.bbrc.2019.01.131. [DOI] [PubMed] [Google Scholar]
- 173.Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet. 2020;11:962. doi: 10.3389/fgene.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi: 10.1016/j.gene.2019.144099. [DOI] [PubMed] [Google Scholar]
- 175.Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi: 10.1016/j.bbrc.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 176.Joseph NA, Chiou S-H, Lung Z, Yang C-L, Lin T-Y, Chang H-W, et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J Hematol Oncol. 2018;11(1):1–14. doi: 10.1186/s13045-018-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang C, Zhang B, Yuan B, Chen C, Zhou Y, Zhang Y, et al. RNA-Seq profiling of circular RNAs in human small cell lung cancer. Epigenomics. 2020;12(8):685–700. doi: 10.2217/epi-2019-0382. [DOI] [PubMed] [Google Scholar]
- 178.Luo Y-H, Zhu X-Z, Huang K-W, Zhang Q, Fan Y-X, Yan P-W, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 179.Liu G, Shi H, Deng L, Zheng H, Kong W, Wen X, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212. doi: 10.1016/j.bbrc.2019.03.213. [DOI] [PubMed] [Google Scholar]
- 180.Xiao Y, Gu S, Yao W, Qin L, Luo J. Circ_0047921 acts as the sponge of miR-1287-5p to stimulate lung cancer progression by regulating proliferation, migration, invasion, and glycolysis of lung cancer cells. World J Surg Oncol. 2022;20(1):1–13. doi: 10.1186/s12957-021-02466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu Q, Shi J, Zhang L, Sheng Y, Zhang Y, Chu D, et al. Circ_0006006 facilitates non-small cell lung cancer progression by modulating miR-924/SRSF7 axis. J Gene Med. 2022;24(5):e3411. doi: 10.1002/jgm.3411. [DOI] [PubMed] [Google Scholar]
- 182.Wang H, Tang Z, Duan J, Zhou C, Xu K, Mu H. Cancer-released exosomal circular RNA circ_0008717 promotes cell tumorigenicity through microRNA-1287-5p/P21-activated kinase 2 (PAK2) axis in non-small cell lung cancer. Bioengineered. 2022;13(4):8937–8949. doi: 10.1080/21655979.2022.2056822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guo F, Li S, Guo C, Xu X, Zhou X, Ma D, et al. Circular RNA circMAGI3 accelerates the glycolysis of non-small cell lung cancer through miR-515-5p/HDGF. Am J transl Res. 2020;12(7):3953. [PMC free article] [PubMed] [Google Scholar]
- 184.She Y, Han Y, Zhou G, Jia F, Yang T, Shen Z. hsa_circ_0062389 promotes the progression of non-small cell lung cancer by sponging miR-103a-3p to mediate CCNE1 expression. Cancer Genet. 2020;241:12–19. doi: 10.1016/j.cancergen.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 185.Cao F, Liu S, Li Z, Meng L, Sang M, Shan B. Activation of circ_0072088/miR-1261/PIK3CA pathway accelerates lung adenocarcinoma progression. Thorac Cancer. 2022 doi: 10.1111/1759-7714.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ning Z, Tian Y, Li Y, Zhao X, Zhang J, Wang C, et al. Exosomal circ_0007385 enhances non-small cell lung cancer cell proliferation and stemness via regulating miR-1253/FAM83A axis. Anticancer Drugs. 2022;33(1):61–74. doi: 10.1097/CAD.0000000000001103. [DOI] [PubMed] [Google Scholar]
- 187.Yang J, Jia Y, Wang B, Yang S, Du K, Luo Y, et al. Circular RNA TUBA1C accelerates the progression of non-small-cell lung cancer by sponging miR-143-3p. Cell Signal. 2020;74:109693. doi: 10.1016/j.cellsig.2020.109693. [DOI] [PubMed] [Google Scholar]
- 188.Li W, Yang P, Zhong C, Shen X, Shi X, Li X. The circ-PITX1 promotes non-small cell lung cancer development via the miR-30e-5p/ITGA6 axis. Cell Cycle. 2022;21(3):304–321. doi: 10.1080/15384101.2021.2020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res. 2019;25(4):1302–1317. doi: 10.1158/1078-0432.CCR-18-1447. [DOI] [PubMed] [Google Scholar]
- 190.Yang B, Teng F, Chang L, Wang J, Liu D-L, Cui Y-S, et al. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging. 2021;13(9):13264. doi: 10.18632/aging.203011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19(1):1–16. doi: 10.1186/s12943-017-0753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293. doi: 10.2147/OTT.S243214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Song J, Shi W, Gao Z, Liu X, Wang W. Downregulation of circRNA_100876 inhibited progression of NSCLC in vitro via targeting miR-636. Technol Cancer Res Treat. 2020;19:1533033820951817. doi: 10.1177/1533033820951817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang W, Lin Y, Zhang G, Shi G, Jiang Y, Hu W, et al. Circ_0002346 suppresses non-small-cell lung cancer progression depending on the regulation of the miR-582–3p/STXBP6 axis. Int J Genom. 2021;2021(1):20. doi: 10.1155/2021/1565660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xu X, Zhou X, Gao C, Cui Y. Hsa_circ_0018818 knockdown suppresses tumorigenesis in non-small cell lung cancer by sponging miR-767-3p. Aging. 2020;12(9):7774. doi: 10.18632/aging.103089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H, et al. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv sci. 2019;6(2):1800654. doi: 10.1002/advs.201800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, et al. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39(8):981–992. doi: 10.1093/carcin/bgy061. [DOI] [PubMed] [Google Scholar]
- 198.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–2090. doi: 10.1080/15384101.2018.1515553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17(1):1–18. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503(3):1503–1509. doi: 10.1016/j.bbrc.2018.07.070. [DOI] [PubMed] [Google Scholar]
- 201.Chi Y, Zheng W, Bao G, Wu L, He X, Gan R, et al. Circular RNA circ_103820 suppresses lung cancer tumorigenesis by sponging miR-200b-3p to release LATS2 and SOCS6. Cell Death Dis. 2021;12(2):1–13. doi: 10.1038/s41419-021-03472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang Y, Zheng F, Wang Z, Lu J, Zhang H. Circular RNA circ-SLC7A6 acts as a tumor suppressor in non-small cell lung cancer through abundantly sponging miR-21. Cell Cycle. 2020;19(17):2235–2246. doi: 10.1080/15384101.2020.1806449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xian J, Su W, Liu L, Rao B, Lin M, Feng Y, et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non–small-cell lung cancer in the chinese population. J Mol Diagn. 2020;22(8):1096–1108. doi: 10.1016/j.jmoldx.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 205.Huo S, Dou D. Circ_0056285 regulates proliferation, apoptosis and glycolysis of osteosarcoma cells via miR-1244/TRIM44 axis. Cancer Manag Res. 2021;13:1257. doi: 10.2147/CMAR.S290645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen F, Huang C, Wu Q, Jiang L, Chen S, Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J Cell Biochem. 2020;121(3):2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 207.Drula R, Braicu C, Harangus A, Nabavi SM, Trif M, Slaby O, et al. Critical function of circular RNAs in lung cancer. Wiley Interdiscip Rev RNA. 2020;11(5):e1592. doi: 10.1002/wrna.1592. [DOI] [PubMed] [Google Scholar]
- 208.Chen Y, Lou C, Ma X, Zhou C, Zhao X, Li N, et al. Serum exosomal hsa_circ_0069313 has a potential to diagnose more aggressive non-small cell lung cancer. Clin Biochem. 2022;102:56–64. doi: 10.1016/j.clinbiochem.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 209.Huang L, Rong Y, Tang X, Yi K, Wu J, Wang F. Circular RNAs are promising biomarkers in liquid biopsy for the diagnosis of non-small cell lung cancer. Front Mol Biosci. 2021;8:625722. doi: 10.3389/fmolb.2021.625722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bian W-X, Xue F, Wang L-Y, Xing X-F. Circular RNA CircCDYL regulates proliferation and apoptosis in non-small cell lung cancer cells by sponging miR-185-5p and upregulating TNRC6A. Cancer Manag Res. 2021;13:633. doi: 10.2147/CMAR.S280315. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 211.Fang K, Chen X, Qiu F, Xu J, Xiong H, Zhang Z. Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small cell lung cancer through the miR-638/FAM83F Axis. Cancer Biother Radiopharm. 2022;37(2):96–110. doi: 10.1089/cbr.2019.3534. [DOI] [PubMed] [Google Scholar]
- 212.Shi Q, Ji T, Ma Z, Tan Q, Liang J. Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother Radiopharm. 2021 doi: 10.1089/cbr.2020.4490. [DOI] [PubMed] [Google Scholar]
- 213.Xu L, Liao W-L, Lu Q-J, Zhang P, Zhu J, Jiang G-N. Hypoxic tumor-derived exosomal circular RNA SETDB1 promotes invasive growth and EMT via the miR-7/Sp1 axis in lung adenocarcinoma. Mol Ther-Nucleic Acids. 2021;23:1078–1092. doi: 10.1016/j.omtn.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 214.Wang X, Wang H, Jiang H, Qiao L, Guo C. Circular RNAcirc_0076305 promotes cisplatin (DDP) resistance of non-small cell Lung cancer cells by regulating ABCC1 through miR-186–5p. Cancer Biothe & Radiopharm. 2021 doi: 10.1089/cbr.2020.4153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wang L, Ma H, Kong W, Liu B, Zhang X. 2019. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. [DOI] [PMC free article] [PubMed] [Retracted]
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.